Abstract
Background
Gut microbiota mainly function in the biotransformation of primary ginsenosides into bioactive metabolites. Herein, we investigated the effects of three prebiotic fibers by targeting gut microbiota on the metabolism of ginsenoside Rb1 in vivo.
Methods
Sprague Dawley rats were administered with ginsenoside Rb1 after a two-week prebiotic intervention of fructooligosaccharide, galactooligosaccharide, and fibersol-2, respectively. Pharmacokinetic analysis of ginsenoside Rb1 and its metabolites was performed, whilst the microbial composition and metabolic function of gut microbiota were examined by 16S rRNA gene amplicon and metagenomic shotgun sequencing.
Results
The results showed that peak plasma concentration and area under concentration time curve of ginsenoside Rb1 and its intermediate metabolites, ginsenoside Rd, F2, and compound K (CK), in the prebiotic intervention groups were increased at various degrees compared with those in the control group. Gut microbiota dramatically responded to the prebiotic treatment at both taxonomical and functional levels. The abundance of Prevotella, which possesses potential function to hydrolyze ginsenoside Rb1 into CK, was significantly elevated in the three prebiotic groups (P < 0.05). The gut metagenomic analysis also revealed the functional gene enrichment for terpenoid/polyketide metabolism, glycolysis, gluconeogenesis, propanoate metabolism, etc.
Conclusion
These findings imply that prebiotics may selectively promote the proliferation of certain bacterial stains with glycoside hydrolysis capacity, thereby, subsequently improving the biotransformation and bioavailability of primary ginsenosides in vivo.
Keywords: Ginsenoside, Prebiotic, Gut microbiota, Biotransformation, Bioavailability
Abbreviations: ANOVA, analysis of variance; AUC, area under the concentration-time curve; Cmax, peak plasma concentration; CAT, CAZymes Analysis Toolkit; CAZymes, carbohydrate active enzymes; CK, compound K; FDR, false discovery rate; FOS, fructooligosaccharide; GOS, galactooligosaccharide; IS, internal standard; KEGG, the Kyoto Encyclopaedia of Genes and Genomes; LCA, lowest common ancestor; LDA, linear discriminant analysis; LEfSe, LDA effect size; LLOQs, lower limits of quantifications; MANOVA, multivariate ANOVA; MRM, multiple reaction monitoring; NMDS, non-metric multidimensional scaling; PCA, principal component analysis; PCoA, principal coordinates analysis; SD, Sprague Dawley; SRA, Sequence Read Archive; UPLC-ESI-QqQ-MS/MS, ultra-high pressure liquid chromatography coupled to an electrospray ionization source and a triple-quadrupole mass spectrometer; Tmax, time of maximum plasma concentration
1. Introduction
Panax ginseng Meyer, as an herbal medicine, has been used for clinical practice and healthcare nourishing for several millennia in China, Japan, Korea, and other East Asian countries [1]. Modern pharmacological research has confirmed that ginsenosides are the major bioactive compounds in P. ginseng and possess multiple therapeutic activities, including antitumor, antihypertension, antivirus, and immune-regulation [2]. Under in vivo condition, primary naturally occurring ginsenosides (e.g. ginsenosides Rb1, Rb2, and Rc) are deglycosylated stepwise by glycoside hydrolases (e.g. β-glucosidase, α-rhamnosidase, and xylosidase), which are present in mammalian gut microbiota, into active secondary ginsenosides such as 20-O-β-D-glucopyranosyl-20(S)-protopanaxadiol (ginsenoside compound K (CK)) [3,4]. The secondary ginsenosides, especially ginsenoside CK, are considered as the major bioactive metabolites attributed to their stronger pharmacological activities than the precursors [5,6]. Furthermore, the prototypical ginsenoside Rb1 has an extremely low oral bioavailability of 0.28–1.18%, while ginsenoside CK has a substantially increased oral bioavailability of 1.8–35.0% [7]. However, the availability of primary ginsenosides for ginsenoside CK manufacturing is compromised by the long-term cultivation periods (5-7 years) and crop rotation cycles (more than 5 years) of Panax; moreover, the industrial production of ginsenoside CK through total synthesis is still impractical [8]. Considering the crucial role of gut microbiota in promoting the metabolic conversion of primary ginsenosides into active secondary ginsenosides, modulating the metabolic function of gut microbiota could be a potential strategy to enhance the pharmacological efficacy of orally administered ginseng [3,4,9].
A three-week intervention of a fermented dairy product that contains two probiotics, Lactobacillus acidophilus and Bifidobacterium bifidum, improved the activity of fecal β-glucosidase in healthy volunteers [10]. Furthermore, a soluble prebiotic fiber called NUTRIOSE was demonstrated to increase the formation and subsequent absorption of ginsenosides Rd and CK following an oral administration of ginseng in rats [11]. Therefore, enhancing the abundances or glucosidase activities of specific gut microbes through probiotic [10,12] and/or prebiotic [11] intervention could potentially increase the formation and subsequent absorption of ginsenoside CK.
However, due to the system complexity and functional redundancy of gut microbiota, the direct link between probiotic or prebiotic intervention and the consistent bacterial composition, enzymes, and metabolic pathways of gut microbiota has not been established. Furthermore, fecal specimen-based approaches, such as the fecal lysate fermentation and in vitro anaerobic culturing, can only partially reflect the physiological condition or the holistic function of gut microbiota in vivo. Meanwhile, the individually specific gut microbiota with differential α/β glucosidase activities and microbial biotransformation potentials contribute to the physiological variability in humans [13]. Therefore, the effect of prebiotics in the whole spectrum of modulating the abundances and metabolic activities of gut microbiota requires in-depth investigation based on a consistent biological baseline. In our current study, the effects of three fibers, namely, fructooligosaccharide (FOS), galactooligosaccharide (GOS), and fibersol-2, which are recognized as prebiotic ingredients, on the pharmacokinetic characteristics of ginsenoside Rb1 and its metabolites, as well as on the gut microbial composition and potential metabolic function in rats were investigated.
2. Materials and methods
2.1. Materials
FOS (purity ≥95%, with ≤5% moisture) and GOS (purity ≥90%, with ≤5% moisture, ≤8.5% anhydrous lactose and ≤1.5% anhydrous glucose) were purchased from Quantum Hi-Tech (China) Biological Co., Ltd. (Jiangmen, Guangdong, China), and fibersol-2 (purity ≥90%, with ≤6% moisture) was obtained from Matsutani Chemical Industry Co., Ltd. (Itami, Hyogo, Japan). Methanol and acetonitrile were provided by Sigma-Aldrich (St. Louis, MO, USA). Acetone and other analytical chemical solvents were obtained from Beijing Chemical Factory (Beijing, China).
The compounds of ginsenosides Rb1, Rd, F2, CK, and the internal standard (IS) saikosaponin A were supplied by the National Institute for the Control of Biological and Pharmaceutical Products (Beijing, China). The hydrophobic polytetrafluoroethylene (0.22 μm) used for organic solution filtration was obtained from Millipore (Millipore, Germany).
2.2. Experimental animals
Male Sprague Dawley (SD) rats (weight 200 ± 20 g, N = 32) were purchased from Beijing Weitong Lihua Experimental Animal Technical Co., Ltd (Beijing, China). These SD rats were housed in a controlled environment with a temperature of 25 °C and 12-h light–dark cycle. The commercial feed (Beijing Keao Co., Beijing, China) and water were autoclaved before providing to the rats, allowing ad libitum. Blank plasma samples were collected from the rats before prebiotic intervention. All the studies were performed in accordance with the proposals approved by the animal ethics committee.
2.3. Prebiotic intervention and sample collection
After one-week acclimation, a total of 32 SD rats were randomly and equally divided into control, FOS, GOS and fibersol-2 groups (N = 8 for each group). To guarantee a consistent prebiotic intake, a temperate and common dose of 5 g/kg (body weight) according to the literature was orally gavaged into the rats. Thus, assuming the average body weight of the six-week-old rats as approximately 200 g, they were gavaged with sterile water (10 ml/kg), FOS, GOS, and fibersol-2 at the dose of 1.0 g/d/rat, respectively, for two weeks.
After an overnight fast, blood samples (approximately 500 μl) were collected from ophthalmic artery plexus of the rats at 0.25, 0.5, 1, 1.5, 2, 3, 4, 6, 8, 10, 12, 24, and 48 h after intragastric administration of ginsenoside Rb1 (100 mg/kg) into capillary tubes. The plasma fraction was separated by centrifugation and then stored at −80 °C until further analysis. One fresh fecal sample was collected from each rat into a sterile tube, directly from the anus to avoid contact with their skin or urine. A total of 32 stool samples were collected from the SD rats in the four groups at each sampling time point (−14 d, 0 h and 48 h) and immediately stored at −80 °C for subsequent analysis.
2.4. Pharmacokinetics
Approximately 100 μl IS of saikosaponin A (141.2 ng/ml) was evaporated under gentle nitrogen stream. Then, 100 μl plasma sample and 400 μl acetone were added, followed by vortex mixing for 5 min, and centrifuging at 12,000 g for 5 min to harvest the supernatant. After evaporating at 40 °C, methanol (100 μl, 50% v/v) was added for re-dissolving, followed by vortex mixing and centrifuging. The supernatant was purified by passing through 0.22 μm Millipore filter before subsequent analysis.
The ultra-high pressure liquid chromatography coupled with an electrospray ionization source and a triple-quadrupole mass spectrometer (UPLC-ESI-QqQ-MS/MS), which contains an Agilent 1290 UPLC system with an Agilent 6460 triple quadrupole mass spectrometry series (Agilent, USA), was used for separating and quantifying the analytes. A reverse-phase Agilent XDB-C18 column (1.8 μm, 3.0 mm × 50 mm) coupled with an Agilent ZORBAX SB-C18 guard column (5 μm, 4.6 mm × 12.5 mm) was also used. The elution gradient was carried out by a binary solvent system consisted of water (A) and MS-grade methanol (B) at a flow rate of 0.2 ml/min and a column temperature of 40 °C. The gradient elution program was presented as follows: 0-8.5 min, 76% B; 8.5-13 min, 82% B; 13-13.1 min, 76% B; 13.1-16 min, 76% B. Electrospray ionization (ESI) of MS was performed in positive mode by using nitrogen as the drying agent. For the positive mode, MS conditions were given as follows: gas temperature, 300 °C; gas flow, 5 L/min; nebulizer, 45 psi; sheath gas temperature, 250 °C; nozzle voltage, 500 V; sheath gas flow, 9 L/min; capillary, 3500 V; HV voltage, 4000 KV; and delta EMV, 200 V. The collision gas of helium was used for collision-induced dissociation, and multiple reaction monitoring (MRM) was used for quantifying.
Under the optimized UPLC-ESI-QqQ-MS/MS method for ginsenosides detection, peak plasma concentration (Cmax) and time of maximum plasma concentration (Tmax) were measured. The formula of area under the concentration-time curve (AUC), namely AUC0-∞ = AUC0-t + Ct/Ke, was applied to calculate the AUC from time zero to infinity (AUC0-∞).
2.5. Gut microbiota structural profiling
Total bacterial genomic DNA was extracted from the fecal sample by using the E.Z.N.A.® stool DNA Kit (Omega Bio-tek, Norcross, GA, USA). The V3/V4 region of 16S rRNA gene with a 468 bp inserting size were amplified with the primers of 338F5′-barcode-ACTCCTACGGGAGGCAGCA)-3′ and 806R5′-GGACTACHVGGGTWTCTAAT-3’ [14]. Purified amplicons from each sample were pooled for paired-end sequencing (PE300). The raw reads were deposited in the Sequence Read Archive (SRA) database at the NCBI (accession number: PRJNA518000).
Quality control of raw fastq files was performed by using QIIME (version 1.17) to trim adapter and remove low-quality sequences, as described by the criteria [15]. The operational taxonomic units (OTUs) were classified at a threshold of 97% sequence similarity by using UPARSE (version 7.1 http://drive5.com/uparse/). RDP Classifier (http://rdp.cme.msu.edu/) was used for the taxonomic assignment of each 16S rRNA gene sequence at a confidence threshold of 70%.
2.6. Metagenome sequencing, assembly, and annotation
Bacterial genomic DNA was fragmented by Covaris M220 focused-ultrasonicator (Covaris Inc., MA, USA) to the size of about 350 bp. Sequencing libraries were constructed by using the NEBNext® Ultra™ DNA Library Prep Kit from Illumina (NEB Inc., MA, USA). After cluster generation in the cBot Cluster Generation System (Illumina, CA, USA), the libraries were sequenced on an Illumina HiSeq platform at the Novogene Bioinformatics Institute (Beijing, China). The metagenome dataset was deposited in the SRA database at the NCBI under accession number PRJNA518011.
The raw reads were screened to generate clean data by removing adaptor contamination, host sequences, and low-quality reads [16]. After assembling with SOAP denovo 2.21 (http://soap.genomics.org.cn/soapdenovo.html) [17], scaffolds were split into scaftigs by removing the Ns. The MetaGeneMark 2.10 (http://topaz.gatech.edu/GeneMark) [18] was used to predict the open reading frames, which were further clustered in CD-HIT 4.5.8 (http://www.bioinformatics.org/cd-hit/) [19] to generate raw gene catalogue. The abundances of Unigenes were evaluated by mapping the corresponding clean data to the gene catalogue by SoapAligner 2.21 (http://soap.genomics.org.cn/soapaligner.html) [20]. After comparing the NR datasets of the NCBI with microbial reference genomes, the Unigenes were identified by the lowest common ancestor (LCA) algorithm for taxonomic analysis [21]. Metabolic function analysis was performed by the Kyoto Encyclopaedia of Genes and Genomes (KEGG) [22] and Carbohydrate Active Enzymes (CAZymes) Analysis Toolkit (CAT) (http://cricket.ornl.gov/cgi-bin/cat.cgi) [23].
2.7. Statistical analysis
The plasma concentrations of ginsenosides Rb1, Rd, F2, and CK in each individual rat were calculated by non-compartment model using DAS 3.2.6 software (BioGuider Co., Shanghai, China). Statistical comparisons were performed by one-way analysis of variance (ANOVA) with the Bonferroni post-test. P value < 0.05 was determined as statistical significance. The overview shift of gut microbiota composition and metabolic function was characterized by principal component analysis (PCA) and principal coordinates analysis (PCoA). Multivariate ANOVA (MANOVA) with PCA or PCoA accounting for approximately 85% of the total variations was conducted for microbial clustering. Significant OTUs or metabolic functions/enzymes and linear discriminant analysis effect size that can distinguish various treatment groups were selected by Metastats. The levels of statistical significance were set at ∗ P < 0.05 and ∗∗P < 0.01.
3. Results
3.1. Effects of prebiotics on pharmacokinetics of ginsenoside Rb1 and its metabolites
The MRM chromatograms of ginsenosides Rb1, Rd, F2, and CK from rat plasma samples and IS were optimized for quantitative analysis using [M+Na]+-MS/MS[Q3]+ paired ions at m/z 1131.6-365.1, 969.6-789.4, 807.6-203.2, 645.4-203.1, and 803.5-331, respectively. Another paired ion ([M+Na]+-MS/MS[Q3]+) was used for the qualitative analysis at m/z 1131.6-789.4, 969.6-365, 807.6-637, 645.4-465.3, and 803.5-203 for ginsenosides Rb1, Rd, F2, and CK, as well as IS, respectively (Supplementary Table S1). We obtained the retention times at 3.7, 5.7, 11.3, 13.6, and 5.2 min for ginsenosides Rb1, Rd, F2, and CK, as well as IS, respectively (Supplementary Fig. S1). No interfering endogenous substance was observed during the detection of the four ginsenosides and IS in the blank control plasma samples (Supplementary Fig. S2). Calibration curve of the four compounds showed good linearity with the available standards (R2 > 0.999), and the lower limits of quantifications (LLOQs) for the four ginsenosides ranged from 0.039 to 0.19 ng/ml (Supplementary Table S2), indicating sufficient sensitivities of the quantitative evaluation.
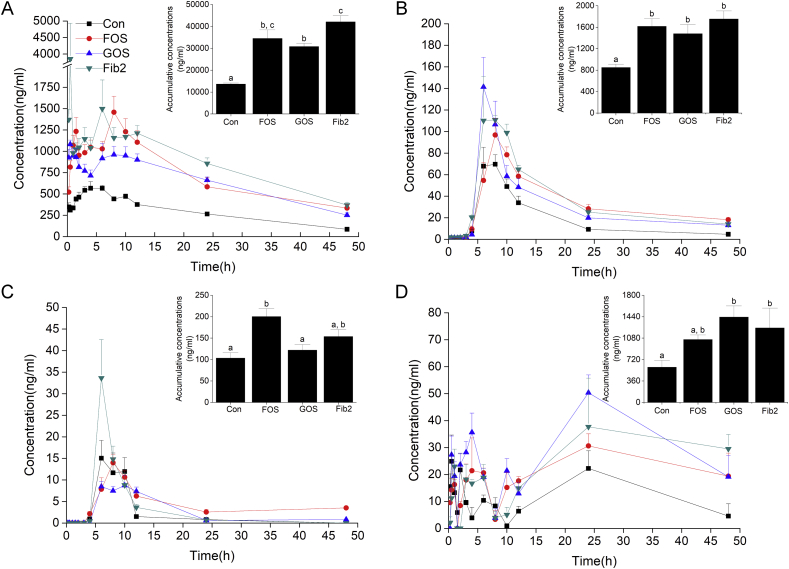
Fig. 1 and Table 1 summarized the pharmacokinetic parameters of the four ginsenosides. The AUCs of ginsenosides Rb1 (Fig. 1A), Rd (Fig. 1B), and F2 (Fig. 1C) were significantly increased by 2.52, 1.91, and 1.94 times respectively comparing with the control group by FOS intervention (P < 0.05). The similar tendency was also observed in the GOS group, wherein the AUCs of ginsenosides Rb1 (Fig. 1A) and Rd (Fig. 1B) were significantly improved by 2.25 and 1.74 times respectively in comparison with the control group (P < 0.05). These parameters of ginsenosides Rb1 and Rd were also increased by 3.07 and 2.06 times by fibersol-2 intervention (P < 0.05). Furthermore, the Cmax values of ginsenosides Rb1, Rd, F2, and CK were increased by various levels after the treatment of the three fibers (Table 1).
Fig. 1.
Integrated concentration-time profile and accumulative concentrations of ginsenoside Rb1 and its metabolites. After the intragastric administration of (A) ginsenoside Rb1 (100 mg/kg), its intermediate metabolites, (B) ginsenoside Rd and (C) ginsenoside F2, and the final metabolite (D) ginsenoside CK in the plasma of the rats in control, fructooligosaccharide (FOS), galactooligosaccharide (GOS), and fibersol-2 groups (mean ± SE) are summarized.
Table 1.
Pharmacokinetic parameters of ginsenosides Rb1, Rd, F2, and CK following the oral administration of ginsenoside Rb1 in rats treated with placebo, fructooligosaccharide (FOS), galactooligosaccharide (GOS), and fibersol-2, respectively
| Groups | Parameters | Control | FOS | GOS | Fibersol-2 |
|---|---|---|---|---|---|
| Rb1 | Tmax1) (h) | 4.40 ± 1.52a4) | 6.30 ± 2.82a | 3.2 ± 3.55ab | 1.00 ± 1.12b |
| Cmax2) (ng/ml) | 657.75 ± 128.02a | 1527.93 ± 342.81ab | 1206.83 ± 125.70b | 3883.77 ± 2369.26ab | |
| AUC3) (ng/ml∗h) | 13695.33 ± 1077.54a | 34494.77 ± 8770.73bc | 30813.79 ± 3334.99b | 42110.46 ± 6452.33c | |
| T1/2 (h) | 15.52 ± 1.58ab | 19.39 ± 4.44b | 19.73 ± 2.06b | 24.23 ± 4.81bc | |
| Rd | Tmax (h) | 7.60 ± 1.67a | 7.60 ± 0.89a | 6.40 ± 0.89a | 7.60 ± 1.67a |
| Cmax (ng/ml) | 88.98 ± 24.30a | 109.93 ± 25.05ab | 156.25 ± 38.96b | 145.06 ± 67.24ab | |
| AUC (ng/ml∗h) | 848.74 ± 140.63a | 1616.98 ± 321.59b | 1479.93 ± 381.35b | 1754.98 ± 335.76b | |
| T1/2 (h) | 11.93 ± 7.54a | 15.20 ± 5.17a | 18.63 ± 18.01a | 10.35 ± 3.79a | |
| F2 | Tmax (h) | 8.00 ± 2.00a | 8.00 ± 1.41a | 7.60 ± 1.67a | 6.80 ± 1.09a |
| Cmax (ng/ml) | 20.26 ± 6.35a | 15.23 ± 2.49a | 11.44 ± 3.44a | 35.27 ± 18.29b | |
| AUC (ng/ml∗h) | 103.40 ± 28.53a | 200.38 ± 41.50b | 121.97 ± 29.35a | 153.78 ± 36.00ab | |
| T1/2 (h) | 3.06 ± 0.29a | 23.22 ± 37.78a | 4.52 ± 3.73a | 2.67 ± 0.95a | |
| CK | Tmax (h) | 1.20 ± 0.76a | 24.80 ± 15.60b | 15.30 ± 11.98ab | 29.00 ± 19.72b |
| Cmax (ng/ml) | 37.91 ± 10.25a | 42.94 ± 12.08a | 57.46 ± 7.17a | 53.38 ± 25.17a | |
| AUC (ng/ml∗h) | 591.98 ± 256.78a | 1056.88 ± 173.22ab | 1434.71 ± 415.30b | 1251.99 ± 740.67b | |
| T1/2 (h) | 3.38 ± 0.26a | 92.12 ± 158.19a | 77.35 ± 129.14a | 50.86 ± 65.79a |
Values (mean ± SE) marked by different letters in the same row indicate a significant difference (P < 0.05).
Tmax, maximum drug concentration time.
Cmax, maximum plasma concentration.
AUC, area under the blood concentration curve from 0 to 48 h.
Analysis of variance is used to detect the differences in means.
The accumulative concentrations of ginsenoside CK (Fig. 1D), the major metabolite of PPD-typed ginsenosides with potent pharmacological activity, were significantly improved by GOS or fibersol-2 intervention (P < 0.05) (Table 1). The AUCs of ginsenoside CK after FOS, GOS and fibersol-2 intervention were 1.76, 2.42, and 2.11 times greater than that of the control group, respectively.
3.2. Structural modulation of gut microbiota after prebiotic intervention
A bar-coded amplicon sequencing based on 16S rRNA genes was performed to profile the overall structural shift of gut microbiota induced by prebiotic intervention. A total of 1,646,123 raw and 1,153,475 high-quality sequences were obtained (37,209 ± 4,272 reads per sample). Approximately 8,634 OTUs were generated at the 97% similarity threshold level (279 ± 59 OTUs per sample). Rarefaction curves and Shannon index (H′) revealed that most of the microbial diversity had already been captured at the current sequencing depth (Supplementary Fig. S3). The bacterial richness and diversity of gut microbiota were tended to decrease after prebiotic intervention, especially after FOS treatment (P < 0.05) (Supplementary Fig. S4).
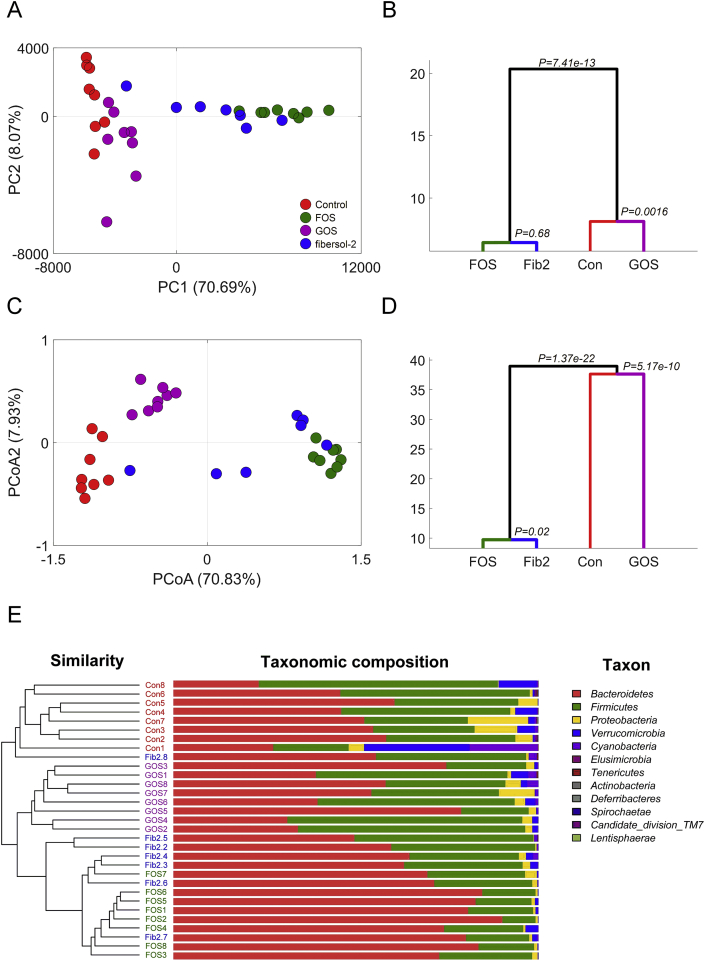
The composition of gut microbiota showed a clear shift on principal component 1 (PC1), indicated by PCA based on OTUs, which accounted for 70.69% of the total microbial variations after the two-week prebiotic intervention (Fig. 2A). The MANOVA derived from PCA scores (the first four PCs that account for 86.34% of the total variations) confirmed the statistically significant (FOS/fibersol-2 vs. control, P = 7.41e−13; GOS vs. control, P = 0.0016) difference between the control and prebiotic intervention groups (Fig. 2B). The structural shift of gut microbiota between the control and GOS groups was also distributed on PC2 (8.07%), indicating different changing patterns between the GOS and FOS/fibersol-2 groups (Fig. 2A). The PCoA and MANOVA (the first four PCs that account for 85.01% of the total variations) analyses based on Bray-Curtis distance further revealed the difference among the four groups (Fig. 2C and D). These findings were also confirmed by the outcomes of hierarchical clustering based on Bray-Curtis distance (Fig. 2E).
Fig. 2.
Structural modulation of gut microbiota by prebiotics intervention. Principal component analysis (PCA) (A), multivariate analysis of variance (MANOVA) of PCA (B), principal coordinates analysis (PCoA) (C), MANOVA analysis of PCoA (D), and hierarchical clustering (E) of taxonomic composition in control, fructooligosaccharide (FOS), galactooligosaccharide (GOS), and fibersol-2 groups.
3.3. Key phylotypes responding to prebiotic intervention
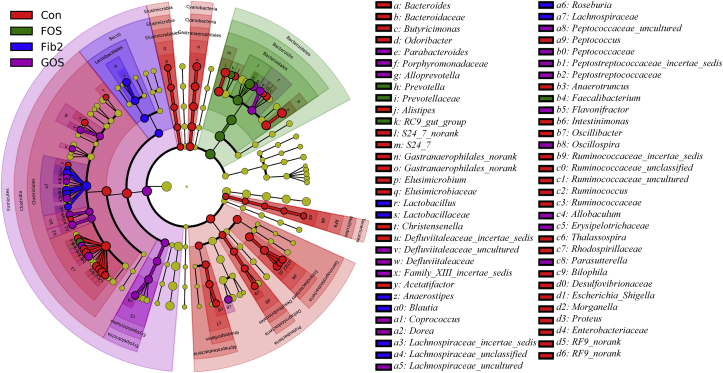
The linear discriminant analysis (LDA) effect size (LEfSe) analysis was performed on the basis of RDP taxonomic data to identify the distinguishing taxa in microbial communities that responded to prebiotic intervention. As shown in Fig. 3, the distinguishing taxa of fecal microbiota between the control and prebiotic intervention groups varied significantly at both phylum and genus levels. The most differentially abundant bacterial phyla of fecal microbiota in the control group were Cyanobacteria, Elusimicrobia, Proteobacteria, and Tenericutes, whereas Bacteroidetes and Firmicutes were dominant in the fecal microbiota in the FOS and GOS groups, respectively. At the genus level, Prevotella was increased dramatically in the gut of the SD rats after FOS treatment. The histogram of the LDA scores (Supplementary Fig. S5) illustrated a clear difference in the composition of 89 differentially abundant microbial clades among the control and the three prebiotic treatment groups (logarithmic LDA score > 3).
Fig. 3.
Taxonomic representations of the fecal microbiome of the rats. The differentially abundant taxa are presented with designated colors based on linear discriminant analysis effect size. The taxa from control, fructooligosaccharide (FOS), galactooligosaccharide (GOS), and fibersol-2 groups are colored in red, green, blue, and purple, respectively. The taxa with insignificant changes between control and prebiotic treatment groups are colored in yellow. The diameter of each small circle represents the taxon abundance.
At the genus level, Prevotella (P = 0.000; false discovery rate, FDR = 0.000) was dramatically and significantly increased by prebiotic treatments, from 4.178 ± 3.593% (control group) to 69.962 ± 8.684% (FOS group), 12.173 ± 4.153% (GOS group), and 44.583 ± 16.482% (fibersol-2 group), respectively (Table 2). Meanwhile, we found that multiple key phylotypes were suppressed by prebiotic intervention. For example, the abundances of Bacteroides (P = 0.013; FDR = 0.065), Escherichia/Shigella (P = 0.024; FDR = 0.109), Oscillibacter (P = 0.002; FDR = 0.020), Proteus (P = 0.004; FDR = 0.033), and S24_7 norank (P = 0.001; FDR = 0.013) were significantly decreased (Table 2).
Table 2.
Significantly different genera a groups by taxon-based comparisons
| Genus | Control | FOS1) | GOS2) | Fibersol-2 | P | FDR |
|---|---|---|---|---|---|---|
| Acetanaerobacterium | 0.002 ± 0.005 | 0.002 ± 0.002 | 0.014 ± 0.018 | 0.002 ± 0.003 | 0.030 | 0.117 |
| Alistipes | 1.077 ± 0.881a | 0.243 ± 0.092b | 1.067 ± 0.413a | 0.503 ± 0.371ab | 0.007 | 0.044 |
| Allobaculum | 0.464 ± 0.604a | 0.081 ± 0.164a | 1.496 ± 1.196b | 0.318 ± 0.279a | 0.002 | 0.020 |
| Alloprevotella | 6.428 ± 6.951ab | 1.828 ± 0.81a | 13.992 ± 11.235b | 3.728 ± 1.885a | 0.008 | 0.044 |
| Anaerostipes | 0±0a | 0.022 ± 0.025ab | 0.024 ± 0.025ab | 0.035 ± 0.032b | 0.049 | 0.140 |
| Anaerotruncus | 0.446 ± 0.365a | 0.112 ± 0.081b | 0.292 ± 0.138ab | 0.313 ± 0.185ab | 0.044 | 0.129 |
| Bacteroides | 15.545 ± 9.961a | 1.81 ± 1.055b | 10.47 ± 8.753ab | 5.733 ± 9.231ab | 0.013 | 0.065 |
| Blautia | 3.666 ± 3.219ab | 1.858 ± 1.328a | 4.172 ± 5.266ab | 8.022 ± 5.045b | 0.044 | 0.129 |
| Butyricimonas | 1.153 ± 0.561a | 0.148 ± 0.081b | 0.872 ± 0.44ac | 0.382 ± 0.248bc | 0.000 | 0.000 |
| Defluviitaleaceae incertae sedis | 0.213 ± 0.171a | 0±0b | 0.038 ± 0.055b | 0.033 ± 0.055b | 0.001 | 0.013 |
| Defluviitaleaceae uncultured | 0.119 ± 0.134a | 0.063 ± 0.037a | 0.497 ± 0.313b | 0.133 ± 0.099a | 0.000 | 0.000 |
| Dorea | 0.019 ± 0.01ab | 0.001 ± 0.002a | 0.03 ± 0.036b | 0.002 ± 0.003a | 0.013 | 0.065 |
| Elusimicrobium | 0.196 ± 0.225a | 0.006 ± 0.011b | 0.021 ± 0.036b | 0.003 ± 0.005b | 0.006 | 0.040 |
| Escherichia/Shigella | 2.652 ± 3.398a | 0.017 ± 0.018b | 0.553 ± 1.124ab | 0.046 ± 0.055ab | 0.024 | 0.109 |
| Faecalibacterium | 0±0a | 1.306 ± 0.921b | 0.042 ± 0.115a | 0.35 ± 0.859a | 0.001 | 0.013 |
| Family XIII incertae sedis | 0.101 ± 0.074ab | 0.037 ± 0.027ab | 0.111 ± 0.065a | 0.025 ± 0.025b | 0.005 | 0.036 |
| Holdemania | 0.008 ± 0.01a | 0.011 ± 0.011ab | 0.031 ± 0.024b | 0.014 ± 0.014ab | 0.033 | 0.117 |
| Intestinimonas | 1.808 ± 0.754a | 0.126 ± 0.078b | 1.066 ± 1.102ab | 0.634 ± 0.489b | 0.001 | 0.013 |
| Lachnospiraceae incertae sedis | 0.309 ± 0.187a | 0.578 ± 0.531ab | 1.673 ± 1.19b | 1.734 ± 1.236b | 0.005 | 0.036 |
| Lachnospiraceae uncultured | 0.888 ± 0.33ab | 0.308 ± 0.259a | 1.988 ± 1.723b | 1.563 ± 1.47ab | 0.034 | 0.117 |
| Lactobacillus | 0.455 ± 0.38a | 1.145 ± 0.584ab | 0.755 ± 0.409ab | 1.726 ± 1.475b | 0.031 | 0.117 |
| Morganella | 0.127 ± 0.157a | 0.001 ± 0.002b | 0.007 ± 0.012b | 0.001 ± 0.002b | 0.008 | 0.044 |
| Odoribacter | 0.077 ± 0.094a | 0±0b | 0.016 ± 0.044ab | 0.013 ± 0.025ab | 0.037 | 0.123 |
| Oscillibacter | 6.59 ± 5.31a | 0.031 ± 0.031b | 1.621 ± 3.197b | 1.032 ± 1.094b | 0.002 | 0.020 |
| Parabacteroides | 1.002 ± 0.901ab | 0.41 ± 0.361a | 2.395 ± 2.104b | 1.269 ± 0.734ab | 0.025 | 0.109 |
| Parasutterella | 0.326 ± 0.383a | 0.749 ± 0.788ab | 1.168 ± 0.679b | 0.505 ± 0.319ab | 0.044 | 0.129 |
| Peptostreptococcaceae incertae sedis | 0.117 ± 0.12a | 0.007 ± 0.015a | 0.879 ± 0.465b | 0.417 ± 0.673ab | 0.001 | 0.013 |
| Phascolarctobacterium | 4.765 ± 2.686 | 5.516 ± 2.488 | 10.581 ± 6.589 | 5.323 ± 3.359 | 0.033 | 0.117 |
| Prevotella | 4.178 ± 3.593a | 69.962 ± 8.684b | 12.173 ± 4.153a | 44.583 ± 16.482c | 0.000 | 0.000 |
| Proteus | 0.566 ± 0.657a | 0.001 ± 0.003b | 0.017 ± 0.024b | 0.007 ± 0.01b | 0.004 | 0.033 |
| Ruminococcaceae incertae sedis | 1.832 ± 0.882a | 0.315 ± 0.102b | 0.75 ± 0.657ab | 1.573 ± 1.251a | 0.003 | 0.027 |
| Ruminococcaceae unclassified | 0.93 ± 0.856a | 0.172 ± 0.096b | 0.386 ± 0.311ab | 0.505 ± 0.401ab | 0.038 | 0.123 |
| Ruminococcaceae uncultured | 11.753 ± 9.126a | 3.275 ± 2.887b | 8.441 ± 5.499ab | 4.445 ± 3.359ab | 0.030 | 0.117 |
| S24_7 norank | 12.453 ± 9.008a | 2.281 ± 1.069b | 4.057 ± 1.794b | 3.237 ± 1.813b | 0.001 | 0.013 |
| Thalassospira | 0.807 ± 1.032a | 0.037 ± 0.035b | 0.115 ± 0.098ab | 0.058 ± 0.077ab | 0.020 | 0.095 |
Values (mean ± SE) marked by different letters in the same row indicate a significant difference (P < 0.05).
FOS, fructooligosaccharide.
GOS, galactooligosaccharide.
3.4. Metabolic function of gut microbiota for ginsenoside hydrolysis
Metagenome analysis was performed based on the representative samples randomly selected from each group (N = 12, three samples per group) to identify the metabolic potential of gut microbiota responding to prebiotic treatment. A total of 67,218.75 Mbp raw data with an average of 5,601.56 ± 441.77 Mbp data per sample were obtained. After the quality control, the total and average clean data were 67,108.54 and 5,592.38 Mbp, respectively (Supplementary Table S3). On average, 37.34 ± 2.95 M reads per sample (Supplementary Table S4) were used for de novo assembly (Supplementary Table S5) and gene prediction (Supplementary Table S6), which constructed 545,353 non-redundant microbial genes. Similarly to the 16S rRNA results, hierarchical clustering based on Bray-Curtis distance and non-metric multidimensional scaling (NMDS) analysis of the relative species abundance (Supplementary Fig. S6) from the metagenomic data also confirmed the structural difference in gut microbiota induced by prebiotic intervention.
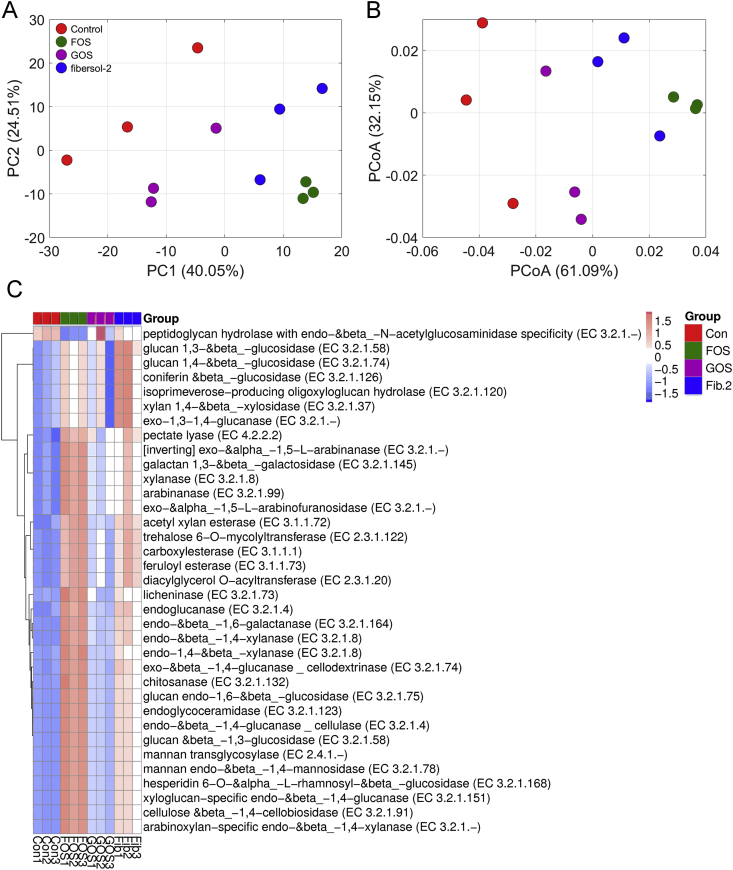
The administration of non-digestible carbohydrates potentially enhanced the fermentation metabolism of gut microbiota and contributed to ginsenoside hydrolysis. From a total number of 545,353 gene catalogues, 20,347 genes encoding CAZymes were detected, accounting for 3.73% of the total assembled gene catalogues (Supplementary Table S7). These CAZyme-encoding genes were distributed in modules at six main categories, namely, 12,226 glycoside hydrolases; 5,291 glycosyl transferases; 2,442 carbohydrate-binding modules; 1,160 carbohydrate esterases; 369 polysaccharide lyases; and 25 auxiliary activities (Supplementary Fig. S7).
At the category level of CAZymes, their compositions were different among the control and prebiotic intervention groups, revealing an overall shift of carbohydrate metabolism (Supplementary Fig. S8). Through LEfSe analysis, 48 CAZyme families responding to prebiotic intervention were statistically (P < 0.05) identified (Supplementary Figs S9, S10). Metastats analysis showed that the prebiotic intervention group contained 35 CAZymes with a higher abundance than that in the control group (Fig. 4). For example, glucan 1,3-β-glucosidase (EC 3.2.1.58), coniferin β-glucosidase (EC 3.2.1.126), xylan 1,4-β-xylosidase (EC 3.2.1.37), exo-1,3-1,4-glucanase (EC 3.2.1.-), and pectate lyase (EC 4.2.2.2) were significantly enriched by prebiotic intervention.
Fig. 4.
Metabolic function of gut microbiota for ginsenoside hydrolysis. Principal component analysis (PCA) (A), principal coordinates analysis (PCoA) (B) score plot, and relative abundance of functional annotation on CAZy database (C).
4. Discussion
Accumulating evidences indicated that gut microbiota play pivotal roles in human health and disease. However, their connection with an individual's response to specific drugs had not been investigated until the past decade [24]. Recently, the term ‘pharmacomicrobiomics’ has been proposed to describe the effect of microbiome variations on drug disposition, which is crucial for the novel establishment of personalized medicine [25,26]. Multiple drugs exhibit prominently varied efficacy and toxicity in individuals, which compromises the treatment on patients and causes additional financial burden [27]. More than 60% of drug responses are related to individual-specific gut microbiota, through microbial biotransformation or modulating host enzymes for drug metabolism [28,29]. As an emerging research field, pharmacomicrobiomics focuses on the interplay between gut microbiota variation and drug response/disposition [26]. Therefore, gut microbiota become an attractive target for enhancing drug efficacy and safety due to the plasticity of their microbial composition and metabolic functions.
Ginseng is frequently used as a crude drug that is administered orally for the treatment of several diseases. To improve its treatment efficacy, we targeted to examine the effectiveness of the active components of ginseng, namely ginsenosides, which are responsible for its pharmacological activities [30]. Rossi et al [31] summarized the potential effect of consuming probiotic on the bioactivity of two glycoconjugates, isoflavones and lignans. However, the correlation between probiotic consumption and isoflavone or lignan metabolite at the urinary and/or plasma levels has not been established [31]. The colonization of these probiotic strains was speculated to inhibit the growth and metabolic activities of microbial species involved in lignan transformation [32]. However, the presence of prebiotic GOS could alleviate such inhibitory effect by modulating the relative abundances of diverse microbes [32]. Additional studies also suggested that transient colonization of different probiotics can alter the metabolic activities of gut microbiota, but it is difficult to predict their specific mechanism [[33], [34], [35]]. Therefore, three prebiotic ingredients, including FOS, GOS, and fibersol-2, were applied in the current study.
Based on the pharmacokinetic results, we speculate that secondary ginsenosides, especially ginsenoside CK, are the major active metabolites generated from the hydrolysis of primary ginsenosides by gut microflora, and fiber intervention probably enhances its bioconversion and bioavailability. As shown in Table 1, we also noticed that the Tmax of ginsenoside CK in the control group was much longer than those in the three fiber groups. Meanwhile, the Cmax of ginsenoside CK was increased by fiber treatment to a certain extent but without statistical significance. We speculated that facilitated by intestinal flora, ginsenoside Rb1 was gradually and persistently metabolized to ginsenoside CK with a higher Cmax.
Prebiotics involve in the progressive nutritional selection by specific bacterial strains, and they were recognized to effectively improve the formation of secondary ginsenosides, especially ginsenoside CK, after the oral administration of ginsenoside Rb1. In the current study, Prevotella, which can hydrolyze ginsenoside Rb1 into CK, was significantly elevated in the prebiotic groups (P < 0.05). The dramatic increase of Prevotella, especially in the FOS and fibersol-2 groups, was reasonable since Prevotella sp. are among the most abundant culturable microbes in the rumen and hind gut of cattle and sheep, where they contribute to break down carbohydrates [36]. Hasegawa et al [37] isolated Prevotella oris strains from fecal specimens of human subjects and identified their metabolic potential to hydrolyze ginsenoside Rb1 into CK. After colimycin treatment (20 mg/kg/day), the ginsenoside Rb1-hydrolysing activity by intestinal microflora was decreased from 22.1 ± 1.2% to 4.7 ± 2.7%, while it was restored to 30.7 ± 3.7% by the inoculation of P. oris isolates [37]. Therefore, we speculate that selective enrichment of Prevotella partially contributes to the metabolism of ginsenoside Rb1 in the FOS and fibersol-2 intervention groups, in the current study. Probiotic lactobacilli and bifidobacteria, which were reported to release aglycones, can produce several glycosyl-hydrolases and thus utilize indigestible oligosaccharides and polysaccharides as their carbon sources [31]. However, no Bifidobacterium in the control and prebiotic treatment groups was detected, while Lactobacillus was slightly increased by the prebiotic intervention (Table 2). Niu et al [30] determined the changes of intestinal bacterial abundances in A/J mice after ginsenoside Rb1 administration; they found that Lactobacillaceae was reduced to be less than 1% and Bifidobacterium was even non-detectable. Kim et al [13] compared the fecal bacterial genera between subjects with different ginsenoside Rb1 metabolizing activities but found no significant differences in the abundances of Prevotella and Bifidobacterium (P > 0.05). The inconsistency of the bacterial responses to prebiotic treatment is partially attributed to the differences between using pure bacterial cultures and fecal specimens in these in vitro studies [38]. It could also be explained by the variation of gut microbes that involve in the intestinal metabolism of ginsenoside Rb1, which determines the responses of gut microbiota to nutritional intervention or xenobiotic metabolism at the biologically functional level, instead of the taxonomic level. In addition, P. oris and Eubacterium sp. A-44 were also demonstrated to transform ginsenoside Rb1 into CK [37,39,40]. Nicholson et al [41] proposed a ‘Pachinko model’ to explain the idiosyncratic reactions and variations in drug interactions based on probabilistic interactions between the host genome and the indigenous microbiome.
Although these observational or interventional studies reported inconsistent changes of gut microbial species, the relative metabolic activities of gut microbiota probably played essential roles in ginsenoside hydrolysis [13,42,43]. Mammalian cells are not capably of hydrolyzing ginsenoside, therefore, biotransformation is required for activating these ginsenosides in mammalian systems [44,45]. In the colonic ecosystem, multiple gut bacterial strains, especially probiotic lactobacilli and bifidobacteria, have evolved to produce several glycosyl-hydrolases, including β-glucosidases, and contribute to release aglycones from glycol-conjugated ginsenosides [31]. Kim et al [11] compared the fecal microbial activities for metabolizing p-nitrophenyl-β-D-glucopyranoside, p-nitrophenyl-β-D-glucuronide, p-nitrophenyl-β-D-galactopyranoside, p-nitrophenyl-α-L-rhamnopyranoside, and ginsenoside Rb1 in ten Koreans. They further found that prebiotic intake could promote glycosidase activity and ginsenoside CK formation in rat intestinal contents based on in vitro culturing [46]. In the current study, prebiotic intervention enriched 35 CAZymes, such as glucan 1,3- & β-glucosidase (EC 3.2.1.58), coniferin & β-glucosidase (EC 3.2.1.126), xylan 1,4- & β-xylosidase (EC 3.2.1.37), exo-1,3-1,4-glucanase (EC 3.2.1.-), and pectate lyase (EC 4.2.2.2) (Fig. 4). The profiling of metabolic capability based on metagenomic analysis would further present the entire spectrum of the metabolic potential of intestinal microbiome.
We also observed the differences in ginsenoside Rb1 metabolism and gut microbiota composition among these three prebiotic intervention groups. The dose-effect relationship was not well-addressed since a uniform dose of these prebiotics was adopted in the study [38]. Moreover, initial bacterial species and abundances can intensively influence the responses of gut microbiota towards dietary intervention, which implies the necessity to homogenize gut microbiota composition at a uniform baseline [47]. In addition to FOS, GOS, and fibersol-2, previous study indicated that soluble prebiotic fiber NUTRIOSE® [11,46,48], ginseng polysaccharides [42], and traditional medicine Daikenchuto (Da-Jian-Zhong-Tang) [43] could also shape gut microbiota architecture and enhance systemic exposure. These findings suggest that the overall efficacy of natural compounds and traditional medicine can be optimized by targeting the metabolic function of gut microbiota.
Declaration of competing interest
The authors declare no conflicts of interest.
Acknowledgments
This research was funded by the National Science and Technology Major Project (No. 2017ZX09301060-012; 2018ZX09201-011), the Youth Program of National Natural Science Foundation of China (No. 81503469) and the Emergency Management Project of National Natural Science Foundation of China (No. 81741060). We would like to thank Zhiying Huang, Xiuting Zhang, Pengpeng Tian and Junqiu Liu for assistance in generating data.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2020.08.001.
Contributor Information
Jiang Xu, Email: jxu@icmm.ac.cn.
Shuiming Xiao, Email: smxiao@icmm.ac.cn.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
Multimedia component 1
References
- 1.Hemmerly T.E. A ginseng farm in Lawrence County, Tennessee. Econ Bot. 1977;31:160–162. [Google Scholar]
- 2.Leung K.W., Wong S.T. Pharmacology of ginsenosides: a literature review. Chin Med. 2010;5:20. doi: 10.1186/1749-8546-5-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Akao T., Kanaoka M., Kobashi K. Appearance of compound K, a major metabolite of ginsenoside Rb1 by intestinal bacteria, in rat plasma after oral administration--measurement of compound K by enzyme immunoassay. Biol Pharm Bull. 1998;21:245–249. doi: 10.1248/bpb.21.245. [DOI] [PubMed] [Google Scholar]
- 4.Akao T., Kida H., Kanaoka M., Hattori M., Kobashi K. Intestinal bacterial hydrolysis is required for the appearance of compound K in rat plasma after oral administration of ginsenoside Rb1 from Panax ginseng. J Pharm Pharmacol. 1998;50:1155–1160. doi: 10.1111/j.2042-7158.1998.tb03327.x. [DOI] [PubMed] [Google Scholar]
- 5.Hasegawa H. Proof of the mysterious efficacy of ginseng: basic and clinical trials: metabolic activation of ginsenoside: deglycosylation by intestinal bacteria and esterification with fatty acid. J Pharmacol Sci. 2004;95:153–157. doi: 10.1254/jphs.fmj04001x4. [DOI] [PubMed] [Google Scholar]
- 6.Wang C.Z., Du G.J., Zhang Z., Wen X.D., Calway T., Zhen Z., Musch M.W., Bissonnette M., Chang E.B., Yuan C.S. Ginsenoside compound K, not Rb1, possesses potential chemopreventive activities in human colorectal cancer. Int J Oncol. 2012;40:1970–1976. doi: 10.3892/ijo.2012.1399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kim D.H. Chemical diversity of Panax ginseng, Panax quinquifolium, and Panax notoginseng. J Gins Res. 2012;36:1–15. doi: 10.5142/jgr.2012.36.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yan X., Fan Y., Wei W., Wang P., Liu Q., Wei Y., Zhang L., Zhao G., Yue J., Zhou Z. Production of bioactive ginsenoside compound K in metabolically engineered yeast. Cell Res. 2014;24:770–773. doi: 10.1038/cr.2014.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee J., Lee E., Kim D., Lee J., Yoo J., Koh B. Studies on absorption, distribution and metabolism of ginseng in humans after oral administration. J Ethnopharmacol. 2009;122:143–148. doi: 10.1016/j.jep.2008.12.012. [DOI] [PubMed] [Google Scholar]
- 10.Marteau P., Pochart P., Flourie B., Pellier P., Santos L., Desjeux J.F., Rambaud J.C. Effect of chronic ingestion of a fermented dairy product containing Lactobacillus acidophilus and Bifidobacterium bifidum on metabolic activities of the colonic flora in humans. Am J Clin Nutr. 1990;52:685–688. doi: 10.1093/ajcn/52.4.685. [DOI] [PubMed] [Google Scholar]
- 11.Kim K.A., Yoo H.H., Gu W., Yu D.H., Jin M.J., Choi H.L., Yuan K., Guerin-Deremaux L., Kim D.H. Effect of a soluble prebiotic fiber, NUTRIOSE, on the absorption of ginsenoside Rd in rats orally administered ginseng. J Gins Res. 2014;38:203–207. doi: 10.1016/j.jgr.2014.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Goossens D., Jonkers D., Russel M., Stobberingh E., Bogaard A.V.D., Stockbrügger R. The effect of Lactobacillus plantarum 299v on the bacterial composition and metabolic activity in faeces of healthy volunteers: a placebo-controlled study on the onset and duration of effects. Aliment Pharmacol Ther. 2003;18:495–505. doi: 10.1046/j.1365-2036.2003.01708.x. [DOI] [PubMed] [Google Scholar]
- 13.Kim K.A., Jung I.H., Park S.H., Ahn Y.T., Huh C.S., Kim D.H. Comparative analysis of the gut microbiota in people with different levels of ginsenoside Rb1 degradation to compound K. PLoS One. 2013;8 doi: 10.1371/journal.pone.0062409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Caporaso J.G., Lauber C.L., Walters W.A., Berg-Lyons D., Huntley J., Fierer N., Owens S.M., Betley J., Fraser L., Bauer M. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6:1621–1624. doi: 10.1038/ismej.2012.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen Y., Xiao S., Gong Z., Zhu X., Yang Q., Li Y., Gao S., Dong Y., Shi Z., Wang Y. Wuji Wan formula ameliorates diarrhea and disordered colonic motility in post-inflammation irritable bowel syndrome rats by modulating the gut microbiota. Front Microbiol. 2017;8:2307. doi: 10.3389/fmicb.2017.02307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liu Z., Sun Y., Zhang Y., Feng W., Lai Z., Fa K., Qin S. Metagenomic and 13C tracing evidence for autotrophic atmospheric carbon absorption in a semiarid desert. Soil Biol Biochem. 2018;125:156–166. [Google Scholar]
- 17.Luo R., Liu B., Xie Y., Li Z., Huang W., Yuan J., He G., Chen Y., Pan Q., Liu Y. SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience. 2012;1:18. doi: 10.1186/2047-217X-1-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Li J., Jia H., Cai X., Zhong H., Feng Q., Sunagawa S., Arumugam M., Kultima J.R., Prifti E., Nielsen T. An integrated catalog of reference genes in the human gut microbiome. Nat Biotechnol. 2014;32:834–841. doi: 10.1038/nbt.2942. [DOI] [PubMed] [Google Scholar]
- 19.Fu L., Niu B., Zhu Z., Wu S., Li W. Cd-Hit: Accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28:3150–3152. doi: 10.1093/bioinformatics/bts565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li R., Yu C., Li Y., Lam T.W., Yiu S.M., Kristiansen K., Wang J. SOAP2: an improved ultrafast tool for short read alignment. Bioinformatics. 2009;25:1966–1967. doi: 10.1093/bioinformatics/btp336. [DOI] [PubMed] [Google Scholar]
- 21.Huson D.H., Mitra S., Ruscheweyh H.J., Weber N., Schuster S.C. Integrative analysis of environmental sequences using MEGAN4. Genome Res. 2011;21:1552–1560. doi: 10.1101/gr.120618.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kanehisa M., Goto S., Kawashima S., Okuno Y., Hattori M. The KEGG resource for deciphering the genome. Nucleic Acids Res. 2004;32:D277. doi: 10.1093/nar/gkh063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Park B.H., Karpinets T.V., Syed M.H., Leuze M.R., Uberbacher E.C. CAZymes Analysis Toolkit (CAT): web service for searching and analyzing carbohydrate-active enzymes in a newly sequenced organism using CAZy database. Glycobiology. 2010;20:1574. doi: 10.1093/glycob/cwq106. [DOI] [PubMed] [Google Scholar]
- 24.Wilson I.D., Nicholson J.K. The role of gut microbiota in drug response. Curr Pharm Des. 2009;15:1519–1523. doi: 10.2174/138161209788168173. [DOI] [PubMed] [Google Scholar]
- 25.Saad R., Rizkallah M.R., Aziz R.K. Gut Pharmacomicrobiomics: the tip of an iceberg of complex interactions between drugs and gut-associated microbes. Gut Pathog. 2012;4:16. doi: 10.1186/1757-4749-4-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Doestzada M., Vila A.V., Zhernakova A., Koonen D.P.Y., Weersma R.K., Touw D.J., Kuipers F., Wijmenga C., Fu J. Pharmacomicrobiomics: a novel route towards personalized medicine? Protein Cell. 2018;9:432–445. doi: 10.1007/s13238-018-0547-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Classen D.C., Pestotnik S.L., Evans R.S., Lloyd J.F., Burke J.P. Adverse drug events in hospitalized patients: excess length of stay, extra costs, and attributable mortality. J Am Med Assoc. 1997;277:301–306. [PubMed] [Google Scholar]
- 28.Haiser H.J., Turnbaugh P.J. Is it time for a metagenomic basis of therapeutics? Science. 2012;336:1253–1255. doi: 10.1126/science.1224396. [DOI] [PubMed] [Google Scholar]
- 29.Lhoste E.F., Ouriet V., Bruel S., Flinois J.P., Brézillon C., Magdalou J., Caroline C., Lionelle N.B. The human colonic microflora influences the alterations of xenobiotic-metabolizing enzymes by catechins in male F344 rats. Food Chem Toxicol. 2003;41:695–702. doi: 10.1016/s0278-6915(03)00010-3. [DOI] [PubMed] [Google Scholar]
- 30.Niu T., Smith D.L., Yang Z., Gao S., Yin T., Jiang Z.H., You M., Gibbs R.A., Petrosino J.F., Hu M. Bioactivity and bioavailability of ginsenosides are dependent on the glycosidase activities of the A/J mouse intestinal microbiome defined by pyrosequencing. Pharm Res. 2013;30:836–846. doi: 10.1007/s11095-012-0925-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rossi M., Amaretti A., Leonardi A., Raimondi S., Simone M., Quartieri A. Potential impact of probiotic consumption on the bioactivity of dietary phytochemicals. J Agric Food Chem. 2013;61:9551–9558. doi: 10.1021/jf402722m. [DOI] [PubMed] [Google Scholar]
- 32.Kekkonen R.A., Holma R., Hatakka K., Suomalainen T., Poussa T., Adlercreutz H., Korpela R. A probiotic mixture including galactooligosaccharides decreases fecal β-glucosidase activity but does not affect serum enterolactone concentration in men during a two-week intervention. J Nutr. 2011;141:870–876. doi: 10.3945/jn.110.137703. [DOI] [PubMed] [Google Scholar]
- 33.Larkin T.A., Price W.E., Astheimer L.B. Increased probiotic yogurt or resistant starch intake does not affect isoflavone bioavailability in subjects consuming a high soy diet. Nutrition. 2007;23:709–718. doi: 10.1016/j.nut.2007.06.010. [DOI] [PubMed] [Google Scholar]
- 34.Nettleton J.A., Greany K.A., Thomas W., Wangen K.E., Adlercreutz H., Kurzer M.S. The effect of soy consumption on the urinary 2:16-hydroxyestrone ratio in postmenopausal women depends on equol production status but is not influenced by probiotic consumption. J Nutr. 2005;135:603–608. doi: 10.1093/jn/135.3.603. [DOI] [PubMed] [Google Scholar]
- 35.Cohen L.A., Crespin J.S., Wolper C., Zang E.A., Pittman B., Zhao Z., Holt P.R. Soy isoflavone intake and estrogen excretion patterns in young women: effect of probiotic administration. In Vivo. 2007;21:507–512. [PubMed] [Google Scholar]
- 36.Avgustin G., Ramsak A., Peterka M. Systematics and evolution of ruminal species of the genus Prevotella. Folia Microbiol. 2001;46:40–44. doi: 10.1007/BF02825882. [DOI] [PubMed] [Google Scholar]
- 37.Hasegawa H., Sung J.H., Benno Y. Role of human intestinal Prevotella oris in hydrolyzing ginseng saponins. Planta Med. 1997;63:436–440. doi: 10.1055/s-2006-957729. [DOI] [PubMed] [Google Scholar]
- 38.Gibson G.R., Probert H.M., Loo J.V., Rastall R.A., Roberfroid M.B. Dietary modulation of the human colonic microbiota: updating the concept of prebiotics. Nutr Res Rev. 2004;17:259–275. doi: 10.1079/NRR200479. [DOI] [PubMed] [Google Scholar]
- 39.Xu J., Chen H.B., Li S.L. Understanding the molecular mechanisms of the interplay between herbal medicines and gut microbiota. Med Res Rev. 2017;37:1140–1185. doi: 10.1002/med.21431. [DOI] [PubMed] [Google Scholar]
- 40.Yang L., Akao T., Kobashi K. Purification and characterization of a geniposide-hydrolyzing β-glucosidase from Euhacterium sp. A-44, a strict anaerobe from human feces. Biol Pharm Bull. 1995;18:1175–1178. doi: 10.1248/bpb.18.1175. [DOI] [PubMed] [Google Scholar]
- 41.Nicholson J.K., Holmes E., Wilson I.D. Gut microorganisms, mammalian metabolism and personalized health care. Nat Rev Microbiol. 2005;3:431–438. doi: 10.1038/nrmicro1152. [DOI] [PubMed] [Google Scholar]
- 42.Zhou S.S., Xu J., Zhu H., Wu J., Xu J.D., Yan R., Li X.Y., Liu H.H., Duan S.M., Wang Z. Gut microbiota-involved mechanisms in enhancing systemic exposure of ginsenosides by coexisting polysaccharides in ginseng decoction. Sci Rep. 2016;6:22474. doi: 10.1038/srep22474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hasebe T., Ueno N., Musch M.W., Nadimpalli A., Kaneko A., Kaifuchi N., Watanabe J., Yamamoto M., Kono T., Inaba Y. Daikenchuto (TU-100) shapes gut microbiota architecture and increases the production of ginsenoside metabolite compound K. Pharma Res Per. 2016;4 doi: 10.1002/prp2.215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Henrissat B., Bairoch A. New families in the classification of glycosyl hydrolases based on amino acid sequence similarities. Biochem J. 1993;293:781–788. doi: 10.1042/bj2930781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Park C.S., Yoo M.H., Noh K.H., Oh D.K. Biotransformation of ginsenosides by hydrolyzing the sugar moieties of ginsenosides using microbial glycosidases. Appl Microbiol Biotechnol. 2010;87:9–19. doi: 10.1007/s00253-010-2567-6. [DOI] [PubMed] [Google Scholar]
- 46.Kim K.A., Yoo H.H., Gu W., Yu D.H., Jin M.J., Choi H.L., Kim D.H., Yuan K., Guerin-Deremaux L. A prebiotic fiber increases the formation and subsequent absorption of compound K following oral administration of ginseng in rats. J Gins Res. 2015;39:183–187. doi: 10.1016/j.jgr.2014.11.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Roberfroid M.B., Loo J.A., Van, Gibson G.R. The bifidogenic nature of chicory inulin and its hydrolysis products. J Nutr. 1998;128:11–19. doi: 10.1093/jn/128.1.11. [DOI] [PubMed] [Google Scholar]
- 48.Lefranc-Millot C., Guérin-Deremaux L., Wils D., Neut C., Miller L., Saniez-Degrave M. Impact of a resistant dextrin on intestinal ecology: how altering the digestive ecosystem with NUTRIOSE®, a soluble fibre with prebiotic properties, may be beneficial for health. J Int Med Res. 2012;40:211–224. doi: 10.1177/147323001204000122. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Multimedia component 1