Abstract
Osteosarcoma (OS) is a rare type of tumor and mostly occurs in children and adolescents. Approximately 10–25% of patients with OS have lung metastases, and lung damage caused by lung metastasis is the main cause of mortality. Therefore, studying the growth and metastasis of OS is key in reducing OS mortality and improving prognosis. The expression of long non-coding RNA (lncRNA) cancer susceptibility 15 (CASC15) in OS patients or OS cell lines were quantified by reverse transcription-quantitative polymerase chain reaction (RT-qPCR). The expression of vimentin, E-cadherin, N-cadherin, and cyclin D were detected by RT-qPCR and western blotting. Mice were injected with OS cell lines via the tail vein to observe tumor formation in the lung. CCK-8 and EdU assays were utilized to evaluate cell proliferation. Both Ttranswell assay and cell scratch test detected cell migration. The results revealed that lncRNA-CASC15 was highly expressed in clinical samples and OS cells. In vitro verification experiments revealed that CASC15 promoted the growth of OS cells. Rescue experiments demonstrated that CASC15 affected the cell cycle by activating the Wnt/β-catenin pathway, thereby promoting cell proliferation. Furthermore, the transfection dose test indicated that lentiviruses expressing various doses of CASC15-overexpression (oe-CASC15) altered the proliferation and migration status of OS cells. CASC15 promoted OS cell metastasis both in vivo and in vitro. The overexpression of CASC15 revealed that the occurrence of metastasis was also related to the Wnt/β-catenin pathway. The western blotting results revealed that CASC15 could lead to β-catenin entering the nucleus via the Wnt pathway to promote the epithelial-mesenchymal transition (EMT) of OS cells. To sum up, CASC15 promoted the proliferation of OS cells in vitro and the growth of OS xenograft tumors in vivo. Moreover, CASC15 promoted the entry of β-catenin into the nucleus, thus activating the Wnt pathway and subsequently promoting the EMT of OS cells.
Keywords: long non-coding RNA cancer susceptibility 15, osteosarcoma, Wnt/β-catenin, epithelial-mesenchymal transition
Introduction
Osteosarcoma (OS) is a rare tumor type, which mostly occurs in children and adolescents (1). OS usually occurs at the fixed end of long bones. Several studies have found that OS occurs mostly in fast-growing bones (2,3). Chemical substances such as methylcholanthrene or SV40 virus may promote the generation of OS (4,5). Approximately 10–25% of patients with OS have lung metastases, and lung injury caused by lung metastasis is the main cause of mortality (6). Osteosarcoma treatment typically involves chemotherapy and surgery. Some common drug treatments include doxorubicin, methotrexate, and vincristine. However, prolonged use of these drugs adversely affects liver function in patients, and their survival cannot be guaranteed. Surgical treatment affects the body of patients (7,8–11). Therefore, it is urgent to understand and explore the pathogenesis of OS and develop effective drugs for the treatment of OS.
Normal cell proliferation depends on a complete and effective cell cycle. The regulation of cell proliferation mainly occurs in the G1 phase cell cycle (12,13). In malignant cells, cell cycle imbalance is an early step in tumor development (14). A large number of signaling pathways and growth factors affect whether the cells enter the S phase for DNA replication (15,16). Of these pathways, cyclin D transfers extracellular mitogenic signals to activate the G1/S phase transition. A variety of signaling pathways can affect the expression of cyclin D, such as the classic Ras/Raf/MEK/ERK (MAPK), Rac, NF-κB, Wnt, and Notch signaling pathways (15). The Wnt/β-catenin signaling pathway involves the stabilization and translocation of β-catenin induced by extracellular Wnt. When β-catenin in the cytoplasm accumulates to a certain level, it transfers to the nucleus and forms β-catenin-TCF/LEF transcription complex, which initiates the transcriptional regulation of cyclin D and c-Myc during the G1-S period of the cell cycle, causing the cells to enter the S phase for DNA replication (17). Previous studies have reported that β-catenin can increase OS cell proliferation and the number of OS cells in the S phase, and promote the invasion and migration of OS cells (18–21). Therefore, the Wnt/β-catenin signaling pathway is a critical pathway in OS.
Most components of the transcriptome that do not encode proteins have traditionally been considered as ‘transcription junk’. However, with the development of high-throughput technologies such as next-generation sequencing in the past decade, researchers have an unprecedented understanding of non-coding genomes. An lncRNA is a type of non-coding RNA over 200 bp (22). In recent years, studies have revealed that there are numerous lncRNAs aberrantly expressed in OS, such as TUG1, UCAI, BCAR4, and HULC, which can be regarded as prognostic indicators of OS (23–26). Other lncRNAs, such as DANCR (27) and FOXC2-AS1 (28), also play important roles in the progression and metastasis of OS. Long non-coding RNA (lncRNA) cancer susceptibility candidate 15 (CASC15) is a long intergenic non-coding RNA (lincRNA) located at chromosome 6p22.3 (29). It has been reported that lncRNA CASC15 regulated the expression of SOX4 in acute leukemia to promote its occurrence (30) and regulated the Wnt/β-catenin signaling pathway through miR-4310 in colon cancer to promote cancer cell proliferation and metastasis (31). The latest research has reported that lncRNA CASC15 is upregulated in OS plasma exosomes. Knockout of lncRNA CASC15 inhibited the proliferation of OS cells and the growth of osteosarcoma in xenograft models (32). In addition, lncRNA CASC15 downregulated the expression of E-cadherin and upregulated the expression of N-cadherin, indicating that lncRNA CASC15 can affect the metastasis of OS cells (32). However, it remains unknown whether lncRNA CASC15 affects the proliferation and metastasis of osteosarcoma by regulating the Wnt/β-catenin signaling pathway.
Materials and methods
Clinical samples
A total of 30 patients with OS treated at the First People's Hospital of Shangqiu (Shangqiu, China) were selected as the observation group, and their OS tissues were obtained as biological samples, and the paired adjacent normal tissues were used as controls. The basic information of OS patients is presented in Table I. The present study was approved by the Ethics Committee of the First People's Hospital of Shangqiu, and the patients provided signed informed consent.
Table I.
Basic information of OS patients.
| OS (n=30) | ||
|---|---|---|
| Characteristics | No. of patients | % |
| Sex | ||
| Male | 17 | 56.7 |
| Female | 13 | 43.3 |
| Tumor size | ||
| ≤9 cm | 14 | 46.7 |
| ≥9 cm | 16 | 53.3 |
| Age at diagnosis | ||
| Mean | – | 22.2 |
| SD | – | 9.2 |
| Minimum | – | 8 |
| Maximum | – | 40 |
None of the patients had received any anticancer therapy prior to tumor resection or had been diagnosed with any additional malignancies. Patients were recruited between January 2017 and December 2019.
Cell culture and transfection
The hFOB1.19 cell line, purchased from American type culture collection (ATCC), was cultured in a 37°C incubator with 5% CO2. Then, the cells were cultured in high-glucose DMEM/F12 (1:1) (Cytiva) supplemented with 10% fetal bovine serum (FBS; Thermo Fisher Scientific, Inc.) and 0.3% G418 (Sigma-Aldrich; Merck KGaA). UMR-106 cells were purchased from ATCC and cultured in complete α-minimum essential medium (α-MEM) containing 15% FBS. The cells were placed in an incubator at 37°C and 5% CO2. Human OS cell line U-2OS (Shanghai Chinese Academy of Sciences Cell Bank) was cultured in RPMI-1640 medium containing 10% FBS at 37°C and 5% CO2. Human osteogenic sarcoma cells Saos-2 were purchased from China Infrastructure of Cell Line Resources. The cells were cultured with a DMEM containing 10% FBS, 100 U/l penicillin, and 100 mg/l streptomycin in a culture flask at 5% CO2 and 37°C.
Lentiviruses were produced using a second-generation lentiviral system in 293T cells. The recombinant lentiviruses expressing overexpressed (oe)-CASCS15, short hairpin (sh)-CASCS15, and their negative controls (oe-NC and sh-NC) were constructed and packaged by Shanghai GenePharma Co., Ltd. The ratio of lentiviral plasmid: psPAX2: pMD2.G was 2:2:1. U-2OS cells were infected with a total of 5 µg lentiviruses expressing oe-CASCS15, sh-CASCS15, and their negative controls (oe-NC and sh-NC) for 48 h using RNAi-Mate transfection reagent (MDBio, Inc.). The multiplicity of infection was 80. Cells were incubated in 5% CO2 at 37°C for 4 days.
Establishment of xenograft model
A total of 15 (female, 4–6 weeks old; weight between 15–20 g; BALb/c; Harlan Sprague Dawley, Inc.) nude mice (n=5 for each group) were used to establish a xenograft model. All mice were housed at a 12-h light/dark cycle with a temperature of 25±2°C and a humidity of 55±5°C. Food and drinking water were provided ad libitum. All animal experiments were approved by the Animal Experimentation Ethics Committee of First People's Hospital of Shangqiu. U-2OS, UMR-106, or Saos-2 cells were inoculated under the skin of the back of nude mice for 4 weeks. Each mouse was inoculated with ~2×106 cells. After 16 days, the mice were euthanized by cervical dislocation following anesthesia with ketamine (50 mg/kg)/xylazine (5 mg/kg), and tumors were harvested. Tumor size was measured by calipers and calculated using the following formula: Volume (mm3) = L (length) × W (width)2/2.
Metastasis assays
A mouse model of pulmonary metastasis was established using tail vein injection (33). When the cells reached 80–90% confluency, U-2OS, Saos-2, UMR-106, and hFOB1.19 cells were separated with 0.2 mmol/l EDTA in Hanks' Balanced Salt Solution (HBSS) without Mg+2-Ca+2-NaHC03. After counting, the cells (2.5×106/ml) were resuspended in ice-cold HBSS. Female athymic mice (4 weeks old; weight ~15 g; BALb/c; Harlan Sprague Dawley, Inc.) were injected with 0.2 ml of the cell suspension through the tail vein. A total of 20 mice was divided into 4 groups (n=5 per group). At 10 weeks after inoculation, mice were euthanized by cervical dislocation following anesthesia with ketamine (50 mg/kg)/xylazine (5 mg/kg). All organs were examined for metastasis formation macroscopically. Lung tissues were harvested and fixed in a mixture of Bouin's fixative and neutral buffered formalin (1:5, v/v). Metastatic nodules in the lungs were counted using an MZ16F dissecting microscope (Leica). Mice were housed at a 12-h light/dark cycle with a temperature of 25±2°C and humidity of 55±5°C. Food and drinking water were provided ad libitum. All animal experiments were approved by the Animal Experimentation Ethics Committee of First People's Hospital of Shangqiu.
Cell Counting Kit-8 (CCK-8) assay
U-2OS cells (4×103 cells/well) were inoculated into a 96-well plate and 8 multiple wells were set up in each group. Cells were cultured in a cell incubator. After lentiviral transfection, the cells were cultured for 48 h. The freshly prepared CCK-8 detection solution (10 µl) was added to each well and then cultured in the incubator for 4 h at 37°C. Then, the OD value was detected at 450 nm with a microplate reader. The experiment was repeated 3 times, and the average value of the experimental results was considered as the final experimental results. The growth inhibition rate was calculated according to the following formula: Cell growth inhibition rate=[(OD control group-OD experimental group)/OD control group] × 100%.
5-Ethynyl-2′-deoxyuridine (EdU) assay
The Cell-Light EdU DNA Cell Proliferation Kit (Guangzhou RiboBio Co., Ltd.) was used to detect cell proliferation by EdU assay. The cells in the logarithmic growth period were seeded into a 96-well plate with ~4×103−1×105 cells/well, and cells were cultured to the normal growth stage. EdU medium was added and incubated with cells for 2 h at 37°C. Cells were washed and incubated with 100 µl of cell immobilization solution (PBS containing 4% paraformaldehyde) in each well for 30 min at room temperature. Glycine (2 mg/ml) was added and incubated on a decolorizing shaking bed for 5 min. Then, the glycine solution was discarded, and cells were washed with PBS for 5 min. Then, 1X Apollo® dyeing reaction solution (100 µl) was added, and the reaction continued for 30 min. Subsequently, the cells were washed with 0.5% Triton X-100 in PBS. After adding 100 µl 1X Hoechst 33342 reaction solution, the reaction solution was discarded, and cells were washed with PBS after incubating on a light-free, room temperature, decolorizing shaking bed for 30 min. Images were obtained using an FSX100 fluorescence microscope (Olympus Corporation) at an ×200 magnification. The cell proliferation rate was evaluated according to the manufacturer's instructions.
Transwell assay
The cells (1×105 cells/well) were suspended in serum-free medium and inoculated into the upper chamber of an 8-µΜ pore Transwell chamber (EMD Millipore). A medium containing 20% FBS was added to the lower chamber, following incubation at 37°C for 24 h. The cells were carefully wiped away in the upper chamber with a cotton swab. Cells that migrated to the lower chamber were fixed with 4% formaldehyde solution and stained with 0.1% crystal violet (Solarbio Life Sciences) for 15 min at 37°C. Finally, an inverted microscope (magnification, ×100; Olympus IX 70; Olympus Corporation) was used to count the number of migrating cells. The migration activity was quantified by counting the migrated cells.
Cell scratch assay
A scratch wound assay was also used to evaluate the cell migration of OS cells. In short, 1×105 U-2OS cells were seeded in 12-well plates. When the cell density reached 80% confluence, the cell monolayer was scraped with a sterile pipette tip. After washing away cell debris with PBS, cells were maintained in serum-free medium. Images were obtained at 0 and 24 h after the scratch. Wound width was measured using ImageJ software version 1.8.0 (National Institutes of Health), and migration was expressed as wound closure fraction.
Cell cycle assay using flow cytometry
Approximately 2×105 cells were collected and washed with PBS, fixed in 70% cold ethanol at 4°C for 2 h, and then filtered through a 70-µm cell strainer (BD Biosciences) to obtain a single-cell suspension. Subsequently, the cells were incubated with RNase A at 37°C for 30 min and stained with PI for 30 min at 4°C (Cell Cycle Detection Kit; BD Biosciences). Flow cytometric analysis was performed on a FACSCalibur flow cytometer (BD Biosciences), and data were analyzed using CellQuest software (version 5.1; BD Biosciences).
Subcellular fractionation
U-2OS Cells were processed using Cytoplasmic Extraction and NE-PER Nuclear kit (Thermo Fisher Scientific, Inc.) according to the manufacturer's protocol. For quantification of β-catenin in indicated fractions, lamin A+C (dilution, 1:200; product code ab40567; Abcam) and actin (dilution, 1:1,000; cat. no. sc-8432; Santa Cruz Biotechnology, Inc.) were employed as the fractionation indicators.
Reverse transcription-quantitative polymerase chain reaction (RT-qPCR)
The RT-qPCR was performed for gene expression analysis in hFOB1.19, U-2OS, Saos-2, UMR-106 cells, and OS tissues. TRIzol reagent (Thermo Fisher Scientific, Inc.) was used to extract total RNA from these cell lines and grated tissues. Nanodrop technology (Thermo Fisher Scientific, Inc.) was used to detect the concentration of RNA, and the ratio of A260/280 was between 1.9 and 2.1. Total RNA was reverse-transcribed using Prime Script™ RT reagent Kit with gDNA Eraser (Takara Biotechnology Co., Ltd.) to reverse RNA to cDNA on a PCR instrument (ABI; Thermo Fisher Scientific, Inc.). The obtained cDNA was mixed with TB Green™ Premix Ex Taq™ II (Takara Biotechnology Co., Ltd.) for RT-qPCR detection, with a Real-time PCR instrument (Roche LightCycler® 480; Roche Diagnostics). The PCR thermal cycling conditions were 95°C for 5 min, and then 45 cycles at 95°C for 5 sec and 60°C for 30 sec. Primers were designed and synthesized by TSINGKE Biological Technology Co., Ltd, as presented in Table SI. β-actin was used as the internal control. The relative expression was calculated using the 2−∆∆Cq method (34).
Western blotting
RIPA lysis buffer (Bio-Rad Laboratories, Inc.) was used to lyse the cells and extract soluble proteins. U-2OS cells were lysed with RIPA lysis buffer. This process lasted 30 min on ice. The obtained system was centrifuged at 12,000 × g for 10 min at 4°C. The supernatant was collected in an EP tube, and the protein concentration was measured with a BCA protein detection kit (Thermo Fisher Scientific, Inc.). The extracted protein (20 µg) was subjected to 7.5% SDS-PAGE electrophoresis to separate the protein. Furthermore, 5% BSA or skimmed milk was prepared in advance, and the membrane was then placed in 5% BSA or skimmed milk, and it was blockedfor 2 h on the shaker at room temperature. The incubated primary antibody was dissolved in 5% BSA or skimmed milk, and the blots were probed overnight at 4°C with antibodies against Wnt1 (dilution 1:1,000; product code ab15251; Abcam), β-catenin (dilution 1:1,000; product code ab68183; Abcam), cyclin D1 (dilution 1:1,000; product no. 55506S; Cell Signaling Technology, Inc.), E-cadherin (dilution 1:50; product code ab1416; Abcam), N-cadherin (dilution 1:10,000; product code ab76011; Abcam), vimentin (dilution 1:5,000; product code ab92547; Abcam), actin (dilution 1:1,000; cat. no. sc-8432; Santa Cruz Biotechnology, Inc.), and lamin A + C (dilution 1:200; product code ab40567; Abcam) followed by incubation for 1 h at room temperature with a horseradish peroxidase-conjugated secondary antibody (cat. no. sc-2357; Santa Cruz Biotechnology, Inc.; or product code ab205719; Abcam). Enhanced chemiluminescence (ECL) substrate (Pierce; Thermo Fisher Scientific, Inc.) was used as the visualization reagent. The film was soaked in the luminescent liquid for 5 min. Then, the film was removed and placed in the luminescence imager, and the luminescent liquid was added dropwise again to collect the images.
Statistical analysis
All data were analyzed with SPSS (version 22.0; IBM Corp.), and the quantitative data were expressed as the mean ± standard deviation (SD). The differences between two groups were compared using unpaired Student's t-test, and one-way analysis of variance (ANOVA) followed by post hoc test Tukey-Kramer was carried out to compare the differences between multiple groups. P<0.05 was considered to indicate a statistically significant difference.
Results
CASC15 is highly expressed in OS cell lines and clinical samples
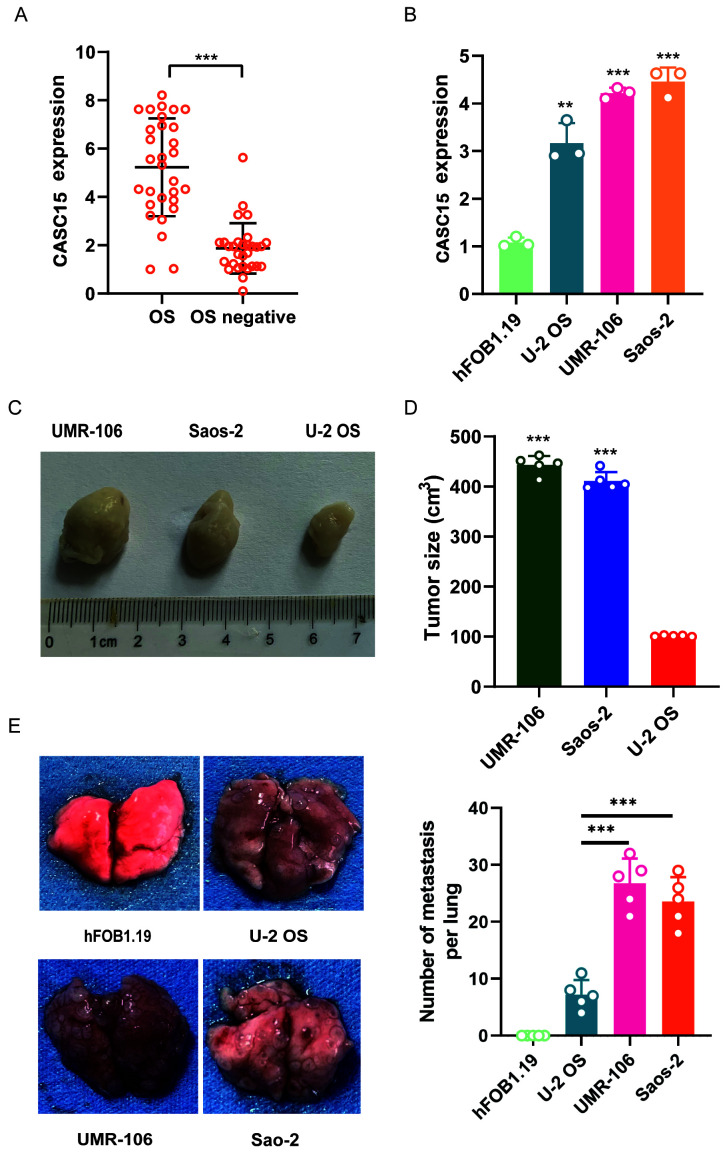
RT-qPCR was used to detect the expression of CASC15 in clinical samples of OS from 30 patients and tissues samples with negative OS biopsy (n=30). The results revealed that the expression of CASC15 in OS biopsy samples was significantly higher than that in biopsy samples without cancer (Fig. 1A). To further explore the expression of CASC15 in OS, different OS cell lines were selected, namely, U-2OS, Saos-2, and UMR-106 as well as normal cell line hFOB1.19, to detect the expression of CASC15. The RT-qPCR results demonstrated that the expression of CASC15 in U-2OS, UMR-106, and Saos-2 was significantly higher than that in hFOB1.19, and the expression of CASC15 in U-2OS was significantly lower than that in Saos-2 and UMR-106 cells (Fig. 1B). U-2OS, Saos-2, and UMR-106 cells in the logarithmic growth phase were inoculated subcutaneously into the back of nude mice (n=5) for 4 weeks at 2×106 cells for each mouse, and tumor formation was observed after 4 weeks. The results revealed that compared with U-2OS nude mice, the tumors of Saos-2 and UMR-106 nude mice were significantly enlarged (Fig. 1C and D). The tail vein injection of U-2OS, Saos-2, UMR-106, and hFOB1.19 in nude mice (n=5) was used to observe tumor formation in the lungs of mice. The results indicated that the nude mice injected with U-2OS had fewer metastatic tumors than the mice injected with Saos-2 and UMR-106 (Fig. 1E).
Figure 1.
Expression of CASC15 in OS cell lines and clinical samples. The OS cell lines used in the present study were hFOB1.19, U-2OS, Saos-2, UMR-106. (A) qPCR was used to detect the expression of CASC15 in clinical samples from 30 patients. (B) qPCR was used to detect the expression of CASC15 in different OS cell lines and a normal cell line. (C and D) Comparison of xenograft tumor formation in nude mice with different cell lines. (E) Nude mice were respectively injected with hFOB1.19, U-2OS, Saos-2, UMR-106 into the tail vein to observe lung tumor formation. **P<0.01 and ***P<0.001 vs. OS, hFOB1.19 or U-2OS. CASC15, cancer susceptibility 15; OS, osteosarcoma; qPCR, quantitative polymerase chain reaction.
CASC15 regulates the cell cycle and promotes the proliferation of OS cells in vitro
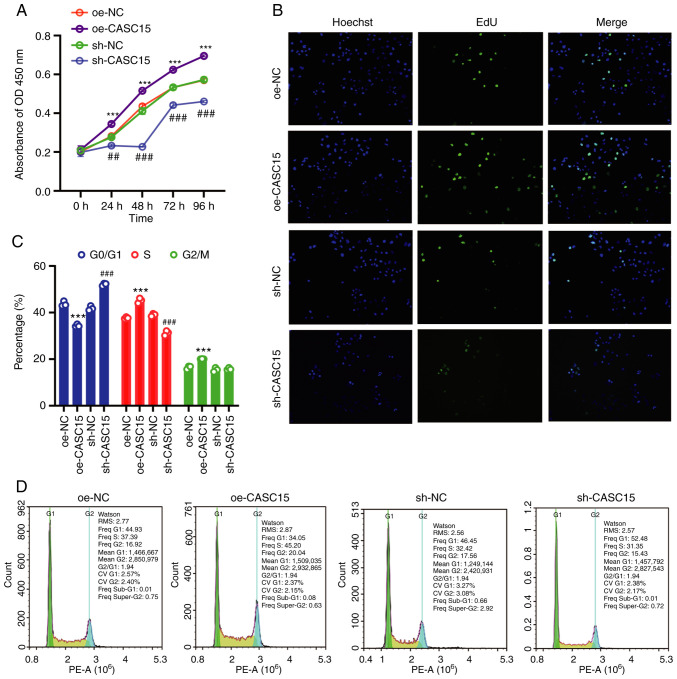
Since CASC15 could promote cancer progression and cancer cell proliferation in leukemia and bowel cancer (30,31,35), and higher expression of CASC15 had been revealed in OS cell lines, transplanted tumors, and clinical samples, whether CASC15 could promote cell proliferation was further studied. Therefore, the U-2OS cell line with low expression of CASC15 was used for the next experiments. The results of CCK-8 and Edu assays demonstrated that compared with the respective control (oe-NC or sh-NC) group, the cell proliferation of the oe-CASC15 group was increased, while the cell proliferation of the sh-CASC15 group was decreased (Fig. 2A and B). Flow cytometry was used to detect the number of cells in the G0/G1, S, and G2/M phases of the cell cycle. The results indicated that compared with the control group, S-phase cells were increased in the oe-CASC15 group, whereas S-phase cells were decreased in the sh-CASC15 group (Fig. 2C and D).
Figure 2.
CASC15 regulates the cell cycle, promotes the proliferation of OS cells in vitro, and promotes the growth of OS. U-2OS cells were transfected with oe-CASC15, sh-CASC15, and their negative controls (oe-NC and sh-NC). (A) Cell Counting Kit-8 assay was used to detect cell proliferation. (B) 5-Ethynyl-2′-deoxyuridine assay was used to detect cell proliferation. (C and D) Flow cytometry was used to detect the cell cycle. ***P<0.001 vs. oe-NC; ##P<0.01 and ###P<0.001 vs. sh-NC. CASC15, cancer susceptibility 15; OS, osteosarcoma; oe, overexpressed; NC, negative control; sh, short hairpin.
CASC15 impacts the proliferation and cell cycle of OS cells by regulating the Wnt/β-catenin pathway
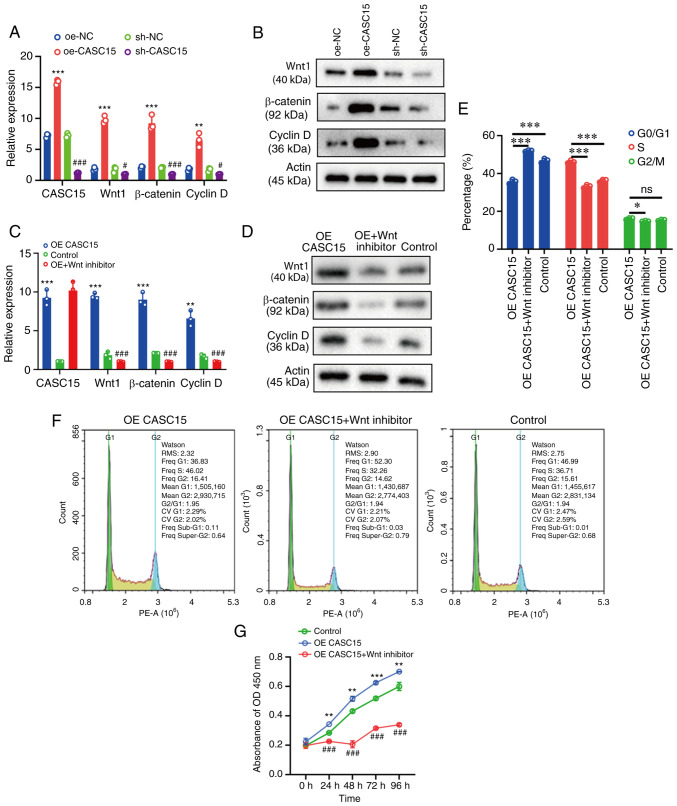
Due to the changes in proliferation and cycle distribution revealed in the present study, RT-qPCR and western blotting were then used to detect the expression of Wnt, β-catenin, and cyclin D, respectively. The transfected U-2OS cells were grouped into oe-CASC15, sh-CASC15, oe-NC and sh-NC. The RT-qPCR results revealed that compared with the respective control (oe-NC or sh-NC) group, the expression of Wnt, β-catenin, and cyclin D was increased in the oe-CASC15 group, while it was decreased in the sh-CASC15 group (Fig. 3A). The western blot results revealed that compared with the control group, the expression of Wnt, β-catenin, and cyclin D was increased in the oe-CASC15 group, whereas it was decreased in the sh-CASC15 group (Fig. 3B). In the rescue experiment, oe-CASC15, oe-CASC15/Wnt inhibitor, or control were transfected into U-2OS cells, and then RT-qPCR and western blotting were used to detect the expression of Wnt, β-catenin, and cyclin D, respectively. The RT-qPCR and western blot results indicated that compared with the control group, the expression of Wnt, β-catenin, and cyclin D was increased in the oe-CASC15 group, whereas it was decreased in the oe-CASC15/Wnt inhibitor group (Fig. 3C and D). The flow cytometric results indicated that compared with the control group, S-phase cells were increased in the oe-CASC15 group, whereas they were decreased in the oe-CASC15/Wnt inhibitor group (Fig. 3E and F). Furthermore, the results of CCK-8 cell proliferation demonstrated that the oe-CASC15 group had increased cell proliferation compared with the control group. The oe-CASC15 + Wnt inhibitor group inhibited cell proliferation, and the proliferation was slightly lower than that of the control group (Fig. 3G).
Figure 3.
CASC15 can activate the Wnt/β-catenin signaling pathway to regulate the cell cycle and interfere with the proliferation of osteosarcoma cells. (A and B) U-2OS cells were transfected with oe-CASC15, sh-CASC15, and their respective negative controls (oe-NC and sh-NC), and RT-qPCR and western blotting were used to detect the expression of Wnt, β-catenin, and cyclin D, respectively. (C and D) U-2OS cells were transfected with oe-CASC15, oe-CASC15 + Wnt inhibitor, and control, and RT-qPCR and western blotting were used to detect the expression of Wnt, β-catenin, and cyclin D, respectively. (E and F) The cell cycle was detected in U-2OS cells which were transfected with oe-CASC15, oe-CASC15 + Wnt inhibitor, and control. (G) A Cell Counting Kit-8 assay was performed in U-2OS cells which were transfected with oe-CASC15, oe-CASC15 + Wnt inhibitor, and control. *P<0.05, **P<0.01 and ***P<0.001 vs. oe-NC, control or oe-CASC15; #P<0.05 and ###P<0.001 vs. sh-NC or control. CASC15, cancer susceptibility 15; oe, overexpressed; NC, negative control; sh, short hairpin; ns, nonstatistically significant difference.
High CASC15 expression promotes the metastasis of OS cells
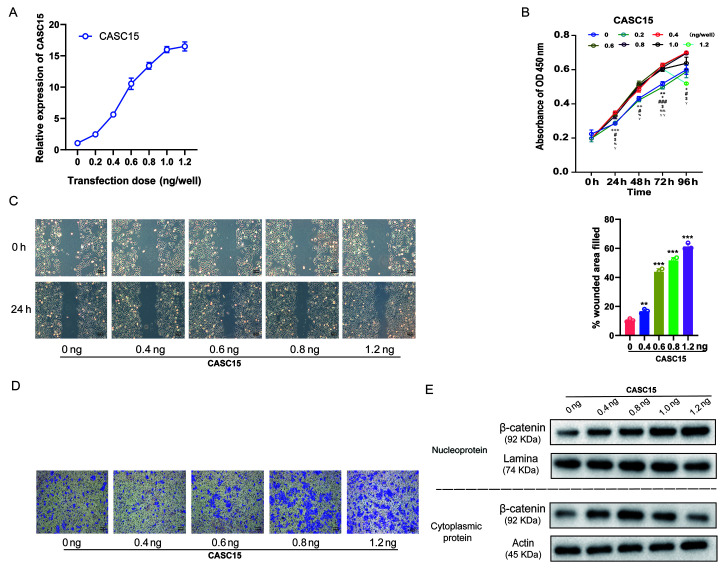
In previous studies, it was revealed that cells with high CASC15 expression were more prone to metastasis, and that CASC15 expression in metastatic tissues was higher (Fig. 1), while promoting cell proliferation via the Wnt/β-catenin signaling pathway (Fig. 3). Considering that the Wnt/β-catenin signaling pathway also promoted the EMT process (36), it was further explored whether the expression level of CASC15 could affect cell proliferation and metastasis. U-2OS cells were transfected with lentiviruses expressing various doses [0.2, 0.4, 0.6, 0.8, 1.0, and 1.2 ng/well (24 wells)] of oe-CASC15. The RT-qPCR results revealed that with the increasing transfection dose, the expression of CASC15 was gradually increased (Fig. 4A). Furthermore, the results of the CCK-8 assay revealed that with the increase in the expression of CASC15, cell proliferation quickly increased, however, when the transfection dose exceeded 0.6 ng/well, the increase of proliferation was insignificant and slightly decreased (Fig. 4B). The results of the cell scratch experiments also indicated that with the increase of CASC15 expression, low-dose transfection did not improve the healing rate, whereas with high-dose transfection it was significantly improved (Fig. 4C). Moreover, the results of the Transwell cell migration experiments revealed that with the increase of CASC15 expression, low-dose transfection did not affect cell migration ability, whereas high-dose transfection significantly increased cell migration (Fig. 4D). Next, the influence of various CASC15 expression levels on the cellular location of β-catenin was also assessed. The western blot results revealed that with the increased expression of CASC15, low-dose transfection increased the expression of β-catenin in the cytoplasm, whereas high-dose transfection increased the expression of β-catenin in the nucleus and decreased its expression in the cytoplasm (Fig. 4E).
Figure 4.
Effect of lncRNA CASC15 on osteosarcoma cells is dose-dependent, and the increase in lncRNA expression gradually exhibits metastatic characteristics. Lentiviruses expressing different doses of overexpressed-CASC15 [0.2, 0.4, 0.6, 0.8, 1.0, 1.2 ng/well (24 wells)] were transfected into U-2OS cells. (A) The expression of CASC15 was detected by reverse transcription-quantitative polymerase chain reaction. (B) Cell proliferation was detected by Cell Counting Kit-8 experiment. **P<0.01: 0.2 vs. 0 ng/well; +P<0.01, ++P<0.01 and +++P<0.001: 0.4 vs. 0 ng/well; #P<0.05 and ###P<0.001: 0.6 vs. 0 ng/well; $P<0.05: 0.8 vs. 0 ng/well; %P<0.05 and %%P<0.01: 1 vs. 0 ng/well; YP<0.05 and YYP<0.01: 1.2 vs. 0 ng/well. (C) The cell scratch test was detected as the speed of healing. **P<0.01 and ***P<0.001 vs. 0 ng/well. (D) The Transwell cell migration assay detected cell migration. (E) The influence of various CASC15 expression levels on the cellular location of β-catenin. lncRNA, long non-coding RNA; CASC15, cancer susceptibility 15.
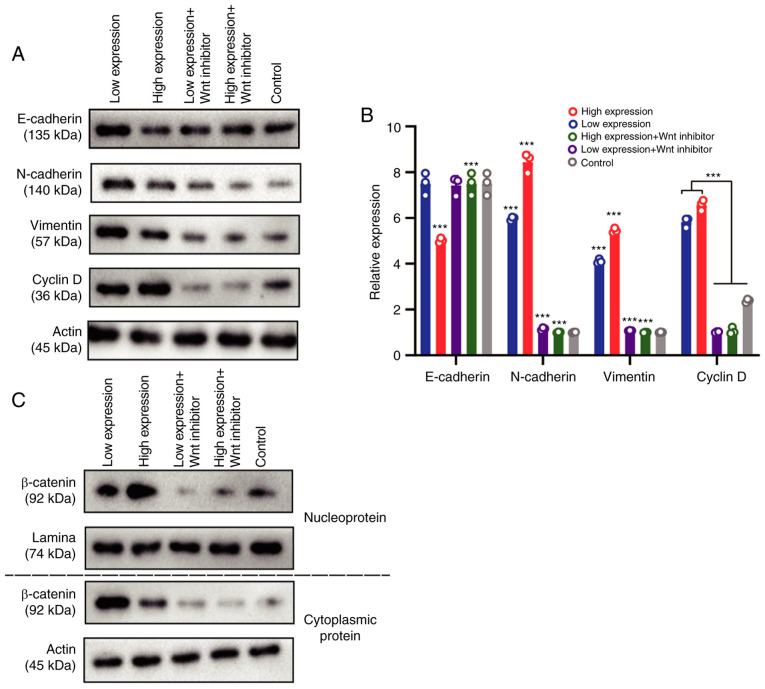
CASC15 promotes metastasis by inducing EMT of OS cells through activation of the Wnt/β-catenin pathway
Since in Fig. 4, the cell proliferation was not obvious whereas cell migration was after the transfection amount was >0.6 ng/well, a dose of 1 ng/well was used as the high-expression dose, and 0.6 ng/well was used as the low-expression dose. Grouping of U-2OS-transfected cells was as follows: high expression of the CASC15 group, low expression of the CASC15 group, high expression of CASC15 + Wnt inhibitor group, low expression of CASC15 + Wnt inhibitor group, and the control group. Western blotting and RT-qPCR were used to detect the expression levels of vimentin, E-cadherin, N-cadherin, and cyclin D. The results revealed that compared with the low-expression CASC15 group, the expression of E-caherin in OS cells was downregulated in the high-expression CASC15 group, while the expression levels of N-cadherin, vimentin and cyclin D were upregulated. After transfection of the Wnt inhibitor, the expression of E-caherin was upregulated in OS cells, whereas the expression levels of N-cadherin, vimentin and cyclin D were downregulated (Fig. 5A and B). After nuclear and cytoplasmic separation, western blotting was used to detect the expression changes of β-catenin among the aforementioned groups. Western blot analysis indicated that when CASC15 was in low concentration, the expression of β-catenin was higher in the cytoplasm than that in the nucleus, indicating that β-catenin was primarily expressed in the cytoplasm; when the concentration of CASC15 was 1 ng/well (high expression), β-catenin was predominantly expressed in the nucleus, indicating that high expression levels of CAS15 could promote the entry of β-catenin into the nucleus, thereby causing the occurrence of EMT in the nucleus (Fig. 5C).
Figure 5.
CASC15 promotes the epithelial-mesenchymal transition of osteosarcoma cells by Wnt/β-catenin pathway. U-2OS cells were transfected with 1 ng/well of oe-CASC15 as the high expression dose, and 0.6 ng/well as the low expression dose. The transfection groups were as follows: high expression, low expression, high expression + Wnt inhibitor, and low expression + Wnt inhibitor. (A and B) Reverse transcription-quantitative polymerase chain reaction and western blotting were used to detect the expression of E-cadherin, N-cadherin, vimentin, and cyclin D, respectively. (C) After nuclear and cytoplasmic separation, western blotting was used to detect the expression changes of β-catenin among the aforementioned groups. ***P<0.01 vs. low expression or control. CASC15, cancer susceptibility 15.
Discussion
EMT was first proposed by Elizabeth Hay. In 1980, she observed that epithelial cells could downregulate epithelial tissue characteristics to obtain mesenchymal tissue characteristics (37). EMT reveals that epithelial cells have a certain degree of plasticity. In the process of EMT, epithelial cells lose their apical-basal polarity and the connection mode between cells, the trefoil factor 3 (TFF3) signaling pathway is involved and altered, and the shape of epithelial cells presents with slender morphology (38). This increases the ability of individual cells to develop their ability to infiltrate into tissues during the period of transformation (39,40). The EMT process is an important step in the formation of gastrulation and neural crest (41). However, EMT occurs again in wound healing, fibrosis, and cancer progression (42). Cancer invasion and metastasis are similar to normal embryonic development (43). Recent studies have revealed that the process of cancer invasion is a recurrence of the EMT process of embryonic development (44,45). During EMT, the expression of E-cadherin and cytokeratin that are specifically expressed in epithelial tissues decrease, whereas the expression of mesenchymal cytoskeleton proteins such as vimentin increase. When epithelial cells undergo the EMT process, they usually secrete fibronectin produced by mesenchymal cells, such as fibroblasts, and concurrently express fibroblast marker proteins such as N-cadherin to replace the marker protein E in epithelial tissues. Cancer cells expressing N-cadherin undergo the EMT process to enhance their affinity with supporting cells, thereby causing cell infiltration and metastasis in normal tissues (46). In the present study, it was revealed that when the expression of CASC15 increased, it could promote the proliferation of OS cells. However, after it increased to a certain level, it did not further promote cell proliferation but appeared to induce cell migration. Therefore, it was considered whether CASC15 activates the Wnt signaling pathway in order for a large amount of β-catenin to enter the nucleus to promote EMT. To this aim, it was first explored whether different doses would affect the EMT process, and it was revealed that different doses of lncRNA CASC15 could alter the expression of β-catenin in the nucleus and cytoplasm. It was revealed that CASC15 activated the Wnt signaling pathway. It was surmised that the formation of the possible migration ability was related to the EMT process caused by β-catenin entering the nucleus. Therefore, the effect of various doses of CASC15 on the content of β-catenin inside and outside the nucleus was detected, and it was revealed that CASC15 could lead to β-catenin entering the nucleus in large quantities via the Wnt pathway to promote the EMT of OS cells.
The Wnt signaling pathway was first discovered in the study of Drosophila wingless genes (47). Later, more Wnt family genes were discovered during embryonic development (48). The discovery of the Wnt1 gene linked the Wnt signaling pathway to cancer. Nusse and Varmus revealed that overexpression of the Wnt1 gene could cause mouse mammary tumors (49). The Wnt signaling pathway is mainly divided into the classic Wnt signaling pathway, namely the Wnt/β-catenin signaling pathway and the non-canonical Wnt signaling pathway (50). The classic Wnt signaling pathway is mainly composed of a Wnt-secreted protein, a transmembrane receptor frizzled protein, a loose protein, β-catenin, glycogen synthesis kinase 3β, and T cell factor/lymph enhancer factor (51). This pathway mainly regulates cell behavior through the transcription properties of DNA-binding proteins of the TCF/LEF family. The key is the stable β-catenin in the cytoplasm (52). In Wnt signaling, β-catenin is mostly bound by E-cadherin, which extends from the cell membrane into the cell. When the Wnt signaling pathway is activated, the Wnt protein secreted by the cell binds to the Frz and LRP5/6 receptor complex on the cell surface to activate the Dsh protein in the cytoplasm. The activated Dsh can inhibit GSK3-β and stimulate β-catenin activity in the cytoplasm. When β-catenin accumulates to a certain level, it transfers to the nucleus and competitively binds to the transcription factor TCF/LEF such as P300 in the nucleus to form a β-catenin-TCF/LEF transcription complex to regulate target gene expression (53). The present study revealed that lncRNA CASC15 triggered the Wnt signaling pathway and led to the downstream β-catenin in the nucleus. β-catenin plays a role in the nucleus. When β-catenin begins to appear in the nucleus, it first regulates the expression of downstream cyclin D and triggers cell proliferation. When there is excessive β-catenin in the nucleus, part of it regulates downstream EMT (54,55). In the present study a competitive binding mechanism of β-catenin with the lncRNA likely occurred, and this mechanism should be further explored in future studies. In addition, in-depth research should be conducted as to how CASC15 regulates the Wnt/β-catenin signaling pathway and even regulates the expression of β-catenin inside and outside the nucleus. It is theorized that CASC15 is likely to inhibit the phosphorylation β-catenin by binding to the β-catenin protein in OS cells (56,57), leading to the accumulation of β-catenin and its entering the nucleus to bind to the LEF/TCF transcription factor family and activate transcription of downstream target genes (such as cyclin D1).
In conclusion, the present study explored the function and effect of lncRNA CASC15 on the proliferation, invasion, and metastasis of OS cells and revealed the preliminary mechanism of the expression level of lncRNA CASC15 on the progression and metastasis of OS, thus providing a new treatment for OS.
Supplementary Material
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HW proposed and designed the study, collected the clinical samples, performed the experiments in vivo, and prepared the manuscript. PZ conducted the experiments in vitro, and analyzed the data. Both authors read and approved the final manuscript.
Ethics approval and consent to participate
The present study was approved by the Ethics Committee of the First People's Hospital of Shangqiu, and the patients provided signed informed consent. All animal experiments were approved by the Animal Experimentation Ethics Committee of First People's Hospital of Shangqiu.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Messerschmitt PJ, Garcia RM, Abdul-Karim FW, Greenfield EM, Getty PJ. Osteosarcoma. J Am Acad Orthop Surg. 2009;17:515–527. doi: 10.5435/00124635-200908000-00005. [DOI] [PubMed] [Google Scholar]
- 2.Schiller AL. Orthopaedic pathology. Semin Diagn Pathol. 2011;28:1–3. doi: 10.1053/j.semdp.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2015. CA Cancer J Clin. 2015;65:5–29. doi: 10.3322/caac.21254. [DOI] [PubMed] [Google Scholar]
- 4.Mendoza SM, Konishi T, Miller CW. Integration of SV40 in human osteosarcoma DNA. Oncogene. 1998;17:2457–2462. doi: 10.1038/sj.onc.1202179. [DOI] [PubMed] [Google Scholar]
- 5.Mazabraud A. Experimental production of bone sarcomas in the rabbit by a single local injection of beryllium. Bull Cancer. 1975;62:49–58. (In French) [PubMed] [Google Scholar]
- 6.Miller BJ, Cram P, Lynch CF, Buckwalter JA. Risk factors for metastatic disease at presentation with osteosarcoma: An analysis of the SEER database. J Bone Joint Surg Am. 2013;95:e89. doi: 10.2106/JBJS.L.01189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Fabbri A, Motta E, Ferrari S, Longhi C, Marchi E, Bacci G, Figus E, Marchesini G. High-dose methotrexate treatment and liver function in patients with osteosarcoma. J Intern Med. 1994;236:209–214. doi: 10.1111/j.1365-2796.1994.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 8.Lin F, Wang Q, Yu W, Tang L, Zheng S, Sun Y, Shen Z, Yao Y, Dong Y. Clinical analysis of Chinese limb osteosarcoma patients treated by two combinations of methotrexate, cisplatin, doxorubicin and ifosfamide. Asia-Pac J Clin Oncol. 2011;7:270–275. doi: 10.1111/j.1743-7563.2011.01406.x. [DOI] [PubMed] [Google Scholar]
- 9.Bacci G, Lari S. Adjuvant and neoadjuvant chemotherapy in osteosarcoma. Chir Organi Mov. 2001;86:253–268. (Article in English, Italian) [PubMed] [Google Scholar]
- 10.Biermann JS, Baker LH. The future of sarcoma treatment. Semin Oncol. 1997;24:592–597. [PubMed] [Google Scholar]
- 11.La Quaglia MP. Osteosarcoma. Specific tumor management and results. Chest Surg Clin N Am. 1998;8:77–95. [PubMed] [Google Scholar]
- 12.Yue Z, Guan X, Chao R, Huang C, Li D, Yang P, Liu S, Hasegawa T, Guo J, Li M. Diallyl disulfide induces apoptosis and autophagy in human osteosarcoma MG-63 cells through the PI3K/Akt/mTOR pathway. Molecules. 2019;24:2665. doi: 10.3390/molecules24142665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kurowska P, Mlyczyńska E, Dawid M, Opydo-Chanek M, Dupont J, Rak A. In vitro effects of vaspin on porcine granulosa cell proliferation, cell cycle progression, and apoptosis by activation of GRP78 receptor and several kinase signaling pathways including MAP3/1, AKT, and STAT3. Int J Mol Sci. 2019;20:5816. doi: 10.3390/ijms20225816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Li B, Zhou P, Xu K, Chen T, Jiao J, Wei H, Yang X, Xu W, Wan W, Xiao J. Metformin induces cell cycle arrest, apoptosis and autophagy through ROS/JNK signaling pathway in human osteosarcoma. Int J Biol Sci. 2020;16:74–84. doi: 10.7150/ijbs.33787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Duronio RJ, Xiong Y. Signaling pathways that control cell proliferation. Cold Spring Harb Perspect Biol. 2013;5:a008904. doi: 10.1101/cshperspect.a008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cavallaro G, Cucina A, Coluccia P, Petramala L, Cotesta D, Polistena A, Zinnamosca L, Letizia C, Rosato L, Cavallaro A, De Toma G. Role of growth factors on human parathyroid adenoma cell proliferation. World J Surg. 2010;34:48–54. doi: 10.1007/s00268-009-0294-x. [DOI] [PubMed] [Google Scholar]
- 17.Kaldis P, Pagano M. Wnt signaling in mitosis. Dev Cell. 2009;17:749–750. doi: 10.1016/j.devcel.2009.12.001. [DOI] [PubMed] [Google Scholar]
- 18.Huang Q, Shi SY, Ji HB, Xing SX. LncRNA BE503655 inhibits osteosarcoma cell proliferation, invasion/migration via Wnt/β-catenin pathway. Biosci Rep. 2019;39:BSR20182200. doi: 10.1042/BSR20182200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keremu A, Maimaiti X, Aimaiti A, Yushan M, Alike Y, Yilihamu Y, Yusufu A. NRSN2 promotes osteosarcoma cell proliferation and growth through PI3K/Akt/MTOR and Wnt/β-catenin signaling. Am J Cancer Res. 2017;7:565–573. [PMC free article] [PubMed] [Google Scholar]
- 20.Zhao H, Hou W, Tao J, Zhao Y, Wan G, Ma C, Xu H. Upregulation of lncRNA HNF1A-AS1 promotes cell proliferation and metastasis in osteosarcoma through activation of the Wnt/β-catenin signaling pathway. Am J Transl Res. 2016;8:3503–3512. [PMC free article] [PubMed] [Google Scholar]
- 21.Jin H, Luo S, Wang Y, Liu C, Piao Z, Xu M, Guan W, Li Q, Zou H, Tan QY, et al. miR-135b stimulates osteosarcoma recurrence and lung metastasis via Notch and Wnt/β-catenin signaling. Mol Ther Nucleic Acids. 2017;8:111–122. doi: 10.1016/j.omtn.2017.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fu X, Duanmu J, Li T, Jiang Q. A 7-lncRNA signature associated with the prognosis of colon adenocarcinoma. PeerJ. 2020;8:e8877. doi: 10.7717/peerj.8877. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang Y, Huang Y, Xiang P, Tian W. LncRNA expression and implication in osteosarcoma: A systematic review and meta-analysis. Onco Targets Ther. 2017;10:5355–5361. doi: 10.2147/OTT.S149889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Xie C, Chen B, Wu B, Guo J, Cao Y. LncRNA TUG1 promotes cell proliferation and suppresses apoptosis in osteosarcoma by regulating miR-212-3p/FOXA1 axis. Biomed Pharmacother. 2018;97:1645–1653. doi: 10.1016/j.biopha.2017.12.004. [DOI] [PubMed] [Google Scholar]
- 25.Ju L, Zhou YM, Yang GS. Up-regulation of long non-coding RNA BCAR4 predicts a poor prognosis in patients with osteosarcoma, and promotes cell invasion and metastasis. Eur Rev Med Pharmacol Sci. 2016;20:4445–4451. [PubMed] [Google Scholar]
- 26.Kong D, Wang Y. Knockdown of lncRNA HULC inhibits proliferation, migration, invasion, and promotes apoptosis by sponging miR-122 in osteosarcoma. J Cell Biochem. 2018;119:1050–1061. doi: 10.1002/jcb.26273. [DOI] [PubMed] [Google Scholar]
- 27.Jiang N, Wang X, Xie X, Liao Y, Liu N, Liu J, Miao N, Shen J, Peng T. lncRNA DANCR promotes tumor progression and cancer stemness features in osteosarcoma by upregulating AXL via miR-33a-5p inhibition. Cancer Lett. 2017;405:46–55. doi: 10.1016/j.canlet.2017.06.009. [DOI] [PubMed] [Google Scholar]
- 28.Zhang CL, Zhu KP, Ma XL. Antisense lncRNA FOXC2-AS1 promotes doxorubicin resistance in osteosarcoma by increasing the expression of FOXC2. Cancer Lett. 2017;396:66–75. doi: 10.1016/j.canlet.2017.03.018. [DOI] [PubMed] [Google Scholar]
- 29.Wu Q, Xiang S, Ma J, Hui P, Wang T, Meng W, Shi M, Wang Y. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol Oncol. 2018;12:799–813. doi: 10.1002/1878-0261.12187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fernando TR, Contreras JR, Zampini M, Rodriguez-Malave NI, Alberti MO, Anguiano J, Tran TM, Palanichamy JK, Gajeton J, Ung NM, et al. The lncRNA CASC15 regulates SOX4 expression in RUNX1-rearranged acute leukemia. Mol Cancer. 2017;16:126. doi: 10.1186/s12943-017-0692-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jing N, Huang T, Guo H, Yang J, Li M, Chen Z, Zhang Y. LncRNA CASC15 promotes colon cancer cell proliferation and metastasis by regulating the miR-4310/LGR5/Wnt/β-catenin signaling pathway. Mol Med Rep. 2018;18:2269–2276. doi: 10.3892/mmr.2018.9191. [DOI] [PubMed] [Google Scholar]
- 32.Zhang H, Wang J, Ren T, Huang Y, Yu Y, Chen C, Huang Q, Guo W. LncRNA CASC15 is upregulated in osteosarcoma plasma exosomes and CASC15 knockdown inhibits osteosarcoma progression by regulating miR-338-3p/RAB14 axis. Onco Targets Ther. 2020;13:12055–12066. doi: 10.2147/OTT.S282053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hurst DR, Edmonds MD, Scott GK, Benz CC, Vaidya KS, Welch DR. Breast cancer metastasis suppressor 1 up-regulates miR-146, which suppresses breast cancer metastasis. Cancer Res. 2009;69:1279–1283. doi: 10.1158/0008-5472.CAN-08-3559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 35.Wang B, Xu W, Cai Y, Guo C, Zhou G, Yuan C. CASC15: A tumor-associated long non-coding RNA. Curr Pharm Des. 2021;27:127–134. doi: 10.2174/1381612826666200922153701. [DOI] [PubMed] [Google Scholar]
- 36.Zhang J, Cai H, Sun L, Zhan P, Chen M, Zhang F, Ran Y, Wan J. LGR5, a novel functional glioma stem cell marker, promotes EMT by activating the Wnt/β-catenin pathway and predicts poor survival of glioma patients. J Exp Clin Cancer Res. 2018;37:225. doi: 10.1186/s13046-018-0864-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hay ED. An overview of epithelio-mesenchymal transformation. Acta Anat (Basel) 1995;154:8–20. doi: 10.1159/000147748. [DOI] [PubMed] [Google Scholar]
- 38.Li P, Pan X, Zheng Z, Sun Y, Han Y, Dong J. LINC00271 inhibits epithelial-mesenchymal transition of papillary thyroid cancer cells by downregulating trefoil factor 3 expression. Aging Pathobiol Ther. 2020;2:78–85. doi: 10.31491/APT.2020.06.018. [DOI] [Google Scholar]
- 39.Thiery JP, Sleeman JP. Complex networks orchestrate epithelial-mesenchymal transitions. Nat Rev Mol Cell Biol. 2006;7:131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 40.Thiery JP, Acloque H, Huang RY, Nieto MA. Epithelial-mesenchymal transitions in development and disease. Cell. 2009;139:871–890. doi: 10.1016/j.cell.2009.11.007. [DOI] [PubMed] [Google Scholar]
- 41.Acloque H, Adams MS, Fishwick K, Bronner-Fraser M, Nieto MA. Epithelial-mesenchymal transitions: The importance of changing cell state in development and disease. J Clin Invest. 2009;119:1438–1449. doi: 10.1172/JCI38019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kalluri R, Weinberg RA. The basics of epithelial-mesenchymal transition. J Clin Invest. 2009;119:1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Li A, Machesky LM. Melanoblasts on the move: Rac1 sets the pace. Small GTPases. 2012;3:115–119. doi: 10.4161/sgtp.19494. [DOI] [PubMed] [Google Scholar]
- 44.Jayanthi P, Varun BR, Selvaraj J. Epithelial-mesenchymal transition in oral squamous cell carcinoma: An insight into molecular mechanisms and clinical implications. J Oral Maxillofac Pathol. 2020;24:189. doi: 10.4103/jomfp.JOMFP_334_19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu X, Qiao B, Zhao T, Hu F, Lam AK, Tao Q. Sox2 promotes tumor aggressiveness and epithelial-mesenchymal transition in tongue squamous cell carcinoma. Int J Mol Med. 2018;42:1418–1426. doi: 10.3892/ijmm.2018.3742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Weber CE, Li NY, Wai PY, Kuo PC. Epithelial-mesenchymal transition, TGF-β, and osteopontin in wound healing and tissue remodeling after injury. J Burn Care Res. 2012;33:311–318. doi: 10.1097/BCR.0b013e318240541e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Sharma RP, Chopra VL. Effect of the Wingless (wg1) mutation on wing and haltere development in Drosophila melanogaster. Dev Biol. 1976;48:461–465. doi: 10.1016/0012-1606(76)90108-1. [DOI] [PubMed] [Google Scholar]
- 48.Yamaguchi TP. Heads or tails: Wnts and anterior-posterior patterning. Curr Biol. 2001;11:R713–R724. doi: 10.1016/S0960-9822(01)00417-1. [DOI] [PubMed] [Google Scholar]
- 49.Nusse R, Varmus HE. Many tumors induced by the mouse mammary tumor virus contain a provirus integrated in the same region of the host genome. Cell. 1982;31:99–109. doi: 10.1016/0092-8674(82)90409-3. [DOI] [PubMed] [Google Scholar]
- 50.Shetti D, Zhang B, Fan C, Mo C, Lee BH, Wei K. Low dose of paclitaxel combined with XAV939 attenuates metastasis, angiogenesis and growth in breast cancer by suppressing Wnt signaling. Cells. 2019;8:892. doi: 10.3390/cells8080892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Saito-Diaz K, Chen TW, Wang X, Thorne CA, Wallace HA, Page-McCaw A, Lee E. The way Wnt works: Components and mechanism. Growth Factors. 2013;31:1–31. doi: 10.3109/08977194.2012.752737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.de Sousa EM, Vermeulen L, Richel D, Medema JP. Targeting Wnt signaling in colon cancer stem cells. Clin Cancer Res. 2011;17:647–653. doi: 10.1158/1078-0432.CCR-10-1204. [DOI] [PubMed] [Google Scholar]
- 53.Jubb AM, Chalasani S, Frantz GD, Smits R, Grabsch HI, Kavi V, Maughan NJ, Hillan KJ, Quirke P, Koeppen H. Achaete-scute like 2 (ascl2) is a target of Wnt signalling and is upregulated in intestinal neoplasia. Oncogene. 2006;25:3445–3457. doi: 10.1038/sj.onc.1209382. [DOI] [PubMed] [Google Scholar]
- 54.Rashid MS, Mazur T, Ji W, Liu ST, Taylor WR. Analysis of the role of GSK3 in the mitotic checkpoint. Sci Rep. 2018;8:14259. doi: 10.1038/s41598-018-32435-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Xiao C, Wu CH, Hu HZ. LncRNA UCA1 promotes epithelial-mesenchymal transition (EMT) of breast cancer cells via enhancing Wnt/beta-catenin signaling pathway. Euro Rev Med Pharmacol Sci. 2016;20:2819–2824. [PubMed] [Google Scholar]
- 56.Yue B, Liu C, Sun H, Liu M, Song C, Cui R, Qiu S, Zhong M. A positive feed-forward loop between LncRNA-CYTOR and Wnt/β-Catenin signaling promotes metastasis of colon cancer. Mol Ther. 2018;26:1287–1298. doi: 10.1016/j.ymthe.2018.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu J, Han Z, Sun Z, Wang Y, Zheng M, Song C. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through β-catenin-dependent Wnt pathway. J Exp Clin Cancer Res. 2018;37:222. doi: 10.1186/s13046-018-0896-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and analyzed during the current study are available from the corresponding author on reasonable request.