Abstract
Nature, which remains a central drug discovery pool, is always looked upon to find a putative druggable lead. The natural products and phytochemical derived from plants are essential during a global health crisis. This class represents one of the most practical and promising approaches to decrease pandemic's intensity owing to their therapeutic potential. The present manuscript is therefore kept forth to give the researchers updated information on undergoing research in allied areas of natural product-based drug discovery, particularly for Covid-19 disease. The study briefly shreds evidence from in vitro and in silico researches done so far to find a lead molecule against Covid-19. Following this, we exhaustively explored the concept of chemical space and molecular similarity parameters for the drug discovery about the lead(s) generated from in silico-based studies. The comparison was drawn using FDA-approved anti-infective agents during 2015–2020 using key descriptors to evaluate druglike properties. The outcomes of results were further corroborated using Molecular Dynamics studies which suggested the outcomes in alignment with chemical space ranking. In a nutshell, current research work aims to provide a holistic strategic approach to drug design, keeping in view the identified phytochemicals against Covid-19.
Keywords: Covid-19, SARS-CoV-2, Natural products, Druggable targets, In silico studies, Chemical space, Molecular similarity
Graphical abstract
List of abbreviations
- ACE2
Angiotensin-Converting Enzyme 2
- ADMET
Absorption, Distribution, Metabolism, Elimination and Toxicity
- AlogP
Implication of Partition Coefficient
- Ang1-7
Angiotensin 1-7
- ARDS
Acute Respiratory Distress Syndrome
- Covid-19
Coronavirus disease
- CSR
Cumulative Scaffold Recovery
- CYPs
Cytochromes P450
- DFT
Density-Functional Theory
- ECFP4
Extended-Connectivity Fingerprints
- EGCG
Epigallocatechin Gallate
- ERGIC
Endoplasmic Reticulum -Golgi Intermediate Compartment
- GCMS
Gas Chromatography–Mass Spectrometry
- HCQ
Hydroxychloroquine;
- HEL
Helicase
- HIV
Human Immunodeficiency Viruses
- HTVS
High-Throughput Virtual Screening
- ISGs
Interferons Stimulating Genes
- KDS
Known Drug Space
- MERS-CoV
Middle East respiratory syndrome coronavirus
- MW
Molecular Weight
- nHBAcc
Hydrogen Bond Acceptors
- nHBDon
Hydrogen Bond Donors
- NMR
Nuclear Magnetic Resonance
- Nsp
Non-Structural Protein
- nROTB
Number of Rotational Bonds
- PUMA
Platform for Unified Molecular Analysis
- RBD
Receptor-Binding Domain
- RNA
Ribonucleic acid
- SARS-CoV
Severe Acute Respiratory Syndrome Coronavirus
- SARS-CoV-2
Severe Acute Respiratory Syndrome Coronavirus 2
- SPCI
Structural and Physiochemical Interpretation
- TMPRSS2
Transmembrane serine protease 2
- TPSA
Topological polar surface area
- QSAR
Quantitative Structure Activity Relationship
1. Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 or SARS-CoV-2 is responsible for Covid-19 diseases and seventh coronavirus strain overall infecting Homo sapiens (Poduri et al., 2020). As of now, Covid-19 disease is the second leading cause of mortality after cardiovascular disease. The situation is even critical as drug candidates to target the SARS-CoV-2 are still not available for clinical use (Thakur et al., 2020). Although vaccines are now being launched, still their safety and efficacy in the population are awaited. Among other therapeutics options, natural and naturally derived products are a vast source of potential drug molecules. Nature provides an immense source of active ingredients yet to be discovered to treat diseases. Historically, clinically important 80% of the drug developments are still inspired by these naturally derived entities. Therefore, naturally originated products or phytochemicals have continuously served humankind as a noble source of therapeutically important moieties (Huang et al., 2020). Phytochemicals are of diversified ranges, including essential oils, secondary metabolites, marine entities, microbes, fungi, making their mark and enabling the development of selective and efficacious chemotherapies against SARS-CoV-2 (da Silva Antonio et al., 2020). Furthermore, natural products become immensely important during a global health crisis and represent one of the most practical and promising approaches to decrease the intensity of pandemics with their therapeutic potential (Christy et al., 2020). Therefore, research and development from phytochemicals and their vitality in drug development are booming to find their place in this competitive league and prove their worth against Covid-19 (da Silva Antonio et al., 2020).
As it is widely known now that small molecules, particularly natural products, share a significant chunk of the commercially available drugs (Christy et al., 2020). Most of these drugs are marketed for antiviral or for use in other infections (Ospanov et al., 2020). Moreover, it has been established that SARS-CoV-2 shares approximately 79.5% genomic similarity with SARS-CoV and 50% similarity with MERS-CoV (Zhu et al., 2020). Therefore, the natural products and related drug candidates with promising effects against the previous pandemics could be repurposed in a current pandemic as well. Few relevant examples from previous pandemics include the use of Allium porrum (Alliacease), Nicotania tobacco (Solanaceae), Urtica diocia (Urticaceae), Hippeastrum hybrid (Amaryllis), etc., active against SARS-CoV and kaempferol derivatives (Viola odorata L), Epigallocatechin gallate, Gallocatechin gallate, quercetin-3-β-d-glucoside, isobavachalchole, herbacetin, and helichrysetin having potential against MERS-CoV. Whereas, tetranderine, fangchinoline, and cepharanthine were found active against HcoV-OC43 and glycyrrhizin and saikosaponin B2 potrayed their potential against HCoV-229E.
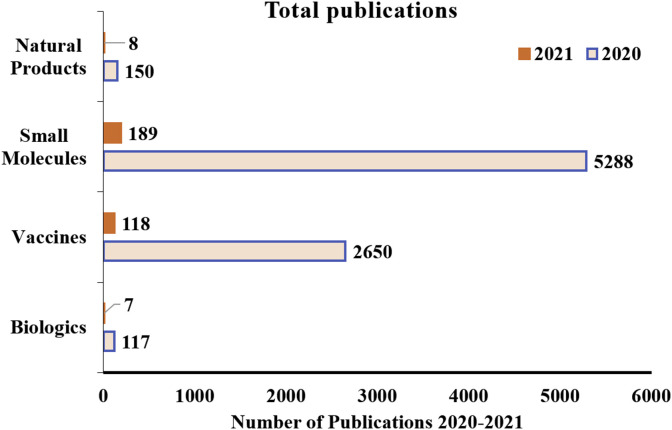
However, a detailed Scopus search (Fig. 1 ) revealed that not many significant studies have been made concerning the evaluation of phytochemicals against Covid-19. The Scopus search with the keyword “Natural products” AND “Covid-19″ AND “SARS-CoV-2″ gave 158 search results, replacing “natural products” in search with “small molecules” gave 5478 entries, with “vaccines” gave 2768 results and “biologics” gave 124 results as on January 8, 2020. There are only a handful of studies pertaining to natural products and their in vitro evaluation against Covid-19. Many studies are undergoing to identify the active leads based on the affinity of naturally derived leads against key druggable targets of SARS-Cov-2 using methodologies of virtual screening, molecular docking and dynamics. However, this approach fastens the drug discovery, but often leads to the identification of molecules deprived of druglikeness. Thus, it is essential to analyze the vital druggable parameters by exploring their chemical space and molecular similarity in the light of already approved drugs.
Fig. 1.
The bar graph quantitatively showcases undergoing research on the basis of total publications in various areas including natural product in a quest to identify a putative lead against SARS-CoV-2 (the data was retrieved from Scopus, last assessed on January 8, 2020).
The current work is therefore kept forth to give the researchers updated information on undergoing research in allied areas of natural product-based drug discovery, particularly for Covid-19. The work briefly put forth evidence from in vitro and in silico studies done so far to find a lead molecule against Covid-19. The work also extends to incorporate the critical druggable targets in SARS-CoV-2 that are explored to identify the putative natural drug leads. Along with this, a discussion on in silico peculiarities to identify the natural lead compounds is also highlighted. Further, we have exhaustively discussed the concept of chemical space and molecular similarity parameters to determine the druggable leads from the ongoing research in the natural products-based drug discovery arena against Covid-19. The concept of chemical space application is further corroborated using molecular dynamics simulations, suggesting proof of concepts. Current work thus aims to provide a holistic strategic background on drug design and discovery form natural derived compounds keeping in view the identified phytochemicals derived lead(s) for Covid-19.
2. Exploring the host-virus interaction SARS-COV-2: Key druggable targets involved
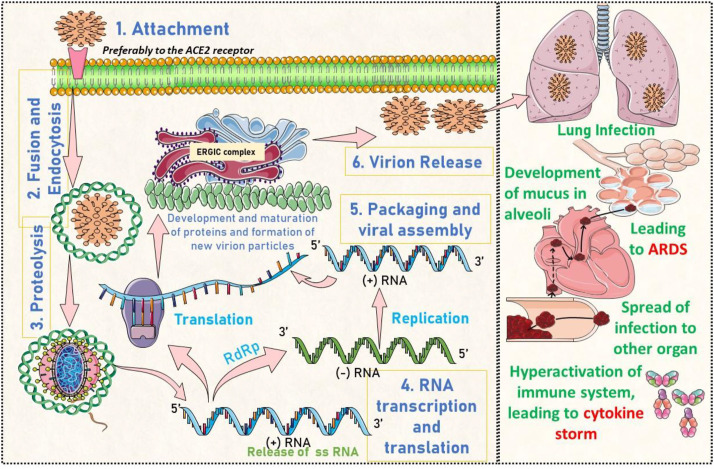
Although covered this aspect in detail by our group (reviewed in(Poduri et al., 2020; Thakur et al., 2020)), briefly SARS-CoV-2 possess a pleomorphic shape with an approximate size of ~125 nm, along with (+)-RNA as the genetic material of 30 kb Genome. The virus consists of four crucial protein which includes Membrane Protein (M), Spike (S) protein, Nucleocapsid (N) protein Envelope (E) and Non-structural protein complexes (Nsp). As per scientific evidence, S protein assists in the attachment of SARS-CoV-2 inside the host cell via its interaction with the host Angiotensin-converting enzyme ACE2 or by employing the Neuropilin-1 receptor, which is lesser-known (Cantuti-Castelvetri et al., 2020). This is followed by TMPRSS2 mediated cleavage of spike protein (also called activation), which allows virus endocytosis (also called membrane fusion) and release of genomic content into the host cell (called proteolysis). This leads to translation and replication of virus (+)-RNA using the host cell machinery, final allowing the synthesis of important Nsp to catalyse these essential reactions followed by synthesis of new virus particles. These newly developed virus particles then evade (virion release) another host cell and increase the severity leading to the rampage of inflammatory pathways called cytokine storm followed by ARDS, multiorgan failure, and eventually death in severe cases. The mechanism is pictorially represented in Fig. 2 .
Fig. 2.
Pictorial representation of crucial pathways of SARS-CoV-2 prognosis and development in the host cell [Reprinted and modified by permission from Ref (Thakur et al., 2020): ©Elsevier Ltd. 2020].
The figure highlights the interaction of the S1 protein of SARS-CoV-2 with the host's ACE2 (attachment), followed by fusion and endocytosis via endosome. The next step, proteolysis, allows the virus to release its genomic ((+)-RNA) content inside the host cell and produces PLpro and 3CLpro that modifies and cleaves pp1a and pp1ab, further transforming into mature Nsp 1–16. All these proteins assemble and prepare the virus for replication and transcription, utilizing these Nsps in the ERGIC complex. These virion particles assemble and release via the exocytosis process to either infect other cells and precipitate conditions like ARDS and cytokine storms associated with the severe form of Covid-19 disease. The key targets are discussed in Table 1 , with a brief discussion on the druggable pocket and their 3D protein representation in Fig. 3 . The key targets are classified broadly as virus-based targets and host-based targets. The critical viral target includes S protein along with Nsps (Nsp3, Nsp7, Nsp8, Nsp12, Nsp13, Nsp10/Nsp16, Nsp14-16) and human targets that include ACE2 and TMPRSS2.
Table 1.
Critical proteins of SARS-CoV-2 and their mechanistic role (Poduri et al., 2020; Thakur et al., 2020).
| Type of protein | Mechanistic role |
|---|---|
| SARS-COV-2 protein | |
| S protein | Glycosylated transmembrane fusion protein (type I) is made of 1160–1400 amino acids. Involved in the attachment to the host cell via the ACE2 receptor. Cleaved by TMPRSS2 at the junction of hydrophobic fusion peptide into two subunits, S1 and S2. |
| S1 subunit | Also known as N-terminal or upper domain. It possesses a receptor-binding domain (RBD) responsible for ACE2 receptor binding. S1 domain is considered to be highly vulnerable to undergo mutation due to evolutionary pressure and its contact with the immune system of the host. This domain is further subdivided into SD-1 and SD-2 which induces conformational modifications in the C-terminal domain once bound to ACE2 receptors. |
| S2 subunit | Also called as C-terminal or lower domain. This domain is much more stable with the evolutionary process and thus less susceptible to undergo mutation. This domain consists of fusion machinery involved in the fusion of virus envelope with the host cell membrane |
| M protein | This is the most abundant protein in SARS-CoV-2 and responsible for eliciting a virus-specific humoral response and neutralizes developed antibodies against the virus inside the host cell |
| N protein | The protein is responsible for encapsulating and protecting (+)-RNA containing the genome of SARS-CoV-2. Also, assist in virion release by allowing a favourable orientation for virion at perinuclear membranes that further help in the release of virus particles. |
| E protein | This is a transmembrane protein acting as an ionophore. It assists in the release of viral genomic material inside a host cell in conjugation with human protease. |
| Non-structural proteins (Nsp 1–16) | The NSP complex is involved and associated with the central dogma of the viral genome. The complex assists and catalyzes viral RNA synthesis, proofreading, capping, and ultimately translation to allow synthesis of polyproteins (pp1a and pp1ab). Only necessary NSPs are discussed herewith |
| Nsp3 | Also called papain-like protease (PLpro). This enzyme possesses the deubiquitinase property and suppresses the innate immune system of the host to allow damage to the virus. These enzymes are also involved in the cleavage of pp1a and pp1ab, thus catalyzing numerous functional and effector proteins. |
| Nsp7-Nsp8 | This is also called a primase complex, involved in de novo initiation and primer extension during translation of viral mRNA |
| Nsp12 | Important Nsp also called RNA dependent RNA polymerase; RdRp. It catalyzes the transcription and replication of RNA strand complementary to a given RNA template of the virus, allowing the synthesis of sgRNA along with structural, and accessory proteins |
| Nsp13 | Also called zinc-binding helicase (HEL). This is involved in the initiation of replication, allowing the unwinding of duplex RNA/DNA with 5′ss tail in 5′–3′ direction |
| Nsp10/Nsp16 complex | This category of Nsp is a type of 2′O-methyltransferases enzymes. Nsp14 is involved with N7-(guanine)-methyltransferase, and Nsp16 is found to link with 2′O-methyltransferases activity, and both are involved with the modification of RNA cap structure. |
| Nsp14-16 | This complex is involved with RNA proofreading. |
| Host proteins | |
| ACE2 | Widely distributed in the human body. Exist as soluble (cleaved) and insoluble (full length) form, both of which contain two domains, a protease domain and the catalytic domain. The protease domain is known to be involved with the S1 binding of SARS-CoV-2, and the catalytic domain is known to play a physiological role (formation of form Ang1-7). The binding of spike protein with protease domain of ACE2 allows its downregulation and thereby diminishes its protective role, which is further deteriorated at latter stages of Covid-19 that involves ACE2 internalization |
| TMPRSS2 | It is a transmembrane serine protease (type II) enzyme known to cleaves the S protein at the hydrophobic monobasic site (685/686 junction) into two domains, S1 and S2. |
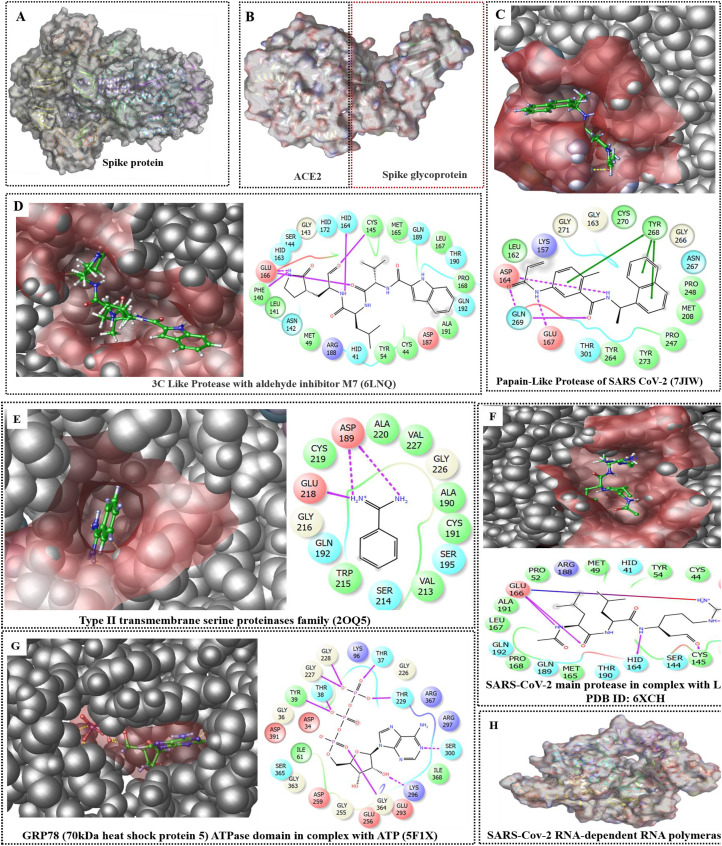
Fig. 3.
(A–H). Pictorial representation of key druggable targets involving both host and virus protein explored for identifying lead molecules using various drug design approaches. Herein, A represents the 3D structure of the spike protein, whereas B represents the binding pattern of viral spike protein with the ACE-2 receptor of the host cells. The interaction of spike protein with host ACE2 protein is critical. It facilitates the viral genome's entry within the host cell and helps in viral propagation and so, the Covid-19 disease. Therefore, designing molecules to prevent spike-ACE2 interaction seems to be a valuable strategy to prevent the propagation of the Covid-19 disease. C represents the papain-like protease of SARS-CoV-2, whereas the co-crystallized ligand's position represents one of the protein's druggable cavity. For the same protein, a 2D protein-ligand interaction profile was also provided, and it represents the critical amino acid residues involved there. Similarly, the druggable cavities of the other essential target proteins viz (D) 3CLPro, (E) type II transmembrane serine proteases, (F) main protease with leupeptin, (G) GRP78 ATPase domain, along with its 3D and 2D protein-ligand interaction profile was provided here. Finally, the 3D structural features of the RNA-dependent RNA polymerase (H) are also provided. All these proteins are important targets for developing drugs against SARS-CoV-2 infection to combat Covid-19 disease.
These targets become the critical basis of natural products-based drug discoveries to identity a putative lead against them. Some crucial studies about natural products and their role against SARS-CoV-2 are briefly compiled in section 3.
3. Studies based on natural products or naturally derived drugs against SARS-CoV-2: An in vitro and in silico approach
Natural products are a rich source of active compounds; their research and development may play a potential role in innovative drug discovery. Amidst the Covid-19 outbreak, few studies have been conducted on medicinal plant and phytochemicals derived from them to explore their potential against SARS-CoV-2. Some of the important in vitro studies are briefly summarized and discussed herein.
Kanjanasirirat et al. screened 122 natural compounds isolated from the Thai plant and assessed them using Vero E6 cells in vitro for their potential against SARS-CoV-2 infection. Among all 114 medicinal plant extract and 8 purified compounds, the Boesenbergia rotunda extract and its phytochemical panduratin-A exhibited very potent activity against SARS-CoV-2 with IC50 of 3.62 μg/ml (CC50 = 28.06 μg/ml) and IC50 0.81 μM (CC 50 = 14.71 μM) compared with the reference standard of two FDA approved drugs HCQ with IC50 of 5.08 μM (CC50 > 100 μM) and Ivermectin with IC50 of 12.68 μM (CC50 of 31.68 μM) respectively (Kanjanasirirat et al., 2020). In another research by Wang et al., the group reported in vitro activity of cholesterol 25-hydroxylase against SARS-CoV-2 infection possessing the potential for preventing membrane fusion. The cholesterol 25-hydroxylase is one of the interferons stimulating genes (ISGs), which convert cholesterol to 25-hydroxycholesterol and possess broad antiviral activity against several viruses, including HIV, Ebola virus, Niptha virus, porcine viruses, reovirus, norovirus, and vesicular stomatitis. The group used two lung epithelial cell lines, Calu3, and A549 cells with ACE2 expression and infected by SARS-CoV-2 (USA-WA1/2020) and examined the inhibition by cholesterol 25-hydroxylase in a dose-dependent manner with IC50 = 550 nM. Additionally, 25-hydroxycholesterol induced the depletion of cholesterol from the plasma membrane by activating ER-localised acyl-CoA, i.e., cholesterol acyltransferase (Wang et al., 2020).
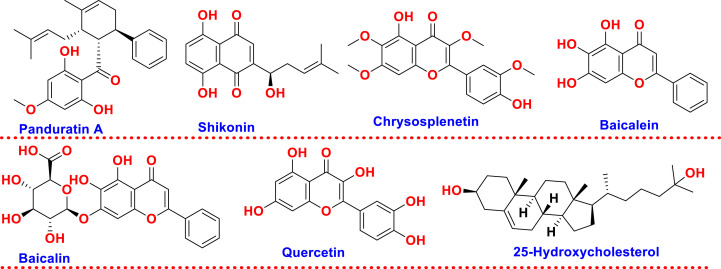
Further, research by Runfeng et al. reported antiviral activity against SARS-CoV-2 by using a herbal mixture of Chinese plants known as Lianhuaqingwen, widely used to treat cough, fever, fatigue, influenza, bronchitis, measles in the initial stage, and pneumonia. Lianhuaqingwen consists of 11 mixtures of Chinese medicinal species, mineral medicine gypsum, and methanol, water, and ethanol used a solvent to prepare the extract. The herbal mixture activity was evaluated in Vero E6 cells by performing plaque reduction and cytopathic effect inhibition assays with IC50 of 411.2 μg/ml that followed a dose-dependent response. The herbal mixture is used in Phase II clinical trials in the USA and is recommended by the Chinese National Health Commission in the treatment of Covid-19 (Li et al., 2020). In a similar context, Jin et al. reported the activity of shikonin, an active ingredient found in Lithospermum erythrorhizon against SARS-CoV-2 3CLpro with IC50 of 15.75 ± 8.22 μmol/L (Jin et al., 2020). Additionally, research by Ebada and group was focused on studying the methanol extract of Crepis sancta, a medicinal plant with activity against SARS-CoV-2 MPro in Covid-19. For phytochemical screening, 10 compounds were isolated, which consist of four eudesmane sesquiterpene lactone, i.e., (6S,7S,10R)-3-oxo-di-nor-eudesm-4-en-6α-hydroxy-11-oic acid, (6S,7S,10R)-3-oxo-di-nor-eudesm-4-en-6α-hydroxy-γ-costic acid, other two coneger 3-oxo-γ-costic acid and its methyl ester, fifth (6S,9R)-roseoside and other methylated flavonols i.e., jaceidin, kumatakenin, penduletin, pachypodol and chrysosplenetin. Among all chrysosplenetin possess a high binding affinity and potential target towards SARS-CoV-2 also was found to be a good candidate as an antiallergic and anti-inflammatory agent (Ebada et al., 2020). Another research done by Su et al. reported traditional Chinese medicine, i.e., Shuanghuanglian, and found it activity against SARS-CoV-2 3 CLpro in Vero E6 cells. The two main ingredients present in Shuanghuanglian were Baicalin and Baicalein. An oral liquid preparation of Shuanghuanglian showed dose-dependent inhibition of SARS-CoV-2 3 CLpro with IC50 of 0.090, 0.064, and 0.076 μl/ml by using FRET-based protease assay (Su et al., 2020). Abian et al., involving biophysical characterization and structural stability along with catalytic activity, performed screening of 150 natural compounds against SARS-CoV-2 3CLpro and found quercetin to be the most active inhibitor against SARS-CoV-2 3CLpro with Ki = 7 μM (Abian et al., 2020). The important chemical structures identified from in vitro based research against Covid-19 are compiled in Fig. 4 and summarization of in vitro studies is made in Table 2 . In contrast to biological determination of the potential of natural products against SARS-CoV-2, many studies have disclosed the identification of putative leads from natural drug databases utilizing in silico based studies which have been dealt in details in subsequent subsections.
Fig. 4.
Chemical structures of molecules identified for their biological potential against SARS-CoV-2 based on in vitro based studies.
Table 2.
Summarization of in vitro based studies undertaken in the quest to identify putative phytochemical(s) against SARS-CoV-2.
| S.No | Plant product | Part Used | Cell Lines | Mechanism of action | Inhibitory Concentration |
|---|---|---|---|---|---|
| 1. | Boesenbergia rotunda (Finger root) | Extract | Vero E6 cells and Calu-3 | Inhibits replication at both pre-entry and post-infection phases | 3.62 μg/mL |
| 2. | Panduratin A | Phytoconstituent | Vero E6 cells and Calu-3 | Inhibits replication at both pre-entry and post-infection phases | Pre-entry IC50: 5.30 μM; Post infection IC50: 0.81 μM |
| 3. | Cholesterol 25-hydroxylase | Interferons stimulating genes | Calu3, and A549-ACE2 | preventing membrane fusion | IC50 : 550 nM |
| 4. | Lianhuaqingwen | Extract | Vero E6 cells | Not known | IC50 : 411.2 μg/ml |
| 5. | Lithospermum erythrorhizon | Phytoconstituent | SARS-CoV-2-infected Vero cells | Inhibits the activity of 3CLpro | IC50 : 15.75 ± 8.22 μmol/L |
| 6. | Crepis sancta, | Methanol extract | mucosal mast-cell from rat basophilic leukaemia (RBL-2H3) | Inhibiting activity MPro in Covid-19 | Neutrophil elastase release inhibition (IC50: 6.66 ± 1.03) and superoxide anion generation inhibition (IC50: 4.32 ± 0.57 μM) |
| 7. | Shuanghuanglian | Oral liquid formulation | Vero E6 cells | Inhibits the activity of 3CLpro | IC50 : 0.090, 0.064, and 0.076 μl/ml |
Exploring medicinal plants and their inherent chemical constituents is a task with both broad accuracy and utmost precision. In work by Yi et al. (2018), the group put forth the idea of identifying the therapeutic significance of plant under three heads, namely i. natural plants with complex mechanisms and their biological screening (Random approach); ii. plants with ethnic use but with less scientific evidence based on phytochemistry and pharmacology (Ethnopharmacology approach); and iii. plants with traditional use and having the potential of repositioning. This is followed by data collection allowing the construction of a natural products-based database followed by their druglike analysis. After refinement, the databases are subjected to in silico application, including virtual screening utilizing the critical parameters of pharmacophore theory, identifying molecular similarity followed by molecular docking. The results thus obtained are analysed and corroborated with extensive biological support to find a putative lead(s) (Ghosh and Gemma, 2014; Yi et al., 2018).
Owing to this, advancements in computer technology has allowed in silico drug and lead identification with minimal effort and at lower cost and time. Simultaneously, computer-aided drug design is vital, as it reduces the experimental use of animals, allows the design of safer drugs with improved druglikeness properties, drug repositioning and hit optimization.
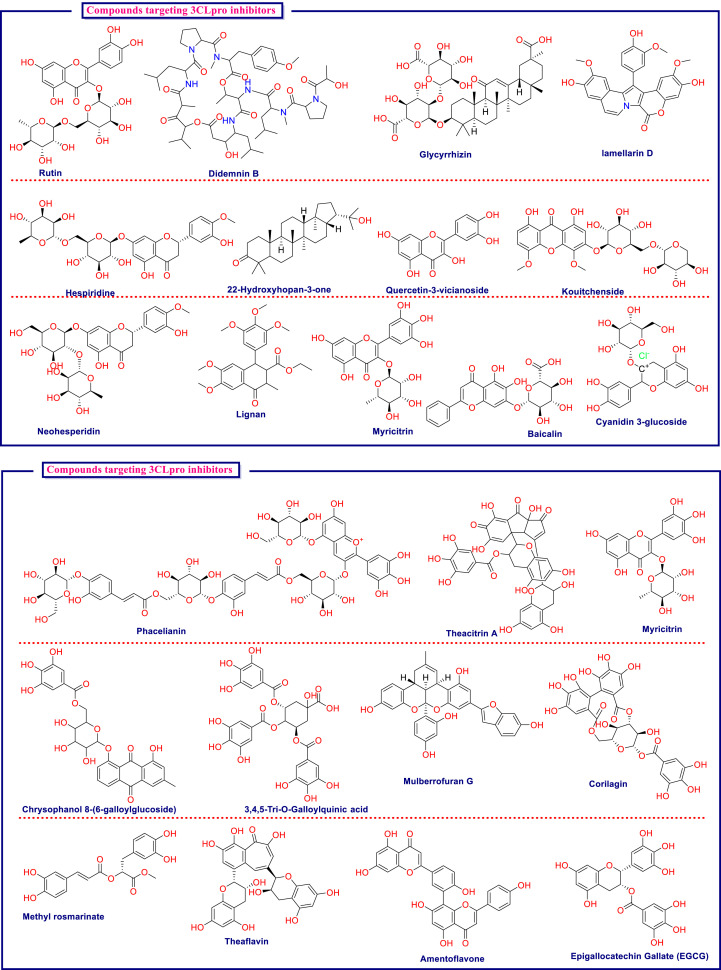
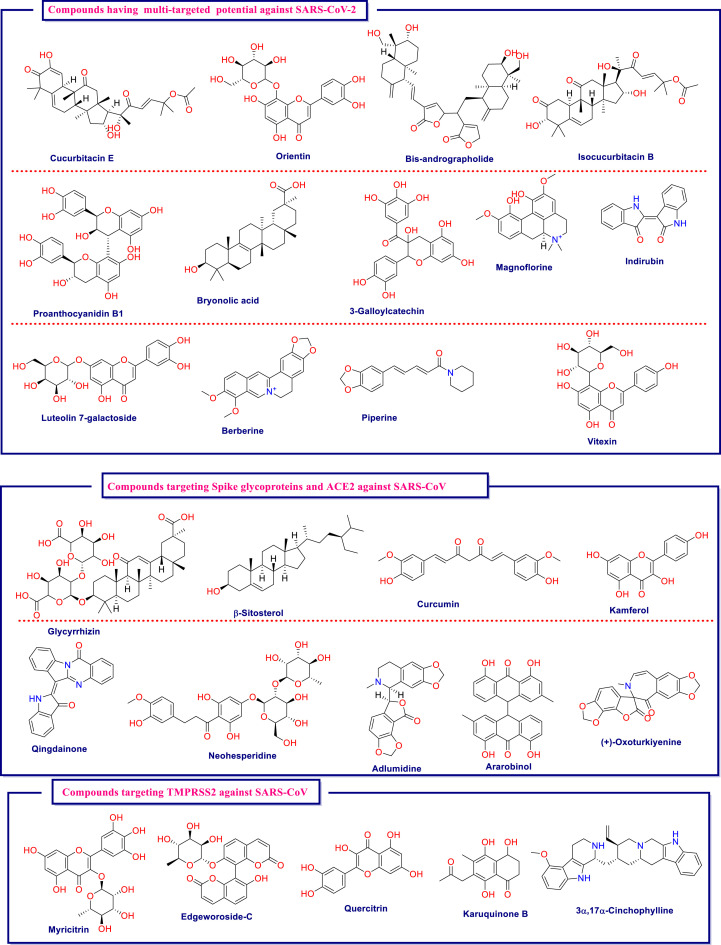
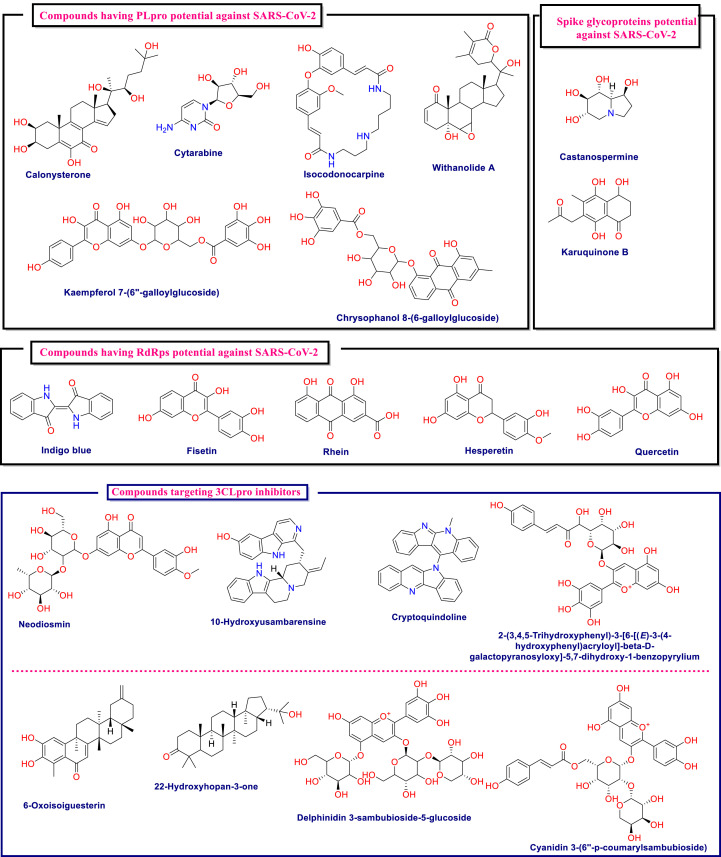
In this process of lead identification against SARS-CoV-2 from natural products, they are well explored by various in silico means like virtual screening, molecular docking, and dynamics. This has led to identification of numerous natural derived drugs and derivatives that may be tested further for their potential against Covid-19. The important in silico-based research pertaining to this area is briefly compiled in Table 3 . The table highlights the essential protein from the virus or host as a drug target, followed by the type of natural-based compounds used for screening, along with the best lead identified from analysis. The chemical structures of the key structures identified are compiled in Fig. 5 .
Table 3.
Compilation of key in silico-based studies undertaken to identify the best leads from the natural products arena against SARS-CoV-2.
| Group [Ref] | Target | Database/natural compounds (n) | Methodology | Outcome (best in silico leads) |
|---|---|---|---|---|
| Ghosh et al., (Ghosh et al., 2021) | Mpro | 113 Compounds of natural origin inhibiting Mpro in SARS-CoV were retrieved, and 88 compounds were further considered for current work on the basis of available IC50 and binding affinity data | QSAR based data mining followed by structural and physiochemical interpretation (SPCI) analysis | Rutin, Hespiridine, 22-Hydroxyhopan-3-one, Oolonghomobisflavan-A, Theasinensin-D, Quercetin, 3-vicianoside, Deacetylcentapicrin, Kouitchenside, Neohesperidin, Lignan, Myricitrin, Baicalin, Cyanidin 3-glucoside |
| Cheke et al., (Cheke et al., 2020) | Spike glycoproteins and ACE2 | 12 Natural compounds with reported antiviral attributes | Molecular docking | Indigo blue, glycyrrhizin, β-sitosterol, indirubin, bicylogermacrene, curcumin, hesperetin, rhein, and berberine |
| Sahin et al., (Sahin et al., 2021) | Mpro | Didemnins A, B and C | Molecular docking | Didemnin B |
| Monajjemi et al., (Monajjemi et al., 2020) | Protease | Vidarabine, Cytarabine, Gemcitabine, and Matrine extracted from Gillan's leaves plants. | Docking simulation and NMR investigation | Cytarabine |
| Saeed et al., (Saeed et al., 2020) | Endoribonuclease NSP15 | 1624 Natural compounds (NuBBE database) | Virtual screening and molecular docking, followed by molecular dynamics for top leads | NuBBE-1970 and NuBBE-242 |
| Caruso et al., (Caruso et al., 2020) | Mpro | Quinone derivatives | Molecular docking and DFT | Embelin |
| Basu et al., (Basu et al., 2020) | Spike protein and human ACE2 | 5 (Flavonoid and anthraquinone subclass) | Homology modelling, Molecular docking | Hesperidin |
| Gopinath et al., (Gopinath et al., 2020) | SARS-CoV-2–ACE2 Receptor Interface | ZINC biogenic (206,800 compounds) FooDB (18,477 compounds), Molport Natural Compound and Natural-Like Compound Database (119,054 compounds), and Super Natural II database (267,762) | Molecular dynamics simulations (MixMD) with high-throughput virtual screening (HTVS) | ZINC000002128789 |
| Al-Sehemi et al., (Al-Sehemi et al., 2020) | Spike glycoprotein | 31000 NPASS library | Virtual screening and molecular docking | Castanospermine and karuquinone B |
| Ibrahim et al., (Ibrahim et al., 2020b) | Mpro | 32 Natural spices (Isolated from 14 cooking seasonings) | Molecular docking, dynamics and Born surface area energy calculations | Salvianolic acid A |
| Kumar et al., (Chidambaram et al., 2020) | Mpro | 11 Natural coumarin analogues | Molecular docking | Toddacoumaquinone |
| Khalifa et al., (Khalifa et al., 2020) | CLpro | 10 Poly-acylated anthocyanins derivatives | Molecular docking | Phacelianin |
| Naik et al., (Naik et al., 2020) | Helicase, Endoribonuclease, Exoribonuclease, RNA-dependent RNA Polymerase, Methyltransferase, 3C-like proteinase |
3963 Compounds from the NPASS database | Virtual screening, molecular docking and Dynamics | NPC214620, NPC52382, and NPC270578 |
| Narkhede et al., (Narkhede et al., 2020) | Mpro | 12 Compounds from the PubChem database | Molecular docking | Glycyrrhizin and Rhein |
| Abdelrheem et al., (Abdelrheem et al., 2020) | Mpro | 10 Compounds from plants or marine algae | Molecular docking and dynamics | Caulerpin |
| Ananth et al., (Vivek-Ananth et al., 2020) | TMPRSS2 and cathepsin L | 14,011 Phytochemicals produced by Indian medicinal plants | Virtual screening, Molecular docking and dynamics | TMPRSS2: Qingdainone, Edgeworoside-C and Adlumidine; CathepsinL: Ararobinol, (+)-oxoturkiyenine and 3α,17α-cinchophylline. |
| Rakib et al., (Rakib et al., 2020) | Mpro | 309 Compounds eluted from methanol extract of T. crispa in GCMS, Lipinski rule applied to 309 compounds and 56 were selected for in silico study | Molecular docking | imidazolidin-4-one, 2-imino-1-(4-methoxy-6-dimethylamino-1,3,5-triazin-2-yl) |
| Alamri et al., (Alamri et al., 2020) | RdRp; 3CLpro; PLpro | ~1000 Compounds available in the in-house database from traditional Saudi medicinal plants | Virtual screening, Molecular docking |
RdRp: Luteolin 7-rutinoside, chrysophanol 8-(6-galloylglucoside) and kaempferol 7-(6″-galloylglucoside); 3CLpro: Chrysophanol, 3,4,5-tri-O-galloylquinic acid and mulberrofuran G; PLpro: Withanolide A, isocodonocarpine and calonysterone |
| Qamar et al., (Tahir Ul Qamar et al., 2020) | 3CLpro | 32,297 Potential antiviral phytochemicals/traditional Chinese medicinal compounds | Virtual screening, molecular docking and dynamics | Myricitrin and methyl rosmarinate |
| Mazzini et al., (Mazzini et al., 2020) | Mpro | 135 Natural and nature-inspired compounds | Virtual screening, Molecular docking | Camptothecin, leopolic acid, and lamellarin D |
| Sayed et al., (Sayed et al., 2020) | Mpro | >24,000 Compounds from a library of natural microbial products | Hyphenated pharmacophoric-based and structural-based virtual screening followed by molecular dynamics | Citriquinochroman, Holyrine B, Proximicin C, Pityriacitrin B, (+)-anthrabenzoxocinone, penimethavone A |
| Olubiyi et al., (Olubiyi et al., 2020) | 3CLpro | 3200 Natural compounds from Nigerian plants | Virtual screening and molecular docking | Theacitrin A, corilagin, theaflavin, amentoflavone, epigallocatechin gallate (EGCG), and neodiosmin |
| Subbaiyan et al., (Subbaiyan et al., 2020) | S-protein | 12 Ligands of herbal origin | Molecular docking | Epigallocatechin |
| Owis et al., (Owis et al., 2020) | Mpro | 11 Flavonoid glycosides were identified from S. persica aerial parts (stem and leaves) on the basis of metabolic profiling (secondary metabolites) | Molecular docking | Narcissin, Kaempferol-3-O-α-l-rhamnopyranosyl-(1 → 6)-β-d-glucopyranoside |
| Rahman et al., (Rahman et al., 2020) | TMPRSS2 | 30,927 Compounds from NPASS library | Ligand-based pharmacophore approach and molecular docking | NPC306344 |
| Gentile et al., (Gentile et al., 2020) | Mpro | 14,064 Marine dataset molecules retrieved from http://docking.umh.es/downloaddb | Pharmacophore filter, virtual screening, molecular dynamics and redocking | Phlorotannin, 1,3,5-trihydroxybenzene |
| Fakhar et al., (Fakhar et al., 2020) | 3CL hydrolase protein | 3435 Anthocyanin derivatives from PubChem database | Virtual screening, molecular docking and dynamics | PubChem IDs 44256891, 44256921, 102452140, 131751762, 131831710 and 139031086 |
| Selvaraj et al., (Selvaraj et al., 2020) | NSP14 | 22122 Compounds from TCM Database | Virtual screening, molecular docking and dynamics | TCM 57025, TCM 3495, TCM 5376, TCM 20111, and TCM 31007 |
| Joshi et al., (Joshi et al., 2020) | Mpro | ~7100 Phytochemicals belonging to alkaloids, flavonoids, glucosinolates, phenolics, terpenes and terpenoid category | Phylogenetic analysis by maximum likelihood (ML) method followed by virtual screening and molecular docking | δ-Viniferin, myricitrin, chrysanthemin, taiwanhomoflavone A, Lactucopicrin 15-oxalate, nympholide A, afzelin, biorobin, hesperidin and phyllaemblicin B |
| Kumar et al., (Kumar et al., 2020) | Mpro | 274,363 Compounds, among which 120,720 belong to Zinc natural database and 14,064 to Marine Natural Products (MNP) database | Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach | SN00293542 and SN00382835 |
| Ibrahim et al., (Ibrahim et al., 2020a) | Mpro | 113,756 Natural and natural-like products from MolPort database | Molecular docking, followed by molecular dynamics and MM-GBSA binding energy calculations | MolPort-004-849-765, MolPort-000-708-794, MolPort-002-513-915 and MolPort-000-702-646 |
| Sharma et al., (Sharma and Shanavas, 2020) | Mpro and ACE2 | 60 Compounds, among which 30 were natural secondary metabolites | Molecular docking, followed by MM-GBSA binding energy calculations | Delphinidin 3,5-diglucoside, Scutellarein 7-glucoside, Avicularin and 3,5-Di-O-galloylshikimic acid |
| Chidambaram et al., (Chidambaram et al., 2021) | Mpro | 10 Compounds, Calanolide A, Cardatolide A, Collinin, Inophyllum A, Mesuol, Isomesuol, Pteryxin, Rutamarin, Seselin, Suksdorin, | Molecular docking | Inophyllum A |
| Khan et al., (Khan et al., 2020) | Mpro | 5 Marine compounds | Molecular docking and dynamics | CID 11170714 |
| Chikhale et al., (Chikhale et al., 2020) | TMPRSS2 | 2230 Natural-based compounds from the Selleckchem database | Homology modelling, virtual screening and molecular dynamics | Neohesperidine, Myricitrin, Quercitrin, Naringin, and Icariin |
| Pandey et al., (Pandey et al., 2020) | Spike protein | 11 Natural compounds | Molecular docking and dynamics | Kaempferol, quercetin, and fisetin |
| Sepay et al., (Sepay et al., 2020) | Mpro | 50 Natural products derived compounds | DFT, molecular docking and dynamics | Terpenoid (T3) from marine sponge Cacospongia mycofijiensis |
| Gahlawat et al., (Gahlawat et al., 2020) | Mpro | Three datasets belonging to natural products isolated from diverse families of plants, Mpro inhibitors from the literature and the FDA approved drugs were employed | Virtual screening, molecular docking and dynamics | The literature of Mproinhibitor: lithospermic acid B, Rutin, neonuezhenide Natural product database: chebulinic acid, delphinidin-3,5-diglucoside, cyanidin-3,5-diglucoside, acteoside |
| Iheagwam et al., (Iheagwam and Rotimi, 2020) | PLpro, 3CLpro), RdRp, 2OMT, S-RBD, ACE2, and TMPRSS2 | 65 African natural products from the ZINC database | Molecular docking | 3-galloylcatechin, proanthocyanidin B1, and luteolin 7-galactoside |
| Lakshmi et al., (Alagu Lakshmi et al., 2020) | Mpro, spike protein and ACE2 | 47 Ligands from 10 ethnomedicine plant of Indian origin | Molecular docking and dynamics | Cucurbitacin E, Orientin, Bis-andrographolide, Cucurbitacin B, Isocucurbitacin B, Vitexin, Berberine, Bryonolic acid, Piperine and Magnoflorine |
| Gyebi et al., (Gyebi et al., 2020) | 3CLpro | 62 Bioactive alkaloids and 100 terpenoids African plants | Molecular docking |
Alkaloids: 10-Hydroxyusambarensine, and Cryptoquindoline; Terpenoids: 6-Oxoisoiguesterin and 22-Hydroxyhopan-3-one |
| Majumdar et al., (Majumder and Mandal, 2020) | Mpro | Initial search for Mpro lead was done from the Sigma-Aldrich plant profiler chemical library. This search led to the identification of Rutin, and 40 similar pharmacophores were retrieved from the SwissSimilarity web tool | Molecular docking and dynamics | Peonidin 3-O-glucoside, Kaempferol 3-O-β–rutinoside, 4-(3,4-Dihydroxyphenyl)-7-methoxy-5-[(6-O-β-D-xylopyranosyl-β-D-glucopyranosyl)oxy]-2H-1-benzopyran-2-one, Quercetin-3-D-xyloside, and Quercetin 3-O-α-L-arabinopyranoside |
Fig. 5.
Chemical structures of key molecules identified for their biological potential against SARS-CoV-2 on the basis of numerous in silico-based studies.
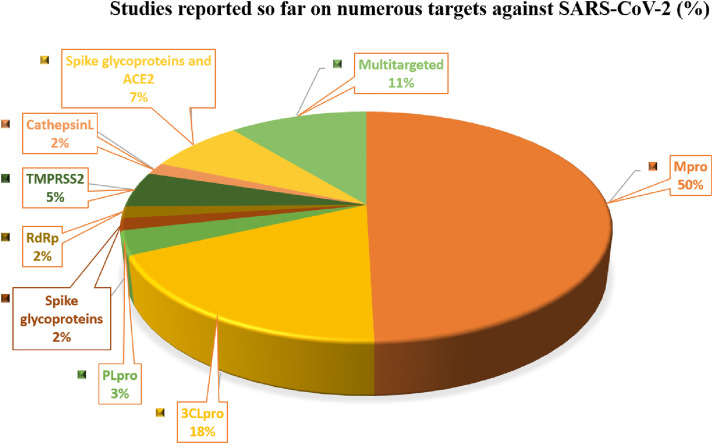
The thorough compilation and analysis suggest that the most crucial target that has been explored to identify lead from natural origin is Mpro (50%) followed by 3CLpro (18%). Moreover, multitargeted ligands have been also identified to target PLpro, 3CLpro, RdRp, 2-OMT, S-RBD, ACE2, and TMPRSS2 simultaneously occupies 11% of total drug discovery efforts from phytochemicals (Fig. 6 ).
Fig. 6.
Identified druggable targets and their share in drug discovery efforts from phytochemicals against SARS-CoV-2.
The analysis revealed the main protease (Mpro) as an attractive drug target because of numerous peculiarities (Ullrich and Nitsche, 2020), important one includes, i. no host cell proteases are known with homologous substrate specificity as Mpro, thus minimizing the probability of off-target effects; ii. Mpro is the only enzyme recognized to cleave pp1a and pp1ab polyproteins to functional proteins, inhibition of which hampers the functioning of other Nsp's; iii. ~99% and ~96% similarity with BatCoV RaTG13 Mpro and SARS-CoV Mpro, respectively; iv. The catalytic dyad (cysteine and histidine) lies within its active centre in Mpro; this is in contrast to other proteases that possess a buried water molecule within the active site, thus hampering efficient drug binding.
However, in comparison to small molecules, in silico approaches, with respect to phytochemicals require a slightly different approach. This ambiguity exists since herbs or natural products as a whole are enriched with multiple pharmacological activities depending upon their chemical entity. This suggests developing phytochemicals as drug candidates require an integrated approach of modern scientific techniques followed by regulatory harmonization. Moreover, the significant challenges in phytochemical drug development are attributed to the presence of greater numbers of chiral centres giving the possibility of generation of a large number of enantiomers (2n, where n is the number of chiral centres present); molecular rigidity; steric intricacy; higher number of hydrogen bond donor and acceptors (Lipinski's rule violation); broader variation in molecular weight, partition coefficient, the topological polar surface area along with divergence of molecular space (Koehn and Carter, 2005). These complexities and technical limitations associated with phytochemicals actually affect their development by pharmaceutical industries, arising the challenges in new drug discovery from natural resources. Further, the advancements in High-throughput screening (HTS), combinatorial chemistry and molecular biology has also declined the traditional area of research in phytochemicals. Considering the hurdles, there is an urge to revisit the scientific enthusiasm concerning phytochemicals and their inclusion in drug discovery and development programs (Katiyar et al., 2012). The research in the past few decades has led us to deduce that computational algorithms and similar methodologies like hit expansion, SAR exploitation, and scaffold hopping in drug design have not seen much success in fetching a putative drug(s). Moreover, studies involving virtual screening and molecular docking only reveals about interaction or affinity of a chemical moiety with the biological target. These studies fail to categories the leads for their pharmacodynamics attribute. The pharmacodynamic parameters that computational studies can detect include the binding energy within the target protein's binding cavity. This represents the protein-ligand binding affinity and not the intrinsic activity of the ligand molecules. Nevertheless, binding affinity and intrinsic activity are the two different pharmacodynamic terms and the biological outcome of the drug majorly depends on the intrinsic activity of the same. However, still, for producing intrinsic activity, the ligand needs to binds efficiently to the target protein, and for that region, binding affinity calculations are considered necessary in the case of drug development.
Further, if we closely look at the reported in silico studies to identify putative lead from phytochemicals against SARS-CoV-2, there is a lack of integration between the reported studies and correlation of their outcomes with the concept of chemical space and molecular similarity for identification of ‘druglike’ candidate. To address this ambiguity, we thought to analyze the chemical space and molecular similarity parameters against the identified in silico phytochemicals in quest to explore druglike lead against SARS-CoV-2. Chemical space is considered as one of the vital concepts in drug discovery. Druglike chemical space is defined as identifying those compounds possessing enough physiochemical attributes to survive human phase trial I completion. It encompasses the compounds that have the potential to become a drug (Naveja and Medina-Franco, 2019). The approach includes a comparison of bioactive or approved drug candidates for molecular similarity against unknown drug candidates. The technique involves multiple descriptors including, Molecular weight (MW); Topological polar surface area (TPSA), number of rotational bonds (nROTB); hydrogen bond donors (nHBDon) and acceptors (nHBAcc) and implication of partition coefficient (AlogP). The detailed exploration of chemical space and molecular similarity using these descriptors is discussed in subsequent sections.
4. Exploring the chemical space and molecular similarities of identified natural leads: A cheminformatics approach
It is now widely accepted that though natural products are biologically active and may possess suitable pharmacokinetic profile, they may or may not satisfy the essential criteria of drug-likenesses. The need of the hour is, therefore, to identify and develop a physiochemical tweaked phytochemicals library in parallel to the lead generation. The quest to find a potential pharmacologically active lead although restricts to the Lipinski rule in general, which is defined by four critical descriptors that include MW; nROTB; nHBDon and cLogP. Among them, MW is an important criterion that should not exceed 500 g/mol as per the Lipinski rule. Other chemical additives, including chemical elements (C, S, H, O, and N) in a molecule, branching, and a maximum of 4 rings in a drug molecule drastically affect the MW, which is further elevated by the use of halogens and radioisotope functionality in a drug candidate. MW is considered to be a substitute for molecular properties of drugs, including molecular rigidity. An increase in MW decreases the oral bioavailability and, at the same time, increases the lipophilicity of the drug candidate (Matsson and Kihlberg, 2017; Veber et al., 2002). Trends also suggest that larger molecular weight compounds often limits target binding and also involves the complex molecular design and tedious chemical synthesis, thus allowing a relatively lower population of drugs with MW more than 500 g/mol. In contrast, the high affinity of drug candidates often overshadows lead-like characteristics during hit selection (Veber et al., 2002). Considering the mean MW of approved USFDA drugs (the year 2020), the mean MW is approximately 438.53 g/mol (513.97 for the anti-infective category) (Bhutani et al., 2021). However, when considering phytochemicals, there existed many phytochemical inspired drugs, including paclitaxel, where if a similar rule was applicable, it would never become a blockbuster. To address this, many variants of Lipinski rules have been considered, the important being Veber's rule. It takes into consideration the TPSA and nROTB, that assist to differentiate between orally active and inactive drugs. According to this rule, compounds possessing ≤140 Å2 TPSA and 10 or fewer rotatable bonds are assumed to possess good oral bioavailability, which, if exceeded, could harm intestinal absorption (Veber et al., 2002). TPSA takes account of metrics of the drug ability to permeate the cells and tissues and at the same time quantify the solvent accessible surface shielded by oxygen, nitrogen and hydrogens. TPSA analysis is focused to allow influence of drug like candidate on oral absorption, cellular potency, peripheral circulation and BBB permeability (Clark, 2011a). Molecules having a TPSA of ≤140 Å2 possess poor cell permeability. The TPSA analysis on approved FDA drugs over the last five years have suggested a mean TPSA of 101.95 Å2 (145.50 Å2 for the anti-infective category) (Bhutani et al., 2021). The next parameter is nROTB, which gives an estimation of drug candidate flexibility and assist in determining oral bioavailability. Thermodynamically, an increase in entropy (ΔS>0) allows strong binding; therefore loss in rotational bond leads to a decrease in entropy, allowing the increase in free energy and consequently loss in binding strength (Zhong et al., 2013). The nROTB analysis approved FDA drugs over the last five years has suggested a mean nROTB of 7.36 (7.78 for the anti-infective drug category) (Bhutani et al., 2021). Further Lipinski rule also focus on the availability of a number of nHBDon and nHBAcc) in a drug candidate. The cut-off includes less than 5 for nHBDon and less than 10 for nHBAcc and play a vital role in drug binding to the receptor. Analysis of FDA approved drugs suggested a mean nHBDon value of 2.30 and nHBAcc value of 6.89 (2.95 and 9.13, respectively, for the anti-infective drug category) (Bhutani et al., 2021). The next parameter is cLogP or its better assessment marker AlogP considers the partition of drug candidate between aqueous and organic phase and is a deciding parameter in drug pharmacokinetics. AlogP in contrast to cLogP, also takes into account of local and molecular hydrophobicity maps along with hydrophobic interactions common in drug-receptor complexes. The acceptable range lies in between −0.4 to +5.6 range. It is considered that drug candidates possessing high AlogP are more vulnerable to CYPs metabolism, therefore reducing LogP or AlogP improves the metabolic stability and consequently ADMET properties of the drug candidate (Gleeson, 2008). The last five-year USFDA approved drugs suggested a trend of −0.32 as LogP (Bhutani et al., 2021).
In the quest to identify drug-likeness of identified leads from in silico studies on phytochemicals by various research groups against SARS-CoV-2, we computed chemical space parameters using the mentioned descriptors and explored druggable leads. The analysis was done using PUMA ((Platform for Unified Molecular Analysis) online server version 1 (Gonzalez-Medina and Medina-Franco, 2017). The comparison was drawn between identified leads with a. the FDA-approved drugs for the antiviral/anti-infective category in the last 5 years (Category A); b. FDA approved drugs derived from natural sources (Category B).
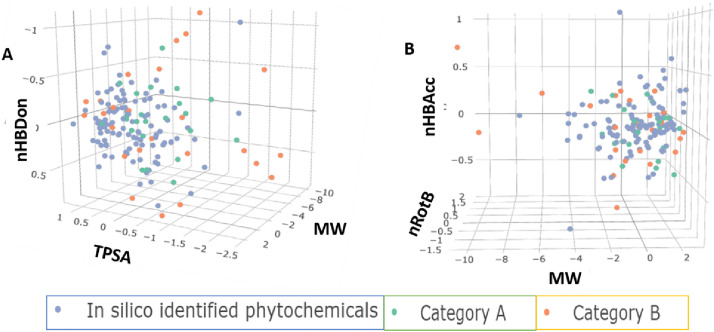
The thorough analysis revealed (in comparison to category A and B) that most natural drugs affecting reported drug targets, as discussed in Table 4 , suggested a deviation in mean MW among various drug targets. Mpro inhibitors identified were found to possess a mean MW of 551.96 g/mol, with 3CLpro inhibitors identified with a mean MW of 642.26 g/mol and S protein inhibitors with the lowest mean MW 226.59 g/mol, followed by a broad statistical variance among all the reported categories (mean MW for category A: 513.97 g/mol and B: 603.95 g/mol). The next parameter we employed was TPSA (Table 4 and Fig. 7 ) which consider the bioavailability of a drug candidate. The analysis on identified phytochemical revealed that the lead compounds intended as PLpro (118.08 Å2), S protein (89.49 Å2), Cathepsin L (82.38 Å2), and multitargeted inhibitors (126.32 Å2), including S Protein and ACE2 (109.83 Å2) follow the TPSA rule efficiently. However, sharing 50% of the total in silico-based drug discovery effort, Mpro inhibitors identified so far do not meet this threshold of TPSA (190.75 Å2), signifying plausibility of decreased or no cellular potency or oral absorption along with the setback of peripheral circulation during the stages of preclinical or clinical development (mean TPSA for category A: 145.50 Å2 and B: 195.61 Å2) (Clark, 2011b). However, a deeper analysis of FDA drugs (Table 5 ) revealed that mainstream anti-infectives launched in the last five years possessed a high value of <140 Å2 TPSA (mean of 131.9 Å2 in 5 years) though the majority were intended with the intravenous or topical route of administration. Considering nROTB next, which give an estimation of drug flexibility, revealed that identified leads belonging to Mpro, 3CLpro, and RdRp defined better mean nROTB scores of 6.38, 6.77 and 6.5, respectively. However, lead belonging to other categories of inhibitors possesses high to medium rigidity (mean 1–3 nROTB) in contrast to category A: 7.78 and B: 10.538. Next, we analysed the trend for nHBDon (cutoff ≤ 5) and nHBAcc (≤10), which suggested identified ligands belonging to Mpro, 3CLpro, RdRp categories are the potential outliers to normal cut-offs as per the Lipinski rule. The next parameter we measured was AlogP that revealed that inhibitors for Cathepsin L and S Protein and ACE2 dual inhibitor obliged this parameter well.
Table 4.
The table illustrates median, mean and standard deviation (SD) values for molecular descriptors employed against reported druggable targets identified using in silico-based studies for natural products.
| Target | MW (g/mol) |
TPSA (Å2) |
nROTB |
nHBDon |
nHBAcc |
AlogP |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | Mean | SD | Median | Mean | SD | Median | Mean | SD | Median | Mean | SD | Median | Mean | SD | Median | Mean | SD | |
| Mpro | 494.12 | 551.96 | 191.8 | 184.98 | 190.75 | 90.49 | 6 | 6.38 | 4.30 | 6 | 6.22 | 3.84 | 11 | 11.94 | 5.27 | −2.24 | −2.03 | 1 |
| 3CLpro | 588.14 | 642.26 | 222.3 | 227.4 | 234.42 | 129.58 | 5.5 | 6.77 | 5.69 | 8.5 | 8.63 | 4.98 | 13 | 13.95 | 7.59 | −2.55 | −2.31 | 8.02 |
| PLpro | 467.74 | 413.71 | 113.8 | 118.76 | 118.08 | 18.98 | 2 | 2.5 | 1.73 | 4 | 4 | 1.63 | 7.5 | 7.25 | 0.95 | −1.29 | −1.11 | 2.94 |
| S protein | 226.59 | 226.59 | 53.03 | 89.49 | 89.49 | 7.54 | 1 | 1 | 1.41 | 3.5 | 3.5 | 0.70 | 5 | 5 | 0 | −1.72 | −1.72 | 2.34 |
| RdRp | 584.11 | 584.11 | 22.62 | 236.82 | 236.82 | 23.06 | 6.5 | 6.5 | 0.70 | 8 | 8 | 1.41 | 14 | 14 | 1.41 | −2.13 | −2.13 | 2.88 |
| TMPRSS2 | 415.60 | 429.44 | 110.1 | 139.71 | 143.25 | 73.87 | 2 | 3 | 3.63 | 4.5 | 4.5 | 3.61 | 8.5 | 9.33 | 3.72 | −1.14 | −1.48 | 3.83 |
| Cathepsin | 466.27 | 441.16 | 54.09 | 83.53 | 82.38 | 33.26 | 1 | 1.66 | 2.08 | 3 | 2.33 | 2.08 | 6 | 6.33 | 1.52 | 0.28 | 0.41 | 1.19 |
| S Protein and ACE2 | 302.04 | 369.69 | 176.4 | 107.22 | 109.83 | 67.27 | 1 | 3 | 3.08 | 3 | 3.55 | 2.06 | 6 | 6.44 | 4 | −0.65 | −0.37 | 1.60 |
| Multitargeted | 448.10 | 448.10 | 120.1 | 138.2 | 126.32 | 67.54 | 3 | 3.53 | 2.18 | 3 | 4.30 | 3.30 | 8 | 7.53 | 3.35 | −1.25 | −0.99 | 3.44 |
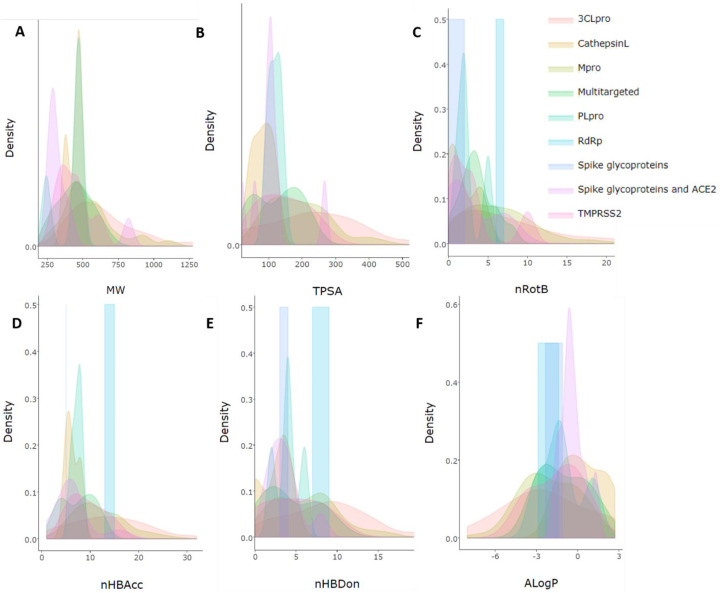
Fig. 7.
Density plot suggesting the deviation between numerous drug targets identified from in silico-based studies for natural products. The essential parameters include A. Molecular weight (MW); B. Topological polar surface area (TPSA); C. number of rotational bonds (nROTB); D. hydrogen bond donors (nHBDon); E. acceptors (nHBAcc); and F. implication of partition coefficient (AlogP).
Table 5.
The table illustrates median, mean and standard deviation (SD) values for molecular descriptors employed against reported USFDA approved drugs under anti-infective category during year 2015–20.
| FDA anti-infective | MW (g/mol) |
TPSA (Å2) |
nROTB |
nHBDon |
nHBAcc |
AlogP |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Median | Mean | SD | Median | Mean | SD | Median | Mean | SD | Median | Mean | SD | Median | Mean | SD | Median | Mean | SD | |
| Year 2015 | 631.67 | 610.44 | 142.53 | 173.29 | 176.61 | 49.88 | 13 | 11.5 | 6.45 | 3 | 3 | 0.81 | 11 | 11 | 2.58 | −1.60 | −1.59 | 1.97 |
| Year 2017 | 311.62 | 345.13 | 168.06 | 81.82 | 82.33 | 4.126 | 5 | 5.25 | 2.21 | 1 | 1.25 | 0.5 | 5 | 5.25 | 2.21 | 0.13 | 0.13 | 1.51 |
| Year 2018 | 564.66 | 556.74 | 99.60 | 149.90 | 153.09 | 69.10 | 6 | 6.62 | 3.77 | 4 | 4.25 | 3.65 | 10.5 | 10 | 3.77 | −0.31 | −1.12 | 3.00 |
| Year 2019 | 433.18 | 493.86 | 185.30 | 110.79 | 146.86 | 111.29 | 6 | 7.5 | 5.19 | 2 | 2.5 | 2.64 | 5.5 | 7 | 4.83 | 0.71 | 0.27 | 3.35 |
| Year 2020 | 583.158 | 523.18 | 120.76 | 187.05 | 166.23 | 58.16 | 10 | 9.66 | 4.50 | 2 | 2.33 | 1.52 | 14 | 12.3 | 3.78 | −3.21 | −2.02 | 2.13 |
Further, the density plots sketched in Fig. 7 also revealed high statistical distribution (high standard deviation) for numerous drug categories, importantly for MW and TPSA which involves a broader median range.
Further, a neck-to-neck comparison of various parameters is made for the in silico based identified phytochemicals and category A and B drugs in Table 6 . Fig. 8 is in a similar context, portrays density variation among these parameters and depicts the outlier regions.
Table 6.
Comparative analysis of reported in silico based identified phytochemicals and category A and B drugs on the basis of key molecular descriptors employed. The values represent mean values for the molecular descriptors.
| Parametersa | Possible leads identified from in silico-based studies from natural sources against Covid-19 | FDA-approved drugs for the antiviral/anti-infective category in the last 5 years (Category A) | FDA approved drugs derived from natural sources (Category B) |
|---|---|---|---|
| MW (g/mol) | 456.33 | 513.97 | 603.95 |
| TPSA (Å2) | 147.92 | 145.50 | 195.61 |
| nROTB | 3.815 | 7.78 | 10.53 |
| nHBDon | 5.003 | 2.95 | 5.26 |
| nHBAcc | 9.085 | 9.13 | 12.38 |
| AlogP | −1.30 | −0.86 | 2.14 |
Represents mean value.
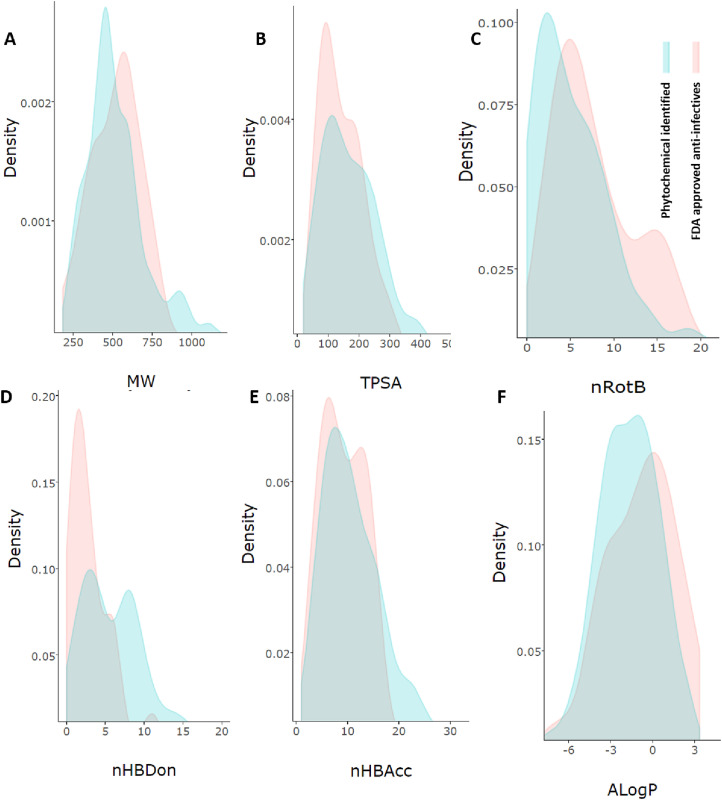
Fig. 8.
Density plot suggesting the complementarity (overlapping area) and dissimilarity between leads from natural products identified from in silico-based studies and reported USFDA approved drugs under anti-infective category during the year 2015–20 using various key molecular descriptor employed. The descriptors include A. Molecular weight (MW); B. Topological polar surface area (TPSA); C. number of rotational bonds (nROTB); D. hydrogen bond donors (nHBDon); E. acceptors (nHBAcc); and F. implication of partition coefficient (AlogP).
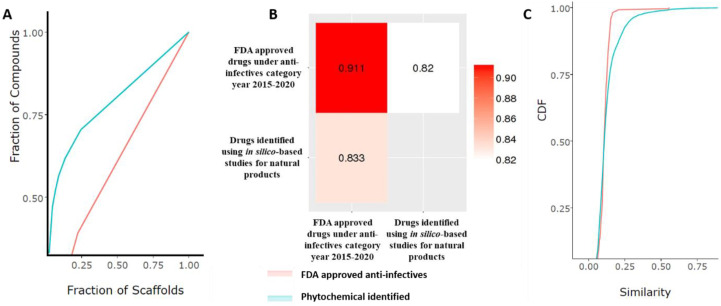
As it is evident that natural product-derived drug discovery would open up many opportunities for drug development involving high structural or chemical complementarity. To deduce the complementarity, we also went on to predict the molecular similarity, if any, between FDA approved anti-infectives and identified natural ligands using cumulative scaffold recovery (CSR) curves methodology. The CSR curves assisted in determining molecular similarity by the plot distribution curves. The diagonal plot is considered to portray high scaffold diversities, whereas gradients or steeper curves suggest low scaffold diversities (Yongye et al., 2012). The analysis revealed (Fig. 9 A) that ligands identified in the natural products category possessed a low molecular diversity (F50: 0.06) among themselves as compared to the FDA-approved (F50: 0.33) drugs suggesting a low molecular overlap and complementarity. Further, molecular similarity using the Tanimoto coefficient (T) was also deduced. The pair of chemical structures are considered similar if they possesses Tanimoto coefficient (T) > 0.85 (Patterson et al., 1996). The analysis revealed that 55.23% identified phytochemicals ligands exhibited Tanimoto coefficient (T) > 0.85 with FDA approved infectives, whereas 44.76 were found to possess a value of T < 0.85, signifying dissimilarity in substructures (Fig. 9B). This was further corroborated with the Extended-connectivity fingerprints (ECFP4) approach (Fig. 9C) to identify molecular features relevant to molecular activity (Rogers and Hahn, 2010). The method gives an analysis of complementarity by identifying compounds with similar bioactivity.
Fig. 9.
The illustration suggests output of descriptive analysis for A. cumulative scaffold recovery (CSR) curves methodology; B. Tanimoto coefficient; and C. Extended-connectivity fingerprints (ECFP4) approach. The research was performed for reported USFDA approved drugs under the anti-infective category during the year 2015–20 and drugs identified using in silico-based studies for natural products.
5. Outcomes from chemical space and molecular similarity analysis and corroboration using molecular dynamics
The thorough analysis of chemical space and molecular similarity parameters led us to identify the leads subset among reported in silico studies. The selection was based on the basis of leads satisfying Lipinski's and Veber's rules and possessed good molecular similarity with reported drugs (Category A and B). We selected the top 10 best scoring molecules that possess a high probability of drug-likeness and may be validated biologically against SARS-CoV-2 to find a putative drug candidate. The compilation of these top-scoring molecules made in Table 7 . The analysis also suggested most ligand-target the essential SARS-CoV-2 protein Mpro, thus conferring it with the status of most prominent drug target from the natural arena as discussed previously. To corroborate the findings, we performed Molecular Docking and Molecular Dynamics (MD) simulation on top identified ligand (Rutin); ligand with a moderate score (narcissin) and ligand with the lowest score (Luteolin 7-galactoside). The docking interaction (with Mpro) is reported in Fig. 10 .
Table 7.
Top 10 most scoring ligands identified among drugs identified using in silico-based studies for natural products against various drug targets of SARS-CoV-2 and host. The identified leads possess a high probability of drug-likeness and may be tested biologically against SARS-CoV-2 for a better corroboration of current studies made.
| Sr No. | Identified ligandsa | Recognized target |
|---|---|---|
| 1. | Rutin | Mpro |
| 2. | Quercetin-3-vicianoside | Mpro and spike protein |
| 3. | Lignan | Mpro |
| 4. | Embelin | Mpro |
| 5. | Toddacoumaquinone | Mpro |
| 6. | Rhein | Spike glycoproteins and ACE2 |
| 7. | Caulerpin | Mpro |
| 8. | imidazolidin-4-one, 2-imino-1-(4-methoxy-6-dimethylamino-1,3,5-triazin-2-yl) | Mpro |
| 9. | Camptothecin | Mpro |
| 10. | Lamellarin D | Mpro |
Only top 10 identified ligand in order of their drug-likeness are illustrated.
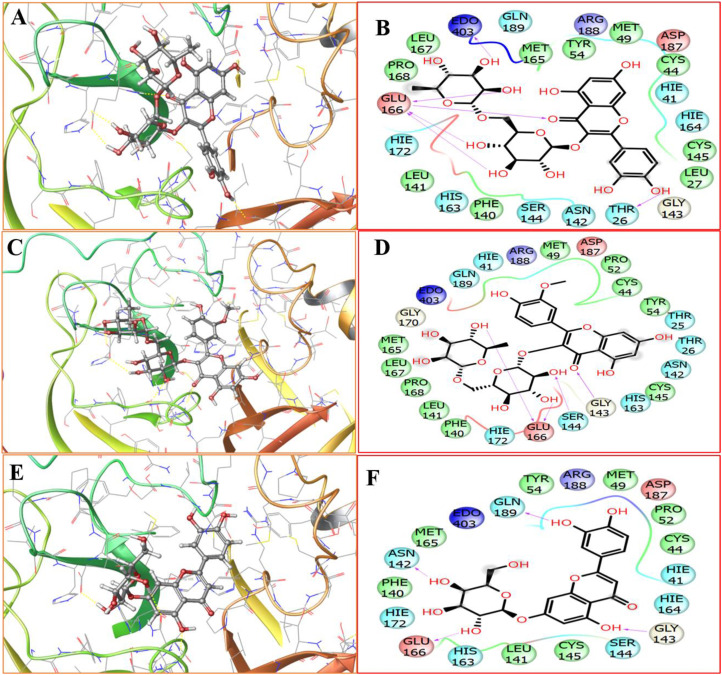
Fig. 10.
Molecular docking study against SARS-CoV-2 major druggable target, MPro, where A and B represents the 3D and 2D docked pose for Rutin; C and D represent the 3D, and 2D docked pose for narcissin and; E and F represent the 3D, and 2D docked pose for Luteolin 7-galactoside.
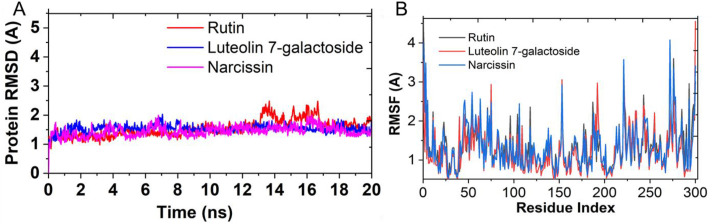
We obtained a docking score of −11.277, −8.939, and −7.957 Kcal/mol, respectively, for Rutin, narcissin, and Luteolin 7-galactoside, respectively, against the Mpro target. Based on the docking score, we have concluded Rutin to be the best, followed by narcissin and Luteolin 7-galactoside with the least binding potential against the Mpro protein. If we compare the trend with the results obtained from the chemical space and molecular similarities results a very high correlation in both models' prediction was observed. To further explore the stability of the protein-ligand complex for Rutin, narcissin, and luteolin 7-galactoside, MD simulation studies were performed considering the protein-ligand RMSD of the ligands with Mpro protein. The studies highlight the RMSD pattern (Fig. 11 A and B) that suggests the Rutin shows comparatively minimal RMSD fluctuations till 13 ns as compared to the other two ligand molecules. However, a section of comparatively higher fluctuations was observed for Rutin that is from 13.5 to 16.5 ns of time frames and thereafter, its RMSD was again found stabilizing after 16.5 ns of a time interval. If we compare the region of 0–13.5 ns the RMSD of other tow ligands viz narcissin and luteolin 7-galactoside, we observed comparatively more fluctuations then that of rutin molecule. In a similar manner, if we compare the RMSF, Rutin was again found to show the least fluctuation compared to the other two molecules. Maximum RMSF fluctuation was observed for luteolin 7-galactoside in this case. Therefore, considering the docking score as well as RMSD and RMSF trends, we can conclude that the protein-ligand binding is best for Rutin, followed by narcissin and luteolin 7-galactoside, respectively.
Fig. 11.
A. RMSD (Ca) for and Ligand- Mpro complex for Rutin, narcissin and Luteolin 7-galactoside as obtained from MD simulation studies. (B) Sidechain RMSF for and Ligand- Mpro complex for Rutin, narcissin and Luteolin 7-galactoside as obtained from MD simulation studies.
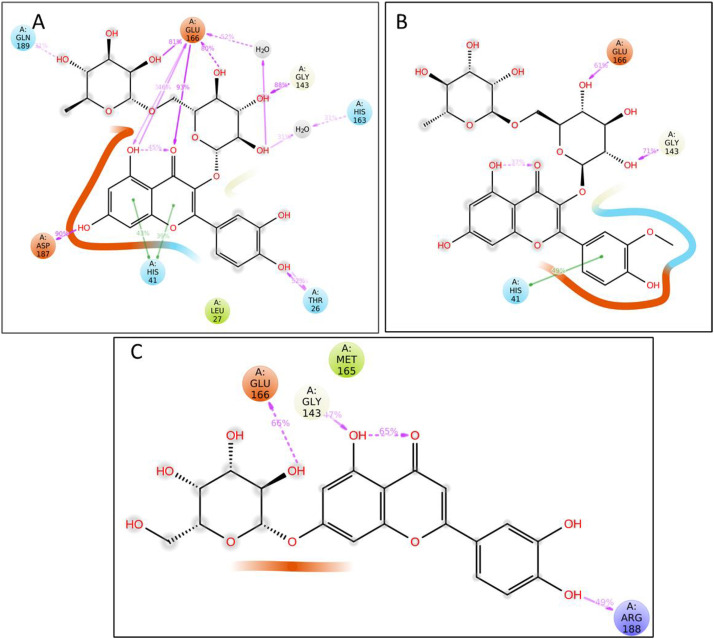
To further investigate the protein-ligand binding strength, the interaction pattern as obtained from the MD-Simulation studies was also studies (Fig. 12 ). Herein, for Rutin, multiple hydrogen-bonding interactions for Thr 26, His 41, Gly 143, His 163, Glu 166, and Asp 186, has been observed. In the case of narcissin, only three of such type of interactions viz His 41, Gly 143 and Glu 166 have been observed. If we compare the structure of both the compounds, there is a high structural similarity, but still, the impact on the interaction profile is enormous. These observations further compliance the applicability of the our chemical space and molecular similarities results that we have discussed in the previous section, as that also revealed Rutin to be the best plausible drug candidate compared to the other two entities. Similarly, only three types of interactions were also visible in the case of our third ligand, luteolin 7-galactoside, suggesting lower interaction. Thus, our study suggested a strong correlation of identified phytochemicals on the basis of chemical space and molecular similarity parameters, which the functionalities of molecular docking and dynamics.
Fig. 12.
Interaction patterns obtained for protein-ligand MD simulation studies, A. Rutin; B. Narcissin; C. Luteolin-7-galactoside.
Thus, the current study is a data-driven effort that assigns the required knowledge about active molecules for the specific targets of SARS-CoV-2. The corroboration of chemical space parameters portrays a criticality to overcome the biases in optimizing chemical space and at the same time allowed thoughtful exploration of chemical space focusing chiefly on target-relevant phytochemical identification.
6. Conclusion
The recent outbreak of Covid-19, caused by SARS-CoV-2, has put the world on red alert. Therefore, putting a strong urge to identify drugs that are capable of curing the infection. Natural product and the phytochemicals derived from them become immensely important during a global health crisis and represent one of the most practical and promising approaches to decrease the intensity of pandemics with their therapeutic potential. Considering the high genomic similarity between SARS-CoV-2 with SARS-CoV and MERS-CoV and use of phytochemicals for treatment or prophylaxis of later has led to research from natural products to identify a wonder drug lead for SARS-CoV-2. Considering research in this area, there are only a handful of studies published that have deduced the effect of phytochemicals against SARS-CoV-2 only under in vitro conditions. Therefore, an exigent need to identify and design drugs could assist in facilitating lead drug discovery by using modern drug discovery techniques than traditional drug discovery. The traditional drug discovery is a costly and time-consuming affair and would delay an urgency to find a druggable lead in pandemic scenarios. Therefore, the latest technologies used to combat disease. Considering this, the majority of publications are based on in silico-based studies to identify leads from phytochemicals on the basis of affinity against key druggable targets of SARS-CoV-2. The release of the detailed 3D structure of viral protein along with critical druggable targets helped in understanding the structural aspects of various molecular targets of SARS-CoV-2 and identification of novel lead molecules. This could be an effective strategy to accelerate the drug discovery and development process, which shorten the research period and low the expenditures involved in drug development for Covid −19. It involves various tools such as homology modelling, molecular docking, QSAR, QSPR, QSTR descriptors and molecular dynamics by using the structural information of either ligands or target to identifying potent lead. Though this approach fastens the drug discovery but many a time, lead identification of molecules is deprived of druglikeness. Since in silico-based methodologies represents the protein-ligand binding affinity and not the intrinsic activity of the ligand molecules. The binding affinity and intrinsic activity are the two different pharmacodynamic terms, and the biological outcome of the drug majorly depends on the intrinsic activity of the same.
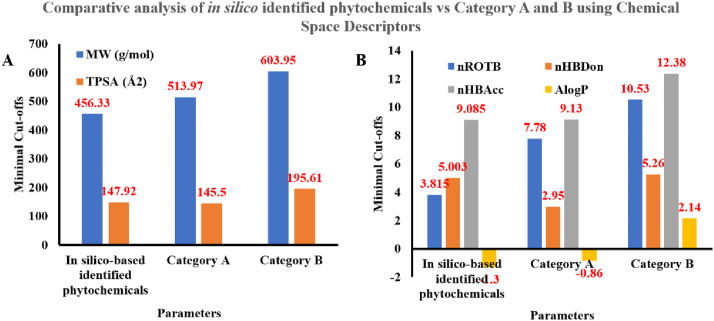
As it is widely accepted that though natural products are biologically active and may possess suitable pharmacokinetic profile, they may or may not satisfies the essential criteria's of ‘drug-likenesses. The need of hour is therefore to identify and develop physiochemical tweaked phytochemicals library in parallel to the lead generation. Considering this, our current research was extended to explore the key druggable parameters by exploring their chemical space and molecular similarity in the light of already identified phytochemicals in Covid-19. The chemical space was explored using key descriptors that includes MW, TPSA, number of rotational bonds (nROTB), hydrogen bond donors (nHBDon), and acceptors (nHBAcc) and implication of partition coefficient (AlogP). The thorough analysis of these parameters in comparison to FDA approved anti-infectives (category A) and FDA approved drugs from natural products (category B) revealed in silico identified leads are well correlated with FDA approved anti-infectives and those with natural products derived FDA approved drugs (Fig. 13 ). The mean MW was found to relatively lower than the mean of category A and B drug but well under Lipinski criteria (≤500 g/mol). The TPSA was found slightly higher than recommended values (≤140 Å2) but was in good accordance with FDA approved anti-infective, which is required since they selectively affect viral or bacterial protein as a drug target, thus conferring selectivity. For nROTB, in silico identified leads were found to be more rigid in comparison to category A and B, whereas nHBAcc and nHBDon were found in accordance with the Lipinski rule. Another essential criterion, i.e., AlogP, was well found to be lower in line with FDA approved anti-infective. High AlogP values are majorly associated with rapid CYPs assisted metabolism leading to a shorter duration of action.
Fig. 13.
Bar graph represents a comparative analysis of chemical space descriptors (A. MW, TPSA; B. nROTB, nHBDon, nHBAcc and AlogP) between in silico identified phytochemicals with category A and B entities.
Furthermore, a good correlation was observed between in silico identified leads with FDA approved infectives. The correlation was observed between MW vs HBDon vs TPSA (Fig. 14 A) and MW vs nRotB vs nHBAcc (Fig. 14B). The natural FDA approved (orange dots, category B) were mainly found scattered in the plot as an outlier with minimal overlapping with former categories. Although studied lead(s) possess a good correlation in terms of physiochemical parameters and other overlapping descriptors, however, ambiguity was noted in term of molecular similarity. Only 55.23% molecular similarity was observed between in silico identified phytochemicals with FDA approved anti-infectives as deduced by the Tanimoto coefficient. This pattern was further correlated with CSR distribution curves which again suggested low molecular diversity as compared to the FDA-approved anti-infective (F50: 0.33) and within themselves (F50: 0.06).
Fig. 14.
Three-dimensional representation highlighting the chemical space parameter comparison A. MW vs HBDon vs TPSA and B. MW vs nRotB vs nHBAcc of in silico identified phytochemicals with those of category A and Category B bioactive. The analysis and graphs were generated using PUMA online server v.1.
The thorough analysis led us to identify the ranking system of in silico leads in order of their drug-likeness among which 10 ligands are highlighted in Table 6. Further, the corroboration of chemical space parameters was done using molecular docking and dynamics simulation. The study suggested the ranking in alignment with the findings. Current work thus aims to provide a holistic strategic background on drug design and discovery form natural derived compounds keeping in view the identified phytochemicals derived lead(s) for Covid-19. Moreover, as per Medgadget, molecular modelling market is likely to become forerunner in new era of modern drug discovery. The Market Research Future (MRFR) predicts CAGR to be 15.46% during year 2017–2023. Thus synchronization of molecular modelling aspects with drug likeness is highly recommend in silico lead identification in future.
In a nutshell, nature provides us with an immense source of active ingredients yet to be discovered to treat diseases. Almost 50% of drugs in the market are originated from nature. Some blockbusters in this category include, Antimalarial drugs Quinine (Cinchona spp.) and Artemisinin (Artemisia annua); anticancer drug Vinblastine (Catharanthus roseus), and Taxol (Taxus brevifolia), etc., to name a few. Apart from natural drugs, whole plant extracts without isolation of bioactive are sometimes used in the form of traditional medicine and produce a better therapeutic effect as compared to the individual compound. Further, the use of computer-assisted drug discovery has brought a revolution in the natural products drug discovery. This includes a handful of successful examples, such as the discovery of dozamide, imatinib, dasatinib, ponatinib, and many more. However, considering the current pandemic of Covid-19, authors believe that natural products have been underexplored for their utility against SARS-CoV-2. The thorough compilation of this work led us to only a handful of studies where the biological mechanism of natural drugs has been elucidated against SARS-CoV-2. Although much effort has been made on in silico-based lead identification, their biological corroboration is still awaited. To get the likelihood of the most putative druglike lead from current research, we did chemical space and molecular similarity analysis. The analysis led us to identify a few leads that possess druglike properties and maybe a strong contender in the quest to identify a suitable lead drug molecule from the natural arena. Further, in the current global pandemic, to date, there is no specific and reliable drug candidate available. Thus, considering the richness of natural products as a rich source of the active compounds and believing that SARS-CoV-2 is not the last virus belonging to the coronavirus family focused research on identifying anti-coronavirus drug from this rich source is highly recommended.
Author contributions
The manuscript has been prepared and drafted with the understanding and contributions of all the authors.
Funding source
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
CRediT authorship contribution statement
Gaurav Joshi: Conceptualization, Data curation, Writing – original draft, preparation, Writing – review & editing. Jayant Sindhu: Methodology, Writing – original draft, preparation, Writing – review & editing. Shikha Thakur: Methodology, Writing – original draft, preparation, Writing – review & editing. Abhilash Rana: Methodology, Writing – original draft, preparation. Geetika Sharma: Methodology, Writing – review & editing. Mayank: Conceptualization. Ramarao Poduri: Conceptualization, Writing – review & editing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
All the authors thank their respective affiliated working institute and universities for providing the necessary infrastructure and support to carry out the current work.
Handling editor: Dr. Jose Luis Domingo
References
- Abdelrheem D.A., Ahmed S.A., Abd El-Mageed H.R., Mohamed H.S., Rahman A.A., Elsayed K.N.M., Ahmed S.A. The inhibitory effect of some natural bioactive compounds against SARS-CoV-2 main protease: insights from molecular docking analysis and molecular dynamic simulation. J. Environ. Sci. Health A. 2020;55:1373–1386. doi: 10.1080/10934529.2020.1826192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abian O., Ortega-Alarcon D., Jimenez-Alesanco A., Ceballos-Laita L., Vega S., Reyburn H.T., Rizzuti B., Velazquez-Campoy A. Structural stability of SARS-CoV-2 3CLpro and identification of quercetin as an inhibitor by experimental screening. Int. J. Biol. Macromol. 2020;164:1693–1703. doi: 10.1016/j.ijbiomac.2020.07.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Sehemi A.G., Olotu F.A., Dev S., Pannipara M., Soliman M.E., Carradori S., Mathew B. Natural products database screening for the discovery of naturally occurring SARS-cov-2 spike glycoprotein blockers. Chemistry. 2020;5:13309–13317. doi: 10.1002/slct.202003349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alagu Lakshmi S., Shafreen R.M.B., Priya A., Shunmugiah K.P. Ethnomedicines of Indian origin for combating COVID-19 infection by hampering the viral replication: using structure-based drug discovery approach. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1778537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alamri M.A., Altharawi A., Alabbas A.B., Alossaimi M.A., Alqahtani S.M. Structure-based virtual screening and molecular dynamics of phytochemicals derived from Saudi medicinal plants to identify potential COVID-19 therapeutics. Arab. J. Chem. 2020;13:7224–7234. doi: 10.1016/j.arabjc.2020.08.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basu A., Sarkar A., Maulik U. Molecular docking study of potential phytochemicals and their effects on the complex of SARS-CoV2 spike protein and human ACE2. Sci. Rep. 2020;10:1–15. doi: 10.1038/s41598-020-74715-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhutani P., Joshi G., Raja N., Bachhav N., Rajanna P.K., Bhutani H., Paul A.T., Kumar R. U.S. FDA approved drugs from 2015–june 2020: a perspective. J. Med. Chem. 2021;64:2339–2381. doi: 10.1021/acs.jmedchem.0c01786. [DOI] [PubMed] [Google Scholar]
- Cantuti-Castelvetri L., Ojha R., Pedro L.D., Djannatian M., Franz J., Kuivanen S., van der Meer F., Kallio K., Kaya T., Anastasina M., Smura T., Levanov L., Szirovicza L., Tobi A., Kallio-Kokko H., Osterlund P., Joensuu M., Meunier F.A., Butcher S.J., Winkler M.S., Mollenhauer B., Helenius A., Gokce O., Teesalu T., Hepojoki J., Vapalahti O., Stadelmann C., Balistreri G., Simons M. Neuropilin-1 facilitates SARS-CoV-2 cell entry and infectivity. Science. 2020;370:856–860. doi: 10.1126/science.abd2985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caruso F., Rossi M., Pedersen J.Z., Incerpi S. Computational studies reveal mechanism by which quinone derivatives can inhibit SARS-CoV-2. Study of embelin and two therapeutic compounds of interest, methyl prednisolone and dexamethasone. J. Infect. Public Health. 2020;13:1868–1877. doi: 10.1016/j.jiph.2020.09.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheke R.S., Narkhede R.R., Shinde S.D., Ambhore J.P., Jain P.G. Natural product emerging as potential sars spike glycoproteins-ace2 inhibitors to combat COVID-19 attributed by in-silico investigations. Biointerface Res. Appl. Chem. 2020;11:10628–10639. doi: 10.33263/BRIAC113.1062810639. [DOI] [Google Scholar]
- Chidambaram S.K., Ali D., Alarifi S., Radhakrishnan S., Akbar I. In silico molecular docking: evaluation of coumarin based derivatives against SARS-CoV-2. J. Infect. Public Health. 2020;13:1671–1677. doi: 10.1016/j.jiph.2020.09.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chidambaram S., El-Sheikh M.A., Alfarhan A.H., Radhakrishnan S., Akbar I. Synthesis of novel coumarin analogues: investigation of molecular docking interaction of SARS-CoV-2 proteins with natural and synthetic coumarin analogues and their pharmacokinetics studies. Saudi J. Biol. Sci. 2021;28:1100–1108. doi: 10.1016/j.sjbs.2020.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chikhale R.V., Gupta V.K., Eldesoky G.E., Wabaidur S.M., Patil S.A., Islam M.A. Identification of potential anti-TMPRSS2 natural products through homology modelling, virtual screening and molecular dynamics simulation studies. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1798813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Christy M.P., Uekusa Y., Gerwick L., Gerwick W.H. Natural products with potential to treat RNA virus pathogens including SARS-CoV-2. J. Nat. Prod. 2020;84:161–182. doi: 10.1021/acs.jnatprod.0c00968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark D.E. What has polar surface area ever done for drug discovery? 2011;3:469–484. doi: 10.4155/fmc.11.1. [DOI] [PubMed] [Google Scholar]
- Clark D.E. What has polar surface area ever done for drug discovery? Future Med. Chem. 2011;3:469–484. doi: 10.4155/fmc.11.1. [DOI] [PubMed] [Google Scholar]
- da Silva Antonio A., Wiedemann L.S.M., Veiga-Junior V.F. Natural products' role against COVID-19. RSC Adv. 2020;10:23379–23393. doi: 10.1039/d0ra03774e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebada S.S., Al-Jawabri N.A., Youssef F.S., El-Kashef D.H., Knedel T.-O., Albohy A., Korinek M., Hwang T.-L., Chen B.-H., Lin G.-H. Anti-inflammatory, antiallergic and COVID-19 protease inhibitory activities of phytochemicals from the Jordanian hawksbeard: identification, structure–activity relationships, molecular modeling and impact on its folk medicinal uses. RSC Adv. 2020;10:38128–38141. doi: 10.1039/d0ra04876c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fakhar Z., Faramarzi B., Pacifico S., Faramarzi S.J., Dynamics Anthocyanin derivatives as potent inhibitors of SARS-CoV-2 main protease: an in-silico perspective of therapeutic targets against COVID-19 pandemic. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1801510. [DOI] [PubMed] [Google Scholar]
- Gahlawat A., Kumar N., Kumar R., Sandhu H., Singh I.P., Singh S., Sjostedt A., Garg P. Structure-based virtual screening to discover potential lead molecules for the SARS-CoV-2 main protease. J. Chem. Inf. Model. 2020;60:5781–5793. doi: 10.1021/acs.jcim.0c00546. [DOI] [PubMed] [Google Scholar]
- Gentile D., Patamia V., Scala A., Sciortino M.T., Piperno A., Rescifina A. Putative inhibitors of SARS-CoV-2 main protease from A library of marine natural products: a virtual screening and molecular modeling study. Mar. Drugs. 2020;18:225. doi: 10.3390/md18040225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghosh A.K., Gemma S. John Wiley & Sons, WILEY‐VCH Verlag GmbH; 2014. Structure-based Design of Drugs and Other Bioactive Molecules: Tools and Strategies. [Google Scholar]
- Ghosh K., Amin S.A., Gayen S., Jha T. Chemical-informatics approach to COVID-19 drug discovery: exploration of important fragments and data mining based prediction of some hits from natural origins as main protease (Mpro) inhibitors. J. Mol. Struct. 2021;1224 doi: 10.1016/j.molstruc.2020.129026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleeson M.P. Generation of a set of simple, interpretable ADMET rules of thumb. J. Med. Chem. 2008;51:817–834. doi: 10.1021/jm701122q. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Medina M., Medina-Franco J.L. Platform for unified molecular analysis: PUMA. J. Chem. Inf. Model. 2017;57:1735–1740. doi: 10.1021/acs.jcim.7b00253. [DOI] [PubMed] [Google Scholar]
- Gopinath K., Jokinen E.M., Kurkinen S.T., Pentikainen O.T. Screening of natural products targeting SARS-CoV-2-ACE2 receptor interface - a MixMD based HTVS pipeline. Front. Chem. 2020;8:589769. doi: 10.3389/fchem.2020.589769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gyebi G.A., Ogunro O.B., Adegunloye A.P., Ogunyemi O.M., Afolabi S.O. Potential inhibitors of coronavirus 3-chymotrypsin-like protease (3CL(pro)): an in silico screening of alkaloids and terpenoids from African medicinal plants. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1764868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang J., Tao G., Liu J., Cai J., Huang Z., Chen J.X. Current prevention of COVID-19: natural products and herbal medicine. Front. Pharmacol. 2020;11:588508. doi: 10.3389/fphar.2020.588508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M.A.A., Abdeljawaad K.A.A., Abdelrahman A.H.M., Hegazy M.F. Natural-like products as potential SARS-CoV-2 M(pro) inhibitors: in-silico drug discovery. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1790037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim M.A.A., Abdelrahman A.H.M., Hussien T.A., Badr E.A.A., Mohamed T.A., El-Seedi H.R., Pare P.W., Efferth T., Hegazy M.F. In silico drug discovery of major metabolites from spices as SARS-CoV-2 main protease inhibitors. Comput. Biol. Med. 2020;126:104046. doi: 10.1016/j.compbiomed.2020.104046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iheagwam F.N., Rotimi S.O. Computer-aided analysis of multiple SARS-CoV-2 therapeutic targets: identification of potent molecules from african medicinal plants. Sci. Tech. Rep. 2020;2020:1878410. doi: 10.1155/2020/1878410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin Z., Du X., Xu Y., Deng Y., Liu M., Zhao Y., Zhang B., Li X., Zhang L., Peng C. Structure of M pro from SARS-CoV-2 and discovery of its inhibitors. Nature. 2020;582:289–293. doi: 10.1038/s41586-020-2223-y. [DOI] [PubMed] [Google Scholar]
- Joshi R.S., Jagdale S.S., Bansode S.B., Shankar S.S., Tellis M.B., Pandya V.K., Chugh A., Giri A.P., Kulkarni M.J. Discovery of potential multi-target-directed ligands by targeting host-specific SARS-CoV-2 structurally conserved main protease. J. Mol. Graph. Model. 2020:1–16. doi: 10.1080/07391102.2020.1760137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanjanasirirat P., Suksatu A., Manopwisedjaroen S., Munyoo B., Tuchinda P., Jearawuttanakul K., Seemakhan S., Charoensutthivarakul S., Wongtrakoongate P., Rangkasenee N. High-content screening of Thai medicinal plants reveals Boesenbergia rotunda extract and its component Panduratin A as anti-SARS-CoV-2 agents. Sci. Rep. 2020;10:1–12. doi: 10.1038/s41598-020-77003-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katiyar C., Gupta A., Kanjilal S., Katiyar S. Drug discovery from plant sources: an integrated approach. Ayu. 2012;33:10. doi: 10.4103/0974-8520.100295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalifa I., Nawaz A., Sobhy R., Althwab S.A., Barakat H. Polyacylated anthocyanins constructively network with catalytic dyad residues of 3CL(pro) of 2019-nCoV than monomeric anthocyanins: a structural-relationship activity study with 10 anthocyanins using in-silico approaches. J. Mol. Graph. Model. 2020;100:107690. doi: 10.1016/j.jmgm.2020.107690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.T., Ali A., Wang Q., Irfan M., Khan A., Zeb M.T., Zhang Y.J., Chinnasamy S., Wei D.Q. Marine natural compounds as potents inhibitors against the main protease of SARS-CoV-2-a molecular dynamic study. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1769733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koehn F.E., Carter G.T. The evolving role of natural products in drug discovery. Nat. Rev. Drug Discov. 2005;4:206–220. doi: 10.1038/nrd1657. [DOI] [PubMed] [Google Scholar]
- Kumar B.K., Faheem, Sekhar K., Ojha R., Prajapati V.K., Pai A., Murugesan S. Pharmacophore based virtual screening, molecular docking, molecular dynamics and MM-GBSA approach for identification of prospective SARS-CoV-2 inhibitor from natural product databases. J. Biomol. Struct. Dyn. 2020:1–24. doi: 10.1080/07391102.2020.1824814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li R., Hou Y., Huang J. Lianhuaqingwen exerts anti-viral and anti-inflammatory activity against novel coronavirus (SARS-CoV-2) Pharmacol. Res. 2020;156 doi: 10.1016/j.phrs.2020.104761. 156:104761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majumder R., Mandal M. Screening of plant-based natural compounds as a potential COVID-19 main protease inhibitor: an in silico docking and molecular dynamics simulation approach. J. Biomol. Struct. Dyn. 2020:1–16. doi: 10.1080/07391102.2020.1817787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsson P., Kihlberg J. How big is too big for cell permeability? J. Med. Chem. 2017;60:1662–1664. doi: 10.1021/acs.jmedchem.7b00237. [DOI] [PubMed] [Google Scholar]
- Mazzini S., Musso L., Dallavalle S., Artali R. Putative SARS-CoV-2 M(pro) inhibitors from an in-house library of natural and nature-inspired products: a virtual screening and molecular docking study. Molecules. 2020;25:3745. doi: 10.3390/molecules25163745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Monajjemi M., Esmkhani R., Mollaamin F., Shahriari S. Prediction of proteins associated with COVID-19 based ligand designing and molecular modeling. Sci. Rep. 2020;125:907–926. [Google Scholar]
- Naik B., Gupta N., Ojha R., Singh S., Prajapati V.K., Prusty D. High throughput virtual screening reveals SARS-CoV-2 multi-target binding natural compounds to lead instant therapy for COVID-19 treatment. Int. J. Biol. Macromol. 2020;160:1–17. doi: 10.1016/j.ijbiomac.2020.05.184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Narkhede R.R., Pise A.V., Cheke R.S., Shinde S.D. Recognition of natural products as potential inhibitors of COVID-19 main protease (Mpro): in-silico evidences. Nat. Prod. Bioprospect. 2020;10:297–306. doi: 10.1007/s13659-020-00253-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naveja J.J., Medina-Franco J.L. Finding constellations in chemical space through core analysis. Front. Chem. 2019;7:510. doi: 10.3389/fchem.2019.00510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olubiyi O.O., Olagunju M., Keutmann M., Loschwitz J., Strodel B. High throughput virtual screening to discover inhibitors of the main protease of the coronavirus SARS-CoV-2. Molecules. 2020;25 doi: 10.3390/molecules25143193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ospanov M., Leon F., Jenis J., Khan I.A., Ibrahim M.A. Challenges and future directions of potential natural products leads against 2019-nCoV outbreak. Curr. Plant Biol. 2020;24:100180. doi: 10.1016/j.cpb.2020.100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Owis A.I., El-Hawary M.S., El Amir D., Aly O.M., Abdelmohsen U.R., Kamel M.S. Molecular docking reveals the potential of Salvadora persica flavonoids to inhibit COVID-19 virus main protease. RSC Adv. 2020;10:19570–19575. doi: 10.1039/d0ra03582c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey P., Rane J.S., Chatterjee A., Kumar A., Khan R., Prakash A., Ray S. Targeting SARS-CoV-2 spike protein of COVID-19 with naturally occurring phytochemicals: an in silico study for drug development. J. Biomol. Struct. Dyn. 2020:1–11. doi: 10.1080/07391102.2020.1796811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patterson D.E., Cramer R.D., Ferguson A.M., Clark R.D., Weinberger L.E. Neighborhood behavior: a useful concept for validation of “molecular diversity” descriptors. J. Med. Chem. 1996;39:3049–3059. doi: 10.1021/jm960290n. [DOI] [PubMed] [Google Scholar]
- Poduri R., Joshi G., Jagadeesh G. Drugs targeting various stages of the SARS-CoV-2 life cycle: exploring promising drugs for the treatment of Covid-19. Cell. Signal. 2020;74:109721. doi: 10.1016/j.cellsig.2020.109721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rahman N., Basharat Z., Yousuf M., Castaldo G., Rastrelli L., Khan H. Virtual screening of natural products against type II transmembrane serine protease (TMPRSS2), the priming agent of coronavirus 2 (SARS-CoV-2) Molecules. 2020;25:2271. doi: 10.3390/molecules25102271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rakib A., Paul A., Chy M.N.U., Sami S.A., Baral S.K., Majumder M., Tareq A.M., Amin M.N., Shahriar A., Uddin M.Z., Dutta M., Tallei T.E., Emran T.B., Simal-Gandara J. Biochemical and computational approach of selected phytocompounds from tinospora crispa in the management of COVID-19. Molecules. 2020;25:3936. doi: 10.3390/molecules25173936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogers D., Hahn M. Extended-connectivity fingerprints. J. Chem. Inf. Model. 2010;50:742–754. doi: 10.1021/ci100050t. [DOI] [PubMed] [Google Scholar]
- Saeed M., Saeed A., Alam M.J., Alreshidi M. Identification of persuasive antiviral natural compounds for COVID-19 by targeting endoribonuclease NSP15: a structural-bioinformatics approach. Molecules. 2020;25:5657. doi: 10.3390/molecules25235657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sahin S., Calapoğlu F., Ozmen I. Didemnins inhibit COVID-19 main protease (Mpro) Biointerface Res. Appl. Chem. 2021;11:8204–8209. [Google Scholar]
- Sayed A.M., Alhadrami H.A., El-Gendy A.O., Shamikh Y.I., Belbahri L., Hassan H.M., Abdelmohsen U.R., Rateb M.E. Microbial natural products as potential inhibitors of SARS-CoV-2 main protease (Mpro) Microorganisms. 2020;8:970. doi: 10.3390/microorganisms8070970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selvaraj C., Dinesh D.C., Panwar U., Abhirami R., Boura E., Singh S.K. Structure-based virtual screening and molecular dynamics simulation of SARS-CoV-2 Guanine-N7 methyltransferase (nsp14) for identifying antiviral inhibitors against COVID-19. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1778535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sepay N., Sekar A., Halder U.C., Alarifi A., Afzal M. Anti-COVID-19 terpenoid from marine sources: a docking, ADMET and molecular dynamics study. J. Mol. Struct. 2020:129433. doi: 10.1016/j.molstruc.2020.129433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma P., Shanavas A. Natural derivatives with dual binding potential against SARS-CoV-2 main protease and human ACE2 possess low oral bioavailability: a brief computational analysis. J. Biomol. Struct. Dyn. 2020:1–12. doi: 10.1080/07391102.2020.1794970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Su H.X., Yao S., Zhao W.F., Li M.J., Liu J., Shang W.J., Xie H., Ke C.Q., Hu H.C., Gao M.N., Yu K.Q., Liu H., Shen J.S., Tang W., Zhang L.K., Xiao G.F., Ni L., Wang D.W., Zuo J.P., Jiang H.L., Bai F., Wu Y., Ye Y., Xu Y.C. Anti-SARS-CoV-2 activities in vitro of Shuanghuanglian preparations and bioactive ingredients. Acta Pharmacol. Sin. 2020;41:1167–1177. doi: 10.1038/s41401-020-0483-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Subbaiyan A., Ravichandran K., Singh S.V., Sankar M., Thomas P., Dhama K., Malik Y.S., Singh R.K., Chaudhuri P. In silico molecular docking analysis targeting SARS-CoV-2 spike protein and selected herbal constituents. J. Pure Appl. Microbiol. 2020;14:989–998. doi: 10.22207/JPAM.14.SPL1.37. [DOI] [Google Scholar]
- Tahir Ul Qamar M., Alqahtani S.M., Alamri M.A., Chen L.L. Structural basis of SARS-CoV-2 3CL(pro) and anti-COVID-19 drug discovery from medicinal plants. J. Pharm. Anal. 2020;10:313–319. doi: 10.1016/j.jpha.2020.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thakur S., Sarkar B., Ansari A.J., Khandelwal A., Arya A., Poduri R., Joshi G. Exploring the magic bullets to identify Achilles' heel in SARS-CoV-2: delving deeper into the sea of possible therapeutic options in Covid-19 disease: an update. Food Chem. Toxicol. 2020;147:111887. doi: 10.1016/j.fct.2020.111887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich S., Nitsche C. The SARS-CoV-2 main protease as drug target. Bioorg. Med. Chem. Lett. 2020;30:127377. doi: 10.1016/j.bmcl.2020.127377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veber D.F., Johnson S.R., Cheng H.-Y., Smith B.R., Ward K.W., Kopple K.D.J. Molecular properties that influence the oral bioavailability of drug candidates. 2002;45:2615–2623. doi: 10.1021/jm020017n. [DOI] [PubMed] [Google Scholar]
- Vivek-Ananth R.P., Rana A., Rajan N., Biswal H.S., Samal A. In silico identification of potential natural product inhibitors of human proteases key to SARS-CoV-2 infection. Molecules. 2020;25:3822. doi: 10.3390/molecules25173822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., Li W., Hui H., Tiwari S.K., Zhang Q., Croker B.A., Rawlings S., Smith D., Carlin A.F., Rana T.M. Cholesterol 25-Hydroxylase inhibits SARS-CoV-2 and other coronaviruses by depleting membrane cholesterol. EMBO J. 2020;39 doi: 10.15252/embj.2020106057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi F., Li L., Xu L.J., Meng H., Dong Y.M., Liu H.B., Xiao P.G. In silico approach in reveal traditional medicine plants pharmacological material basis. Chin. Med. 2018;13:33. doi: 10.1186/s13020-018-0190-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yongye A.B., Waddell J., Medina-Franco J.L. Molecular scaffold analysis of natural products databases in the public domain. Chem. Biol. Drug Des. 2012;80:717–724. doi: 10.1111/cbdd.12011. [DOI] [PubMed] [Google Scholar]
- Zhong H.A., Mashinson V., Woolman T.A., Zha M.J.C. vol. 13. 2013. pp. 1290–1307. (Understanding the Molecular Properties and Metabolism of Top Prescribed Drugs). [DOI] [PubMed] [Google Scholar]
- Zhu Z., Lian X., Su X., Wu W., Marraro G.A., Zeng Y. From SARS and MERS to COVID-19: a brief summary and comparison of severe acute respiratory infections caused by three highly pathogenic human coronaviruses. Respir. Res. 2020;21:224. doi: 10.1186/s12931-020-01479-w. [DOI] [PMC free article] [PubMed] [Google Scholar]