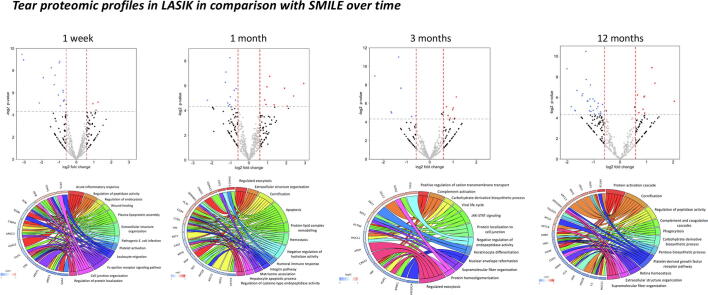
Graphical abstract
Keywords: Tear, Proteomics, Neuromediators, Small incision lenticule extraction, Laser-assisted in-situ keratomileusis, Dry eye
Abstract
Introduction
The tear proteomics and neuromediators are associated with clinical dry eye parameters following refractive surgery.
Purpose
To investigate and compare the tear proteomic and neuromediator profiles following small incision lenticule extraction (SMILE) versus laser-assisted in-situ keratomileusis (LASIK).
Methods
In this randomized controlled trial with paired-eye design, 70 patients were randomized to receive SMILE in one eye and LASIK in the other eye. Tear samples were collected preoperatively, and 1 week, 1, 3, 6 and 12 months postoperatively, and were examined for protein concentration changes using sequential window acquisition of all theoretical fragment ion mass spectrometry (SWATH-MS). The data were analyzed with DAVID Bioinformatics Resources for enriched gene ontology terms and over-represented pathways. Tear neuromediators levels were correlated with clinical parameters.
Results
Post-SMILE eyes had significantly better Oxford staining scores and tear break-up time (TBUT) than post-LASIK eyes at 1 and 3 months, respectively. Tear substance P and nerve growth factor levels were significantly higher in the LASIK group for 3 months and 1 year, respectively. SMILE and LASIK shared some similar biological responses postoperatively, but there was significant up-regulation in leukocyte migration and wound healing at 1 week, humoral immune response and apoptosis at 1 month, negative regulation of endopeptidase activity at 3 to 6 months, and extracellular structure organization at 1 year in the post-LASIK eyes. Tear mucin-like protein 1 and substance P levels were significantly correlated with TBUT (r = -0.47, r = -0.49, respectively).
Conclusion
Significant differences in the tear neuromediators and proteomics were observed between SMILE and LASIK, even though clinical dry eye signs have subsided and became comparable between 2 procedures.
Introduction
Small incision lenticule extraction (SMILE) has become a viable alternative to laser-assisted in situ keratomileusis (LASIK) for the refractive correction of myopia and myopic astigmatism [1]. Due to the small incision in SMILE, as opposed to a circumferential flap in LASIK, the impact on the ocular surface is expected to be less. A systematic review reported that the tear breakup time (TBUT) and ocular surface disease index (OSDI) scores were significantly better in SMILE than LASIK 6 months postoperatively [2]. A non-randomized comparative study showed that at 6 months postoperatively, 80% of patients in the SMILE group did not use any topical lubricants versus 57% in the LASIK group, with 20% of the LASIK group needing daily and frequent use of tear substitutes or even gels versus none of the patients in the SMILE group [3].
The risk factors for dry eye disease following refractive surgery are multifactorial. Pathophysiological mechanisms include corneal denervation, goblet cell loss, meibomian gland dysfunction, and ocular surface inflammation [4]. Ocular surface damage triggers an inflammatory response and subsequent production of cytokines such as tumor necrosis factor, interleukin (IL)-1, growth factors, free radicals, adhesion molecules and proteolytic enzymes [5]. Zhong et al. evaluated the early changes of tear mediators after SMILE versus femtosecond lenticule extraction (FLEx) and found that the SMILE group had lower expression of tear tumor growth factor (TGF)-β1 and IL-1α at 1 month postoperatively [6]. The same authors also investigated the tear mediators in SMILE versus LASIK, and lower IL-6 tear level was noted in SMILE at 1 month [7]. These studies, however, are limited by their short follow-up time (up to postoperative 3 months), small patient number, and data from non-paired eyes that would introduce inter-subject variability.
Tear neurotransmitters and neuropeptides that mediate neurogenic inflammation also play an important role in postoperative dry eye diseases. Surgical incisions and laser exposure activate stromal keratocytes to undergo a wound healing response and trigger neurogenic inflammation [8]. The key mediators of neurogenic inflammation include tachykinins, calcitonin gene-related peptide (CGRP), substance P, and proinflammatory cytokines especially IL-6. CGRP and substance P act directly on vascular endothelial and smooth muscle cells, and induce vasodilation as well as increased vascular permeability, resulting in the recruitment and attraction of immune cells to the corneal wound site [9]. In patients with chronic idiopathic dry eyes, the tear CGRP level was decreased, and this was associated with increased corneal staining [10]. Nerve growth factor (NGF) is another neuropeptide that can accelerate corneal epithelial healing and tract repair, as well as induce keratocyte migration and facilitate corneal nerve regeneration. Increased tear NGF levels have been observed in LASIK patients at 6 months postoperatively [11].
To decipher the changes in protein expression in a biological process, an approach using advanced quantitative proteomics, such as isobaric tags for relative and absolute quantitation (iTRAQ) or sequential window acquisition of all theoretical fragment mass spectra (SWATH-MS), has been used recently [12]. The advantages of SWATH-MS include its higher reproducibility especially when dealing with a large cohort, the ability to pick up signals from low abundant peptides or proteins, and the capacity to allow for comparisons across multiple time points, especially when the sample quantity is limited, such as tear samples [13].
In the present study, we investigated the longitudinal changes of tear proteome and neuromediators over 1-year, using the SWATH technique, in patients who have undergone SMILE and LASIK. A randomized controlled trial (RCT) with paired-eye design was used to ensure more precise comparisons, by eliminating the inter-individual and inter-eye variation. The results of tear proteomics and neuromediators were also correlated with clinical dry eye parameters.
Methods
Patients and surgical procedures
This RCT (NCT01216475) included 70 patients randomized to undergo SMILE in one eye and femtosecond laser-assisted LASIK in the other eye at the Singapore National Eye Center between August 2012 and November 2016, with the inclusion and exclusion criteria detailed in Supplementary Table 1. The random allocation sequence was be generated by a computer with no blocks or restrictions, and implemented by concealing the number-coded surgery within sealed envelopes until just before the procedure. The right eye was always operated first, hence this was the eye to which the random allocation applied [14]. SMILE procedure was performed as previously described [1], [14]. In brief, a S-sized curved interface cone was applied, and the femtosecond laser (Visumax; Carl Zeiss, Jena, Germany) with the power set at 145 nJ, was used to create the following planes: posterior surface of the lenticule, anterior surface of the lenticule, which extended beyond the posterior lenticule diameter by 0.5 mm to form the anterior cap, and a 2.8 mm vertical circumferential incision placed at 120° [15]. The cap thickness was 120 μm, cap diameter was 7.5 mm, optical zone was 6.5 mm, and the side-cut angle was at 90°. A SMILE dissector (ASICO, Westmont, IL, USA) was used for the dissection of the anterior and posterior planes of lenticule, followed by the removal of the lenticule. For the LASIK procedure, a superiorly-hinged, 120 μm- thick flap was created using the Visumax femtosecond laser. Excimer laser ablation was then performed the WaveLight EX500 excimer laser (Alcon Laboratories, Inc, Fort Worth, TX). After ablation, the flap was carefully repositioned. All the procedures were performed under topical anesthesia by the same refractive surgeon. The postoperative regimen for SMILE and LASIK groups were identical, consisting of topical preservative-free dexamethasone and moxifloxacin, each 3 hourly for a week then 4 times a day for 2 weeks. Artificial tears (Tears Naturale Free; AlconLaboratories, Kaysersberg, France) were also prescribed, and the dosage was the same for the first 3 months for both eyes, afterwards they were applied ad libitum. Approval for the study was granted by the institutional review board of SingHealth, Singapore (2011/109/A), and the study was conducted in accordance to the Declaration of Helsinki.
Dry eye assessments
Dry eye assessments were performed preoperatively, and 1 week, and 1, 3, 6, 12 months postoperatively for all patients. The assessments were comprised of Schirmer’s I test (without anesthesia, mm/5 min), ocular surface fluorescein staining (Oxford score; 0: absent, 5: severe), and TBUT. In Schirmer’s test, after the wetted area of Schirmer’s strip was measured, the strips were stored at −80 °C until the day of analysis. For TBUT, the right eye was evaluated first followed by the left eye, by placing a single fluorescein strip over the inferior tear meniscus after shaking off a small drop of normal saline. The time between eye opening and the first appearance of a dry spot on the cornea was recorded as the TBUT [16]. Three consecutive tests were performed and the average value was taken.
Quantitative proteomics
Thirty pairs of tear samples selected randomly for proteomics analysis. Tear proteins from finely cut Schirmer’s strip were extracted by ammonium bicarbonate (50 mM, 100 μl per strip; Sigma-Aldrich, St. Louis, MO, USA) at 1300 rpm, room temperature (23 °C) for 1 h, and the protein concentration was measured. Protein samples (100 μg) were applied for SWATH-MS and a spectral library was established as previously described [17], [18]. The digested samples were reconstituted in loading buffer (0.1% formic acid, 2% acetonitrile in water), and iRT standard (Biognosys, Switzerland) was spiked. Sample analysis was performed using an Ultimate 3000 nanoLC system (Dionex, Thermo Fisher Scientific, MA, USA) coupled with AB Sciex 5600 triple TOF (AB Sciex, Framingham, MA, USA) as previously reported [18], [19].
Enzyme-linked immunosorbent assay (ELISA)
Forty patients were randomly selected for neuromediator analysis. Tear proteins from each Schirmer strip sample were eluted by submerging finely cut Schirmer’s strips into 200 µl elution buffer containing 0.55 M NaCl, 0.33% Tween-20, 0.55% bovine serum albumin and protease inhibitor, with brief sonication and agitation at 450 rpm for 17 h at 4 °C. The elutes were centrifuged and clear supernatants were collected for protein assay, using a Micro BCA Protein Assay Kit (Pierce Biotechnology, Inc. MA, USA). ELISA for CGRP, NGF and Substance P were performed per manufacturers’ instructions (CGRP ELISA kit from Phoenix Pharmaceuticals, Runcorn, UK; NGF and Substance P ELISA kits from R&D Systems, Minneapolis, USA). Each eluted tear sample was diluted 4x, 4x, and 1.5x for Substance P, CGRP and NGF, respectively, with individual assay diluent to a final volume of 50 µl per well. All samples were evaluated in duplicate per analyte per run. Optical density (OD) was read at 450 nm using a microplate reader, and OD reading at 540 nm was set as background. The concentrations were interpolated from the standard curve.
Differential protein expression and pathway analyses
SWATH proteomics data were processed with SpectronautTM (Biognosys, Schlieren, Switzerland). The relative quantitation data in terms of ratio was generated, and they included (1) post-SMILE data at different time points versus preoperative data; (2) post-LASIK data at different time points versus preoperative data; and (3) pairwise LASIK versus SMILE ratios at different time points. The differentially expressed proteins were determined with linear models for microarray data (LIMMA) algorithm [20] with fold changes (FC) > 1.50 or < 0.67, i.e. the log2 FC > 0.58 or < -0.58, and P values < 0.05. Only proteins that exhibited consistent changes among all 30 sample pairs were reported and used for pathway analyses. Proteins with statistically significant changes (P < 0.05) without the concordant changes in all 30 SMILE/LASIK pairs were excluded. Gene Ontology (GO) term analysis to identify enriched biological themes was done by inputting database to web-based DAVID Bioinformatics Resources v6.8 (NIAID, NIH, USA) [21]. Adjusted Benjamini P values were calculated by Modified Fisher Exact test with the smaller P values representing higher chance of enrichment. Bioinformatics analysis was performed using Metascape [22]. The differentially regulated proteins were also analysed using Reactome Knowledgebase [23] to explore the functional relationship and their contribution in the biological process network. Custom R scripts with R 64-bit version 3.4.0 [24] was used to generate the plots to illustrate the results of protein expression and GO analysis.
Statistical analysis
All data were expressed as mean ± standard deviation. The required sample size was calculated based on the pilot data of the clinical parameter, which is the primary outcome of the study (6-months TBUT). A sample size of 62 eyes per arm, with a power of ≥ 80% and at a 5% significance, was sufficient to detect the difference between the SMILE and LASIK groups. Considering a dropout rate of 10%, a total of 70 patients were recruited. A paired t-test was used to compare the SMILE and LASIK groups, as well as preoperative and postoperative data. The correlation analysis was performed with a Spearman (ordinal variables) or Pearson test (continuous variables). The statistical analysis was carried out using STATA (STATACorp, College Station, TX, USA), and a P < 0.05 without adjusting for multiplicity was considered as significant.
Results
Patient characteristics
The mean age at the time of surgery was 27.6 ± 5.1 years (female: male = 45:25). The preoperative spherical equivalent was −5.3 ± 1.8 D and −5.2 ± 1.7 D for the SMILE and LASIK groups, respectively (P = 0.88). All the patients completed their follow-up visits for postoperative 3 months, and there were 4 and 6 patients who lost the follow up at 6 and 12 months, respectively.
Clinical dry eye assessments
All dry eye parameters were statistically comparable between 2 groups before the surgery. After surgery, the SMILE group had significantly better TBUT than the LASIK group at 1 week (8.4 ± 3.2 versus 6.1 ± 2.7 s; P = 0.0028), 1 month (9.3 ± 2.6 versus 7.0 ± 2.3 s; P < 0.001) and 3 months (10.0 ± 2.6 versus 8.3 ± 3.2 s; P = 0.011). The TBUT was recovered to the preoperative level at 6 months in post-SMILE eyes, whereas impaired TBUT was still noted at 12 months in post-LASIK eyes (Supplementary Fig. 1A). The oxford score was significantly worse in the LASIK than SMILE group at 1 week (0.51 ± 0.28 versus 0.87 ± 0.33; P = 0.03) and 1 month (0.24 ± 0.19 versus 0.48 ± 0.11; P = 0.04), but not thereafter (Supplementary Fig. 1B). There was no significant difference between 2 groups in the Schirmer I test at all time points (Supplementary Fig. 1C). For the ocular surface staining and Schirmer’s test evaluation, a significant difference was noted at 1 week in post-SMILE eyes, and at 1 week and 1 month in post-LASIK eyes, in comparison with preoperative level.
Proteomic analysis
SWATH analysis identified and quantified an average of 541 ± 46 proteins with 95% confidence for the whole dataset.
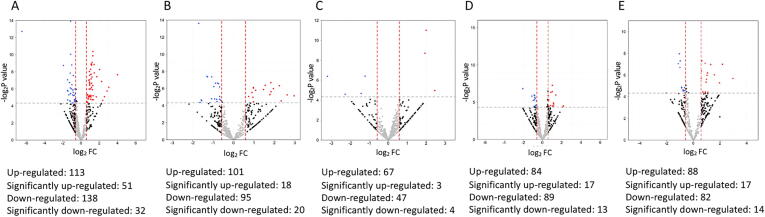
Post-SMILE tear protein changes
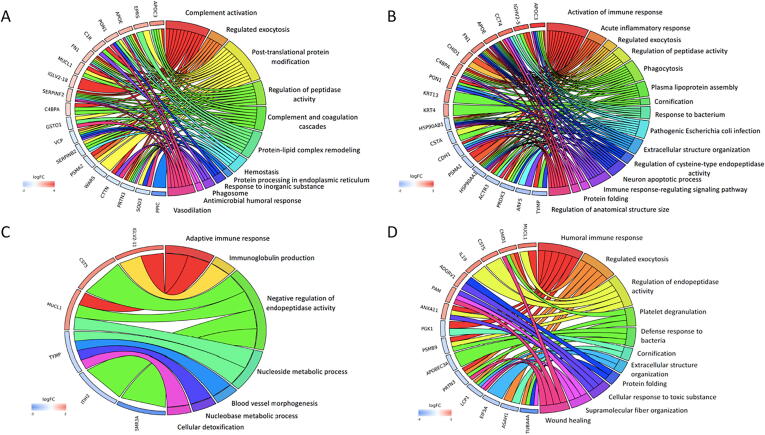
The number and fold change of differentially abundant proteins following SMILE over time was illustrated in Fig. 1. In the early postoperative period (1 week and 1 month), immunoglobulin lambda variable 2–18 (IGLV2-18) that is involved in complement activation (log2FC = 1.73; 1 week), apolipoprotein C3 (APOC3) that is involved in complement and coagulation cascades (log2FC = 3.99 at 1 week and log2FC = 2.97 at 1 month), APOE (log2FC = 2.85 at 1 week, log2FC = 2.24 at 1 month) and fibronectin 1 (FN1, log2FC = 2.26 at 1 week, log2FC = 1.95 at 1 month) that are related to acute inflammation, phagocytosis, and fibrotic responses, were significantly up-regulated (Fig. 2A, B; Table 1). At 3 months, up-regulated IGLV2-11 (log2FC = 2.46), cystatin D (CST5, log2FC = 2.00), and mucin-like 1 (MUCL1, log2FC = 1.94) were detected, and they are linked to the process of adaptive immune, immunoglobulin production and endopeptidase activities (Fig. 2C). Elevated APOC3 level was still noted at 6 months (log2FC = 6.13), as well as serum amyloid A4 (SAA4; log2FC = 2.15) and immunoglobulin heavy constant gamma 3 (IGHG3; log2FC = 1.52) that all relate to chronic immunological responses. The complement responses subsided thereafter, and proteins involved in the endopeptidase activity (MUCL1, CST5 and interleukin (IL)-19), wound healing (chitinase domain-containing protein 1, CHID1), protein folding, and response to the toxic substance were significantly up-regulated (Fig. 2). Submaxillary gland androgen-regulated protein 3A (SMR-3A) was significantly down-regulated (log2FC = −3.21) at 3 months. Table 1 lists the differentially regulated proteins over time after SMILE.
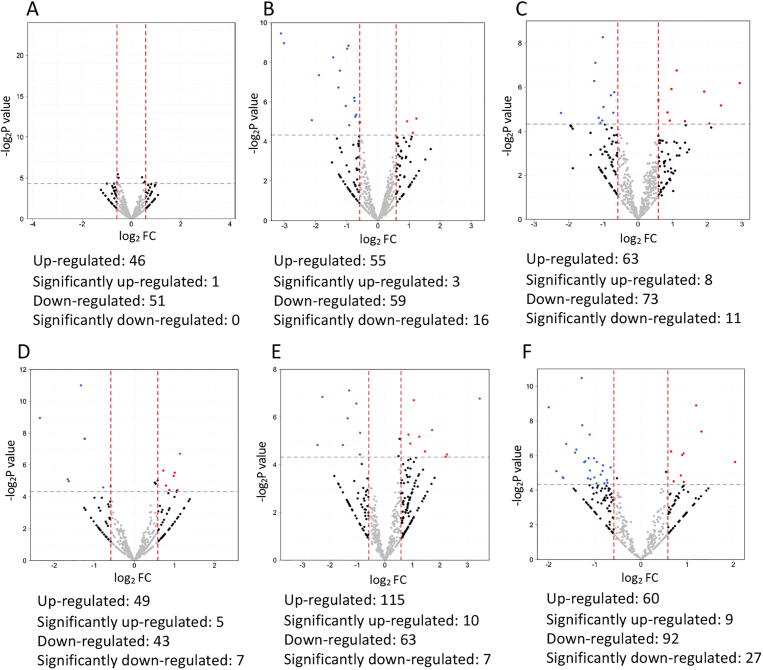
Fig. 1.
Volcano plot showing the fold changes and p values comparing the proteomic data of different postoperative time points (A: 1 week, B: 1 month, C: 3 months, D: 6 months, E: 12 months) with that of baseline in SMILE. Red dots indicate significantly up-regulated proteins (FC > 1.5 and P < 0.05, i.e. log2 FC > 0.58 and -log2P > 4.3), and blue dots indicate significantly down-regulated proteins (FC < 0.67 and P < 0.05, i.e. log2 FC < -0.58 and -log2P > 4.3).
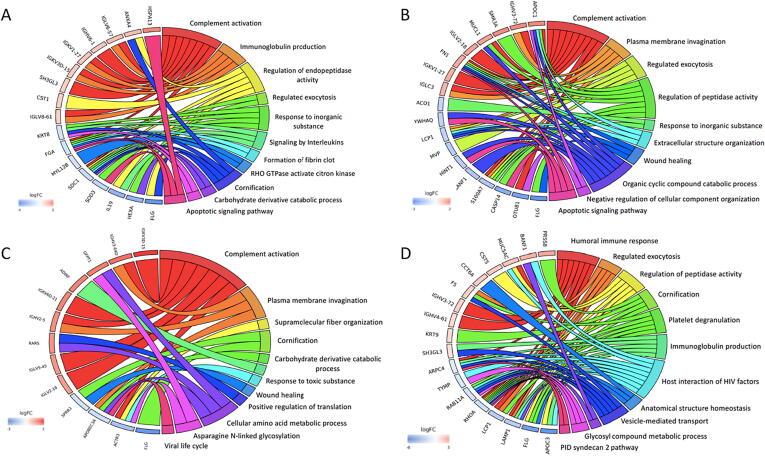
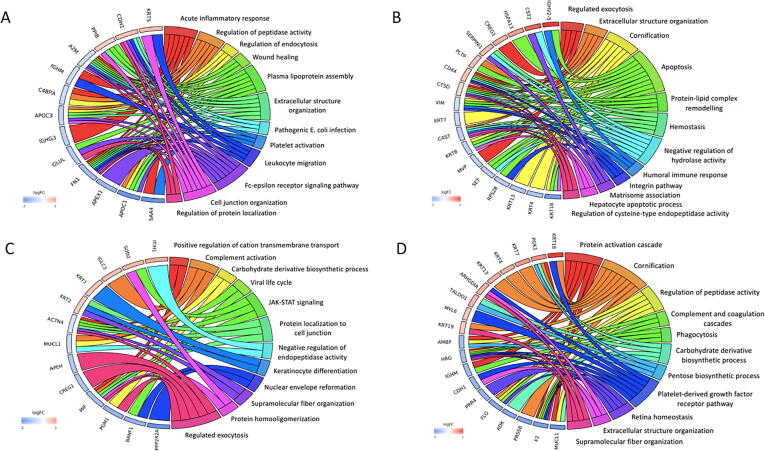
Fig. 2.
Chord plots illustrating GO analysis of top 10 up-regulated and down-regulated proteins and associated pathways in SMILE surgery at 1 week (A), 1 month (B), 3 months (C) and 12 months (D), in comparison to the baseline. Chord plots represent a circular dendrogram of the clustering of the expression profiles. Log FC: log2 (fold-changes).
Table 1.
Top 10 up-regulated and down-regulated tear proteins in post-SMILE eyes at different time points postoperatively.
| 1 week |
1 month |
3 months |
6 months |
12 months |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein Name | log2 FC | P Value | Protein Name | log2 FC | P Value | Protein Name | lo g 2 FC | P Value | Protein Name | log2 FC | P Value | Protein Name | log2 FC | P Value | |
| Up-regulated proteins | |||||||||||||||
| 1 | Apolipoprotein C3 (APOC3) | 3.9943 | 0.0050 | Apolipoprotein C3 (APOC3) | 2.9685 | 0.0284 | Ig lambda variable 2–11 (IGLV2-11) | 2.4571 | 0.0323 | Apolipoprotein C3 (APOC3) | 6.1280 | 0.0000 | Mucin-like protein 1 (MUCL1) | 3.3505 | 0.0017 |
| 2 | Glutamate/proline-tRNA ligase (EPRS) | 3.0457 | 0.0138 | Ig heavy variable2–5 (IGHV2-5) | 2.6279 | 0.0264 | Cystatin D (CST5) | 2.0036 | 0.0005 | Cathepsin H (CTSH) | 2.1736 | 0.0442 | Chitinase domain Containing 1 (CHID1) | 3.0661 | 0.0177 |
| 3 | Apolipoprotein E (APOE) | 2.8546 | 0.0187 | Chaperonin containingTCP1 subunit 4 (CCT4) | 2.3345 | 0.0430 | Mucin-like protein 1 (MUCL1) | 1.9420 | 0.0024 | Serum amyloid A4 (SAA4) | 2.1507 | 0.0458 | Cystatin D (CST5) | 2.9637 | 0.0193 |
| 4 | Serum paraoxonase (PON1) | 2.5651 | 0.0297 | Apolipoprotein E (APOE) | 2.2432 | 0.0126 | Ceroid-lipofuscinosis neuronal protein 5 (CLN5) | 2.1010 | 0.0497 | Interleukin 19 (IL19) | 2.8668 | 0.0210 | |||
| 5 | Complement C1r (C1R) | 2.3810 | 0.0399 | Fibronectin 1 (FN1) | 1.9528 | 0.0159 | Ig heavy constant γ3 (IGHG3) | 1.5210 | 0.0129 | Adhesion G protein-coupled receptor V1 (ADGRV1) | 2.4898 | 0.0154 | |||
| 6 | Fibronectin 1 (FN1) | 2.2581 | 0.0086 | Chitinase domaincontaining 1 (CHID1) | 1.8373 | 0.0204 | Peptidyl-glycine α-amidating monooxygenase (PAM) | 1.3885 | 0.0161 | Peptidylglycine alpha-amidating monooxygenase (PAM) | 2.3752 | 0.0340 | |||
| 7 | Mucin-like protein 1 (MUCL1) | 2.0267 | 0.0032 | C4b-binding proteinα chain (C4BPA) | 1.8304 | 0.0098 | C4b-binding protein α chain (C4BPA) | 1.3818 | 0.0307 | Annexin A11 (ANXA11) | 2.2104 | 0.0406 | |||
| 8 | Ig lambda variable2–18 (IGLV2-18) | 1.7282 | 0.0059 | Serum paraoxonase (PON1) | 1.7841 | 0.0256 | Inter-α-trypsin inhibitor heavy chain H1 (ITIH1) | 1.1360 | 0.0340 | ||||||
| 9 | α2-antiplasmin (SERPINF2) | 1.6716 | 0.0300 | Keratin 13 (KRT13) | 1.7798 | 0.0260 | |||||||||
| x | C4b-binding proteinα chain (C4BPA) | 1.4591 | 0.0043 | Keratin 4 (KRT4) | 1.5519 | 0.0121 | |||||||||
| Down-regulated proteins | |||||||||||||||
| 1 | Peptidyl-prolyl cis–trans isomerase C (PPIC) | −6.4759 | 0.0001 | Thymidine Phosphorylase (TYMP) | −1.7024 | 0.0001 | Submaxillary gland androgen regulated protein 3A (SMR3A) | −3.2100 | 0.0120 | Phosphoglucomutase 2 (PGM2) | −2.1769 | 0.0439 | Tubulin α4a (TUBA4A) | −4.4151 | 0.0066 |
| 2 | Superoxide dismutase 3 (SOD3) | −1.9301 | 0.0024 | ADP ribosylation Factor 5 (ARF5) | −1.6447 | 0.0490 | Inter-α-trypsin inhibitor heavy chain 2 (ITIH2) | −1.4295 | 0.0396 | Aldo-keto reductase 7A2 (AKR7A2) | −2.0444 | 0.0089 | N-Acylsphingosine amidohydrolase 1 (ASAH1) | −2.5089 | 0.0296 |
| 3 | Myeloblastin (PRTN3) | −1.4818 | 0.0180 | Peroxiredoxin 3 (PRDX3) | −1.5867 | 0.0397 | Thymidine phosphorylase (TYMP) | −1.2192 | 0.0119 | Histone H1.4 (HIST1H1E) | −1.4605 | 0.0364 | Eukaryotic translation initiation factor 5A (EIF5A) | −2.0286 | 0.0499 |
| 4 | Src substrate cortactin (CTTN) | −1.4813 | 0.0075 | Actin related protein 3 (ACTR3) | −1.3065 | 0.0059 | Actin-related protein 3 (ACTR3) | −1.4155 | 0.0206 | Lymphocyte cytosolic protein 1 (LCP1) | −1.8740 | 0.0113 | |||
| 5 | Tryptophan--tRNA ligase (WARS) | −1.4618 | 0.0384 | Heat shock protein 90 αA member 1 (HSP90AA1) | −1.2742 | 0.0061 | Tumor protein D52 (TPD52) | −1.3124 | 0.0103 | Proteinase 3 (PRTN3) | −1.5471 | 0.0209 | |||
| 6 | Proteasome subunit α2 (PSMA2) | −1.4296 | 0.0189 | Proteasome subunit α1 (PSMA1) | −1.1553 | 0.0496 | Actin-related protein 2/3 complex subunit 4 (ARPC4) | −1.0978 | 0.0179 | Apolipoprotein B mRNA editing enzyme catalytic subunit 3A (APOBEC3A) | −1.5370 | 0.0287 | |||
| 7 | Plasminogen activator inhibitor 2 (SERPINB2) | −1.3320 | 0.0256 | Cadherin-1 (CDH1) | −1.0734 | 0.0363 | Protein disulfide-isomerase A3 (PDIA3) | −0.9677 | 0.0266 | Proteasome subunit β9 (PSMB9) | −1.4406 | 0.0038 | |||
| 8 | Transitional endoplasmic reticulum ATPase (VCP) | −1.2211 | 0.0413 | Cystatin-A (CSTA) | −1.0685 | 0.0062 | Transitional endoplasmic reticulum ATPase (VCP) | −0.8789 | 0.0457 | Phosphoglycerate kinase 1 (PGK1) | −1.2959 | 0.0371 | |||
| 9 | Glutathione S-transferase omega-1 (GSTO1) | −1.1882 | 0.0425 | Heat shock protein 90β (HSP90AB1) | −0.9649 | 0.0135 | Pyruvate kinase PKM (PKM) | −0.8758 | 0.0161 | ||||||
| 10 | Thymidine phosphorylase (TYMP) | −0.8306 | 0.0253 | ||||||||||||
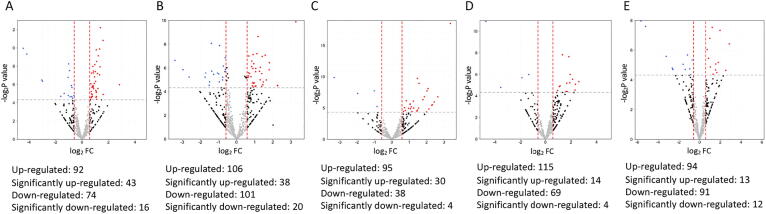
Post-LASIK tear protein changes
The number and fold change of up-regulated and down-regulated proteins following LASIK over time were illustrated in Fig. 3. In the early post-LASIK period (1 week and 1 month), various immunoglobulin proteins (IGLV6-57, IGLV8-61, immunoglobulin kappa variable (IGKV)1–27, IgKV3D-15) were significantly up-regulated, indicating complement activation and immunoglobulin production (Table 2). Apoptotic signalling pathway (heat shock protein (HSP)A13; log2FC = 2.82) and endopeptidase activity (APOC1, log2FC = 2.27; CST1, log2FC = 1.21; FN1, log2FC = 1.65) were also observed (Fig. 4A, B). At 3 months, proteins playing a role in complement (IGKV3D-15, IGKV6D-21, IGLV5-45, and IGLV2-18) and wound healing (arginine-tRNA ligase (RARS), log2FC = 2.08) were expressed at a significantly higher level than baseline. At 6 months, significantly up-regulated proteins included APOB (log2FC = 2.32) and various types of immunoglobulins (Table 2). At 1 year, immunoglobulin production reaction was still observed, and significantly altered proteins associated with endopeptidase activity (MUC5AC, log2FC = 1.97; CST5, log2FC = 1.80) and platelet degranulation (coagulation factor-V (F5), log2FC = 1.57) were emerged (Fig. 4). Filaggrin (FLG) was significantly downregulated in all time points after LASIK (Table 2).
Fig. 3.
Volcano plot presenting the fold changes and p values comparing the proteomic data of different postoperative time points (A: 1 week, B: 1 month, C: 3 months, D: 6 months, E: 12 months) with that of baseline in LASIK. Red dots indicate significantly up-regulated proteins (FC > 1.5 and P < 0.05, i.e. log2 FC > 0.58 and -log2P > 4.3), and blue dots indicate significantly down-regulated proteins (FC < 0.67 and P < 0.05, i.e. log2 FC < -0.58 and -log2P > 4.3).
Table 2.
Top 10 up-regulated and down-regulated tear proteins in post-LASIK eyes at different time points postoperatively.
| 1 week |
1 month |
3 months |
6 months |
12 months |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein Name | Log2 FC | P Value | Protein Name | Log2 FC | P Value | Protein Name | Log2 FC | P Value | Protein Name | Log 2FC | P Value | Protein Name | Log2 FC | P Value | |
| Up-regulated proteins | |||||||||||||||
| 1 | Heat shock protein A13 (HSPA13) | 2.8282 | 0.0160 | Apolipoprotein C1 (APOC1) | 2.2720 | 0.0436 | Ig kappa variable 3D-15 (IGKV3D-15) | 3.3696 | 0.0000 | Ig kappa variable 1–6 (IGKV1-6) | 2.6160 | 0.0251 | Serine protease 8 (PRSS8) | 2.8660 | 0.0117 |
| 2 | Annexin A4 (ANXA4) | 1.6041 | 0.0335 | Ig heavy variable 3–72 (IGHV3-72) | 2.2617 | 0.0442 | Ig heavy variable 3-64D (IGHV3-64D) | 2.6444 | 0.0091 | Ig kappa variable 2–24 (IGKV2-24) | 2.3451 | 0.0298 | BAF nuclear assembly Factor 1 (BANF1) | 2.5272 | 0.0402 |
| 3 | Ig lambda variable 6–57 (IGLV6-57) | 1.5958 | 0.0006 | Submaxillary gland androgen regulated protein 3A (SMR3A) | 2.2576 | 0.0445 | Glutamine-fructose-6-phosphate transaminase 1 (GFPT1) | 2.4423 | 0.0159 | Apolipoprotein B (APOB) | 2.3173 | 0.0433 | Mucin 5AC (MUC5AC) | 1.9723 | 0.0062 |
| 4 | Ig heavy variable 6–1 (IGHV6-1) | 1.3898 | 0.0002 | Mucin-like protein 1 (MUCL1) | 1.8144 | 0.0115 | Adipogenesis regulatory factor (ADIRF) | 2.3268 | 0.0216 | Ig lambda Like polypeptide 5 (IGLL5) | 2.1285 | 0.0216 | Cystatin D (CST5) | 1.8037 | 0.0276 |
| 5 | Ig kappa variable 1–27 (IGKV1-27) | 1.3662 | 0.0041 | Ig lambda variable 2–18 (IGLV2-18) | 1.8029 | 0.0174 | Ig kappa variable 6D-21 (IGKV6D-21) | 2.3009 | 0.0231 | Ig heavy variable 6–1 (IGHV6-1) | 1.9338 | 0.0158 | Chaperonin containing TCP1 subunit 6A (CCT6A) | 1.6666 | 0.0448 |
| 6 | Ig kappa variable 3D-15 (IGKV3D-15) | 1.3271 | 0.0076 | Fibronectin 1 (FN1) | 1.6470 | 0.0318 | Ig heavy variable 2–5 (IGHV2-5) | 2.1339 | 0.0351 | Ig lambda Like polypeptide 1 (IGLL1) | 1.9081 | 0.0271 | Coagulation factor V (F5) | 1.5672 | 0.0298 |
| 7 | Endophilin-A3 (SH3GL3) | 1.2315 | 0.0018 | Ig kappa variable 1–27 (IGKV1-27) | 1.5837 | 0.0074 | Arginine-tRNA ligase (RARS) | 2.0840 | 0.0036 | Ig kappa variable 3D-15 (IGKV3D-15) | 1.8353 | 0.0050 | Ig heavy variable 3–72 (IGHV3-72) | 1.5448 | 0.0395 |
| 8 | Cystatin-SN (CST1) | 1.2112 | 0.0213 | Ig lambda constant 3 (IGLC3) | 1.4769 | 0.0358 | Ig lambda variable 5–45 (IGLV5-45) | 2.0257 | 0.0047 | Ig lambda constant 2 (IGLC2) | 1.7103 | 0.0474 | Ig heavy variable 4–61 (IGHV4-61) | 1.3004 | 0.0481 |
| 9 | Ig lambda variable 8–61 (IGLV8-61) | 1.1692 | 0.0068 | Ig lambda variable 2–18 (IGLV2-18) | 1.8719 | 0.0090 | Ig heavy variable 3–7 (IGHV3-7) | 1.6702 | 0.0276 | Keratin 9 (KRT9) | 1.2178 | 0.0054 | |||
| 10 | Ig heavy constant mu (IGHM) | 1.6130 | 0.0469 | Endophilin-A3 (SH3GL3) | 1.0415 | 0.0344 | |||||||||
| Down-regulated proteins | |||||||||||||||
| 1 | Filaggrin (FLG) | −4.1638 | 0.0016 | Filaggrin (FLG) | −3.3967 | 0.0100 | Filaggrin (FLG) | −3.3168 | 0.0011 | Filaggrin (FLG) | −4.6876 | 0.0005 | Apolipoprotein C3 (APOC3) | −5.6508 | 0.0040 |
| 2 | Hexosaminidase subunit alpha (HEXA) | −3.0295 | 0.0112 | OTU deubiquitinase, ubiquitin aldehyde binding 1 (OTUB1) | −2.9543 | 0.0173 | Actin related protein 3 (ACTR3) | −1.9630 | 0.0061 | Apolipoprotein C3 (APOC3) | −3.5306 | 0.0365 | Filaggrin (FLG) | −5.1985 | 0.0052 |
| 3 | Interleukin 19 (IL19) | −2.9780 | 0.0122 | Caspase 14 (CASP14) | −2.6177 | 0.0269 | Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A (APOBEC3A) | −1.0129 | 0.0047 | Coronin 1A (CORO1A) | −1.8676 | 0.0196 | Lysosomal associated membrane protein 1 (LAMP1) | −2.6320 | 0.0361 |
| 4 | Superoxide dismutase 3 (SOD3) | −1.5974 | 0.0384 | S100 calcium binding protein A7 (S100A7) | −1.7365 | 0.0270 | Small proline-rich protein 3 (SPRR3) | −0.8491 | 0.0266 | Proteinase 3 (PRTN3) | −1.3437 | 0.0159 | Lymphocyte cytosolic protein 1 (LCP1) | −2.5816 | 0.0380 |
| 5 | Syndecan-1 (SDC1) | −1.3405 | 0.0308 | BAF nuclear assembly factor 1 (BANF1) | −1.6998 | 0.0211 | Transforming protein RhoA (RHOA) | −1.5814 | 0.0302 | ||||||
| 6 | Myosin regulatory light chain 12B (MYL12B) | −1.0435 | 0.0091 | Histidine triad nucleotide-binding protein 1 (HINT1) | −1.5126 | 0.0434 | Ras-related protein Rab-11A (RAB11A) | −1.3830 | 0.0385 | ||||||
| 7 | Fibrinogen α chain (FGA) | −1.0334 | 0.0361 | Major vault protein (MVP) | −1.4863 | 0.0356 | Thymidine phosphorylase (TYMP) | −1.1653 | 0.0196 | ||||||
| 8 | Keratin 8 (KRT8) | −1.0017 | 0.0033 | Plastin-2 (LCP1) | −1.3899 | 0.0037 | Actin-related protein 2/3 complex subunit 4 (ARPC4) | −0.9512 | 0.0375 | ||||||
| 9 | 14–3-3 protein theta (YWHAQ) | −1.3646 | 0.0188 | ||||||||||||
| 10 | Cytoplasmic aconitate hydratase (ACO1) | −1.2620 | 0.0229 | ||||||||||||
Fig. 4.
Chord plots demonstrating GO analysis of top 10 up-regulated and down-regulated proteins in LASIK procedure when comparing the profiles at 1 week (A), 1 month (B), 3 months (C) and 12 months (D) to those of the baseline. Chord plots represent a circular dendrogram of the clustering of the expression profiles. Log FC: log2 (fold-changes).
Tear protein comparison between LASIK and SMILE
Before surgery, the tear protein profiles were comparable between the SMILE and LASIK groups (Fig. 5A). The comparisons of tear proteomic profiles following LASIK and SMILE are summarized in Table 3 and Fig. 5,6. In the early postoperative period, the post-LASIK eyes contained significantly upregulated peptidyl-prolyl cis–trans isomerase B (PPIB, log2FC = 0.94) that is involved in wound healing, HSPA13 (log2FC = 1.36) in apoptosis, IGHV2-5 (log2FC = 2.39) in humoral immune reaction, cadherin 1 (CDH1, log2FC = 1.12) in leukocyte migration, and CST2 (log2FC = 2.06) in negative regulation of hydrolase activity. Thereafter, superoxide dismutase protein (superoxide dismutase (SOD2), log2FC = 1.01 at 3 months; log2FC = 1.46 at 6 months) and endoplasmic reticulum resident protein 29 (ERP-29, log2FC = 3.45 at 6 months) were significantly up-regulated in the LASIK group. At 1 year, the LASIK eyes had a significantly higher level of Rho GDP-dissociation inhibitor 1 (log2FC = 0.90) involved in extracellular structure organization, while FLG, MUCL1 and a lacrimal gland-abundant protein, proline-rich 4 (PRR4) were significantly lower (log2FC = −1.61, log2FC = -2.00 and log2FC = −1.43, respectively). In particular, the fold changes of tear MUCL1 level were negatively correlated with TBUT (r = -0.47, P = 0.04) (see Fig. 6.).
Fig. 5.
Volcano plot presenting the fold changes and p values comparing the proteomic data between LASIK and SMILE at different time points (A: before surgery, B: 1 week, C: 1 month, D: 3 months, E: 6 months, F: 12 months). Red dots indicate significantly up-regulated proteins (FC > 1.5 and P < 0.05, i.e. log2 FC > 0.58 and -log2P > 4.3) and blue dots indicate significantly down-regulated proteins (FC < 0.67 and P < 0.05, i.e. log2 FC < -0.58 and -log2P > 4.3). Tear proteomic profiles were comparable between LASIK and SMILE group before surgery (A).
Table 3.
Top 10 up-regulated and down-regulated tear proteins comparing post-LASIK versus post-SMILE eyes at different time points postoperatively.
| 1 week |
1 month |
3 months |
6 months |
12 months |
|||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Protein Name | Log2 FC | P Value | Protein Name | Log2 FC | P Value | Protein Name | Log2 FC | P Value | Protein Name | Log2 FC | P Value | Protein Name | Log2 FC | P Value | |
| Up-regulated proteins | |||||||||||||||
| 1 | Keratin 5 (KRT5) | 1.2390 | 0.0280 | Ig heavy variable 2–5 (IGHV2-5) | 2.3930 | 0.0278 | Inter-alpha-trypsin inhibitor heavy chain H1 (ITIH1) | 1.1398 | 0.0096 | Endoplasmic reticulum resident protein 29 (ERP29) | 3.4489 | 0.0091 | Keratin 18 (KRT18) | 2.0409 | 0.0204 |
| 2 | Cadherin-1 (CDH1) | 1.1240 | 0.0462 | Cystatin-SA (CST2) | 2.0569 | 0.0491 | Superoxide dismutase (SOD2) | 1.0080 | 0.0220 | S-methyl-5′-thioadenosine phosphorylase (MTAP) | 2.2605 | 0.0464 | Phosphoglycerate kinase 1 (PGK1) | 1.3122 | 0.0059 |
| 3 | Peptidyl-prolyl cis–trans isomerase B (PPIB) | 0.9430 | 0.0309 | Heat shock 70 kDa protein 13 (HSPA13) | 1.3550 | 0.0456 | Ig lambda constant 3 (IGLC3) | 0.8751 | 0.0467 | Plastin-2 (LCP1) | 2.2162 | 0.0495 | Keratin 7 (KRT7) | 1.1975 | 0.0021 |
| 4 | Cellular repressor of E1A stimulated gene 1 (CREG1) | 1.1145 | 0.0093 | Keratin 7 (KRT7) | 0.7884 | 0.0385 | Tumor protein D52 (TPD52) | 1.7152 | 0.0228 | Keratin 4 (KRT4) | 0.9291 | 0.0450 | |||
| 5 | Neuroserpin (SERPINI1) | 0.9644 | 0.0166 | Keratin 2 (KRT2) | 0.7235 | 0.0201 | Superoxide dismutase 2 (SOD2) | 1.4622 | 0.0424 | Keratin 13 (KRT13) | 0.9256 | 0.0143 | |||
| 6 | Phospholipid transfer protein (PLTP) | 0.9164 | 0.0446 | Glutaredoxin-1 (GLRX) | 1.4557 | 0.0424 | Rho GDP-dissociation inhibitor 1 (ARHGDIA) | 0.8959 | 0.0154 | ||||||
| 7 | CD44 antigen (CD44) | 0.8495 | 0.0347 | Alcohol dehydrogenase class 4 mu/sigma chain (ADH7) | |||||||||||
| 1.2539 | 0.0277 | Transaldolase (TALDO1) | 0.8651 | 0.0349 | |||||||||||
| 8 | Cathepsin D (CTSD) | 0.5876 | 0.0235 | Aldehyde dehydro-genase 3A1 (ALDH3A1) | 1.0541 | 0.0096 | Myosin light polypeptide 6 (MYL6) | 0.7116 | 0.0441 | ||||||
| 9 | Heat shock protein 90β1 (HSP90B1) | 0.9227 | 0.0338 | Keratin 19 (KRT19) | 0.6566 | 0.0132 | |||||||||
| 10 | Cytosolic non-specific dipeptidase (CNDP2) | 0.8565 | 0.0260 | ||||||||||||
| Down-regulated tear proteins | |||||||||||||||
| 1 | Serum amyloid A-4 protein (SAA4) | −3.1164 | 0.0014 | Keratin 18 (KRT18) | −2.2235 | 0.0352 | Serine/threonine-protein phosphatase 2A 55 kDa regulatory subunit B α isoform (PPP2R2A) | −2.3558 | 0.0020 | Filaggrin (FLG) | −2.4520 | 0.0351 | Mucin-like protein 1 (MUCL1) | −1.9951 | 0.0023 |
| 2 | Apolipoprotein C-I (APOC1) | −3.0195 | 0.0020 | Keratin 4 (KRT4) | −1.2640 | 0.0129 | Barrier-to-autointegration factor (BANF1) | −1.6611 | 0.0293 | Fibronectin (FN1) | −1.5303 | 0.0352 | Prothrombin (F2) | −1.8304 | 0.0293 |
| 3 | DNA-(apurinic or apyrimidinic site) lyase (APEX1) | −2.1237 | 0.0297 | Keratin 13 (KRT13) | −1.2336 | 0.0073 | Phosphoglucomutase-1 (PGM1) | −1.6352 | 0.0319 | Ig lambda constant 3 (IGLC3) | −1.3581 | 0.0162 | Prostasin (PRSS8) | −1.6934 | 0.0376 |
| 4 | Fibronectin (FN1) | −1.8946 | 0.0061 | 40S ribosomal protein S28 (RPS28) | −1.1354 | 0.0410 | Macrophage migration inhibitory factor (MIF) | −1.3305 | 0.0005 | Inter-α-trypsin inhibitor heavy chain H1 (ITIH1) | −1.3023 | 0.0072 | Adenosine kinase (ADK) | −1.6797 | 0.0386 |
| 5 | Glutamine synthetase (GLUL) | −1.4370 | 0.0033 | Protein SET (SET) | −1.0877 | 0.0482 | Protein CREG1 (CREG1) | −1.2379 | 0.0049 | Plasminogen (PLG) | −1.0351 | 0.0106 | Filaggrin (FLG) | −1.6161 | 0.0097 |
| 6 | Ig heavy constant γ3 (IGHG3) | −1.2682 | 0.0094 | Major vault protein (MVP) | −1.0333 | 0.0446 | Acylamino-acid-releasing enzyme (APEH) | −1.2353 | 0.0050 | Ig heavy constant gamma 4 (IGHG4) | −0.9052 | 0.0465 | Proline-rich protein 4 (PRR4) | −1.4252 | 0.0139 |
| 7 | Apolipoprotein C-III (APOC3) | −1.2207 | 0.0052 | Keratin 8 (KRT8) | −1.0166 | 0.0033 | Mucin-like protein 1 (MUCL1) | −0.7758 | 0.0418 | Inter-α-trypsin inhibitor heavy chain H2 (ITIH2) | −0.8991 | 0.0247 | Cadherin-1 (CDH1) | −1.3985 | 0.0122 |
| 8 | C4b-binding protein alpha chain (C4BPA) | −1.0333 | 0.0180 | Calpastatin (CAST) | −0.9974 | 0.0291 | α-Actinin-4 (ACTN4) | −0.5929 | 0.0396 | Ig heavy constant mu (IGHM) | −1.2822 | 0.0007 | |||
| 9 | Ig heavy constant mu (IGHM) | −0.9886 | 0.0024 | Keratin 7 (KRT7) | −0.7841 | 0.0202 | Histidine-rich glycoprotein (HRG) | −1.2697 | 0.0046 | ||||||
| 10 | α2-Macroglobulin (A2M) | −0.9468 | 0.0022 | Vimentin (VIM) | −0.7130 | 0.0350 | Protein AMBP (AMBP) | −1.2277 | 0.0205 |
Fig. 6.
Chord plots demonstrating GO analysis of top 10 up-regulated and down-regulated protein profiles when comparing LASIK versus SMILE procedure at 1 week (A), 1 month (B), 3 months (C) and 12 months (D). Chord plots show the clustering of the expression profiles. Log FC: log2 (fold-changes).
Pathway analysis using the Reactome database showed similar findings: LASIK surgery was associated with significantly enriched biological processes involving the immune system, DNA replication, cell cycle, programmed cell death, hemostasis, and protein metabolism, compared to SMILE (Supplementary Figs. 2–4 and Supplementary Tables 2–4).
Tear neuromediator profiles
After LASIK surgery, the NGF level was elevated throughout the study period of 1 year, whereas this elevation was only observed for the initial 1 month after SMILE. There was a significant difference in the tear NGF level between the 2 groups at all time points (all P < 0.05). The substance P level was increased postoperatively in the LASIK group, whereas it did not change significantly in the SMILE patients. A significant difference was noted at 1 and 3 months (P = 0.02 for both; Table 4). The postoperative CGRP levels remained at a constant range for both surgeries. Overall, there was a significant but only moderate correlation between the tear substance P level and TBUT (r = -0.49, P = 0.02). There was no significant correlation between other clinical dry eye outcomes and tear neuromediators.
Table 4.
Tear neuromediator concentration in the SMILE and LASIK groups over postoperative 1 year.
| Pre-op | 1 week | 1 month | 3 months | 6 months | 12 months | |
|---|---|---|---|---|---|---|
| NGF(pg/ml) | ||||||
| SMILE | 36.8 ± 16.3 | 37.1 ± 18.2 | 42.5 ± 14.7 | 37.6 ± 23.4 | 38.0 ± 19.8 | 36.2 ± 11.6 |
| LASIK | 35.9 ± 15.6 | 40.5 ± 18.5 | 54.4 ± 17.2 | 44.0 ± 19.4 | 45.2 ± 20.8 | 41.4 ± 24.7 |
| P value | 0.77 | 0.047 | <0.01 | 0.04 | <0.01 | 0.01 |
| Substance P (pg/ml) | ||||||
| SMILE | 1940.0 ± 647.3 | 1900.4 ± 773.5 | 1858.3 ± 828.4 | 1833.1 ± 890.7 | 1890.3 ± 693.0 | 1956.4 ± 818.5 |
| LASIK | 1874.0 ± 652.4 | 1962.0 ± 618.5 | 2157.1.1 ± 792.5 | 2156.4 ± 803.5 | 1998.2 ± 682.0 | 2114.9 ± 756.2 |
| P value | 0.68 | 0.78 | 0.04 | 0.04 | 0.74 | 0.88 |
| CGRP (ng/ml) | ||||||
| SMILE | 3.7 ± 1.3 | 3.6 ± 0.9 | 3.6 ± 1.2 | 3.5 ± 1.4 | 3.7 ± 0.9 | 3.9 ± 1.1 |
| LASIK | 3.8 ± 1.4 | 3.6 ± 1.2 | 3.7 ± 1.0 | 3.9 ± 1.5 | 4.0 ± 1.8 | 4.0 ± 0.9 |
| P value | 0.79 | 0.85 | 0.84 | 0.74 | 0.71 | 0.75 |
Discussion
In the present study, we characterized detailed tear proteomic profiles following SMILE and LASIK over a 1-year postoperative period. The tear neuromediator profiles were also investigated and correlated to clinical dry eye outcome measures. Significant differences in the TBUT and tear substance P level were observed between 2 groups for the initial 3 months after surgery, whereas the differences in the tear NGF level and levels of other proteins, such as proteins involved in complement and coagulative cascades and extracellular structural organization, between 2 groups, were still noted at 1 year. To our knowledge, this is the first study comparing SMILE and LASIK from a proteomic perspective, using a high-sensitivity SWATH-MS technique. The results give a better understanding of the underlying pathophysiological alterations on a molecular level accounting for the clinical ocular surface changes postoperatively. The randomized trial design, as well as the use of data of paired eyes from the same patient, provided more accurate comparison by minimizing the inter-individual and inter-eye variation as well as selection bias.
The SWATH-MS technology is emerging as a preferred tool for clinical proteomic studies with a large number of samples involved, especially when analyzing limited volume of samples for multiple time points [13]. Our group has previously used this method to document the tear protein changes in patients with thyroid eye diseases and keratoconus [17], [18]. Our data showed that during the first month postoperatively, the complement activation and immunoglobulin production were the dominant responses shown in both SMILE and LASIK. From postoperative 3 months onwards, while the immunological response persisted, cellular metabolic processes were also up-regulated. A higher level of apolipoproteins (APOC1, APOC3, and APOE), immunoglobulin family, proteins involved in complement cascade and FN1 were detected in the operated eyes in both groups, compared to the preoperative baseline. Apolipoproteins participate in lipoprotein metabolism and have immune-regulatory functions [25]. APOC1 is a pro-inflammatory protein and positively correlates with IL-6 level [25]. APOE regulates macrophage function and enhances lipid antigen presentation by CD1 molecules to natural killer T cells [26]. Higher expression of APOE and APOC3 in aqueous has been reported in patients with open-angle glaucoma or exfoliative glaucoma due to more disrupted blood-aqueous barrier compared to normal subjects [27]. It is not surprising that tear FN1 also was elevated following surgery, as it has an important role in wound healing, depositing at the incision site and providing a guide for cell attachment and migration [28].
Three to six months following surgery, up-regulated expression of different types of immunoglobulins was observed in both SMILE and LASIK groups, whereas FLG and SMR3A were the top down-regulated proteins in both groups. FLG is expressed in the basal layer of corneal epithelium [29] and its function is to compensate for ocular surface stress [30]. Reduced expression of FLG has been reported in patients with atopic dermatitis-related keratitis due to the chronic ocular surface inflammation [29]. Similarly, the down-regulation of FLG in our study could be the consequence of surgery-induced inflammation. Similar results of reduced SMR3A in post-SMILE eyes were observed in the analysis of salivary glands in patients with primary Sjogren syndrome [31]. At 1 year, proteins modulating tear endopeptidase function, including goblet cell-derived mucin proteins (MUCL1 and MUC5AC) and cystatin D protein (CST5), were expressed at significantly higher levels than the baseline in both groups. Tissue endopeptidase are involved in wound remodeling in response to tissue stress [32], [33], and its expression has been shown to be increased in dry eyes [32]. The tear film on the ocular surface epithelia is maintained by mucins on its surface as well as by membrane-associated mucins in the apical cell surface [34]. MUC5AC has been considered as a potential marker for dry eye [34]. Its level in tears was positively correlated with the corneal staining in contact lens users and post-menopausal dry eye patients (r = 0.37–0.53)[35], [36]. High expression of MUCL1 was found in patients with dry eye or meibomian gland dysfunction (MGD), with correlation observed between its concentration and corneal staining as well as dry eye symptom questionnaire [36], [37]. The elevated tear MUCL1 and MUC5AC levels presented in our study could be a compensatory response and attributable to post-refractive surgery-related neurogenic keratopathy. MUCL1 plays a major role in ocular surface lubrication, apical surface barrier, and osmo-sensing [38]. We found that the tear MUCL1 level was significantly correlated with TBUT. The potential of the use of MUCL1 as an indicator for the evaluation of tear film stability after refractive surgery warrants further investigation.
We subsequently compared the tear proteomes between SMILE and LASIK procedure (Table 3). In the first month, the differentially regulated tear proteins in LASIK showed a significantly greater humoral immune response, wound healing, apoptosis, negative regulation of hydrolase activity, and leukocyte migration. We found that the protein contents had significant variation at 1 week after surgery, and this could be due to the use of postoperative topical steroids that affected the ocular response and tear protein profiles. At 1 month, CST2 and HSPA13 were among the top 5 up-regulated tear proteins observed in LASIK compared to SMILE. The CST family are cysteine protease inhibitors and exert direct immunomodulatory properties [39]. Previous studies have observed up-regulation of CST1 in eyes following femtosecond laser-assisted LASIK at 1 week [40], and down-regulation of CST4 in patients with dry eye and MGD [41]. CST5 was down-regulated in hard and soft contact lens users [42], but was significantly increased in patients after brain injury, similar to the change in the present study that CST5 levels increased after surgical injury [43], regardless of the surgical types. HSPA13 belongs to HSP70 family [44], which is ubiquitously expressed during cell stress, and is involved in T cell regulation in various inflammatory conditions, such as the retinal laser photocoagulation [44]. Given the nature of the larger incision and excimer photoablation involved in LASIK, it could be anticipated that the activation of HSP would be more intensive in response to more extensive tissue reaction in LASIK than SMILE [45]. Significantly higher expression of SOD2 was observed following LASIK after 3 months. The increase in SOD2 could be a feedback response to the greater tissue reshaping from LASIK, to facilitate the clearance of reactive oxygen species and inflammatory cytokines [46].
In the later postoperative period (6 to 12 months), ERP29 and KRT18 were the two most over-expressed proteins in the LASIK compared to SMILE group, while the clinical dry eye parameters had become comparable between 2 groups. Up-regulation of ERP29 facilitates DNA and tight junction repair, extracellular matrix homeostasis and oxidative stress clearance, and ERP29 may also act as a neuroprotectant [47]. We speculate that ERP29 up-regulation might be feedback to the greater tissue modification and response in LASIK. PRR4 is a lacrimal gland-secreted protein and was down-regulated in the post-LASIK eyes, compared to post-SMILE eyes, at 1 year. PRR4 was found to be decreased in patients with dry eye syndrome, and this decrease was correlated with the severity of clinical symptoms [48]. The down-regulation of PRR4 in the LASIK group may imply that the lacrimal neural arc reflex originating from the corneal nerves was still impaired at 1 year, and had greater impairment in the LASIK group, although the clinical dry eye signs had returned to the preoperative level and were comparable between 2 procedures. Similarly, a previous study on the evaluation of the effects of LASIK on tear proteomes showing that eyes with a larger flap were associated with reduced expression of lacrimal gland proteins due to greater corneal nerve damage [40].
We noted that the tear proteomic expression was somewhat different from that in patients with idiopathic dry eye syndrome [41], [49], [50]. This might be due to two reasons. Firstly, the pathogenesis and ocular surface changes resulting from refractive surgery and subsequent wound healing are neurogenic and neuro-inflammatory reaction-related, which are essentially different from those of idiopathic dry eye. This was also reported in our previous study showing that the tear proteome after LASIK was different from that reported in patients with idiopathic dry eye [40]. Secondly, the postoperative regimen, consisting of corticosteroids and antibiotics, would also alter the initial protein expression. These eye drops were given during the early postoperative period, when the most pronounced pathophysiological process occurred.
We were also interested in the changes of neuromediator profiles following surgery as these are related to the postoperative corneal reinnervation and have not been investigated in SMILE. The neuromediators are released by corneal nerves and maintain the function and integrity of corneal epithelial and stromal keratocytes [51], and they in turn regulate the maintenance, regeneration and integration of nerve function [51], [52]. The effects of NGF on promoting corneal wound healing have drawn considerable attention. NGF participates in ocular surface homeostasis by producing neurotrophins and facilitating sensory dependent corneal and tearing reflex [51]. Our results showed that, in the same patient, the NGF level in the post-LASIK eyes was significantly higher than in the post-SMILE eyes even 1 year after surgery. The level returned to the preoperative range at 1 month in SMILE, while it was persistently high in LASIK. After surgery, the pro-inflammatory and inflammatory cytokines amplify the expression of NGF to be released from the corneal epithelial cells and keratocytes [53]. The more extensive disruption on the cornea by LASIK results in more severe tissue stress, inflammation and greater wound healing responses, which have been demonstrated in our SWATH-MS proteomic results and our previous work comparing these two procedures using animal models [45], [54]. The differences in the tissue responses could explain the differences in the NGF level. Similarly, Lee et al. presented the tear NGF concentrations were significantly higher in photorefractive keratectomy than LASIK [11].
Substance P and CGRP are the most abundant neuropeptides in corneas and are considered as the main triggers of neurogenic inflammation [55]. We observed that the tear substance P level increased significantly for 3 months following LASIK but did not in the SMILE group. This is in agreement with a study reporting an increase in substance P level in post-LASIK eyes at 3 months [56], but we further demonstrated this increase lasted for 1 year. In addition, our results showed the tear CGRP concentration did not change after either LASIK or SMILE. This is consistent with a previous study presenting the CGRP level did not differ from the baseline to 3 months [56]. However, another study reported that the CGRP level was significantly higher in LASIK than controls at 1 year [8]. This disparity may come from the femtosecond laser system used in the surgery. The Intralase system (Abott Medical Optics, Santa Ana, CA, USA) employs a lower repetition rate but higher laser energy, while the Visumax system, the platform used in the present study, uses a higher repetition rate and lower laser energy [40]. The former applies the suction on the conjunctiva and sclera, which may stimulate the goblet cells to release CGRP [51], whereas the latter generates the suction only on the cornea via a smaller cone. Following SMILE, the substance P and CGRP concentrations did not change significantly.
Our RCT data on dry eye variables showed that the difference between the 2 procedures was noted for the initial 1 month for corneal staining, and 3 months for the TBUT, with more favorable outcomes in SMILE. There was no difference in the Schirmer’s test results. A systematic review has also demonstrated similar trends, although some studies reported differences in TBUT might be present until 6 months [2], [3]. A significant but only moderate correlation was observed between the substance P level and TBUT, and no correlation was found between other dry eye parameters and neuromediators. This suggests that the current clinical assessment for dry eye may be too coarse and not sensitive enough to reflect the underlying biological changes. Similarly, previous evaluation on the correlation between the tear NGF concentrations and ocular surface parameters such as OSDI and non-invasive TBUT, in FLEx or SMILE procedure, have shown only moderate correlation existed (r = 0.35–0.59) [6]. In another study on LASIK, tear substance P level was positively but also only moderately correlated with dry eye symptoms [8]. Studies in contact lens wearers also found that tear NGF levels correlated significantly with clinical dry eye severity grading, ocular surface fluorescein staining and conjunctival hyperemia, while CGRP levels showed opposite changes compared to those of NGF, in which CGRP was correlated inversely with clinical dry eye severity. However, all these correlations presented were only weak to moderate [52].
The merit of this study is its RCT and paired-eye design to eliminate the inter-individual and inter-eye variation, especially when the high-sensitivity proteomic techniques are used to detect fine protein changes. Due to limited tear protein amount, the SWATH-MS and ELISA experiments were conducted in two different cohorts. Hence, we could not explore the link between the tear neuromediator profiles and SWATH-MS data in this study. Tear collection using a microcapillary, rather than a Schirmer’s strip, may ensure a higher protein concentration to be collected. For studies on surgical techniques, the inherent limitation is a double-masked study design is not possible as the surgeons performed the surgery and investigators could differentiate the surgical type with slit lamp during the assessments, for example, during the dry eye assessments in this study. Subjective evaluation on dry eye symptoms, such as questionnaires, would be considered to improve the study objectivity. Validation experiments on selected up- and down-regulated proteins using an additional cohort should be considered. Future studies will include analysis on the corneal nerve plexus imaged by in-vivo confocal microscopy to illustrate the relationship between the tear neuromediators and corneal nerve metrics, and to identify potential biomarkers for the severity of corneal denervation and the status of nerve regeneration following refractive surgery [57].
Conclusions
We have identified the proteomic response following SMILE and LASIK. Over a one-year postoperative period, there were some similar responses observed in these two procedures. However, there was significantly higher tear NGF levels, as well as up-regulated leukocyte migration, humoral immune response wound healing, apoptotic, and endopeptidase activities, and down-regulated lacrimal gland-abundant protein, in the post-LASIK eyes. These results indicate that different proteomic profiles were still observed even though clinical dry eye evaluation findings became comparable between these 2 procedures. Tear protein science is an evolving area, and current evidence has suggested that tear proteins have a role in modifying clinical practice [34]. This is the first study to demonstrate the detailed proteome changes of SMILE and LASIK. Our data provide new insights into the biological responses on the ocular surface after refractive surgery.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
Acknowledgement
Nil.
Financial Support
This work was supported by the HREF grant, Singapore National Eye Centre, Singapore (R1568/67/2018).
Footnotes
Peer review under responsibility of Cairo University.
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jare.2020.11.001.
Appendix A. Supplementary material
The following are the Supplementary data to this article:
References
- 1.Liu Y.C., Rosman M., Mehta J.S. Enhancement after small-incision lenticule extraction: Incidence, risk factors, and outcomes. Ophthalmology. 2017;124(6):813–821. doi: 10.1016/j.ophtha.2017.01.053. [DOI] [PubMed] [Google Scholar]
- 2.Kobashi H., Kamiya K., Shimizu K. Dry eye after small incision lenticule extraction and femtosecond laser-assisted LASIK: Meta-analysis. Cornea. 2017;36(1):85–91. doi: 10.1097/ICO.0000000000000999. [DOI] [PubMed] [Google Scholar]
- 3.Denoyer A., Landman E., Trinh L., Faure J.F., Auclin F., Baudouin C. Dry eye disease after refractive surgery: comparative outcomes of small incision lenticule extraction versus LASIK. Ophthalmology. 2015;122(4):669–676. doi: 10.1016/j.ophtha.2014.10.004. [DOI] [PubMed] [Google Scholar]
- 4.Fuest M., Mehta J.S. Iatrogenic dry eye following cataract and refractive surgical procedures. In: He M.G., editor. Ocular surface diseases, disorders, & dysfunctions. Thorofare; NJ, Healio: 2016. pp. 3–6. [Google Scholar]
- 5.Liu Y.C., Tan D.T.H., Mehta J.S. Wound healing after ReLEx surgery. In: Sekundo W., editor. small incision lenticule extraction: Principles, techniques, complication management and future concepts. NY, Springer; New York: 2015. pp. 13–26. [Google Scholar]
- 6.Zhang C., Ding H., He M., Liu L., Liu L., Li G. Comparison of early changes in ocular surface and inflammatory mediators between femtosecond lenticule extraction and small-incision lenticule extraction. PLoS ONE. 2016;11(3) doi: 10.1371/journal.pone.0149503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gao S., Li S., Liu L., Wang Y., Ding H., Li L. Early changes in ocular surface and tear inflammatory mediators after small-incision lenticule extraction and femtosecond laser-assisted laser in situ keratomileusis. PLoS ONE. 2014;9(9) doi: 10.1371/journal.pone.0107370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chao C., Golebiowski B., Zhao X., Chen S., Zhou S., Stapleton F. Long-term effects of LASIK on corneal innervation and tear neuropeptides and the associations with dry eye. J Refract Surg. 2016;32(8):518–524. doi: 10.3928/1081597X-20160603-01. [DOI] [PubMed] [Google Scholar]
- 9.Chiu I.M., von Hehn C.A., Woolf C.J. Neurogenic inflammation and the peripheral nervous system in host defense and immunopathology. Nat Neurosci. 2012;15(8):1063–1067. doi: 10.1038/nn.3144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Solomon R., Donnenfeld E.D., Perry H.D. The effects of LASIK on the ocular surface. Ocul Surf. 2004;2(1):34–44. doi: 10.1016/s1542-0124(12)70022-8. [DOI] [PubMed] [Google Scholar]
- 11.Lee H.K., Lee K.S., Kim H.C., Lee S.H., Kim E.K. Nerve growth factor concentration and implications in photorefractive keratectomy vs laser in situ keratomileusis. Am J Ophthalmol. 2005;139(6):965–971. doi: 10.1016/j.ajo.2004.12.051. [DOI] [PubMed] [Google Scholar]
- 12.Zhou L., Beuerman R.W. Tear analysis in ocular surface diseases. Prog Retin Eye Res. 2012;31(6):527–550. doi: 10.1016/j.preteyeres.2012.06.002. [DOI] [PubMed] [Google Scholar]
- 13.Jylha A., Nattinen J., Aapola U., Mikhailova A., Nykter M., Zhou L. Comparison of iTRAQ and SWATH in a clinical study with multiple time points. Clin Proteomics. 2018;15:24. doi: 10.1186/s12014-018-9201-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ang M., Tan D., Mehta J.S. Small incision lenticule extraction (SMILE) versus laser in-situ keratomileusis (LASIK): study protocol for a randomized, non-inferiority trial. Trials. 2012;13:75. doi: 10.1186/1745-6215-13-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ang M., Farook M., Htoon H.M., Mehta J.S. Randomized clinical trial comparing femtosecond LASIK and Small-incision lenticule extraction. Ophthalmology. 2020;127(6):724–730. doi: 10.1016/j.ophtha.2019.09.006. [DOI] [PubMed] [Google Scholar]
- 16.Yi Teo C.H., Ong H.S., Liu Y.C., Tong L. Meibomian gland dysfunction is the primary determinant of dry eye symptoms: Analysis of 2346 patients. Ocul Surf. 2020;(20):30105–30111. doi: 10.1016/j.jtos.2020.06.008. S1542–0124. [DOI] [PubMed] [Google Scholar]
- 17.Chng C.L., Seah L.L., Yang M., Shen S.Y., Koh S.K., Gao Y. Tear proteins calcium binding protein A4 (S100A4) and prolactin induced protein (PIP) are potential biomarkers for thyroid eye disease. Sci Rep. 2018;8(1):16936. doi: 10.1038/s41598-018-35096-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yam G.H., Fuest M., Zhou L., Liu Y.C., Deng L., Chan A.S. Differential epithelial and stromal protein profiles in cone and non-cone regions of keratoconus corneas. Sci Rep. 2019;9(1):2965. doi: 10.1038/s41598-019-39182-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yawata N., Selva K.J., Liu Y.C., Tan K.P., Lee A.W., Siak J. Dynamic change in natural killer cell type in the human ocular mucosa in situ as means of immune evasion by adenovirus infection. Mucosal Immunol. 2016;9(1):159–170. doi: 10.1038/mi.2015.47. [DOI] [PubMed] [Google Scholar]
- 20.Kammers K., Cole R.N., Tiengwe C., Ruczinski I. Detecting significant changes in protein abundance. EuPA Open Proteom. 2015;7:11–19. doi: 10.1016/j.euprot.2015.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dennis G., Jr., Sherman B.T., Hosack D.A., Yang J., Gao W., Lane H.C. DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 2003;4:P3. [PubMed] [Google Scholar]
- 22.Zhou Y., Zhou B., Pache L., Chang M., Khodabakhshi A.H., Tanaseichuk O. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10(1):1523. doi: 10.1038/s41467-019-09234-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jassal B., Matthews L., Viteri G., Gong C., Lorente P., Fabregat A. The reactome pathway knowledgebase. Nucleic Acids Res. 2020;48(D1):D498–D503. doi: 10.1093/nar/gkz1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Team RC. R: A language and environment for statistical com‐puting. R Foundation for Statistical Computing. 2017;Vienna, Austria.
- 25.Ko H.L., Wang Y.S., Fong W.L., Chi M.S., Chi K.H., Kao S.J. Apolipoprotein C1 (APOC1) as a novel diagnostic and prognostic biomarker for lung cancer: A marker phase I trial. Thorac Cancer. 2014;5(6):500–508. doi: 10.1111/1759-7714.12117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Allan L.L., Hoefl K., Zheng D.J., Chung B.K., Kozak F.K., Tan R. Apolipoprotein-mediated lipid antigen presentation in B cells provides a pathway for innate help by NKT cells. Blood. 2009;114(12):2411–2416. doi: 10.1182/blood-2009-04-211417. [DOI] [PubMed] [Google Scholar]
- 27.Inoue T., Kawaji T., Tanihara H. Elevated levels of multiple biomarkers of Alzheimer's disease in the aqueous humor of eyes with open-angle glaucoma. Invest Ophthalmol Vis Sci. 2013;54(8):5353–5358. doi: 10.1167/iovs.13-12245. [DOI] [PubMed] [Google Scholar]
- 28.Pankov R., Yamada K.M. Fibronectin at a glance. J Cell Sci. 2002;115(20):3861–3863. doi: 10.1242/jcs.00059. [DOI] [PubMed] [Google Scholar]
- 29.Lapp T., Auw-Haedrich C., Reinhard T., Evans R., Rodriguez E., Weidinger S. Analysis of filaggrin mutations and expression in corneal specimens from patients with or without atopic dermatitis. Int Arch Allergy Immunol. 2014;163(1):20–24. doi: 10.1159/000355965. [DOI] [PubMed] [Google Scholar]
- 30.Tong L., Corrales R.M., Chen Z., Villarreal A.L., De Paiva C.S., Beuerman R. Expression and regulation of cornified envelope proteins in human corneal epithelium. Invest Ophthalmol Vis Sci. 2006;47(5):1938–1946. doi: 10.1167/iovs.05-1129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang L., Xu P., Wang X., Zhang Z., Zhao W., Li Z. Identification of differentially expressed genes in primary Sjogren's syndrome. J Cell Biochem. 2019;120(10):17368–17377. doi: 10.1002/jcb.29001. [DOI] [PubMed] [Google Scholar]
- 32.Stevenson W., Chauhan S.K., Dana R. Dry eye disease: an immune-mediated ocular surface disorder. Arch Ophthalmol. 2012;130(1):90–100. doi: 10.1001/archophthalmol.2011.364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim R.R., Tan A., Liu Y.C., Barathi V.A., Mohan R.R., Mehta J.S. ITF2357 transactivates Id3 and regulate TGFbeta/BMP7 signaling pathways to attenuate corneal fibrosis. Sci Rep. 2016;6:20841. doi: 10.1038/srep20841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.D'Souza S., Tong L. Practical issues concerning tear protein assays in dry eye. Eye Vis (Lond) 2014;1:6. doi: 10.1186/s40662-014-0006-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berry M., Pult H., Purslow C., Murphy P.J. Mucins and ocular signs in symptomatic and asymptomatic contact lens wear. Optom Vis Sci. 2008;85(10):E930–E938. doi: 10.1097/OPX.0b013e318188896b. [DOI] [PubMed] [Google Scholar]
- 36.Gipson I.K., Spurr-Michaud S.J., Senchyna M., Ritter R., 3rd, Schaumberg D. Comparison of mucin levels at the ocular surface of postmenopausal women with and without a history of dry eye. Cornea. 2011;30(12):1346–1352. doi: 10.1097/ICO.0b013e31820d852a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ni Q., Zhao J., Gao Y., Qin D., Chen X., Ainiwaer X. Prediction of potential drugs and targets based on meibomian gland dysfunction module classification to guide individualized treatment. J Cell Biochem. 2019;120(9):14813–14821. doi: 10.1002/jcb.28742. [DOI] [PubMed] [Google Scholar]
- 38.Corrales R.M., Narayanan S., Fernandez I., Mayo A., Galarreta D.J., Fuentes-Paez G. Ocular mucin gene expression levels as biomarkers for the diagnosis of dry eye syndrome. Invest Ophthalmol Vis Sci. 2011;52(11):8363–8369. doi: 10.1167/iovs.11-7655. [DOI] [PubMed] [Google Scholar]
- 39.Fabian T.K., Hermann P., Beck A., Fejerdy P., Fabian G. Salivary defense proteins: their network and role in innate and acquired oral immunity. Int J Mol Sci. 2012;13(4):4295–4320. doi: 10.3390/ijms13044295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.D'Souza S., Petznick A., Tong L., Hall R.C., Rosman M., Chan C. Comparative analysis of two femtosecond LASIK platforms using iTRAQ quantitative proteomics. Invest Ophthalmol Vis Sci. 2014;55(6):3396–3402. doi: 10.1167/iovs.14-14113. [DOI] [PubMed] [Google Scholar]
- 41.Soria J., Duran J.A., Etxebarria J., Merayo J., Gonzalez N., Reigada R. Tear proteome and protein network analyses reveal a novel pentamarker panel for tear film characterization in dry eye and meibomian gland dysfunction. J Proteomics. 2013;78:94–112. doi: 10.1016/j.jprot.2012.11.017. [DOI] [PubMed] [Google Scholar]
- 42.Manicam C., Perumal N., Wasielica-Poslednik J., Ngongkole Y.C., Tschabunin A., Sievers M. Proteomics unravels the regulatory mechanisms in human tears following acute renouncement of contact lens use: A Comparison between hard and soft lenses. Sci Rep. 2018;8(1):11526. doi: 10.1038/s41598-018-30032-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hill L.J., Di Pietro V., Hazeldine J., Davies D., Toman E., Logan A. Cystatin D (CST5): An ultra-early inflammatory biomarker of traumatic brain injury. Sci Rep. 2017;7(1):5002. doi: 10.1038/s41598-017-04722-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lyu Q., Ludwig I.S., Kooten P.J.S., Sijts A., Rutten V., van Eden W. Leucinostatin acts as a co-inducer for heat shock protein 70 in cultured canine retinal pigment epithelial cells. Cell Stress Chaperones. 2020;25(2):235–243. doi: 10.1007/s12192-019-01066-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu Y.C., Ang H.P., Teo E.P., Lwin N.C., Yam G.H., Mehta J.S. Wound healing profiles of hyperopic-small incision lenticule extraction (SMILE) Sci Rep. 2016;6:29802. doi: 10.1038/srep29802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Becuwe P., Ennen M., Klotz R., Barbieux C., Grandemange S. Manganese superoxide dismutase in breast cancer: from molecular mechanisms of gene regulation to biological and clinical significance. Free Radic Biol Med. 2014;77:139–151. doi: 10.1016/j.freeradbiomed.2014.08.026. [DOI] [PubMed] [Google Scholar]
- 47.McLaughlin T., Falkowski M., Wang J.J., Zhang S.X. Molecular Chaperone ERp29: A potential target for cellular protection in retinal and neurodegenerative diseases. Adv Exp Med Biol. 2018;1074:421–427. doi: 10.1007/978-3-319-75402-4_52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Aluru S.V., Agarwal S., Srinivasan B., Iyer G.K., Rajappa S.M., Tatu U. Lacrimal proline rich 4 (LPRR4) protein in the tear fluid is a potential biomarker of dry eye syndrome. PLoS ONE. 2012;7(12) doi: 10.1371/journal.pone.0051979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Jung J.H., Ji Y.W., Hwang H.S., Oh J.W., Kim H.C., Lee H.K. Proteomic analysis of human lacrimal and tear fluid in dry eye disease. Sci Rep. 2017;7(1):13363. doi: 10.1038/s41598-017-13817-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Zhou L., Beuerman R.W., Chan C.M., Zhao S.Z., Li X.R., Yang H. Identification of tear fluid biomarkers in dry eye syndrome using iTRAQ quantitative proteomics. J Proteome Res. 2009;8(11):4889–4905. doi: 10.1021/pr900686s. [DOI] [PubMed] [Google Scholar]
- 51.Al-Aqaba M.A., Dhillon V.K., Mohammed I., Said D.G., Dua H.S. Corneal nerves in health and disease. Prog Retin Eye Res. 2019;73 doi: 10.1016/j.preteyeres.2019.05.003. [DOI] [PubMed] [Google Scholar]
- 52.Lambiase A., Micera A., Sacchetti M., Cortes M., Mantelli F., Bonini S. Alterations of tear neuromediators in dry eye disease. Arch Ophthalmol. 2011;129(8):981–986. doi: 10.1001/archophthalmol.2011.200. [DOI] [PubMed] [Google Scholar]
- 53.Lambiase A., Manni L., Bonini S., Rama P., Micera A., Aloe L. Nerve growth factor promotes corneal healing: structural, biochemical, and molecular analyses of rat and human corneas. Invest Ophthalmol Vis Sci. 2000;41(5):1063–1069. [PubMed] [Google Scholar]
- 54.Mohamed-Noriega K., Riau A.K., Lwin N.C., Chaurasia S.S., Tan D.T., Mehta J.S. Early corneal nerve damage and recovery following small incision lenticule extraction (SMILE) and laser in situ keratomileusis (LASIK) Invest Ophthalmol Vis Sci. 2014;55(3):1823–1834. doi: 10.1167/iovs.13-13324. [DOI] [PubMed] [Google Scholar]
- 55.He J., Bazan H.E. Neuroanatomy and neurochemistry of mouse cornea. Invest Ophthalmol Vis Sci. 2016;57(2):664–674. doi: 10.1167/iovs.15-18019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chao C., Stapleton F., Zhou X., Chen S., Zhou S., Golebiowski B. Structural and functional changes in corneal innervation after laser in situ keratomileusis and their relationship with dry eye. Graefes Arch Clin Exp Ophthalmol. 2015;253(11):2029–2039. doi: 10.1007/s00417-015-3120-1. [DOI] [PubMed] [Google Scholar]
- 57.Liu Y.C., Jung A.S.J., Chin J.Y., Yang L.W.Y., Mehta J.S. Cross-sectional Study on Corneal Denervation in Contralateral Eyes Following SMILE Versus LASIK. J Refract Surg. 2020;36(10):653–660. doi: 10.3928/1081597X-20200730-01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.