Abstract
Globally, stroke is a leading cause of death and disability. Traditional risk factors like hypertension, diabetes, and obesity do not fully account for all stroke cases. Recent infection is regarded as changes in systemic immune signaling, which can increase thrombosis formation and other stroke risk factors. We have previously shown that administration of lipopolysaccharide (LPS) 30-minutes prior to stroke increases in infarct volume. In the current study, we found that animals intermittently exposed to LPS have larger cortical infarcts when compared to saline controls. To elucidate the mechanism behind this phenomenon, several avenues were investigated. We observed significant upregulation of tumor necrosis factor-alpha (TNF-α) mRNA, especially in the ipsilateral hemisphere of both saline and LPS exposed groups compared to sham surgery animals. We also observed significant reductions in expression of genes involved in autophagy in the ipsilateral hemisphere of LPS stroke animals. In addition, we assessed DNA methylation of autophagy genes and observed a significant increase in the ipsilateral hemisphere of LPS stroke animals. Intermittent exposure to LPS increases cortical infarct volume, downregulates autophagy genes, and induces hypermethylation of the corresponding CpG islands. These data suggest that intermittent immune activation may deregulate epigenetic mechanisms and promote neuropathological outcomes after stroke.
Keywords: autophagy, DNA methylation, LPS, stroke, TNF-α
Introduction
Stroke is the second leading cause of death and the leading cause of disability globally (Gorelick, 2019). Traditionally recognized risk factors for stroke and other acute vascular events include hypertension, diabetes, obesity, and tobacco use (Yang et al., 2017). In addition, recent (7-10 days) bacterial or viral infections are also recognized as potential stroke risk factors (Grau et al., 1995; Bova et al., 1996; Paganini-Hill et al., 2003; Grau et al., 2005). Systemic infection can alter thrombosis formation by increasing platelet aggregation (Zeller et al., 2005; Elkind et al., 2011), and can also increase circulating cytokine concentrations (Wilson et al., 1998; Guedes et al., 2016) which has been shown to play a major role in fibronectin accumulation (Pawluczyk and Harris, 1998) and thereby contribute to thrombosis formation (Maurer et al., 2010). Additionally, systemic inflammation can induce changes in endothelial cell function (Stern et al., 1988) which may contribute to the development of atherosclerosis (Sessa et al., 2014; Campbell and Rosenfeld, 2015).
An increasing number of research reports show that pathogens can change the chromatin structure and transcriptional program in the host cells by manipulating the epigenetic machinery, including DNA methylation (Day, 2014). In fact, many bacteria (or their products) can selectively silence the expression of specific host genes and/or host signaling cascades epigenetically to decrease the inflammatory response and contribute to its tolerance by host cells (Day, 2014).
DNA methylation is the key player in epigenetic regulation of gene expression in mammalian cells and plays a significant role in carcinogenesis, fibrosis, and inflammation. DNA methylation is mediated by DNA methyl transferases (DNMT) at the 5-position of cytosine in CpG islands of the promoter; the addition of a methyl group here can lead to gene silencing (Gujar et al., 2019). In humans, there are four DNMTs responsible for DNA methylation; DNMT1, -3a, -3b, and 3L, which are ubiquitously expressed in fetal and adult tissues. DNMT1 is the most abundant and is involved in the maintenance of methylation, 3a, and 3b, function as de novo methylation enzymes, and DNMT3L lacks methylation capacities (Aapola et al., 2000). Many studies have shown that hypermethylation is associated with bacterial infection, or the bacterial product lipopolysaccharide (LPS) (Medzhitov and Horng, 2009; Bayarsaihan, 2011). Bacterial-induced DNA methylation can regulate various genes involved in cell proliferation, pro-inflammatory suppression, and pathogen persistence (Zhang et al., 2010; Yin and Chung, 2011; Tolg et al., 2011). This immunosuppressive response results in increased susceptibility of the individual to further infection for extended periods of time, and as well as LPS tolerance (McCall et al., 2010; Carson et al., 2011).
Interestingly, a recent study has shown that activation of TLR4 by LPS impairs autophagy in microglia (Lee et al., 2019). Autophagy is a lysosomal-mediated intracellular degradation pathway that maintains cellular homeostasis by turning over cellular components, waste, old or damaged organelles, and microbes. These components are sequestered into double-membrane vesicles (autophagosomes) which bind to lysosomes, resulting in degradation to their basic components to be reutilized for anabolic pathways and ATP generation. Thus, autophagy plays a vital role in organelle quality control, defense, homeostasis, and energy production (Feng et al., 2014). Energy depletion induces autophagy and upregulates formation of autophagosomes through inhibition of the mechanistic target of rapamycin (mTOR) (Jung et al., 2010). The formation of the autophagosome is mediated by many molecules, importantly by 2 ubiquitin-like conjugation systems: microtubule-associated protein 1, light chain 3 (LC3) and the autophagy protein, autophagy-related gene 5–12 (ATG5-12). The conversion of LC3-I (unconjugated cytosolic form) to LC3-II (autophagosomal membrane-associated phosphatidylethanolamine-conjugated form) is a symbol of autophagosome formation and autophagy flux (Feng et al., 2014). Therefore, the transformation of LC3-I to LC3-II denotes autophagy stimulation and autophagosome formation (Kang et al., 2011). Beclin1 is a phylogenetically conserved protein that is considered as a central regulator of autophagy initiation including the nucleation of phagohore. It is expressed in different human and murine tissues and is primarily localized within cytoplasmic structures. Many studies have shown that Beclin-1 is regulated by transcription factors or epigenetic regulators (e.g. miRNAs) independently (Kusama et al., 2009). Accumulating evidences indicate that the autophagy pathway is activated upon ischemic stroke in several cell types in the brain, with minimum explanation as to how it contributes to stroke and facilitating pathogen infection in the long term (Wang et al., 2018).
Previous work from our laboratory has found that exposure to LPS 30 minutes prior to experimental stroke significantly increases infarct volumes when compared to vehicle injected controls (Doll, Engler-Chiurazzi et al., 2015). In the current study, we sought to expand these findings and determine if intermittent LPS exposure could be additive over time, increasing infection burden of the organism, and lead to significantly larger stroke infarcts by inhibiting autophagy. In an effort to mimic the rate of illness experienced by an adult human, we induced an immune response in mice every two weeks (akin to ∼2 years in a human), with ample time to fully recover between insults. Despite animals being fully recovered from the last LPS injection at the time of surgery, we observed a significant increase in infarct size in the LPS-treated group when compared to the saline controls, which corresponded to a significant reduction in the expression of key autophagy genes, and increased methylation of their associated promoters.
Materials and Methods
The data that support the findings of this study are available from the corresponding author upon reasonable request.
Animals
Twenty four male C57BL/6J mice (12 weeks of age) were acquired from Jackson Laboratory (Bar Harbor, ME). Animals were housed in sterile micro-isolator cages with sterile bedding, and provided food and water ad libitum in the XX University Health Science Center animal facility. All the steps in the animal procedures, experiments and imaging were conducted in strict accordance with the Guide for the Care and Use of Laboratory Animals published by the National Institute of Health (NIH, USA 2018) and were evaluated and approved by XXX University ethics and Animal Care and Use Committees (XX-ACUC, protocol number 1604002043).
Intermittent Immune Activation Paradigm
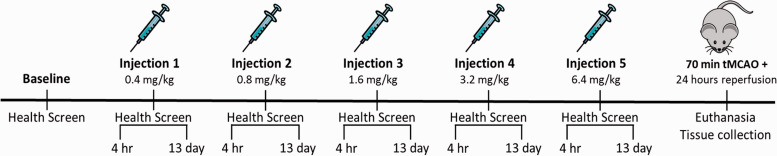
Lipopolysaccharide (LPS; Escherichia coli 055:B5, Sigma, St. Louis, MO) was reconstituted in sterile, injectable saline (B. Braun Medical Inc, Irvine, CA). Animals were administered LPS (or an equal volume of saline) via intraperitoneal (i.p.) injection. To ensure that animals elicited an immune response, the concentration of LPS was doubled with each injection; i.e. injection 1: 0.4 mg/kg, injection 2: 0.8 mg/kg, injection 3: 1.6 mg/kg, injection 4: 3.2 mg/kg, and injection 5: 6.4 mg/kg (Figure 1). Each injection was separated by a two-week recovery period to allow for animals’ health to return to baseline levels.
Figure 1.
Experimental Timeline and Study Design. Prior to the start of the LPS (or saline) injection paradigm, baseline health scores were obtained. Four hours post-injection, health screen scores were measured to ensure animals exhibited sickness behaviors. To confirm recovery between LPS exposures, thirteen days post-injection (i.e. one day prior to the next injection) the health screen was again administered. LPS concentrations were doubled with each injection. Thirteen days after injection 5, the health screen was administered once more to verify that animals were no longer exhibiting sickness behavior. The following day, animals were subjected to a 70 minute tMCAO surgery, followed by a 24-hour reperfusion period, at the conclusion of which, animals were euthanized and tissues were collected.
Sickness Behavior Monitoring
To measure animals’ sickness behavior prior to, and after each LPS (or saline) injection, a 0-20 point scale of overall health was implemented (Doll et al., 2015). For the majority of the health screen, animals were observed in their home cage to assess behavior across several parameters, including general appearance, posture, respiration, and spontaneous locomotion/social interaction. Animals were removed from the cage to then assess their body condition, temperature, and weight. A score was assigned for each parameter, which were then summed to provide an overall assessment of the animal’s health.
Prior to the beginning of the injection paradigm, baseline health screen measurements were obtained, which were then used for determining changes from the norm for each individual animal. Four hours post-injection, the health screen was administered to ensure animals were exhibiting a sickness response due to LPS exposure. Additionally, 13 days post-injection the health screen was conducted to ensure animals recovered to baseline levels of health prior to undergoing transient middle cerebral artery occlusion (tMCAO) (Figure 1).
Transient Middle Cerebral Artery Occlusion and Sham Surgery
Surgical anesthesia was induced with 4-5% isoflurane and maintained with 1-2% isoflurane via face-mask in oxygen-enriched air. tMCAO model was performed with a 6.0 monofilament suture (Doccol, Sharon, Massachusetts) for seventy minutes, with rectal body temperature maintained at 37°C ± 0.5°C during surgery with a warm blanket (Stoelting Co, Wood Dale, IL). To confirm successful occlusion (>70% decrease in blood flow), regional cerebral blood flow was detected using a Laser Speckle Imager (Moor instruments, United Kingdom). All surgeries were performed by one surgeon who was blinded to pre-treatments.
Exclusion Criteria for Animal Experiments
Several exclusion criteria for tMCAO were implemented in this experiment, as we have previously described (Doll et al., 2015; Sun et al., 2016). (1) A decrease of regional cerebral blood flow < 70% during occlusion as detected by laser Doppler flowmetry; (2) surgery time lasting > 70 minutes; (3) neurological score 0, indicating no neurologic deficit 3 hours after tMCAO; (4) no infarction in the MCA territory as indicated by TTC staining; (5) subarachnoid hemorrhage upon post-mortem examination; (6) substantial ambient temperature changes in the animal housing facility; and (7) body temperature at or below 32°C.
In this study, 3 mice were excluded: 1 mouse (saline treated) because the Laser Speckle Imager did not reach 70% reduction during the occlusion, and 2 mice (LPS treated) because body temperatures were below 32°C.
Analysis of Cerebral Infarct Volume
All animals were euthanized 24 hours post-ischemia via cardiac puncture. Animals were then perfused with 1X phosphate buffered saline (PBS). The remaining brains were removed and cut into 2-mm sections with a mouse brain matrix (World Precision Instruments). Sections were stained with 2% 2,3,5-triphenyltetrazolium chloride (TTC; Sigma, Saint Louis, Missouri) in PBS solution at 37°C for 15 minutes, and digitally photographed.
After imaging TTC stained brain slices, brain tissue was dissected into four groups. The main infarcted tissue was dissected from each of the slices and pooled together as the ipsilateral core, while the remaining tissue from the ipsilateral hemisphere was pooled together as the ipsilateral penumbra. Mirroring sections were taken from the contralateral hemisphere and termed contralateral core and contralateral penumbra. Tissue was snap frozen in liquid nitrogen and stored at –80°C. Each pooled sample was homogenized for RNA analysis as an independent sample.
Digitized images of each brain section were analyzed using computerized image analysis software (ImageJ, RRID:SCR_003070) in a blinded manner. Brain and infarct volumes for right (ipsilateral) and left (contralateral) hemispheres were calculated. Volumes were calculated as a percentage of contralateral cortex, striatum, and total hemisphere to reduce the effect of brain edema.
RNA Isolation and qPCR
Total RNA was isolated from brain tissue via Trizol extraction (Invitrogen) as per the manufacturer’s instructions and as previously done (Dakhlallah et al., 2013). RNA concentrations were measured with a NanoDrop One Microvolume UV-Vis Spectrophotometer (ThermoFisher), and 1 µg was reverse transcribed using the SuperScript™ III First-Strand Synthesis SuperMix for qRT-PCR (Invitrogen) as per the manufacturer’s instructions. The resulting cDNA was used for qPCR for 40 cycles, using the ABI 7900HT real-time PCR system in combination with SYBR Green PCR Master mix (Life Technologies, Grand Island, NY) in addition to 10μM of RT2 qPCR Assay primer mix (primers listed below) in a 10 µl reaction. Expression of TNF-α, Beclin-1, ATG5, LC3a, LC3b, DNMT1, DNMT3a, and DNMT3b mRNA was normalized to verified specific PrimeTime qPCR primers for Ribosomal Protein L4 (RPL4) and adenylate cyclase-associated protein 1(CAP1), which were used as internal controls (Integrated DNA Technologies, Coralville, IA). Quantification of PCR amplified mRNA specific cDNA was done by comparative cycle threshold CT method (2^-ΔΔCT). Two independent values were obtained for each hemisphere (core and penumbra) for all animals, which were then pooled together for analysis as the ipsilateral and contralateral hemispheres. Fold changes of target gene expression in the ipsilateral and contralateral hemispheres of saline or LPS exposed stroke animals were relative to that of the corresponding hemisphere of the sham animals. cDNA could not be synthesized from samples with low RNA yield (<1 µg total RNA), and were excluded from analysis as previously described (Dakhlallah et al., 2013). Significant five outliers total, as determined by GraphPad Prism 8.0 (GraphPad Prism, RRID:SCR_002798), were excluded from analysis.
DNA Methylation
We isolated genomic DNA using the DNeasy Blood and Tissue Kit (Qiagen, Valencia, CA) specific to the manufacturer’s protocol. DNA concentration was measured with the NanoDrop (ND-1000) spectrophotometer (Thermo Scientific Nanodrop 1000 Spectrophotometer, RRID:SCR_016517). Two independent values were obtained for each hemisphere (core and penumbra) for all animals, which were then pooled together for analysis as the ipsilateral and contralateral hemispheres. The DNA methylation status of autophagy gene promoters’ CpG island was evaluated using the EpiTect Methyl II PCR Primer Assay, specific for each gene (Qiagen, Valencia, CA) according to manufactures instructions, and as previously described (Dakhlallah et al., 2013).
Statistical Analyses
All quantitative data were assessed for significance using either a one-way ANOVA with Dunnett’s or Tukey’s post hoc test, or Student’s t test. All results were analyzed by GraphPad Prism 8.0 software (GraphPad Software, La Jolla, CA). Results from the experiments are reported as means ± SEM. A p value < 0.05 was used to establish significance; significance presented as *p<0.05, **p<0.01, ***p<0.001, and ****p<0.0001.
Results
LPS Injections Significantly Alter Animal Health Status
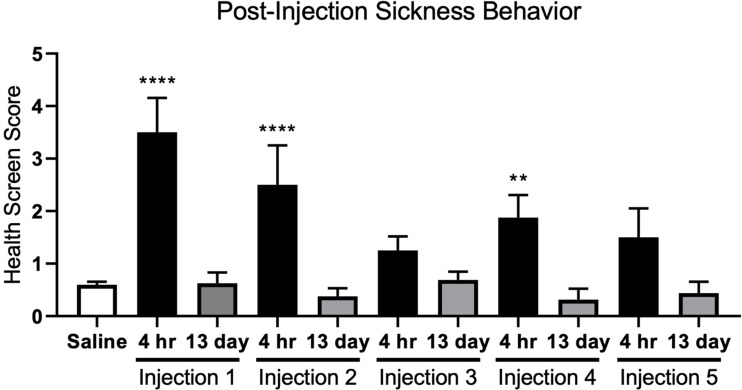
By using a health screening tool previously developed in our laboratory (Doll et al., 2015), we assessed the overall health of the animals at both four hours and thirteen days post-LPS exposure. As assessed 4 hours post-injection, LPS induces an acute sickness response, from which animals recovered (Figure 1). Animals exposed to LPS exhibited significant sickness behaviors four hours post-injection after injections 1, 2, and 4, while injection 5 trended towards significance. Across all five injections, at 13 days post-injection, health screen scores averaged < 1, indicating that animals returned to normal health status between injections (Figure 1). Throughout the duration of the experiment, there were no significant changes in health screen scores for animals exposed to saline; thus all health screen data for these animals was grouped together.
Intermittent LPS Exposure Significantly Increases Cortical Infarct Volume
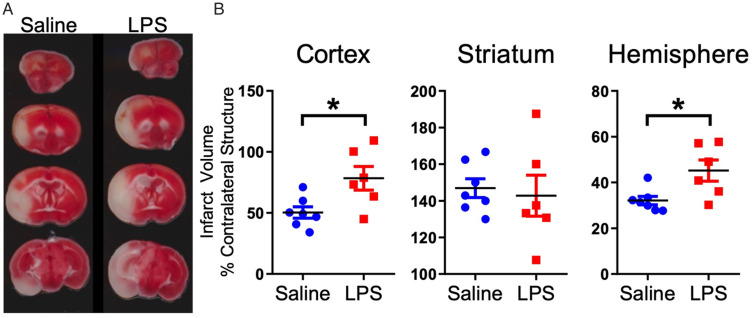
By day 13-post injection 5, all animals exhibited a full behavioral recovery. Despite this, we observed a significant increase in stroke infarct volume in animals that were intermittently injected with LPS, compared to the saline-injected controls (Figure 2). There were significantly larger infarct volumes in the cortex and total ipsilateral hemisphere in the animals that were intermittently injected with LPS compared to saline; however, there were no significant differences in striatal infarct volume between the two groups (Figure 3A and B)
Figure 2.
LPS Exposure Induces an Acute Sickness Response, From Which Animals Recovered. Health screen scores for animals exposed to saline were grouped together as they did not significantly differ across time. There was a significant effect of LPS exposure on sickness behavior when compared to saline injected animals (F10,157 = 12.69, p < 0.0001). Dunnett’s post-hoc analysis indicated that when compared to saline-treated animals, animals exposed to LPS exhibited significant sickness behavior four hours post-injections 1, 2, and 4, while injection 5 trended towards significance (p = 0.0667). Further, health sickness scores at the 13 day time point did not significantly differ from those in the saline-injected group, indicating that animals returned to normal health status. Data are expressed as mean ± SEM; one-way ANOVA. **p < 0.01 and ****p < 0.0001.
Figure 3.
Repeated LPS Exposure Significantly Increases Stroke Infarct Volume. A: Representative TTC stained coronal slices used to analyze infarct volume 24 hours post-stroke. B: Animals repeatedly exposed to LPS had significantly larger infarct volume than saline exposed mice in cortex and total hemisphere. Data are expressed as mean ± SEM; brackets connect statistically significant groups; Student t test. *p < 0.05.
TNF-α mRNA Transcription Is Significantly Upregulated 24 Hours Post-Stroke
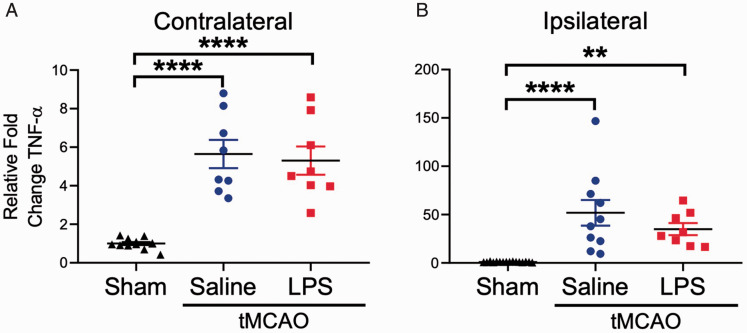
Tumor necrosis factor-alpha (TNF-α) is a proinflammatory cytokine which is thought to play an important role in the post-stroke immune response (Barone et al., 1997). We measured TNF-α mRNA in the contralateral and ipsilateral hemispheres of saline or LPS-injected animals that underwent tMCAO, as well as sham surgery controls (Figure 4). We found a significant increase in TNF-α mRNA expression in both hemispheres of the animals subjected to tMCAO when compared to the sham surgery group. Interestingly, TNF-α mRNA expression increased in both saline and LPS groups ∼ 5-fold in the contralateral hemisphere (Figure 4A), while in the ipsilateral hemisphere, TNF-α mRNA was increased ∼ 50-fold in the saline group, but only ∼ 35-fold in the LPS group (Figure 4B).
Figure 4.
Stroke Induces Significant Upregulation of TNF-α mRNA Expression Across the Entire Brain. A: Compared to sham animals, TNF-α mRNA was significantly upregulated in the contralateral hemisphere of animals subjected to a 70 minute tMCAO, regardless of prior saline or LPS exposure (F2, 25 = 28.33, p < 0.0001). B: Similarly, expression of TNF-α mRNA was significantly upregulated in the ipsilateral hemisphere of animals subjected to a 70 minute tMCAO when compared to sham (F2, 31 = 14.92, p < 0.0001). Data are expressed as mean ± SEM; one-way ANOVA with Dunnett’s post hoc test; brackets connect statistically significant groups;**p < 0.01 and ****p < 0.0001.
LPS Exposure Significantly Alters Post-Stroke Autophagy-Associated mRNAs
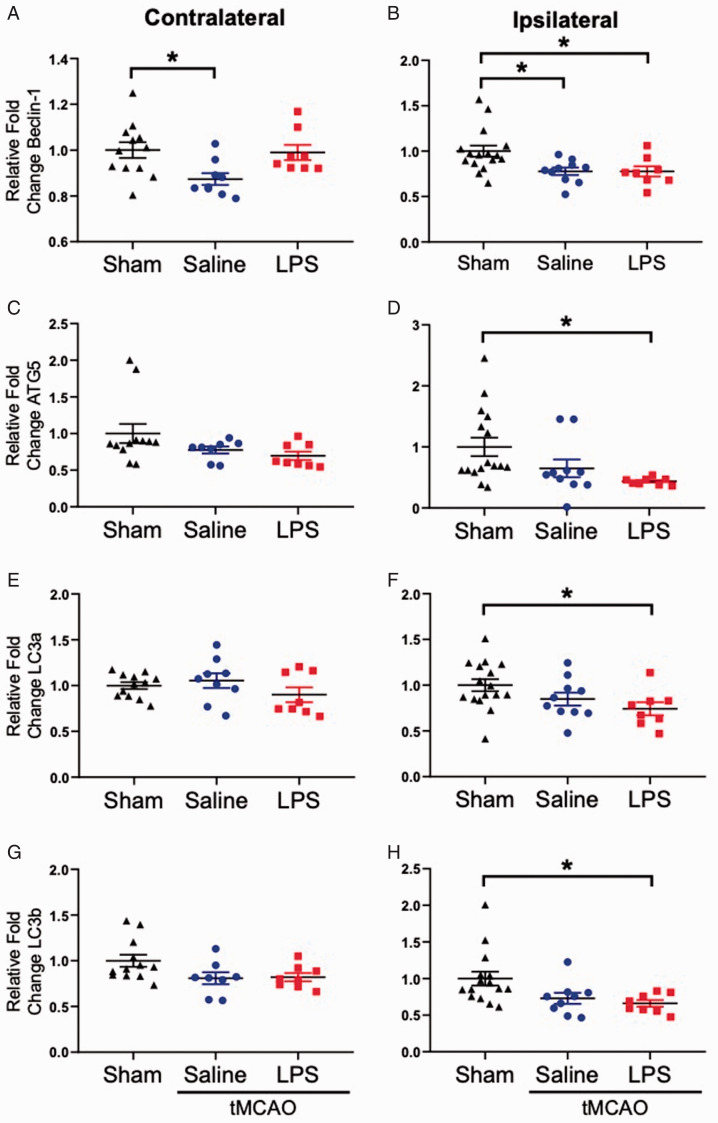
As changes in TNF-α mRNA expression did not appear to be the driving force leading to increased infarct size after repeated LPS exposure, we examined expression of key genes associated with autophagy that have CpG islands in their promoter (Beclin, Atg5, LC3a and LC3b). As shown in Figure 5, we observed significant reductions in all four autophagy genes in the ipsilateral hemispheres of at 24-hours post-stroke in the LPS exposed animals when compared to shams (Figure 5A to H). Beclin-1 was also significantly reduced in both hemispheres of the saline exposed stroke animals when compared to sham (Figure 5A and B).
Figure 5.
Autophagy-Related Genes Are Downregulated in the Ipsilateral Hemisphere of LPS Exposed Stroke Animals. A: In the contralateral hemisphere, only saline injected animals had a significant reduction in Beclin-1 mRNA expression when compared to sham controls (F2, 26 = 4.644, p = 0.0189). B: Both saline and LPS injected animals expressed significantly lower levels of Beclin-1 in the ipsilateral hemisphere when compared to sham controls (F2, 31 = 5.457, p < 0.0093). In the contralateral hemisphere, no significant differences in (C) ATG5, (E) LC3a, or (G) LC3b mRNA expression were observed. In the ipsilateral hemisphere, significant downregulation of (D) ATG5 (F2, 31 = 3.972, p = 0.0291), (F) LC3a (F2, 31 = 3.393, p = 0.0465), and (H) LC3b mRNA expression was observed in the LPS exposed stroke animals when compared to sham controls. Data are expressed as mean ± SEM; one-way ANOVA with Dunnett’s post hoc test; brackets connect statistically significant groups; *p < 0.05.
DNA Methylation of Autophagy Genes Is Significantly Increased After Stroke
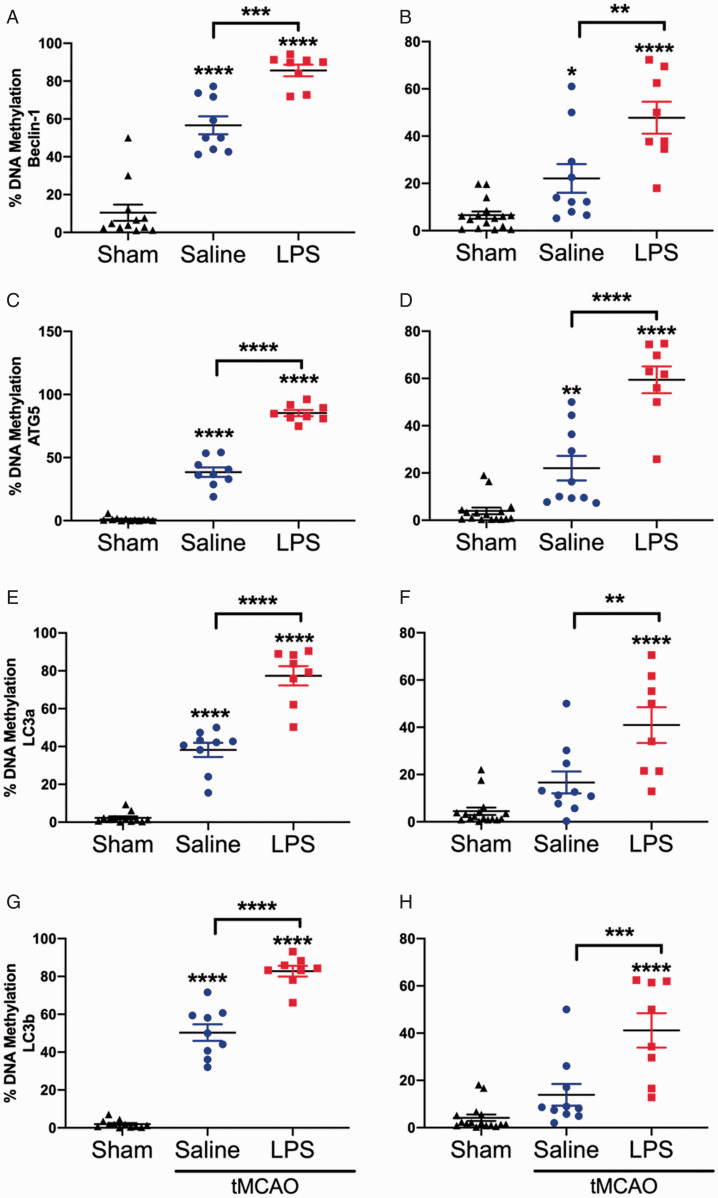
mRNA expression of the autophagy genes, Beclin-1, ATG5, LC3a, and LC3b was significantly reduced in the ipsilateral hemispheres of the LPS exposed stroke animals, so we next sought to understand the potential mechanism by which this phenomenon occurs. The methylation status of all four genes was significantly increased in the contralateral (Figure 6A, C, E, and G) and ipsilateral (Figure 6B, D, F and H) hemispheres of both saline and LPS exposed stroke animals. Further, we also observed significant differences between saline and LPS exposed stroke animals, with LPS exposed animals having significantly higher methylation than the saline exposed animals (Figure 6).
Figure 6.
DNA Methylation of Autophagy Genes Is Increased Post-Stroke. DNA methylation of Beclin-1 was significantly increased in both the (A) contralateral (F2, 26 = 81.77, p < 0.0001) and (B) ipsilateral (F2, 31 = 21.76, p < 0.0001) hemispheres of both the saline and LPS exposed stroke animals, when compared to the sham controls. Similarly, DNA methylation of ATG5 was significantly increased in both the (C) contralateral (F2, 26 = 322.5, p < 0.0001) and (D) ipsilateral (F2, 31 = 53.58, p < 0.0001) hemispheres of both the saline and LPS exposed stroke animals, when compared to the sham controls. DNA methylation of LC3a was significantly increased in the (E) contralateral (F2, 26 = 140.4, p < 0.0001) hemisphere of both saline and LPS exposed stroke animals, while it was significantly upregulated in only the (F) ipsilateral (F2, 31 = 19.32, p < 0.0001) hemispheres LPS exposed stroke animals, when compared to the sham controls. DNA methylation of LC3b was also significantly increased in the (G) contralateral (F2, 26 = 229.7, p < 0.0001) hemisphere of both saline and LPS exposed stroke animals, while it was significantly upregulated in only the (H) ipsilateral (F2, 31 = 21.45, p < 0.0001) hemispheres LPS exposed stroke animals, when compared to the sham controls. For all assessed genes, LPS exposed stroke animals had significantly higher levels of DNA methylation than the saline exposed stroke animals (A-H). Data are expressed as mean ± SEM; one-way ANOVA with Tukey’s post hoc test; brackets connect statistically significant groups; *p < 0.05, **p < 0.01, **p < 0.001, and ****p < 0.0001.
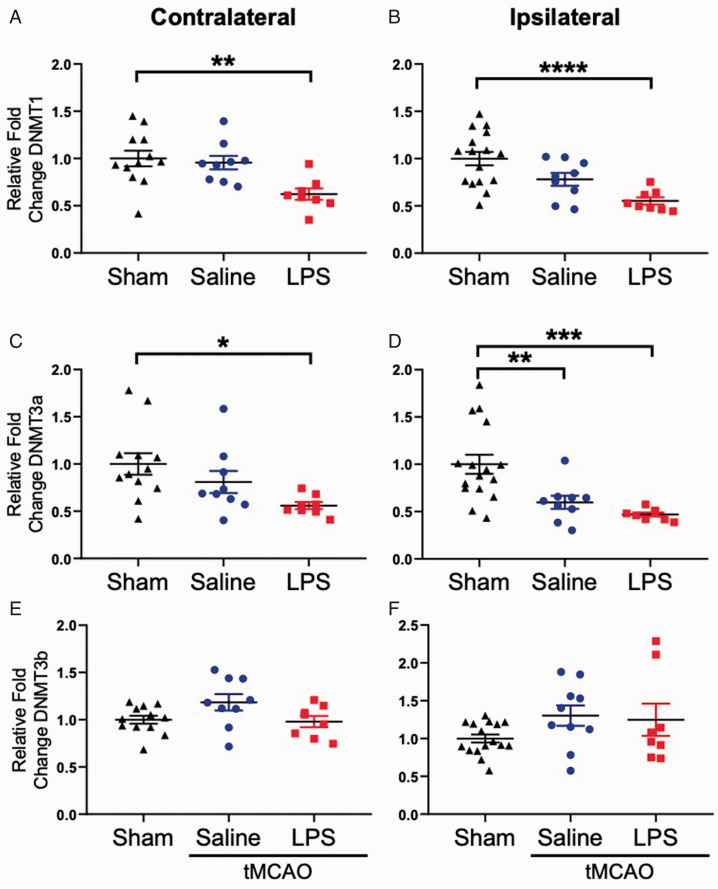
LPS Exposure Prior to Stroke Significantly Reduces DNMT mRNA Expression
De novo DNA methylation occurs via DNMT3a and DNMT3b, while DNMT1 functions to maintain methylation (Okano et al., 1999). To our surprise, we observed a significant downregulation of DNMT1 mRNA expression in both the contralateral (Figure 7A) and ipsilateral (Figure 7B) hemispheres of stroke animals exposed to LPS when compared to sham controls. We also observed significant downregulation of DNMT3a mRNA expression in the contralateral hemisphere (Figure 7C) of LPS exposed stroke animals, and downregulation in the ipsilateral hemispheres (Figure 7D) of both saline and LPS exposed stroke animals. We observed no significant changes in DNMT3b expression (Figure 7E and F).
Figure 7.
DNMT mRNA Expression Is Reduced Post-Stroke. DNMT1 mRNA expression was significantly reduced in both the (A) contralateral (F2, 26 = 6.713, p = 0.0045) and (B) ipsilateral (F2, 30 = 10.02, p = 0.0005) hemispheres of the LPS exposed stroke animals, when compared to sham controls. Similarly, DNMT3a mRNA expression was significantly reduced in the (C) contralateral hemisphere of LPS stroke animals (F2, 26 = 4.388, p = 0.0228), and in the (D) ipsilateral hemisphere of both saline and LPS stroke animals (F2, 30 = 9.661, p = 0.0006). No significant changes in DNMT3b mRNA expression were observed (E, F). Data are expressed as mean ± SEM; one-way ANOVA with Dunnett’s post hoc test; brackets connect statistically significant groups; **p < 0.01, ***p < 0.001, and ****p < 0.0001.
Discussion
In this study, we found that intermittent immune activation with the gram-negative bacterial wall component, LPS, induces significant sickness behaviors 4 hours post-injection, and that animals recover from this effect in < 13 days. Despite making a full recovery after the last injection of LPS, animals subjected to tMCAO had significantly larger infarct volumes at 24-hours reperfusion when compared to saline injected animals. We investigated several avenues to elucidate the mechanism behind this phenomenon. Consistent with the literature, we found significant upregulation of TNF-α mRNA in the brains of stroke animals, when compared to sham controls. We also assessed expression of autophagy related mRNAs and found significant reductions in expression within the ipsilateral hemispheres of the LPS exposed stroke mice. Reduced autophagic function can lead to the accumulation of damaged/unfunctional proteins and organelles, leading to cellular senescence, apoptosis and necrosis, and worsened stroke outcomes. Although we did not observe statistically significant differences between saline and LPS exposed stroke mice, we did observe significantly lower gene expression levels in the LPS stroke animals compared to sham, an effect that was not observed in the saline exposed stroke animals, with the exception of Beclin-1. These data indicate that intermittent LPS exposure prior to stroke does significantly reduce expression of key autophagy genes, however future experiments should focus on further elucidating the molecular mechanisms involved in these processes. We next assessed changes in DNMT expression which may alter the methylation status of the autophagy genes. Our data show significant decreases in DNMT1 and DNMT3a mRNA expression in LPS exposed stroke animals, with concurrent hypermethylation of autophagy gene promoters. This inverse relationship has been shown in other studies as well (Sharma et al., 2007; LaPlant et al., 2010; Shukla et al., 2014; Khalil et al., 2016), suggesting that DNMT mRNA are likely translated into proteins under regulation of posttranslational modifications, which in turns methylate the promoter of autophagy genes (Bayraktar and Kreutz, 2018). Similar to our observations of autophagy gene expression, we did not observe significant differences in DNMT gene expression between saline and LPS exposed stroke animals. With the exception of DNMT3a in the ipsilateral hemisphere, LPS exposed animals had significantly lower DNMT expression compared to sham, while the saline exposed animals did not. Interestingly, we did observe significant differences in methylation status between saline and LPS exposed animals, with LPS exposed stroke animals having significantly higher levels of methylation than the saline exposed stroke animals. Future studies should assess changes in gene expression and DNA methylation across different post-stroke timepoints and include assessments of protein expression to better understand the mechanisms by which LPS is imparting its negative post-stroke effects.
At the transcriptional level, TNF-α mRNA is significantly upregulated in post-stroke brains (Pan and Kastin, 2007), and protein expression in blood plasma of stroke patients 24 hours post-insult has been positively correlated with infarct volume (Zaremba and Losy, 2001). Administering exogenous TNF-α prior to experimental stroke significantly exacerbates infarct volume, while blocking endogenous TNF-α signaling post-stroke has shown to be neuroprotective (Barone et al., 1997). Previous work has also demonstrated that systemic administration of LPS can induce TNF-α mRNA expression (Buttini et al., 1997). Further, it is well established that systemic inflammation activates the sympathetic nervous system (Pongratz and Straub, 2014), and can lead to enhanced transcription of TNF-α mRNA within the brain (Zielinski et al., 2013). We hypothesized that our observation of LPS-induced increases in stroke infarct volume may be mediated by increased expression of TNF-α 24 hours post-stroke. Despite observing significant post-stroke upregulation of TNF-α mRNA expression, we observed no significant differences in TNF-α mRNA expression between saline and LPS exposed stroke animals, indicating that another mechanism is likely underlying this phenomenon.
Although not significant, the saline exposed stroke animals expressed higher levels of TNF-α mRNA (∼ 50-fold) than the LPS exposed stroke animals (∼ 35-fold). A recent study described an interesting LPS-mediated effect of immune tolerance within the brain. Two consecutive injections of LPS were required to initiate an immune response within the brain, however this effect was attenuated by the fourth injection (Wendeln et al., 2018). We may have observed reduced TNF-α mRNA in the LPS group, compared to the saline, due to immune tolerance, as these animals already had several experiences with brain-immune activation, while the animals injected with saline were experiencing a significant inflammatory event for the first time (i.e. the stroke). We also observed attenuation of sickness behavior with each subsequent LPS exposure, further strengthening the notion of LPS-tolerance after intermittent exposure.
TNF-α signaling can activate the transcription factor NF-κB, leading to the activation of mTOR, a prominent inhibitor of autophagy (Djavaheri-Mergny et al., 2006). We hypothesized that chronic LPS exposure would significantly upregulate TNF-α expression and potentially inhibit autophagy, leading to the promotion of apoptosis and necrosis, thus resulting in significantly larger infarct sizes. While this may occur in the general pathology of stroke, there do not appear to be significant differences in TNF-α expression between the saline and LPS exposed stroke animals.
Autophagy is generally regarded as a self-protective mechanism, which breaks down and recycles long-lived, damaged, or misfolded proteins and organelles (Wang et al., 2018). Oxygen and nutrient deprivation can activate autophagy, and several studies have found that activation of autophagy during cerebral ischemia has neuroprotective benefits by degrading damaged structures and thereby limiting apoptosis (Wang et al., 2012; Papadakis et al., 2013). We assessed mRNA expression of several genes involved in the autophagy pathway; Beclin-1, ATG5, LC3a, and LC3b, all of which have important functions in the formation and maturation of the autophagosome (Wang et al., 2018). If post-stroke autophagy is inhibited in the ipsilateral hemisphere of LPS exposed animals, damaged organelles and proteins would not be degraded. The accumulation of such damaged cellular contents can trigger apoptosis, or induce necrosis in these cells. This could contribute to the enlarged infarct we observed in the LPS exposed animals.
DNA methylation is an epigenetic modification that can be inherited, as well as influenced by environmental factors. In recent years a growing interest in stroke-induced alterations of DNA methylation has surfaced (Qureshi and Mehler, 2010; Soriano-Tárraga et al., 2014; Soriano-Tárraga et al., 2017). Segments (≥500 base pairs) of DNA which contain > 55% cytosine and guanine nucleotides, known as CpG islands, can be epigenetically modified via the methylation of cytosine residues. This mechanism is mediated by DNA methyl transferases (DNMTs), and inhibits gene transcription by preventing transcription factor binding (Krupinski et al., 2018). Aging, which is characterized by low-grade chronic inflammation (Franceschi et al., 2018), is the main risk factor for stroke, and is associated with global DNA hypomethylation, but CpG island hypermethylation (Johnson et al., 2012). Inflammation has also been directly associated with changes in DNA methylation (Abu-Remaileh et al., 2015; Maiuri et al., 2018). In animals chronically exposed to LPS, we observed a significant increase in DNA methylation of key autophagy genes as well as repression of their mRNA expression in the ischemic hemisphere post-stroke, while DNMT mRNA expression was decreased. These observations suggest that DNMT mRNA was utilized to synthesize DNMT proteins responsible for the observed DNA hypermethylation. Unfortunately, to our knowledge, no study has thoroughly investigated the role of posttranscriptional regulation of DNMT protein activity and DNA methylation status of the genome after intermittent exposures to LPS alone, as well as post-stroke.
In conclusion, this study has shown that chronic systemic inflammation induced by intermittent injections of LPS can significantly increase cortical infarcts in an experimental stroke model. We show that in the brain, TNF-α mRNA expression is significantly elevated post-stroke, regardless of prior saline or LPS exposure. Further, we show that mRNA expression of autophagy genes is significantly downregulated in the ischemic hemisphere of LPS exposed mice. We propose that the mechanism by which significant increases in infarct volume occurs in the LPS exposed mice is by de-regulated autophagy, leading to significantly increased neuronal apoptosis. Additionally, in view of our observation of downregulated DNMT mRNA expression in the ipsilateral hemisphere of LPS exposed stroke animals, we also propose that changes in DNA methylation may also significantly contribute to the increased infarct volume observed in this group.
Summary Statement
In the current study, we have found that intermittent exposure to an immune activator (lipopolysaccharide) increases cortical infarct size and impairs autophagy. Our data suggest that these observations may be mediated through deregulated epigenetic mechanisms.
Authors' Contributions
AER, DD, XR, and JWS designed the experiments. AER, JZC, AR, MV, AHD, HH, and DD performed the experiments. AA intellectual contribution. AER, DD, CBM, and JWS wrote the manuscript.
Supplementary Material
Footnotes
Ethical Approval: Approved by West Virginia University.
Declaration of Conflicting Interests: The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Funding: The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by NIH grants P20 GM109098, P01 AG027956, U54 GM104942, T32 AG052375, AHA SDG (16SDG31170008), and NSF 1008182R.
ORCID iDs: Candice M. Brown https://orcid.org/0000-0001-5845-0221
Duaa Dakhlallah https://orcid.org/0000-0002-6288-4927
Supplemental material: Supplemental material for this article is available online.
References
- Aapola U., Kawasaki K., Scott H. S., Ollila J., Vihinen M., Heino M., Shintani A., et al. (2000). Isolation and initial characterization of a novel zinc finger gene, DNMT3L, on 21q22.3, related to the cytosine-5-methyltransferase 3 gene family. Genomics, 65, 293–298. [DOI] [PubMed] [Google Scholar]
- Abu-Remaileh M., Bender S., Raddatz U, Ansari I., Cohen D., Gutekunst J., Musch T., et al. (2015). Chronic Inflammation Induces a Novel EpigeneticProgram That Is Conserved in Intestinal Adenomasand in Colorectal Cancer. Tumor and Stem Cell Biology. Advance online publication. 10.1158/0008-5472.CAN-14-3295. [DOI] [PubMed]
- Barone F. C., Arvin B., White R. F., Miller A., Webb C. L., Willette R. N., Lysko P. G., Feuerstein G. Z. (1997). Tumor necrosis factor-alpha. A mediator of focal ischemic brain injury. Stroke, 28, 1233–1244. [DOI] [PubMed] [Google Scholar]
- Bayarsaihan D. (2011). Epigenetic mechanisms in inflammation. J Dent Res, 90, 9–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bayraktar G., Kreutz M. R. (2018). Neuronal DNA methyltransferases: Epigenetic mediators between synaptic activity and gene expression? SAGE Publications Inc. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bova I. Y., Bornstein N. M., Korczynl A. D. (1996). Acute infection as a risk factor for ischemic stroke. Stroke, 27, 2204–2206. [DOI] [PubMed] [Google Scholar]
- Buttini M., Mir A., Appel K., Wiederhold K. H., Limonta S., Gebicke-Haerter P. J., Boddeke H. W. G. M. (1997). Lipopolysaccharide induces expression of tumour necrosis factor alpha in rat brain: inhibition by methylprednisolone and by rolipram, Br J Pharmacol, 122, 1483–1489. [DOI] [PMC free article] [PubMed]
- Campbell L. A., Rosenfeld M. E. (2015). Infection and atherosclerosis development. Arch Med Res, 46, 339–350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carson W. F., Cavassani K. A., Dou Y., Kunkel S. L. (2011). Epigenetic regulation of immune cell functions during post-septic immunosuppression. Epigenetics, 6, 273–283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dakhlallah D., Batte K., Wang Y., Cantemir-Stone C. Z., Yan P., Nuovo G., Mikhail A., et al. (2013). Epigenetic regulation of miR-17∼92 contributes to the pathogenesis of pulmonary fibrosis. Am J Respir Crit Care Med, 187, 397–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Day J. J. (2014). New approaches to manipulating the epigenome. Dialogues Clin Neurosci, 16, 345–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Djavaheri-Mergny M., Amelotti M., Mathieu J., Besançon F., Bauvy C., Souquère S., Pierron G., Codogno P. (2006). NF-kappaB activation represses tumor necrosis factor-alpha-induced autophagy. J Biol Chem, 281, 30373–30382. [DOI] [PubMed] [Google Scholar]
- Doll D. N., Engler-Chiurazzi E. B., Lewis S. E., Hu H., Kerr A. E., Ren X., Simpkins J. W. (2015). Lipopolysaccharide exacerbates infarct size and results in worsened post-stroke behavioral outcomes. Behav Brain Funct, 11, 32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doll D. N., Hu H., Sun J., Lewis S. E., Simpkins J. W., Ren X. (2015). Mitochondrial crisis in cerebrovascular endothelial cells opens the blood–brain barrier. Stroke, 46, 1681–1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkind M. S. V, Carty C. L., O’Meara E. S., Lumley T., Lefkowitz D., Kronmal R. A., Longstreth W. T., Jr.(2011). Hospitalization for infection and risk of acute ischemic stroke: the Cardiovascular Health Study. Stroke, 42, 1851–1856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng Y., He D., Yao Z., Klionsky D. J. (2014). The machinery of macroautophagy. Cell Res, 24, 24–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Franceschi C., Garagnani P., Parini P., Giuliani C., Santoro A. (2018). Inflammaging: A new immune–metabolic viewpoint for age-related diseases. Nat Rev Endocrinol, 14, 576–590. [DOI] [PubMed] [Google Scholar]
- Gorelick P. B. (2019). The global burden of stroke: persistent and disabling. Lancet Publishing Group. [DOI] [PubMed] [Google Scholar]
- Grau A. J., Buggle F., Heindl S., Steichen-Wiehn C., Banerjee T., Maiwald M., Rohlfs M., Suhr H., Fiehn W., Becher H. (1995). Recent infection as a risk factor for cerebrovascular ischemia. Stroke, 26, 373–379. [DOI] [PubMed] [Google Scholar]
- Grau A. J., Fischer B., Barth C., Ling P., Lichy C., Buggle F. (2005). Influenza vaccination is associated with a reduced risk of stroke. Stroke, 36, 1501–1506. [DOI] [PubMed] [Google Scholar]
- Guedes P. M. M., de Andrade C. M., Nunes D. F., de Sena Pereira N., Queiroga T. B. D., Machado-Coelho G. L. L., Nascimento M. S. L., et al. (2016). Inflammation enhances the risks of stroke and death in chronic chagas disease patients. PLoS Negl Trop Dis, 10, e0004669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gujar H., Weisenberger D. J., Liang G. (2019). The roles of human DNA methyltransferases and their isoforms in shaping the epigenome. Genes (Basel), 10, 172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A. A., Akman K., Calimport , S. R. G., Wuttke D., Stolzing A., de Magalhães J. P. (2012) The role of DNA methylation in aging, rejuvenation, and age-related disease. Rejuvenation Res, 15, 483–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung C. H., Ro S.-H., Cao J., Otto N. M., Kim D.-H. (2010. ). mTOR regulation of autophagy. FEBS Lett, 584, 1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang R., Zeh H. J., Lotze M. T., Tang D. (2011). The Beclin 1 network regulates autophagy and apoptosis. Cell Death Differ, 18, 571–580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil H., Tazi M., Caution K., Ahmed A., Kanneganti A., Assani K., Kopp B., Marsh C., Dakhlallah D., Amer A. O. (2016). Aging is associated with hypermethylation of autophagy genes in macrophages. Epigenetics, 11, 381–388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krupinski J., Carrera C., Muiño E., Torres N., Al-Baradie R., Cullell N., Fernandez-Cadenas I. (2018). DNA methylation in stroke. Update of latest advances. Comput Struct Biotechnol J, 16, 1–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kusama Y., Sato K., Kimura N., Mitamura J., Ohdaira H., Yoshida K. (2009). Comprehensive analysis of expression pattern and promoter regulation of human autophagy-related genes. Apoptosis, 14, 1165–1175. [DOI] [PubMed] [Google Scholar]
- LaPlant Q., Vialou V., Covington H. E., Dumitriu D., Feng J., Warren B. L., Maze I., et al. (2010). Dnmt3a regulates emotional behavior and spine plasticity in the nucleus accumbens. Nat Neurosci, 13, 1137–1143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J.-W., Nam H., Kim L. E., Jeon Y., Min H., Ha S., Lee Y., et al. (2019). TLR4 (toll-like receptor 4) activation suppresses autophagy through inhibition of FOXO3 and impairs phagocytic capacity of microglia. Autophagy, 15, 753–770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiuri A. R., Li H., Stein B. D., Tennessen J. M., O’Hagan H. M. (2018). Inflammation-induced DNA methylation of DNA polymerase gamma alters the metabolic profile of colon tumors. Cancer Metab, 6, 9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer L. M., Tomasini-Johansson B. R., Mosher D. F. (2010). Emerging roles of fibronectin in thrombosis. Thromb Res, 125, 287–291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCall C. E., Yoza B., Liu T., El Gazzar M. (2010). Gene-specific epigenetic regulation in serious infections with systemic inflammation. J Innate Immun, 2, 395–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R., Horng T. (2009). Transcriptional control of the inflammatory response. Nat Rev Immunol, 9, 692–703. [DOI] [PubMed] [Google Scholar]
- Okano M., Bell D. W., Haber D. A., Li E. (1999). DNA methyltransferases Dnmt3a and Dnmt3b are essential for de novo methylation and mammalian development. Cell, 99, 247–257. [DOI] [PubMed] [Google Scholar]
- Paganini-Hill A., Lozano E., Fischberg G., Perez Barreto M., Rajamani K., Ameriso S. F., Heseltine P. N. R., Fisher M. (2003). Infection and risk of ischemic stroke. Stroke, 34, 452–457. [DOI] [PubMed] [Google Scholar]
- Pan W., Kastin A. J. (2007). Tumor necrosis factor and stroke: Role of the blood-brain barrier. Prog Neurobiol, 83, 363–374. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papadakis M., Hadley G., Xilouri M., Hoyte L. C., Nagel S., McMenamin M. M., Tsaknakis G., et al. (2013). Tsc1 (hamartin) confers neuroprotection against ischemia by inducing autophagy. Nat Med, 19, 351–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawluczyk I. Z., Harris K. P. (1998). Cytokine interactions promote synergistic fibronectin accumulation by mesangial cells. Kidney Int, 54, 62–70. [DOI] [PubMed] [Google Scholar]
- Pongratz G., Straub R. H. (2014). The sympathetic nervous response in inflammation. Arthritis Res Ther, 16, 504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qureshi I. A., Mehler M. F. (2010). Emerging role of epigenetics in stroke: part 1: DNA methylation and chromatin modifications. Arch Neurol, 67, 1316–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessa R., Di P. M., Filardo S., Turriziani O. (2014). Infectious burden and atherosclerosis: A clinical issue. World J Clin Cases, 2, 240–249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma R. P., Tun N., Grayson D. R. (2007). Depolarization induces downregulation of DNMT1 and DNMT3a in primary cortical cultures. Epigenetics, 3, 74–80. [DOI] [PubMed] [Google Scholar]
- Shukla S., Patric I. R. P., Patil V., Shwetha S. D., Hegde A. S., Chandramouli B. A., Arivazhagan A., Santosh V., Somasundaram K. (2014). Methylation silencing of ULK2, an autophagy gene, is essential for astrocyte transformation and tumor growth. J Biol Chem, 289, 22306–22318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano-Tárraga C., Jiménez-Conde J., Giralt-Steinhauer E., Mola M., Ois Á., Rodríguez-Campello A., Cuadrado-Godia E., et al. (2014). Global DNA methylation of ischemic stroke subtypes. PLoS One, 9, e96543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soriano-Tárraga C., Mola-Caminal M., Giralt-Steinhauer E., Ois A., Rodríguez-Campello A., Cuadrado-Godia E., Gómez-González A., et al. (2017). Biological age is better than chronological as predictor of 3-month outcome in ischemic stroke. Neurology, 89, 830–836. [DOI] [PubMed] [Google Scholar]
- Stern D. M., Kaiser E., Nawroth P. P. (1988). Regulation of the coagulation system by vascular endothelial cells. Pathophysiol Haemost Thromb, 18, 202–214. [DOI] [PubMed] [Google Scholar]
- Sun J., Hu H., Ren X., Simpkins J. W. (2016). Tert-butylhydroquinone compromises survival in murine experimental stroke. Neurotoxicol Teratol, 54, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolg C., Sabha N., Cortese R., Panchal T., Ahsan A., Soliman A., Aitken K. J., Petronis A., Bägli D. J. (2011). Uropathogenic E. coli infection provokes epigenetic downregulation of CDKN2A (p16INK4A) in uroepithelial cells. Lab Investig, 91, 825–836. [DOI] [PubMed] [Google Scholar]
- Wang P., Guan Y.-F., Du H., Zhai Q.-W., Su D.-F., Miao C.-Y. (2012). Induction of autophagy contributes to the neuroprotection of nicotinamide phosphoribosyltransferase in cerebral ischemia. Autophagy, 8, 77–87. [DOI] [PubMed] [Google Scholar]
- Wang P., Shao B.-Z., Deng Z., Chen S., Yue Z., Miao C.-Y. (2018). Autophagy in ischemic stroke. Prog Neurobiol, 163–164, 98–117. [DOI] [PubMed] [Google Scholar]
- Wendeln A.-C., Degenhardt K., Kaurani L., Gertig M., Ulas T., Jain G., Wagner J., et al. (2018). Innate immune memory in the brain shapes neurological disease hallmarks. Nature, 556, 332–338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson M., Seymour R., Henderson B. (1998). Bacterial perturbation of cytokine networks. Infect Immun, 66, 2401–2409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Q., Tong X., Schieb L., Vaughan A., Gillespie C., Wiltz J. L., King S. C., et al. (2017). Vital signs: Recent trends in stroke death rates — United States, 2000–2015. MMWR Morb Mortal Wkly Rep, 66, 933–939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yin L., Chung W. O. (2011). Epigenetic regulation of human β-defensin 2 and CC chemokine ligand 20 expression in gingival epithelial cells in response to oral bacteria. Mucosal Immunol, 4, 409–419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaremba J., Losy J. (2001). Early TNF-alpha levels correlate with ischaemic stroke severity. Acta Neurol Scand, 104, 288–295. [DOI] [PubMed] [Google Scholar]
- Zeller J. A., Lenz A., Eschenfelder C. C., Zunker P., Deuschl G. (2005). Platelet-leukocyte interaction and platelet activation in acute stroke with and without preceding infection. Arterioscler Thromb Vasc Biol, 25, 1519–1523. [DOI] [PubMed] [Google Scholar]
- Zhang Y., Liu D., Chen X., Li J., Li L., Bian Z., Sun F., et al. (2010). Secreted monocytic miR-150 enhances targeted endothelial cell migration. Mol Cell, 39, 133–144. [DOI] [PubMed] [Google Scholar]
- Zielinski M. R., Dunbrasky D. L., Taishi P., Souza G., Krueger J. M. (2013). Vagotomy attenuates brain cytokines and sleep induced by peripherally administered tumor necrosis factor-α and lipopolysaccharide in mice. Sleep, 36, 1227–1238, 1238A. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.