Abstract
Background
Dysregulation of circular RNAs (circRNAs) is associated with bladder cancer progression. Nevertheless, the mechanisms of circRNA centrosomal protein 128 (circCEP128) underlying bladder cancer progression remain poorly understood.
Methods
The levels of circCEP128, microRNA-515-5p (miR-515-5p) and syndecan-1 (SDC1) were determined via reverse transcription-quantitative polymerase chain reaction or Western blot. The effects of circCEP128, miR-515-5p and SDC1 on bladder cancer progression were investigated via MTT and colony formation assays, flow cytometry and transwell analysis and subcutaneous xenograft experiments. The interactions between miR-515-5p and circCEP128 or SDC1 were examined through bioinformatics prediction and luciferase reporter assay.
Results
circCEP128 and SDC1 were highly expressed and miR-515-5p was low expressed in bladder cancer tissues and cells. circCEP128 knockdown hindered cell proliferation, migration and invasion and promoted cell apoptosis in bladder cancer. circCEP128 loss increased miR-515-5p expression through direct interaction in bladder cancer cells. MiR-515-5p depletion mitigated the influences of circCEP128 knockdown on bladder cancer cell phenotypes. SDC1 was a direct target of miR-515-5p. circCEP128 positively regulated SDC1 expression via miR-515-5p. MiR-515-5p restrained the malignant progression of bladder cancer cells by decreasing SDC1 expression. circCEP128 knockdown hindered the growth of bladder cancer xenograft tumors by up-regulating miR-515-5p and down-regulating SDC1.
Conclusion
circCEP128 knockdown hampered the tumorigenesis and progression of bladder cancer by regulating miR-515-5p/SDC1 axis in vitro and in vivo, deepening our understanding on the molecular mechanisms of circCEP128 in bladder cancer.
Keywords: bladder cancer, circCEP128, microRNA-515-5p, SDC1
Introduction
Bladder cancer is a urinary malignancy with higher morbidity in men than women.1 Although great advances have been made in the management of bladder cancer, the clinical outcomes of patients with advanced disease remain poor.2 Thus, there is an urgent need to deeply investigate the pathogenesis of bladder cancer and identify novel molecular therapeutic targets for bladder cancer.
Circular RNAs (circRNAs) are a type of covalently closed circular RNA molecules that are formed by the back-splicing process.3 Over the past decades, accumulating circRNAs have been found to be implicated in the carcinogenesis and progression of bladder cancer.4 For example, circRNA forkhead box O3 (circFOXO3) suppressed cell proliferation, migration and invasion via controlling miR-9-5p and transforming growth factor-beta receptor II (TGFBR2) expression in bladder cancer.5 CircBC048201 knockdown hampered bladder cancer cell proliferation, migration and invasion via modulating the expression of miR-1184 and integrin alpha 3 (ITGA3).6 CircRNA CEP128 (circCEP128, hsa_circ_0032821), a circRNA derived from CEP128 gene, has been identified as an oncogenic factor in some malignancies such as glioma and gastric cancer.7,8 Moreover, circCEP128 was found to be highly expressed in bladder cancer tissues, and circCEP128 knockdown hindered the tumorigenesis and progression of bladder cancer in vitro and in vivo.9,10 In this study, other molecular targets or regulatory pathways involved in mediating circCEP128 functions were explored in bladder cancer.
MicroRNAs (miRNAs), a class of short noncoding RNAs, have been documented as crucial players in the tumorigenesis and progression of bladder cancer.11 Previous studies demonstrated that miR-515-5p could hamper cell proliferation and metastasis in bladder cancer.12,13 However, it is unknown whether miR-515-5p is involved in mediating the oncogenic effects of circCEP128 in bladder cancer. Syndecan-1 (SDC1) also have been found to be closely linked with tumorigenesis and progression of malignancies by regulating tumor cell phenotypes such as proliferation, apoptosis, invasion and migration.14 Furthermore, previous studies showed that the aberrant expression of SDC1 was associated with the prognosis of bladder cancer patients.15,16 Bioinformatics prediction analysis suggested that miR-515-5p might interact with circCEP128 or SDC1. However, the interaction between miR-515-5p and circCEP128 or SDC1 has not been previously reported.
In this work, the roles of circCEP128 in bladder cancer progression were further examined by in vitro cell experiments and in vivo xenograft experiments. Additionally, we further investigated whether circCEP128 could exert its functions by regulating miR-515-5p/SDC1 axis in bladder cancer.
Materials and Methods
Bioinformatics Analysis
The genomic location of circCEP128 was analyzed via circBase database (http://www.circbase.org/). CircCEP128 was visualized using the circView software. The differential expression profiles of circRNAs in bladder cancer tissues versus normal tissues were identified according to the GES92675 dataset (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE92675). MiRNAs with the possibility to interact with circCEP128 were predicted by circBank (http://www.circbank.cn/) and circinteractome (https://circinteractome.nia.nih.gov/) and RNAhybrid 2.2 (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid). The binding site of miR-515-5p and SDC1 was analyzed via starBase (http://starbase.sysu.edu.cn/).
Patient Tissues
The tumor tissues and adjacent normal tissues were harvested from 39 bladder cancer patients via surgery at Henan Provincial People’s Hospital. Bladder cancer patients suffered from any other treatment before surgery were excluded. The cancer and non-cancer regions were identified by two experienced pathologists and then stored in liquid nitrogen. The written informed consents were obtained from every subject. This research was approved by the Ethics Committee of Henan Provincial People’s Hospital. Our project was conducted in accordance with the Declaration of Helsinki.
Cell Culture and Transfection
Human bladder cancer cells (T24 and 5637), and normal urothelial cells (SV-HUC-1) were purchased from Procell Life Science & Technology Co., Ltd. (Wuhan, China), and maintained in Roswell Park Memorial Institute (RPMI)-1640 medium (Procell) supplemented with 10% fetal bovine serum (FBS) (Solarbio, Beijing, China) and 1% antibiotics (Procell) at 37°C and 5% CO2.
The full-length sequence of SDC1 coding region was constructed into pcDNA3.1 vector (Thermo Fisher Scientific, Waltham, MA, USA) to generate pcDNA3.1-SDC1 overexpression plasmid (SDC1) with pcDNA3.1 empty vector as the negative control. siRNA targeting circCEP128 (si-circCEP128), siRNA negative control (si-NC), miR-515-5p mimic, negative control of mimic (miR-NC), miR-515-5p inhibitor (anti-miR-515-5p), and negative control of inhibitor (anti-miR-NC) were obtained from Genomeditech (Shanghai, China). The oligonucleotide sequences are displayed in Table 1. T24 and 5637 cells at 60–70% confluence were transfected with 1 μg vectors or 30 nM oligos using Lipofectamine 2000 (Solarbio Technology Co., ltd., Beijing, China).
Table 1.
The Sequences for Transfection in This Study
| Name | Sequence (5ʹ-3ʹ) |
|---|---|
| si-circCEP128 | AUGUUUUUCCUCUCUUCUCA |
| si-NC | AAGACAUUGUGUGUCCGCCTT |
| miR-515-5p mimic | UUCUCCAAAAGAAAGCACUUUCUG |
| miR-NC | CGAUCGCAUCAGCAUCGAUUGC |
| anti-miR-515-5p | CAGAAAGUGCUUUCUUUUGGAGAA |
| anti-miR-NC | CUAACGCAUGCACAGUCGUACG |
Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
RNA was extracted using Trizol (Solarbio) according to the instructions of manufacturer. To purify circRNA or analyze circRNA stability, extracted RNA was further treated with 3U/μg RNase R for 20 min. The complementary DNA (cDNA) was generated from 800 ng RNA using the GoScript reverse transcription kit (Promega, Madison, WI, USA). Next, cDNA was amplified and quantified using SYBR (Thermo Fisher Scientific) and quantitative primer pairs. Real Time-quantitative PCR was performed using the following procedures: 95°C for 5 min, and 40 cycles of 95°C for 10 s, 58°C for 20 s and 72°C for 1 min. The specific quantitative primers synthesized by Sangon (Shanghai, China) were displayed in Table 2. U6 or GAPDH was regarded as the negative control. Relative RNA abundance was analyzed according to 2−ΔΔCt method.17
Table 2.
The Primer Sequences for RT-qPCR in This Study
| Name | Sequence (5ʹ-3ʹ) | |
|---|---|---|
| Forward | Reverse | |
| miR-515-5p | GCCGAGTTCTCCAAAAGAAAGC | CAGTGCGTGTCGTGGAGT |
| U6 | GCTCGCTTCGGCAGCACA | GAGGTATTCGCACCAGAGGA |
| circCEP128 | GCAGGAGCAGCTTCTGGATGA | ATCTGGCCTCTTCCATCCTCCT |
| linearCEP128 | GATCTCTGCAGGACCGTGTA | AAGCGACTGTGATCCTGCC |
| SDC1 | CGAGCTGAAAGGCCGGGAA | CAGGGGTTGAGGTCTCATGG |
| GAPDH | GAAAGCCTGCCGGTGACTAA | TTCCCGTTCTCAGCCTTGAC |
Western Blot
Protein was extracted from bladder cancer cells and xenograft tumors with radio-immunoprecipitation assay buffer (Solarbio), and then quantified using the bicinchoninic acid assay kit (Beyotime, Shanghai, China). Subsequently, 20 μg of protein was separated via 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis and transferred on nitrocellulose membranes (Solarbio). After blocked with 3% bovine serum albumin (Solarbio), the membranes were incubated with primary antibodies against SDC1 (ab60199, 1:3000 dilution, Abcam, Cambridge, UK), and β-actin (ab8227, 1:2000 dilution, Abcam) overnight at 4°C, and secondary antibody labeled with horseradish peroxidase (ab205718, 1:8000 dilution, Abcam) for 2 h at room temperature. Next, the membranes were developed using the BeyoECL Plus kit (Beyotime). Relative protein abundance was determined via Image J software (NIH, Bethesda, MD, USA). β-actin served as the house-keeping gene to normalize the expression of SDC1.
MTT
Cell proliferation activity was determined via MTT assay. T24 and 5637 cells (1 × 104) were placed into 96-well plates. After 0, 24, 48 or 72 h of culture, 10 μL of 0.5% MTT solution (Solarbio) was added into each well and incubated for 4 h. Next, formazan complex was dissolved using dimethyl sulfoxide (Sigma-Aldrich, St. Louis, MO, USA), and the optical density (OD) values were detected at 570 nm with a microplate reader (Allsheng, Hangzhou, China).
Colony Formation Assay
Cell proliferative ability was also assessed by colony formation assay. Briefly, transfected cells at the logarithmic growth phase (1000 cells/dish) were seeded into 10 cm culture dishes. After 2 weeks of culture, cells were fixed with methanol for 15 min and stained with 0.1% crystal violet (Solarbio) for 20 min. The number of colonies containing more than 20 cells was counted under a microscope.
Flow Cytometry
Cell apoptotic rate was examined via flow cytometry (Agilent, Santa Clara, CA, USA) using an Annexin V-fluorescein isothiocyanate (FITC) apoptosis detection kit (Thermo Fisher Scientific) following the manufacturer’s instructions. Briefly, 2 × 105 T24 and 5637 cells were inoculated into 12-well plates. After 72 h of culture, cells were harvested and resuspended in Annexin V binding buffer. Next, cells were dyed with Annexin V-FITC and propidium iodide (PI) for 10 min. Cell apoptotic patterns were analyzed using a flow cytometry. Cell apoptotic rate was represented as the percentage of cells at early (Annexin V-FITC+ and PI−) and late (Annexin V-FITC+ and PI+) apoptotic phases.
Transwell Assay
Cell migratory and invasive abilities were tested with 24-well transwell chambers (Corning, New York, NY, USA). For migration analysis, 2 × 105 T24 and 5637 cells suspended in RPMI-1640 medium without serum were placed into the upper chambers, and 600 μL of medium containing 10% FBS was added into the low chambers. After 24 h of incubation, migrated cells were dyed using 0.5% crystal violet (Solarbio), imaged with a microscope (Olympus, Tokyo, Japan), and counted via Image J software. For invasion analysis, 5 × 105 T24 and 5637 cells were seeded into the upper chambers precoated with Matrigel (Solarbio). The other procedures in the invasion experiments were the same as those of migration analysis.
Dual-Luciferase Reporter Analysis
The wild-type (WT) sequence of circCEP128 or SDC1 3ʹ UTR with miR-515-5p complementary site was cloned into pGL3-Basic vector (Promega) to generate the luciferase reporter vectors (circCEP128 WT or SDC1 3ʹUTR WT). The mutant (MUT) vectors (circCEP128 MUT and SDC1 3ʹUTR MUT) were also constructed via mutating corresponding binding site of miR-515-5p. T24 and 5637 cells were transfected with 1 μg of vector and 30 nM of miR-515-5p mimic or miR-NC. The luciferase activities were examined via a dual-luciferase assay kit (Solarbio) with Renilla luciferase activity as the normalization control.
Tumor Xenograft Analysis
The BALB/c nude mice (male, 4-week-old) were obtained from Charles River Laboratories (Beijing, China). Mice were randomly divided into sh-NC or sh-circCEP128 group (n=5 in each group). T24 cells cannot generate the subcutaneous xenograft tumors in nude mice according to the information from Procell. Therefore, 5637 cells were used to establish the xenograft model. The sh-NC or sh-circCEP128 lentiviral particles carrying circCEP128 targeting sequences or control sequences were constructed via Ribobio, and transduced into 5637 cells. The stably transduced cells were screened out via 5 μg/mL of puromycin (Thermo Fisher Scientific). After resuspended in phosphate buffer saline (PBS), 5 × 106 5637 cells stably transduced with sh-NC or sh-circCEP128 were subcutaneously injected into mice in the corresponding group. Tumor volume was determined every 7 days and calculated using the formula: volume = (length × width2)/2. After 35 days, the mice were euthanized and tumors were dissected out and weighed. The expression levels of circCEP128, miR-515-5p and SDC1 in tumor tissues were determined by RT-qPCR and Western blot assays. The animal experiments were conducted with the approval of the Animal Care and Use Committee of Henan Provincial People’s Hospital and the Guide for the Care and Use of Laboratory Animals (8th edition) by the National Research Council of the National Academies (The National Academies Press).
Statistical Analysis
The results were shown as mean ± standard deviation (SD). The in vitro experiments were conducted 3 times. Data were analyzed using the SPSS 20.0 software (SPSS, Chicago, IL, USA). Difference analysis was performed using Student’s t-test or one-way analysis of variance (ANOVA) followed by Tukey post hoc test. The difference was considered as statistically significant when P values were less than 0.05.
Results
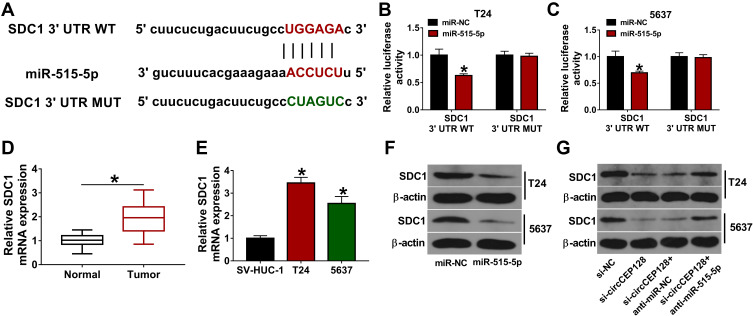
circCEP128 is Highly Expressed in Bladder Cancer Tissues and Cell Lines
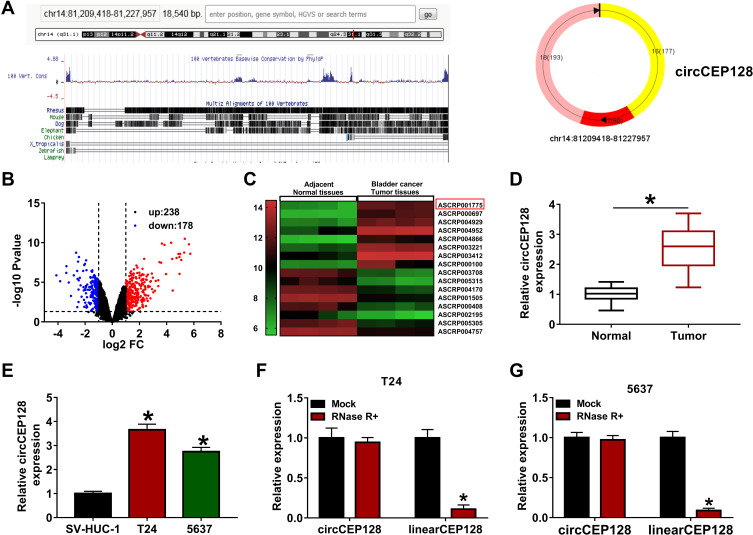
The circBase and circView showed that circCEP128 (hsa_circ_0032821, genome position: chr14: 81,209,418–81,227,957) was derived from the exon 16–18 of CEP128 gene (Figure 1A). To examine whether circCEP128 was aberrantly expressed in bladder cancer, circRNA expression profiles of GSE92675 dataset were downloaded from Gene Expression Omnibus (GEO) database. Differential expression analysis for GSE92675 dataset revealed that 416 circRNAs (238 up-regulated and 178 down-regulated) were differentially expressed in bladder cancer tissues (n=4) compared to adjacent normal tissues (n=4) (Figure 1B). Moreover, circCEP128 (ASCRP001775) expression level was markedly increased in bladder cancer tumor tissues (n=4) relative to adjacent normal tissues (n=4) (Figure 1C). To further validate the overexpression of circCEP128 in bladder cancer, 39 pairs of bladder cancer tumor tissues and adjacent normal tissues were collected from our hospital and then circCEP128 level in these tissues was measured by RT-qPCR assay. As expected, circCEP128 expression was higher in bladder cancer tumor tissues (n=39) compared to their normal counterparts (n=39) (Figure 1D). In addition, circCEP128 was highly expressed in bladder cancer cell lines (T24 and 5637) than that in an immortalized human bladder epithelial cell line (SV-HUC-1) (Figure 1E). Next, total RNA isolated from T24 and 5637 cells was treated with or without RNase R to further validate the circular characteristic of circCEP128. As presented in Figure 1F and G, circCEP128 was resistant to RNase R digestion, suggesting that circCEP128 was indeed a circRNA. Considering the overexpression of circRNA circCEP128 in bladder cancer tissues and cells, we supposed that circCEP128 might play vital roles in the development and progression of bladder cancer.
Figure 1.
circCEP128 is highly expressed in bladder cancer tissues and cell lines. (A) The schematic diagram of circCEP128 genomic location. (B and C) The volcano plot and heat map of differentially expressed circRNAs in bladder cancer tissues and adjacent normal tissues based on the dataset of GSE92675. (D) CircCEP128 level in bladder cancer tissues (n=39) and adjacent normal tissues (n=39) was examined via RT-qPCR assay. (E) CircCEP128 level was measured through RT-qPCR assay in T24, 5637 and SV-HUC-1 cells. (F and G) CircCEP128 and linearCEP128 abundances were detected via RT-qPCR assay in T24 and 5637 cells treated with or without RNase R. *P<0.05.
circCEP128 Knockdown Suppresses Bladder Cancer Cell Proliferation, Migration, Invasion and Colony Formation, and Induces Bladder Cancer Cell Apoptosis
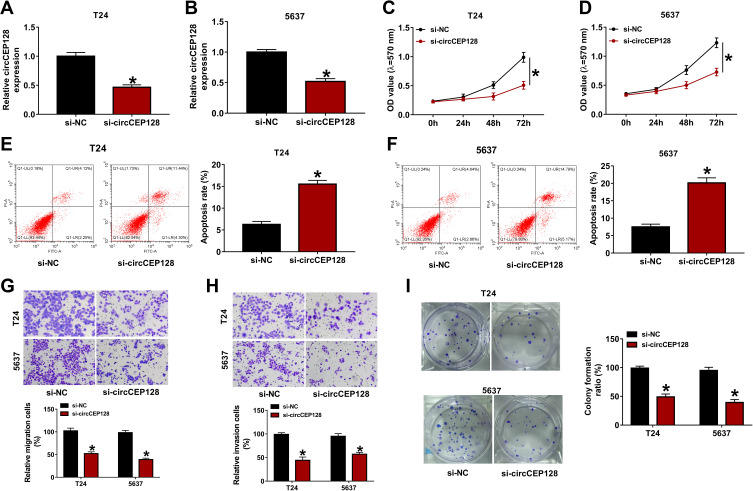
To study the functions of circCEP128 in bladder cancer cell progression, si-circCEP128 and si-NC were synthesized. Knockdown efficiency analysis revealed that the transfection of si-circCEP128 effectively reduced circCEP128 abundance in T24 and 5637 cells (Figure 2A and B). MTT assays displayed that circCEP128 knockdown markedly weakened the proliferative ability of T24 and 5637 cells (Figure 2C and D). Also, flow cytometry results showed that circCEP128 loss led to the notable increase of cell apoptotic rate in T24 and 5637 cells (Figure 2E and F). Moreover, Transwell and colony formation assays showed that circCEP128 depletion noticeably curbed cell migration, invasion and colony formation in T24 and 5637 cells (Figure 2G–I). These data suggested that circCEP128 knockdown hampered bladder cancer progression in vitro.
Figure 2.
circCEP128 knockdown suppresses cell proliferation, migration and invasion and induces cell apoptosis in bladder cancer. (A and B) circCEP128 abundance was measured via RT-qPCR in cells transfected with si-NC or si-circCEP128. (C and D) Cell proliferation activity was detected via MTT assays in cells transfected with si-NC or si-circCEP128. (E and F) Cell apoptotic rate was measured via flow cytometry in cells after transfection with si-NC or si-circCEP128. (G and H) Cell migratory and invasive abilities were determined using transwell assay in cells after transfection with si-NC or si-circCEP128. (I) Colony formation abilities were determined using transwell assay in cells after transfection with si-NC or si-circCEP128. *P<0.05.
miR-515-5p is a Target of circCEP128
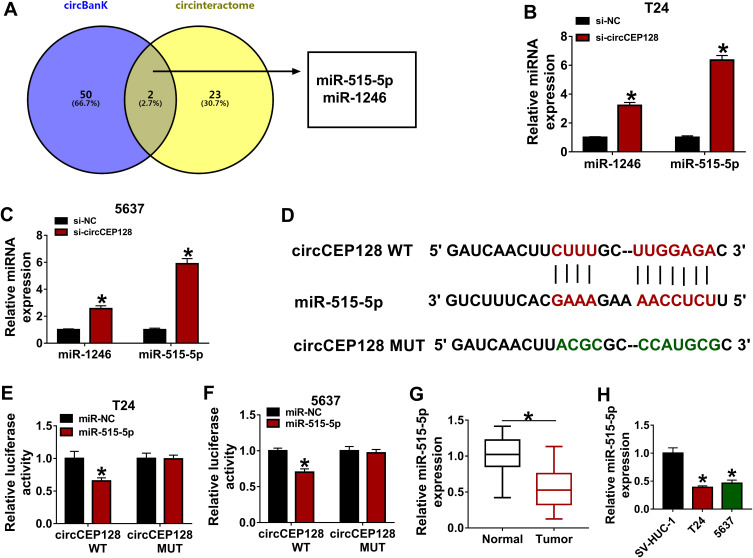
Next, miRNAs that had the possibility to interact with circCEP128 were predicted by circinteractome, circBank databases. Combined with the prediction outcomes of circinteractome, circBank databases, two common miRNAs (miR-515-5p and miR-1246) were screened out by Venn analysis (Figure 3A). Subsequent RNAhybrid analysis also verified the target interaction between circCEP128 and miR-515-5p. Moreover, the effect of circCEP128 loss on the expression of miR-515-5p and miR-1246 was examined in T24 and 5637 cells. As displayed in Figure 3B and C, circCEP128 knockdown markedly increased miR-515-5p and miR-1246 levels in T24 and 5637 cells. Considering the stronger influence of circCEP128 loss on miR-515-5p expression in bladder cancer cells, the interaction between circCEP128 and miR-515-5p was further investigated by luciferase reporter assay. The binding sites between circCEP128 and miR-515-5p were also validated by RNAhybrid website (Figure 3D). Luciferase reporter assay showed that the introduction of miR-515-5p mimic conspicuously reduced the luciferase activity of circCEP128 WT reporter, but did not alter the luciferase activity of circCEP128 MUT reporter with mutant miR-515-5p binding sites in T24 and 5637 cells (Figure 3D–F), suggesting that circCEP128 could bind with miR-515-5p by putative complementary sites. Next, RT-qPCR assay disclosed that miR-515-5p abundance was markedly decreased in bladder cancer tumor tissues (n=39) and cell lines relative to their counterparts (Figure 3G and H). These findings suggested that miR-515-5p was a target of circCEP128 in bladder cancer cells.
Figure 3.
miR-525-5p is a target of circCEP128. (A) MiRNAs that had the possibility to interact with circCEP128 were predicted by circinteractome and circBank database. Common miRNAs in the prediction outcomes of circinteractome and circBank were screened out by Venn analysis. (B and C) miR-1246 and miR-515-5p abundances were examined via RT-qPCR in cells transfected with si-NC or si-circCEP128. (D) Putative binding sequences of circCEP128 and miR-515-5p and mutant sequences in circCEP128 MUT reporter. (E and F) Luciferase activities were detected in T24 and 5637 cells transfected with miR-NC or miR-515-5p mimic and circCEP128 WT or circCEP128 MUT reporter. (G) MiR-515-5p level was examined via RT-qPCR in bladder cancer tumor tissues (n=39) and adjacent normal tissues (n=39). (H) miR-515-5p abundance was examined via RT-qPCR in T24, 5637 and SV-HUC-1 cells. *P<0.05.
The Introduction of miR-515-5p Inhibitor Mitigates the Effects of circCEP128 Knockdown on Bladder Cancer Cell Proliferation, Migration, Invasion and Apoptosis
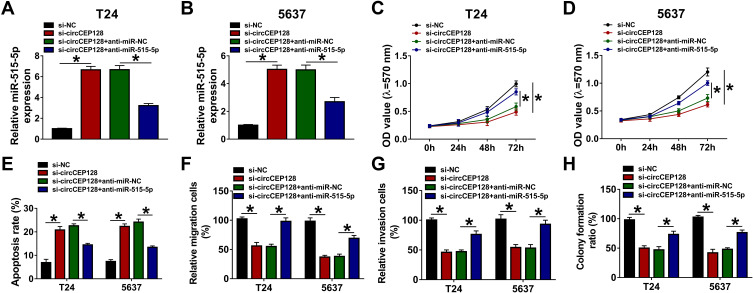
To analyze whether miR-515-5p was involved in mediating the functions of circCEP128 in bladder cancer, T24 and 5637 cells were transfected with si-NC, si-circCEP128, si-circCEP128+anti-miR-NC or si-circCEP128+anti-miR-515-5p. RT-qPCR assay confirmed that the introduction of anti-miR-515-5p inhibited the increase of miR-515-5p expression induced by circCEP128 depletion in T24 and 5637 cells (Figure 4A and B). Subsequent experiments further demonstrated that miR-515-5p down-regulation weakened si-circCEP128-mediated anti-proliferative, pro-apoptotic, anti-migratory, anti-invasive, and anti-colony formation effects in T24 and 5637 cells (Figure 4C–H). These results showed that circCEP128 knockdown inhibited bladder cancer cell progression through up-regulating miR-515-5p.
Figure 4.
The introduction of miR-515-5p inhibitor weakens the influences of circCEP128 knockdown on cell proliferation, migration, invasion and apoptosis in bladder cancer. T24 and 5637 cells were transfected with si-NC, si-circCEP128, si-circCEP128 + anti-miR-NC or si-circCEP128 + anti-miR-515-5p, followed by the measurement of miR-515-5p expression (A and B), cell proliferative potential (C and D), apoptotic rate (E), migratory ability (F), invasive capacity (G), and colony formation (H). *P<0.05.
circCEP128 Promotes SDC1 Expression by Acting as a Molecular Sponge of miR-515-5p in Bladder Cancer Cells
Next, potential targets of miR-515-5p were searched by starBase database. Among candidate targets, SDC1 was selected for further investigations due to its close link with the pathogenesis of cancers including bladder cancer. Subsequent luciferase reporter assay demonstrated that miR-515-5p overexpression markedly reduced the luciferase activity of SDC1 3ʹUTR WT reporter, but did not influence the luciferase activity of SDC1 3ʹUTR MUT reporter carrying mutant miR-515-5p complementary sites in T24 and 5637 cells (Figure 5A–C). Moreover, higher SDC1 expression was observed in bladder cancer tumor tissues (n=39) and cell lines than that in their counterparts (Figure 5D and E). Additionally, the ectopic expression of miR-515-5p triggered the noticeable down-regulation of SDC1 protein level in T24 and 5637 cells (Figure 5F). To further examine whether SDC1 was a downstream mediator of circCEP128/miR-515-5p axis in bladder cancer, the effects of circCEP128 depletion alone or in combination with miR-515-5p loss on SDC1 expression were measured by Western blot assay in T24 and 5637 cells. The outcomes showed that circCEP128 knockdown triggered the notable down-regulation of SDC1 protein level, while the introduction of miR-515-5p inhibitor markedly weakened the inhibitory effect of circCEP128 depletion on SDC1 expression in T24 and 5637 cells (Figure 5G). These results indicated that circCEP128 could regulate SDC1 expression indirectly via miR-515-5p in bladder cancer cells.
Figure 5.
circCEP128 positively regulates SDC1 expression by miR-525-5p. (A) The binding site of miR-515-5p and SDC1 3ʹUTR was predicted by starBase database. Mutant site in SDC1 3ʹUTR MUT reporter. (B and C) Luciferase activities were examined in T24 and 5637 cells transfected with miR-NC or miR-515-5p mimic and SDC1 3ʹUTR WT or SDC1 3ʹUTR MUT. (D) SDC1 abundance was examined via RT-qPCR in bladder cancer tumor tissues (n=39) and adjacent normal tissues (n=39). (E) SDC1 mRNA expression level was examined via RT-qPCR in T24, 5637 and SV-HUC-1 cells. (F) SDC1 expression was determined via Western blot in cells transfected with miR-NC or miR-515-5p mimic. (G) SDC1 protein level was measured by Western blot in cells transfected with si-NC, si-circCEP128, si-circCEP128 + anti-miR-NC or si-circCEP128 + anti-miR-515-5p. *P<0.05.
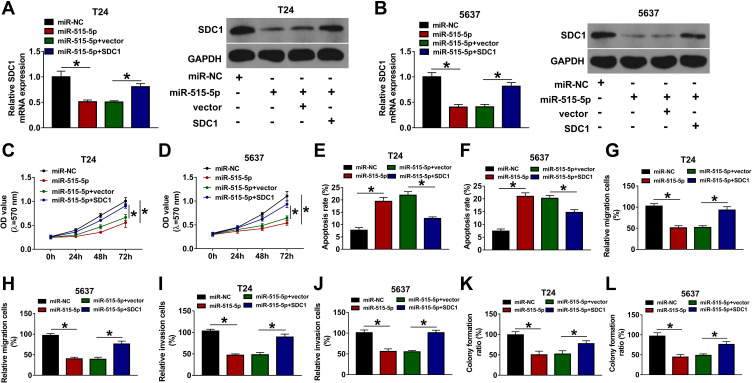
miR-515-5p Overexpression Weakens Cell Proliferative, Migratory and Invasive Capacities and Elevated Cell Apoptotic Rate by Targeting SDC1 in Bladder Cancer
Next, we further investigated whether miR-515-5p could regulate the phenotypes of bladder cancer cells by targeting SDC1. RT-qPCR and Western blot assays validated that the introduction of miR-515-5p mimic led to the noticeable reduction of SDC1 mRNA and protein levels and SDC1 overexpression partially rescued the inhibitory effect of miR-515-5p on SDC1 expression in T24 and 5637 cells (Figure 6A and B). The functional analyses showed that miR-515-5p overexpression hindered cell proliferation (Figure 6C and D), migration (Figure 6G and H), invasion (Figure 6I and J), colony formation (Figure 6K and L) and facilitated cell apoptosis (Figure 6E and F) in T24 and 5637 cells. Restoration experiments demonstrated that SDC1 up-regulation notably lessened the detrimental effects of miR-515-5p on cell proliferation, migration, invasion, colony formation and curbed the increase of cell apoptotic rate induced by miR-515-5p in T24 and 5637 cells (Figure 6C–L). These results suggested that miR-515-5p hindered bladder cancer progression via targeting SDC1 in vitro.
Figure 6.
miR-515-5p regulates cell proliferation, migration, invasion and apoptosis via decreasing SDC1 level in bladder cancer. SDC1 expression (A and B), cell proliferation (C and D), apoptosis (E and F), migration (G and H), invasion (I and J), and colony formation (K and L) were detected in T24 and 5637 cells transfected with miR-NC, miR-515-5p mimic, miR-515-5p mimic + vector or miR-515-5p mimic + SDC1 overexpression vector. *P<0.05.
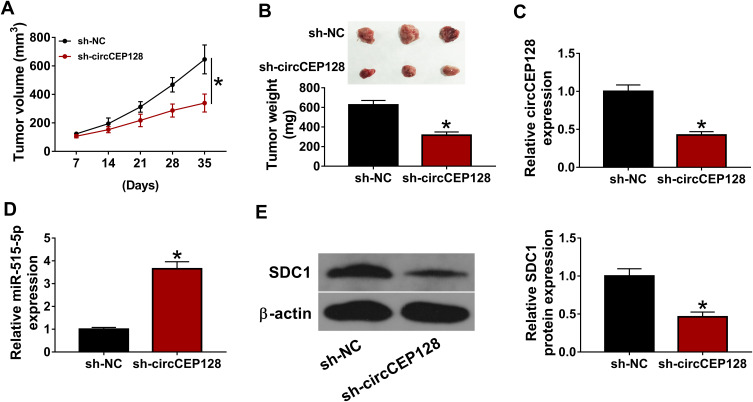
circCEP128 Knockdown Hampers the Growth of Bladder Cancer Xenograft Tumors by Regulating miR-515-5p/SDC1 Axis
Next, in vivo experiments demonstrated that circCEP128 knockdown markedly impeded the growth of bladder cancer xenograft tumors, as evidenced by the noticeable reduction of tumor volume and weight in sh-circCEP128 group versus sh-NC group (Figure 7A and B). Moreover, RT-qPCR and Western blot assays further verified that CEP128 and SDC1 expression levels were markedly decreased and miR-515-5p level was notably increased in xenograft tumors of sh-circCEP128 group relative to sh-NC group (Figure 7C–E). These results indicated that circCEP128 depletion inhibited the tumorigenesis of bladder cancer by up-regulating miR-515-5p and down-regulating SDC1 in vivo.
Figure 7.
circCEP128 knockdown inhibits the growth of bladder cancer xenograft tumors by up-regulating miR-515-5p and down-regulating SDC1. The xenograft model was established using 5637 cells stably transduced with sh-NC or sh-circCEP128 lentiviruses. (A and B) Tumor volume and weight were examined. (C–E) circCEP128, miR-515-5p and SDC1 abundances in xenograft tumors were determined by RT-qPCR and Western blot assays. *P<0.05.
Discussion
Bladder cancer is a prevalent cancer with about 75% of new diagnostic cases as non-muscle-invasive bladder cancer and approximately 25% of new cases as muscle-invasive bladder cancer.18 The 5-year overall survival rate for patients with non-muscle-invasive or muscle-invasive bladder cancer is about 90% or 60–70%, respectively.2 However, bladder cancer patients with metastatic disease have a 5-year overall survival rate of approximately 15%.18 Therefore, it is imperative to have a deep insight into the molecular regulatory mechanisms implicated in the tumorigenesis and progression of bladder cancer, which contributes to the development of better therapeutic schedules and the improvement of clinical outcomes.
Based on the GSE92675 dataset, we found that circCEP128 expression was markedly up-regulated in bladder cancer tissues relative to normal tissues. Also, analyses for clinical tissue specimens and cell lines further validated that circCEP128 was highly expressed in bladder cancer tumor tissues and cells. Consistent with previous studies,9,10 our data also revealed that cicCEP128 knockdown markedly weakened cell proliferative, migratory and invasive capacities and facilitated cell apoptosis in vitro and hindered the growth of xenograft tumors in vivo in bladder cancer.
Over the past decades, emerging circRNAs have been found to be involved in the regulation of cancer progression by acting as the molecular sponges of miRNAs.4,19 Prior studies also showed that circCEP128 could exert its oncogenic effect by sponging miR-145-5p in bladder cancer.9,10 In this study, we demonstrated that circCEP128 could negatively regulate miR-515-5p expression by direct interaction in bladder cancer cells. MiR-515-5p has been reported to be a potential tumor suppressor in multiple malignancies such as gastric, breast and hepatocellular cancers.20–22 In addition, Gong et al demonstrated that miR-515-5p could inhibit cell proliferation in bladder cancer.12 Dai et al revealed that miR-515-5p hindered cell proliferation, migration and invasion by regulating Go‐Ichi‐Ni‐San (GINS) complex subunit 2 (GINS2) in bladder cancer.13 In line with these reports,12,13 our outcomes also presented that miR-515-5p overexpression lessened cell proliferative, migratory and invasive abilities and elevated cell apoptotic rate in bladder cancer. Moreover, we further demonstrated that circCEP128 knockdown could hamper the tumorigenesis and progression of bladder cancer in vitro and in vivo by negatively regulating miR-515-5p expression. In other words, miR-515-5p as another miRNA target of circCEP128 was identified in bladder cancer.
It is well known to us that miRNAs can exert their functions by regulating the expression of target genes at post-transcriptional levels.23 Hence, potential target genes of miR-515-5p were searched by bioinformatics analysis. Among these targets, SDC1 was screened out for further investigations due to its vital roles in the tumorigenesis and progression of malignancies.14,24 For instance, SDC1 was reported to be an oncogene in glioma, renal cell cancer and pancreatic cancer.25–27 However, some studies also showed that SDC1 functioned as a tumor suppressor in colorectal cancer, oral cancer and gallbladder cancer.28–30 These data suggested that SDC1 might exert specific roles in different cancers. Moreover, Szarvas et al suggested that SDC1 abundance was elevated in bladder cancer, and SDC1 could function as an independent risk factor for the poor survival of bladder cancer patients.15 In addition, Wu et al revealed that SDC1 facilitated bladder cancer cell proliferation, migration and invasion,31 hinting the potential oncogenic effect of SDC1 in bladder cancer. Consequently, we further investigated whether SDC1 was a downstream mediator of circCEP128/miR-515-5p signaling and functions in bladder cancer. In this text, we demonstrated that SDC1 was a target of miR-515-5p and circCEP128 could positively regulate SDC1 expression by sequestering miR-515-5p in bladder cancer cells. Additionally, miR-515-5p exerted its anti-proliferative, pro-apoptotic, anti-migratory and anti-invasive effects by targeting SDC1 in bladder cancer cells. The subcutaneous xenograft model is one of important models for studying the pathogenesis of bladder cancer.32 Here, the anti-tumor function of circCEP128 loss was further validated in a xenograft model of bladder cancer. Moreover, our data disclosed that circCEP128 knockdown curbed the growth of bladder cancer xenograft tumors by up-regulating miR-515-5p and down-regulating SDC1.
In conclusion, circCEP128 knockdown suppressed the tumorigenesis and progression of bladder cancer by regulating miR-515-5p/SDC1 axis in vitro and in vivo. This is the first study to demonstrate that circCEP128 can exert its oncogenic effects by regulating miR-515-5p/SDC1 axis in bladder cancer. The elucidation of another molecular regulatory mechanisms of circCEP128 might deepen our understanding on pathogenesis of bladder cancer. Moreover, the aberrant expression of circCEP128, miR-515-5p, and SDC1 in bladder cancer tissues and cells indicated the potential diagnostic values of these molecules in bladder cancer. Furthermore, our study might contribute to the better management of bladder cancer considering the regulatory effects of these molecules on disease progression.
Disclosure
The authors declare that they have no conflicts of interest.
References
- 1.Sanli O, Dobruch J, Knowles MA, et al. Bladder cancer. Nat Rev Dis Primers. 2017;3:17022. [DOI] [PubMed] [Google Scholar]
- 2.Patel VG, Oh WK, Galsky MD. Treatment of muscle-invasive and advanced bladder cancer in 2020. CA Cancer J Clin. 2020;70(5):404–423. doi: 10.3322/caac.21631 [DOI] [PubMed] [Google Scholar]
- 3.Patop IL, Wust S, Kadener S. Past, present, and future of circRNAs. EMBO J. 2019;38(16):e100836. doi: 10.15252/embj.2018100836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li C, Fu X, He H, Chen C, Wang Y, He L. The biogenesis, functions, and roles of circRNAs in bladder cancer. Cancer Manag Res. 2020;12:3673–3689. doi: 10.2147/CMAR.S245233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y, Qiao L, Zang Y, Ni W, Xu Z. Circular RNA FOXO3 suppresses bladder cancer progression and metastasis by regulating MiR-9-5p/TGFBR2. Cancer Manag Res. 2020;12:5049–5056. doi: 10.2147/CMAR.S253412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Jiang P, Zhu Y, Xu Z, Chen X, Xie L. Interference with circBC048201 inhibits the proliferation, migration, and invasion of bladder cancer cells through the miR-1184/ITGA3 axis. Mol Cell Biochem. 2020. doi: 10.1007/s11010-020-03835-2 [DOI] [PubMed] [Google Scholar]
- 7.Hua L, Huang L, Zhang X, Feng H, Shen B. Knockdown of circular RNA CEP128 suppresses proliferation and improves cytotoxic efficacy of temozolomide in glioma cells by regulating miR-145-5p. Neuroreport. 2019;30(18):1231–1238. doi: 10.1097/WNR.0000000000001326 [DOI] [PubMed] [Google Scholar]
- 8.Jiang Y, Zhang Y, Chu F, Xu L, Wu H. Circ_0032821 acts as an oncogene in cell proliferation, metastasis and autophagy in human gastric cancer cells in vitro and in vivo through activating MEK1/ERK1/2 signaling pathway. Cancer Cell Int. 2020;20(1):74. doi: 10.1186/s12935-020-1151-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wu Z, Huang W, Wang X, et al. Circular RNA CEP128 acts as a sponge of miR-145-5p in promoting the bladder cancer progression via regulating SOX11. Mol Med. 2018;24(1):40. doi: 10.1186/s10020-018-0039-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sun M, Zhao W, Chen Z, et al. Circular RNA CEP128 promotes bladder cancer progression by regulating Mir‐145‐5p/Myd88 via MAPK signaling pathway. Int J Cancer. 2019;145:2170–2181. doi: 10.1002/ijc.32311 [DOI] [PubMed] [Google Scholar]
- 11.Li Q, Wang H, Peng H, et al. MicroRNAs: key players in bladder cancer. Mol Diagn Ther. 2019;23:579–601. [DOI] [PubMed] [Google Scholar]
- 12.Gong P, Xu R, Zhuang Q, He X. A novel circular RNA (hsa_circRNA_102336), a plausible biomarker, promotes the tumorigenesis by sponging miR-515-5p in human bladder cancer. Biomed Pharmacother. 2020;126:110059. doi: 10.1016/j.biopha.2020.110059 [DOI] [PubMed] [Google Scholar]
- 13.Dai G, Huang C, Yang J, et al. LncRNA SNHG3 promotes bladder cancer proliferation and metastasis through miR-515-5p/GINS2 axis. J Cell Mol Med. 2020;24(16):9231–9243. doi: 10.1111/jcmm.15564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Palaiologou M, Delladetsima I, Tiniakos D. CD138 (syndecan-1) expression in health and disease. Histol Histopathol. 2014;29:177–189. doi: 10.14670/HH-29.177 [DOI] [PubMed] [Google Scholar]
- 15.Szarvas T, Reis H, Kramer G, et al. Enhanced stromal syndecan-1 expression is an independent risk factor for poor survival in bladder cancer. Hum Pathol. 2014;45(4):674–682. doi: 10.1016/j.humpath.2013.10.036 [DOI] [PubMed] [Google Scholar]
- 16.Kim JH, Park J. Prognostic significance of heme oxygenase-1, S100 calcium-binding protein A4, and syndecan-1 expression in primary non–muscle-invasive bladder cancer. Hum Pathol. 2014;45:1830–1838. doi: 10.1016/j.humpath.2014.04.020 [DOI] [PubMed] [Google Scholar]
- 17.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262 [DOI] [PubMed] [Google Scholar]
- 18.Nadal R, Bellmunt J. Management of metastatic bladder cancer. Cancer Treat Rev. 2019;76:10–21. [DOI] [PubMed] [Google Scholar]
- 19.Cong L, Yang Q, Hu C, Yu Q, Hao S, Li D. Current status of functional studies on circular RNAs in bladder cancer and their potential role as diagnostic and prognostic biomarkers: a review. Med Sci Monit. 2019;25:3425–3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang D, Liu K, Chen E. LINC00511 promotes proliferation and invasion by sponging miR-515-5p in gastric cancer. Cell Mol Biol Lett. 2020;25:4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huang FJ, Dang JQ, Zhang S, Cheng ZY. Circular RNA hsa_circ_0008039 promotes proliferation, migration and invasion of breast cancer cells through upregulating CBX4 via sponging miR-515-5p. Eur Rev Med Pharmacol Sci. 2020;24:1887–1898. [DOI] [PubMed] [Google Scholar]
- 22.Ni JS, Zheng H, Ou YL, et al. miR-515-5p suppresses HCC migration and invasion via targeting IL6/JAK/STAT3 pathway. Surg Oncol. 2020;34:113–120. [DOI] [PubMed] [Google Scholar]
- 23.Saliminejad K, Khorram Khorshid HR, Soleymani Fard S, Ghaffari SH. An overview of microRNAs: biology, functions, therapeutics, and analysis methods. J Cell Physiol. 2019;234:5451–5465. [DOI] [PubMed] [Google Scholar]
- 24.Gharbaran R. Advances in the molecular functions of syndecan-1 (SDC1/CD138) in the pathogenesis of malignancies. Crit Rev Oncol Hematol. 2015;94:1–17. [DOI] [PubMed] [Google Scholar]
- 25.Shi S, Zhong D, Xiao Y, et al. Syndecan-1 knockdown inhibits glioma cell proliferation and invasion by deregulating a c-src/FAK-associated signaling pathway. Oncotarget. 2017;8:40922–40934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang J, Cao Z, Ding X, et al. The lncRNA XIST regulates the tumorigenicity of renal cell carcinoma cells via the miR-302c/SDC1 axis. Int J Clin Exp Pathol. 2017;10:7481–7491. [PMC free article] [PubMed] [Google Scholar]
- 27.Chen X, Zhao H, Chen C, et al. The HPA/SDC1 axis promotes invasion and metastasis of pancreatic cancer cells by activating EMT via FGF2 upregulation. Oncol Lett. 2020;19:211–220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang S, Zhang X, Wang G, et al. Syndecan-1 suppresses cell growth and migration via blocking JAK1/STAT3 and Ras/Raf/MEK/ERK pathways in human colorectal carcinoma cells. BMC Cancer. 2019;19:1160. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 29.Wang X, He J, Zhao X, Qi T, Zhang T, Kong C. Syndecan-1 suppresses epithelial-mesenchymal transition and migration in human oral cancer cells. Oncol Rep. 2018;39:1835–1842. [DOI] [PubMed] [Google Scholar]
- 30.Liu Z, Jin H, Yang S, et al. SDC1 knockdown induces epithelial-mesenchymal transition and invasion of gallbladder cancer cells via the ERK/Snail pathway. J Int Med Res. 2020;48:300060520947883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu JP, Zhang GY, Sun XZ. LncRNA ZNFX1-AS1 targeting miR-193a-3p/SDC1 regulates cell proliferation, migration and invasion of bladder cancer cells. Eur Rev Med Pharmacol Sci. 2020;24:4719–4728. [DOI] [PubMed] [Google Scholar]
- 32.Ringuette-Goulet C, Bolduc S, Pouliot F. Modeling human bladder cancer. World J Urol. 2018;36:1759–1766. [DOI] [PubMed] [Google Scholar]