Abstract
An outbreak of a new coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), began in December 2019. Accurate, rapid, convenient, and relatively inexpensive diagnostic methods for SARS-CoV-2 infection are important for public health and optimal clinical care. The current gold standard for diagnosing SARS-CoV-2 infection is reverse transcription-polymerase chain reaction (RT-PCR). However, RTPCR assays are designed for use in well-equipped laboratories with sophisticated laboratory infrastructure and highly trained technicians, and are unsuitable for use in under-equipped laboratories and in the field. In this study, we report the development of an accurate, rapid, and easy-to-implement isothermal and nonenzymatic signal amplification system (a catalytic hairpin assembly (CHA) reaction) coupled with a lateral flow immunoassay (LFIA) strip-based detection method that can detect SARSCoV-2 in oropharyngeal swab samples. Our method avoids RNA isolation, PCR amplification, and elaborate result analysis, which typically takes 6–8 h. The entire CHA-LFIA detection method, from nasopharyngeal sampling to obtaining test results, takes less than 90 min. Such methods are simple and require no expensive equipment, only a simple thermostatically controlled water bath and a fluorescence reader device. We validated our method using synthetic oligonucleotides and clinical
samples from 15 patients with SARS-CoV-2 infection and 15 healthy individuals. Our detection method provides a fast, simple, and sensitive (with a limit of detection (LoD) of 2000 copies/mL) alternative to the SARS-CoV-2 RT-PCR assay, with 100 % positive and negative predictive agreements.
Keywords: sars-cov-2, Nucleic acid test, Rapid diagnostic test, Catalytic hairpin assembly reaction, Lateral flow immunoassay strip
1. Introduction
In December 2019, there was an outbreak of a new coronavirus, severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which causes coronavirus disease 2019 (COVID-19) [1,2]. SARS-CoV-2 infection is a serious global public health issue that has caused serious economic losses worldwide, leading the World Health Organization to characterize the COVID-19 outbreak as a public health emergency of international concern. In general, vaccination is the most effective method to prevent virus infection. Since humans typically acquire infection through the inhalation of contaminated aerosols [3], self-quarantine is one of the most effective infection prevention and control measures before effective vaccines become available [4]. To make matters worse, mild and asymptomatic cases of COVID-19 have been reported [5,6]. Asymptomatic patients have positive detection of SARS-CoV-2 nucleic acids; however, they have no typical clinical symptoms or signs, and no apparent abnormalities by imaging, including by lung computed tomography [[7], [8], [9], [10]]. The majority of people with asymptomatic infections are unaware of their infection status and remain infectious to others [11]. This situation increases the difficulty of current global SARS-CoV-2 infection prevention and control efforts. A reliable, quick, inexpensive, and accurate diagnostic test for suspected patients would play a significant role in COVID-19 case identification, infection control, and disease management.
Clinical and laboratory-based methods for the diagnosis of SARS-CoV-2 infection involve isolating the virus and detecting its RNA [12,13]. Isolation is the gold standard for diagnosing viral infections; however, it is tedious, time-consuming, labor-intensive, and expensive, and requires specialized equipment and trained technicians that are not available in many healthcare institutions. Thus, this technology is difficult to use in routine clinical examinations. The current gold standard for detecting SARS-CoV-2 infection is reverse transcription-polymerase chain reaction (RT-PCR) [14]. After the publication of the complete genome of SARS-CoV-2, public health laboratories, clinical laboratories, and diagnostic companies in different countries developed numerous RT-PCR methods for SARS-CoV-2 detection, targeting different genes and genomic regions (Orf1ab, RdRp, N, E, etc.) [[14], [15], [16]]. However, RT-PCR also has many shortcomings in practical application, especially during a public health emergency such as the COVID-19 outbreak. Although RT-PCR assays are easier to perform than viral isolation, they still require highly skilled laboratory technicians and sophisticated laboratory infrastructure, which are often located in a central laboratory (biosafety level 2 or above), precluding its use in on-site detection. Therefore, in remote regions with limited resources, sample transportation is inevitable, which is time-consuming and increases the likelihood of viral transmission in the meantime. Moreover, RT-PCR assays for SARS-CoV-2 detection have high false negative rates (30–50 %) for a variety of reasons [17,18], which remains an issue that cannot be neglected. Thus, researchers have been searching for an alternative to RT-PCR for SARS-CoV-2 virus nucleic acid detection.
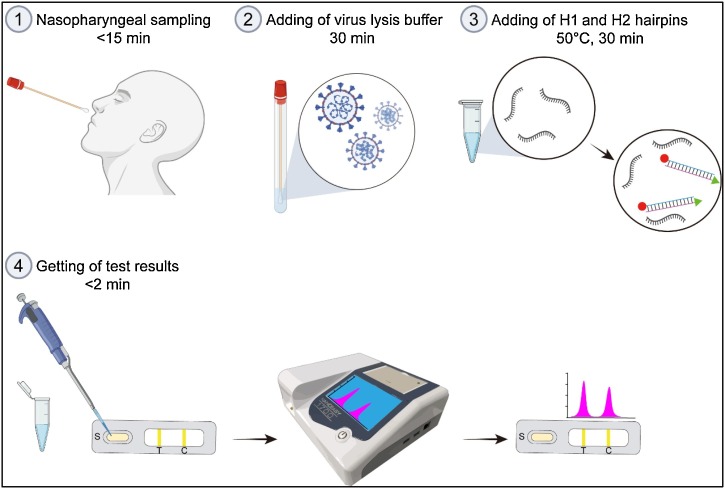
In this work, we report a catalytic hairpin assembly (CHA) reaction-based signal amplification system coupled with a lateral flow immunoassay (LFIA) strip for the rapid and high-sensitivity detection of SARS-CoV-2 RNA. This method requires neither RNA extraction nor RT-PCR, making it well suited for use in on-site clinical settings. As shown in Fig. 1 , the CHA-LFIA detection method consists of two steps: (1) signal amplification by CHA reaction, and (2) ultra-sensitive detection by LFIA. The CHA reaction is an isothermal and enzyme-free amplification technique, which was first proposed by Yin et al. in 2008 [19]. Because of its multiple advantages, such as high efficiency, easy operation, and great versatility, the CHA amplification system can be used to detect various nucleic acids [[20], [21], [22], [23], [24]]. However, to the best of our knowledge, this study is the first to apply CHA to viral detection. The entire procedure, from nasopharyngeal sampling to obtaining test results, takes less than 90 min. Our method exhibits excellent performance, such as easy operation, short detection time and better meet the needs for rapid detection, is expected to provide a simple and faster alternative to the traditional SARS-CoV-2 RT-PCR assay.
Fig. 1.
Overview of the CHA-LFIA method for SARS-CoV-2 viral RNA detection.
2. Experimental Section
2.1. Chemicals, reagents and samples
The oligonucleotides (Table S1) used in this work were synthesized and purified (HPLC) by Sangon Biotech. Co., Ltd. (Shanghai, China). Native PAGE gel was purchased from Sangon Biotech. Co., Ltd. (Shanghai, China). SARS-Cov-2 nucleic acid detection kit (fluorometric PCR) was purchased from BioGerm Medical Technology Co., Ltd. (Shanghai, China). SARS-CoV-2 RNA reference material was obtained from the National Institute of Metrology, China (Beijing, China). Except where indicated, all reagents were obtained from Sigma (Shanghai, China). All Clinical samples were obtained from Affiliated Zhongda Hospital of Southeast University.
2.2. Identification of specificity sequence for SARS-CoV-2 virus and design of DNA hairpins
The full genome sequence data of the SARS-CoV-2 were obtained from the NCBI Gene Bank (http://www.ncbi.nlm.nih.gov/genbank/) and GISAID databases (https://www.gisaid.org/) for identification of the common regions among all retrieved sequence of the SARS-CoV-2. The multiple sequence alignments were carried out by using ClustalW (http://www.ebi.ac.uk/Tools/msa/clustalw2/). Consensus sequences were determined by selecting the most common base at each nucleotide site.
DNA probes for detection selected RNA sequences of SARS-CoV-2 were designed according to the principles of DNA hairpins design. The secondary structure of DNA probe was carried out NUPACK software package (www.nupack.org) [25]. DNA hairpins were diluted with TAE/Mg2+ buffer (40 mM Tris-Acetate, 1 mM EDTA, and 12.5 mM magnesium acetate, pH 8.0) to reach a final concentration of 100 μM, and annealed over a temperature gradient from 95 to 25 °C in 90 min to create the hairpin structure, followed by preservation at −20 °C for standby application.
2.3. Theoretical calculation and modelling
For theoretical calculation and modelling, the free-energy changes of each reaction were estimated by NUPACK software package [25]. The predicted secondary structures of the DNA hairpins with Gibbs-free energy (ΔG) at different temperatures were shown in Figure S1. And the Gibbs-free energy (ΔG) of each hybridization reaction product at different reaction temperatures were shown in Table S2. The change of the free energy (ΔG°) for hybridization reaction are calculated using the Van’t Hoff formula (Formula 1).
| (1) |
Where ΔH and ΔS are the changes in enthalpy and entropy of binding, respectively. T is the temperature (K), R is the universal gas constant (8.314 J/mol K, and Keq represents the equilibrium constant.
Thus, the following holds at equilibrium (Formula 2) for a dilute solution containing strands A (H1 probe) and B (H2 probe) that can interact to form complex AB (H1-H2 hybrids).
| (2) |
Where for each complex, i, xi is the mole fraction, [i] is the concentration (e.g. in units of mol/L), ΔGi is the free energy, and ρH2O is the concentration of water.
The CHA reaction consists of two steps of hybridization reaction: 1) H1 probe hybridizes with the target RNA to form an H1-Tartget RNA hybrid complexes, and 2) H2 probe compete with Target RNA to bind to H1 probe to form an H1-H2 hybrid complexes. Therefore, the can be readily calculated the equilibrium concentration for H1-H2 hybrid complexes.
2.4. Native-polyacrylamide gel electrophoresis
The feasibility of CHA reaction was evaluated by using 12 % native polyacrylamide gel electrophoresis. The details of electrophoretic samples were shown in Table S3. The reaction was conducted at 50 °C during 2 h. Gel electrophoresis was performed at 120 V for 1 h. TAE buffer was used as the electrophoresis buffer. Finally, Gels were stained with 10 mg/mL ethidium bromide (EB) for 20 min and visualized under UV light at 280 nm.
2.5. Optimization of reaction conditions of the CHA reaction
Different parameters should be optimized, including the concentration ratio between hairpins DNA H1 probe and H2 probe, reaction temperature and time, to drive the equilibrium of the hairpin DNA cascade reaction to yield more H1-H2 hybrid duplexes. In addition, the concentration of hairpins DNA H1 probe and H2 probe also need to be optimized to improve limits of detection. Since the RNA concentrations of swab samples vary from 10^3 to 10^7 copies/mL, we therefore optimize the reaction conditions with the concentration of target RNA of 1 fM (approximately 6 ^5 copies/mL).
First, we optimized the concentration ratio between hairpins DNA H1 probe and H2 probe. Briefly, the concentration of H1 probe was held constant at 50 nM in the reactions while the concentration of H2 probe were varied from 50 to 200 nM in 50 nM steps, i.e., 50 nM, 100 nM, 150 nM and 200 nM. The total volume of each reaction mixture (H1 probe, H2 probe and targeted RNA (or non-targeted RNA) in 200 μL of TAE buffer) was 200 μL. Reaction mixture was incubated in water bath at 50 °C for 2 h. After the reaction, 80 μL of the reaction mixture was taken and placed in the sample well of the nucleic acid test strip, and the fluorescence intensity values of the T line and C line were recorded (the same below).
Based on the results of previous optimum concentration between H1 probe and H2 probe, a further investigation on optimal reaction temperature was performed. The optimal temperature was determined by incubating the reaction mixture for 2 h at different temperatures ranging from 20 to 70 °C (20,30,40,50,60and 70 °C).
And then, experiments with different reaction time were performed under the conditions optimized above. The reaction mixture was incubated for different incubation times (10 min, 20 min, 30 min, 1 h, 3 h and 6 h).
Finally, a further investigation on optimal concentration of hairpins DNA H1 probe and H2 probe were performed. We compared the effects of different concentrations of H1 and H2 probes (1pM:3pM; 50pM:150pM; 100pM:300pM; 1nM:3nM; 50 nM:150 nM; 100 nM:300nM) on the detection results.
2.6. Sensitivity and specificity of the CHA-LFIA method
In this study, we evaluated the sensitivity of the CHA-LFIA method (i.e. the minimum detectable concentration of targeted RNA) in oropharyngeal swab samples of healthy people that confirmed to be free of SARS-CoV-2 or other types of human coronaviruses infection. Each swab was stored in a sterile EP tube with 5 mL Hanks balanced salt solution. To estimate the minimum detectable concentration of targeted RNA, a series of low concentrations of targeted RNA from 50 nM∼1 aM in oropharyngeal swab samples reaction mediums under optimal conditions according to previous work.
To verify the sequence specificity of the CHA-LFIA method, we examined whether mismatched sequences affect detection result in CHA-LFIA method. Non-target RNA (random sequence generated from target RNA), single- and double-mismatched target RNA sequences (see Table S1 for the sequences) with concentration of 1 fM were assessed with CHA-LFIA method against the fully matched RNA target. The fluorescence value of fully matched target RNA was set to 100 %(F0) to calculate the relative fluorescence values (ΔF/F0) of non-target RNA, single- and double-mismatched target RNA.
2.7. RNA extraction and reverse transcription-PCR
Total RNA was extracted using total RNA extraction kit (Invitrogen) according to the instructions recommended by the manufacturer. And the nucleic acid was eluted in 50 μL of nuclease-free water and stored at −80 °C until use. rRT-PCR was performed using commercial kits (Biogerm, Shanghai, China) according to the manufacturer's instructions. Orf1ab (FAM reporter) and N (HEX/VIC reporter) genes of SARS-CoV-2 were detected. Samples were positive when PCR gave rise to reliable signals (Ct<38) for either or both genes
3. Results and discussion
3.1. Principle of the CHA-LFIA method
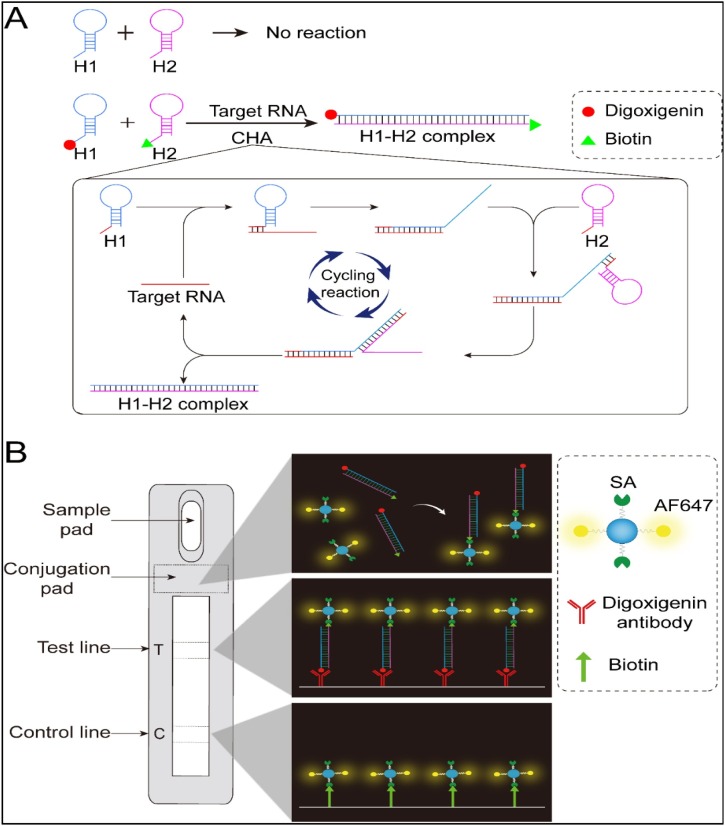
The mechanism of the CHA-LFIA detection method is illustrated in Fig. 2 . The CHA amplification reaction system consists of two complementary single-stranded hairpin DNA probes (termed the H1 and H2 probes). Although these probes are complementary, they maintain their stable hairpin structure and stably coexist in solution, as the complementary domains are caged within the hairpin stems (Fig. 2A). However, the probes are opened by the target RNA, which acts as a toehold switch, yielding a stable hybrid duplex through a free-energy-driven, isothermal, autonomous process [19,26]. In this reaction, the target RNA sequence plays the role of the enzyme that would be required to catalyze H1-H2 hybridization, without being consumed in the process. As the reaction proceeds, an increasing number of H1-H2 hybrid duplexes is generated, enabling signal amplification. Thus, the presence of the target RNA can be confirmed by detecting H1-H2 hybrid duplexes. The 5′ ends of the H1 and H2 probes are labeled with digoxigenin and biotin, respectively, for further processing and subsequent detection. A lateral flow immunoassay (LFIA) strip is used to detect digoxigenin- and biotin-double-labeled H1-H2 hybrid duplexes, as shown in Fig. 2B. The LFIA strip is composed of a sample pad, a conjugation pad, a nitrocellulose membrane with one test (T) line and one control (C) line, and an absorbent pad. The conjugation pad was prepared by spraying it with polyethylene (PE) nanoparticles doubly labeled with streptavidin and the fluorophore Alexa Fluor 647. The test line was prepared by spraying it with mouse anti-digoxin/digoxigenin monoclonal antibodies, and the control line was formed by spreading biotin on it. The amplification product to be detected (digoxigenin-biotin double-labeled H1-H2 hybrids) is added to the sample pad, which soaks up the specimen fluid. The fluid then migrates to the conjugation pad, which contains the doubly labeled PE nanoparticles. Here, H1-H2 hybrid-PE nanoparticle complexes form via biotin-streptavidin interactions. The complexes then migrate further along the strip until they reach the T line, where they bind the anti-digoxigenin antibodies coating it. Uncomplexed PE nanoparticles continue to migrate toward the C line, where they are captured by biotin. Finally, the fluorescence intensities of the T and C lines are quantified using a fluorescence-detecting device.
Fig. 2.
Schematic of the CHA-LFIA method for SARS-CoV-2 viral RNA detection.
(A) The CHA reaction without and with addition of target RNA; (B) The LFIA strip used to detect digoxigenin-biotin double-labeled H1-H2 hybrid duplexes. SA, streptavidin; AF647, Alexa Fluor 647.
3.2. Identification of specific target sequences for SARS-CoV-2 and DNA hairpin design
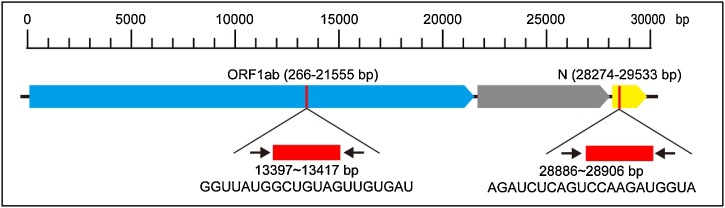
DNA hairpins were designed to detect conserved regions in SARS-CoV-2′s ORF1ab and N genes. Although overall, variation in the open reading frames of SARS-CoV-2 is low, sites of variation in the ORF1ab and N regions have been reported by several studies. [[27], [28], [29]] These variable regions should be avoided when designing DNA hairpins. To eliminate the impact of variation in the ORF1ab and N regions, and to minimize false negative results, we performed multiple sequence alignments of the ORF1ab and N genes to identify sequences common to all samples (as of March 15, 2020, 97 sequences were available for analysis). We identified common regions among all retrieved full genome sequences of SARS-CoV-2, and selected target sequences without variation sites. Selected sequences for specific SARS-CoV-2 detection and their relative positions in the ORF1ab and N genes are shown in Fig. 3 . These two DNA hairpin pairs (see Table S1 for their sequences) were termed the O-H1 and O-H2 probes (for ORF1ab), and the N-H1 and N-H2 probes (for N). The predicted secondary structures of these hairpins are shown in Figure S1. As controls, a random RNA sequences generated from the target RNA sequence produced by a random RNA sequence generator served (i.e. non-target RNA sequence) (Table S1).
Fig. 3.
Selected target sequences for specific SARS-CoV-2 detection and their relative positions.
3.3. Feasibility of the CHA reaction
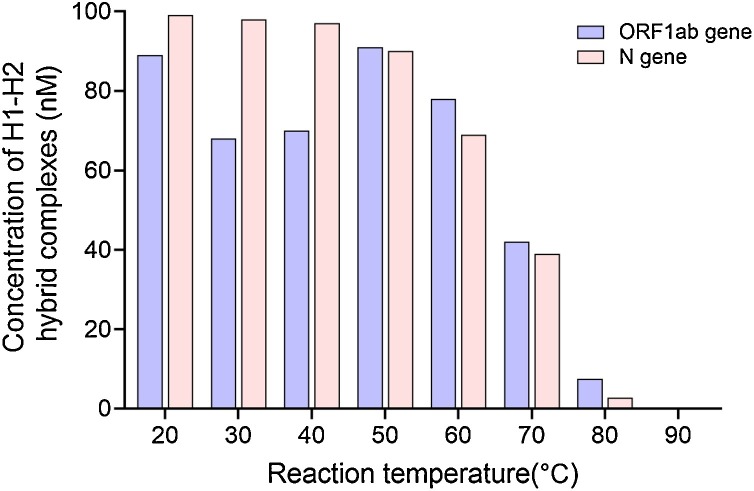
The equilibrium concentrations of H1-H2 hybrid complexes at different temperatures are shown in Fig. 4 , assuming equal initial concentrations of the H1 probe, H2 probe, and target RNA (100 nM each). Over 60 % of the H1 probe hybridized to the H2 probe to form H1-H2 hybrid complexes at 20–60 °C. These results suggest that the CHA reaction is thermodynamically feasible.
Fig. 4.
Thermodynamic analysis of the equilibrium concentrations of H1-H2 hybrid complexes at different temperatures with equal initial concentrations (100 nM) of the H1 probe, H2 probe, and target RNA.
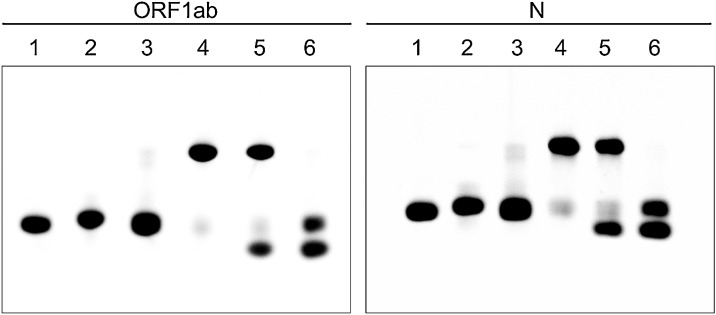
The practical feasibility of the CHA reaction was monitored by native polyacrylamide gel electrophoresis (PAGE), as shown in Fig. 5 . The two hairpin DNAs (H1 and H2) stably coexisted when target RNA strands were not present (lanes 3 and 6). However, once the target RNA strands (the lowest band in lane 5) were added, hybridization of the H1 and H2 probes was initiated, resulting in stable H1-H2 hybrid duplexes (the upper band in lane 5). More importantly, the hybridization reaction between the H1 and H2 probes triggered by the target RNA was comparable to that of the annealed hairpin H1 and H2 probes (lane 4). These results were consistent with the thermodynamic calculations.
Fig. 5.
Analysis of the CHA reaction by native PAGE.
Lane 1: H1 probe; lane 2: H2 probe; lane 3: H1 probe and H2 probe; lane 4: annealed H1 and H2; lane 5: H1 probe, H2 probe, and target RNA; lane 6: H1, H2, and non-target RNA. The concentration of all components was 50 nM.
3.4. Optimization of CHA reaction conditions
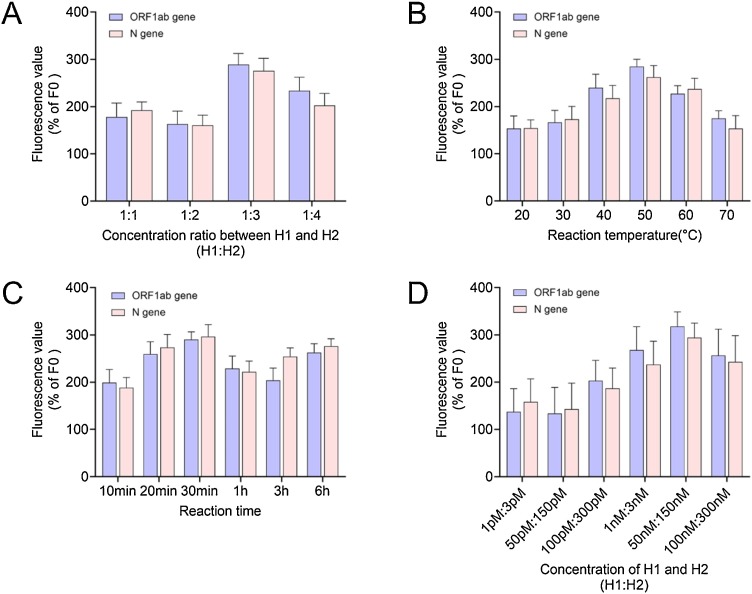
To optimize the H1:H2 concentration ratio, the fluorescence value of non-target RNA with concentration of 1 fM was set to 100 %(F0), and the relative fluorescence values (ΔF/F0) of reactions with different concentration ratios were calculated. The results, showing the average of three independent experiments, are shown in Fig. 6 A. The relative fluorescence value at 1:3 was highest. Therefore, we concluded that the optimal H1:H2 concentration ratio was 1:3. Using this ratio, we next optimized the reaction temperature. Similarly, the fluorescence value of non-target RNA with concentration of 1 fM was set to 100 %(F0), and calculated the relative fluorescence values (ΔF/F0) of reactions conducted at different temperatures. As shown in Fig. 6B, the relative fluorescence value at 50 °C was highest. Therefore, we concluded that the optimal reaction temperature was 50 °C. Importantly, our method can be performed efficiently at a constant temperature without the requirement for a thermal cycler, which is an advantage compared to RT-PCR. Using the reaction conditions determined above, we next investigated the optimal CHA reaction time. As shown in Fig. 6C, the relative fluorescence value at 30 min was highest. Finally, optimal concentration of hairpins DNA H1 probe and H2 probe were performed. As shown in Fig. 6D, the relative fluorescence value of concentration of hairpins DNA H1 probe and H2 probe with 50 nM:150 nM was highest.
Fig. 6.
Optimization of the CHA reaction.
(A) Optimization of the H2 concentration (50–200 nM) at a constant H1 concentration (50 nM) in a 2 h reaction at 50 °C; (B) Reaction temperature optimization using a 2 h reaction with 50 nM of H1 and 150 nM of H2; (C) Optimization of the CHA reaction time at 50 °C with 50 nM of H1 and 150 nM of H2; (D) Optimization of the concentrations of H1 and H2 in a 30 min reaction at 50 °C.
3.5. Sensitivity of the CHA-LFIA method
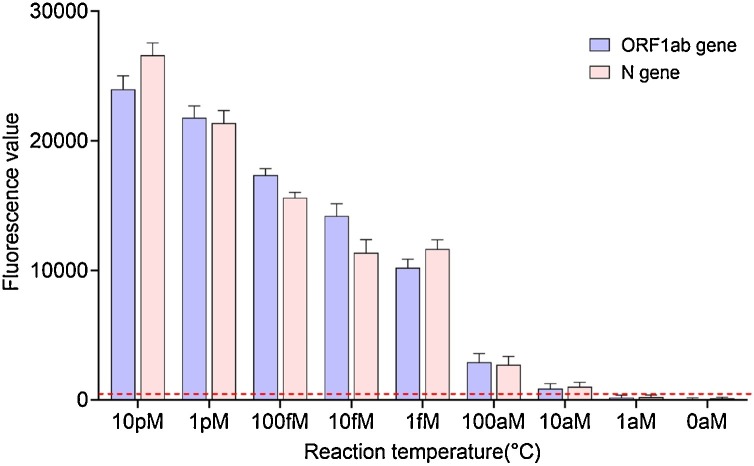
As previous studies have verified the sensitivity of the CHA system, [22,[30], [31], [32], [33], [34]] we focused on verifying the sensitivity of the combined CHA-LFIA method. In addition, previously reported measurements of CHA sensitivity were performed using TAE buffer, a relatively pure reaction medium. However, the CHA-LFIA method was designed to detect the presence of the SARS-CoV-2 genome in complex biological samples, including oropharyngeal swab specimens, sputum, saliva, and possibly even serum, plasma, and urine. The complexity of biological samples may have a greater impact on the detection outcome than TAE. Thus, we spiked an oropharyngeal swab sample solution from healthy people with target RNA, and determined the minimum detectable concentration of target RNA using the optimal conditions described above. As expected, the fluorescence value decreased as the target RNA concentration decreased (Fig. 7 ). The average fluorescence values of the control (non-target) RNA) were 88.24 ± 25.76 for ORF1ab gene and 129.12 ± 30.91 for N gene, respectively. Using a fluorescence value of 300(over the mean+ 3SD) as a threshold, the CHA-LFIA detection method had a minimum detectable target RNA concentration of 10 aM, consistent with previous studies [22,[31], [32], [33]].
Fig. 7.
Fluorescence kinetics of the CHA-LFIA method with various concentrations of target RNA in an oropharyngeal swab sample healthy person.
3.6. Specificity of the CHA-LFIA method
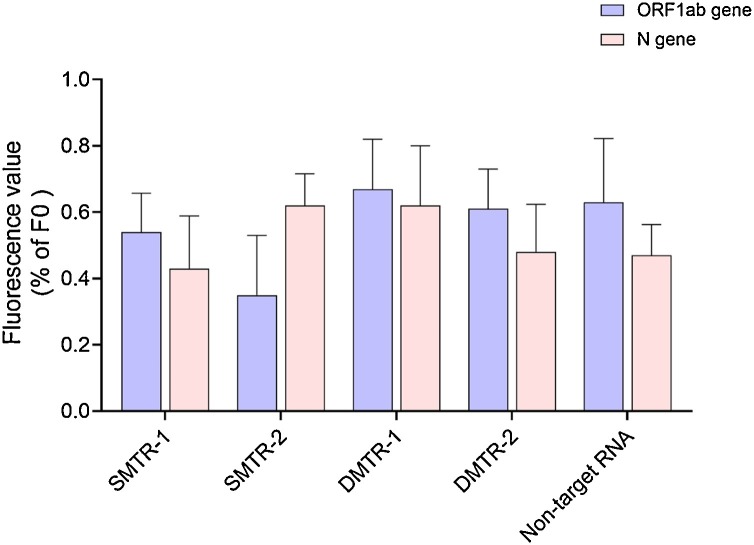
To verify the sequence specificity of the CHA-LFIA method, we examined whether mismatched sequences affected the detection results. Non-target RNA and single- and double-mismatched target RNA sequences (Table S1) were assessed by the CHA-LFIA method alongside the fully matched RNA target. The fluorescence values of the fully matched target RNAs for each gene were set to 100 % (F0) to calculate the relative fluorescence values (ΔF/F0) of the non-target RNA and single- and double-mismatched target RNA (Fig. 8 ). There are no significant differences among the non-target RNA, single- and double-mismatched target RNA groups. The fluorescence values of the non-target RNA, single- and double-mismatched target RNA less than 1% of target RNA. This suggests very high sequence specificity for the CHA reaction.
Fig. 8.
Sequence specificity analysis of the CHA-LFIA method.
The results are the average of three independent trials. SM, single-mismatched target RNA; DM, double-mismatched target RNA.
3.7. Detection of SARS-CoV-2 RNA reference material using the CHA-LFIA method
To determine the sensitivity of the CHA-LFIA method, we tested a five-fold dilution series of SARS-CoV-2 RNA reference material (Table 1 ). We set the cutoff value as 300 based on the fluorescence value of control (0 copies/mL), the lower limit of the detection sensitivity for the CHA-LIFA method is 4000 copies/mL. These results indicated that the sensitivity of detection of SARS-CoV-2 RNA reference material by CHA-LFIA method is approximately half an order of magnitude lower than that of RT-PCR. In view of a mean viral load of 1.4 × 106 copies/mL in the oropharyngeal swabs of patients with COVID-19 [[35], [36], [37]], our CHA-LFIA method is sufficiently sensitive to detect SARS-CoV-2 at an early stage of infection using oropharyngeal swab specimens.
Table 1.
Comparison of CHA-LFIA and RT-PCR for SARS-CoV-2 RNA reference material detection.
| Copies/mL | RT-PCR |
CHA-LFIA fluorescence values |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Result | Ct Value |
Orf1ab |
N |
||||||
| Orf1ab | N | Test 1 | Test 2 | Test 3 | Test 1 | Test 2 | Test 3 | ||
| 500,000 | Positive | 18.25 | 17.33 | 11,960 | 8950 | 8997 | 9463 | 8215 | 10,448 |
| 100,000 | Positive | 20.43 | 18.38 | 5669 | 4430 | 5401 | 4688 | 5805 | 3752 |
| 20,000 | Positive | 21.68 | 19.99 | 2319 | 2800 | 1921 | 1707 | 1695 | 2725 |
| 4000 | Positive | 29.62 | 28.50 | 892 | 598 | 734 | 523 | 817 | 825 |
| 800 | Positive | 36.59 | 35.63 | 192 | 131 | 72 | 167 | 100 | 198 |
| 160 | Negative | 39.16 | 38.32 | 180 | 125 | 123 | 102 | 99 | 186 |
| 0 | Negative | 0 | 0 | 95 | 69 | 79 | 93 | 133 | 140 |
3.8. Detection of SARS-CoV-2 in clinical samples using the CHA-LFIA method
We next determined whether the CHA-LFIA method was suitable for SARS-CoV-2 detection in clinical samples. Thirty oropharyngeal swab samples were tested using the CHA-LFIA method. Of these, 15 were confirmed to be SARS-CoV-2-positive and 15 as SARS-CoV-2 negative by RT-PCR (RT-PCR data of oropharyngeal swab samples are detailed in the supplemental material). Three independent tests were performed, and the results are shown in Table S4. At a fluorescence value threshold of 300, the CHA-LFIA results completely agreed with the RT-PCR results (Table 2 ).
Table 2.
Comparison of CHA-LFIA and RT-PCR for SARS-CoV-2 detection in 30 clinical samples.
| Samples | CHA-LFIA for Orf1ab |
CHA-LFIA for N |
||
|---|---|---|---|---|
| Positive | Negative | Positive | Negative | |
| RT-PCR-positive samples (n = 15) | 15 (100 %) | 0 (100 %) | 15 (100 %) | 0 (100 %) |
| RT-PCR-negative samples (n = 15) | 0 (100 %) | 15 (100 %) | 0 (100 %) | 15 (100 %) |
4. Conclusion
In summary, the present study describes a simple, highly sensitive, fast, and reliable method for the detection of SARS-CoV-2 RNA. By exploiting the signal amplification of the CHA reaction and the sensitivity of the lateral flow immunoassay strip for the detection of digoxigenin-biotin double-labeled double-stranded DNA, we achieved rapid and sensitive RNA detection with a low background signal. Our method avoids RNA isolation, PCR amplification, and elaborate data analysis, which typically takes 6–8 h. In contrast, the entire CHA-LFIA method, from nasopharyngeal sampling to obtaining test results, takes less than 90 min. Although a limited number of clinical samples were used in the present study, it demonstrates that the proposed strategy could be a convenient tool for detecting SARS-CoV-2 infection, as it requires minimal laboratory facilities and is relatively simple and inexpensive to perform. Such CHA-LFIA method can be very helpful in providing standardized and cost-effective screening at a larger scale screening of SARS-CoV-2 infection.
CRediT authorship contribution statement
Mingyuan Zou: Methodology, Investigation, Visualization, Writing - original draft. Feiya Su: Methodology, Data curation. Rui Zhang: Validation. Xinglu Jiang: Software, Validation, Formal analysis. Han Xiao: Validation, Formal analysis. XueJiao Yan: Funding acquisition, Methodology, Formal analysis. Chuankun Yang: Methodology. Xiaobo Fan: Conceptualization, Resources, Funding acquisition, Methodology, Formal analysis. Guoqiu Wu: Conceptualization, Resources, Funding acquisition, Project administration, Supervision, Writing - review & editing.
Declaration of Competing Interest
The authors declare no conflict of interest.
Acknowledgements
This work was supported by grants from the National Science and Technology Major Project (No. 2020ZX09201015), National Natural Science Foundation of China (No. 81773624 & 81603016 & 81600415 & 81900453), National Natural Science Foundation of Jiangsu Province (No. BE2017746 & BK20160706), and the Fundamental Research Funds for the Central Universities (2242020K40130 & 2242020K10020).
Biographies
Mingyuan Zou is a Ph.D candidate under the supervision of Prof. Guoqiu Wu. His research interests include the Pathogen detection and Biotechnology.
Feiya Su received her Bachelor degree in 2019 at Southeast University in China. She is currently a master candidate under the supervision of Prof Guoqiu Wu and Dr Xiaobo Fan.
Rui Zhang received her Bachelor degree in 2018 at Henan University of Science and Technology in China. She is currently a master candidate under the supervision of Prof Guoqiu Wu and Dr Xiaobo Fan.
Xinglu Jiang is a Ph.D candidate under the supervision of Prof. Guoqiu Wu. His research interests include the development of therapeutic agents for clinic pathogens and cancer.
Han Xiao: is a Ph.D candidate under the supervision of Prof. Guoqiu Wu. Her research interests include the development of therapeutic agents for clinic pathogens and Nanomedicine.
XueJiao Yan received her Ph.D degree in 2016 under the supervision of Prof. Naifeng Liu. She is a physician at The Affiliated Changzhou No. 2 People's Hospital of Nanjing Medical University. Her research interests include nanomaterials and virus detection.
Chuankun Yang is technician in Center of Clinical Laboratory Medicine of Zhongda Hospital of Southeast University in China. His research interests include virus detection and biomedical engineering.
Xiaobo Fan received his Ph.D degree in 2014 under the supervision of Prof. Michael Wink at the University of Heidelberg in Germany. He is currently a lecturer at Southeast University. His research interests include the development of probes and therapeutic agents for clinic pathogens.
Guoqiu Wu received her Ph.D degree in 2003 under the supervision of Prof. Zilong Shen at the China Pharmaceutical University in China. He is currently a professor at Southeast University and the director of diagnostics department at ZhongDa Hospital. His research interests include the development of probes and therapeutic agents for clinic pathogens and tumor.
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.snb.2021.129899.
Appendix A. Supplementary data
The following is Supplementary data to this article:
References
- 1.Lauxmann M.A., Santucci N.E., Autran-Gomez A.M. The SARS-CoV-2 coronavirus and the COVID-19 outbreak. Int. Braz. J. Urol. 2020;46 doi: 10.1590/S1677-5538.IBJU.2020.S101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Buder F., Hitzenbichler F., Ehrenstein B., et al. The outbreak of COVID-19 in China. Internist (Berl) 2020 doi: 10.1007/s00108-020-00833-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tang S., Mao Y., Jones R.M., et al. Aerosol transmission of SARS-CoV-2? Evidence, prevention and control. Environ. Int. 2020;144 doi: 10.1016/j.envint.2020.106039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Koo J.R., Cook A.R., Park M., et al. Interventions to mitigate early spread of SARS-CoV-2 in Singapore: a modelling study. Lancet Infect. Dis. 2020;20(6):678–688. doi: 10.1016/S1473-3099(20)30162-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ooi E.E., Low J.G. Asymptomatic SARS-CoV-2 infection. Lancet Infect. Dis. 2020 doi: 10.1016/S1473-3099(20)30460-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gao M., Yang L., Chen X., et al. A study on infectivity of asymptomatic SARS-CoV-2 carriers. Respir. Med. 2020;169 doi: 10.1016/j.rmed.2020.106026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng Y., Fu J., Yu X., et al. Should computed tomography (CT) be used as a screening or follow-up tool for asymptomatic patients with SARS-CoV-2 infection? Quant. Imaging Med. Surg. 2020;10(5):1150–1152. doi: 10.21037/qims.2020.04.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu R., Du M., Li L., et al. CT imaging of one extended family cluster of corona virus disease 2019 (COVID-19) including adolescent patients and "silent infection". Quant. Imaging Med. Surg. 2020;10(3):800–804. doi: 10.21037/qims.2020.02.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang Y.X.J.A. Call for caution in extrapolating chest CT sensitivity for COVID-19 derived from hospital data to patients among general population. Quant. Imaging Med. Surg. 2020;10(3):798–799. doi: 10.21037/qims.2020.03.04. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bernheim A., Mei X., Huang M., et al. Chest CT findings in coronavirus Disease-19 (COVID-19): relationship to duration of infection. Radiology. 2020;295(3) doi: 10.1148/radiol.2020200463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wu J., Liang J., Zhou H., et al. Clinical features and outcomes of asymptomatic cases of SARS-CoV-2 infection. J. Infect. 2020;81(1):e102–e103. doi: 10.1016/j.jinf.2020.04.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang C., Wen T., Shi F.J., et al. Rapid detection of IgM antibodies against the SARS-CoV-2 virus via colloidal gold nanoparticle-based lateral-flow assay. ACS Omega. 2020;5(21):12550–12556. doi: 10.1021/acsomega.0c01554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mathuria J.P., Yadav R., Rajkumar Laboratory diagnosis of SARS-CoV-2 - A review of current methods. J. Infect. Public Health. 2020 doi: 10.1016/j.jiph.2020.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shih H.I., Wu C.J., Tu Y.F., et al. Fighting COVID-19: a quick review of diagnoses, therapies, and vaccines. Biomed. J. 2020 doi: 10.1016/j.bj.2020.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Waggoner J.J., Stittleburg V., Pond R., et al. Triplex real-time RT-PCR for severe acute respiratory syndrome coronavirus 2. Emerg. Infect. Dis. 2020;26(7) doi: 10.3201/eid2607.201285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hogan C.A., Sahoo M.K., Huang C., et al. Comparison of the Panther Fusion and a laboratory-developed test targeting the envelope gene for detection of SARS-CoV-2. J. Clin. Virol. 2020;127 doi: 10.1016/j.jcv.2020.104383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramdas K., Darzi A., Jain S. ’Test, re-test, re-test’: using inaccurate tests to greatly increase the accuracy of COVID-19 testing. Nat. Med. 2020 doi: 10.1038/s41591-020-0891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li Y., Yao L., Li J., et al. Stability issues of RT-PCR testing of SARS-CoV-2 for hospitalized patients clinically diagnosed with COVID-19. J. Med. Virol. 2020;92(7):903–908. doi: 10.1002/jmv.25786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yin P., Choi H.M., Calvert C.R., et al. Programming biomolecular self-assembly pathways [J] Nature. 2008;451(7176):318–322. doi: 10.1038/nature06451. [DOI] [PubMed] [Google Scholar]
- 20.Bi S., Yue S., Wu Q., et al. Triggered and catalyzed self-assembly of hyperbranched DNA structures for logic operations and homogeneous CRET biosensing of microRNA. Chem Commun. 2016;52(31):5455–5458. doi: 10.1039/c6cc01308b. [DOI] [PubMed] [Google Scholar]
- 21.Zang Y., Lei J., Hao Q., et al. CdS/MoS2 heterojunction-based photoelectrochemical DNA biosensor via enhanced chemiluminescence excitation. Biosens. Bioelectron. 2016;77:557–564. doi: 10.1016/j.bios.2015.10.010. [DOI] [PubMed] [Google Scholar]
- 22.Duan Z., Li Z., Dai J., et al. Nucleotide base analog pyrrolo-deoxycytidine as fluorescent probe signal for enzyme-free and signal amplified nucleic acids detection. Talanta. 2017;164:34–38. doi: 10.1016/j.talanta.2016.10.079. [DOI] [PubMed] [Google Scholar]
- 23.Karunanayake Mudiyanselage A., Yu Q., Leon-Duque M.A., et al. Genetically encoded catalytic hairpin assembly for sensitive RNA imaging in live cells. J. Am. Chem. Soc. 2018;140(28):8739–8745. doi: 10.1021/jacs.8b03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Zhang H., Wang K., Bu S., et al. Colorimetric detection of microRNA based on DNAzyme and nuclease-assisted catalytic hairpin assembly signal amplification. Mol. Cell. Probes. 2018;38:13–18. doi: 10.1016/j.mcp.2018.02.002. [DOI] [PubMed] [Google Scholar]
- 25.Zadeh J.N., Steenberg C.D., Bois J.S., et al. NUPACK: analysis and design of nucleic acid systems. J. Comput. Chem. 2011;32(1):170–173. doi: 10.1002/jcc.21596. [DOI] [PubMed] [Google Scholar]
- 26.Liu J., Zhang Y., Xie H., et al. Applications of catalytic hairpin assembly reaction in biosensing. Small. 2019;15(42) doi: 10.1002/smll.201902989. [DOI] [PubMed] [Google Scholar]
- 27.Demir A.B., Benvenuto D., Abacioglu H., et al. Identification of the nucleotide substitutions in 62 SARS-CoV-2 sequences from Turkey. Turk. J. Biol. 2020;44(3):178–184. doi: 10.3906/biy-2005-69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ugurel O.M., Ata O., Turgut-Balik D. An updated analysis of variations in SARS-CoV-2 genome. Turk. J. Biol. 2020;44(3):157–167. doi: 10.3906/biy-2005-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wang C., Liu Z., Chen Z., et al. The establishment of reference sequence for SARS-CoV-2 and variation analysis. J. Med. Virol. 2020;92(6):667–674. doi: 10.1002/jmv.25762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wu C., Cansiz S., Zhang L., et al. A nonenzymatic hairpin DNA cascade reaction provides high signal gain of mRNA imaging inside live cells. J. Am. Chem. Soc. 2015;137(15):4900–4903. doi: 10.1021/jacs.5b00542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yao J., Zhang Z., Deng Z., et al. An enzyme free electrochemical biosensor for sensitive detection of miRNA with a high discrimination factor by coupling the strand displacement reaction and catalytic hairpin assembly recycling. Analyst. 2017;142(21):4116–4123. doi: 10.1039/c7an01224a. [DOI] [PubMed] [Google Scholar]
- 32.Dai W., Zhang J., Meng X., et al. Catalytic hairpin assembly gel assay for multiple and sensitive microRNA detection. Theranostics. 2018;8(10):2646–2656. doi: 10.7150/thno.24480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu X., Fan J., Duan B., et al. Single-molecule catalytic hairpin assembly for rapid and direct quantification of circulating miRNA biomarkers. Anal. Chim. Acta. 2018;1042:109–115. doi: 10.1016/j.aca.2018.08.037. [DOI] [PubMed] [Google Scholar]
- 34.Yin C., Wu Y., Li X., et al. Highly selective, Naked-Eye, and trace discrimination between perfect-match and mismatch sequences using a plasmonic nanoplatform. Anal. Chem. 2018;90(12):7371–7376. doi: 10.1021/acs.analchem.8b00756. [DOI] [PubMed] [Google Scholar]
- 35.Mawaddah A., Gendeh H.S., Lum S.G., et al. Upper respiratory tract sampling in COVID-19. Malays. J. Pathol. 2020;42(1):23–35. [PubMed] [Google Scholar]
- 36.Huang Y., Chen S., Yang Z., et al. SARS-CoV-2 viral load in clinical samples from critically ill patients. Am. J. Respir. Crit. Care Med. 2020;201(11):1435–1438. doi: 10.1164/rccm.202003-0572LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan Y., Zhang D., Yang P., et al. Viral load of SARS-CoV-2 in clinical samples. Lancet Infect. Dis. 2020;20(4):411–412. doi: 10.1016/S1473-3099(20)30113-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.