Abstract
Partial replacement of α-amino acid residues with β-amino acid residues has been established as a strategy for preserving target-engagement by helix-forming polypeptides while altering other properties. The impact of β-residue incorporation within polypeptides that adopt less regular conformations, however, has received less attention. The HRC domains of fusion glycoproteins from pathogenic paramyxoviruses contain a segment that must adopt an extended conformation in order to co-assemble with the HRN domain in the post-fusion state and drive merger of the viral envelope with a target cell membrane. Here we examine the impact of single α-to-β substitutions within this extended N-terminal segment of an engineered HRC peptide designated VIQKI. Stabilities of hexameric co-assemblies formed with the native HPIV3 HRN have been evaluated, the structures of five co-assemblies have been determined, and antiviral efficacies have been measured. Many sites within the extended segment show functional tolerance of α-to-β substitution. These results offer a basis for future development of paramyxovirus infection inhibitors with novel biological activity profiles, possibly including resistance to proteolysis.
Keywords: alpha/beta-peptide, beta amino acid, HPIV3, entry inhibitor, fusion glycoprotein, viral entry
The disruption of interactions within or between proteins is an attractive basis for drug design. This strategy requires molecules that can engage large surfaces on target proteins, a goal that is generally difficult to achieve with small molecules (MW<1000).1-3 Clinically useful inhibitors of pathogenic protein-protein interactions are typically engineered proteins or large peptides. Antibodies and other large proteins are expensive to produce.4 Smaller polypeptides offer the advantage of chemical synthesis but suffer from rapid degradation by endogenous proteases.1-3 One strategy for retaining the benefits of polypeptides while minimizing proteolytic susceptibility is to incorporate non-natural subunits into the peptidic backbone.5-12 The challenge inherent in this approach is to introduce a sufficient number and distribution of backbone modifications to hinder protease action without loss of target protein engagement. We have shown that the recognition properties of an α-helix can be retained after periodic replacement of α-amino acid residues with β residues.13-22 The resulting “α/β-peptides” adopt an α-helix-like conformation but resist enzymatic degradation. Less is known about the impact of β residue incorporation in non-helical segments.
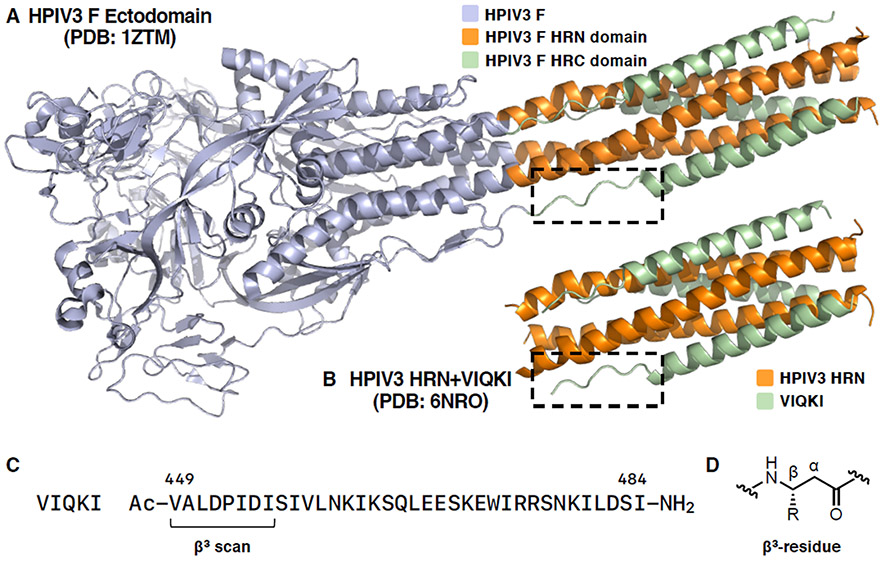
Paramyxoviruses, such as parainfluenza, measles, Nipah, and Hendra viruses, are negative-strand RNA viruses that negatively impact global human health. Here, we describe fundamental studies of α-to-β modifications in the context of an extended segment that plays a critical role in a paramyxovirus infection mechanism. We focus on an eight-residue portion of the fusion (F) glycoprotein of human parainfluenza virus 3 (HPIV3). The F protein orchestrates fusion of the HPIV3 envelope and the outer membrane of a target cell. This process involves a complex set of structural transitions within an F trimer that ultimately leads to the formation of a “six-helix bundle” (6HB) assembly between N-terminal heptad repeat (HRN) and C-terminal heptad repeat (HRC) domains of F.23-25 In the post-fusion 6HB state, the N-terminal portion of the HRC domain (residues 449-456) adopts an extended conformation, while the majority of the HRC domain residues (457-484) form an α-helix (Figure 1A).26
Figure 1.
(A)HPIV3 F ectodomain in postfusion conformation (PDB 1ZTM), (B) VIQKI co-assembled with HPIV3-HRN (PDB 6NRO), (C) sequence of VIQKI, (D) structure of a β3-residue.
The few previous evaluations of α-to-β modification in extended peptide ligands had variable outcomes. For example, three groups have explored systematic “β scans” of short peptide antigens (8 or 9 residues) that bind to class I major histocompatibility complexes (MHC I).27-29 In each system, MHC binding was retained for β substitution at some positions, but most of the resulting peptide-MHC complexes were not effective in stimulating T cells that respond to the natural antigen. A subsequent study found greater functional tolerance of single α-to-β replacements in a 17-mer antigen that binds to a class II MHC.30 A systematic α-to-β analogue series was recently reported for a 10-residue phosphopeptide that binds in an extended conformation to 14-3-3 proteins.31 β substitution near either end caused modest declines in affinity (~10-fold), but more severe losses in affinity were observed for substitutions closer to the center of the peptide.
Based on these precedents, it was not clear whether α-to-β substitutions would be structurally or functionally tolerated within the extended portion of an HPIV3 inhibitor. We were motivated to proceed, however, by two considerations. First, peptide binding to an MHC or a 14-3-3 protein involves multiple H-bonds between the backbone of the peptide ligand and the protein partner. In contrast, there are relatively few H-bonds between the extended segment of the HPIV3 HRC domain and the HRN trimer core in the post-fusion 6HB state of F. Second, coronaviruses (CoVs) feature even longer extended segments within the post-fusion HRC domains of their fusion glycoproteins (S) than do paramyxoviruses, but the backbones of these extended CoV segments form relatively few H-bonds with the HRN trimer core.32-34 α-to-β replacement seems likely to disrupt backbone H-bonding in an extended conformation; therefore, we predicted that this type of modification might be better tolerated in an extended segment of a viral fusion protein HRC domain than in other linear peptide epitopes.
METHODS
Our study began with a previously reported peptide derived from the HRC domain of HPIV3, designated VIQKI (Figure 1C), which blocks infection by both HPIV3 and respiratory syncytial virus (RSV).35-37 These two viruses are leading causes of fatal lower respiratory infections in young children.38-40 A crystal structure of VIQKI co-assembled with HPIV3-HRN (Figure 1B) revealed a 6HB assembly similar to the HRC/HRN assembly in the post-fusion conformation of the HPIV3 F protein (Figure 1A).35 As expected, two distinct conformations were observed within the VIQKI peptide, an α-helix spanning residues corresponding to 457–484 of the F protein, and an extended conformation for residues corresponding to 449–456. Our studies focused on backbone modifications in the extended portion of VIQKI (Figure 1, dashed boxes).
A set of VIQKI analogues was synthesized in which each of the eight N-terminal residues was replaced individually with the homologous β3-amino acid residue (Figure 1D). Thus, each replacement preserved the side chain found in VIQKI but introduced an extra CH2 unit into the backbone. The effect of each single α-to-β3 substitution on assembly stability with HPIV3-HRN was examined using variable-temperature circular dichroism (VT-CD) spectroscopy. When combined with a 51-residue peptide corresponding to the HPIV3-HRN domain, each α/β-VIQKI variant formed a helical assembly, as detected by CD minima at 208 and 222 nm. Each assembly exhibited a cooperative thermal transition at 222 nm, which allowed us to determine apparent thermal denaturation (Tm,app) values, based on the mid-point of each transition (Table 1). For each α/β-VIQKI variant, a ΔTm,app value was calculated based on comparison with Tm,app for VIQKI itself [ΔTm,app = Tm,app (HPIV3-HRN+VIQKI β3-variant) – Tm,app (HPIV3-HRN+VIQKI)].
Table 1.
Apparent thermal denaturation temperatures (Tm,app) of co-assemblies formed between HPIV3-HRN+VIQKI β3-variant pairs
| Peptide | Tm,app (°C)a | ΔTm,appb |
|---|---|---|
| VIQKI | 88.6±0.4 | |
| VIQKI-V449(β3V) | 89.0±0.8 | +0.4 |
| VIQKI-A450(β3A) | 89.2±1.0 | +0.6 |
| VIQKI-L451 (β3L) | 88.7±0.8 | +0.1 |
| VIQKI-D452(β3D) | 89.1±0.4 | +0.5 |
| VIQKI-P453(β3P) | 86.1±0.9 | −2.5 |
| VIQKI-I454(β3I) | 86.7±1.0 | −1.9 |
| VIQKI-D455(β3D) | 81.5±0.5 | −7.1 |
| VIQKI-I456(β3I) | 86.2±1.1 | −2.4 |
1:1 mixture of Inhibitor (50 μM) and HRN (50 μM) peptides. Tm,app values were determined as mean values of three replicates ± standard deviation.
ΔTm.app = Tm,app (HPIV3-HRN+VIQKI β3-variant) - Tm,app (HPIV3-HRN+VIQKI)
The VT-CD analysis revealed that most α-to-β replacements in the N-terminal region of VIQKI caused little or no change in ΔTm,app. Since omission of the three N-terminal residues (N449-L451) has little effect HRC-HRN assembly stability,35 it is perhaps not surprising that the β3 residue was well tolerated at each of these positions. The D452-I456 segment, on the other hand, was shown by truncation studies to be crucial for assembly stability,35 and it was therefore not expected that only one α-to-β replacement among these five residues, D455(β3D), would be highly destabilizing.
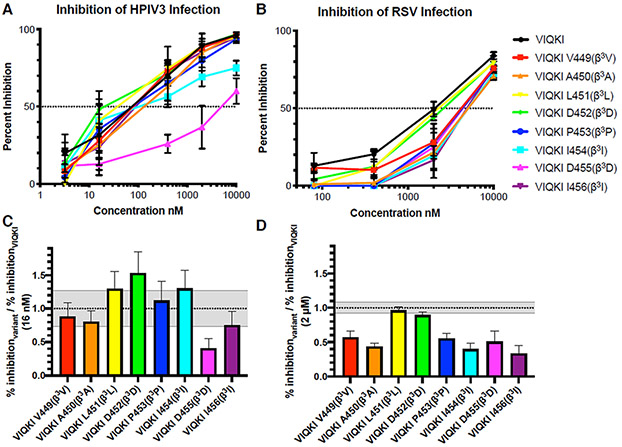
To assess the functional impact of each single α-to-β substitution, we evaluated inhibitory activity toward HPIV3 infection using plaque reduction assays. Hep2 cell monolayers were infected with engineered recombinant HPIV3 that expresses green fluorescent protein (GFP) in the presence of various concentrations of each VIQKI variant, and the extent of infection was determined by quantification of fluorescence. The results are provided in two formats. Figure 2A shows extent of inhibition of infection as a function of VIQKI variant concentration. Figure 2C shows the ratio of extent of inhibition for each variant relative to inhibition for VIQKI itself at the experimental concentration closest to the IC50 of VIQKI (16 nM). Consistent with the VT-CD results, the functional data show that seven of the eight β-substituted peptides display substantial antiviral activity. Activity was lowest for the substitution that was most destabilizing to HRC-HRN assembly, D455(β3D).
Figure 2.
Anti-viral efficacy of VIQKI β3-variants against HPIV3 or RSV. Peptide activity against HPIV3 (left) or RSV (right) were determined by plaque reduction assay in infected Hep-2 cell monolayers. Data are shown as (A and B) percent inhibition of infection as a function of inhibitor concentration and (C and D) a ratio of the percent inhibition of each β3 variant with the percent inhibition of VIQKI. Gray zones represent the standard deviation of the VIQKI control. Data are expressed as mean ± standard deviation (n=3 separate experiments).
The high sensitivity of D455 to β residue replacement, in terms of both assembly stability and antiviral activity, was intriguing because of a critical role suggested by previous structural studies for the D455 side chain. The carboxylate of D455 in the post-fusion 6HB form of the HPIV3 F protein engages in a H-bond with the backbone amide N-H of S457.26 The same H-bond is seen in the co-crystal structure of VIQKI with the HPIV3 HRN domain.35 This i,i+2 interaction appears to serve as an N-terminal cap41 for the α-helical secondary structure that starts at S457 and extends to the C-terminus.
To try to gain insight into the structural impact of the α-to-β substitutions, we co-crystallized five VIQKI variants with the HPIV3-HRN peptide, D452(β3D), P453(β3P), I454(β3I), D455(β3D) and I456(β3I). Each structure was solved by molecular replacement using a search model based on the HPIV3-HRN+VIQK1 structure (PDB 6NRO).35 Chains A and B from 6NRO were truncated to include residues 153–173 (chain A) and 460–480 (chain B). All hydrogen atoms and side chains were removed prior to molecular replacement. Crystallographic models were refined to 1.70 Å [HPIV3-HRN+VIQKI-D452(β3D), (PDB 6PZ6)]; 1.87 Å [HPIV3-HRN+VIQKI-P453(β3P), (PDB 6PRL)]; 1.49 Å [HPIV3-HRN+VIQKI-I454(β3I), (PDB 6VAS)]; 1.79 Å [HPIV3-HRN+VIQKI-D455(β3D), (PDB 6PYQ)]; and 2.17 Å [HPIV3-HRN+VIQKI-I456(β3I), (PDB 6V3V)].
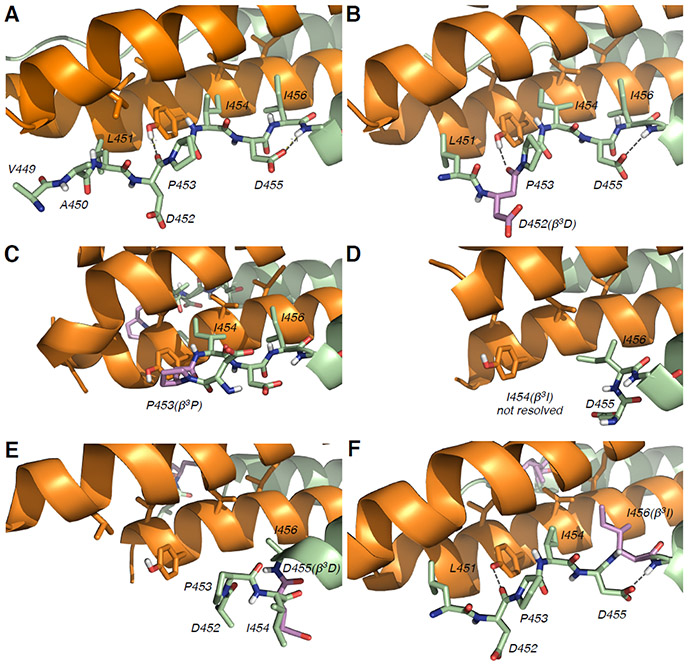
In each of the five new structures, the single β3 residue substitution within the extended N-terminal segment did not affect the α-helical conformation of residues 457–484. However, the impact of α-to-β3 substitution on the conformation of the N-terminal segment differed among the five variants (Figure 3).
Figure 3.
X-ray crystal structures of VIQKI and β3-variants co-assembled with HPIV3-HRN. (A) HPIV3-HRN+VIQKI, PDB:6NRO; (B) HPIV3-HRN+VIQKI-D452(β3D), (PDB:6PZ6); (C) HPIV3-HRN+VIQKI-P453(β3P), (PDB:6PRL); (D) HPIV3-HRN+VIQKI-I454(β3I), (PDB:6VAS); (E) HPIV3-HRN+VIQKI-D455(β3D), (PDB:6PYQ); (F) HPIV3-HRN+VIQKI-I456(β3I), (PDB:6V3V). HPIV3-HRN (orange), VIQKI and β3-variants (green) with β3-residues highlighted in violet.
In three of the five co-crystal structures, those containing P453(β3P), I454(β3I), or D455(β3D) substitutions, the backbone modification resulted in a significant remodeling of the extended conformation in the N-terminal region relative to VIQKI. For VIQKI-P453(β3P), contacts involving the I454 and I456 side chains and hydrophobic residues from the HPIV3-HRN trimer were retained, as was the helix capping interaction of D455. However, this substitution nucleated a hairpin turn that reoriented residues 449–452, with loss of contacts observed between these residues in VIQKI and HPIV3-HRN (Figures 3C, 4B). Since previous work showed that contacts involving residues 449–451 do not significantly contribute to six-helix bundle stability,35 this distortion may reflect effects of packing with lattice neighbors. For VIQKI-I454(β3I), only residues 455–484 could be resolved. No significant electron density was observed for the β3I residue or residues 449–453.
Figure 4.
2mFo-DFc weighted electron density maps for β3-residues in (A) HPIV3-HRN+VIQKI-D452(β3D), (B) HPIV3-HRN+VIQKI-P453(β3P), (C) HPIV3-HRN+VIQKI-D455(β3D), (D) HPIV3-HRN+VIQKI-I456(β3I). β3-residues are shown in violet.
In the co-assembly of VIQKI-D455(β3D) and HPIV3-HRN, residues 452–484 of the VIQKI variant were resolved. However, the transition from extended to α-helical conformation, which occurred at S457 in the HPIV3-HRN+VIQKI structure, was not observed for the D455(β3D) variant. Instead, VIQKI-D455(β3D) adopted a helical conformation from residue 453 to 484 (Figures 3E, 4C). We hypothesize that the additional backbone carbon atom between the carboxylate side chain of β3D455 and the backbone amide N-H of S457 in this variant, relative to VIQKI itself, interferes with the helix-capping interaction involving the side chain of D455 in VIQKI. This conformational rearrangement in the N-terminal segment may explain the loss of 6HB stability (Table 1) and diminished antiviral activity (Figure 2) observed for VIQKI-D455(β3D).
In the crystal structures of VIQKI-D452(β3D) and VIQKI-I456(β3I) co-assembled with HPIV3-HRN, the extended conformation observed for VIQKI itself was retained (Figures 3B, 3F), although in both variants the two terminal residues, V449 and A450, could not be resolved. In both new structures, key interactions between the N-terminal segment of the inhibitor and the HPIV3-HRN core were preserved. These interactions include contacts of side chains from L451, I454, and I456 (or β3I456) with hydrophobic residues in the HRN trimer, as well as H-bonds between the backbone carbonyl of D452 with the side chain hydroxyl of HRN residue Y178. The helix capping interaction was present in both of these VIQKI variants.
Although assessing activity of the β-substituted analogues of VIQKI toward RSV was tangential to the main goals of this work, we evaluated these peptides for the ability to inhibit RSV infection of Hep-2 cells. Figures 2B,D show that the effect of α-to-β substitutions on efficacy against RSV was more pronounced than the effect on efficacy against HPIV3. Most VIQKI variants were significantly less active relative to VIQKI itself. However, variants L451(β3L) and D452(β3D) seemed to match VIQKI in efficacy against RSV infection. The N-terminal segment of VIQKI adopts different conformations when co-assembled with HPIV3-HRN vs. RSV-HRN.30 Residues P453-I456 are part of the extended segment in the assembly with HPIV3-HRN, but these residues have been incorporated into the α-helix in the assembly with RSV-HRN. It is noteworthy that only two variants, L451(β3L) and D452(β3D), maintained inhibition of infection by both viruses.
We have used a peptide that inhibits HPIV3 infection via a well-characterized mode of target engagement to explore the effects of replacing α-amino acid residues by homologous β3 residues within a segment that is extended in the bioactive conformation. This modification preserves the side chain but alters the backbone via introduction of a CH2 unit. Based on previous studies involving β replacements in extended peptide ligands, it could not have been predicted that most sites in the extended portion of the α-peptide inhibitor VIQKI would tolerate α-to-β3 substitution while retaining the ability to inhibit HPIV3 infection. We hypothesize that the extended segments of viral HRC domains in the 6HB assemblies of post-fusion forms may generally prove to be tolerant of backbone modifications because the backbone amide groups in these segments form few H-bonds with HRN domain trimers. This work offers a step toward development of protease-resistant dual HPIV3/RSV inhibitors that might address life-threatening child-hood infections for which there are no current drugs. (We have not examined the effects of proteases on the β-substituted peptides discussed here, because, with only one backbone modification, these 36-mers will undoubtedly be susceptible at most sites of potential cleavage.) In addition, given the presence of long extended segments in the post-fusion HRC/HRN assemblies formed by coronavirus fusion glycoproteins,32-34 these results encourage application of the backbone modification strategy to inhibitor development directed toward these pathogens.
Supplementary Material
ACKNOWLEDGMENT
This work was supported by NIH/NIAID R01AI114736 to AM and NIH/NIAID R01AI121349 to MP. VKO was funded in part by a Ruth L. Kirschstein National Research Service Award from the NIH (F32GM122263). This research used resources of the Advanced Photon Source, a U.S. Department of Energy (DOE) Office of Science User Facility operated for the DOE Office of Science by Argonne National Laboratory under Contract No. DE-AC02-06CH11357. Use of the LS-CAT Sector 21 was supported by the Michigan Economic Development Corporation and the Michigan Technology Tri-Corridor (Grant 085P1000817). We thank Craig Bingman of the University of Wisconsin Crystallography Core facility for organization of x-ray data collection.
Footnotes
Supporting Information
The Supporting Information is available free of charge on the ACS Publications website at DOI: 10.1021/acsinfecdis.0c00385
Additional figures, synthetic procedures, and characterization data for all synthesized peptides, as well as protocols for plaque reduction and thermal denaturation assays (PDF)
REFERENCES
- (1).Fosgerau K; Hoffmann T Peptide Therapeutics: Current Status and Future Directions. Drug Discov. Today 2015, 20 (1), 122–128. [DOI] [PubMed] [Google Scholar]
- (2).Lau JL; Dunn MK Therapeutic Peptides: Historical Perspectives, Current Development Trends, and Future Directions. Bioorg. Med. Chem 2018, 26 (10), 2700–2707. [DOI] [PubMed] [Google Scholar]
- (3).Henninot A; Collins JC; Nuss JM The Current State of Peptide Drug Discovery: Back to the Future? J. Med. Chem 2018, 61 (4), 1382–1414. [DOI] [PubMed] [Google Scholar]
- (4).Elgundi Z; Reslan M; Cruz E; Sifniotis V; Kayser V The State-of-Play and Future of Antibody Therapeutics. Advanced Drug Delivery Reviews 2017, 122 (C), 2–19. [DOI] [PubMed] [Google Scholar]
- (5).Nowick JS; Lam KS; Khasanova TV; Kemnitzer WE; Maitra S; Mee HT; Liu R An Unnatural Amino Acid That Induces Beta-Sheet Folding and Interaction in Peptides. J. Am. Chem. Soc 2002, 124 (18), 4972–4973. [DOI] [PubMed] [Google Scholar]
- (6).Wang D; Chen K; Kulp Iii JL; Arora PS Evaluation of Biologically Relevant Short Alpha-Helices Stabilized by a Main-Chain Hydrogen-Bond Surrogate. J. Am. Chem. Soc 2006, 128 (28), 9248–9256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (7).Goodman CM; Choi S; Shandler S; DeGrado WF Foldamers as Versatile Frameworks for the Design and Evolution of Function. Nat. Chem. Biol 2007, 3 (5), 252–262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (8).Henchey LK; Jochim AL; Arora PS Contemporary Strategies for the Stabilization of Peptides in the A-Helical Conformation. Curr. Opin. Chem. Biol 2008, 12 (6), 692–697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (9).Azzarito V; Long K; Murphy NS; Wilson AJ Inhibition of A-Helix-Mediated Protein–Protein Interactions Using Designed Molecules. Nat. Chem 2013, 5 (3), 161–173. [DOI] [PubMed] [Google Scholar]
- (10).Barnard A; Long K; Martin HL; Miles JA; Edwards TA; Tomlinson DC; Macdonald A; Wilson AJ Selective and Potent Proteomimetic Inhibitors of Intracellular Protein-Protein Interactions. Angew. Chem. Int. Ed. 2015, 54 (10), 2960–2965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (11).Wu H; Qiao Q; Teng P; Hu Y; Antoniadis D; Zuo X; Cai J New Class of Heterogeneous Helical Peptidomimetics. Org. Lett 2015, 17 (14), 3524–3527. [DOI] [PubMed] [Google Scholar]
- (12).Bolarinwa O; Zhang M; Mulry E; Lu M; Cai J Sul-fono-ɣ-AA Modified Peptides That Inhibit HIV-1 Fusion. Org. Biomol. Chem 2018, 16 (42), 7878–7882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (13).Horne WS; Johnson LM; Ketas TJ; Klasse PJ; Lu M; Moore JP; Gellman SH Structural and Biological Mimicry of Protein Surface Recognition by Alpha/Beta-Peptide Foldamers. Proc. Natl. Acad. Sci. USA 2009, 106 (35), 14751–14756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (14).Johnson LM; Horne WS; Gellman SH Broad Distribution of Energetically Important Contacts Across an Extended Protein Interface. J. Am. Chem. Soc 2011, 133 (26), 10038–10041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (15).Boersma MD; Haase HS; Peterson-Kaufman KJ; Lee EF; Clarke OB; Colman PM; Smith BJ; Horne WS; Fairlie WD; Gellman SH Evaluation of Diverse α/β-Backbone Patterns for Functional α-Helix Mimicry: Analogues of the Bim BH3 Domain. J. Am. Chem. Soc 2012, 134 (1), 315–323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Johnson LM; Mortenson DE; Yun HG; Horne WS; Ketas TJ; Lu M; Moore JP; Gellman SH Enhancement of α-Helix Mimicry by an α/β-Peptide Foldamer via Incorporation of a Dense Ionic Side-Chain Array. J. Am. Chem. Soc 2012, 134 (17), 7317–7320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (17).Smith BJ; Lee EF; Checco JW; Evangelista M; Gellman SH; Fairlie WD Structure-Guided Rational Design of α/β-Peptide Foldamers with High Affinity for BCL-2 Family Prosurvival Proteins. ChemBioChem 2013, 14 (13), 1564–1572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (18).Cheloha RW; Maeda A; Dean T; Gardella TJ; Gellman SH Backbone Modification of a Polypeptide Drug Alters Duration of Action in Vivo. Nat. Biotechnol 2014, 32 (7), 653–655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Johnson LM; Barrick S; Hager MV; McFedries A; Homan EA; Rabaglia ME; Keller MP; Attie AD; Saghatelian A; Bisello A; Gellman SH A Potent α/β-Peptide Analogue of GLP-1 with Prolonged Action in Vivo. J. Am. Chem. Soc 2014, 136 (37), 12848–12851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (20).Checco JW; Kreitler DF; Thomas NC; Belair DG; Rettko NJ; Murphy WL; Forest KT; Gellman SH Targeting Diverse Protein-Protein Interaction Interfaces with α/β-Peptides Derived From the Z-Domain Scaffold. 2015, 112 (15), 4552–4557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Peterson-Kaufman KJ; Haase HS; Boersma MD; Lee EF; Fairlie WD; Gellman SH Residue-Based Preorganization of BH3-Derived α/β-Peptides: Modulating Affinity, Selectivity and Proteolytic Susceptibility in α-Helix Mimics. ACS Chem. Biol 2015, 10 (7), 1667–1675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Olson KE; Kosloski-Bilek LM; Anderson KM; Diggs BJ; Clark BE; Gledhill JM; Shandler SJ; Mosley RL and Gendelman HE Selective VIP Receptor Agonists Facilitate Immune Transformation for Dopaminergic Neuroprotection in MPTP-Intoxicated Mice. J. Neurosci 2015, 35 (50), 16463–16478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (23).Harrison SC Viral Membrane Fusion. Nat Struct Mol Biol 2008, 15 (7), 690–698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (24).Chang A; Dutch RE Paramyxovirus Fusion and Entry: Multiple Paths to a Common End. Viruses 2012, 4 (12), 613–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (25).Kielian M Mechanisms of Virus Membrane Fusion Proteins. Annual Review of Virology 2014, 1 (1), 171–189. [DOI] [PubMed] [Google Scholar]
- (26).Yin H-S; Paterson RG; Wen X; Lamb RA; Jardetzky TS Structure of the Uncleaved Ectodomain of the Paramyxovirus (hPIV3) Fusion Protein. Proc Natl Acad Sci USA 2005, 102 (26), 9288–9293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (27).Guichard G, Zerbib A, Le Gal FA, Hoebeke J, Connan F, Choppin J, Briand JP, and Guillet JG Melanoma peptide MART-1(27–35) analogues with enhanced binding capacity to the human class I histocompatibility molecule HLA-A2 by introduction of a β-amino acid residue: Implications for recognition by tumor- infiltrating lymphocytes. J. Med. Chem 2000, 43, 3803–3808. [DOI] [PubMed] [Google Scholar]
- (28).Reinelt S, Marti M, Dedier S, Reitinger T, Folkers G, de Castro JAL, and Rognan D β-Amino acid scan of a Class I major histocompatibility complex-restricted allore-active T-cell epitope. J. Biol. Chem 2001, 276, 24525–24530. [DOI] [PubMed] [Google Scholar]
- (29).Webb AI; Dunstone MA; Williamson NA; Price JD; de Kauwe A; Chen WS; Oakley A; Perlmutter P; McCluskey J; Aguilar MI; Rossjohn J; Purcell AW T cell determinants incorporating β-amino acid residues are protease resistant and remain immunogenic in vivo. J. Immunol 2005, 175, 3810–3818. [DOI] [PubMed] [Google Scholar]
- (30).Cheloha RW; Woodham AW; Bousbaine D; Wang T; Liu S; Sidney J; Sette A; Gellman SH; Ploegh HL Recognition of Class II MHC Peptide Ligands That Contain B-Amino Acids. J. Immunol 2019, 203 (6), 1619–1628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (31).Andrei SA; Thijssen V; Brunsveld L; Ottmann C; Milroy L-G A Study on the Effect of Synthetic α-to-β 3-Amino Acid Mutations on the Binding of Phosphopeptides to 14-3-3 Proteins. Chem. Commun 2019, 55 (98), 14809–14812. [DOI] [PubMed] [Google Scholar]
- (32).Duquerroy S; Vigouroux A; Rottier PJM; Rey FA; Jan Bosch B Central Ions and Lateral Asparagine/Glutamine Zippers Stabilize the Post-Fusion Hairpin Conformation of the SARS Coronavirus Spike Glycoprotein. Virology 2005, 335 (2), 276–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Lu L; Liu Q; Zhu Y; Chan K-H; Qin L; Li Y; Wang Q; Chan JF-W; Du L; Yu F; Ma C; Ye S; Yuen KY; Zhang R; Jiang S Structure-Based Discovery of Middle East Respiratory Syndrome Coronavirus Fusion Inhibitor. Nat. Commun 2014, 5 (1), 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Xia S; Yan L; Xu W; Agrawal AS; Algaissi A; Tseng C-TK; Wang Q; Du L; Tan W; Wilson IA; Jiang S; Yang B; Lu L A Pan-Coronavirus Fusion Inhibitor Targeting the HR1 Domain of Human Coronavirus Spike. Sci. Adv 2019, 5 (4). [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Outlaw VK; Bottom-Tanzer S; Kreitler DF; Gellman SH; Porotto M; Moscona A Dual Inhibition of Human Parainfluenza Type 3 and Respiratory Syncytial Virus Infectivity with a Single Agent. J. Am. Chem. Soc 2019, 141 (32), 12648–12656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (36).Outlaw VK; Lemke JT; Zhu Y; Gellman SH; Porotto M; Moscona A Structure-Guided Improvement of a Dual HPIV3/RSV Fusion Inhibitor. J. Am. Chem. Soc 2020. 142 (5), 2140–2144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (37).Marcink TC; Yariv E; Rybkina K; Más V; Bovier FT; Georges des, A.; Greninger AL; Alabi CA; Porotto M; Ben-Tal N; Moscona A Hijacking the Fusion Complex of Human Parainfluenza Virus as an Antiviral Strategy. mBio 2020, 11 (1), 1969–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (38).Nair H; Nokes DJ; Gessner BD; Dherani M; Madhi SA; Singleton RJ; O’Brien KL; Roca A; Wright PF; Bruce N; Chandran A; Theodoratou E; Sutanto A; Sedyaningsih ER; Ngama M; Munywoki PK; Kartasasmita C; Simões EA; Rudan I; Weber MW; Campbell H Global Burden of Acute Lower Respiratory Infections Due to Respiratory Syncytial Virus in Young Children: a Systematic Review and Meta-Analysis. The Lancet 2010, 375 (9725), 1545–1555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (39).Englund JA; Moscona A Paramyxoviruses: Parainfluenza viruses. In Viral Infections of Humans; Kaslow RA, Stanberry LR, Le Duc JW, Eds.; Springer: New York, 2014; pp 579–600. [Google Scholar]
- (40).Weinberg GA; Hall CB; Iwane MK; Poehling KA; Edwards KM; Griffin MR; Staat MA; Curns AT; Erdman DD; Szilagyi PG New Vaccine Surveillance Network. Parainfluenza Virus Infection of Young Children: Estimates of the Population-Based Burden of Hospitalization. J. Pediatr 2009, 154 (5), 694–699. [DOI] [PubMed] [Google Scholar]
- (41).Serrano L; Fersht AR Capping and A-Helix Stability. Nature 1989, 342 (6247), 296–299. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.