Extended Data Fig. 2 |. Factors that affect the formation of a gap junction.
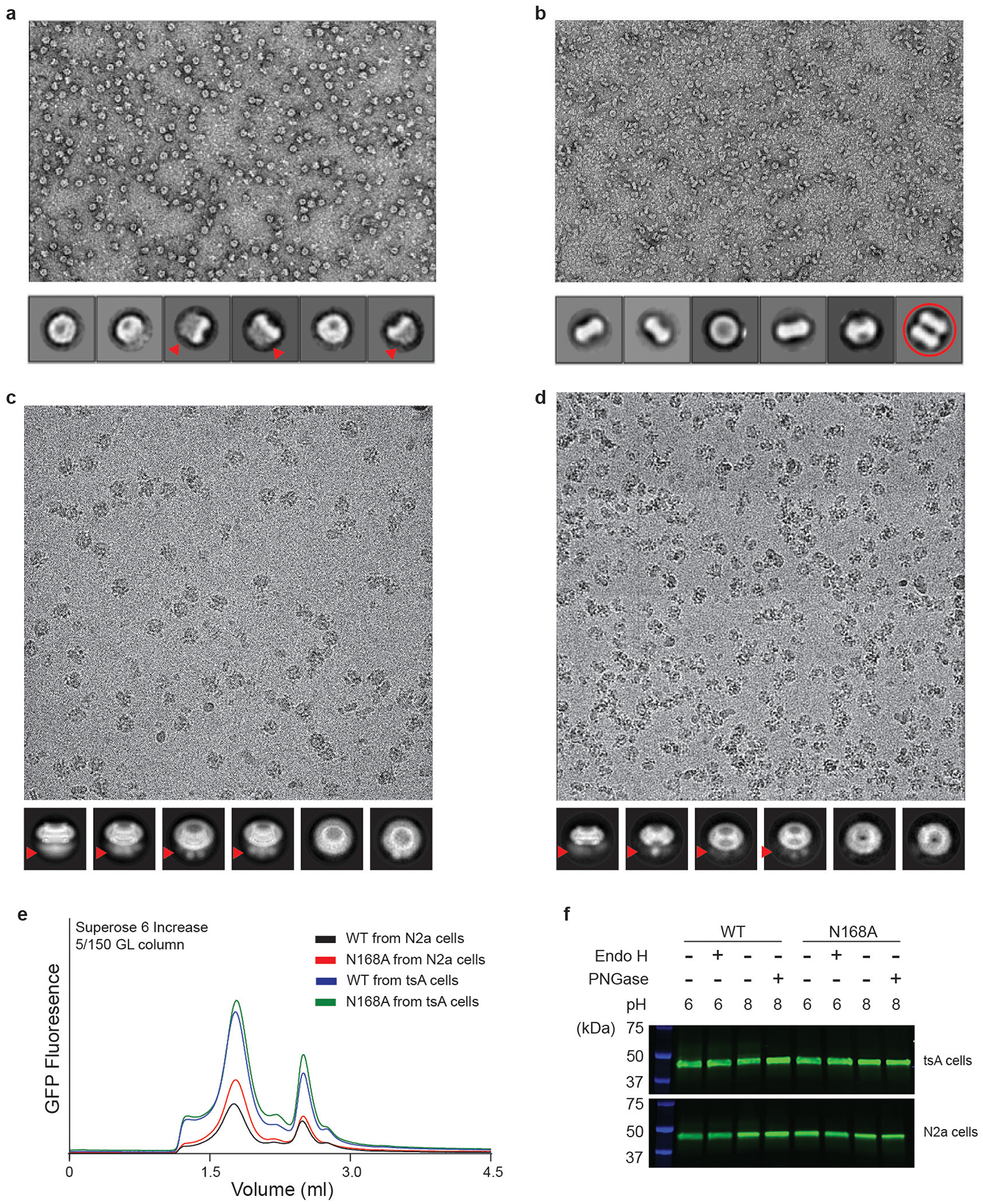
a, A representative micrograph of purified CALHM2N by negative-stain electron microscopy; selected 2D classes are shown below. Fuzzy areas (indicated by red arrows) are caused by flexible GFP tags; this is also implied in c and d. Only hemichannels, and not gap junctions, appear in the micrograph. b, A representative micrograph of purified CALHM2C (from which the GFP tag has been cleaved) by negative-stain electron microscopy. The fuzzy areas in a, c, d disappeared in the 2D classes, which confirms that they are indeed due to the GFP tag. One of the 2D class averages represents a gap junction (red circle). c, A representative micrograph of purified CALHM2N by cryo-EM in the presence of 1 mM EDTA, collected using the Titan Krios; selected 2D classes are shown below. Only hemichannels, and not gap junctions, are observed. d, A representative micrograph of purified CALHM2C by cryo-EM using the Talos Arctica; selected 2D classes are shown below. Only hemichannels, and not gap junctions, are seen. e, FSEC profile of wild-type CALHM2 and the CALHM2(N168A) mutant, expressed in tsA 201 and N2a cells. This experiment was repeated three times, each yielding similar results. f, The wild-type CALHM2 ran at the same height in SDS gel before and after treatment using 250 U of Endo H and 250 U PNGase at optimum pH. Moreover, the CALHM2(N168A) mutant ran at the same height as the wild-type CALHM2. These data suggest CALHM2 is not glycosylated. This experiment was repeated multiple times (Endo H, n = 6; PNGase, n = 3), all yielding non-glycosylation phenotypes.
