Abstract
A self-transcribing and replicating RNA (STARR)-based vaccine (LUNAR-COV19) has been developed to prevent SARS-CoV-2 infection. The vaccine encodes an alphavirus-based replicon and the SARS-CoV-2 full-length spike glycoprotein. Translation of the replicon produces a replicase complex that amplifies and prolongs SARS-CoV-2 spike glycoprotein expression. A single prime vaccination in mice led to robust antibody responses, with neutralizing antibody titers increasing up to day 60. Activation of cell-mediated immunity produced a strong viral antigen-specific CD8+ T lymphocyte response. Assaying for intracellular cytokine staining for interferon (IFN)γ and interleukin-4 (IL-4)-positive CD4+ T helper (Th) lymphocytes as well as anti-spike glycoprotein immunoglobulin G (IgG)2a/IgG1 ratios supported a strong Th1-dominant immune response. Finally, single LUNAR-COV19 vaccination at both 2 μg and 10 μg doses completely protected human ACE2 transgenic mice from both mortality and even measurable infection following wild-type SARS-CoV-2 challenge. Our findings collectively suggest the potential of LUNAR-COV19 as a single-dose vaccine.
Keywords: SARS-CoV-2, conventional mRNA, self-amplifying RNA, STARR, LUNAR-COV19, COVID-19, vaccine, coronavirus
Graphical abstract
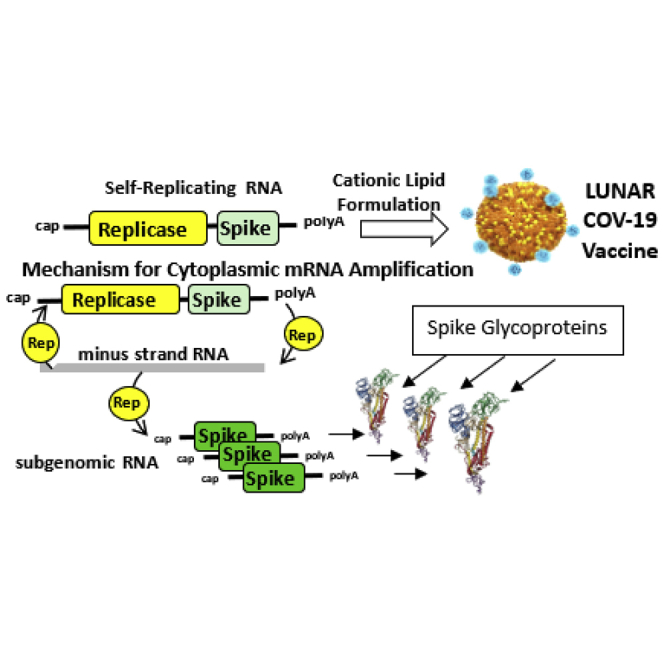
The LUNAR-COV19 SARS-CoV-2 vaccine is a self-replicating RNA-based vaccine that increases antigen expression and also duration of expression. This increased antigen expression combined with the self-activation of the innate immune system produces a low-single-dose vaccine that yields protective immunity against SARS-CoV-2 virus infection.
Introduction
The pandemic of coronavirus disease 2019 (COVID-19) has afflicted tens of millions of people, of which over 2.5 million have died from severe respiratory dysfunction and other complications of this disease.1 The etiological agent of COVID-19 is the severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), which may have first emerged from a zoonotic source to then spread from person to person until global dissemination.1 Current control measures to curb the pandemic, such as national lockdowns, closure of work places and schools, and reduction of international travel, are threatening to draw the world into a global economic recession of unprecedented scale.2 Ending the pandemic will likely require the manufacture of a variety of vaccines to generate the billions of doses required to vaccinate the global population.3 Encouragingly, hundreds of different vaccine development efforts are currently in progress, currently 16 of which have even entered phase III clinical trials.4,5
Of these candidates, two vaccines have been granted emergency use authorization in several countries based on early phase III results demonstrating nearly 95% protection from developing COVID-19. Both candidates, one from Moderna and the other from Pfizer-BioNTech, mediate immunity by a prime and boost regimen through intramuscular (i.m.) injection of a messenger RNA (mRNA) encoding the SARS-CoV-2 S antigen encased by a lipid nanoparticle (LNP). The use of adjuvants is obviated by the ability of the LNP itself to activate an innate immune response.6,7 The swift development of these RNA vaccines from design to demonstrated efficacy in humans highlights the advantages of such a system.5 The cell-free, rapidly scalable techniques used to manufacture these vaccines provide an additional advantage.8
Despite these benefits, the need to manufacture the billions of vaccine doses required to immunize the world’s population remains a challenging problem.9 This issue is compounded by the need to immunize individuals with two doses of the currently approved vaccines, as a single dose does not elicit robust immunity likely needed for protection.10, 11, 12 Although some have demonstrated robust anti-SARS-CoV-2 immunity in a small animal model with a single dose of mRNA vaccine, self-replicating mRNA (replicon) vaccines may offer dual advantages of protective immunity with a single low-dose administration.8 A single-dose vaccine would not only avoid logistics and compliance challenges associated with multi-dose vaccines but also allow vaccination of more individuals with each batch.9
Herein, the immunogenicity of and host response to a self-replicating RNA vaccine using Arcturus’ proprietary self-transcribing and replicating RNA (STARR technology) against SARS-CoV-2 was examined. Our findings indicate the added benefit of self-replication in amplifying the immunogenicity of RNA vaccine against COVID-19.
Results
Design and expression of LUNAR-COV19
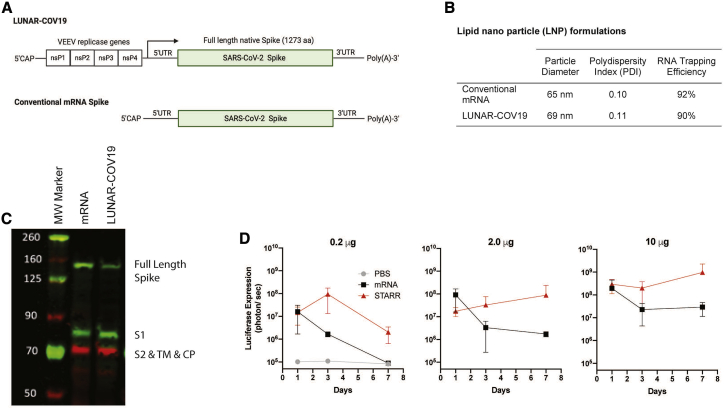
LUNAR-COV19 was designed to encode the full-length, unmodified, SARS-CoV-2 spike (S) protein (1,273 aa) as well as the Venezuelan equine encephalitis virus (VEEV) replicase genes, nsP1, nsP2, nsP3, and nsP4, required for self-amplification (Figure 1A). To determine the usefulness of including the VEEV replicase gene complex in the same positive RNA strand bearing the S gene, the LUNAR-COV19 vaccine candidate was tested against a conventional mRNA control bearing the S gene only. Both sets of RNAs were produced by in vitro transcription and then formulated with the same LUNAR LNP lipid formulation. The mRNA contained a N1-methyl-pseudouridine (N1-Me-PU) substitution.
Figure 1.
Design and expression of a SARS-COV-2 vaccine with conventional mRNA and self-transcribing and replicating RNA (STARR) platforms
(A) Schematic diagram of the SARS-CoV-2 self-replicating STARR RNA (LUNAR-COV19) and conventional mRNA vaccine constructs. The STARR construct encodes for the four non-structural proteins, ns1–ns4, from Venezuelan equine encephalitis virus (VEEV) and the unmodified full-length pre-fusion spike protein of SARS-CoV-2. The mRNA construct also codes for the same SARS-CoV-2 full-length spike protein. (B) Physical characteristics and RNA trapping efficiency of the LNPs encapsulating conventional mRNA and LUNAR-COV19 vaccines. (C) Western blot detection of SARS-CoV-2 Spike protein following transfection of Hep3b cells with LUNAR-COV19 and conventional mRNA. (D) In vivo comparison of protein expression following i.m. administration of LNPs containing luciferase-expressing STARR RNA or conventional mRNA. BALB/c mice (n = 3/group) were injected i.m. with 0.2 μg, 2.0 μg, or 10.0 μg of STARR RNA or conventional mRNA formulated with the same LNPs. Luciferase expression was measured by in vivo bioluminescence on days 1, 3, and 7 post-i.m. administration. Results are shown as mean with standard deviation error bars. S1, S domain 1; S2, S domain 2; TM, transmembrane domain; CP, cytoplasmic domain; aka, also known as.
Physical chemical characterization of the LUNAR-COV19 vaccine candidate and LNP formulated mRNA showed that the LNP diameters, polydispersity indexes, and RNA trapping efficiencies were very similar (Figure 1B), despite the LUNAR-COV19 RNA transcript being ~4 times longer than the mRNA. In vitro expression of the LUNAR-COV19 and conventional mRNA control was confirmed in cell lysates 24 h post-transfection through positive western blot detection of full-length and furin-cleaved S protein (Figure 1C).
Prior to testing immunogenicity, the impact of the VEEV replicase on duration of in vivo protein expression was examined with STARR that expressed a luciferase reporter and compared with a luciferase mRNA control that contained the N1-Me-PU substitution. Increased luciferase signal in BALB/c mice receiving an i.m. injection of the STARR construct was compared to i.m. administration of equivalent doses of conventional mRNA control at all time points and tested doses (Figure 1D). These data support the potential of STARR for greater and prolonged antigen expression in vivo.
Immune gene expression following LUNAR-COV19 vaccination
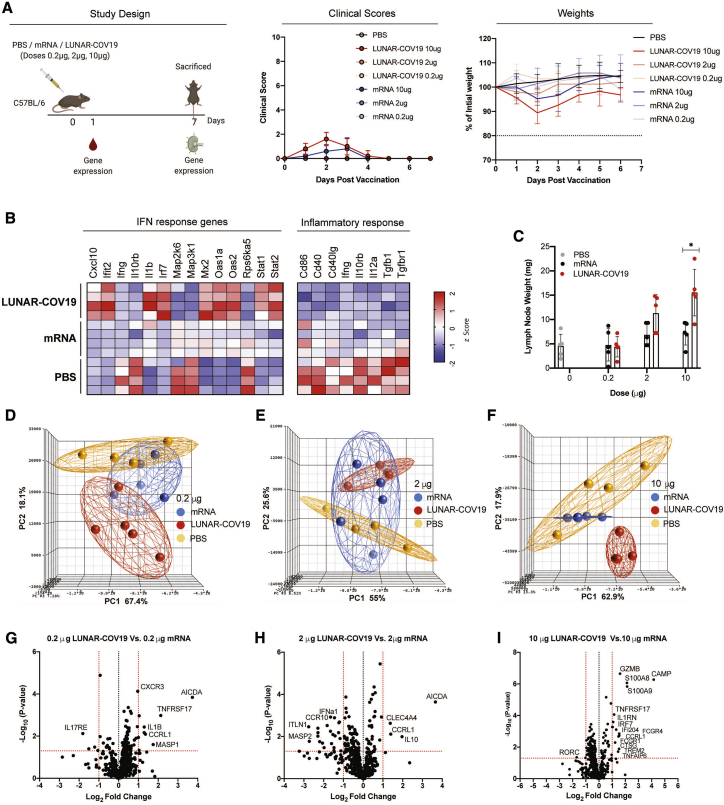
To determine how the self-replicating property of LUNAR-COV19 translates to immunogenicity, we inoculated C57BL/6J mice with LUNAR-COV19 at 0.2 μg, 2 μg, and 10 μg doses and compared these to equivalent doses of conventional mRNA controls. A negative PBS control was also included. No significant mean loss in animal weight occurred over the first 4 days, except for those that received 10 μg of LUNAR-COV19 (Figure 2A). However, apart from weight loss, there were few other clinical signs, as indicated by the minimal differences in clinical scores. Both weight and clinical scores improved after day 3 post-vaccination.
Figure 2.
Clinical scores, mouse weights, and transcriptomic analysis of immune genes after vaccination with LUNAR-COV19
(A) C57BL/6 mice (n = 5/group) were immunized with either PBS, mRNA, or LUNAR-COV19 (doses 0.2 μg, 2 μg, or 10 μg), had weight and clinical scores assessed every day, were bled at day 1 post-immunization, and were sacrificed at 7 days post-vaccination, and lymph nodes were harvested. Gene expression of inflammatory genes and immune genes were measured in whole blood (at day 1) and lymph nodes (at day 7), respectively. (B) Expression of IFN and inflammatory response genes in whole blood presented as heatmap of Z scores. (C) Lymph node weights at 7 days post-vaccination. (D–F) Principal-component analysis (PCA) of immune gene expression following vaccination with LUNAR-COV19 or conventional mRNA control at doses 0.2 μg (D), 2 μg (E), and 10 μg (F). (G–I) Volcano plots of fold change of LUNAR-COV19 versus conventional mRNA control (x axis) and log10 p value of LUNAR-COV19 versus conventional mRNA control (y axis) for doses 0.2 μg (G), 2 μg (H), and 10 μg (I). Study design schematic diagram created with BioRender.com. Weights of lymph nodes were compared between groups with a two-tailed Mann-Whitney U test; ∗0.05 < p < 0.01.
The innate immune response, particularly the type I interferon (IFN) response, has previously been shown to be associated with vaccine immunogenicity following yellow fever vaccination.13, 14, 15 Furthermore, reactive oxygen species-driven pro-inflammatory response-underpinned systemic adverse events in yellow fever vaccination have also been observed.16,17 Based on these previous observations, the expression of innate immune and pro-inflammatory genes from whole blood of C57BL/6 mice inoculated with LUNAR-COV19 or controls was tested. Genes in the type I IFN pathway were the most highly expressed in animals inoculated with LUNAR-COV19 compared with either conventional mRNA or PBS controls (Figure 2B; Figure S1). By contrast, expression of genes associated with pro-inflammatory responses were mostly reduced after LUNAR-COV19 vaccination relative to both conventional mRNA and PBS controls (Figure 2B; Figure S1).
Since adaptive immune responses develop in germinal centers in the draining lymph nodes, the draining lymph nodes at day 7 post-inoculation were tested (study schematic in Figure 2A). The inguinal lymph nodes of mice inoculated with LUNAR-COV19 but not conventional mRNA controls showed a dose-dependent increase in weight (Figure 2C). Principal-component analysis (PCA) of immune gene expression showed clustering of responses to each of the 3 doses of LUNAR-COV19 away from the conventional mRNA and PBS controls (depicted as red and orange spheres in Figures 2D–2F), indicating clear differences in immune gene expression following LUNAR-COV19 vaccination.
Differentially expressed genes were examined in the lymph nodes of mice injected with LUNAR-COV19 compared with those administered a similar dose of conventional mRNA control. Volcano plot analysis identified significant upregulation of several innate, B and T cell genes in LUNAR-COV19-immunized animals (Figures 2G–2I). Some of the most highly differentially expressed genes included GZMB (required for target cell killing by cytotoxic immune cells),18 S100A8 and S100A9 (factors that regulate immune responses via TLR4),19 TNFRSF17 (also known as BCMA; regulates humoral immunity),20 CXCR3 (chemokine receptor involved in T cell trafficking and function),21 and AICDA (mediates antibody class switching and somatic hypermutation in B cells).22 These findings collectively suggest the stimulation and development of adaptive immune responses in the draining lymph nodes of mice inoculated with LUNAR-COV19.
LUNAR-COV19 induced robust T cell responses
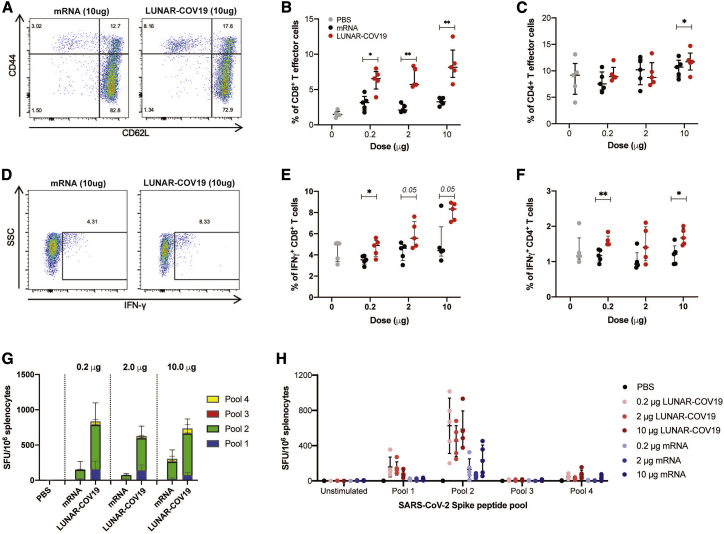
The cellular immune response following vaccination of C57BL/6 mice (n = 5 per group) with LUNAR-COV19 was investigated. At day 7 post-vaccination, spleens were harvested and assessed for CD8 and CD4 T cells by flow cytometry. The CD8+ T cell CD44+CD62L− effector/memory subset was significantly expanded in LUNAR-COV19-vaccinated mice compared with animals inoculated with PBS or conventional mRNA controls (Figures 3A and 3B). However, these same animals showed only a minimal difference in the proportion of CD4+ T effector cells (Figure 3C). IFNγ+ CD8+ T cells (with 2 μg and 10 μg doses) and IFNγ+ CD4+ T cells (in 0.2 μg and 10 μg) were proportionately higher in animals inoculated with LUNAR-COV19 compared with conventional mRNA controls (Figures 3D–3F).
Figure 3.
Cellular immune responses following vaccination with LUNAR-COV19
C57BL/6 mice (n = 5 per group) were immunized i.m. with 0.2 μg, 2.0 μg, or 10.0 μg of LUNAR-COV19 or conventional mRNA control and sacrificed at day 7 post-vaccination, and spleens were analyzed for cellular T cell responses by flow cytometry and ELISpot. (A–C) CD8+ (A and B) and CD4+ (C) T effector cells were assessed in vaccinated animals with surface staining for T cell markers and flow cytometry. (D–F) IFNγ+ CD8+ T cells (D and E) and ratio of IFNγ+ IL-4+ CD4+ T cells (F) in spleens of immunized mice were assessed after ex vivo stimulation with PMA/IO and intracellular staining. (G and H) SARS-CoV-2 Spike protein-specific responses to pooled Spike protein peptides were assessed with IFNγ ELISpot assays after vaccination with conventional mRNA controls or LUNAR-COV19. Percentage of CD8+ cells, CD4+ cells, and IFNγ- and IL-4-producing T cells were compared between groups with two-tailed Mann-Whitney U test; ∗0.05 < p < 0.01; ∗∗0.01 < p < 0.001.
SARS-CoV-2-specific cellular responses were assessed in vaccinated animals by ELISpot. A set of 15-mer peptides covering the full-length SARS-CoV-2 S were divided into 4 pools and tested for IFNγ+ responses in splenocytes of mice inoculated with LUNAR-COV19, conventional mRNA, or PBS control. SARS-CoV-2-specific cellular responses (displayed as IFNγ+ spot forming units [SFU]/106 cells) were higher across all doses in animals inoculated with LUNAR-COV19 compared with conventional mRNA controls (Figures 3G and 3H). Collectively, these findings indicate the potential of LUNAR-COV19 in eliciting SARS-CoV-2-specific cytotoxic cellular immune responses.
LUNAR-COV19 induced humoral immune responses
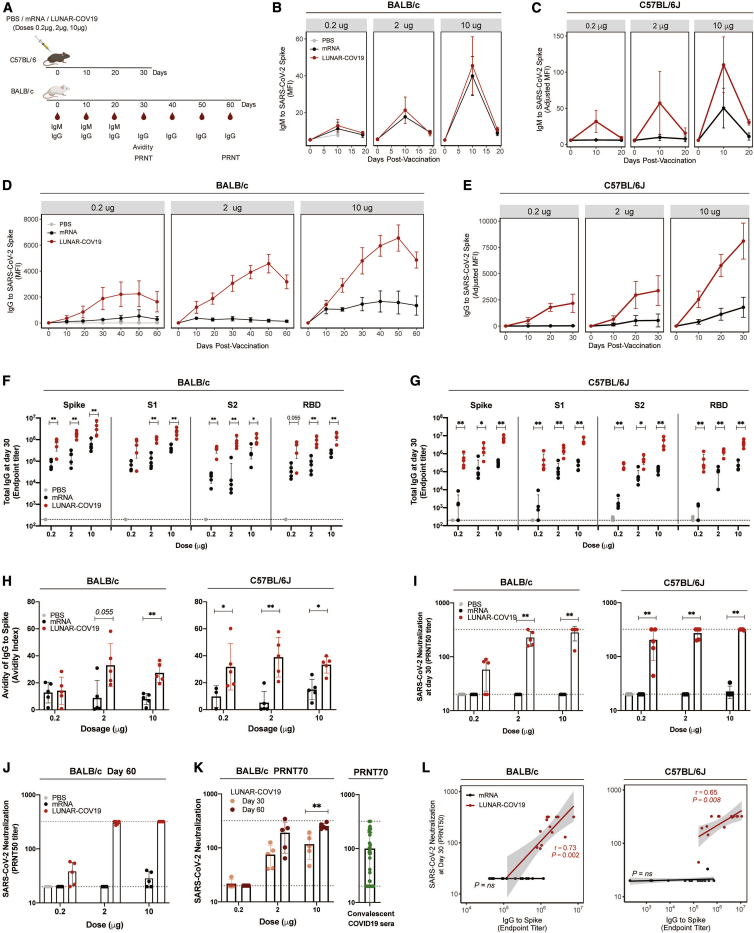
SARS-CoV-2-specific humoral responses following a single inoculation were characterized in two different mouse models, BALB/c and C57BL/6. Female mice (n = 5 per group) were inoculated at day 0 and bled every 10 days, up to day 60 for BALB/c and day 30 for C57BL/6 (Figure 4A). SARS-CoV-2 S-specific immunoglobulin M (IgM) responses were tested at 1:2,000 serum dilution with an in-house Luminex immuno-assay. All tested doses of LUNAR-COV19 produced detectable S-specific IgM responses in both mouse models (Figures 4B and 4C). Likewise, LUNAR-COV19-vaccinated BALB/c and C57BL/6 mice also produced SARS-CoV-2 S-specific IgG (at 1:2,000 serum dilution) levels. The levels of these IgG antibodies were universally higher from day 20 onward compared with conventional mRNA controls (Figures 4D and 4E). Immunized BALB/c mice were also monitored for an extended 60-day period; C57BL/6 mice were only monitored for 30 days post-inoculation. The SARS-CoV-2 S-specific IgG levels continued to increase until day 50 in mice inoculated with LUNAR-COV19 but not the equivalently dosed conventional mRNA control (Figure 4D).
Figure 4.
Antibody response following LUNAR-COV19 vaccination
(A) BALB/c and C57BL/6J mice were i.m. immunized with 0.2 μg, 2 μg, or 10 μg of LUNAR-COV19 or conventional mRNA control (n = 5/group). Blood sampling was conducted at baseline and days 10, 19, 30, 40, 50, and 60 post-vaccination for BALB/c and days 10, 20, and 30 for C57BL/6J. (B–E) IgM (B and C) and IgG (D and E) against the SARS-CoV-2 Spike protein over time, assessed with insect cell-derived whole Spike protein in a Luminex immuno-assay (measured as MFI). (F and G) IgG endpoint titers to mammalian-derived whole Spike protein and S1, S2, and RBD proteins to mammalian-derived whole Spike protein at day 30 post-vaccination were assessed in BALB/c (F) and C57BL/6J (G). (H) Avidity of SARS-CoV-2 Spike protein-specific IgG at day 30 post-immunization was measured with 8 M urea washes. (I) Neutralizing antibody (PRNT50 titers) at day 30 post-vaccination against a clinically isolated live SARS-CoV-2 virus measured in both BALB/c and C57BL/6J. Gray dashed lines depict the serum dilution range (i.e., from 1:20 to 1:320) tested by PRNT. (J and K) PRNT50 (J) and PRNT70 (K) of SARS-CoV-2 neutralization at day 30 and day 60 post-vaccination in BALB/c and convalescent sera from COVID-19 patients. (L) Correlation analysis of Spike-specific IgG endpoint titers against SARS-CoV-2 neutralization (PRNT50). Antibody data were compared between groups with a two-tailed Mann-Whitney U test; ∗0.05 < p < 0.01; ∗∗0.01 < p < 0.001.
An in-depth characterization was conducted of the SARS-CoV-2-specific IgG response at day 30 post-inoculation with LUNAR-COV19 to determine which regions of S were targeted. IgG endpoint titers were estimated to full ectodomain S protein and S domain 1 (S1), S2, and receptor binding domain (RBD) regions. As expected, although high IgG endpoint titers were also detected to S2 protein, the majority of SARS-CoV-2-specific IgG recognized S1, which contains the RBD—an immunodominant site targeted by neutralizing antibodies23,24 (Figures 4F and 4G). LUNAR-COV19-elicited IgG endpoint titers were universally and significantly higher compared with conventional mRNA controls (Figures 4F and 4G).
To complete the characterization of the antibody immune response, the binding strength (avidity) and the neutralizing ability of the antibody response elicited by LUNAR-COV19 were assessed. Serum IgG avidity was measured at day 30 post-vaccination with a modified Luminex immuno-assay with 8 M urea washes. LUNAR-COV19 elicited high-avidity S-specific IgG in both mouse models at all tested doses compared with conventional mRNA controls (Figure 4H). These differences were observed, with the exception of 0.2 μg in BALB/c, across all doses (Figure 4H), indicating that LUNAR-COV19 vaccination elicited favorable B cell affinity maturation.
Neutralization of live SARS-CoV-2 by serum from vaccinated animals was assessed by the plaque reduction neutralization test (PRNT). At day 30, LUNAR-COV19-vaccinated BALB/c mice showed a clear dose-dependent elevation in PRNT50 titers; 4 out of 5 (80%) of mice in the 10 μg LUNAR-COV19 group showed PRNT50 titers >320, which was the upper limit of the dilution (Figure 4I). Similar dose-dependent trends in PRNT50 titers were also found in C57BL/6 mice, although PRNT50 titers of several animals exceeded 320 even with the lowest 0.2 μg dose vaccination (Figure 4I). In sharp contrast, except for one C57BL/6J mouse that received the 10 μg dose, PRNT50 titers in animals inoculated with the conventional mRNA controls were all <20 (Figure 4I). Unexpectedly but encouragingly, PRNT50 and PRNT70 titers continued to rise between day 30 and day 60 in LUNAR-COV19-vaccinated animals (Figures 4J and 4K) for doses ≥2.0 μg with titers comparable to PRNT70 titers for sera from convalescent COVID-19 patients (Figure 4K; Table S1).
Further analysis revealed that the S-specific IgG titers positively correlated with PRNT50 titers with LUNAR-COV19 but not the conventional mRNA control (Figure 4L). Similar correlative findings were also observed with IgG against S1 and RBD (Figure S2). Taken collectively, the findings suggest that the SARS-CoV-2 neutralization activities of antibodies produced from LUNAR-COV19 inoculation not only are strongly associated with the amount of IgG produced but are also of a higher quality with regard to binding avidity.
LUNAR-COV19 vaccination showed a Th1-dominant response
A safety concern for a coronavirus vaccine is the risk of vaccine-associated immune enhancement of respiratory disease (VAERD).25 Indeed, SARS-CoV and MERS-CoV vaccine development has highlighted the importance of T helper (Th)1-skewed responses in mitigating the risk of vaccine-induced immune enhancement.26,27 Therefore, the Th1/Th2 balance elicited by vaccination with both conventional mRNA and LUNAR-COV19 was investigated. The IgG subclass fate of plasma cells is highly influenced by Th cells.28
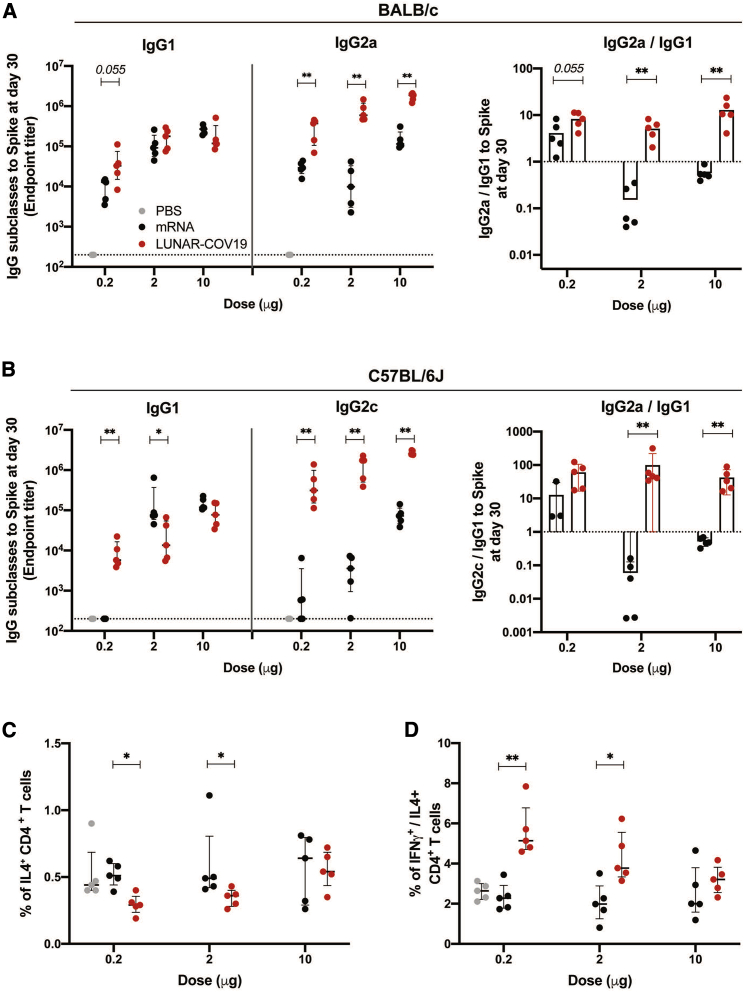
To determine whether LUNAR-COV19 showed skewing of Th1 over Th2 responses, measured Th1-associated IgG subclasses—IgG2a (BALB/c) and IgG2c (C57BL/6)—against the Th2-associated IgG1 were measured. In both strains of mice, all doses of LUNAR-COV19 vaccination induced Th1-skewed IgG subclass responses compared with conventional mRNA control-inoculated animals (Figures 5A and 5B). Similarly, ICS investigation of CD4+ T cells in the spleen of LUNAR-COV19-vaccinated C56BL/6J mice also showed a greater than unity ratio of IFNγ (Th1 cytokine) to interleukin (IL)-4 (Th2 cytokine) across all doses (Figures 5C and 5D). These findings collectively indicate that the self-replicating LUNAR-COV19 elicits Th1-dominant immune responses.
Figure 5.
LUNAR-COV19 elicits Th1-biased immune responses
(A and B) SARS-CoV-2 spike-specific IgG subclasses and the ratio of IgG2a/c/IgG1 at 30 days post-vaccination with LUNAR-COV19 compared with conventional mRNA control in BALB/c (A) and C57BL/6J (B) mice (n = 5/group). (C and D) Th2 cytokine and Th1/Th2 skew in CD4 T cells at day 7 post-vaccination in C57BL/6J mice measured by ICS as percentage of IL-4+ CD4 T cells (C) and ratio of IFNγ+/IL-4+ CD4+ T cells (D). Antibody titers and T cell data were compared between groups with a two-tailed Mann-Whitney U test; ∗0.05 < p < 0.01; ∗∗0.01 < p < 0.001.
Single dose of LUNAR-COV19 protects from a lethal infection of SARS-CoV-2
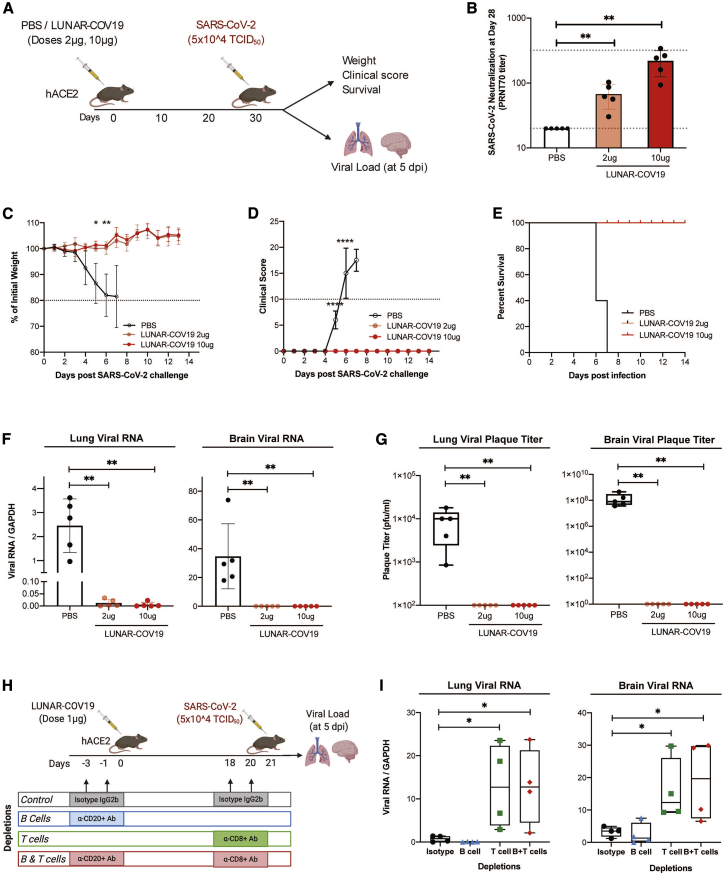
The protection against a lethal virus challenge of SARS-CoV-2 following a single dose of LUNAR-COV19 was tested in a human ACE2 (hACE2) transgenic C57BL/6 mouse model. K18-hACE2 transgenic mice immunized with either 2 μg or 10 μg of LUNAR-COV19 were intranasally challenged with a clinical isolate of SARS-CoV-2 (5 × 104 TCID50) at day 30 post-vaccination. The lethality of SARS-CoV-2 was controlled for by using PBS as a placebo control. Mice were then divided into two groups: one group was tracked for weight, clinical scores, and survival; a second group of mice was euthanized at 5 days post-injection (dpi), and viral loads were assessed in the respiratory tract (trachea to lung) and brain (Figure 6A). Measurement of PRNT70 titers confirmed the generation of neutralizing antibodies in LUNAR-COV19-vaccinated hACE2 mice (Figure 6B). Irrespective of tested dosages, mice that received the LUNAR-COV19 vaccine showed unchanged weight and no clinical sign. In contrast, negative control mice that received PBS showed significant drop in weight and increased clinical scores upon challenge with wild-type SARS-CoV-2 (Figures 6C and 6D). LUNAR-COV19 vaccination at both 2 μg and 10 μg doses fully protected hACE2 mice from an otherwise 100% mortality at day 7 post-challenge (Figure 6E). Assessment of tissue viral load at day 5 post-challenge found minimal to no SARS-CoV-2 RNA as well as no infectious viral particles in both the lung and brain of vaccinated compared with unvaccinated animal controls (Figures 6F and 6G).
Figure 6.
Single dose of LUNAR-COV19 protects hACE2 mice against a lethal challenge of SARS-CoV-2 virus
(A) hACE2 transgenic mice were immunized with a single dose of either PBS or 2 μg or 10 μg of LUNAR-COV19 (n = 5 per group), then challenged with live SARS-CoV-2 at 30 days post-vaccination, and either assessed for survival (with daily weights and clinical scores) or sacrificed at day 5 post-challenge for measurement of lung and brain tissue viral loads. (B) Live SARS-CoV-2 neutralizing antibody titers (PRNT70) measured at 28 days post-vaccination. (C–E) Weight (C), clinical score (D), and survival (E) were estimated after challenge with a lethal dose (5 × 105 TCID50) of live SARS-CoV-2 virus. (F and G) Viral RNA (F) and infectious virus (G) in the lungs and brain of challenged mice were measured with qRT-PCR and plaque assay, respectively. (H) The role of B cell and T cell depletion in LUNAR-COV19-vaccinated mice was studied after challenge with SARS-CoV-2 virus. (I) At 5 dpi, viral RNA was assessed in both mouse lungs and brain. Study design schematic diagrams were created with BioRender.com. PRNT70 and viral titers (RNA and plaque titers) were compared across groups with the non-parametric Mann-Whitney U test. Weights and clinical scores at different time points were compared between PBS and 10 μg LUNAR-COV19-immunized mice with multiple t tests. ∗0.05 < p < 0.01; ∗∗0.01 < p < 0.001; ∗∗∗0.001 < p < 0.0001; ∗∗∗∗p < 0.00001.
Finally, the contribution of protection against a SARS-CoV-2 virus challenge by vaccine-engendered B and T cell responses was assessed in the hACE2 transgenic mouse model. The antibody response to vaccination was reduced by depleting CD20+ B cells with an anti-CD20 antibody prior to LUNAR-COV19 inoculation. Depletion of B cells after vaccination would hinder the generation of IgG. Impairment of CD8+ T cell response was achieved by treating the vaccinated animals with anti-CD8 antibody prior to SARS-CoV-2 challenge (Figure S3). Treatment of vaccinated animals with matched isotype antibodies served as control. A reduced RNA dose of 1 μg of LUNAR-COV19 was used to minimize lymphocyte count expansion from vaccination (Figure 6H). Depletion of CD8+ T cells or both CD8+ T cells and CD20+ B cells resulted in breakthrough SARS-CoV-2 infection. In contrast, neither isotype-treated nor anti-CD20 only antibody-treated animals developed breakthrough SARS-CoV-2 infection (Figure 6I).
Collectively, these data show that a single dose of LUNAR-COV19 vaccine is able to elicit adaptive immunity that protects against SARS-CoV-2 challenge.
Discussion
The pandemic of COVID-19 has necessitated rapid development of vaccines. Encouragingly, after less than a year of development, several vaccines have received emergency use authorization, and more are entering first-in-human trials. However, the capacity to manufacture vaccines for billions of people globally has presented a challenge. The majority of vaccine candidates require two or more doses for sufficient adaptive immune responses, consequently complicating compliance rates in mass vaccination campaigns and resulting in fewer subjects vaccinated per manufactured lot. Vaccines that generate immunity after a single dose or with lower doses than those currently approved may lead to more efficient and rapid immunization of the global populace.
The proposed mechanism for self-replicating RNAs to enhance the immune response starts with delivery of the RNA to the cytosol; the RNA encoding the replicase is translated as a polyprotein. The nsP4 RNA-dependent RNA polymerase in combination with the polyprotein 1-2-3 makes a complementary strand of the RNA, producing a double-stranded RNA (dsRNA) replication intermediate. Multiple copies of the S glycoprotein are then transcribed by nsP4 from the complementary negative strand, and nsP1 and nsP2 facilitate 5′ capping and the addition of a poly A tail to the amplified RNA, thus increasing expression of the S glycoprotein.29 Additionally, the dsRNA replication intermediate has the potential to interact with RIG-I and/or MDA5, inducing type I IFN production,30 thus activating an innate immune response that can transition into an adaptive immune response.
Several studies have shown RNA vaccines to be immunogenic.11,31, 32, 33 While RNA vaccines have made unprecedented progress in clinical development, self-replicating RNA vaccine may elicit dose-sparing and other useful properties in the global effort to bring the COVID-19 pandemic under control.34 This study thus explored the ability of a STARR construct to safely confer improved immunogenicity of an RNA vaccine against COVID-19, specifically reducing the required RNA dose and potentially achieving protective immunity following a single vaccination. An unmutated SARS-CoV-2 S glycoprotein was expressed by STARR and mRNA. All variables were minimized to provide a direct comparison of a self-replicating RNA-based vaccine with a non-self-replicating RNA vaccine. There are reports in the literature showing that conventional mRNA vaccines can yield neutralizing antibody titers after a single priming vaccination.35, 36, 37 However, direct comparison of assay results are difficult because of differences in S glycoprotein quaternary structure, LNP formulation composition, and virus neutralization assays. However, trends in results can be compared. One study in which the furin cleavage site was inactivated produced similar anti-S glycoprotein IgG titers at 4 and 9 weeks post-vaccination and neutralizing antibody titers 9 weeks post-vaccination.35,36 However, this was achieved at RNA doses at least 10-fold greater than the lowest effective LUNAR-COV19 RNA dose (2 μg), and no virus challenge study was conducted to demonstrate protective immunity. Another study with Moderna’s mRNA-1273 vaccine, which contains two point mutations to stabilize the prefusion conformation of the S glycoprotein, also yielded neutralizing antibody titers 14 days post-prime vaccination at RNA doses comparable to LUNAR-COV19. In addition, significant reduction in lung viral RNA was observed following challenge with a mouse-adapted virus (SARS-CoV-2 MA).37 However, unlike the lethal challenge model in the present study, the virus RNA titers with the mouse-adapted strain peaked at day 2 post-infection and were cleared by day 4 post-infection, whereas K18-hACE2 (hACE2) mice used for the lethal challenge began to lose weight by day 4 post-infection along with development of clinical signs, and all mice were dead by day 7. Not only was there 100% survival 14 days post-virus challenge after a single vaccination of 2 μg, there was no weight loss, no development of clinical signs, and no change in overall behavior compared with their behavior prior to vaccination.
These preclinical results confirmed that LUNAR-COV19 vaccination elicited innate immune responses, in both the blood and draining lymph nodes of mice, similar to those elicited by a mRNA vaccine lacking the VEEV replicase but at doses that were 5–50 times lower. In terms of adaptive immunity, LUNAR-COV19 was able to elicit SARS-CoV-2 S-specific Th1-predominant humoral and cellular immune responses. Critically, it was also observed that even under circumstances where B cell responses to LUNAR-COV19 vaccination are compromised, cellular immunity generated by this vaccine could protect against SARS-CoV-2 infection. Our findings thus highlight the utility of self-replicating RNA vaccine against COVID-19.
The balance between immunogenic and reactogenic responses is important. Replication of LUNAR-COV19 results in the formation of a negative-strand template for production of more positive-strand mRNA and sub-genomic mRNA expressing the S transgene. Interaction between the negative and positive strands forms a dsRNA intermediate, which would interact with TLR3 and RIG-I-like receptors to stimulate type I IFN responses,38, 39, 40 which we and others have previously shown to correlate with superior adaptive immune responses.13, 14, 15 Our data show a 10% weight loss in mice receiving the highest dose, an unfortunate consequence of increased innate immune responses. However, weight loss is reduced at the 2 μg dose, comparable to that of the (nucleoside modified) mRNA at 10 μg, while showing significant increases in adaptive immune responses.
It is unclear whether the VEEV nsP1-4 forming the replication complex contain any immunogenic properties, although mutations in the nsP proteins have been shown to affect the induction of type I IFN.41 Although unexplored in our current study, VEEV replicons have also been shown to adjuvant immune responses at mucosal sites,42 further justifying the use of the STARR platform to develop a COVID-19 vaccine.
In conclusion, the STARR vaccine platform, as exemplified by LUNAR-COV19, offers an approach to simulate key immunogenic properties of live virus vaccination and offers the potential for an effective single-shot vaccination against COVID-19.
Materials and methods
Vaccine plasmid constructs and design
A human codon-optimized S glycoprotein gene of SARS-CoV-2 (GenBank: YP_009724390) was cloned into plasmids pARM2922 and pARM2379 for generation of SARS-CoV-2 S-expressing STARR and conventional mRNA, respectively. The STARR plasmid also encoded the VEEV non-structural proteins nsP1, nsP2, nsP3, and nsP4 (GenBank: MH891622), which together form the replicase complex that binds to the VEEV 26 S sub-genomic promoter placed right before the S protein sequence (sequences will be made available upon request). The cloned portions of all plasmid constructs were verified by DNA sequencing. The pARM2922 and pARM2379 DNA plasmids were linearized immediately after the poly(A) stretch and used as a template for in vitro transcription reaction with T7 RNA polymerase. For LUNAR-COV19 vaccine, the reaction for RNA was performed as previously described,43 with proprietary modifications to allow highly efficient co-transcriptional incorporation of a proprietary Cap1 analog and to achieve a high-quality RNA molecule >11,000 nt long. RNA was then purified through silica column (Macherey-Nagel) and quantified by UV absorbance. For the conventional mRNA vaccine, the RNA was also synthesized as previously described43 but with 100% substitution of UTP with N1-Me-PU. For both LUNAR-COV19 and conventional mRNA vaccines, the RNA quality and integrity were verified by 0.8%–1.2% non-denaturing agarose gel electrophoresis as well as Fragment Analyzer (Advanced Analytical). The purified RNAs were stored in RNase-free water at −80°C until further use.
Vaccine LNPs
LUNAR nanoparticles encapsulating STARR were prepared by mixing an ethanolic solution of lipids with an aqueous solution of STARR. Lipid excipients (Arcturus Therapeutics proprietary ionizable lipid, DSPC, cholesterol, and PEG2000-DMG) are dissolved in ethanol at a mole ratio of 50:10:38.5:1.5 or 50:13:35.5:1.5. An aqueous solution of the vaccine RNA is prepared in citrate buffer pH 4.0. The lipid mixture is then combined with the vaccine RNA solution at a flow rate ratio of 1:3 (V/V) via a proprietary mixing module. Nanoparticles thus formed are stabilized by dilution with phosphate buffer followed by HEPES buffer, pH 8.0. Ultrafiltration and diafiltration (UF/DF) of the nanoparticle formulation is then performed by tangential flow filtration (TFF) using modified PES hollow-fiber membranes (100 kDa MWCO) and HEPES pH 8.0 buffer. After UF/DF, the formulation is filtered through a 0.2 μm PES filter. An in-process RNA concentration analysis is then performed. Concentration of the formulation is adjusted to the final target RNA concentration, followed by filtration through a 0.2 μm PES sterilizing-grade filter. After sterile filtration, bulk formulation is aseptically filled into glass vials, stoppered, capped, and frozen at −70°C ± 10°C. Analytical characterization included measurement of particle size and polydispersity by dynamic light scattering (ZEN3600, Malvern Instruments), pH, osmolality, RNA, content and encapsulation efficiency by a fluorometric assay using RiboGreen RNA reagent, RNA purity by capillary electrophoresis using fragment analyzer (Advanced Analytical), and lipid content by high-performance liquid chromatography (HPLC).
In vitro transfection and immunoblot detection of S protein
Hep3b cells (seeded in 6-well plates at a density of 7 × 105 cells/well a day before) were transfected with purified in vitro transcripts (IVTs) (2.5 μg conventional mRNA control and 2.5 μg STARR) by Lipofectamine MessengerMax transfection reagent (Thermo Fisher Scientific) according to the manufacturer’s instructions. The cells were harvested the next day with a hypotonic buffer (10 mM Tris-HCl, 10 mM NaCl supplemented with protease inhibitor cocktail [Roche]), followed by sonication. Samples were deglycosylated, followed by treatment with PNGase F (New England Biolabs) according to the manufacturer’s instructions.
The protein lysate (10 μg) was resolved on a 7.5% NuPAGE Tris-acetate gel (Thermo Fisher Scientific), and the S protein expression was analyzed by the LI-COR Quantitative Western Blot system using a rabbit antibody detecting S1 (40150-T62-COV2, Sino Biological) and a mouse antibody for S2 region (GTX632604, GeneTex) along with appropriate secondary antibodies (goat anti-rabbit 800 and goat anti-mouse 680).
Animal studies
BALB/c studies
All BALB/c mouse studies were approved by the Explora BioLabs Institutional Animal Care and Use Committee (IACUC) and performed under Animal Care and Use protocol number EB-17-004-003. To examine protein expression in vivo, the STARR platform was designed to express a luciferase reporter gene. BALB/c mice (Jackson Laboratory) were immunized i.m. in the rectus femoris with STARR at doses of 0.2, 2, and 10 μg and compared to conventional non-replicating mRNA vaccine expressing the same luciferase reporter gene (n = 3 mice/group). Luciferase expression was measured at days 1, 3, and 7 post-inoculation by imaging the mice for bioluminescence.
Humoral responses following vaccination with LUNAR-COV19 or conventional mRNA controls, both containing the same S gene, were tested in female BALB/c mice (Jackson Laboratory) aged 8–10 weeks by i.m. immunization of the rectus femoris at doses of 0.2 μg, 2 μg, and 10 μg (n = 5 mice/group). Mice were bled at baseline and at 10, 19, 30, 40, 50, and 60 days post-vaccination to assess SARS-CoV-2-specific humoral immune responses.
C57BL/6
All C57BL/6 mouse studies were performed in accordance with protocols approved by the IACUC at Singapore Health Services, Singapore (ref no.: 2020/SHS/1554). C57BL/6 mice purchased from inVivos were housed in a BSL-2 animal facility at Duke-NUS Medical School. Groups of 6- to 8-week-old wild-type C57BL/6 female mice were vaccinated i.m. with either LUNAR-COV19 or conventional mRNA controls bearing the same SARS-CoV-2 S gene at doses of 0.2 μg, 2 μg, and 10 μg. For transcriptomic and T cell studies, submandibular bleeds were performed for whole blood at 24 h post-vaccination. Day 7 post-immunization, mice were sacrificed, and inguinal lymph nodes and spleens were harvested for investigation of immune gene expression and T cell responses, respectively. Splenocyte suspensions for measuring T cell responses were obtained by crushing spleen through a 70 μm cell strainer (Corning). Red blood cells were removed by lysis using BD PharmLyse reagent. For antibody studies, another set of vaccinated 6- to 8-week-old mice were bled at baseline and at 10, 20, and 30 days post-vaccination.
SARS-CoV-2 challenge studies
SARS-CoV-2 challenge experiments were conducted with female B6;SJL-Tg(K18-hACE2)2Prlmn/J (hACE2) mice purchased from Jackson Laboratory. Groups of 6- to 8-week-old wild-type C57BL/6 female mice were vaccinated i.m. with 100 μL of LUNAR-COV19 at doses of 2 μg and 10 μg. Submandibular bleeds were performed for serum isolation to determine antibody titers via PRNT 28 days post-vaccination. Animals were infected with 5 × 104 TCID50 in 50 μL via the intranasal route. Daily weight measurements and clinical scores were obtained. Mice were sacrificed when exhibiting >20% weight loss or clinical score of 10. To assess organ viral loads, mice were sacrificed 5 days post-infection and harvested organs were frozen at −80°C. Whole lungs and brains were homogenized with MP lysing matrix A and F according to manufacturer’s instructions in 1 mL of PBS. Homogenate was used to assess both plaque titers and RNA extraction with TRIzol LS (Invitrogen). No blinding was done for animal studies.
B cell and CD8+ T cell depletion studies
All CD20+ B and CD8+ T cell depletion studies followed by wild-type SARS-CoV-2 challenge were carried out by adapting a previously published approach. B cells were depleted from hACE2 transgenic mice through the intraperitoneal (i.p.) administration of 50 μg of anti-CD20 antibody (BioLegend SA271G2) at 72 h (−3 days) and 24 h (−1 day) before vaccination with LUNAR-COV19. For CD8+ T cell depletion, 5 μg of anti-CD8+ antibody (Bio X Cell, Cat#BE0061) was injected i.p. at −3 days and −1 day before challenge with SARS-CoV-2. For control mice, isotype IgG2b mouse control antibody (Bio X Cell, Cat#BE0090) was similarly injected. Successful depletions of CD20+ B and/or CD8+ T cells were confirmed via flow cytometry. Briefly, 24 h after the second injection of anti-CD20+ and/or anti-CD8+ antibodies, sub-mandibular blood was stained with AF488-conjugated anti-mouse CD45R (BD, Cat#557669), APC-conjugated anti-mouse CD3 (BD, Cat#565643), and BUV395-conjugated anti-mouse CD8b antibodies (BD, Cat#74027).
Gene expression of immune and inflammatory genes
Whole blood collected 1 day post-vaccination was lysed with BD PharmLyse reagent, and RNA was extracted with the QIAGEN RNAeasy kit. Mouse lymph nodes collected from 7 days post-vaccination were homogenized, and RNA was extracted with TRIzol LS. RNA (50 ng) from whole blood cells and lymph nodes were hybridized to the NanoString nCounter mouse inflammation and immunology v2 panels (NanoString Technologies), respectively. As previously described,17,44 RNA was hybridized with reconstituted CodeSet and ProbeSet. Reactions were incubated for 24 h at 65°C and ramped down to 4°C. Hybridized samples were then immobilized onto a nCounter cartridge and imaged on a nCounter SPRINT (NanoString Technologies). Data were analyzed with nSolver Analysis software (NanoString Technologies) and Partek Genomics Suite. For normalization, samples were excluded when percentage field of vision registration was <75, binding density outside the range 0.1–1.8, positive control R2 value was <0.95, and 0.5 fM positive control was ≤2 SD above the mean of the negative controls. Background subtraction was performed by subtracting estimated background from the geometric means of the raw counts of negative control probes. Probe counts less than the background were floored to a value of 1. The geometric mean of positive controls was used to compute positive control normalization parameters. Samples with normalization factors outside 0.3–3.0 were excluded. The geometric mean of housekeeping genes was used to compute the reference normalization factor. Samples with reference factors outside the 0.10–10.0 range were also excluded. Hierarchical clustering was performed with Partek Genomics Suite v.6 on gene sets and Z score values by Euclidean dissimilarity and average linkage.
To identify DEGs between groups, Partek Genomics Suite Analysis v.7 software was used to analyze variance (ANOVA) with a cutoff of p < 0.05. Log2 fold changes generated were used for volcano plots constructed with Prism v.8.1.0 software. DEGs were identified by a fold change cutoff of 2. Unsupervised PCA was performed to visualize variability between vaccinated and non-vaccinated animals with Partek Genomics Suite Analysis v.7 software. PCA ellipsoids were drawn with a maximum density and 3 subdivisions.
T cell assessment using flow cytometry
Surface staining was performed on freshly isolated splenocytes with the following panel of antibodies and reagents: B220 (RA3-6B2), CD3 (17A2), CD4 (RM4-5), CD8α (53-6.7), CD44 (IM7), CD62L (MEL-14) and DAPI. Intracellular cytokine staining was performed by stimulating freshly isolated splenocytes with 50 ng/mL phorbol myristate acetate (PMA) and 500 ng/mL ionomycin (IO) in the presence of GolgiPlug (BD) for 6 h. After stimulation, surface staining of CD3, CD4, and CD8a was performed, followed by intracellular staining of IFNγ (XMG1.2) and IL-4 (11B11). Data acquisition was performed on a BD LSRFortessa and analyzed with FlowJo.
ELISpot
ELISpot was performed with the Mouse IFN-γ ELISpotBASIC Kit (Mabtech). A similar protocol has been used for human SARS-CoV-2 samples.45. In brief, 4 × 105 freshly isolated splenocytes were plated into polyvinylidene fluoride (PVDF)-coated 96-well plates containing IFNγ capture antibody (AN18). Cells were stimulated with a 15-mer peptide library covering part of the S protein. One hundred forty-three total peptides were divided into four pools and used at a final concentration of 1 μg/mL per peptide. Negative control wells contained no peptide. After overnight stimulation, plates were washed and sequentially incubated with biotinylated IFNγ detection antibody (R4-6A2), streptavidin-ALP, and finally BCIP/NBT. Plates were imaged with ImmunoSpot Analyzer and quantified with ImmunoSpot software.
Luminex immuno-assay
Longitudinal assessment of binding antibody
Longitudinal IgM and IgG responses in BALB/c and C57BL/6 were measured using an in-house Luminex immuno-assay. Similar Luminex immuno-assays have been previously described for antibody detection against SARS-CoV-2 antigens.46,47 Briefly, MAGPIX Luminex beads were covalently conjugated to insect-derived HIS-tagged SARS-CoV-2 whole S protein (Sino Biological) with the ABC coupling kit (Thermo) as per manufacturer’s instructions. Beads were then blocked with 1% BSA, followed by incubation with serum (diluted at 1:2,000 in block) for 1 h at 37°C. Beads are then washed, and SARS-CoV-2 S-specific mouse antibodies were detected using the relevant biotinylated secondary antibody (i.e., anti-mouse IgM-biotin and anti-mouse IgG-biotin; SouthernBiotech) for IgM and IgG assessment, respectively) with streptavidin-PE (SouthernBiotech). Antibody binding to S was then measured on a MAGPIX instrument as median fluorescence intensity (MFI). S antigen quantity on beads was also probed with anti-6xHIS-PE antibodies, and all MFI values were then corrected to S antigen quantity to account for experiment-to-experiment variation.
IgG and IgG subclass endpoint titers
IgG endpoint titers to mammalian-derived SARS-CoV-2 S, S1, S2 and RBD in sera at day 30 post-immunization were measured by Luminex immuno-assay. Assay was conducted as described above, with the modification of serially diluting serum 10-fold from 200 to 2 × 108. Similarly, IgG subclass endpoint titers (i.e., IgG1 and IgG2a in BALB/c and IgG1 and IgG2c in C57BL/6) were measured against mammalian-derived SARS-CoV-2 S protein, using serially diluted mouse sera (5-fold from 200 to 3.1 × 106) and secondary antibodies anti-IgG1-biotin, anti-IgG2a-biotin, or anti-IgG2b-biotin (SouthernBiotech). Four parameter logistic (4PL) curves were fitted to the measured MFI data from serially diluted sera, and three times the background (i.e., 3 × MFI with no serum) was used as a threshold cutoff to estimate endpoint titers.
IgG avidity
Avidity index of IgG to SARS-CoV-2 S protein in sera at day 30 post-immunization was estimated with the Luminex immuno-assay. Assay was conducted as described above, with the minor modification of following bead incubation with serum (diluted at 1:2,000) with either a 10 min PBS or 8 M urea wash. Avidity index was estimated by subtracting background MFI from all sample values and then dividing MFI with 8 M urea wash by MFI with PBS wash.
PRNT
Neutralization of live SARS-CoV-2 was measured by PRNT at day 30 post-vaccination in both BALB/c and C57BL/6 mice. Similar protocols have been published previously for SARS-CoV-2.48 Briefly, mouse sera were serially diluted from 1:20 to 1:320 in culture media and incubated with a clinical isolate of SARS-CoV-2,BetaCoV/Singapore/2/2020 (GISAID: EPI_ISL_406973), for 1 h at 37°C. Virus-antibody mixtures were then added to Vero-E6 cells in 24-well plates, incubated for 1–2 h, and then overlaid with carboxymethyl cellulose (CMC) and incubated at 37°C and 5% CO2. At 5 days, cells were washed and stained with crystal violet, and the number of plaques was counted visually. The serum dilution leading to neutralization of 50% of virus, i.e., PRNT50, was then calculated.
Acknowledgments
We thank the Economic Development Board of Singapore for initiating this collaboration and for funding the development of LUNAR-COV19. We also thank The Hour Glass for its philanthropic support for the development of several assays reported here. R.d.A. received support from the National Medical Research Council (NMRC) Young Investigator Award. NMRC provided salary support for E.E.O. and J.G.H.L. through the Clinician-Scientist Award and S.K. through the Transition Award. ViREMiCS was established through a generous gift from the Tanoto Foundation.
Author contributions
R.d.A., E.S.G., S.C., S.M.S., and E.E.O. designed the studies. R.d.A., Y.S.L., S.L.Z., and H.C.T. conducted the antibody assays. S.C. conducted the T cell studies. E.S.G. and C.Y. conducted the C57BL/6 and K18-hACE mouse studies in Singapore. S.P., M.S., and J.A.G. conducted the BALB/c mouse immunogenicity studies in San Diego. S.M.S., D.M., K.T., H.B., E.C.A., and P.C. designed, constructed, and tested LUNAR-COV19 and mRNA expression. K.-J.J.P., M.A., J.D., and A.D. manufactured and characterized the LUNAR-COV19 and conventional mRNA RNAs. P.K., Y.B., B.C., J.V., and S.R. manufactured and characterized LUNAR COV-19 and mRNA LNPs. J.G.H.L. and S.K. obtained written informed consent from the COVID-19 cases and collected their convalescent sera. S.M.S., S.G.H., P.C., and E.E.O. reviewed the data. R.d.A., E.S.G., S.C., S.M.S., B.M.S., and E.E.O. wrote the manuscript.
Declaration of interests
D.M., E.C.A., P.H., K.-J.J.P., M.A., H.B., A.D., Y.B., B.C., J.V., S.R, J.A.G., M.S., R.Y., W.T., K.T., S.P., P.K., J.D., S.M.S., B.M.S, S.G.H., and P.C. are employees of Arcturus Therapeutics, Inc. E.E.O is an unpaid member of the Scientific Advisory Board of Arcturus Therapeutics, Inc.
Footnotes
Supplemental information can be found online at https://doi.org/10.1016/j.ymthe.2021.04.001.
Supplemental information
References
- 1.WHO . Dashboard; 2020. WHO Coronavirus Disease (COVID-19) [Google Scholar]
- 2.World Bank . 2020. The Global Economic Outlook During the COVID-19 Pandemic: A Changed World.https://www.worldbank.org/en/news/feature/2020/06/08/the-global-economic-outlook-during-the-covid-19-pandemic-a-changed-world [Google Scholar]
- 3.Randolph H.E., Barreiro L.B. Herd Immunity: Understanding COVID-19. Immunity. 2020;52:737–741. doi: 10.1016/j.immuni.2020.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.WHO . 2020. Draft landscape of COVID-19 candidate vaccines.https://www.who.int/publications/m/item/draft-landscape-of-covid-19-candidate-vaccines [Google Scholar]
- 5.Thanh Le T., Andreadakis Z., Kumar A., Gómez Román R., Tollefsen S., Saville M., Mayhew S. The COVID-19 vaccine development landscape. Nat. Rev. Drug Discov. 2020;19:305–306. doi: 10.1038/d41573-020-00073-5. [DOI] [PubMed] [Google Scholar]
- 6.Hassett K.J., Benenato K.E., Jacquinet E., Lee A., Woods A., Yuzhakov O., Himansu S., Deterling J., Geilich B.M., Ketova T. Optimization of Lipid Nanoparticles for Intramuscular Administration of mRNA Vaccines. Mol. Ther. Nucleic Acids. 2019;15:1–11. doi: 10.1016/j.omtn.2019.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zeng C., Zhang C., Walker P.G., Dong Y. Formulation and Delivery Technologies for mRNA Vaccines. Curr. Top. Microbiol. Immunol. 2020 doi: 10.1007/82_2020_217. Published online June 2, 2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Jackson N.A.C., Kester K.E., Casimiro D., Gurunathan S., DeRosa F. The promise of mRNA vaccines: a biotech and industrial perspective. NPJ Vaccines. 2020;5:11. doi: 10.1038/s41541-020-0159-8. 32047656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lurie N., Saville M., Hatchett R., Halton J. Developing Covid-19 Vaccines at Pandemic Speed. N. Engl. J. Med. 2020;382:1969–1973. doi: 10.1056/NEJMp2005630. [DOI] [PubMed] [Google Scholar]
- 10.Folegatti P.M., Ewer K.J., Aley P.K., Angus B., Becker S., Belij-Rammerstorfer S., Bellamy D., Bibi S., Bittaye M., Clutterbuck E.A. Safety and immunogenicity of the ChAdOx1 nCoV-19 vaccine against SARS-CoV-2: a preliminary report of a phase 1/2, single-blind, randomised controlled trial. Lancet. 2020;396:467–478. doi: 10.1016/S0140-6736(20)31604-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jackson L.A., Anderson E.J., Rouphael N.G., Roberts P.C., Makhene M., Coler R.N., McCullough M.P., Chappell J.D., Denison M.R., Stevens L.J. An mRNA Vaccine against SARS-CoV-2 - Preliminary Report. N. Engl. J. Med. 2020;383:1920–1931. doi: 10.1056/NEJMoa2022483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhu F.C., Li Y.H., Guan X.H., Hou L.H., Wang W.J., Li J.X., Wu S.P., Wang B.S., Wang Z., Wang L. Safety, tolerability, and immunogenicity of a recombinant adenovirus type-5 vectored COVID-19 vaccine: a dose-escalation, open-label, non-randomised, first-in-human trial. Lancet. 2020;395:1845–1854. doi: 10.1016/S0140-6736(20)31208-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kasturi S.P., Skountzou I., Albrecht R.A., Koutsonanos D., Hua T., Nakaya H.I., Ravindran R., Stewart S., Alam M., Kwissa M. Programming the magnitude and persistence of antibody responses with innate immunity. Nature. 2011;470:543–547. doi: 10.1038/nature09737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Querec T.D., Akondy R.S., Lee E.K., Cao W., Nakaya H.I., Teuwen D., Pirani A., Gernert K., Deng J., Marzolf B. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat. Immunol. 2009;10:116–125. doi: 10.1038/ni.1688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chan K.R., Wang X., Saron W.A.A., Gan E.S., Tan H.C., Mok D.Z.L., Zhang S.L., Lee Y.H., Liang C., Wijaya L. Cross-reactive antibodies enhance live attenuated virus infection for increased immunogenicity. Nat. Microbiol. 2016;1:16164. doi: 10.1038/nmicrobiol.2016.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chan C.Y., Chan K.R., Chua C.J., Nur Hazirah S., Ghosh S., Ooi E.E., Low J.G. Early molecular correlates of adverse events following yellow fever vaccination. JCI Insight. 2017;2:e96031. doi: 10.1172/jci.insight.96031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chan K.R., Gan E.S., Chan C.Y.Y., Liang C., Low J.Z.H., Zhang S.L., Ong E.Z., Bhatta A., Wijaya L., Lee Y.H. Metabolic perturbations and cellular stress underpin susceptibility to symptomatic live-attenuated yellow fever infection. Nat. Med. 2019;25:1218–1224. doi: 10.1038/s41591-019-0510-7. [DOI] [PubMed] [Google Scholar]
- 18.Salti S.M., Hammelev E.M., Grewal J.L., Reddy S.T., Zemple S.J., Grossman W.J., Grayson M.H., Verbsky J.W. Granzyme B regulates antiviral CD8+ T cell responses. J. Immunol. 2011;187:6301–6309. doi: 10.4049/jimmunol.1100891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ehrchen J.M., Sunderkötter C., Foell D., Vogl T., Roth J. The endogenous Toll-like receptor 4 agonist S100A8/S100A9 (calprotectin) as innate amplifier of infection, autoimmunity, and cancer. J. Leukoc. Biol. 2009;86:557–566. doi: 10.1189/jlb.1008647. [DOI] [PubMed] [Google Scholar]
- 20.Yu G., Boone T., Delaney J., Hawkins N., Kelley M., Ramakrishnan M., McCabe S., Qiu W.R., Kornuc M., Xia X.Z. APRIL and TALL-I and receptors BCMA and TACI: system for regulating humoral immunity. Nat. Immunol. 2000;1:252–256. doi: 10.1038/79802. [DOI] [PubMed] [Google Scholar]
- 21.Groom J.R., Luster A.D. CXCR3 in T cell function. Exp. Cell Res. 2011;317:620–631. doi: 10.1016/j.yexcr.2010.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Conticello S.G., Ganesh K., Xue K., Lu M., Rada C., Neuberger M.S. Interaction between antibody-diversification enzyme AID and spliceosome-associated factor CTNNBL1. Mol. Cell. 2008;31:474–484. doi: 10.1016/j.molcel.2008.07.009. [DOI] [PubMed] [Google Scholar]
- 23.Premkumar L., Segovia-Chumbez B., Jadi R., Martinez D.R., Raut R., Markmann A., Cornaby C., Bartelt L., Weiss S., Park Y. The receptor binding domain of the viral spike protein is an immunodominant and highly specific target of antibodies in SARS-CoV-2 patients. Sci. Immunol. 2020;5:eabc8413. doi: 10.1126/sciimmunol.abc8413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Rogers T.F., Zhao F., Huang D., Beutler N., Burns A., He W.T., Limbo O., Smith C., Song G., Woehl J. Isolation of potent SARS-CoV-2 neutralizing antibodies and protection from disease in a small animal model. Science. 2020;369:956–963. doi: 10.1126/science.abc7520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.de Alwis R., Chen S., Gan E.S., Ooi E.E. Impact of immune enhancement on Covid-19 polyclonal hyperimmune globulin therapy and vaccine development. EBioMedicine. 2020;55:102768. doi: 10.1016/j.ebiom.2020.102768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Honda-Okubo Y., Barnard D., Ong C.H., Peng B.H., Tseng C.T., Petrovsky N. Severe acute respiratory syndrome-associated coronavirus vaccines formulated with delta inulin adjuvants provide enhanced protection while ameliorating lung eosinophilic immunopathology. J. Virol. 2015;89:2995–3007. doi: 10.1128/JVI.02980-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hashem A.M., Algaissi A., Agrawal A.S., Al-Amri S.S., Alhabbab R.Y., Sohrab S.S., S Almasoud A., Alharbi N.K., Peng B.H., Russell M. A Highly Immunogenic, Protective, and Safe Adenovirus-Based Vaccine Expressing Middle East Respiratory Syndrome Coronavirus S1-CD40L Fusion Protein in a Transgenic Human Dipeptidyl Peptidase 4 Mouse Model. J. Infect. Dis. 2019;220:1558–1567. doi: 10.1093/infdis/jiz137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Higgins B.W., McHeyzer-Williams L.J., McHeyzer-Williams M.G. Programming Isotype-Specific Plasma Cell Function. Trends Immunol. 2019;40:345–357. doi: 10.1016/j.it.2019.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rupp J.C., Sokoloski K.J., Gebhart N.N., Hardy R.W. Alphavirus RNA synthesis and non-structural protein functions. J. Gen. Virol. 2015;96:2483–2500. doi: 10.1099/jgv.0.000249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Loo Y.M., Fornek J., Crochet N., Bajwa G., Perwitasari O., Martinez-Sobrido L., Akira S., Gill M.A., García-Sastre A., Katze M.G., Gale M., Jr. Distinct RIG-I and MDA5 signaling by RNA viruses in innate immunity. J. Virol. 2008;82:335–345. doi: 10.1128/JVI.01080-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anderson E.J., Rouphael N.G., Widge A.T., Jackson L.A., Roberts P.C., Makhene M., Chappell J.D., Denison M.R., Stevens L.J., Pruijssers A.J. Safety and Immunogenicity of SARS-CoV-2 mRNA-1273 Vaccine in Older Adults. N. Engl. J. Med. 2020;383:2427–2438. doi: 10.1056/NEJMoa2028436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Walsh E.E., Frenck R.W., Jr., Falsey A.R., Kitchin N., Absalon J., Gurtman A., Lockhart S., Neuzil K., Mulligan M.J., Bailey R. Safety and Immunogenicity of Two RNA-Based Covid-19 Vaccine Candidates. N. Engl. J. Med. 2020;383:2439–2450. doi: 10.1056/NEJMoa2027906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Polack F.P., Thomas S.J., Kitchin N., Absalon J., Gurtman A., Lockhart S., Perez J.L., Marc G.P., Moreira E.D., Zerbini C. Safety and Efficacy of the BNT162b2 mRNA Covid-19 Vaccine. N. Engl. J. Med. 2020;383:2603–2615. doi: 10.1056/NEJMoa2034577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McKay P.F., Hu K., Blakney A.K., Samnuan K., Brown J.C., Penn R., Zhou J., Bouton C.R., Rogers P., Polra K. Self-amplifying RNA SARS-CoV-2 lipid nanoparticle vaccine candidate induces high neutralizing antibody titers in mice. Nat. Commun. 2020;11:3523. doi: 10.1038/s41467-020-17409-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Laczkó D., Hogan M.J., Toulmin S.A., Hicks P., Lederer K., Gaudette B.T., Castaño D., Amanat F., Muramatsu H., Oguin T.H., 3rd A Single Immunization with Nucleoside-Modified mRNA Vaccines Elicits Strong Cellular and Humoral Immune Responses against SARS-CoV-2 in Mice. Immunity. 2020;53:724–732.e7. doi: 10.1016/j.immuni.2020.07.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lederer K., Castaño D., Gómez Atria D., Oguin T.H., 3rd, Wang S., Manzoni T.B., Muramatsu H., Hogan M.J., Amanat F., Cherubin P. SARS-CoV-2 mRNA Vaccines Foster Potent Antigen-Specific Germinal Center Responses Associated with Neutralizing Antibody Generation. Immunity. 2020;53:1281–1295.e5. doi: 10.1016/j.immuni.2020.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Corbett K.S., Edwards D.K., Leist S.R., Abiona O.M., Boyoglu-Barnum S., Gillespie R.A., Himansu S., Schäfer A., Ziwawo C.T., DiPiazza A.T. SARS-CoV-2 mRNA vaccine design enabled by prototype pathogen preparedness. Nature. 2020;586:567–571. doi: 10.1038/s41586-020-2622-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.von Herrath M.G., Bot A. Immune responsiveness, tolerance and dsRNA: implications for traditional paradigms. Trends Immunol. 2003;24:289–293. doi: 10.1016/s1471-4906(03)00121-2. [DOI] [PubMed] [Google Scholar]
- 39.Jin B., Sun T., Yu X.H., Liu C.Q., Yang Y.X., Lu P., Fu S.F., Qiu H.B., Yeo A.E. Immunomodulatory effects of dsRNA and its potential as vaccine adjuvant. J. Biomed. Biotechnol. 2010;2010:690438. doi: 10.1155/2010/690438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Pepini T., Pulichino A.M., Carsillo T., Carlson A.L., Sari-Sarraf F., Ramsauer K., Debasitis J.C., Maruggi G., Otten G.R., Geall A.J. Induction of an IFN-Mediated Antiviral Response by a Self-Amplifying RNA Vaccine: Implications for Vaccine Design. J. Immunol. 2017;198:4012–4024. doi: 10.4049/jimmunol.1601877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Maruggi G., Shaw C.A., Otten G.R., Mason P.W., Beard C.W. Engineered alphavirus replicon vaccines based on known attenuated viral mutants show limited effects on immunogenicity. Virology. 2013;447:254–264. doi: 10.1016/j.virol.2013.07.021. [DOI] [PubMed] [Google Scholar]
- 42.Thompson J.M., Whitmore A.C., Konopka J.L., Collier M.L., Richmond E.M., Davis N.L., Staats H.F., Johnston R.E. Mucosal and systemic adjuvant activity of alphavirus replicon particles. Proc. Natl. Acad. Sci. USA. 2006;103:3722–3727. doi: 10.1073/pnas.0600287103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ramaswamy S., Tonnu N., Tachikawa K., Limphong P., Vega J.B., Karmali P.P., Chivukula P., Verma I.M. Systemic delivery of factor IX messenger RNA for protein replacement therapy. Proc. Natl. Acad. Sci. USA. 2017;114:E1941–E1950. doi: 10.1073/pnas.1619653114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gan E.S., Tan H.C., Le D.H.T., Huynh T.T., Wills B., Seidah N.G., Ooi E.E., Yacoub S. Dengue virus induces PCSK9 expression to alter antiviral responses and disease outcomes. J. Clin. Invest. 2020;130:5223–5234. doi: 10.1172/JCI137536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Le Bert N., Tan A.T., Kunasegaran K., Tham C.Y.L., Hafezi M., Chia A., Chng M.H.Y., Lin M., Tan N., Linster M. SARS-CoV-2-specific T cell immunity in cases of COVID-19 and SARS, and uninfected controls. Nature. 2020;584:457–462. doi: 10.1038/s41586-020-2550-z. [DOI] [PubMed] [Google Scholar]
- 46.Ayouba A., Thaurignac G., Morquin D., Tuaillon E., Raulino R., Nkuba A., Lacroix A., Vidal N., Foulongne V., Le Moing V. Multiplex detection and dynamics of IgG antibodies to SARS-CoV2 and the highly pathogenic human coronaviruses SARS-CoV and MERS-CoV. J. Clin. Virol. 2020;129:104521. doi: 10.1016/j.jcv.2020.104521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Atyeo C., Fischinger S., Zohar T., Slein M.D., Burke J., Loos C., McCulloch D.J., Newman K.L., Wolf C., Yu J. Distinct early serological signatures track with SARS-CoV-2 survival. Immunity. 2020;53:524–532.e4. doi: 10.1016/j.immuni.2020.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.GeurtsvanKessel C.H., Okba N.M.A., Igloi Z., Bogers S., Embregts C.W.E., Laksono B.M., Leijten L., Rokx C., Rijnders B., Rahamat-Langendoen J. An evaluation of COVID-19 serological assays informs future diagnostics and exposure assessment. Nat. Commun. 2020;11:3436. doi: 10.1038/s41467-020-17317-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.