Abstract
It has been suggested that a few methanogens are capable of extracellular electron transfers. For instance, Methanosarcina barkeri can directly capture electrons from the coexisting microbial cells of other species. Methanothrix harundinacea and Methanosarcina horonobensis retrieve electrons from Geobacter metallireducens via direct interspecies electron transfer (DIET). Recently, Methanobacterium, designated strain YSL, has been found to grow via DIET in the co-culture with Geobacter metallireducens. Methanosarcina acetivorans can perform anaerobic methane oxidation and respiratory growth relying on Fe(III) reduction through the extracellular electron transfer. Methanosarcina mazei is capable of electromethanogenesis under the conditions where electron-transfer mediators like H2 or formate are limited. The membrane-bound multiheme c-type cytochromes (MHC) and electrically-conductive cellular appendages have been assumed to mediate the extracellular electron transfer in bacteria like Geobacter and Shewanella species. These molecules or structures are rare but have been recently identified in a few methanogens. Here, we review the current state of knowledge for the putative extracellular electron transfers in methanogens and highlight the opportunities and challenges for future research.
Keywords: extracellular electron transfer, methanogenic archaea, c-type cytochrome, archaellum, direct interspecies electron transfer, direct electron transfer
Introduction
Methanogens are important to the carbon biogeochemical cycle and global methane emissions. Approximately 1 billion tons of methane is generated annually by methanogens in natural and man-made anoxic environments, as a consequence, about half of that is emitted into the atmosphere (Thauer, 1998; Thauer et al., 2008). Methanogens belong to members of the archaeal domain and occur mostly in the phylum Euryarchaeota. The species found to date fall into seven orders that differ both in energy conservation and ecological niches (Liu and Whitman, 2008; Thauer et al., 2008). Methanogenesis can be generally performed through the hydrogenotrophic, aceticlastic, methylotrophic, or methyl-reducing pathways depending on the substrates available in environments. Methanogenesis from syntrophic oxidation of short-chain fatty acids and alcohols is the key process during the anaerobic decomposition of complex organic matter. In this process, H2 and formate are usually used as electron transfer mediators. (Stams and Plugge, 2009). An increasing of observations addressing some methanogens perform extracellular electron transfers, however, have questioned this knowledge of methanogenesis.
The extracellular electron transfer (EET) has been well studied in some bacteria regarding Geobacter and Shewanella species (Shi et al., 2016). EET pathways contain the direct electron transfer (DET) pathway in which solid abiotic materials, such as iron minerals or electrodes, can function as terminal electron acceptors/donors, and the alternative pathway is the direct interspecies electron transfer (DIET) where living microbes can serve as terminal electron acceptors or donors. The strongest evidence for DIET is from a co-culture of Geobacter metallireducens and Geobacter sulfurreducens, which can oxidize ethanol with the reduction of fumarate to succinate (Summers et al., 2010; Lovley, 2017). DIET or DET cannot be established with the Geobacter mutants deprived of the electrically conductive pili (e-pili) or the outer-membrane multi-heme c-type cytochromes (MHC); these components are thus critical to extracellular electron transfer (Summers et al., 2010; Shrestha et al., 2013; Shi et al., 2016; Lovley, 2017).
The DIET pathway has also been proposed in methanogenic aggregates (Morita et al., 2011) and was initially demonstrated in the co-cultures of G. metallireducens with Methanothrix harundinacea or Methanosarcina barkeri (Rotaru et al., 2014a,b). Supplementation of iron oxides (hematite and magnetite) or other conductive materials, such as granular activated carbon, can facilitate the syntrophic growth of co-cultures consuming acetate or ethanol (Kato et al., 2012; Liu et al., 2012). Magnetite-facilitated syntrophic oxidation of propionate and butyrate was detected in methanogenic enrichments (Li et al., 2015; Zhang and Lu, 2016; Xia et al., 2019); consequently, the DIET pathway has been proposed to be an alternate strategy for syntrophic metabolisms in methanogenic environments (Kato et al., 2012; Liu et al., 2012; Li et al., 2015). The discovery of DIET in the co-cultures of Geobacter and methanogens (Rotaru et al., 2014a,b; Yee and Rotaru, 2020) supports a long-standing hypothesis that some methanogens can directly obtain electrons from the outside (Dinh et al., 2004). The DET pathways of methanogens have been proposed to occur with electrodes or minerals serving as electron sources (Cheng et al., 2009; Beese-Vasbender et al., 2015; Soo et al., 2016; Yan et al., 2018). Putative DIET and DET in methanogens greatly expand our understanding of methanogen’s roles in biogeochemistry, and future research shall re-assess the contribution of different methanogens to address global methane emission challenges. The mechanisms of extracellular electron transfers from methanogenic archaea, however, have yet to be resolved. The purpose of this review is to summarize the current understanding of putative extracellular electron transfers from diverse methanogens and highlight the challenges of future research.
Diverse Strategies in Methanosarcinales
Methanosarcina barkeri
M. barkeri is metabolically versatile and capable of hydrogenotrophic (H2/CO2), methylotrophic (methanol, methylamine), methyl-reducing (H2 and methanol/methylamine), and aceticlastic (acetate) methanogenesis (Welander and Metcalf, 2005; Thauer et al., 2008; Welte and Deppenmeier, 2014). When performing hydrogenotrophic methanogenesis, M. barkeri employs the energy-converting ferredoxin-dependent hydrogenase (Ech), F420-reducing hydrogenase (Frh), and membrane-bound methanophenazine-dependent hydrogenase (Vht; Thauer et al., 2008; Kulkarni et al., 2009; Welte and Deppenmeier, 2014). In the pathway of methylotrophic and aceticlastic methanogenesis of M. barkeri, Ech, Frh, and Vht also participate (Deppenmeier et al., 1995; Meuer et al., 2002; Welander and Metcalf, 2005; Kulkarni et al., 2009, 2018; Mand et al., 2018). Hydrogenases from M. barkeri can catalyze the H2 production of electrodes in electrochemical reactors. Cathodes inoculated with Ms. barkeri, with the set potential of –0.6 V vs. the standard hydrogen electrode (SHE), could produce H2 at a rate of 120 ± 18 nmol d-1 ml-1, and about half of this rate was detected with the cell extracts of M. barkeri (Yates et al., 2014). Recently, it was shown that hydrogenases in combination with ferredoxin (Fd) and F420 from M. barkeri could attach to the electrode surface and catalyze the formation of H2. Then, the produced H2 could be consumed rapidly by M. barkeri, resulting in a low or undetectable level of H2 accumulation (Rowe et al., 2019).
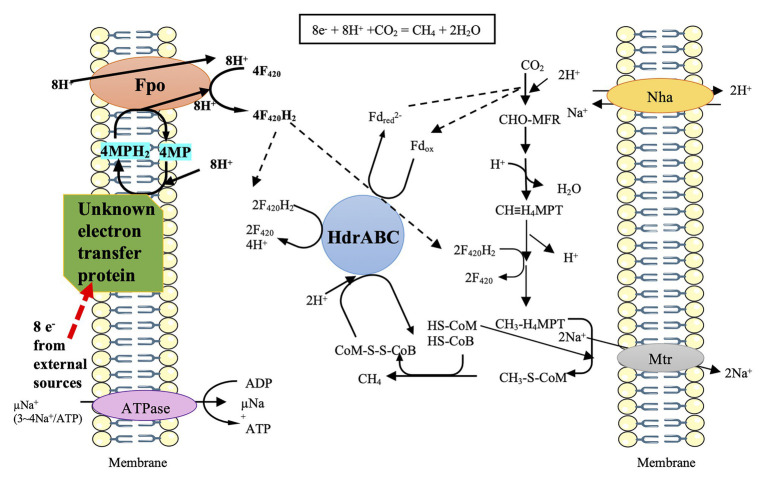
However, a hydrogenase deletion mutant of M. barkeri still exhibited the ability of electromethanogenesis with the cathode potential poised at –0.484 V vs. SHE, indicating a hydrogenase-independent mechanism to facilitate the cathodic activity (Rowe et al., 2019). Furthermore, M. barkeri is capable of conducting DIET to accept electrons from syntrophic growth with G. metallireducens on ethanol (Rotaru et al., 2014a; Holmes et al., 2018). The stoichiometric conversion of ethanol to methane (1.5 CH4 per ethanol) in the co-culture of G. metallireducens and M. barkeri indicated that M. barkeri not only metabolized the acetate produced by G. metallireducens but also used the electrons released from ethanol oxidation. The transcriptome was compared between the co-culture of G. metallireducens/M. barkeri and the co-culture of Pelobacter carbinolicus/M. barkeri (H2 was used as the electron transfer mediator) (Holmes et al., 2018). It showed the significant upregulation of gene expression of the most subunits of the membrane-bound F420-dehydrogenase (Fpo) in the co-culture of G. metallireducens/M. barkeri. In addition, the expression of nine genes predicted to be involved in ubiquinone/menaquinone biosynthesis and those genes coding for HdrA1B1C1, HdrA2, and HdrB2 were also upregulated in the co-culture of G. metallireducens/M. barkeri. Therefore, a model for the electron and proton flux of the CO2 reduction to CH4 in M. barkeri during DIET-based growth has been postulated based on the above transcriptome comparison (Figure 1; Holmes et al., 2018). M. barkeri may obtain electrons from an unknown electron carrier and donate the electrons to methanophenazine (MP), a membrane-bound electron carrier analogous to ubiquinones. Then, the membrane-bound, proton-pumping F420-dehydrogenase (Fpo) may transfer electrons from MPH2 to F420, resulting in the formation of F420H2. Half of the F420H2 is proposed to serve as a reductant in the CO2 reduction pathway, while the remaining F420H2 donates electrons to HdrABC. With participation of electron bifurcation, HdrABC may transfer electrons to Fdox and CoM-S-S-CoB, respectively (Holmes et al., 2018).
Figure 1.
Prediction model of direct electron uptake in Methanosarcina barkeri, cited (Holmes et al., 2018). The black arrows represent the possible transfers of electrons via F420 and Fd. The red arrow represents the possible route for electron uptake from the outside. The unknown electron transfer proteins may gain 8 e— from external sources and then channel these electrons via MP/MPH2 to Fpo. Fpo can utilize F420/F420H2 to deliver electrons to the process of CO2 to CH4.
M. barkeri does not contain MHC, and Fpo has no active sites on the outer surface of the membrane (Welte and Deppenmeier, 2014); how the external electrons are channeled into Fpo thus remains an important question (Figure 1). Future research shall focus on the alternate redox-active proteins in M. barkeri that can potentially aid in direct electron uptake. Moreover, a few studies have shown that the membrane-bound methanophenazine greatly contributes to the electrical conductivity of the membrane of Methanosarcina acetivorans growing on methanol (Duszenko and Buan, 2017; Yan et al., 2018). Further research is necessary to identify the effect of methanophenazine on the possible augmentation of membrane conductivity in Ms. barkeri performing direct electron uptake.
Methanothrix harundinacea
Both Methanosarcina and Methanothrix are known as the aceticlastic methanogens, while Methanothrix species are the specialists having a much lower threshold concentration for acetate metabolism (Jetten et al., 1992; Welte and Deppenmeier, 2014). Methanothrix species have no genes coding for the hydrogenases like Ech, Frh, and Vht (Welte and Deppenmeier, 2011). Although Methanothrix species are restricted to acetate degradation, the gene repository for CO2 reduction exists in their genome (Rotaru et al., 2014b). Afterwards, Mt. harundinacea has been suggested to perform DIET with G. metallireducens (Rotaru et al., 2014b; Yee and Rotaru, 2020). The co-culture of G. metallireducens and M. harundinacea converted ethanol to methane in a stoichiometry of ca. 1.5 moles CH4 per mole ethanol. The inability of G. metallireducens to generate H2 or formate and the inability of M. harundinacea to metabolize H2 or formate ruled out the possibility of electron transfer via mediated electron carrier. So, the finding strongly suggested that DIET occurrence in the co-culture of G. metallireducens and M. harundinacea (Rotaru et al., 2014b). In addition, the genes coding for CO2 reduction were highly expressed in Mt. harundinacea from the co-culture (Zhu et al., 2012; Rotaru et al., 2014b). This methanogen, however, cannot utilize the cathode as the sole electron donor (Rotaru et al., 2014b; Yee and Rotaru, 2020).
The specific electron transfer route for DIET remains elusive in M. harundinacea. The cell surface of Methanothrix genera consists of a protein sheet that is thought to be composed of amyloid proteins. The amyloid proteins can cluster together binding peptides and metal irons, which may facilitate the direct electron uptake from external electron donors (Maji et al., 2009; Viles, 2012; Dueholm et al., 2015; Yee and Rotaru, 2020). However, this is highly speculative and the experimental evidence has yet to be obtained.
Methanosarcina horonobensis
M. horonobensis has a relatively narrow substrate spectrum compared with M. mazei and M. barkeri, only growing on methanol, dimethylamine, and acetate but not on H2/CO2 (Shimizu et al., 2011). M. horonobensis is able to retrieve electrons from G. metallireducens via DIET. Specifically, the co-culture of G. metallireducens and M. horonobensis converted 8.8 ± 0.4 mM ethanol to 13.1 ± 0.8 mM CH4. Therefore, each mole of ethanol yielded ca. 1.5 moles of CH4, indicating complete conversion of the added ethanol to CH4 (Yee et al., 2019). Similar to the co-culture of M. harundinecea/G. metallireducens, the inability of G. metallireducens to generate H2 or formate and the inability of M. horonobensis to metabolize H2 ruled out the possibility of electron transfer via the mediated electron carrier. However, M. horonobensis failed to perform electromethanogenesis at the cathode potential poised at –0.4 V vs. SHE (Yee et al., 2019; Yee and Rotaru, 2020). Further research needs to elucidate why M. horonobensis can accept electrons from electroactive microbes (typically G. metallireducens) rather than from electrodes. M. horonobensis has the membrane-bound MHC (Yee and Rotaru, 2020), and future efforts to study DIET of the co-culture of G. metallireducens and M. horonobensis must be intensified and eventually provide the clarified membrane-bound electron transport chain and MHC expression response to DIET.
Methanosarcina mazei and Methanothrix soehngenii
M. mazei is closely related to M. barkeri in the phylogenetic relationship and can consume a wide range of substrates, including H2/CO2, methanol, methylamine, and acetate (Welte and Deppenmeier, 2014). It is possible that hydrogenases are coupled with ferredoxin (Fd) and F420 from M. mazei to capture electrons from cathodes to form H2, which is then consumed for the CH4 production. However, the experimental evidence for this speculation has yet to be revealed (Welte and Deppenmeier, 2014; Rowe et al., 2019). M. soehngenii is a strict non-hydrogenotrophic methanogen. A recent study showed that both M. mazei and M. soehngenii could pair with G. metallireducens (Yee and Rotaru, 2020). However, only 7.7 ± 0.7 mM CH4 was produced from 10 mM ethanol in the co-culture of M. mazei and G. metallireducens, and only 1.8 ± 1.0 mM CH4 was produced from 20 mM ethanol in the co-culture of M. soehngenii and G. metallireducens (Yee and Rotaru, 2020), indicating incomplete conversion of the added ethanol to CH4. Therefore, laboratory study should further verify if M. mazei and M. soehngenii can establish DIET with G. metallireducens or other Geobacter species.
Methanosarcina acetivorans
M. acetivorans does not possess Ech and Vht hydrogenases and hence is incapable of H2-dependent methanogenesis (Ollivier et al., 1984; Sowers et al., 1984; Ferry, 2020). The presence of the membrane-bound Rnf complex (homolog of rhodobacter nitrogen fixation complex) can oxidize Fdred or hydroquinone of flavodoxin A (FldAhq) to Fdox or semiquinone of flavodoxin A (FldAsq), which distinguishes M. acetivorans from all the H2-utilizing methanogens among Methanosarcina (Li et al., 2006; Wang et al., 2011; Schlegel et al., 2012; Prakash et al., 2019b). It is worth noting that the Rnf genes in M. acetivorans cluster with the gene coding for a c-type cytochrome with multiheme-binding motifs (MmcA) (Galagan et al., 2002; Li et al., 2006; Schlegel et al., 2012). The ν mmcA mutant strain of M. acetivorans, however, still grows on acetate, indicating MmcA is unnecessary for the acetotrophic growth (Holmes et al., 2019).
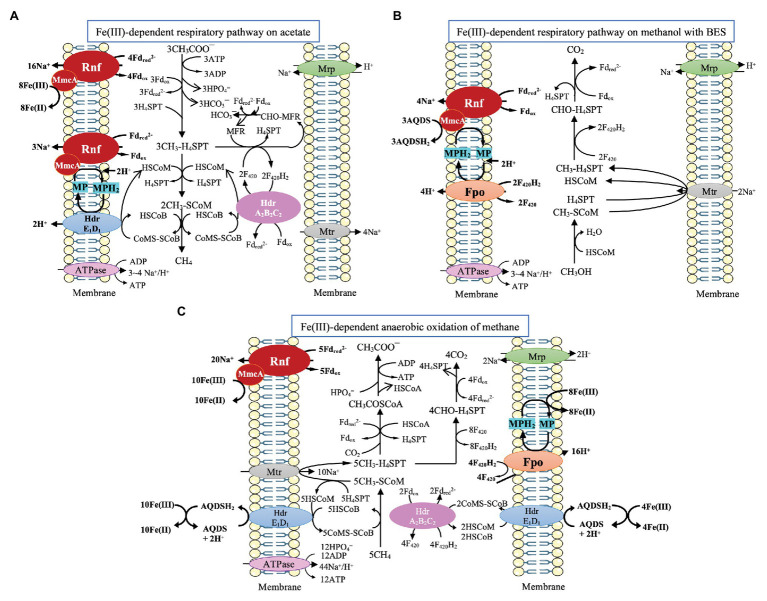
M. acetivorans has been shown to perform Fe(III)-dependent respiratory growth on acetate with the simultaneous reduction of Fe(III) to Fe(II) and production of CH4. The relevant pathway is illustrated in Figure 2A (Prakash et al., 2019a). One-carbon transformations leading to CH4 are the same as its aceticlastic pathway of methanogenesis where Fdred can be generated. By reversal of reactions of the CO2 reduction pathway, the respiratory electron transport occurs through oxidation of methyl-tetrahydrosarcinapterin (CH3-H4SPT) to formyl-methanofuran (CHO-MFR) and then to HCO3¯ along with the generation of F420H2 and Fdred (Lessner et al., 2006; Prakash et al., 2019a; Ferry, 2020). Then, through the combination of Rnf enzyme complex with MmcA, electrons are transferred from Fdred for the reduction of Fe(III) to Fe(II). Two Na+ are translocated for each Fe(III) reduced to Fe(II) in this process (Yan et al., 2018; Prakash et al., 2019a). Given that the expression of Fpo is down-regulated, it is postulated that the reoxidation of F420H2 occurs through the electron bifurcation performing by HdrA2B2C2 (Prakash et al., 2019a; Ferry, 2020). Importantly, the Fe(III)-dependent respiratory growth showed higher acetate consumption, a greater ratio of ATP/ADP, and a higher growth rate, indicating the improved energy conservation (Prakash et al., 2019a; Ferry, 2020). Interestingly, Ms. acetivorans can also perform respiratory growth on methanol with AQDS (anthraquinone-2,6-disulfonate, an analog of humic substances in the environment) as the external electron acceptor in the presence of methanogenesis inhibitor 2-biomoethanesulfonate (BES; Figure 2B; Holmes et al., 2019). F420H2 and Fdred generated from the oxidization of methanol are probably reoxidized by Fpo and Rnf complex, respectively, and electrons are channeled via either MP/MPH2 or MmcA for the external reduction of AQDS. Fpo and Rnf can pump H+ and Na+, respectively. Notably, the ν mmcA mutant strain is incapable of methanol-dependent respiratory growth, indicating the importance of MmcA in the external electron transfer (Holmes et al., 2019).
Figure 2.
Pathway proposed for Fe(III)-dependent respiratory pathway and CH4 oxidation by Ms. acetivorans. (A) Fe(III)-dependent respiratory pathway on acetate, cited (Prakash et al., 2019a). (B) Fe(III)-dependent respiratory pathway on methanol with 2-bromoethanesulfonate (BES), cited (Holmes et al., 2019). (C) Fe(III)-dependent CH4 oxidation, cited (Yan et al., 2018).
M. acetivorans is also capable of Fe(III)-dependent anaerobic oxidation of methane through the reversal of aceticlastic and CO2-reducing methanogenesis (Figure 2C; Moran et al., 2005, 2007; Soo et al., 2016; Yan et al., 2018). Methane is assumed to be oxidized by the methyl-coenzyme M methyl reductase (Mcr) to yield methyl-coenzyme M (CH3-SCoM), and the methyl group is transferred through Mtr to tetrahydrosarcinapterin (H4SPT). During the oxidation of CH3-H4SPT to CO2, Fdred and F420H2 are formed. Fdred is probably used for the reduction of Fe(III) through the combination of the Rnf complex with MmcA. The Na+ gradient generated by the Rnf complex can power the transfer of the methyl group of CH3-SCoM to H4SPT (Yan et al., 2018). F420H2 is probably reoxidized by HdrA2B2C2 through electron bifurcation for the coupling reduction of CoM-S-S-CoB and Fdox. The produced HSCoM and HSCoB are postulated to be reoxidized by the membrane-bound HdrE1D1, driving the AQDS mediated reduction of Fe(III) to Fe(II) (Yan et al., 2018). Additionally, MP is postulated to gain electrons via Fpo from F420H2, and the produced MPH2 is reoxidized for the reduction of Fe(III) (Yan et al., 2018).
Notably, though MHC (also MmcA here) in Ms. acetivorans is analogous to MHC in Shewanella and Geobacter species (Yan et al., 2018; Holmes et al., 2019), the exact route for the MHC-mediated electron transfer has yet to be elucidated. Up to now, there has been no evidence indicating that Ms. acetivorans can gain electrons directly from electrodes or materials. It is also unknown whether this methanogen can develop an electrical connection with other microbes like Geobacter species. In addition, the deletion of the MHC gene does not obviously impair the growth of Ms. acetivorans on acetate and especially does not influence the expression of Rnf genes, indicating MHC gene might be independent of Rnf (Holmes et al., 2019). This independence, however, may promise the flexibility of MHC to interact with other enzymes like Fpo and make M. acetivorans adaptable to varying environmental conditions.
Strategies in Hydrogenotrophic Methanogens
Methanospirillum hungatei With Electrically Conductive Protein Filaments
Methanospirillum species belong to the members of Methanospirillaceae within the order of Methanomicrobiales and represent a group of methanogenic archaea utilizing hydrogen or formate. Methanospirillum hungatei JF1 is the first isolated strain of Methanospirillum genera (Gunsalus et al., 2016). The potential c-type cytochrome in M. hungatei is predicted to be located in the cytoplasm, indicating that it may not take part in the extracellular electron transfer (Yee and Rotaru, 2020). A unique trait of M. hungatei is the synthesis of extracellular filaments, called archaella, that can drive cellular motility, promote biofilm formation, and participate in cellular adhesion (Schopf et al., 2008; Jarrell et al., 2011; Jarrell and Albers, 2012; Albers and Jarrell, 2015). The archaella more resemble the bacterial type IV pili in terms of evolution and structure than the bacterial flagella (Faguy et al., 1994; Thomas et al., 2001; Jarrell and Albers, 2012; Albers and Jarrell, 2015, 2018). The type IV pili in the Geobacter species have been found to be electrically conductive (hence named e-pili in short), mediating the long-distance extracellular electron transfer (Malvankar et al., 2012; Shi et al., 2016). So far, the e-pili have been explored mainly in bacteria, which raises the question of whether such conductive protein filaments have ever been evolved in archaea.
Initial screening of the relative conductivity of bacterial pili is typically determined with conductive atomic force microscopy (Reguera et al., 2005; Steidl et al., 2016; Sure et al., 2016; Liu et al., 2019), so a similar method was applied to the M. hungatei archaella (Walker et al., 2019). To avoid chemical alteration of the archaellum structure and determine the conductivity of hydrated archaella, 100 μl of a culture of M. hungatei grown in low-phosphate medium to induce archaellum expression (Faguy et al., 1993) was drop-cast onto highly oriented pyrolytic graphite (HOPG), washed, dried, and equilibrated at 40% relative humidity. Then, a conductive tip serving as a translatable top electrode was used for conductivity measurements. The above process could mimic physiologically relevant conditions (Walker et al., 2019). The local conductive imaging showed that the archaella of M. hungatei were electrically conductive. The linear-like current response to applied voltage was revealed in the point-mode current-voltage spectroscopy. The conductance estimated from this response curve was 16.9 ± 3.9 nS for the archaella of M. hunagtei, compared with 4.5 ± 0.3 nS for the pili of wild-type Geobacter sulfurreducens and 0.004 ± 0.002 nS for the pili of G. sulfurreducens strain Aro-5 designed for producing pili with poor conductivity (Reguera et al., 2005; Walker et al., 2019). The atomic model from the cryo-electron microscopy structure of archaella at 3.4 Å resolution revealed that the archaella of M. hungatei possessed a core of closely packed phenylalanines (Poweleit et al., 2017). This amino acid arrangement was considered as the key to the electrical conductivity of M. hungatei archaella (Walker et al., 2019).
The function of the electrically conductive archaella of M. hungatei in nature remains elusive. An earlier study showed that M. hungatei could reduce extracellular electron acceptors in which H2 was used as the electron-transfer mediator (Cervantes et al., 2002). Electrically conductive archaella of M. hungatei may merely facilitate cell attachment by dissipating charge barriers between cells and minerals/electrodes (Walker et al., 2019). Whether the archaella of M. hungatei can be used as conduits for the extracellular electron transfer remains an open question. It will be a great challenge to clarify mechanisms of archaellum-mediated electron transport and determine how this electron transport process is coordinated with the catabolic electron flux during growth of M. hungatei.
Methanococcus maripaludis Capable of Extracellular Enzyme-Dependent Electron Uptake
Methanococcus maripaludis, which belongs to the order Methanococcales, is often used as a model for genetic investigation (Leigh et al., 2011). This methanogen utilizes H2 and formate as the electron donor to reduce CO2 to methane (Brileya et al., 2014). M. maripaludis possesses cytoplasmic heterodisulfide reductase (HdrABC) but does not contain membrane-bound HdrDE. The cytoplasmic HdrABC and F420-nonreducing hydrogenase (Vhu) form a complex which can perform the flavin-based electron bifurcation, driving the endergonic reduction of oxidized ferredoxin (Fdox) by coupling with the exergonic reduction of heterodisulfide. The reduced ferredoxin (Fdred) is used for the first step of CO2 reduction by the formyl-methanofuran dehydrogenase (Fwd; Thauer et al., 2008). When formate is supplied, the formate dehydrogenase (Fdh) can be activated and incorporated into the complex of Vhu, Hdr, and Fwd (Thauer et al., 2008; Costa et al., 2010; Kaster et al., 2011). F420-reducing hydrogenase (Frh) in the cytoplasm consumes H2 to produce F420H2 feeding electrons into the pathway of methanogenesis (Thauer et al., 2008). Overall, M. maripaludis contains six catabolic hydrogenases and an additional energy-converting ferredoxin-dependent hydrogenase (Eha).
Analysis of bioelectrochemical performance revealed that the methane formation rate of the hydrogenase-deprived MM1284 strain of M. maripaludis was only about one-tenth of the rate of the wild-type strain (Lohner et al., 2014). The MM1284 mutant carries markerless in-frame deletions of all five catabolic hydrogenase genes except Eha, which is essential to reduce ferredoxin for anabolic reactions (Lie et al., 2012; Costa et al., 2013a; Lohner et al., 2014). The significantly reduced rate of methane production of the MM1284 strain suggested that most of the cathodic electrons used for methane production in the wild type M. maripaludis were derived from a hydrogenase-dependent mechanism (Lohner et al., 2014). It was further showed that the hydrogenases and other redox enzymes (like formate dehydrogenases) from cells could precipitate on Fe(0) or the electrode surface. Meanwhile, these enzymes catalyzed the formation of H2, or formate, which was then consumed by M. maripaludis cells (Deutzmann et al., 2015). In the early view, it was thought that H2 or formate was produced from electrodes via the abiotic way in the cathode chamber with methanogens. While the above studies showed that H2 or formate production was mainly catalyzed by hydrogenase, or formate dehydrogenase from methanogens (biotic way), resulting in the high conversion efficiency of current to methane (Cheng et al., 2009). And this process of electron uptake from electrodes can be defined as surface-associated redox enzyme-dependent electron uptake in methanogens (Figure 3).
Figure 3.
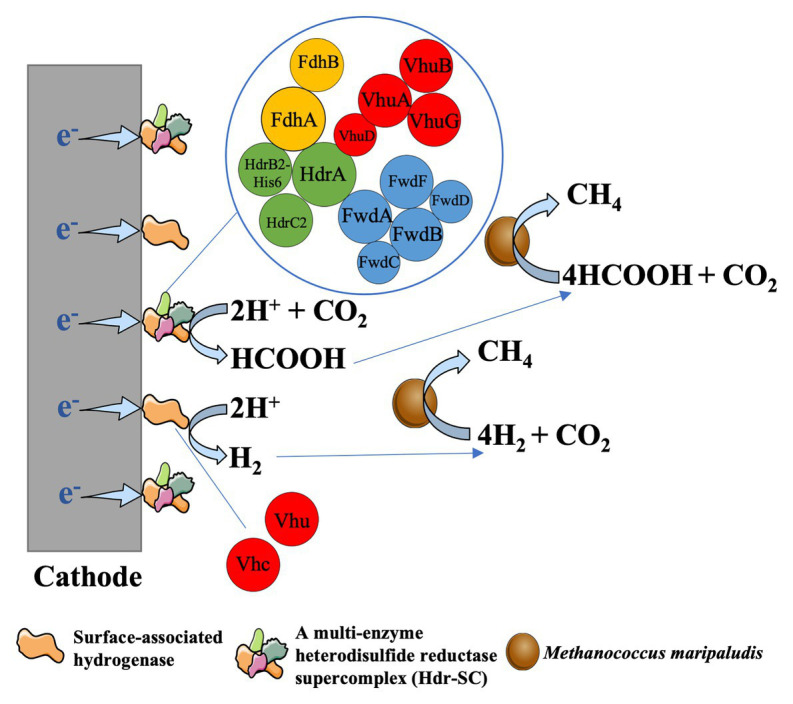
Enzyme-dependent external electron uptake in Methanococcus maripaludis, cited (Lienemann et al., 2018). Hydrogenase and formte dehydrogenase can be released from the living or dead cells of methanogens and then are absorbed on the cathode surface. These surface-associated enzymes can catalyze the formation of H2 or formate, which was then rapidly consumed by M. maripaludis cells to produce CH4 (Deutzmann et al., 2015).
The composition of these cell-free and surface-associated redox enzymes however might be complicated. Many studies have shown that hydrogenases and formate dehydrogenases are electroactive and capable of generating hydrogen, or formate by consuming electrons from electrodes (Cracknell et al., 2008; Parkin et al., 2008; Reda et al., 2008; Armstrong et al., 2009; Lojou, 2011; Baffert et al., 2012; Sakai et al., 2016; Chica et al., 2017; Hu et al., 2018; Lienemann et al., 2018; Yuan et al., 2018; Cordas et al., 2019). Recently it has been shown that the M. maripaludis-derived NiFeSe hydrogenase and the NiFe hydrogenase, when immobilized at a cathode in a cobaltocene-functionalized polyallylamine (Cc-PAA) redox polymer, could mediate the rapid and efficient hydrogen evolution (Ruth et al., 2020). In addition, a multi-enzyme heterodisulfide reductase supercomplex (Hdr-SC) of M. maripaludis was purified. In Fe(0) corrosion experiments, hydrogen formation rates from Fe(0) in the crude lysate amended and purified Hdr-SC vials were 0.14 ± 0.04 μmol d−1 μl−1 and 0.007 ± 0.03 μmol d−1 μl−1 lysate equivalent, respectively. The formate formation rate from Fe(0) by cell lysate was 0.62 ± 0.03 μmol d−1 μl−1, and a quarter of this activity (0.15 ± 0.01 μmol d−1 μl−1 lysate equivalent) was recovered from purified Hdr-SC (Lienemann et al., 2018). The electrocatalytic activity of purified Hdr-SC was also examined. Upon applying a potential of –0.6 V vs. SHE, the electrochemical reactors with 60 μg of purified Hdr-SC accumulated formate and hydrogen at initial rates of 266 μmol h−1 L−1 catholyte and 17 μmol h−1 L−1 catholyte, respectively (Lienemann et al., 2018). Therefore, the hydrogen formation of cell lysate was more likely to be catalyzed by a non-Hdr-SC hydrogenase activity, while Hdr-SC was the main component catalyzing formate production from Fe(0)-derived and cathode-derived electrons. The Hdr-SC in M. maripaludis consists of a heterodisulfide reductase (HdrABC), a formate dehydrogenase (FdhAB), and a NiFe-hydrogenase (VhuABDG; Costa et al., 2010, 2013b; Lienemann et al., 2018). In a recent study, homodimeric Hdr complexes containing either (Vhu)2 or (Fdh)2 have been identified and purified (Milton et al., 2018). Although the structure and function of flavin-based electron bifurcation of Hdr-SC have been documented, it remains unclear why the Hdr-SC deposited on Fe(0) or the cathode surface tends to produce more formate than hydrogen.
It is worth noting that, albeit at a slow rate, the bioelectrochemical methane formation in the hydrogenase-deficient MM1284 strain of M. maripaludis was detected (Lohner et al., 2014). Lowering the cathode potential did not increase the rate of methanogenesis, and no formate was detected in the reactors containing MM1284 cells, indicating that a direct electron uptake might occur in the MM1284 strain (Lohner et al., 2014). However, the absence of a detectable level of electron-carrying mediators cannot rule out the possibility of rapid cycling of these redox mediators in the electrochemical reactors. For instance, the cell extracts from MM1284 strain can catalyze formate formation on cathodes (Deutzmann et al., 2015). A recent study designed a combined method using a hydrogen microsensor system and cyclic voltammetry (CV) to determine in situ hydrogen concentration within the cathodic biofilm (Cai et al., 2020). A similar method can be explored to detect other in situ redox mediators, like formate, within the cathodic biofilm of reactors with MM1284 strain.
Future Perspectives
M. barkeri has been demonstrated to perform DIET and utilize electrons from electrodes. M. harundinacea and M. horonobensis can retrieve electrons from G. metallireducens via DIET but cannot perform electromethanogenesis. M. acetivorans can perform Fe(III)-dependent respiratory growth and anaerobic oxidation of methane. Ms. mazei is capable of electromethanogenesis under the potential of –0.4 V vs. SHE. A strain of Methanobacterium, designated strain YSL, can establish DIET with G. metallireducens (Zheng et al., 2020), indicating that the DIET pathway is more broadly distributed among methanogens than previously thought.
However, the external electron-acquisition/donation mechanisms have remained unclear. The external electron-acquisition/donation processes need to coordinate with the internal energy metabolism. Different methanogens perform different energy conservation; as a consequence, the possible external electron-acquisition/donation gadgets may show a high diversity among methanogens. Some methanogens contain MHC, or electrically conductive archaella (M. hungatei), while others may have unknown electron-acquisition/donation gadgets.
Many puzzles on the extracellular electron transfer of methanogens remain to be resolved. It warrants further research to figure out why some methanogens can accept electrons from other microbes but cannot utilize electrons from electrodes. Additionally, it remains unclear whether the electrically conductive archaella of M. hungatei can help M. hungatei electrically interact with other microbes, minerals, or electrodes. It is also unknown whether M. acetivorans can gain electrons directly from the outside. Moreover, given the diversity of Methanosarcina species, different kinds of external electron-acquisition/donation gadgets may exist and are worthy to be further explored. The novel technologies, such as metagenomics, metatranscriptomics, and high-resolution cryo-electron microscopy, may help us identify more methanogens that may perform DIET or DET in nature methanogenic communities. The biochemical approaches, for example, using the washed everted membrane vesicles to study Fe(III)-dependent anaerobic oxidation of methane in M. acetivorans (Yan et al., 2018), can also be employed to explore the mechanisms of DIET/DET in other methanogens.
Author Contributions
KG and YL conceived the study and wrote the manuscript. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We would like to thank Dr. Zhen Yang for her assistance in correcting orthographic and grammatical errors.
Footnotes
Funding. This study was financially supported by the National Natural Science Foundation of China (No. 41630857; 91951206).
References
- Albers S.-V., Jarrell K. F. (2015). The archaellum: how archaea swim. Front. Microbiol. 6:23. 10.3389/fmicb.2015.00023, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albers S.-V., Jarrell K. F. (2018). The Archaellum: an update on the unique archaeal motility structure. Trends Microbiol. 26, 351–362. 10.1016/j.tim.2018.01.004, PMID: [DOI] [PubMed] [Google Scholar]
- Armstrong F. A., Belsey N. A., Cracknell J. A., Goldet G., Parkin A., Reisner E., et al. (2009). Dynamic electrochemical investigations of hydrogen oxidation and production by enzymes and implications for future technology. Chem. Soc. Rev. 38, 36–51. 10.1039/B801144N, PMID: [DOI] [PubMed] [Google Scholar]
- Baffert C., Sybirna K., Ezanno P., Lautier T., Hajj V., Meynial-Salles I., et al. (2012). Covalent attachment of FeFe hydrogenases to carbon electrodes for direct electron transfer. Anal. Chem. 84, 7999–8005. 10.1021/ac301812s, PMID: [DOI] [PubMed] [Google Scholar]
- Beese-Vasbender P. F., Grote J. P., Garrelfs J., Stratmann M., Mayrhofer K. J. J. (2015). Selective microbial electrosynthesis of methane by a pure culture of a marine lithoautotrophic archaeon. Bioelectrochemistry 102, 50–55. 10.1016/j.bioelechem.2014.11.004, PMID: [DOI] [PubMed] [Google Scholar]
- Brileya K. A., Connolly J. M., Downey C., Gerlach R., Fields M. W. (2014). Taxis toward dydrogen gas by Methanococcus maripaludis. Sci. Rep. U.K. 3:3140. 10.1038/srep03690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cai W. W., Liu W. Z., Wang B., Yao H., Guadie A., Wang A. J. (2020). Semiquantitative detection of hydrogen-associated or hydrogen-free electron transfer within methanogenic biofilm of microbial electrosynthesis. Appl. Environ. Microbiol. 86:e01056–20. 10.1128/aem.01056-20, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cervantes F. J., de Bok F. A. M., Duong-Dac T., Stams A. J. M., Lettinga G., Field J. A. (2002). Reduction of humic substances by halorespiring, sulphate-reducing and methanogenic microorganisms. Environ. Microbiol. 4, 51–57. 10.1046/j.1462-2920.2002.00258.x, PMID: [DOI] [PubMed] [Google Scholar]
- Cheng S. A., Xing D. F., Call D. F., Logan B. E. (2009). Direct biological conversion of electrical current into methane by electromethanogenesis. Environ. Sci. Technol. 43, 3953–3958. 10.1021/es803531g, PMID: [DOI] [PubMed] [Google Scholar]
- Chica B., Wu C. H., Liu Y., Adams M. W. W., Lian T., Dyer R. B. (2017). Balancing electron transfer rate and driving force for efficient photocatalytic hydrogen production in CdSe/CdS nanorod-[NiFe] hydrogenase assemblies. Energy Environ. Sci. 10, 2245–2255. 10.1039/C7EE01738C [DOI] [Google Scholar]
- Cordas C. M., Campanico M., Baptista R., Maia L. B., Moura I., Moura J. J. G. (2019). Direct electrochemical reduction of carbon dioxide by a molybdenum-containing formate dehydrogenase. J. Inorg. Biochem. 196:110694. 10.1016/j.jinorgbio.2019.110694, PMID: [DOI] [PubMed] [Google Scholar]
- Costa K. C., Lie T. J., Jacobs M. A., Leigh J. A. (2013a). H2-independent growth of the hydrogenotrophic methanogen Methanococcus maripaludis. MBio 4:e00062–13. 10.1128/mBio.00062-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa K. C., Lie T. J., Xia Q., Leigh J. A. (2013b). VhuD facilitates electron flow from H2 or formate to heterodisulfide reductase in Methanococcus maripaludis. J. Bacteriol. 195, 5160–5165. 10.1128/jb.00895-13, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa K. C., Wong P. M., Wang T. S., Lie T. J., Dodsworth J. A., Swanson I., et al. (2010). Protein complexing in a methanogen suggests electron bifurcation and electron delivery from formate to heterodisulfide reductase. Proc. Natl. Acad. Sci. U. S. A. 107, 11050–11055. 10.1073/pnas.1003653107, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cracknell J. A., Vincent K. A., Armstrong F. A. (2008). Enzymes as working or inspirational electrocatalysts for fuel cells and electrolysis. Chem. Rev. 108, 2439–2461. 10.1021/cr0680639, PMID: [DOI] [PubMed] [Google Scholar]
- Deppenmeier U., Blaut M., Lentes S., Herzberg C., Gottschalk G. (1995). Analysis of the vhoGAC and vhtGAC operons from Methanosarcina mazei strain Gö1, both encoding a membrane-bound hydrogenase and a cytochrome b. Eur. J. Biochem. 227, 261–269. 10.1111/j.1432-1033.1995.tb20383.x, PMID: [DOI] [PubMed] [Google Scholar]
- Deutzmann J. S., Sahin M., Spormann A. M. (2015). Extracellular enzymes facilitate electron uptake in biocorrosion and bioelectrosynthesis. MBio 6:e00496–15. 10.1128/mBio.00496-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dinh H. T., Kuever J., Mussmann M., Hassel A. W., Stratmann M., Widdel F. (2004). Iron corrosion by novel anaerobic microorganisms. Nature 427, 829–832. 10.1038/nature02321, PMID: [DOI] [PubMed] [Google Scholar]
- Dueholm M. S., Larsen P., Finster K., Stenvang M. R., Christiansen G., Vad B. S., et al. (2015). The tubular sheaths encasing Methanosaeta thermophila filaments are functional amyloids. J. Biol. Chem. 290, 20590–20600. 10.1074/jbc.M115.654780, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duszenko N., Buan N. R. (2017). Physiological evidence for isopotential tunneling in the electron transport chain of methane-producing archaea. Appl. Environ. Microbiol. 83:e00950–17. 10.1128/aem.00950-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faguy D. M., Jarrell K. F., Kuzio J., Kalmokoff M. L. (1994). Molecular analysis of archaeal flagellins: similarity to the type IV pilin - transport superfamily widespread in bacteria. Can. J. Microbiol. 40, 67–71. 10.1139/m94-011, PMID: [DOI] [PubMed] [Google Scholar]
- Faguy D. M., Koval S. F., Jarrell K. F. (1993). Effect of changes in mineral composition and growth temperature on filament length and flagellation in the archaeon Methanospirillum hungatei. Arch. Microbiol. 159, 512–520. 10.1007/BF00249028 [DOI] [Google Scholar]
- Ferry J. G. (2020). Methanosarcina acetivorans: a model for mechanistic understanding of aceticlastic and reverse methanogenesis. Front. Microbiol. 11:1806. 10.3389/fmicb.2020.01806, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galagan J. E., Nusbaum C., Roy A., Endrizzi M. G., Macdonald P., Fitzhugh W., et al. (2002). The genome of M. acetivorans reveals extensive metabolic and physiological diversity. Genome Res. 12, 532–542. 10.1101/gr.223902, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunsalus R. P., Cook L. E., Crable B., Rohlin L., Mcdonald E., Mouttaki H., et al. (2016). Complete genome sequence of Methanospirillum hungatei type strain JF1. Stand. Genomic Sci. 11:2. 10.1186/s40793-015-0124-8, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. E., Rotaru A.-E., Ueki T., Shrestha P. M., Ferry J. G., Lovley D. R. (2018). Electron and proton flux for carbon dioxide reduction in Methanosarcina barkeri during direct interspecies electron transfer. Front. Microbiol. 9:3109. 10.3389/fmicb.2018.03109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmes D. E., Ueki T., Tang H.-Y., Zhou J., Smith J. A., Chaput G., et al. (2019). A membrane-bound cytochrome enables Methanosarcina acetivorans to conserve energy from extracellular electron transfer. MBio 10:e00789–19. 10.1128/mBio.00789-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Harris D. F., Dean D. R., Liu T. L., Yang Z. Y., Seefeldt L. C. (2018). Electrocatalytic CO2 reduction catalyzed by nitrogenase MoFe and FeFe proteins. Bioelectrochemistry 120, 104–109. 10.1016/j.bioelechem.2017.12.002, PMID: [DOI] [PubMed] [Google Scholar]
- Jarrell K. F., Albers S.-V. (2012). The archaellum: an old motility structure with a new name. Trends Microbiol. 20, 307–312. 10.1016/j.tim.2012.04.007, PMID: [DOI] [PubMed] [Google Scholar]
- Jarrell K. F., Stark M., Nair D. B., Chong J. P. J. (2011). Flagella and pili are both necessary for efficient attachment of Methanococcus maripaludis to surfaces. FEMS Microbiol. Lett. 319, 44–50. 10.1111/j.1574-6968.2011.02264.x, PMID: [DOI] [PubMed] [Google Scholar]
- Jetten M. S. M., Stams A. J. M., Zehnder A. J. B. (1992). Methanogenesis from acetate: a comparison of the acetate metabolism in Methanothrix soehngenii and Methanosarcina spp. FEMS Microbiol. Lett. 88, 181–197. 10.1111/j.1574-6968.1992.tb04987.x [DOI] [Google Scholar]
- Kaster A. K., Moll J., Parey K., Thauer R. K. (2011). Coupling of ferredoxin and heterodisulfide reduction via electron bifurcation in hydrogenotrophic methanogenic archaea. Proc. Natl. Acad. Sci. U. S. A. 108, 2981–2986. 10.1073/pnas.1016761108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato S., Hashimoto K., Watanabe K. (2012). Methanogenesis facilitated by electric syntrophy via (semi)conductive iron-oxide minerals. Environ. Microbiol. 14, 1646–1654. 10.1111/j.1462-2920.2011.02611.x, PMID: [DOI] [PubMed] [Google Scholar]
- Kulkarni G., Kridelbaugh D. M., Guss A. M., Metcalf W. W. (2009). Hydrogen is a preferred intermediate in the energy-conserving electron transport chain of Methanosarcina barkeri. Proc. Natl. Acad. Sci. U. S. A. 106, 15915–15920. 10.1073/pnas.0905914106, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni G., Mand T. D., Metcalf W. W. (2018). Energy conservation via hydrogen cycling in the methanogenic archaeon Methanosarcina barkeri. MBio 9:e01256–18. 10.1128/mBio.01256-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leigh J. A., Albers S.-V., Atomi H., Allers T. (2011). Model organisms for genetics in the domain archaea: methanogens, halophiles, Thermococcales and Sulfolobales. FEMS Microbiol. Rev. 35, 577–608. 10.1111/j.1574-6976.2011.00265.x, PMID: [DOI] [PubMed] [Google Scholar]
- Lessner D. J., Li L., Li Q., Rejtar T., Andreev V. P., Reichlen M., et al. (2006). An unconventional pathway for reduction of CO2 to methane in CO-grown Methanosarcina acetivorans revealed by proteomics. Proc. Natl. Acad. Sci. U. S. A. 103, 17921–17926. 10.1073/pnas.0608833103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H. J., Chang J. L., Liu P. F., Fu L., Ding D. W., Lu Y. H. (2015). Direct interspecies electron transfer accelerates syntrophic oxidation of butyrate in paddy soil enrichments. Environ. Microbiol. 17, 1533–1547. 10.1111/1462-2920.12576, PMID: [DOI] [PubMed] [Google Scholar]
- Li Q. B., Li L. Y., Rejtar T., Lessner D. J., Karger B. L., Ferry J. G. (2006). Electron transport in the pathway of acetate conversion to methane in the marine archaeon Methanosarcina acetivorans. J. Bacteriol. 188, 702–710. 10.1128/JB.188.2.702-710.2006, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lie T. J., Costa K. C., Lupa B., Korpole S., Whitman W. B., Leigh J. A. (2012). Essential anaplerotic role for the energy-converting hydrogenase Eha in hydrogenotrophic methanogenesis. Proc. Natl. Acad. Sci. U. S. A. 109, 15473–15478. 10.1073/pnas.1208779109, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lienemann M., Deutzmann J. S., Milton R. D., Sahin M., Spormann A. M. (2018). Mediator-free enzymatic electrosynthesis of formate by the Methanococcus maripaludis heterodisulfide reductase supercomplex. Bioresour. Technol. 254, 278–283. 10.1016/j.biortech.2018.01.036, PMID: [DOI] [PubMed] [Google Scholar]
- Liu F. H., Rotaru A. E., Shrestha P. M., Malvankar N. S., Nevin K. P., Lovley D. R. (2012). Promoting direct interspecies electron transfer with activated carbon. Energy Environ. Sci. 5, 8982–8989. 10.1039/c2ee22459c [DOI] [Google Scholar]
- Liu X., Wang S. W., Xu A. M., Zhang L., Liu H. S., Ma L. Z. (2019). Biological synthesis of high-conductive pili in aerobic bacterium Pseudomonas aeruginosa. Appl. Microbiol. Biotechnol. 103, 1535–1544. 10.1007/s00253-018-9484-5, PMID: [DOI] [PubMed] [Google Scholar]
- Liu Y. C., Whitman W. B. (2008). Metabolic, phylogenetic, and ecological diversity of the methanogenic archaea. Ann N Y Acad. Sci. 1125, 171–189. 10.1196/annals.1419.019, PMID: [DOI] [PubMed] [Google Scholar]
- Lohner S. T., Deutzmann J. S., Logan B. E., Leigh J., Spormann A. M. (2014). Hydrogenase-independent uptake and metabolism of electrons by the archaeon Methanococcus maripaludis. Isme J. 8, 1673–1681. 10.1038/ismej.2014.82, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lojou E. (2011). Hydrogenases as catalysts for fuel cells: strategies for efficient immobilization at electrode interfaces. Electrochim. Acta 56, 10385–10397. 10.1016/j.electacta.2011.03.002 [DOI] [Google Scholar]
- Lovley D. R. (2017). Syntrophy goes electric: direct interspecies electron transfer. Annu. Rev. Microbiol. 71, 643–664. 10.1146/annurev-micro-030117-020420, PMID: [DOI] [PubMed] [Google Scholar]
- Maji S. K., Perrin M. H., Sawaya M. R., Jessberger S., Vadodaria K., Rissman R. A., et al. (2009). Functional amyloids as natural storage of peptide hormones in pituitary secretory granules. Science 325, 328–332. 10.1126/science.1173155, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malvankar N. S., Tuominen M. T., Lovley D. R. (2012). Lack of cytochrome involvement in long-range electron transport through conductive biofilms and nanowires of Geobacter sulfurreducens. Energy Environ. Sci. 5, 8651–8659. 10.1039/c2ee22330a [DOI] [Google Scholar]
- Mand T. D., Kulkarni G., Metcalf W. W. (2018). Genetic, biochemical, and molecular characterization of Methanosarcina barkeri mutants lacking three distinct classes of hydrogenase. J. Bacteriol. 200:e01256–18. 10.1128/JB.00342-18, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meuer J., Kuettner H. C., Zhang J. K., Hedderich R., Metcalf W. W. (2002). Genetic analysis of the archaeon Methanosarcina barkeri Fusaro reveals a central role for Ech hydrogenase and ferredoxin in methanogenesis and carbon fixation. Proc. Natl. Acad. Sci. U. S. A. 99, 5632–5637. 10.1073/pnas.072615499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Milton R. D., Ruth J. C., Deutzmann J. S., Spormann A. M. (2018). Methanococcus maripaludis employs three functional heterodisulfide reductase complexes for flavin-based electron bifurcation using hydrogen and formate. Biochemistry 57, 4848–4857. 10.1021/acs.biochem.8b00662, PMID: [DOI] [PubMed] [Google Scholar]
- Moran J. J., House C. H., Freeman K. H., Ferry J. G. (2005). Trace methane oxidation studied in several Euryarchaeota under diverse conditions. Archaea 1, 303–309. 10.1155/2005/650670, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran J. J., House C. H., Thomas B., Freeman K. H. (2007). Products of trace methane oxidation during nonmethyltrophic growth by Methanosarcina. J. Geophys. Res. Biogeo. 112:G02011. 10.1029/2006JG000268 [DOI] [Google Scholar]
- Morita M., Malvankar N. S., Franks A. E., Summers Z. M., Giloteaux L., Rotaru A. E., et al. (2011). Potential for direct interspecies electron transfer in methanogenic wastewater digester aggregates. MBio 2:e00159–11. 10.1128/mBio.00159-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ollivier B., Lombardo A., Garcia J. L. (1984). Isolation and characterization of a new thermophilic Methanosarcina strain (strain MP). Annales De Microbiologie 135, 187–198. 10.1016/s0769-2609(84)80026-5 [DOI] [PubMed] [Google Scholar]
- Parkin A., Goldet G., Cavazza C., Fontecilla-Camps J. C., Armstrong F. A. (2008). The difference a Se makes? Oxygen-tolerant hydrogen production by the NiFeSe -hydrogenase from Desulfomicrobium baculatum. J. Am. Chem. Soc. 130, 13410–13416. 10.1021/ja803657d, PMID: [DOI] [PubMed] [Google Scholar]
- Poweleit N., Ge P., Nguyen H. H., Loo R. R. O., Gunsalus R. P., Zhou Z. H. (2017). CryoEM structure of the Methanospirillum hungatei archaellum reveals structural features distinct from the bacterial flagellum and type IV pilus. Nat. Microbiol. 2:16222. 10.1038/nmicrobiol.2016.222, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash D., Chauhan S. S., Ferry J. G. (2019a). Life on the thermodynamic edge: respiratory growth of an acetotrophic methanogen. Sci. Adv. 5:eaaw9059. 10.1126/sciadv.aaw9059, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prakash D., Iyer P. R., Suharti S., Walters K. A., Santiago-Martinez M. G., Golbeck J. H., et al. (2019b). Structure and function of an unusual flavodoxin from the domain Archaea. Proc. Natl. Acad. Sci. U. S. A. 116, 25917–25922. 10.1073/pnas.1908578116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reda T., Plugge C. M., Abram N. J., Hirst J. (2008). Reversible interconversion of carbon dioxide and formate by an electroactive enzyme. Proc. Natl. Acad. Sci. U. S. A. 105, 10654–10658. 10.1073/pnas.0801290105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reguera G., Mccarthy K. D., Mehta T., Nicoll J. S., Tuominen M. T., Lovley D. R. (2005). Extracellular electron transfer via microbial nanowires. Nature 435, 1098–1101. 10.1038/nature03661, PMID: [DOI] [PubMed] [Google Scholar]
- Rotaru A. E., Shrestha P. M., Liu F., Markovaite B., Chen S., Nevin K. P., et al. (2014a). Direct interspecies electron transfer between Geobacter metallireducens and Methanosarcina barkeri. Appl. Environ. Microbiol. 80, 4599–4605. 10.1128/aem.00895-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rotaru A. E., Shrestha P. M., Liu F. H., Shrestha M., Shrestha D., Embree M., et al. (2014b). A new model for electron flow during anaerobic digestion: direct interspecies electron transfer to Methanosaeta for the reduction of carbon dioxide to methane. Energy Environ. Sci. 7, 408–415. 10.1039/c3ee42189a [DOI] [Google Scholar]
- Rowe A. R., Xu S., Gardel E., Bose A., Girguis P., Amend J. P., et al. (2019). Methane-linked mechanisms of electron uptake from cathodes by Methanosarcina barkeri. MBio 10:12. 10.1128/mBio.02448-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ruth J. C., Milton R. D., Gu W., Spormann A. M. (2020). Enhanced electrosynthetic hydrogen evolution by hydrogenases embedded in a redox-active hydrogel. Chem. Eur. J. 26, 7323–7329. 10.1002/chem.202000750, PMID: [DOI] [PubMed] [Google Scholar]
- Sakai K., Kitazumi Y., Shirai O., Takagi K., Kano K. (2016). Efficient bioelectrocatalytic CO2 reduction on gas-diffusion-type biocathode with tungsten-containing formate dehydrogenase. Electrochem. Commun. 73, 85–88. 10.1016/j.elecom.2016.11.008 [DOI] [Google Scholar]
- Schlegel K., Welte C., Deppenmeier U., Muller V. (2012). Electron transport during aceticlastic methanogenesis by Methanosarcina acetivorans involves a sodium-translocating Rnf complex. FEBS J. 279, 4444–4452. 10.1111/febs.12031, PMID: [DOI] [PubMed] [Google Scholar]
- Schopf S., Wanner G., Rachel R., Wirth R. (2008). An archaeal bi-species biofilm formed by Pyrococcus furiosus and Methanopyrus kandleri. Arch. Microbiol. 190, 371–377. 10.1007/s00203-008-0371-9, PMID: [DOI] [PubMed] [Google Scholar]
- Shi L., Dong H. L., Reguera G., Beyenal H., Lu A. H., Liu J., et al. (2016). Extracellular electron transfer mechanisms between microorganisms and minerals. Nat. Rev. Microbiol. 14, 651–662. 10.1038/nrmicro.2016.93, PMID: [DOI] [PubMed] [Google Scholar]
- Shimizu S., Upadhye R., Ishijima Y., Naganuma T. (2011). Methanosarcina horonobensis sp nov., a methanogenic archaeon isolated from a deep subsurface miocene formation. Int. J. Syst. Evol. Microbiol. 61, 2503–2507. 10.1099/ijs.0.028548-0, PMID: [DOI] [PubMed] [Google Scholar]
- Shrestha P. M., Rotaru A.-E., Summers Z. M., Shrestha M., Liu F., Lovley D. R. (2013). Transcriptomic and genetic analysis of direct interspecies electron transfer. Appl. Environ. Microbiol. 79, 2397–2404. 10.1128/AEM.03837-12, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soo V. W. C., Mcanulty M. J., Tripathi A., Zhu F., Zhang L., Hatzakis E., et al. (2016). Reversing methanogenesis to capture methane for liquid biofuel precursors. Microb. Cell Fact. 15:11. 10.1186/s12934-015-0397-z, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sowers K. R., Baron S. F., Ferry J. G. (1984). Methanosarcina acetivorans sp. nov., an acetotrophic methane-producing bacterium isolated from marine sediments. Appl. Environ. Microbiol. 47, 971–978. 10.1128/AEM.47.5.971-978.1984, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stams A. J. M., Plugge C. M. (2009). Electron transfer in syntrophic communities of anaerobic bacteria and archaea. Nat. Rev. Microbiol. 7, 568–577. 10.1038/nrmicro2166, PMID: [DOI] [PubMed] [Google Scholar]
- Steidl R. J., Lampa-Pastirk S., Reguera G. (2016). Mechanistic stratification in electroactive biofilms of Geobacter sulfurreducens mediated by pilus nanowires. Nat. Commun. 7:12217. 10.1038/ncomms12217, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Summers Z. M., Fogarty H. E., Leang C., Franks A. E., Malvankar N. S., Lovley D. R. (2010). Direct exchange of electrons within aggregates of an evolved syntrophic coculture of anaerobic bacteria. Science 330, 1413–1415. 10.1126/science.1196526, PMID: [DOI] [PubMed] [Google Scholar]
- Sure S., Ackland M. L., Torriero A. a. J., Adholeya A., Kochar M. (2016). Microbial nanowires: an electrifying tale. Microbiol-Sgm 162, 2017–2028. 10.1099/mic.0.000382 [DOI] [PubMed] [Google Scholar]
- Thauer R. K. (1998). Biochemistry of methanogenesis: a tribute to Marjory Stephenson. Microbiol. U. K. 144, 2377–2406. 10.1099/00221287-144-9-2377 [DOI] [PubMed] [Google Scholar]
- Thauer R. K., Kaster A.-K., Seedorf H., Buckel W., Hedderich R. (2008). Methanogenic archaea: ecologically relevant differences in energy conservation. Nat. Rev. Microbiol. 6, 579–591. 10.1038/nrmicro1931, PMID: [DOI] [PubMed] [Google Scholar]
- Thomas N. A., Bardy S. L., Jarrell K. F. (2001). The archaeal flagellum: a different kind of prokaryotic motility structure. FEMS Microbiol. Rev. 25, 147–174. 10.1111/j.1574-6976.2001.tb00575.x, PMID: [DOI] [PubMed] [Google Scholar]
- Viles J. H. (2012). Metal ions and amyloid fiber formation in neurodegenerative diseases. Copper, zinc and iron in Alzheimer’s, Parkinson’s and prion diseases. Coord. Chem. Rev. 256, 2271–2284. 10.1016/j.ccr.2012.05.003 [DOI] [Google Scholar]
- Walker D. J. F., Martz E., Holmes D. E., Zhou Z. M., Nonnenmann S. S., Lovley D. R. (2019). The archaellum of Methanospirillum hungatei is electrically conductive. MBio 10:e00579–19. 10.1128/mBio.00579-19, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Tomb J.-F., Ferry J. G. (2011). Electron transport in acetate-grown Methanosarcina acetivorans. BMC Microbiol. 11:165. 10.1186/1471-2180-11-165, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welander P. V., Metcalf W. W. (2005). Loss of the mtr operon in Methanosarcina blocks growth on methanol, but not methanogenesis, and reveals an unknown methanogenic pathway. Proc. Natl. Acad. Sci. U. S. A. 102, 10664–10669. 10.1073/pnas.0502623102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte C., Deppenmeier U. (2011). Membrane-bound electron transport in Methanosaeta thermophila. J. Bacteriol. 193, 2868–2870. 10.1128/JB.00162-11, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Welte C., Deppenmeier U. (2014). Bioenergetics and anaerobic respiratory chains of aceticlastic methanogens. BBA-Bioenergetics 1837, 1130–1147. 10.1016/j.bbabio.2013.12.002, PMID: [DOI] [PubMed] [Google Scholar]
- Xia X. X., Zhang J. C., Song T. Z., Lu Y. H. (2019). Stimulation of Smithella-dominating propionate oxidation in a sediment enrichment by magnetite and carbon nanotubes. Environ. Microbiol. Rep. 11, 236–248. 10.1111/1758-2229.12737, PMID: [DOI] [PubMed] [Google Scholar]
- Yan Z., Joshi P., Gorski C. A., Ferry J. G. (2018). A biochemical framework for anaerobic oxidation of methane driven by Fe(III)-dependent respiration. Nat. Commun. 91642. 10.1038/s41467-018-04097-9, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yates M. D., Siegert M., Logan B. E. (2014). Hydrogen evolution catalyzed by viable and non-viable cells on biocathodes. Int J Hydrogen Energ. 39, 16841–16851. 10.1016/j.ijhydene.2014.08.015 [DOI] [Google Scholar]
- Yee M. O., Rotaru A.-E. (2020). Extracellular electron uptake in Methanosarcinales is independent of multiheme c-type cytochromes. Sci. Rep. U. K. 10:372. 10.1038/s41598-019-57206-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yee M. O., Snoeyenbos-West O. L., Thamdrup B., Ottosen L. D. M., Rotaru A.-E. (2019). Extracellular electron uptake by two Methanosarcina Species. Front. Energy Res. 7:29. 10.3389/fenrg.2019.00029 [DOI] [Google Scholar]
- Yuan M., Sahin S., Cai R., Abdellaoui S., Hickey D. P., Minteer S. D., et al. (2018). Creating a low-potential redox polymer for efficient electroenzymatic CO2 reduction. Angew. Chem. Int. Edit. 57, 6582–6586. 10.1002/anie.201803397 [DOI] [PubMed] [Google Scholar]
- Zhang J. C., Lu Y. H. (2016). Conductive Fe3O4 nanoparticles accelerate syntrophic methane production from butyrate oxidation in two different lake sediments. Front. Microbiol. 7:1316. 10.3389/fmicb.2016.01316, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S. L., Liu F. H., Wang B. C., Zhang Y. C., Lovley D. R. (2020). Methanobacterium capable of direct interspecies electron transfer. Environ. Sci. Technol. 54, 15347–15354. 10.1021/acs.est.0c05525, PMID: [DOI] [PubMed] [Google Scholar]
- Zhu J. X., Zheng H. J., Ai G. M., Zhang G. S., Liu D., Liu X. L., et al. (2012). The Genome characteristics and predicted function of methyl-group oxidation pathway in the obligate aceticlastic methanogens, Methanosaeta spp. PLoS One 7:e36756. 10.1371/journal.pone.0036756, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]