Abstract
Bladder cancer (BC) is one of the most commonly diagnosed cancers globally. Recently, circular RNAs (circRNAs) have been revealed to participate in BC progression with diverse mechanisms. However, mechanisms of circ_100984 in BC have not been determined. Here, we found that circ_100984 and YBX‐1 were high presented, while miR‐432‐3p was low presented in BC. Silencing of circ_100984 and YBX‐1 repressed BC tumor growth, migration, and invasion in vitro and in vivo. Mechanistically, we revealed that circ_100984 served as a competing endogenous RNA that sponged miR‐432‐3p to indirectly regulate YBX‐1 and epithelial‐mesenchymal transition (EMT)‐related molecules. Moreover, we confirmed that YBX‐1 or c‐Jun acted as a transcription regulatory factor for β‐catenin or YBX‐1, respectively, in BC cells. Knockdown of YBX‐1 inhibited the expression of β‐catenin and c‐Jun, whereas downregulated c‐Jun inversely repressed the expression of YBX‐1 and β‐catenin. Our results suggested that circ_100984‐miR‐432‐3p axis regulated c‐Jun/YBX‐1/β‐catenin feedback loop promotes BC progression, providing a potential therapeutic axis for BC progression.
Keywords: β‐catenin, bladder cancer, circ_100984, c‐Jun, epithelial‐mesenchymal transition, miR‐432‐3p, YBX‐1
circ_100984 sponges miR‐432‐3p to form c‐Jun/YBX‐1/β‐catenin feedback loop and promotes bladder cancer progression, providing a potential therapeutic axis for BC progression.
1. INTRODUCTION
Bladder cancer (BC) is the most common tumor that occurs in the urinary system, ranking as the fourth most frequent tumor in Europe and the United States. 1 , 2 According to an estimation from the American Cancer Society, over 81 400 new BC cases and 17 980 deaths will occur in 2020 in the United States. 3 At this time, surgical resection has been adopted as the mainstay therapeutic option for BC patients, and chemotherapy and radiation therapy are used as effective adjunctive therapies. 4 However, due to its high recurrence rate and metastasis, as well as the deficiency in effective early screen biomarkers, the 5‐y survival rate of BC patients remains dismal. 5 Understanding the molecular mechanisms of BC tumorigenesis may contribute to the development of promising diagnosis biomarkers and effective therapeutic targets for BC. Y box binding protein‐1 (YBX‐1) is a key subtype of the cold‐shock protein family that has been proposed to bind to DNA and RNA to manipulate DNA transcription and repair, as well as mRNA translation and packaging. 6 , 7 Due to its important regulatory activity in gene expression, YBX‐1 dysregulation is known to be closely linked to unfavorable clinical outcomes. 8 Dysregulation of YBX‐1 has been found recently in BC tissues, and it has been reported to be a potential prognostic factor of BC. 9 It was also found to facilitate BC tumor progression by elevating glycolysis. 10 Therefore, the study of YBX‐1 regulation in BC might be a valuable research direction.
Circular RNAs (circRNAs) are known as important members of the non‐coding RNA family and contains a unique close loop structure. 11 It is presented differentially in numerous cancerous tissues, and is closely associated with the status and prognosis of tumors. 12 , 13 For their mechanisms, circRNAs have been proposed to regulate tumor occurrence by acting as microRNAs (miRNAs) sponges and preventing miRNA‐induced mRNA cleavage of target genes. 14 , 15 In a circRNA microarray analysis in 2018, circ_100984 was found to be upregulated in BC tissues compared with matched normal tisues, 16 however its role and mechanism in BC remain undetermined.
The Wnt/β‐catenin cascade participates in numerous cellular biological events including cell proliferation, metastasis, and apoptosis. 17 Inhibition of the Wnt/β‐catenin cascade has been found to repress BC tumor cell proliferation and promote BC cell apoptosis. 18 , 19 YBX‐1 was reported to promote the Wnt/β‐catenin cascade by upregulation of β‐catenin in hepatocellular carcinoma. 20 Knockdown of YBX‐1 could repress triple‐negative breast cancer cell invasiveness by regulating MMP1 and β‐catenin expression. 21 Moreover, 1 previous study indicated that β‐catenin positively regulated the expression of c‐Jun in breast cancer. 22 These findings implied that c‐Jun/YBX‐1/β‐catenin might function as an axis in cancer progression, however it is still unknown if this axis exists and plays a role during BC tumorigenesis.
Therefore, in this study we aimed to investigate whether circ_100984 could regulate BC tumor progression through the Jun/YBX‐1/β‐catenin axis.
2. MATERIALS AND METHODS
2.1. BC tissue samples and cell lines
Twenty pairs of BC and the corresponding non‐cancer tissues were collected from BC patients (12 men, 8 women, aged 42–65 years, mean 52.43 ± 5.18) who were treated in Xiangya Hospital of Central South University from January 2017 to January 2019; tissues were maintained at −80°C until use. There were 15 cases of superficial BC and 2 cases of invasive BC. Pathological grades were: 10 cases of Grade G1, 6 cases of Grade G2, and 4 cases of Grade G3. Clinical stages were: 5 cases of Ta, 4 cases of T1, 5 cases of T2, 3 cases of T3, and 3 cases of T4. There were 3 cases of lymph node metastasis. Subjects were firstly diagnosed as BC patients using pathological diagnosis and cytological examination. Patients all received surgical resection treatment without any chemotherapy or radiotherapy and their expected survival time was greater than 3 mo. Subjects with following conditions were excluded: (a) with surgical contraindications; (b) with other tumors; (c) complicated with infectious diseases; (d) combined with autoimmune diseases; (e) serious functional insufficiency of the heart, liver, kidney and other important organs; and (f) suffering from mental illness or disorder of consciousness. This study was approved by the Ethics Committee of Xiangya Hospital of Central South University and informed consent documentation was provided to each patient. BC cell lines (HT‐1376, HTB9, 253J, BT‐B, Biu‐87 and 5637) were purchased from ATCC and a normal human urothelial cell line (SV‐HUC‐1) was supplied by Procell Life Science & Technology Co., Ltd. Cells were kept in DMEM containing 10% fetal calf serum (FBS, Sigma) and 1% ampicillin/streptomycin, and maintained in an incubator containing 95% O2 and 5% CO2 at 37°C.
2.2. Quantitative real‐time PCR (qRT‐PCR) analysis
Total RNAs of BC tissues and cells were separated after TRIzol (Invitrogen) incubation. After examining its quality and concentration, RNAs were transcribed into cDNA using the Gibco BRL kit (Life Technologies), and qRT‐PCR was completed following standard procedures for the SYBR Green PCR kit (Takara) on an ABI Prism system (PE Applied Biosystems). Sequences of primers are shown in Table 1. The relative levels of RNA normalized to β‐actin or U6 snRNA were calculated using the 2−ΔΔCt method.
TABLE 1.
Oligonucleotide primer sets for qRT‐PCR
| Name | Sequence (5′–3′) | Length |
|---|---|---|
| circ_100984 F | GAGCCACCACATTGGACTTC | 20 |
| circ_100984 R | CAGAGGTGCTCCTCAATTCC | 20 |
| YBX‐1 F | GATAAATTTAAACCTGAAAA | 20 |
| YBX‐1 R | ATCTTGTTTCTATCTTCCAA | 20 |
| miR‐432‐3p F | GCGGCGGCTGGATGGCTCCTCC | 22 |
| miR‐432‐3p R | CAGTGCGTGTCGTGGAGT | 18 |
| U6 F | GCTTCGGCAGCACATATACTAAAA | 24 |
| U6 R | CGCTTCACGAATTTGCGTGTCAT | 23 |
| β‐actin F | ACACTGTGCCCATCTACG | 18 |
| β‐actin R | TGTCACGCACGATTTCC | 17 |
Abbreviations: F, forward; R, reverse; YBX‐1, Y box binding protein 1.
2.3. Western blot analysis
Total proteins of BC tissues and cells were extracted by high‐speed centrifugation at 10 000 g/min for 10 min after lysis in RIPA buffer (Beyotime). Proteins (50 μg) were loaded into and isolated by 10% SDS‐PAGE. Isolated proteins were transferred onto nitrocellulose membrane and incubated with 5% low‐fat milk for non‐specific site blocking. Afterwards, membranes were probed with primary antibodies against YBX‐1 (Rabbit, 1:1000, ab12148, Abcam), Slug‐1 (Rabbit, 1:1000, ab27568, Abcam), Vimentin (Rabbit, 1:3000, ab137321, Abcam), N‐cadherin (Rabbit, 1:1000, ab18203, Abcam), E‐cadherin (Rabbit, 1:10 000, ab40772, Abcam), β‐catenin (Rabbit, 1:4000, ab6302, Abcam), c‐Jun (Rabbit, 1:1000, ab131497, Abcam), and β‐actin (Rabbit, 1:5000, ab179467, Abcam). After overnight incubation, membranes were incubated for 2 h with the indicated secondary antibodies. Bands were visualized using enhanced chemiluminescence reagent (EMD, Millipore).
2.4. RNA transfection
Biu‐87 and HTB9 cells were infected with lentivirus containing specific shRNAs against circ_100984 (sh‐circ_100984‐1, sh‐circ_100984‐2), YBX‐1 (sh‐YBX‐1, sh‐YBX‐2), and β‐catenin (sh‐β‐catenin‐1, sh‐β‐catenin‐2), or circ_100984, and c‐Jun gene. miR‐432‐3p mimics, as well as their negative control, were transduced into Biu‐87 and HTB9 cells using Lipofectamine 3000 reagent (Thermo Fisher).
2.5. Cell proliferation viability assessment
Proliferation viability was estimated by Cell Counting Kit‐8 (CCK‐8) and colony formation experiments. For CCK‐8, treated BC cells (2 × 104 cells/well) were added to 96‐well plates containing DMEM. Next, CCK‐8 solution (10 μL) was added into each well and wells were incubated at 37°C for 2 h. Absorbance in each well was determined at 450 nm. For colony formation, 1000 treated BC cells were harvested and added into 6‐well plates. After incubation in a cell incubator for 2 wk, visualized colonies were fixed in methanol and stained with crystal violet. The numbers of colony were counted manually.
2.6. Cell migration estimation
Cell migratory ability was estimated using a wound‐healing assay. Treated BC cells were collected and seeded into 35‐mm Petri dishes and then cultured at 37°C until 100% confluency. A scratch was made by a pipette tip in the BC cell surface and images were taken after 0 and 24 h after scratching.
2.7. Cell invasion estimation
Cell invasive ability was estimated by transwell chambers with Matrigel (8 μm pore size, Corning Incorporated). Treated BC cells (2 × 105) were collected and re‐suspended in 500 μL serum‐free DMEM, which was then added into the upper chamber. The lower chamber was filled with 500 µL DMEM containing 10% FBS. After 24 h of incubation, cells that remained in the upper chamber were discarded and those on the undersurface were fixed and stained with 5% crystal violet. Invasive cell number was counted under a microscope.
2.8. Immunofluorescence analysis
Treated BC cells (1 × 104) were collected and seeded into 96‐well plates, and then cells were fixed with 4% paraformaldehyde overnight after washing with PBS 3 times. Cells permeabilization was carried out in Triton X‐100 at 4°C for 20 min, followed by blocking with 5% bovine serum albumin for 1 h. Next, cells were probed with primary antibodies against Vimentin (Rabbit, 1:1000, ab137321, Abcam) and E‐cadherin (Rabbit, 1:500, ab40772, Abcam), followed by secondary antibodies incubation for 2 h. DAPI was applied to stain nuclei. Imaging was performed using a FV1000 confocal microscope and relative fluorescence intensity was analyzed using Image J software.
2.9. Mouse xenografts
Nude mice (8 wk old, male) were supplied by the Human SJA Laboratory Animal Co., Ltd and kept in animal rooms under a 12 h/12 h, day/night cycle and with free access to food and water. Animal manipulation was approved by the Ethics Committee of Central South University. The subcutaneous tumor animal model was established by injecting 2 × 104 treated Biu‐87 and HTB9 cells into the back of the nude mice. These animals were sacrificed 35 d after inoculation and tumors were excised for further study. For in vivo metastasis evaluation, 2 × 104 treated Biu‐87 cells were inoculated using the tail vein into animals. These nude mice were sacrificed after 35 d of inoculation, and liver and lung tissues were collected for metastatic nodules analysis. After fixation in 4% paraformaldehyde, liver and lung tissues were sliced into 5‐μm sections followed by hematoxylin and eosin (H&E) staining.
2.10. Dual‐luciferase reporter assay
To validate the interplay between YBX‐1 and miR‐432‐3p, recombinant luciferase reporter plasmids were established by inserting wild type (WT) or mutant (MUT) predicted miR‐432‐3p sequences of YBX‐1 into pGL3 vector. Recombinant plasmids were named YBX‐1‐WT or YBX‐1‐MUT. Next, miR‐432‐3p mimics or NC mimics and YBX‐1‐WT or YBX‐1‐MUT were co‐transfected into Biu‐87 and HTB9 cells, followed by detection of luciferase activity of Biu‐87 and HTB9 cells using the Dual‐Luciferase Reporter Assay System (Promega). Validation of the interaction between miR‐432‐3p and circ_100984, as well as between YBX‐1 and c‐Jun, was again performed as above.
2.11. RNA immunoprecipitation (RIP)
Biu‐87 and HTB9 cells were co‐transfected with MS2bs‐Rluc, MS2bs‐circ_100984 and MS2bs‐circ_100984‐MUT, followed by performance of the RIP assay after 48 h of co‐transfection, using the Magna RIP RNA‐Binding Protein Immunoprecipitation Kit (Millipore) in accordance with the manufacturer’s instructions.
2.12. ChIP assay
ChIP analysis was conducted using a ChIP assay kit (Thermo Scientific) following the instructions supplied by the manufacturer. DNA fragments were obtained after isolation and micrococcal nuclease digestion of chromatin. Anti‐YBX‐1/c‐Jun or IgG was added into the reaction system for immunoprecipitation. Subsequently, qRT‐PCR was adopted to detect DNA enrichment after elution and purification.
2.13. Electrophoretic mobility shift assay (EMSA)
Binding activities of YBX‐1 or c‐Jun on the promoter area of β‐catenin or YBX‐1 were assessed using an EMSA kit (Thermo Scientific) in a reaction system including nuclear extracts and specific biotin probes in accordance with manufacturer’s procedures.
2.14. Statistical analysis
Data are shown as mean ± standard deviation (SD) and were analyzed using Student t test or one‐way ANOVA and GraphPad Prism software (Version 7.0, USA). A P‐value <.05 was considered significant.
3. RESULTS
3.1. circ_100984, YBX‐1, and miR‐432‐3p were dysregulated in BC
To determine whether circ_100984, YBX‐1, and miR‐432‐3p played a role during BC tumorigenesis, we firstly examined by qRT‐PCR their expression levels in 20 pairs of BC and adjacent non‐cancer tissues. The results illustrated that circ_100984 and YBX‐1 were dramatically increased, whereas miR‐432‐3p was obviously decreased in BC tissues, compared with that in non‐cancer control tissues (Figure 1A). Consistently, we found that circ_100984 and YBX‐1 were dramatically increased, whereas miR‐432‐3p was obviously decreased in BC cell lines HT‐1376, HTB9, 253J, BT‐B, Biu‐87 and 5637 compared with the SV‐HUC‐1 cell line (Figure 1B). In addition, western blot analysis showed that YBX‐1 protein expression was significantly upregulated in BC cell lines HT‐1376, HTB9, 253J, BT‐B, Biu‐87 and 5637 compared with the SV‐HUC‐1 cell line (Figure 1C). These results implied that circ_100984, YBX‐1, and miR‐432‐3p might exert a vital effect during BC tumorigenesis.
FIGURE 1.
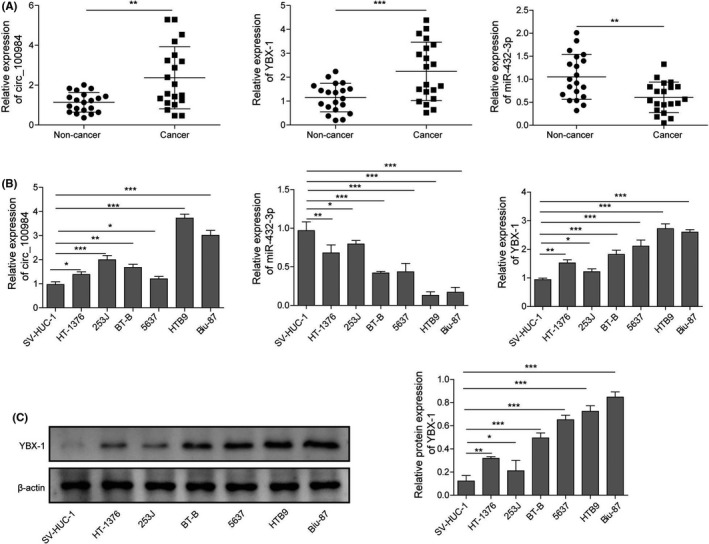
circ_100984, YBX‐1, and miR‐432‐3p were dysregulated in BC. A, Relative expression levels of circ_100984, YBX‐1 mRNA, and miR‐432‐3p of BC tissues were determined by qRT‐PCR, matched non‐cancer tissues were applied as the control group, n = 20. B, Relative expression levels of circ_100984, YBX‐1 mRNA and miR‐432‐3p of 6 BC cell lines (HT‐1376, HTB9, 253J, BT‐B, Biu‐87 and 5637) and 1 normal human urothelial cell line (SV‐HUC‐1). C, Protein expression level of YBX‐1 was evaluated by western blot in BC cell lines, SV‐HUC‐1 was used as the control group. *P < .05; **P < .01; ***P < .001
3.2. circ_100984 silencing repressed BC cell proliferation, migration, invasion, and EMT in vitro and in vivo
To elucidate the biological functions of circ_100984 in BC, we introduced circ_100984 shRNAs (sh‐circ_100984‐1, sh‐circ_100984‐2) into Biu‐87 and HTB9 cells and observed a significantly downregulation in circ_100984 expression in both Biu‐87 and HTB9 cells compared with the matched control cells (Figure 2A). In vitro functional assays revealed that knockdown of circ_100984 significantly repressed cell viability (Figure 2B), proliferation (Figure 2C), migration (Figure 2D) and invasion (Figure 2E) activities of Biu‐87 and HTB9 cells. Moreover, Vimentin downregulation and E‐cadherin upregulation were observed in circ_100984 silenced Biu‐87 and HTB9 cells (Figure 2F), indicating that circ_100984 knockdown repressed the EMT process of BC cells. The in vivo oncogenic effect of circ_100984 was then evaluated in mouse models. Tumors derived from circ_100984 silenced Biu‐87 and HTB9 cells were obviously smaller compared with those from control cells (Figure S1A). Tumor weight and volume of sh‐circ_100984‐1 and sh‐circ_100984‐2 groups were significantly reduced compared with that of sh‐NC group (Figure S1B,C). Moreover, the pulmonary and hepatic metastasis models indicated that, in contrast with metastatic nodules in the sh‐NC group, less metastatic nodules were found in mice injected with circ_100984 shRNAs (Figure S1D,E). Histopathologic analysis showed that circ_100984 knockdown obviously alleviated the pathological lesion of lung and liver (Figure S1F). In addition, we found that protein expression levels of YBX‐1, N‐cadherin, Vimentin, and Slug‐1 were significantly downregulated while E‐cadherin was upregulated in the tumors derived from circ_100984 silenced Biu‐87 and HTB9 cells (Figure 2G). These findings suggested that circ_100984 silencing repressed BC cell proliferation, migration, invasion, and EMT in vitro and in vivo.
FIGURE 2.
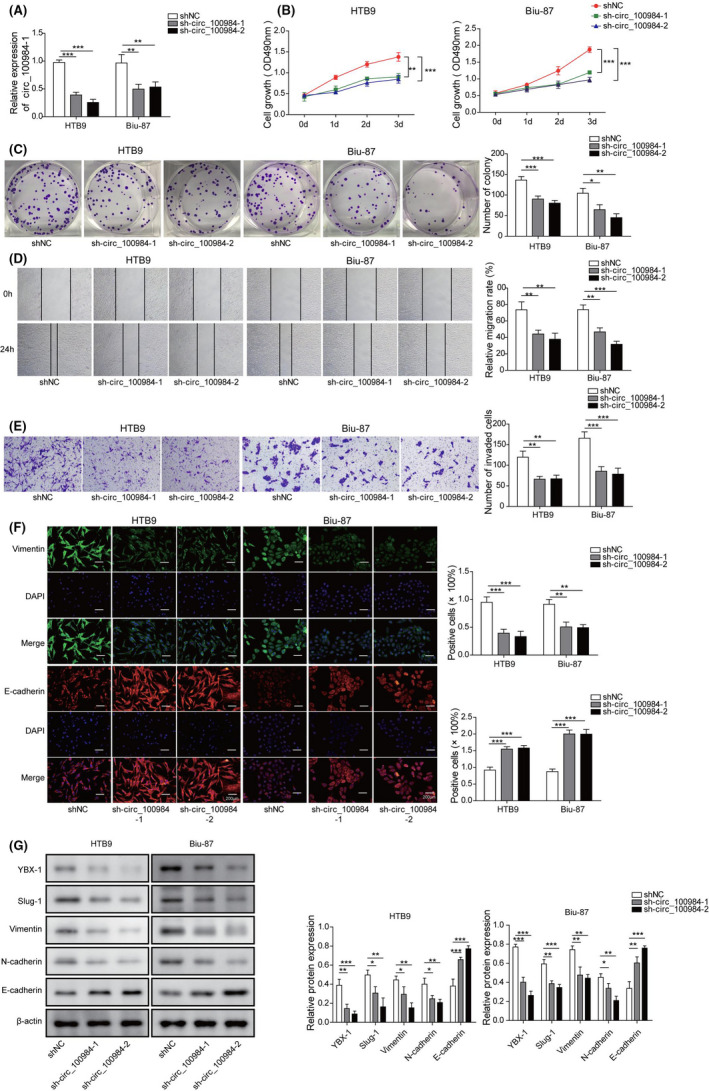
circ_100984 silencing repressed BC cell proliferation, migration, invasion, and EMT in vitro and in vivo. A, Knockdown efficiencies of sh‐circ_100984‐1 and sh‐circ_100984‐2 were examined by qRT‐PCR in Biu‐87 and HTB9 cells. B, After 0, 1, 2 and 3 d of transfection with sh‐NC, sh‐circ_100984‐1 and sh‐circ_100984‐2, Biu‐87, and HTB9 cells were subjected to cell viability analysis using CCK‐8 assay. C, Colony formation assay was performed to assess the proliferation of Biu‐87 and HTB9 cells after transfection with sh‐NC, sh‐circ_100984‐1 and sh‐circ_100984‐2. Effects of circ_100984 knockdown on cell (D) migration and (E) invasion were evaluated by wound‐healing and transwell assay, respectively. F, Immunofluorescence comparing circ_100984 silenced Biu‐87 and HTB9 cells with their respective cells are shown for relative expression of Vimentin and E‐cadherin. G, Protein expression levels of YBX‐1, Slug‐1, Vimentin, N‐cadherin, and E‐cadherin in the tumors of nude mice following sh‐NC, sh‐circ_100984‐1 and sh‐circ_100984‐2 transfection were examined by western blot. *P < .05; **P < .01; ***P < .001
3.3. YBX‐1 silencing repressed BC cell proliferation, migration, invasion, and EMT in vitro, and in vivo
The same strategy was applied to study the effects of YBX‐1 knockdown on BC cell viability, proliferation, migration, invasion, and EMT in vitro and in vivo. Transfection of YBX‐1 shRNAs (sh‐YBX‐1‐1 and sh‐YBX‐1‐2) caused a significant YBX‐1 downregulation in Biu‐87 and HTB9 cells compared with sh‐NC transfection (Figure 3A). In vitro functional analysis demonstrated that YBX‐1 silencing resulted in a significant inhibition effect on cell viability (Figure 3B), proliferation (Figure 3C), migration (Figure 3D) and invasion (Figure 3E) of Biu‐87 and HTB9 cells. YBX‐1 silencing also caused a significant Vimentin downregulation and E‐cadherin upregulation in Biu‐87 and HTB9 cells (Figure 3F). An in vivo xenograft tumor assay was then adopted to further evaluate the effects of YBX‐1 silencing on BC tumor growth, metastasis, and EMT. Tumors derived from YBX‐1 silenced Biu‐87 and HTB9 cells were obviously smaller compared with those from control cells (Figure S2A). Tumor weight and volume of YBX‐1 silenced groups were significantly reduced compared with that of sh‐NC group (Figure S2B,C). Moreover, the pulmonary and hepatic metastasis models indicated that in contrast with metastatic nodules in the sh‐NC group, less metastatic nodules were found in mice injected with YBX‐1 shRNAs (Figure S2D,E). Histopathologic analysis showed that YBX‐1 knockdown obviously alleviated the pathological lesions of lung and liver (Figure S2F). In addition, we found that protein expression levels for N‐cadherin, Vimentin, and Slug‐1 were significantly downregulated, while E‐cadherin was significantly upregulated in the tumors derived from YBX‐1 silenced Biu‐87 and HTB9 cells (Figure 3G). These findings suggested that YBX‐1 silencing repressed BC cell proliferation, migration, invasion, and EMT in vitro, and in vivo.
FIGURE 3.
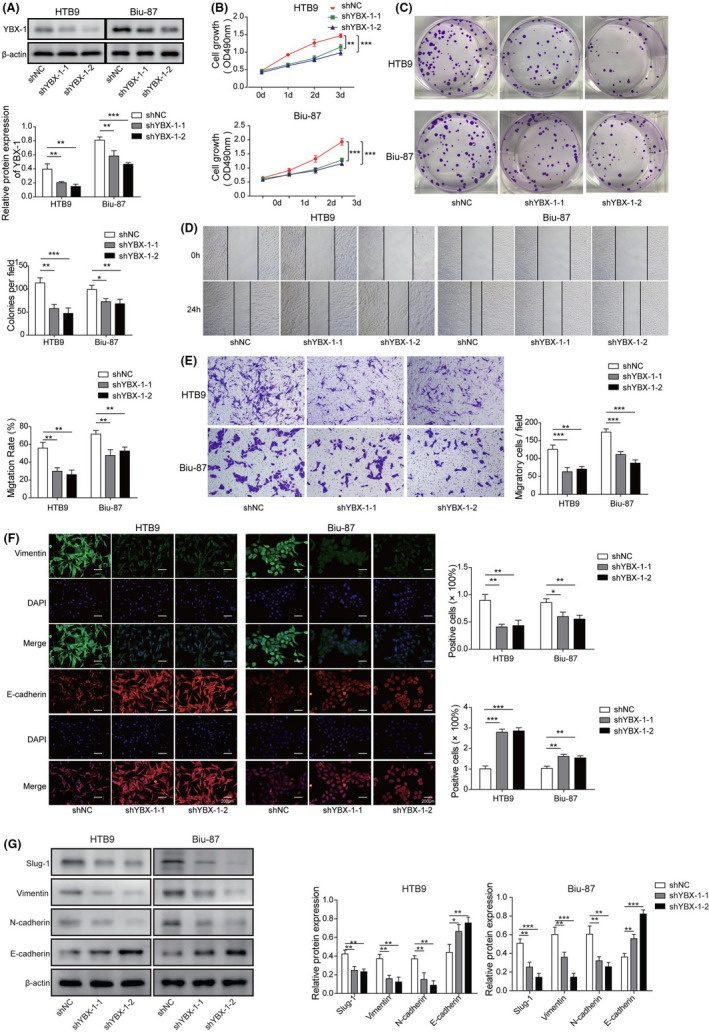
YBX‐1 silencing repressed BC cell proliferation, migration, invasion, and EMT in vitro and in vivo. A, Knockdown efficiencies of sh‐YBX‐1‐1, and sh‐YBX‐1‐2 were examined by western blot in Biu‐87 and HTB9 cells. B, After 0, 1, 2 and 3 d of transfection with sh‐NC, sh‐YBX‐1‐1, and sh‐YBX‐1‐2, Biu‐87, and HTB9 cells were subjected to cell viability analysis using CCK‐8 assay. C, Colony formation assay was performed to assess the proliferation of Biu‐87 and HTB9 cells after transfection with sh‐NC, sh‐YBX‐1‐1, and sh‐YBX‐1‐2. Effects of YBX‐1 knockdown on cell (D) migration and (E) invasion were evaluated by wound‐healing and transwell assay, respectively. F, Immunofluorescence comparing YBX‐1 silenced Biu‐87 and HTB9 cells with their respective cells are shown for relative expression of Vimentin and E‐cadherin. G, Protein expression levels of YBX‐1, Slug‐1, Vimentin, N‐cadherin, and E‐cadherin in the tumors of nude mice with sh‐NC, sh‐YBX‐1‐1, and sh‐YBX‐1‐2 following transfection were examined by western blot. *P < .05; **P < .01; ***P < .001
3.4. miR‐432‐3p regulated EMT‐related molecules by targeting YBX‐1 in BC
To investigate the relationship between miR‐432‐3p and YBX‐1, we evaluated the expression of YBX‐1 using western blot in miR‐432‐3p mimics transfected Biu‐87 and HTB9 cells. Transfection of miR‐432‐3p mimics caused a marked upregulation of miR‐432‐3p in both Biu‐87 and HTB9 cells compared with NC mimics transfection (Figure 4A). Overexpression of miR‐432‐3p dramatically decreased the protein expression of YBX‐1 in Biu‐87 and HTB9 cells (Figure 4B). Based on StarBase prediction showing the complementary sequence between YBX‐1 and miR‐432‐3p (Figure 4C), a dual‐luciferase reporter assay was employed to verify their interaction. The results indicated that miR‐432‐3p mimics, but not NC mimics, transfection could attenuate the luciferase activity of Biu‐87 and HTB9 cells driven by YBX‐1 WT, although those cells driven by YBX‐1 MUT were not affected by either NC or miR‐432‐3p mimics (Figure 4D). Furthermore, results from the RIP assay showed that miR‐432‐3p could be significantly enriched by MS2bs‐YBX‐1 3′UTR, but not MS2bs‐YBX‐1 3′UTR MUT (Figure 4E). In addition, we demonstrated that miR‐432‐3p mimics transfection dramatically decreased the expression of YBX‐1, Slug‐1, Vimentin, and N‐cadherin, although it increased the expression of E‐cadherin in Biu‐87 and HTB9 cells. YBX‐1 overexpression exhibited opposite effects, moreover co‐transfection of YBX‐1 and miR‐432‐3p mimics reversed the dysregulation caused by miR‐432‐3p mimics (Figure 4F). These findings indicated that miR‐432‐3p regulated EMT‐related molecules by targeting and negatively regulating YBX‐1 in BC.
FIGURE 4.
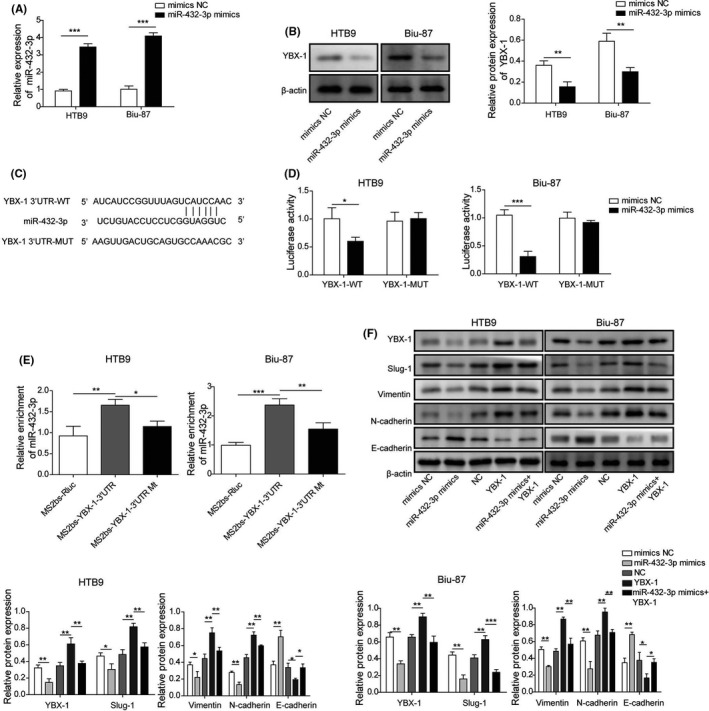
miR‐432‐3p regulates EMT‐related molecules by targeting YBX‐1 in BC. A, Relative expression of miR‐432‐3p was measured in Biu‐87 and HTB9 cells transfected with NC and miR‐432‐3p mimics. B, YBX‐1 protein expression of miR‐432‐3p overexpressed Biu‐87 and HTB9 cells was examined by western blot. C, StarBase software analysis showing the predicted consequential pairing of the target regions of YBX‐1 and miR‐432‐3p. The interplay between YBX‐1 and miR‐432‐3p was verified by (D) dual‐luciferase reporter and (E) RIP assay. F, Protein expression levels of YBX‐1, Slug‐1, Vimentin, N‐cadherin and E‐cadherin of Biu‐87 and HTB9 cells were evaluated by western blot after 24 h of transfection of miR‐432‐3p mimics or YBX‐1 plasmid. *P < .05; **P < .01; ***P < .001
3.5. circ_100984 regulated YBX‐1 and EMT‐related molecules by targeting miR‐432‐3p in BC
StarBase prediction also predicted the complementary sequence between miR‐432‐3p and circ_100984 (Figure 5A). In the dual‐luciferase assay, we found that miR‐432‐3p mimics transfection reduced the luciferase intensity of Biu‐87 and HTB9 cells driven by circ_100987 WT (Figure 5B). In the RIP assay, miR‐432‐3p was significantly enriched by MS2bs‐circ_100984 but not MS2bs‐circ_100984 MUT (Figure 5C). Additionally, we found that co‐transfection of circ_100984 and miR‐432‐3p could also reverse the dysregulation of YBX‐1, Slug‐1, Vimentin, N‐cadherin and E‐cadherin caused by miR‐432‐3p in Biu‐87 and HTB9 cells (Figure 5D). These results showed that circ_100984 regulated YBX‐1 and EMT‐related molecules by targeting miR‐432‐3p in BC.
FIGURE 5.
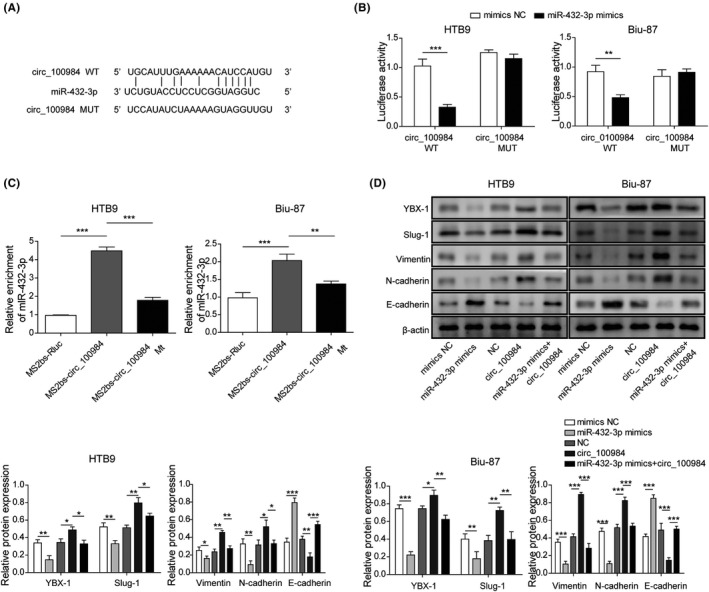
circ_100984 regulates YBX‐1 and EMT‐related molecules by targeting miR‐432‐3p in BC. A, StarBase software analysis showing the predicted consequential pairing of target region of circ_100984 and miR‐432‐3p. The interplay between circ_100984 and miR‐432‐3p was verified by (B) dual‐luciferase reporter and (C) RIP assay. D, Protein expression levels of YBX‐1, Slug‐1, Vimentin, N‐cadherin and E‐cadherin of Biu‐87 and HTB9 cells were evaluated by western blot after 24 h of transfection of miR‐432‐3p mimics or circ_100984. *P < .05; **P < .01; ***P < .001
3.6. YBX‐1, β‐catenin and c‐Jun acted as a feedback loop to regulate EMT‐related molecules
Using western blot, we found that transfection of sh‐YBX‐1‐1 and sh‐YBX‐1‐2 significantly decreased the expression levels of β‐catenin and c‐Jun in Biu‐87 and HTB9 cells (Figure 6A). Moreover, transfection of sh‐β‐catenin‐1 and sh‐β‐catenin‐2 resulted in a significant downregulation of YBX‐1, β‐catenin, c‐Jun, Slung‐1, Vimentin and N‐cadherin and a significant upregulation of E‐cadherin in Biu‐87 and HTB9 cells (Figure 6B). Effects of c‐Jun overexpression on the expression levels of YBX‐1, β‐catenin, c‐Jun, Slung‐1, Vimentin, N‐cadherin and E‐cadherin were opposite to that of β‐catenin knockdown. Meanwhile, we found that co‐transfection of c‐Jun and sh‐β‐catenin reversed the effects of β‐catenin knockdown in both Biu‐87 and HTB9 cells (Figure 6C). These results implied that YBX‐1, β‐catenin and c‐Jun acted as a feedback loop to regulate EMT‐related molecules.
FIGURE 6.
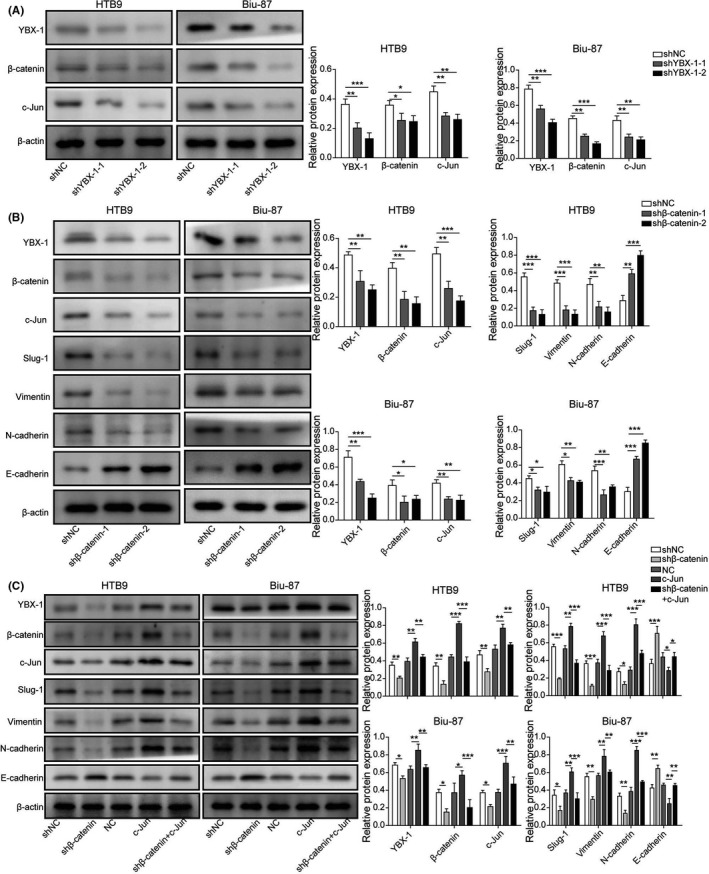
YBX‐1, β‐catenin and c‐Jun act as a feedback loop to regulate EMT‐related molecules. A, Western blot analysis was carried out in Biu‐87 and HTB9 cells transfected with sh‐NC, sh‐YBX‐1‐1, and sh‐YBX‐1‐2 to analyze the expression of YBX‐1, β‐catenin and c‐Jun. B, Protein expression levels of YBX‐1, β‐catenin, c‐Jun, Slug‐1, Vimentin, N‐cadherin and E‐cadherin of Biu‐87 and HTB9 cells were detected after β‐catenin knockdown. C, Protein expression levels of YBX‐1, β‐catenin, c‐Jun, Slug‐1, Vimentin, N‐cadherin and E‐cadherin were detected in Biu‐87 and HTB9 cells transfected with sh‐β‐catenin or c‐Jun. *P < .05; **P < .01; ***P < .001
3.7. YBX‐1 regulated β‐catenin activity by binding to its promoter
According to the analysis results of the JASPAR (http://jaspar.genereg.net/), the β‐catenin promoter area contains 1 putative YBX‐1 binding site (from 1749 to 1757 bp) (Figure 7A). Further EMSA analysis demonstrated that YBX‐1 formed a DNA‐protein complex with the native probe containing the putative binding sequence (Figure 7B). Meanwhile, ChIP‐PCR analysis showed that YBX‐1 knockdown resulted in a significant downregulation of enrichment of YBX‐1 in the β‐catenin promoter (Figure 7C). These results indicated that YBX‐1 might regulate β‐catenin activity by binding to its promoter.
FIGURE 7.
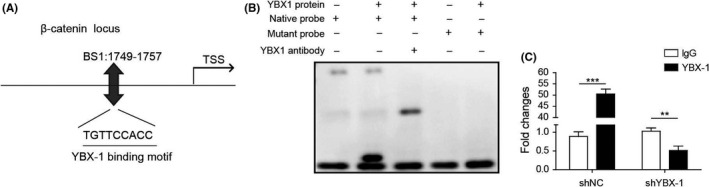
YBX‐1 regulates β‐catenin activity by binding to its promoter. A, JASPAR analysis showing the binding sequences of YBX‐1 at the promoter area of β‐catenin. B, Protein‐DNA interplay between YBX‐1 protein and β‐catenin promoter was verified by EMSA assay. C, ChIP‐PCR analysis was used to evaluate YBX‐1 binding to β‐catenin promoter in YBX‐1 blocked Biu‐87 cells. ***P < .001
3.8. c‐Jun regulated β‐catenin activity by binding to YBX‐1 promoter
Transfections of sh‐c‐Jun‐1 and sh‐c‐Jun‐2 were revealed to cause a significant inhibitory effect on the expression of c‐Jun, YBX‐1 and β‐catenin compared with sh‐NC transfection (Figure 8A). In addition, JASPAR prediction indicated that the YBX‐1 promoter region contained 3 putative c‐Jun binding sites (BSs) (BS1, from 405 to 418; BS2, from 565 to 578; BS3, from 1020 to 1033) (Figure 8B). Next, ChIP assay indicated that c‐Jun preferentially bound to BS2, but not to BS1 or BS3 in the YBX‐1 promoter region (Figure 8C). Moreover, EMSA assay demonstrated that c‐Jun protein could form a DNA‐protein complex with native probe containing BS2 (Figure 8D). Moreover, we adopted a dual‐luciferase reporter assay to evaluate the interaction between c‐Jun and YBX‐1 directly. The results indicated that transfection of c‐Jun caused a significant downregulation of luciferase activity of cells driven by mut‐BS2, but not mut‐BS1 and mut‐BS3 (Figure 8E). We also detected the protein expression of β‐catenin in BS1‐, BS2‐, and BS3‐mutant BC cells. Compared with BS2‐mutant cells, protein expression of β‐catenin was significantly increased in BS1‐ and BS3‐mutant BC cells (Figure 8F). These findings indicated that c‐Jun regulated β‐catenin activity by binding to the YBX‐1 promoter.
FIGURE 8.
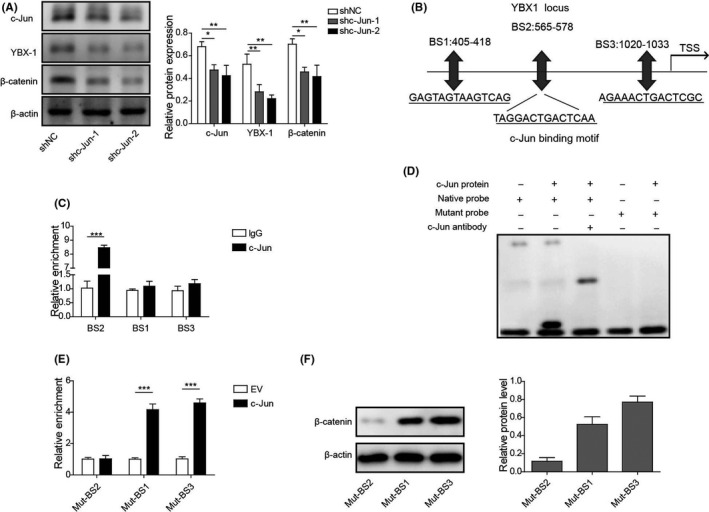
c‐Jun regulates β‐catenin activity by binding to the YBX‐1 promoter. A, Protein expression levels of c‐Jun, YBX‐1, and β‐catenin were examined by western blot in c‐Jun silenced BC cell. B, JASPAR analysis showing the binding sequences of c‐Jun at the promoter area of YBX‐1. C, ChIP‐PCR analysis was used to evaluate c‐Jun binding to YBX‐1 promoter in c‐Jun blocked Biu‐87 cells. D, Protein‐DNA interplay between c‐Jun protein and YBX‐1 promoter was verified by EMSA assay. E, Dual‐luciferase reporter assay was performed to examine the interaction between YBX‐1 and c‐Jun in BC cell. F, Protein expression of β‐catenin was evaluated by western blot in BS1‐, BS2‐, and BS3‐mutant BC cells. *P < .05; **P < .01; ***P < .001
4. DISCUSSION
YBX‐1 provides diverse pro‐oncogenic roles in tumors, including tumor growth, invasion, metastasis, chemo‐resistance, and angiogenesis within tumors. 7 , 23 Its expression has been regarded as a prognostic factor in multiple tumors such as breast cancer, ovarian cancer, and prostate cancer. 24 , 25 , 26 YBX‐1 was also identified in 2014 by Song et al 9 as a promising prognostic factor of BC and its nuclear expression was revealed to be correlated in 2017 27 with resistance to multiple chemotherapeutic drugs in TP53‐mutated BC. In addition, YBX‐1 was found to facilitate BC tumor progression by increasing glycolysis levels through the regulation of c‐Myc and HIF‐1α. 10 circ_100984 was previously identified to be upregulated in BC in a circRNAs microarray analysis, however its role and mechanism remain unknown. Consistently, we found that expression levels of circ_100984 and YBX‐1 were upregulated in BC. Moreover, silencing of circ_100984 and YBX‐1 repressed BC tumor progression in vitro and in vivo. Taken together, these findings suggest that circ_10084 and YBX‐1 act as an oncogene of BC.
Regulation of gene expression is considered to be the most important property of miRNAs, and circRNAs have been demonstrated to indirectly regulate gene expression by functioning as sponges of miRNAs. 28 The circRNAs‐miRNAs‐mRNAs axis has been found to be one of the most common molecular mechanisms in human cancer. 29 As a well documented oncogene, YBX‐1 has been indirectly modulated by several circRNAs through miRNAs‐mediated action in human cancers. 30 , 31 In this study, circ_100984 was demonstrated to indirectly regulate the expression of YBX‐1 by functioning as a sponge of miR‐432‐3p.
EMT is considered to be a key process that promotes cancer cell aggressiveness. Repression of EMT has been well demonstrated to slow BC progression, 32 , 33 and circRNAs was revealed to be a critical regulator of EMT process in BC through an miRNAs/mRNA mechanism. 16 , 34 , 35 , 36 Canonical Wnt/β‐catenin cascade facilitates the EMT process by elevating several transcription factors including Slug, Twist, and YBX‐1. 21 , 37 , 38 In hepatocellular carcinoma, YBX‐1 was not only found to activate EMT process, but also to modulate the expression of Wnt ligands and β‐catenin. 20 Moreover, YBX‐1 and β‐catenin could form a transcriptional complex with Kindlin‐2 that binds to the epidermal growth factor receptor (EGFR) promoter and promote its transcription. 39 c‐Jun has been reported to be a Wnt/β‐catenin downstream positive modulator, and is known as a key transcription factor in mammals that involves regulating diverse cellular physiological functions such as proliferation, metastasis, and death. 40 , 41 , 42 A prior study demonstrated that c‐Jun could be induced by MYH9 via Wnt/β‐catenin cascade, and c‐Jun, MYH9, β‐catenin and miR‐6089 act as a negative feedback loop to regulate ovarian cancer progression. 43 Additionally, β‐catenin could promote c‐Jun expression and form a feedback loop with miR‐5188 and FOXO1 in breast cancer cell. 22 In this study, YBX‐1 was found to regulate β‐catenin expression by targeting its promoter, in addition c‐Jun was demonstrated to regulate YBX‐1 expression by binding to its promoter.
Taken together, c‐Jun/YBX‐1/β‐catenin formed a feedback loop in BC, and circ_100984 promoted BC progression and EMT process by modulating this loop through miR‐432‐3p. Our findings offer a novel target axis for BC treatment that might contribute to the development of an efficient therapeutic strategy.
DISCLOSURE
This work has no conflict of interest.
Supporting information
Fig S1
Fig S2
ACKNOWLEDGMENTS
This work was supported by the National Natural Science Foundation of China (No. 81974397).
Tong L, Yang H, Xiong W, Tang G, Zu X, Qi L. circ_100984‐miR‐432‐3p axis regulated c‐Jun/YBX‐1/β‐catenin feedback loop promotes bladder cancer progression. Cancer Sci.2021;112:1429–1442. 10.1111/cas.14774
REFERENCES
- 1. Borden LS Jr, Clark PE, Hall MC. Bladder cancer. Curr Opin Oncol. 2004;16:257‐262. [DOI] [PubMed] [Google Scholar]
- 2. Leung HY, Griffiths TR, Neal DE. Bladder cancer. Postgrad Med J. 1996;72:719‐724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7‐30. [DOI] [PubMed] [Google Scholar]
- 4. Huang J, Chen X, Lin TX. [Strategies for diagnosis and treatment of bladder cancer in precise times]. Zhonghua Wai Ke Za Zhi. 2016;54:734‐737. [DOI] [PubMed] [Google Scholar]
- 5. Racioppi M, D’Agostino D, Totaro A, et al. Value of current chemotherapy and surgery in advanced and metastatic bladder cancer. Urol Int. 2012;88:249‐258. [DOI] [PubMed] [Google Scholar]
- 6. El‐Naggar AM, Veinotte CJ, Cheng H, et al. Translational activation of HIF1alpha by YB‐1 promotes sarcoma metastasis. Cancer Cell. 2015;27:682‐697. [DOI] [PubMed] [Google Scholar]
- 7. Eliseeva IA, Kim ER, Guryanov SG, Ovchinnikov LP, Lyabin DN. Y‐box‐binding protein 1 (YB‐1) and its functions. Biochemistry (Mosc). 2011;76:1402‐1433. [DOI] [PubMed] [Google Scholar]
- 8. Kuwano M, Oda Y, Izumi H, et al. The role of nuclear Y‐box binding protein 1 as a global marker in drug resistance. Mol Cancer Ther. 2004;3:1485‐1492. [PubMed] [Google Scholar]
- 9. Song YH, Shiota M, Yokomizo A, et al. Twist1 and Y‐box‐binding protein‐1 are potential prognostic factors in bladder cancer. Urol Oncol. 2014;32(31):e1‐e7. [DOI] [PubMed] [Google Scholar]
- 10. Xu L, Li H, Wu L, Huang S. YBX1 promotes tumor growth by elevating glycolysis in human bladder cancer. Oncotarget. 2017;8:65946‐65956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Memczak S, Jens M, Elefsinioti A, et al. Circular RNAs are a large class of animal RNAs with regulatory potency. Nature. 2013;495:333‐338. [DOI] [PubMed] [Google Scholar]
- 12. Li J, Yang J, Zhou P, et al. Circular RNAs in cancer: novel insights into origins, properties, functions and implications. Am J Cancer Res. 2015;5:472‐480. [PMC free article] [PubMed] [Google Scholar]
- 13. Liu L, Wu SQ, Zhu X, et al. Analysis of ceRNA network identifies prognostic circRNA biomarkers in bladder cancer. Neoplasma. 2019;66:736‐745. [DOI] [PubMed] [Google Scholar]
- 14. Rong D, Sun H, Li Z, et al. An emerging function of circRNA‐miRNAs‐mRNA axis in human diseases. Oncotarget. 2017;8:73271‐73281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Liu Z, Yu Y, Huang Z, et al. CircRNA‐5692 inhibits the progression of hepatocellular carcinoma by sponging miR‐328‐5p to enhance DAB2IP expression. Cell Death Dis. 2019;10:900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen X, Chen R‐X, Wei W‐S, et al. PRMT5 circular RNA promotes metastasis of urothelial carcinoma of the bladder through sponging miR‐30c to induce epithelial‐mesenchymal transition. Clin Cancer Res. 2018;24:6319‐6330. [DOI] [PubMed] [Google Scholar]
- 17. Nusse R, Clevers H. Wnt/beta‐catenin signaling, disease, and emerging therapeutic modalities. Cell. 2017;169:985‐999. [DOI] [PubMed] [Google Scholar]
- 18. Li J, Li Y, Meng F, Fu L, Kong C. Knockdown of long non‐coding RNA linc00511 suppresses proliferation and promotes apoptosis of bladder cancer cells via suppressing Wnt/beta‐catenin signaling pathway. Biosci Rep. 2018;38(4):BSR20171701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Yuan H, Yu S, Cui Y, et al. Knockdown of mediator subunit Med19 suppresses bladder cancer cell proliferation and migration by downregulating Wnt/beta‐catenin signalling pathway. J Cell Mol Med. 2017;21:3254‐3263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Chao HM, Huang HX, Chang PH, Tseng KC, Miyajima A, Chern E. Y‐box binding protein‐1 promotes hepatocellular carcinoma‐initiating cell progression and tumorigenesis via Wnt/beta‐catenin pathway. Oncotarget. 2017;8:2604‐2616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lim JP, Nair S, Shyamasundar S, et al. Silencing Y‐box binding protein‐1 inhibits triple‐negative breast cancer cell invasiveness via regulation of MMP1 and beta‐catenin expression. Cancer Lett. 2019;452:119‐131. [DOI] [PubMed] [Google Scholar]
- 22. Zou Y, Lin X, Bu J, et al. Timeless‐stimulated miR‐5188‐FOXO1/beta‐Catenin‐c‐Jun feedback loop promotes stemness via ubiquitination of beta‐catenin in breast cancer. Mol Ther. 2020;28:313‐327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kuwano M, Shibata T, Watari K, Ono M. Oncogenic Y‐box binding protein‐1 as an effective therapeutic target in drug‐resistant cancer. Cancer Sci. 2019;110:1536‐1543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Ferreira AR, Bettencourt M, Alho I, et al. Serum YB‐1 (Y‐box binding protein 1) as a biomarker of bone disease progression in patients with breast cancer and bone metastases. J Bone Oncol. 2017;6:16‐21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Heumann A, Kaya Ö, Burdelski C, et al. Up regulation and nuclear translocation of Y‐box binding protein 1 (YB‐1) is linked to poor prognosis in ERG‐negative prostate cancer. Sci Rep. 2017;7:2056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Rohr I, Braicu EI, En‐Nia A, et al. Y‐box protein‐1/p18 as novel serum marker for ovarian cancer diagnosis: a study by the Tumor Bank Ovarian Cancer (TOC). Cytokine. 2016;85:157‐164. [DOI] [PubMed] [Google Scholar]
- 27. Yamashita T, Higashi M, Momose S, Morozumi M, Tamaru JI. Nuclear expression of Y box binding‐1 is important for resistance to chemotherapy including gemcitabine in TP53‐mutated bladder cancer. Int J Oncol. 2017;51:579‐586. [DOI] [PubMed] [Google Scholar]
- 28. Xiong D‐D, Dang Y‐W, Lin P, et al. A circRNA‐miRNA‐mRNA network identification for exploring underlying pathogenesis and therapy strategy of hepatocellular carcinoma. J Transl Med. 2018;16:220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Yin Y, Long J, He Q, et al. Emerging roles of circRNA in formation and progression of cancer. J Cancer. 2019;10:5015‐5021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Fang J, Hong H, Xue X, et al. A novel circular RNA, circFAT1(e2), inhibits gastric cancer progression by targeting miR‐548g in the cytoplasm and interacting with YBX1 in the nucleus. Cancer Lett. 2019;442:222‐232. [DOI] [PubMed] [Google Scholar]
- 31. Wang L, Wang P, Su X, Zhao B. Circ_0001658 promotes the proliferation and metastasis of osteosarcoma cells via regulating miR‐382‐5p/YB‐1 axis. Cell Biochem Funct. 2020;38:77‐86. [DOI] [PubMed] [Google Scholar]
- 32. Chen X, Li S, Yu Z, Tan W. Yes‐associated protein 1 promotes bladder cancer invasion by regulating epithelial‐mesenchymal transition. Int J Clin Exp Pathol. 2019;12:1070‐1077. [PMC free article] [PubMed] [Google Scholar]
- 33. Lei Y, Yang L, Hongwei J, Hongyuan Y, Tao L. TIS111D can affect bladder cancer cells by regulating epithelial‐mesenchymal transition. Life Sci. 2019;235:116832. [DOI] [PubMed] [Google Scholar]
- 34. Li Y, Wan B, Liu L, Zhou L, Zeng Q. Circular RNA circMTO1 suppresses bladder cancer metastasis by sponging miR‐221 and inhibiting epithelial‐to‐mesenchymal transition. Biochem Biophys Res Commun. 2019;508:991‐996. [DOI] [PubMed] [Google Scholar]
- 35. Zhong Z, Huang M, Lv M, et al. Circular RNA MYLK as a competing endogenous RNA promotes bladder cancer progression through modulating VEGFA/VEGFR2 signaling pathway. Cancer Lett. 2017;403:305‐317. [DOI] [PubMed] [Google Scholar]
- 36. Zhang M, Wang S, Tang LU, et al. Downregulated circular RNA hsa_circ_0067301 regulates epithelial‐mesenchymal transition in endometriosis via the miR‐141/Notch signaling pathway. Biochem Biophys Res Commun. 2019;514:71‐77. [DOI] [PubMed] [Google Scholar]
- 37. DiMeo TA, Anderson K, Phadke P, et al. A novel lung metastasis signature links Wnt signaling with cancer cell self‐renewal and epithelial‐mesenchymal transition in basal‐like breast cancer. Cancer Res. 2009;69:5364‐5373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wang ZY, Hu M, Dai MH, et al. Upregulation of the long non‐coding RNA AFAP1‐AS1 affects the proliferation, invasion and survival of tongue squamous cell carcinoma via the Wnt/beta‐catenin signaling pathway. Mol Cancer. 2018;17:3. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 39. Ou Y, Zhao Z, Zhang W, et al. Kindlin‐2 interacts with beta‐catenin and YB‐1 to enhance EGFR transcription during glioma progression. Oncotarget. 2016;7:74872‐74885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Liang Z, Liu Z, Cheng C, et al. VPS33B interacts with NESG1 to modulate EGFR/PI3K/AKT/c‐Myc/P53/miR‐133a‐3p signaling and induce 5‐fluorouracil sensitivity in nasopharyngeal carcinoma. Cell Death Dis. 2019;10:305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhao M, Xu P, Liu Z, et al. Dual roles of miR‐374a by modulated c‐Jun respectively targets CCND1‐inducing PI3K/AKT signal and PTEN‐suppressing Wnt/beta‐catenin signaling in non‐small‐cell lung cancer. Cell Death Dis. 2018;9:78. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42. Liu Y, Jiang Q, Liu X, et al. Cinobufotalin powerfully reversed EBV‐miR‐BART22‐induced cisplatin resistance via stimulating MAP2K4 to antagonize non‐muscle myosin heavy chain IIA/glycogen synthase 3beta/beta‐catenin signaling pathway. EBioMedicine. 2019;48:386‐404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Liu L, Ning Y, Yi J, et al. miR‐6089/MYH9/beta‐catenin/c‐Jun negative feedback loop inhibits ovarian cancer carcinogenesis and progression. Biomed Pharmacother. 2020;125:109865. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Fig S1
Fig S2


