Graphical abstract
Keywords: Ion channel receptors, Synaptic anchoring, Protein-protein interaction, Glycine receptor alpha1 subunit, Polyproline II helix, Collybistin, Gephyrin, SH3 domains, Pleckstrin homology domains
Abstract
Introduction
The inhibitory glycine receptor (GlyR), a mediator of fast synaptic inhibition, is located and held at neuronal synapses through the anchoring proteins gephyrin and collybistin. Stable localization of neurotransmitter receptors is essential for synaptic function. In case of GlyRs, only beta subunits were known until now to mediate synaptic anchoring.
Objectives
We identified a poly-proline II helix (PPII) in position 365–373 of the intra-cellular TM3-4 loop of the human GlyRα1 subunit as a novel potential synaptic anchoring site. The potential role of the PPII helix as synaptic anchoring site was tested.
Methods
Glycine receptors and collybistin variants were generated and recombinantly expressed in HEK293 cells and cultured neurons. Receptor function was assessed using patch-clamp electrophysiology, protein-protein interaction was studied using co-immuno-precipitation and pulldown experiments.
Results
Recombinantly expressed collybistin bound to isolated GlyRα1 TM3-4 loops in GST-pulldown assays. When the five proline residues P365A, P366A, P367A, P369A, P373A (GlyRα1P1-5A) located in the GlyRα1-PPII helix were replaced by alanines, the PPII secondary structure was disrupted. Recombinant GlyRα1P1-5A mutant subunits displayed normal cell surface expression and wildtype-like ion channel function, but binding to collybistin was abolished. The GlyRα1-collybistin interaction was independently confirmed by o-immunoprecipitation assays using full-length GlyRα1 subunits. Surprisingly, the interaction was not mediated by the SH3 domain of collybistin, but by its Pleckstrin homology (PH) domain. The mutation GlyRα1P366L, identified in a hyperekplexia patient, is also disrupting the PPII helix, and caused reduced collybistin binding.
Conclusion
Our data suggest a novel interaction between α1 GlyR subunits and collybistin, which is physiologically relevant in vitro and in vivo and may contribute to postsynaptic anchoring of glycine receptors.
Introduction
The inhibitory glycine receptor (GlyR), a pentameric ion channel belonging to the Cys-loop receptor family is predominantly expressed in mammalian spinal cord and brain stem [1], [2]. Each GlyR subunit is characterized by a large extracellular domain, four transmembrane domains (TM1-4) and a large TM3-4 loop followed by a short extracellular C-terminus. GlyR alpha subunits (α1-4) can form functional, homomeric channels, while the beta subunit is associated with intracellular anchoring [1], [2]. The GlyR beta subunit was shown to bind gephyrin, a synaptic protein that anchors GlyRs and GABAARs to the subsynaptic cytoskeleton and mediates tethering of glycine [3], [4], [5], [6] and GABAA [7], [8], [9], [10] receptors at postsynaptic membranes. Since GlyR α subunits failed to bind recombinant gephyrin, it was concluded that the beta subunit is necessary for successful clustering [3], [7], [11], [12]. Screening approaches to identify gephyrin-binding proteins revealed the brain-specific GDP/GTP exchange factor collybistin as a gephyrin binding partner [4], [13], [14], [15]. Gephyrin was shown in vitro to bind simultaneously to both collybistin and the GlyR β-subunit binding motif [16], however direct interactions between gephyrin and GlyR alpha subunits have not been observed [14], although mutations in the GlyRα1 subunit were shown to affect receptor surface expression [17], [18].
Subunit dependent GABAAR binding to gephyrin is weak to intermediate, while the interaction between gephyrin and GlyRβ was shown to be strong [11], [19]. Formation of complexes between GABAAR α2 subunits and collybistin was reported [20], and the role of gephyrin in the stabilization of GABAergic and glycinergic synapses is well documented [3], [7], [10], [21]. A critical interaction partner of gephyrin at inhibitory synapses is the GTP/GDP exchange factor collybistin [14], [15], [22]. A recent study defines a collybistin-based network of protein interactions that controls the gephyrin content of inhibitory postsynapses [14], [23], [24]. There, collybistin can adopt open/active or closed/inactive conformations to act as a switchable adaptor that links gephyrin to plasma membrane phosphoinositides [23], [24]. This function of collybistin is regulated by binding of the adhesion protein neuroligin-2, which stabilizes the open/active conformation of collybistin at the postsynaptic plasma membrane [24].
A polyproline II (PPII)-helix in the GlyRα1 subunit TM3-4 loop was discovered by secondary structure analysis using circular dichroism (CD) spectroscopy of reconstituted [25] and recombinant [26] GlyRs. While GlyRα3 subunits lack the PPII motif, GlyRβ as well as α2 subunits both contain a polyproline motif of related sequence at this position. PPII conformations consisting of a homopolymer of proline residues were first discovered in 1968 [27]. They resemble single-stranded collagen triple helices, comprising 4–8 proline, hydroxyproline and glycine residues. The PPII helix structure is characterized by the lack of any intra- or intermolecular hydrogen bonds that are present in α-helices and β-sheets [28]. In general, PPII helical structures play an essential role in mediating protein-protein interactions [29], [30], [31]. Binding partners of PPII helices include SH3 [32], and EVH-1 domains [31], [33].
Here, we show that a PPII helix in the glycine receptor α1 intracellular loop interacts with collybistin. An isolated TM3-4 polypeptide is sufficient to pull down the heterologously expressed collybistin splice variants 1 (SH3+) and 2 (SH3-) indicating that this interaction is not mediated via the SH3 domain. Replacement of five neighbouring proline residues (P365A, P366A, P367A, P369A, P373A) by alanine, generating the GlyRα1P1-5A mutant, interfered with collybistin binding. Immunoprecipitation experiments from HEK293 cells expressing GFP-CB2 or an isolated GFP-CB-PH domain confirmed GlyRα1 - collybistin association, suggesting the C-terminal PH domain as critical binding site. Notably, expression of full length GlyRα1, carrying the GlyRα1P1-5A group mutation did not affect subcellular localization or functional GlyR properties. Our data suggest that the PPII motif may play a relevant role in synaptic anchoring of α1 GlyRs.
Materials and methods
Constructs and mutagenesis
Single-nucleotide exchanges were introduced by PCR-mediated site-directed mutagenesis using an overlap extension PCR approach. Mutagenesis primers (MWG, Ebersberg, Germany) contained nucleotides encoding for specific amino acid exchanges. PCRs were set up as previously described [34]. For GlyRα1 mutations GlyRα1P1-5A and α1P366L [18], PCR products were inserted into the plasmid pRK5 using EcoRI and PstI restriction sites. For bacterial expression and purification of GlyRα1 TM3-4 loop constructs, PCR products were inserted into the plasmid pET30a (Novagen, Darmstadt, Germany) using BamHI and HindIII restriction sites. All clones were verified by DNA sequencing (MWG, Ebersberg, Germany). The cDNA of HA-GlyRα1 fusion protein was generated by insertion of annealed oligonucleotides including a Bsp1407I overhang into pCIS- GlyRα1. The cDNAs encoding full-length collybistin II or the collybistin PH-domain were amplified and subcloned as XhoI fragment or EcoRI -XhoI fragment, respectively (pAcGFP-C vector, Clontech, Mountain View, CA, USA).
Expression and purification of fusion proteins in E. coli
After transformation and expressi-on of GlyRα1 TM3-4 loop constructs, BL21 cells (Novagen, Darmstadt, Germany) were harvested by centrifugation, resuspended in 50 mM Tris-HCl, 2.5 mM EDTA, pH 7.4, treated with lysozyme (0.1 mg/ml, 30 min, 0 °C) and sonicated on ice. Sonification was repeated and the supernatant collected for purification. Native purification was performed using a Ni-agarose column by washing and eluting with increasing imidazole concentrations, followed by size exclusion chromatography. Successful purification was verified on Coomassie stained SDS PAGE gel electrophoresis.
CD Spectroscopy
Purified intracellular GlyR domains, expressed in E. coli were buffered in 10 mM K-Pi pH 7.4 and subjected to CD analysis. Measurements were performed on a JASCO-J810 spectrometer (JASCO, Gross-Umstadt, Germany) in a 0.1 cm analytical cell chamber. All spectra were baseline corrected by subtracting buffer runs. Eight individual scans were taken at 22 °C in a range of 320 to 185 nm, with a 0.5 nm step size and averaged. Protein concentrations were determined by measuring the absorbance at 280 nm using the equation c = A280 nm/(e*L). The path length L was 1 cm, the extinction coefficients were calculated from the protein sequence [26]. While α-helical structure elements lead to distinct CD spectra with negative maxima at 222 nm and 208 nm, spectra obtained from β-sheets reveal a broad negative maximum at 215 nm [35]. The left-handed type II polyproline (PPII) helix yields spectra with an intense negative band at 204 nm [35], [36], [37].
Cell Culture, transfection and membrane preparation
HEK293 cells were grown in 10 cm tissue culture Petri dishes in Minimum Essential Medium (MEM, Sigma, Deisenhofen, Germany) supplemented with 10% FBS (Invitrogen, Karlsruhe, Germany) and Penicillin/Streptomycin at 5% CO2 and 37 °C in a water saturated atmosphere. For electrophysiological experiments, cells were plated on poly-lysine treated glass coverslips in 6 cm dishes. For Western blot analysis, cells were prepared in 10 cm dishes. Transfection was performed 1 day after cell passage using the Ca2+ phosphate method. 10 µg plasmid DNA, 10 µg GFP vector, 430 µl H2O and 50 µl CaCl2 (2.5 M) were mixed and 500 µl HBS-buffer (50 mM HEPES, 12 mM Glucose, 10 mM KCl, 280 mM NaCl, 1.5 mM Na2HPO4 × 12 H2O, pH 6.95) was added drop wise. After a 20 min incubation period, the transfection mixture was added to the cells. Cells were washed 1 day later with MEM full medium and harvested 24–48 h later. Briefly, cells were taken in ice cold PBS buffer, centrifuged (10 min, 2000 g, 4 °C) and pellets were taken in buffer H (20 mM K-phosphate pH 7.4, 5 mM EDTA, 5 mM EGTA, complete protease inhibitor cocktail (Roche, Penzberg, Germany)) and homogenized. Resulting suspension supernatants were centrifuged (20 min, 20,000 rpm, 4 °C), the pellets resuspended in Buffer H, containing 200 mM KCl and further processed for Western blotting.
Western blot analysis
After SDS PAGE gel electrophoresis proteins were transferred onto nitrocellulose membrane using the semi-dry method. Membranes were blocked with TBB solution (50 mM Tris-HCl pH 8.0, 150 mM NaCl, 0.5% TritonX-100, 3% bovine serum albumin (BSA)). Primary antibodies were applied for 60 min. Secondary antibodies were incubated for 30 min in the dark. After washing, proteins labelled by the secondary antibody were detected on a Storm 860 Fluoroimager (Molecular Dynamics, Krefeld, Germany). Primary antibodies: monoclonal mouse anti GlyR mAB4a (hybridoma cells supernatant 1:1); secondary antibody: goat anti mouse IgG-Cy3 (1:200) (Dianova, Hamburg, Germany).
Immunocytochemistry on HEK293 cells
All steps were performed at room temperature. Fixation: coverslips containing transfected HEK293 cells were transferred into a 24 well plate containing 500 μl medium per well. The medium was exchanged by a 4% paraformaldehyde-PBS solution. After a 10 min incubation period, cells were washed twice with cold PBS buffer. Staining of surface proteins: cells were blocked with PBS buffer including 5% BSA for 30 min; intracellular staining: cells were blocked with PBS buffer including 5% BSA and 0.1% Triton X-100. After blocking, primary antibodies (mAb4a dilution 1:200 in PBS, cat. no. 146011; Synaptic Systems, Göttingen, Germany) were applied for 60 min. Cells were washed with PBS. The secondary antibody (goat-anti-mouse IgG-Cy3, 1:250 in PBS, Dianova, Germany) was applied for 60 min. After washing, cells were analysed using a Zeiss LSM5 Pascal confocal laser scan microscope (Carl Zeiss, Jena, Germany), and Zeiss LSM Image Browser software (Carl Zeiss, Jena, Germany).
Hippocampal cultures
Hippocampal neurons were prepared from mouse embryos of the CD1 strain at stage E18 (male and female embryos were taken) and grown in neurobasal medium (Thermo Fischer, Darmstadt, Germany) containing 5 ml of L-glutamine (200 mM) and B27 supplement (Thermo Fischer, Darmstadt, Germany) with an exchange of 50% medium after 7 days in culture. Experiments were authorized by the local veterinary authority and Committee on the Ethics of Animal Experiments (Regierung von Unterfranken).
Hippocampal neurons were infected with 1 µg lentiviral low copy vector FUVal-GFP (provided by R. Blum) containing the cDNA of GlyRα1 wildtype or GlyRα1P366L after seeding and cultured for 14 days (days in vitro = DIV). At DIV15 live cell stainings of surface GlyRs were performed using an α1-specific mAb2b antibody (recognizes a native epitope in the N-terminus of GlyRα1 (residues 1–10 of mature protein; cat. no.146111, SYSY, Göttingen, Germany) for 2 hrs at 4 °C. After 20 min of fixation (4% paraformaldehyde, 4% sucrose) cells were blocked and permeabilized with 5% goat serum and 0.1% Triton for 30 min. Endogenous collybistin was stained overnight with the rabbit anti-collybistin antibody at 4 °C. Subsequently, cells were washed and incubated with goat-anti-mouse-Cy3 and goat-anti-rabbit Cy5 secondary antibodies (Dianova, Hamburg, Germany) for 2 hrs at 21 °C. Before mounting of the coverslips with mowiol, nuclear staining using DAPI was performed.
Confocal microscopy, image acquisition and analysis on Hippocampal Neurons
An inverted Olympus IX81 microscope equipped with an Olympus FV1000 confocal laser scanning system, a FVD10 SPD spectral detector and diode lasers of 495 nm (Alexa488) and 550 nm (Cy3) (Olympus, Tokyo, Japan) was used to acquire confocal images. To take images an Olympus UPLSAPO 60x (oil, numerical aperture: 1.35) objective was used. The whole cell collybistin signal intensity was analyzed from infected cells (GFP positive) and discriminated between cell soma and neurite using the Open View software [38]. The mAb2 channel was used as a mask (cluster) and the collybistin signal intensity was determined within the mAb2b clusters. All immunofluorescence analysis is shown as means ± standard errors of the mean (SEM). Calculated signal intensities were compared using ANOVA with a probability of error of p < 0.05 considered significant. The images were further developed and organized by Adobe Photoshop (Adobe, San Jose, CA, USA) or ImageJ (1.51)/Fiji (https://imagej.net/ImageJ).
Electrophysiological recordings and data analysis
HEK293 cells were transfected 2–4 days prior to electrophysiological recordings. Current responses were measured at room temperature (21–23 °C) at a holding potential of −50 mV. Whole-cell recordings were performed using a HEKA EPC10 amplifier (HEKA Electronics, Lambrecht, Germany) controlled by Pulse software (HEKA Electronics). Recording pipettes were pulled from borosilicate glass (World Precision Instruments, Berlin, Germany) using a Sutter P-97 horizontal puller (Sutter, Novato, CA). Solutions were applied using an Octaflow system (NPI electronics, Tamm, Germany), where cells were bathed in a laminar flow of buffer, giving a time resolution for solution exchange and re-equilibration of about 100 ms. The external buffer consisted of 135 mM NaCl, 5.5 mM KCl, 2 mM CaCl2, 1.0 mM MgCl2, and 10 mM Hepes (pH 7.4, NaOH); the internal buffer was 140 mM CsCl, 1.0 mM CaCl2, 2.0 mM MgCl2, 5.0 mM EGTA, and 10 mM Hepes (pH 7.2, CsOH). Using a nonlinear algorithm in Microcal Origin (Additive, Friedrichsdorf, Germany), dose-response data were fitted to the Hill equation where IGly is the current amplitude at a given glycine concentration, Isat is the current amplitude at saturating concentrations of glycine, EC50 is the glycine concentration at half-maximal current responses, and nH is the Hill coefficient. In all experiments EC50 values were determined for each individual cell from a non-linear fit of dose response data to the logistic equation (above). Differences between EC50 values recorded for wildtype and GlyRα1P1-5A mutant receptor or co-expression of wildtype and collybistin were compared using one-way ANOVA with p ≤ 0.05 taken as significant. Significance levels are indicated with p- and F-values. An unweighed average ± standard error (SEM) was calculated from all individual EC50 values, without considering the fitting errors.
Fusion constructs and GST Pulldown assays
HEK293 cells were transfected with HA-tagged collybistin I or II cDNAs [14] using the calcium phosphate precipitation method. 16 hrs after transfection cells were washed with PBS and harvested in 1 ml PBS supplemented with 1% Triton X-100 and 1 mM PMSF. After 30 min incubation on ice, the lysate was centrifuged at 1,000 × g for 5 min and the supernatant was retained. GST-fusion proteins of the isolated GlyRα1 wildtype or mutant GlyRα1P1-5A TM3-4 loop were recombinantly expressed in E. coli BL21 cells (New England Biolabs Inc. Ipswich, MA). Cells were pelletized and lysed by sonification in protein extraction buffer (400 mM NaCl, 100 mM HEPES (pH 7.9 KOH), 10% (v/v) glycerol). After centrifugation at 10,000g for 30 min, bacterial lysates were coupled to glutathione sepharose beads (Thermo Scientific, Dreieich, Germany) for 3 hrs. Beads were washed 3 times with washing buffer (50 mM Tris (pH 7.5), 150 mM NaCl, 5 mM MgCl2) and subsequently incubated with HEK293 cell lysates for 10–12 hrs. Beads were then washed 5 times with washing buffer for 5 min, resolved with PBS and boiled in SDS sample buffer. After SDS PAGE, proteins were transferred onto PVDF membranes using the wet blot method. Membranes were blocked with rabbit anti-HA antibody (Sigma-Aldrich, Taufkirchen, Germany) at 4 °C in milk/TBSB. The secondary antibody (donkey anti rabbit HRP, (Dianova, Hamburg, Germany)) was incubated for 1 h at room temperature in milk/TBST. After incubation with ECL substrate, signals were detected by ChemoCam ECL detection system (Intas GmbH, Göttingen, Germany).
Co-immunoprecipitation
GlyRα1 – GFP-CB2/ GFP-PH domain
For co-immunoprecipitation experiments, HEK-293 cells were cotransfected with GFP-CB2 or GFP-PH domain, respectively, together with pCIS-HA-GlyRα1 using the calcium phosphate method. 48 hrs after transfection, HEK293 cells were washed with PBS and harvested in 2 ml PBS, supplemented with 1% Triton, complete protease inhibitor cocktail (Roche, Mannheim, Germany) and 1 mM PMSF (Carl Roth, Karlsruhe, Germany). After 30 min incubation lysates were clarified by centrifugation at 1000g for 5 min. After coupling magnetic Protein G beads (Thermo Scientific, Dreieich, Germany) with 5 µg mouse anti-HA antibody (Roche, Mannheim, Germany) or mouse IgG (Sigma-Aldrich, Taufkirchen, Germany) for 4 hrs, HEK293 cell lysates were incubated with antibody coupled beads overnight at 4 °C. Beads were washed six times with IP wash buffer (50 mM Tris, 500 mM NaCl, 5 mM MgCl2, 1% Triton X-100, 1% Nonidet NP-40, pH 7.5), resolved in PBS and boiled in SDS sample buffer. After SDS PAGE proteins were transferred onto PVDF membrane using wet blot method. Membranes were blocked with milk/TBST. Secondary antibody goat anti chicken HRP (Dianova, Hamburg, Germany) was incubated 1 h at room temperature in milk/TBST. After incubation with ECL substrate signals were detected by ChemoCam ECL detection system.
GlyRα1 – CB2; GlyRα1-β – CB2 – Gephyrin
HEK293 cells were grown on 10-cm dishes and used 48 hrs after transfection. Cells were transfected with (i) GlyRα1 together with collybistin I (CB1) or collybistin II (CB2) in a ratio of 1:1, (ii) GlyRα1 with GlyRβ, CB2, and gephyrin in a ratio of 1:2:2:2 (or mutant GlyRα1P366L was used instead of GlyRα1 wildtype; same ratio transfected) (Fig. 5C). GFP transfections were used as mock control. After washing the cells once with PBS, 1 ml of lysis buffer (Cytobuster Protein Extraction Reagent, Merck Millipore, Darmstadt, Germany) was added to each plate to scrape off the cells following the manufacturer’s protocol. After centrifugation (5 min, 4 °C, 13500 × g) the supernatant (lysate) was used as input for co-immunoprecipitation experiments. Protein concentrations were measured with the Bradford protein assay (Bio-Rad Laboratories, Munich, Germany). To 300 µg of the respective protein solution, mAb2b antibody (1:200; cat. no.146111, Synaptic Systems, Göttingen, Germany) and 50 µl of protein A-sepharose beads (1:1, GE Healthcare, Freiburg, Germany) were added and agitated overnight at 4 °C. Following removal of the supernatant, beads were washed three times with lysis buffer. Proteins were eluted from beads by incubating in 30 µl of 2× SDS sample buffer at 95 °C for 7 min. Proteins were separated on 11% SDS gels and stained after Western blotting with the polyclonal rabbit anti-collybistin antibody (1:1000, cat. no. 261003, Synaptic Systems, Germany) to identify coprecipitated collybistin. Input controls were performed for collybistin, gephyrin and GlyRα1 (polyclonal gephyrin antibody 1:500, cat. no. 147008; polyclonal rabbit anti-GlyRα1 antibody 1:500, cat. no. 146003, both from Synaptic Systems, Göttingen, Germany). GAPDH (glyceraldehyde-3-phosphate dehydrogenase) was used as housekeeping protein (1:1000 monoclonal anti-GAPDH antibody, cat. no. CB1001, EMD Millipore, San Diego, CA, USA). For statistical analysis collybistin signals were normalized to GlyRα1 input signals and adjusted to wildtype GlyRα1. Calculated data were compared using ANOVA with a probability of error of p < 0.05 considered significant.
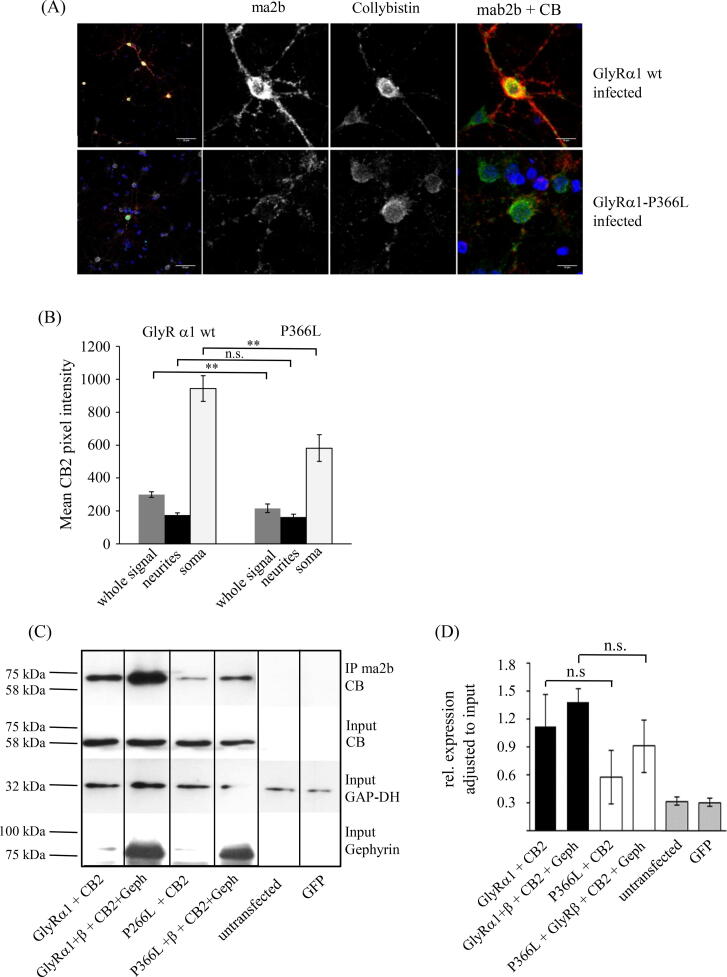
Fig. 5.
Endogenous collybistin colocalizes with GlyRα1 in primary murine neurons. (A) Hippocampal neurons were infected with a lentivirus encoding either GlyRα1 wildtype or a pathological GlyRα1P366L variant carrying a mutation in the PPII helix. At DIV15, cells were co-stained for GlyRα1 (mAb2b, 1:500, red) and endogenous collybistin (polyclonal rabbit anti-collybistin antibody, 1:500, green). Note, infected cells were controlled by bidirectional GFP expression. Hence, collybistin was stained with the secondary goat-anti-rabbit Cy5 antibody and is shown in false color. Nuclear DAPI staining is shown in blue. White bar in left overview panels refers to 50 µm, white bar in zoomed pictures (second to fourth lane) refers to 10 µm. (B) Quantification of the collybistin intensities determined in mAb2b (GlyRα1) clusters. Mean intensities are shown for whole cells, neurites and soma. P-values to represent level of significance are indicated **p < 0.01, n.s. = non-significant. (C) Co-immunoprecipitation of the GlyRα1 wildtype and the pathological GlyRα1P366L variant with collybistin following overexpression in HEK293 cells. Both variants were expressed with CB2 (ratio 1:1) only to detect direct interaction and co-transfected with the GlyRβ subunit and gephyrin (ratio 1:2:2:2) to detect if these structural GlyR complex proteins promote interaction with collybistin. Precipitated collybistin is shown at the appropriate molecular weight of 60 kDa in the upper panel, collybistin input second panel, GAP-DH (32 kDa) input control third panel, gephyrin (93 kDa) expression lower panel. Note, although the collybistin expression is similar (input), collybistin precipitated more efficiently in the presence of GlyRβ and gephyrin (abbreviation Geph). (D) Quantification of relative collybistin expression in the presence of collybistin only or together with GlyRβ and gephyrin. Four independent experiments have been performed and were used for analysis; n.s. non-significant.
Experimental design and statistical analysis
All experiments were performed as at least three independent biological replicates (unless stated otherwise). The number of experiments or cells recorded for analyses are presented in the result session referring to the appropriate experiments.
Quantification of GlyR and GlyR-derived protein: mAb4A signals for each clone were normalized to pan-cadherin signals and adjusted to α1 wildtype (wt = non-mutated full-length GlyRα1 protein). Calculated data were compared using one-way ANOVA (Microcal Origin, Additive, Karlsruhe, Germany) with a probability of error of p < 0.05 considered significant. For statistical analysis of binding data from infected hippocampal neurons, signals were normalized to GlyRα1 input signals and adjusted to wildtype. Calculated data were compared using ANOVA with a probability of error of p < 0.05 considered significant.
Quantification of data obtained from electrophysiological recordings: when datasets were compared one-way ANOVA was performed with a probability of error of p < 0.05 considered significant.
Results
GlyRα1 subunits carrying a proline to alanine group mutation in the PPII helix of the TM3-4 loop lose PPII helix conformation but retain cell surface membrane expression
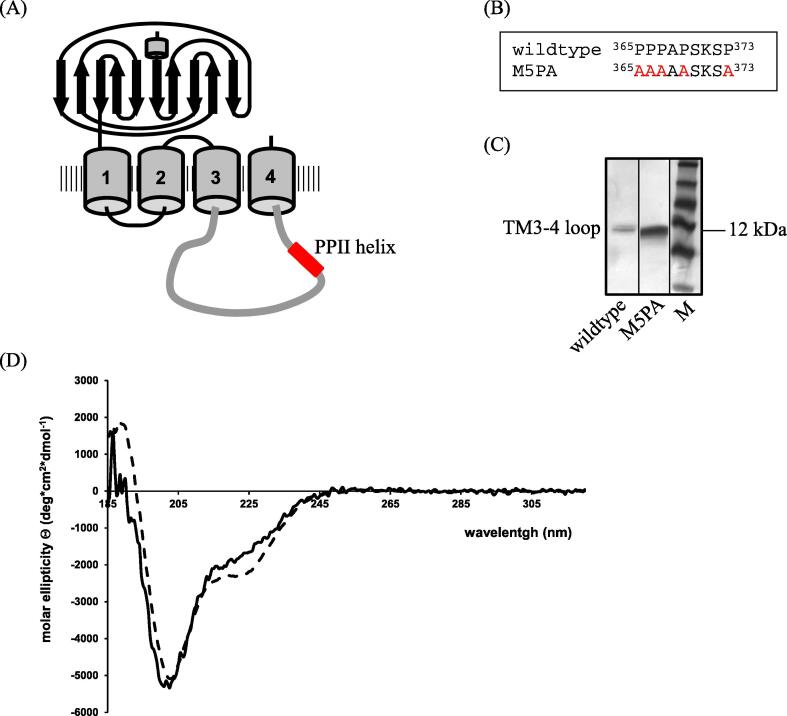
A polyproline II (PPII) helix within the GlyRα1 TM3-4 loop (Fig. 1A) has been demonstrated via CD spectroscopy [26], [28], [30]. Within this loop the motif 365PPPAPSKSP373 is thought to mediate its secondary structure (Fig. 1B). To investigate the influence of the PPII helix on receptor targeting and localization at the cell surface, proline residues located between amino acid positions 365 and 373 were systematically substituted to alanine. Mutant GlyRα1P1-5A is characterized by the simultaneous exchange of five proline to alanine residues within the helix motif: P365A, P366A, P367A, P369A and P373A (Fig. 1B).
Fig. 1.
Localization and characterization of the GlyRα1 PPII helix. (A) Model of GlyRα1 with its predicted PPII helix in the TM3-4 loop (adapted from [44]. (B) Sequence of wildtype GlyR in comparison with group mutant at position 365–373. (C) SDS-PAGE of GlyRα1 as well as GlyRα1P1-5A mutant TM3-4 loops after over-expression in E. coli cells and purification via Ni-NTA column and gel filtration chromatography. Lane 1: α1 wildtype TM3-4 loop; lane 2: GlyRα1P1-5A TM3-4 loop; lane 3: protein standard. Position of TM3-4 loop and 12 kDa is indicated. (D) CD spectra of α1-wt (solid line) and GlyRα1P1-5A (dashed line). All spectra were measured in 10 mM K-phosphate, pH 7.4 in a 1 cm cuvette at 22 °C. Eight single spectra were summed and the reference spectrum (10 mM K-phosphate, pH 7.4) was subtracted.
Structural studies of the PPII helix required pure protein samples of the isolated TM3-4 loop. Expression of the wildtype or mutant hs α1 TM3-4 loop polypeptide in E. coli cells and subsequent purification using a Ni-agarose column and size exclusion chromatography yielded pure protein samples, as visualized by SDS PAGE Coomassie Blue staining (Fig. 1C). Purified TM3-4 loops of mutant and wildtype receptor were subjected to CD analysis. Wildtype spectra displayed a minimum at 205 and a shoulder at 225 nm, typical for structures containing β-sheets and PPII helices (Fig. 1D), and consistent with previous reports [28], [36]. The GlyRα1P1-5A mutant, in contrast, had the minimum shifted to 208 nm, and the shoulder in the 225–228 nm region became more pronounced, presenting as a new minimum. These differences are in agreement with a loss of the PPII helical conformation and an increase of the α-helical content of the protein.
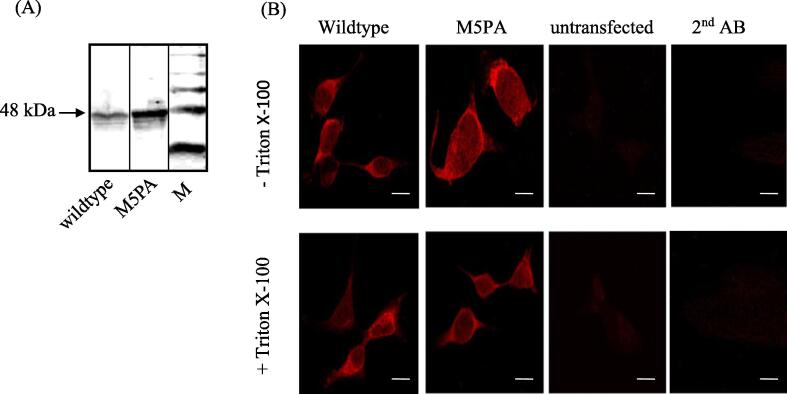
We then examined plasma membrane targeting of the full length GlyR group mutant GlyRα1P1-5A upon expression in HEK293 cells, using Western blot analysis (Fig. 2A). Here, the mutant receptor was present in the membrane fractions enriched for the plasma membrane. Immunocytochemistry also confirmed the delivery of mutant receptors to the cell surface similar to the wildtype (Fig. 2B), suggesting normal cell surface delivery of GlyRα1P1-5A mutant receptors.
Fig. 2.
Expression and cellular distribution of GlyRα1 wildtype and GlyRα1P1-5A mutant subunits. (A) Western blot analysis of GlyRα1 subunits. 20 μg of membrane preparation was loaded per lane, primary antibody was mAb4a supernatant. Lane 1: α1 wildtype; lane 2: GlyRα1P1-5A; lane 3: protein standard; 48 kDa is indicated. (B) Immunofluorescence: HEK293 cells were transfected on cover slides and treated for immunocytochemistry with mAb4a and goat anti mouse Cy3. For surface expression no Triton X-100 was added. To detect the intracellular protein distribution, the cell membrane was permeabilized with Triton X-100. Controls: untransfected cells or GlyRα1 transfected cells treated with secondary antibody only are shown. Pictures were taken at 400× magnification. Scale bar indicates 10 µm.
GlyRα1 subunits carrying a proline to alanine group mutation in the PPII helix of the TM3-4 loop display normal glycinergic currents
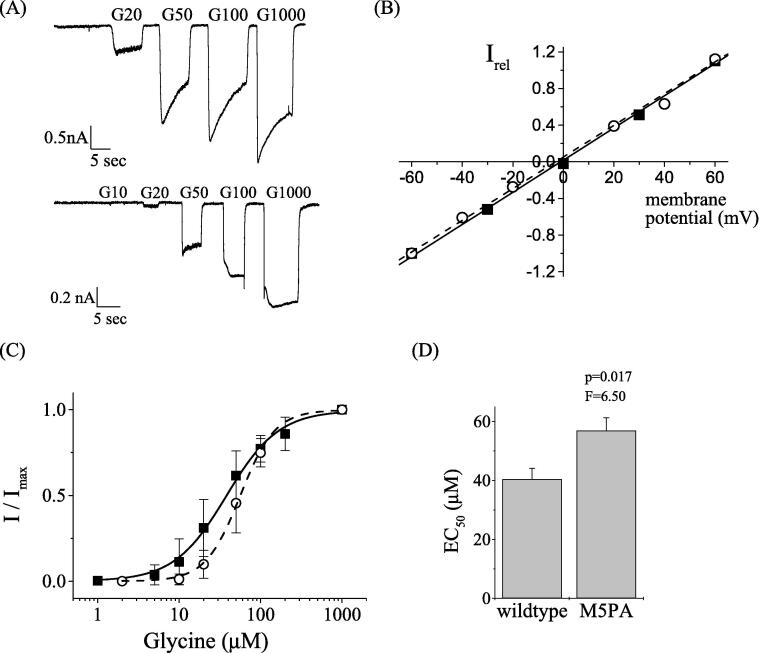
To study the role of the mutant GlyRα1P1-5A on receptor function, electrophysiological experiments were performed on homomeric full length GlyRs carrying the wildtype GlyRα1 subunit, as compared to GlyRs carrying the α1 group mutant GlyRα1P1-5A (Fig. 1B). To this end HEK293 cells were transfected with the respective constructs and current responses analysed at increasing glycine concentrations of 10–1000 µM. GlyRα1 wildtype receptors showed EC50 values of 38.4 ± 3.4 μM (n = 20), while receptors carrying the GlyRα1P1-5A group revealed EC50 values of 56.9 ± 4.4 μM (n = 9) (Fig. 3A-B, D). These data show that exchanging all proline residues to alanine in the intracellular PPII helix does not affect channel functions of homomeric α1 GlyRs. Although the increase of EC50 was statistically significant (p = 0.016), a physiologically active mutant channel was retained. We therefore compared the dependency of current responses to the transmembrane voltage for both subunit species at saturating concentrations of glycine (2 mM) in the range of −60 mV to +60 mV (Fig. 3C). Both, wildtype and mutant receptors displayed a linear I-V relationship, as expected for inhibitory GlyRs. These data suggest that ion conductance of the GlyR channel is independent of the polyproline motif located in the GlyRα1 subunit TM3-4 loop.
Fig. 3.
Patch-clamp electrophysiological characterization of homomeric GlyRα1 wildtype and GlyRα1P1-5A mutant receptors. (A) Current responses of human wildtype GLRA1 (hs GlyRα1-wt) and mutant receptors GlyRα1P1-5A. Whole-cell patch clamp currents were recorded from transfected HEK293 cells at glycine concentrations between 10 and 1000 μM and a membrane potential of −50 mV. (B) Dose response curve of α1-wt (solid squares, solid line) and GlyRα1P1-5A (open circles and dashed line). EC50 values were 38.4 ± 3.4 μM for α1-wt (n = 20) and 56.9 ± 4.4 μM (n = 9) for the mutant receptor. Hill constants were 1.9 ± 0.1 and 2.0 ± 0.1 for wildtype and mutant, respectively. (C) Current potential curves were measured at 2 mM glycine in a potential range of −60 mV to +60 mV. Current responses were normalized to maximal currents at −60 mV. (D) Comparison of wildtype and mutant receptor dose responses; the difference was statistically significant (p = 0.016).
Proline residues in the PPII helix of an isolated GlyRα1 TM3-4 loop are critical for interaction with collybistin
The PPII motif of the GlyR belongs to the class II type of polyproline 2 helices. Some PPII helices have been described to preferentially interact with SH3 domains of specific target proteins [30], [39], [40]. We therefore applied GST assays to select for different candidate SH3 domains containing class II PPII helices that have been described in the literature and may be potential GlyR-interacting domains. In addition, a search for potential SH3 domains was performed using the iSPOT database (http://cbm.bio.uniroma2.it/ispot) [41]. Nine promising candidates (T7-CRK-A, T7-SRC8, T7-YES, T7-PI3K, T7-Grb2-A, T7-Grb2-B, T7-NCK1-A, T7-NCK1-B, T7-NCK1-C) for binding to the GlyR motif 365PPPAPSKSP373 were tested, however, none did lead to detectable interactions (data not shown).
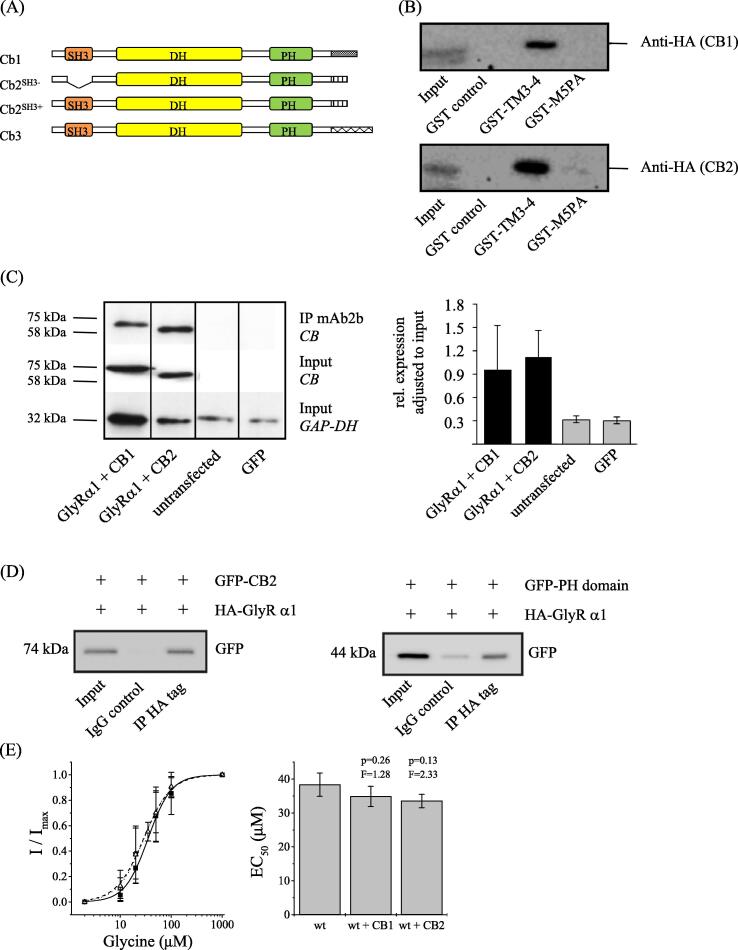
Further GST-pulldown assays with the isolated wildtype GlyRα1 TM3-4 loop revealed interaction with heterologously expressed full length collybistin I (CB1SH3+), a protein located at inhibitory postsynaptic sites, which is known to harbour an N-terminal SH3 domain (Fig. 4A-B). However, as a collybistin II splice variant lacking the SH3 domain (CB2SH3-), was also pulled-down (Fig. 4B) it is likely that other protein domains, rather than the SH3 motif mediate binding to the GlyR loop.
Fig. 4.
Interaction between collybistin and the GlyRα1 TM3-4 loop. (A) Schematic structure of collybistin variants 1–3. (B) Binding studies between α1-GlyR and collybistin I-II using GST pull down assay upon HEK293 cell transfection with CB1SH+-HA and CB2SH--HA. GST-fusion proteins of the isolated GlyRα1 wildtype or GlyRα1P1-5A TM3-4 loop were recombinantly expressed in E. coli BL21 cells, coupled to glutathione-sepharose beads and finally incubated with HEK293 cell lysates. Results are shown after Western Blot analysis. Lane 1: input, lane 2: GST control, lane 3: α1-wt TM3-4 loop, lane 4: GlyRα1P1-5A TM3-4 loop. C Co-immunoprecipitation of glycine receptors and collybistin splice variants. The GlyRα1-specific antibody mAb2b was used for co-precipitation. Lane 1: GlyRα1 + CB1 (1:1); lane 2: GlyRα1 + CB2 (1:1); lane 3: GlyRα1 + GlyRβ + CB2 + gephyrin (1:2:2:2); lane 4: untransfected cells; lane 5: GFP = mock control (left panel). Collybistin was detected at the appropriate molecular weight of 60 kDa (precipitated with GlyRα1 – see IP, upper panel and input control expression of collybistin - see input second panel; gephyrin at 93 kDa, and GAP-DH at 32 kDa. The observed shift in molecular weight between CB1 and CB2 is due to the presence of the SH3 domain in CB1 but not CB2. Right panel: Quantification of the relative collybistin expression normalized to GAPDH. At least 4 independent experiments have been performed and were used for quantification analysis. (D) Co-immunoprecipitation of GFP-CB2 or GFP-PH domain together with pCIS-HA-GlyRα1. Beads were coupled with mouse anti-HA antibody or mouse IgG, HEK293 cell lysates were incubated with antibody coupled beads (see methods). Samples were subjected to SDS PAGE and Western Blotting. Lane 1: input; lane 2: IgG control; lane 3: IP HA tag. Left panel: GFP-CB2; right panel: GFP-PH domain. (E) Electrophysiological data after co-expression of GlyRα1 with CB1 or CB2. Left panel: EC50 curve of GlyRα1 (solid squares, solid line); GlyRα1 co-expressed with CB1 (open circles, dashed line) and GlyRα1 co-expressed with CB2 (open triangle, dotted line). Right panel: comparison of GlyRα1 and co-transfection with CB1 and CB2. Differences were not significant (p > 0.05, one-way ANOVA).
Notably, HEK293 cell pulldowns using mutant GlyRα1P1-5A TM3-4 loop (compare with Fig. 1B) abolished binding of both collybistin variants, indicating a critical involvement of the GlyRα1 polyproline helix. In spite of endogenous expression of gephyrin in HEK293 cell, cells are unlikely to endogenously express a GlyRβ subunit. Our data therefore suggest that GlyRα1-collybistin interactions may occur in addition to GlyRβ- gephyrin interactions or may be an alternative way to anchor homomeric glycine receptors. Next, we investigated the binding of full length GlyRα1 wildtype using immunoprecipitation upon HEK293 cell expression. Co-expression experiments of GlyRα1 and CB1 or CB2 displayed strong collybistin signals detected in Western blot analysis, confirming the interaction of GlyRα1 and collybistin variants CB1 and CB2 (Fig. 4C). GAPDH was used as a loading control.
We then tested whether the C-terminal collybistin PH domain would be sufficient for GlyRα1-collybistin binding. Therefore, we heterologously co-expressed full-length wildtype HA-GlyRα1 with either GFP-CB2 or an isolated CB2-PH domain fused to GFP. HA specific antibodies led to co-IP of full length CB2. In addition, an isolated CB2-PH domain fusion protein also revealed interaction with the full-length wildtype HA-GlyRα1 subunit (Fig. 4D), suggesting PH domain residues as critical mediators of GlyRα1 binding.
The influence of collybistin on glycinergic currents was tested in patch-clamp experiments. To this end, GlyRα1 subunits were co-expressed with either CB1 or CB2 in HEK293 cells. Whole cell recordings displayed no significant differences of half maximum currents. EC50 values were 34.8 ± 3.0 μM and 31.5 ± 2.2 μM for cells co-expressing CB1 or CB2, respectively, as compared to 38.4 ± 3.4 μM in wildtype controls in the absence of collybistin (Fig. 4E).
Finally, we compared a pathological GlyRα1 mutant, GlyRα1P366L, which had been identified in a hyperekplexia patient [18] to the wildtype receptor. Immunocytochemistry experiments on hippocampal neurons (DIV15) revealed co-assembly of GlyRα1 and collybistin, which was reduced in mutant GlyRα1P366L receptors (Fig. 5A). Collybistin intensities adjusted to a GlyRα1 (mAb2b mask) were determined for whole cells and differentiated between signal in neurites and soma (Fig. 5B). While no significant difference was detected between wildtype and mutant receptors in neurites, differences in whole cell signal and more dominant in soma were detected (**p < 0.01), indicating that exchange of only one proline residue in the PPII helix motif into the bulky leucine is sufficient to weaken the binding between GlyRα1 and collybistin (Fig. 5B). In addition, co-immunoprecipitation experiments were performed on wildtype GlyRα1 and GlyRα1P366L receptors. Co-expression of GlyRα1 and CB2 revealed strong CB signals in Western blot analysis upon immunoprecipitation with mAb2b, which was only slightly increased in co-expression experiments with GlyRα1, GlyRβ, CB2 and gephyrin (Fig. 5C-D). When co-expression experiments were performed on GlyRα1P366L receptors, the relative signal adjusted to input was reduced to 0.6 for the mutant compared to 1.1 for wildtype GlyRα1. Co-expression of GlyRα1, GlyRβ, CB2 and gephyrin resulted in a reduction of similar extent of relative signal intensity for the GlyRα1P366L mutant compared to wildtype (Fig. 5D).
In summary, we conclude that the PPII motif located in the GlyRα1 TM3-4 loop mediates interaction with the guanine nucleotide exchange factor collybistin. Mutagenesis in the PPII motif neither alters GlyR surface targeting nor receptor function, but appears to affect synaptic anchoring of GlyRα1 subunits.
Discussion
The large intracellular loop of the inhibitory GlyR contains numerous motifs for intracellular interaction [2], [4], [17], [18], [42], [43], [44], [45] and modulation of receptor function [7], [17], [34], [46], [47]. Here, we investigated the function of a proline-rich motif, 365PPPAPSKSP373, which forms a PPII helix that is located in the intracellular TM3-4 loop of the human GlyRα1 subunit. Proline residues of this motif were replaced by alanine, resulting in the group mutant P365A, P366A, P367A, P369A and P373A, termed GlyRα1P1-5A. The isolated TM3-4 loop of wildtype GlyRα1 displayed robust binding to the anchoring protein collybistin, binding both splice isoforms, collybistin ISH3+ and collybistin IISH3-, equally well. Thus, binding between collybistin and the GlyRα1 subunit was independent of the N-terminal SH3 domain, while other SH3 domains, notably that of syndapin, do bind to the PPII helix motif [18]. Here, the C-terminal PH domain of collybistin was shown to mediate GlyRα1 binding via the PPII helix, since disruption of the PPII helix motif in the group mutant GlyRα1P1-5A, or a hyperekplexia mutation GlyRα1P366L [18] abolished collybistin binding, While indirect binding through one or several linker proteins cannot be ruled out completely, our data strongly suggest that a specific interaction between the PPII motif and the PH domain of collybistin is present. Our data suggest a direct interaction between GlyRα1 subunits and collybistin.
The isolated TM3-4 loops of GlyRα1 wildtype and GlyRα1P1-5A revealed distinct changes of secondary structure, as determined by CD spectroscopy. Spectra of the wildtype agreed with the presence of a PPII helix, while that of the GlyRα1P1-5A mutant was consistent with a loss of the PPII helix, and an increase in α-helical structure. Complete replacement of all prolines by alanine seems to destroy the PPII structure, but replacement of a single residue (as found in the rat α1 sequence) may be tolerated. The loss of the observed interaction between the polyproline helix and collybistin in the GlyRα1P1-5A mutant indicates that the prolines in the region 365–373 play an important role for the interaction between GlyRα1 and the binding partner collybistin. The interaction between the pleckstrin homology (PH) domain [48], [49], [50] of collybistin and the GlyRα1 PPII helix – initially surprising – could be rationalized from the protein structures. The PH domain of collybistin shows structural similarity to Enabled/VASP Homology 1 (EVH-1) domains, which are known to bind to PPII domains. Here, the backbone conformation is defined by the PPII helix, while the specific surface, formed by proline side chains is required for the binding to EVH1 [31], [33]. The sequence identity between the PH domain of collybistin and human EVH-1 is only ~ 8%, yet the PPII-binding regions of both proteins overlay very well, to a RMSD of 2.2 Å, as evident from a comparison between the pdb structures of rat collybistin [51] (pdb file 2DFK) and a murine EVH-1 domain [33] (pdb file 1EVH).
In structural and functional tests, GlyRα1P1-5A mutant receptors were expressed, targeted to the cell surface and showed wildtype-like GlyR currents. Half maximum concentrations were slightly but significantly increased in the mutant receptor compared to wildtype. Large variations (up to 10-fold) of recombinant wildtype GlyR EC50 values have been described previously [52], [53] and were compatible with regular “wildtype” function of the receptor. Thus, the 1.5-fold increase in EC50 of the GlyRα1P1-5A mutant is not expected to be physiologically relevant. After co-expression of wildtype GlyRα1 and CB1 or CB2 we observed a slight decrease in the EC50 of glycine, which was not statistically significant. Taken together, we conclude that the GlyRα1P1-5A mutant receptor is active and that absence or presence of collybistin I or II has no relevant influence on wildtype channel activity of the receptor.
To date, GlyRs are known to cluster via β-subunits that directly interact with the sub-synaptic scaffolding protein gephyrin at inhibitory postsynaptic sites [3], [5], [6], [7], [11], [12], [54]. Since we tested the binding mediated by the α1 subunit alone, we mostly expressed homomeric α1 receptors (all experiments in Fig. 2, Fig. 3, Fig. 4 were in the absence of GlyRβ expression), thus avoiding possible interactions of the GlyRβ subunit with gephyrin in the recombinant system. Homomeric α1 GlyRs have never been reported to interact with gephyrin, in fact, absence of this interaction has been suggested [14]. It is presently unknown whether (and how) homomeric α1 glycine receptors could attach and cluster at postsynaptic plasma membranes. Expression of homomeric GlyRα1 or α3 subunits in neurons is mostly unknown, as also demonstrated by electrophysiological investigation of neuronal glycine receptor channels, where only conductance levels of αβ GlyRs were observed [2].
Conductance levels consistent with homomeric α1 or α3 GlyRs have seldom been reported, suggesting that these are not expressed in significant amounts. There is, however, sporadic evidence that homomeric GlyRs are expressed in non-somatic locations on presynaptic nerve terminals of central neurons [2], [55]. A study performed on neurons of the rat supraoptic nucleus showed a differential distribution of heteromeric and homomeric GlyRs, where former are distributed exclusively on the soma and dendrites and homomeric GlyRs are found exclusively on distal axonal regions [56]. A recent study showed that α1 subunits can also affect the GlyR-gephyrin interaction despite the absence of direct binding [57].
Our data demonstrate that the intracellular TM3-4 loop of GlyRα1 subunit is able to bind collybistin in GST pull down assays and suggest that the α1 subunit itself may be able to mediate membrane attachment. Since binding of CB1 or CB2 to the GlyRα1P1-5A TM3-4 loop was absent in HEK293 cells, we conclude that the PPII motif mediates this interaction. To validate the finding of pulldown experiments, we performed immunoprecipitation studies of the GlyRα1 and collybistin or the isolated CB-PH domain. Our results confirmed the binding of GlyRα1 to CB1 and CB2 as well as the isolated PH domain, indicating that the PH domain mediates the observed interaction. Our data extend the present ensemble of glycine receptor – scaffolding proteins interaction, suggesting alternative collybistin binding determinants in different GlyR subunits.
Interaction between collybistin and gephyrin [13], [14], [16], [23] occurs near the N-terminus of collybistin, in a linker region between SH3 domain and the DH domain [16] while GABAAR binding sites are located inside the N-terminal SH3 domain [20]. Hence, in contrast to the GlyRα1 subunit, the GABAAR interaction requires the presence of the SH3 domain. Interactions of collybistin with both, gephyrin or GABAAR are independent of the PH domain which contains the binding site of the here discovered GlyRα1-CB interaction. Interestingly, Harvey and colleagues reported that gephyrin clustering in recombinant systems and cultured neurons required both, CB-gephyrin interactions and an intact collybistin PH domain [13]. Also, in hippocampus, substitution of residues in the PH domains of CB2 abolished clustering of gephyrin [58]. Thus, the PH domain of collybistin plays an important role in synaptic clustering.
In contrast to glycine receptors, direct interactions between GABAAR subunits α2 and α3 and collybistin are known [19], [20], [22], [59], [60]. Although GlyRs and GABAARs are structurally similar, both member being of the cysteine loop superfamily of ligand-gated ion channel [1], [2], the collybistin binding motif ‘AYAVAVANYA’ of the GABAAR α2 subunit is situated in the TM3-4 loop upstream of the binding motif ‘PPPAPSKSP’ of the glycine receptor. Collybistin binds the α1 glycine receptor via its PH domain (this study) while the GABAA α2 receptor binds via its SH3 domain [20]. Thus, location and contributing domains of collybistin binding are different between glycine and GABAA receptors.
It has been reported that GlyR clusters remained largely unaffected in collybistin knockout mice in contrast to GABAARs which were reduced following collybistin depletion [60]. Binding between gephyrin and GlyRβ subunits in neurons is much stronger than that of gephyrin and GABAAR α1, α2 or α3 subunits, which is reported to be moderate or even absent [11], [19]. Thus, the rather weak binding of GABAA receptors to gephyrin may require support via collybistin whereas GlyRβ-gephyrin interactions may be sufficient for effective synaptic clustering, independent of collybistin. Recently, subunit-specific clustering of glycine receptors was studied, showing that clustering of α1, but not α3 glycine receptors was sensitive to interleukin-1β and suggesting that alpha subunits alone can affect GlyR-gephyrin binding at synapses without need for β-subunits [57]. Collybistin expression is regulated by excitatory synaptic input [61] and driven by neuronal proteins such as neuroligin-2 [23]. The direct interaction between GlyRα1 subunits and collybistin described here may be relevant for the regulation of synaptic glycine receptor clustering.
Our data indicate a direct binding of collybistin to the GlyRα1 subunit which would enable α1 homomers to cluster at synapses without involvement of the GlyRβ subunit. This adds to the variety of interactions that contribute to synaptic anchoring of glycine receptors. The role of the collybistin-GlyRα1 interaction in synaptic clustering requires further investigation.
Author contributions
U.B. and K.W. performed mutagenesis and patch-clamp electrophysiology experiments, K.W. and R.E. planned and performed Y2H tests, Y.P. performed pulldown experiments, G.L. performed immunofluorescence and immunoprecipitation, H.S. performed structural analysis, C.M.B., U.B., and H.G.B. initiated project, U.B., H.G.B., M.K., and C.V. performed data analysis, U.B. and H.G.B. designed and supervised project and wrote manuscript.
Compliance with Ethics Requirements
The work described here did not involve human probands or animals. All relevant ethical and scientific standards were observed.
Declaration of Competing Interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgements
U.B. and H.G.B. were supported by German Academic Exchange Service DAAD research collaboration travel grants. We thank R. Blum for the lentiviral FUVa1-GFP vector, Finn Bauer, Section Biopolymers, University Bayreuth, Germany, for help with the CD spectroscopic analysis, and Mousa Abdalla Mousa for technical assistance.
Footnotes
Peer review under responsibility of Cairo University.
References
- 1.Breitinger HG. Glycine Receptors. Chichester: eLS. John Wiley & Sons Ltd; 2014.
- 2.Lynch J.W. Native glycine receptor subtypes and their physiological roles. Neuropharmacology. 2009;56:303–309. doi: 10.1016/j.neuropharm.2008.07.034. [DOI] [PubMed] [Google Scholar]
- 3.Grunewald N., Jan A., Salvatico C., Kress V., Renner M., Triller A. Sequences flanking the gephyrin-binding site of GlyRbeta tune receptor stabilization at synapses. eNeuro. 2018;5 doi: 10.1523/ENEURO.0042-17.2018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kneussel M., Betz H. Receptors, gephyrin and gephyrin-associated proteins: novel insights into the assembly of inhibitory postsynaptic membrane specializations. J Physiol. 2000;525(Pt 1):1–9. doi: 10.1111/j.1469-7793.2000.t01-4-00001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Ogino K., Yamada K., Nishioka T., Oda Y., Kaibuchi K., Hirata H. Phosphorylation of Gephyrin in Zebrafish Mauthner cells governs glycine receptor clustering and behavioral desensitization to sound. J Neurosci. 2019;39:8988–8997. doi: 10.1523/JNEUROSCI.1315-19.2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prior P., Schmitt B., Grenningloh G., Pribilla I., Multhaup G., Beyreuther K. Primary structure and alternative splice variants of gephyrin, a putative glycine receptor-tubulin linker protein. Neuron. 1992;8:1161–1170. doi: 10.1016/0896-6273(92)90136-2. [DOI] [PubMed] [Google Scholar]
- 7.Kasaragod V.B., Schindelin H. Structure-function relationships of glycine and GABAA receptors and their interplay with the scaffolding protein gephyrin. Front Mol Neurosci. 2018;11:317. doi: 10.3389/fnmol.2018.00317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kasaragod V.B., Schindelin H. Structure of heteropentameric GABAA receptors and receptor-anchoring properties of gephyrin. Front Mol Neurosci. 2019;12:191. doi: 10.3389/fnmol.2019.00191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kneussel M., Brandstätter J.H., Laube B., Stahl S., Müller U., Betz H. Loss of postsynaptic GABA(A) receptor clustering in gephyrin-deficient mice. J Neurosci. 1999;19:9289–9297. doi: 10.1523/JNEUROSCI.19-21-09289.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pizzarelli R., Griguoli M., Zacchi P., Petrini E.M., Barberis A., Cattaneo A. Tuning GABAergic inhibition: gephyrin molecular organization and functions. Neuroscience. 2020;439:125–136. doi: 10.1016/j.neuroscience.2019.07.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Maric H.M., Mukherjee J., Tretter V., Moss S.J., Schindelin H. Gephyrin-mediated gamma-aminobutyric acid type A and glycine receptor clustering relies on a common binding site. J Biol Chem. 2011;286:42105–42114. doi: 10.1074/jbc.M111.303412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer G., Kirsch J., Betz H., Langosch D. Identification of a gephyrin binding Motif on the glycine receptor beta subunit. Neuron. 1995;15:563–572. doi: 10.1016/0896-6273(95)90145-0. [DOI] [PubMed] [Google Scholar]
- 13.Harvey K., Duguid I.C., Alldred M.J., Beatty S.E., Ward H., Keep N.H. The GDP-GTP exchange factor collybistin: an essential determinant of neuronal gephyrin clustering. J Neurosci. 2004;24:5816–5826. doi: 10.1523/JNEUROSCI.1184-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kins S., Betz H., Kirsch J. Collybistin, a newly identified brain-specific GEF, induces submembrane clustering of gephyrin. Nat Neurosci. 2000;3:22–29. doi: 10.1038/71096. [DOI] [PubMed] [Google Scholar]
- 15.Papadopoulos T., Soykan T. The role of collybistin in gephyrin clustering at inhibitory synapses: facts and open questions. Front Cell Neurosci. 2011;5:11. doi: 10.3389/fncel.2011.00011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grosskreutz Y., Hermann A., Kins S., Fuhrmann J.C., Betz H., Kneussel M. Identification of a gephyrin-binding motif in the GDP/GTP exchange factor collybistin. Biol Chem. 2001;382:1455–1462. doi: 10.1515/BC.2001.179. [DOI] [PubMed] [Google Scholar]
- 17.Breitinger U., Bahnassawy L.M., Janzen D., Roemer V., Becker C.M., Villmann C. PKA and PKC modulators affect ion channel function and internalization of recombinant Alpha1 and Alpha1-beta glycine receptors. Front Mol Neurosci. 2018;11:154. doi: 10.3389/fnmol.2018.00154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Langlhofer G., Schaefer N., Maric H.M., Keramidas A., Zhang Y., Baumann P. A novel glycine receptor variant with startle disease affects Syndapin I and glycinergic inhibition. J Neurosci. 2020;40:4954–4969. doi: 10.1523/JNEUROSCI.2490-19.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tretter V., Kerschner B., Milenkovic I., Ramsden S.L., Ramerstorfer J., Saiepour L. Molecular basis of the gamma-aminobutyric acid A receptor alpha3 subunit interaction with the clustering protein gephyrin. J Biol Chem. 2011;286:37702–37711. doi: 10.1074/jbc.M111.291336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Saiepour L., Fuchs C., Patrizi A., Sassoe‘-Pognetto M., Harvey R.J., Harvey K. Complex role of collybistin and gephyrin in GABAA receptor clustering. J Biol Chem. 2010;285:29623–29631. doi: 10.1074/jbc.M110.121368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Panzanelli P., Gunn B.G., Schlatter M.C., Benke D., Tyagarajan S.K., Scheiffele P. Distinct mechanisms regulate GABAA receptor and gephyrin clustering at perisomatic and axo-axonic synapses on CA1 pyramidal cells. J Physiol. 2011;589:4959–4980. doi: 10.1113/jphysiol.2011.216028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Chiou T.T., Bonhomme B., Jin H., Miralles C.P., Xiao H., Fu Z. Differential regulation of the postsynaptic clustering of gamma-aminobutyric acid type A (GABAA) receptors by collybistin isoforms. J Biol Chem. 2011;286:22456–22468. doi: 10.1074/jbc.M111.236190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kilisch M., Mayer S., Mitkovski M., Roehse H., Hentrich J., Schwappach B. A GTPase-induced switch in phospholipid affinity of collybistin contributes to synaptic gephyrin clustering. J Cell Sci. 2020;133 doi: 10.1242/jcs.232835. [DOI] [PubMed] [Google Scholar]
- 24.Soykan T., Schneeberger D., Tria G., Buechner C., Bader N., Svergun D. A conformational switch in collybistin determines the differentiation of inhibitory postsynapses. Embo J. 2014;33:2113–2133. doi: 10.15252/embj.201488143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cascio M., Shenkel S., Grodzicki R.L., Sigworth F.J., Fox R.O. Functional reconstitution and characterization of recombinant human alpha 1-glycine receptors. J Biol Chem. 2001;276:20981–20988. doi: 10.1074/jbc.M010968200. [DOI] [PubMed] [Google Scholar]
- 26.Breitinger U., Breitinger H.G., Bauer F., Fahmy K., Glockenhammer D., Becker C.-M. Conserved high affinity ligand binding and membrane association in the native and refolded extracellular domain of the human glycine receptor alpha1-subunit. J Biol Chem. 2004;279:1627–1636. doi: 10.1074/jbc.M303811200. [DOI] [PubMed] [Google Scholar]
- 27.Isemura T., Okabayashi H., Sakakibara S. Steric structure of L-proline oligopeptides. I. Infrared absorption spectra of the oligopeptides and poly-L-proline. Biopolymers. 1968;6:307–321. doi: 10.1002/bip.1968.360060306. [DOI] [PubMed] [Google Scholar]
- 28.Sreerama N., Woody R.W. Molecular dynamics simulations of polypeptide conformations in water: A comparison of alpha, beta, and poly(pro)II conformations. Proteins. 1999;36:400–406. [PubMed] [Google Scholar]
- 29.Kelly M.A., Chellgren B.W., Rucker A.L., Troutman J.M., Fried M.G., Miller A.F. Host-guest study of left-handed polyproline II helix formation. Biochemistry. 2001;40:14376–14383. doi: 10.1021/bi011043a. [DOI] [PubMed] [Google Scholar]
- 30.Adzhubei A.A., Sternberg M.J., Makarov A.A. Polyproline-II helix in proteins: structure and function. J Mol Biol. 2013;425:2100–2132. doi: 10.1016/j.jmb.2013.03.018. [DOI] [PubMed] [Google Scholar]
- 31.Peterson F.C., Volkman B.F. Diversity of polyproline recognition by EVH1 domains. Front Biosci (Landmark Ed). 2009;14:833–846. doi: 10.2741/3281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kay B.K., Williamson M.P., Sudol M. The importance of being proline: the interaction of proline-rich motifs in signaling proteins with their cognate domains. Faseb J. 2000;14:231–241. [PubMed] [Google Scholar]
- 33.Prehoda K.E., Lee D.J., Lim W.A. Structure of the enabled/VASP Homology 1Domain–Peptide Complex: A Key Component in the Spatial Control of Actin Assembly. Cell. 1999;97:471–480. doi: 10.1016/s0092-8674(00)80757-6. [DOI] [PubMed] [Google Scholar]
- 34.Breitinger H.G., Villmann C., Melzer N., Rennert J., Breitinger U., Schwarzinger S. Novel regulatory site within the TM3-4 loop of human recombinant alpha3 glycine receptors determines channel gating and domain structure. J Biol Chem. 2009;284:28624–28633. doi: 10.1074/jbc.M109.043174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Greenfield N.J. Methods to estimate the conformation of proteins and polypeptides from circular dichroism data. Anal Biochem. 1996;235:1–10. doi: 10.1006/abio.1996.0084. [DOI] [PubMed] [Google Scholar]
- 36.Schweimer K., Hoffmann S., Bauer F., Friedrich U., Kardinal C., Feller S.M. Structural investigation of the binding of a herpesviral protein to the SH3 domain of tyrosine kinase. Biochemistry. 2002;41:5120–5130. doi: 10.1021/bi015986j. [DOI] [PubMed] [Google Scholar]
- 37.Tiffany M.L., Krimm S. Circular dichroism of poly-L-proline in an unordered conformation. Biopolymers. 1968;6:1767–1770. doi: 10.1002/bip.1968.360061212. [DOI] [PubMed] [Google Scholar]
- 38.Tsuriel S., Geva R., Zamorano P., Dresbach T., Boeckers T., Gundelfinger E.D. Local sharing as a predominant determinant of synaptic matrix molecular dynamics. PLoS Biol. 2006;4 doi: 10.1371/journal.pbio.0040271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mayer B.J. SH3 domains: complexity in moderation. J Cell Sci. 2001;114:1253–1263. doi: 10.1242/jcs.114.7.1253. [DOI] [PubMed] [Google Scholar]
- 40.Sparks A.B., Rider J.E., Hoffman N.G., Fowlkes D.M., Quilliam L.A., Kay B.K. Distinct ligand preferences of Src homology 3 domains from Src, Yes, Abl, Cortactin, p53bp2, PLCgamma, Crk, and Grb2. Proc Natl Acad Sci U S A. 1996;93:1540–1544. doi: 10.1073/pnas.93.4.1540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brannetti B., Helmer-Citterich M. iSPOT: A web tool to infer the interaction specificity of families of protein modules. Nucleic Acids Res. 2003;31:3709–3711. doi: 10.1093/nar/gkg592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kneussel M., Betz H. Clustering of inhibitory neurotransmitter receptors at developing postsynaptic sites: the membrane activation model. Trends Neurosci. 2000;23:429–435. doi: 10.1016/s0166-2236(00)01627-1. [DOI] [PubMed] [Google Scholar]
- 43.Langlhofer G., Villmann C. The intracellular loop of the glycine receptor: It's not all about the size. Front Mol Neurosci. 2016;9:41. doi: 10.3389/fnmol.2016.00041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Villmann C., Oertel J., Melzer N., Becker C.M. Recessive hyperekplexia mutations of the glycine receptor alpha1 subunit affect cell surface integration and stability. J Neurochem. 2009;111:837–847. doi: 10.1111/j.1471-4159.2009.06372.x. [DOI] [PubMed] [Google Scholar]
- 45.Zhang Z.Y., Guo Z., Li H.L., He Y.T., Duan X.L., Suo Z.W. Ubiquitination and inhibition of glycine receptor by HUWE1 in spinal cord dorsal horn. Neuropharmacology. 2019;148:358–365. doi: 10.1016/j.neuropharm.2019.02.002. [DOI] [PubMed] [Google Scholar]
- 46.Breitinger U., Breitinger H.G. Modulators of the inhibitory glycine receptor. ACS Chem Neurosci. 2020;11:1706–1725. doi: 10.1021/acschemneuro.0c00054. [DOI] [PubMed] [Google Scholar]
- 47.Moraga-Cid G., San Martin V.P., Lara C.O., Munoz B., Marileo A.M., Sazo A. Modulation of glycine receptor single-channel conductance by intracellular phosphorylation. Sci Rep. 2020;10:4804. doi: 10.1038/s41598-020-61677-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaw G. The pleckstrin homology domain: an intriguing multifunctional protein module. BioEssays. 1996;18:35–46. doi: 10.1002/bies.950180109. [DOI] [PubMed] [Google Scholar]
- 49.Scheffzek K., Welti S. Pleckstrin homology (PH) like domains - versatile modules in protein-protein interaction platforms. FEBS Lett. 2012;586:2662–2673. doi: 10.1016/j.febslet.2012.06.006. [DOI] [PubMed] [Google Scholar]
- 50.Naveenkumar N., Sowdhamini R., Srinivasan N. Specialized structural and functional roles of residues selectively conserved in subfamilies of the pleckstrin homology domain family. FEBS Open Bio. 2019;9:1848–1859. doi: 10.1002/2211-5463.12725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xiang S., Kim E.Y., Connelly J.J., Nassar N., Kirsch J., Winking J. The crystal structure of Cdc42 in complex with collybistin II, a gephyrin-interacting guanine nucleotide exchange factor. J Mol Biol. 2006;359:35–46. doi: 10.1016/j.jmb.2006.03.019. [DOI] [PubMed] [Google Scholar]
- 52.Breitinger U., Raafat K.M., Breitinger H.G. Glucose is a positive modulator for the activation of human recombinant glycine receptors. J Neurochem. 2015;134:1055–1066. doi: 10.1111/jnc.13215. [DOI] [PubMed] [Google Scholar]
- 53.De Saint Jan D, David-Watine B., Korn H., Bregestovski P. Activation of human alpha1 and alpha2 homomeric glycine receptors by taurine and GABA. J Physiol. 2001;535:741–755. doi: 10.1111/j.1469-7793.2001.t01-1-00741.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Maric H.M., Kasaragod V.B., Hausrat T.J., Kneussel M., Tretter V., Stromgaard K. Molecular basis of the alternative recruitment of GABA(A) versus glycine receptors through gephyrin. Nat Commun. 2014;5:5767. doi: 10.1038/ncomms6767. [DOI] [PubMed] [Google Scholar]
- 55.Hruskova B., Trojanova J., Kulik A., Kralikova M., Pysanenko K., Bures Z. Differential distribution of glycine receptor subtypes at the rat calyx of Held synapse. J Neurosci. 2012;32:17012–17024. doi: 10.1523/JNEUROSCI.1547-12.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deleuze C., Runquist M., Orcel H., Rabié A., Dayanithi G., Alonso G. Structural difference between heteromeric somatic and homomeric axonal glycine receptors in the hypothalamo-neurohypophysial system. Neuroscience. 2005;135:475–483. doi: 10.1016/j.neuroscience.2005.05.024. [DOI] [PubMed] [Google Scholar]
- 57.Patrizio A., Renner M., Pizzarelli R., Triller A., Specht C.G. Alpha subunit-dependent glycine receptor clustering and regulation of synaptic receptor numbers. Sci Rep. 2017;7:10899. doi: 10.1038/s41598-017-11264-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Reddy-Alla S., Schmitt B., Birkenfeld J., Eulenburg V., Dutertre S., Bohringer C. PH-domain-driven targeting of collybistin but not Cdc42 activation is required for synaptic gephyrin clustering. Eur J Neurosci. 2010;31:1173–1184. doi: 10.1111/j.1460-9568.2010.07149.x. [DOI] [PubMed] [Google Scholar]
- 59.Körber C., Richter A., Kaiser M., Schlicksupp A., Mukusch S., Kuner T. Effects of distinct collybistin isoforms on the formation of GABAergic synapses in hippocampal neurons. Mol Cell Neurosci. 2012;50:250–259. doi: 10.1016/j.mcn.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 60.Papadopoulos T., Korte M., Eulenburg V., Kubota H., Retiounskaia M., Harvey R.J. Impaired GABAergic transmission and altered hippocampal synaptic plasticity in collybistin-deficient mice. Embo J. 2007;26:3888–3899. doi: 10.1038/sj.emboj.7601819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Horn M.E., Nicoll R.A. Somatostatin and parvalbumin inhibitory synapses onto hippocampal pyramidal neurons are regulated by distinct mechanisms. Proc Natl Acad Sci USA. 2018;115:589–594. doi: 10.1073/pnas.1719523115. [DOI] [PMC free article] [PubMed] [Google Scholar]