Summary
T cell development is a major branch of lymphoid development and a key output of hematopoiesis, especially in early life, but the molecular requirements for T-cell potential have remained obscure. Considerable advances have now been made toward solving this problem through single-cell transcriptome studies, interfaced with in vitro differentiation assays that monitor potential efficiently at the single cell level. This review focuses on a series of recent reports studying mouse and human early T cell precursors, both in the developing fetus and in stringently purified postnatal samples of intrathymic and prethymic T-lineage precursors. Cross-comparison of results shows a robustly conserved core program in mouse and human, but with some informative and provocative variations between species and between ontogenic states. Repeated findings are the multipotent progenitor regulatory signature of thymus-seeding cells and the proximity of the T cell program to dendritic cell programs, especially to plasmacytoid dendritic cells in humans.
Keywords: T lymphocyte development, Single-cell transcriptome, Thymus, Lineage choice, Regulatory factors
Graphical abstract
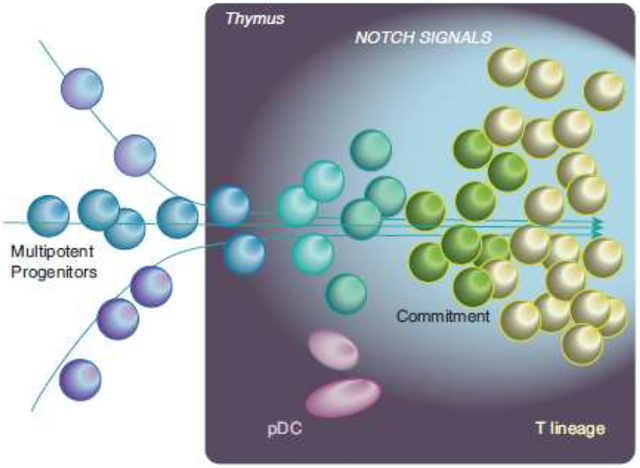
Introduction
T and B lymphocyte developmental processes have enjoyed particularly in-depth characterization [1–6]. However, relatively little knowledge concerning T cells has been integrated into current discussions of broader hematopoietic differentiation. Most single-cell transcriptome analyses of multilineage hematopoietic stem and progenitor cells, while powerful and sophisticated [7–12], have undercharacterized lymphoid precursors. Cells starting the B cell and natural killer (NK) cell programs are detected in bone marrow and are often labeled as general “lymphoid” precursors [13–15], but benchmarks for initiation of the T-cell program have been lacking. This is a problem because the pathway toward T-cell development is intrinsically different from pathways to B and NK cells. First, it depends on the microenvironment of the thymus. Also, at least some T cell precursors diverge from B cell precursors prethymically, in mice and humans [14, 16, 17]. Recently, multiple single-cell transcriptome publications have clarified the earliest stages of T cell development. These insights provide an opportunity to link the T cell developmental pathway better to the rest of hematopoiesis.
Several issues contribute to the poor characterization of T-cell precursors in bone marrow and cord blood. First, although abundant in blood, T cells are long-lived, and they undergo extensive proliferation both within the thymus during development and also as part of their function after they are mature. Thus, their population homeostasis does not depend on continuous input of large numbers of precursors. In young mice, the thymus can generate ~5×107 cells/day, exporting ~106 mature cells/day, from inputs of just ~10 cells/day [18]. Thus, any dedicated T-lymphoid precursors could be rare in postnatal BM and still supply the thymus in steady state. Second, the thymus is a formidable black box between transcriptional states in bone marrow and blood populations. The cells coming into the thymus should resemble not mature T cells but the most immature thymocytes, which are rare even within the thymus and unfamiliar to most hematologists and immunologists alike. Third, T-cell development initiation depends on intense, multi-day exposure to Notch signals that inhibit other lymphoid and myeloid fates as they push T-cell differentiation forward [19, 20]. This greatly complicates attempts to test single precursors for T-cell potential simultaneously with other potentials. While assay systems have been designed [13, 21, 22], they all require caveats.
Mice and humans also differ in the timing of their first waves of T cell development relative to gestation. In mice, with a 20-day gestation period, T cell development starts at ~ d13 and the thymus makes its first cohort of mature-type TCRαβ+ T cells only just before birth [23]. In humans, on the other hand, T cell development starts by 8–9 weeks of gestation and the first T cells are mature in the thymus by week 15, less than halfway through gestation [24]. Neonatal thymectomy is therefore lethal for mice, but not for humans.
Mouse-human differences are relevant because most understanding of lymphocyte development has come from complex genetic manipulations and in vivo cell transfers in mice. In contrast, much of the recent single-cell analysis of hematopoietic precursors has focused on human samples. The cell-surface markers that enrich or define particular developmental stages are also different between mouse and human. Now, however, using the common platform of single-cell transcriptomics, it has become possible to bring together insights from both biological systems to elicit general principles about the hematopoietic roots of T-cell development.
Overview of T-cell development
T cell development is outlined in Fig. 1, in the framework of the mouse thymus [5, 6, 18, 20, 25, 26](for the human system, see [19]). In each cohort of precursors, cells between entry into the thymus and the first functional expression of T-cell receptor complexes (TCR) are called “pro-T cells”, and their development depends on Notch signaling. The stages can be subdivided into thymus-settling precursors (TSP), “Early T-cell Precursors” (ETP), DN2 and DN3 (each with subdivisions)(Fig. 1). T cell identity is determined during the pro-T cell stages, before TCR expression. The process involved, as recently illuminated by single-cell analysis, is the focus of this review.
Figure 1. Stages of mouse T-cell development.
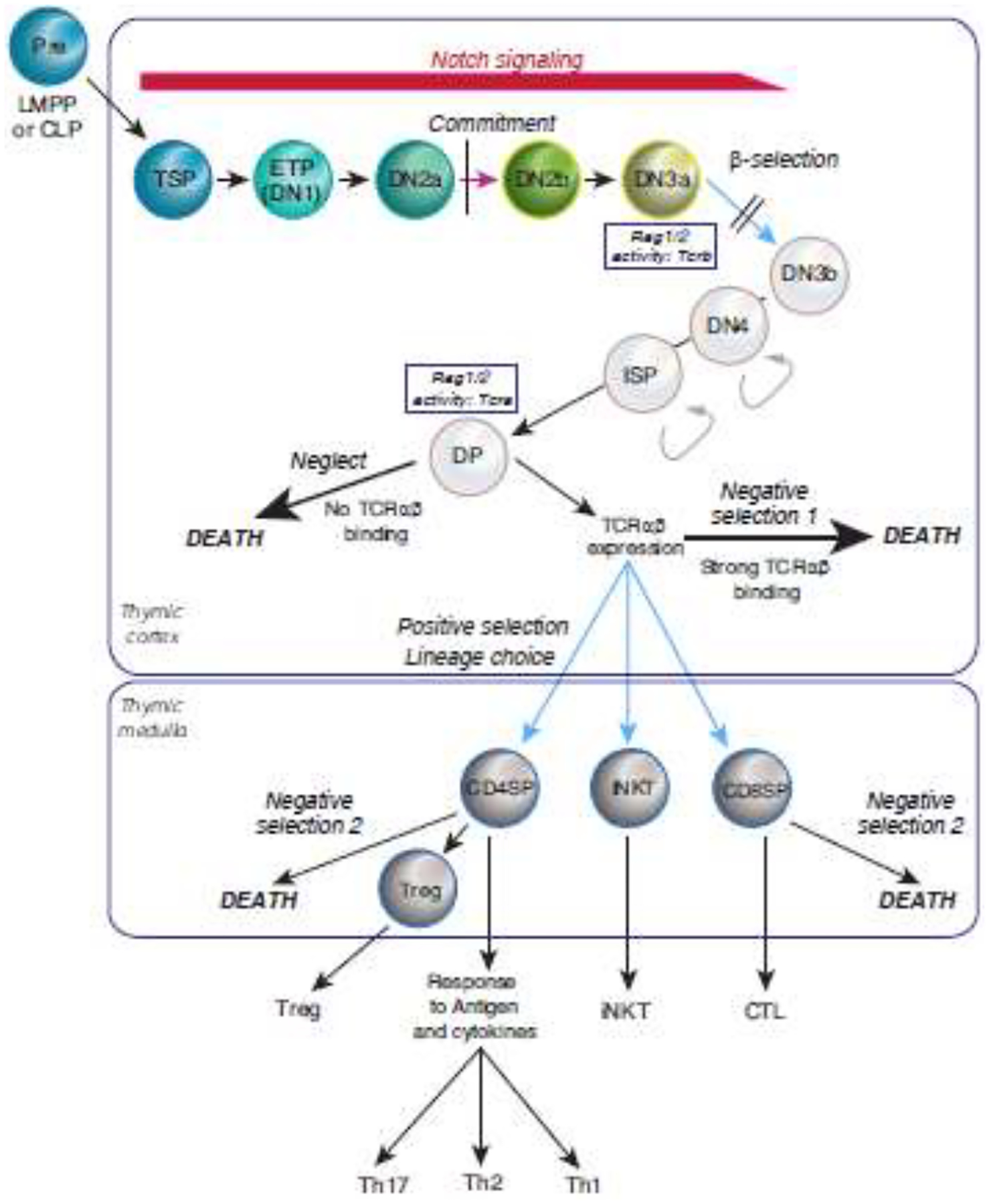
Stages of early T cell development in the postnatal mouse are shown, from TSP to DN3a, in the context of the full span of intrathymic T cell development. Color highlights “hematopoietic” stages of T cell development, the focus of this review. Intrathymic Notch signaling (red) triggers and sustains proliferation and drives changes in surface phenotype from ETP (early T-cell precursor) to DN3a (DN= “Double negative”, lacking both CD4 and CD8). Substage markers are shown in Table 1 and Fig. 2 (also see refs [25, 28]). Commitment occurs between DN2a and DN2b (magenta arrow). At DN3a stage, cells recombine T-cell receptor (TCR) coding gene(s), β as shown (Rag1/2 activity: Tcrb) or γ and δ (not shown), enabling TCR protein(s) to be expressed if the recombination creates an intact protein reading frame. Signaling through a TCR complex containing TCRβ is required (blue arrow, dependent on TCR) for cells to pass “β-selection”, a stringent developmental checkpoint. Successful cells proliferate intensely (curved arrows), lose sensitivity to Notch signaling, and progress through DN3b, DN4, and ISP intermediates to become DP cells (CD4, CD8 “double positive”), then do not proliferate further. DP cells recombine the TCRα coding gene (Rag1/2 activity: Tcra), enabling expression of the complete TCRαβ complex. DP cells are selected for survival based on whether their TCRαβ complex has an appropriate affinity interaction with ligands expressed by the thymic epithelium. The great majority of DP cells are killed intrathymically, due to TCRαβ complex expression failure or failure to bind these ligands at all (neglect), or due to binding too strongly, triggering apoptosis (negative selection 1). TCR complexes showing appropriate affinity enable cells to survive and become positively selected (blue arrows, dependent on TCR). Positive selection causes cells to diverge toward functionally different destinies (lineage choice) and move from the thymic cortex to the medulla. TCR interaction with additional microenvironmental elements is re-tested in the medulla, with another round of negative selection for strongly autoreactive cells (negative selection 2). A subset of autoreactive cells is shunted to a Treg fate rather than death. Surviving cells finally mature and exit the thymus. CD4 SP, CD4 single-positive cells, mostly helpers. iNKT, invariant NK-like T cells, mostly rapid cytokine producers. CD8 SP, mostly precursors of killer T cells (CTL). Treg, regulatory T cells, immunosuppressive. Th1, Th2, Th17 cells, quasi-lineage subtypes of cytokine-producing helper T cells.
T cells are closely related both to B cells and to Innate Lymphoid Cells (ILC), including both helper-type ILCs and natural killer (NK) cells. The similarities are modular [3]. T cells share with B cells their mechanisms for generating clonally unique antigen receptor sequences (immunoglobulin for B cells, TCR for T cells), through RAG1-RAG2-mediated somatic recombination. They share with ILCs nearly all their effector functions (cytolytic mechanisms and secretion of particular cytokine combinations) and transcriptional regulators, although ILCs are RAG independent. Thus, a priori, it is hard to predict whether prethymic precursors should be more like B cells, more like ILC, or more like multipotent precursors with broader potentials.
T cell identity involves both positive activation of T-cell gene expression and loss of access to alternative pathways. From population studies in the mouse, it is known that the T-cell transcriptional regulatory program begins in the ETP and DN2 stages, substantially before the TCR gene recombination which occurs in DN3 stage [27–29], and this pattern is shared in humans (see below)[30]. Important landmarks are known from purified subpopulation studies. Mouse T cell precursors begin expressing T-cell transcription factors, GATA3 and TCF1 (encoded by Tcf7), soon after entering the thymus, while other transcription factors, including IKAROS and RUNX family factors and basic helix-loop-helix “E proteins” like E2A, are expressed even before entry. Some genes shared between T and B lineage cells, such as Dntt (encoding Terminal Deoxynucleotidyl Transferase), may be expressed in TSP as well. However, T-cell lineage-specific genes, like those encoding the CD3 chains that provide membrane anchoring and signaling function for the TCR, are only upregulated in the late DN2 stage, and the Rag1, Rag2 recombinase-coding genes turn on even later, in DN3 stage.
Within the thymus, the strong Notch ligand presentation by the environment makes it overwhelmingly likely that immigrant precursors will generate T-cell progeny; however, the cells themselves are not intrinsically committed to do this when they arrive. Instead, for many cell cycles, their fate as T cells remains completely environment-dependent [21, 31–34]. If ETP or DN2 cells are removed from the thymus and placed in conditions without Notch signaling, the cells develop into non-T cells instead. In contrast, normal DN3 cells can no longer make this shift, but have become intrinsically committed. Further work has located the transition from multipotency to commitment (assayed at the single-cell level) within the mouse DN2 stage(s), when the transcription factor BCL11B is upregulated [35–37] and multipotent progenitor-associated transcription factors are downregulated [25](Fig. 1). Thus, although the undisturbed thymic microenvironment enforces a T cell fate on nearly all progenitors that enter it, the cells themselves do not internalize this pressure into an intrinsically committed state until after multiple rounds of proliferation in the thymic microenvironment [37–40].
Single cell transcriptome analysis has recently shed light on multiple questions about T cell development. It has refined the program of regulatory change seen at population level, and while showing conservation between mouse and human, it has identified some intriguing differences. It has also illuminated unexpectedly distinct types of precursors that can enter the T-cell program.
Single-cell analyses of early T-cell development in mice and humans
T cell developmental progressions have now been studied by single-cell transcriptome analysis using two kinds of material, both in mouse and in human. First, the initial wave of T-cell development has been tracked through gestational time in whole organ thymus populations from fetal mouse [41] and first-trimester human [24, 42], following both developing hematopoietic cells and the epithelial and mesenchymal cells. Second, the most immature T cell precursors have been studied in highly enriched pro-T cell populations from postnatal mouse and human thymus, using cell sorting deliberately to exclude non-hematopoietic cells and mature hematopoietic cells of all lineages, including mature T cells. In the mouse, Lin− Kithi expression criteria were used [43], and in human thymus Lin− CD34+ [44]or Lin− CD34+ CD1a− criteria were used [45] to enrich for cells in the continuum of the earliest T lineage-defining stages.
The whole-organ embryological studies [24, 41, 42] followed thymic cell populations through successive gestational ages after arrival of the first hematopoietic immigrants. Data from ref. [41] were also used in a sophisticated computational model to relate differentiation and population dynamics in the mouse fetal thymus [46]. The stages beyond the first TCR-dependent checkpoint (β-selection or γδ-selection) were characterized as they appeared in later gestation [24, 41, 42], with special depth in ref. [24], and transcriptome program divergences between αβ and γδ lineage T cells were highlighted [24, 42]. One postnatal human cell study [44], too, identified new molecular markers for β-selection, although notably, these TCR-dependent stages were deliberately excluded by the sorting strategies used in other postnatal studies [43, 45]. The postnatal studies focused on relationships of specific transcriptome subsets within the T-lineage developmental progression, enriching the sequencing data by in vitro developmental potential assays [43–45], both for T-cell fate and for developmental alternative branches [43–45]. In the postnatal mouse system, the pseudotime-predicted precursor-product relationships were validated by real-time kinetic tests in clonal developmental assays [43].
Taken together, these studies established that pro-T cell developmental programs have a highly consistent conserved regulatory core in the two species. This conservation is noteworthy because the cell surface markers previously used to distinguish stages are different between mouse and human (Fig. 2, top). Population designations have not used a consistent nomenclature, even in the same species (Table 1). At a deeper level, transcriptome relationships emerging from these studies also suggest that the well-used surface markers may not perfectly coincide with the underlying changes in regulatory state. While general state changes roughly correlated with surface change, cells with generally similar transcriptomes, defined by cluster, often cut across surface marker-defined boundaries (Table 1). Single-cell transcriptome analyses generally may present interpretation issues, including limited sensitivity, inadequate clustering of samples containing too few cells, and treatment of subpopulations with different cell cycle distributions. Nevertheless, despite ontogenic and species differences, central features of the intrathymic T-cell developmental pathway were concordant in all five studies.
Figure 2. A common core program of regulatory gene expression change in mouse and human.
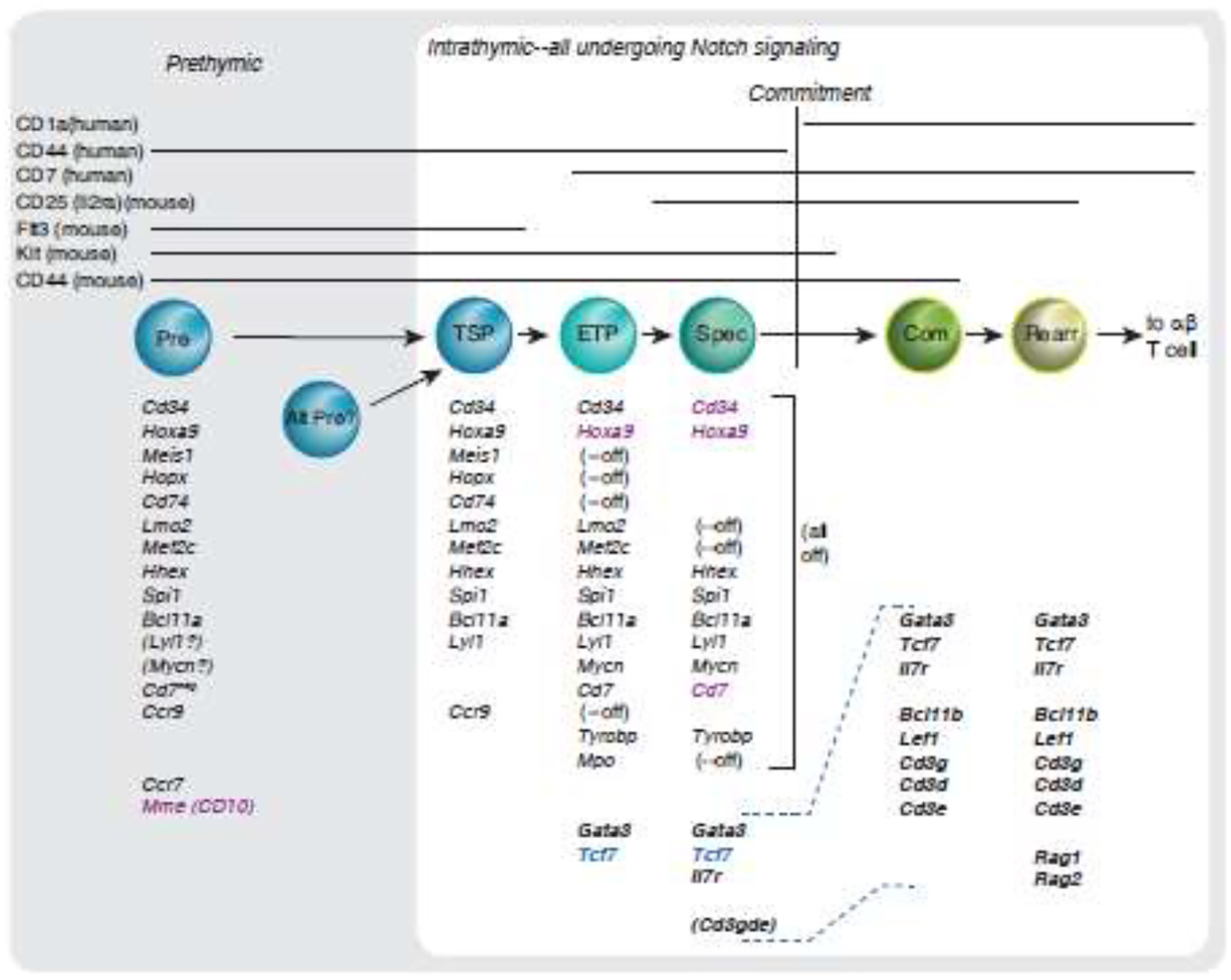
A consensus conserved gene expression program is shared by early T cells, in postnatal mice and humans. The figure shows key regulatory and signaling genes expressed at the corresponding indicated stages in both mouse and human [43–45]. The framework of stages shown is rooted in the TSP1 precursor trajectory with early intrathymic upregulation of Cd7 [44, 45] and the mouse stage equivalents. Stage markers are shown at the top and aligned as in Table 1. Note that mouse and human marker expression patterns are slightly different relative to underlying transcriptomes even when the same marker is tracked in both. Where possible, each gene in a group is listed in the same row across the figure. (--off), silencing of the gene in this row expressed in previous stages. Gene names in bold: T-cell program genes activated in T-lineage precursors by thymic microenvironment. Gene names in black: genes expressed in common between mouse and human cells at corresponding stages. Gene names in magenta: genes expressed in indicated stages persist later than in mouse, or lack a mouse equivalent. Gene name in blue: gene expressed at this stage is activated earlier than in human lineage. (Cd3gde), slight upregulation of the Cd3g, Cd3d, Cd3e gene cluster detectable before commitment, although greatly enhanced after commitment. (Lyl1), (Mycn) in Prethymic: probable expression but not reported directly. Evidence for gene expression in the prethymic mouse precursor is based on LMPP phenotypes and evidence from genetic studies of Ccr9 and Ccr7 impact on thymic immigration [98, 99]. Note that these lists are approximate, not comprehensive, and quantitative changes in expression are not represented. Patterns are gleaned from data shown in the figures and tables of the references as published [43–45] and not from reanalysis of all genomic datasets with consistent statistical criteria; comparative levels of genes shown were not always available across all populations. Population abbreviations: TSP, stage 0 in Table 1. ETP, stage 1. Spec, specified (stage 2). Com, committed (stage 3). Rearr, TCR gene rearranging (stage 4). Dashed lines in gene lists from Spec to Com aid to align gene groups.
TABLE 1.
ALIGNMENTS BETWEEN EARLY T CELL PRECURSOR NOMENCLATURES
| Stage | Mouse postnatal | Casero, Ha | Lavaert | Le | Zeng | Kernfeld |
|---|---|---|---|---|---|---|
| Stage 0 | (TSP ≈ Flt3+ ETP) | Thy1 | TSP1 and TSP2 | Cluster 1 | Embryonic ETP | Tconv1 |
| Stage 1 | Flt3− ETP | Thy1-Thy2 | ETP | Cluster 2; Thy2a, 2b | Fetal ETP1?; TSP-like ETP | Tconv1 |
| Stage 2 | DN2a | Thy2 | Specified | Cluster2–3; Thy2a, 2b | Fetal ETP1; proliferating ETP | Tconv1; Tconv2 |
| COMMITMENT | ||||||
| Stage 3 | DN2b | Thy3 | Committed | Thy3; Cluster3–4 | Fetal ETP2, γδ T cell precursor | Tconv2; Tconv3 |
| Stage 4 | DN3a | Thy4 | Rearranging | Clusters 5–11 | αβ T cell precursor2 | Tconv3 |
| References | [27, 28, 35, 37, 70, 96] | [30, 97] | [45] | [44] | [42] | [41] |
Approximate correspondences based on consensus of measured developmental potentials and single-cell transcriptome similarities in postnatal T-cell precursors.
Key cell-surface marker landmarks: mouse precursors starting Kithigh, Flt3+, CD44high, CD25−, Thy1−, IL7R−, CD4− CD8− TCR−; human precursors starting CD34+, (Flt3, Kit?), CD7−, CD1a−, CD44+, CD4− CD8− TCR−. Note that the surface marker distinguishing one “stage” from the next may not be precisely representative of the timing of the main transcriptome cluster shift
Mouse, transition stage 0 to 1: Flt3 downregulation, reduced access to DC fates.
Human, transition stage 0 to 1: onset of CD7 expression. Note that this definition only characterizes a state transition where the starting point is a classic TSP1 (CD34+ CD7−). CD7+ CD34-low TSP2 cells [45], likely to represent a separate precursor stream, could appear as an IRF8+ CD123+ minor subset within the Thy2+ population [44].
Mouse, transition stage 1–2: onset of CD25, Thy1, IL7R expression (CD7 is turned on within ETP compartment in mice, then off). More pronounced transcriptome change than in human.
Human, transition stage 1–2: increased expression of T-cell signaling genes, e.g.
Mouse, commitment: onset of Bcl11b expression (fluorescent reporter), Kit starts to go down
Human, commitment: onset of CD1a expression (no Cd1a exists in mice), CD7 and CD44 go down
Mouse, advent of rearrangement: CD44 and Kit off, RAG1 and RAG2 on
Human, advent of rearrangement: CD34 downregulated, RAG1 and RAG2 on
A conserved mouse-human developmental trajectory for pro-T cells
All studies showed that the earliest hematopoietic cells in the thymus, including the cells identified as likely thymus-settling precursors, expressed a consistent core of multipotent progenitor-associated regulatory genes (Fig. 2), including Hoxa9, Meis1, Hopx, Lmo2, Mef2c, Hhex, Spi1 (PU.1), Bcl11a, and Lyl1. These regulatory genes, except Hopx, have previously been identified as signature features of the pre-commitment “Phase 1” ETP and DN2a stages in the mouse, based on population RNA analyses [25]. Expression of these genes in human was similarly confined to the early intrathymic stages [44, 45], associated with cell-surface phenotypes of populations that are not yet T-lineage committed [47, 48]. Both in mouse and human, they were downregulated before or during commitment, with Hopx, Lmo2, and Meis1 among the earliest downregulated and Hhex, Bcl11a, and Spi1 among the last.
The distinctive T-lineage specific gene expression profile was activated from specification to commitment, in human and mouse precursors alike (Fig. 2), with upregulation of the genes encoding the invariant CD3 chains of the TCR complex Cd3g, Cd3d, and Cd3e, and the transcription factor coding genes Bcl11b and Lef1. Activation of the Rag genes, to enable TCR gene locus recombination, occurred at the next stage. Similar to results using a fluorescent Bcl11b expression indicator in the mouse system [37], the onset of Bcl11b expression in individual human cells [44, 45] corresponded closely to the phenotypic transition associated with T-cell lineage commitment (CD44+ to CD44− in human)[47]. Most remaining Phase 1 transcription factor expression was downregulated, and the functional impact of loss of the Phase 1 transcription factors could be seen in global motif analysis of the promoters of genes changing expression in human pro-T cells [45]. This agreed with previous mouse work showing the epigenetic closure of many sites as the Phase 1 factor PU.1 (encoded by Spi1) was turned off [49–51].
The Phase 1 genes are usually considered part of non-T cell programs, and many are associated with T-lineage oncogenesis if they are expressed after commitment (reviewed in [25, 52, 53]) [54, 55]. However, single-cell analyses confirmed that they were indeed conserved physiological components of T cell development, detectable in the absolute majority of individual cells categorized as ETPs or TSPs in both human and mouse [43, 44]. Importantly, the mouse TSP-ETP populations that expressed these genes showed high T-lineage precursor frequencies, ~90% among clonogenic cells [43]. Coexpression analysis in mouse and human further verified that the same individual cells turning on the T-cell program genes were also expressing Phase 1 factors [43, 44]. When higher-sensitivity methods were used to overcome the problem of false negatives [43], the absolute transcript counts from multiple genes in single cells confirmed that multipotent progenitor-associated genes are normally continuing expression in the great majority of cells as they start the T-cell program [40].
The early Phase 1 compartment was not homogeneous. First, the cells represented all cell cycle states, and second, they comprised more than one developmental stage (roughly, “TSP” to “Specified”, Fig. 2). Pseudotime models in both the murine [43] and the human systems [44, 45] indicated several different regulatory processes. Post-TSP ETPs in mice transiently upregulated “non-T lineage” genes including Tyrobp, Hhex and Mpo, often associated with myeloid fate, suggesting multilineage priming [43]. Similar myeloid-like gene expression was also found within the early intrathymic human precursors [44, 45]. In the mouse system, the order of events predicted by the pseudotime model was verified by in vitro differentiation races between sets of 50–60 single-cell clones with founders sorted from distinct pseudotime subsets [43]. The results indicated that cells upregulating these “myeloid” genes were indeed intermediates on the T-cell pathway. These results strongly suggest that the thymic microenvironment drives several transient regulatory gene expression changes even before T-cell lineage commitment.
Variations: Species differences and changes in ontogeny
These strong overarching similarities did not mean that the mouse and human regulatory programs were identical. Earlier population-level measurements had identified differences in activation of the Notch target Dtx1 [56, 57], in regulation of the ILC-promoting regulatory gene Id2 [30], and in the impacts of strong vs. weak Notch signaling on the TCRαβ vs. TCRγδ lineage choice [58–61]. Single-cell analysis showed that the important T-cell transcription factor gene, TCF7, was activated slightly later in human, accompanying rather than preceding BCL11B as in the mouse [44, 45]. HOXA9 expression in human appeared to persist slightly longer in precommitment human cells, and IRF8 was also more conspicuously expressed in Phase 1 human pro-T cells, while FLT3 and KIT were more prominent in mouse. One study [44] emphasized several additional timing or expression differences in gene expression.
T-cell precursors in the mouse thymus are also different in the first ontogenic wave of embryonic T cell development than in all subsequent waves [23, 62], and may even arise from precursors that precede appearance of definitive stem cells or from the very earliest intra-embryonic stem cells [63–66]. First-wave mouse ETPs (E13 in the mouse) differ from second-wave ETPs (E18) in their more restricted developmental plasticity and more limited potential for intrathymic proliferation before they reach differentiation endpoints [67]. In vivo, they progress through intrathymic stages of T-cell development faster than any later cohorts of precursors [68, 69] and uniquely give rise to specific skin-homing and mucosal subsets of TCRγδ cells [62]. Interestingly, in the single-cell studies, the earliest prenatal Phase 1 thymocytes were harder to separate into sequential gene-expression substages than the Phase 1 cells from postnatal samples in both mouse and human [24, 41, 42]. Phase 1 genes were expressed, and then were downregulated, but the stages resolved (re-averaged from the single-cell clusters) appeared to suggest increased activation of specification and commitment genes while Phase 1 genes were still active [41, 42]. Possibly, this could reflect the statistical problem of subclustering such rare cells. However, this might indicate that in cells of earlier fetal cohorts, regulatory changes that otherwise occur sequentially in later waves of T-cell development are mechanistically accelerated to overlap (cf. [70]).
Both murine and human single-cell RNA-seq analyses of first-wave embryonic/fetal thymic populations also concurred in showing populations initially rich in NK and innate lymphoid cell precursors [24, 41, 42], followed by a wave of γδ lineage T cells, before αβ lineage T cells became dominant. Recently, first-wave (E13) mouse ETPs were also reported to be uniquely bipotent to generate T and Lymphoid Tissue Inducer cells, a specific subset of group 3 ILCs [66]. This could confirm an ETP origin for ILC3-type cells that have been found in the mouse [71] and early human [24] fetal thymuses, but which disappear later in ontogeny. As gestation progressed, all these ILC and “nonconventional” T cell types became less prominent [24, 41, 42], becoming rare in pro-T cell populations after birth, as analyzed in the single-cell studies [43–45].
Linking T cells to other hematopoietic lineages: precursors and alternatives
Individual T cell precursors retain intrinsic access to alternative myeloid and dendritic-cell fates for many cell cycles after arriving in the thymus [21, 32–34, 72–75], albeit losing B cell potential much earlier [33, 34, 74]. Their myeloid or DC competence is readily demonstrated by removing them from the thymus and from Notch signaling. Thus, they appear to arise from lymphomyeloid precursors [38, 76–78], or else from common lymphoid precursors that somehow regain myeloid potential transiently within the thymus. Within the thymus, despite this potential, Notch signaling inhibits their myeloid differentiation efficiently [75, 79–81], and the thymus is poor in myeloid-supporting cytokines (in contrast to its abundance of lymphoid-supporting IL-7).
A strongly debated question has been whether any of the same precursors that generate T cells normally generate myeloid or dendritic cells also, within the thymus microenvironment [82, 83]. Granulocytic-committed precursors appear consistently as a minority within mouse ETP populations [43, 75], and may be generated in vivo from some ETPs [75]. Much evidence has accumulated to suggest that thymic dendritic cells could emerge, at least in part, from the same thymic immigrants as T cells [21, 84, 85]. Plasmacytoid dendritic cells (pDC) are particularly prominent, and share with T-cell precursors their strong use of E protein transcription factors [86], leading to some expression of “cross-lineage” genes [87, 88]. The potential for pDC development has long been noted: when Notch1 is mutationally disrupted specifically within T-cell precursors, pDC emerge from these precursors within the mouse thymus [81], and human thymocytes cultured without Notch signaling also generate pDC [87]. A lower-intensity Notch-signaling niche has now been described in the human thymus which appears to be naturally permissive for pDC development [89].
Single-cell analyses confirm that pDC are reproducibly found in postnatal human thymus [44, 45], and some uncommitted early pro-T cells in the postnatal human thymus are apparently transcriptionally pre-primed for a pDC developmental path [44]. One pro-T cell subset distinctively expressing the IL-3 receptor subunit CD123 and the transcription factor IRF8 showed a higher ability to generate pDC cells than any other non-T alternative [44], supporting earlier evidence for some pDC-biased precursors in the thymus [89]. Evidence discussed below indicates that a separate prethymic precursor stream [45] might be the source of the CD123+ pro-T cells. Thus, a pre-ordained pDC bias may characterize one particular stream of precursors supplying the human thymus.
If thymus-seeding cells have granulocytic and pDC potential, then where should they fall in the spectra of bone marrow, cord blood, and fetal liver hematopoietic precursors? In human, prior studies had used varied combinations of CD38, CD45RA, ITGB7, CD7, CD10, CD127 (IL-7 receptor α chain), and/or CD2 for enrichment and fractionation of possible T cell precursors among CD34+ cells [11, 13–15], but it was not clear whether all assays were identifying the same precursors. In the mouse, an elegant barcode-based lineage-tracing analysis (“LARRY”) identified transcriptomes of cells with the highest precursor activity for T cell generation in vivo, finding them close to DC precursors and sharing with them enriched expression of Flt3, Ighm, and Dntt [12]. Other important datasets came previously from bulk microarray and RNA-seq measurements and single-cell analyses of finely dissected hematopoietic progenitors [8, 9, 13, 14, 17, 76], but explicit quantitative comparison to early intrathymic populations was lacking until the recent studies, in which single-cell transcriptomes of primitive intrathymic cells were matched with single-cell clusters from blood or fetal liver to find TSP candidates based on similarity [24, 42, 44, 45]. In human fetuses, the most immature fetal thymocyte transcriptomes were found to adjoin a continuum of states between lymphomyeloid and CLP-like fetal liver cells [24]. On deeper analysis, the results suggested three possible types of cells that may seed the human thymus: at least two different precursor types for postnatal thymus, and a third, probably distinct type for early fetal thymus.
In postnatal human blood, two different thymus-seeding candidates were identified [45], each with a counterpart within the thymus. Both expressed multiple progenitor transcription factors and CD74, but they were otherwise distinct. One of the precursors, TSP1, was CD10+, CD7−, and ITGB7−, lacked expression of Notch-inducible genes, and expressed a stem/progenitor-like set of genes. The other, TSP2, was CD10− CD7+, and ITGB7+, high in interferon response gene and IRF8 expression, lower in expression of HOXA9 and CD34, and expressed Notch target genes (CD7, CD3E) even before reaching the thymus [45]. While their CD7 and CD3E expression resembled features of most intrathymic “specified” cells, their other properties did not. The possibility of prethymic Notch priming was consistent with some previous evidence from mouse and human alike [15, 90]. These IRF8-high TSP2 cells might be the source of pro-T cells with high pDC potential. Despite their differences, however, TSP1 and TSP2 were functionally indistinguishable in T cell potential [45](Fig. 3). Both effectively generated T lineage cells in T-cell conditions in stromal culture, and both could similarly repopulate fetal thymus organ cultures in a G-protein coupled receptor-dependent way, consistent with the known CCR7- or CCR9- dependent mechanism in vivo [45]. A third apparent TSP-like population was found in the very early fetal liver (8 pcw)[42]. These cells were TSP2-like, with high IRF8 and CD123 (IL3RA) expression. However, they also expressed IL7R [42], like the distinctive first-wave T-cell precursors in mice [16, 23], but unlike postnatal TSP1 and TSP2, and contrasting with cord blood, where IL7R reportedly marks precursors preferring a B-cell fate [14]. If these were indeed the earliest-wave fetal TSP source, then they should give rise to special fetal-restricted ILC and γδ cell lineages, as well as conventional T cells and possible pDCs.
Figure 3. Potential alternative origins for T cell precursors.
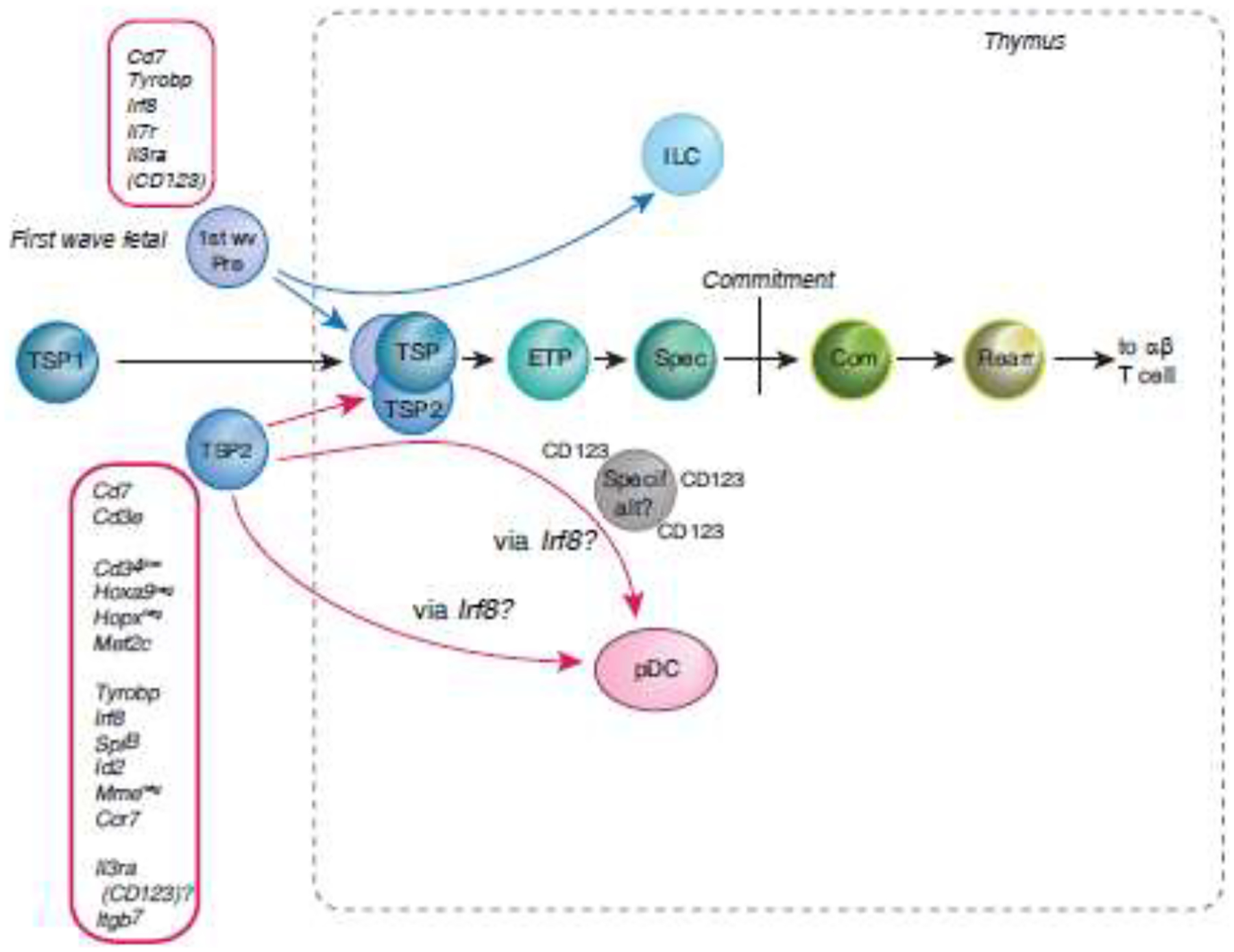
Shown are distinct properties of two alternative sources of prethymic TSPs, for contrast with the TSP1 cells shown in Fig. 2. Below main sequence: TSP2 cells. TSP1 cells have more readily detectable counterparts in mouse and human ETP populations, but TSP2-like cells [14] have been proposed as T cell precursors in the bone marrow because of their apparently T-lineage affiliated CD7+ phenotype [14, 15]. They lack the CD10 (MME), Hoxa9, and Hopx expression of TSP1 cells, and express regulatory genes that seemingly clash with the T cell program (Irf8, SpiB, Id2), yet rapidly develop into T-lineage cells in assays in vitro and in humanized mice [14, 15]. The figure shows the distinct transcriptomes of the TSP2 cells from [45] and their high propensity to act as bipotent T-pDC precursors [44, 45]. Above main sequence: first-wave fetal TSPs (1st wave Pre). Functional studies in mice [16, 17] as well as single-cell transcriptome alignments in human [42] indicate that first-wave prethymic precursors express Il7r (CD127) and show preferential ability to generate ILCs as well as T cells [24, 41, 42, 66]. These cells are rare, but transcriptome analysis shows TSP2-like Irf8, Tyrobp, and Il3ra (CD123) expression.
Thus, two or more distinct regulatory states are consistent with an ability to migrate to the thymus and give rise to T cells once in the thymus, and they are likely to come with distinct repertoires of alternative lineage potentials.
Concluding remarks: Single cell insights into T cell development in its hematopoietic context
This review has focused on aspects of T-cell development that link it to the rest of hematopoiesis. In fact, the recent studies discussed here extend well beyond this focus. For example, refs. [24, 41, 42] also characterize intrathymic stromal cell types (also see ref. [91] for pioneering detail on epithelial cell types) and signaling ligand/receptor pairs potentially mediating communication of microenvironmental cells with T-cell precursors [24, 42]. Some focus on T-cell receptor repertoire selection in detail [24], and on the regulatory divergences at the split between TCRαβ and TCRγδ lineages [24, 42, 44]. These topics interface with much larger immunological literatures, and also with examination of gene regulatory mechanisms controlling developmental progressions, including roles of transcription factors individually (rev. in [4]) and epigenetic changes [49, 50, 92, 93].
Other recent studies, moreover, have now used single-cell transcriptome analyses to demonstrate the crucial role of the TCF1 transcription factor (encoded by Tcf7) both in early intrathymic T-cell development [94] and in the corresponding stages of ILC2 development in the bone marrow [95]. Lack of TCF1 was shown to divert DN1 and DN2/3 pro-T cells into altered, distinct gene expression states, prone to leukemic transformation [94], while TCF1 was found to be indispensable for ILC precursors to undergo transition to lineage commitment, a regulatory shift homologous in many details to commitment in pro-T cells [95]. The similarities between the ILC and T cell programs remain a fascinating challenge to facile pictures of how the thymus triggers T cell development.
The next challenge is to link the origins of the T-cell program more accurately to the diverse span of progenitor states in the bone marrow [11, 13, 15]. While factors like SOX4, ID2 and SATB1 have been correlated with lymphoid precursor activity [13, 15], the evidence reviewed here suggests a different set of genes distinctive for thymus-seeding precursors, and the genuine possibility that more than one such prethymic source exists. The new richness of single-cell transcriptome evidence suggests fresh ways to identify the sources of T-cell precursors, and to reveal what determines their T-lineage competence.
HIGHLIGHTS.
Single-cell transcriptome analyses show homology of mouse and human T cell programs
T-cell precursors begin T program from multipotent progenitor-like regulatory state
Progenitor-like regulatory genes are turned off, BCL11B is turned on at commitment
T cell precursors maintain close linkage to plasmacytoid dendritic cell program
Transcriptionally distinct precursor types can convergently develop into T cells
Acknowledgments
The author thanks Ido Amit, Avinash Bhandoola, Gay Crooks, Ana Cumano, Kenneth Dorshkind, Berthold Göttgens, Muzlifah Haniffa, Andreas Krueger, Chintan Parekh, Hans-Reimer Rodewald, Tom Taghon, Sarah Teichmann, and Wen Zhou for sharing insightful discussions about work before it was published and about general principles of the field. Relevant support for work in the author’s lab was from USPHS grants R01HL119102, R01HD076915, and R01HD100039, with support for E.V.R. from the Albert Billings Ruddock Professorship of Biology.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest
E.V.R. is a member of the Scientific Advisory Board of Century Therapeutics and has consulted for A2 Biotherapeutics.
REFERENCES
- [1].Boller S, Grosschedl R. The regulatory network of B-cell differentiation: a focused view of early B-cell factor 1 function. Immunol Rev. 2014;261:102–115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Somasundaram R, Prasad MA, Ungerbäck J, Sigvardsson M. Transcription factor networks in B-cell differentiation link development to acute lymphoid leukemia. Blood. 2015;126:144–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Rothenberg EV. Programming for T-lymphocyte fates: modularity and mechanisms. Genes Dev. 2019;33:1117–1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Hosokawa H, Rothenberg EV. How transcription factors drive choice of the T cell fate. Nat Rev Immunol. 2020: doi: 10.1038/s41577-020-00426-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].De Obaldia ME, Bhandoola A. Transcriptional regulation of innate and adaptive lymphocyte lineages. Annu Rev Immunol. 2015;33:607–642. [DOI] [PubMed] [Google Scholar]
- [6].Seo W, Taniuchi I. Transcriptional regulation of early T-cell development in the thymus. Eur J Immunol. 2016;46:531–538. [DOI] [PubMed] [Google Scholar]
- [7].Paul F, Arkin Y, Giladi A, et al. Transcriptional heterogeneity and lineage commitment in myeloid progenitors. Cell. 2015;163:1663–1677. [DOI] [PubMed] [Google Scholar]
- [8].Zheng S, Papalexi E, Butler A, Stephenson W, Satija R. Molecular transitions in early progenitors during human cord blood hematopoiesis. Mol Syst Biol. 2018;14:e8041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Velten L, Haas SF, Raffel S, et al. Human haematopoietic stem cell lineage commitment is a continuous process. Nat Cell Biol. 2017;19:271–281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Giladi A, Paul F, Herzog Y, et al. Single-cell characterization of haematopoietic progenitors and their trajectories in homeostasis and perturbed haematopoiesis. Nat Cell Biol. 2018;20:836–846. [DOI] [PubMed] [Google Scholar]
- [11].Pellin D, Loperfido M, Baricordi C, et al. A comprehensive single cell transcriptional landscape of human hematopoietic progenitors. Nat Commun. 2019;10:2395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Weinreb C, Rodriguez-Fraticelli A, Camargo FD, Klein AM. Lineage tracing on transcriptional landscapes links state to fate during differentiation. Science. 2020;367:aaw3381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Karamitros D, Stoilova B, Aboukhalil Z, et al. Single-cell analysis reveals the continuum of human lympho-myeloid progenitor cells. Nat Immunol. 2018;19:85–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Alhaj Hussen K, Vu Manh TP, Guimiot F, et al. Molecular and Functional Characterization of Lymphoid Progenitor Subsets Reveals a Bipartite Architecture of Human Lymphopoiesis. Immunity. 2017;47:680–696.e8. [DOI] [PubMed] [Google Scholar]
- [15].Laurenti E, Doulatov S, Zandi S, et al. The transcriptional architecture of early human hematopoiesis identifies multilevel control of lymphoid commitment. Nat Immunol. 2013;14:756–763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Masuda K, Kubagawa H, Ikawa T, et al. Prethymic T-cell development defined by the expression of paired immunoglobulin-like receptors. EMBO J. 2005;24:4052–4060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Berthault C, Ramond C, Burlen-Defranoux O, et al. Asynchronous lineage priming determines commitment to T cell and B cell lineages in fetal liver. Nat Immunol. 2017;18:1139–1149. [DOI] [PubMed] [Google Scholar]
- [18].Krueger A, Ziętara N, Łyszkiewicz M. T Cell Development by the Numbers. Trends Immunol. 2017;38:128–139. [DOI] [PubMed] [Google Scholar]
- [19].Famili F, Wiekmeijer AS, Staal FJ. The development of T cells from stem cells in mice and humans. Future Sci OA. 2017;3:FSO186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Shah DK, Zúñiga-Pflücker JC. An overview of the intrathymic intricacies of T cell development. J Immunol. 2014;192:4017–23. [DOI] [PubMed] [Google Scholar]
- [21].Shen HQ, Lu M, Ikawa T, et al. T/NK bipotent progenitors in the thymus retain the potential to generate dendritic cells. J Immunol. 2003;171:3401–3406. [DOI] [PubMed] [Google Scholar]
- [22].Heinzel K, Benz C, Martins VC, Haidl ID, Bleul CC. Bone marrow-derived hemopoietic precursors commit to the T cell lineage only after arrival in the thymic microenvironment. J Immunol. 2007;178:858–868. [DOI] [PubMed] [Google Scholar]
- [23].Cumano A, Berthault C, Ramond C, et al. New Molecular Insights into Immune Cell Development. Annu Rev Immunol. 2019;37:497–519. [DOI] [PubMed] [Google Scholar]
- [24].Park JE, Botting RA, Dominguez Conde C, et al. A cell atlas of human thymic development defines T cell repertoire formation. Science. 2020;367:aay3224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Yui MA, Rothenberg EV. Developmental gene networks: a triathlon on the course to T cell identity. Nat Rev Immunol. 2014;14:529–545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Love PE, Bhandoola A. Signal integration and crosstalk during thymocyte migration and emigration. Nat Rev Immunol. 2011;11:469–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Zhang JA, Mortazavi A, Williams BA, Wold BJ, Rothenberg EV. Dynamic transformations of genome-wide epigenetic marking and transcriptional control establish T cell identity. Cell. 2012;149:467–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Mingueneau M, Kreslavsky T, Gray D, et al. The transcriptional landscape of αβ T cell differentiation. Nat Immunol. 2013;14:619–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Johnson JL, Georgakilas G, Petrovic J, et al. Lineage-Determining Transcription Factor TCF-1 Initiates the Epigenetic Identity of T Cells. Immunity. 2018;48:243–257.e10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Ha VL, Luong A, Li F, et al. The T-ALL related gene BCL11B regulates the initial stages of human T-cell differentiation. Leukemia. 2017;31:2503–2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Schmitt TM, Ciofani M, Petrie HT, Zúñiga-Pflücker JC. Maintenance of T cell specification and differentiation requires recurrent Notch receptor-ligand interactions. J Exp Med. 2004;200:469–479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Shortman K, Wu L. Early T lymphocyte progenitors. AnnuRevImmunol. 1996;14:29–47. [DOI] [PubMed] [Google Scholar]
- [33].Bell JJ, Bhandoola A. The earliest thymic progenitors for T cells possess myeloid lineage potential. Nature. 2008;452:764–767. [DOI] [PubMed] [Google Scholar]
- [34].Wada H, Masuda K, Satoh R, et al. Adult T-cell progenitors retain myeloid potential. Nature. 2008;452:768–772. [DOI] [PubMed] [Google Scholar]
- [35].Yui MA, Feng N, Rothenberg EV. Fine-scale staging of T cell lineage commitment in adult mouse thymus. J Immunol. 2010;185:284–293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Masuda K, Kakugawa K, Nakayama T, Minato M, Katsura Y, Kawamoto H. T cell lineage determination precedes the initiation of TCRβ rearrangement. J Immunol. 2007;179:3699–3706. [DOI] [PubMed] [Google Scholar]
- [37].Kueh HY, Yui MA, Ng KKH, et al. Asynchronous combinatorial action of four regulatory factors activates Bcl11b for T cell commitment. Nat Immunol. 2016;17:956–965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Lu M, Tayu R, Ikawa T, et al. The earliest thymic progenitors in adults are restricted to T, NK, and dendritic cell lineage and have a potential to form more diverse TCRβ chains than fetal progenitors. J Immunol. 2005;175:5848–5856. [DOI] [PubMed] [Google Scholar]
- [39].Manesso E, Chickarmane V, Kueh HY, Rothenberg EV, Peterson C. Computational modelling of T-cell formation kinetics: output regulated by initial proliferation-linked deferral of developmental competence. J R Soc Interface. 2013;10:20120774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Porritt HE, Gordon K, Petrie HT. Kinetics of steady-state differentiation and mapping of intrathymic-signaling environments by stem cell transplantation in nonirradiated mice. J Exp Med. 2003;198:957–962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Kernfeld EM, Genga RMJ, Neherin K, Magaletta ME, Xu P, Maehr R. A single-cell transcriptomic atlas of thymus organogenesis resolves cell types and developmental maturation. Immunity. 2018;48:1258–1270.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Zeng Y, Liu C, Gong Y, et al. Single-cell RNA sequencing resolves spatiotemporal development of pre-thymic lymphoid progenitors and thymus organogenesis in human embryos. Immunity. 2019;51:930–948.e6. [DOI] [PubMed] [Google Scholar]
- [43].Zhou W, Yui MA, Williams BA, et al. Single-cell analysis reveals regulatory gene expression dynamics leading to lineage commitment in early T cell development. Cell Syst. 2019;9:321–337.e9.s [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Le J, Park JE, Ha VL, et al. Single-Cell RNA-Seq Mapping of Human Thymopoiesis Reveals Lineage Specification Trajectories and a Commitment Spectrum in T Cell Development. Immunity. 2020;52:1105–1118.e9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Lavaert M, Liang KL, Vandamme N, et al. Integrated scRNA-Seq Identifies Human Postnatal Thymus Seeding Progenitors and Regulatory Dynamics of Differentiating Immature Thymocytes. Immunity. 2020;52:1088–1104.e6. [DOI] [PubMed] [Google Scholar]
- [46].Fischer DS, Fiedler AK, Kernfeld EM, et al. Inferring population dynamics from single-cell RNA-sequencing time series data. Nat Biotechnol. 2019;37:461–468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Canté-Barrett K, Mendes RD, Li Y, et al. Loss of CD44dim Expression from Early Progenitor Cells Marks T-Cell Lineage Commitment in the Human Thymus. Front Immunol. 2017;8:32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Hao QL, George AA, Zhu J, et al. Human intrathymic lineage commitment is marked by differential CD7 expression: identification of CD7- lympho-myeloid thymic progenitors. Blood. 2008;111:1318–1326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Yoshida H, Lareau CA, Ramirez RN, et al. The cis-Regulatory Atlas of the Mouse Immune System. Cell. 2019;176:897–912.e20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Roels J, Kuchmiy A, De Decker M, et al. Distinct and temporary-restricted epigenetic mechanisms regulate human αβ and γδ T cell development. Nat Immunol. 2020;21:1280–1292. [DOI] [PubMed] [Google Scholar]
- [51].Ungerbäck J, Hosokawa H, Wang X, et al. Pioneering, chromatin remodeling, and epigenetic constraint in early T-cell gene regulation by SPI1 (PU.1). Genome Res. 2018;28:1508–1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Hoang T, Lambert JA, Martin R. SCL/TAL1 in hematopoiesis and cellular reprogramming. Curr Top Dev Biol. 2016;118:163–204. [DOI] [PubMed] [Google Scholar]
- [53].Haydu JE, Ferrando AA. Early T-cell precursor acute lymphoblastic leukaemia. Curr Opin Hematol. 2013;20:369–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [54].Smith S, Tripathi R, Goodings C, et al. LIM domain only-2 (LMO2) induces T-cell leukemia by two distinct pathways. PLoS One. 2014;9:e85883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Rosenbauer F, Owens BM, Yu L, et al. Lymphoid cell growth and transformation are suppressed by a key regulatory element of the gene encoding PU.1. Nat Genet. 2006;38:27–37. [DOI] [PubMed] [Google Scholar]
- [56].Van de Walle I, Dolens AC, Durinck K, et al. GATA3 induces human T-cell commitment by restraining Notch activity and repressing NK-cell fate. Nat Commun. 2016;7:11171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Van de Walle I, De Smet G, De Smedt M, et al. An early decrease in Notch activation is required for human TCR-αβ lineage differentiation at the expense of TCR-γδ T cells. Blood. 2009;113:2988–2998. [DOI] [PubMed] [Google Scholar]
- [58].Garcia-Peydro M, de Yebenes VG, Toribio ML. Sustained Notch1 signaling instructs the earliest human intrathymic precursors to adopt a γδ T cell fate in fetal thymus organ culture. Blood. 2003;102:2444–2451. [DOI] [PubMed] [Google Scholar]
- [59].Taghon T, Rothenberg EV. Molecular mechanisms that control mouse and human TCR-αβ and TCR-γδ T cell development. Semin Immunopathol. 2008;30:383–398. [DOI] [PubMed] [Google Scholar]
- [60].Joachims ML, Chain JL, Hooker SW, Knott-Craig CJ, Thompson LF. Human αβ and γδ thymocyte development: TCR gene rearrangements, intracellular TCR β expression, and γδ developmental potential--differences between men and mice. J Immunol. 2006;176:1543–1552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Dolens AC, Durinck K, Lavaert M, et al. Distinct Notch1 and BCL11B requirements mediate human γδ/αβ T cell development. EMBO Rep. 2020;21:e49006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Ikuta K, Kina T, MacNeil I, et al. A developmental switch in thymic lymphocyte maturation potential occurs at the level of hematopoietic stem cells. Cell. 1990;62:863–874. [DOI] [PubMed] [Google Scholar]
- [63].Boïers C, Carrelha J, Lutteropp M, et al. Lymphomyeloid contribution of an immune-restricted progenitor emerging prior to definitive hematopoietic stem cells. Cell Stem Cell. 2013;13:535–548. [DOI] [PubMed] [Google Scholar]
- [64].Beaudin AE, Boyer SW, Perez-Cunningham J, et al. A Transient Developmental Hematopoietic Stem Cell Gives Rise to Innate-like B and T Cells. Cell Stem Cell. 2016;19:768–783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Yoshimoto M, Porayette P, Glosson NL, et al. Autonomous murine T-cell progenitor production in the extra-embryonic yolk sac before HSC emergence. Blood. 2012;119:5706–5714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Elsaid R, Meunier S, Burlen-Defranoux O, et al. A wave of bipotent T/ILC-restricted progenitors shapes the embryonic thymus microenvironment in a time-dependent manner. Blood. 2020:blood.2020006779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Ramond C, Berthault C, Burlen-Defranoux O, et al. Two waves of distinct hematopoietic progenitor cells colonize the fetal thymus. Nat Immunol. 2014;15:27–35. [DOI] [PubMed] [Google Scholar]
- [68].Kawamoto H, Ohmura K, Hattori N, Katsura Y. Hemopoietic progenitors in the murine fetal liver capable of rapidly generating T cells. J Immunol. 1997;158:3118–3124. [PubMed] [Google Scholar]
- [69].Scripture-Adams DD, Damle SS, Li L, et al. GATA-3 dose-dependent checkpoints in early T cell commitment. J Immunol. 2014;193:3470–3491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Belyaev NN, Biro J, Athanasakis D, Fernandez-Reyes D, Potocnik AJ. Global transcriptional analysis of primitive thymocytes reveals accelerated dynamics of T cell specification in fetal stages. Immunogenetics. 2012;64:591–604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Jones R, Cosway EJ, Willis C, et al. Dynamic changes in intrathymic ILC populations during murine neonatal development. Eur J Immunol. 2018;48:1481–1491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Balciunaite G, Ceredig R, Rolink AG. The earliest subpopulation of mouse thymocytes contains potent T, significant macrophage, and natural killer cell but no B-lymphocyte potential. Blood. 2005;105:1930–1936. [DOI] [PubMed] [Google Scholar]
- [73].Franco CB, Scripture-Adams DD, Proekt I, et al. Notch/Delta signaling constrains reengineering of pro-T cells by PU.1. Proc Natl Acad Sci U S A. 2006;103:11993–11998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Allman D, Sambandam A, Kim S, et al. Thymopoiesis independent of common lymphoid progenitors. Nat Immunol. 2003;4:168–174. [DOI] [PubMed] [Google Scholar]
- [75].De Obaldia ME, Bell JJ, Bhandoola A. Early T-cell progenitors are the major granulocyte precursors in the adult mouse thymus. Blood. 2013;121:64–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [76].Luc S, Luis TC, Boukarabila H, et al. The earliest thymic T cell progenitors sustain B cell and myeloid lineage potential. Nat Immunol. 2012;13:412–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Masuda K, Itoi M, Amagai T, Minato N, Katsura Y, Kawamoto H. Thymic anlage is colonized by progenitors restricted to T, NK, and dendritic cell lineages. J Immunol. 2005;174:2525–2532. [DOI] [PubMed] [Google Scholar]
- [78].Doulatov S, Notta F, Eppert K, Nguyen LT, Ohashi PS, Dick JE. Revised map of the human progenitor hierarchy shows the origin of macrophages and dendritic cells in early lymphoid development. NatImmunol. 2010;11:585–593. [DOI] [PubMed] [Google Scholar]
- [79].Del Real MM, Rothenberg EV. Architecture of a lymphomyeloid developmental switch controlled by PU.1, Notch and Gata3. Development. 2013;140:1207–1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].De Obaldia ME, Bell JJ, Wang X, et al. T cell development requires constraint of the myeloid regulator C/EBP-α by the Notch target and transcriptional repressor Hes1. Nat Immunol. 2013;14:1277–1284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Feyerabend TB, Terszowski G, Tietz A, et al. Deletion of Notch1 converts pro-T cells to dendritic cells and promotes thymic B cells by cell-extrinsic and cell-intrinsic mechanisms. Immunity. 2009;30:67–79. [DOI] [PubMed] [Google Scholar]
- [82].Schlenner SM, Madan V, Busch K, et al. Fate mapping reveals separate origins of T cells and myeloid lineages in the thymus. Immunity. 2010;32:426–436. [DOI] [PubMed] [Google Scholar]
- [83].Richie Ehrlich LI, Serwold T, Weissman IL. In vitro assays misrepresent in vivo lineage potentials of murine lymphoid progenitors. Blood. 2011;117:2618–2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [84].Ardavin C, Wu L, Li CL, Shortman K. Thymic dendritic cells and T cells develop simultaneously in the thymus from a common precursor population. Nature. 1993;362:761–763. [DOI] [PubMed] [Google Scholar]
- [85].Corcoran L, Ferrero I, Vremec D, et al. The lymphoid past of mouse plasmacytoid cells and thymic dendritic cells. J Immunol. 2003;170:4926–4932. [DOI] [PubMed] [Google Scholar]
- [86].Cisse B, Caton ML, Lehner M, et al. Transcription factor E2–2 is an essential and specific regulator of plasmacytoid dendritic cell development. Cell. 2008;135:37–48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Dontje W, Schotte R, Cupedo T, et al. Delta-Like1-induced Notch1 signalling regulates the human plasmacytoid dendritic cell versus T cell lineage decision through control of GATA-3 and Spi-B. Blood. 2006;107:2446–2452. [DOI] [PubMed] [Google Scholar]
- [88].Shigematsu H, Reizis B, Iwasaki H, et al. Plasmacytoid dendritic cells activate lymphoid-specific genetic programs irrespective of their cellular origin. Immunity. 2004;21:43–53. [DOI] [PubMed] [Google Scholar]
- [89].Martín-Gayo E, González-García S, García-León MJ, et al. Spatially restricted JAG1-Notch signaling in human thymus provides suitable DC developmental niches. J Exp Med. 2017;214:3361–3379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Chen ELY, Thompson PK, Zúñiga-Pflücker JC. RBPJ-dependent Notch signaling initiates the T cell program in a subset of thymus-seeding progenitors. Nat Immunol. 2019;20:1456–1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Bornstein C, Nevo S, Giladi A, et al. Single-cell mapping of the thymic stroma identifies IL-25-producing tuft epithelial cells. Nature. 2018;559:622–626. [DOI] [PubMed] [Google Scholar]
- [92].Hu G, Cui K, Fang D, et al. Transformation of accessible chromatin and 3D nucleome underlies lineage commitment of early T cells. Immunity. 2018;48:227–242.e8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Vigano MA, Ivanek R, Balwierz P, et al. An epigenetic profile of early T-cell development from multipotent progenitors to committed T-cell descendants. Eur J Immunol. 2014;44:1181–1193. [DOI] [PubMed] [Google Scholar]
- [94].Wang F, Qi Z, Yao Y, et al. Exploring the stage-specific roles of Tcf-1 in T cell development and malignancy at single-cell resolution. Cell Mol Immunol. 2020: 10.1038/s41423-020-00527-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [95].Harly C, Kenney D, Ren G, et al. The transcription factor TCF-1 enforces commitment to the innate lymphoid cell lineage. Nat Immunol. 2019;20:1150–1160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Arenzana TL, Schjerven H, Smale ST. Regulation of gene expression dynamics during developmental transitions by the Ikaros transcription factor. Genes Dev. 2015;29:1801–1816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Casero D, Sandoval S, Seet CS, et al. Long non-coding RNA profiling of human lymphoid progenitor cells reveals transcriptional divergence of B cell and T cell lineages. Nat Immunol. 2015;16:1282–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Calderon L, Boehm T. Three chemokine receptors cooperatively regulate homing of hematopoietic progenitors to the embryonic mouse thymus. Proc Natl Acad Sci U S A. 2011;108:7517–7522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Zlotoff DA, Sambandam A, Logan TD, Bell JJ, Schwarz BA, Bhandoola A. CCR7 and CCR9 together recruit hematopoietic progenitors to the adult thymus. Blood. 2010;115:1897–1905. [DOI] [PMC free article] [PubMed] [Google Scholar]


