Abstract
Background and Objective:
Gastric cancer (GC) is a common tumor malignancy with high incidence and poor prognosis. Laminin is an indispensable component of basement membrane and extracellular matrix, which is responsible for bridging the internal and external environment of cells and transmitting signals. This study mainly explored the association of the LAMB1 expression with clinicopathological characteristics and prognosis in gastric cancer.
Methods:
The expression data and clinical information of gastric cancer patients were downloaded from The Cancer Genome Atlas (TCGA) and Asian Cancer Research Group (ACRG). And we analyzed the relationship between LAMB1 expression and clinical characteristics through R. CIBERSORTx was used to calculate the absolute score of immune cells in gastric tumor tissues. Then COX proportional hazard models and Kaplan-Meier curves were performed to evaluate the role of LAMB1 and its influence on prognosis in gastric cancer patients. Finally, GO and KEGG analysis were applied for LAMB1-related genes in gastric cancer, and PPI network was constructed in Cytoscape software.
Results:
In the TCGA cohort, patients with gastric cancer frequently generated LAMB1 gene copy number variation, but had little effect on mRNA expression. Both in the TCGA and ACRG cohorts, the mRNA expression of LAMB1 in gastric cancer tissues was higher than it in normal tissues. All patients were divided into high expression group and low expression group according to the median expression level of LAMB1. The elevated expression group obviously had more advanced cases and higher infiltration levels of M2 macrophages. COX proportional hazard models and Kaplan-Meier curves revealed that patients with enhanced expression of LAMB1 have a worse prognosis. GO/KEGG analysis showed that LAMB1-related genes were enriched in PI3K-Akt signaling pathway, focal adhesion, ECM-receptor interaction, etc.
Conclusions:
The high expression of LAMB1 in gastric cancer is related to the poor prognosis of patients, and it may be related to microenvironmental changes in tumors.
Keywords: the β1 subunit of laminin, LAMB1, gastric cancer, GC, prognosis, tumor microenvironment, TME, CIBERSORTx, survival, therapeutic target, tumor-infiltrating immune cells, TICs
Introduction
According to the report from the World Health Organization (WHO: http://www.who.int/cancer/en/), Gastric cancer (GC) is one of the most common cancers world-wide.1 The incidence of gastric cancer (GC) has been declining in the past few decades worldwide. However, GC is still the fifth most frequently diagnosed cancer and the third leading cause of cancer-related death.2 Despite tremendous advances in surgery, radiotherapy, chemotherapy and targeted molecular therapy, the overall effectiveness of treatment is low, with a poor median overall survival (OS) which is shorter than 1 year. Many biomarkers had been developed for the prognosis prediction of patients with advanced GC and the early diagnosis of gastric cancer, but few biomarkers can be used in clinical practice.3-5 Moreover, as multiple clinical trials of targeted drugs for gastric cancer have failed, there is an urgent need to explore more sensitive and specific GC-related biomarkers as diagnostic and therapeutic targets.
Laminin is a large molecular weight glycoprotein assembled by 3 disulfide-bonded polypeptides (α, β and γ chains).6,7 It is an indispensable component for cells to bridge the internal and external environments of cells and carry out signal transmission. It is also a basement membrane and an essential component of the extracellular matrix. Including 5 alpha chains (LAMA1, LAMA2, LAMA3, LAMA4 and LAMA5), 4 beta chains (LAMB1, LAMB2, LAMB3 and LAMB4), and 3 gamma chains (LAMC1, LAMC2 and LAMC3), the human genome encodes for 12 different laminin chains varying in expression and distribution within the tissues. The β1 subunit (LAMB1) is ubiquitously expressed in skin, kidneys, lungs, intestine, bladder and stomach. And in addition to their role in maintaining structural integrity of tissues, the laminin-binding β1 also involved in the function of bidirectional signaling. Current studies have shown that LAMB1 plays an important role in a variety of tumors, such as8 prostate cancer,9 hepatocellular carcinoma,10 breast cancer and11 glioblastoma multiforme. However, there is limited systematic research investigating the associations between LAMB1 mRNA expression and patients’ with gastric cancer clinicopathological characteristics and prognosis. Therefore, based on The Cancer Genome Atlas database (TCGA) and Asian Cancer Research Group (ACRG) dataset, this study retrospectively investigated the transcriptome and genome of LAMB1 in gastric cancer and its impact on the prognosis of gastric cancer. Furthermore, the genomic changes and functional networks in GC related to LAMB1 had been analyzed.
Materials and Methods
Data Resource and Description
Demographic information and clinical data, as well as expression data (genomic data was included in TCGA cohort) of gastric cancer was selected form the Asian Cancer Research Group (ACRG) study and The Cancer Genome Atlas (TCGA) dataset, The ACRG cohort (GSE66229) containing gastric cancer expression data of tumor and non-tumor samples were obtained from the National Center for Biotechnology Information’s (NCBI) Gene Expression Omnibus (GEO, https://www.ncbi.nlm.nih.gov/geo/). In TCGA cohort, all of the publicly available gastric cancer RNA-Seq data, copy number variation (CNV) data and genomic data information were downloaded from TCGA official website (https://cancergenome.nih.gov/). TCGA RNA-seq data was comprised of 414 tumor samples and 36 non-tumor samples. GSE66229 was comprised of 300 tumor samples and 100 non-tumor samples (details in Tables 1 and 2).
Table 1.
Patients in TCGA Dataset.
| TCGA | P | |||||
|---|---|---|---|---|---|---|
| Total | Low | High | ||||
| Age | 67(58-73) | 67(58-73) | 67(57-72) | 13538 | 0.6215 | |
| Gender | Female | 146 | 81 | 65 | 2.3806 | 0.1228 |
| Tstage | T1 | 19 | 15 | 4 | 22.444 | 0.0001635 |
| T2 | 81 | 44 | 37 | |||
| T3 | 167 | 87 | 80 | |||
| T4 | 106 | 35 | 71 | |||
| TX | 41 | 26 | 15 | |||
| Nstage | N0 | 98 | 59 | 52 | 3.0078 | 0.5565 |
| N1 | 105 | 53 | 52 | |||
| N2 | 73 | 32 | 41 | |||
| N3 | 74 | 34 | 40 | |||
| NX | 51 | 29 | 22 | |||
| Mstage | M0 | 380 | 192 | 188 | 0.364 | |
| M1 | 30 | 12 | 18 | |||
| MX | 4 | 3 | 1 | |||
| Grade | G1 | 12 | 4 | 8 | 0.06371 | |
| G2 | 148 | 76 | 72 | |||
| G3 | 245 | 126 | 119 | |||
| GX | 9 | 1 | 8 | |||
| Gastric cardia adenocarcinoma (GCA) | 89 | 46 | 43 | 0.057252 | 0.8109 | |
| Type | Intestinal type | 177 | 83 | 94 | 3.5851 | 0.1665 |
| Diffuse type | 80 | 36 | 44 | |||
| Mix or other | 157 | 88 | 69 | |||
| Race | Asian | 86 | 42 | 44 | 4.7432 | 0.1916 |
| Black | 12 | 9 | 3 | |||
| White | 260 | 124 | 136 | |||
| Other | 56 | 32 | 24 | |||
| Total | 414 | 207 | 207 | |||
Table 2.
Patients in ACRG Dateset.
| ACRG | P | |||||
|---|---|---|---|---|---|---|
| Total | Low | High | ||||
| Age | 64(55-70) | 65(57-70) | 62(52-70) | 12690 | 0.05529 | |
| Gender | Female | 101 | 54 | 47 | 0.53734 | 0.4635 |
| Male | 199 | 96 | 103 | |||
| Tstage | T1 | 0 | 0 | 0 | 11.174 | 0.003746 |
| T2 | 188 | 108 | 80 | |||
| T3 | 91 | 34 | 57 | |||
| T4 | 21 | 8 | 13 | |||
| Nstage | N0 | 38 | 21 | 17 | 10.254 | 0.01652 |
| N1 | 131 | 76 | 55 | |||
| N2 | 80 | 36 | 44 | |||
| N3 | 51 | 17 | 34 | |||
| Mstage | M0 | 273 | 138 | 135 | 0.1628 | 0.6866 |
| M1 | 27 | 12 | 15 | |||
| Gastric cardia adenocarcinoma (GCA) | 32 | 15 | 17 | 0.034981 | 0.8516 | |
| Subtype | MSI | 68 | 50 | 18 | 37.87 | 3.012e-08 |
| EMT | 46 | 7 | 39 | |||
| TP53- | 107 | 51 | 56 | |||
| TP53+ | 79 | 42 | 37 | |||
| Type | Intestinal type | 146 | 83 | 63 | 8.6851 | 0.013 |
| Diffuse type | 135 | 55 | 80 | |||
| Mix or other | 19 | 12 | 7 | |||
| Total | 300 | 150 | 150 | |||
Bioinformatics Analysis for Identifying LAMB1 Expression
Raw CEL files of the micro array of each GEO dataset were normalized by the quantile method of Robust Multichip Analysis (RMA) from the R12 affy package and the normalized gene expression levels were presented as log2-transformed values by RMA. The copy number variation (CNV) v3 data of 406 gastric cancer patients (TCGA) were annotated by13 annovar. And the absolute value of segment mean >0.3 will be defined as gain or loss.
TICs Profile
CIBERSORTx was used to calculate the absolute score of tumor-infiltrating immune cells (TICs) in gastric tumor tissues, with reference to14 LM22 gene signature. The CIBERSORTx is an analytical tool to impute gene expression profiles and provide an estimation of the abundances of member cell types in a mixed cell population, using gene expression data. LM22 defines 22 subtypes of immune cells referring to the annotated gene signature matrix, downloaded from the CIBERSORTx website portal (https://cibersortx.stanford.edu/). The 22 immune cells contain 2 subtypes of B cells, 7 subtypes of T cells, 2 subtypes of NK cells, 3 subtypes of Macrophages, 2 subtypes of Dendritic cells, 2 subtypes of Mast cells, Monocytes, Eosinophils and Neutrophils. Wilcoxon rank-sum test was performed to analyze the differential abundances of infiltrating immune cells between low- and high-LAMB1 level groups, which were visualized using the “ggplot2” package.
Survival Analysis
Patients, defining the median of LAMB1 expression values as the cutoff point, were classified into a low expression group and high expression group to analyze the correlation between LAMB1 expression with survival rates and clinical pathological characteristics. The survivorship curve was plotted by R package15,16 survival and r-base.
Identification of LAMB1-Related Genes
The Pearson’s correlation coefficient between the mRNA expression value of LAMB1 and other gene were calculated. If any genes’ absolute value of Pearson’s correlation coefficient > 0.6 and adj. P-value < 0.05, it were defined as LAMB1-related genes.
KEGG/GO Biological Process Enrichment
The R package17 clusterProfiler was used to Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway enrichment analysis and Gene Ontology (GO) enrichment analysis, including biological process (BP), cellular components (CC), and molecular function (MF). Adjusted P-value < 0.05 was considered as the threshold.
Protein-Protein Interaction (PPI) Network Construction
Protein-protein interaction (PPI) networks were analyzed using the Search Tool for the Retrieval of Interacting Genes/Proteins (STRING; http://www.string-db.org/), an online database comprising comprehensive known and predicted interactions, to determine the interactive relationships among the LAMB1-related genes.18 Then, the PPI pairs were inputted into19 Cytoscape software version 3.8.0 (http://www.cytoscape.org) to construct the PPI network, and the cytoscape plug-in cytoHuba were used to calculate the top 10 central genes.
Statistical Analyses
All analyses were carried out using the R language,20 version 3.6.3, and nonparametric rank sum tests and t tests revealed that the LAMB1 mRNA expression differences in different clinical variables were visualized through the21 ggplot2 package. The Mann-Kendall test were performed to uncover the trend of change by R package22 trend. Chi-square test or Fisher exact test were used for enumeration data.
Results
LAMB1 Expression and Mutation in Gastric Cancer
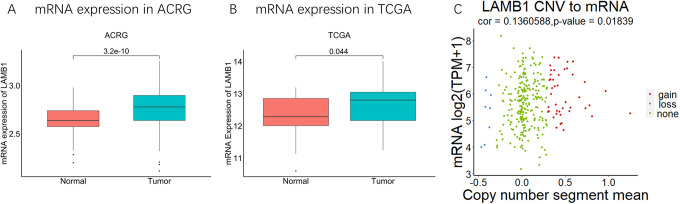
As illustrated in Figure 1, the expression of LAMB1 in tumor samples was higher than non-tumor samples in ACRG (P = 3.2e-10, Figure 1A). The similar tendency was observed in TCGA cohort (P = 0.04398065, Figure 1B). Then we inspected somatic mutation and copy number variation of LAMB1. The TCGA cohort showed more frequent copy number variation compared with somatic mutation.30 patients generated somatic mutation in 406 patients, but most of these mutations belong to (21/30,70%) synonymous variant or missense variant.64 patients generated copy number variation in 406 patients, include 55 gains and 9 losses, but, in those patients with both mRNA data and CNV data, little correlation is showed (cor = 0.1360588, P-value = 0.01839, Figure 1C).
Figure 1.
The expression of LAMB1 in gastric cancer. A-B. Comparison of LAMB1 mRNA expression between tumor and normal samples in ACRG (A) and TCGA-STAD (B). C. The relationship between mRNA of LAMB1 and copy number variation of LAMB1 in TCGA-STAD.
Association Between LAMB1 and Clinicopathological Features in Gastric Cancer Patients
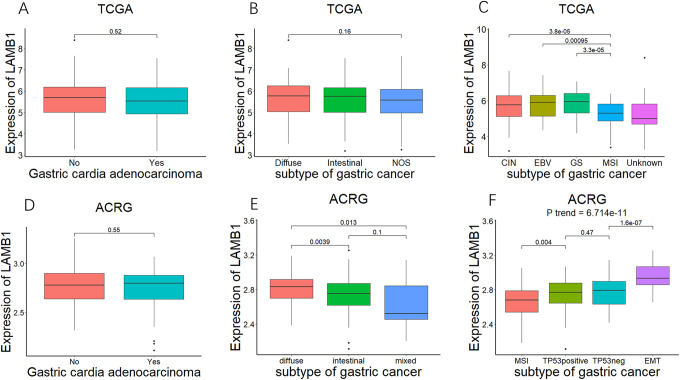
In order to clarify the relationship between LAMB1 mRNA expression and the clinical characteristics of gastric cancer samples, we respectively divided TCGA and ACRG cohorts into high expression groups and low expression groups based on the median of LAMB1 mRNA expression. As delineated in Tables 1 and 2, both in ACRG and TCGA, age, gender and distant metastasis status were not showing significantly difference between LAMB1 expression low group and high group (P > 0.05). Both in the TCGA and ACRG cohorts, there was a significant increase in T3 and T4 cases in the LAMB1 high expression group (P = 0.0001635 in TCGA and P = 0.003746 in ACRG). The LAMB1 high expression group in the GEO cohort seems to have more N2, N3 cases (P = 0.01652) and more diffuse gastric cancer (P = 0.013), but this was not found in the TCGA cohorts (P > 0.05). Therefore, we investigated the expression of LAMB1 in different gastric cancer subtypes and TNM stages. In the TCGA cohort, the expression of LAMB1 in T2 stage gastric cancer was significantly higher than it in T1 stage and the expression of LAMB1 in T3 stage tumors was significantly higher than it in T2 stage. LAMB1 mRNA expression showed a significant increasing trend with the increase of tumor T stage (P trend < 0.05, Figure 2A), and a similar trend was observed in ACRG (Figure 2D). More patients, in ACRG, accompanied with lymph node metastasis in the LAMB1 high expression group, but there is no similar situation in TCGA (Tables 1 and 2). Therefor we separately analyzed the relationship between the expression of LAMB1 and lymph node metastasis in different dataset. The results, both in ACRG and TCGA, showed a tendency of LAMB1 expression increased with N stages (Figure 2B, P trend = 0.02739 in TCGA, Figure 2E, P trend = 0.00453 in ACRG). In addition, the expression of LAMB1 in patients with distant metastasis tended to be higher than that in patients without metastasis, but there was no significant statistical difference (Figure 2C, P = 0.16 in TCGA, Figure 2E, P = 0.11 in ACRG), which may be due to lower proportion of ACRG and TCGA distant metastases. Subsequently, we analyzed the LAMB1 expression of different subtypes of gastric cancer, and the results suggested that the expression of LAMB1 in gastric cancer has nothing to do with the tumor location (P > 0.5, Figure 3A and Figure 3D). The expression of LAMB1 in diffuse gastric cancer in ACRG was significantly higher than it in intestinal gastric cancer or mixed gastric cancer (Figure 3E), but there was no significant difference in the expression of LAMB1 in the 3 gastric cancers in the TCGA cohort (Figure 3B). Finally, we analyzed the expression of LAMB1 in different subtypes in the molecular typing proposed by the TCGA project and the ACRG project. In TCGA, the expression of microsatellite unstable (MSI) LAMB1 was significantly lower than that of other subtypes (Figure 3C). The expression of LAMB1 of MSI subtype in ACRG was also lower than that of other subtypes (Figure 3F). At the same time, as the prognosis of the subtypes deteriorated, the expression of LAMB1 showed obviously increasing trend (P trend < 0.05, Figure 3F).
Figure 2.
The mRNA expression of LAMB1 in different TNM stage of gastric cancer. A-F. The association between LAMB1 mRNA expression and different T stages (A&D), different N stages (B&E), different distant metastasis status (C&F).
Figure 3.
The mRNA expression of LAMB1 in different subtype of gastric cancer. A-F. Comparison of LAMB1 mRNA expression between gastric cardia adenocarcinoma and gastric non-cardia adenocarcinoma (A&D), different histological subtype (B&E), and different molecular subtypes in TCGA-STAD (C) and ACRG (F).
Associations Between LAMB1 and Survival in Gastric Cancer Patients
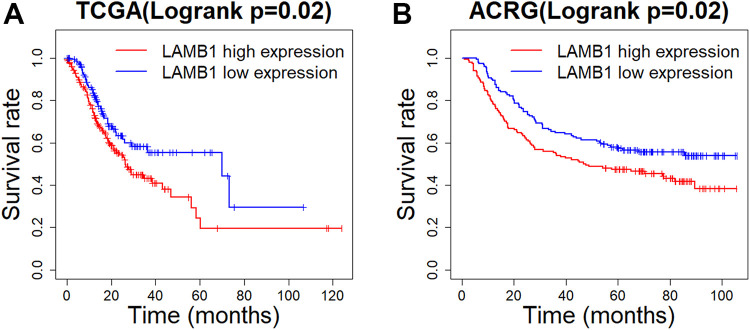
Considering that the expression of LAMB1 is closely related to the T staging of gastric cancer and the MSI subtype of gastric cancer, Kaplan-Meier curves, along with log-rank test, evaluated the association between LAMB1 mRNA and prognosis of patients with gastric cancer. It was found that increased mRNA expression of LAMB1 in gastric tumor tissues was considerably associated with poor overall survival (Figure 4A and Figure 4B) in patients with gastric cancer. Then univariate and multivariate Cox proportional hazards models with age, gender and other factors as covariates were performed, and the factors with a P value of less than 0.1 in the univariate COX proportional hazard model are included in the multivariate COX proportional hazard model. Regardless of TCGA or ACRG, the LAMB1 expression, just like TNM stages, has a significant impact on the prognosis of gastric cancer patients (Table 3).
Figure 4.
Differences in overall survival between LAMB1 low expression group and LAMB1 high expression group in TCGA (A) and ACRG (B).
Table 3.
COX Proportional Hazard Model.
| Univariate | Multivariate | ||||
|---|---|---|---|---|---|
| Cohort | Variable | HR (95%CI) | P-value (logrank) | HR (95%CI) | P-value (wald) |
| ACRG | Stage (III&IV VS.I&II) | 3.41(2.34-4.96) | 1.53e-10 | 3.24(2.20-4.76) | 2.29e-09 |
| LAMB1(mRNA level) | 4.59(1.95-10.81) | 0.00048 | 4.41(1.79- 10.88) | 0.001280 | |
| Age(per decade after 50 years old) | 1.16(0.98-1.37) | 0.0831 | 1.37(1.15-1.64) | 0.000432 | |
| Diffused type(compare with intestinal type) | 1.68(1.20-2.35) | 0.00257 | 1.44(1.02-2.03) | 0.000432 | |
| Mixed type(compare with intestinal type) | 2.13(1.17-3.88) | 0.01370 | 2.15(1.16-3.98) | 0.014463 | |
| Gastric cardia adenocarcinoma | 1.55(0.98-2.46) | 0.0624 | |||
| TCGA | Stage (III&IV VS.I&II) | 2.08(1.47-2.93) | 3.08e-05 | 2.13(1.50-3.01) | 2.21e-05 |
| Age(per decade after 50 years old) | 1.19(1.02-1.39) | 0.0256 | 1.27(1.08-1.49) | 0.00325 | |
| LAMB1(mRNA level) | 1.23(1.03-1.46) | 0.023 | 1.41(1.02-1.95) | 0.03838 | |
| MSI subtype(Yes VS.No) | 0.64(0.46-1.0) | 0.0514 | |||
Correlation of LAMB1 With Immune Signatures in Gastric Cancer Patients
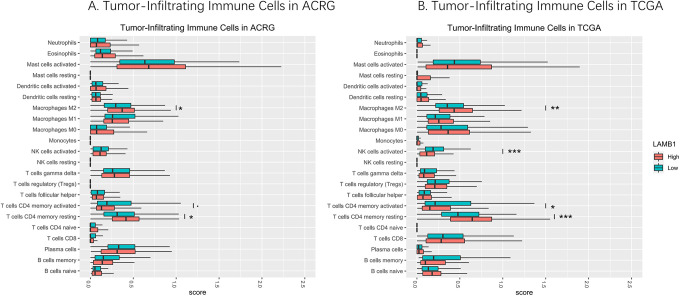
CIBERSORTx algorithm calculated the absolute score of 22 infiltrating immune cells in the gastric cancer samples (Figure 5). Then, Wilcoxon rank-sum test was used to reveal the difference of infiltrating immune cells between the low- and high-LAMB1 expression level samples. Both in the TCGA and ACRG cohorts, high-LAMB1 level group presented a significantly higher infiltration levels of resting CD4+ memory T cells and M2 macrophages than the low-LAMB1 group (Figure 5). On the other hand, the low-LMB1 level group showed a higher infiltration level of activated NK cells and activated CD4+ memory T cells in the TCGA cohorts (Figure 5B). And a similar trend was observed in ACRG, although the difference in the trend is not significant (Figure 5A).
Figure 5.
The distribution of infiltrating immune cells in the low- and high-LAMB1 expression level samples of ACRG (A) and TCGA-STAD (B) cohort (*P < 0.05,**P < 0.01,***P < 0.001).
KEGG/GO Biological Process Enrichment
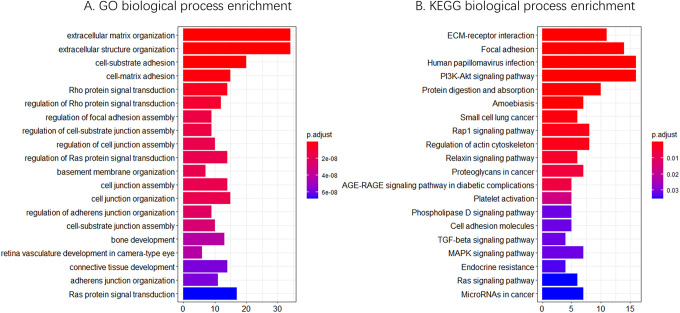
The GO analysis of LAMB1-related genes showed that the most of enriched pathways were closely related to extracellular matrix (ECM) and cancer progression, such as extracellular matrix organization, extracellular structure organization, cell-substrate adhesion, Ras protein signal transduction, cell-substrate junction assembly (Figure 6A). The results from KEGG analysis indicated that among the pathways in which these genes were particularly enriched, many were closely related to cancer progression, such as the PI3K-Akt signaling pathway, focal adhesion, and ECM-receptor interaction (Figure 6B).
Figure 6.
GO (A) and KEGG (B) biological process enrichment of LAMB1-related genes.
Protein-Protein Interaction (PPI) Network Construction
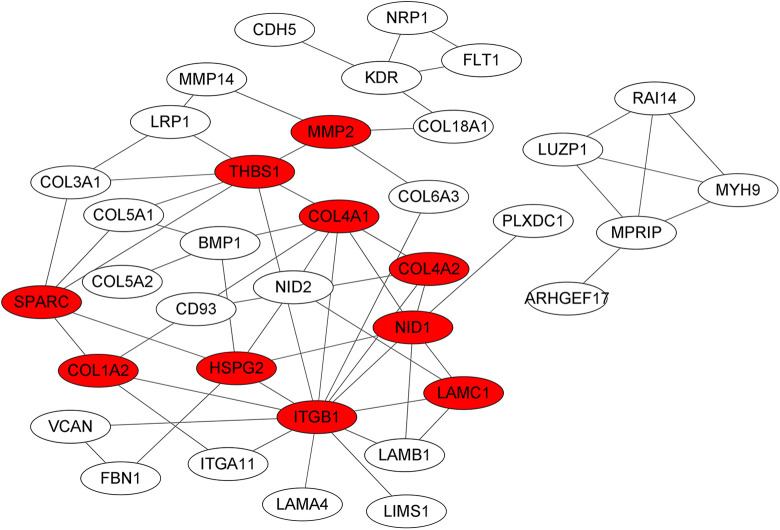
Using the STRING online database and Cytoscape software, 36 genes of the 113 LAMB1-related genes were filtered into the PPI network complex, containing 61 edges in their intricate network (Figure 7), and the top 10 central genes, calculated by the cytoscape plug-in cytoHuba, were ITGB1, THBS1, COL4A1, NID1, HSPG2, SPARC, COL4A2, COL1A2, LAMC1 and MMP2.
Figure 7.
Protein-protein interaction (PPI) network of LAMB1-related genes, the top 10 central genes were marked by red.
Discussion
In addition to tumor cells, cancer also includes a complex ecosystem composed of peripheral blood vessels, extracellular matrix (ECM), other non-malignant cells and signaling molecules.23 These non-tumor cell components together constitute the tumor microenvironment (TME). In recent decades,24 cancer research has undergone an overturning shift from focusing exclusively on a seemingly obvious target in malignant cells toward appreciation of key roles of the tumor microenvironment (TME) in cancer progression and therapy.6,23,25 The tumor microenvironment (TME), including laminins, in the tumor microenvironment is important for tumor invasion, progression and chemoresistance. This is further confirmed by numerous advances in tumor microenvironment research.26 Many subtypes of laminins have been described to promote cell adhesion and migration via ITG interaction. Studies have confirmed that LAMB1 is associated with tumor progression in11 glioma and10 breast cancer.8 In addition, LAMB1, in prostate cancer, participate in cell movement and is involved in tumor invasion into ECM.9 And study shows that LAMB1 has a crucial role in the invasion and metastasis of human HCC. To gain more detailed insights into the potential functions of LAMB1 in GC and its regulatory network, bioinformatics analysis of public sequencing data was performed.
More than 600 GC clinical samples including TCGA and ACRG revealed that the mRNA level of LAMB1 in GC was slightly higher than it in normal gastric mucosa (Figure 1). Despite the fact that the increase in the copy number of LAMB1 occurs frequently in gastric cancer tissues, the up-regulation of copy number of LAMB1 has relatively little effect on mRNA levels. It seems to indicate that LAMB1 is more regulated by its related genes. In addition, the high expression of LAMB1 is significantly related to low survival rate and high T stage. The Mann-Kendall test showed that ACRG and TCGA cases shared a significant increasing trend in LAMB1 mRNA expression with the increase of tumor T stage and N stage.27 Another study in stomach cancer pointed out that LAMB1 can promote tumor growth, cell invasion and migration of gastric cancer cells, which is in good agreement with our results.
Tumor-infiltrating immune cells as integral component of the tumor microenvironment are associated with tumor progress, prognosis and responses to immunotherapy. Therefore, we studied the relationship between LAMB1 and the tumor immune micro-environment of gastric cancer in TCGA and ACRG. The study found that, in tissues with high expression of LAMB1, M2 macrophages and resting CD4+ memory T cells increased significantly, while the activated CD4+ memory T cells decreased. Memory B cells, plasma cells and NK activated cells all have a downward trend, although not all of them have significant statistical differences. In gastric cancer,28 the activated CD4+ memory T cells and plasma cells,29 contrary to resting CD4+ memory T cells, has been known as a protective factor. And for macrophages,30 the 2 main polarization-based subtypes have more or less opposite functions in tumor: M1 macrophages are believed to exert anti-tumor effect by promoting the Th1 immune response; M2 macrophages favor the Th2 immune response, which facilitates tumor progression. This discovery that a high level of LAMB1 expression in gastric cancer predicted an increase in immune cells with a poor prognosis and a decrease in cells with a good prognosis is highly consistent with our previous results. It seems to indicate that LAMB1 may be involved in the activation of CD4+ memory T cells and the polarization or recruitment of macrophages. After using the COX proportional hazard model to correct the influence of age, gender and other factors, high expression of LAMB1 still predicts a poorer prognosis. Consistent with previous findings in many other tumors, our results suggest that the elevated expression of LAMB1 is associated with poor prognosis in gastric cancer patients, indicating an important role of LAMB1 in gastric cancer development and progression.
Next, GO/KEGG analysis of genes with medium to high intensity correlation with LAMB1 expression showed that LAMB1 was significantly related to signal pathways such as extracellular matrix, PI3K-Akt and Ras.31,32 These signal pathways had been shown to be involved in tumor progression, EMT and other biological processes.33 The PI3K-Akt pathway regulates macrophage survival, migration, and proliferation but also contributes to macrophage polarization and activation.34 And the MAPK signaling pathway, a crucial driver of tumorigenesis, is associated with activation of T cells. Based on the current evidence, we speculate that LAMB1 may have the ability to affect the prognosis by interacting with immune cells. Analysis of co-expression and PPI network showed that ITGB1, THBS1, COL4A1, NID1, HSPG2, SPARC, COL4A2, COL1A2, LAMC1, MMP2 and other molecules connect the interaction of multiple proteins and have a strong correlation with LAMB1.35-38 A lot of studies had shown that the integrin family, Collagen family, and MMP family are related to the tumor occurrence and development, EMT, poor prognosis, and even resistance to some chemotherapeutic drugs. For example,37 enhanced SPARC expression in GC led to a worse clinical outcome of patients and might induce Adriamycin (Adr) sensitivity of GC cells.36 And a meta-analysis indicated that abnormal MMP2 expression strongly correlated with poor prognosis in patients with GC. Therefore, we have more reason to believe that LAMB1 presents a comprehensive biological effect of promoting progression of tumor in gastric cancer, which may be utilized as a potential diagnostic or prognostic marker and therapeutic targets and it is valuable to study more.
Unfortunately, we have not yet conducted experimental studies to explore the potential mechanism of LAMB1 in the development of gastric cancer. This will be done in our subsequent research. However, combining previous reports and the findings of this study, we can still put forward the conclusion that GC patients with high expression of LAMB1 have a poor prognosis. Moreover, it may be related to microenvironmental changes in tumors.
Acknowledgments
The authors thank TCGA and ACRG for their contributions to human tumor research and the establishment of open database. The authors also thank Chongqing Health and Family Planning Commission for their financial support.
Authors’ Note: All of the authors have read and approved the manuscript. Our study did not require an ethical board approval because it did not contain human or animal trials.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by Chongqing Health and Family Planning Commission [Grant Number: 2017ZDXM010].
ORCID iD: Tao Ran, MD  https://orcid.org/0000-0001-5742-745X
https://orcid.org/0000-0001-5742-745X
References
- 1. Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68(6):394–424. doi:10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2. Van Cutsem E, Sagaert X, Topal B, Haustermans K, Prenen H. Gastric cancer. Lancet Lond Engl. 2016;388(10060):2654–2664. doi:10.1016/S0140-6736(16)30354-3 [DOI] [PubMed] [Google Scholar]
- 3. Hecht JR, Bang Y-J, Qin SK, et al. Lapatinib in combination with capecitabine plus oxaliplatin in human epidermal growth factor receptor 2-positive advanced or metastatic gastric, esophageal, or gastroesophageal adenocarcinoma: TRIO-013/LOGiC—a randomized phase III trial. J Clin Oncol Off J Am Soc Clin Oncol. 2016;34(5):443–451. doi:10.1200/JCO.2015.62.6598 [DOI] [PubMed] [Google Scholar]
- 4. Adelstein DJ, Rodriguez CP, Rybicki LA, Ives DI, Rice TW. A phase II trial of gefitinib for recurrent or metastatic cancer of the esophagus or gastroesophageal junction. Invest New Drugs. 2012;30(4):1684–1689. doi:10.1007/s10637-011-9736-z [DOI] [PubMed] [Google Scholar]
- 5. Tebbutt NC, Price TJ, Ferraro DA, et al. Panitumumab added to docetaxel, cisplatin and fluoropyrimidine in oesophagogastric cancer: ATTAX3 phase II trial. Br J Cancer. 2016;114(5):505–509. doi:10.1038/bjc.2015.440 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Qin Y, Rodin S, Simonson OE, Hollande F. Laminins and cancer stem cells: partners in crime? Semin Cancer Biol. 2017;45:3–12. doi:10.1016/j.semcancer.2016.07.004 [DOI] [PubMed] [Google Scholar]
- 7. Ramovs V, Te Molder L, Sonnenberg A. The opposing roles of laminin-binding integrins in cancer. Matrix Biol J Int Soc Matrix Biol. 2017;57-58:213–243. doi:10.1016/j.matbio.2016.08.007 [DOI] [PubMed] [Google Scholar]
- 8. Alinezhad S, Väänänen R-M, Mattsson J, et al. Validation of novel biomarkers for prostate cancer progression by the combination of bioinformatics, clinical and functional studies. PloS One. 2016;11(5):e0155901. doi:10.1371/journal.pone.0155901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Govaere O, Petz M, Wouters J, et al. The PDGFRα-laminin B1-keratin 19 cascade drives tumor progression at the invasive front of human hepatocellular carcinoma. Oncogene. 2017;36(47):6605–6616. doi:10.1038/onc.2017.260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Qiu X, Tan H, Fu D, Zhu Y, Zhang J. Laminin is over expressed in breast cancer and facilitate cancer cell metastasis. J Cancer Res Ther. 2018;14(Supplement):S1170–S1172. doi:10.4103/0973-1482.191035 [DOI] [PubMed] [Google Scholar]
- 11. Chen Q, Lu G, Cai Y, et al. MiR-124-5p inhibits the growth of high-grade gliomas through posttranscriptional regulation of LAMB1. Neuro-Oncol. 2014;16(5):637–651. doi:10.1093/neuonc/not300 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Gautier L, Cope L, Bolstad BM, Irizarry RA. Affy—analysis of affymetrix genechip data at the probe level. Bioinformatics 2004;20(3):307–315. [DOI] [PubMed] [Google Scholar]
- 13. Wang K, Li M, Hakonarson H. ANNOVAR: Functional annotation of genetic variants from next-generation sequencing data. Nucleic Acids Res 2010;38(16):E164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Newman AM, Liu CL, Green MR, et al. Robust enumeration of cell subsets from tissue expression profiles. Nat Methods. 2015;12(5):453–457. doi:10.1038/nmeth.3337 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Therneau T. _A Package for Survival Analysis in R_. R Package Version 3.2-3. 2020. Updated July 08, 2020. Accessed March 10, 2020. <URL: Https://CRAN.R-Project.Org/Package=survival>
- 16. Therneau TM, Grambsch PM. Modeling Survival Data: Extending the Cox Model. Springer; 2000. ISBN 0-387-98784-3. [Google Scholar]
- 17. Yu G, Wang LG, Han Y, He QY. Cluster profiler: an R package for comparing biological themes among gene clusters. OMICS. 2012;16(5):284–287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Szklarczyk D, Morris JH, Cook H, et al. The STRING database in 2017: quality-controlled protein–protein association networks, made broadly accessible. Nucleic Acids Res. 2017;45(Database issue):D362–D368. doi:10.1093/nar/gkw937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Killcoyne S, Carter GW, Smith J, Boyle J. Cytoscape: a community-based framework for network modeling. Methods Mol Biol Clifton NJ. 2009;563:219–239. doi:10.1007/978-1-60761-175-2_12 [DOI] [PubMed] [Google Scholar]
- 20. R Core Team. R: A Language and Environment for Statistical Computing. 2020. R Foundation for Statistical Computing. Updated March 10, 2020. Accessed February 29, 2020. URL Https://Www.R-Project.Org/ [Google Scholar]
- 21. Wickham H. Ggplot2: Elegant Graphics for Data Analysis. Springer-Verlag; 2016. [Google Scholar]
- 22. Pohlert T. Trend: non-parametric trend tests and change-point detection. R Package Version 1.1.4. 2020. Updated October 01, 2020. Accessed September 17, 2020. Https://CRAN.R-Project.Org/Package=trend
- 23. Hui L, Chen Y. Tumor microenvironment: sanctuary of the devil. Cancer Lett. 2015;368(1):7–13. doi:10.1016/j.canlet.2015.07.039 [DOI] [PubMed] [Google Scholar]
- 24. Siemann DW. Ed. Tumor Microenvironment. John Wiley & Sons, Ltd; 2011. [Google Scholar]
- 25. Cukierman E, Bassi DE. The mesenchymal tumor microenvironment: a drug-resistant niche. Cell Adhes Migr. 2012;6(3):285–296. doi:10.4161/cam.20210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Patarroyo M, Tryggvason K, Virtanen I. Laminin isoforms in tumor invasion, angiogenesis and metastasis. Semin Cancer Biol. 2002;12(3):197–207. doi:10.1016/S1044-579X(02)00023-8 [DOI] [PubMed] [Google Scholar]
- 27. Lee H, Kim W-J, Kang H-G, et al. Upregulation of LAMB1 via ERK/c-Jun axis promotes gastric cancer growth and motility. Int J Mol Sci. 2021;22(2):626. doi:10.3390/ijms22020626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Xie S, Mo P, Li N, et al. Tumor-infiltrating lymphocyte-based risk score for predicting prognosis in gastric cancer. Front Oncol. 2020;10:2111. doi:10.3389/fonc.2020.522015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Li F, Sun Y, Huang J, Xu W, Liu J, Yuan Z. CD4/CD8 + T cells, DC subsets, Foxp3, and IDO expression are predictive indictors of gastric cancer prognosis. Cancer Med. 2019;8(17):7330–7344. doi:10.1002/cam4.2596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Najafi M, Hashemi Goradel N, Farhood B, et al. Macrophage polarity in cancer: a review. J Cell Biochem. 2019;120(3):2756–2765. doi:10.1002/jcb.27646 [DOI] [PubMed] [Google Scholar]
- 31. Fresno Vara JA, Casado E, de Castro J, Cejas P, Belda-Iniesta C, González-Barón M. PI3K/Akt signalling pathway and cancer. Cancer Treat Rev. 2004;30(2):193–204. doi:10.1016/j.ctrv.2003.07.007 [DOI] [PubMed] [Google Scholar]
- 32. Bos JL. Ras oncogenes in human cancer: a review. Cancer Res. 1989;49(17):4682–4689. [PubMed] [Google Scholar]
- 33. Vergadi E, Ieronymaki E, Lyroni K, Vaporidi K, Tsatsanis C. Akt signaling pathway in macrophage activation and M1/M2 polarization. J Immunol Baltim Md 1950. 2017;198(3):1006–1014. doi:10.4049/jimmunol.1601515 [DOI] [PubMed] [Google Scholar]
- 34. Marquis M, Boulet S, Mathien S, et al. The non-classical MAP kinase ERK3 controls T cell activation. PloS One. 2014;9(1):e86681. doi:10.1371/journal.pone.0086681 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Kashihara H, Shimada M, Yoshikawa K, et al. Correlation between thrombospondin-1 expression in non-cancer tissue and gastric carcinogenesis. Anticancer Res. 2017;37(7):3547–3552. doi:10.21873/anticanres.11724 [DOI] [PubMed] [Google Scholar]
- 36. Wang H-L, Zhou P-Y, Zhang Y, Liu P. Relationships between abnormal MMP2 expression and prognosis in gastric cancer: a meta-analysis of cohort studies. Cancer Biother Radiopharm. 2014;29(4):166–172. doi:10.1089/cbr.2014.1608 [DOI] [PubMed] [Google Scholar]
- 37. Li Z, Li A-D, Xu L, et al. SPARC expression in gastric cancer predicts poor prognosis: results from a clinical cohort, pooled analysis and GSEA assay. Oncotarget. 2016;7(43):70211–70222. doi:10.18632/oncotarget.12191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Xu S, Xu H, Wang W, et al. The role of collagen in cancer: from bench to bedside. J Transl Med. 2019;17(1):309. doi:10.1186/s12967-019-2058-1 [DOI] [PMC free article] [PubMed] [Google Scholar]