Abstract
Herpesviruses encode multiple glycoproteins required for different stages of viral attachment, fusion, and envelopment. The protein encoded by the human cytomegalovirus (HCMV) open reading frame UL116 forms a stable complex with glycoprotein H that is incorporated into virions. However, the function of this complex remains unknown. Herein, we characterize R116, the rat CMV (RCMV) putative homolog of UL116. Two R116 transcripts were identified in fibroblasts with three proteins expressed with molecular weights of 42, 58, and 82 kDa. R116 is N-glycosylated, expressed with late viral gene kinetics, and is incorporated into the virion envelope. RCMV lacking R116 failed to result in productive infection of fibroblasts and siRNA knockdown of R116 substantially reduced RCMV infectivity. Complementation in trans of an R116-deficient virus restored ability of the virus to infect fibroblasts. Finally, UL116 knockdown also decreased HCMV infectivity indicating that R116 and UL116 both contribute to viral infectivity.
Keywords: Cytomegalovirus, glycoproteins, viral entry
Introduction
Human cytomegalovirus (HCMV) is ubiquitous in the human population, with primary infections resulting in life-long infection. Although HCMV is generally mild or asymptomatic in immunocompetent individuals, immunocompromised patients suffer severe acute disease and in utero infection can result in permanent neurological injuries [1]. In addition, HCMV replication within transplanted tissues promotes chronic rejection and affects patient and graft survival despite use of prophylactic anti-CMV strategies [2–5]. Therefore, the development of an effective HCMV vaccine remains a high-priority [6]. Previous vaccine candidates have targeted CMV envelope glycoproteins with varying efficacy [7–10].
CMVs encode approximately 19 structural glycoproteins that are incorporated into the mature virion. However, not all of these glycoproteins directly participate in the viral entry process [11]. Glycoprotein B (gB), gH, gL, gM, gN, gO, UL128, UL130, and UL131A are the most well characterized for their roles in virion assembly and the virus entry process. These glycoproteins form several identified complexes (gB homotrimers, gM/gN, gH/gL/gO, and gH/gL/UL128/UL130/UL131A) that promote entry through either pH-independent entry by macropinocytosis or membrane fusion, or pH-dependent entry via endocytosis or macropinocytosis. Many cellular receptors have been proposed as having a role in these HCMV entry processes, but further work remains to be done to detail the mechanisms through which they promote entry [12,13].
The HCMV gB complex is a functional trimer that mediates viral membrane fusion with the host membrane [14,15]. This glycoprotein complex has also been implicated in binding of PDGFRα and several integrins to help facilitate entry [16–18], although no follow-up studies have validated these interactions. The glycoproteins gN and gM form the most abundant glycoprotein complex on the virion surface [11]. The precise role of the gM/gN complex is not yet known; however, the complex has been identified as binding heparan sulfate proteoglycans, suggesting a role in the initial tethering step [19]. Interestingly, viral mutants containing a gM C’ terminal deletion result in unstable gM proteins and fail to produce viable virus, and similarly mutations in the structural domains of gM generate replication-deficient virus [20]. C’ terminal deletion mutants of gN are also replication-deficient and fail to be enveloped, suggesting a further role for the gM/gN complex in viral assembly [21].
The gH and gL glycoproteins form a scaffold for the two known HCMV entry complexes: the gH/gL/gO heterotrimer and the pentameric complex consisting of gH/gL/UL128/UL130/UL131A [13,22–24]. These two complexes compete for the same binding region on the gH/gL scaffold [22]. The levels of trimer and pentamer incorporated into viral particles are influenced by HCMV UL148 [25]. Disruption of UL148 leads to a loss of mature trimer and promotes infection of epithelial cells, whereas rescue of UL148 expression decreases levels of the pentamer and decreases infection of epithelial cells. The trimeric gH/gL/gO complex is essential for entry into fibroblasts via binding of PDGFRα and for entry into epithelial cells [26–30]. However, the precise role of trimer in entry into epithelial cells and endothelial cells is unclear, as PDGFRα is not highly expressed on these cell types whereas viruses lacking pentamer have impaired entry efficiency into these cells [26,31–33]. In addition to being necessary for entry into endothelial and epithelial cells, the pentamer is also essential for entry into dendritic cells and monocytes [34,35]. The Integrin/Src/Paxillin signaling pathway is activated in pentamer associated entry processes [35]. Furthermore, Neuropilin-2 and OR14I1 were recently identified as cellular receptors for the pentamer [36,37].
An additional gH complex has been recently shown to exist that contains the viral glycoprotein UL116. UL116 is incorporated into the HCMV viral envelope complexed with gH [38]; however, the function of this complex and influence on viral infectivity remain unknown. Proteomics analysis also indicated the M116 was also present in mouse CMV (MCMV) particle preparations [39]. To better define the role of UL116, we described the role of a putative homolog of UL116 in rat cytomegalovirus (RCMV), and we have previously reported that the R116 gene is highly expressed in many different rat tissues following infection with rat cytomegalovirus (RCMV) [40]. In the present study, we investigated R116 expression, virion association and requirement for entry. We demonstrate that R116 is a virion-associated glycoprotein that is required for efficient viral infection of fibroblasts, and trans-complementation of R116 restored infectivity. Similarly, we also demonstrate here that HCMV UL116 is required for efficient production of infectious HCMV.
Results
R116 in vitro and in vivo expression profiles
R116 transcription kinetics were determined by Northern blot analysis using a double stranded DNA probe to identify all transcripts sharing the R116 sequence. Two transcripts, measuring 1kb and 3kb in length, were detectable at 48 hours post-infection (hpi), with a faint band detected at 24 hpi. R116 mRNA expression was blocked by treatment with the viral DNA synthesis inhibitor Foscarnet, suggesting that R116 is expressed with late viral gene expression kinetics (Figure 1a). Using a cDNA library generated from RCMV infected fibroblasts harvested at 48 hpi, we observed that the 3 kb transcript contained R116, R115 (gL), and R114, but that the 1 kb transcript contained only R116 sequences. A truncated form of R116 (880bp) was additionally identified at 24 hpi, corresponding in size to the shifted northern bands observed at 24 hpi. Analysis of these three R116 transcripts revealed a common splice event such that 84 base pairs are removed from 355 to 436 bp of the gene (Figure 1e). A recombinant R116 protein based on the mid-sized transcript and encoding amino acid residues 19–222 of the protein, excluding the predicted signal peptide (residues 1–18), was used to generate a rabbit polyclonal α-R116 antibody. The predicted molecular weight of R116 is 42 kDa; however, four different molecular weight R116 protein species were detected under denaturing conditions with this polyclonal serum (Figure 1b). The lowest molecular weight band was detected by 24 hpi corresponding to the small transcript, and expression of the low molecular weight version was unaltered by Foscarnet treatment at 48 hpi. In contrast, the larger R116 isoforms (42, 58, and 82 kDa) were expressed with late viral expression kinetics, as their expression was first detectable at 48 hpi and was sensitive to Foscarnet. Interestingly, western blot analysis on solubilized salivary gland tissue from mock-infected and RCMV-infected rats at 28 days post-infection (dpi) revealed that only the two higher molecular weight species of R116 were detected in salivary gland lysates (Figure 1c). The 82 kDa R116 band was the most abundant in vivo, whereas the 58 kDa band was the most abundant in vitro. The presence of R116 in the salivary glands was confirmed by immunofluorescence staining of frozen sections from rats infected with RCMV-GFP at 21 dpi (Figure 1d).
Figure 1. R116 expression profiles differ between in vitro and in vivo infections.
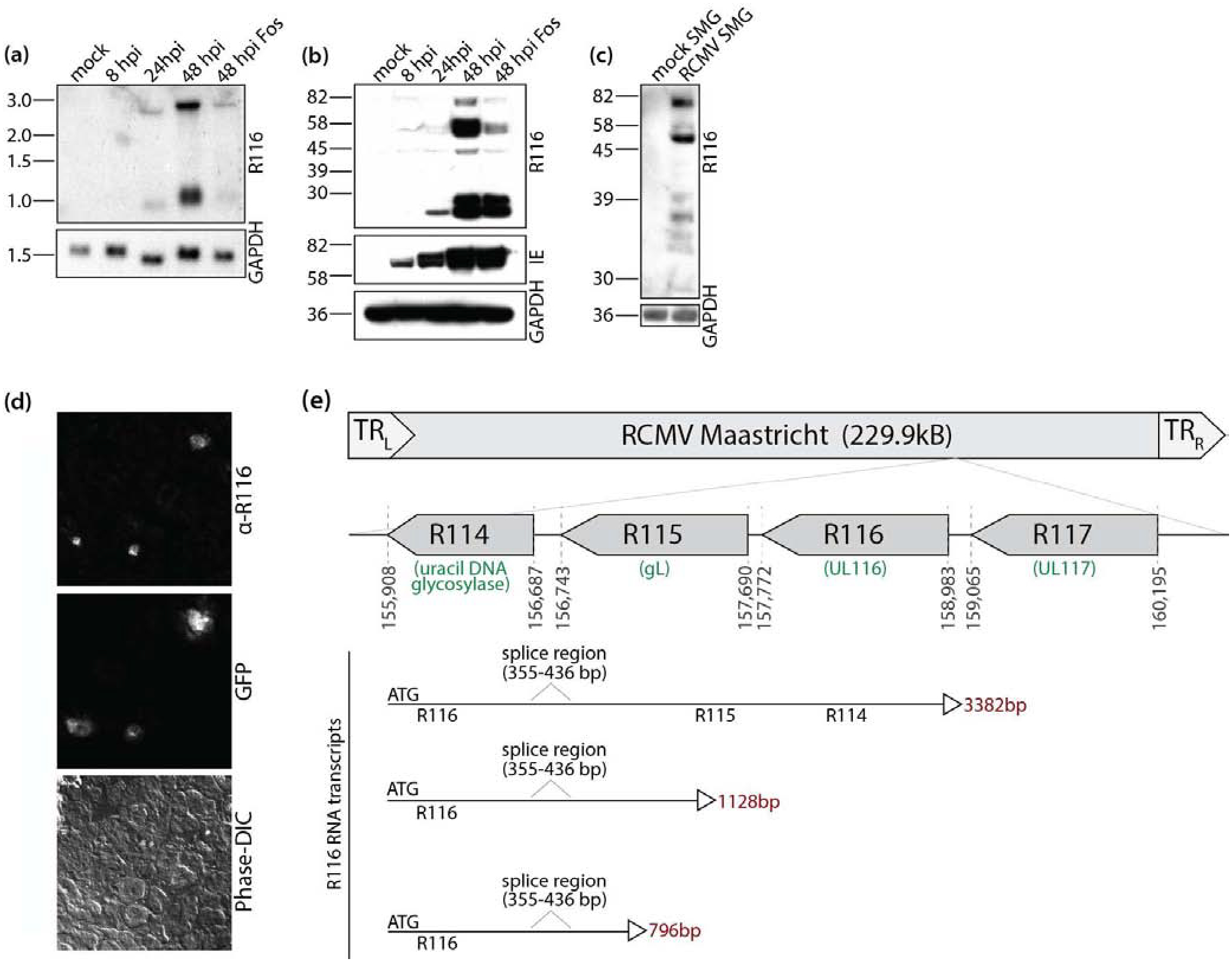
(a) R116 is a spliced message expressed on two viral transcripts with late viral gene expression kinetics. Rat fibroblasts were mock infected or infected with RCMV (MOI=1) and harvested at 8, 24, and 48 hpi. One infected cell sample was treated with Foscarnet (100 μg/ml) and harvested at 48 hpi. RNA was separated by electrophoresis utilizing a 1% formaldehyde agarose gel. RNA was transferred to nitrocellulose membranes and probed for R116. The blot was then stripped and re-probed for GAPDH. The Northern blots were visualized by autoradiography. (b) R116 protein is expressed with early and late viral kinetics in vitro. Rat RFL6 fibroblasts were mock infected or infected with RCMV (MOI=1) and harvested at 8, 24, and 48 hpi in Laemmli’s sample buffer. An additional infected cell sample was treated with Foscarnet (100 μg/ml) upon infection and harvested at 48 hpi. The blot was probed with antibodies directed against the RCMV proteins R116 and immediate early proteins 1 and 2 (IE1&2) as well as GAPDH. (c) Only high molecular weight R116 proteins are expressed in the salivary gland of RCMV infected rats. Salivary glands were harvested from RCMV infected rats at 28 dpi. Salivary glands from uninfected rats served as a control. The salivary glands were homogenized in RIPA buffer, analyzed by SDS-PAGE and probed using the anti-R116 polyclonal antibody. Equal loading was confirmed by staining for the cellular protein GAPDH. Western blots were visualized by autoradiography. (d) R116 protein expression is limited to glandular infected cells. Salivary glands were harvested from rats infected with RCMV-GFP RCMV at 21 dpi. Embedded frozen tissues were cut and sections were fixed with 2% PFA, washed and stained with antibodies directed against R116. Infected cells were detected by GFP expression. Deconvolution microscopy was used to visualize the stained cells. Mag=60X. (e) The RCMV R116 gene is expressed as three transcript variants with the largest containing transcripts for R115 (gL) and R114. A short transcript corresponding to the low molecular weight protein identified by western blots in (b) is shown.
RCMV R116 localizes to the virion assembly compartment
The cellular localization of R116 was identified using immunofluorescence microscopy by co-staining RCMV-infected RFL6 fibroblasts (48hpi) for R116 and either the endoplasmic reticulum (ER) marker KDEL, the trans-Golgi network (TGN) marker TGN-38, or the lysosomal marker LAMP-1. R116 protein localized to the TGN, but not within the ER or lysosomes in infected fibroblasts (Figure 2a). In order to chase R116 to its natural final compartment, cells infected for 48 hours were treated with cycloheximide and fixed and stained at 0, 1, 2, or 4 hours post treatment. Virion-associated glycoprotein gB translocates from the surface into the assembly compartment within 4–6 hours (Figure 2b). While R116 does not relocate to the same extent as gB, by 6 hours a portion of the cellular R116 protein localizes with gB to a perinuclear site, reminiscent of the virion assembly compartment.
Figure 2. R116 localizes with the trans-Golgi network marker TGN-38 and the viral glycoprotein gB.
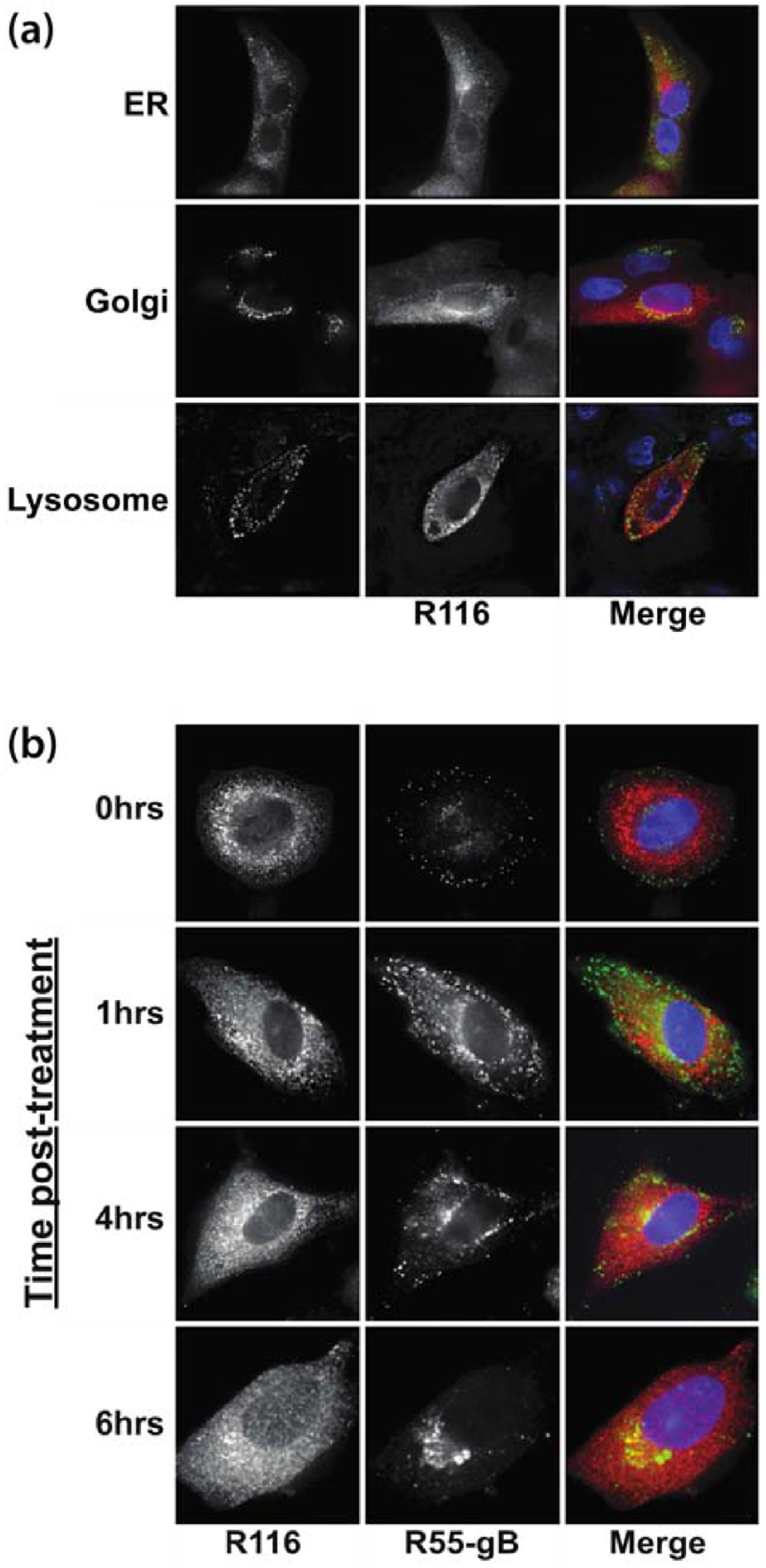
Rat fibroblasts were infected with RCMV (MOI=1). (a) At 48 hpi the cells were fixed with 2% paraformaldehyde (PFA) diluted in PBS. The fixed cells were stained for R116 (red) and antibodies to the cellular ER, TGN or lysosomal compartments using antibodies directed against KDEL, LAMP-1 or TGN-38 (green), and DAPI (blue). (b) To confirm the cellular localization of R116 (red) in relationship to the viral glycoprotein gB (green), RCMV infected fibroblasts were fixed in 2% PFA at 0, 1, 2, 4, and 6 hrs post treatment with Cycloheximide (100 ug/ml) that was initiated at 48 hpi. Deconvolution microscopy was used to visualize the stained cells. Mag=60x.
RCMV R116 is a virion associated glycoprotein
Since R116 localized to the virus assembly compartment, it was possible that R116 was incorporated into RCMV particles. Western blot analysis of gradient purified virions indicated that the high molecular weight species of R116 are preferentially enriched in RCMV virions, whereas the lowest molecular weight species was excluded from the virions (Figure 3a). RCMV IE and GAPDH are present in infected cell lysates but not incorporated into virions, and were used as controls. To determine whether R116 was incorporated into the virion as part of the viral envelope, aliquots of gradient purified RCMV virions were treated with either 2% NP40 or PBS and centrifuged to separate the virion envelope from the capsid/tegument fraction (Figure 3b). The pellet and supernatant fractions were analyzed by western blot with α-R116 and α-gB antibodies. Similar to the viral glycoprotein gB, R116 protein was present in the pellet fraction of the PBS-treated virus preparation and in the supernatant fraction when treated with NP40 (Figure 3c). This finding suggests that R116 is part of the viral envelope. In addition, R116 in gradient purified virions was sensitive to trypsin degradation indicating that R116 was present on the surface of the viral particle (Figure 3c).
Figure 3. R116 is a virion surface envelope glycoprotein.
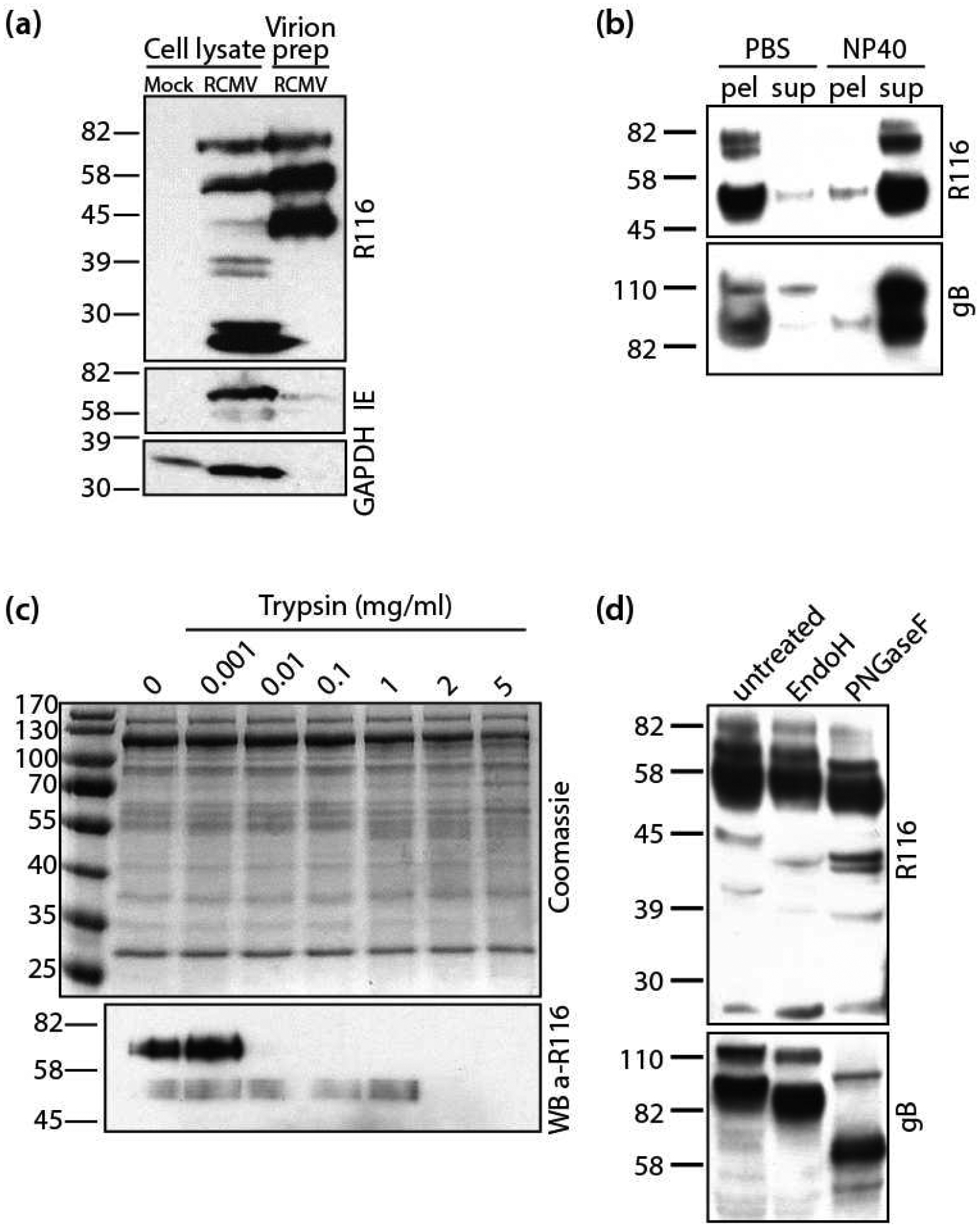
RCMV virus particles were purified over a discontinuous 10-to-50% Nycodenz gradient and then resuspended and pelleted through a 10% sorbitol cushion. (a) The RCMV virion preparation, infected fibroblast lysate, and lysate from mock-infected fibroblasts were analyzed by SDS-PAGE for R116, IE1&2 and GAPDH by Western blot. (b) RCMV virion preparation was split into two samples. One sample was treated with 1% NP-40 and the control sample with an equal volume of PBS. Both samples were pelleted at 100,000×g for 30 minutes. The pellet and supernatant fractions were analyzed by SDS-PAGE and probed for RCMV R116 and gB. (c) RCMV virion preparation was subjected to increasing concentrations of trypsin (0–5μg) for 15 minutes at 37°C. The samples were analyzed on two separate SDS-PAGE gels; one was stained with Coomassie Brilliant Blue and the other was probed for R116. (d) RCMV particles was treated with either EndoH or PNGaseF and analyzed by SDS-PAGE. The Western blot was stained for RCMV R116 and gB and visualized by chemiluminescence.
Examination of the R116 amino acid sequence indicates that the protein contains three predicted N-glycosylation sites. Virion associated R116 and gB were treated with endoglycosidase H (Endo H) and Peptide N Glycosidase F (PNGaseF) to trim off sugar moieties. Endo H removes high mannose and some hybrid types of N-linked carbohydrates, whereas PNGaseF cleaves all N-linked carbohydrates without regard to type. Similar to gB, both enzymes reduced the molecular weight of R116, from the 82kDa and 58 kDa bands to 42 kDa (Figure 3d). The predicted size of R116 without glycosylation is 42 kDa. However, PNGase F had a greater effect than Endo H, suggesting that R116 is N-linked glycosylated with a mixture of high mannose or hybrid and complex glycans.
Loss of R116 impairs RCMV infectivity
Since R116 is a virion envelope glycoprotein, we hypothesized that R116 plays a role in virus entry. A bacterial artificial chromosome (BAC) was generated containing a GFP reporter and the RCMV genome. BAC recombineering was then used to generate a mutant virus containing a 2xSTOP mutation at the beginning of the R116 coding sequence. RFL6 cells transfected with recombinant BACs containing the R116 2xSTOP mutation produced GFP and the viral IE proteins indicating successful transfection of the BAC DNA and viral gene expression. However, exhaustive transfection experiments with multiple BAC clones and preparations failed to rescue infectious virus (data not shown).
As an alternative approach, R116 knockdown experiments were performed in WT RCMV-infected fibroblasts using two different siRNAs specific for R116. Western blot analysis of cell lysates validated that both R116 specific siRNAs, but not a control siRNA, reduced R116 protein production (Figure 4a). Titration of supernatant virus from this experiment revealed that knockdown of R116 reduced RCMV infectivity by nearly 10-fold when compared to the negative control siRNA or non-transfected controls (Figure 4b). To determine whether R116 knock-down affects viral genome replication or the release of genome containing particles, viral DNA levels in supernatants and cell pellets from siRNA treated cells were analyzed by quantitative PCR. Knockdown of R116 did not decrease viral genomic DNA levels in the infected cells indicating that the R116 knockdown did not decrease the production of infectious virus by preventing DNA replication (Figure 4c). Furthermore, knockdown of R116 did not decrease the release of viral genome containing particles into the supernatant suggesting that R116 deficient viruses display a reduced infectivity per particle ratio, rather than a reduction in the production of viral particles (Figure 4c).
Figure 4. R116 is necessary for viral entry into fibroblasts.
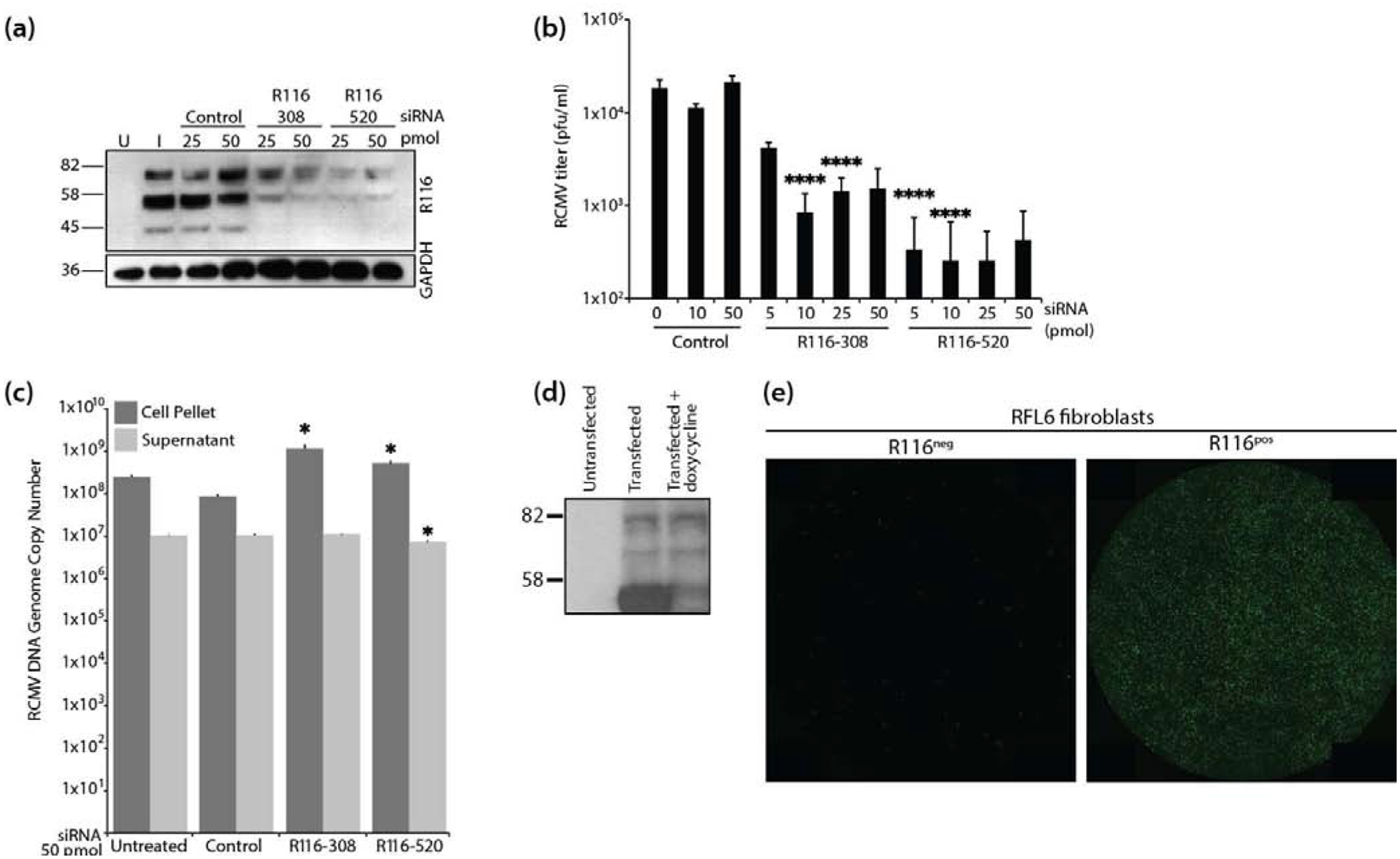
(a-c) In triplicate wells RFL6 fibroblasts were transfected with a control siRNA or siRNAs targeting sequences present with the R116 gene. At 24 hours post transfection, the cells were infected with RCMV (MOI=1.0) and allowed to incubate for 4 hours at which time the virus inocula were removed and the cells were washed twice with fresh medium. At 48 hpi, supernatants and cell pellets were collected and analyzed by: (a) Western blotting for R116 and GAPDH; (b) Plaque assay for the presence of infectious virus. Statistical significance was determined by Student’s t-test [control vs R116 siRNA at 10 and 50pmol ****p<0.001, n=3]; and (c) Quantitative PCR for the presence of viral DNA genomes. Statistical significance was determined by Student’s t-test [control vs. siRNA R116–308 cell pellet p=0.02 and supernatant p=0.33, n=3; control vs. siRNA R116–520 cell pellet and supernatant p=0.01, *p<0.05, n=3]. (d-e) R116 expression by RFL6 cells restores infectivity of ΔR116-RCMV. (d) RFL6 fibroblasts were harvested in lysis buffer containing protease inhibitor, analyzed by SDS-PAGE and probed using the anti-R116 polyclonal antibody. Western blots were visualized by autoradiography. (e) GFP expression 5 days after infection with ΔR116-RCMV in triplicate wells wild-type RFL6 fibroblasts (left) or R116-expressing RFL6 fibroblasts (right). Representative images are shown.
We next investigated whether the reduced infectivity per particle ratio was due to a defect in an entry process or a post-entry step. For these experiments, cells were treated with polyethylene glycol (PEG) following infection with WT RCMV collected from cells treated with the anti-R116 or control siRNA. PEG is a fusogenic substance, known to promote viral and cell membrane fusion [41,42]. PEG restored infectivity of the R116-deficient virus to PEG-treated control siRNA levels (Table 1). While PEG treatment increased the infectivity of virus from both control and R116 siRNA treated cells, the effect of PEG was greatest in cells infected with the R116-deficient virus, wherein PEG increases the infectivity by 10–12-fold. This result indicates there were similar numbers of RCMV virions present in the R116 knockdown supernatants compared to controls and confirms our findings of equal viral genome levels in the R116-deficient virions. Thus, R116-deficient viral particles have a reduced infectivity per particle ratio and lack the necessary entry machinery to complete early entry steps.
Table 1.
PEG treatment restores infectivity of RCMV lacking R116.
| # plaques | Fold increase with PEGa | ||
|---|---|---|---|
| No treatment | PEG | ||
| Control siRNA | 725 +/− 199 | 2417 +/− 736 | 3.3 |
| R116–308 | 158 +/− 143b | 1950 +/− 421c | 12.3 |
| R116–520 | 208 +/− 174d | 2183 +/− 640e | 10.5 |
=PEG treatment increases infectivity for all siRNA (p<0.005)
=siRNA to R116 decreases infectivity vs. control (p<0.0003, p<0.0008 respectively)
=PEG restores infectivity of RCMV lacking R116 to control levels (p=0.21, p=0.57 respectively)
Since R116 is necessary for virus entry into fibroblasts, we hypothesized that trans-complementation may permit the rescue of recombinant RCMV lacking expression of R116. For this experiment, fibroblasts were constructed that express R116 in a doxycycline-inducible manner using a Tet OFF system. R116 expression was confirmed by western blot of lysates from transfected fibroblasts (Figure 4d). To evaluate whether trans-complementation was possible for viruses lacking R116, these cells expressing R116 were transfected with the RCMV-GFP-116-2xStop BAC, in the absence of doxycycline. Supernatants were collected at 7 days post transfection and titered on fibroblasts by GFP expression. Importantly, virus was recovered from the R116 expressing cells, which was used to infect WT or R116 expressing cells to assess viral entry and support of viral replication. Cells were infected with a low multiplicity of infection (MOI = 0.1) and visualized fluorescence microscopy at 5 dpi for the presence of virus as indicated by GFP. R116 complementation restored infectivity of RCMV (Figure 4e) and the RCMV-GFP-116-2xStop virus was capable of spreading in the cells expressing R116 but not in cells that did not express R116. This data confirms that R116 is required for the production of infectious virus.
UL116 knockdown impairs HCMV infectivity
Previously it has been reported that UL116 binds to gH but the function of this complex remains unknown [38]. To examine whether UL116 is required for HCMV infectivity, we utilized a UL116 knockdown technique in WT HCMV infected cells, which was similar to our approach for R116. For this experiment, UL116 was knocked-down with 2 unique siRNAs targeting different regions of the UL116 gene and then infected with HCMV-TB40EgfpUL116-HiBiT. This virus contains a C’terminal in-frame fusion of UL116 with an 11 amino acid epitope that binds to Large BiT in trans for luminescence detection (Figure 5a). Multistep growth curves in NHDFs infected with HCMV WT and UL116 HiBiT viruses demonstrated that the HiBiT-tag did not affect viral replication (Figure 5b).
Figure 5. HCMV UL116 is required for entry into fibroblasts.
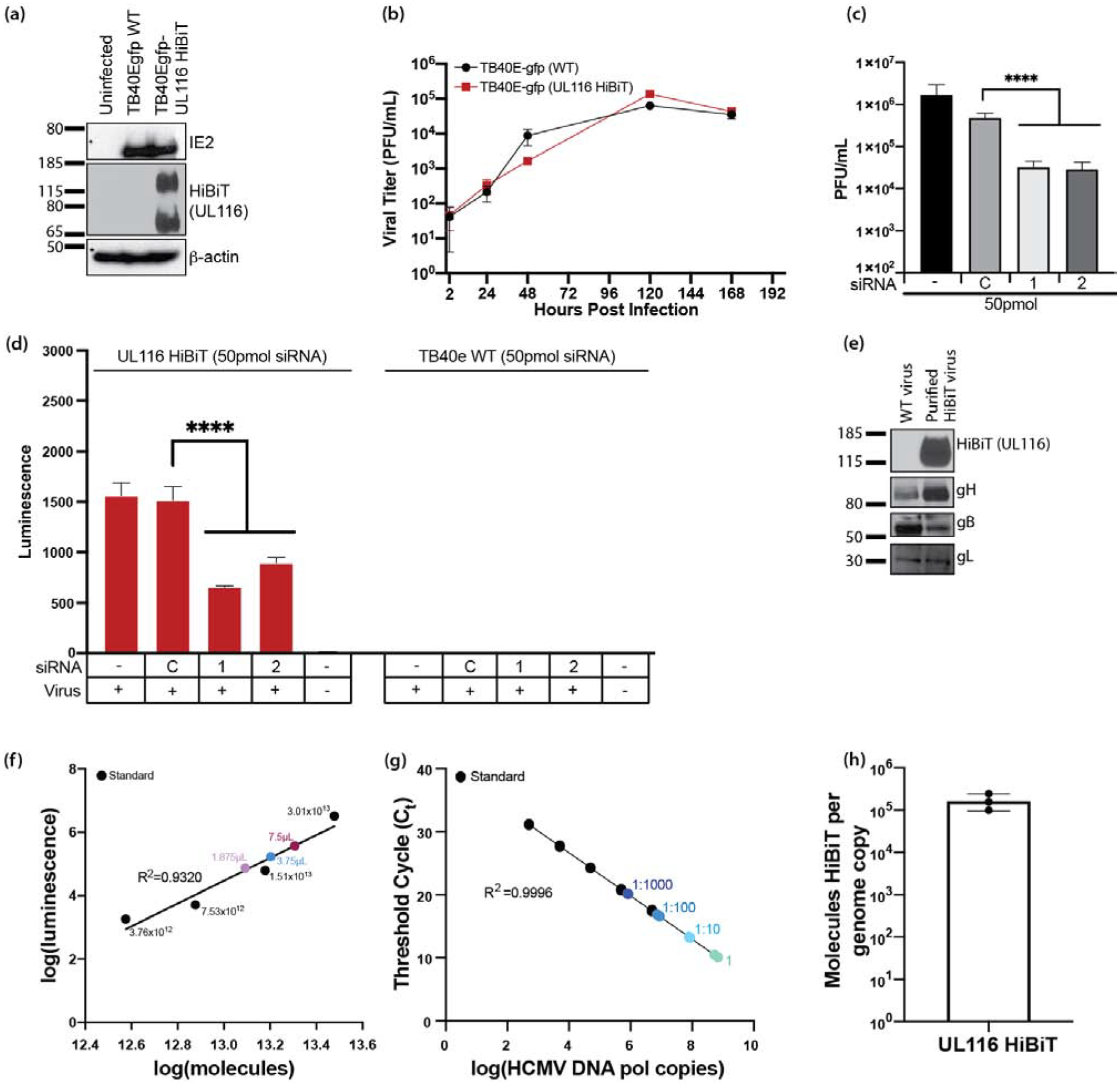
HCMV UL116 was molecularly tagged with the HiBiT tag using BAC recombineering in a TB40e background. (a) NHDF fibroblasts were infected with TB40e GFP HCMV or TB40e GFP UL116-HiBiT HCMV and were harvested in lysis buffer. Western blots were performed for IE2, HiBiT, and β-actin to confirm expression of UL116-HiBiT. (b) NHDF fibroblasts were infected at a MOI=0.3 with HCMV TB40egfp-UL116-HiBiT or WT virus. At 2 hours post infection, the virus inoculums were removed, cells were washed with PBS, and then fresh culture media was added. Supernatants were collected at 24, 48, 120, and 168 hpi and titered using NHDF fibroblasts. (c) NHDF fibroblasts were plated in 24-well plates and treated in quadruplicates. NHDF fibroblasts were transfected with a control siRNA or siRNAs targeting sequences present within the UL116 gene at 50pmol/well. At 24 hours post transfection, the cells were infected with HCMV (MOI=0.3) and incubated for 2 hours at which time the virus inoculums were removed and the cells were washed twice with fresh medium. At 5 days post-infection, supernatants and cell pellets were collected and analyzed by plaque assay for the presence of infectious virus. Statistical significance was determined by one-way ANOVA with Dunnett’s correction for multiple comparisons [****p<0.0001, n=4]. (d) NHDF fibroblasts were seeded in 96-well black plates and transfected with control siRNA or siRNAs targeting sequences present within the UL116 gene. Cells were infected at a MOI=0.3 using a UL116-HiBiT tagged HCMV or TB40e WT HCMV for 2 hours, at which time the virus inoculums were removed and cells were washed with PBS and fresh media was added. At 3 days post-infection cells were harvested and analyzed by HiBiT lytic detection assay. Statistical significance was determined by two-way ANOVA with Dunnett’s correction for multiple comparisons [****p<0.0001]. (e) gH, gB, and gL are incorporated into viral particles of UL116-HiBiT HCMV. WT and UL116-HiBiT HCMV were expanded over NHDF fibroblasts. Viruses were harvested at the time of maximum cytopathic effect and pelleted through a 20% sorbitol cushion. UL116-HiBiT viral particles were further purified over a 10–50% histodenz gradient. Purified viral particles were run on SDS-PAGE gels and western blots for gH, gB, gL, and HiBiT were performed. (f-h) UL116 is incorporated into HCMV virions at 1.06×105 molecules/HCMV genome. HCMV UL116 HiBiT viral particles were isolated over a histodenz gradient and molecules of HiBiT was determined against a standard curve by HiBiT lytic detection assay (f) and genome copies were determined by qPCR for HCMV DNA polymerase (g). HiBiT control protein (Promega) and known genome copies of HCMV TB40e were used as respective assay controls. Ratio of UL116-HiBiT molecules over genome copies was calculated (h).
For siRNA knockdown experiments, culture supernatants were collected at 5 days post-infection and titered to determine viral infectivity. siRNA knockdown of UL116 with 50pmol siRNA significantly decreased viral loads at 5 days post-infection with HCMV TB40e UL116-HiBiT (MOI=0.3) in NHDF fibroblasts (Figure 5c). Knock-down of UL116-HiBiT in this experiment was confirmed in supernatants from the infected cells using the NanoGlo HiBiT lytic detection system (Figure 5d), which detects HiBiT-tagged proteins following treatment with a membrane-dissociation buffer. These data indicate that UL116 contributes to the production of infectious HCMV.
Quantification of UL116 incorporation into HCMV
To determine the number of UL116 molecules incorporated into viral particles, we isolated UL116-HiBiT tagged virus particles by serial centrifugation over a 20% sorbitol cushion, a discontinuous 10–50% histodenz gradient, and a 20% sorbitol cushion. Gradient purified HCMV incorporation of UL116-HiBiT was detected by western blot analysis and compared to pelleted HCMV TB40e WT virus (Figure 5e). gH, gL, and gB were all incorporated into the viral particle of UL116-HiBiT HCMV (Figure 5e). Molecules of HiBiT tagged protein in three different volumes of virus were determined by HiBiT lytic detection assay relative to a tagged control protein purchased from Promega (Figure 5f). Viral genome copies in the virus preparations were determined by qPCR using primers and probes directed against UL54 gene sequences and this data was used to calculate the number of viral genomes per μL of the virus preparation (Figure 5g). By directly comparing the number of HCMV genome copies with HiBiT molecules per volume, it was calculated that, on average, 1.06×105 molecules of UL116-HiBiT per viral genome are incorporated into virus particles (Figure 5h).
Discussion
The UL116 proteins are incorporated into multiple CMVs including HCMV, MCMV and RCMV. In this report, we demonstrate that RCMV ORF R116 is a virion surface envelope glycoprotein required for an early step of virus entry. R116 protein localizes to the TGN in a compartment that co-stains with RCMV glycoprotein gB. From this data and given the incorporation of R116 into the virion, we predict that this compartment is the virion assembly compartment. Transfection with a RCMV BAC containing a disrupted R116 gene was unable to spread in fibroblasts in vitro. Similarly, siRNA knockdown of R116 decreased RCMV infectivity of fibroblasts by approximately a log but did not alter viral genome replication or release of viral genome containing particles. Polyethylene glycol treatment promotes the fusion of membranes and as such restored the infectivity of virus lacking R116, indicating that the R116-deficient particles are otherwise intact. R116 complementation restored RCMV infectivity in fibroblasts. Finally, knockdown of UL116 decreased HCMV infectivity in human fibroblasts by approximately a log. Importantly, we determined that UL116 is incorporated at 1.06×105 molecules per genome copy. Taken together the data presented in this manuscript demonstrate that CMV ORF 116 encodes a virion glycoprotein involved in the production of infectious virus.
Despite a low identity (18%) with UL116, the RCMV R116 protein shares many structural characteristics with its HCMV homologue. Both proteins contain predicted 18 amino-acid signal peptides in their N-terminal regions, with a cleavage site between position 18 and 19 for R116 (www.cbs.dtu.dk/services/SignalP/). Additionally, both proteins are expressed with late viral gene expression, as observed for UL116 [38]. Multiple variants of UL116 have been described, with a 35 kDa, a 76 kDa, and a 125 kDa form accumulating after 3 dpi [38]. Similarly, we found two distinct R116 forms of about 82 and 58 kDa expressed in vivo. R116 is glycosylated, and post-translational modification prediction reveals 3 N-linked glycosylation sites at positions 98, 134 and 335 and more than 60 O-glycosylation sites (http://www.cbs.dtu.dk/services/NetNGlyc), while UL116 contains 14 N-glycosylation sites and numerous O-glycosylation sites. Therefore, we suggest that the function of ORF 116 might be conserved between RCMV, MCMV and HCMV. Notably, treatment of cell lysates with PNGase F decreased the size of R116, but a large proportion of R116 was not reduced to the un-glycosylated size of 42kDa. Since PNGase F treatment had significantly more effect on reducing the size at which gB migrates, this suggests that additional post-translational modifications beyond N-linked glycosylation occur to R116. The requirement for R116 and UL116 in the production of infectious virus suggests that either the protein encoded by ORF116 is a member of a putative entry complex or is involved in regulation of other entry complexes. A putative entry complex containing UL116 has been proposed previously for HCMV, consisting of gH/UL116 [38]. So far, gH/gL/gO has been described as the primary gH based complex for entry into fibroblasts, and is believed to function with gB to create the core fusion machinery for entry [13]. Importantly, the decrease in production of infectious CMV via UL116/R116 knockdown may alternatively be due to effects on an mRNA also containing UL115/R115 (gL); however, our trans-complementation of RCMV deficient of R116 would indicate that expression of only R116 is sufficient to regain infectivity.
Interaction between gH and gB to form gB:gH/gL complexes is essential to induce cell-to-cell fusion [43,44]. The crystal structure for HCMV gH was recently described in complex with the other pentamer components [45]. This crystal structure showed substantial similarities to previous descriptions of gH for Epstein-Barr virus (EBV) and Herpes Simplex virus-2 (HSV-2) [45–49]. Based on mechanistic studies involving different neutralizing antibodies, some studies determined that binding of gH/gL to gB depends on N-terminal domain H1 of gH [47–49], which is supported by the inability of MSL-109 (an anti-CMV neutralizing antibody binding domain H2 of gH) to prevent the generation of gB:gH/gL [46]. Here, we found that gB and R116 are colocalized, but formation of a gB:gH/R116 (or gB:gH/UL116) complex remains to be explored.
Numerous receptors of CMV envelope glycoproteins have been identified, conferring a broad cell tropism to CMV, including fibroblasts, endothelial cells, epithelial cells and myeloid cells [16–18,37,50]. Platelet-derived growth factor alpha is known as the main receptor of gH/gL/gO, while neuropilin-2 and OR14I1 are known as the main receptors of the pentameric complex [26,36,37]. Calo et al. showed that UL116 competes with gL for gH binding, leading to the formation of a stable gH/UL116 complex, which is transported to the plasma membrane [38]. How UL116 interacts with gH remains unknown, but UL116 does not contain the necessary cysteine residues to form disulfide bonds with gH. Further work remains to be done to identify binding partners of the gH/UL116 complex and to determine whether UL116 limits CMV cell tropism beyond fibroblasts. Additional studies are required to determine whether UL116/R116 acts a chaperone promoting gH complex formation. In conclusion, ORF 116 encodes a CMV envelope glycoprotein essential for production of infectious virus in both HCMV and RCMV, and should be considered as an attractive therapeutic target when developing vaccines and monoclonal antibodies against CMV.
Materials and Methods
Antibodies.
Rabbit polyclonal antibodies were generated to ORF R116 by immunizing rabbits with a HIS tag R116 (19–222aa) fusion protein. Rabbit anti-RCMV-IE polyclonal antibody was previously described [51]. A rat anti-RCMV gB monoclonal antibody was produced by Dan Cawley at the OHSU-VGTI Monoclonal Antibody Facility from splenocytes derived from RCMV-infected rats. Mouse monoclonal antibodies directed against GAPDH (ab8245), KDEL (ab12223), LAMP-1 (ab13523), and TGN-38 (ab16059) were purchased from AbCAM. Secondary anti-mouse and anti-rabbit horseradish peroxidase (HRP)-conjugated antibodies (NA934V and NA931V) were purchased from Amersham and rabbit anti-rat HRP (6180-05) was purchased from Southern Biotech. Secondary anti-mouse (A11020), anti-rabbit (A11046) and anti-rat (A21470) fluorescently tagged antibodies were purchased from BioSource International.
RCMV.
The RCMV Maastricht strain genome was captured as a Bacterial Artificial Chromosome (BAC) using homologous recombination by replacing ORFs r144-r146 with a BAC cassette expressing eGFP under the HCMV major immediate early promoter [52]. Two-step recombination was used to mutate RCMV open reading frames in the creation of the R116 2xSTOP mutant. The first recombination step incorporated a gene cassette expressing galK and Kanamycin resistance genes into the RCMV BAC genome at the genomic site for introduction of the mutation, via PCR amplification of a 100bp homology region flanking the galK/Kan gene cassette. The amplified DNA was treated with Dpn1, purified, and then electroporated into competent RCMV-BAC SW102 cells. Clones were positively selected for gain of resistance against Kanamycin, screened by PCR and sequenced to identify clones with the proper insertion. PCR was used to generate DNA fragments containing RCMV sequences with the desired mutation. This amplicon was electroporated into competent bacteria and negatively selected for galK replacement on 2-deoxy-galactose-1-phosphate (DOG) negative selection plates. BAC clones were verified by PCR and the fragments were sequenced for correct incorporation of mutations. DNA from correct clones was transfected into RFL6 fibroblasts (RFL6, ATCC) to rescue the virus. RCMV mutants and WT were then expanded on RFL6 fibroblasts and purified over a 10% sorbitol cushion by ultracentrifugation at 25,000RPM (SW32) for 1 hour. Purified virus was then titered over RFL6 fibroblasts in serial dilutions to determine viral titer as measured in PFU/mL.
HCMV.
In order to evaluate the efficiency of UL116 knockdown using specific siRNA, we modified a GFP-expressing BAC of wild type HCMV clinical isolate TB40E using galK positive/negative selection as described for RCMV. The HiBiT tag (5’-GTGAGCGGCTGGCGGCTGTTCAAGAAGATTAGC-3’) was inserted between nucleotides 1026 and 1027 in the UL116 gene, corresponding to the C-terminal region of the protein. Correct insertion of the tag was confirmed by sequencing.
Identification of RCMV R116 transcripts.
An RCMV cDNA library was constructed from rat RFL6 fibroblasts infected with RCMV at a multiplicity of infection (MOI) of 1 for 24 and 48 hours post infection (hpi) using the Superscript Plasmid System with Gateway Technology for cDNA synthesis and cloning (Invitrogen). The cDNA was ligated into the plasmid pSPORT and screened by Southern blotting using a R116 DNA probe. The R116-positive clones were sequenced using oligonucleotides corresponding to Sp6 and T7 binding sites present in the plasmid flanking the cDNA insert. The predicted splicing of R116 was confirmed by RT-PCR using flanking primers. The products were analyzed by gel electrophoresis and the products were sequenced.
Northern blot.
RFL6 fibroblasts plated on 10cm dishes were infected with RCMV (MOI=1). At 8, 24 and 48 hpi the cells were washed and lysed with Trizol for 5 min at room temperature. Subsequently, the samples were scraped and stored at −80°C. RNA was isolated per the manufacturer’s instructions and electrophoresed through a 1% agarose/formaldehyde gel and transferred to GeneScreen Plus nylon membranes (Dupont/NEN). The blots were hybridized with probes specific for R116 and GAPDH generated from 500bp BamHI fragments of plasmids containing R116 or GAPDH using Roche Random Prime Labeling kit. Alternatively, single stranded probes were made by end labeling DNA oligonucleotides complementary for R116 or GAPDH sequences using T4 polynucleotide kinase (New England Bio). The Northern blots were hybridized in Express Hybe (Clontech) and washed with low stringency wash (2xSSC with 0.05% SDS) followed by high stringency wash (0.1xSSC with 0.1% SDS). The blots were exposed to autoradiography film (Kodak Biomax MS) using intensifying screens at −80°C, developed, and visualized.
Western blots.
Cells were placed on ice, washed once with 4°C PBS, and 1x cell lysis buffer (cell signaling) containing HALT protease inhibitor (ThermoFisher) was added. Cells were incubated on ice for 15 minutes and scraped. Cell nuclei were pelleted out at 10,000RPM for 10 minutes at 4°C and clarified supernatants were transferred to new Eppendorf tubes. NuPage loading buffer with 2% BME was added to cell lysates at a final concentration of 2x. Samples were boiled for 5 minutes and loaded on 4–12% SDS-PAGE protein gels. Proteins were transferred using a semi-dry transfer system to PVDF membranes and membranes were probed with the appropriate antibodies with membrane blocking in 5% milk-TBST. HiBiT blots on PVDF membranes were washed in TBST (10mM Tris, pH 7.2, 100mM sodium chloride, 0.1% Tween-20) for 15 minutes at room temperature, incubated with 1:200 LgBiT in 1x HiBiT blotting buffer for 1 hour at room temperature, and HiBiT substrate was added at 1:500 for 5 minutes at room temperature. Blots were then developed by chemiluminescence.
Salivary gland protein isolation.
R116 in vivo protein expression was performed on approximately 100μg of protein extracted from flash frozen salivary gland tissue from uninfected control rats and rats infected with RCMV for 28 days (n=3). Tissues were homogenized in RIPA buffer (50mM Tris HCl pH7.4, 150mM NaCl, 5mM MgCl2, 0.5% NP-40, 1mM PMSF and protease inhibitor cocktail) using a Precellys tissue homogenizer. Lysates were analyzed by SDS-PAGE and Western blotted for R116 and GAPDH as described above.
Cellular localization of R116 assessed by immunofluorescence microscopy.
Fibroblasts grown in 4-well chamber slides were infected with RCMV (MOI=0.5) for 48 hpi and then treated with cycloheximide (100μg/ml) for an additional 0, 1, 2, 4 and 6 hpi to block new protein synthesis. At the indicated time points, cells were fixed in 2% paraformaldehyde in PBS and then permeabilized with Saponin buffer (0.2% saponin, 1%BSA, PBS) blocking with normal goat serum. Localization of viral and cellular proteins was determined by immunofluorescence (IF) microscopy utilizing primary antibodies directed against R116, gB, KDEL, LAMP-1, and TGN-38. Samples were then incubated with appropriate fluorescently-labeled species-specific secondary antibodies and DAPI DNA stain. Frozen, thin-sections of rat salivary glands from rats infected with RCMV-GFP at 21 days post-infection were fixed and stained with antibodies directed against R116 using the same protocols described above. Deconvolution microscopy was used to visualize the stained cells in the 4-well chamber slides and tissue sections (mag=60X).
Virion localization and glycosylation of R116.
The protein composition of the pelleted virus was analyzed by Western blotting to determine whether R116 was incorporated into virions. We analyzed samples of RFL6 fibroblasts infected with RCMV (MOI=1.0), salivary gland tissue homogenates (supplementary data), or different preparations of virus pellets. RCMV virus particles were purified by layering over a discontinuous 10-to-50% Nycodenz gradient and centrifuged at 110,000×g for 2 hours at 4°C. Virus banded at 20–30% Nycodenz was brought up in PBS and pelleted at 59,439×g for 1 hr through a 10% sorbitol cushion. The virus pellet was resuspended in a minimal volume of PBS. First, to determine whether RCMV R116 was an envelope protein, the resuspended viral pellet was split into 2 samples. One sample was treated with 1% NP-40 in PBS added in a 1:1 ratio and the second half of the original sample was treated with an equivalent volume of PBS. The samples were incubated for 30 minutes at 4°C and centrifuged in a mini-ultracentrifuge at 100,000 ×g for 30 minutes to acquire both the pellet and supernatant fractions. Second, to determine whether R116 was on the virion surface, an additional preparation of gradient banded virus particles was incubated with increasing concentrations of trypsin and incubated at 37°C for 15 minutes. Laemmli’s sample buffer was added to the virus/protease mixture to stop the reaction and the samples were analyzed by SDS-PAGE in combination with either Coomassie brilliant blue staining or Western blotting for R116. Third, to determine whether virion-associated R116 was glycosylated, RCMV particles were treated with PNGaseF (NEB) or EndoH (Roche). The treated virus samples were analyzed by SDS PAGE and Western blotting for R116 and gB.
siRNA transfection and infection.
Two R116-specific siRNAs (R116 ORF positions 308–326 nt and 520–538 nt) and a control siRNA were purchased from Dharmacon. The sequences of the R116 siRNA were: R116–308 5’-CTACATTACCCTCGCAAAT-3’ and siRNA R116–520 5’-GCGACGAGGCGATACGTTT-3’. Two UL116-specific siRNA were purchased from Horizon having the following sequences: UL116-1 5’-ACAAGAAACACAAGGAAUAUU-3’ and UL116-2 5’-CCGUCAUCGUCGCGGGUAAUU-3’. RFL6 fibroblasts and NHDF fibroblasts seeded in 24-well plates were transfected twice with each siRNA using Lipofectamine RNAiMAX Reagent (Invitrogen) according to the manufacturer’s protocol. At 24 hours after the initial transfection step, the cells were infected with RCMV (MOI=1) or HCMV (MOI=0.3). The cells and supernatants were collected and titered using a standard plaque assay at 2 and 5 dpi for RCMV and at 5 dpi for HCMV. For HCMV, protein levels were determined by HiBiT lytic detection, as described below. For RCMV, the triplicate samples of cells were lysed in either DNAzol (viral DNA analysis), Trizol (mRNA analysis) or Laemmli’s buffer (viral protein analysis). Host cell production of the viral mRNAs was quantified by RT-PCR Taqman. R116 protein expression levels were determined by Western blotting. Viral genomic DNA was analyzed by Real Time PCR for R54 (viral DNA polymerase) as described below.
Plaque Assays.
Serial dilutions (10−1 to 10−6) of viral supernatant were performed in complete DMEM and 100μL were added to confluent 24-well plates of rat fibroblasts or NHDF fibroblasts as appropriate. Viral solutions were incubated for 2 hours at 37°C. Carboxy-methyl-cellulose (CMC; 500μL per well) was overlaid onto the cells and cells were incubated for 7 days for RCMV or 14 days for HCMV at 37°C. Titer plates were fixed in 3.7% PFA and stained with methylene blue and plaques were counted using a dissecting microscope. HCMV titers were counted by GFP fluorescence microscopy.
Viral Entry Assays.
A 1:10 dilution of supernatant from siRNA treated cells infected with RCMV was allowed to bind for 1 hour and then treated with the fusogenic substance polyethylene glycol (PEG) 6000 (Calbiochem) diluted in PBS (44%) for 30 seconds at 37°C using a protocol designed by Ryckman et al. for the study of HCMV entry [41,42]. Following PEG treatment, the cells were washed once with PBS, twice with media and then overlaid with CMC and incubated for 5 days to allow for plaquing of virus. Cells were then fixed with 3.7% paraformaldehyde and stained with methylene blue and plaques were counted.
Quantitative PCR detection of RCMV genomic DNA and viral gene expression.
For the quantification of RCMV DNA in virus particles and infected cells treated with siRNA, total genomic DNA was extracted from cells and supernatants using DNAzol (Invitrogen). A total of 0.5 μg of DNA was analyzed using a Taqman probe/primer set recognizing a RCMV R54 DNA polymerase sequence as previously described [40,53]. PCR reactions were set up using the TaqMan Universal PCR Master Mix (Applied Biosystems). Following thermal activation of AmpliTaq Gold (10 min. at 95°C), a total of 40 cycles were performed (15 sec. at 95°C and 1 min. at 58°C) using an ABI StepOnePlus Real Time PCR machine. The sensitivity of detection for this assay was <100 copies.
Real-time (RT)-PCR was used to quantify expression levels of RCMV R114, R115 and R116 from infected cells collected following treatment with siRNA [40]. cDNA was generated from mRNA using Superscript III RT (Invitrogen) and analyzed by real-time PCR techniques using primer sets recognizing RCMV gene sequences. RT-PCRs were performed using the SYBR Green PCR Master Mix (Applied Biosystems) as previously described using an ABI StepOnePlus Real Time PCR machine [40,51]. Plasmid clones containing each gene fragment were used as positive controls and quantification standards. The sensitivity of detection of this assay was <100 plasmid copies for all of the tested RCMV genes. Quantitative PCR data were analyzed by ANOVA and student’s t-test.
Fibroblast R116 transfection and infection.
The entire R116 gene was ligated into the plasmid pGEM®-T easy (PROMEGA). Briefly, we combined 22ng of amplified modified R116 gene (forward primer 5’-TCCACGAACACACATACGTA-3’, reverse primer 5’-TTCGACATCTGTTGGCGAAT-3’), 50 ng of pGEM®-T easy vector and 5μl of rapid ligation buffer in a total volume of 10 μL. After incubation for 1 hour at room temperature, we carried out transformation of high-efficiency competent cells using 10μL of ligation reactions. Mix was successively incubated for 20 minutes on ice, heat-shocked for 45 seconds at 42 °C, and chilled for 2 minutes on ice. Transformed competent cells were then recovered in 500 μl of LB media for 1 hour at 37°C with shaking (300 RPM) and transformants were selected on carbenicillin-containing 2xYT agar plates. Colonies were PCR screened and positive colonies were expanded in carbenicillin-containing 2xYT broth. DNA was purified from pGEM-T easy R116 positive clones and empty pB vector and digested with Kpn1 and Xho1. Purified DNA was then isolated by gel electrophoresis, followed by gel extraction. The R116 gene was then ligated into the pB expression vector and transformed into chemically competent cells as above. Colonies were screened by PCR and sequenced to confirm R116 insertion.
Sequence-confirmed constructs were transfected into rat RFL6 fibroblasts. The pB vector is a TET-Off system, so treatment with doxycycline (250μg/ml) was used to silence R116 gene expression. R116 production in absence of doxycycline was confirmed by western blot. R116-expressing RFL6 cells were infected with RCMV-116–2xStop to produce a R116 complemented RCMV. Spreading of infection was monitored by GFP detection.
Virion purification of UL116 HiBiT.
HCMV UL116 HiBiT was expanded over NHDF fibroblasts. Supernatants were pelleted at 76,755×g for 2 hours over a 20% sorbitol cushion. Viral particles were then resuspended in TNE buffer (50mM Tris [pH 7.4], 100mM NaCl, 10mM EDTA) and layered over a discontinuous 10-to-50% Histodenz gradient and centrifuged at 111,132 ×g for 2 hours at 4°C. Virus banded at 20–30% Histodenz was brought up in PBS and pelleted at 76,755 ×g for 2 hours through a 20% sorbitol cushion. The virus pellet was resuspended in a minimal volume of PBS.
qPCR for genome copies of UL116 HiBiT:
DNA was extracted from histodenz-purified virus with the GeneJet viral DNA and RNA purification kit (ThermoFisher). Dilutions of DNA were prepared (Undiluted, 1:10, 1:100, 1:1000) and qPCR against HCMV DNA pol (UL54) was performed in triplicate. HCMV TB40e of known genome copies was used to create a standard curve in triplicate. UL54 primers and probe were as follows: (P1: CGCTGCGAGTCCACCTTATAC, P2: ACCGGTTACAACATCAACTCTTTTG, Probe: CGCGTGAGGATGTACT). Genome copies per μL of the histodenz-purified virus preparation was determined for use in calculating UL116-HiBiT molecules/genome copy.
HiBiT lytic detection assay for UL116-HiBiT.
For infection of NHDF with UL116 HiBiT, supernatants were removed from cells and 25μL per well of fresh media was added to cells. For detection of virion incorporated proteins, three quantities of histodenz-purified HCMV UL116 HiBiT (7.5μL, 3.75μL, 1.875μL) were diluted with PBS to 25μL in a black 96-well plate and assayed in triplicate. A standard curve was generated using HiBiT control protein (ProMega) from 1000 to 15.625 nM. For all assays, 25μL per well of HiBiT lytic detection reagent with LgBiT and substrate was then added to wells as per the manufacturer’s protocol. Plates were incubated at room temperature for 10 minutes on an orbital shaker. Luminescence was read on a 96-well plate reader with a gain setting of 135.
Table 2.
Primer and probe sets for qRT-PCR.
| Probe | |||
|---|---|---|---|
| R54 | 5’-CCTCACGGGCTACAACATCA-3’ | 5’-GAGAGTTGACGAAGAACCGACC-3’ | 5’-VIC-CGGCTTCGATATCAAGTATCTCCTGCACC-TAMRA-3’ |
| R114 | 5’-ACCTTTACGGAACCGGAGTTG-3’ | 5’-ACGGACAAGGTCGATAGGGA-3’ | 5’-FAM-ACTCCAACGGTGCACGT-MGB-3’ |
| R115 | 5’-GAGCAACTCCGCGTCTTACTCA-3’ | 5’-TACCTGGCCGGATACAACGA-3’ | 5’-FAM-TCATCCAATCCTCTCC-MGB-3’ |
| R116 | 5’-TCCGGCTGAATAAGACCTCG-3’ | 5’-CCCATCCTCAACAGCACACA-3’ | 5’-FAM-CATAGACCCGAAGATCTGTA-MGB-3’ |
Highlights.
Rat cytomegalovirus R116 is a virion-associated glycoprotein
R116 is required for production of infectious virus
Human cytomegalovirus UL116 is required for the production of infectious virus
Acknowledgments
The authors would like to thank Drs. Jeremy P. Kamil (LSU) and Brent Ryckman (MSU) for their critical reading of the manuscript and constructive discussion over the course of these studies. This work was supported by a grant from the National Institutes of Health NIAID R01 AI116633. IJ was supported by the OHSU Molecular Microbiology and Immunology Interactions at the Microbe/Host Interface training grant NIH 2T32 AI007472 and NIH F31 1F311AI145193-01.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Declaration of competing interest
Authors declare no competing interest.
References
- 1.Boeckh M; Geballe AP Cytomegalovirus: pathogen, paradigm, and puzzle. J. Clin. Invest 2011, 121, 1673–1680, doi: 10.1172/JCI45449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Dzabic M; Rahbar A; Yaiw K-C; Naghibi M; Religa P; Fellstrom B; Larsson E; Soderberg-Naucler C Intragraft Cytomegalovirus Protein Expression Is Associated With Reduced Renal Allograft Survival. Clin. Infect. Dis 2011, 53, 969–976, doi: 10.1093/cid/cir619. [DOI] [PubMed] [Google Scholar]
- 3.Gatault P; Halimi J; Forconi C; Thibault G; Barbet C; Mérieau E; Graffin-Gaudy C; Marlière J-F; Goudeau A; Bruyère F; et al. CMV Infection in the Donor and Increased Kidney Graft Loss : Impact of Full HLA-I Mismatch and Posttransplantation CD8+ Cell Reduction. Am. J. Transplant 2013, 2119–2129, doi: 10.1002/ajt.12298. [DOI] [PubMed] [Google Scholar]
- 4.Leeaphorn N; Garg N; Thamcharoen N; Khankin EV; Cardarelli F; Pavlakis M Cytomegalovirus mismatch still negatively affects patient and graft survival in the era of routine prophylactic and preemptive therapy: A paired kidney analysis. Am. J. Transplant 2018, 19, ajt.15183, doi: 10.1111/ajt.15183. [DOI] [PubMed] [Google Scholar]
- 5.Helanterä I; Koskinen P; Finne P; Loginov R; Kyllönen L; Salmela K; Grönhagen-Riska C; Lautenschlager I Persistent cytomegalovirus infection in kidney allografts is associated with inferior graft function and survival. Transpl. Int 2006, 19, 893–900, doi: 10.1111/j.1432-2277.2006.00364.x. [DOI] [PubMed] [Google Scholar]
- 6.Committee to Study Priorities for Vaccine Development; Division of Health Promotion and Disease Prevention; Institute of Medicine Vaccines for the 21st Century; Stratton KR, Durch JS, Lawrence RS, Eds.; National Academies Press: Washington, D.C., 2000; ISBN 978-0-309-05646-5. [PubMed] [Google Scholar]
- 7.Macagno A; Bernasconi NL; Vanzetta F; Dander E; Sarasini A; Revello MG; Gerna G; Sallusto F; Lanzavecchia A Isolation of Human Monoclonal Antibodies That Potently Neutralize Human Cytomegalovirus Infection by Targeting Different Epitopes on the gH/gL/UL128–131A Complex. J. Virol 2010, 84, 1005–1013, doi: 10.1128/JVI.01809-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Shen S; Wang S; Britt WJ; Lu S DNA vaccines expressing glycoprotein complex II antigens gM and gN elicited neutralizing antibodies against multiple human cytomegalovirus (HCMV) isolates. Vaccine 2007, 25, 3319–3327, doi: 10.1016/j.vaccine.2007.01.011. [DOI] [PubMed] [Google Scholar]
- 9.Shimamura M; Mach M; Britt WJ Human Cytomegalovirus Infection Elicits a Glycoprotein M (gM)/gN-Specific Virus-Neutralizing Antibody Response. J. Virol 2006, 80, 4591–4600, doi: 10.1128/JVI.80.9.4591-4600.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wussow F; Chiuppesi F; Contreras H; Diamond D Neutralization of Human Cytomegalovirus Entry into Fibroblasts and Epithelial Cells. Vaccines 2017, 5, 39, doi: 10.3390/vaccines5040039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Varnum SM; Streblow DN; Monroe ME; Smith P; Auberry KJ; Paša-Tolić L; Wang D; Camp DG; Rodland K; Wiley S; et al. Identification of Proteins in Human Cytomegalovirus (HCMV) Particles: the HCMV Proteome. J. Virol 2004, 78, 10960–10966, doi: 10.1128/JVI.78.20.10960-10966.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gardner TJ; Tortorella D Virion Glycoprotein-Mediated Immune Evasion by Human Cytomegalovirus: a Sticky Virus Makes a Slick Getaway. Microbiol. Mol. Biol. Rev 2016, 80, 663–677, doi: 10.1128/MMBR.00018-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nguyen CC; Kamil JP Pathogen at the gates: Human cytomegalovirus entry and cell tropism. Viruses 2018, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sharma S; Wisner TW; Johnson DC; Heldwein EE HCMV gB shares structural and functional properties with gB proteins from other herpesviruses. Virology 2013, 435, 239–249, doi: 10.1016/j.virol.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wille PT; Wisner TW; Ryckman B; Johnson DC Human Cytomegalovirus (HCMV) Glycoprotein gB Promotes Virus Entry In Trans Acting as the Viral Fusion Protein Rather than as a Receptor-Binding Protein. MBio 2013, 4, e00332–13, doi: 10.1128/mbio.00332-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Feire AL; Koss H; Compton T Cellular integrins function as entry receptors for human cytomegalovirus via a highly conserved disintegrin-like domain. Proc. Natl. Acad. Sci. U. S. A 2004, 101, 15470–5, doi: 10.1073/pnas.0406821101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Feire AL; Roy RM; Manley K; Compton T The Glycoprotein B Disintegrin-Like Domain Binds Beta 1 Integrin To Mediate Cytomegalovirus Entry. J. Virol 2010, 84, 10026–10037, doi: 10.1128/JVI.00710-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Soroceanu L; Akhavan A; Cobbs CS Platelet-derived growth factor-α receptor activation is required for human cytomegalovirus infection. Nature 2008, 455, 391–395, doi: 10.1038/nature07209. [DOI] [PubMed] [Google Scholar]
- 19.Kari B; Gehrz R A human cytomegalovirus glycoprotein complex designated gC-II is a major heparin-binding component of the envelope. J. Virol 1992, 66, 1761–1764, doi: 10.1128/JVI.66.3.1761-1764.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krzyzaniak M; Mach M; Britt WJ The Cytoplasmic Tail of Glycoprotein M (gpUL100) Expresses Trafficking Signals Required for Human Cytomegalovirus Assembly and Replication. J. Virol 2007, 81, 10316–10328, doi: 10.1128/JVI.00375-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mach M; Osinski K; Kropff B; Schloetzer-Schrehardt U; Krzyzaniak M; Britt W The Carboxy-Terminal Domain of Glycoprotein N of Human Cytomegalovirus Is Required for Virion Morphogenesis. J. Virol 2007, 81, 5212–5224, doi: 10.1128/JVI.01463-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ciferri C; Chandramouli S; Donnarumma D; Nikitin PA; Cianfrocco MA; Gerrein R; Feire AL; Barnett SW; Lilja AE; Rappuoli R; et al. Structural and biochemical studies of HCMV gH/gL/gO and Pentamer reveal mutually exclusive cell entry complexes. Proc. Natl. Acad. Sci. U. S. A 2015, 112, 1767–1772, doi: 10.1073/pnas.1424818112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Huber MT; Compton T The Human Cytomegalovirus UL74 Gene Encodes the Third Component of the Glycoprotein H-Glycoprotein L-Containing Envelope Complex. J. Virol 1998, 72, 8191–8197, doi: 10.1128/JVI.72.10.8191-8197.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li L; Nelson JA; Britt WJ Glycoprotein H-related complexes of human cytomegalovirus: identification of a third protein in the gCIII complex. J. Virol 1997, 71, 3090–3097, doi: 10.1128/JVI.71.4.3090-3097.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li G; Nguyen CC; Ryckman BJ; Britt WJ; Kamil JP A viral regulator of glycoprotein complexes contributes to human cytomegalovirus cell tropism. Proc. Natl. Acad. Sci 2015, 112, 4471–4476, doi: 10.1073/pnas.1419875112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kabanova A; Marcandalli J; Zhou T; Bianchi S; Baxa U; Tsybovsky Y; Lilleri D; Silacci-Fregni C; Foglierini M; Fernandez-Rodriguez BM; et al. Platelet-derived growth factor-α receptor is the cellular receptor for human cytomegalovirus gHgLgO trimer. Nat. Microbiol 2016, 1, 16082, doi: 10.1038/nmicrobiol.2016.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stegmann C; Abdellatif MEA; Laib Sampaio K; Walther P; Sinzger C Importance of Highly Conserved Peptide Sites of Human Cytomegalovirus gO for Formation of the gH/gL/gO Complex. J. Virol 2017, 91, 1–22, doi: 10.1128/JVI.01339-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Vanarsdall AL; Chase MC; Johnson DC Human Cytomegalovirus Glycoprotein gO Complexes with gH/gL, Promoting Interference with Viral Entry into Human Fibroblasts but Not Entry into Epithelial Cells. J. Virol 2011, 85, 11638–11645, doi: 10.1128/jvi.05659-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu Y; Prager A; Boos S; Resch M; Brizic I; Mach M; Wildner S; Scrivano L; Adler B Human cytomegalovirus glycoprotein complex gH/gL/gO uses PDGFR-α as a key for entry. PLoS Pathog. 2017, 13, 1–24, doi: 10.1371/journal.ppat.1006281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhou M; Lanchy J-M; Ryckman BJ Human Cytomegalovirus gH/gL/gO Promotes the Fusion Step of Entry into All Cell Types, whereas gH/gL/UL128–131 Broadens Virus Tropism through a Distinct Mechanism. J. Virol 2015, 89, 8999–9009, doi: 10.1128/JVI.01325-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adler B; Scrivano L; Ruzcics Z; Rupp B; Sinzger C; Koszinowski U Role of human cytomegalovirus UL131A in cell type-specific virus entry and release. J. Gen. Virol 2006, 87, 2451–2460, doi: 10.1099/vir.0.81921-0. [DOI] [PubMed] [Google Scholar]
- 32.Hahn G; Revello MG; Patrone M; Percivalle E; Campanini G; Sarasini A; Wagner M; Gallina A; Milanesi G; Koszinowski U; et al. Human Cytomegalovirus UL131–128 Genes Are Indispensable for Virus Growth in Endothelial Cells and Virus Transfer to Leukocytes. J. Virol 2004, 78, 10023–10033, doi: 10.1128/JVI.78.18.10023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Patrone M; Secchi M; Fiorina L; Ierardi M; Milanesi G; Gallina A Human cytomegalovirus UL130 protein promotes endothelial cell infection through a producer cell modification of the virion. J. Virol 2005, 79, 8361–8373, doi: 10.1128/JVI.79.13.8361-8373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gerna G; Percivalle E; Lilleri D; Lozza L; Fornara C; Hahn G; Baldanti F; Revello MG Dendritic-cell infection by human cytomegalovirus is restricted to strains carrying functional UL131–128 genes and mediates efficient viral antigen presentation to CD8+ T cells. J. Gen. Virol 2005, 86, 275–284, doi: 10.1099/vir.0.80474-0. [DOI] [PubMed] [Google Scholar]
- 35.Nogalski MT; Chan GCT; Stevenson EV; Collins-McMillen DK; Yurochko AD The HCMV gH/gL/UL128–131 Complex Triggers the Specific Cellular Activation Required for Efficient Viral Internalization into Target Monocytes. PLoS Pathog. 2013, 9, e1003463, doi: 10.1371/journal.ppat.1003463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Xiaofei E; Meraner P; Lu P; Perreira JM; Aker AM; McDougall WM; Zhuge R; Chan GC; Gerstein RM; Caposio P; et al. OR14I1 is a receptor for the human cytomegalovirus pentameric complex and defines viral epithelial cell tropism. Proc. Natl. Acad. Sci. U. S. A 2019, 116, 7043–7052, doi: 10.1073/pnas.1814850116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martinez-Martin N; Marcandalli J; Huang CS; Arthur CP; Perotti M; Foglierini M; Ho H; Dosey AM; Shriver S; Payandeh J; et al. An Unbiased Screen for Human Cytomegalovirus Identifies Neuropilin-2 as a Central Viral Receptor. Cell 2018, 174, 1158–1171.e19, doi: 10.1016/j.cell.2018.06.028. [DOI] [PubMed] [Google Scholar]
- 38.Caló S; Cortese M; Ciferri C; Bruno L; Gerrein R; Benucci B; Monda G; Gentile M; Kessler T; Uematsu Y; et al. The Human Cytomegalovirus UL116 Gene Encodes an Envelope Glycoprotein Forming a Complex with gH Independently from gL. J. Virol 2016, 90, 4926–4938, doi: 10.1128/jvi.02517-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kattenhorn LM; Mills R; Wagner M; Lomsadze A; Makeev V; Borodovsky M; Ploegh HL; Kessler BM Identification of Proteins Associated with Murine Cytomegalovirus Virions. J. Virol 2004, 78, 11187–11197, doi: 10.1128/JVI.78.20.11187-11197.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Streblow DN; van Cleef KWR; Kreklywich CN; Meyer C; Smith P; Defilippis V; Grey F; Fruh K; Searles R; Bruggeman C; et al. Rat Cytomegalovirus Gene Expression in Cardiac Allograft Recipients Is Tissue Specific and Does Not Parallel the Profiles Detected In Vitro. J. Virol 2007, 81, 3816–3826, doi: 10.1128/JVI.02425-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ryckman BJ; Chase MC; Johnson DC HCMV gH/gL/UL128–131 interferes with virus entry into epithelial cells: evidence for cell type-specific receptors. Proc. Natl. Acad. Sci. U. S. A 2008, 105, 14118–23, doi: 10.1073/pnas.0804365105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ryckman BJ; Rainish BL; Chase MC; Borton JA; Nelson JA; Jarvis MA; Johnson DC Characterization of the human cytomegalovirus gH/gL/UL128–131 complex that mediates entry into epithelial and endothelial cells. J. Virol 2008, 82, 60–70, doi: 10.1128/JVI.01910-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schulz U; Solidoro P; Müller V; Szabo A; Gottlieb J; Wilkens H; Enseleit F CMV Immunoglobulins for the Treatment of CMV Infections in Thoracic Transplant Recipients. Transplantation 2016, 100, S5–S10, doi: 10.1097/TP.0000000000001097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Vanarsdall AL; Ryckman BJ; Chase MC; Johnson DC Human Cytomegalovirus Glycoproteins gB and gH/gL Mediate Epithelial Cell-Cell Fusion When Expressed either in cis or in trans. J. Virol 2008, 82, 11837–11850, doi: 10.1128/JVI.01623-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chandramouli S; Malito E; Nguyen T; Luisi K; Donnarumma D; Xing Y; Norais N; Yu D; Carfi A Structural basis for potent antibody-mediated neutralization of human cytomegalovirus. Sci. Immunol 2017, 2, eaan1457, doi: 10.1126/sciimmunol.aan1457. [DOI] [PubMed] [Google Scholar]
- 46.Fouts AE; Comps-Agrar L; Stengel KF; Ellerman D; Schoeffler AJ; Warming S; Eaton DL; Feierbach B Mechanism for neutralizing activity by the anti-CMV gH/gL monoclonal antibody MSL-109. Proc. Natl. Acad. Sci 2014, 111, 8209–8214, doi: 10.1073/pnas.1404653111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chowdary TK; Cairns TM; Atanasiu D; Cohen GH; Eisenberg RJ; Heldwein EE Crystal structure of the conserved herpesvirus fusion regulator complex gH–gL. Nat. Struct. Mol. Biol 2010, 17, 882–888, doi: 10.1038/nsmb.1837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Omerović J; Lev L; Longnecker R The Amino Terminus of Epstein-Barr Virus Glycoprotein gH Is Important for Fusion with Epithelial and B Cells. J. Virol 2005, 79, 12408–12415, doi: 10.1128/JVI.79.19.12408-12415.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Plate AE; Smajlović J; Jardetzky TS; Longnecker R Functional Analysis of Glycoprotein L (gL) from Rhesus Lymphocryptovirus in Epstein-Barr Virus-Mediated Cell Fusion Indicates a Direct Role of gL in gB-Induced Membrane Fusion. J. Virol 2009, 83, 7678–7689, doi: 10.1128/JVI.00457-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Wang X; Huong S-M; Chiu ML; Raab-Traub N; Huang E-S Epidermal growth factor receptor is a cellular receptor for human cytomegalovirus. Nature 2003, 424, 456–461, doi: 10.1038/nature01818. [DOI] [PubMed] [Google Scholar]
- 51.Streblow DN; Kreklywich CN; Andoh T; Moses AV; Dumortier J; Smith PP; Defilippis V; Fruh K; Nelson JA; Orloff SL The Role of Angiogenic and Wound Repair Factors During CMV-Accelerated Transplant Vascular Sclerosis in Rat Cardiac Transplants. Am. J. Transplant 2008, 8, 277–287, doi: 10.1111/j.1600-6143.2007.02062.x. [DOI] [PubMed] [Google Scholar]
- 52.Vomaske J; Denton M; Kreklywich C; Andoh T; Osborn JM; Chen D; Messaoudi I; Orloff SL; Streblow DN Cytomegalovirus CC chemokine promotes immune cell migration. J. Virol 2012, 86, 11833–11844, doi: 10.1128/JVI.00452-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Streblow DN; Kreklywich C; Yin Q; De La Melena VT; Corless CL; Smith PA; Brakebill C; Cook JW; Vink C; Bruggeman CA; et al. Cytomegalovirus-mediated upregulation of chemokine expression correlates with the acceleration of chronic rejection in rat heart transplants. J. Virol 2003, 77, 2182–94, doi: 10.1128/jvi.77.3.2182-2194.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]


