Abstract
Senescent cells that gradually accumulate during aging are one of the leading causes of aging. While senolytics can improve aging in humans as well as mice by specifically eliminating senescent cells, the effect of the senolytics varies in different cell types, suggesting variations in senescence. Various factors can induce cellular senescence, and the rate of accumulation of senescent cells differ depending on the organ. In addition, since the heterogeneity is due to the spatiotemporal context of senescent cells, in vivo studies are needed to increase the understanding of senescent cells. Since current methods are often unable to distinguish senescent cells from other cells, efforts are being made to find markers commonly expressed in senescent cells using bulk RNA-sequencing. Moreover, single-cell RNA (scRNA) sequencing, which analyzes the transcripts of each cell, has been utilized to understand the in vivo characteristics of the rare senescent cells. Recently, transcriptomic cell atlases for each organ using this technology have been published in various species. Novel senescent cells that do not express previously established marker genes have been discovered in some organs. However, there is still insufficient information on senescent cells due to the limited throughput of the scRNA sequencing technology. Therefore, it is necessary to improve the throughput of the scRNA sequencing technology or develop a way to enrich the rare senescent cells. The in vivo senescent cell atlas that is established using rapidly developing single-cell technologies will contribute to the precise rejuvenation by specifically removing senescent cells in each tissue and individual.
Keywords: aging, cellular senescence, heterogeneity, single-cell RNA sequencing, transcriptomics
INTRODUCTION
Various types of damages, such as those in tissues, cells, and molecules, accumulate during aging and often lead to cellular senescence (Soares et al., 2014; Zhang et al., 2015). Senescent cells are secretory cells that are still metabolically active, although their cell cycle is stably stopped (Dörr et al., 2013). Senescent cells accumulate with age and have a significant impact on the health and longevity of an individual (Biran et al., 2017; Burd et al., 2013; Jeyapalan et al., 2007; Zhu et al., 2014). Cellular senescence is induced by various stresses. For example, telomere shortening occurs due to repetitive cell division, leading to a kind of senescence called replicative senescence, or Hayflick’s limit (Biran et al., 2017; Burd et al., 2013; Jeyapalan et al., 2007; Zhu et al., 2014). Other stresses, including the activation of oncogenes such as Ras, the inhibition of tumor suppressor genes such as p53, the accumulation of DNA damage, chromatin disruption, and reactive oxygen species, can trigger senescence.
However, cellular senescence can also occur in specific physiological conditions, such as development and wound healing (Demaria et al., 2014; Storer et al., 2013). Senescent cells affect the surrounding cells by secreting senescence-associated secretory phenotypes (SASPs), including cytokines, chemokines, growth factors, and matrix metalloproteases (Acosta et al., 2008; Coppé et al., 2008; Krtolica et al., 2001). Depending on the physiological context, SASPs can be involved in diverse mechanisms, such as immune cell recruitment, inflammation, and extracellular matrix (ECM) remodeling (Borodkina et al., 2018; Coppé et al., 2008; Xu et al., 2015). For example, SASPs can result in immune activation, growth arrest, and differentiation, processes that are essential for tissue restoration. SASPs can also result in cell growth, migration, and invasion, which have adverse effects on cancer. This context difference also exists in young and old tissues. In young tissues, the effects of SASPs are temporary because senescent cells can be eliminated by the immune cells recruited by pro-inflammatory SASPs (Demaria et al., 2014; Kang et al., 2011; Xue et al., 2007); thus, senescent cells facilitate wound healing, tumor suppression, and the restoration of tissue homeostasis (Campisi, 2001; Demaria et al., 2014; Storer et al., 2013). Conversely, as we get older, the removal of senescent cells becomes slower, resulting in the gradual accumulation of senescent cells. In other words, the effect of SASPs persists, likely resulting in chronic inflammation, aging-related deterioration or disease, and tumorigenesis (Chen et al., 2020; Coppé et al., 2008; Krizhanovsky et al., 2008; Nathan and Ding, 2010). It remains unclear whether the accumulation of senescent cells inhibits the immune surveillance system or if the aging of the immune surveillance system leads to the accumulation of senescent cells (Ray and Yung, 2018).
While senescence has a beneficial aspect in facilitating development and repairing damage, it has another aspect that induces organ dysfunction and aging. Therefore, there have been various attempts to reverse aging by regulating the accumulation of senescent cells and SASPs (Paez‐Ribes et al., 2019). Among these efforts, senolytics attempt to improve aging by selectively eliminating senescent cells. For example, when p16-expressing senescent cells are specifically eliminated in aged mice (p16-3MR, INK/ATTAC, etc.), aging-related dysfunctions are improved, and healthspan is extended (Pajvani et al., 2005). Most senolytic drugs induce the death of apoptosis-resistant senescent cells via a pro-apoptotic mechanism (Lozano-Torres et al., 2019). Recently, senolytics have been extended to chimeric antigen receptor (CAR) T-cell therapy that precisely target surface molecules expressed on various senescent cells. CAR T-cell therapy could inhibit liver fibrosis and restored liver function in old mice (Amor et al., 2020).
However, this strategy does not work effectively against all types of senescent cells since cellular senescence is induced by various mechanisms to have different transcription signatures (Hernandez-Segura et al., 2017). In particular, p16, a marker for senescent cells, is not only universally expressed in various senescent cell types but is also expressed after the cell cycle is temporarily stopped (Marthandan et al., 2014). In addition, the representative senolytic drugs, such as dasatinib, quercetin, and navitoclax, had different effects depending on the cell type (Zhu et al., 2015; 2016). These data suggest that some degree of heterogeneity exists in senescent cells. Therefore, it would be essential to consider the heterogeneity of senescent cells to eliminate senescent cells more efficiently.
HETEROGENEITY OF SENESCENT CELLS
When irreparable DNA damage occurs in a cell, it dies through mechanisms such as apoptosis or necrosis or activates senescence (Childs et al., 2014). The cell cycle of senescent cells is arrested through the DNA-damage-response dependent p53/p21 pathway or the p16INK4a/RB pathway (Serrano et al., 1993). The factors determining the choice between apoptosis and senescence remain elusive. Still, given that the pro-apoptotic factors PUMA and BIM are upregulated in senescent cells (Baar et al., 2017), the senescent cell is likely committed to apoptosis but unable to enter apoptosis. Accordingly, in senescent cells, pro-inflammatory SASP components, such as interleukin (IL)-6, IL-8, and IL-1, suppress apoptosis via NF-κB by regulating the expression of anti-apoptotic B cell lymphoma proteins (Bcl) such as Bcl-2 and Bcl-Xl (Catz and Johnson, 2001; Gabellini et al., 2008; Liu et al., 2016; Lozano-Torres et al., 2019). Additionally, the increase in the translational burden of the SASPs in senescent cells leads to the unfolded protein response, which is one of the major causes of senescence-specific morphological changes, including flattened shape, enlarged nuclear and nucleolus, and accumulated lysosomal senescence-associated β-galactosidase (SA-β-gal) (Druelle et al., 2016; Muñoz-Espín and Serrano, 2014).
Various methods have been developed to detect senescent cells based on their characteristics. The SA-β-gal staining assay and quantitative analysis of biomarkers, such as p16, p21, and p53, are commonly used (Dimri et al., 1995; Krishnamurthy et al., 2004; Ressler et al., 2006; Sharpless and Sherr, 2015). However, these methods also detect non-senescent cells. For example, both quiescent cells, which are induced by serum starvation or pH change in a culture environment, and confluent cells are also stained positive in the SA-β gal staining assay (Yang and Hu, 2005). In addition, p16 is highly expressed in non-senescent macrophages and pRb-negative cells (Benassi et al., 1999; Hall et al., 2017; Hara et al., 1996). Furthermore, while premature senescence, induced by the telomere deprotection, involves p53/p21 and p16 pathways in human cells, it only involves the p53/p21 pathway in mouse cells (Smogorzewska and de Lange, 2002). In humans, during aging, p16 and p21 show tissue-specific expression patterns (Casella et al., 2019; Idda et al., 2020). Therefore, the current detection method is not only ineffective in distinguishing between the senescent and non-senescent cells but also unable to illustrate the molecular mechanisms of senescence between species. Further understanding of senescent cells is required to develop methods that detect senescent cells specifically.
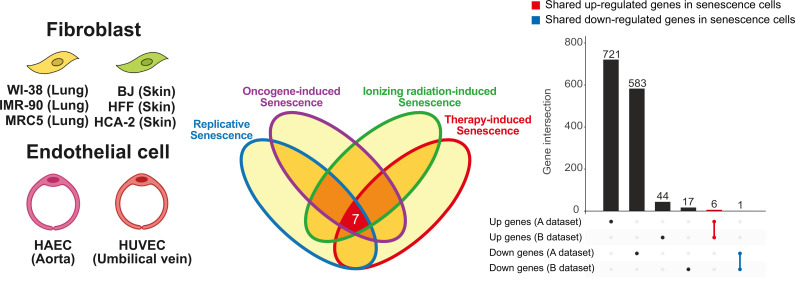
Several studies have been tried to identify common markers observed in all senescent cells through unbiased transcriptomic in vitro analysis (Fig. 1, Tables 1 and 2). The differentially expressed genes (DEGs) analysis of various human fibroblasts induced by different stimuli revealed senescence transcriptomic signatures, which were commonly changed in all fibroblasts and did not overlap with the transcriptomic signatures of quiescent cells. The core senescence signature was defined by comparing the signatures between fibroblast and the three other cell types from DEG analyses. Interestingly, this signature did not include classical senescence marker genes, such as p16, p21, LMNB1, and members of SASP, but rather genes related to DNA damage checkpoints and mitosis, such as PLK3, CCND1, DYNLT3, and CHMP5 (Hernandez-Segura et al., 2017).
Fig. 1. Transcriptonal heterogentiy of in vitro senescent cells.
Transcriptomic changes induced by various types of senescence are highly heterogeneous. The expression of only 7 genes is commonly changed in six fibroblasts and two endothelial cells during senescence induced by ionizing radiation, doxorubicin, oncogene activation, and telomere shortening. After senescence is induced, 727 genes are upregulated and 584 genes are downregulated in dataset A (Hernandez-Segura et al., 2017), while 50 genes are upregulated and 18 genes are downregulated in dataset B (Casella et al., 2019).
Table 1.
List of commonly changed genes in senescent cells
| Ensembl ID | Gene symbol | Status | Description | GO: Cellular component | Cell type | No. of RNA-seq |
|---|---|---|---|---|---|---|
| ENSG00000033100 | CHPF2 | Up | Chondroitin polymerizing factor 2 | Membrane | Fibroblast, endothelial cells | 17 |
| ENSG00000083444 | PLOD1 | Up | Procollagen-lysine,2-oxoglutarate 5-dioxygenase 1 | Membrane | Fibroblast, endothelial cells | 17 |
| ENSG00000084444 | FAM234B | Up | Family with sequence similarity 234 member B | Membrane | Fibroblast, endothelial cells | 17 |
| ENSG00000112697 | TMEM30A | Up | Transmembrane protein 30A | Membrane | Fibroblast, endothelial cells | 17 |
| ENSG00000186866 | POFUT2 | Up | Protein O-fucosyltransferase 2 | Endoplasmic reticulum membrane | Fibroblast, endothelial cells, melanocytes, keratinocytes, astrocytes | 20 |
| ENSG00000197077 | KIAA1671 | Up | Unknown protein coding | Unknown | Fibroblast, endothelial cells | 17 |
| ENSG00000143815 | LBR | Down | Lamin B receptor | Membrane | Fibroblast, endothelial cells | 17 |
Fibroblast cells: WI-38, IMR90, HCA-2, BJ, HFF, MRC-5. Endothelial cells: HUVEC, HAEC.
Table 2.
Multiple cell types bulk RNA-seq studies (in vitro, senescence-specific)
| Hernandez-Segura et al., (2017) | Casella et al., (2019) | |
|---|---|---|
| Cell lines | 6 different fibroblast strains (BJ, IMR90, HFF, MRC5, WI38, and HCA2), human neonatal foreskin epidermal melanocytes, keratinocytes, human fetal astrocytes | Human diploid fibroblast from fetal lung (WI-38, IMR-90), human aortic endothelial cells (HAECs), human umbilical vein endothelial cells (HUVECs) |
| Induction stimuli | RS, OIS, IRIS, oxidative stress | RS, IRIS, OIS, doxorubicin |
| Platform | Illumina Hiseq 2000 | Illumina Hiseq 4000 |
| No. of common DEGs | 55 | 68 |
| p16 in DEGs | No | No |
RS, replicative senescence; OIS, oncogene-induced senescence; IRIS, ionic radiation-induced senescence.
In addition, in another independently performed transcriptomic analysis, universally expressed genes were found in various cell lines, such as lung fibroblast, umbilical vein endothelial, and alveolar endothelial cells, using various cellular senescence induction stimuli, such as replicative, oncogene-induced, ionizing radiation-induced, or doxorubicin-induced senescence. According to this analysis, the expression level of senescence markers, such as p16 and p21, slightly or did not commonly change (Casella et al., 2019). When comparing these in vitro senescence transcriptomes, only 7 genes were consistently changed regardless of cell type or senescence induction method (Fig. 1, Table 1). These analyses suggest that previously known senescence markers might be insufficient as common markers.
IN VIVO TRANSCRIPTOMIC STUDY OF SENESCENT CELL
Interestingly, bulk RNA expression was found to be highly correlated to the cell type more than the method of inducing senescence. In other words, regardless of the senescence inducer, such as telomere shortening, ionizing radiation, doxorubicin treatment, and the epxression of the HRASG12V oncogene, the endothelial cells (HUVECs, HAECs) and fibroblasts (WI-38, IMR-90) are grouped, respectively (Casella et al., 2019). This observation suggests that cellular senescence is likely cell-type-specific. Furthermore, even within the same cell type, a transcriptomic feature related to the origin of cell types is more prominent than the senescence-related transcriptomic changes. These results suggest that the in vivo transcriptomic signature of cellular senescence, i.e., the occurrence of senescence in specific cell types in specific tissues, may be more intricate and diverse depending on the spatial diversity.
Two main factors likely lead to the in vivo heterogeneity of senescent cells. First, during the RNA-sequencing analysis of gene expression changes in various mouse organs, most aging-related changes were grouped by organ (Zhou et al., 2020). However, the different organs have varying epigenetic programs and microenvironments and undergo diverging aging processes (Ashapkin et al., 2017). As a result, the senescence in an organ appears heterogeneous.
Second, senescent cells are not static; their transcriptomic and epigenomic states may change over time (Fig. 2). By tracking transcriptomic changes over time, the senescence induced by ionizing radiation was divided into three stages: early senescence, after 4 days of senescence induction; intermediate senescence, after 10 days of senescence induction; and late senescence, after 20 days of senescence induction. The early, intermediate, and late stages were enriched for transcript related to DNA damage responses, signaling mediated by p38-MAPK, and cell cycle arrest, respectively. In addition, the SASP-encoding genes also significantly changed over time (Hernandez-Segura et al., 2017). Therefore, studies that consider the effects of temporal and spatial context are required to understand in vivo cellular senescence.
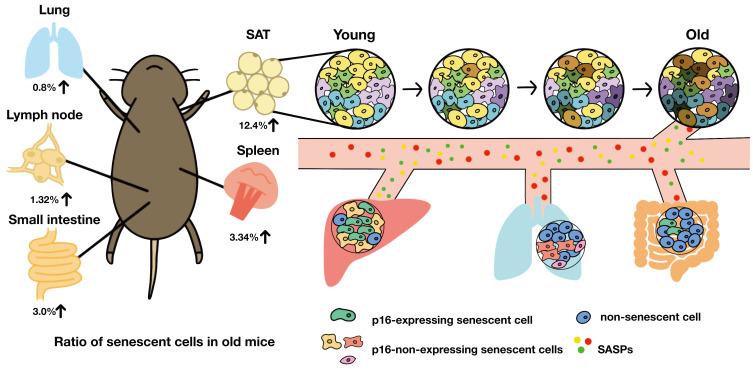
Fig. 2. Accumulation of senescent cells in vivo.
The composition of the senescent cell is different for each organ. Due to aging, heterogeneity between cells increases, and some cells become senescent. Although senescent cells are a rare population in tissue, SASP secreted by senescent cells not only affects neighboring cells but also travels through the blood to affect other tissues. The percentage of senescent cells is high in adipose tissue and low in the lung. SAT, subcutaneous adipose tissue.
Since senescent cells rarely exist in organs, it has been challenging to track and analyze their transcription signatures in vivo (Fig. 2) (Biran et al., 2017). The SASPs secreted by senescent cells vary depending on the senescence induction stimuli and cell types (Basisty et al., 2020). In vivo SASPs mediate senescent cell clearance by recruiting immune cells, maintaining tissue integrity, i.e., preventing excessive fibrosis by ECM degradation enzymes, and reprogramming adjacent cells through an NFκB-driven secretome like IL-6 (Chiche et al., 2017; Kang et al., 2011; Krizhanovsky et al., 2008; Mosteiro et al., 2016; Xue et al., 2007). Also, the persistence of the effects of SASPs could create a chronic inflammatory microenvironment that is favorable to cancer formation and senescence induction (Chen et al., 2005; Coppé et al., 2006; Perez-Mancera et al., 2014). Moreover, SASPs can travel through the blood and systemically drive aging-related dysfunctions (Fig. 2) (Baar et al., 2017; Childs et al., 2016; Wiley et al., 2019). Therefore, the contents of SASPs are likely more diverse in vivo than in vitro, adding to the complexity in analyzing senescent cells in vivo. It will be difficult to fully explain the complexity of in vivo senescent cells with only in vitro studies.
Studies based on bulk RNA-sequencing have been used to track the longitudinal changes in transcriptomes in various mice tissues and expand the understanding of in vivo aging. For example, (Schaum et al., 2019) tracked aging-related transcriptomic changes in 17 murine organs. At 12 months (mid-life), the transcripts in specific organs, such as subcutaneous and gonadal adipose tissues, changed more rapidly than in other organs. The changes in adipose tissue during aging can lead to inflammation in various tissues (Palmer and Kirkland, 2016). Indeed, genes related to the immune response continued to be activated throughout the organs from mid-life. The temporal clustering of the changes in genes showed that genes related to the extracellular matrix, mitochondrion, and protein folding were decreased with age, and the genes related to stress and immune responses were increased with age. However, these changes were not commonly detected in whole organs. In particular, no changes in the immune response genes were detected in bones, the pancreas, or the skin. No changes in genes related to protein folding were detected in white blood cells, the marrow, and the intestine. Since various transcriptomic changes, such as those in genes related to the extracellular matrix, protein folding, and immune response, were highly enriched in adipose tissues, adipose tissue is likely not only a source of inflammation but also an organ undergoing the most changes during aging.
SINGLE-CELL RNA SEQUENCING IN THE STUDY OF AGING
It is unclear whether the transcriptomic changes in each organ during aging are due to a cell-intrinsic change or a change in cellular composition. Indeed, tracking changes in the cell populations, such as the accumulation of senescent cells, the infiltration of immune cells, and the reduction of stem cells, which can significantly influence changes in aging transcriptome, are challenging in bulk RNA-sequencing. Single-cell RNA-sequencing (scRNA-seq) technology, which can identify intra-organ and inter-organ heterogeneity by tracking changes in the transcriptome of each individual cell in the aging process, could be applied to aging research (Fig. 3).
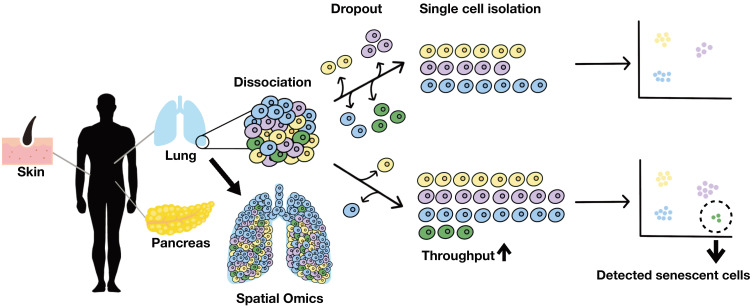
Fig. 3. The single-cell RNA-sequencing of human tissue.
scRNA-seq has been performed only in a limited number of human tissues. Due to the morphology and rarity of senescent cells, senescence can be underestimated by scRNA-seq. Since senescent cells may be lost occur during organ dissociation and library construction, it is necessary to enrich senescent cells or increase throughput to understand senescent cells thoroughly.
Unlike bulk RNA-seq, scRNA-seq needs to distinguish every single cell and molecule from a mixed sample. Most scRNA-seq methods enrich the mRNA using poly(T) oligonucleotides with barcodes that can tag distinct cells and molecules. Specifically, the cell barcode identifies each cell to facilitate multiplexing. The unique molecule identifier (UMI) is a barcode that can identify each molecule, thereby correcting amplification biases. Meanwhile, template switching (TS) is used in the reverse transcription step to obtain full-length transcripts. When a cDNA molecule reaches the 5′-end of an mRNA molecule, a few nucleotides are added to the 3′-end of the cDNA by a specific reverse transcriptase. Then, a unique TS oligo binds to this site and uses it as a template, so that cDNA synthesis continues to the 5′-end of TS oligo. Afterward, the cDNA molecule is amplified by polymerase chain reaction or in vitro transcription and can be subject to the full-length or 3′- or 5′-end mRNA sequencing.
Various microfluidic- and plate-based methods have been rapidly developed based on this process. For example, plate-based methods, such as Smart-seq2, STRT-seq, and Quartz-seq, have the advantage of higher cell capture rate and are applied in full-length or 3′- or 5′-end transcript sequencing. On the other hand, InDrops and Drop-seq, which are microfluidic system-based on single-cell encapsulation in droplets, can easily scale up the number of cells, thus increasing the possibility of detecting rare cells (Lafzi et al., 2018). Since each method has strengths and weaknesses, the appropriate method should be selected while considering the cell type, costs, efficiency, and scalability.
Recently, the Tabula Muris Senis (Mouse Aging Cell Atlas), which analyzes the aging in mice with scRNA-seq, has been released (Almanzar et al., 2020) (Table 3). In this study, 23 organs from 19 male and 11 female mice have been analyzed at 6 time points. The study observed the appearance of aging-specific cell types in various tissues, such as the liver and bladder, and the immune infiltration of various tissues. In addition, the number of T and B cells in gonadal adipose tissues increased with age, and the B cell cluster highly expressing immunoglobulin J was found specifically in old mice. Therefore, scRNA-seq enables the monitoring of the changes in cell population and the discovery of new rare populations associated with aging.
Table 3.
Multiple cell types scRNA-seq studies (in vivo, senescence non-specific)
| Kimmel et al., (2019) | Almanzar et al. (2020) | |
|---|---|---|
| Tissue | Kidney, lung, spleen | Bladder, bone marrow, brain (cerebellum, cortex, hippocampus and striatum), fat (brown, gonadal, mesenteric and subcutaneous), heart and aorta, kidney, large intestine, limb muscle and diaphragm, liver, lung, mammary gland, pancreas, skin, spleen, thymus, tongue and trachea |
| Mice age (mo) | 7, 22-23 | 1, 3, 18, 21, 24, 30 |
| Method | 10× Genomics | 10× Genomics, Smart-seq2 |
| Capture format | Droplets | Droplets, plate |
| Transcript coverage | 3’ end | 3’ end Full length |
| No. of cells | 55,293 | 529,823 |
However, scRNA-seq has difficulty detecting senescent cells, which account for only 2% of specific in vivo tissues (Biran et al., 2017). In the Tabula Muris Senis dataset, the p16-expressing cell fraction at 24 months was doubled of that in 3 months, and the expression level of p16 also increased by approximately 2-fold. However, according to a murine scRNA-seq study of 3 organs at 2 time points conducted by Calico, the fraction of p16-expressing cells and the average p16 expression level did not increase during aging (Kimmel et al., 2019) (Table 3). This inconsistency suggests that p16-expressing senescent cells are quite rare and are likely to be lost during tissue dissociation and sequencing library construction. Alternatively, the accumulation pattern of p16-expressing senescent cells may differ between individuals.
Due to ethical issues, scRNA-seq analysis has been performed only in a limited number of human tissues (Table 4 and references therein). In human lung tissues obtained from donors and pulmonary fibrosis patients, rare cell populations, such as senescent cells, appeared when pulmonary fibrosis progressed with age (Reyfman et al., 2019). Interestingly, senescence markers such as CDKN2A, IL6, SERPINE1, and GLB1, were not specifically expressed, but the transcriptomic signature of replicative senescence was enriched in senescent cells. These data suggest that in vivo, human senescent cells may have a different transcriptomic signature from in vitro senescent cells. Therefore, more in vivo longitudinal studies are needed to elucidate the transcriptomic signature of senescent cells and their effects on the surrounding tissues and organismal aging.
Table 4.
Single organ scRNA-seq studies (in vivo, senescence non-specific)
| Enge et al. (2017) | Hammond et al. (2019) | Yi et al. (2020) | Zheng et al. (2020) | De Micheli et al. (2020) | Zou et al. (2021) | |
|---|---|---|---|---|---|---|
| Species | Human | Mouse | Human, primate | Human | Human | Human |
| Organ | Pancreas | Brain (microglia) | Retina | Peripheral blood mononuclear cells (PBMCs) | Muscle | Eyelid skin |
| Age | Juvenile (1 mo, 5 y, 6 y), young adult (21 y, 22 y), adult/middle aged (38 y, 44 y, 54 y) | Embryonic day 14.5 (E14.5), early postnatal day 4/5 (P4/P5), late juvenile stage (P30), adulthood (P100), old age (P540), injury (P100) |
Human: infant (8 days), adult (35-87 y) Macaque: juvenile (2 y), adults (4-23 y) |
Cohort-1: young healthy adults (YA) (20-45 y), aged healthy adults (AA) (≥ 60 y) Cohort-2: comprising young adults (30-45 y) (YH), aged healthy adults (AH) (≥ 60 y), young COVID-19 onset patients (30-50 y), aged COVID-19 onset patients (≥ 70 y) Cohort-3: comprising YH AH, young recovered COVID-19 patients, aged recovered COVID-19 patients |
Donors (range, 41-81 y) | Young, middle aged, old* |
| Method | Smart-seq2 | 10× Genomics | 10× Genomics | 10× Genomics | 10× Genomics | 10× Genomics |
| Capture format | Plate | Droplets | Droplets | Droplets | Droplets | Droplets |
| Transcript coverage | Full length | 3’ end | 3’ end | 5’ end | 3’ end | 3’ end |
| No. of cells | 2,544 | 76,149 | 119,520 | 166,609 | Over 22,000 | 35,678 |
| Remark | An age-dependent mutational signature of endocrine cells is attributed to guanine oxidation selectively induced by reactive oxygen species. | Two microglia clusters are enriched in aging mice; one clustered has 2-4 times more microglia expressing inflammatory signals, such as Ccl4, Il1b, and Ccr5. | Human retinal aging occurs in the foveal region earlier. MYO9A− rods and the horizontal cell subtype, reduced in aging retina, are vulnerable to aging. | Age-induced immune cell polarization and expression of inflammation-related genes, such as FOS, DUSP1, IL1B, and cellular senescence-related genes, such as the CDKN family, are associated with vulnerability to COVID-19. | The muscle stem/progenitor cell (MuSC) population consists of MuSC1 and MuSC2 subpopulations. MuSC2 is enriched for inflammation markers, including CCL2, CXCL1, IL32, and TNFRSF12/FN14, that may constitute a marker set for MuSC variation in chronic muscle inflammation. | The cell-type specific downregulation of key TFs, such as KLF6 in keratinocytes and HES1 in dermal fibroblast, promote senescence phenotypes including increased SA-β-gal-positive cells and increased inflammation. |
*Specific ages are not defined.
PERSPECTIVE
Senescent cells, which have heterogeneous transcriptomic signature, are accumulated with aging. Moreover, the senescent cells can affect the surrounding cells and the whole body by SASPs. We need to understand not only the features but also the ecosystem of the senescent cells. However, the throughput of scRNA-seq, which has only captured approximately 10,000 cells to date, needs to be improved to capture the rare population of senescent cells thoroughly (Fig. 3). Also, a technology that enriches senescent cells needs to be developed. In addition, it will be necessary to establish an organ-specific, in vivo senescent cell marker gene other than p16, which may not be a suitable marker for every context.
Furthermore, the degree of heterogeneity of senescent cells in vivo and the effect of SASPs on cells located in neighbor and other tissues remain elusive. Utilizing single-cell spatial transcriptomics will help elucidate the origin of the senescent cells and the range of cells are affected by SASP. (Fig. 2). In addition, utilizing single-cell spatial transcriptomics to study various tissues longitudinally can help expand the understanding of the systemic effects of senescent cells (Roy et al., 2020).
A detailed understanding of senescent cells in vivo will enable the discovery of new targets that can be used to specifically eliminate senescent cells and the establishment of new strategies to reverse aging.
ACKNOWLEDGMENTS
This work was supported by the National Research Foundation of Korea (NRF-2020R1C1C101220611) and KRIBB Research Initiative Program.
Footnotes
AUTHOR CONTRIBUTIONS
S.K. and C.K. wrote the manuscript.
CONFLICT OF INTEREST
The authors have no potential conflicts of interest to disclose.
REFERENCES
- Acosta J.C., O'Loghlen A., Banito A., Guijarro M.V., Augert A., Raguz S., Fumagalli M., Da Costa M., Brown C., Popov N., et al. Chemokine signaling via the CXCR2 receptor reinforces senescence. Cell. 2008;133:1006–1018. doi: 10.1016/j.cell.2008.03.038. [DOI] [PubMed] [Google Scholar]
- Almanzar N., Antony J., Baghel A.S., Bakerman I., Bansal I., Barres B.A., Beachy P.A., Berdnik D., Bilen B., Brownfield D., et al. A single-cell transcriptomic atlas characterizes ageing tissues in the mouse. Nature. 2020;583:590–595. doi: 10.1038/s41586-020-2496-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amor C., Feucht J., Leibold J., Ho Y.J., Zhu C., Alonso-Curbelo D., Mansilla-Soto J., Boyer J.A., Li X., Giavridis T., et al. Senolytic CAR T cells reverse senescence-associated pathologies. Nature. 2020;583:127–132. doi: 10.1038/s41586-020-2403-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashapkin V.V., Kutueva L.I., Vanyushin B.F. Aging as an epigenetic phenomenon. Curr. Genomics. 2017;18:385–407. doi: 10.2174/1389202918666170412112130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baar M.P., Brandt R.M., Putavet D.A., Klein J.D., Derks K.W., Bourgeois B.R., Stryeck S., Rijksen Y., van Willigenburg H., Feijtel D.A., et al. Targeted apoptosis of senescent cells restores tissue homeostasis in response to chemotoxicity and aging. Cell. 2017;169:132–147.e16. doi: 10.1016/j.cell.2017.02.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basisty N., Kale A., Jeon O.H., Kuehnemann C., Payne T., Rao C., Holtz A., Shah S., Sharma V., Ferrucci L., et al. A proteomic atlas of senescence-associated secretomes for aging biomarker development. PLoS Biol. 2020;18:e3000599. doi: 10.1371/journal.pbio.3000599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benassi M.S., Molendini L., Gamberi G., Ragazzini P., Sollazzo M.R., Merli M., Asp J., Magagnoli G., Balladelli A., Bertoni F., et al. Alteration of pRb/p16/cdk4 regulation in human osteosarcoma. Int. J. Cancer. 1999;84:489–493. doi: 10.1002/(sici)1097-0215(19991022)84:5<489::aid-ijc7>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- Biran A., Zada L., Abou Karam P., Vadai E., Roitman L., Ovadya Y., Porat Z., Krizhanovsky V. Quantitative identification of senescent cells in aging and disease. Aging Cell. 2017;16:661–671. doi: 10.1111/acel.12592,. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borodkina A., Deryabin P., Giukova A.A., Nikolsky N. “Social Life” of senescent cells: what is SASP and why study it? Acta Naturae. 2018;10:4–14. [PMC free article] [PubMed] [Google Scholar]
- Burd C.E., Sorrentino J.A., Clark K.S., Darr D.B., Krishnamurthy J., Deal A.M., Bardeesy N., Castrillon D.H., Beach D.H., Sharpless N.E. Monitoring tumorigenesis and senescence in vivo with a p16INK4a-luciferase model. Cell. 2013;152:340–351. doi: 10.1016/j.cell.2012.12.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campisi J. Cellular senescence as a tumor-suppressor mechanism. Trends Cell Biol. 2001;11:S27–S31. doi: 10.1016/s0962-8924(01)02151-1. [DOI] [PubMed] [Google Scholar]
- Casella G., Munk R., Kim K.M., Piao Y., De S., Abdelmohsen K., Gorospe M. Transcriptome signature of cellular senescence. Nucleic Acids Res. 2019;47:7294–7305. doi: 10.1093/nar/gkz555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Catz S.D., Johnson J.L. Transcriptional regulation of bcl-2 by nuclear factor κB and its significance in prostate cancer. Oncogene. 2001;20:7342–7351. doi: 10.1038/sj.onc.1204926. [DOI] [PubMed] [Google Scholar]
- Chen X., Xu H., Hou J., Wang H., Zheng Y., Li H., Cai H., Han X., Dai J. Epithelial cell senescence induces pulmonary fibrosis through Nanog-mediated fibroblast activation. Aging (Albany NY) 2020;12:242. doi: 10.18632/aging.102613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Z., Trotman L.C., Shaffer D., Lin H.K., Dotan Z.A., Niki M., Koutcher J.A., Scher H.I., Ludwig T., Gerald W., et al. Crucial role of p53-dependent cellular senescence in suppression of Pten-deficient tumorigenesis. Nature. 2005;436:725–730. doi: 10.1038/nature03918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiche A., Le Roux I., von Joest M., Sakai H., Aguín S.B., Cazin C., Salam R., Fiette L., Alegria O., Flamant P., et al. Injury-induced senescence enables in vivo reprogramming in skeletal muscle. Cell Stem Cell. 2017;20:407–414.e4. doi: 10.1016/j.stem.2016.11.020. [DOI] [PubMed] [Google Scholar]
- Childs B.G., Baker D.J., Kirkland J.L., Campisi J., Van Deursen J.M. Senescence and apoptosis: dueling or complementary cell fates? EMBO Rep. 2014;15:1139–1153. doi: 10.15252/embr.201439245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Childs B.G., Baker D.J., Wijshake T., Conover C.A., Campisi J., Van Deursen J.M. Senescent intimal foam cells are deleterious at all stages of atherosclerosis. Science. 2016;354:472–477. doi: 10.1126/science.aaf6659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coppé J.P., Kauser K., Campisi J., Beauséjour C.M. Secretion of vascular endothelial growth factor by primary human fibroblasts at senescence. J. Biol. Chem. 2006;281:29568–29574. doi: 10.1074/jbc.M603307200. [DOI] [PubMed] [Google Scholar]
- Coppé J.P., Patil C.K., Rodier F., Sun Y., Muñoz D.P., Goldstein J., Nelson P.S., Desprez P.Y., Campisi J. Senescence-associated secretory phenotypes reveal cell-nonautonomous functions of oncogenic RAS and the p53 tumor suppressor. PLoS Biol. 2008;6:2853–2868. doi: 10.1371/journal.pbio.0060301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demaria M., Ohtani N., Youssef S.A., Rodier F., Toussaint W., Mitchell J.R., Laberge R.M., Vijg J., Van Steeg H., Dollé M.E., et al. An essential role for senescent cells in optimal wound healing through secretion of PDGF-AA. Dev. Cell. 2014;31:722–733. doi: 10.1016/j.devcel.2014.11.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Micheli A.J., Spector J.A., Elemento O., Cosgrove B.D. A reference single-cell transcriptomic atlas of human skeletal muscle tissue reveals bifurcated muscle stem cell populations. Skelet. Muscle. 2020;10:19. doi: 10.1186/s13395-020-00236-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dimri G.P., Lee X., Basile G., Acosta M., Scott G., Roskelley C., Medrano E.E., Linskens M., Rubelj I., Pereira-Smith O. A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U. S. A. 1995;92:9363–9367. doi: 10.1073/pnas.92.20.9363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dörr J.R., Yu Y., Milanovic M., Beuster G., Zasada C., Däbritz J.H.M., Lisec J., Lenze D., Gerhardt A., Schleicher K., et al. Synthetic lethal metabolic targeting of cellular senescence in cancer therapy. Nature. 2013;501:421–425. doi: 10.1038/nature12437. [DOI] [PubMed] [Google Scholar]
- Druelle C., Drullion C., Deslé J., Martin N., Saas L., Cormenier J., Malaquin N., Huot L., Slomianny C., Bouali F., et al. ATF6α regulates morphological changes associated with senescence in human fibroblasts. Oncotarget. 2016;7:67699–67715. doi: 10.18632/oncotarget.11505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enge M., Arda H.E., Mignardi M., Beausang J., Bottino R., Kim S.K., Quake S.R. Single-cell analysis of human pancreas reveals transcriptional signatures of aging and somatic mutation patterns. Cell. 2017;171:321–330.e14. doi: 10.1016/j.cell.2017.09.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabellini C., Castellini L., Trisciuoglio D., Kracht M., Zupi G., Del Bufalo D. Involvement of nuclear factor‐kappa B in bcl‐xL‐induced interleukin 8 expression in glioblastoma. J. Neurochem. 2008;107:871–882. doi: 10.1111/j.1471-4159.2008.05661.x. [DOI] [PubMed] [Google Scholar]
- Hall B.M., Balan V., Gleiberman A.S., Strom E., Krasnov P., Virtuoso L.P., Rydkina E., Vujcic S., Balan K., Gitlin I.I., et al. p16 (Ink4a) and senescence-associated β-galactosidase can be induced in macrophages as part of a reversible response to physiological stimuli. Aging (Albany NY) 2017;9:1867–1884. doi: 10.18632/aging.101268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammond T.R., Dufort C., Dissing-Olesen L., Giera S., Young A., Wysoker A., Walker A.J., Gergits F., Segel M., Nemesh J., et al. Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity. 2019;50:253–271.e6. doi: 10.1016/j.immuni.2018.11.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara E., Smith R., Parry D., Tahara H., Stone S., Peters G. Regulation of p16CDKN2 expression and its implications for cell immortalization and senescence. Mol. Cell. Biol. 1996;16:859–867. doi: 10.1128/mcb.16.3.859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Segura A., de Jong T.V., Melov S., Guryev V., Campisi J., Demaria M. Unmasking transcriptional heterogeneity in senescent cells. Curr. Biol. 2017;27:2652–2660.e4. doi: 10.1016/j.cub.2017.07.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Idda M.L., McClusky W.G., Lodde V., Munk R., Abdelmohsen K., Rossi M., Gorospe M. Survey of senescent cell markers with age in human tissues. Aging (Albany NY) 2020;12:4052–4066. doi: 10.18632/aging.102903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeyapalan J.C., Ferreira M., Sedivy J.M., Herbig U. Accumulation of senescent cells in mitotic tissue of aging primates. Mech. Ageing Dev. 2007;128:36–44. doi: 10.1016/j.mad.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang T.W., Yevsa T., Woller N., Hoenicke L., Wuestefeld T., Dauch D., Hohmeyer A., Gereke M., Rudalska R., Potapova A., et al. Senescence surveillance of pre-malignant hepatocytes limits liver cancer development. Nature. 2011;479:547–551. doi: 10.1038/nature10599. [DOI] [PubMed] [Google Scholar]
- Kimmel J.C., Penland L., Rubinstein N.D., Hendrickson D.G., Kelley D.R., Rosenthal A.Z. Murine single-cell RNA-seq reveals cell-identity-and tissue-specific trajectories of aging. Genome Res. 2019;29:2088–2103. doi: 10.1101/gr.253880.119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krishnamurthy J., Torrice C., Ramsey M.R., Kovalev G.I., Al-Regaiey K., Su L., Sharpless N.E. Ink4a/Arf expression is a biomarker of aging. J. Clin. Invest. 2004;114:1299–1307. doi: 10.1172/JCI22475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krizhanovsky V., Yon M., Dickins R.A., Hearn S., Simon J., Miething C., Yee H., Zender L., Lowe S.W. Senescence of activated stellate cells limits liver fibrosis. Cell. 2008;134:657–667. doi: 10.1016/j.cell.2008.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krtolica A., Parrinello S., Lockett S., Desprez P.Y., Campisi J. Senescent fibroblasts promote epithelial cell growth and tumorigenesis: a link between cancer and aging. Proc. Natl. Acad. Sci. U. S. A. 2001;98:12072–12077. doi: 10.1073/pnas.211053698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lafzi A., Moutinho C., Picelli S., Heyn H. Tutorial: guidelines for the experimental design of single-cell RNA sequencing studies. Nat. Protoc. 2018;13:2742–2757. doi: 10.1038/s41596-018-0073-y. [DOI] [PubMed] [Google Scholar]
- Liu Z., Wild C., Ding Y., Ye N., Chen H., Wold E.A., Zhou J. BH4 domain of Bcl-2 as a novel target for cancer therapy. Drug Discov. Today. 2016;21:989–996. doi: 10.1016/j.drudis.2015.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozano-Torres B., Estepa-Fernández A., Rovira M., Orzáez M., Serrano M., Martínez-Máñez R., Sancenón F. The chemistry of senescence. Nat. Rev. Chem. 2019;3:426–441. [Google Scholar]
- Marthandan S., Priebe S., Hemmerich P., Klement K., Diekmann S. Long-term quiescent fibroblast cells transit into senescence. PLoS One. 2014;9:e115597. doi: 10.1371/journal.pone.0115597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mosteiro L., Pantoja C., Alcazar N., Marión R.M., Chondronasiou D., Rovira M., Fernandez-Marcos P.J., Muñoz-Martin M., Blanco-Aparicio C., Pastor J., et al. Tissue damage and senescence provide critical signals for cellular reprogramming in vivo. Science. 2016;354:aaf4445. doi: 10.1126/science.aaf4445. [DOI] [PubMed] [Google Scholar]
- Muñoz-Espín D., Serrano M. Cellular senescence: from physiology to pathology. Nat. Rev. Mol. Cell Biol. 2014;15:482–496. doi: 10.1038/nrm3823. [DOI] [PubMed] [Google Scholar]
- Nathan C., Ding A. Nonresolving inflammation. Cell. 2010;140:871–882. doi: 10.1016/j.cell.2010.02.029. [DOI] [PubMed] [Google Scholar]
- Paez-Ribes M., González-Gualda E., Doherty G.J., Muñoz-Espín D. Targeting senescent cells in translational medicine. EMBO Mol. Med. 2019;11:e10234. doi: 10.15252/emmm.201810234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajvani U.B., Trujillo M.E., Combs T.P., Iyengar P., Jelicks L., Roth K.A., Kitsis R.N., Scherer P.E. Fat apoptosis through targeted activation of caspase 8: a new mouse model of inducible and reversible lipoatrophy. Nat. Med. 2005;11:797–803. doi: 10.1038/nm1262. [DOI] [PubMed] [Google Scholar]
- Palmer A.K., Kirkland J.L. Aging and adipose tissue: potential interventions for diabetes and regenerative medicine. Exp. Gerontol. 2016;86:97–105. doi: 10.1016/j.exger.2016.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Mancera P.A., Young A.R., Narita M. Inside and out: the activities of senescence in cancer. Nat. Rev. Cancer. 2014;14:547–558. doi: 10.1038/nrc3773. [DOI] [PubMed] [Google Scholar]
- Ray D., Yung R. Immune senescence, epigenetics and autoimmunity. Clin. Immunol. 2018;196:59–63. doi: 10.1016/j.clim.2018.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ressler S., Bartkova J., Niederegger H., Bartek J., Scharffetter-Kochanek K., Jansen-Dürr P., Wlaschek M. p16INK4A is a robust in vivo biomarker of cellular aging in human skin. Aging Cell. 2006;5:379–389. doi: 10.1111/j.1474-9726.2006.00231.x. [DOI] [PubMed] [Google Scholar]
- Reyfman P.A., Walter J.M., Joshi N., Anekalla K.R., McQuattie-Pimentel A.C., Chiu S., Fernandez R., Akbarpour M., Chen C.I., Ren Z., et al. Single-cell transcriptomic analysis of human lung provides insights into the pathobiology of pulmonary fibrosis. Am. J. Respir. Crit. Care Med. 2019;199:1517–1536. doi: 10.1164/rccm.201712-2410OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy A.L., Sierra F., Howcroft K., Singer D.S., Sharpless N., Hodes R.J., Wilder E.L., Anderson J.M. A blueprint for characterizing senescence. Cell. 2020;183:1143–1146. doi: 10.1016/j.cell.2020.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaum N., Lehallier B., Hahn O., Pálovics R., Hosseinzadeh S., Lee S.E., Sit R., Lee D.P., Losada P.M., Zardeneta M.E. Ageing hallmarks exhibit organ-specific temporal signatures. Nature. 2020;583:596–602. doi: 10.1038/s41586-020-2499-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serrano M., Hannon G.J., Beach D. A new regulatory motif in cell-cycle control causing specific inhibition of cyclin D/CDK4. Nature. 1993;366:704–707. doi: 10.1038/366704a0. [DOI] [PubMed] [Google Scholar]
- Sharpless N.E., Sherr C.J. Forging a signature of in vivo senescence. Nat. Rev. Cancer. 2015;15:397–408. doi: 10.1038/nrc3960. [DOI] [PubMed] [Google Scholar]
- Smogorzewska A., de Lange T. Different telomere damage signaling pathways in human and mouse cells. EMBO J. 2002;21:4338–4348. doi: 10.1093/emboj/cdf433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soares J.P., Cortinhas A., Bento T., Leitão J.C., Collins A.R., Gaivão I., Mota M.P. Aging and DNA damage in humans: a meta-analysis study. Aging (Albany NY) 2014;6:432–439. doi: 10.18632/aging.100667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Storer M., Mas A., Robert-Moreno A., Pecoraro M., Ortells M.C., Di Giacomo V., Yosef R., Pilpel N., Krizhanovsky V., Sharpe J., et al. Senescence is a developmental mechanism that contributes to embryonic growth and patterning. Cell. 2013;155:1119–1130. doi: 10.1016/j.cell.2013.10.041. [DOI] [PubMed] [Google Scholar]
- Wiley C.D., Liu S., Limbad C., Zawadzka A.M., Beck J., Demaria M., Artwood R., Alimirah F., Lopez-Dominguez J.A., Kuehnemann C., et al. SILAC analysis reveals increased secretion of hemostasis-related factors by senescent cells. Cell Rep. 2019;28:3329–3337.e5. doi: 10.1016/j.celrep.2019.08.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu M., Palmer A.K., Ding H., Weivoda M.M., Pirtskhalava T., White T.A., Sepe A., Johnson K.O., Stout M.B., Giorgadze N., et al. Targeting senescent cells enhances adipogenesis and metabolic function in old age. Elife. 2015;4:e12997. doi: 10.7554/eLife.12997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue W., Zender L., Miething C., Dickins R.A., Hernando E., Krizhanovsky V., Cordon-Cardo C., Lowe S.W. Senescence and tumour clearance is triggered by p53 restoration in murine liver carcinomas. Nature. 2007;445:656–660. doi: 10.1038/nature05529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang N.C., Hu M.L. The limitations and validities of senescence associated-β-galactosidase activity as an aging marker for human foreskin fibroblast Hs68 cells. Exp. Gerontol. 2005;40:813–819. doi: 10.1016/j.exger.2005.07.011. [DOI] [PubMed] [Google Scholar]
- Yi W., Lu Y., Zhong S., Zhang M., Sun L., Dong H., Wang M., Wei M., Xie H., Qu H., et al. A single-cell transcriptome atlas of the aging human and macaque retina. Natl. Sci. Rev. 2020 doi: 10.1093/nsr/nwaa179. 2020 Aug 25 [Epub]. https://doi.org/10.1101/2020.07.17.207977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Chen H.Z., Liu D.P. The four layers of aging. Cell Syst. 2015;1:180–186. doi: 10.1016/j.cels.2015.09.002. [DOI] [PubMed] [Google Scholar]
- Zheng Y., Liu X., Le W., Xie L., Li H., Wen W., Wang S., Ma S., Huang Z., Ye J., et al. A human circulating immune cell landscape in aging and COVID-19. Protein Cell. 2020;11:740–770. doi: 10.1007/s13238-020-00762-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Q., Wan Q., Jiang Y., Liu J., Qiang L., Sun L. A landscape of murine long non-coding RNAs reveals the leading transcriptome alterations in adipose tissue during aging. Cell Rep. 2020;31:107694. doi: 10.1016/j.celrep.2020.107694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Armstrong J.L., Tchkonia T., Kirkland J.L. Cellular senescence and the senescent secretory phenotype in age-related chronic diseases. Curr. Opin. Clin. Nutr. Metab. Care. 2014;17:324–328. doi: 10.1097/MCO.0000000000000065. [DOI] [PubMed] [Google Scholar]
- Zhu Y., Tchkonia T., Fuhrmann-Stroissnigg H., Dai H.M., Ling Y.Y., Stout M.B., Pirtskhalava T., Giorgadze N., Johnson K.O., Giles C.B., et al. Identification of a novel senolytic agent, navitoclax, targeting the Bcl‐2 family of anti‐apoptotic factors. Aging Cell. 2016;15:428–435. doi: 10.1111/acel.12445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Tchkonia T., Pirtskhalava T., Gower A.C., Ding H., Giorgadze N., Palmer A.K., Ikeno Y., Hubbard G.B., Lenburg M., et al. The Achilles’ heel of senescent cells: from transcriptome to senolytic drugs. Aging Cell. 2015;14:644–658. doi: 10.1111/acel.12344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zou Z., Long X., Zhao Q., Zheng Y., Song M., Ma S., Jing Y., Wang S., He Y., Esteban C.R., et al. A single-cell transcriptomic atlas of human skin aging. Dev. Cell. 2021;56:383–397.e8. doi: 10.1016/j.devcel.2020.11.002. [DOI] [PubMed] [Google Scholar]