Abstract
Accumulating studies have suggested that microRNAs (miRs) play a significant role in lung cancer development and progression, especially in non-small cell lung cancer (NSCLC). The present study aimed to investigate the associations between miR-454-3p and NSCLC progression. qPCR assay was applied to examine the expression of miR-454-3p and transforming growth factor-β2 (TGFB2) in tissues and cell lines. CCK-8 and EdU assays were used to detect cell proliferation. Wound-healing and Transwell assays were conducted to assess cell migration and invasion. Western blotting assay was performed to explore the protein levels of epithelial-mesenchymal transition (EMT) markers. The interaction between miR-454-3p and TGFB2 was investigated with a luciferase reporter assay. miR-454-3p was downregulated in NSCLC tissues and NSCLC cell lines. miR-454-3p overexpression led to the suppression of proliferation, migration, and invasion in A549 and NCI-H1650 cells. In addition, the overexpression of miR-454-3p in A549 and NCI-H1650 cells significantly inhibited EMT. TGFB2 was revealed to be a direct target of miR-454-3p by using TargetScan database and luciferase reporter assay. TGFB2 was observed to be upregulated in NSCLC tissues and cell lines. Further mechanistic studies revealed that the inhibitory effects of miR-454-3p on NSCLC were reversed upon overexpression of TGFB2. These findings provided strong evidence that miR-454-3p suppressed NSCLC cell proliferation and metastasis by targeting TGFB2. The study suggests that targeting miR-454-3p could be a promising strategy for treating NSCLC.
Keywords: non-small cell lung cancer, microRNA-454-3p, transforming growth factor-β2, proliferation, epithelial-mesenchymal transition, metastasis
Introduction
Lung cancer, one of the most common globally diagnosed cancers, has a high mortality rate. The percentage of non-small cell lung cancer (NSCLC) is estimated at 85% among all lung cancers (1). Although clinical diagnosis and therapeutic strategies of NSCLC have advanced over the past three decades, the 5-year overall survival rate remains <16% (2). Among the reasons, tumor metastasis is one of the main causes of clinical treatment failure and recurrence (3,4). Therefore, it is necessary to elucidate the underlying mechanisms of NSCLC cell growth and metastasis, which may shed light on potential therapies for NSCLC.
MicroRNAs (miRNAs) belong to a class of small non-coding RNAs which regulating gene expression and protein-coding genes (5). Increasing studies have revealed that miRNAs are closely associated with tumor cell proliferation, migration, invasion, drug resistance and apoptosis (6,7). For instance, miR-410 was revealed to promote epithelial-mesenchymal transition (EMT) and radio-resistance by targeting the PI3K/mTOR axis in NSCLC (8). miR-219 functioned as a tumor suppressor, which inhibited NSCLC and ovarian cancer cell growth and metastasis (9,10).
miR-454-3p is a member of the miR-130-3p/301-3p/454-3p miRNA cluster, which was revealed to play a critical role in the progression and malignancies of various cancers (11). At present, some studies have revealed that miR-454-3p functioned as a suppressive miRNA in several cancers, including glioblastoma (12), lung cancer (13), bladder cancer (14) and pancreatic ductal adenocarcinoma (15). However, the detailed functions of miR-454-3p in NSCLC proliferation and metastasis remain to be characterized.
Transforming growth factor-β2 (TGFB2) is a protein coding gene of TGF-β2, and it has been revealed to regulate tumor cell growth, proliferation and metastasis (16,17). Emerging evidence has revealed that TGF-β2 was upregulated in numerous types of cancer (18). Bruna et al revealed that TGF-β2 was abundant in glioma and correlated with poor prognosis (19). Yang et al have reported that TGF-β2 contributed to EMT and tumor mutation burden in gastric cancer (17). EMT is a key biological process that induces malignant tumor cell migration and invasion (20,21). TGF-β is regarded as the most crucial factor for EMT (22). Consequently, targeting TGF-β2 could be a promising treatment strategy for cancer. Whether the expression of TGF-β2 is related with the expression level of miR-454-3p in NSCLC remains to be elucidated.
In the present study, the expression of miR-454-3p and TGFB2 was investigated in NSCLC tissues and cell lines. Furthermore, it was explored how miR-454-3p and TGFB2 contribute to the progression of NSCLC and the underlying mechanisms were also investigated.
Materials and methods
Clinical specimens
The present study was carried out with a total number of 56 human NSCLC tissue and 56 adjacent non-tumor tissue samples. These patients were between 25–75 years old, and the ratio of males to females was 1.15:1. Patients were confirmed to have no other serious diseases except NSCLC. The 56 NSCLC patients were clinically diagnosed and undergoing surgery (had never received any neo-adjuvant treatment) at the Yuebei People's Hospital of Shaoguan from March 2013 to October 2018. Patients provided written informed consent for their participation in the present study. These clinical samples were stored in liquid nitrogen until their use in experiments. The research was approved and carried out according to the ethical standards of the Ethics Committee of Yuebei People's Hospital (approval no. DD-KY-2018310).
Cells and cell culture
The normal lung cell line BEAS-2B and NSCLC cell lines A549, NCI-H1299, NCI-H1650, NCI-H460, NCI-H1975 were purchased from American Type Culture Collection (ATCC). All the cells were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10% fetal bovine serum (FBS; Shanghai ExCell Biotech Co., Ltd.) and 1% penicillin/streptomycin (Gibco; Thermo Fisher Scientific, Inc.) in 5% CO2 at 37°C. By separately using an STR Multi-Amplification Kit (Microreader 21 ID System; Suzhou Microread Genetics) and Mycoplasma Detection Set (M&C Gene Technology), all the cell lines were identified with no cross contamination with other cell lines and negative for mycoplasma.
Reverse transcription-quantitative (RT-q)PCR
E.Z.N.A.® Total RNA Kit I (Omega Bio-Tek) was used to extract total RNA from tissues and cultured cells in accordance with the manufacturer's protocol. Then, cDNAs were synthesized by All-in-One cDNA Synthesis SuperMix (Bimake) on a PCR instrument (C1000; Bio-Rad Laboratories, Inc.). The thermocycling conditions were as follows: 25°C/10 min; 42°C/30 min; and 85°C/5 min. Next, real-time qPCR was performed using 2X SYBR Green qPCR Master Mix (Bimake) on a LightCycler 480 System (Roche Diagnostics). The thermocycling conditions were as follows: Pre-incubation at 95°C for 5 min; then 40 cycles of amplification at 95°C for 10 sec, 56°C for 20 sec, 72°C for 20 sec; melting curve, 1 cycle at 95°C for 5 sec, 65°C for 1 min and continuously at 97°C; finally cooling at 95°C for 10 sec. miRNA and mRNA expression were defined based on the quantification cycle (Cq), and respectively normalized to U6 and GAPDH levels. The relative expression levels were analyzed by using 2−ΔΔCq method (23). The sequence of all primers applied in the present study are listed as follows: miR-454-3p forward, 5′-ACCCTATCAATATTGTCTCTGC-3′ and reverse, 5′-GCGAGCACAGAATTAATACGAC-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′; TGFB2 forward, 5′-GTTCGATTTGACGTCTCAGCAAT-3′ and reverse, 5′-CAATCCGTTGTTCAGGCACTCT-3′; GAPDH forward, 5′-TGCACCACCAACTGCTTAGC-3′ and reverse, 5′-GGCATGGACTGTGGTCATGAG-3′.
Cell transient transfection
A549 and NCI-H1650 cells were seeded in 6-well plates (2.1×104 cells/well) and incubated overnight. After cell fusion reached 80%, the cells were transfected with i) negative control (NC) or miR-454-3p mimics. The sequence of the miR-454-3p mimic was 5′-UAGUGCAAUAUUGCUUAUAGGGU-3′ and that of the negative control 5′-UCACAACCUCCUAGAAAGAGUAGA-3′; ii) NC with Vector (pCMV3), pCMV3-TGFB2 plasmid, or pCMV3-TGFB2 plasmid with miR-454-3p mimics using Lipofectamine™ 3000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. The concentration of NC and miR-454-3p mimics was 100 pmol and the mass of pCMV3 and pCMV3-TGFB2 was 2 µg. miR-454-3p mimics and negative control (NC) oligonucleotides were synthesized by Shanghai GenePharma Co., Ltd. Vector (pCMV3) and pCMV3-TGFB2 were purchased from Origene Technologies, Inc.. After incubation with transfection reagent for 6 h, the cells were cultured using complete medium for another 48 h in 5% CO2 at 37°C. Then, the transfected cells were used to perform subsequent experiments.
Bioinformatics analysis
Bioinformatics analysis was conducted to predict the assumed targets of miR-454-3p using TargetScan (version 6.0; www.targetscan.org).
Luciferase reporter assay
Luciferase reporter plasmids, pmirGLO-TGFB2-3′-UTR wild-type (WT), and pmirGLO-TGFB2-3′-UTR mutant (Mut) were synthesized by Shanghai GenePharma Co., Ltd. A549 and NCI-H1650 cells were seeded in 96-well plates (7.0×103 cells/well) and cultured for 24 h. Then, the pmirGLO-TGFB2-3′-UTR-WT or pmirGLO-TGFB2-3′-UTR-Mut plasmid was co-transfected with NC or miR-454-3p mimics into the cells using Lipofectamine™ 3000 transfection reagent (Invitrogen; Thermo Fisher Scientific, Inc.). After transfection for 48 h, a Dual Luciferase Reporter Assay kit (Promega Corporation) was used to measure the luciferase signals and the Renilla luciferase activity was used for normalization.
Cell proliferation assay
A549 and NCI-H1650 cells were plated into 96-well plates (5.0×103 cells/well) and incubated overnight. After transfection, the cells were cultured for another 24 h. Cell proliferation was analyzed by Cell Counting Kit-8 (CCK-8) assay (Beyotime Institute of Biotechnology) at 0, 24, 48 and 72 h, whereupon 10 µl CCK-8 solution was added to each well. Following incubation at 37°C for another 1 h, the optical density (OD) at 450 nm was measured using a microplate reader (Bio-Rad Laboratories, Inc.).
5-Ethynyl-2-deoxyuridine (EdU) proliferation assay
Transfected cells were cultured in 96-well plates (6.0×103 cells/well). After a 24-h incubation, the cells were subjected to the iClick™ EdU Andy Fluor 555 Imaging Kit (GeneCopoeia, Inc.) in accordance with the indicated protocol. The images were acquired with a fluorescence microscope (magnification, ×10) (Zeiss AG) and the EdU-positive cells were calculated by Image-Pro Plus 6.0 software (Media Cybernetics, Inc.).
Wound healing assay
A wound-healing assay can be used to evaluate cell migration capacity (24). A549 and NCI-H1650 cells were transfected with miRNA mimics and/or plasmid in 12-well plates (1.5×104 cells/well). After incubation for another 24 h, the cells were starved with serum-free DMEM for 6 h and then cell monolayers were scratched with a 10-µl pipette tip. Next, the cells were washed gently with phosphate-buffered saline (PBS) to remove the cell debris and then cultured with total medium. Images were captured at 0 and 12 h after the scratch by an optical microscope (magnification, ×5) (Zeiss AG). The wound widths were analyzed with ImageJ 1.38 software (National Institutes of Health).
Invasion assays
The invasion assays were conducted with 24-well Boyden chambers (8 µm pore size; Corning, Inc.). First, the upper chambers were pre-coated with diluted Matrigel (Corning, Inc.) and allowed to settle for 30 min at 37°C. The transfected cells (1.0×105 cells/ml) were resuspended in serum-free DMEM medium and seeded into the upper chamber (100 µl/well). The bottom chamber was filled with DMEM supplemented with 10% FBS (600 µl/well). After incubation for 24 h, the invasive cells were fixed with 4% paraformaldehyde for 30 min and then stained with 0.1% crystal violet for 15 min at room temperature. A cotton swab was used to rub away the non-invading cells on the inner side of upper chamber. The invasive cells in the lower surface were photographed with an inverted microscope (magnification, ×10) (Olympus Corporation) and counted by Image-Pro Plus 6.0 software.
Western blotting
A549 and NCI-H1650 cells after transfection were collected and the total protein of cells was extracted using RIPA buffer (Thermo Fisher Scientific, Inc.) containing proteinase inhibitors (Roche Diagnostics). A Pierce™ Bicinchoninic Acid assay kit (Thermo Fisher Scientific, Inc.) was used to detect the protein concentrations. Equal amounts of protein (30 µg) were separated by 10% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) and then transferred onto 4.5 µm polyvinylidene fluoride (PVDF) membranes (EMD Millipore). Subsequently, 5% skim milk in TBS containing 0.1% Tween-20 (TBST) was used to block the membranes for 1 h at room temperature. The membranes were incubated with primary antibodies against E-cadherin (cat. no. 3195), N-cadherin (cat. no. 4061) and GAPDH (cat. no. 5174S), which were purchased from Cell Signaling Technology, Inc.. The dilution of the primary antibodies was 1:1,000. After washing with TBST for three times, the horseradish peroxidase (HRP)-conjugated secondary antibody (goat anti-rabbit IgG; cat. no. sc-2004; 1:1,000; Santa Cruz Biotechnology, Inc.) was probed with the blots for 1 h at room temperature. After washing with TBST, the protein bands were visualized by an enhanced chemiluminescence (ECL; Pierce; Thermo Fisher Scientific, Inc.) solutions.
Statistical analysis
The experimental data were statistically analyzed with GraphPad Prism 8.0 software (GraphPad Software, Inc.). Each experiment was repeated at least three times and the results are expressed as the mean ± SD. The unpaired Student's t-test was conducted for comparisons between two groups and one-way ANOVA followed by the Tukey's post hoc test was used to analyze the significance among more than two groups. Survival analysis was investigated using Kaplan-Meier method followed by the Gehan-Breslow-Wilcoxon test which was used to analyze the significance. The correlation between miR-454-3p and TGFB2 expression were evaluated using Spearman's analysis. P<0.05 was considered to indicate a statistically significant difference.
Results
miR-454-3p is downregulated in NSCLC tissues and cell lines
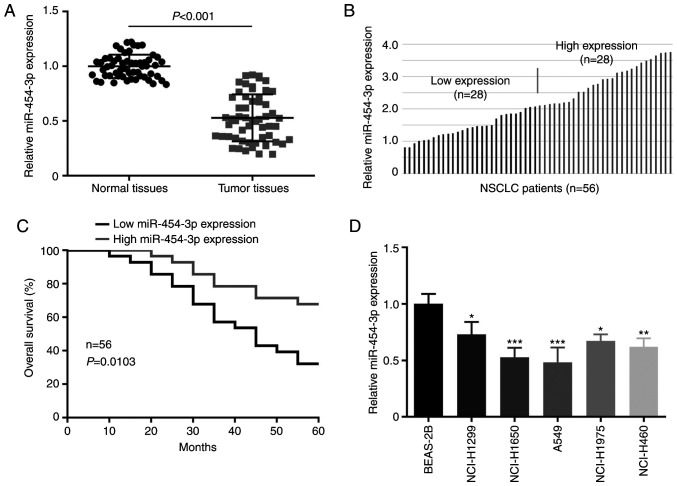
To understand the role of miR-454-3p in NSCLC progression, the expression level of miR-454-3p in NSCLC tissues and cells was examined by RT-qPCR assay. As presented in Fig. 1A, miR-454-3p was downregulated in NSCLC tissues by comparison with adjacent normal tissues. In the clinicopathological analysis of NSCLC patients, miR-454-3p was significantly associated with tumor size, pathological stage and tumor metastasis, but not with age and sex (Table I). Then, the association between overall survival and miR-454-3p expression in NSCLC patients was analyzed. The 56 NSCLC tissues were divided into a low-miR-454-3p expression group and a high-miR-454-3p expression group (Fig. 1B). Kaplan-Meier analysis revealed that the low miR-454-3p expression was associated with worse overall survival in patients with NSCLC (Fig. 1C). The expression of miR-454-3p in NSCLC cells was also detected. The RT-qPCR assay revealed that miR-454-3p expression was significantly lower in 5 NSCLC cell lines (NCI-H1299, NCI-H1650, A549, NCI-H1975 and NCI-H460) than in normal lung cell line BEAS-2B (Fig. 1D).
Figure 1.
Expression of miR-454-3p is downregulated in NSCLC tissues and cell lines. (A) The relative expression level of miR-454-3p in 56 NSCLC tissues and paired adjacent normal tissues. (B) NSCLC tissues (n=56) were divided into two groups (miR-454-3p low expression and high expression) based on the median value. (C) The relationship between the overall survival and miR-454-3p expression in NSCLC patients was analyzed by Kaplan-Meier method. (D) RT-qPCR analysis for miR-454-3p expression in BEAS-2B and 5 NSCLC cell lines. Data are presented as the mean ± SD. *P<0.05, **P<0.01 and ***P<0.001 compared with BEAS-2B cells. miR, microRNA; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR.
Table I.
Association between miR-454-3p and clinicopathological characteristics of patients with NSCLC.
| Expression level of miR-454-3p | ||||
|---|---|---|---|---|
| Pathological parameters | n | High | Low | P-value |
| Age (years) | 0.7825 | |||
| <50 | 21 | 10 | 11 | |
| ≥50 | 35 | 18 | 17 | |
| Sex | ||||
| Male | 30 | 14 | 16 | 0.5920 |
| Female | 26 | 14 | 12 | |
| Tumor size | 0.0331a | |||
| <3 cm | 27 | 17 | 10 | |
| ≥3 cm | 29 | 10 | 19 | |
| Pathological stage | 0.0044b | |||
| Well | 20 | 14 | 6 | |
| Poor | 36 | 11 | 25 | |
| Metastasis | 0.0014b | |||
| Negative | 31 | 22 | 9 | |
| Positive | 25 | 7 | 18 | |
P<0.05, the expression of miR-454-3p was significantly associated with tumor size
P<0.01, the expression of miR-454-3p was significantly associated with pathological stage and tumor metastasis. miR, microRNA; NSCLC, non-small cell lung cancer.
miR-454-3p inhibits the proliferation of NSCLC cells
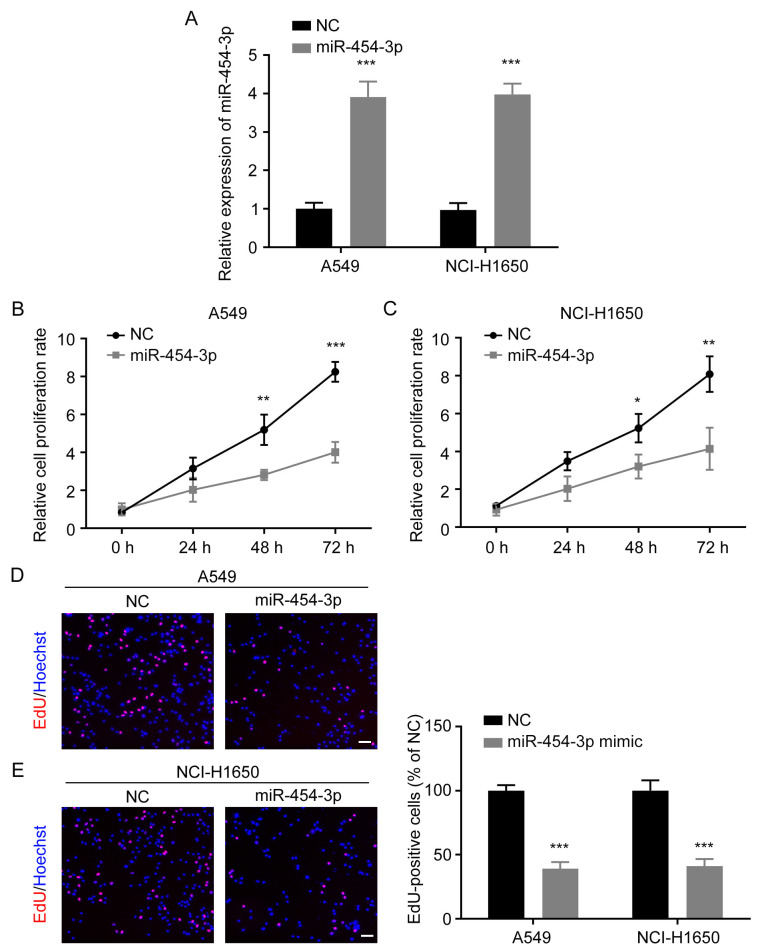
A549 and NCI-H1650 cells were transfected with NC or miR-454-3p mimics. The transfection efficiency was determined by RT-qPCR and it was revealed that the expression of miR-454-3p was significantly increased in both A549 and NCI-H1650 cells after transfection with miR-454-3p mimics (Fig. 2A). Then, CCK-8 and EdU cell proliferation assays were performed to investigate the effect of miR-454-3p overexpression on NSCLC cell proliferation. The CCK-8 assay revealed that miR-454-3p overexpression significantly suppressed the proliferation rate of A549 and NCI-H1650 cells (Fig. 2B and C). EdU assay also revealed that the overexpression of miR-454-3p markedly reduced the number of EdU-positive cells (Fig. 2D and E). These results provided evidence that miR-454-3p functions as a tumor suppressor and inhibits the proliferation of NSCLC cells.
Figure 2.
Ectopic expression of miR-454-3p suppresses NSCLC cell proliferation. (A) The expression of miR-454-3p in NSCLC cells transfected with miR-454-3p mimics was evaluated by qPCR assay. (B and C) A549 and NCI-H1650 cells were transfected with miR-454-3p mimics for 24 h, and the cell proliferation rate was determined by CCK-8 assay. (D and E) The effect of miR-454-3p on A549 and NCI-H1650 cell proliferation was detected by EdU cell proliferation kit. EdU-positive cells were photographed under an inverted fluorescence microscope. Scale bar, 100 µm. Data are presented as the mean ± SD, n=3. *P<0.05, **P<0.01 and ***P<0.001 vs. cells transfected with the NC-mimic group. miR, microRNA; NSCLC, non-small cell lung cancer; qPCR, quantitative PCR; CCK-8, Cell Counting Kit-8; EdU, 5-ethynyl-2-deoxyuridine; NC, negative control mimics; miR-454-3p, miR-454-3p mimics.
miR-454-3p attenuates metastases and suppresses EMT in NSCLC cells
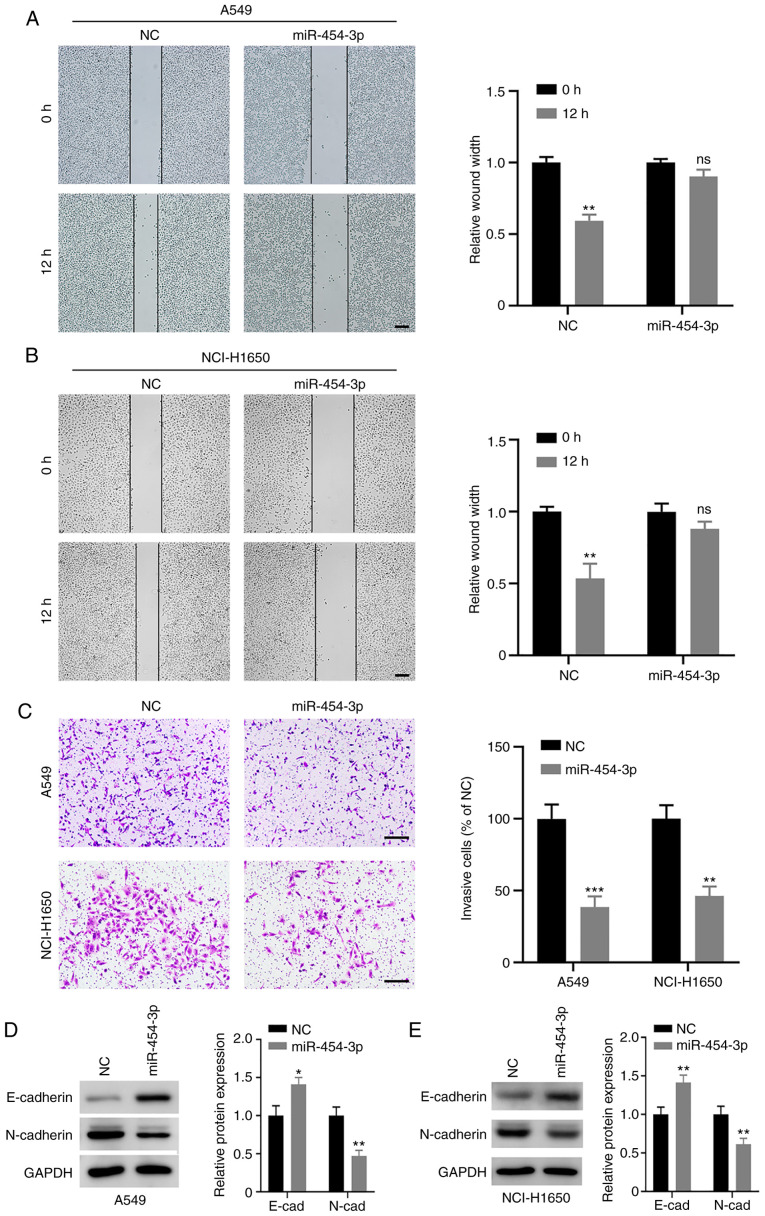
To further determine the biological effect of miR-454-3p on NSCLC cell metastasis, wound-healing and Transwell invasion assays were conducted. The wound-healing assay revealed that the migratory capability of A549 and NCI-H1650 cells was attenuated when miR-454-3p was overexpressed (Fig. 3A and B). Furthermore, miR-454-3p significantly decreased the number of invasive A549 and NCI-H1650 cells (Fig. 3C). Next, the role of miR-454-3p in EMT processes was evaluated. The present results revealed that the overexpression of miR-454-3p led to a decrease in mesenchymal marker N-cadherin and increased the expression of epithelial marker E-cadherin (Fig. 3D and E). Collectively, these data indicated that miR-454-3p may block EMT to inhibit NSCLC cell metastasis.
Figure 3.
Effect of miR-454-3p on migration, invasion, and EMT in NSCLC cells. (A and B) A549 and NCI-H1650 cells were transfected with NC or miR-454-3p mimics, and the migration capacity of cells was assessed with a wound healing assay. Scale bar, 200 µm. (C) The effect of miR-454-3p overexpression on NSCLC cell invasion was examined using a Transwell invasion assay. Scale bar, 200 µm. (D and E) The protein levels of EMT markers in A549 and NCI-H1650 cells after transfection with NC or miR-454-3p mimics were determined by western blotting. Data are presented as the mean ± SD, n=3. *P<0.05, **P<0.01 and ***P<0.001 compared with the NC group. miR, microRNA; EMT, epithelial-mesenchymal transition; NSCLC, non-small cell lung cancer; NC, negative control mimics; ns, not significant; miR-454-3p, miR-454-3p mimics.
TGFB2 is a direct target of miR-454-3p in NSCLC
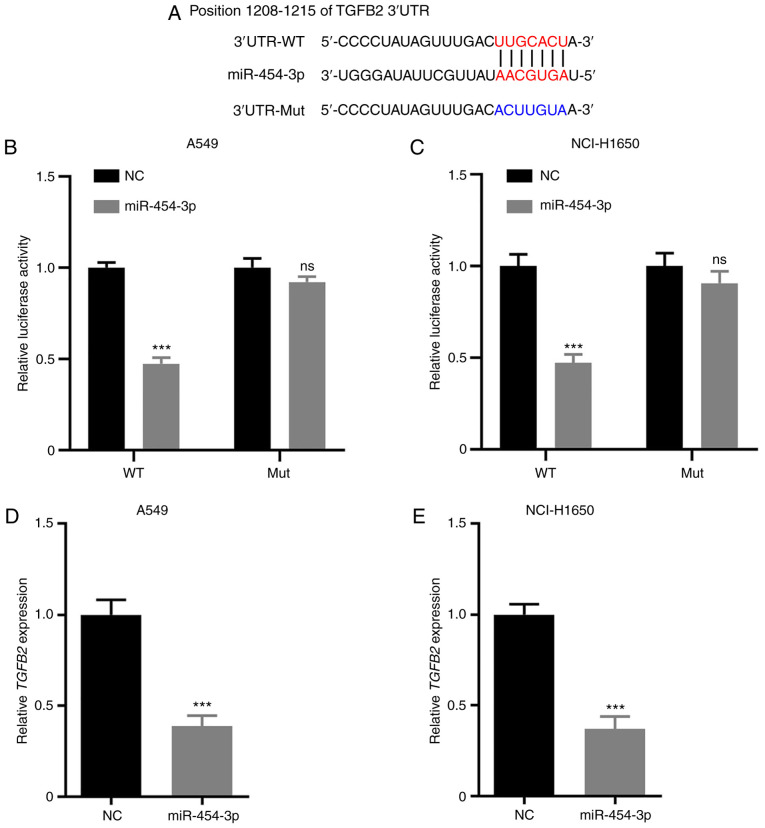
TargetScan was selected for bioinformatics analysis to identify a potential target gene of miR-454-3p. The results suggested that TGFB2 is a putative target of miR-454-3p (Fig. 4A). To confirm whether the 3′-UTR of TGFB2 could be directly targeted by miR-454-3p, a luciferase reporter assay was performed. The results revealed that miR-454-3p overexpression resulted in significantly decreased the luciferase activity in the WT 3′-UTR of TGFB2, but not the Mut reporter (Fig. 4B and C). In addition, RT-qPCR assays were performed to investigate the mRNA levels of TGFB2 in A549 and NCI-H1650 cells transfected with NC or miR-454-3p mimics. As revealed in Fig. 4D and E, overexpression of miR-454-3p significantly attenuated TGFB2 mRNA expression. Consequently, these results demonstrated that TGFB2 is a direct target of miR-454-3p in NSCLC cells.
Figure 4.
TGFB2 is a direct target of miR-454-3p in NSCLC cells. (A) The predicted seed region of miR-454-3p at the 3′UTR of TGFB2 was analyzed with TargetScan software and the mutated version is presented. (B and C) A549 and NCI-H1650 cells were co-transfected with miR-454-3p mimics and TGFB2-WT or TGFB2-Mut for 48 h. Then, the luciferase activity was detected using the Dual Luciferase Reporter Assay kit. (D and E) The TGFB2 mRNA expression in A549 and NCI-H1650 cells transfected with NC or miR-454-3p mimics was assessed by RT-qPCR. Data are expressed as the mean ± SD, n=3. ***P<0.001 compared with NC group. TGFB2, transforming growth factor-β2; miR, microRNA; NSCLC, non-small cell lung cancer; WT, wild-type; Mut, mutant; NC, negative control mimics; RT-qPCR, reverse transcription-quantitative PCR; ns, not significant; miR-454-3p, miR-454-3p mimics.
miR-454-3p is negatively correlated with TGFB2 expression in NSCLC tissues
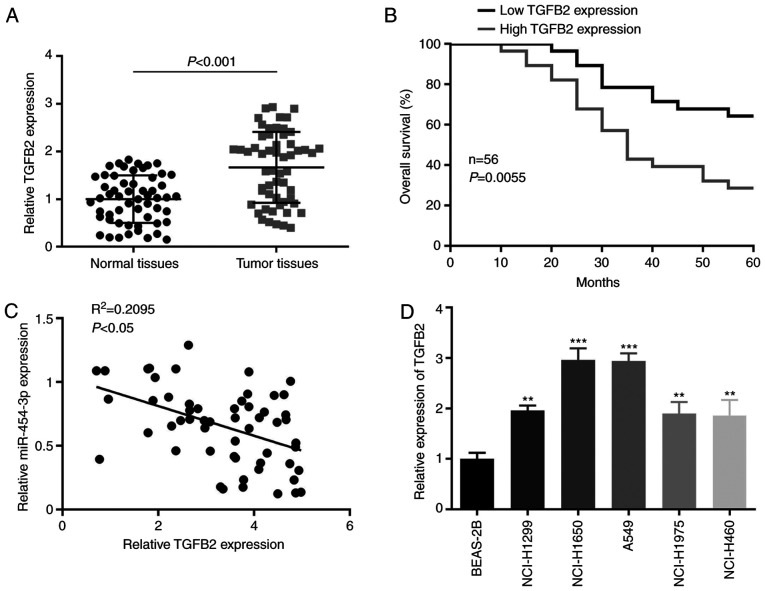
To determine the association between miR-454-3p and TGFB2 in NSCLC tissues, the expression of TGFB2 in NSCLC tissues and adjacent non-cancerous tissues was examined. It was revealed that the expression level of TGFB2 was significantly higher in NSCLC tissues than in non-tumor tissues (Fig. 5A) and high TGFB2 expression indicated poor prognosis in patients with NSCLC (Fig. 5B). In addition, it was observed that the expression of miR-454-3p had a negative correlation with TGFB2 expression in NSCLC tissues (Fig. 5C). Furthermore, when compared with normal lung cell line BEAS-2B, TGFB2 expression was significantly higher in all five NSCLC cell lines (NCI-H1299, NCI-H1650, A549, NCI-H1975 and NCI-H460) (Fig. 5D). Collectively, the present results revealed that TGFB2 expression was negatively correlated with miR-454-3p in NSCLC tissues and cells, which may confirm that TGFB2 could be a target of miR-454-3p.
Figure 5.
TGFB2 is upregulated in NSCLC tissues and cells. (A) The expression of TGFB2 was significantly higher in NSCLC tissues than in adjacent non-cancerous tissues. (B) The association between TGFB2 expression level and overall survival in 56 NSCLC patients was assessed by Kaplan-Meier analysis. (C) The correlation between TGFB2 and miR-454-3p in NSCLC tissues was analyzed by Spearman's analysis. (D) RT-qPCR analysis of the TGFB2 expression level in NSCLC cells. Data are presented as the mean ± SD, n=3. **P<0.01 and ***P<0.001 vs. the BEAS-2B group. TGFB2, transforming growth factor-β2; NSCLC, non-small cell lung cancer; miR, microRNA; RT-qPCR, reverse transcription-quantitative PCR.
TGFB2 overexpression abrogates miR-454-3p-mediated inhibition of NSCLC cell proliferation
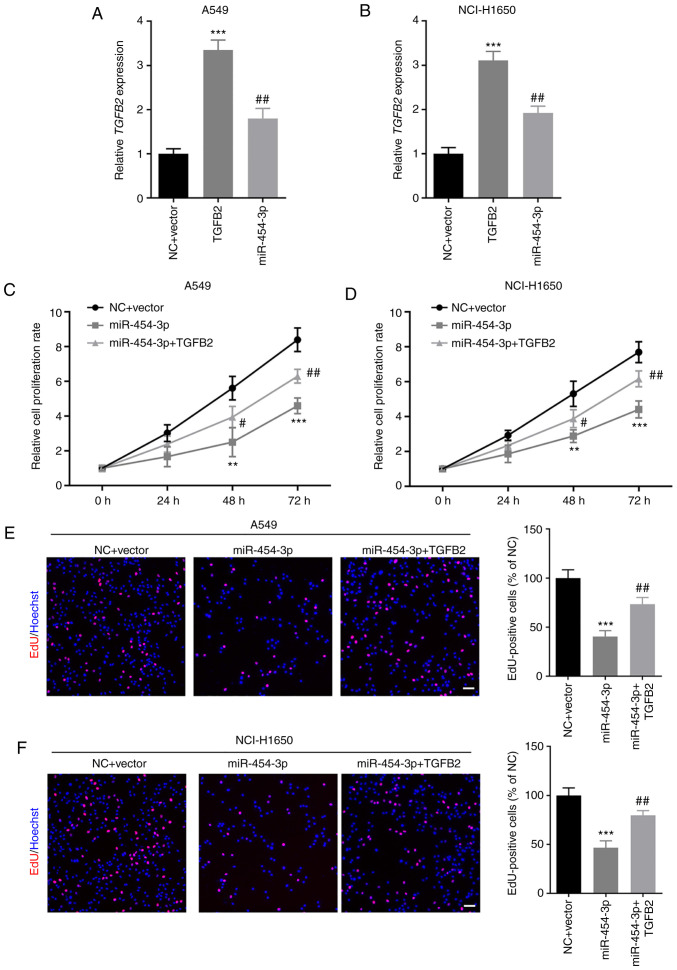
Next, the effect of TGFB2 and miR-454-3p on NSCLC cell proliferation was investigated. First, A549 and NCI-H1650 cells were co-transfected with TGFB2-overexpressing plasmid and miR-454-3p mimics. RT-qPCR assays revealed that TGFB2 was significantly upregulated in A549 and NCI-H1650 cells after transfection with TGFB2 overexpression plasmid, which could be significantly inhibited by miR-454-3p mimics (Fig. 6A and B). Then, CCK-8 and EdU cell proliferation assays were performed to examine the proliferation of NSCLC cells. The cell proliferation rate was significantly increased in NSCLC cells co-transfected with TGFB2 plasmid and miR-454-3p mimics compared with cells transfected with miR-454-3p mimics alone (Fig. 6C and D). Furthermore, the number of EdU-positive A549 and NCI-H1650 cells in the co-transfection groups was significantly higher than cells transfected with miR-454-3p mimics alone (Fig. 6E and F). The results indicated that the overexpression of TGFB2 abrogated the miR-454-3p-mediated inhibitory effect on NSCLC cell proliferation.
Figure 6.
TGFB2 overexpression counteracts the miR-454-3p-mediated suppressive effects on NSCLC cell proliferation. (A and B) A549 and NCI-H1650 cells were co-transfected with TGFB2-overexpressing plasmid and miR-454-3p mimics and then the expression of TGFB2 was determined by RT-qPCR assay. (C-F) The effect of miR-454-3p and TGFB2 on the proliferation of A549 and NCI-H1650 cells was examined by (C and D) CCK-8 assay and (E and F) EdU cell proliferation assay. Scale bar, 100 µm. Data are presented as the mean ± SD, n=3. **P<0.01 and ***P<0.001 compared with cells transfected with the NC + Vector group. #P<0.05 and ##P<0.01 compared with cells transfected with miR-454-3p mimics. TGFB2, transforming growth factor-β2; miR, microRNA; NSCLC, non-small cell lung cancer; RT-qPCR, reverse transcription-quantitative PCR; CCK-8, Cell Counting Kit-8; EdU, 5-ethynyl-2-deoxyuridine; NC, negative control mimics; miR-454-3p, miR-454-3p mimics.
TGFB2 reverses miR-454-3p-mediated inhibition of metastasis and EMT in NSCLC cells
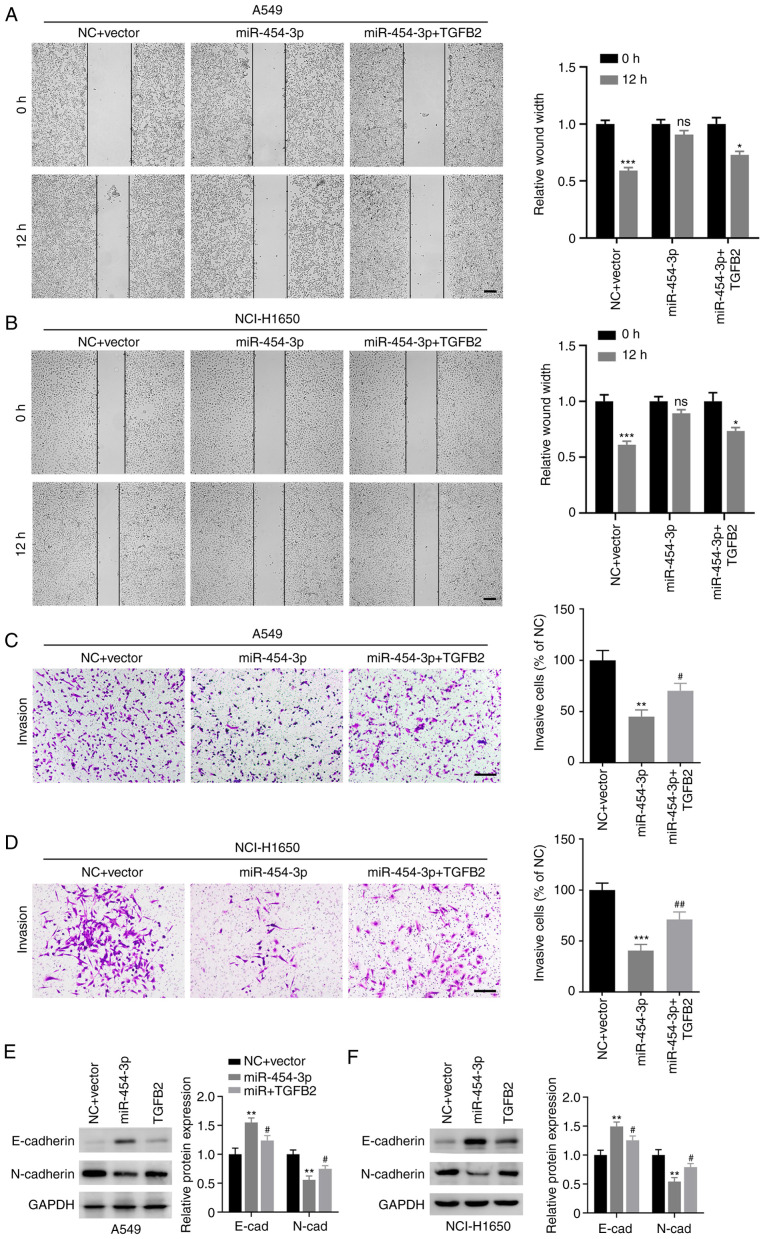
Subsequently, the role of TGFB2 in miR-454-3p-mediated NSCLC cell metastasis and EMT were evaluated. As revealed in Fig. 7A-D, the number of migrated and invasive cells was markedly decreased in A549 and NCI-H1650 cells transfected miR-454-3p mimics, which could be reversed by overexpression of TGFB2. In addition, TGFB2 overexpression decreased the expression E-cadherin and increased the expression of N-cadherin in A549 and NCI-H1650 cells (Fig. 7E and F). Collectively, these data demonstrated that miR-454-3p inhibited the metastasis and EMT of NSCLC cells via antagonizing TGFB2 expression.
Figure 7.
TGFB2 reverses miR-454-3p-mediated inhibition of migration, invasion and EMT in NSCLC cells. A549 and NCI-H1650 cells were transfected with TGFB2-overexpressing plasmid and miR-454-3p mimics. (A and B) A wound healing assay was performed to evaluate cell migration capacity. Scale bar, 200 µm. (C and D) Transwell invasion assay was conducted to investigate cell invasion capacity. Scale bar, 200 µm. (E and F) Western blot analysis was used to reveal the expression of E-cadherin and N-cadherin in cells after the indicated transfections. Data are presented as the mean ± SD, n=3. *P<0.05, **P<0.01 and ***P<0.001 vs. cells transfected with the NC + Vector group. #P<0.05 and ##P<0.01 vs. cells transfected with miR-454-3p mimics. TGFB2, transforming growth factor-β2; miR, microRNA; EMT, epithelial-mesenchymal transition; NSCLC, non-small cell lung cancer; NC, negative control mimics; miR-454-3p, miR-454-3p mimics; ns, no significance.
Discussion
NSCLC is closely associated with high cancer-related incidence and fatality rate (25). Challenges remain due to failure of early diagnosis of the cancer, metastasis or drug resistance (26–28). The last two decades have witnessed the rapid developments of tumor epigenetic alternations (29,30). miRNAs play an important role in tumorigenesis and development (31,32). It is urgent to determine the expression, functions and potential targets of dysregulated miRNAs in NSCLC, which could provide effective strategies to improve the treatment and prognosis of patients.
It has been reported that miR-454-3p was downregulated in various cancers and its ectopic expression impaired tumor malignant behaviors (12,33). For example, miR-454-3p suppressed cell proliferation and induced apoptosis by directly downregulating nuclear factor of activated T-cells c2 (NFATc2) in glioblastoma (GBM) (12). It also targeted cytoplasmic polyadenylation element-binding protein 1 (CPEB1) to inhibit GBM cell proliferation and mobility (34). For these reasons, it may be regarded as a tumor suppressor and a novel biomarker for GBM therapy. In addition, low miR-454-3p expression in bladder cancer tissues, predicted poor prognosis of bladder cancer patients (14). Moreover, miR-454-3p functioned as a tumor inhibitor of esophageal cancer by inhibiting the ERK/AKT signaling pathways (35). With regard to NSCLC, the expression of miR-454-3p was downregulated in NSCLC tissues and cell lines (13). Further study suggested that aberrant expression of miR-454-3p significantly restrained NSCLC cell proliferation and accelerated cell apoptosis by downregulating calbindin 1 (CALB1) (13). These studies indicated that miR-454-3p could be a tumor-suppressive miRNA as revealed in a great number of cancer types. Conversely, it was observed to be upregulated in breast cancer and further enhanced cell metastasis, leading to a shorter relapse-free survival of patients with breast cancer (36). Song et al expounded that miR-454-3p depletion reduced cervical cancer cell growth, facilitating cancer cell apoptosis (37). Thus, miR-454-3p can function as either a tumorigenic or tumor suppressive miRNA, which may depend on the type of cancer. In present study, the expression of miR-454-3p was revealed to be decreased in NSCLC tissues and cells. Further mechanistic studies suggested that its aberrant expression attenuated migration and invasion and had an impact on E-cadherin and N-cadherin expression in NSCLC cells.
To gain insight into the mechanism of miR-454-3p in NSCLC, TargetScan database was applied to predict the potential target genes of miR-454-3p. Among the candidate genes, a complementary miR-454-3p site was present in the 3′-UTR of TGFB2 mRNA. Previous evidence has revealed the involvement of TGFB2 in numerous malignancies (38,39). As a result, TGFB2 was selected for subsequent investigations. Dual luciferase reporter assays revealed that TGFB2 could directly bind to miR-454-3p in NSCLC cells. TGFB2 is known as an isoform of the TGFB family. TGFBs could function as oncogenic genes and bind with a range of TGFB receptors and activate transcription factors such as the SMAD superfamily members which regulate cell homeostasis (39–41). TGFBs (TGFB1, TGFB2, and TGFB3) were upregulated in gastric carcinoma, however, only high expression of TGFB2 was associated with poor prognosis of gastric carcinoma patients (42). Schlingensiepen et al also reported that the high expression of TGFB2 was revealed in high-grade gliomas and indicated an unfavorable prognosis of glioma patients (43). Moreover, TGFB2 could induce EMT and promote tumor metastasis in nasopharyngeal carcinoma (44) and gallbladder carcinoma (45). The major biological function of miRNAs is to regulate the expression of genes by post-transcriptional mRNA regulation (31). Some miRNAs have been reported to regulate TGFB2 in malignant tumors. miR-200a suppressed renal cell carcinoma development by directly targeting TGFB2 (46). A recent study revealed that TGFB2 downregulation mediated by miR-324-5p mimics could inhibit gallbladder carcinoma cell migration, invasion and EMT, which contributed to suppression of gallbladder carcinoma metastatic behaviors (45). In the present study, the results revealed that TGFB2 was markedly upregulated in NSCLC tissues and cell lines. Moreover, it was negatively correlated with miR-454-3p in NSCLC tissues. Further research corroborated that TGFB2 reversed miR-454-3p-mediated migration, invasion and E-cadherin and N-cadherin expression in NSCLC cells.
TGFB2 is a hallmark in various malignant tumors, and treatment of cancer based on the specific inhibition of TGFB2 is currently being developed (17). The mode of action, effectuality and tolerability in vitro and in vivo of Trabedersen (AP-12009), an antisense oligodeoxynucleotide inhibitor targeting TGFB2, has been confirmed (47). Furthermore, AP-12009 is being evaluated in a phase I/II dose escalation study with high-grade glioma, metastatic colorectal carcinoma, metastatic melanoma, and advanced pancreatic carcinoma (47,48). In present study, the regulatory effect of miR-454-3p/TGFB2 concatenation on NSCLC malignant behaviors in vitro was confirmed. However, the study of miR-454-3p and TGFB2 in vivo remains to be further investigated. In addition, studies using TGFB2 inhibitors can be carried out in vitro and in vivo, which may provide a more rationale and potential treatment target for NSCLC patients.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets generated and analyzed during the study are available from the corresponding author upon reasonable request.
Authors' contributions
HL and RW conceived and designed this study. YL and LK collected the samples and performed the experiments. YX and TY analyzed and verified the data. HL wrote the manuscript. RW revised the manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The human tissue used in the present study were obtained from Yuebei People's Hospital of Shaoguan (Shaoguan, China). The data were published after internal ethical approval and patient consent were obtained. The proposed experiments and procedures were reviewed and approved by the Ethics Committee of Yuebei People's Hospital. The present study was performed according to the Declaration of Helsinki of 1975 and its succeeding amendments.
Patient consent for publication
Not applicable.
Competing interests
The author declare that they have no competing interests.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70:7–30. doi: 10.3322/caac.21601. [DOI] [PubMed] [Google Scholar]
- 2.Didkowska J, Wojciechowska U, Mańczuk M, Łobaszewski J. Lung cancer epidemiology: Contemporary and future challenges worldwide. Ann Transl Med. 2016;4:150. doi: 10.21037/atm.2016.03.11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jonna S, Subramaniam DS. Molecular diagnostics and targeted therapies in non-small cell lung cancer (NSCLC): An update. Discov Med. 2019;27:167–170. [PubMed] [Google Scholar]
- 4.Foster CC, Pitroda SP, Weichselbaum RR. Beyond palliation: The rationale for metastasis-directed therapy for metastatic non-small cell lung cancer. J Thorac Oncol. 2019;14:1510–1512. doi: 10.1016/j.jtho.2019.05.025. [DOI] [PubMed] [Google Scholar]
- 5.Galagali H, Kim JK. The multifaceted roles of microRNAs in differentiation. Curr Opin Cell Biol. 2020;67:118–140. doi: 10.1016/j.ceb.2020.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee TJ, Yuan X, Kerr K, Yoo JY, Kim DH, Kaur B, Eltzschig HK. Strategies to modulate MicroRNA functions for the treatment of cancer or organ injury. Pharmacol Rev. 2020;72:639–667. doi: 10.1124/pr.119.019026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu M, Wang G, Tian W, Deng Y, Xu Y. miRNA-based therapeutics for lung cancer. Curr Pharm Des. 2018;23:5989–5996. doi: 10.2174/1381612823666170714151715. [DOI] [PubMed] [Google Scholar]
- 8.Yuan Y, Liao H, Pu Q, Ke X, Hu X, Ma Y, Luo X, Jiang Q, Gong Y, Wu M, et al. miR-410 induces both epithelial-mesenchymal transition and radioresistance through activation of the PI3K/mTOR pathway in non-small cell lung cancer. Signal Transduct Target Ther. 2020;5:85. doi: 10.1038/s41392-020-0182-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun X, Xu M, Liu H, Ming K. MicroRNA-219 is downregulated in non-small cell lung cancer and inhibits cell growth and metastasis by targeting HMGA2. Mol Med Rep. 2017;16:3557–3564. doi: 10.3892/mmr.2017.7000. [DOI] [PubMed] [Google Scholar]
- 10.Xing F, Song Z, He Y. miR-219-5p inhibits growth and metastasis of ovarian cancer cells by targeting HMGA2. Biol Res. 2018;51:50. doi: 10.1186/s40659-018-0199-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shao Y, Liang B, Long F, Jiang SJ. Diagnostic MicroRNA biomarker discovery for non-small-cell lung cancer adenocarcinoma by integrative bioinformatics analysis. Biomed Res Int. 2017;2017:2563085. doi: 10.1155/2017/2563085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zuo J, Yu H, Xie P, Liu W, Wang K, Ni H. miR-454-3p exerts tumor-suppressive functions by down-regulation of NFATc2 in glioblastoma. Gene. 2019;710:233–239. doi: 10.1016/j.gene.2019.06.008. [DOI] [PubMed] [Google Scholar]
- 13.Jin C, Lin T, Shan L. Downregulation of calbindin 1 by miR-454-3p suppresses cell proliferation in nonsmall cell lung cancer in vitro. Cancer Biother Radiopharm. 2019;34:119–127. doi: 10.1089/cbr.2018.2598. [DOI] [PubMed] [Google Scholar]
- 14.Wang S, Zhang G, Zheng W, Xue Q, Wei D, Zheng Y, Yuan J. miR-454-3p and miR-374b-5p suppress migration and invasion of bladder cancer cells through targetting ZEB2. Biosci Rep. 2018;38:BSR20181436. doi: 10.1042/BSR20181436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fan Y, Shi C, Li T, Kuang T. microRNA-454 shows anti-angiogenic and anti-metastatic activity in pancreatic ductal adenocarcinoma by targeting LRP6. Am J Cancer Res. 2017;7:139–147. [PMC free article] [PubMed] [Google Scholar]
- 16.Abraham CG, Ludwig MP, Andrysik Z, Pandey A, Joshi M, Galbraith MD, Sullivan KD, Espinosa JM. ΔNp63α suppresses TGFB2 expression and RHOA activity to drive cell proliferation in squamous cell carcinomas. Cell Rep. 2018;24:3224–3236. doi: 10.1016/j.celrep.2018.08.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yang B, Bai J, Shi R, Shao X, Yang Y, Jin Y, Che X, Zhang Y, Qu X, Liu Y, Li Z. TGFB2 serves as a link between epithelial-mesenchymal transition and tumor mutation burden in gastric cancer. Int Immunopharmacol. 2020;84:106532. doi: 10.1016/j.intimp.2020.106532. [DOI] [PubMed] [Google Scholar]
- 18.Dave H, Trivedi S, Shah M, Shukla S. Transforming growth factor beta 2: A predictive marker for breast cancer. Indian J Exp Biol. 2011;49:879–887. [PubMed] [Google Scholar]
- 19.Bruna A, Darken RS, Rojo F, Ocaña A, Peñuelas S, Arias A, Paris R, Tortosa A, Mora J, Baselga J, Seoane J. High TGFbeta-Smad activity confers poor prognosis in glioma patients and promotes cell proliferation depending on the methylation of the PDGF-B gene. Cancer Cell. 2007;11:147–160. doi: 10.1016/j.ccr.2006.11.023. [DOI] [PubMed] [Google Scholar]
- 20.Mittal V. Epithelial mesenchymal transition in aggressive lung cancers. Adv Exp Med Biol. 2016;890:37–56. doi: 10.1007/978-3-319-24932-2_3. [DOI] [PubMed] [Google Scholar]
- 21.Singh M, Yelle N, Venugopal C, Singh SK. EMT: Mechanisms and therapeutic implications. Pharmacol Ther. 2018;182:80–94. doi: 10.1016/j.pharmthera.2017.08.009. [DOI] [PubMed] [Google Scholar]
- 22.Chang H, Brown CW, Matzuk MM. Genetic analysis of the mammalian transforming growth factor-beta superfamily. Endocr Rev. 2002;23:787–823. doi: 10.1210/er.2002-0003. [DOI] [PubMed] [Google Scholar]
- 23.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 24.Rodriguez LG, Wu X, Guan JL. Wound-healing assay. Methods Mol Biol. 2005;294:23–29. doi: 10.1385/1-59259-860-9:023. [DOI] [PubMed] [Google Scholar]
- 25.Bray F, Ferlay J, Soerjomataram I, Siegel RL, Torre LA, Jemal A. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin. 2018;68:394–424. doi: 10.3322/caac.21492. [DOI] [PubMed] [Google Scholar]
- 26.Hung JJ, Jeng WJ, Hsu WH, Chou TY, Huang BS, Wu YC. Predictors of death, local recurrence, and distant metastasis in completely resected pathological stage-I non-small-cell lung cancer. J Thorac Oncol. 2012;7:1115–1123. doi: 10.1097/JTO.0b013e31823b564a. [DOI] [PubMed] [Google Scholar]
- 27.Lee WC, Reuben A, Hu X, McGranahan N, Chen R, Jalali A, Negrao MV, Hubert SM, Tang C, Wu CC, et al. Multiomics profiling of primary lung cancers and distant metastases reveals immunosuppression as a common characteristic of tumor cells with metastatic plasticity. Genome Biol. 2020;21:271. doi: 10.1186/s13059-020-02175-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lim ZF, Ma PC. Emerging insights of tumor heterogeneity and drug resistance mechanisms in lung cancer targeted therapy. J Hematol Oncol. 2019;12:134. doi: 10.1186/s13045-019-0818-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Diaz-Lagares A, Mendez-Gonzalez J, Hervas D, Saigi M, Pajares MJ, Garcia D, Crujerias AB, Pio R, Montuenga LM, Zulueta J, et al. A novel epigenetic signature for early diagnosis in lung cancer. Clin Cancer Res. 2016;22:3361–3371. doi: 10.1158/1078-0432.CCR-15-2346. [DOI] [PubMed] [Google Scholar]
- 30.Duruisseaux M, Esteller M. Lung cancer epigenetics: From knowledge to applications. Semin Cancer Biol. 2018;51:116–128. doi: 10.1016/j.semcancer.2017.09.005. [DOI] [PubMed] [Google Scholar]
- 31.Iqbal MA, Arora S, Prakasam G, Calin GA, Syed MA. MicroRNA in lung cancer: Role, mechanisms, pathways and therapeutic relevance. Mol Aspects Med. 2019;70:3–20. doi: 10.1016/j.mam.2018.07.003. [DOI] [PubMed] [Google Scholar]
- 32.Wu KL, Tsai YM, Lien CT, Kuo PL, Hung AJ. The roles of MicroRNA in lung cancer. Int J Mol Sci. 2019;20:1611. doi: 10.3390/ijms20071611. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jiang D, Li H, Xiang H, Gao M, Yin C, Wang H, Sun Y, Xiong M. Long chain non-coding RNA (lncRNA) HOTAIR knockdown increases miR-454-3p to suppress gastric cancer growth by targeting STAT3/cyclin D1. Med Sci Monit. 2019;25:1537–1548. doi: 10.12659/MSM.913087. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 34.Hui X, Zhang S, Wang Y. miR-454-3p suppresses cell migration and invasion by targeting CPEB1 in human glioblastoma. Mol Med Rep. 2018;18:3965–3972. doi: 10.3892/mmr.2018.9386. [DOI] [PubMed] [Google Scholar]
- 35.Yan A, Wang C, Zheng L, Zhou J, Zhang Y. MicroRNA-454-3p inhibits cell proliferation and invasion in esophageal cancer by targeting insulin-like growth factor 2 mRNA-binding protein 1. Oncol Lett. 2020;20:359. doi: 10.3892/ol.2020.12223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ren L, Chen H, Song J, Chen X, Lin C, Zhang X, Hou N, Pan J, Zhou Z, Wang L, et al. miR-454-3p-mediated Wnt/β-catenin signaling antagonists suppression promotes breast cancer metastasis. Theranostics. 2019;9:449–465. doi: 10.7150/thno.29055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Song Y, Guo Q, Gao S, Hua K. miR-454-3p promotes proliferation and induces apoptosis in human cervical cancer cells by targeting TRIM3. Biochem Biophys Res Commun. 2019;516:872–879. doi: 10.1016/j.bbrc.2019.06.126. [DOI] [PubMed] [Google Scholar]
- 38.Bartel DP. MicroRNAs: Target recognition and regulatory functions. Cell. 2009;136:215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Strauss J, Heery CR, Schlom J, Madan RA, Cao L, Kang Z, Lamping E, Marté JL, Donahue RN, Grenga I, et al. Phase I trial of M7824 (MSB0011359C), a bifunctional fusion protein targeting PD-L1 and TGFβ, in advanced solid tumors. Clin Cancer Res. 2018;24:1287–1295. doi: 10.1158/1078-0432.CCR-17-2653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Derynck R, Akhurst RJ, Balmain A. TGF-beta signaling in tumor suppression and cancer progression. Nat Genet. 2001;29:117–129. doi: 10.1038/ng1001-117. [DOI] [PubMed] [Google Scholar]
- 41.Moustakas A, Heldin CH. Mechanisms of TGFβ-induced epithelial-mesenchymal transition. J Clin Med. 2016;5:63. doi: 10.3390/jcm5070063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Vagenas K, Spyropoulos C, Gavala V, Tsamandas AC. TGFbeta1, TGFbeta2, and TGFbeta3 protein expression in gastric carcinomas: Correlation with prognostics factors and patient survival. J Surg Res. 2007;139:182–188. doi: 10.1016/j.jss.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 43.Schlingensiepen KH, Schlingensiepen R, Steinbrecher A, Hau P, Bogdahn U, Fischer-Blass B, Jachimczak P. Targeted tumor therapy with the TGF-beta 2 antisense compound AP 12009. Cytokine Growth Factor Rev. 2006;17:129–139. doi: 10.1016/j.cytogfr.2005.09.002. [DOI] [PubMed] [Google Scholar]
- 44.Zhao L, Lin L, Pan C, Shi M, Liao Y, Bin J, Liao W. Flotillin-2 promotes nasopharyngeal carcinoma metastasis and is necessary for the epithelial-mesenchymal transition induced by transforming growth factor-β. Oncotarget. 2015;6:9781–9793. doi: 10.18632/oncotarget.3382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang X, Zhang L, Chen M, Liu D. miR-324-5p inhibits gallbladder carcinoma cell metastatic behaviours by downregulation of transforming growth factor beta 2 expression. Artif Cells Nanomed Biotechnol. 2020;48:315–324. doi: 10.1080/21691401.2019.1703724. [DOI] [PubMed] [Google Scholar]
- 46.Lu R, Ji Z, Li X, Qin J, Cui G, Chen J, Zhai Q, Zhao C, Zhang W, Yu Z. Tumor suppressive microRNA-200a inhibits renal cell carcinoma development by directly targeting TGFB2. Tumour Biol. 2015;36:6691–6700. doi: 10.1007/s13277-015-3355-9. [DOI] [PubMed] [Google Scholar]
- 47.Jaschinski F, Rothhammer T, Jachimczak P, Seitz C, Schneider A, Schlingensiepen KH. The antisense oligonucleotide trabedersen (AP 12009) for the targeted inhibition of TGF-β2. Curr Pharm Biotechnol. 2011;12:2203–2213. doi: 10.2174/138920111798808266. [DOI] [PubMed] [Google Scholar]
- 48.Bogdahn U, Hau P, Stockhammer G, Venkataramana NK, Mahapatra AK, Suri A, Balasubramaniam A, Nair S, Oliushine V, Parfenov V, et al. Targeted therapy for high-grade glioma with the TGF-β2 inhibitor trabedersen: Results of a randomized and controlled phase IIb study. Neuro Oncol. 2011;13:132–142. doi: 10.1093/neuonc/noq142. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets generated and analyzed during the study are available from the corresponding author upon reasonable request.