Abstract
Background and Purpose :
Malassezia yeasts are lipophilic normal flora of the skin in humans and other warm-blooded vertebrates. This genus includes 18 species and is responsible for dermatological disorders, such as pityriasis versicolor, atopic dermatitis, seborrheic dermatitis, folliculitis, and dandruff. The aim of the present study was to identify the etiologic agents of Malassezia infections among the patients referring to the Referral Dermatology Clinic of Al-Zahra Hospital, Isfahan, Iran, during 2018-2019.
Materials and Methods:
For the purpose of the study, clinical specimens, including skin scrapings and dandruff, were collected and subjected to direct microscopy, culture, and polymerase chain reaction (PCR) sequencing. Direct PCR was performed on the clinical samples to amplify the D1/D2 region of 26S rDNA, using specific primers; subsequently, the amplicons were sent for sequencing.
Results:
This study was conducted on 120 patients with suspected pityriasis versicolor and seborrheic dermatitis, who referred to the Referral Dermatology Clinic of Al-Zahra Hospital, Isfahan, Iran, during 2018-2019. Out of this population, 50 (41.7%), 26 (52%), and 24 (48%) cases had Malassezia infection, pityriasis versicolor, and seborrheic dermatitis, respectively. Malassezia globosa was found to be the most prevalent species (n=29, 58%), followed by M. restricta (n=20, 40%), and M. arunalokei (n=1, 2%).
Conclusion:
The epidemiologic study was indicative of the frequency of some Malassezia species, such as M. globosa and M. restricta, in Isfahan, Iran. It can be concluded that direct PCR on clinical samples could be used as a simple, precise, effective, fast, and affordable method for research and even routine medical mycology laboratory studies.
Keywords: Malassezia species, Pityriasis versicolor, Seborrheic dermatitis, 26S rDNA sequencing
Introduction
Malassezia species are lipophilic commensal yeasts found as normal skin flora in 70-95% of healthy individuals. This species is also responsible for dermatological conditions, such as pityriasis versicolor (PV), atopic dermatitis, seborrheic dermatitis (SD), folliculitis, and dandruff [ 1 ]. Malassezia genus consists of 18 species, among which M. furfur, M. obtuse, M. globosa, M. slooffiae, M. sympodialis, and M. restricta frequently cause human infection. Additionally, M. pachydermatis, a zoophilic specie, and M. dermatis have been occasionally isolated from human diseases [ 2 ]. These animal-associated species can cause complicated disseminated fungal infections mainly in neonates, immunocompromised infants, and young children candidate for indwelling catheter [ 3 ].
For the first time, Gaitanis et al. [ 4 ] extracted Malassezia DNA from skin scrapings using hexadecyltrimethylammonium bromide and identified Malassezia species directly from the clinical samples by ITS primers. Since M. restricta and M. obtusa are fastidious, the isolation of these species from culture media cannot be indicative of real normal flora. Several epidemiological studies on Malassezia-related infections have been performed in Iran. For example, Zarei Mahmoudabadi et al. [ 5 ] determined the frequency of the prevalent Malassezia species in patients with SD and PV using the nested polymerase chain reaction (PCR) method, in Ahwaz, Iran. They reported M. obtusa as the most common species (51.3%), followed by M. globosa (35.2%) and M. restricta. With this background in mind, the present study was performed to identify the etiologic agents of PV and SD among the patients referring to the Referral Dermatology Clinic of Al-Zahra Hospital, Isfahan, Iran, during 2018-2019.
Materials and Methods
Patients
Patients with hypo- or hyper-pigmented scaly patches, red skin, and dandruff involving the oily regions of the body, who were suspected to have PV or SD were included in the study during June 2018 to November 2019. On the other hand, those with disseminated skin inflammation or skin peeling or the individuals who had taken antifungal agents within the last 2 months were excluded from the study. The study was approved by the Ethics Committee of Isfahan University of Medical Sciences, Isfahan, Iran (IR.MUI.MED.REC.1397.054).
Sampling
Skin scrapings and dandruff were collected from the included patients and then subjected to direct microscopic examination with potassium hydroxide 10%, culture on Dixon's agar (HiMedia, India), and direct PCR sequencing. In addition, a concentrated suspension of positive cultures was prepared and kept at -20°C as a source of specimen.
Direct polymerase chain reaction for the amplification of D1/D2 region of 26S rDNA
Genomic DNA was extracted from the skin scrapings and dandruff samples using glass bead and phenol/chloroform techniques [ 6 , 7 ]. Briefly, a loopful of the skin scale or dandruff was transferred to a 2-mL Eppendorf tube, containing 300 μL lysis buffer (200 mM Tris/HCl with a pH of 7.5, 25 mM EDTA, 0.5% SDS, and 250 mM NaCl) and 300 μL glass beads. Subsequently, the samples were centrifuged for 1 min at 6,000 rpm for three times and then added with 300 μL phenol/chloroform, followed by vortexing and centrifugation for 5 min at 5,000 rpm. In the next stage, the supernatant was transferred to a new tube, added with the same amount of chloroform, and centrifuged for 5 min at 5,000 rpm. After transferring the superna- tant to a new tube and adding alcohol (2.5 times) and 3 M sodium acetate (1/10 volume), the final mixture was stored at -20°C for 1 h and then centrifuged for 5 min at 10,000. Following the removal of the supernatant, 500 μL alcohol 70% was added to the pellet, which was then centrifuged for 10 min at 10,000 rpm. At this stage, the supernatant was thrown away again, and 50 μL double distilled water was added and kept at -20°C. The PCR amplification was performed at a final volume of 50 µl. Each PCR reaction included 5 µl of 10×PCR buffer, 0.2 mM of each deoxynucleoside triphosphate, 0.5 mM of each forward (5΄- TAACAAGGATTCCCCTAGTA-3΄) and reverse (5΄- ATTACGCCAGCATCCTAAG-3΄) primers [ 8 ], 1.25 U of Taq polymerase, and 2 µl template DNA. The PCR conditions consisted of an initial denaturation step at 95ºC for 5 min, followed by 32 cycles of denaturation at 95ºC for 45 sec, annealing at 55ºC for 45 sec, and extension at 72ºC for 1 min, with a final extension step of 72ºC for 7 min [ 8 ]. The amplified products were visualized by 1.5% (w/v) agarose gel electrophoresis in TBE buffer, stained with ethidium bromide (0.4 µg/ml), and photographed under ultraviolet transilluminator (UVITEC, UK).
Polymerase chain reaction sequencing
All PCR products were subjected to sequence analysis. The amplicons were purified using the ethanol purification method; furthermore, cycle sequencing reactions were performed in a forward direction (Bioneer, South Korea). The sequencing products were analyzed with Chromas 2.4 (https://chromas.software.informer.com/2.4/) and then evaluated using the NCBI BLAST searches against fungal sequences existing in DNA databases (https://blast.ncbi.nlm.nih.gov/Blast.cgi).
Statistical analysis
The results were analyzed by the Chi-square and Fisher’s exact tests in the SPSS software, version 23 (Armonk, NY: IBM Corp). A p-value less than 0.05 was considered statistically significant.
Results
A total of 120 patients suspected of PV and SD referred to the Referral Dermatology Clinic of Al- Zahra Hospital from June 2018 to November 2019. Out of 120 subjects, 50 (41.6%), 26 (52%), and 24 (48%) cases had Malassezia-related infection, PV, and SD, respectively. The age range of the patients was between 9 and 60 years with the median age of 30.2 years. The age ranges of 21-30 (44%) and 0-10 (2%) years had the highest and lowest frequency, respectively. The male to female ratio of the study participants was 35/15. With regard to the occupational status, the majority of the patients were shopkeepers (22%), students (18%), housekeepers (18%), employees (12%), and construction workers (10%).
The clinical specimens were obtained from dandruff (28%), lesions on the neck (24%), lesions on the chest (12%), flakes behind the ears (8%), lesions on the upper back (6%), flakes around the sides of the nose (6%), lesions on the arms (4%), lesions of the armpits (4%), flakes of the eyebrows (4%), groin (2%), and flakes of the beard (2%; Figure 1). The predisposing factors were identified as hyperhidrosis (24%), stress (14%), wearing tight clothes (10%), poor hygiene (8%), solid organ transplantation (2%), and overweight (2%; Table 1). In addition, Malassezia globosa was found to be the most prevalent species (n=29, 58%), followed by M. restricta (n=20, 40%), and M. arunalokei (n=1, 2%; Table 1, Figure 2). The results of the Fisher’s exact test showed that the association between the clinical sample type and Malassezia species was not statistically significant (P=0.87).
Figure 1.
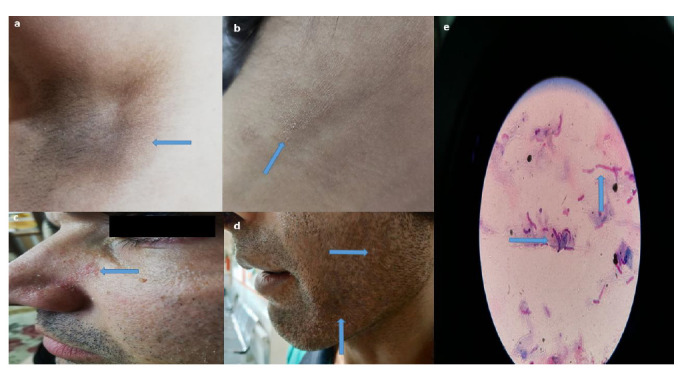
Clinical signs of infection among patients; a) lesions of the armpits, b) lesions on the neck, c) flakes around the sides of the nose, d) flakes of beard, and e) hyphae and globose yeast cells (spaghetti and meatballs) (typical microscopy for Malassezia species, Gram stain, original magnification ×100)
Table 1.
Demographic and clinical characteristics of patients with different types of Malassezia infection
| No. | Gender | Age (years) | Occupation | Primary diagnosis | Predisposing factors | Seasonal distribution | Malassezia spp. | Accession number |
|---|---|---|---|---|---|---|---|---|
| 1 | Male | 35 | Construction worker | SD | Unknown | Summer | M. restricta | MT645538 |
| 2 | Female | 28 | Housekeeper | PV | Hyperhidrosis | Summer | M. globosa | MT645539 |
| 3 | Male | 26 | Shopkeeper | SD | Poor hygiene | Summer | M. restricta | MT645540 |
| 4 | Male | 26 | Shopkeeper | PV | Poor hygiene | Summer | M. globosa | MT645541 |
| 5 | Female | 30 | Employee | PV | Stress | Summer | M. globosa | MT645542 |
| 6 | Male | 32 | Construction worker | PV | Hyperhidrosis | Summer | M. restricta | MT645543 |
| 7 | Male | 35 | Shopkeeper | SD | Wearing tight clothes | Summer | M. restricta | MT645544 |
| 8 | Male | 25 | Free-lancer | SD | Stress | Summer | M. restricta | MT645545 |
| 9 | Male | 30 | Shopkeeper | PV | Hyperhidrosis | Summer | M. restricta | MT645546 |
| 10 | Male | 20 | Athlete | PV | Wearing tight clothes | Summer | M. globosa | MT645547 |
| 11 | Male | 22 | Shopkeeper | SD | Unknown | Summer | M. restricta | MT645548 |
| 12 | Male | 31 | Shopkeeper | SD | Wearing tight clothes | Summer | M. restricta | MT645549 |
| 13 | Male | 35 | Construction worker | SD | Unknown | Spring | M. restricta | MT645550 |
| 14 | Male | 40 | Employee | SD | Stress | Summer | M. globosa | MT645551 |
| 15 | Female | 38 | Housekeeper | PV | Wearing tight clothes | Spring | M. restricta | MT645552 |
| 16 | Male | 28 | Student | SD | Stress | Spring | M. globosa | MT645553 |
| 17 | Female | 30 | Housekeeper | SD | Unknown | Fall | M. restricta | MT645554 |
| 18 | Female | 29 | Housekeeper | SD | Unknown | Summer | M. globosa | MT645555 |
| 19 | Male | 17 | Student | SD | Unknown | Summer | M. restricta | MT645556 |
| 20 | Male | 13 | Student | PV | Stress | Summer | M. restricta | MT645557 |
| 21 | Male | 54 | Shopkeeper | PV | Unknown | Summer | M. globosa | MT645558 |
| 22 | Female | 32 | Housekeeper | SD | Unknown | Spring | M. restricta | MT645559 |
| 23 | Male | 27 | Free-lancer | SD | Unknown | Fall | M. globosa | MT645560 |
| 24 | Male | 23 | Student | SD | Stress | Fall | M. arunalokei | MT645561 |
| 25 | Male | 27 | Employee | SD | Unknown | Summer | M. restricta | MT645562 |
| 26 | Female | 24 | Student | SD | Unknown | Summer | M. globosa | MT645563 |
| 27 | Male | 29 | Shopkeeper | PV | Unknown | Fall | M. restricta | MT645564 |
| 28 | Male | 25 | Unemployed | SD | Hyperhidrosis | Spring | M. globosa | MT645565 |
| 29 | Male | 60 | Retired | PV | Solid organ transplantation | Spring | M. globosa | MT645566 |
| 30 | Female | 31 | Employee | PV | Hyperhidrosis | Spring | M. globosa | MT645567 |
| 31 | Male | 35 | Shopkeeper | SD | Unknown | Spring | M. restricta | MT645568 |
| 32 | Male | 30 | Free-lancer | SD | Unknown | Summer | M. globosa | MT645569 |
| 33 | Male | 26 | Unemployed | SD | Unknown | Summer | M. globosa | MT645570 |
| 34 | Male | 34 | Construction worker | PV | Poor hygiene | Winter | M. globosa | MT645571 |
| 35 | Male | 13 | Student | SD | Stress | Fall | M. restricta | MT645572 |
| 36 | Male | 27 | Driver | SD | Unknown | Fall | M. restricta | MT645573 |
| 37 | Female | 33 | Housekeeper | PV | Unknown | Summer | M. globosa | MT645574 |
| 38 | Female | 30 | Employee | PV | Unknown | Spring | M. globosa | MT645575 |
| 39 | Female | 32 | Housekeeper | PV | Hyperhidrosis | Spring | M. globosa | MT645576 |
| 40 | Male | 38 | Driver | PV | Unknown | Winter | M. globosa | MT645577 |
| 41 | Male | 39 | Shopkeeper | PV | Hyperhidrosis | Summer | M. globosa | MT645578 |
| 42 | Female | 38 | Housekeeper | PV | Overweight | Winter | M. globosa | MT645579 |
| 43 | Male | 38 | Construction worker | PV | Hyperhidrosis | Fall | M. globosa | MT645580 |
| 44 | Female | 23 | Student | PV | Hyperhidrosis | Fall | M. globosa | MT645581 |
| 45 | Male | 53 | Shopkeeper | PV | Wearing tight clothes | Fall | M. globosa | MT645582 |
| 46 | Female | 32 | Sports Coach | PV | Hyperhidrosis | Fall | M. globosa | MT645583 |
| 47 | Male | 9 | Student | PV | Poor hygiene | Spring | M. globosa | MT645584 |
| 48 | Female | 32 | Housekeeper | PV | Hyperhidrosis | Spring | M. globosa | MT645585 |
| 49 | Male | 30 | Employee | PV | Hyperhidrosis | Spring | M. globosa | MT645586 |
| 50 | Male | 15 | Student | SD | Unknown | Spring | M. restricta | MT645587 |
PV: Pityriasis versicolor; SD: Seborrheic dermatitis
Figure 2.
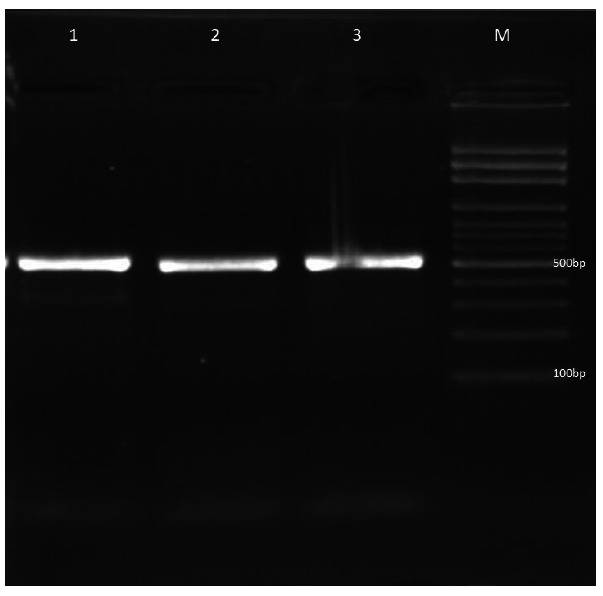
Gel electrophoresis of 26S rDNA-polymerase chain reaction amplicons of Malassezia species; lanes 1-3) M. restricta and lane M) 100-bp DNA size marker
Discussion
Malassezia yeasts are lipophilic normal microbial flora needing specific lipids, such as oleic acid, for their growth. Colony formation begins quickly after birth and remarkably increments with the increase of neonatal age. Skin colonization by Malassezia species is increased from 7% at the first week to 40% at 3-5 weeks of life [ 9 ]. Malassezia genus has undergone many taxonomic classifications during the last 2 decades. Up to 1995, three Malassezia species, containing M. obtusa, M. sympodialis, and M. pachydermatis, had been identified [ 10 ].
Guého et al. [ 11 ] revealed a taxonomic revision that resulted in four new Malassezia species, including M. obtuse, M. globosa, M. slooffiae, and M. restricta, based on ultrastructure, physiology, morphology, and also molecular information of rDNA sequencing. Thereafter, other new species were isolated and identified from humans (M. japonica [ 12 ], M. dermatis [ 13 ], M. yamatoensis) [ 14 ], as well as animals (M. nana [ 15 ], M. equine [ 16 ], M. caprae [ 16 ], and M. cuniculi [ 17 ]). Furthermore, four undifferentiated phylotypes were offered in psoriatic and healthy individuals [ 18 , 19 ].
Recently, new Malassezia species have been isolated from opossum and parrots based on multilocus sequence analysis [ 20 ], suggesting that Malassezia genus may comprise more species than presently known. Similar to the present study, Prohic et al. [ 21 ] and Rodoplu et al.
reported M. globosa as the predominant Malassezia species isolated from PV lesions in India (52%) and Turkey (65%), respectively. In the same vein, Rasi et al.
reported M. globosa as the most prevalent Malassezia species in Tehran, Iran, from 2006 to 2007. In the mentioned study, M. obtusa was found to be the second most frequent species; however, Malassezia furfur was not isolated from Isfahan samples in the current study. Rasi et al. used phenotypic tests, such as Tween assimilation test, splitting of esculin, and catalase reaction, for the identification of the clinical isolates. In the mentioned research, 30.2% of the Malassezia culture was negative, which is similar to the result obtained in the present study (32%).
Malassezia restricta and M. globosa are the species most repeatedly found on the skin of healthy and infected humans. However, other species, such as M. obtusa and M. sympodialis, have been also associated with different human skin disorders [ 24 , 25 ]. Malassezia globosa is found more regularly on the arms and chest, whereas M. restricta is reported to be more common on the forehead and scalp [ 21 , 26 ]. Furthermore, M. restricta has been isolated more frequently in teenagers and young people, while M. globosa is the most prevalent species in individuals over 50 years of age [ 27 ]. In many studies, Malassezia species are identified based on microscopic characteristics, colony morphology, and complicated and time-consuming tests (e.g., esculin hydrolysis tests, assimilation of Tween 20, 40, and 80, and Cremophor El) [ 28 , 29 ].
Nevertheless, for the first time, Gaitanis et al. [ 4 ] used the nested PCR, PCR-restriction fragment length polymorphism (RFLP), and PCR sequencing for the identification of Malassezia species isolated from skin scrapings. They extracted Malassezia DNA from 45.5% of samples; however, in the present research, the successful extraction rate from clinical samples was obtained as 100%. The PCR-based method is less costly and more rapid in comparison to the phenotypic methods. Moreover, the traditional methods for Malassezia identification are complex and require experienced specialists.
Tarazooie et al. [ 30 ] compared the distribution of Malassezia species isolated from PV lesions and those isolated from healthy skins by phenotypic tests. Similar to our study, they reported M. globose as the most prevalent species; however, contrary to our study, they did not isolate any M. restricta from the infected patients. The PV is uncommon in children and the elderly. Accordingly, in the present study, there were just one case of PV in a child aged < 10 years and two cases of PV in the subjects aged > 50 years. This result is completely in line with those obtained by Tarazooie et al. [ 30 ].
In another study, Berenji et al. [ 31 ] obtained a prevalence of 58.1% for the superficial fungal infections caused by Malassezia species in Mashhad, from 2000 to 2011. In the mentioned study, PV and SD were observed in 19% and 58.4% of the cases, respectively. In the present research, one M. arunalokei isolate was isolated from the scalp of a 23-year-old student. He was very stressful because of his lessons and had a long history of dandruff. Malassezia arunalokei was described as a new species by Honnavar et al. [ 32 ] in 2016. They observed sequence divergence in the D1/D2 domain, intergenic spacer 1 region of rDNA, ITS region, and the TEF1 gene of new species. Similar to their findings, in the present study,
M. arunalokei was isolated from a patient with SD. To the best of our knowledge, the current study is the first report on the isolation of this species from Iran.
Conclusion
As the findings indicated, the two-step molecular technique adopted in this study could facilitate the identification of all Malassezia species without any need for cultivating the clinical isolates. This technique only involved one PCR reaction and a sequencer to identify the clinical isolates at the species level. Accordingly, it can be concluded that this technique can be used as a simple, precise, effective, fast, and affordable method for research and even routine medical mycology laboratory studies.
Acknowledgement
This study was supported by the Isfahan University of Medical Sciences, Isfahan, Iran (Number: 397113), also contributing to the design of the investigation.
Author’s contribution
R. M. designed the study. M. G. and F. M. performed data collection. R. M., M. G., and F. M. carried out data analysis. R. M. prepared the manuscript. All authors read and approved the final manuscript.
Conflict of Interest: The authors declare that there is no conflict of interest.
Financial disclosure
The authors declare no financial disclosure.
References
- 1.Angiolella L, Carradori S, Maccallini C, Giusiano G, Supuran TC. Targeting Malassezia species for novel synthetic and natural antidandruff agents. Curr Med Chem. 2017; 24(22):2392–412. doi: 10.2174/0929867324666170404110631. [DOI] [PubMed] [Google Scholar]
- 2.Theelen B, Cafarchia C, Gaitanis G, Bassukas ID, Boekhout T, Dawson TL Jr. Malassezia ecology, pathophysiology, and treatment. Med Mycol. 2018; 56(Suppl 1):S10–25. doi: 10.1093/mmy/myx134. [DOI] [PubMed] [Google Scholar]
- 3.Diongue K, Kébé O, Faye M, Samb D, Diallo M, Ndiaye M, et al. MALDI-TOF MS identification of Malassezia species isolated from patients with pityriasis versicolor at the Seafarers’ Medical Service in Dakar, Senegal. J Mycol Med. 2018; 28(4):590–3. doi: 10.1016/j.mycmed.2018.09.007. [DOI] [PubMed] [Google Scholar]
- 4.Gaitanis G, Velegraki A, Frangoulis E, Mitroussia A, Tsigonia A, Tzimogianni A, et al. Identification of Malassezia species from patient skin scales by PCR-RFLP. Clin Microbiol Infect. 2002; 8(3):162–73. doi: 10.1046/j.1469-0691.2002.00383.x. [DOI] [PubMed] [Google Scholar]
- 5.Zarei Mahmoudabadi A, Zarrin M, Azish M. Detection of Malassezia species isolated from patients with pityriasis versicolor and seborrheic dermatitis using Nested-PCR. Jentashapir J Health Res. 2015; 5(6):e26683. [Google Scholar]
- 6.Yamada Y, Makimura K, Merhendi H, Ueda K, Nishiyama Y, Yamaguchi H, et al. Comparison of different methods for extraction of mitochondrial DNA from human pathogenic yeasts. Jpn J Infect Dis. 2002; 55(4):122–5. [PubMed] [Google Scholar]
- 7.Gupta A, Kohli Y, Summerbell R. Molecular differentiation of seven Malassezia species. J Clin Microbiol. 2000; 38(5):1869–75. doi: 10.1128/jcm.38.5.1869-1875.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mirhendi H, Makimura K, Zomorodian K, Yamada T, Sugita T, Yamaguchi H. A simple PCR-RFLP method for identification and differentiation of 11 Malassezia species. J Microbiol Methods. 2005; 61(2):281–4. doi: 10.1016/j.mimet.2004.11.016. [DOI] [PubMed] [Google Scholar]
- 9.Ayhan M, Sancak B, Karaduman A, Arikan S, Sahin S. Colonization of neonate skin by Malassezia species: relationship with neonatal cephalic pustulosis. J Am Acad Dermatol. 2007; 57(6):1012–8. doi: 10.1016/j.jaad.2007.02.030. [DOI] [PubMed] [Google Scholar]
- 10.1Ashbee HR. Update on the genus Malassezia. Med Mycol. 2007; 45(4):287–303. doi: 10.1080/13693780701191373. [DOI] [PubMed] [Google Scholar]
- 11.Guého E, Midgley G, Guillot J. The genus Malassezia with description of four new species. Antonie Van Leeuwenhoek. 1996; 69(4):337–55. doi: 10.1007/BF00399623. [DOI] [PubMed] [Google Scholar]
- 12.Sugita T, Takashima M, Kodama M, Tsuboi R, Nishikawa A. Description of a new yeast species, Malassezia japonica, and its detection in patients with atopic dermatitis and healthy subjects. J Clin Microbiol. 2003; 41(10):4695–9. doi: 10.1128/JCM.41.10.4695-4699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sugita T, Takashima M, Shinoda T, Suto H, Unno T, Tsuboi R, et al. New yeast species, Malassezia dermatis, isolated from patients with atopic dermatitis. J Clin Microbiol. 2002; 40(4):1363–7. doi: 10.1128/JCM.40.4.1363-1367.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sugita T, Tajima M, Takashima M, Amaya M, Saito M, Tsuboi R, et al. A new yeast, Malassezia yamatoensis, isolated from a patient with seborrheic dermatitis, and its distribution in patients and healthy subjects. Microbiol Immunol. 2004; 48(8):579–83. doi: 10.1111/j.1348-0421.2004.tb03554.x. [DOI] [PubMed] [Google Scholar]
- 15.Hirai A, Kano R, Makimura K, Duarte ER, Hamdan JS, Lachance MA, et al. Malassezia nana sp. nov., a novel lipid- dependent yeast species isolated from animals . Int J Syst Evol Microbiol. 2004; 54(Pt 2):623–7. doi: 10.1099/ijs.0.02776-0. [DOI] [PubMed] [Google Scholar]
- 16.Cabañes FJ, Theelen B, Castellá G, Boekhout T. Two new lipid- dependent Malassezia species from domestic animals. FEMS Yeast Res. 2007; 7(6):1064–76. doi: 10.1111/j.1567-1364.2007.00217.x. [DOI] [PubMed] [Google Scholar]
- 17.Cabañes F, Vega S, Castellá G. Malassezia cuniculi sp. nov., a novel yeast species isolated from rabbit skin. Med Mycol. 2011; 49(1):40–8. doi: 10.3109/13693786.2010.493562. [DOI] [PubMed] [Google Scholar]
- 18.Paulino LC, Tseng CH, Strober BE, Blaser MJ. Molecular analysis of fungal microbiota in samples from healthy human skin and psoriatic lesions. J Clin Microbiol. 2006; 44(8):2933–41. doi: 10.1128/JCM.00785-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paulino LC, Tseng CH, Blaser MJ. Analysis of Malassezia microbiota in healthy superficial human skin and in psoriatic lesions by multiplex real-time PCR. FEMS Yeast Res. 2008; 8(3):460–71. doi: 10.1111/j.1567-1364.2008.00359.x. [DOI] [PubMed] [Google Scholar]
- 20.Castellá G, Coutinho SD, Cabañes FJ. Phylogenetic relationships of Malassezia species based on multilocus sequence analysis. Med Mycol. 2014; 52(1):99–105. doi: 10.3109/13693786.2013.815372. [DOI] [PubMed] [Google Scholar]
- 21.Prohic A, Simic D, Sadikovic TJ, Krupalija-Fazlic M. Distribution of Malassezia species on healthy human skin in Bosnia and Herzegovina: correlation with body part, age and gender. Iran J Microbiol. 2014; 6(4):253–62. [PMC free article] [PubMed] [Google Scholar]
- 22.Rodoplu G, Saracli M, Gümral R, Yildiran ST. Distribution of Malassezia species in patients with pityriasis versicolor in Turkey. J Mycol Med. 2014; 24(2):117–23. doi: 10.1016/j.mycmed.2014.02.001. [DOI] [PubMed] [Google Scholar]
- 23.Rasi A, Naderi R, Heshmatzade Behzadi A, Falahati M, Farehyar S, Honarbakhsh Y, et al. Malassezia yeast species isolated from Iranian patients with pityriasis versicolor in a prospective study. Mycoses. 2010; 53(4):350–5. doi: 10.1111/j.1439-0507.2009.01727.x. [DOI] [PubMed] [Google Scholar]
- 24.Difonzo E, Faggi E, Bassi A, Campisi E, Arunachalam M, Pini G, et al. Malassezia skin diseases in humans. G Ital Dermatol Venereol. 2013; 148(6):609–19. [PubMed] [Google Scholar]
- 25.Jagielski T, Rup E, Ziółkowska A, Roeske K, Macura AB, Bielecki J. Distribution of Malassezia species on the skin of patients with atopic dermatitis, psoriasis, and healthy volunteers assessed by conventional and molecular identification methods. BMC Dermatol. 2014; 14:3. doi: 10.1186/1471-5945-14-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kaur M, Narang T, Bala M, Gupte S, Aggarwal P, Manhas A. Study of the distribution of Malassezia species in patients with pityriasis versicolor and healthy individuals in Tertiary Care Hospital, Punjab. Indian J Med Microbiol. 2013; 31(3):270–4. doi: 10.4103/0255-0857.115636. [DOI] [PubMed] [Google Scholar]
- 27.Lee Y, Yim S, Lim S, Choe Y, Ahn K. Quantitative investigation on the distribution of Malassezia species on healthy human skin in Korea. Mycoses. 2006; 49(5):405–10. doi: 10.1111/j.1439-0507.2006.01239.x. [DOI] [PubMed] [Google Scholar]
- 28.Guillot J, Gueho E, Lesourd M, Midgley G, Chevrier G, Dupont B. Identification of Malassezia species. A practical approach. J Mycol Med. 1996; 6(3):103–10. [Google Scholar]
- 29.Mayser P, Haze P, Papavassilis C, Pickel M, Gruender K, Gueho E. Differentiation of Malassezia species: selectivity of Cremophor El, castor oil and ricinoleic acid for M. furfur. Br J Dermatol. 1997; 137(2):208–13. doi: 10.1046/j.1365-2133.1997.18071890.x. [DOI] [PubMed] [Google Scholar]
- 30.Tarazooie B, Kordbacheh P, Zaini F, Zomorodian K, Saadat F, Zeraati H, et al. Study of the distribution of Malassezia species in patients with pityriasis versicolor and healthy individuals in Tehran, Iran. BMC Dermatol. 2004; 4:5. doi: 10.1186/1471-5945-4-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Berenji F, Mahdavi Sivaki M, Sadabadi F, Andalib Aliabadi Z, Ganjbakhsh M, Salehi M. A retrospective study of cutaneous fungal infections in patients referred to Imam Reza Hospital of Mashhad, Iran during 2000-2011. Curr Med Mycol. 2016; 2(1):20–3. doi: 10.18869/acadpub.cmm.2.1.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Honnavar P, Prasad GS, Ghosh A, Dogra S, Handa S, Rudramurthy SM. Malassezia arunalokei sp. nov., a novel yeast species isolated from seborrheic dermatitis patients and healthy individuals from. India J Clin Microbiol. 2016; 54(7):1826–34. doi: 10.1128/JCM.00683-16. [DOI] [PMC free article] [PubMed] [Google Scholar]


