Abstract
Long non-coding RNAs (lncRNAs) are non-coding transcripts that have emerged as one of the largest and diverse RNA families that regulate gene expression. Accumulating evidence has suggested a number of lncRNAs are involved in diabetes mellitus (DM) pathogenesis. However, results about lncRNA expressions in DM patients are still inconclusive. Thus, we performed a systematic review of the literature on the subject followed by bioinformatics analyses to better understand which lncRNAs are dysregulated in DM and in which pathways they act. Pubmed, Embase, and Gene Expression Omnibus (GEO) repositories were searched to identify studies that investigated lncRNA expression in cases with DM and non-diabetic controls. LncRNAs consistently dysregulated in DM patients were submitted to bioinformatics analysis to retrieve their target genes and identify potentially affected signaling pathways under their regulation. Fifty-three eligible articles were included in this review after the application of the inclusion and exclusion criteria. Six hundred and thirty-eight lncRNAs were differentially expressed between cases and controls in at least one study. Among them, six lncRNAs were consistently dysregulated in patients with DM (Anril, Hotair, Malat1, Miat, Kcnq1ot1, and Meg3) compared to controls. Moreover, these six lncRNAs participate in several metabolism-related pathways, evidencing their importance in DM. This systematic review suggests six lncRNAs are dysregulated in DM, constituting potential biomarkers of this disease.
Keywords: lncRNAs (long non-coding RNAs), type 1 diabetes mellitus (DM1), type 2 diabetes mellitus (T2DM), systematic review, target prediction
Introduction
Diabetes mellitus (DM) is a group of metabolic disorders that have in common the chronic hyperglycemia, which results from defects in insulin secretion, insulin action, or both (1). Accordingly to the International Diabetes Federation Atlas 2019, an estimated 463 million adults are currently living with DM (9.3% of the world population), and this number is projected to reach 700 million by 2045 (2). Thus, DM has achieved epidemic proportions worldwide, being associated with increased morbidity and mortality rates due to its specific micro- and macrovascular complications (1, 2).
Type 1 DM (T1DM) accounts for 5–10% of all DM cases and usually appears in people younger than 30 years (1, 2). T1DM is an autoimmune disease caused by the progressive destruction of pancreatic beta-cells by macrophages and T lymphocytes, making patients insulin-dependent for life (1, 3). Type 2 DM (T2DM) comprises 90–95% of worldwide diabetic cases and generally arises in subjects older than 40 years and with obesity. Hyperglycemia in T2DM patients is caused by insulin resistance associated with different degrees of a relative beta-cell failure (1, 2). It is well known that susceptibility for both T1DM and T2DM is triggered by a multifaceted interaction among several environmental, genetic, and epigenetic factors (4–8).
Epigenetic factors regulate the complex crosstalk between genes and environmental factors without altering the DNA sequence and include DNA methylation, histone posttranslational modifications, and non-coding RNAs (ncRNAs) (7, 8). NcRNAs are regulatory RNAs that typically lack protein-coding capacity and play key roles in both physiological and pathological processes (9, 10). According to their length and functions, ncRNAs can be classified into different subtypes, including the long ncRNAs (lncRNAs), which are those ncRNAs with >200 nucleotides in length (10, 11).
LncRNAs can be located in the nucleus or cytoplasm and exhibit more specific expression profiles than mRNAs, being expressed in cell/tissue-, developmental stage-, or disease state-specific manners (10, 12, 13). A number of studies have suggested lncRNAs participate in several molecular processes involved in gene regulation, including epigenetic, transcriptional, and post-transcriptional regulation, through interaction with chromatin-remodeling complexes, binding to transcription factors or regulation of mRNA-binding proteins and microRNAs (another class of ncRNAs) (10, 14–16).
In this context, growing evidence has shown lncRNAs play key roles in regulating beta-cell function, apoptosis, insulin secretion, glucose metabolism, and insulin resistance (10, 17–22). Accordingly, a number of studies have reported changes in lncRNA expressions in patients with DM or in murine models of T1DM or T2DM (10, 23–29). Thus, lncRNAs are likely to be novel potential biomarkers for early diagnosis and prognosis of T1DM or T2DM (10, 29). For example, Carter et al. showed GAS5 might be a prognostic biomarker for T2DM since this lncRNA was decreased in serum of patients with DM from a US military veterans cohort (23). Individuals with lower GAS5 expression were almost 12× more likely to have T2DM (23). Li et al. reported ENST00000550337.1 upregulation in blood had high diagnostic value for identifying pre-DM and T2DM in patients from a Chinese cohort (25).
Therefore, to further investigate which lncRNAs may be involved in DM pathogenesis and used as potential biomarkers of this disease, we performed a systematic review of the literature on the subject. Moreover, bioinformatics analyses were performed to investigate the regulatory and functional roles of dysregulated lncRNAs in DM pathogenesis.
Materials and Methods
Search Strategy, Eligibility of Studies, and Data Extraction
This systematic review was designed and described in accordance with current guidelines (30, 31), and its protocol was registered at PROSPERO (http://www.crd.york.ac.uk/PROSPERO), under the identification: CRD42019124368. PubMed and EMBASE repositories were searched to retrieve all articles that investigated lncRNA expressions in T1DM or T2DM patients compared to non-diabetic controls. The research question was constructed based on the PICOS strategy (31), as follows: P (Population): patients with T1DM or T2DM; I (Intervention): lncRNA expression; C (Comparators): healthy control groups; O (Outcomes): DM; S (Study designs): case–control study, cross-sectional or cohort. The following medical subject headings (MeSH) were used: (“diabetes mellitus” OR “diabetes mellitus, type 1” OR “diabetes mellitus, type 2”) AND (“RNA, long noncoding” OR “untranslated RNA”). The search was restricted to English, Spanish, or Portuguese language papers and was finished on April 2020. Reference lists from all included articles were also manually reviewed in order to identify other relevant citations. Moreover, studies were also searched in the GEO database (https://www.ncbi.nlm.nih.gov/geo/).
We included original articles that analyzed lncRNA expressions in patients with T1DM or T2DM (cases) and subjects without DM (controls). Studies that did not have an appropriate control group were excluded. Two researchers (CD and NL) independently reviewed titles and abstracts of all articles to evaluate if they were eligible for inclusion in this systematic review.
Results were independently collected by two investigators (CD and NL) using a standardized abstraction form (31). Discrepancies between investigators were solved by discussion between them and, when necessary, a third reviewer (DC) was consulted. The following information was collected from each study included in this review: 1) characteristics of studies and samples; 2) information regarding lncRNA expressions, quantification method, analyzed tissue, and number of lncRNAs investigated; and 3) lncRNA expression profile in case and control groups.
Evaluation of lncRNA Putative Target Genes and Functional Enrichment Analysis
Potential target genes for the consistently dysregulated lncRNAs in DM were searched using lncRNA2Target v2.0 (32) and starBase (33). The criteria for selecting the consistently dysregulated lncRNAs were: 1) lncRNAs with concordant results in ≥75% of the studies in which they were analyzed; and 2) lncRNAs analyzed in at least three studies. Statistical significances were reported after Benjamini–Hochberg (q-value) corrections for multiple comparisons (34). To better understand the biological relevance of lncRNA target genes, a network analysis was executed using PathDIP (accessed 23th April 2020) (35). The nomenclature of mRNAs and lncRNAs were unified based on HUGO gene nomenclature committee (HGNC) and LNCipedia v5.2, respectively.
Results
Literature Search and Characteristics of Eligible Studies
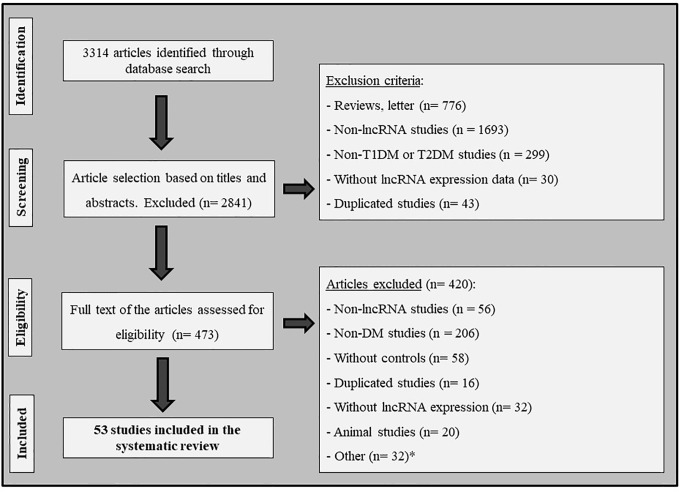
Figure 1 shows the flowchart illustrating the strategy used to identify and select articles for inclusion in this systematic review. Following the search criteria, a total of 3,314 publications were retrieved from databases; however, after careful full text analysis, only 53 articles fulfilled the eligibility criteria and were included in the present review. The main characteristics of these studies are shown in Table 1 and the Supplementary Table 1.
Figure 1.
Flowchart illustrating the search strategy used to identify studies that investigated the association between lncRNAs and diabetes mellitus. *Other: articles excluded due to lack of important information; studies with cell lines; and studies written in other idioms (not English, Spanish or Portuguese).
Table 1.
Characteristics of studies included in the systematic review.
| Author, year [Reference] | Sample size Case/Control | Tissue | Method | Total number of studied lncRNAs | Statistically significant lncRNAs | |
|---|---|---|---|---|---|---|
| Upregulated | Downregulated | |||||
| Akerman et al. 2017 (17) | 10 T2DM patients/50 controls | Pancreatic islets | RNA-seq and qPCR | 2,373 | 0 | 16 |
| Alikhah et al. 2018 (36) | 18 T2DM patients/18 controls | PBMCs | qPCR | 1 | 0 | 0 |
| Carter et al. 2015 (23) | 5 T2DM patients/5 controls 47 T2DM patients/49 controls (validation) |
Serum | Microarray and qPCR | 84 | 0 | 1 |
| Chen et al. 2019 (37) | 25 DM patients/20 controls | Serum | qPCR | 1 | 0 | 0 |
| Chen et al. 2018 (38) | 27 DM patients/17 controls | Serum | qPCR | 1 | 0 | 0 |
| Cheng et al. 2019 (39) | 30 DM patients/30 controls | Peripheral blood | qPCR | 1 | 1 | 0 |
| Dai et al. 2020 (40) | 60 T2DM patients/60 controls | Plasma | qPCR | 1 | 0 | 0 |
| Das et al. 2018 (41) | 5 T2DM patients/5 controls | CD14+ monocytes | qPCR | 1 | 1 | 0 |
| De Gonzalo-Calvo et al. 2016 (42) | 48 T2DM patients/12 controls | Serum | qPCR | 12 | 1 | 3 |
| Erfanian Omidvar et al. 2019 (24) | 100 T2DM patients/100 controls | PBMCs | qPCR | 2 | 0 | 2 |
| Esguerra et al. 2020 (43) | 9 T2DM patients/10 controls | Pancreatic islets | qPCR | 1 | 1 | 0 |
| Fadista et al. 2014 (44) | 12 T2DM patients/51 controls | Pancreatic islets | RNA-seq | 493 | NA | NA |
| Fawzy et al. 2020 (45) | 53 T2DM patients/110 controls | Plasma | qPCR | 2 | 1 | 1 |
| Gao et al. 2014 (46) | 5 T2DM patients/4 controls | Lateral quadriceps muscle biopsy | qPCR | 1 | 0 | 1 |
| Jiao et al. 2019 (47) | 43 DM patients/48 controls | Serum | qPCR | 1 | 1 | 0 |
| Kameswaran et al. 2014 (48) | 4 T2DM patients/3 controls | Pancreatic islets | qPCR | 1 | 0 | 1 |
| Li et al. 2018 (49) | 10 T2DM patients/10 controls | Liver biopsy | qPCR | 1 | 1 | 0 |
| Li et al. 2019 (50) | 56 T2DM patients/40 controls | Serum | qPCR | 1 | 0 | 0 |
| Li et al. 2018 (51) | 63 DM patients/56 controls | Plasma | qPCR | 1 | 0 | 0 |
| Li et al. 2018 (25) | 6 T2DM patients/6 controls 20 T2DM patients/20 controls (validation) |
Peripheral blood | Microarray and qPCR | 41,000 | 14 | 3 |
| Liu et al. 2019 (52) | 90 T2DM patients/30 controls | Serum | qPCR | 1 | 1 | 0 |
| Luo et al. 2018 (53) | 6 T2DM patients/6 controls 26 T2DM patients/26 controls (validation) |
PBMCs | Microarray and qPCR | NA | 316 | 126 |
| Ma et al. 2020 (54) | 5 T2DM patients/5 controls 122 T2DM patients/125 controls (validation) |
PBMCs | Array and qPCR | 41,000 | 44 | 24 |
| Mansoori et al. 2018 (26) | 100 T2DM patients/100 controls | PBMCs | qPCR | 2 | 0 | 2 |
| Mohamadi et al. 2019 (55) | 100 T2DM patients/100 controls | PBMCs | qPCR | 2 | 0 | 0 |
| Móran et al. 2012 (56) | 16 T2DM patients/19 controls | Pancreatic islets | qPCR | 13 | 1 | 1 |
| Motterle et al. 2017 (57) | 10 T2DM patients/10 controls | Pancreatic islets | qPCR | 1 | 0 | 1 |
| Pengyu et al. 2020 (58) | 4 T2DM patients/4 controls | Serum | RNAseq and qPCR | NA | 68763 | 28523 |
| Pradas-Juni et al. 2020 (59) | 4 T2DM patients/4 controls | Liver | RNAseq | 13,805 | 126 | 384 |
| Reddy et al. 2014 (60) | 4 T2DM patients/4 controls | Monocytes | qPCR | 1 | 1 | 0 |
| Ren et al. 2019 (61) | 178 T2DM patients/44 controls | Plasma | qPCR | 1 | 0 | 0 |
| Ruan et al. 2018 (19) | 3 T2DM patients/3 controls 30 T2DM patients/30 controls (validation) |
Blood | Microarray and qPCR | 40,914 | 2269 | |
| 30 T2DM patients/30 controls | Exosome serum/exosome-free serum | qPCR | 1 | 1 | 0 | |
| Saeidi et al. 2018 (27) | 100 T2DM patients/100 controls | PBMCs | qPCR | 2 | 0 | 2 |
| Sathishkumar et al. 2018 (21) | 30 T2DM patients/32 controls | PBMCs | qPCR | 17 | 13 | 2 |
| Shaker et al. 2019 (62) | 30 T2DM patients/81 controls | Blood | qPCR | 2 | 2 | 0 |
| Toraih et al. 2019 (63) | 55 T2DM patients/108 controls | Plasma | qPCR | 4 | 4 | 0 |
| Wan et al. 2020 (64) | 32 T2DM patients/32 controls | Serum | qPCR | 1 | 1 | 0 |
| Wang et al. 2018 (65) | 296 T2DM patients/56 controls | Serum | qPCR | 1 | 0 | 0 |
| Wang et al. 2018 (66)* | 2 T2DM patients/2 controls | Blood | Microarray and qPCR | NA | NA | NA |
| Wang et al. 2017 (28) | 6 T2DM patients/6 controls 60 T2DM patients/60 controls (validation) |
Peripheral blood | Microarray and qPCR | NA | 39 | 16 |
| Wang et al. 2020 (67) | 156 T2DM/100 controls | Peripheral blood | qPCR | 3 | 3 | 0 |
| Yang et al. 2018 (68) | 8 DM patients/8 controls | Serum | qPCR | 1 | 1 | 0 |
| Yang et al. 2018 (69) | 6 DM patients/6 controls | Serum | qPCR | 1 | 1 | 0 |
| Yang et al. 2018 (70) | 36 DM patients/41 controls | Serum | qPCR | 1 | 0 | 0 |
| Yang et al. 2019 (71) | DM patients/controls | Serum | Array | 30,586 | 245 | 680 |
| Yin et al. 2019 (72) | 62 DM patients/48 controls | Plasma | qPCR | 1 | 0 | 0 |
| Zha et al. 2019 (73) | 244 T2DM patients/126 controls | Plasma | qPCR | 1 | 0 | 1 |
| Zhang et al. 2018 (74) | 28 DM patients/30 controls | Serum | qPCR | 1 | 0 | 1 |
| Zhang et al. 2020 (75) | 99 T2DM patients/50 controls | Serum | qPCR | 1 | 0 | 1 |
| Zhang et al. 2017 (76) | 30 DM patients/28 controls | Plasma | Microarray | NA | NA | NA |
| Zhang et al. 2019 (77) | 24 T2DM patients/26 controls | Serum | qPCR | 1 | 1 | 0 |
| Zhang et al. 2019 (78) | 244 T2DM patients/102 controls | Plasma | qPCR | 1 | 0 | 0 |
| Zhang et al. 2019 (79) | 60 DM patients/60 controls | Plasma | qPCR | 1 | 0 | 0 |
*Abstract from congress. DM, diabetes mellitus; NA, information not available; PBMCs, Peripheral blood mononuclear cells; qPCR, quantitative real time PCR; RNA seq, RNA sequencing; T2DM, type 2 diabetes mellitus.
The number of lncRNAs differentially expressed between case and control groups from the different included studies varied from 1 (23, 39, 41, 43, 46–49, 52, 57, 60, 64, 68, 69, 73–75, 77) to 97,286 (58), and the sample sizes ranged from 4 (66) to 370 (73). Among the 53 studies included in this systematic review, 74% of them analyzed T2DM patients, while 26% did not report which DM type patients had. The tissues most analyzed were serum, plasma, and peripheral blood mononuclear cells (PBMCs).
Differentially Expressed lncRNAs in DM
As shown in the Supplementary Table 2, 623 lncRNAs were reported as being dysregulated in patients with DM from one study (17, 21, 24–28, 41, 42, 44, 47, 54, 55, 57–60, 64, 73, 75), while only seven were dysregulated in cases in two studies (ENST00000550337.1, Pluto, LncRNAp3134, n335556, n336109, n342533, and Pvt1) (17, 19, 21, 25, 28, 63, 66, 67). Eight lncRNAs were dysregulated in patients from three or more studies, being chosen for further evaluation (Supplementary Table 2 and Table 2). Among these eight lncRNAs, those showing concordant results in more than 75% of the studies were considered consistently dysregulated in DM. Thus, as shown in Table 2, six lncRNAs were consistently dysregulated in patients with DM (upregulated: Anril, Hotair, Malat1, Miat, and Kcnq1ot1; downregulated: Meg3) compared to controls. GAS5 and H19 were upregulated in patients from some studies and downregulated in others, which could be explained by differences in the tissue types analyzed (serum, pancreatic islets, liver, plasma, and PBMCs) (Table 2).
Table 2.
LncRNAs differentially expressed in at least three studies included in the systematic review.
| LncRNA | Reference | Samples | Tissue | Change of expression |
|---|---|---|---|---|
| ANRIL | Sathishkumar et al. (21) | T2DM patients | PBMCs | Up |
| Toraih et al. (63) | T2DM patients | Plasma | Up | |
| Zhang and Wang (77) | T2DM patients | Serum | Up | |
| GAS5 | Carter et al. (23) | T2DM patients | Serum | Down |
| Esguerra et al. (43) | T2DM patients | Pancreatic islets | Up | |
| Sathishkumar et al. (21) | T2DM patients | PBMCs | Up | |
| H19 | Cheng et al. (39) | T2DM patients | Peripheral blood | Up |
| Fawzy et al. (45) | T2DM patients | Plasma | Up | |
| Gao et al. (46) | T2DM patients | Muscle | Down | |
| HOTAIR | Li et al. (49) | T2DM patients | Liver | Up |
| Sathishkumar et al. (21) | T2DM patients | PBMCs | Up | |
| Shaker et al. (62) | T2DM patients | Blood | Up | |
| Kcnq1ot1 | Móran et al. (56) | T2DM patients | Pancreatic islets | Up |
| Yang et al. (68) | DM patients | Serum | Up | |
| Yang et al. (69) | DM patients | Serum | Up | |
| MALAT1 | Liu et al. (52) | T2DM patients | Serum | Up |
| Luo et al. (53) | T2DM patients | Blood | Up | |
| Sathishkumar et al. (21) | T2DM patients | PBMCs | Up | |
| Shaker et al. (62) | T2DM patients | Blood | Up | |
| Toraih et al. (63) | T2DM patients | Plasma | Up | |
| MEG3 | Kameswaran et al. (48) | T2DM patients | Pancreatic islets | Down |
| Luo et al. (53) | T2DM patients | Blood | Down | |
| Sathishkumar et al. (21) | T2DM patients | PBMCs | Up | |
| Zhang et al. (74) | DM patients | Serum | Down | |
| MIAT | De Gonzalo-Calvo et al. (42) | T2DM patients | Serum | Up |
| Sathishkumar et al. (21) | T2DM patients | PBMCs | Up | |
| Toraih et al. (63) | T2DM patients | Plasma | Up |
DM, diabetes mellitus; PBMCs, Peripheral blood mononuclear cells; T2DM, type 2 diabetes mellitus.
Putative Target Genes and Enrichment Pathway Analysis of the Six Differentially Expressed lncRNAs in Human Samples
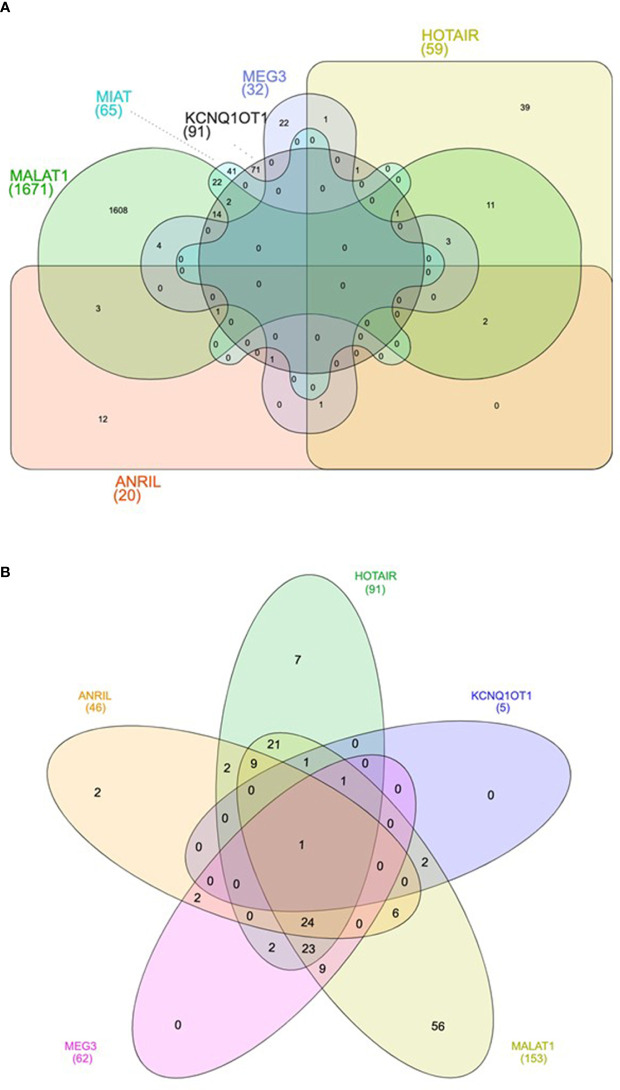
Bioinformatics analyses were carried out to find putative targets and biological pathways regulated by the six lncRNAs (Anril, Hotair, Malat1, Miat, Kcnq1ot1, and Meg3) consistently dysregulated in samples of DM patients. These six lncRNAs regulate together the expression of 1,860 unique target genes (Supplementary Table 3). Malat1 has the largest number of target genes (1,671), followed by Kcnq1ot1 (91), Miat (65), and Hotair (59), while Meg3 and Anril have the lowest number of targets (32 and 20, respectively) (Figure 2A and Supplementary Table 3). Among the 1,860 target genes, 1,307 were protein coding genes, 287 were pseudogenes, 100 were small nuclear RNAs (snRNAs), and 225 were other type of ncRNAs, including microRNAs, rRNA, tRNA, and mitochondrial RNA (mtRNA) (Supplementary Table 3).
Figure 2.
Venn diagram showing the shared target genes (A) and pathways (B) of the six lncRNAs consistently dysregulated in DM.
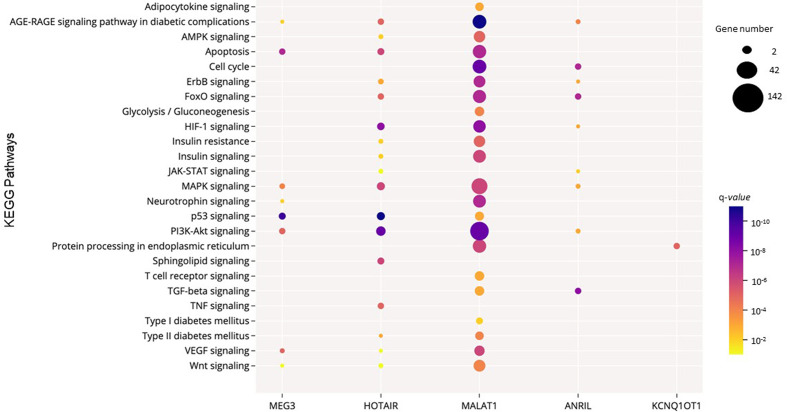
Next, to further explore the functional consequences of the dysregulation of the six lncRNAs of interest, we performed functional enrichment analysis of their protein-encoding target genes using pathways maps from the KEGG repository. As a result, a total of 168 unique pathways were enriched for lncRNA target genes (Supplementary Table 4). Moreover, as demonstrated in Figure 2B, only one pathway is shared among the five lncRNAs (Anril, Hotair, Malat1, Kcnq1ot1, and Meg3): Kaposi sarcoma-associated herpes virus infection. Many of the 168 pathways are well established to be involved in DM pathogenesis, such as PI3K/Akt, MAPK, apoptosis, AGE/RAGE, and FoxO (Figure 3 and Supplementary Table 4). Of note, we could not find any significant KEGG pathway for Miat.
Figure 3.
Significant KEGG pathways potentially regulated by the consistently dysregulated lncRNAs in DM. The size and the color of the dots represent the gene number and the range of the pathway’s q-value, respectively. The y-axis represents the KEGG pathways, and the x-axis shows the five lncRNAs that participated in each selected pathway. MIAT was not significantly enriched in these selected pathways. Q-values: P-values corrected for multiple tests using the Benjamini–Hochberg method.
Discussion
Currently, several studies have reported the association between epigenetic mechanisms and DM development [reviewed in (6, 7, 80, 81)]. In this context, lncRNAs are a class of ncRNAs that appear to be involved in DM pathogenesis (10). Thus, here, we performed a systematic review to further investigate which lncRNAs are mainly associated with DM. Our results demonstrated six lncRNAs were consistently dysregulated in patients with DM. Anril, Hotair, Kncq1ot1, Malat1, and Miat were consistently upregulated, while Meg3 was downregulated in diabetic cases compared to controls.
Malat1 (metastasis‐associated lung adenocarcinoma transcript 1, also known as Neat2) is one of the most analyzed lncRNAs in T2DM samples. Here, our qualitative analysis shows this lncRNA is upregulated in serum, plasma, and PBMCs of T2DM patients (21, 52, 53, 62, 63). Moreover, studies performed in animal models of DM indicate that the expression of Malat1 is increased in liver, macrophages, and serum of different murine models of T2DM compared to controls (20, 27, 52). Malat1 is a highly conserved nuclear lncRNA initially identified as a predictor of lung cancer metastasis (82). Several studies have reported the involvement of this lncRNA in signaling pathways related to DM pathogenesis, such as PI3K/Akt (83), NF-κB (84), MAPK/ERK (85, 86), and Wnt/β-catenin (87). Accordingly, our in silico analysis shows Malat1 is involved in a number of pathways involved in DM and its complications that, besides PI3K/Akt, MAPK, and Wnt, include apoptosis, insulin, cell cycle, AMPK, FoxO, ErbB, HIF-1, AGE/RAGE, adipocytokines, and protein processing in endoplasmic reticulum. In agreement with Malat1 upregulation in T2DM, its expression was also increased in human umbilical vein endothelial cells (HUVECs) cultured with high-glucose (HG) and positively correlated with inflammatory cytokine (IL6 and TNF) levels (88). Additionally, this lncRNA was upregulated in mice with diabetic retinopathy (DR) compared to control animals (89).
Hotair was also consistently upregulated in liver, blood, and PBMCs of patients with T2DM (21, 38, 62). Accordingly, Li et al. reported this lncRNA was upregulated in liver of two T2DM murine models (db/db and C57BL/6J mice) treated with high-fat diet (49). Hotair is located within the HOMEOBOX C (HOXC) gene cluster on chromosome 12q13.13 and is involved in cellular proliferation, inhibition of apoptosis, genomic instability, angiogenesis, and metastasis (90–92). Moreover, Hotair upregulation promotes hepatic insulin resistance via the Akt/GSK pathway (38), which might partially explain its association with T2DM. Our in silico analysis demonstrates the involved of Hotair in several DM-related pathways, such apoptosis, PI3K-Akt, MAPK, HIF-1, TNF, and FoxO. This lncRNA seems also to be involved in the pathogenesis of diabetic chronic complications. Hotair was upregulated in serum of patients with different degrees of DR compared to healthy controls, and its expression was able to distinguish patients with non-proliferative DR from those with proliferative DR (62). Increased expression of Hotair was also found in kidney of patients with diabetic kidney disease (DKD) and in kidneys of db/db and STZ-induced diabetic mice (93). Accordingly, mouse podocytes cultured under HG conditions also expressed high levels of Hotair (93).
In addition to Malat1 and Hotair, the lncRNA Anril was also increased in PBMCs, plasma, or serum of patients with T2DM compared to controls (21, 63, 77). This lncRNA has been associated with several types of cancer, such as gliomas, breast, lung, liver, colon, and thyroid cancers [reviewed in (94)]. Anril seems also to be involved in DR pathogenesis, since its expression was upregulated in human retinal endothelial cells (HRECs) cultured under HG conditions and in retinal tissue of STZ-induced diabetic mice (95). Blockade of Anril prevented HG-induced VEGF upregulation in HRECs, which is a key angiogenic factor in DR pathogenesis (95, 96). In line with these findings, Zhang et al. showed Anril overexpression in diabetic rats complicated with cerebral infarction upregulated VEGF and improved angiogenesis through activation of the NF-κB pathway (97). Our in silico analysis indicates that Anril is also involved in the TGFβ, PI3K-Akt, MAPK, cell cycle, FoxO, and AGE/RAGE pathways, which are known pathways related to DM and its chronic complications.
Kcnq1ot1 is another lncRNA consistently upregulated in islets and serum of patients with T2DM (56, 68, 69). Kcnq1ot1 is an antisense lncRNA that seems to regulate the expression of both neighboring or distant genes (98), including the CDKN1C, a known regulator of beta-cell development (99). Interestingly, a meta-analysis study, including 51,075 DM cases and 10,6134 controls, demonstrated the association between the rs231362 polymorphism in the Kcnq1ot1 gene and risk for T2DM [OR 1.10 (95% CI 1.06–1.15), P < 10−4] (100). Our in silico analysis indicates this lncRNA regulates genes from the protein processing in endoplasmic reticulum stress pathway.
Miat was also consistently upregulated in serum, plasma, or PBMCs of T2DM patients compared to controls (21, 42, 63). This lncRNA seems to act as a regulator of several signaling pathways related to cellular function, such as proliferation and apoptosis and as a competitive endogenous RNA (101). Additionally, Miat seems to be involved in diabetic complications (102). Miat was upregulated in the myocardium of diabetic rats, while its knockdown inhibited apoptosis in cardiomyocytes exposed to HG (103). In contrast, in renal tubuli of diabetic rats, Miat was downregulated compared to control rats and negatively correlated to serum creatinine levels (104). Growing evidence has also shown Miat dysregulation in a number of diseases, such as myocardial infarction, age-related cataract, different cancers, and ischemic stroke [reviewed in (101)]. Here, we were not able to find any significant KEGG pathway for Miat; therefore, how this lncRNA is involved in DM and other diseases still needs to be clarified.
Our systematic review indicates Meg3 is downregulated in islets, whole blood, and serum of patients with DM (48, 53, 74). Accordingly, this lncRNA was downregulated in islets of db/db mice (105) and in serum of diabetic patients with DR compared to controls (74). However, it was upregulated in liver or primary hepatocytes of different T2DM murine models (59, 106). In a murine beta-cell line (MIN6), Meg3 suppression led to increased apoptosis due to caspase-3 and Bax upregulation and Bcl2 downregulation (105). In addition, Meg3 seems to regulate insulin synthesis and secretion since its blockade in murine beta-cells decreased the expression of key transcription factors involved in insulin synthesis (Pdx-1 and mafA); thus, decreasing insulin gene transcription (105). Besides apoptosis, our in silico analysis suggests this lncRNA is involved in PI3K/Akt, VEGF, and MAPK pathways.
Of note, our bioinformatics analysis also demonstrated that Anril, Hotair, Malat1, Kcnq1ot1, and Meg3 regulate genes from the Kaposi sarcoma-associated herpes virus infection (KSHV) pathway. KSHV, also known as human herpesvirus 8, is a human tumor virus associated with the pathogenesis of Kaposi’s sarcoma, primary effusion lymphoma, and Multicentric Castleman’s disease. The KSHV pathway contains genes related to IFN antiviral response, inflammatory cytokines, and cell proliferation pathways [https://www.genome.jp/kegg/kegg2.html]. Interestingly, the association between KSHV and DM was previously reported by observational studies (107, 108). Cui et al. described that patients with T2DM had an elevated risk of KSHV (107). Accordingly, Piras et al. showed 58% of T2DM patients were seropositive for KSHV vs. 27% of the healthy subjects (108). Even though the mechanisms behind this association are unknown, this virus causes metabolic changes that might lead to altered insulin uptake and accumulation of neutral lipids in cells and also induce an impairment of the immune system [review in (109)], which are mechanisms related to DM pathogenesis.
Even though this systematic review indicates a group of lncRNAs consistently associated with DM and the pathways possible regulated by them, it has few limitations. First, there is no official nomenclature for lncRNAs; thus, we cannot exclude the possibility that we have lost some information. Second, some studies, especially those using RNAseq and microarrays technologies, did not inform which were the differentially expressed lncRNAs or their expression pattern (up- or downregulation) (19, 25, 44, 53, 54, 58, 66, 71, 76). Third, studies used different techniques to quantify lncRNA expressions and usually did not provide the expression values, only the pattern of expression of the dysregulated lncRNAs; therefore, making impossible to perform a reliable quantitative analysis of the data (meta-analysis). Fourth, most of the studies investigated lncRNAs in patients with T2DM or did not inform the type of DM, evidencing the lack of studies in T1DM population. In this context, four of the dysregulated lncRNAs found in this study were analyzed only in T2DM patients (Anril, Hotair, Malat1, and Miat). Thus, our results are more representative of this type of DM. Fifth, although six lncRNAs were consistently dysregulated in patients with DM compared to controls, it was not possible to perform a stratified analysis by tissue type since the number of studies that evaluated the same lncRNA in a given tissue is very small. Lastly, as commented above, Anril, Hotair, Kcnq1ot1, Malat, Meg3, and Miat lncRNAs seem to be dysregulated in patients with DR and DKD. However, most of the studies included in this systematic review did not report the percentage of patients with these diabetic chronic complications. Thus, here, it was impossible to evaluate if presence of diabetic chronic complications is impacting our results. Further studies are required to clarify this point.
In conclusion, our systematic review indicates that six lncRNAs are consistently dysregulated in DM, especially in patients with T2DM. This study also contributes to enlighten the pathways regulated by these lncRNAs and involved in the DM pathogenesis, such as PI3K/Akt, MAPK, apoptosis, AGE/RAGE, and FoxO. Although this systematic review included 53 studies which analyzed lncRNA expression in DM-related tissues, further studies are necessary to better understand the involvement of lncRNAs in the pathogenesis of this complex disease and its chronic complications. As much as lncRNAs seem to be good candidates as biomarkers and therapeutic targets for DM, further investigations on organ-specific distribution of these regulatory molecules may be useful to clarify their role in DM.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
CD designed the study, researched data, performed the analysis, and wrote the manuscript. NL researched data, performed the analysis, and reviewed the manuscript. NC researched data and reviewed the manuscript. TA researched data, performed the bioinformatics analyses, contributed to discussion, and reviewed the manuscript. DC designed the study, contributed to the discussion, and wrote and reviewed the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This study was partially supported by grants from the Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), Fundo de Incentivo à Pesquisa e Eventos (FIPE, number 2018-0470) at Hospital de Clínicas de Porto Alegre, Fundação de Amparo à Pesquisa do Estado do Rio Grande do Sul (FAPERGS) (Edital FAPERGS/CNPq 12/2014 PRONEX - Processo n° 16/2551 - 0000483-8), and Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES). DC is recipient of scholarships from CNPq, while CD and TSA are recipients from scholarships from CAPES, and NL is recipient of scholarships from FAPERGS.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fendo.2021.602597/full#supplementary-material
References
- 1. American Diabetes Association - ADA . 2. Classification and Diagnosis of Diabetes: Standards of Medical Care in Diabetes-2019. Diabetes Care (2019) 42:S13–28. 10.2337/dc19-S002 [DOI] [PubMed] [Google Scholar]
- 2. International Diabetes Federation - IDF . IDF Diabetes Atlas. 9th edn. Brussels, Belgium: International Diabetes Federation; (2019). [Google Scholar]
- 3. Katsarou A, Gudbjornsdottir S, Rawshani A, Dabelea D, Bonifacio E, Anderson BJ, et al. Type 1 diabetes mellitus. Nat Rev Dis Primers (2017) 3:17016. 10.1038/nrdp.2017.16 [DOI] [PubMed] [Google Scholar]
- 4. Ilonen J, Lempainen J, Veijola R. The heterogeneous pathogenesis of type 1 diabetes mellitus. Nat Rev Endocrinol (2019) 15:635–50. 10.1038/s41574-019-0254-y [DOI] [PubMed] [Google Scholar]
- 5. Nyaga DM, Vickers MH, Jefferies C, Perry JK, O’Sullivan JM. The genetic architecture of type 1 diabetes mellitus. Mol Cell Endocrinol (2018) 477:70–80. 10.1016/j.mce.2018.06.002 [DOI] [PubMed] [Google Scholar]
- 6. Zhang H, Pollin TI. Epigenetics Variation and Pathogenesis in Diabetes. Curr Diabetes Rep (2018) 18:121. 10.1007/s11892-018-1091-4 [DOI] [PubMed] [Google Scholar]
- 7. Dhawan S, Natarajan R. Epigenetics and Type 2 Diabetes Risk. Curr Diabetes Rep (2019) 19:47. 10.1007/s11892-019-1168-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Loh M, Zhou L, Ng HK, Chambers JC. Epigenetic disturbances in obesity and diabetes: Epidemiological and functional insights. Mol Metab (2019) 27S:S33–41. 10.1016/j.molmet.2019.06.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet (2006) 15(Spec No 1):R17–29. 10.1093/hmg/ddl046 [DOI] [PubMed] [Google Scholar]
- 10. Guo J, Liu Z, Gong R. Long noncoding RNA: an emerging player in diabetes and diabetic kidney disease. Clin Sci (Lond) (2019) 133:1321–39. 10.1042/CS20190372 [DOI] [PubMed] [Google Scholar]
- 11. St Laurent G, Wahlestedt C, Kapranov P. The Landscape of long noncoding RNA classification. Trends Genet (2015) 31:239–51. 10.1016/j.tig.2015.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res (2012) 22:1775–89. 10.1101/gr.132159.111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet (2016) 17:47–62. 10.1038/nrg.2015.10 [DOI] [PubMed] [Google Scholar]
- 14. Chen X, Sun Y, Cai R, Wang G, Shu X, Pang W. Long noncoding RNA: multiple players in gene expression. BMB Rep (2018) 51:280–9. 10.5483/BMBRep.2018.51.6.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Sun M, Kraus WL. Minireview: Long noncoding RNAs: new “links” between gene expression and cellular outcomes in endocrinology. Mol Endocrinol (2013) 27:1390–402. 10.1210/me.2013-1113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Paraskevopoulou MD, Hatzigeorgiou AG. Analyzing MiRNA-LncRNA Interactions. Methods Mol Biol (2016) 1402:271–86. 10.1007/978-1-4939-3378-5_21 [DOI] [PubMed] [Google Scholar]
- 17. Akerman I, Tu Z, Beucher A, Rolando DMY, Sauty-Colace C, Benazra M, et al. Human Pancreatic beta Cell lncRNAs Control Cell-Specific Regulatory Networks. Cell Metab (2017) 25:400–11. 10.1016/j.cmet.2016.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Jin F, Wang N, Zhu Y, You L, Wang L, De W, et al. Downregulation of Long Noncoding RNA Gas5 Affects Cell Cycle and Insulin Secretion in Mouse Pancreatic β Cells. Cell Physiol Biochem (2018) 43:2062–73. 10.1159/000484191 [DOI] [PubMed] [Google Scholar]
- 19. Ruan Y, Lin N, Ma Q, Chen R, Zhang Z, Wen W, et al. Circulating LncRNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet β-Cell Function. Cell Physiol Biochem (2018) 46:335–50. 10.1159/000488434 [DOI] [PubMed] [Google Scholar]
- 20. Yan C, Chen J, Chen N. Long noncoding RNA MALAT1 promotes hepatic steatosis and insulin resistance by increasing nuclear SREBP-1c protein stability. Sci Rep (2016) 6:22640. 10.1038/srep22640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sathishkumar C, Prabu P, Mohan V, Balasubramanyam M. Linking a role of lncRNAs (long non-coding RNAs) with insulin resistance, accelerated senescence, and inflammation in patients with type 2 diabetes. Hum Genomics (2018) 12:41. 10.1186/s40246-018-0173-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Feng SD, Yang JH, Yao CH, Yang SS, Zhu ZM, Wu D, et al. Potential regulatory mechanisms of lncRNA in diabetes and its complications. Biochem Cell Biol (2017) 95:361–7. 10.1139/bcb-2016-0110 [DOI] [PubMed] [Google Scholar]
- 23. Carter G, Miladinovic B, Patel AA, Deland L, Mastorides S, Patel NA. Circulating long noncoding RNA GAS5 levels are correlated to prevalence of type 2 diabetes mellitus. BBA Clin (2015) 4:102–7. 10.1016/j.bbacli.2015.09.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Erfanian Omidvar M, Ghaedi H, Kazerouni F, kalbasi S, Shanaki M, Miraalamy G, et al. Clinical significance of long noncoding RNA VIM-AS1 and CTBP1-AS2 expression in type 2 diabetes. J Cell Biochem (2019) 120:9315–23. 10.1002/jcb.28206 [DOI] [PubMed] [Google Scholar]
- 25. Li X, Zhao Z, Gao C, Rao L, Hao P, Jian D, et al. The Diagnostic Value of Whole Blood lncRNA ENST000005503371 for Pre-Diabetes and Type 2 Diabetes Mellitus. Exp Clin Endocrinol Diabetes (2017) 125:377–83. 10.1055/s-0043-100018 [DOI] [PubMed] [Google Scholar]
- 26. Mansoori Z, Ghaedi H, Sadatamini M, Vahabpour R, Rahimipour A, Shanaki M, et al. Downregulation of long non-coding RNAs LINC00523 and LINC00994 in type 2 diabetes in an Iranian cohort. Mol Biol Rep (2018) 45:1227–33. 10.1007/s11033-018-4276-7 [DOI] [PubMed] [Google Scholar]
- 27. Saeidi L, Ghaedi H, Sadatamini M, Vahabpour R, Rahimipour A, Shanaki M, et al. Long non-coding RNA LY86-AS1 and HCG27_201 expression in type 2 diabetes mellitus. Mol Biol Rep (2018) 45(6):2601–8. 10.1007/s11033-018-4429-8 [DOI] [PubMed] [Google Scholar]
- 28. Wang X, Chang X, Zhang P, Fan L, Zhou T, Sun K. Aberrant Expression of Long Non-Coding RNAs in Newly Diagnosed Type 2 Diabetes Indicates Potential Roles in Chronic Inflammation and Insulin Resistance. Cell Physiol Biochem (2017) 43:2367–78. 10.1159/000484388 [DOI] [PubMed] [Google Scholar]
- 29. He X, Ou C, Xiao Y, Han Q, Li H, Zhou S. LncRNAs: Key players and novel insights into diabetes mellitus. Oncotarget (2017) 8:71325–41. 10.18632/oncotarget.19921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Stroup DF, Berlin JA, Morton SC, Olkin I, Williamson GD, Rennie D, et al. Meta-analysis of observational studies in epidemiology: a proposal for reporting. Meta-analysis Of Observational Studies in Epidemiology (MOOSE) group. JAMA (2000) 283:2008–12. 10.1001/jama.283.15.2008 [DOI] [PubMed] [Google Scholar]
- 31. Moher D, Liberati A, Tetzlaff J, Altman DG, Group P. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. BMJ (2009) 339:b2535. 10.1136/bmj.b2535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cheng L, Wang P, Tian R, Wang S, Guo Q, Luo M, et al. LncRNA2Target v2.0: a comprehensive database for target genes of lncRNAs in human and mouse. Nucleic Acids Res (2019) 47:D140–4. 10.1093/nar/gky1051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Li JH, Liu S, Zhou H, Qu LH, Yang JH. starBase v2.0: decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA interaction networks from large-scale CLIP-Seq data. Nucleic Acids Res (2014) 42:D92–7. 10.1093/nar/gkt1248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Benjamini Y, Hochberg Y. Controlling the False Discovery Rate: A Practical and Powerful Approach to Multiple Testing. J R Stat Soc.: Ser B (Methodol) (1995) 57:289–300. 10.1111/j.2517-6161.1995.tb02031.x [DOI] [Google Scholar]
- 35. Rahmati S, Abovsky M, Pastrello C, Jurisica I. pathDIP: an annotated resource for known and predicted human gene-pathway associations and pathway enrichment analysis. Nucleic Acids Res (2017) 45:D419–26. 10.1093/nar/gkw1082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Alikhah A, Pahlevan Kakhki M, Ahmadi A, Dehghanzad R, Boroumand MA, Behmanesh M. The role of lnc-DC long non-coding RNA and SOCS1 in the regulation of STAT3 in coronary artery disease and type 2 diabetes mellitus. J Diabetes Its Complications (2018) 32:342–8. 10.1016/j.jdiacomp.2017.12.001 [DOI] [PubMed] [Google Scholar]
- 37. Chen SZ, Zhong HM, Wang Y, Wang ZH, Liang XQ, Li SQ, et al. The clinical significance of long non-coding RNA ANRIL level in diabetic retinopathy. Acta Diabetol (2019) 57:409–18. 10.1007/s00592-019-01442-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Chen Y, Tan S, Liu M, Li J. LncRNA TINCR is downregulated in diabetic cardiomyopathy and relates to cardiomyocyte apoptosis. Scandinavian Cardiovasc J (2018) 52:335–9. 10.1080/14017431.2018.1546896 [DOI] [PubMed] [Google Scholar]
- 39. Cheng XW, Chen ZF, Wan YF, Zhou Q, Wang H, Zhu HQ. Long Non-coding RNA H19 Suppression Protects the Endothelium Against Hyperglycemic-Induced Inflammation via Inhibiting Expression of miR-29b Target Gene Vascular Endothelial Growth Factor a Through Activation of the Protein Kinase B/Endothelial Nitric Oxide Synthase Pathway. Front Cell Dev Biol (2019) 7:11. 10.3389/fcell.2019.00263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Dai R, Sun Z, Qian Y, Zhang B, Han Y, Deng G. LncRNA LUADT1 inhibits cell apoptosis in diabetic retinopathy by regulating miR-383/peroxiredoxin 3 axis. Arch Physiol Biochem (2020) 1–6. 10.1080/13813455.2020.1716016 [DOI] [PubMed]
- 41. Das S, Reddy MA, Senapati P, Stapleton K, Lanting L, Wang M, et al. Diabetes Mellitus-Induced Long Noncoding RNA Dnm3os Regulates Macrophage Functions and Inflammation via Nuclear Mechanisms. Arteriosclerosis Thrombosis Vasc Biol (2018) 38:1806–20. 10.1161/ATVBAHA.117.310663 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. de Gonzalo-Calvo D, Kenneweg F, Bang C, Toro R, van der Meer RW, Rijzewijk LJ, et al. Circulating long-non coding RNAs as biomarkers of left ventricular diastolic function and remodelling in patients with well-controlled type 2 diabetes. Sci Rep (2016) 6:37354. 10.1038/srep37354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Esguerra JLS, Ofori JK, Nagao M, Shuto Y, Karagiannopoulos A, Fadista J, et al. Glucocorticoid induces human beta cell dysfunction by involving riborepressor GAS5 LincRNA. Mol Metab (2020) 32:160–7. 10.1016/j.molmet.2019.12.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Fadista J, Vikman P, Laakso EO, Mollet IG, Esguerra JL, Taneera J, et al. Global genomic and transcriptomic analysis of human pancreatic islets reveals novel genes influencing glucose metabolism. Proc Natl Acad Sci U S A (2014) 111:13924–9. 10.1073/pnas.1402665111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Fawzy MS, Abdelghany AA, Toraih EA, Mohamed AM. Circulating long noncoding RNAs H19 and GAS5 are associated with type 2 diabetes but not with diabetic retinopathy: A preliminary study. Bosnian J Basic Med Sci (2020) 20(3):365–71. 10.17305/bjbms.2019.4533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Gao Y, Wu F, Zhou J, Yan L, Jurczak MJ, Lee HY, et al. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res (2014) 42:13799–811. 10.1093/nar/gku1160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Jiao H, Xie D, Qiao Y. LncRNA PRINs is involved in the development of nephropathy in patients with diabetes via interaction with smad7. Exp Ther Med (2019) 17:3203–8. 10.3892/etm.2019.7307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Kameswaran V, Bramswig NC, McKenna LB, Penn M, Schug J, Hand NJ, et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab (2014) 19:135–45. 10.1016/j.cmet.2013.11.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Li M, Guo Y, Wang XJ, Duan BH, Li L. HOTAIR participates in hepatic insulin resistance via regulating SIRT1. Eur Rev Med Pharmacol Sci (2018) 22:7883–90. 10.26355/eurrev_201811_16414 [DOI] [PubMed] [Google Scholar]
- 50. Li P, Zhang N, Ping F, Gao Y, Cao L. LncRNA SCAL1 inhibits inducible nitric oxide synthase in lung cells under high-glucose conditions. Exp Ther Med (2019) 18:1831–6. 10.3892/etm.2019.7729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wen X, Han XR, Wang YJ, Wang S, Shen M, Zhang ZF, et al. Down-regulated long non-coding RNA ANRIL restores the learning and memory abilities and rescues hippocampal pyramidal neurons from apoptosis in streptozotocin-induced diabetic rats via the NF-κB signaling pathway. J Cell Biochem (2018) 119:5821–33. 10.1002/jcb.26769 [DOI] [PubMed] [Google Scholar]
- 52. Liu SX, Zheng F, Xie KL, Xie MR, Jiang LJ, Cai Y. Exercise Reduces Insulin Resistance in Type 2 Diabetes Mellitus via Mediating the lncRNA MALAT1/MicroRNA-382-3p/Resistin Axis. Mol Ther Nucleic Acids (2019) 18:34–44. 10.1016/j.omtn.2019.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Luo L, Ji LD, Cai JJ, Feng M, Zhou M, Hu SP, et al. Microarray Analysis of Long Noncoding RNAs in Female Diabetic Peripheral Neuropathy Patients. Cell Physiol Biochem (2018) 46:1209–17. 10.1159/000489071 [DOI] [PubMed] [Google Scholar]
- 54. Ma Q, Wang L, Yang Y, Su Y, Wang T, Hou Q, et al. Association between lncRNA and GCKR gene in type 2 diabetes mellitus. Clin Chim Acta (2020) 501:66–71. 10.1016/j.cca.2019.10.004 [DOI] [PubMed] [Google Scholar]
- 55. Mohamadi M, Ghaedi H, Kazerouni F, Erfanian Omidvar M, Kalbasi S, Shanaki M, et al. Deregulation of long noncoding RNA SNHG17 and TTC28-AS1 is associated with type 2 diabetes mellitus. Scandinavian J Clin Lab Invest (2019) 79:519–23. 10.1080/00365513.2019.1664760 [DOI] [PubMed] [Google Scholar]
- 56. Morán I, Akerman I, Van De Bunt M, Xie R, Benazra M, Nammo T, et al. Human β cell transcriptome analysis uncovers lncRNAs that are tissue-specific, dynamically regulated, and abnormally expressed in type 2 diabetes. Cell Metab (2012) 16:435–48. 10.1016/j.cmet.2012.08.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Motterle A, Gattesco S, Peyot ML, Esguerra JLS, Gomez-Ruiz A, Laybutt DR, et al. Identification of islet-enriched long non-coding RNAs contributing to β-cell failure in type 2 diabetes. Mol Metab (2017) 6:1407–18. 10.1016/j.molmet.2017.08.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Pengyu Z, Yan Y, Xiying F, Maoguang Y, Mo L, Yan C, et al. The Differential Expression of Long Noncoding RNAs in Type 2 Diabetes Mellitus and Latent Autoimmune Diabetes in Adults. Int J Endocrinol (2020) 2020:12. 10.1155/2020/9235329 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Pradas-Juni M, Hansmeier NR, Link JC, Schmidt E, Larsen BD, Klemm P, et al. A MAFG-lncRNA axis links systemic nutrient abundance to hepatic glucose metabolism. Nat Commun (2020) 11:644. 10.1038/s41467-020-14323-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Reddy MA, Chen Z, Park JT, Wang M, Lanting L, Zhang Q, et al. Regulation of inflammatory phenotype in macrophages by a diabetes-induced long noncoding RNA. Diabetes (2014) 63:4249–61. 10.2337/db14-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ren S, Zhang Y, Li B, Bu K, Wu L, Lu Y, et al. Downregulation of lncRNA-SRA participates in the development of cardiovascular disease in type II diabetic patients. Exp Ther Med (2019) 17:3367–72. 10.3892/etm.2019.7362 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Shaker OG, Abdelaleem OO, Mahmoud RH, Abdelghaffar NK, Ahmed TI, Said OM, et al. Diagnostic and prognostic role of serum miR-20b, miR-17-3p, HOTAIR, and MALAT1 in diabetic retinopathy. IUBMB Life (2019) 71:310–20. 10.1002/iub.1970 [DOI] [PubMed] [Google Scholar]
- 63. Toraih EA, Abdelghany AA, Abd El Fadeal NM, Al Ageeli E, Fawzy MS. Deciphering the role of circulating lncRNAs: RNCR2, NEAT2, CDKN2B-AS1, and PVT1 and the possible prediction of anti-VEGF treatment outcomes in diabetic retinopathy patients. Graefes Arch Clin Exp Ophthalmol (2019) 257:1897–913. 10.1007/s00417-019-04409-9 [DOI] [PubMed] [Google Scholar]
- 64. Wan W, Wan W, Long Y, Li Q, Jin X, Wan G, et al. Physcion 8-O-β-glucopyranoside exerts protective roles in high glucose-induced diabetic retinopathy via regulating lncRNA NORAD/miR-125/STAT3 signalling. Artif Cells Nanomed Biotechnol (2020) 48:463–72. 10.1080/21691401.2019.1709861 [DOI] [PubMed] [Google Scholar]
- 65. Wang L, Su N, Zhang Y, Wang G. Clinical significance of serum lncRNA cancer susceptibility candidate 2 (CASC2) for chronic renal failure in patients with type 2 diabetes. Med Sci Monitor (2018) 24:6079–84. 10.12659/MSM.909510 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wang W, Liu H, Zhou S, Yu P. Differential expression of lncRNA MEG3 in patients with different glycometabolism and analysis of related functions. Diabetes/Metabol Res Rev (2018) 34:1–2. [Google Scholar]
- 67. Wang X, Cheng Y, Zhang P, Chang X, Sun K. Potentials of long non-coding RNAs to differentiate latent autoimmune diabetes in adults from type 2 diabetes. Int J Clin Exp Med (2020) 13:742–9. [Google Scholar]
- 68. Yang F, Qin Y, Wang Y, Li A, Lv J, Sun X, et al. LncRNA KCNQ1OT1 Mediates Pyroptosis in Diabetic Cardiomyopathy. Cell Physiol Biochem Int J Exp Cell Physiol Biochem Pharmacol (2018) 50:1230–44. 10.1159/000494576 [DOI] [PubMed] [Google Scholar]
- 69. Yang F, Qin Y, Lv J, Wang Y, Che H, Chen X, et al. Silencing long non-coding RNA Kcnq1ot1 alleviates pyroptosis and fibrosis in diabetic cardiomyopathy. Cell Death Dis (2018) 9:13. 10.1038/s41419-018-1029-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Yang H, Kan QE, Su Y, Man H. Long Non-Coding RNA CASC2 Improves Diabetic Nephropathy by Inhibiting JNK Pathway. Exp Clin Endocrinol Diabetes (2018) 127(8):533–7. 10.1055/a-0629-9958 [DOI] [PubMed] [Google Scholar]
- 71. Yang Y, Lv X, Fan Q, Wang X, Xu L, Lu X, et al. Analysis of circulating lncRNA expression profiles in patients with diabetes mellitus and diabetic nephropathy: Differential expression profile of circulating lncRNA. Clin Nephrol (2019) 92:25–35. 10.5414/CN109525 [DOI] [PubMed] [Google Scholar]
- 72. Yin L, Sun Z, Ren Q, Su X, Zhang D. Long non-coding RNA BANCR is overexpressed in patients with diabetic retinopathy and promotes apoptosis of retinal pigment epithelial cells. Med Sci Monitor (2019) 25:2845–51. 10.12659/MSM.913359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Zha T, Su F, Liu X, Yang C, Liu L. Role of Long Non-Coding RNA (LncRNA) LINC-PINT Downregulation in Cardiomyopathy and Retinopathy Progression Among Patients with Type 2 Diabetes. Med Sci Monit (2019) 25:8509–14. 10.12659/MSM.918358 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Zhang D, Qin H, Leng Y, Li X, Zhang L, Bai D, et al. LncRNA MEG3 overexpression inhibits the development of diabetic retinopathy by regulating TGF−β1 and VEGF. Exp Ther Med (2018) 16:2337–42. 10.3892/etm.2018.6451 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Zhang FF, Liu YH, Wang DW, Liu TS, Yang Y, Guo JM, et al. Obesity-induced reduced expression of the lncRNA ROIT impairs insulin transcription by downregulation of Nkx6.1 methylation. Diabetologia (2020) 63:811–24. 10.1007/s00125-020-05090-y [DOI] [PubMed] [Google Scholar]
- 76. Zhang L, Li R, He J, Yang Q, Wu Y, Huang J, et al. Co-expression analysis among microRNAs, long non-coding RNAs, and messenger RNAs to understand the pathogenesis and progression of diabetic kidney disease at the genetic level. Methods (2017) 124:46–56. 10.1016/j.ymeth.2017.05.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Zhang L, Wang YM. Expression and function of lncRNA ANRIL in a mouse model of acute myocardial infarction combined with type 2 diabetes mellitus. J Chin Med Assoc (2019) 82:685–92. 10.1097/JCMA.0000000000000182 [DOI] [PubMed] [Google Scholar]
- 78. Zhang X, Zou X, Li Y, Wang Y. Downregulation of lncRNA BANCR participates in the development of retinopathy among diabetic patients. Exp Ther Med (2019) 17:4132–8. 10.3892/etm.2019.7444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Zhang X, Shi E, Yang L, Fu W, Hu F, Zhou X. LncRNA AK077216 is downregulated in diabetic retinopathy and inhibited the apoptosis of retinal pigment epithelial cells by downregulating miR-383. Endocrine J (2019) 66:1011–6. 10.1507/endocrj.EJ19-0080 [DOI] [PubMed] [Google Scholar]
- 80. Kato M, Natarajan R. Epigenetics and epigenomics in diabetic kidney disease and metabolic memory. Nat Rev Nephrol (2019) 15:327–45. 10.1038/s41581-019-0135-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rosen ED, Kaestner KH, Natarajan R, Patti ME, Sallari R, Sander M, et al. Epigenetics and Epigenomics: Implications for Diabetes and Obesity. Diabetes (2018) 67:1923–31. 10.2337/db18-0537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Ji P, Diederichs S, Wang W, Boing S, Metzger R, Schneider PM, et al. MALAT-1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early-stage non-small cell lung cancer. Oncogene (2003) 22:8031–41. 10.1038/sj.onc.1206928 [DOI] [PubMed] [Google Scholar]
- 83. Dong Y, Liang G, Yuan B, Yang C, Gao R, Zhou X. MALAT1 promotes the proliferation and metastasis of osteosarcoma cells by activating the PI3K/Akt pathway. Tumour Biol J Int Soc Oncodevelopmental Biol Med (2015) 36:1477–86. 10.1007/s13277-014-2631-4 [DOI] [PubMed] [Google Scholar]
- 84. Zhao G, Su Z, Song D, Mao Y, Mao X. The long noncoding RNA MALAT1 regulates the lipopolysaccharide-induced inflammatory response through its interaction with NF-kappaB. FEBS Lett (2016) 590:2884–95. 10.1002/1873-3468.12315 [DOI] [PubMed] [Google Scholar]
- 85. Chen L, Feng P, Zhu X, He S, Duan J, Zhou D. Long non-coding RNA Malat1 promotes neurite outgrowth through activation of ERK/MAPK signalling pathway in N2a cells. J Cell Mol Med (2016) 20:2102–10. 10.1111/jcmm.12904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Liu S, Yan G, Zhang J, Yu L. Knockdown of Long Noncoding RNA (lncRNA) Metastasis-Associated Lung Adenocarcinoma Transcript 1 (MALAT1) Inhibits Proliferation, Migration, and Invasion and Promotes Apoptosis by Targeting miR-124 in Retinoblastoma. Oncol Res (2018) 26:581–91. 10.3727/096504017X14953948675403 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 87. Liang J, Liang L, Ouyang K, Li Z, Yi X. MALAT1 induces tongue cancer cells’ EMT and inhibits apoptosis through Wnt/beta-catenin signaling pathway. J Oral Pathol Med (2017) 46:98–105. 10.1111/jop.12466 [DOI] [PubMed] [Google Scholar]
- 88. Puthanveetil P, Chen S, Feng B, Gautam A, Chakrabarti S. Long non-coding RNA MALAT1 regulates hyperglycaemia induced inflammatory process in the endothelial cells. J Cell Mol Med (2015) 19:1418–25. 10.1111/jcmm.12576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Liu JY, Yao J, Li XM, Song YC, Wang XQ, Li YJ, et al. Pathogenic role of lncRNA-MALAT1 in endothelial cell dysfunction in diabetes mellitus. Cell Death Dis (2014) 5:10. 10.1038/cddis.2014.466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Toy HI, Okmen D, Kontou PI, Georgakilas AG, Pavlopoulou A. HOTAIR as a Prognostic Predictor for Diverse Human Cancers: A Meta- and Bioinformatics Analysis. Cancers (2019) 11:17. 10.3390/cancers11060778 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Tang Q, Hann SS. HOTAIR: An Oncogenic Long Non-Coding RNA in Human Cancer. Cell Physiol Biochem (2018) 47:893–913. 10.1159/000490131 [DOI] [PubMed] [Google Scholar]
- 92. Hajjari M, Salavaty A. HOTAIR: an oncogenic long non-coding RNA in different cancers. Cancer Biol Med (2015) 12:1–9. 10.7497/j.issn.2095-3941.2015.0006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Majumder S, Hadden MJ, Thieme K, Batchu SN, Niveditha D, Chowdhury S, et al. Dysregulated expression but redundant function of the long non-coding RNA HOTAIR in diabetic kidney disease. Diabetologia (2019) 62(11):2129–42. 10.1007/s00125-019-4967-1 [DOI] [PubMed] [Google Scholar]
- 94. Kong Y, Hsieh CH, Alonso LC. ANRIL: A lncRNA at the CDKN2A/B Locus With Roles in Cancer and Metabolic Disease. Front Endocrinol (2018) 9:405. 10.3389/fendo.2018.00405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Thomas AA, Feng B, Chakrabarti S. ANRIL: A Regulator of VEGF in Diabetic Retinopathy. Invest Ophthalmol Visual Sci (2017) 58:470–80. 10.1167/iovs.16-20569 [DOI] [PubMed] [Google Scholar]
- 96. Ruiz MA, Feng B, Chakrabarti S. Polycomb repressive complex 2 regulates MiR-200b in retinal endothelial cells: potential relevance in diabetic retinopathy. PLoS One (2015) 10:e0123987. 10.1371/journal.pone.0123987 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97. Zhang B, Wang D, Ji TF, Shi L, Yu JL. Overexpression of lncRNA ANRIL up-regulates VEGF expression and promotes angiogenesis of diabetes mellitus combined with cerebral infarction by activating NF-kappaB signaling pathway in a rat model. Oncotarget (2017) 8:17347–59. 10.18632/oncotarget.14468 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 98. Mitsuya K, Meguro M, Lee MP, Katoh M, Schulz TC, Kugoh H, et al. LIT1, an imprinted antisense RNA in the human KvLQT1 locus identified by screening for differentially expressed transcripts using monochromosomal hybrids. Hum Mol Genet (1999) 8:1209–17. 10.1093/hmg/8.7.1209 [DOI] [PubMed] [Google Scholar]
- 99. Kassem SA, Ariel I, Thornton PS, Hussain K, Smith V, Lindley KJ, et al. p57(KIP2) expression in normal islet cells and in hyperinsulinism of infancy. Diabetes (2001) 50:2763–9. 10.2337/diabetes.50.12.2763 [DOI] [PubMed] [Google Scholar]
- 100. Liu J, Wang F, Wu Y, Huang X, Sheng L, Xu J, et al. Meta-analysis of the effect of KCNQ1 gene polymorphism on the risk of type 2 diabetes. Mol Biol Rep (2013) 40:3557–67. 10.1007/s11033-012-2429-7 [DOI] [PubMed] [Google Scholar]
- 101. Sun C, Huang L, Li Z, Leng K, Xu Y, Jiang X, et al. Long non-coding RNA MIAT in development and disease: a new player in an old game. J BioMed Sci (2018) 25:23. 10.1186/s12929-018-0427-3 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Yan B, Yao J, Liu JY, Li XM, Wang XQ, Li YJ, et al. lncRNA-MIAT regulates microvascular dysfunction by functioning as a competing endogenous RNA. Circ Res (2015) 116:1143–56. 10.1161/CIRCRESAHA.116.305510 [DOI] [PubMed] [Google Scholar]
- 103. Zhou X, Zhang W, Jin M, Chen J, Xu W, Kong X. lncRNA MIAT functions as a competing endogenous RNA to upregulate DAPK2 by sponging miR-22-3p in diabetic cardiomyopathy. Cell Death Dis (2017) 8:e2929. 10.1038/cddis.2017.321 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Zhou L, Xu DY, Sha WG, Shen L, Lu GY, Yin X. Long non-coding MIAT mediates high glucose-induced renal tubular epithelial injury. Biochem Biophys Res Commun (2015) 468:726–32. 10.1016/j.bbrc.2015.11.023 [DOI] [PubMed] [Google Scholar]
- 105. You L, Wang N, Yin D, Wang L, Jin F, Zhu Y, et al. Downregulation of Long Noncoding RNA Meg3 Affects Insulin Synthesis and Secretion in Mouse Pancreatic Beta Cells. J Cell Physiol (2016) 231:852–62. 10.1002/jcp.25175 [DOI] [PubMed] [Google Scholar]
- 106. Zhu X, Wu YB, Zhou J, Kang DM. Upregulation of lncRNA MEG3 promotes hepatic insulin resistance via increasing FoxO1 expression. Biochem Biophys Res Commun (2016) 469:319–25. 10.1016/j.bbrc.2015.11.048 [DOI] [PubMed] [Google Scholar]
- 107. Cui M, Fang Q, Zheng J, Shu Z, Chen Y, Fan Y, et al. Kaposi’s sarcoma-associated herpesvirus seropositivity is associated with type 2 diabetes mellitus: A case-control study in Xinjiang, China. Int J Infect Dis (2019) 80:73–9. 10.1016/j.ijid.2019.01.003 [DOI] [PubMed] [Google Scholar]
- 108. Piras E, Madeddu MA, Palmieri G, Angius F, Contini P, Pompei R, et al. High Prevalence of Human Herpesvirus 8 Infection in Diabetes Type 2 Patients and Detection of a New Virus Subtype. Adv Exp Med Biol (2017) 973:41–51. 10.1007/5584_2016_73 [DOI] [PubMed] [Google Scholar]
- 109. Pompei R. The Role of Human Herpesvirus 8 in Diabetes Mellitus Type 2: State of the Art and a Medical Hypothesis. Adv Exp Med Biol (2016) 901:37–45. 10.1007/5584_2015_5014 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.