Graphical abstract
Keywords: Genome editing, CRISPR, Cas, Crop improvement, Gene function, PAM
Abstract
Background
It is a long-standing goal of scientists and breeders to precisely control a gene for studying its function as well as improving crop yield, quality, and tolerance to various environmental stresses. The discovery and modification of CRISPR/Cas system, a nature-occurred gene editing tool, opens an era for studying gene function and precision crop breeding.
Aim of Review
In this review, we first introduce the brief history of CRISPR/Cas discovery followed the mechanism and application of CRISPR/Cas system on gene function study and crop improvement. Currently, CRISPR/Cas genome editing has been becoming a mature cutting-edge biotechnological tool for crop improvement that already used in many different traits in crops, including pathogen resistance, abiotic tolerance, plant development and morphology and even secondary metabolism and fiber development. Finally, we point out the major issues associating with CRISPR/Cas system and the future research directions.
Key Scientific Concepts of Review: CRISPR/Cas9 system is a robust and powerful biotechnological tool for targeting an individual DNA and RNA sequence in the genome. It can be used to target a sequence for gene knockin, knockout and replacement as well as monitoring and regulating gene expression at the genome and epigenome levels by binding a specific sequence. Agrobacterium-mediated method is still the major and efficient method for delivering CRISPR/Cas regents into targeted plant cells. However, other delivery methods, such as virus-mediated method, have been developed and enhanced the application potentials of CRISPR/Cas9-based crop improvement. PAM requirement offers the CRISPR/Cas9-targted genetic loci and also limits the application of CRISPR/Cas9. Discovering new Cas proteins and modifying current Cas enzymes play an important role in CRISPR/Cas9-based genome editing. Developing a better CRISPR/Cas9 system, including the delivery system and the methods eliminating off-target effects, and finding key/master genes for controlling crop growth and development is two major directions for CRISPR/Cas9-based crop improvement.
Introduction
Since crops were domesticated, scientists and breeders have been attempting to improve different traits for making crops better in growth, yield and quality as well as response to different environmental factors, including pests and other pathogens as well as various abiotic stress, such as drought and salinity. At the earliest stage, people just selected the good characters from the population growing in the field although they do not know what causes the difference. Until 1800 s, Gregor Mendel discovered the fundamental laws of inheritance, human started to clearly know that each crop trait is controlled by genes and that the combination of genes from both sides of parents controls the phenotype of an individual plant. Mendel's Law of Inheritance provides the foundation for traditional breeding and then for molecular and transgenic breeding. For a long period, breeders combine the good traits through cross and backcross breeding. Then, as the development of gene cloning and transformation technology, transgenic technology was become a method for improving specific trait in plants by inducing a foreign gene. Although great progress has been made and many transgenic plants have been widely adopted around the world that are also generating huge economic and environmental benefits, the majority of transgenic crops used in the field are insect- and/or herbicide-resistant crops due to the limitation of desirable foreign genes [1], [2]. Additionally, inserting these foreign genes into plant genome is always random and it may also affect other genes that results in gene silencing or altered expression [3]. Thus, more precision way is warranted to edit the genes for both gene function study and crop improvement.
Genome editing is a dream method for accurately and precisely recognizing and locating a specific sequence within a genome, and then altering the targeted sequences for various purposes. Unlike traditional transgenic technology that randomly inserts genetic materials into a genome, genome editing technology works on a specific genome sequence within the genome. To achieve this, genome editing requires a specific restriction nuclease that can recognize the specific sequences; these nucleases not only locate the genome position but also serve as molecule scissors to cut the specific sequences. Up to now, there are four major families of nucleases used for genome editing; these nucleases were developed at different time and they are meganucleases, zinc finger nucleases (ZFNs), transcription activator-like effector-based nucleases (TALEN), and the clustered regularly interspaced short palindromic repeats associated nucleases (CRISPR/Cas) [4]. Meganucleases, discovered in the late 1980 s, are the first nucleases used for genome editing, followed by discovering ZFNs and TALENs for genome editing [4], [5], [6]. All three technologies, particularly ZFNs and TALENs, have been extensively studied and applied to edit a range of genes in both plants and animals, and even generated some commercial products or used in clinical trials in treating human genetic diseases [7]. However, all these systems are difficult to use and all of them are lab- and cost-consuming. Later, CRISPR/Cas system caught scientists' attention and now become a popular method for editing a genome/gene.
CRISPR is an extensive class of short palindromic repeat sequences, which are widely existed in many prokaryotes, including most bacteria and Archaea. In prokaryotes, these short repeat sequences complimentary with some foreign DNA sequences, such as virus DNA, that invade bacteria or Archaea. When viruses infect bacteria, bacteria will produce this type of DNA and bind to virus DNAs; with working with one nuclease, called Cas, the Cas enzyme will cut the invaded DNA into pieces. Thus, CRISPR/Cas is a type of acquired immune defense mechanism for prokaryotes against virus. It is also a naturally occurred genome editing tool.
The first CRISPR was cloned from E. coli by accident in 1987 [8]. Later the similar CRISPR sequences were also cloned from other bacteria and Archaea [9], [10], [11], [12]. However, at the first almost one decade, the scientists did not know the function of these special repeated sequences and just thought they are a special sequence among different species and used them as strain typing [13]. In 1995, Mojica and colleagues constructed plasmid containing fragment of CRISPR sequences and then transformed into halophilic Archaea Haloferax volcanii; their results demonstrated that extra copies of CRISPR sequences resulted in the altered distribution of H. volcanii genome [14]. This is the first time that shows the incompatibility between external plasmid CRISPR and Archaea. However, their real function was unclear until that scientists found CRISPR-associated protein (Cas) genes [12] and their functions in bacterial defense mechanisms [15]. Since then, we understand that CRISPR/Cas system is a naturally occurred and acquired immune defense mechanism in bacteria and Archaea to against foreign DNA material infection, majorly the viruses. This also allows scientists to think a new way to edit a gene using this naturally occurred immune defense system. In 2012, two independent research laboratories reported that CRISPR/Cas system can be reconstructed in vitro and the in vitro reconstructed CRISPR/Cas systems had biological functions and were capable to cut an individual DNA sequence [16], [17]. This provides foundation for using CRISPR/Cas as a genome editing tool. Their studies and others also show that, to make CRISPR/Cas system function on genome editing, there are three required parts of components that are Cas enzyme, crRNA and tracRNA. Jinek and colleagues (2012) show that CRISPR RNA (crRNA) and trans-activating crRNA (tracRNA) can be constructed together to form a single chimeric synthetic RNA molecule, called single guide RNA (sgRNA) that work perfectly on the genome editing activities [17]. This research is significant to simply the DNA construction for making CRISPR/Cas system as a biotechnological tool for genome editing, that is always very simple compared with previously used genome editing tools, such as ZFNs and TALENs. Thus, these two studies opened the door and lead to the next levels for using CRISPR/Cas system to edit genome sequences for a variety of purposes. Followed these two studies, CRISPR/Cas systems were immediately employed to test the feasible genome editing in mammalian genomes. Cong and colleagues (2013) successfully knockout several genes in both human and mouse cell lines by constructing CRISPR/Cas systems using S. pyogenes Cas9 (SpCas9). Mali and colleagues (2013) also used Cas9 to knockout functional genes in several human cell lines. Both studies modified the Cas9 enzyme to allow its high function in human and mouse cells using a single crRNA-tracRNA fusion sgRNA transcript. Both studies also introduced multiple gRNAs for targeting multiple genes simultaneously and show the flexibility of multiplex editing [18], [19]. Following these two studies, CRISPR/Cas system have been quickly adopted by the scientific community to edit, regulate, and/or monitor genes in bacteria, animals, and plants.
CRISPR/Cas system working mechanisms and CRISPR/Cas family
The commonly used Cas endonucleases, Cas9, contain two nuclease domains, RuvC and HNH, and a PAM-interacting domain (PI) [20], [21]. RuvC and HNH domains clave the double-strand DNA and form a double-strand break (DSB). However, the function of RuvC and HNH domains requires the Cas nuclease to bind a specific DNA location that are protospacer adjacent motif (PAM) dependent. After Cas cuts the DNAs, DSBs will be repaired using the cell own DNA repair mechanisms. There are two repair pathways, one is non-homologous end joining (NHEJ) repair and another one is homology-directed repair (HDR). HDR usually requires a template DNA for DSB repair, that can be a foreign single- or double-strand exogenous DNA, the homologous chromosome, the sister chromatid, or even some sequences that are highly related to the sequences with DSBs.
There are two reasons for that Cas nuclease can precisely cut a specific sequence within the genome. One is that CRISPR/Cas system needs one gRNA that contains both crRNA and tracRNA, in which crRNA guides the Cas nuclease to the target sequence. In an engineered CRISPR/Cas system, the gRNA or sgRNA is usually a short synthetic RNA containing a scaffold tracRNA sequence and a spacer with about 20 nucleotides. The tracRNA serves as scaffold for Cas enzyme binding. The spacer is a sequence complementary with the target site, which is user-designed sequence and commonly called gRNA when we design the CRISPR/Cas system for gene editing. gRNA can target either strand of a gene due to the Cas enzyme have two nuclease domains, one cuts sense strand and another one cuts the anti-sense strand. However, not all sequences can serve as a CRISPR/Cas target site; this is because all target sites require a PAM sequence immediately adjacent to its target. PAM is required for Cas enzyme function and PAM sequence serves as a binding signal for Cas nuclease; without a PAM sequence, Cas enzyme does not know where to bind and where to cut the sequence. Thus, PAM sequence is required for all current CRISPR/Cas systems used for genome editing.
CRISPR/Cas systems are a big family; according to a recent study, it can be divided into 2 classes, 6 types and 33 subtypes based on several criteria, including sequence similarity, distinct features of the components and phylogenetic analysis [22]. As more and more new CRISPR/Cas are discovered, this classification will be expended in the future. The major difference between class 1 and class 2 CRISPR/Cas system is that, in the Class 1 CRISPR/Cas system, multiple Cas proteins work together to form a functional complex for degrading DNA sequences in which the complicate complex binds crRNA and target together for function. However, in class 2 CRISPR/Cas system, although there may be additional Cas proteins, only one larger Cas protein is needed to play the same function as the multiple Cas proteins in the class 1 system. Thus, for CRISPR/Cas system as a genome editing tool, class 1 CRISPR/Cas system is hard to be constructed and transformed into plant or animal cells, it is rarely used as a genome editing tool. Currently, all CRISPR/Cas genome editing is using the class 2 CRISPR/Cas system because it is easy to construct with straightforward protocol in which two components (one single Cas protein gene and a single gRNA including the tracRNA and crRNA) are only needed. Actually, for most commercially CRISPR/Cas systems, the only thing one research lab do is to design a gRNA and insert it to the construct. Thus, it is easy and simple with high successful potentials.
The class 2 CRISPR/Cas system currently contains 3 types (II, V and VI) and 17 subtypes. Because of its usage, this class of CRISPR/Cas systems has quickly expanded in the last 5 years from 2 types and 4 subtypes in 2015 to 3 types and 17 subtypes in 2020 [22]. Among these Cas enzymes, there are two special ones, one is Cas13 and another is Cas14. Cas 13 targets RNA sequences instead of DNA sequence [23] and Cas14 target ssDNA sequences instead of dsDNA sequences [24]. Cas14 is a family of Cas variants which enable high-fidelity SNP genotyping instead of genome editing [24]. Although there are many differences among these three types, including CRISPR sequences, as considering the aspect of genome editing tool, the major difference is that different type contains different Cas proteins. This has been well reviewed in a recent paper [22]. If you are interested in, please refer to that paper.
Different Cas enzymes, even closely related Cas enzymes from different species, recognizes different PAM sequences (Table 1) and show their specificity. For example, the commonly used SpCas9 only requires NGG PAM sequences, where N represent any nucleotide [16], [17]. However, the S. thermophilus Cas9 (StCas9) recognizes NNAGAAW PAM sequences, where W is A or T [25], [26]. Additionally, different CRISPR/Cas system also requires the PAM sitting at different direction of the target DNA sequence. For the majority of Cas enzymes, the required PAMs sit at the 3′ end of the target DNA sequences; however, for certain Cas enzymes, such as Cas12 (previous also known as cpf1), they require that the PAMs are located at the 5′ end of the targeted DNA sequences. Thus, when selecting a CRISPR/Cas system for genome editing, people need to pay attention which Cas they will employ and then pay more attention on guide RNA design to make sure that they target the right location and the designed gRNA works well. Due to the fact that sgRNAs are solely responsible for recruiting Cas9 and targeting on a specific genomic location, it is steamy important to optimize sgRNA design for successful gnome editing experiments. For designing a good sgRNA, the DNA sequences must contain an appropriate PAM and immediately adject to the gRNAs designed. Other strong criteria for designing gRNAs with high activity include: gRNAs are about 20 bp in length, gRNAs containing intermediate GC content is better than high or low GC content, and that a purine in the most PAM-proximal position can enhance Cas9 cutting efficacy [27]. The good thing is that there currently are many web-based computational programs we can use to design gRNAs in which the program will automatically search the right DNA sequences with right PAM requirement.
Table 1.
Major naturally occurring and genetically modified Cas enzymes used for genome editing.
| Name | Cas | Resources | CRISPR/Cas | PAM* | PAM location | Reference |
|---|---|---|---|---|---|---|
| SpCas9 | Cas9 | Streptococcus pyogenes | Type II | NGG | 3′ | [16], [17] |
| SaCas9 | Cas9 | Streptococcus aureus | Type II | NNGRRT | 3′ | [130] |
| FnCas9 | Cas9 | Francisella Novicida | Type II | NGG | 3′ | [131] |
| NmCas9 | Cas9 | Neisseria meningitidis | Type II | NNNNGATT | 3′ | [132] |
| CjCas9 | Cas9 | Campylobacter jejuni | Type II | NNNNRYAC | 3′ | [133] |
| St1Cas9 | Cas9 | Streptococcus thermophilus | Type II | NNAGAAW | 3′ | [25], [26] |
| St1Cas9 | Cas9 | Streptococcus thermophilus | Type II | NGGNG | 3′ | [25], [26] |
| AsCas12a | Cas12a(cpf1) | Acidaminococcus sp. | Type II | TTTV | 5′ | [134] |
| LbCas12a | Cas12a(cpf1) | Lachnospiraceae bacterium | Type II | TTTV | 5′ | [134] |
| FnCas12a | Cas12a(cpf1) | Francisella Novicida | Type II | TTTN or YTN | 5′ | [134], [135] |
| LsCas13# | Cas13 (C2c2) | Leptotrichia shahii | Type VI | [23] | ||
| Cas14& | Cas14 | Archaea | [24] | |||
| FnCas9 variant | Cas9 | Modified FnCas9 | Type II | YG | 3′ | [131] |
| Modified SpCas9 | Cas9 | Engineered SpCas9 | Type II | NGA or NAG | 3′ | [136] |
| SaCas9-KKH | Cas9 | Engineered SaCas9 | Type II | NNNRRT | 3′ | [29] |
| SpCas9-HF | Cas9 | Engineered SpCas9 | Type II | NGG | 3′ | [137] |
| eSpCas9 | Cas9 | Engineered SpCas9 | Type II | NGG | 3′ | [138] |
| SpCas9-NG | Cas9 | Engineered SpCas9 | Type II | NG | 3′ | [28] |
| xCas9 | Cas9 | Engineered SpCas9 | Type II | NG | 3′ | [139] |
| Sniper-Cas9 | Cas9 | Engineered SpCas9 | Type II | NGG | 3′ | [140] |
| evoCas9 | Cas9 | Mutated SpCas9 | Type II | NGG | 3′ | [141] |
| HypaCas9 | Cas9 | Mutated SpCas9-HF | Type II | NGG | 3′ | [142] |
| Cas9-NRNH | Cas9 | Engineered SpCas9 | Type II | NRNH | 3′ | [143] |
| SpG | Cas9 | Engineered SpCas9 | Type II | NGN | 3′ | [31] |
| SpRY | Cas9 | Engineered SpCas9 | Type II | NRN or NYN | 3′ | [31] |
* N is any nucleotide. R is A or G. H is A, C or T. Y is C or T. W is A or T. # Cas13 targets RNA sequences instead of DNA; $ Cas14 targets ssDNAs instead of dsDNAs, which doesnot require a PAM.
PAM requirements limited the DNA sequence accessible for genome editing. To edit genome sequences as more as possible, scientists have been attempted to expand the PAM sequences so we can make more genome sequence accessible (Table 1). Although different strategies have been employed, they can be divided into two major approaches: 1) discovering new resources with different PAM requirements, this strategy majorly focus on identifying new Cas proteins from new organisms, this is also the major contribution for the expanding Cas family. Due to this, the CRISPR/Cas system has been quickly expanding in the past couple of years [22]. Except the Cas binds to DNA sequences, the Cas binding to and editing RNA sequences is also discovered. 2) Modifying current Cas proteins, particularly on the currently commonly used SpCas9 proteins, to relax the PAM requirement. Nishimasu and colleagues (2008) structurally engineered SpCas9 and allowed the Cas9 loss of the base-specific interaction with the third nucleotide of PAMs, and then developed the engineered SpCas9-NG variant; this new variant relaxed the third nucleotide of SpCas9 PAM to become NG PAM recognition [28]; their study also show that SpCas9-NG can be used to edit DNA sequences with NG PAMs and induce indel mutations in human cell lines [28]. Other formed PAM modifications were also ready to use; studies show that the newly modified Cas9, such as SpCas9-NG and SpCas9-VQR can be accessible to the new DNA sequences that WT Cas9 can not [28], [29], [30]. Recently, Walton and colleagues (2020) employed structure-guided engineering approach to further modify the SpCas9 enzyme to SpG and SpRY [31]. Their study show that the modified SpRY relaxed all the three nucleotide requirement of PAM and allowed almost all DNA sequences accessible for CRISPR/Cas editing without change the Cas enzyme activities, including gene editing and base editing [31]. This modified CRISPR/Cas system will boost the application of CRISPR/Cas genome editing in both foundational and applied research [32].
CRISPR/Cas delivery methods
CRISPR/Cas-based genome editing is based on genetic transformation, in which we need to deliver the Cas enzymes and gRNAs into the cells and let them work together to edit the DNA sequences. Thus, plant transformation is a necessary and an important and first step for genome editing process. Thus, any traditionally used methods for transgenic plants can be used to deliver Cas and gRNAs.
Traditionally Agrobacterium-mediated gene transformation
As traditional transgenic technology, Agrobacterium-mediated gene transformation is also the dominant method for delivering both Cas protein genes and gRNAs into a plant cell, in which both Cas gene and gRNAs are inserted into the Ti plasmid with different promoters. To make sure both Cas and gRNAs are highly expressed in the targeted cells, CaMV 35S promoter is commonly used promoter to express Cas enzymes whereas U6 is commonly used promoter for gRNAs. However, other promoters are also used to express Cas proteins and/or gRNAs. To make each gene with high expression, the promoters isolated from protein-coding genes (e.g. CaMV 35S) are generally used for expressing Cas proteins; the promoters isolated from small RNA genes (e.g. U6) are commonly used for gRNAs majorly due to that gRNA genes are usually small sequences. Although most people use the promoters from model plant species, such as Arabidopsis, some studies also show that cloning the promoters from the target plant species may be better for high expression of the gRNAs with increase in genome editing efficiency.
Due to the fact that the majority of CRISPR/Cas-based genome editing is based on plant transformation that requires the transformed cells develop into an entire plant. Plant tissue culture and regeneration is still the major challenge for genome editing, which limits the application of genome editing in all plant species. If there is no plant tissue culture and regeneration system has been established for an individual plant species, it is hard to edit a gene in that species no matter how good you are on the genome editing. To overcome this obstacle, some in planta transformation methods have been adopted to obtain genome editing events. A recent study obtained genome editing events by de novo induction of meristems, in which they co-transferred CRISPR/Cas9 system with developmental regulators into tobacco plants [33]. Several studies show that developmental regulators, particularly SHOOT MERISTEMLESS (stm) and WUSCHEL (wus), allow the somatic cells reprogramming and differentiate into shoots [34]. After co-transformation of CRISPR/Cas9 system with developmental regulators in tobacco plants, plant somatic cell can directly differentiate into shoot after the genome editing events [33]. This method opens a new way to obtain genome editing events. However, more studies may need to be performed whether this method can used to obtain genome editing events in other plant species, such as agriculturally important crops. Currently, Agrobacterium-mediated CRISPR/Cas9 genome editing is the most commonly used delivery method; for the majority of plant species, this method can achieve a high efficiency compared with other delivery methods.
RNA and protein-level's delivery enhances the application of CRISPR/Cas system
Traditional transgenic technology, such as Agrobacterium-mediated gene transformation, require inserting the gene (DNA) sequences into plant genome for making it inheritable. For CRISPR/Cas-mediated genome editing, we do not need to deliver the final products into plant cell, we just need to deliver the molecular tool (both gRNAs and Cas protein) into the plant cells. Thus, it is not required to let the gRNAs and Cas stably expressed in the plant genomes. Thus, it becomes possible for scientists to directly deliver functional products (gRNAs and Cas RNA and/or protein) of gRNA genes and Cas genes for making CRISPR/Cas genome editing.
RNAs have been directly delivered into plant cells. Both Cas proteins and sgRNAs can be in vitro transcribed and even synthesized, then the RNAs are ready to be delivered into plant cells using direct methods, including protoplast transformation and particle bombardment-mediated transformation. Zhang and colleagues first coated golden particles using in vitro synthesized Cas9 RNAs and sgRNA, then they employed gene gun to direct deliver the RNAs into wheat immature embryos and successfully obtained genome-edited mutants [35]. Some studies also tried to deliver the Cas9 DNA/RNAs and sgRNAs using the plant virus vector and successfully obtained genome-editing mutants in different plant species [36], [37]. Cody and colleagues (2017) employed tobacco mosaic virus-derived vector to test the flexibility of delivering sgRNAs into plant cells and obtained genome editing events for all tested genes [38]. Mei and colleagues (2019) explored foxtail mosaic virus (FoMV) vector for delivering sgRNAs into corn, tobacco and other plant cell and also obtained genome editing events [36]. Recently, significant progress has been achieved in CRISPR/Cas9 genome editing using engineered virus to directly deliver Cas9 and gRNAs into plant cells by virus infection in planta. Ma and colleagues (2020) reported for the first time the delivery of the entire CRISPR/Cas9 cassette into plant cells by virus infection by an engineered plant negative-strand RNA virus, sonchus yellow net rhabdovirus (SYNV), and then they obtained single, multiplex mutagenesis, and chromosome deletions with a high frequency in tobacco [39]. Additionally, Ellison and colleagues (2020) used RNA viruses and mobile single guide RNAs to generate mutant progeny in the second generation of tobacco plants at frequencies ranging from 65 to 100% [40]. These two studies represent important advances in developing virus-mediated CRISPR/Cas reagent delivery methods. Combined these two methods may revolutionize CRISPR/Cas9 genome editing in plants [41]. The DNA-free delivery of CRISPR/Cas reagents by engineered virus vectors represents a promising approach for genome/gene editing.
In 2015, Woo and colleagues attempted to deliver the preassembled CRISPR/Cas9 ribonucleoprotein (RNP) complexes of purified Cas9 protein and guide RNA into plant cells by protoplast transformation and successfully obtained genome editing events on several plant species, including Arabidopsis, tobacco, rice, and lettuce [42]. Following, Malnoy and colleagues (2016) also investigated the possibility of direct delivery of purified CRISPR/Cas9 ribonucleoproteins into grape and apple cells using protoplast transformation treated by PEG6000; they successfully edited several disease-related genes, including mlo-7 in grape and DIPM-1, DIPM-2, and DIPM-4 in apple tree [43]. At almost the same time, Svitashew and colleagues (2016) successfully delivered pre-assembled Cas9–gRNA ribonucleoproteins into corn embryo cells by using gene gun direct transformation technology; because embryo cells have high potentials of plant regeneration, it is easy for them to obtain genome editing corn plants after they derived the CRISPR/Cas9 RNP complex into corn embryo cells [44]. This method was also successfully employed to obtain CRISPR/Cas-edited wheat [45], potato [46], Brassica [47], and rice [48].
Except gene knockout, RNP-mediated genome editing combing with donor DNA was also successfully employed to obtain knockin mutants by HDR. These studies and others suggest that RNP-mediated genome editing is a robust method for obtaining genome editing events in plants. One of the major advantage of RNP-mediated genome editing is that all genome editing event are transgene-free, that eliminates a lot of approval process and issues in which all transgenic products need to be federally approved before they move to the field and markets. Another advantage of RNP-mediated genome editing is that RNP transformation may reduce the off-target effects of CRISPR/Cas system compared with the Agrobacterium-mediated CRISPR/Cas genome editing [45]. Due to the short lifespan time of RNPs, the CRISPR/Cas system more likely produce low possibility of cutting other DNA locations except the real targeted sites.
Applications of CRISPR/Cas system on gene function study
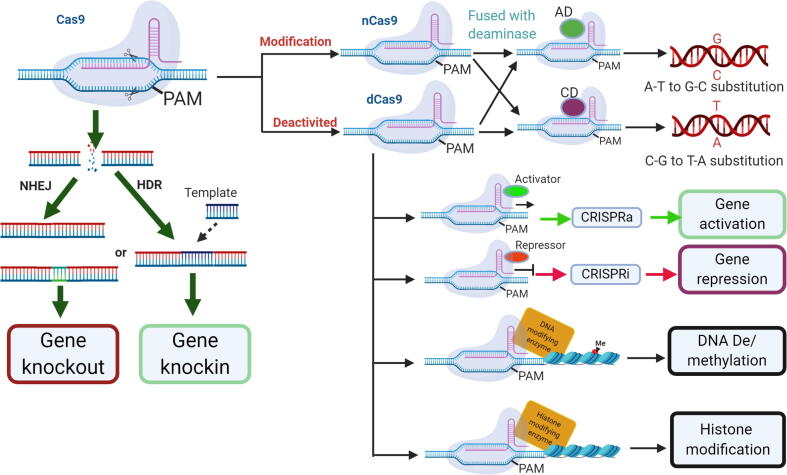
CRISPR/Cas-mediated genome editing has many potential applications, depending on the Cas enzyme activities and DSB repair mechanism. DSB repairs may cause several different consequences that lead to different applications (Fig. 1).
Fig. 1.
Application of CRISPR/Cas genome editing in gene functional study. CRISPR/Cas system has a diversity of application in gene functional study. Based the DNA double strand break (DSB) repair mechanism, CRISPR can directly cause gene knockout (silencing) by insertion or deletion of a couple of nucleotides and repaired by non-homologous end join (EHEJ); however, if the homologue-directed repair (HDR) happed, with a DNA donor, CRISPR/Cas genome editing can be used to replace an undesirable gene or over express (knockin) and an individual gene. If deactivating the Cas9 enzyme, and with transcription effector or other enzymes fused with the dCas9, CRISPR/Cas system also can be used to base editing, epigenome editing and imaging.
Gene knockout for silencing an individual gene
Gene knockout/silencing is the dominant application of CRISPR/Cas-based genome editing. Since its usability was recognized [16], [17], CRISPR/Cas system has been widely used in knockout of an individual gene in many plant species. The major reason is because of its gene editing and repair mechanisms. The most commonly used CRISPR/Cas9 and other CRSIPR/Cas systems usually cut the double-stranded DNA and generate a DSB. In the most cases, a DSB will be repaired by NHEJ. Due to that fact that random one or more nucleotides are deleted or added, it will interrupt the expression of the targeted gene because of the base frame shift if it is a protein-coding gene, genome editing usually results in gene silencing or gene knockout. Compared to T-DNA mutagenesis, CRISPR/Cas-caused gene silencing has many advantages. The most advantage is the specificity for CRISPR/Cas-based gene editing; it can be targeted on an individual gene without other side effects. Thus, CRISPR/Cas system can quickly and precisely silence a gene on purpose. This is particularly useful for studying a gene function and/or removing some undesirable traits controlling by a specific gene. Thus, since this method was developed, CRISPR/Cas system has been widely employed to silence an individual gene. However, this may majorly be used to study the function of a protein-coding gene because base frame shift will cause nonsense mutation and/or code a different protein. For non-coding RNA genes, particularly for small RNAs, such as microRNAs (miRNAs), an extensive class of small RNAs with versatile functions in almost all biological and metabolic processes in both plant and animal kingdoms [49], it is a little bit of difficult to silence them majorly due to that base frame shift generally did not affect the targeting of a miRNAs on a protein-coding gene. Thus, there are few studies on CRISPR/Cas9-based on genome editing although designing multiple gRNAs still can completely remove the miRNA genes.
Gene knock in for over-expressing an individual gene
Agrobacterium-mediated gene transformation is traditionally used to over express an individual gene in a plant cell through T-DNA insertion that contains the foreign DNA sequences. However, the random insertion of a targeted gene into a plant genome always causes many side impacts, including silencing other functional genes. CRISPR/Cas genome editing can cut the DNA sequence at a specific site and then HDR will insert a foreign DNA sequence (targeted gene) into this cleavage site. Thus, CIRSPR/Cas can be used to precisely insert a foreign gene into a specific location within a genome without interrupting other genes, another word avoiding position-effects. Compared with gene knockout, CRISPR/Cas-based gene knockin is much more challenge and a little bit of complicate with lower genome editing efficiency. By using geminiviral vectors to deliver enough amount of donor template DNAs into rice cells, Wang and colleagues (2017) obtained about 19% of gene knockin efficiency for CRISPR/Cas-based gene editing by using a report gene gfp [50]. Recently, Kim and colleagues (2020) modified the CRISPR/Cas gene knockin delivery system and successfully obtained gene knockin with high efficiency (37%) by using a combination of ribonucleoprotein (RNP) complex and DNA fragment (antibiotics resistance gene) that provided a selection marker for selecting genome editing events [51]. However, CRISPR/Cas9-based targeted insertion or replacement of long sequences and genes is still challenging in plants. Recently, Lu et al. (2020) used chemically modified donor DNA and CRISPR/Cas9 to knock in long DNA sequences and achieved the insertion of up to 2,049 base pairs (bp) into the rice genome at an efficiency of 25% [52]. They also developed a method for gene replacement with up to 130-bp sequences with 6.1% efficiency, which relied on homology-directed repair, chemically modified donor DNA and the presence of tandem repeats at target sites [52]. This study represents a breakthrough in CRISPR/Cas-mediated targeted sequence insertion and replacement in plants. As the modification of CRISPR/Cas-based gene knockin efficiency, CRISPR/Cas-based gene knockin will become another tool for over expressing certain genes without any position effects.
Gene replacement for gene mutations or undesirable genes
Due to the nature of HDR, any sequence can be insert and replace the original genome sequence around the DSB sites. Thus, if the gene sequences have mutations, even point mutations and/or any undesirable sequences, we can use a desirable gene/DNA sequence with homologous arms with the original sequences to replace them. As the same as CRISPR/Cas-based gene knockin, CRISPR/Cas-based gene replacement also remains a lot of challenging with low efficiency for delivering donor DNA repair/placement template and competing with NHEJ. Recently, Li and colleagues (2019) used transcript-template HDR to successfully obtain acetolactate synthase gene (ALS) replacement in rice through delivery of a DNA-free ribonucleoprotein complex [53].
Recently developed prime editing technology opens a new era for gene replacement for gene mutation and any undesirable gene. Prime editing was developed in 2019 by the Liu group at Harvard University [54], in which a reverse transcriptase is fused with Cas9 nickase (nCas9) and a prime editing guide RNA (pegRNA) was used to replace the traditional gRNA. nCas9 will cut the opposite strand of DNA and form a single strand DNA break where the broken DNA sequence will serve as a primer to bind the primer binding site in the pegRNA and synthesize a new DNA sequence using the CRISPR/Cas9-fused reverse transcriptase. Although the prime editing technology was first developed in animal system, it is quickly adopted to edit a specific gene sequence in plants. Currently, prime editing has been successfully to replace genes in several important plant species, including rice [55], [56], [57], [58], wheat [55], tomato [59], and maize [60].
CRISPR/Cas-mediated base editing
Point mutants is a common phenomenon in plant genome and genetics. Many plant traits, including resistance to different diseases, are controlled/regulated by single nucleotide polymorphism (SNP). Thus, it is important and also necessary to develop a genomic tool for precisely changing a single nucleotide within a genome. Although HDR-based CRISPR/Cas9 can be used to induce and/or create a single nucleotide change by inducing a DNA template containing the desirable nucleotide substitution. However, HDR is usually at a very low frequency and the change is very low for repairing the point mutants, particularly in the undivided cells. Due to the huge potentials of CRISPR/Cas9 system and the wide application on single base genome editing, scientists quickly modified CRISPR/Cas system to become a CRISPR/Cas base editor for editing certain bases within a genome. Currently, CRISPR/Cas-based base editing has become one of the major progresses and application of this emerging cutting-edge technology.
The principle of CRISPR/Cas9-based base editing is that gRNA can recognize a specific DNA sequence and then covert a specific base into a different base. To achieve this, Cas9 enzyme can not generate DSBs. Thus, scientists modified the wide type of Cas9 enzymes into nickase Cas9 (nCas9) and later even deactivated Cas9, called dead Cas9 or dCas9. Both nCas9 and dCas9 can bind to the specific DNA sequence with the help of gRNAs. Then, scientists fused a base conversion enzyme to change the specific DNA base. Currently, there are two base covert enzymes used, one is cytidine deaminase (CD) and another one is adenine deaminase (AD). CD converts nucleotide substitution from C to U, and then become T to achieve the substitution from C·G to T·A. AD converts nucleotide substitution from A to I and then become G to achieve the final substitution from A·T to G·C. Cytidine deaminase-mediated base editing (CBE) has been well studied in many plant species, including major crops, such as wheat and corn [61], [62]. CBE is also employed to create herbicide-resistant plants and also for other proposes [63], [64]. Recently, CRISPR/Cas9 base editing is also employed to create knockout mutants by change the sense code to nonsense code [65], [66]. Compared with NHEJ of CRISPR/Cas9-generated knockout, CRISPR/Cas base editor-induced knockout has more advantages, including easy operation and high efficiency.
CRISPR/Cas-mediated gene regulation
The reason of CRISPR/Cas9 system-mediating gene editing is that there are two domains (HNH and RuvC) cutting the targeted double stranded DNA sequences to generate DSBs. If letting one domain loss activities by inducing mutation inside the functional domains, Cas9 enzyme will not work appropriately in which another domain still works and only generate a single strand break of DNA. If both domains loss their cutting functions, there will be no DNA cutting happened. Based on this principle, scientists have been modified Cas enzyme and deactivated Cas9 or dead Cas9 (dCas9) can be used to other purposes, such as regulating gene expression and epigenome editing as well as localization of a gene for imaging.
Deactivating the nuclease function of Cas9 HNH and RuvC does not affect the binding activity of dCas9 to the target DNA sequences. Studies also show that after Cas9 binds to a target DNA sequence, it will block other protein to bind the same DNA sequence. Based on this principle, scientists have developed CRISPR/dCas9 system as a CRISPR/dCas interference (CRISPRi) system to inhibit transcriptional process and used as a gene knockdown approach for regulating gene expression [67]. CRISPRi can be used to knock down a gene expression by blocking transcription initiation and or transcription elongation based on where the CRISPR/dCas system binds to. If the system binds to the promoter region, the CRISPR/dCas system will block transcription factor and RNA polymerase (RNAP) binding to the promoter region and inhibit transcription initiation; otherwise, if the CRISPR/dCas system binds to the coding region, it will block RNAP binding and inhibit transcription elongation [67]. In both cases, CRISPR/dCas will serve as an RNA-guided DNA-binding complex that blocks the gene expression at the transcriptional levels. More excitingly and usefully, when a repressor is fused to the dCas9 protein, the CRISPR/dCas9 system inhibits gene initiation and elongation more strongly [68]. At the same time, if a transcription activator binds to the dCas9 protein, the CRISPR/dCas system will enhance the expression of the targeted genes [68]. Recently, Papikian and colleagues (2019) employed CRISPR/dCa9-SunTag system, in which they fused dCas9 with the transcriptional activator VP64, to drive robust and specific activation of several loci in Arabidopsis; this system worked very well in many genes, including protein coding genes and transposable elements, in diverse chromatin contexts [69]. By fused transcription activator VP64 with dCas9, Lee and colleagues (2019) targeted floral regulator FLOWERING LOCUS T (FT) gene and they obtained Arabidopsis plants with early flowering phenotype [70]. Some editing plants even flowered at five leaf stage. Gene expression analysis show that FT gene was strongly and significantly unregulated by the CRISPR/dCas9-VP64 compared with the wild type of Arabidopsis seedlings [70].
Thus, CRISPR/dCas system is also a great tool of regulating gene expression at the transcription levels, and deep sequencing analysis showed that CRISPR/dCas-based gene regulation is highly gene specific [68].
CRISPR/Cas-mediated epigenome editing
Unlike the classical Mendelian genetics, epigenetic inheritance generally can not be inherited from generations to generations due to the factor that epigenetic change does not alter the DNA sequences but modifying the DNA sequence for controlling their expression levels. More and more evidence show that epigenetics plays an equal important role on controlling plant traits. If it is said that traditional genetics (DNA sequences) determine the traits, then epigenetics can control whether the traits appear. Thus, epigenetic study has been becoming one of the hottest research topics in the last couple of years. There are many layers for epigenetics to regulate the gene expression, which majorly contain DNA methylation, histone modification, and non-coding RNAs.
DNA methylation is a common gene regulation mechanism. It is a nucleotide at a specific location which is methylated or demethylated. Generally speaking, a gene with methylation is usually silenced. In plants, CG (or CpG), CHG, or CHH (H is A, T or C) are three common sequence contexts with high frequency of methylation [71]. Regulating the methylation and demethylation of a gene can be an efficient way to control the expression of this gene and further control the trait controlled by this gene. Thus, as the CRISPR/Cas system was approved to be an efficient genome-editing tool, scientists have been attempting to modify this tool to edit DNA methylation. In this case, dCas9 will be fused with a methyltransferase or a demethylase. Due to dCas9 losing catalyzed function but it still tightly binds to the target site, the enzyme (methyltransferase or demethylase) can methylate or demethylate the nucleotides. By utilizing the CRISPR/dCas-SunTag system with the catalytic domain of the Nicotiana tabacum DRM methyltransferase, Papikian and colleagues (2019) developed a robust CRISPR/Cas9-based methylation targeting system for plants and this system can efficiently target DNA methylation to specific loci in plants [69]. In another study, Devesa-Guerra and colleagues (2020) linked the catalytic domain of Arabidopsis ROS1 5-mrthylcytosine (5-meC) DNA glycosylase to dCas9 and they found that dCas9-ROS1, but not dCas9-TET1, induced partially demethylation of the methylation-silenced genes in a replication-independent manner [72]. They also found dCas9-ROS1 as well as two different CRISPR-based chromatin effectors (dCas9-VP160 and dCas9-p300) generally decreased the methylation density of targeted DNA sequences [72].
Histone posttranslational modification is another major factor epigenetically controlling gene expression. Histone protein is one important part of DNA package and chromatin modeling. The activities and structures of histone proteins significantly affected the activities of genes associating with them in the structures. Methylation and acetylation are two commonly modification of an amino acid within a histone protein. Each modification will affect the chromatin remodeling and further affect the gene function associating with the chromatin structures. Thus, employing CRISPR/Cas system to change the status of histone proteins, including methylation and acetylation, will change the epigenetic and genetic features of chromatin and affect gene functions. Recently, Lee and colleagues (2019) fused p300 with dCas9, which is an H3K27 histone acetyltransferase associating with gene activation. After they expressed both histone-modified constructs in Arabidopsis seedlings, they observed the altered flowering time in transgenic Arabidopsis [70]. Chromatin immunoprecipitation (ChIP) analysis showed that H3K27 acetylation (H3K27ac) was increased by about two-folds in CRISPR/dCas9-p300 plants compared with the wild type controls; however, no FT gene expression change was observed between the CRISPR-edited lines and the wildtype controls [70]. This suggests that CRISPR/dCas system is efficient to modify histone marks and further affect plant traits.
Non-coding RNAs (ncRNAs) are an extensive class of regulatory RNA molecules, which are divided into long non-coding RNAs (lncRNAs) and small non-coding RNAs. There are many classes of small ncRNAs, including well known microRNAs (miRNAs) and small interfering (siRNAs) as well as piwi-interacting RNAs (piRNAs) and repeat associated small interfering RNAs (rasiRNAs). It is already well studied that both small RNAs, particularly miRNAs, and lncRNAs play a versatile role in regulating gene expression in almost all biological and metabolic processes [73]. Unlike protein-coding genes in which deletion and/or addition of several nucleotides will result in base frameshift and further cause gene silencing, CRISPR/Cas-caused deletion or addition do not change too much on the non-coding RNA, particularly do not affect too much on mature miRNA biogenesis which is the functional part of miRNA genes [74]. Thus, there are few successful reports on CRISPR/Cas9-based gene editing for miRNAs. Although there are several reports, the majority of them designed two gRNAs and completely removed the miRNA gene sequences [75], [76]. The major reason is that a miRNA sequence, particularly the seed region of a miRNA, is too short, it is hard to find a PAM sequence that are required for CRISPR/Cas genome editing. As more diversity of Cas proteins are identified and continuously modifying current Cas proteins, the PAM requirement will be relaxed, and more genetic loci will become accessible by CRISPR/Cas system. For example, a current research by Walton and colleagues structurally modified the most commonly used SpCas9 to SpRY that completely relaxes the NGG PAM to near-PAMless NRN or NYN PAM [31]. This modification allows the CRISPR/SpRY system accessible to almost all genetic locations, including miRNA sequences and SNPs [32]. Thus, engineered CRISPR/SpRY will boost genetic research and gene function study for both fundamental and applied research [32].
Except base, gene, and epigenome editing, CRISPR/Cas system also can be used to image the chromatin and localize the genes [71]. As new discovery, more applications will become possible on other gene function study.
Application of CRISPR/Cas on crop improvement
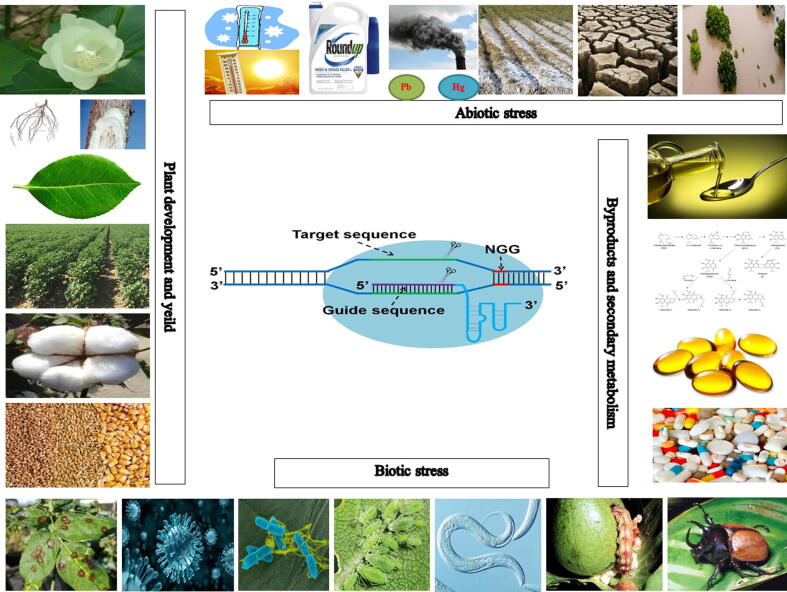
Since it was recognized as a cutting-edge tool for genome/gene editing, CRISPR/Cas system has been quickly attracted attention from both scientific researchers and industries for improving crop breeding with promising achievement. In a short period since it was applied on plants, the CRISPR/Cas-based genome editing systems have been established for almost all major crops, including rice, wheat, and cotton [77]. Currently, the CRISPR/Cas genome editing has become a mature biotechnological tool for improving crop growth and development as well as response to various environmental stress (Fig. 2).
Fig. 2.
Application of CRISPR/Cas genome editing in crop improvement, CRISPR/Cas is a robust tool and have huge potentials on crop improvement. Though targeting an individual DNA sequence, CRISPR/Cas can be used to add and/or change the expression of an individual gene that controls plant tolerance to abiotic and biotic stresses as well as plant growth and development. CRISPR/Cas also can change the secondary metabolism for controlling the biosynthesis of protein, carbohydrate, oil, and functional components. Partial figures are modified from previous publications [74], [108].
CRISPR/Cas genome editing improves plant tolerance to pathogen and insect infection
Pest- and pathogen-caused diseases are a big problem for many plants, including the major crops, such as cotton and wheat. Biotic factors not only affect plant growth and development but also decrease crop yield and quality, even result in complete yield loss in certain cases.
It is a complicated process and mechanism for plant response to various diseases; there are many genes associated with this process. Although transgenic technology has been employed to overexpress certain genes for obtaining transgenic plants with high tolerance to different pathogen, a little progress has been made due to the fact that no single gene plays a significant dominant role in plant response to diseases. However, it seems that knockout of an undesirable gene offers plant tolerance to those pathogens. Thus, since CRISPR/Cas9 was used as a genome editing tool, it has been quickly employed to create genome editing mutants with plant resistance to various diseases, caused by fungus, viruses, and bacteria (Table 2).
Table 2.
CRISPR/Cas-based crop improvement with resistance to virus, bacterial and fungal diseases.
| Crop | Targeted gene | Disease resistance | Results | CRISPR/Cas system | Transformation method | Type 0f mutation | Reference |
|---|---|---|---|---|---|---|---|
| Watermelon | Clpsk1 | Fungus | Resistance to Fusarium oxysporum f.sp. niveum | CRISPR/Cas9 | Agrobacterium | Stable | [83] |
| Arabidopsis | eIF(iso)4E | potyvirus | Resistance to Turnip mosaic virus (TuMV) | CRISPR/Cas9 | Floral dip | Stable | [87] |
| Rice | Os8n3 | Bacteria | Resistance to Xanthomonas oryzae pv. oryzae (Xoo) | CRISPR/Cas9 | Agrobacterium | Stable | [90] |
| Rice | OsERF922 | Fungus | Resistance to Magnaporthe oryzae | CRISPR/Cas9 | Agrobacterium | Stable | [85] |
| Tomato | SlJAZ2 | Bacteria | Resistance to Pseudomonas syringae pv. tomato (Pto) DC3000 | CRISPR/Cas9 | Agrobacterium | Stable | [93] |
| Citrus | CsLOB1 | Bacteria | Resistance to Xanthomonas citri subsp. citri (Xcc) | CRISPR/Cas9 | Agrobacterium | Stable | [91] |
| Orange | CsWRKY22 | Bacteria | Xanthomonas citri subsp. citri (Xcc) | CRISPR/Cas9 | Agrobacterium | Stable | [92] |
| Cucumber | eif4e | Virus | Resistance to Cucumber vein yellowing virus, Zucchini yellow mosaic virus and Papaya ring spot mosaic virus‐W | CRISPR/Cas9 | Agrobacterium | Stable | [88] |
| Cotton | 14–3-3 | Fungus | Resistance to Verticillium dahliae | CRISPR/Cas9 | Agrobacterium | Stable | [86] |
| Wheat | mlo | Fungus | Resistance to powdery mildew | CRISPR/Cas9 | Gene gun and protoplast | Stable | [80] |
| Tomato | SiMlo | Fungus | Resistance to powdery mildew | CRISPR/Cas9 | Agrobacterium | Stable | [81] |
Powdery mildew is a common disease caused by fungal pathogens in many plant species, including agriculturally important crops, such as wheat and tomato [78]. A study shows that MILDEW-RESISTANCE LOCUS (MLO) genes may be responsible to respond powdery mildew infection and inhibition of mlo genes repressed powdery mildew diseases in barley [79]. Just after CRISPR/Cas9 system was applied in plant research, Wang and colleagues (2014) immediately adopted the CRISPR/Cas9 to knockout mlo gene in hexaploid bread wheat and confirmed that transcription activator–like effector nuclease (TALEN) knockout of three mlo genes in wheat confers wheat resistance to powdery mildew disease [80]. This is first report attempting to create disease resistant materials in plants. Since then, more and more researches have been performed by using CRISPR/Cas technology to obtain genome editing materials in a wide variety of plant species. Mlo gene was also CRISPR/Cas9 knocked out in tomato and transgene-free tomato plants showed significantly resistance to powdery mildew without affecting other agronomical traits [81].
Fusarium wilt is another most severe vascular fungal diseases in many plant species, including watermelon, which is caused by Fusarium oxysporum f.sp.niveum (FON). FON can cause up to 30–80% and even more yield loss in watermelon [82]. However, there are currently no FON-resistant germplasm that limits the traditional breeding for breeding FON resistant cultivars [83]. Recently, Zhang and colleagues (2020) successfully employed CRISPR/Cas9 system to knockout Clpsk1 gene that encodes phytosulfokine (PSK) precursor, a disulfated pentapeptide plant hormone regulating plant immunity [84]. Their results show that watermelon with Clpsk1 knockout significantly enhanced plant resistance to Fusarium wilt disease [83]. In rice, Wang and colleagues (2016) silenced ERF transcription factor gene OsERF922 by CRISPR/Cas9, their results show that all tested genome-editing rice plants were significantly enhanced in resistance to rice blast in both seedling and tillering stages [85]. CRISPR/Cas9 knockout of 14–3-3 genes conferred plant tolerance to Verticillium dahliae in cotton with significant decrease in both disease plant rate and disease index; the total fungal biomass on the infected cotton leaves was also significantly decreased by 80% [86].
CRISPR/Cas technology has also been employed to create genome-editing plants with high resistance to viruses. Plant viruses ubiquitously exist in natural environment and frequently infect plants and cause significant effects on plant growth and development. In 2016, Pyott and colleagues employed CRISPR/Cas9 technology and successfully knocked eIF(iso)4E gene out in the model plant species, Arabidopsis. Their results show that eIF(iso)4E knockout mutants resisted to the infection of turnip mosaic virus (TuMV), a major pathogen in field-grown vegetable crops, without affecting other traits, including biomass and flowering time [87]. Chandrasekaran and colleagues (2016) also demonstrated that knockout of eif4e significantly improved cucumber resistance to a broad range of disease, including Cucumber vein yellowing virus (Ipomovirus), potyviruses Zucchini yellow mosaic virus and Papaya ring spot mosaic virus‐W [88].
Many bacteria cause a variety of plant diseases. Such as, bacterial blight is a globally devastating disease in rice, which is caused by Xanthomonas oryzae pv. oryzae (Xoo). Xoo requires a specific class of transcription activator-like (TAL) effectors to bind certain host disease-susceptibility genes for infecting the plants. Rice Os8n3 (also called OsSweet11) gene is one of these susceptive genes that Xoo TAL binds to and makes rice susceptive to Xoo infection and further causes bacterial blight [89]. Kim and colleagues (2019) employed CRISPR/Cas9 to knockout Os8n3 gene in rice, the genome-editing rice plants show significant resistance to Xoo bacterial infection [90]. In citrus, Peng and colleagues (2017) knocked out citrus susceptibility gene CsLOB1 by CRISPR/Cas9 and the genome editing citrus showed the enhanced resistance to citrus canker, caused by Xanthomonas citri subsp. citri (Xcc) [91]. Knockout of transcription factor CsWRKY22 gene also show enhanced resistance to citrus canker in the Wanjincheng orange [92]. Ortigosa and colleagues (2018) silenced SiJaz2 gene in tomato using CRISPR/Cas9 and gene editing plants show resistance to bacterial speck disease but not to the necrotrophic fungal pathogen Botrytis cinerea [93].
Compared with creating disease-resistant plants by using CRISPR/Cas9 technology, there are very few report on the application of CRISPR/Cas technology on creating insect-resistant plants although there are several reports on CRISPR/Cas-based editing of insect genome for pest management [94].
CRISPR/Cas genome editing improves plant tolerance to abiotic stresses
As global warming, abiotic stress is becoming more and more serious for crop growth and development. The most common abiotic stresses include drought, salinity, extreme temperatures as well as environmental pollutions. Each of them significantly affects plant growth and development and further affect plant biomass, yield, and quality.
Lots of structural and regulatory genes, including non-coding RNAs, are involved in plant response to different environmental stresses [95]. All these genes can be the target for improving crop tolerance to abiotic stresses by using traditional transgenic technology and the currently advanced genome editing tools. However, due to the fact that thousands genes may be associated with a single stress condition in a plant and not a single gene plays a dominant role during this process, unlikely as biotic stresses, a little progress has been made in creating genome editing mutants for improving plant tolerance to abiotic stresses by using CRISPR/Cas technology [96]. Recently, CRISPR/Cas9-edited AGROS8 gene enhanced maize tolerance to drought, in which ARGOS8 is a negative regulator of ethylene response [97]. This suggests the potentials of CRISPR/Cas technology on improving plant tolerance to abiotic stresses. SlAGAMOUS-Like 6 (SlAGL6) is an R gene, knockout of this gene by CRISPR/Cas9 technology allowed tomato plant grew better and developed fruit even at the condition of heat stress [98]. Under high temperature treatment that severely hampers fertilization-dependent fruit set in the wild type tomato, genome editing tomato plants demonstrated improved fruiting setting with the capability of fruit production, and tomato sexual reproduction capacity is maintained under heat stress [98]. A recent study shows that CRISPR/Cas-edited Arabidopsis mutants of dpa4-sod7-aitr256 enhanced plant tolerance to drought treatment [99]. Knockdown and knockout of ARF4 transcription factor gene improved the water usage efficiency in tomato and allow the editing plants more tolerance to salinity and osmotic stress [100]. CRISPR/Cas-generated mutants of G protein genes gs3 and dep1 enhanced rice tolerance to various abiotic stresses, especially for salinity stress [101]. Knockout of ppa6 gene enhanced rice tolerance to alkaline stress [102].
Although most studies show that CRISPR-edited plants show improvement of plant tolerance to abiotic stresses, some studies also show the opposite phenotype. For example, CRISPR/Cas9-knockout of nonexpressor of pathogenesis-related gene 1 (npr1) reduced plant tolerance to drought stress in tomato [103]. CRISPR/Cas9 knockout mutants of PtrADA2b-3 also show highly sensitive to drought treatment in Populus trichocarpa [104].
CRISPR/Cas system not only be used to knockout an individual gene, but also can be used to activate the expression of certain genes. In a recent study, Roca Paixao and colleagues (2019) fused a histone acetyltransferase with deactivated CRISPR/dCas9 system to activate the endogenous promoter of abscisic acid (ABA)-responsive element binding protein gene 1 (AREB1), the results show that the genome-edited plant were highly expressed in AREB1 gene and improved drought stress response in Arabidopsis [105].
CRIPSR/Cas genome editing improves plant growth and development
CRISPR/Cas system has been widely adopted to study the gene function on plant growth and development, and it also has been used as a robust tool for improving plant morphology for enhancing plant growth and development. Arginase (ARG) is an important enzyme regulating root development by targeting nitric oxide synthase (NOS). Over expression of ARG inhibited the formation of lateral roots [106]. Recently, Wang and colleagues (2017) employed CRISPR/Cas 9 technology to knockout out the ARG gene in cotton. For T1 generation of genome knockout lines, no matter in high or low nitrogen medium, the ARG knockout out lines show significantly increases in the number of laterals roots by 25% and in the total root surface area by 52% [107]. This suggests that CRISPR/Cas9-edited ARG plants have better root development that will enhance genome-edited plant absorb more water and nutrients from the soil and finally enhance plant growth and development and response to various environmental stresses [108].
CRISPR/Cas9-based genome editing demonstrated the essential regulatory function of MADS box transcription factor gene MADS78 and MADS79 in endosperm cellularization and early seed development in rice [109]. Single knockout mutant of MADS78 or MADS79 showed precocious endosperm cellularization and double mutants inhibited seed development and failed to make viable seeds in rice [109]. Rice hexokinase hxk5 gene mutant created by CRISPR/Cas9 results in male sterility on rice [110]. Knockout of COPII components sarib and saric genes also altered pollen development and caused male sterility in Arabidopsis [111]. Bnspl3 mutants, generated by CRISPR/Cas9 technology, exhibited developmental delay phenotype in allotetraploid oilseed rape [112]. CRISPR/Cas9 knockout of terminal flower1 (tfl1) gene altered the phase change and flowering timing in Brassica napus [113]. Knockout of tfl1 gene altered plant architecture, particularly on plant height and branching in Brassica napus [113]. Inducing mutants in S and SP promoter by CRISPR/Cas9 technology resulted in improved inflorescence and plant architectures in tomato [114].
CRIPSR/Cas genome editing improves crop yield
It is always a long-term goal to improve crop yield through traditional breeding and transgenic technology. However, due to the fact that yield trait is a complicated trait controlling by multiple genes and gene network, scientists and breeders have not made big progress by gene cloning and transgenic approach. Although it is nearly impossible to find a single gene majorly controlling yield, many studies do show that the expression of certain genes negatively affect crop yield. If this type of genes were inhibited and even removed, it may directly increase crop yield. The quickly developed CRISPR/Cas system provides a perfect tool for knockout of genes negatively controlling yield in plants. A study performed by Li and colleagues (2016) shows that knockout of crop yield negative regulators, gn1a, dep1 or gs3 gene, significantly enhanced certain traits associating rice yield, which include larger grain size, improved grain weight, grain number, dense, and erect panicles [115]. Another study, also in rice, Xu and colleagues (2016) employed CRISPR/Cas9 technology to knockout gw2, gw5 and tgw 6, grain weight negative regulators; the genome editing mutants show enhanced grain weight and size [116]. Rice seed weight and size were also controlled by vacuolar invertase2 (vin2) gene; CRISPR/Css9 knockout mutants show reduced seed size and grain weight majorly due to controlling sucrose flux into developing seed grains [117]. Grain weight was improved in wheat by CRISPR/Cas9-edited mutants of GASR7 [118]. Larger fruit size was achieved in tomato by CRISPR/Cas-edited cis-regulatory element CLV-WUS [114].
A field test shows that transgenic corn with CRISPR/Cas-edited AGROS8 gene increased grain yield by five bushels/acre under drought treatment at the flowering stage compared with the controls [97]. In rice, CRISPR/Cas9-edited variants at the 3′-end of OsLOGL5′s coding sequence (CDS) significantly increased grain yield under drought and low nitrogen treatment as well as well-watered and normal nitrogen control condition, this increase was consistence at multiple geographical locations [119]. Knockout of G protein genes gs3 and dep1 allowed rice to grow more preferable agronomic traits, including grain size and number per panicle [101]. This suggests that CRISPR/Cas genome editing is becoming a new approach for precision breeding for improving crop yield.
CRIPSR/Cas genome editing improves secondary metabolism and crop quality
CRISPR/Cas technology was also employed to explore the possibility to modify secondary metabolism in plants for improving crop quality. Protein, carbohydrate, oil, and bioactive compounds are the major functional compounds in food and their composition decides the quality of a crop product. Lycopene is a well-known functional bioactive component for treating several human diseases, including cardiovascular diseases and certain cancers. Recently, Li and colleagues (2018) enriched lycopene biosynthesis and production in tomato fruits by CRISPR/Cas-mediated multiplex genome editing, in which they edited several genes associated with the carotenoid metabolic pathway [120].
Protein is a major nutrient in crop products. However, the composition of protein decides the quality of protein mixture. There are about 1–2% of human population who are hypoimmunogenic sensitive to gluten that are widely existed in wheat and some other products. By employing CRISPR/Cas9 genome editing technology, Sanchez-Leon and colleagues targeted one class of α-gliadin gene and obtained low-gluten transgene-free wheat [121]. CRISPR/Cas-based knockout of d-hordein gene resulted in increased protein matrix surrounding the starch granules in barley [122]. CRISPR/Cas9 genome editing of seed storage protein CRUCIFERIN C (CRUC) gene altered the protein profiles in Camelina sativa seeds [123]; at the same time, amino acid composition was significantly changed in the genome editing plants with an increase in alanine, cysteine, and proline and an decrease in isoleucine, tyrosine, and valine [123].
CRISPR/Cas-based genome editing is also used to modify the carbohydrate production and composition in crops. Grains with high amylose content and resistant starch has human health benefits. CRISPR/Cas9 genome editing shows that knockout of pat2/5 genes resulted in increased accumulation of starch in the mature seeds of allotetraploid rapeseed [124]. Sun and colleagues (2018) employed CRISPR/Cas technology to successfully knockout the starch branching enzyme (SBE) gene SBEIIb and generated genome-edited rice with high content of amylose, in which up to 25% of amylose was generated with higher proportion of long chain in debranched amylopectin in rice seeds [125]. The similar results were also obtained in sweet potato by knockout of SBEII gene using CRISPR/Cas9 while the genome editing plant did not affect total starch contain [126]. Altered starch quality was also achieved in potato by CRISPR/Cas9-editing the granule-bound starch synthase (GBSS) gene [127]. CRISPR/Cas-based knockout of d-hordein gene resulted in decreased size of starch granules and a considerable decrease in the prolamines and an increase in the glutenins in barley [122].
Oil is another major trait that scientists want to improve by using CRISPR/Cas9 genome editing technology. Karunarathna and colleagues (2020) obtained enhanced seed oil content without adverse effects on other agronomic traits, such as seed germination and vigour in rapseed by simultaneous CRISPR knockout of several BnSFAR4 and BnSFAR5 genes [128]. In Brassica napus, knockout of tt8 gene enhanced oil and protein content as well as altered fatty acid (FA) composition in the seed, but the genome editing plant did not cause any serious defects in the yield-related traits [129].
Except the application of CRISPR/Cas technology are mentioned here on crop improvement, CRISPR/Cas genome editing is also employed to study crop domestication and certain unique traits in a specific crop. For example, CRISPR/Cas9 was used to knock out an MYB transcription factor gene and the edited cotton became fibreless [77].
Future perspectives
Although it is a short period since the naturally occurred genome editing tool was recognized, CRISPR/Cas system has become one of the most powerful tools in the history for molecular biology study (Fig. 1). Quick development of CRISPR/Cas-based genome editing holds great promise to both fundamental and applied research, which has been becoming a powerful biotechnological tool for addressing important biological and agricultural issues. CRISPR/Cas-based genome editing will also be becoming one of the major breeding technologies for crop improvement (Fig. 2). In the past 8 years, no technology as the CRISPR/Cas made so quickly developed and applied in biological and biomedical research. However, there are still many questions to be solved for wider application of this powerful system.
Developing a PAM-independent CRISPR/Cas system will enhance its potential application
All currently widely used CRISPR/Cas system requires a specific PAM sequence. PAM sequences guide the editing sites of an individual event and provide a specificity. However, PAM requirement limits the genes that CRISPR/Cas can be accessible for efficient editing. Developing a PAM-independent CRISPR/Cas system will boost the application of CRISPR/Cas on both fundamental and applied research, including gene function study and precision breeding. Although scientists have been attempting to identify new Cas proteins and modifying current Cas enzymes for expanding PAM variants and the DNA sequences that CRISPR/Cas can target are still very limited, more research is warranted. Exploring new Cas resources may be the easiest and most useful way to expand PAM variants. In the past 8 years, most PAM expansion is contributed by finding new Cas resources, including finding CRISPR/Cas can bind to RNA sequences for RNA editing [22]. There are thousands of bacteria and Archaea, currently there are only limited number of them explored; as timing and more research going, it is undoubted that more Cas resources will be identified with different PAM variants. Secondly, it is also a smart idea to continuously modify current Cas variants, particularly the most commonly used ones, including Cas9, Cas12, and Cas13. Many experiments show that modification of Cas enzymes can relax the PAM requirement and create new tool for gene function study, such as deactivated Cas has lots of more application, including gene regulation, epigenome target, and imaging.
Developing an efficient delivery method for CRRISPR/Cas system is needed
No matter what purpose, CRISPR/Cas-based genome editing event is a single cell editing event, it do requires the single cell to become an entire plant for later study and application. Thus, plant regeneration from a single cell is required, even for some in planta transformation. Currently, plant transformation and regeneration system are still the bottleneck for genome editing plants. Thus, development of an efficient delivery and plant regeneration system is important and necessary.
The success of transformation/genome editing mainly depends on the ability of the recipient cells to regenerate plants. At the early stage, it is hard for many plant species for obtaining regenerated plants, such as in rice, corn, and wheat. Later, scientists have attempted to culture immature seeds/embryos that have high potentials to become an entire plant and quickly develop an efficient tissue culture and plant regeneration system in an individual plant species. In the future, we may focus on more immature seeds and even seeds for plant transformation and genome editing [108].
Delivery method of CRISPR/Cas system is also important. It not only affects the transformation efficiency, but also impact the off-target effect and later regulation purpose. Traditional T-DNA-mediated transgenic and genome editing usually inserts the T-DNA and/or targeted genes into plant genome; those genes will be consistently expressed and inherited to the following generations. If we deliver RNAs or proteins of CRISPR/Cas system into a plant cell, unlikely CRISPR/Cas system will be inherited in plant and thus transgene-free genome editing events will be generated. This will avoid the regulation-related issues, which usually take many years for safety test and government approval.
Finding gene targets is the key for CRISPR/Cas-based genetic improvement
Rapid development of CRISPR/Cas-based genome editing provides a new opportunity for crop improvement by using the cutting-edge powerful tool for precision breeding. However, selecting and finding the desired genes for a desired trait is the key and also the major challenge for precisely improving desired crop traits, particularly for the traits with a complicated genetic background, such as crop yield and quality [108]. No matter for what trait, genes, controlling each trait, can be classified into structural and regulatory classes. Structural genes are usually protein-coding gene and directly control crop trait, they usually directly control the unique traits. Thus, finding a structural gene is the best choice for genome editing for improving the trait controlled by those genes. However, for many plant traits are not simply controlled by certain structural genes; there are always lots of regulatory elements involving in that processes. There are many regulatory genes in plants, which include but not only limit to transcription factors and non-coding RNAs. Some transcription factors specifically respond to certain biological processes, such as stress-responsible transcription factors. Thus, targeting on transcription factor genes may control multiple downstream structural genes for certain desirable traits; this is the reason for current genome editing research that focused on transcription factor genes for improving plant tolerance to abotic and biotic stresses. Thus, in the future, scientists need to find the major gene players that control directly or indirectly desirable traits in plant, which will serve as the targets for genome editing. Due to the fact that plant traits are generally controlled by multiple structural genes and regulatory elements, CRISPR/Cas9-mediated multiple gene-editing may be an alternative approach for crop improvement. Currently, there are many successfully reports on editing multiple genes at the same time using CRISPR/Cas9 technology. However, when using this technology, it is better to make sure that no huge fragments are removed from the chromosome that will significantly affect other related traits.
Off-target effect is a big concern for CRISPR/Cas application
Off-target effects are frequently reported and widely existed in genome editing events although it is less happened in plant genome editing than that in animals. In human clinic research and CRISPR/Cas gene therapy, off-target is really a big issue and should be eliminated [7]. Although it is not so serious for the CRISPR/Cas applications in plants, off-target effects may cause undesirable results; thus, eliminating off-target effect is always needed for genome editing.
CRISPR/Cas delivery system affects off-target of genome editing. Direct delivery of the protein or RNAs of Cas enzymes and sgRNAs, such as RNP transformation, can significantly reduce off-target effects. This is because that these functional molecules are not inserted into plant genome; after their functions, they can be quickly degraded; in which they only have a short life time in plant cells, thus have a less chance to target other locations.
Conclusions
CRISPR/Cas9 system is a naturally occurred genome editing tool, which was widely adopted and modified for precisely editing, regulating and monitoring an individual gene in plants and animals as well as micorbes for a wide range of purposes, including improvement of crop growth, development and further yield and quality as well as tolerance to various environmental biotic and abiotic stress. Developing a more efficiency and robust CRISPR/Cas system, including CRISPR/Cas regent delivery system, with less off-target effects is still the currently challenge for the majority of plant species. Current multiple omics, including whole genome sequencing, RNA-seq and small RNA-seq, will help to find more gene targets for CRISPR/Cas9-based crop improvement.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgement
We greatly appreciate the scientific community for making this field so quickly growing. We were trying to cite as more references as possible. However, due to the page limitation, there may be some important works not being cited in this paper, we apologize for this. D.Z. is supported by the National Natural Science Foundation of China (31172257), the Science Research Project in Henan Province of China (192102110174), and the Talent Project of Henan Agriculture University. Z.Z. is supported by the National Natural Science Foundation of China (31571600) and the Key Science and Technology Special Project of Xinxiang City of China (ZD2020004). B.Z. is supported in part by Cotton Incorporated and the National Science Foundation (award 1658709).
Biographies

Dr. Dangquan Zhang is currently working as a distinguished professor at Henan Agricultural University (HAU, China). Dr. Zhang received his PhD from Sun Yat-Sen University in 2004, then work on plant biotechnology and utilization. Dr. Zhang has authored more than 100 publications with more than 2,000 citations. Dr. Zhang frequently reviews manuscripts for many journals, and also serves as an Associate Editor or Guest Editor for 2 journals. He has reviewed proposals for more than 100 funding agents, including NSFC. Dr. Zhang is elected to the High level talents in Henan Province in 2019, Hunan Province new century talent project in 2011, and Academic leader of Henan Education Department in 2020. He won First prize of Hunan province natural science in 2011, First prize of China industry-university-research cooperation innovation award in 2017, and Second prize of Henan province science and technology progress award in 2018.

Dr. Zhiyong Zhang is a full professor at Henan Institute of Science and Technology. He got his Ph.D. degree from China Agricultural University in 2007 and are currently working on plant molecular biology, physiology and biotechnology. His research is supported by “The Science and Technology Innovation Talents Project of Henan Province of China (114100510008)”, “National Natural Science Foundation of China (31271648; 31571600)”, and “Key Science and Technology Special Project of Xinxiang City of China (ZD2020004)”. He has published more than 50 papers in peer-reviewed journals and won the Distinguished Professorship of Henan Province of China in 2020.

Dr. Turgay Unver achieved his PhD degree from the Middle East Technical University (METU; Turkey) in 2008. He became Assistant Professor at Cankiri Karatekin University in 2009 and obtained his Associated Professorship in 2011. His main research areas involve genome and transcriptome analyses, including microRNA. Currently, he works on important biological problems using emerging technologies such as CRISPR Cas9, deep sequencing, and bioinformatics collaboratively. He has published 50 indexed articles with more than 4000 citations. In 2012, he was awarded the TWAS prize. He was selected as outstanding young scientist by the Turkish Academy of Science in 2013. He is currently running two Biotechnology companies and bearing editorial duties for Genomics, BMC Genomics, and PloS One.

Dr. Baohong Zhang is currently working as a distinguished professor at East Carolina University (ECU). Dr. Zhang graduated from China Agricultural University. After received his bachelor degree, he worked on cotton biotechnology at Chinese Academy of Agricultural Sciences. Then he received his PhD from Texas Tech University in 2006. Dr. Zhang has authored more than 200 publications with more than 14,000 citations, many of them are listed as highly cited papers by the Web of Sciences, his h-index is 55. Dr. Zhang frequently reviewed manuscripts for many international journals, including Nature and Nature Biotechnology. He also served as an Associate Editor or Guest Editor for 7 journals, including Scientific Reports and Plant Biotechnology Journal. He has reviewed proposals for more than 40 funding agents, including NSF, US DoE, US USDA, and NSFC. Dr. Zhang is elected to the Fellow of the American Association for the Advancement of Science (AAAS) in 2018. He won the Researcher of the Year 2018 award by the ICAC. In 2018, Dr. Zhang won the THCAS Distinguished Professorship and the Highly Cited Scholar awards. Recently, he also won the Lifetime Research Achievement Award, the top honor for ECU faculty. He also won the Cotton Biotechnology Award in 2020.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Bennett, A.B. et al. (2013) Agricultural Biotechnology: Economics, Environment, Ethics, and the Future. In Annual Review of Environment and Resources, Vol 38 (Gadgil, A. and Liverman, D.M. eds), pp. 249-279.
- 2.Raney T. Economic impact of transgenic crops in developing countries. Curr Opin Biotechnol. 2006;17(2):174–178. doi: 10.1016/j.copbio.2006.02.009. [DOI] [PubMed] [Google Scholar]
- 3.Gelvin S.B. Agrobacterium-mediated plant transformation: the biology behind the “gene-jockeying” tool. Microbiology and molecular biology reviews : MMBR. 2003;67(1):16–37. doi: 10.1128/MMBR.67.1.16-37.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gaj T. ZFN, TALEN, and CRISPR/Cas-based methods for genome engineering. Trends Biotechnol. 2013;31(7):397–405. doi: 10.1016/j.tibtech.2013.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Silva G. Meganucleases and Other Tools for Targeted Genome Engineering: Perspectives and Challenges for Gene Therapy. Curr Gene Ther. 2011;11(1):11–27. doi: 10.2174/156652311794520111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Demirci Y. CRISPR/Cas9: An RNA-guided highly precise synthetic tool for plant genome editing. 2018;233(3):1844–1859. doi: 10.1002/jcp.25970. [DOI] [PubMed] [Google Scholar]
- 7.Zhang, B. (2020) CRISPR/Cas gene therapy. n/a (n/a). 10.1002/jcp.30064. [DOI]
- 8.Ishino Y. Nucleotide sequence of the iap gene, responsible for alkaline phosphatase isozyme conversion in Escherichia coli, and identification of the gene product. J Bacteriol. 1987;169(12):5429–5433. doi: 10.1128/jb.169.12.5429-5433.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nakata A. Unusual nucleotide arrangement with repeated sequences in the Escherichia coli K-12 chromosome. J Bacteriol. 1989;171(6):3553–3556. doi: 10.1128/jb.171.6.3553-3556.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hermans P.W. Insertion element IS987 from Mycobacterium bovis BCG is located in a hot-spot integration region for insertion elements in Mycobacterium tuberculosis complex strains. Infect Immun. 1991;59(8):2695–2705. doi: 10.1128/iai.59.8.2695-2705.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mojica F.J.M. Transcription at different salinities of Haloferax mediterranei sequences adjacent to partially modified PstI sites. Mol Microbiol. 1993;9(3):613–621. doi: 10.1111/j.1365-2958.1993.tb01721.x. [DOI] [PubMed] [Google Scholar]
- 12.Jansen R. Identification of genes that are associated with DNA repeats in prokaryotes. Mol Microbiol. 2002;43(6):1565–1575. doi: 10.1046/j.1365-2958.2002.02839.x. [DOI] [PubMed] [Google Scholar]
- 13.Mojica F.J.M., Montoliu L. On the rrigin of CRISPR-Cas technology: from prokaryotes to mammals. Trends Microbiol. 2016;24(10):811–820. doi: 10.1016/j.tim.2016.06.005. [DOI] [PubMed] [Google Scholar]
- 14.Mojica F.J.M. Long stretches of short tandem repeats are present in the largest replicons of the Archaea Haloferax mediterranei and Haloferax volcanii and could be involved in replicon partitioning. Mol Microbiol. 1995;17(1):85–93. doi: 10.1111/j.1365-2958.1995.mmi_17010085.x. [DOI] [PubMed] [Google Scholar]
- 15.Mojica F.J.M. Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. J Mol Evol. 2005;60(2):174–182. doi: 10.1007/s00239-004-0046-3. [DOI] [PubMed] [Google Scholar]
- 16.Gasiunas G. Cas9-crRNA ribonucleoprotein complex mediates specific DNA cleavage for adaptive immunity in bacteria. Proc Natl Acad Sci USA. 2012;109(39):E2579–E2586. doi: 10.1073/pnas.1208507109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jinek M. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial i9mmunity. Science. 2012;337(6096):816–821. doi: 10.1126/science.1225829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cong L. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339:819–823. doi: 10.1126/science.1231143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mali P. RNA-Guided Human Genome Engineering via Cas9. Science. 2013;339(6121):823–826. doi: 10.1126/science.1232033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Anders C. Structural basis of PAM-dependent target DNA recognition by the Cas9 endonuclease. Nature. 2014;513:569–573. doi: 10.1038/nature13579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Nishimasu H. Crystal structure of Cas9 in complex with guide RNA and target DNA. Cell. 2014;156:935–949. doi: 10.1016/j.cell.2014.02.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Makarova K.S. Evolutionary classification of CRISPR–Cas systems: a burst of class 2 and derived variants. Nat Rev Microbiol. 2020;18(2):67–83. doi: 10.1038/s41579-019-0299-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abudayyeh O.O. C2c2 is a single-component programmable RNA-guided RNA-targeting CRISPR effector. 2016;353(6299):aaf5573. doi: 10.1126/science.aaf5573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harrington L.B. Programmed DNA destruction by miniature CRISPR-Cas14 enzymes. 2018;362(6416):839–842. doi: 10.1126/science.aav4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Garneau J.E. The CRISPR/Cas bacterial immune system cleaves bacteriophage and plasmid DNA. Nature. 2010;468(7320):67–71. doi: 10.1038/nature09523. [DOI] [PubMed] [Google Scholar]
- 26.Magadán A.H. Cleavage of phage DNA by the Streptococcus thermophilus CRISPR3-Cas system. PLoS ONE. 2012;7(7) doi: 10.1371/journal.pone.0040913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang T. Genetic screens in human cells using the CRISPR-Cas9 system. Science. 2014;343(6166):80–84. doi: 10.1126/science.1246981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nishimasu H. Engineered CRISPR-Cas9 nuclease with expanded targeting space. Science. 2018;361:1259–1262. doi: 10.1126/science.aas9129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kleinstiver B.P. Broadening the targeting range of Staphylococcus aureus CRISPR-Cas9 by modifying PAM recognition. Nat Biotechnol. 2015;33(12):1293–1298. doi: 10.1038/nbt.3404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hu J.H. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556:57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Walton R.T. Unconstrained genome targeting with near-PAMless engineered CRISPR-Cas9 variants. Science. 2020;368:290–296. doi: 10.1126/science.aba8853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang, D. and Zhang, B. (2020) SpRY: engineered CRISPR/Cas9 harnesses new genome-editing power. Trends in Genetics. 36(8):546–548. 10.1016/j.tig.2020.05.001. [DOI] [PubMed]
- 33.Maher M.F. Plant gene editing through de novo induction of meristems. Nat Biotechnol. 2020;38(1):84–89. doi: 10.1038/s41587-019-0337-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gallois J.L. Combined SHOOT MERISTEMLESS and WUSCHEL trigger ectopic organogenesis in Arabidopsis. Development. 2002;129(13):3207–3217. doi: 10.1242/dev.129.13.3207. [DOI] [PubMed] [Google Scholar]
- 35.Zhang Y. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. 2016;7:12617. doi: 10.1038/ncomms12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mei Y. Protein expression and gene editing in monocots using foxtail mosaic virus vectors. Plant Direct. 2019;3 doi: 10.1002/pld3.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Alagoz Y. Manipulating the Biosynthesis of Bioactive Compound Alkaloids for Next-Generation Metabolic Engineering in Opium Poppy Using CRISPR-Cas 9 Genome Editing Technology. Sci Rep. 2016;6:30910. doi: 10.1038/srep30910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Cody W.B. Multiplexed Gene Editing and Protein Overexpression Using a Tobacco mosaic virus Viral Vector. Plant Physiol. 2017;175:23–35. doi: 10.1104/pp.17.00411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ma X. Highly efficient DNA-free plant genome editing using virally delivered CRISPR–Cas9. Nat Plants. 2020;6(7):773–779. doi: 10.1038/s41477-020-0704-5. [DOI] [PubMed] [Google Scholar]
- 40.Ellison E.E. Multiplexed heritable gene editing using RNA viruses and mobile single guide RNAs. Nat Plants. 2020;6(6):620–624. doi: 10.1038/s41477-020-0670-y. [DOI] [PubMed] [Google Scholar]
- 41.Liu, H. and Zhang, B. Virus-based CRISPR/Cas9 genome editing in plants. Trends in Genetics 36(11):810-813. 10.1016/j.tig.2020.08.002. [DOI] [PubMed]
- 42.Woo J.W. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat Biotechnol. 2015;33(11):1162–1164. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 43.Malnoy M. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front Plant Sci. 2016;7:1904. doi: 10.3389/fpls.2016.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Svitashev S. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat Commun. 2016;7(1):13274. doi: 10.1038/ncomms13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liang Z. Efficient DNA-free genome editing of bread wheat using CRISPR/Cas9 ribonucleoprotein complexes. Nat Commun. 2017;18:14261. doi: 10.1038/ncomms14261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Andersson M. Genome editing in potato via CRISPR-Cas9 ribonucleoprotein delivery. Physiol Plant. 2018;164:378–384. doi: 10.1111/ppl.12731. [DOI] [PubMed] [Google Scholar]
- 47.Murovec J. DNA-Free Genome Editing of Brassica oleracea and B. rapa Protoplasts Using CRISPR-Cas9 Ribonucleoprotein Complexes. Front. Plant Sci. 2018;9:1594. doi: 10.3389/fpls.2018.01594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Banakar R. High-frequency random DNA insertions upon co-delivery of CRISPR-Cas9 ribonucleoprotein and selectable marker plasmid in rice. Sci Rep. 2019;9:19902. doi: 10.1038/s41598-019-55681-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Zhang B. MicroRNAs and their regulatory roles in animals and plants. J Cell Physiol. 2007;210:279–289. doi: 10.1002/jcp.20869. [DOI] [PubMed] [Google Scholar]
- 50.Wang M. Gene Targeting by Homology-Directed Repair in Rice Using a Geminivirus-Based CRISPR/Cas9 System. Molecular Plant. 2017;10(7):1007–1010. doi: 10.1016/j.molp.2017.03.002. [DOI] [PubMed] [Google Scholar]
- 51.Kim J. Site-specific gene knock-out and on-site heterologous gene overexpression in Chlamydomonas reinhardtii via a CRISPR-Cas9-mediated knock-in method. Front Plant Sci. 2020;11(306) doi: 10.3389/fpls.2020.00306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lu Y. Targeted, efficient sequence insertion and replacement in rice. Nature. 2020 doi: 10.1038/s41587-020-0581-5. Biotechnology. [DOI] [PubMed] [Google Scholar]
- 53.Li S. Precise gene replacement in rice by RNA transcript-templated homologous recombination. Nat Biotechnol. 2019;37(4):445–450. doi: 10.1038/s41587-019-0065-7. [DOI] [PubMed] [Google Scholar]
- 54.Anzalone A.V. Search-and-replace genome editing without double-strand breaks or donor DNA. Nature. 2019;576(7785):149–157. doi: 10.1038/s41586-019-1711-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lin Q. Prime genome editing in rice and wheat. Nat Biotechnol. 2020;38(5):582–585. doi: 10.1038/s41587-020-0455-x. [DOI] [PubMed] [Google Scholar]
- 56.Hua, K. et al. Precision genome engineering in rice using prime editing system. n/a (n/a). [DOI] [PMC free article] [PubMed]
- 57.Xu R. Development of Plant Prime-Editing Systems for Precise Genome Editing. Plant Communications. 2020;1(3) doi: 10.1016/j.xplc.2020.100043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tang X. Plant Prime Editors Enable Precise Gene Editing in Rice Cells. Mol Plant. 2020;13(5):667–670. doi: 10.1016/j.molp.2020.03.010. [DOI] [PubMed] [Google Scholar]
- 59.Veillet, F. et al. (2020) Prime editing is achievable in the tetraploid potato, but needs improvement. 2020.06.18.159111.
- 60.Jiang, Y.-Y. et al. (2020) Prime editing efficiently generates W542L and S621I double mutations in two ALS genes of maize. 2020.07.06.188896. [DOI] [PMC free article] [PubMed]
- 61.Wang M. Targeted base editing in rice with CRISPR/ScCas9 system. Plant Biotechnol J. 2020 doi: 10.1111/pbi.13330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xu W. Multiplex nucleotide editing by high-fidelity Cas9 variants with improved efficiency in rice. BMC Plant Biol. 2019;19(1):511. doi: 10.1186/s12870-019-2131-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Wu J. In press; Plant Biotechnol J: 2020. Engineering herbicide-resistant oilseed rape by CRISPR/Cas9-mediated cytosine base-editing. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kuang Y. Base-editing-mediated artificial evolution of OsALS1 In planta to develop novel herbicide-tolerant rice germplasms. Mol Plant. 2020;13:565–572. doi: 10.1016/j.molp.2020.01.010. [DOI] [PubMed] [Google Scholar]
- 65.Kuscu C. CRISPR-STOP: gene silencing through base-editing-induced nonsense mutations. Nat Methods. 2017;14:710–712. doi: 10.1038/nmeth.4327. [DOI] [PubMed] [Google Scholar]
- 66.Billon P. CRISPR-mediated base editing enables efficient disruption of eukaryotic genes through induction of STOP codons. Mol Cell. 2017;67:1068–1079. doi: 10.1016/j.molcel.2017.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Qi L.S. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013;152:1173–1183. doi: 10.1016/j.cell.2013.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gilbert L.A. CRISPR-mediated modular RNA-guided regulation of transcription in eukaryotes. Cell. 2013;154:442–451. doi: 10.1016/j.cell.2013.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Papikian A. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat Commun. 2019;10(1):729. doi: 10.1038/s41467-019-08736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Lee J.E. CRISPR-based tools for targeted transcriptional and epigenetic regulation in plants. PLoS ONE. 2019;14 doi: 10.1371/journal.pone.0222778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Adli M. The CRISPR tool kit for genome editing and beyond. Nat Commun. 2018;9(1) doi: 10.1038/s41467-018-04252-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Devesa-Guerra I. DNA methylation editing by CRISPR-guided excision of 5-methylcytosine. J Mol Biol. 2020;432:2204–2216. doi: 10.1016/j.jmb.2020.02.007. [DOI] [PubMed] [Google Scholar]
- 73.Li C., Zhang B. MicroRNAs in Control of Plant Development. J Cell Physiol. 2016;231(2):303–313. doi: 10.1002/jcp.25125. [DOI] [PubMed] [Google Scholar]
- 74.Zhang B., Unver T. A critical and speculative review on microRNA technology in crop improvement: Current challenges and future directions. Plant Sci. 2018;274:193–200. doi: 10.1016/j.plantsci.2018.05.031. [DOI] [PubMed] [Google Scholar]
- 75.Bi H. Disruption of miRNA sequences by TALENs and CRISPR/Cas9 induces varied lengths of miRNA production. Plant Biotechnol J. 2019 doi: 10.1111/pbi.13315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Yao X. An Essential Role for miRNA167 in Maternal Control of Embryonic and Seed Development. Plant Physiol. 2019;180(1):453–464. doi: 10.1104/pp.19.00127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Li C. A high-efficiency CRISPR/Cas9 system for targeted mutagenesis in Cotton (Gossypium hirsutum L.) Sci Rep. 2017;7:43902. doi: 10.1038/srep43902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Huckelhoven R., Panstruga R. Cell biology of the plant-powdery mildew interaction. Curr Opin Plant Biol. 2011;14(6):738–746. doi: 10.1016/j.pbi.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 79.Buschges R. The barley Mlo gene: a novel control element of plant pathogen resistance. Cell. 1997;88(5):695–705. doi: 10.1016/s0092-8674(00)81912-1. [DOI] [PubMed] [Google Scholar]
- 80.Wang Y. Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat Biotechnol. 2014;32(9):947–951. doi: 10.1038/nbt.2969. [DOI] [PubMed] [Google Scholar]
- 81.Nekrasov V. Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci Rep. 2017;7(1):482. doi: 10.1038/s41598-017-00578-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Martyn R.D., Netzer D. Resistance to races 0, 1, and 2 of Fusarium wilt of watermelon in Citrullus sp. PI-296341 -FR. 1991;26(4):429. [Google Scholar]
- 83.Zhang M. CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f.sp. niveum. Plant Cell Rep. 2020;39(5):589–595. doi: 10.1007/s00299-020-02516-0. [DOI] [PubMed] [Google Scholar]
- 84.Hammes U.Z. Novel roles for phytosulfokine signalling in plant–pathogen interactions. Plant, Cell Environ. 2015;39(7):1393–1395. doi: 10.1111/pce.12679. [DOI] [PubMed] [Google Scholar]
- 85.Wang F. Enhanced Rice Blast Resistance by CRISPR/Cas9-Targeted Mutagenesis of the ERF Transcription Factor Gene OsERF922. PLoS ONE. 2016;11(4) doi: 10.1371/journal.pone.0154027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Zhang Z. Simultaneous editing of two copies of Gh14-3-3d confers enhanced transgene-clean plant defense against Verticillium dahliae in allotetraploid upland cotton. Front Plant Sci. 2018;9:842. doi: 10.3389/fpls.2018.00842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Pyott D.E. Engineering of CRISPR/Cas9-mediated potyvirus resistance in transgene-free Arabidopsis plants. Mol Plant Pathol. 2016;17(8):1276–1288. doi: 10.1111/mpp.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Chandrasekaran J. Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol Plant Pathol. 2016;17(7):1140–1153. doi: 10.1111/mpp.12375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Yang B. Os8N3 is a host disease-susceptibility gene for bacterial blight of rice. PNAS. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Kim Y.-A. CRISPR/Cas9-targeted mutagenesis of Os8N3 in rice to confer resistance to Xanthomonas oryzae pv. oryzae. Rice. 2019;12(1):67. doi: 10.1186/s12284-019-0325-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Peng A. Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol J. 2017;15(12):1509–1519. doi: 10.1111/pbi.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Wang L. CRISPR/Cas9-mediated editing of CsWRKY22 reduces susceptibility to Xanthomonas citri subsp. citri in Wanjincheng orange (Citrus sinensis (L.) Osbeck) Plant Biotechnology Reports. 2019;13(5):501–510. [Google Scholar]
- 93.Ortigosa A.S. Design of a bacterial speck resistant tomato by CRISPR/Cas9-mediated editing of SlJAZ2. Plant Biotechnol J. 2018;17(3):665–673. doi: 10.1111/pbi.13006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Smagghe G. Targeting female reproduction in insects with biorational insecticides for pest management: a critical review with suggestions for future research. Curr Opin Insect Sci. 2019;31:65–69. doi: 10.1016/j.cois.2018.10.009. [DOI] [PubMed] [Google Scholar]
- 95.Zhang B. MicroRNA: a new target for improving plant tolerance to abiotic stress. J Exp Bot. 2015;66:1749–1761. doi: 10.1093/jxb/erv013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Zafar S.A. Engineering abiotic stress tolerance via CRISPR/ Cas-mediated genome editing. J Exp Bot. 2020;71:470–479. doi: 10.1093/jxb/erz476. [DOI] [PubMed] [Google Scholar]
- 97.Shi J. ARGOS8 variants generated by CRISPR-Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol J. 2017;15:207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Klap C. Tomato facultative parthenocarpy results from SlAGAMOUS-LIKE 6 loss of function. Plant Biotechnol J. 2017;15:634–647. doi: 10.1111/pbi.12662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Chen S. Genome editing to integrate seed size and abiotic stress Ttolerance traits in Arabidopsis reveals a role for DPA4 and SOD7 in the regulation of inflorescence architecture. Int J Mol Sci. 2019;20:2695. doi: 10.3390/ijms20112695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Bouzroud S. Down regulation and loss of auxin response factor 4 function using CRISPR/Cas9 alters plant growth, stomatal function and improves tomato tolerance to salinity and osmotic stress. Genes. 2020;11:272. doi: 10.3390/genes11030272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Cui Y. Heterotrimeric G protein are involved in the regulation of multiple agronomic traits and stress tolerance in rice. BMC Plant Biol. 2020;20:90. doi: 10.1186/s12870-020-2289-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Wang B. Mutagenesis Reveals That the OsPPa6 Gene Is Required for Enhancing the Alkaline Tolerance in Rice. Front Plant Sci. 2019;10:759. doi: 10.3389/fpls.2019.00759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Li R. CRISPR/Cas9-Mediated SlNPR1 mutagenesis reduces tomato plant drought tolerance. BMC Plant Biol. 2019;19:38. doi: 10.1186/s12870-018-1627-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Li S. The AREB1 transcription factor influences histone acetylation to regulate drought responses and tolerance in Populus trichocarpa. Plant Cell. 2019;31(3):663–686. doi: 10.1105/tpc.18.00437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Roca Paixao J.F. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a Histone AcetylTransferase. Sci Rep. 2019;9:8080. doi: 10.1038/s41598-019-44571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Meng Z. Expression of the rice arginase gene OsARG in cotton influences the morphology and nitrogen transition of seedlings. PLoS ONE. 2015;10 doi: 10.1371/journal.pone.0141530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Wang Y. Increased lateral root formation by CRISPR/Cas9-mediated editing of arginase genes in cotton. Science China Life Sciences. 2017;60(5):524–527. doi: 10.1007/s11427-017-9031-y. [DOI] [PubMed] [Google Scholar]
- 108.Peng R.H. From sequencing to genome editing for cotton improvement. Trends Biotechnol. 2020 doi: 10.1016/j.tibtech.2020.09.001. [DOI] [PubMed] [Google Scholar]
- 109.Paul P. MADS78 and MADS79 are essential regulators of early seed development in rice. Plant Physiol. 2020;182:933–948. doi: 10.1104/pp.19.00917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Lee S.K. Deficiency of rice hexokinase HXK5 impairs synthesis and utilization of starch in pollen grains and causes male sterility. J Exp Bot. 2020;71:116–125. doi: 10.1093/jxb/erz436. [DOI] [PubMed] [Google Scholar]
- 111.Liang X. COPII components Sar1b and Sar1c play distinct yet interchangeable roles in pollen development. Plant Physiol. 2020 doi: 10.1104/pp.20.00159. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Li C. An Efficient CRISPR/Cas9 Platform for Rapidly Generating Simultaneous Mutagenesis of Multiple Gene Homoeologs in Allotetraploid Oilseed Rape. Front Plant Sci. 2018;9:442. doi: 10.3389/fpls.2018.00442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Sriboon S. Knock-out of TERMINAL FLOWER 1 genes altered flowering time and plant architecture in Brassica napus. BMC Genet. 2020;21:52. doi: 10.1186/s12863-020-00857-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Rodriguez-Leal D. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171(2):470–480.e8. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 115.Li M. Reassessment of the four yield-related genes Gn1a, DEP1, GS3, and IPA1 in rice using a CRISPR/Cas9 system. Front Plant Sci. 2016;7(377) doi: 10.3389/fpls.2016.00377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Xu R. Rapid improvement of grain weight via highly efficient CRISPR/Cas9-mediated multiplex genome editing in rice. J Genet Genomics. 2016;43:529–532. doi: 10.1016/j.jgg.2016.07.003. [DOI] [PubMed] [Google Scholar]
- 117.Lee D.W. The Role of Rice Vacuolar Invertase2 in Seed Size Control. Mol Cells. 2019;42:711–720. doi: 10.14348/molcells.2019.0109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Zhang Y. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat Commun. 2016;7(1):12617. doi: 10.1038/ncomms12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wang C. A cytokinin-activation enzyme-like gene improves grain yield under various field conditions in rice. Plant Mol Biol. 2020;102(4):373–388. doi: 10.1007/s11103-019-00952-5. [DOI] [PubMed] [Google Scholar]
- 120.Li X. Lycopene Is Enriched in Tomato Fruit by CRISPR/Cas9-Mediated Multiplex Genome Editing. Front Plant Sci. 2018;9(559) doi: 10.3389/fpls.2018.00559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Sanchez-Leon S. Low-gluten, nontransgenic wheat engineered with CRISPR/Cas9. Plant Biotechnol J. 2018;16(4):902–910. doi: 10.1111/pbi.12837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Yang Q. Mutation of the d-hordein gene by RNA-guided Cas9 targeted editing reducing the grain size and changing grain compositions in barley. Food Chem. 2020;311 doi: 10.1016/j.foodchem.2019.125892. [DOI] [PubMed] [Google Scholar]
- 123.Lyzenga W.J. CRISPR/Cas9 editing of three CRUCIFERIN C homoeologues alters the seed protein profile in Camelina sativa. BMC Plant Biol. 2019;19:292. doi: 10.1186/s12870-019-1873-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Zhang K. Effective editing for lysophosphatidic acid acyltransferase 2/5 in allotetraploid rapeseed (Brassica napus L.) using CRISPR-Cas9 system. Biotechnol Biofuels. 2019;12:225. doi: 10.1186/s13068-019-1567-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Sun Y. Generation of High-Amylose Rice through CRISPR/Cas9-Mediated Targeted Mutagenesis of Starch Branching Enzymes. Front Plant Sci. 2017;8:298. doi: 10.3389/fpls.2017.00298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Wang H. CRISPR/Cas9-Based Mutagenesis of Starch Biosynthetic Genes in Sweet Potato (Ipomoea Batatas) for the Improvement of Starch Quality. Int J Mol Sci. 2019;20:4702. doi: 10.3390/ijms20194702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Andersson M. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 2017;36(1):117–128. doi: 10.1007/s00299-016-2062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Karunarathna, N.L. et al. (2020) Elevating seed oil content in a polyploid crop by induced mutations in SEED FATTY ACID REDUCER genes. Plant Biotechnology Journal n/a (n/a), In press. [DOI] [PMC free article] [PubMed]
- 129.Zhai Y. Targeted mutagenesis of BnTT8 homologs controls yellow seed coat development for effective oil production in Brassica napus L. Plant Biotechnol J. 2020;18:1153–1168. doi: 10.1111/pbi.13281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Ran F.A. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–191. doi: 10.1038/nature14299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hirano H. Structure and Engineering of Francisella novicida Cas9. Cell. 2016;164(5):950–961. doi: 10.1016/j.cell.2016.01.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Hou Z. Efficient genome engineering in human pluripotent stem cells using Cas9 from <em>Neisseria meningitidis</em>. 2013;110(39):15644–15649. doi: 10.1073/pnas.1313587110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Kim E. In vivo genome editing with a small Cas9 orthologue derived from Campylobacter jejuni. Nat Commun. 2017;8:14500. doi: 10.1038/ncomms14500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zetsche B. Cpf1 is a single RNA-guided endonuclease of a class 2 CRISPR-Cas system. Cell. 2015;163(3):759–771. doi: 10.1016/j.cell.2015.09.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Fonfara I. The CRISPR-associated DNA-cleaving enzyme Cpf1 also processes precursor CRISPR RNA. Nature. 2016;532(7600):517–521. doi: 10.1038/nature17945. [DOI] [PubMed] [Google Scholar]
- 136.Kleinstiver B.P. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–485. doi: 10.1038/nature14592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Kleinstiver B.P. High-fidelity CRISPR–Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–495. doi: 10.1038/nature16526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Slaymaker I.M. Rationally engineered Cas9 nucleases with improved specificity. 2016;351(6268):84–88. doi: 10.1126/science.aad5227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Hu J.H. Evolved Cas9 variants with broad PAM compatibility and high DNA specificity. Nature. 2018;556(7699):57–63. doi: 10.1038/nature26155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Lee J.K. Directed evolution of CRISPR-Cas9 to increase its specificity. Nat Commun. 2018;9(1):3048. doi: 10.1038/s41467-018-05477-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Casini A. A highly specific SpCas9 variant is identified by in vivo screening in yeast. Nat Biotechnol. 2018;36(3):265–271. doi: 10.1038/nbt.4066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Chen J.S. Enhanced proofreading governs CRISPR–Cas9 targeting accuracy. Nature. 2017;550(7676):407–410. doi: 10.1038/nature24268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Miller S.M. Continuous evolution of SpCas9 variants compatible with non-G PAMs. Nat Biotechnol. 2020;38(4):471–481. doi: 10.1038/s41587-020-0412-8. [DOI] [PMC free article] [PubMed] [Google Scholar]