Abstract
Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) pandemic has been predominantly respiratory. This study aimed to evaluate the presence of virus in non-airborne body fluids as transmission vehicles. Medline, EMBASE, and Cochrane Library databases were searched from December 01, 2019, to July 01, 2020, using terms relating to SARS-CoV-2 and non-airborne clinical sample sources (feces, urine, blood, serum, serum, and peritoneum). Studies in humans, of any design, were included. Risk of bias assessment was performed using the Quality Assessment of Diagnostic Accuracy 2 tool. Preferred Reporting Items for Systematic Reviews & Meta-Analyses) guidelines were used for abstracting data. If ≥5 studies reported proportions for the same non-respiratory site, a meta-analysis was conducted using either a fixed or random-effects model, depending on the presence of heterogeneity. A total of 22 studies with 648 patients were included. Most were cross-sectional and cohort studies. The SARS-CoV-2 RNA was most frequently detected in feces. Detectable RNA was reported in 17% of the blood samples, 8% of the serum, 16% in the semen, but rarely in urine. Prevalence of SARS-CoV-2 in non-airborne sites varies widely with a third of non-airborne fluids. Patients with bowel and non-specific symptoms have persistence of virus in feces for upto 2 weeks after symptom resolution. Although there was a very low detection rate in urine, given the more frequent prevalence in blood samples, the presence of SARS-CoV-2 in patients with disrupted urothelium or undergoing urinary tract procedures, is likely to be higher. Healthcare providers need to consider non-airborne transmission and persistence of SARS-CoV-2 in body fluids to enable appropriate precautions to protect healthcare workers and carers.
Keywords: Body fluids, coronavirus disease 2019, healthcare worker risk, non-airborne, severe acute respiratory syndrome coronavirus 2
Introduction
The severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection was declared a pandemic in early March 2020. Despite the similarity to 2 predecessors of SARS-CoV-2, the severe respiratory syndrome coronavirus (SARS-CoV) and the Middle East respiratory syndrome coronavirus (MERS-CoV)[1]; scientific debate continues on the transmission modalities and viral behavior. Primary transmissibility of SARS-CoV-2 is via an airborne route, such as respiratory droplets when a positive individual sneezes or coughs,[2] with the most common symptoms being fever, fatigue, dry cough, and anosmia.[3] However, other reports suggest SARS-CoV-2 can present with atypical symptoms such as heart palpitations, abdominal pain, diarrhea, headaches,[4] and dysgeusia.[5] Asymptomatic transmission is a characteristic feature.[6] Incubation period is variable (2–15 days), especially in younger patients (<35 years), with symptoms occurring >25 days from exposure in some cases.[7,8]
The coronavirus disease 2019 (COVID-19) tests include detecting SARS-CoV-2 viral RNA using polymerase chain reaction (PCR) based techniques in throat and nasal swabs.[9] Serology assays developed to identify patients with antibodies are the basis for contact tracing, monitoring exposure, and immune status of geographical areas.[10] However, with variability in symptoms and imperfect diagnostic test accuracy, it remains difficult to accurately identify infected patients and subsequently quarantine them. This necessitates additional investigations such as chest CT scans to negate disease characteristics, especially in patients scheduled for medical procedures or surgery.[6,11]
Pathways to improve capacity for patient consultations and surgery while minimizing risk to the patients and healthcare workers are needed. The need to minimize aerosol-generating procedures has resulted in scheduling only urgent surgeries. The unpredictability of disease spread is problematic in planning healthcare capacity recovery. A French hospital study highlighted exhaustive demands and resources utilized for patients with COVID-19, where staff numbers had to be increased 3-fold to meet the demands of the pandemic.[12]
Reports have emerged of the virus being detected in various other bodily fluids.[2,13] Therefore, we conducted a systematic review and meta-analysis of studies evaluating the presence of the SARS-CoV-2 virus in non-airborne bodily fluids. This study aimed to offer information to aid healthcare workers, surgeons, and policymakers to identify operative procedures with potential increased risk on the basis of exposure risk to various body fluids.
Material and methods
Search strategy
This systematic review was conducted and reported with reference to the Preferred Reporting Items for Systematic Reviews & Meta-Analyses guidelines.[14] A systematic search of the literature was conducted using the databases Medline, EMBASE, and the Cochrane Library from December 01, 2019, to July 01, 2020. The search strategy used MeSH terms and keywords relating to the following domains:
SARS-CoV-2; and
Non-airborne clinical sample sources-specifically feces, urine, blood, serum, and peritoneum.
Searches relating to 2 domains were combined with the Boolean operator AND. (An example of the complete search strategy can be found in Appendix 1, Supplementary Material.) Search results were limited to those involving humans and published in the English language. Following de-duplication, titles, abstracts, and full texts were sequentially screened for inclusion by 2 reviewers independently (HJ, MG) with disagreements resolved by mutual discussion. To retrieve additional relevant citations, we hand-searched reference lists of potentially eligible articles. The inter-rater agreement of reviewers for citation selection of abstracts and full texts was summarized using Cohen’s kappa statistic.[15]
Eligibility criteria
All observational or experimental studies, from any country, were eligible for inclusion. Conference proceedings, editorials, opinions, and consensus statements were excluded, as were economic evaluations that did not include previously unreported primary data. The selected studies also needed to satisfy the following criteria:
studied humans (including adults, children of any age, and pregnant woman) with confirmed SARS-CoV-2 infection as defined by a positive PCR test result from an upper respiratory, lower respiratory, or sputum sample[16];
participants were also tested for presence of SARS-CoV-2 RNA in 1 or more of extra-respiratory clinical sites of interest (feces, urine, blood, serum, semen, or peritoneal fluid).
Exclusion criteria consisted of articles that:
did not follow an empirical study design, such as single-case studies or reviews;
were not written in English;
studied convalescent patients with SARS-CoV-2; or
did not assess non-respiratory samples for SARS-CoV-2 RNA.
Data extraction
Relevant information from eligible articles was extracted using a predefined data extraction form by 2 reviewers independently (HJ, MG), with disagreements resolved by mutual discussion. The following data were recorded: year of publication, first author’s name, study design, country, inclusion criteria, exclusion criteria, sex distribution, age distribution, specific symptoms (such as fever, dry cough, fatigue, and gastrointestinal symptoms), number of individuals with a SARS-CoV-2 positive respiratory sample, number of individuals with a SARS-CoV-2 positive non-respiratory sample (recorded separately for feces, urine, blood, serum, semen, and peritoneal fluid), diagnostic test used, test company, and test cycle threshold used to determine positivity.
Quality assessment
The Quality Assessment of Diagnostic Accuracy (QUADAS-2) tool was used to evaluate the risk of bias, applicability of diagnostic accuracy, and level of concurrence with the review question of interest.[17] QUADAS-2 includes the following 4 key domains: patient selection, index test, reference standard, and flow and timing. We defined the index test to be that of non-respiratory samples with the use of respiratory samples as a reference standard test.
Statistical analysis
For each study, the proportion of SARS-CoV-2 respiratory-positive individuals with detectable RNA in each non-respiratory site was calculated. Where 5 or more studies reported proportions for the same non-respiratory site, a meta-analysis was conducted using either a fixed or random-effects model, with the latter performed where significant clinical or methodological heterogeneity was deemed to exist. Summary effect measures were reported by a pooled proportion, with 95% confidence intervals. Statistical heterogeneity was assessed using the I2 statistic. For sites where 10 or more studies existed, visual inspection of funnel plots was performed to investigate possible publication bias as well as an Egger test of funnel plot asymmetry. Statistical analyses were conducted using the Statistical Package for Social Sciences (version 26, IBM Corporation, Armonk, NY, USA) and R (version 3.6)[18] with the metafor package. A p value of <0.05 was deemed to be statistically significant.
Results
Study selection
Of 9,278 unique citations found, 46 articles went through full-text screening. There was a good overall agreement (Cohen’s Kappa) between reviewers for abstract and full-text screening (χ =0.79 and 0.58, respectively).[19] Of the full-text articles reviewed, 24 were excluded for following reasons, in descending order of exclusion hierarchy:
convalescent patients studied (n=4);
negative respiratory swab sample for SARS-CoV-2 RNA (n=7);
non-empirical study design (n=8);
non-airborne sample not assessed for SARS-CoV-2 RNA (n=2); and
not written in English (n=3).
A total of 22 studies were eligible for inclusion in the systematic review and meta-analysis.[20–41] Figure 1 illustrates the study selection process.
Figure 1.
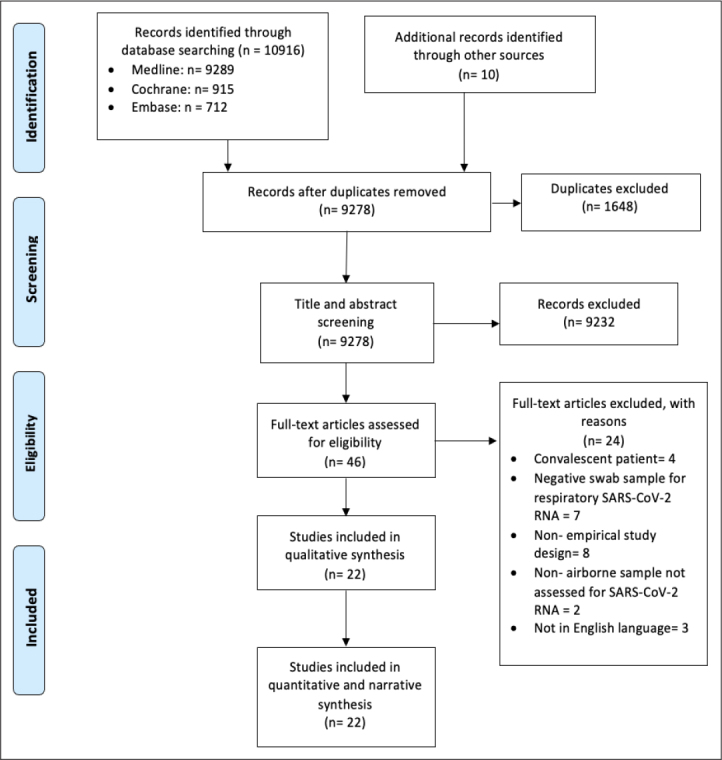
Prisma 2009 Flow Diagram
Study characteristics
Study dates ranged from January 01, 2020, to June 30, 2020. The majority of the studies took place in China (86%) and 1 study each in Singapore, Hong Kong, and the United States. The studies that looked for the presence of SARS-CoV-2 RNA in non-airborne samples were variable, with 21 studies including feces (95%), 8 studies testing urine, 5 studies each testing blood or serum, and 1 study on sperm. A total of 3 reports on peritoneal fluid had single patients, therefore, did not match the inclusion criteria; and the 22 included studies involved 648 patients in total with a mean of 50.6% men (range 25.0%–100%) and an age range from 2 months to 87 years. Mean symptom prevalence, where available, were as follows: fever (80.5%; 14/22 studies), cough (52.5%; 14/22 studies), fatigue (32.6%, 9/22 studies), and gastrointestinal symptoms (25.2%; 14/22 studies). Only 6 studies had interpretable temperature recordings, ranging from 36.1°C to 39.6 °C. A total of 21 studies used PCR as the diagnostic test, and 1 study did not specify the diagnostic test that was implemented. Furthermore, only 8 studies stated the PCR test company brand and 9 studies stated cycle threshold (CT) cut-off value for a positive SARS-CoV-2 RNA result. CT values ranged from <33 to <40. A summary of the characteristics of included studies is found in Table 1.
Table 1.
Characteristics of included studies
| Study, author (year) | Study design | Study dates (2020) | Location | Sample size | Male (%) | Age distribution (years) | Prevalence of symptoms (%) | Diagnostic test | Test company | Cycle threshold | Sample site | No. of positive samples/ No. tested | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Cai et al. [20] (2020) | Case series | 19 Jan–3 Feb | Shanghai, China | 6 | 45% | Mean 6.77 (range, 0.25–10.92) | Fever | 80% | rRT-PCR | NR | <35 | Feces | 5/6 |
| Cough | 60% | Urine | 0/6 | ||||||||||
| Fatigue | NR | Serum | 0/6 | ||||||||||
| GI symptoms | 0% | ||||||||||||
|
| |||||||||||||
| Chen et al.[21] (2020) | Cross-sectional | 20 Jan–9 Feb | Wuhan, China | 42 | 36% | Median 51 (IQR, 42.75–62) | Fever | 85.7 | rRT-PCR | NR | NR | Feces | 28/42 |
| Cough | 52.30% | Urine | 0/10 | ||||||||||
| Fatigue | 52.30% | ||||||||||||
| GI symptoms | 19.10% | ||||||||||||
|
| |||||||||||||
| Cheung et al.[22] (2020) | Cross-sectional | 2 Feb–29 Feb | Hong Kong | 53 | 46% | Median 58.5 (IQR, 43.5–68.0) | Fever | 94.90% | RT-PCR | NR | NR | Feces | 9/59 |
| Cough | 37.30% | ||||||||||||
| Fatigue | 37.30% | ||||||||||||
| GI symptoms | NR | ||||||||||||
|
| |||||||||||||
| Kujawski et al.[23] (2020) | Case series | 20 Jan–5 Feb | USA | 10 | 67% | Median 53 (range, 21–68) | Fever | 58.30% | rRT-PCR | NR | <33 | Feces | 7/9 |
| Cough | 66.70% | Urine | 0/9 | ||||||||||
| Fatigue | 41.70% | Serum | 0/10 | ||||||||||
| GI symptoms | 8.30% | ||||||||||||
|
| |||||||||||||
| Study, author (year) | Study design | Study dates (2020) | Location | Sample size | Male (%) | Age distribution (years) | Prevalence of symptoms (%) | Diagnostic test | Test company | Cycle threshold | Sample site | No. of positive samples/ No. tested | |
|
| |||||||||||||
| Lei et al.[24] (2020) | Cross-sectional | 22 Jan–12 Feb | Guangzhou, China | 7 | 50% | Mean 43.2 (range, 25–64) | Fever | 80% | rRT-PCR | NR | NR | Feces | 4/7 |
| Cough | 11% | ||||||||||||
| Fatigue | 35% | ||||||||||||
| GI symptoms | 25% | ||||||||||||
|
| |||||||||||||
| Li et al.[25] (2020) | Cohort study | 26 Jan–16 Feb | Shangqiu, China | 38 | 100% | Range (20–50) | None reported | rRT-PCR | NR | NR | Semen | 6/38 | |
|
| |||||||||||||
| Lin et al. [26] (2020) | Cross-sectional | 17 Jan–15 Feb | China | 65 | 47% | Mean 45.3 (range, 27–63.6) | GI symptoms | 61% | rRT-PCR | Shanghai ZJ BioTech Co, Ltd, Shanghai, China |
<37 | Feces | 31/65 |
|
| |||||||||||||
| Lo et al.[27] (2020) | Cross-sectional | 21 Jan–16 Feb | Macau SAR, China | 8 | 30% | Median 54 (IQR, 27–64) | Fever | 80% | qRT-PCR | BioGerm, China | ≤35 | Feces | 8/8 |
| Cough | 50% | Urine | 0/8 | ||||||||||
| Fatigue | 30% | ||||||||||||
| GI symptoms | 80% | ||||||||||||
|
| |||||||||||||
| Ma et al. [28] (2020) | Cross-sectional | NR | China | 4 | 25% | Mean 12.2 (range, 0.92–39.0) | Fever | 38% | rRT-PCR | BioGerm, Shanghai, | NR | Feces | 0/4 |
|
| |||||||||||||
| Study, author (year) | Study design | Study dates (2020) | Location | Sample size | Male (%) | Age distribution (years) | Prevalence of symptoms (%) | Diagnostic test | Test company | Cycle threshold | Sample site | No. of positive samples/ No. tested | |
|
| |||||||||||||
| Peng et al.[29] (2020) | Cross-sectional | 19 Feb–unknown | Guangdong, China | 7 | 44% | Mean 38.9 (range, 27.1–40.7) | Fever | 100% | qRT-PCR | SUPI-0509; Supbio | NR | Feces | 2/7 |
| Cough | 67% | Blood | 2/7 | ||||||||||
| Fatigue | 22% | Urine | 1/7 | ||||||||||
| GI symptoms | 11% | ||||||||||||
|
| |||||||||||||
| Tan et al.[30] (2020) | Cross-sectional | 27 Jan–10 Mar | China | 10 | 30% | Mean 7 (range, 1–12) | Fever | NR | rRT-PCR | NR | NR | Feces | 1/10 |
| Cough | 40% | ||||||||||||
| Fatigue | 30% | ||||||||||||
| GI symptoms | NR | ||||||||||||
|
| |||||||||||||
| Wang et al.[31] (2020) | Cross-sectional | 1 Jan–17 Feb | China | 12 | 66% | Mean 44 (range, 5–67) | None reported | rRT-PCR | NR | <40 | Feces | 5/12 | |
| Urine | 0/9 | ||||||||||||
| Blood | 2/5 | ||||||||||||
|
| |||||||||||||
| Wei et al.[32] (2020) | Cross-sectional | 19 Jan–7 Feb | Wuhan, China | 84 | 33% | Median 37 (range, 24–74) | Fever | 86% | rRT-PCR | NR | NR | Feces | 28/84 |
| Cough | 57% | ||||||||||||
| Fatigue | 44% | ||||||||||||
| GI symptoms | 31% | ||||||||||||
|
| |||||||||||||
| Study, author (year) | Study design | Study dates (2020) | Location | Sample size | Male (%) | Age distribution (years) | Prevalence of symptoms (%) | Diagnostic test | Test company | Cycle threshold | Sample site | No. of positive samples/ No. tested | |
|
| |||||||||||||
| Wu et al.[33] (2020) | Cohort study | 16 Jan–15 Mar | China | 41 | NR | NR | None reported | rRT-PCR | NR | NR | Feces | 29/41 | |
|
| |||||||||||||
| Xiao et al.[34] (2020) | Cross-sectional | 1 Feb–14 Feb | China | 73 | 56% | Mean 43 (range, 0.83–78) | Fever | NR | rRT-PCR | NR | NR | Feces | 39/73 |
| Cough | NR | ||||||||||||
| Fatigue | 36% | ||||||||||||
| GI symptoms | NR | ||||||||||||
|
| |||||||||||||
| Xing et al.[35] (2020) | Cross-sectional | 17 Jan–23 Feb | Shanong, China | 3 | 67% | Mean 4.2 (range, 1.5–6) | Fever | 100% | rRT-PCR | NR | <40 | Feces | 2/3 |
| Cough | 33% | ||||||||||||
| Fatigue | NR | ||||||||||||
| GI symptoms | 33% | ||||||||||||
|
| |||||||||||||
| Xu et al.[36] (2020) | Cross-sectional | 22 Jan–20 Feb | Guangdong, China | 10 | 60% | Mean 7.54 (range 0.17–15) | Fever | 70% | rRT-PCR | NR | <37 | Feces | 8/10 |
| Cough | 50% | ||||||||||||
| Fatigue | 30% | ||||||||||||
| GI symptoms | NR | ||||||||||||
|
| |||||||||||||
| Young et al.[37] (2020) | Case series | 23 Jan–3 Feb | Singapore | 12 | 50% | Median 47 | Fever | 72% | rRT-PCR | Qiagen | <38 | Feces | 4/8 |
| Cough | 83% | Blood | 1/12 | ||||||||||
| Fatigue | NR | Urine | 0/10 | ||||||||||
| GI symptoms | 17% | ||||||||||||
|
| |||||||||||||
| Study, author (year) | Study design | Study dates (2020) | Location | Sample size | Male (%) | Age distribution (years) | Prevalence of symptoms (%) | Diagnostic test | Test company | Cycle threshold | Sample site | No. of positive samples/ No. tested | |
|
| |||||||||||||
| Yun et al.[38] (2020) | Cross-sectional | 23 Jan–25 Feb | Changsha, China | 32 | 49% | Median 50 (IQR, 37–66) | None reported | Unspecified | Hunan Shengxiang Biology Co., Ltd | NR | Feces | 8/32 | |
| Blood | 0/32 | ||||||||||||
| Serum | 0/32 | ||||||||||||
|
| |||||||||||||
| Zhang et al.[39] (2020) | Cross-sectional | 27 Jan–10 Feb | Zhejiang, China | 14 | 50% | Median 41 (range, 18–87) | Fever | 93% | rRT-PCR | NR | NR | Feces | 5/14 |
| Cough | 71% | ||||||||||||
| Fatigue | NR | ||||||||||||
| GI symptoms | 0% | ||||||||||||
|
| |||||||||||||
| Zheng et al.[40] (2020) | Cohort | 19 Jan–20 Mar | Zhejiang, China | 96 | 60% | Median 55 (IQR, 44.3–64.8) | Fever | 89% | qRT-PCR | BoJie, Shanghai, China | ≤38.0 | Feces | 55/96 |
| Cough | 56% | Serum | 39/96 | ||||||||||
| Fatigue | 9% | Urine | 1/96 | ||||||||||
| GI symptoms | 10% | ||||||||||||
|
| |||||||||||||
| Zhang et al.[41] (2020) | Cohort | NR | Wuhan, China | 15 | NR | NR | None reported | qRT-PCR | Vazyme Biotech Co., Ltd | NR | Feces | 4/15 | |
| Blood | 6/15 | ||||||||||||
| Serum | 3/15 | ||||||||||||
NR: not reported; GI: gastrointestinal; rRT-PCR: real-time reverse-transcriptase polymerase chain reaction; qRT-PCR: quatitative reverse-transcriptase polymerase chain reaction; no., number
Quality assessment
Most included studies were cross-sectional studies or case series, and an overall high risk of bias was judged to exist across all 4 domains, specifically in patient selection (90.1%), index test for non-respiratory SARS-CoV-2 samples (95.4%), reference test for respiratory SARS-CoV-2 samples (95.4%), and the flow and timing (86.4%, Figure 2 and Table 2). There are various reasons for this high risk of bias. Firstly, the patient selection process across most studies was not documented; for example, it was often not clear whether all the patients who were SARS-CoV-2 positive were included over a given time frame, or if specific exclusion criteria were applied. Secondly, the index test and the standard test was interpreted with limited blinding and without predetermined criteria, along with certain studies lacking the mention of a CT value. Finally, the time interval between the tests was not strictly specified in most studies. However, the applicability concerns of the majority of studies were low across the 3 domains of patient selection, index test, and reference standard (Figure 2).
Figure 2.
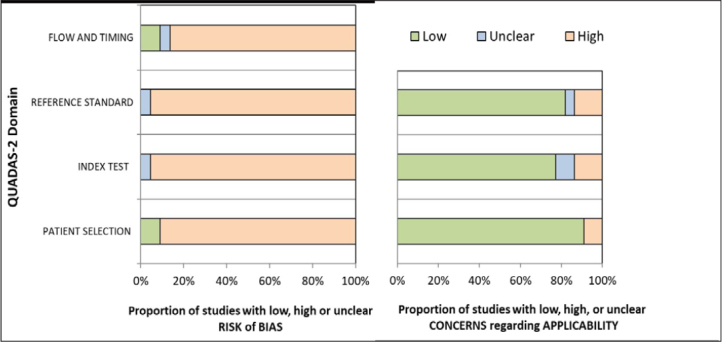
Summary of Percentage of Each Judgement in QUADAS
Table 2.
Quality assessment
| Study (2020) | Applicability | Bias | |||||
|---|---|---|---|---|---|---|---|
|
|
|
||||||
| Patient selection | Index test | Reference standard | Patient selection | Index test | Reference standard | Flow and timing | |
| Cai et al.[20] | Low | Low | Low | Low | High | High | Unclear |
|
| |||||||
| Chen et al.[21] | Low | Unclear | Unclear | High | High | Unclear | High |
|
| |||||||
| Cheung et al.[22] | Low | Unclear | High | High | High | High | High |
|
| |||||||
| Kujawski et al.[23] | Low | Low | Low | High | High | High | High |
|
| |||||||
| Lei et al.[24] | Low | Low | High | High | High | High | High |
|
| |||||||
| Li et al.[25] | Low | High | Low | High | High | High | High |
|
| |||||||
| Lin et al.[26] | Low | Low | Low | High | High | High | High |
|
| |||||||
| Lo et al.[27] | Low | Low | Low | High | High | High | Low |
|
| |||||||
| Ma et al.[28] | Low | Unclear | Low | High | Unclear | High | High |
|
| |||||||
| Peng et al.[29] | Low | Low | Low | High | High | High | High |
|
| |||||||
| Tan et al.[30] | Low | Low | Low | High | High | High | High |
|
| |||||||
| Wang et al.[31] | Low | Low | Low | High | High | High | High |
|
| |||||||
| Wei et al.[32] | Low | Low | Low | High | High | High | High |
|
| |||||||
| Wu et al.[33] | Low | Low | Low | High | High | High | High |
|
| |||||||
| Xiao et al.[34] | Low | Low | Low | Low | High | High | High |
|
| |||||||
| Xing et al.[35] | Low | Low | Low | High | High | High | High |
|
| |||||||
| Xu et al.[36] | Low | Low | Low | High | High | High | High |
|
| |||||||
| Young et al.[37] | Low | Low | Low | High | High | High | Low |
|
| |||||||
| Yun et al.[38] | Low | High | High | High | High | High | High |
|
| |||||||
| Zhang et al.[39] | High | Low | Low | High | High | High | High |
|
| |||||||
| Zhang et al.[40] | Low | Low | Low | High | High | High | High |
|
| |||||||
| Zhang et al.[41] | High | Low | Low | High | High | High | High |
Presence of SARS-CoV-2 in non-respiratory samples
Feces
For the meta-analysis of the proportion of patients who were SARS-CoV-2 respiratory positive with a positive SARS-CoV-2 fecal sample, there were a total of 605 patients from 21 studies.[20–24,26–41] There was significant statistical heterogeneity between studies (I2= 81%, p<0.01). The pooled proportion was 0.48 (95% confidence interval [CI] 0.37–0.59, random effects) for the presence of SARS-CoV-2 viral RNA in fecal samples of non-convalescent COVID-19 patients (Figure 3). There was no evidence of publication bias (Egger’s test p=0.69 and on visual inspection of the funnel plot).
Figure 3.
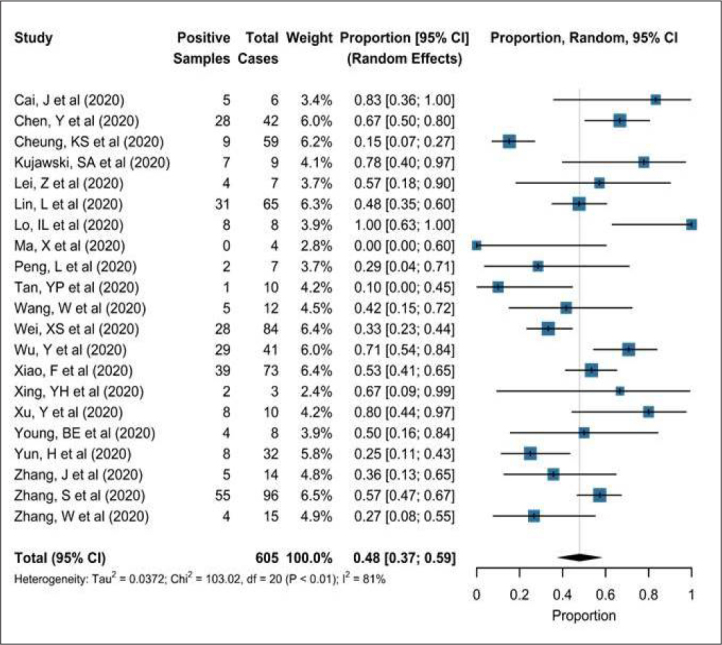
SARS-CoV-2 viral RNA in Faeces
Blood and serum
A total of 71 patients with COVID-19 from 5 studies had results for the presence of SARS-CoV-2 RNA in whole blood samples.[29,31,37,38,40] There was significant statistical heterogeneity between the studies (I2=81%, p<0.01), and the pooled proportion for the presence of SARS-CoV-2 RNA in whole blood samples of non-convalescent patients with COVID-19 were 0.17 (95% CI, 0.00–0.45; random effects, Figure 4).
Figure 4.
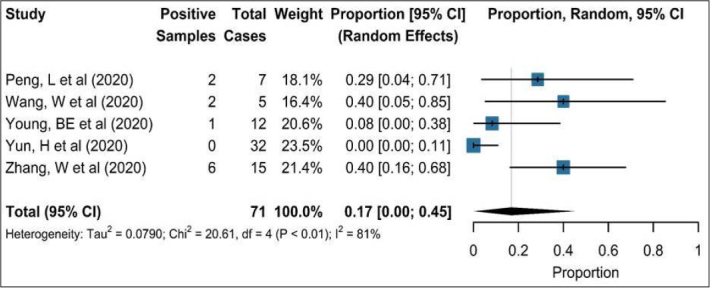
SARS-CoV-2 viral RNA in whole blood samples
A total of 5 studies with 159 patients specifically looked for SARS-CoV-2 RNA in the serum of patients with COVID-19.[20,23,38,40,41] Interestingly, 3 of these studies found no individuals with detectable RNA in serum, whereas the largest of the studies (with 96 patients) found SARS-CoV-2 RNA in the serum of 41% of the patients.[41] The pooled proportion for serum positivity was 0.08 (95% CI, 0.00–0.35, random effects; I2=91%, p<0.01 for heterogeneity.[20,23,38,40,41]
Urine
A total of 8 studies were included with a total of 155 patients,[20,21,23,27,29,31,37,41] of which, 6 found no detectable SARS-CoV-2 RNA in any of their cases, 1 study found it in only 1 of 7 patients, and the largest study of 96 patients also only found detectable RNA in the urine of 1 individual.
Sperm
Only 1 study documented the evidence of SARS-CoV-2 viral RNA in the semen of individuals with SARS-CoV-2 positive respiratory secretions, with 6 (16%) of 38 having detectable viral load in semen.[25] Specifically, they report the presence of SARS-CoV-2 in 26.7% of patients during the acute phase and 8.7% in the recovery phase.
Peritoneal Fluid
None of the included studies examined the peritoneal fluid. However, we are aware of 2 single-case study reports with conflicting results. A positive peritoneal swab was reported in a patient undergoing emergency laparotomy but not in a patient undergoing laparoscopic appendicectomy.[42] In 1 patient failing peritoneal dialysis, SARS-CoV-2 was identified in the peritoneal dialysate, with recovery from COVID-19 correlating with re-effectiveness of peritoneal dialysis.[43]
Discussion
From end of March 2020 till the end of July 2020, there has been a drastic reduction in delivery of medical interventions in the community, primary, and secondary healthcare services, except for emergency or cancer-related procedures. Diagnostic services have also sharply reduced in most urology services with only urgent procedures taking place.
There is a likelihood of increasing hospital referrals and an urgent necessity to enable surgeries that had been put on hold. With the specter of a second wave of the pandemic on the horizon, there is a need to ramp up the work rate and maximize throughput and enabling “catch-up” by optimizing capacity. This would require logistic expansion that would be increasingly difficult with the need for staff to maintain social distancing, staggered patient attendances, and full personal protection equipment (PPE) precautions, including precautions for aerosol-generating procedures. The present scenario predicts a reduced capacity in comparison with the pre-COVID times.
In the community and primary care, there is an urgent need to review and manage those patients who have had attention and care diverted, and the need to identify risk from exposure to body fluids. This would inform the level of PPE required for physicians, nurses, and specialist healthcare workers such as continence and stoma nurses, tissue viability nurses, phlebotomists, and podiatrists.
The current evidence suggests transmission mainly via respiratory droplets and mucosal contact with contaminated material (oral, ocular, and nasal).[44] In the hospital setting, this suggests increased exposure risk in thoracic, head and neck, upper gastrointestinal, dental, and anesthetic procedures. However, viral load and exposure to surgeons and theater staff from various bodily fluids during abdominal and pelvic surgery is poorly understood. This systematic review and meta-analysis of 22 studies with 937 patients with COVID-19 infection have reported on the prevalence of SARS-CoV-2 positivity in various bodily fluids of interest to bowel, urological, vascular, transplant, pelvic, and gynecological surgeons.
Feces
Detection of viral RNA in stool was reported in almost half of the tested patients from 21 studies. Patients presenting with gastrointestinal symptoms were more likely to have a positive stool test for viral RNA.[22] However, the presence of SARS-CoV-2 in feces did not correlate with the severity of gastrointestinal symptoms.[26] Lo et al.[27] have suggested a delay of viral RNA conversion in stool implying infectivity through the fecal-oral route even during convalescence, with implications for follow-up care and further interventions. Intestinal epithelial injury caused by infection with SARS-CoV-2[32] or virus secretion from gastrointestinal cells[34] could explain the high detection rates of viral RNA from feces. Patients with gastrointestinal symptoms have a greater liability to suffer direct damage on gut mucosa owing to increased angiotensin-converting enzyme 2 expression in the small intestine and colon than in the lungs.[45] It has previously been reported that intestinal infections have been observed at a later stage in MERS-CoV.[40] However, the reasons underlying the persistence of viral RNA in feces and its relationship with infectivity has not been elucidated.
It is important for healthcare workers caring for patients with stomas and conduits and performing urological and bowel procedures to be aware of the persistence of the virus within feces, for up to 30 days from a nasopharyngeal swab turning negative,[20] potentially reflecting a persistent risk of fecal-oral transmission. There is a need for extended feces sampling in patients who are convalescing to understand the transmission potential from the patients.[33] Furthermore, knowledge of whether detectable RNA reflects presence of virions is required before considering the patients clear from an infective potential.[34]
Blood and serum
SARS-CoV-2 viremia has been reported in 15% (11/71) of the blood samples and 26% (42/159) of the serum samples across the included studies. Not all the included patients had systemic symptoms suggesting SARS-CoV-2 could infect multiple systems with few symptoms.[29]
Urine and vaginal fluid
Just 2 of the 155 individuals had detectable SARS-CoV-2 RNA in the urine. These were from patients with only respiratory symptoms and an intact urothelium. Systemic viremia in a fifth of the patients with SARS-CoV-2 suggests the potential for infective risk in those with disrupted urothelia, especially after surgical interventions such as resection of bladder tumors and endometrial ablation. Although not included in this review, SARS-CoV-2 has been reported to be absent in vaginal fluid (1 report) by analyzing vaginal swabs.[46]
Peritoneal fluid
There exists a potential for higher viral exposure to dialysis staff, operating department staff, and specialist nurses in the care of open wounds where the peritoneum is breached. This applies to both open surgery and minimally invasive procedures, where there is exposure to smoke and intra-abdominal gas. However, careful measures to mitigate risk in dialysis centers and reduce exposure in operating departments to aerosol generation, blood splatter, and reducing pneumoperitoneum may be effective in minimizing exposure.[47]
Semen
Only 1 study analyzed semen for the presence of SARS-CoV-2 RNA(25), reporting the presence of non-sexually transmitted viruses in the male urinary and reproductive tract. Although the study was limited by a small sample size and short-term follow-up, the possibility of sexual transmission of SARS-CoV-2 should not be discounted especially in those with active systemic or local inflammation. This would be pertinent when evaluating patients for fertility-associated procedures such as assisted conception, vasectomy, and diagnostic urological procedures. A longer follow-up and supplementary semen analysis would have identified duration of viral presence.
Strengths and Limitations
A strength of this review is the broad nature of the search strategy, which is likely to have found a large proportion of the relevant literature. To account for the heterogeneity of the included studies, we reported random-effects pooled proportions only where sufficient numbers of studies existed. Limitations include a lack of larger scale observational studies with clear eligibility criteria. As much of the included studies were small, with a high risk of selection bias, there was a resulting lack of generalizability. Furthermore, the risk of viral RNA detection in non-respiratory sites may vary with other clinical variables, such as age or co-morbidities. Cohort studies with testing at multiple time points would help delineate the trends for viral shedding. Finally, the presence of viral RNA in a sample does not necessarily mean that there are viable virus particles capable of transmission.
Conclusion
The meta-analysis has shown the presence of SARS-CoV-2 in a third of all non-airborne fluids. Almost half of the feces specimens were positive, with continued presence in convalescence suggesting a potential need for continued protection for healthcare workers looking after patients with lower gastrointestinal conditions or stomas. Continued presence of viral RNA for almost 2 weeks after clearance in the airway needs better elucidation. There was a very low detection rate in urine (<2%). However, with over a tenth of the blood samples being positive, the presence of SARS-CoV-2 in patients who have disrupted urothelium or undergoing urinary tract procedures is likely to be higher. With a 16% positive detection rate in semen, the risk of sexual transmission should be considered. Healthcare workers and carers should consider continuing protection, even after patients are symptomatically better and with negative nasopharyngeal swabs. Other modalities of transmission such as fecal-oral and sexual transmission should be considered.
Main Points.
SARS CoV-2 RNA has been reported to be positive in almost half the faeces samples.
Detection of viral RNA has been identified in almost a fifth of patients in semen and blood, suggesting non-airborne transmission risk.
Persistence of viral RNA is reported in faeces of convalescing patients even after negative oro-pharyngeal negative swabs, suggesting infectivity maybe longer; especially for Health care workers caring for patients with gastrointestinal symptoms.
Search Strategy
The following with title, abstract and keyword for Embase (1974–2020) and Medline (1946 – present):
| 1. | SARS-CoV-2 |
| 2. | Coronavirus |
| 3. | corona-virus |
| 4. | 2019-nCoV |
| 5. | nCoV |
| 6. | COVID-19 |
| 7. | Covid19 |
| 8. | SARS-CoV |
| 9. | SARSCov2 |
| 10. | Ncov |
| 11. | 2019 coronavirus |
| 12. | 2019 corona virus |
| 13. | novel corona virus |
| 14. | new corona virus |
| 15. | nouveau corona virus |
The following with title, abstract and keyword for Cochrane Search Manager
| #1 | SARS-CoV-2 |
| #2 | Coronavirus |
| #3 | MeSH descriptor: [Coronavirus] explode all trees |
| #4 | corona-virus |
| #5 | nCoV |
| #6 | COVID-19 |
| #7 | Covid19 |
| #8 | SARS-CoV |
| #9 | MeSH descriptor: [SARS Virus] explode all trees |
| #10 | SARSCov2 |
| #11 | Ncov |
| #12 | 2019 coronavirus |
| #13 | 2019 corona virus |
| #14 | covid19 |
| #15 | novel corona virus |
| #16 | new corona virus |
| #17 | nouveau corona virus |
Footnotes
Peer-review: This manuscript was prepared by the invitation of the Editorial Board and its scientific evaluation was carried out by the Editorial Board.
Author Contributions: Concept – H.H., J.P.; Design – H.J., M.G., S.S., J.P.; Supervision – S.S., J.T., B.R., O.M.A., J.P.; Resources – H.J., M.G., S.S., B.R., H.H., J.P.; Materials – H.J., M.G., O.M.A., H.H., J.P.; Data Collection and/or Processing – H.J., M.G., S.S., B.R., H.H., J.P.; Analysis and/or Interpretation –H.J., M.G., S.S., J.T., B.R., O.M.A., J.P.; Literature Search – H.J., M.G., S.S., J.P.; Writing Manuscript – H.J., M.G., S.S., J.C., B.R., O.M.A., H.H., J.P.; Critical Review – S.S., J.T., B.R., O.M.A., H.H., J.P.
Conflict of Interest: The authors have no conflicts of interest to declare.
Financial Disclosure: The authors declared that this study has received no financial support.
References
- 1.Song Z, Xu Y, Bao L, Zhang L, Yu P, Qu Y, et al. From SARS to MERS, Thrusting Coronaviruses into the Spotlight. Viruses. 2019;11:59. doi: 10.3390/v11010059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang C, Horby PW, Hayden FG, Gao GF. A novel coronavirus outbreak of global health concern. Lancet. 2020;395:470–3. doi: 10.1016/S0140-6736(20)30185-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D, Hu B, Hu C, Zhu F, Liu X, Zhang J, et al. Clinical Characteristics of 138 Hospitalized Patients With 2019 Novel Coronavirus-Infected Pneumonia in Wuhan, China. JAMA. 2020;323:1061–9. doi: 10.1001/jama.2020.1585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Agarwal A, Chen A, Ravindran N, To C, Thuluvath PJ. Gastrointestinal and Liver Manifestations of COVID-19. J Clin Exp Hepatol. 2020;10:263–5. doi: 10.1016/j.jceh.2020.03.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Russell B, Moss C, Rigg A, Hopkins C, Papa S, Van Hemelrijck M. Anosmia and ageusia are emerging as symptoms in patients with COVID-19: What does the current evidence say? Ecancermedicalscience. 2020;14:ed98-ed. doi: 10.3332/ecancer.2020.ed98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rothe C, Schunk M, Sothmann P, Bretzel G, Froeschl G, Wallrauch C, et al. Transmission of 2019-nCoV Infection from an Asymptomatic Contact in Germany. N Engl J Med. 2020;382:970–1. doi: 10.1056/NEJMc2001468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Liao J, Fan S, Chen J, Wu J, Xu S, Guo Y, et al. Epidemiological and clinical characteristics of COVID-19 in adolescents and young adults. medRxiv. 2020 doi: 10.1016/j.xinn.2020.04.001. 2020.03.10.20032136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Montero Feijoo A, Maseda E, Adalia Bartolomé R, Aguilar G, González de Castro R, Gómez-Herreras JI, et al. Practical recommendations for the perioperative management of patients with suspicion or serious infection by coronavirus SARS-CoV. Revista Española de Anestesiología y Reanimación (English Edition) 2020;67:253–60. doi: 10.1016/j.redare.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Carter LJ, Garner LV, Smoot JW, Li Y, Zhou Q, Saveson CJ, et al. Assay Techniques and Test Development for COVID-19 Diagnosis. ACS Cent Sci. 2020;6:591–605. doi: 10.1021/acscentsci.0c00501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Infantino M, Damiani A, Gobbi FL, Grossi V, Lari B, Macchia D, et al. Serological Assays for SARS-CoV-2 Infectious Disease: Benefits, Limitations and Perspectives. Isr Med Assoc J. 2020;22:203–10. [PubMed] [Google Scholar]
- 11.Bai Y, Yao L, Wei T, Tian F, Jin D-Y, Chen L, et al. Presumed Asymptomatic Carrier Transmission of COVID-19. JAMA. 2020;323:1406–7. doi: 10.1001/jama.2020.2565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peiffer-Smadja N, Lucet J-C, Bendjelloul G, Bouadma L, Gerard S, Choquet C, et al. Challenges and issues about organising a hospital to respond to the COVID-19 outbreak: experience from a French reference centre. Clin Microbiol Infect. 2020 doi: 10.1016/j.cmi.2020.04.002. S1198743X(20)30187-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cui J, Li F, Shi ZL. Origin and evolution of pathogenic coronaviruses. Nat Rev Microbiol. 2019;17:181–92. doi: 10.1038/s41579-018-0118-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Moher D, Liberati A, Tetzlaff J, Altman DG. Preferred reporting items for systematic reviews and meta-analyses: the PRISMA statement. PLoS Med. 2009;6:e1000097. doi: 10.1371/journal.pmed.1000097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Orwin R, Cooper I, Hedges L. The handbook of research synthesis. New York, NY: Russell Sage Foundation; 1994. pp. 139–62. [Google Scholar]
- 16.Deeks JJ, Dinnes J, Takwoingi Y, Davenport C, Leeflang MMG, Spijker R, et al. Diagnosis of SARS-CoV-2 infection and COVID-19: accuracy of signs and symptoms; molecular, antigen, and antibody tests; and routine laboratory markers. Cochrane Database of Systematic Reviews. 2020 doi: 10.1002/14651858.CD013596. doi: 10.1002/14651858.CD013596. [DOI] [Google Scholar]
- 17.Whiting PF, Rutjes AW, Westwood ME, Mallett S, Deeks JJ, Reitsma JB, et al. QUADAS-2: a revised tool for the quality assessment of diagnostic accuracy studies. Ann Intern Med. 2011;155:529–36. doi: 10.7326/0003-4819-155-8-201110180-00009. [DOI] [PubMed] [Google Scholar]
- 18.R Core Team. R: A language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing; 2019. [Google Scholar]
- 19.Cicchetti DV, Sparrow SA. Developing criteria for establishing interrater reliability of specific items: applications to assessment of adaptive behavior. Am J Ment Defic. 1981;86:127–37. [PubMed] [Google Scholar]
- 20.Cai J, Xu J, Lin D, Yang Z, Xu L, Qu Z, et al. A Case Series of children with 2019 novel coronavirus infection: clinical and epidemiological features. Clin Infect Dis. 2020;71:1547–51. doi: 10.1093/cid/ciaa198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chen Y, Chen L, Deng Q, Zhang G, Wu K, Ni L, et al. The presence of SARS-CoV-2 RNA in the feces of COVID-19 patients. J Med Virol. 2020;92:833–40. doi: 10.1002/jmv.25825. [DOI] [PubMed] [Google Scholar]
- 22.Cheung KS, Hung IF, Chan PP, Lung KC, Tso E, Liu R, et al. Gastrointestinal Manifestations of SARS-CoV-2 Infection and Virus Load in Fecal Samples from the Hong Kong Cohort and Systematic Review and Meta-analysis. Gastroenterology. 2020 doi: 10.1053/j.gastro.2020.03.065. S00165085(20)30448-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kujawski SA, Wong KK, Collins JP, Epstein L, Killerby ME, Midgley CM, et al. Clinical and virologic characteristics of the first 12 patients with coronavirus disease 2019 (COVID-19) in the United States. Nat Med. 2020;26:861–8. doi: 10.1038/s41591-020-0877-5. [DOI] [PubMed] [Google Scholar]
- 24.Lei Z, Jie Y, Huang Z, Guo X, Chen J, Peng L, et al. A cross-sectional comparison of epidemiological and clinical features of patients with coronavirus disease (COVID-19) in Wuhan and outside Wuhan, China. Travel Med Infect Dis. 2020;35:101664. doi: 10.1016/j.tmaid.2020.101664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li D, Jin M, Bao P, Zhao W, Zhang S. Clinical Characteristics and Results of Semen Tests Among Men With Coronavirus Disease 2019. JAMA Network Open. 2020;3:e208292-e. doi: 10.1001/jamanetworkopen.2020.8292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lin L, Jiang X, Zhang Z, Huang S, Zhang Z, Fang Z, et al. Gastrointestinal symptoms of 95 cases with SARS-CoV-2 infection. Gut. 2020;69:997–1001. doi: 10.1136/gutjnl-2020-321013. [DOI] [PubMed] [Google Scholar]
- 27.Lo IL, Lio CF, Cheong HH, Lei CI, Cheong TH, Zhong X, et al. Evaluation of SARS-CoV-2 RNA shedding in clinical specimens and clinical characteristics of 10 patients with COVID-19 in Macau. Int J Biol Sci. 2020;16:1698–707. doi: 10.7150/ijbs.45357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ma X, Su L, Zhang Y, Zhang X, Gai Z, Zhang Z. Do children need a longer time to shed SARS-CoV-2 in stool than adults? J Microbiol Immunol Infect. 2020;53:373–6. doi: 10.1016/j.jmii.2020.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Peng L, Liu J, Xu W, Luo Q, Chen D, Lei Z, et al. SARS-CoV-2 can be detected in urine, blood, anal swabs and oropharyngeal swabs specimens. J Med Virol. 2020;92:1676–80. doi: 10.1101/2020.02.21.20026179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tan YP, Tan BY, Pan J, Wu J, Zeng SZ, Wei HY. Epidemiologic and clinical characteristics of 10 children with coronavirus disease 2019 in Changsha, China. J Clin Virol. 2020;127:104353. doi: 10.1016/j.jcv.2020.104353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wang W, Xu Y, Gao R, Lu R, Han K, Wu G, et al. Detection of SARS-CoV-2 in Different Types of Clinical Specimens. JAMA. 2020:e203786. doi: 10.1001/jama.2020.3786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Wei XS, Wang X, Niu YR, Ye LL, Peng WB, Wang ZH, et al. Diarrhea is associated with prolonged symptoms and viral carriage in COVID-19. Clin Gastroenterol Hepatol. 2020 doi: 10.1016/j.cgh.2020.04.030. S1542-3565(20)30526-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wu Y, Guo C, Tang L, Hong Z, Zhou J, Dong X, et al. Prolonged presence of SARS-CoV-2 viral RNA in faecal samples. Lancet Gastroenterol Hepatol. 2020;5:434–5. doi: 10.1016/S2468-1253(20)30083-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiao F, Tang M, Zheng X, Liu Y, Li X, Shan H. Evidence for Gastrointestinal Infection of SARS-CoV-2. Gastroenterology. 2020;158:1831–3.e3. doi: 10.1053/j.gastro.2020.02.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xing YH, Ni W, Wu Q, Li WJ, Li GJ, Wang WD, et al. Prolonged viral shedding in feces of pediatric patients with coronavirus disease 2019. J Microbiol Immunol Infect. 2020;53:473–80. doi: 10.1016/j.jmii.2020.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xu Y, Li X, Zhu B, Liang H, Fang C, Gong Y, et al. Characteristics of pediatric SARS-CoV-2 infection and potential evidence for persistent fecal viral shedding. Nat Med. 2020;26:502–5. doi: 10.1038/s41591-020-0817-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Young BE, Ong SWX, Kalimuddin S, Low JG, Tan SY, Loh J, et al. Epidemiologic Features and Clinical Course of Patients Infected With SARS-CoV-2 in Singapore. JAMA. 2020;323:1488–94. doi: 10.1001/jama.2020.3204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yun H, Sun Z, Wu J, Tang A, Hu M, Xiang Z. Laboratory data analysis of novel coronavirus (COVID-19) screening in 2510 patients. Clin Chim Acta. 2020;507:94–7. doi: 10.1016/j.cca.2020.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang J, Wang S, Xue Y. Fecal specimen diagnosis 2019 novel coronavirus-infected pneumonia. J Med Virol. 2020;92:680–2. doi: 10.1002/jmv.25742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang W, Du RH, Li B, Zheng XS, Yang XL, Hu B, et al. Molecular and serological investigation of 2019-nCoV infected patients: implication of multiple shedding routes. Emerg Microbes Infect. 2020;9:386–9. doi: 10.1080/22221751.2020.1729071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zheng S, Fan J, Yu F, Feng B, Lou B, Zou Q, et al. Viral load dynamics and disease severity in patients infected with SARS-CoV-2 in Zhejiang province, China, January–March 2020: retrospective cohort study. BMJ. 2020;369:m1443. doi: 10.1136/bmj.m1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ngaserin SHN, Koh FH, Ong BC, Chew MH. COVID-19 not detected in peritoneal fluid: a case of laparoscopic appendicectomy for acute appendicitis in a COVID-19-infected patient. Langenbeck’s Arch Surg. 2020;405:353–5. doi: 10.1007/s00423-020-01891-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Candellier A, Goffin É. Letter regarding “SARS-CoV-2 in the peritoneal waste in a patient treated with peritoneal dialysis”. Kidney Int. 2020 doi: 10.1016/j.kint.2020.05.034. S0085-2538(20)30660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Montero Feijoo A, Maseda E, Adalia Bartolomé R, Aguilar G, González de Castro R, Gómez-Herreras JI, et al. Practical recommendations for the perioperative management of the patient with suspection or serious infection by coronavirus SARS-CoV. Rev Esp Anestesiol Reanim. 2020;67:253–60. doi: 10.1016/j.redare.2020.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gheblawi M, Wang K, Viveiros A, Nguyen Q, Zhong JC, Turner AJ, et al. Angiotensin-Converting Enzyme 2: SARS-CoV-2 Receptor and Regulator of the Renin-Angiotensin System: Celebrating the 20th Anniversary of the Discovery of ACE2. Circ Res. 2020;126:1456–74. doi: 10.1161/CIRCRESAHA.120.317015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Qiu L, Liu X, Xiao M, Xie J, Cao W, Liu Z, et al. SARS-CoV-2 is not detectable in the vaginal fluid of women with severe COVID-19 infection. Clin Infect Dis. 2020;71:813–7. doi: 10.1093/cid/ciaa375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mintz Y, Arezzo A, Boni L, Baldari L, Cassinotti E, Brodie R, et al. The risk of COVID-19 transmission by laparoscopic smoke may be lower than for laparotomy: a narrative review. Surg Endosc. 2020;34:3298–305. doi: 10.1007/s00464-020-07652-y. [DOI] [PMC free article] [PubMed] [Google Scholar]


