Abstract
Purpose
Long non-coding RNAs (lncRNAs) play critical roles in cancer onset and development, including head and neck squamous cell carcinoma (HNSCC). This study aimed to investigate the biological role of LINC00460 and the mechanisms underlying epithelial-mesenchymal transition (EMT) in HNSCC.
Methods
Aberrantly LINC00460 expression in HNSCC and overall survival outcomes were constructed using the TCGA database. Quantitative real-time polymerase chain reaction (RT-qPCR) was applied to examine the LINC00460 expression level in HNSCC cell lines. The role of LINC00460 knockdown on HNSCC cell growth, migration, invasion, and EMT was investigated in vitro using cell counting kit-8 (CCK-8), colony formation, transwell assay, and Western blot assay. Besides, bioinformatics prediction, dual-luciferase reporter assay, and RNA immunoprecipitation (RIP) were performed to reveal the interaction among LINC00460 and its target genes. The function of the LINC00460/miR-320a/BGN axis in HNSCC cells was clarified by rescue assays. Furthermore, the in vivo effects of LINC00460 on tumor growth were investigated using mice xenograft models.
Results
In this study, LINC00460 was upregulated in HNSCC tissues and cells and was associated with poor clinical prognosis. Further functional analysis showed that LINC00460 knockdown decreased HNSCC cell proliferation, migration, invasion, as well as EMT in vitro. Mechanistic investigation indicated that LINC00460 sponged miR-320a to upregulate Biglycan (BGN) expression, thereby facilitating HNSCC progression and induced EMT. Moreover, knockdown of LINC00460 significantly suppressed the progression of HNSCC cells in vivo.
Conclusion
Taken together, LINC00460 mediates miR-320a/BGN signaling axis to promote cell proliferation, migration, invasion, and induce the EMT process in HNSCC cells. Our findings elucidated a novel mechanism underlying the progression of HNSCC. LINC00460 could serve as a potential therapeutic target for the treatment of HNSCC.
Keywords: head and neck squamous cell carcinoma, LINC00460, miR-320a, BGN, EMT
Introduction
Head and neck squamous cell carcinoma (HNSCC) is a heterogeneous disease and accounts for 90% of all head and neck cancers.1 Annually, about 600,000 new cases are diagnosed in the world.2 Nowadays, the main treatment regimens for HNSCC are surgical resection, adjuvant and neoadjuvant chemotherapy, radiation therapy, and chemoradiotherapy. Despite the development of comprehensive therapy, a high rate of recurrence and metastasis impede the effective treatment of HNSCC. The 5-year survival rates are dissatisfactory, ranging from approximately 25% in hypopharyngeal, roughly 60% in laryngeal cancer, approximately 64% in oral cancer, to roughly 72% in salivary glands cancer based on different anatomical subsites.3 Therefore, the identification of molecular mechanisms of HNSCC is needed to establish novel treatment strategies and to gain potential therapeutic targets.
Epithelial to mesenchymal transition (EMT) is a dynamic process that switching the epithelial phenotype to mesenchymal form in cellular organization.4 EMT determines stem cell behavior, metastasis formation, wound healing, and represents a vital target for the treatment of HNSCC.5 Long non-coding RNA (lncRNA), a class of non-coding RNA more than 200 nucleotides in length, has been reported to be aberrantly expressed and related to diverse malignancy processes in HNSCC.6 Long non-coding RNA 00460 (LINC00460) is an intergenic lncRNA located at 13q33.2. In various malignant tumors, LINC00460 has been determined to be abnormally upregulated, such as gastric cancer,7 lung cancer,8 and glioma.9 Previous studies also reported that LINC00460 is overexpressed and promotes the EMT process in HNSCC.10 However, the underlying mechanism and functional roles and of LINC00460 have not been radically investigated in HNSCC.
In our study, the differences in LINC00460 expression between HNSCC tissues and adjacent non-tumor tissues were investigated, and the survival outcomes of patients with HNSCC were assessed. Besides, silencing LINC00460 inhibited HNSCC cell proliferation, migration, and invasion. Furthermore, we found that LINC00460 played a functional role in EMT promotion by sponging miR-320a expression and then upregulating Biglycan (BGN) expression. Our discovery provides new evidence that LINC00460 exerts a crucial role in HNSCC progression and highlight the potential target for the treatment of HNSCC.
Materials and Methods
Bioinformatics Analysis
The Cancer Genome Atlas (TCGA) (https://portal.gdc.cancer.gov/) and Gene Expression Profiling Interactive Analysis (GEPIA) (http://gepia.cancer-pku.cn) was used to obtain LINC00460 expression in HNSCC tissues and adjacent normal tissues. The prognosis analysis of HNSCC patients was calculated using the Kaplan-Meier method by GEPIA. The Starbase v3.0 database (http://starbase.sysu.edu.cn/) was used to predict the interaction among lncRNA, miRNA, and mRNA and reveal the correlation between the expression of lncRNA/mRNA and miRNA in HNSCC tissues.
Cell Culture and Treatment
Human HNSCC cell line FaDu, Cal-27, SCC4, SCC9, and human immortalized keratinocytes cell line (HaCaT, normal control) were bought from American Type Culture Collection (ATCC, Manassas, VA, USA). FaDu and Cal-27 cells were cultured in Dulbecco’s modified Eagle’s medium (DMEM, Gibco, Grand Island, NY, USA). SCC4 and SCC9 cells were cultured in DMEM/F1:2 (Gibco, Grand Island, NY, USA). HaCaT cells were cultured in RPMI-1640 (Gibco, Grand Island, NY, USA). All the medium was supplemented with 10% fetal bovine serum (FBS, Gibco, Grand Island, NY, USA), and 1% penicillin/streptomycin (Beyotime Biotechnology, Shanghai, China). Cell mediums were maintained in an incubator containing 5% CO2 at 37 °C.
To perform lentivirus-mediated suppression of LINC00460 and BGN in HNSCC cells, the following shRNA and scrambled control shRNA were inserted into the pLVX-tdTomato-Puro vector (GenePharma, Shanghai, China): sh-LINC00460-#1: GCCTCTGAAATGGTGACAATA; sh-LINC00460-#2: GCCATCCACTTCAAAGTATTC; sh-LINC00460-#3: GCGTGGGAAAGAAGACGCATT; and sh-BGN: GCCATTCATGATGAACGATGA. After 72 h of transfection, the medium was mixed with puromycin and cultured for 14 days, the stably stained cells were screened. MiR-320a mimics, inhibitors, and the negative controls (miR-NC, miR-NC inhibitor) were synthesized by RiboBio Company (Guangzhou, China). Cells were transfected using Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific, CA, USA).
RNA Extraction and Quantitative Real-Time Polymerase Chain Reaction (RT-qPCR)
Total RNA was extracted from HNSCC cells using TRIzol reagent (Invitrogen, Carlsbad, CA, USA). RNA integrity and quantity were examined using a Nano-Drop spectrophotometer (Thermo Scientific, Wilmington, DE, USA). Reverse transcription was performed using a Transcriptor First Strand cDNA Synthesis Kit (Roche, Basel, Switzerland) and a TaqMan miRNA reverse transcript kit (Applied Biosystems, Branchburg, NJ, USA). An ABI PRISM 7500 FAST sequence detection system (Applied Biosystems, Foster City, CA, USA) was employed to conduct RT-qPCR. After the reaction cycles, the threshold cycle (Ct) values were determined, and the relative mRNA levels were calculated based on Ct values and normalized to the normalized to housekeeping gene glyceraldehyde-3-phosphate dehydrogenase (GAPDH) or U6 level in each sample. The gene expression fold changes were calculated according to the formula of 2−ΔΔCt. Each test was repeated three times. Specific primer sequences were as follows:
LINC00460 forward: 5ʹ-GCATGCACACTTCTCGGCTA −3ʹ
LINC00460 reverse: 5ʹ-GAATGCGTCTTCTTTCCCACG −3ʹ
BGN forward: 5ʹ- GGGTCTCCAGCACCTCTACGC −3ʹ
BGN reverse: 5ʹ- TGAACACTCCCTTGGGCACCT −3ʹ
GAPDH forward: 5ʹ- GATCATCAGCAATGCCTCCT −3ʹ
GAPDH reverse: 5ʹ- TGAGTCCTTCCACGATACCA −3ʹ
miR-320a forward: 5ʹ- AAAAGCTGGGTTGAGAGGGCGA −3ʹ
miR-320a reverse: 5ʹ- GCGAGCACAGAATTAATACGAC −3ʹ
U6 forward: 5ʹ-CTCGCTTCGGCAGCACA-3ʹ
U6 reverse: 5ʹ-AACGCTTCACGAATTTGCGT-3ʹ
Cell Counting Kit-8 (CCK-8) Assay
After transfection, the cells were plated in 96-well plates (2000 cells/well). Cell proliferation was determined every 24 h for 4 days according to the manufacturer’s instruction. Briefly, the cells were mixed with 10 μL of CCK-8 solution (MedChemExpress, Shanghai, China) per well and incubated for a further 1 h at 37°C. The amount of formazan dye generated by cellular dehydrogenase activity was measured for absorbance at 450 nm using a microplate reader (Multiscan FC, Thermo Scientific, MA, USA).
Colony Formation Assay
After transfection, the cells were plated in 6-well plates (500 cells/well) and incubated at 37°C with 5% CO2 for 2 weeks to form cell colonies. Then, the cells were stained with 0.1% crystal violet, after which the positive colonies were imaged. Positive colonies with more than 50 cells were counted under a microscope.
Transwell Migration and Invasion Assays
Transwell migration and invasion assays were performed using Transwell chambers (8-μm pores, Corning, NY, USA) that coated without or with Matrigel (BD, Biosciences, NJ, USA). Briefly, the cells (5×104) were seeded in the upper chambers and incubated in 500 μL serum-free medium, while 500 μL medium supplemented with 10% FBS was placed in the lower chambers. After incubation for 18 h (migration) or 24 h (invasion) in a 5% CO2 humidified incubator at 37°C, the migrated or invaded cells were fixed with 1% formaldehyde solution, stained with 0.1% crystal violet, and captured under a light microscope (Olympus, Tokyo, Japan). The cell numbers were counted in five random fields of each filter.
Dual-Luciferase Reporter Gene Assay
LINC00460-miR-320a binding sites and miR-320a-BGN binding sites were identified on the bioinformatics prediction website StarBase v3.0 (http://starbase.sysu.edu.cn/). The full-length sequences of LINC00460 cDNA and BGN 3ʹ-UTR were cloned and inserted into a pmirGLO (Promega, Madison, WI, USA) Dual-luciferase vector respectively and were named pLINC00460-wide type (WT) and pBGN-WT. Site-specific mutagenesis was performed on LINC00460-miR-320a binding sites and miR-320a-BGN binding sites to construct pLINC00460-mutant (MUT) type and pBGN-MUT vectors. The pRL-TK vector expressing Renilla luciferase was regarded as the internal reference. The vectors of miR-320a negative control (NC) and miR-320a mimic were co-transfected with luciferase reporter vector into Cal-27 and FaDu cells with the use of Lipofectamine 3000 (Invitrogen, Thermo Fisher Scientific, CA, USA), separately. 48 h following the transfection, the relative luciferase activities were detected by a Luciferase Reporter Assay Kit (Promega, Madison, WI, USA) according to the manufactures’ instructions.
RNA-Binding Protein Immunoprecipitation (RIP) Assay
RIP assay was performed using a Magna RIP RNA-Binding Protein Immunoprecipitation Kit (Millipore, Bedford, MA, USA) according to the manufacturers’ protocol. Cells transiently transfected with miR-320a were isolated and lysed using a RIP lysis buffer and incubated with antibody against Argonaute 2 (AGO2, Abcam, Cambridge, MA, USA) at 4 °C overnight. IgG (Abcam, Cambridge, MA, USA) was used as negative control (NC). The beads were attracted by a magnetic separator, and samples were fixed with proteinase K. The immunoprecipitated RNAs were then extracted by TRIzol and quantified by RT-qPCR.
Western Blotting
The total protein was extracted using RIPA (Applygen, Beijing, China). Protein concentration was determined using a BCA Protein Assay Kit (Beyotime, Shanghai, China). The proteins were electrophoretically separated by SDS-PAGE and transferred onto a PVDF membrane (Millipore, MA, USA). Membrane blockade was performed using 5% bovine serum albumin for 1 h and added with primary antibodies to rabbit anti-human antibodies to GAPDH (1:1000, Cell Signaling Technology, CST), BGN (1:2000, Abcam), N-cadherin (1:1000, Abcam), Vimentin (1:2000, Abcam) at 4°C overnight. The membrane was then incubated with a goat anti-rabbit antibody to IgG (Beyotime, Shanghai, China) for 1 h. Then the protein strips were visualized and detected using a chemiluminescent reagent kit (Thermo Fisher Scientific, MA, USA). Images were acquired by a ChemiDoc MP Imaging System (Bio-Rad, CA, USA) and analyzed by the ImageJ software. GAPDH was used as an internal control. The uncropped images of membranes used for the research were shown in Figure S1.
Tumor Xenografts
Lentiviruses containing shRNA for LINC00460, or scrambled control shRNA lentiviruses, were purchased from GenePharma Company (Shanghai, China). These lentiviruses were mixed with Cal-27 cells, followed by selection with 1 μg/mL puromycin for 14 days. Four-to six-week-old female BALB/c nude mice were obtained from Shanghai SLAC Laboratory Animal Co. Ltd. (Shanghai, China). All of the BALB/c nude mice were maintained under Specific Pathogen Free conditions. 1×106 stably transfected Cal-27 cells were suspended in 100 μL of PBS and subcutaneously injected into the subcutis. The width and length of the tumor were measured every 6 days. The tumor volume was calculated using the following formula: Volume (mm3)=length × width2× 0.5. Moreover, 30 days following the injection, the nude mice were sacrificed, and tumors were photographed and weighed. Animal handling and procedures were approved by the Capital Medical University Institutional Animal Care. The name of the guidelines followed for the welfare of the laboratory animals is “Guide for the Care and Use of Laboratory Animals (8th Edition)”.
Statistical Analysis
SPSS 22.0 software (SPSS Inc., Chicago, IL, USA) was used for data analysis. All measurement data were presented as mean±standard deviation (SD). Survival analysis was carried out using the Log rank test in association with Kaplan-Meier analysis. The comparison between two experimental groups was analyzed by a paired t-test or unpaired t-test. Comparison among multiple groups was analyzed by one-way analysis of variance (ANOVA), followed by Tukey’s post hoc test. P-value <0.05 was considered as statistically significant.
Results
LINC00460 is Highly Expressed in HNSCC and Predicts Poor Overall Survival
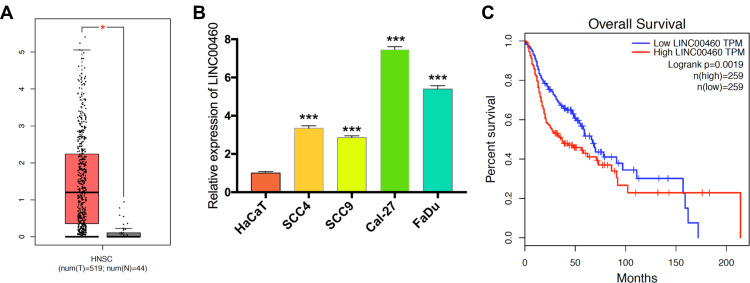
Firstly, we explored the TCGA database and found that LINC00460 expression was upregulated in HNSCC tissues compared with adjacent non-tumor tissues (Figure 1A). Meanwhile, LINC00460 expression levels were elevated in HNSCC cell lines (especially Cal-27 and FaDu cell lines) than human immortalized keratinocytes cells (HaCaT, normal control; Figure 1B). Moreover, according to LINC00460 mRNA expression (median), patients with high LINC00460 expression levels had poorer overall survival (OS) rate than those with low LINC00460 level ones (Figure 1C). These results indicated that LINC00460 might play a pivotal role in HNSCC tumorigenesis and development.
Figure 1.
Levels of LINC00460 were highly expressed in HNSCC and associated with poor prognosis. (A) LINC00460 expression in HNSCC tissues and normal tissues in the TCGA database. (B) The expression of LINC00460 in four HNSCC cell lines and normal control cells HaCaT. (C) The relationship between LINC00460 expression and overall survival time of HNSCC patients analyzed using the Kaplan-Meier method. *P < 0.05, ***P < 0.001.
LINC00460 Silencing Suppresses HNSCC Cell Proliferation, Migration, Invasion, and EMT
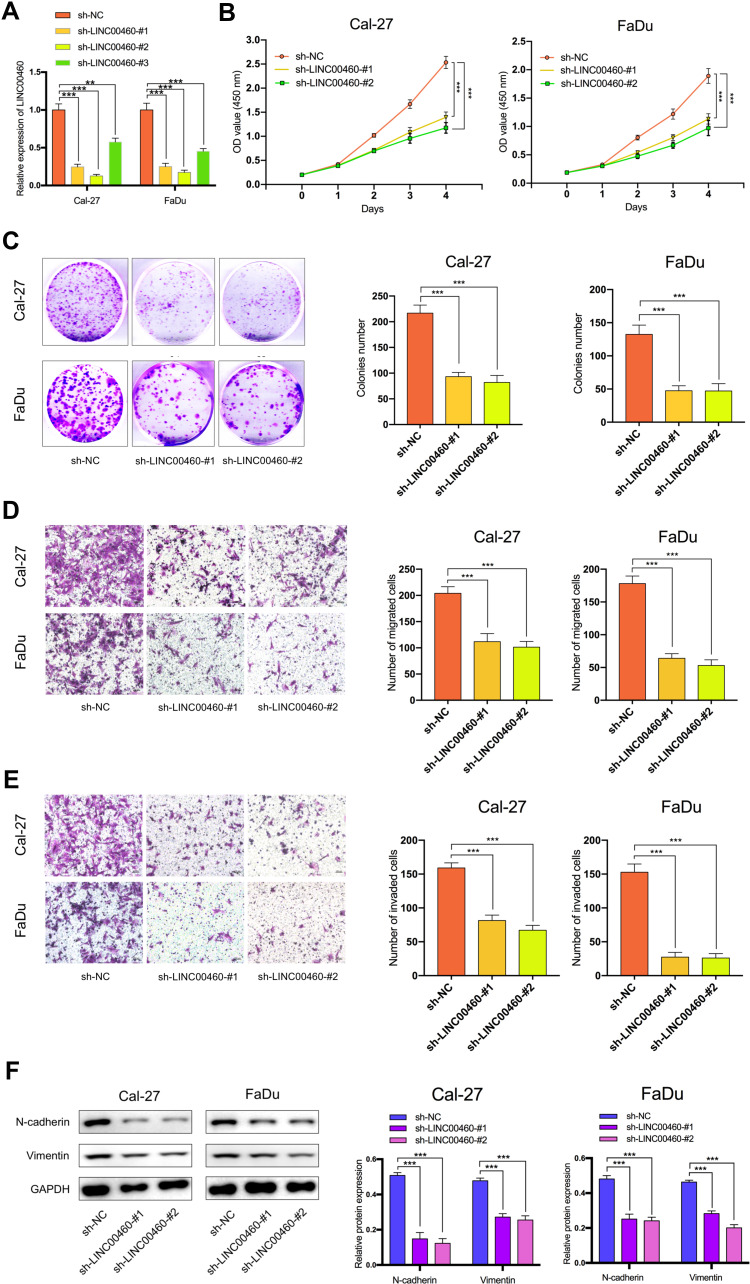
To explore the regulation effect of LINC00460 in HNSCC cells, a total of three shRNAs (sh-LINC00460-#1, sh-LINC00460-#2, sh-LINC00460-#3) targeted to LINC00460, and one scrambled control shRNA (sh-NC) were applied. Cal-27 and FaDu cells were chosen for lentivirus transfection because of highly expressing LINC00460. The efficiency was then determined in stably transfected cells by RT-qPCR and sh-LINC00460-#1/sh-LINC00460-#2 were selected for the following experiments as their better knockdown efficiencies (Figure 2A). CCK-8 assay (Figure 2B) and colony formation assay (Figure 2C) demonstrated that Cal-27 and FaDu cell proliferation was significantly decreased after knockdown of LINC00460. To investigate the effect of LINC00460 on cell migration and invasion in HNSCC cells, the transwell migration and invasion assays were performed. Results showed that migrated and invaded Cal-27 and FaDu cells in transwell assays were reduced following LINC00460 knockdown (Figure 2D and E), which suggested that knockdown of LINC00460 inhibited the migration and invasiveness of HNSCC cells.
Figure 2.
Silencing of LINC00460 attenuated HNSCC cell proliferation, invasion, migration, and EMT. (A) RT-qPCR analysis of LINC00460 mRNA in Cal-27 and FaDu cells treated with sh-LINC00460 (B) CCK-8 assays were applied to detect cell viability of Cal-27 and FaDu cells. (C) Colony formation assays were used to measure cell proliferation ability. (D and E) Representative results of transwell migration assays and invasion assays in Cal-27 and FaDu cells. (F) Relative expression of EMT-related genes in Cal-27 and FaDu cells detected by Western blot analysis. (scale bar: 200 μm for transwell assay). **P < 0.01, ***P < 0.001.
To further investigate the mechanism by which LINC00460 promotes cell progression, invasion, and metastasis of HNSCC, we performed Western blotting to measure the epithelial and mesenchymal markers. As shown in Figure 2F, knockdown of LINC00460 in both Cal-27 and FaDu cells led to decreased expression of N-cadherin and Vimentin. Taken together, LINC00460 silencing suppressed cell progression, migration, invasion, and EMT in HNSCC.
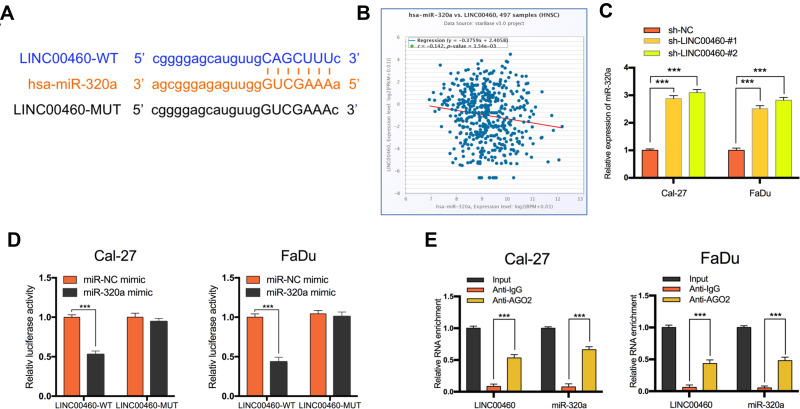
LINC00460 Acts as a Molecular Sponge for miR-320a
Increasing evidence confirmed that many lncRNAs exert their functions by binding to miRNAs and mechanically liberating the target RNA transcripts. To explore the potential mechanisms of LINC00460, we predicted the potential miRNA that may interact with LINC00460 using Starbase v3.0 and observed a complementary sequence to miR-320a (Figure 3A). TCGA database has revealed that miR-320a expression was negatively associated with the expression of LINC00460 in HNSCC tissues (Figure 3B, P=1.54E-3, r=−0.142). Consistent with TCGA analysis results, we observed that after knockdown of LINC00460, the expression of miR-320a was significantly increased in Cal-27 and FaDu cells (Figure 3C). Then the dual-luciferase reporter assays demonstrated that miR-320a significantly inhibited the luciferase activity that carried wild type (wt) but not mutant (mut) 3ʹ-UTR of LINC00460 (Figure 3D). Consistently, results of RIP also confirmed that miR-320a was a target of LINC00460 in HNSCC cells. As presented in Figure 3E, LINC00460 and miR-320a were both substantially enriched in AGO2 antibody-incubated beads, compared with those harboring control IgG in Cal-27 and FaDu cells, indicating that endogenous binding might occur between LINC00460 and miR-320a. In summary, these data indicated that LINC00460 could directly sponge miR-320a in HNSCC.
Figure 3.
LINC00460 negatively regulated the expression and activity of miR-320a in HNSCC cells. (A) Starbase3.0 revealed the predicted miR-320a binding site in the LINC00460 3ʹUTR, wildtype (WT) and mutated (MUT) 3ʹUTR of LINC00460 was also shown. (B) The relationship between LINC00460 and miR-320a expression of HNSCC patients in the TCGA database (C) Expression of miR-320a in Cal-27 and FaDu cells transfected with sh-LINC00460 or sh-NC. (D) miR-320a markedly reduced luciferase activity in LINC00460-wild not in LINC00460 mut in Cal-27 and FaDu cells. (E) The correlations between LINC00460, miR-320a, and AGO2 were detected in the RIP assay with anti-AGO2 in Cal-27 and FaDu cells. ***P < 0.001.
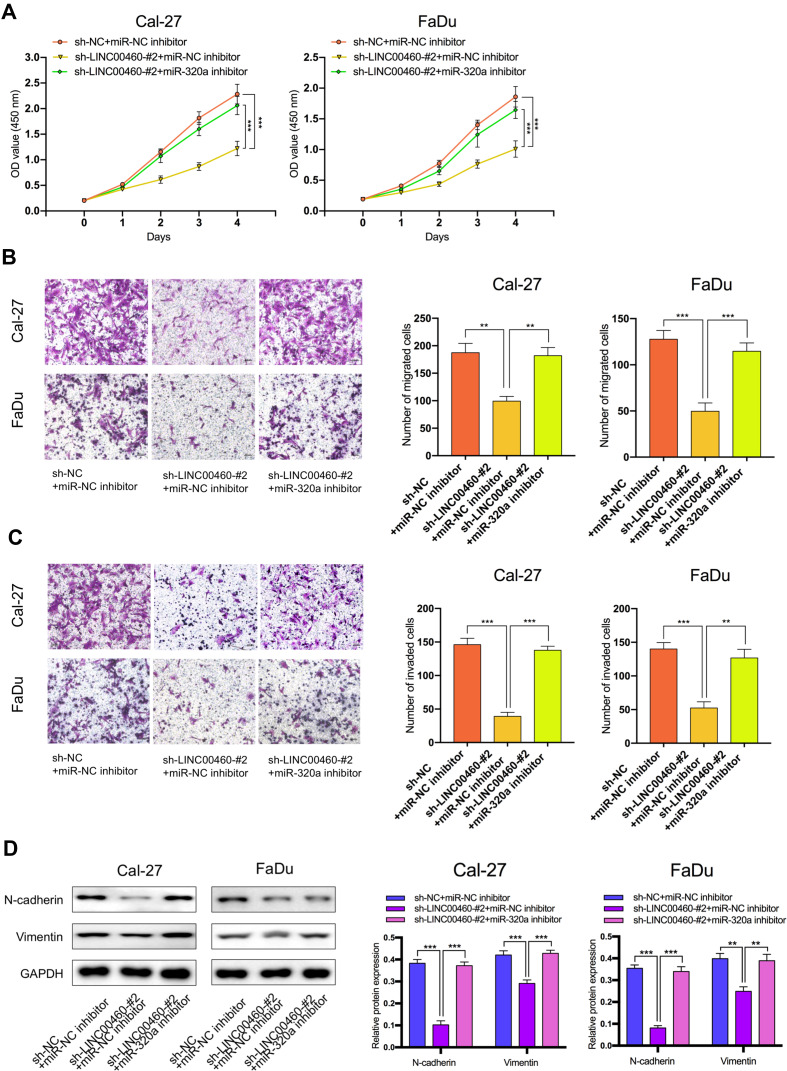
The Effect of LINC00460 on HNSCC Cell Progression and EMT is Mediated by miR-320a
We further explored whether LINC00460 functions through miR-320a. We found that in Cal27 and FaDu cells, silencing of LINC00460 mediated reduction of cell proliferation, migration, and invasion were partially rescued by simultaneous co-transfection with miR-320a inhibitor (Figure 4A–C). Then, we detected the expression of genes associated with EMT by Western blot analysis. LINC00460 reduction reduced N-cadherin and Vimentin expression. This effect was partially rescued by the co-transfection of sh-LINC00460 and the miR-320a inhibitor (Figure 4D). Therefore, the effect of LINC00460 on HNSCC progression and EMT was partially mediated by negative regulation of miR-320a.
Figure 4.
The regulation of LINC00460 on HNSCC cells was mediated by miR-320a. (A–C) CCK-8 assays, transwell migration, and invasion assays were used to detect the cell proliferation ability, cell migration, and invasion ability after transfection Cal-27 and FaDu cells with sh-NC+NC inhibitor, sh-LINC00460-#2+miR-NC inhibitor, or sh-LINC00460-#2+miR-320a inhibitor. (scale bar: 200 μm for transwell assay). (D) Western blot analysis was used to detect the expression of EMT-related genes after transfecting Cal-27 and FaDu cells. **P < 0.01, ***P < 0.001.
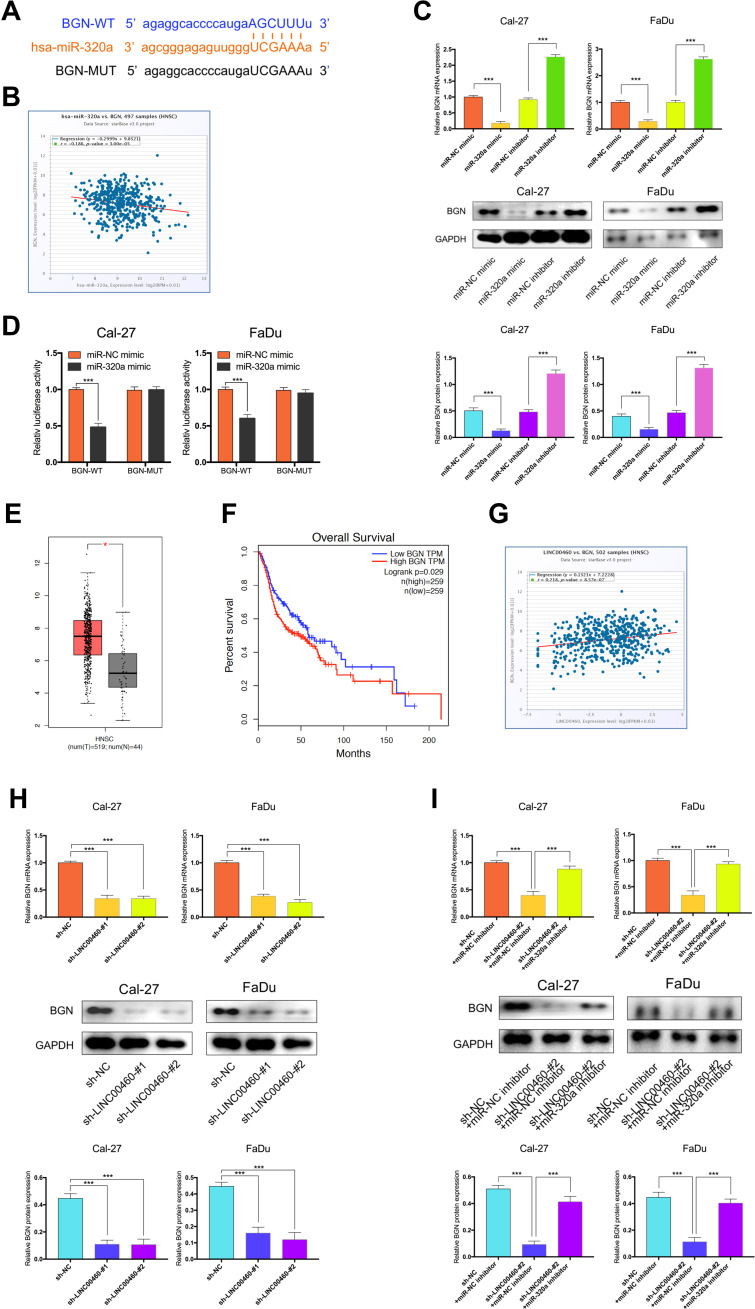
BGN is a Target Gene of miR-320a and Regulated by LINC00460
To explore the potential ceRNA mechanism between LINC00460, miR-320a, and its target genes in HNSCC, we used StarBase v3.0 to predict the candidate target of miR-320a and found that BGN may be the direct target (Figure 5A). According to the TCGA database, we observed that an obvious inverse correlation between the levels of miR-320a and BGN in HNSCC tissues (Figure 5B, P=3.00E-5, r=−0.186). Consistently, our results revealed that the miR-320a inhibitor significantly increased the mRNA and protein expression of BGN in Cal-27 and FaDu cells. By contrast, miR-320a mimic led to BGN depletion, which was opposite to miR-320a reduction (Figure 5C). Dual-luciferase reporter assays confirmed that miR-320a mimic inhibited luciferase activity of wild type (wt) BGN 3ʹ-UTR but not the mutant (mut) BGN 3ʹ-UTR in Cal-27 and FaDu cells (Figure 5D). So, BGN was considered as a direct target of miR-320a. Next, we explored whether LINC00460 could regulate the expression of BGN. TCGA database showed that the BGN was higher expressed in HNSCC tumor tissues compared with adjacent non-tumor tissues (Figure 5E) and patients with high BGN expression levels had poorer OS rate than those with low BGN level ones (Figure 5F). The BGN expression was positively correlated to the LINC00460 expression in HNSCC tissues (Figure 5G, P=8.57E-7, r=0.218). Consistently, we observed that after knockdown of LINC00460, the mRNA and protein expression of BGN was significantly decreased in Cal-27 and FaDu cells (Figure 5H). Besides, we found that silencing of LINC00460 mediated reduction of BGN were partially rescued by downregulation of miR-320a in Cal-27 and FaDu cells (Figure 5I). These results provided evidence that BGN was a direct target of miR-320a and regulated by LINC00460 via miR-320a in HNSCC cells.
Figure 5.
BGN was a target gene of miR-320a and regulated by LINC00460. (A) Starbase3.0 revealed the predicted miR-320a binding site in the BGN 3ʹUTR, wildtype (WT) and mutated (MUT) 3ʹUTR of BGN was also shown. (B) The relationship between miR-320a and BGN expression of HNSCC patients in the TCGA database (C) Relative mRNA and protein levels of BGN in Cal-27 and FaDu cells transfected with miR-NC mimic, miR-320a mimic, miR-NC inhibitor, or miR-320a inhibitor. (D) miR-320a markedly reduced luciferase activity in LINC00460-wild not in LINC00460 mut in Cal-27 and FaDu cells. (E) BGN expression in HNSCC tissues and normal tissues in TCGA database. (F) The relationship between LINC00460 expression and overall survival time of HNSCC patients analyzed using the Kaplan-Meier method. (G) The relationship between LINC00460 and BGN expression of HNSCC patients in the TCGA database (H) Relative mRNA and protein levels of BGN in Cal-27 and FaDu cells transfected with sh-LINC00460 or sh-NC. (I) Relative mRNA and protein levels of BGN in Cal-27 and FaDu cells transfected with sh-NC+miR-NC inhibitor, sh-LINC00460-#2+miR-NC inhibitor, or sh-LINC00460-#2+miR-320a inhibitor. * P< 0.05, ***P < 0.001.
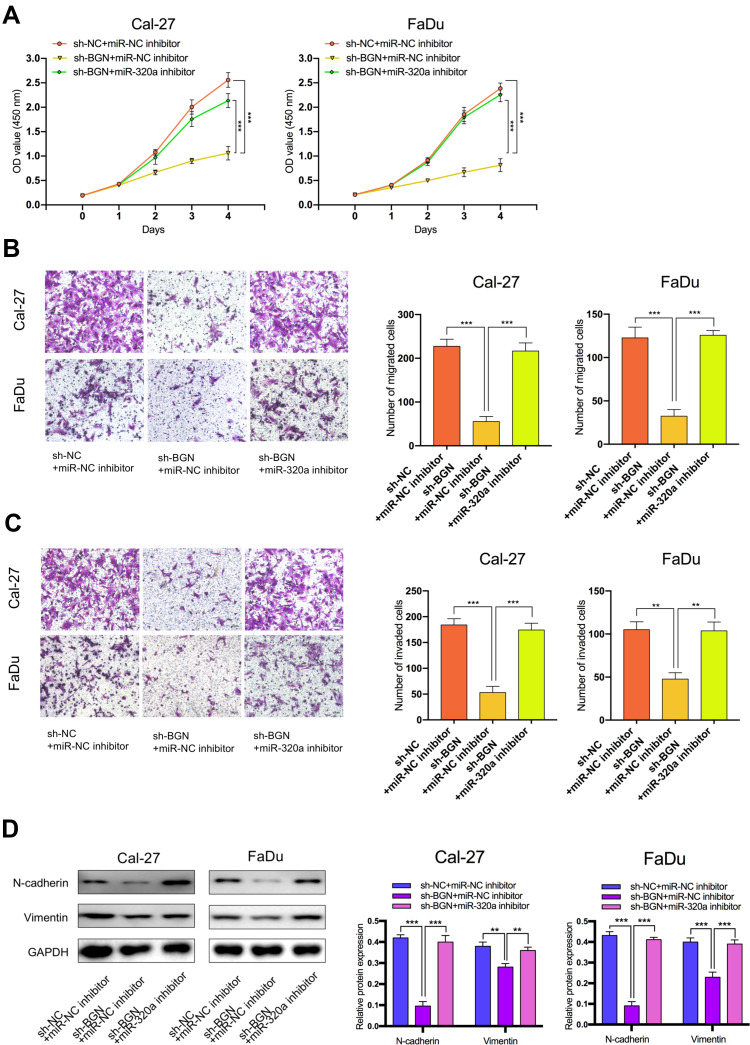
The Effect of BGN on HNSCC Cell Progression and EMT is Regulated by miR-320a
To explore the effect of the miR-320a/BGN axis on HNSCC progression, we adopted CCK-8 assay (Figure 6A), transwell assay (Figure 6B and C) to examine cell proliferation, migration, and invasion in HNSCC. We also used Western blotting analysis to test the levels of mRNAs and proteins linked to EMT (Figure 6D). Results revealed that knockdown of BGN attenuated the proliferation, migration, and invasion of HNSCC cells and diminished N-cadherin, Vimentin expression. Conversely, knockdown of miR-320a resulted in promoting cell proliferation, migration, and invasion of HNSCC and elevation in N-cadherin, Vimentin expression. Depletion of BGN, as well as depletion miR-320a, could partially rescue the suppressive role of BGN in cell proliferation, migration, invasion, as well as EMT of HNSCC. In summary, these results illustrated that the effect of BGN on HNSCC progression and EMT was negatively regulated by miR-320a.
Figure 6.
The effect of BGN on HNSCC cells was regulated by miR-320a. (A–C) CCK-8 assays, transwell migration and invasion assays were used to detect the cell proliferation ability, cell migration and invasion ability after transfection Cal-27 and FaDu cells with sh-NC+NC inhibitor, sh-BGN+miR-NC inhibitor, or sh-BGN+miR-320a inhibitor. (scale bar: 200 μm for transwell assay). (D) Western blot analysis was used to detect the expression of EMT-related genes after transfecting Cal-27 and FaDu cells. **P < 0.01, ***P < 0.001.
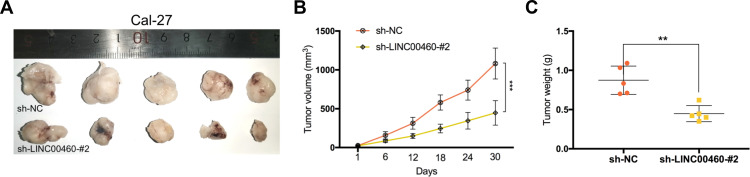
LINC00460 Silencing Inhibits Tumorigenesis of HNSCC in vivo
To explore the effects of LINC00460 on HNSCC tumorigenesis in vivo, we subcutaneously injected Cal-27 cells stably transfected with sh-LINC00460-#2 or their negative control groups into nude mice. Representative images of subcutaneous tumors were shown in Figure 7A. The volume and weight of tumors in the LINC00460 knockdown group were significantly reduced when compared with those in the control group (Figure 7B and C). Overall, these data demonstrated that LINC00460 silencing might inhibit tumorigenesis of HNSCC in vivo.
Figure 7.
Silencing of LINC00460 inhibited the tumor growth of HNSCC in vivo. (A) Subcutaneous tumors in the nude mouse model under different treatments. (B and C) Analysis of tumor weight and size in different groups. **P < 0.01, ***P < 0.001.
Discussion
HNSCC is a group of squamous cell carcinomas arising from the squamous mucosal surfaces of the upper aerodigestive tract with unsatisfactory curative effects in the clinical practice.11 Sustaining proliferation, activating invasion, and metastasis are the hallmarks of cancer, which have closely connected with poor prognosis of HNSCC patients.12 Recent evidence has reported that the EMT process leads to the progression and metastasis of HNSCC.13 However, the regulatory mechanisms of EMT on HNSCC still need to be further explored, especially the effects involving lncRNAs. Numerous lncRNAs play oncogenic roles in multiple cancers.14 Hence, the current study selected an upregulated lncRNA LINC00460 with the help of the TCGA-HNSC database. Survival analysis revealed that a high LINC00460 level was linked to a poor survival rate, which suggested that LINC00460 might be a potential prognostic predictor for HNSCC patients. Consequently, we found that LINC00460 was highly expressed in HNSCC cells comparing with human immortalized keratinocytes cell HaCaT, especially in Cal-27 and FaDu cells. For exploring the role of LINC00460 in HNSCC cells, we performed a knockdown of LINC00460 in Cal-27 and FaDu cells. As a result, knockdown of LINC00460 significantly restrained the proliferation, migration, and invasion of HNSCC cells in vitro. Moreover, we found that LINC00460 facilitated the EMT process in HNSCC cells. These findings demonstrate that LINC00460 plays a pivotal role in HNSCC and needs further mechanistic research.
A growing number of studies confirm that lncRNAs can exert a regulatory effect on the progression of HNSCC by sponging miRNAs and regulate the expression of target genes.15 miRNAs are short, evolutionarily conserved RNAs containing about 20–24 nucleotides.16 In the present study, we performed bioinformatics analysis and experimental assays to confirm that LINC00460 shared a complementary binding site with miR-320a. miR-320a has been identified as an inhibitor of oncogenes in several studies and suppresses tumor progression by multiple mechanisms. For instance, miR-320a suppresses cervical cancer cell proliferation, migration, and invasion by suppressing FoxM1 expression.17 miR-320a inhibits invasion and metastasis in hepatocellular carcinoma by regulating HMGB1.18 Besides, miR-320a could reduce stemness and cisplatin resistance in laryngeal carcinoma cells.19 However, there is a limited understanding of the roles of miR-320a in HNSCC. We found that there was a negative correlation between the expression of LINC00460 and miR-320a in HNSCC tissues and cell lines. Moreover, we demonstrated that miR-320a could partially mediate the tumor-promoting function of LINC00460 in HNSCC progression and EMT.
Aiming to further explore the function of the LINC00460/miR-320a axis, we predicted the potential target of miR-320a with bioinformatics analysis. We found that the 3ʹ-UTR region of BGN contained potential binding sites for miR-320a and verified this finding by experimental assays. BGN has been reported as a classical type of extracellular matrix protein and has been found to play an important role in numerous cancer-related biological behaviors such as cell growth, migration, autophagy, and differentiation of epithelial cells.20 In colon cancer, BGN could mediate epigenetic silencing of immunosuppressive ligands and promote carcinogenesis.21 In patients with prostate cancer, the up-regulation of BGN is associated with poor prognosis and PTEN deletion.22 In the present study, we found that there was a negative correlation between the expression of miR-320a and BGN and a positive correlation between the expression of LINC00460 and BGN. Then, we performed a series of experiments and rescue assays to confirmed that the LINC00460/miR-320a/BGN axis was involved in the cell progression, migration, invasion, and EMT of HNSCC.
In this study, evidence was obtained demonstrating that silencing LINC00460/miR-320a/BGN axis inhibited EMT, which was accompanied by the reductions in the expression of N-cadherin, and Vimentin. EMT is an important process of cancer development that has been widely documented as a key factor in the diagnosis and treatment of cancers.4 Repressed LINC00460 expression has been previously reported to inhibit EMT in colon cancer through the downregulation of the ANXA2/miR-433-3p pathway.23 Furthermore, LINC00460 has been shown to promote EMT in HNSCC by facilitating peroxiredoxin-1 into the nucleus.10 Chou et al asserted that miR-320a could inhibit the expression of the transcription factor FOXM1 and downstream proteins related to EMT, thereby leading to the suppression of liver cancer cell growth and invasion.24 Zhang et al reported that miR-320a inhibits endometrial carcinoma cell metastatic capability by preventing TGF-β1-induced EMT.25 Previous studies also reported that BGN could induce the EMT process of diverse malignancies. Thakur et al demonstrated that BGN is necessary and sufficient to mediate the pro-EMT effect in pancreatic ductal adenocarcinoma.26 Also, BGN is regulated by the TGF-β pathway, a key regulator of the EMT process. Furthermore, BGN is also considered as one of the potential EMT biomarkers in colorectal cancer.27 Our findings demonstrated that knockdown of LINC00460 was able to repress EMT, cell progression, migration, and invasion in HNSCC by increasing miR-320a and decreasing BGN. These results largely expanded the EMT regulating effect of LINC00460, miR-320a, BGN, and provided more supporting evidence for the ceRNA regulatory network. However, a detailed regulatory network of LINC00460 in HNSCC needs further investigation.
Conclusion
In conclusion, our findings are the first to demonstrate that LINC00460 promotes HNSCC progression, migration, invasion, and EMT by sponging miR-320a and further upregulating BGN. The LINC00460/miR-320a/BGN axis may contribute to the development of a new therapeutic target for HNSCC.
Acknowledgments
This work was supported by the Beijing Municipal Administration of Hospitals’ Ascent Plan (No. DFL20180202), Capital Health Development Research Project (No. S2018-2-2054), Beijing Natural Science Foundation Program and Scientific Research Key Program of Beijing Municipal Commission of Education (No. KZ201910025034), and National Key Research and Development Program of China (No. 2020YFB1312805).
Disclosure
The authors declare no conflicts of interest in this work.
References
- 1.Wyss A, Hashibe M, Chuang SC, et al. Cigarette, cigar, and pipe smoking and the risk of head and neck cancers: pooled analysis in the international head and neck cancer epidemiology consortium. Am J Epidemiol. 2013;178(5):679–690. doi: 10.1093/aje/kwt029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leemans CR, Snijders PJF, Brakenhoff RH. The molecular landscape of head and neck cancer. Nat Rev Cancer. 2018;18(5):269–282. doi: 10.1038/nrc.2018.11 [DOI] [PubMed] [Google Scholar]
- 3.Gatta G, Botta L, Sánchez MJ, Anderson LA, Pierannunzio D, Licitra L. Prognoses and improvement for head and neck cancers diagnosed in Europe in early 2000s: the EUROCARE-5 population-based study. Eur J Cancer. 2015;51(15):2130–2143. doi: 10.1016/j.ejca.2015.07.043 [DOI] [PubMed] [Google Scholar]
- 4.Yang J, Antin P, Berx G, et al. Guidelines and definitions for research on epithelial-mesenchymal transition. Nat Rev Mol Cell Biol. 2020;21(6):341–352. doi: 10.1038/s41580-020-0237-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thierauf J, Veit JA, Hess J. Epithelial-to-mesenchymal transition in the pathogenesis and therapy of head and neck cancer. Cancers. 2017;9(12):7. doi: 10.3390/cancers9070076 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kopp F, Mendell JT. Functional classification and experimental dissection of long noncoding RNAs. Cell. 2018;172(3):393–407. doi: 10.1016/j.cell.2018.01.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Yang J, Lian Y, Yang R, et al. Upregulation of lncRNA LINC00460 Facilitates GC Progression through Epigenetically Silencing CCNG2 by EZH2/LSD1 and Indicates Poor Outcomes. Mol Ther Nucleic Acids. 2020;19:1164–1175. doi: 10.1016/j.omtn.2019.12.041 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Li K, Sun D, Gou Q, et al. Long non-coding RNA linc00460 promotes epithelial-mesenchymal transition and cell migration in lung cancer cells. Cancer Lett. 2018;420:80–90. doi: 10.1016/j.canlet.2018.01.060 [DOI] [PubMed] [Google Scholar]
- 9.Feng L, Rao M, Zhou Y, Zhang Y, Zhu Y. Long noncoding RNA 00460 (LINC00460) promotes glioma progression by negatively regulating miR-320a. J Cell Biochem. 2019;120(6):9556–9563. doi: 10.1002/jcb.28232 [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 10.Jiang Y, Cao W, Wu K, et al. LncRNA LINC00460 promotes EMT in head and neck squamous cell carcinoma by facilitating peroxiredoxin-1 into the nucleus. J Exp Clin Cancer Res. 2019;38(1):365. doi: 10.1186/s13046-019-1364-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Budach V, Tinhofer I. Novel prognostic clinical factors and biomarkers for outcome prediction in head and neck cancer: a systematic review. Lancet Oncol. 2019;20(6):e313–e326. doi: 10.1016/S1470-2045(19)30177-9 [DOI] [PubMed] [Google Scholar]
- 12.Hanahan D, Weinberg RA. Hallmarks of cancer: the next generation. Cell. 2011;144(5):646–674. doi: 10.1016/j.cell.2011.02.013 [DOI] [PubMed] [Google Scholar]
- 13.Puram SV, Tirosh I, Parikh AS, et al. Single-cell transcriptomic analysis of primary and metastatic tumor ecosystems in head and neck cancer. Cell. 2017;171(7):1611–1624.e1624. doi: 10.1016/j.cell.2017.10.044 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Esposito R, Bosch N, Lanzós A, Polidori T, Pulido-Quetglas C, Johnson R. Hacking the cancer genome: profiling therapeutically actionable long non-coding RNAs using CRISPR-Cas9 Screening. Cancer Cell. 2019;35(4):545–557. doi: 10.1016/j.ccell.2019.01.019 [DOI] [PubMed] [Google Scholar]
- 15.Zhang XZ, Liu H, Chen SR. Mechanisms of long non-coding RNAs in cancers and their dynamic regulations. Cancers. 2020;12:5. doi: 10.3390/cancers12051245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Treiber T, Treiber N, Meister G. Regulation of microRNA biogenesis and its crosstalk with other cellular pathways. Nat Rev Mol Cell Biol. 2019;20(1):5–20. doi: 10.1038/s41580-018-0059-1 [DOI] [PubMed] [Google Scholar]
- 17.Hong H, Zhu H, Zhao S, et al. The novel circCLK3/miR-320a/FoxM1 axis promotes cervical cancer progression. Cell Death Dis. 2019;10(12):950. doi: 10.1038/s41419-019-2183-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lv G, Wu M, Wang M, et al. miR-320a regulates high mobility group box 1 expression and inhibits invasion and metastasis in hepatocellular carcinoma. Liver Int. 2017;37(9):1354–1364. doi: 10.1111/liv.13424 [DOI] [PubMed] [Google Scholar]
- 19.Yuan Z, Xiu C, Song K, et al. Long non-coding RNA AFAP1-AS1/miR-320a/RBPJ axis regulates laryngeal carcinoma cell stemness and chemoresistance. J Cell Mol Med. 2018;22(9):4253–4262. doi: 10.1111/jcmm.13707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Roedig H, Damiescu R, Zeng-Brouwers J, et al. Danger matrix molecules orchestrate CD14/CD44 signaling in cancer development. Semin Cancer Biol. 2020;62:31–47. [DOI] [PubMed] [Google Scholar]
- 21.Huang HC, Cai BH, Suen CS, et al. BGN/TLR4/NF-B mediates epigenetic silencing of immunosuppressive siglec ligands in colon cancer cells. Cells. 2020;9:2. doi: 10.3390/cells9020397 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jacobsen F, Kraft J, Schroeder C, et al. Up-regulation of biglycan is associated with poor prognosis and PTEN deletion in patients with prostate cancer. Neoplasia. 2017;19(9):707–715. doi: 10.1016/j.neo.2017.06.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hong W, Ying H, Lin F, Ding R, Wang W, Zhang M. lncRNA LINC00460 Silencing Represses EMT in Colon Cancer through Downregulation of ANXA2 via Upregulating miR-433-3p. Mol Ther Nucleic Acids. 2020;19:1209–1218. doi: 10.1016/j.omtn.2019.12.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chou LF, Chen CY, Yang WH, et al. Suppression of hepatocellular carcinoma progression through FOXM1 and EMT Inhibition via Hydroxygenkwanin-Induced miR-320a Expression. Biomolecules. 2019;10:1. doi: 10.3390/biom10010020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhang HH, Li R, Li YJ, et al. eIF4E‑related miR‑320a and miR‑340‑5p inhibit endometrial carcinoma cell metastatic capability by preventing TGF‑β1‑induced epithelial‑mesenchymal transition. Oncol Rep. 2020;43(2):447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Thakur AK, Nigri J, Lac S, et al. TAp73 loss favors Smad-independent TGF-β signaling that drives EMT in pancreatic ductal adenocarcinoma. Cell Death Differ. 2016;23(8):1358–1370. doi: 10.1038/cdd.2016.18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li H, Zhong A, Li S, et al. The integrated pathway of TGFβ/Snail with TNFα/NFκB may facilitate the tumor-stroma interaction in the EMT process and colorectal cancer prognosis. Sci Rep. 2017;7(1):4915. doi: 10.1038/s41598-017-05280-6 [DOI] [PMC free article] [PubMed] [Google Scholar]