Abstract
Background
Prostate carcinoma is the second most common cancer among men worldwide. Developing new therapeutic approaches and diagnostic biomarkers for prostate cancer (PC) is a significant need. The Chinese herbal medicine Panax quinquefolius saponins (PQS) have been reported to show anti-tumor effects. We hypothesized that PQS exhibits anti-cancer activity in human PC cells and we aimed to search for novel biomarkers allowing early diagnosis of PC.
Methods
We used the human PC cell line DU145 and the prostate epithelial cell line PNT2 to perform cell viability assays, flow cytometric analysis of the cell cycle, and FACS-based apoptosis assays. Microarray-based gene expression analysis was used to display specific gene expression patterns and to search for novel biomarkers. Western blot and quantitative real-time PCR were performed to demonstrate the expression levels of multiple cancer-related genes.
Results
Our data showed that PQS inhibited the viability of DU145 cells and induced cell cycle arrest at the G1 phase. A significant decrease in DU145 cell invasion and migration were observed after 24 h treatment by PQS. PQS up-regulated the expression levels of p21, p53, TMEM79, ACOXL, ETV5, and SPINT1 while it down-regulated the expression levels of bcl2, STAT3, FANCD2, DRD2, and TMPRSS2.
Conclusion
PQS promoted cells apoptosis and inhibited the proliferation of DU145 cells, which suggests that PQS may be effective for treating PC. TMEM79 and ACOXL were expressed significantly higher in PNT2 than in DU145 cells and could be novel biomarker candidates for PC diagnosis.
Keywords: Chinese medicinal herbs, Panax quinquefolius, Potential biomarkers, Prostate cancer cells, saponins
Abbreviations: ACOXL, Acyl-CoA oxidase-like protein; bcl2, B-cell lymphoma 2; DRD2, dopamine receptor D2; ETV5, ETS variant 5; FACS, fluorescence-activated cell sorting; FANCD2, fanconi anemia group D2; p21, cyclin-dependent kinase inhibitor p21; p53, tumor suppressor p53; PC, prostate cancer; PQS, Panax quinquefolius saponins; qRT-PCR, quantitative real-time PCR; SPINT1, serine peptidase inhibitor Kunitz type 1; STAT3, signal transducer and activator of transcription 3; TCM, Traditional Chinese Medicine; TMEM79, transmembrane protein 79; TMPRSS2, transmembrane protease serine 2
1. Introduction
Prostate cancer (PC) is currently one of the most common malignancies in males worldwide and the second leading cause of malignancy mortality in developed western countries [[1], [2], [3]]. In Europe, prostate carcinomas are the second most prevalent tumors in men after the age of 50 [[1], [2], [3], [4]]. Prevalent diagnosis and treatment of PC include screening trial, digital rectal examination, prostate-specific antigen (PSA) testing, multi-parametric magnetic resonance imaging, hormone treatment, and radiotherapy [4]. The low specificity of the clinical PSA test often potentially results in overdiagnosis and overtreatment of patients [[4], [5], [6]]. Most of the patients with PC respond to traditional androgen ablation therapy, leading to the regression of prostate tumors [7,8]. However, with disease progression, PC can often develop resistance to androgen ablation therapy. Recent studies have shown that approximately 50% of PC is able to progress from an androgen-dependent to a hormone refractory disease state and will spread from the original tumor to other parts of the body [9]. Immunotherapies and medications with denosumab, abiraterone, bisphosphonates, enzalutamide, and Radium-223 have debatable efficacy for the hormone refractory state of disease [10]. Abiraterone may cause cardiac disorders and hypertension [4,11,12]. Enzalutamide blocks androgen receptor transfer and thus inhibits any possible agonist-like activity [[11], [12], [13]]. Bisphosphonates and denosumab can cause low calcium levels in the blood (hypocalcemia). Toxicity of Radium-223 may lead to gastrointestinal side effects, neutropenia, and thrombocytopenia [14]. Therefore, new alternative therapies that demonstrate efficacy in PC treatment with less associated toxicities and side effects are still in urgent need.
Complementary and alternative medicine has been investigated widely during the past few decades [[15], [16], [17]]. Phytotherapies have been shown to have beneficiary effects as well as they are of low cost; they have mild adverse effects for PC patients [18]. A pilot study indicates that a combination of phytotherapeutic intervention of turmeric, resveratrol, green tea, and broccoli sprouts can significantly alleviate complication symptoms caused by PC and increase patient's quality of life [[18], [19], [20]]. A number of isolated herbal components have been discovered to have inhibitory effects on PC cell growth and to have therapeutic potential for treating PC [[21], [22], [23], [24]]. For example, lycopenes in tomatoes and isoflavones in soybeans might help prevent PC [17,25,26]. Green tea catechins have been reported to reduce the incidence of PC risk and are effective for preventing PC [27]. Chinese herbal medicines have been shown to have various biological effects, including anti-inflammation, anti-oxidation, anti-myocardial infarction, and anti-tumor activities [[28], [29], [30], [31], [32]]. In this study, we used the Chinese medicinal herb Panax quinquefolius, one of the main species of the ginseng genus, which plays an important part in improving the immune system and the treatment and prevention of various diseases in China [33,34]. Clinical evidence of the beneficiary effects P. quinquefolius in cancer has been reported in various recent studies [18,21,[35], [36], [37], [38], [39], [40], [41]].
P. quinquefolius contains common bioactive ingredients including saponins (ginsenosides), flavonoids, polysaccharides, volatile oils, polyenes, fatty acids, and polyacetylenic alcohols, among which ginsenosides are thought to be the most beneficial to human health [[42], [43], [44], [45]]. P. quinquefolius saponins (PQS) have various physiological and pharmacological characteristics including anti-tumorigenic activities [[31], [32], [33], [34],43]. The chemical composition includes ginsenosides Rc, Rd, Re, Rb1, Rg1, Rh1, Rb2, Rf2, Rb3, Rg3, quinquenoside R1, quinquefolans A, B, C, pseudo-ginsenoside F11, essential oils, and rutin [[46], [47], [48], [49]]. Studies indicated that ginsenoside Rg3 has anti-vascularization effects and inhibits colorectal cancer cell growth and metastasis in vivo [50,51]. Besides, Rg3 also suppresses the proliferation of non-small cell lung cancer cells [52] and pancreatic cancer cells [53] in vitro. Ginsenosides Rb3 and Rd reduce the colorectal cancer tumor size and number in mice [54]. In ovarian cancer cells, the epithelial–mesenchymal transition was inhibited by Ginsenoside Rb1 [55,56].
We used isolated PQS, which consist of the eight major ginsenosides Re, Rg1, Rc, Rb1, Rb2, Rb3, Rd, and Rg3 for this study. We aimed to evaluate the anti-cancer activities of PQS and to understand the important role of PQS in regulating cellular mechanisms such as proliferation, cell cycle, differentiation, cytotoxic efficacy, and cell apoptosis using the PC cell line DU145 in comparison with the prostate epithelial cell line PNT2. Cell invasion assays and wound healing assays were carried out to measure the activities of cell migration. The effects of PQS treatment on mRNA-related gene expression were examined in DU145 and PNT2 cells. We used the whole human genome microarray technique to obtain data of PQS treatment-related genes and pathways and to analyze the expression levels of multiple cancer-related genes and gene-related pathways.
Our aim was to investigate a useful alternative medicine, which has anti-cancer properties and low adverse side effects, and to search for new biomarkers allowing for early diagnosis and therapy of PC.
2. Materials and methods
2.1. Sample preparation
The purified PQS extracts were supplied by Jian Yisheng Pharmaceutical Company (Jilin, China). The PQS powder was dissolved in the RPMI-1640 culture medium (10% FBS, 1% Pen/Strep, 1% glutamine, and 1% Hepes, GIBCO, Dublin, Ireland) for cell culture experiments. For UHPLC-QTOF-MS/MS analysis, 0.3 g of PQS extracts was ultrasonicated with 10 mL of 70% formic acid for 30 min at RT. The extract was centrifuged for 20 min at 10,000 rpm. The supernatant was collected and filtered using a 0.22 μm PTFE syringe filter for the UHPLC-QTOF-MS/MS analysis.
2.2. Quality control of PQS using ultra-high performance liquid chromatography (UHPLC) and mass spectrometry
Using a Waters ACQUITY UPLC system (Waters Co., Milford, MA, USA) equipped with an auto-sampler, a PDA detector, and a binary solvent delivery system, we performed the UHPLC for the quality control of PQS extracts. The chromatographic separation was processed by a Waters ACQUITY HSS T3 column (2.1 mm × 100 mm, 1.8 μm). The mobile phases consisted of A (0.1% formic acid in water) and B (acetonitrile containing 0.1% formic acid) phases. Optimized elution conditions were as follows: 5–15% B for 0–1 min, 15–30% B for 1–5 min, 30–38% B for 5–15 min, 38–60% B for 15–15.5 min, 60% B for 15.5–23 min, 60–95% B for 23–23.5 min, 95% B for 23.5–25 min, 95–5% B for 25–25.5 min, and isocratic at 5% B for 25.5–27 min. The flow rate was 0.5 mL/min. The injection volumes of ginsenoside standard solutions and PQS extracts were 1 μL. Mass spectrometry coupled with the electrospray ionization interface was applied on a Waters Synapt G2-S Q-TOF system (Waters MS Technologies, Manchester, UK). The desolvation gas was fixed to 900 L/h at 450°C and the temperature of source was at 100°C. The voltage of capillary and cone were set at 2,500 V and 30 V. The Q-TOF acquisition rate was 0.2 s. The energy for collision-induced dissociation for the precursor ion was 6 V and those for fragmentation information were 30–60 V. The accurate mass and elemental composition for the fragment ions and the precursor ions were analyzed by the MassLynx V4.1 software (Waters Co., Milford, MA, USA).
2.3. Cell lines and cell culture
The human prostate carcinoma cell line DU145 and the human prostate epithelial cell line PNT2 were used for this study. The cells were incubated in RPMI-1640 medium in a 25 cm2 Nunc Nunclon tissue culture flask (Thermo Scientific, Waltham, MA, USA) at 37°C with 5% CO2. The DU145 cells were treated with PQS extracts in different concentrations (0, 1.5, 5, 15, 50, and 150 μg/mL). Control cells PNT2 were treated with RPMI-1640 medium. The medium was changed every 2 days. After washing with PBS, cells were collected by centrifugation at 900 rpm for 8 min at RT. The cells were calculated using a TC20 automated cell counter (BIO-RAD, CA, USA) and divided into a tissue culture flask for the following experiments.
2.4. Cell proliferation and cell viability assay
DU145 and PNT2 cells were seeded in the 96-well plates (Thermo Fisher Scientific, MA, USA). After incubation for 24 h at 37°C, the DU145 cells were treated with different concentrations (0, 5, 15, 50, 100, and 150 μg/mL) of PQS at different time points for 24, 48, and 72 h. PNT2 cells as control group were treated with RPMI medium. Cell proliferation of both cell lines was measured after treatment using the CellTiter-Blue Cell Viability Assay and the ApoTox-Glo Triplex Assay reagents (Promega Co., WI, USA). The absorbance and fluorescence were measured using a Tecan microplate reader (Epoch, Bio-Tek Instruments, Winooski, VT, USA) at 560 nmEx/590 nmEm. The data were calculated as a ratio of treated versus untreated cells (vehicle set at 100%).
A FITC Annexin V Apoptosis Detection Kit with PI (Biolegend, California, USA) was used to detect the apoptosis of DU145 cells after PQS treatment for 24 h and 48 h. DU145 cells (106 cells/mL) were treated with PBS (negative control), 100% ethanol (positive control), and 150 μg/mL of PQS for 24 h and 48 h in a 6-well plate. Subsequently, cells were collected, washed twice with PBS buffer, and centrifuged. Cells were suspended in 200 μL of binding buffer. After adding 10 μL of Annexin V-FITC and 5 μL of PI (Propidium Iodide Solution), and incubating for 15 min at room temperature without exposure to light, 400 μl of binding buffer was added. The stained cells were analyzed using a flow cytometer to detect apoptosis under the excitation wavelength of 488 nm. The experiment was repeated three times to obtain mean values.
2.5. Caspase-3/7 activity assay
DU145 cells were treated with PQS (0, 15, 50, 100, and 150 μg/mL) for 48 h for determining caspase-3/7 activity using the ApoTox-Glo Triplex assay kit. The caspase-3/7 activity was measured using the ApoTox-Glo Triplex Assay reagents and the luminescence of caspase-3/7 activity was detected using a Tecan microplate reader. The data were calculated as a ratio of treated versus untreated cells (vehicle set at 100%) and shown as fold-increased activities. The assays were carried out in three independent plates running in triplicate each time. For each assay, at least 104 cells were counted.
2.6. Western blot analysis of cleaved caspase-3
DU145 cells were collected after treatment for 48 h with PQS (0, 50, 100, and 150 μg/mL) and lysed in cold radio immunoprecipitation assay (RIPA) buffer containing 1% protease inhibitor cocktail. After centrifugation, the supernatant was collected. Samples for SDS-PAGE were denatured for 5 min at 95°C and loaded on 4–15% Mini-PROTEAN TGX gels (Bio-Rad, Hercules, CA, USA). After electrophoresis, proteins were transferred to a polyvinylidene fluoride or polyvinylidene difluoride (PVDF) membrane (Bio-Rad, Hercules, CA, USA) and blocked in tris-buffered saline, 0.1% Tween 20 (TBST) buffer with 5% nonfat milk. Blots were incubated with 1:1,000 diluted cleaved caspase-3 antibodies (Cell Signaling Technology, Massachusetts, USA) overnight at 4°C, followed by incubation for 2 h with goat anti-rabbit IgG (H + L) secondary antibodies. The bound cleaved caspase-3 protein was detected using the BCIP/NBT Color Development Substrate (Promega Co., WI, USA). Beta-actin was used as a control protein.
2.7. LDH cytotoxicity assay
Cytotoxicity was assessed using a LDH Cytotoxicity Assay Kit (Thermo Scientific Pierce, Massachusetts, USA), which measures the release of the cytoplasmic enzyme lactate dehydrogenase (LDH) by damaged cells. DU145 cells cultured in 96-well plates were treated with H2O (spontaneous controls), different concentrations of PQS (0, 15, 50, 100, and 150 μg/mL), and 10× lysis buffer (maximum LDH activity controls). After PQS treatment for 24 h and 48 h, the culture supernatant was collected and incubated with the reaction mixture. The LDH activity was measured at 490 nm and 680 nm absorbance. Percent cytotoxicity was calculated as follows:
2.8. Wound healing assay and cell invasion assay
The mobility and the invasion ability of DU145 cells were measured by the wound healing and the cell invasion assays using the Cyto Select 24-well wound healing assay kit and the cell invasion assay kit (Cell BioLab, CA, USA), respectively. Wound closure was monitored and the percent closure was measured (Percent closure rate = migrated cell surface area/total surface area × 100). PQS treated cells were resuspended in culture medium and incubated for 48 h at 37°C with 5% CO2. The migrated DU145 cells through the membrane were stained and photographed. The invaded cells were quantified at OD 560 nm. Data were interpreted as mean ± SEM in three independent groups.
2.9. Cell cycle assay
For cell cycle assays, 2 × 105 DU145 cells were seeded in the 6-well plates. DU145 cells were treated with PQS extracts (0, 50, 100, and 150 μg/mL) for 48 h. Trypsin was added to collect the adherent cells, which were fixed with 80% ethanol and then stored for 2 h at 4°C. The fixed cells were resuspended in 200 μL of PI/RNase staining buffer (Abcam, Cambridge, UK) and incubated for 30 min at 37°C in the dark. At least 105 cells were counted and measured using a FACS flow cytometer for each measurement.
2.10. Agilent whole genome oligo microarrays
The whole human genome oligo microarray represents all known genes and transcripts in the human genome. Sequences were compiled from a broad source survey and optimized by alignment to the assembled human genome. RNA samples were extracted from the harvested DU145 and PNT cells using the TriZol reagent (Invitrogen, Carlsbad, USA). Total RNA clean-up and RNA quality control were performed using the RNeasy MinElute Cleanup Kit (Qiagen, Hilden, Germany). RNA integrity was evaluated by standard denaturing agarose gel electrophoresis.
Sample labeling and array hybridization were carried out following the protocol of one-color microarray-based gene expression analysis (Agilent Technology, Santa Clara, USA). Total RNA was amplified and labeled with Cy3-UTP. The RNeasy Mini kit (Qiagen, Valencia, CA) was used to purify the labeled cRNAs. The Nano Drop ND-1000 was used to analyze the concentration and specific activity of the labeled cRNAs (pmol Cy3/μg cRNA). Labeled cRNA (1 μg) was fragmented by adding 2.2 μL of 25 × fragmentation buffer and 11 μL of 10 × blocking agent. After incubating for 30 min at 60°C, the labeled cRNA was diluted by adding 55 μL of 2 × GE hybridization buffer. Hybridization solution (100 μL) was used for assembling to the gene expression microarray slide. The slides were washed and scanned using a DNA microarray scanner after incubating for 17 h at 65°C in a hybridization oven. The gene expression levels were identified by setting up the fold-change filters, which were increased or decreased more than two-fold in PQS-treated cells compared to untreated cells. The quantitative real-time PCR (qRT-PCR) analysis was followed to confirm the accuracy of the microarray analyses.
2.11. Validation of mRNA expression using qRT-PCR and western blot analysis
DU145 cells were treated with the different concentrations of PQS (0, 50, 100, and 150 μg/mL) for 48 h. After isolating the total RNA of DU145 cells, the concentration was quantified using a BioPhotometer plus photometer (Eppendorf, Germany). The first strand of cDNA was synthesized using a Super Script IV Reverse Transcriptase kit (Invitrogen, Carlsbad, USA). The fold-change of cancer-related genes (p21, p53, bcl2, STAT3, TMEM79, ACOXL, DRD2 TMPRSS2, SPINT1, ETV5, and FANCD2) after PQS treatment was evaluated using qRT-PCR, respectively.
The reaction mixture for qRT-PCR contained 10 pmol/μL of SYBR Green PCR Master Mix (Applied Biosystems, USA) and each primer. Thermal cycling conditions were as follows: for 10 min at 95°C, followed by 40 cycles for 15 s at 95°C, for 1 min at 60°C, and for 15 s at 95°C. A melting curve analysis of the products was performed after amplification by high-resolution data collection during an incremental temperature increase from 60°C to 95°C. The qRT-PCR analysis was performed on an Applied Biosystems StepOnePlus System and data were analyzed quantitatively using the StepOne software version 2.1 (Applied Biosystems, USA). Experiments were repeated three times. Relative expression levels of the genes were assessed using the 2−ΔΔct method. The methods of western blot are described in Section 2.6. Blots were incubated with selected antibodies p21, p53, bcl2, TMEM79, ACOXL, DRD2, FANCD2, and TMPRSS2 (Abcam, Cambridge, UK). Beta-actin was used as a loading control.
2.12. Statistical analysis
Data are expressed as mean ± SEM/SD of three independent experiments, and one-way ANOVA analysis was conducted using the Graphpad Prism 6 software. Statistically significant results among means were set at ∗p < 0.01, ∗∗p < 0.001, ∗∗∗p < 0.0001, and ∗∗∗∗p < 0.00001.
3. Results
3.1. Quality control of PQS by UHPLC-QTOF-MS/MS
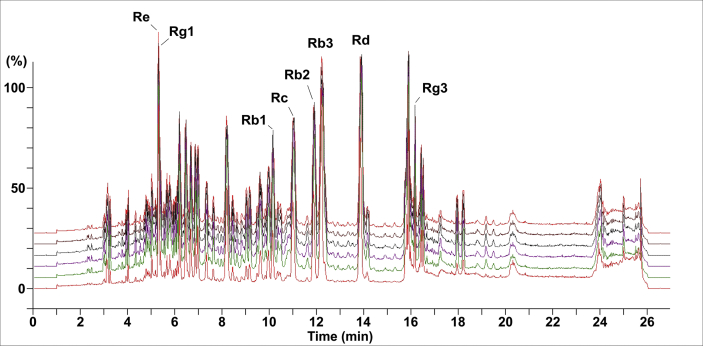
To qualify and quantify ginsenosides in PQS, six different batches of PQS extracts were analyzed with UHPLC-QTOF-MS/MS column chromatography following chromatographic conditions based on previous studies [48]. Eight ginsenosides of PQS were identified by their retention times, accurate mass data of molecular ions, and content of the reference compounds. The characteristic peaks in six batches were consistent (Fig. 1). The retention times of the eight main peaks were 5.28 min (Ginsenoside Re), 5.31min (Ginsenoside Rg1), 10.81 min (Ginsenoside Rc), 9.69 min (Ginsenoside Rb1), 11.68 min (Ginsenoside Rb2), 12.00 min (Ginsenoside Rb3), 13.60 min (Ginsenoside Rd), and 16.16 min (Ginsenoside Rg3), respectively.
Fig. 1.
Quality control of six batches of purified PQS by UHPLC-QTOF-MS/MS. The fingerprints of ginsenosides in the methanol fraction include Re, Rg1, Rc, Rb1, Rb2, Rb3, Rd, and Rg3. The characteristic peaks were consistent among all six batches repeated within one month.
The sum of these saponins in PQS including Ginsenosides Re (330.28 μg/g), Rg1 (105.91 μg/g), Rc (181.12 μg/g), Rb1 (92.64 μg/g), Rb2 (284.01 μg/g), Rb3 (382.12 μg/g), Rd (217.58 μg/g), and Rg3 (118.03 μg/g) accounts for approximately 1,711.69 μg/g, which is calculated using the following formula:
Our results showed that UHPLC-QTOF-MS/MS is a sensitive, rapid, and precise method for simultaneous resolution of the ginsenosides in PQS and a useful technique for the quality control of PQS as well as other Chinese herbal medicines.
3.2. PQS inhibited DU145 cell viability and induced cell apoptosis
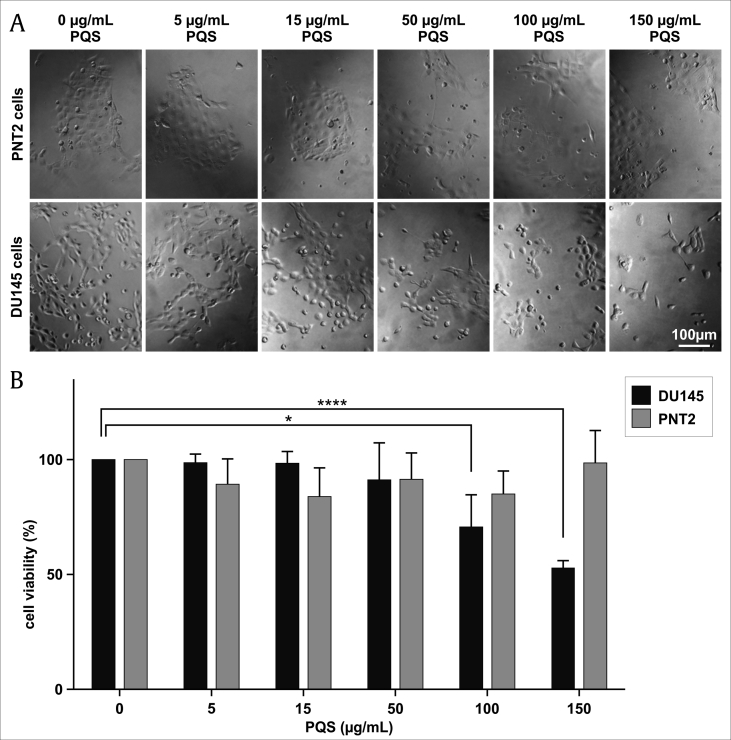
To evaluate the cell growth inhibition, DU145 cells were treated with indicated concentrations (0, 15, 50, 100, and 150 μg/mL) of PQS for 24, 48, and 72 h. The results showed that PQS reduced DU145 cell viability in a time- and concentration-dependent manner compared with untreated DU145 cells. A significant reduction of 47.1% viable DU145 cells was observed at 48 h compared to the control group (Fig. 2A). Furthermore, DU145 and PNT2 cells were treated with 0, 50, 100, and 150 μg/mL of PQS for 48 h. PQS treatment caused a 33.2% decrease of DU145 cell viability compared to a 14.5% decrease of PNT2 cell viability at a concentration of 150 μg/mL (Fig. 2B).
Fig. 2.
PQS inhibited the viability of DU145 cells in a dose-dependent manner. Human PC DU145 cells and prostate epithelial PNT2 cells were treated without (0 μg/mL, vehicle control) or with increasing concentrations of PQS (5, 15, 50, 100, and 150 μg/mL) for 48 h. Cells were photographed using a microscope (A) and cell viability (% of control, B) was measured using a Cell Titer-Blue Cell Viability assay kit. Error bars represent standard deviations (SD) from n ≥ 3 independent experiments in triplicate. ∗ and ∗∗∗∗ represent p < 0.05 and p < 0.0001, as obtained by one-way ANOVA.
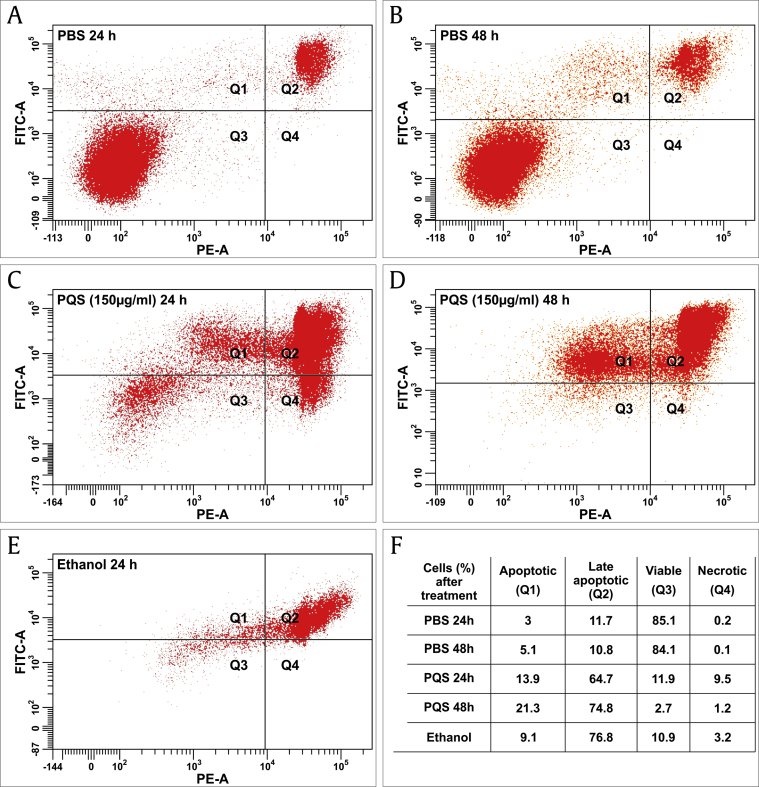
Apoptosis of DU145 cells after PQS treatment for 24 h and 48 h was detected using a FITC Annexin V Apoptosis Detection Kit with PI. Cells treated with PQS exhibited significant apoptosis at both time points (24 h and 48 h). The FACS results showed a significant increase of apoptotic cells after PQS treatment in a time-dependent manner (Fig. 3). After exposure to PBS (negative control) for 24 h, 3% of DU145 cells exhibited an early stage apoptosis and 11.7% a late stage apoptosis, while 85.1% of cells were viable. After 48 h of PBS treatment, 5.1% of DU145 cells exhibited an early stage apoptosis and 10.8% a late stage apoptosis, while 84.1% of cells were viable. After ethanol treatment (positive control), 9.1% of cells exhibited an early stage apoptosis and 76.8% a late stage apoptosis while 10.9% of cells were viable. PQS-treated cells showed an apparently increased apoptosis ranging from 13.9% early stage apoptosis to a 64.7% late stage apoptosis after 24 h treatment, and from 21.3% early stage apoptosis to a 74.8% late stage apoptosis after 48 h treatment. Total apoptosis of PQS-treated cells increased from 78.6% at the time point of 24 h to 96.1% total apoptosis at 48 h. The viable cell percentage was significantly reduced from 11.9% after 24 h treatment to 2.7% at 48 h (Fig. 3).
Fig. 3.
PQS induced the apoptosis of DU145 cells. DU145 cells were treated with PBS as a negative control, 100% ethanol as a positive control, and PQS (150 μg/mL) for 24 and 48 h and measured by FACS using a FITC Annexin V Apoptosis Detection Kit with PI. Cells treated with PQS exhibited a significantly increased apoptosis in a time-dependent manner. Total apoptosis of cells increased from 78.6% at the time point of 24 h to 96.1% total apoptosis after 48 h of PQS treatment. The viable cell percentage was significantly reduced from 11.9% after 24 h to 2.7% after 48 h of treatment.
3.3. PQS induced caspase-3/7 activity and cytotoxicity
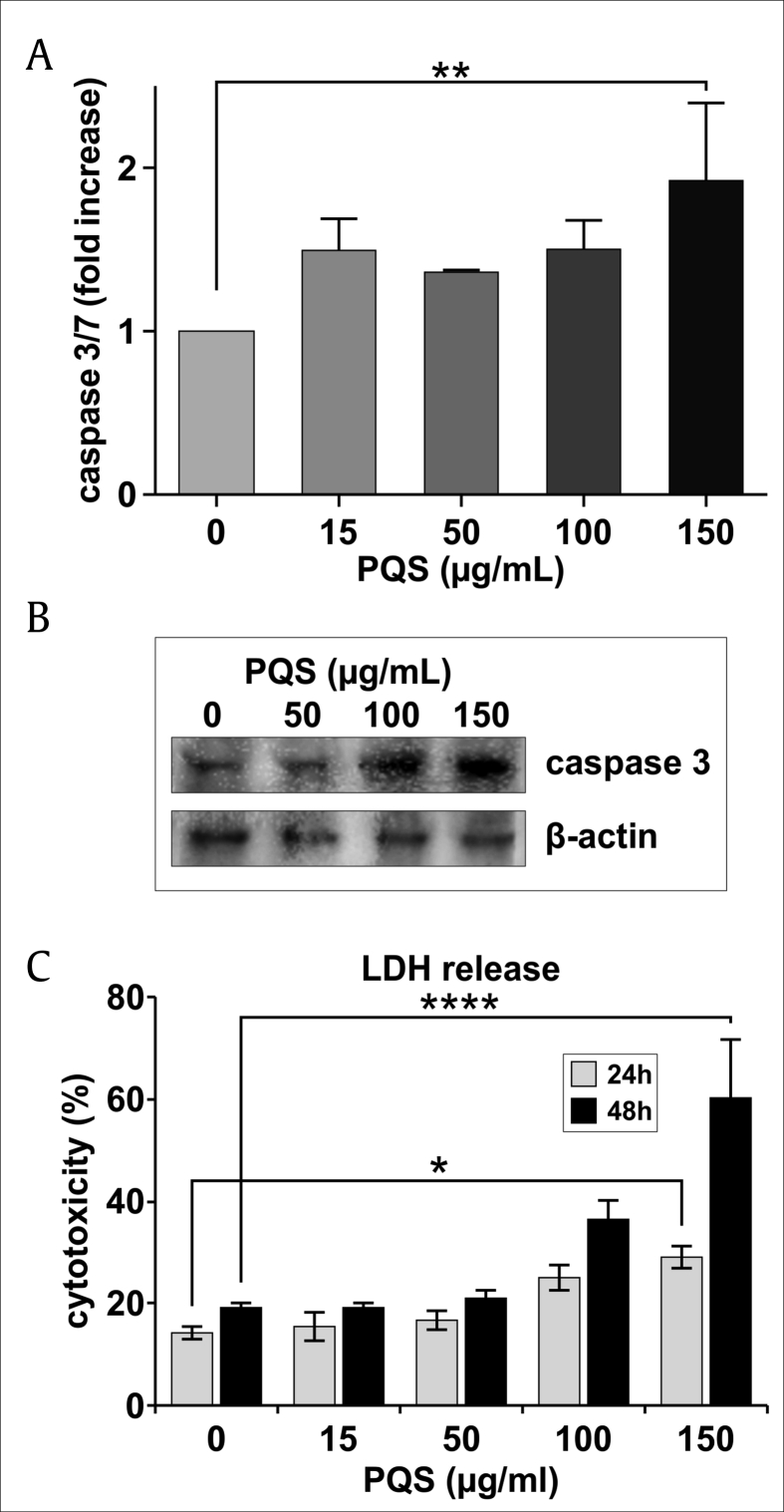
To further evaluate anti-proliferation effects of PQS on DU145 cells, the ApoTox-Glo Triplex assay, which includes the viability and caspase assays, and the LDH cytotoxicity assay, was performed in parallel under the same conditions. In the caspase assay, apoptosis was detected using Caspase-Glo to measure caspase-3/7 activity. Caspase activity is shown as fold increase of treated cells versus untreated cells (vehicle). The results showed that PQS increased significantly caspase activity by 90.54% at 150 μg/mL (Fig. 4A).
Fig. 4.
PQS treatment induced caspase-3/7 activity (A), expression levels of cleaved caspase-3 (B) and cytotoxicity (C). DU145 cells were treated with PQS (0, 15, 50, 100, and 150 μg/mL) for 48 h for the ApoTox-Glo Triplex assay and western blots, and for 24 h and 48 h for LDH cytotoxicity assay. Caspase-3/7 activity was determined by luminescence measurement and is shown as fold increase of treated cells versus untreated cells (vehicle) (A). Western blot analysis of cleaved caspase-3 (B) showed that PQS treatment raised the levels of the cleaved caspase-3 in a dose-dependent manner, which is in agreement with the data of caspase-3/7 activity assay. Cytotoxicity was determined using a LDH release assay by fluorescence measurement at 485Ex/520Em (C). PQS caused a significant, time- and dose-dependent increase of LDH release at concentrations of 15 to 150 μg/mL in comparison with the control group. Error bars represent standard deviations (SD) from n ≥ 3 independent experiments in triplicate. ∗, ∗∗, and ∗∗∗∗ represent p < 0.05, p < 0.01, and p < 0.0001, as obtained by one-way ANOVA (A).
Western blot analysis showed that PQS treatment with different concentrations (50, 100, and 150 μg/mL) raised the expression levels of cleaved caspase-3 significantly in a dose-dependent manner (Fig. 4B), which was in consistence with caspase-3/7 activity data obtained by ApoTox-Glo caspase assays (Fig. 4A, ∗∗p < 0.01).
The cytotoxic effect of different concentrations (0, 15, 50, 100, and 150 μg/mL) of PQS on DU145 cells was determined using a LDH Cytotoxicity Assay Kit. PQS caused a significant, time- and dose-dependent increase in LDH release at concentrations of 15 to 150 μg/mL after treatment for 24 h and 48 h. A concentration of 150 μg/mL of PQS led to a cytotoxicity value of 29.01% after 24 h and to 60.21% after 48 h in comparison with the control group (Fig. 4C, ∗p < 0.01 and ∗∗∗∗p < 0.0001).
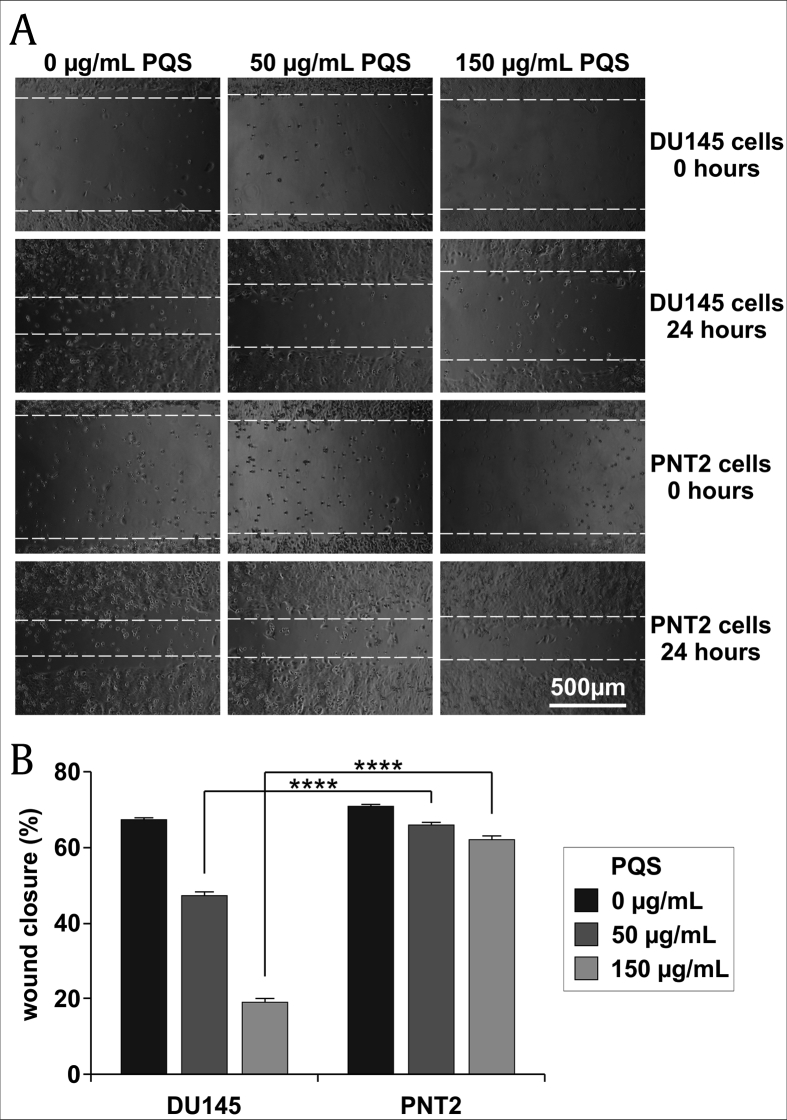
3.4. PQS treatment reduced the migration and invasion of DU145 cells
To gauge the anti-migratory activity of PQS, wound healing assays were carried out with DU145 and PNT2 cells after a 24 h treatment with PQS (150 μg/mL). Wound fields with a defined gap of 0.9 mm were inserted. Wound closures were photographed at 0 and 48 h after wounding and the percent closures were measured (Percent Closure = Migrated Cell Surface Area/Total Surface Area × 100). Fig. 5A and B show that the migration of PQS-treated DU145 cells was significantly inhibited compared to untreated cells. The percent wound closure was 24.8% in DU145 cells whereas it was 72.8% in PNT2 cells after treatment.
Fig. 5.
PQS treatment inhibited the migration of DU145 cells as shown by a wound healing assay. DU145 and PNT2 cells were treated with the indicated concentration of PQS and were tested using the CytoSelect Wound Healing Assay kit. Wound closures were photographed at 0 h and 24 h after wounding (A). The wound closure of DU145 cells was significantly smaller than that of PNT2 cells at concentrations of 50 and 150 μg/mL (B). Error bars represent standard deviations (SD) from n ≥ 3 independent experiments in triplicate. ∗∗∗∗ represents p < 0.0001 as obtained by one-way ANOVA.
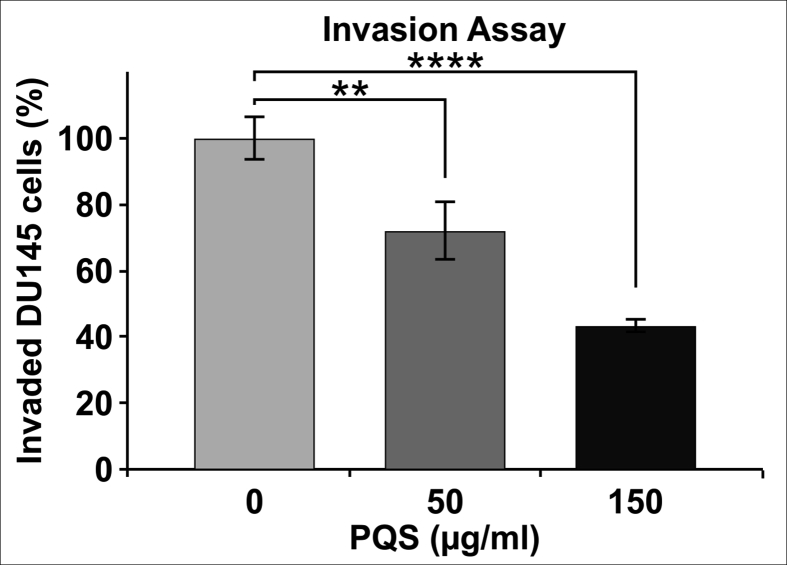
Invasion assays were performed using a CytoSelect Invasion Assay kit. Cells were treated with indicated concentrations of PQS for 24 h. Only 43.28% of DU145 cells were able to invade through the polycarbonate membrane compared to the untreated cells. The results showed that both concentrations of PQS (50 and 150 μg/mL) inhibited DU145 cell invasion (Fig. 6).
Fig. 6.
PQS treatment reduced the invasion of DU145 cells. Cells were treated with different concentrations of PQS (50 and 150 μg/mL) for 24 h. Invasion assays were performed using the CytoSelect Invasion Assay kit. Both concentrations of PQS inhibited DU145 cell invasion compared to untreated cells. Only 43.28% of DU145 cells were able to invade through the polycarbonate membrane after treatment with 150 μg/mL PQS. Error bars represent standard deviations (SD) from n ≥ 3 independent experiments in triplicate. ∗∗ and ∗∗∗∗ represent p < 0.01 and p < 0.0001 as obtained by two-way ANOVA.
3.5. PQS induced the DU145 cell cycle arrest at the G1 phase
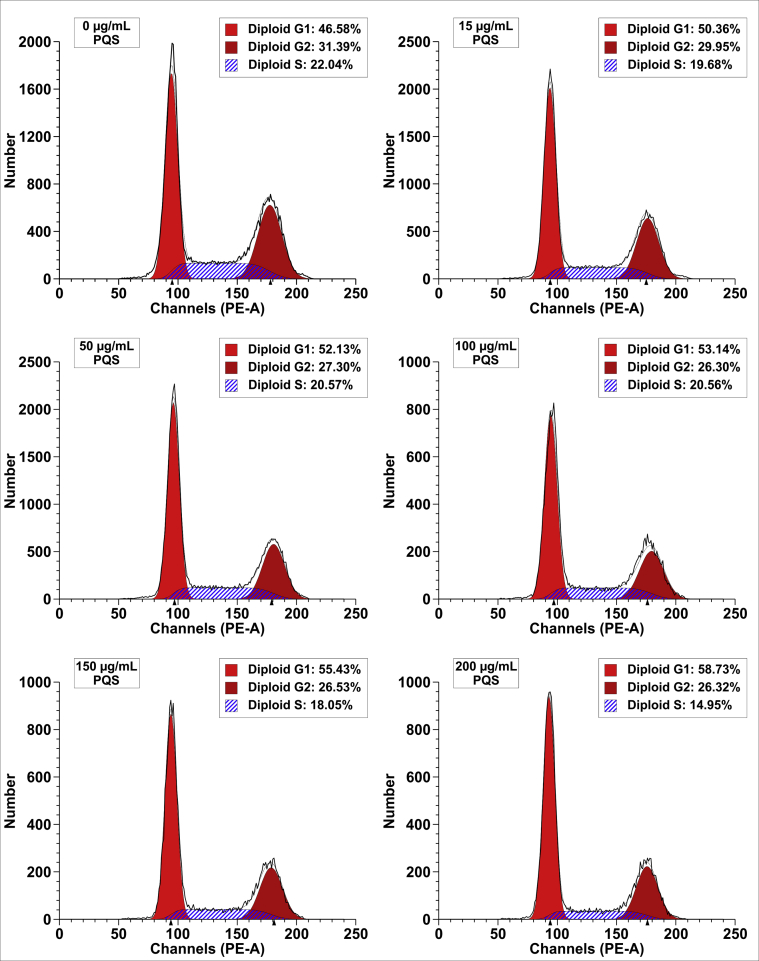
A main characteristic of cancer cells is the deregulation of cell cycle controls. To further define whether the anti-proliferative effect of PQS is due to inhibition of cell cycle progression, we performed a flow cytometric analysis of the cell cycle in DU145 cells. DU145 cells were treated with indicated concentrations of PQS (0, 50, 100, and 150 μg/mL) for 48 h, stained with the PI/RNase staining buffer, and analyzed by FACS. PQS increased the number of DU145 cells from 46.58% to 58.73% in the G1 phase, decreased the number of DU145 cells from 31.39% to 26.32% in the G2 phase, and from 22.04% to 14.95% in the S phase in a dose-dependent manner (Fig. 7). These findings demonstrate that cell cycle arrest in DU145 cells is induced by PQS via alteration of protein levels that influence cell cycle progression in the G1 phase. Examination of the cellular levels of cell cycle regulatory proteins that promote the G1 transition furthermore confirmed the PQS-induced cell cycle arrest of DU145 cells. The overall arrest induced by PQS may account for its inhibitory effect on DU145 cell growth.
Fig. 7.
PQS treatment induced the cell cycle arrest at the G1 phase in DU145 cells. DU145 cells were treated with the indicated concentrations of PQS for 48 h. Propidium iodide was used for FACS analysis of DNA content in fixed cells. PQS increased the number of cells in the G1 phase from 46.58% to 58.73% and decreased the number of cells in the G2 phase from 31.39% to 26.32% and in the S phase from 22.04% to 14.95% in a dose-dependent manner.
3.6. PQS treatment regulated the expression level of multiple cancer-related genes
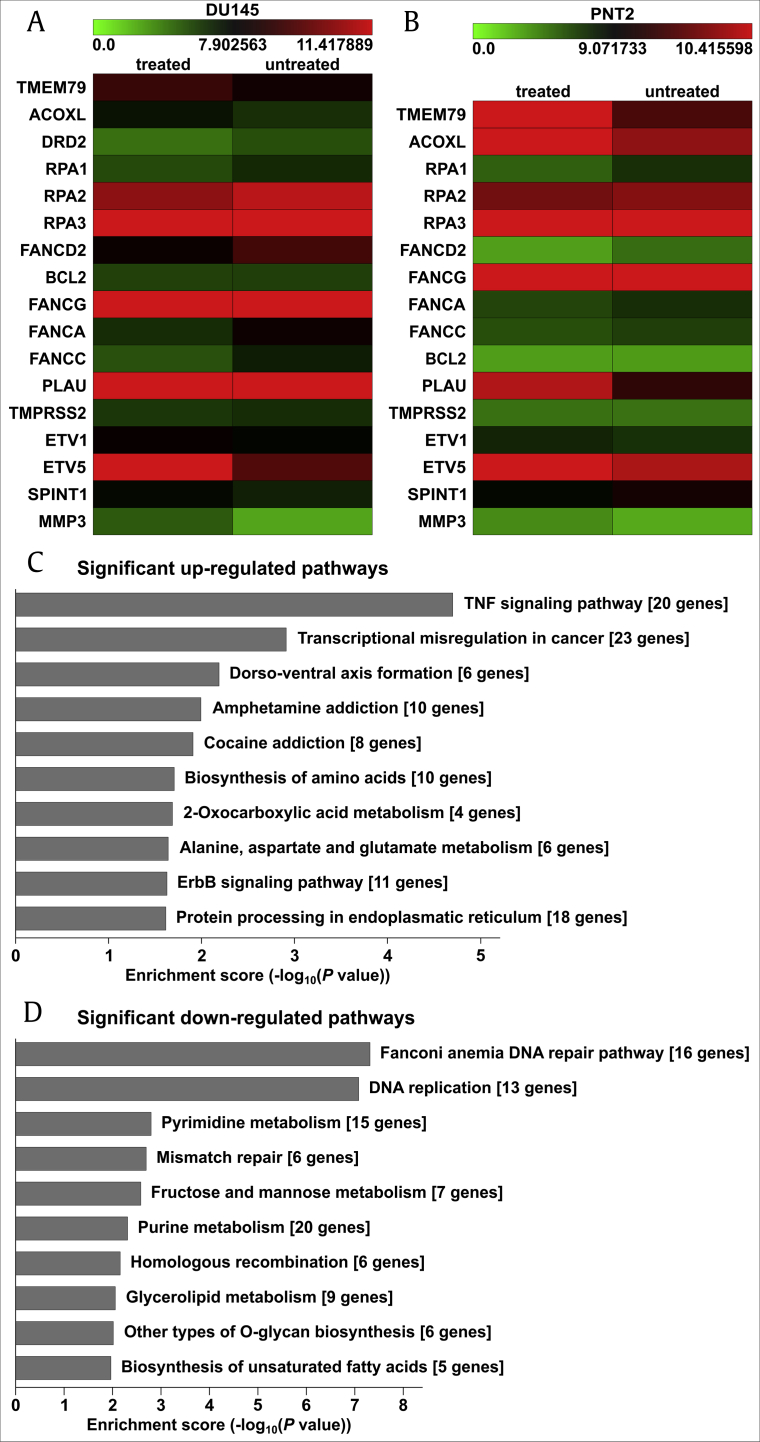
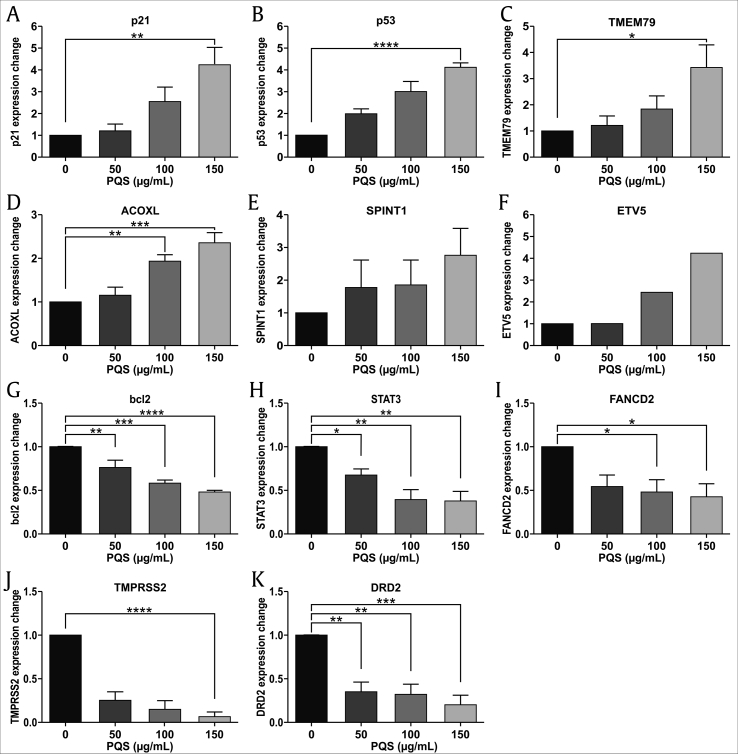
We used the oligonucleotide microarray technique to analyze miRNA expression profiles in DU145 and PNT2 cells after PQS treatment. Differential miRNA expression data were evaluated using a hierarchical cluster analysis software. A total of 28,647 human genes were detected and 3,199 differentially expressed genes were discovered in DU145 cells whereas 25,602 genes were detected and 1,205 differentially expressed genes were identified in PNT2 cells. Differentially expressed genes were identified when their expression was decreased or increased more than two fold in PQS-treated cells. The results showed that the expression of 1,501 genes was down-regulated and 1,698 genes was up-regulated in PQS-treated DU145 cells. Likewise, the expression of 598 genes was down-regulated and 607 genes was up-regulated in PQS-treated PNT2 cells. Among the expressed genes which were the most related to altered migration and cell cycle arrest after PQS treatment, p21, p53, ETV5, SPINT1, ACOXL, and TMEM79 were up-regulated, whereas bcl2, DRD2, TMPRSS2, FANCD2, and STAT3 were down-regulated in our experiments (Fig. 8A and B). The miRNA expression intensity is visualized using a color scale ranging between green (low) and red (high). The top ten significantly altered pathways are listed in Fig. 8C and D.
Fig. 8.
PQS treatment regulated the expression levels of multiple cancer-related genes and pathways. An oligonucleotide microarray was used to analyze the miRNA expression profiles in DU145 and PNT2 cells after PQS treatment (A, B). Hierarchical clustering analysis was employed to evaluate differential miRNA expression between treated and untreated groups. p21, p53, ETV5, SPINT1, ACOXL, and TMEM79 were up-regulated, whereas bcl2, DRD2, TMPRSS2, FANCD2, and STAT3 were down-regulated in the treatment groups. The enrichment scores of ten significant up- and down-regulated pathways are shown (C, D).
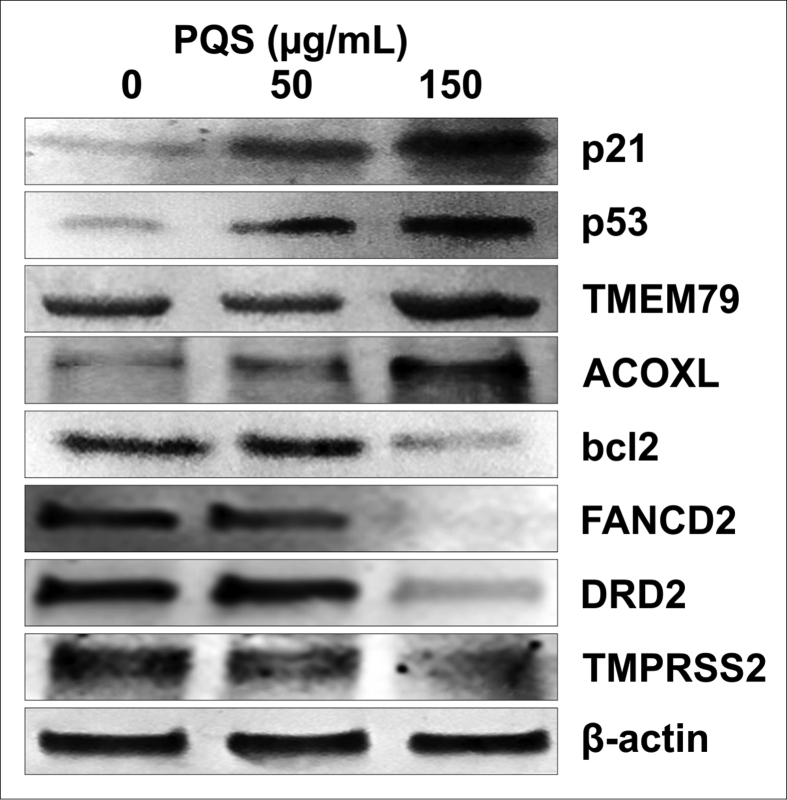
We performed qRT-PCR and western blot analyses to evaluate the altered mRNAs and protein expression levels after PQS treatment. Current studies indicate that p53, p21, STAT3, and bcl2 are the most important genes associated with cell apoptosis and cell cycle arrest [21,22,[57], [58], [59], [60], [61], [62]]. TMPRSS2, ETV5, SPINT1, FANCD2, and DRD2 are involved in tumor migration and invasion [[63], [64], [65], [66], [67], [68], [69]]. TMEM79 and ACOXL have been reported to be novel markers for PC research as well [5,70]. We tested whether PQS would alter the levels of these genes in DU145 cells and thereby would convey its anti-cancer activities. The qRT-PCR results displayed that PQS significantly down-regulated the expression levels of bcl2, DRD2, TMPRSS2, FANCD2, and STAT3 and up-regulated the expression levels of p21, p53, ETV5, SPINT1, ACOXL, and TMEM79 in DU145 cells versus in PNT2 cells in a dose-dependent manner mostly (Fig. 9, ∗p < 0.05, ∗∗p < 0.01, ∗∗∗p < 0.001, ∗∗∗∗p < 0.0001). Representative protein bands of western blots supported the qRT-PCR results. Upon PQS treatment of DU145 cells, we found that the protein levels of bcl2, DRD2, FANCD2, and TMPRSS2 were decreased, whereas those of p53, p21, TMEM79, and ACOXL were increased (Fig. 10). PQS treatment regulated the mRNA and protein expression levels in DU145 cells consistently.
Fig. 9.
PQS treatment regulated the expression of multiple cancer-related genes at the mRNA level. DU145 cells were treated with indicated concentrations of PQS for 48 h. mRNA of cells was extracted for qRT-PCR. The expression levels of p21, p53, TMEM79, ACOXL, SPINT1, and ETV5 were up-regulated after PQS treatment (A–F). The expression levels of bcl2, STAT3, FANCD2, TMPRSS2, and DRD2 were down-regulated after PQS treatment (G–K). The qRT-PCR was performed using a SYBR Green PCR Master Mix kit. Error bars represent standard deviations (SD) from n ≥ 3 independent experiments in triplicate. ∗, ∗∗, ∗∗∗, and ∗∗∗∗ represent p < 0.05, p < 0.01, p < 0.001, and p < 0.0001 as obtained by one-way ANOVA.
Fig. 10.
PQS treatment regulated the expression of multiple cancer-related genes at the protein level. DU145 cells were collected after PQS treatment for 48 h. Cell lysates were prepared for SDS-PAGE and western blotting. β-Actin served as the control protein. The expression levels of p21, p53, TMEM79, and ACOXL were up-regulated and the expression levels of bcl2, FANCD2, TMPRSS2, and DRD2 were down-regulated after PQS treatment in a dose-dependent manner. Proteins were detected using BCIP/NBT color development substrate.
4. Discussion
PC is the most prevalent cancer among men worldwide [70]. Hormone treatment and radiotherapy are generally administered to shrink the tumor, but often are accompanied by different side effects [14,[71], [72], [73], [74], [75], [76], [77]]. Therefore complementary and alternative therapies are receiving more attention and have been applied in oncotherapy in some Asian and European countries due to their relatively low toxicity and fewer side effects [78,79]. The Chinese herbal medicine PQS has been applied for treatment of various diseases including cancer [34,36,37,44,53,55,[80], [81], [82], [83]]. Published studies showed that different single ginsenosides exhibit an inhibitory effect on the proliferation of colorectal cancer cells [50,51,54], non-small cell lung cancer cells [52], pancreatic cancer cells [53], ovarian cancer cells [55,56], and PC cells [18,21,[83], [84], [85], [86], [87]]. To isolate and purify single ginsenosides for the treatment requires complicated separation techniques and the yield is low [48]. We investigated the ginsenosides of PQS, which were identified as Re, Rg1, Rc, Rb1, Rb2, Rb3, Rd, and Rg3, to evaluate their anti-PC efficacy in this study. Our data suggest that PQS have efficacious anti-PC cell activities in agreement with previous published results.
According to the WHO global survey, the efficacy, safety, and quality control of traditional medicine and complementary and alternative medicines have become inportant issues for TCM research [88]. We used the UHPLC-QTOF-MS/MS method to evaluate the stability and quality of different batches of PQS during our study [48,89]. We obtained comparable chromatograms of six batches of PQS measured three times in one month. Our data demonstrated that the PQS extracts were stable during the measurements and have repeatable chromatographs among batches, which were consistent with previous studies [89]. We used quality-controlled PQS by UHPLC-QTOF-MS/MS for our subsequent cellular and immunological experiments.
We compared the cell viability of DU145 cells with PC3 cells, another human PC cell line, and epithelial PNT2 cells. The inhibitory activity of PQS on DU145 cells was found to be similar to that of PC3 cells versus a much reduced effect on PNT2 cells (data not shown). We studied the influence of PQS on cell proliferation, invasion, migration, apoptosis, and cell cycle of both DU145 and PNT2 cells. Our data showed that PQS exhibited a potent inhibitory effect on DU145 cell growth, but apparently did not have a cytotoxic effect on PNT2 cells.
Cell migration is a particular important aspect to investigate in cancer research and it is also widely used in immunological and wound healing studies [55,90]. Because distant metastasis is especially common in PC patients, the inhibition of migratory and invasive abilities of cancer cells is remarkably important [20,65,86,91]. Transwell cell migration and invasion assays as well as wound closure assays show detailed information of cell migratory behaviors and thus may reveal molecular mechanisms of cell migration [90]. Accordingly, cell motility assays were used to determine the invasion capabilities and migration velocity of DU145 and PNT2 cells after PQS treatment, which significantly inhibited the migration distance and suppressed the invasive properties of DU145 cells in a dose-dependent manner.
Biomarker assays for diagnosis, prognosis, and treatment prediction in PC is a rapidly expanding field in recent years. A combination of different markers will be able to guide future clinical decisions rather than a single biomarker [[92], [93], [94]]. Current research indicated that p53, p21, STAT3, and bcl2 are the most important genes related to cancer research [21,22,[57], [58], [59], [60], [61], [62]]. TMPRSS2, ETV5, SPINT1, FANCD2, and DRD2 were found to be related to cell migration and invasion in PC [[63], [64], [65], [66], [67], [68], [69]]. TMEM79 and ACOXL were reported to be novel markers for PC as well [5,70]. Our results suggested that these cancer-related genes associated with restraining cell growth, inhibiting cell proliferation, regulating cell cycle, inducing apoptosis, and suppressing cell migration and invasion were modified by PQS, which likely contributed to its anti-cancer effect. This suggested that PQS not only works on a certain pathway, but also acts on multiple cancer-related pathways, thus inducing apoptosis and inhibiting proliferation.
p53 and bcl2 are two of the earliest known cancer-related genes [58]. The p53 gene represents a key regulator of cellular growth control, which has been reported to have the ability to induce either growth arrest or cell death [95]. Alterations of the p53 protein were mediated via interaction with its downstream effector p21 [57,59,69,96]. Up-regulated expression of p21 following p53 activation in response to DNA damage can cause either cell cycle arrest at the G1 phase or apoptosis, which may be important in the development of PC [57]. We showed that after PQS treatment, the expression levels of p53 and p21 were up-regulated in DU145 cells. This suggested that PQS may affect the cell cycle, which consequently leads to apoptosis in DU145 cells. The proteins of the bcl2 family are important regulators of many signals leading to caspase activation [60,97,98]. The p53 gene inhibits bcl2 transcription and directly impacts bcl2 activity as part of a transcription-independent pathway to cell death [58,99,100]. Overexpression of bcl2 significantly affected the expression or phosphorylation of STAT3 [101], which is believed to be a potential molecular target due to the regulation of different genes related to metastasis and angiogenesis [22,62,102]. Our experiments indicated that PQS down-regulated the expression levels of bcl2 and STAT3, which is eventually leading to the suppression of DU145 cell proliferation, angiogenesis, and metastasis.
TMPRSS2, ETV5, and SPINT1 were found to be associated with a more aggressive form of PC [64,65,92] and have been reported to play a significant role in cell invasion, migration, and proliferation [63,91]. DRD2 was reported to have an effect on cell invasion and migration in human non-functioning pituitary cancer cells and pancreatic ductal adenocarcinoma [68,69]. The FANCD2 protein was found to be involved in the regulation of the downstream DNA-repair events and cell cycle in rhabdomyosarcoma cells, bystander cells, and HeLa cells [66,[103], [104], [105]]. Our data indicated that the expression levels of TMPRSS2, DRD2, and FANCD2 were down-regulated whereas ETV5 and SPINT1 were up-regulated by PQS in PC DU145 cells. This indicated that the anti-PC activity of PQS could be due to the modified regulation of these genes.
Transmembrane proteins play a crucial role as transporters and receptors [106]. Misplaced transmembrane proteins are related to several serious diseases, making transmembrane proteins important targets for biological drugs [107,108]. Recent research showed that TMEM79 was highly expressed in normal prostate tissue but only lowly expressed in PC tissue and indicated a transfer of the proteins from the inside of the membrane into the cytoplasm when the normal epithelium transformed into tumor epithelial cells [5,70]. The potential mechanisms and consequences of low expression levels in PC cells are unclear as the function of TMEM79 is yet to be clarified [70]. It is of importance to continue the further functional analysis, such as comparison of gene expression levels in matched tissues and serum, to examine its clinical impact in disease screening.
ACOXL showed high expression in the granular cytoplasm of the mitochondrial region in most of benign glands and low expression in the prostate tumors, which enables specificity and high sensitivity in recognizing benign prostate glands in tissue samples [70]. Another study showed an enrichment of ACOXL in PC serum indicating that the transformation of normal epithelium to tumor epithelium may lead to reduced levels of ACOXL expression in the epithelium possibly because of ACOXL leaking into the serum [70]. The expression level of TMEM79 and ACOXL in PC tissue was lower than in normal tissue [109]. We tested the expression of these genes in DU145 and PNT2 cells treated with PQS. We found these two genes were always expressed lower in DU145 cells than in PNT2 cells. The expression level of TMEM79 and ACOXL in DU145 cells was up-regulated after PQS treatment, which was consistent with the previous results [70,106]. O'Hurley et al. indicated in 2015 in their study that the observed high sensitivity and specificity of TMEM79 and ACOXL at detecting benign prostate may suggest that these markers could be beneficial to assist pathological diagnosis. The differentiation of the expression levels of TMEM79 and ACOXL genes in DU145 and PNT2 cells proved that both genes might have potential as specific biomarkers in PC research.
In conclusion, we demonstrated that PQS have good stability and quality, which might be beneficiary for an effective herbal remedy playing a complementary or alternative role for treating PC. Our data indicated that PQS suppressed the proliferation of DU145 cells, reduced their cell viability, and exhibited higher cytotoxicity in comparison with PNT2 cells. G1 phase cell cycle arrest and apoptosis were also induced by PQS. A significant decrease in DU145 cell invasion and migration were observed after PQS treatment. PQS up-regulated the expression levels of p21, p53, TMEM79, ACOXL, ETV5, and SPINT1 whereas it down-regulated the expression levels of bcl2, STAT3, FANCD2, DRD2, and TMPRSS2. TMEM79 and ACOXL were expressed significantly higher in PNT2 cells than in DU145 cells and thus could be novel biomarker candidates for PC diagnosis in the future.
Conflicts of interest
All authors have no conflicts of interest to declare.
Acknowledgments
This study was funded by the Austrian Federal Ministry of Science and Research, the Austrian Federal Ministry of Health (GZ 402.000/0006-II/6b/2012), and the Austrian Federal Ministry of Science, Research and Economy (GZ 402.000/0009-WF/V/6/2016) awarded to Yan Ma, and supported by Austrian Eurasia Pacific UNINET Technology Scholarships awarded to Lu Liu (2014) and Lixia Lou (2016) and an Ernst Mach Scholarship awarded to Fanqiao Lyu (2017). The authors would like to thank Professor Enikö Kallay group for supporting the cell lines.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.jgr.2019.12.007.
Appendix A. Supplementary data
The following is the Supplementary data to this article:
References
- 1.Cao B., Bray F., Beltran-Sanchez H., Ginsburg O., Soneji S., Soerjomataram I. Benchmarking life expectancy and cancer mortality: global comparison with cardiovascular disease 1981-2010. BMJ. 2017;357:j2765. doi: 10.1136/bmj.j2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Siegel R.L., Miller K.D., Jemal A. Cancer statistics. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. 2017. [DOI] [PubMed] [Google Scholar]
- 3.Sadeghi-Gandomani H.R., Yousefi M., Rahimi S., Yousefi S., Karimi-Rozveh A., Hosseini S., Mahabadi A.A., Abarqui H.F., Borujeni N.N., Salehiniya H. The incidence, risk factors, and knowledge about the prostate cancer through worldwide and Iran. WCRJ. 2017;4(4):e972. 2017. [Google Scholar]
- 4.Mottet N., Bellmunt J., Briers E., Bolla M., Cornford P., De Santis M., Henry A., Joniau S., Lam T., Mason M.D. European Association of Urology; 2016. EAU - ESTRO - SIOG guidelines on prostate cancer. 2016. [DOI] [PubMed] [Google Scholar]
- 5.Pin E., Henjes F., Hong M.G., Wiklund F., Magnusson P., Bjartell A. Identification of a novel autoimmune peptide epitope of prostein in prostate cancer. J Proteome Res. 2017;16:204–216. doi: 10.1021/acs.jproteome.6b00620. [DOI] [PubMed] [Google Scholar]
- 6.Loeb S., Bjurlin M.A., Nicholson J., Tammela T.L., Penson D.F., Carter H.B. Overdiagnosis and overtreatment of prostate cancer. Eur Urol. 2014;65:1046–1055. doi: 10.1016/j.eururo.2013.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suzuki Y., Kondo Y., Himeno S., Nemoto K., Akimoto M., Imura N. Role of antioxidant systems in human androgen-independent prostate cancer cells. Prostate. 2000;43:144–149. doi: 10.1002/(sici)1097-0045(20000501)43:2<144::aid-pros9>3.0.co;2-h. [DOI] [PubMed] [Google Scholar]
- 8.Feldman B.J., Feldman D. The development of androgen-independent prostate cancer. Nature Reviews Cancer. 2001;1(1):34–45. doi: 10.1038/35094009. [DOI] [PubMed] [Google Scholar]
- 9.Thalmann G.N., Anezinis P.E., Chang S.M., Zhau H.E., Kim E.E., Hopwood V.L. Androgen-independent cancer progression and bone metastasis in the LNCaP model of human prostate cancer. Cancer Res. 1994;54:2577–2581. [PubMed] [Google Scholar]
- 10.Chaturvedi S., Garcia J.A. Novel agents in the management of castration resistant prostate cancer. J Carcinog. 2014;13:5. doi: 10.4103/1477-3163.128185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Antonarakis E.S., Lu C., Wang H., Luber B., Nakazawa M., Roeser J.C. AR-V7 and resistance to enzalutamide and abiraterone in prostate cancer. N Engl J Med. 2014;371:1028–1038. doi: 10.1056/NEJMoa1315815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang T., Zhu J., George D.J., Armstrong A.J. Enzalutamide versus abiraterone acetate for the treatment of men with metastatic castration-resistant prostate cancer. Expert Opin Pharmacother. 2015;16:473–485. doi: 10.1517/14656566.2015.995090. [DOI] [PubMed] [Google Scholar]
- 13.Badrising S., van der Noort V., van Oort I.M., van den Berg H.P., Los M., Hamberg P. Clinical activity and tolerability of enzalutamide (MDV3100) in patients with metastatic, castration-resistant prostate cancer who progress after docetaxel and abiraterone treatment. Cancer. 2014;120:968–975. doi: 10.1002/cncr.28518. [DOI] [PubMed] [Google Scholar]
- 14.Harrison M.R., Wong T.Z., Armstrong A.J., George D.J. Radium-223 chloride: a potential new treatment for castration-resistant prostate cancer patients with metastatic bone disease. Cancer Manag Res. 2013;5:1–14. doi: 10.2147/CMAR.S25537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Domnick M., Domnick M., Wiebelitz K.R., Beer A.M. Evaluation of the effectiveness of a multimodal complementary medicine program for improving the quality of life of cancer patients during adjuvant radiotherapy and/or chemotherapy or outpatient Aftercare. Oncology. 2017;93:83–91. doi: 10.1159/000468939. [DOI] [PubMed] [Google Scholar]
- 16.van Die M.D., Williams S.G., Emery J., Bone K.M., Taylor J.M., Lusk E. A placebo-controlled double-blinded randomized pilot study of combination phytotherapy in biochemically recurrent prostate cancer. Prostate. 2017;77:765–775. doi: 10.1002/pros.23317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paur I., Lilleby W., Bohn S.K., Hulander E., Klein W., Vlatkovic L. Tomato-based randomized controlled trial in prostate cancer patients: effect on PSA. Clin Nutr. 2017;36:672–679. doi: 10.1016/j.clnu.2016.06.014. [DOI] [PubMed] [Google Scholar]
- 18.Park J.Y., Choi P., Kim H.K., Kang K.S., Ham J. Increase in apoptotic effect of Panax ginseng by microwave processing in human prostate cancer cells: in vitro and in vivo studies. J Ginseng Res. 2016;40:62–67. doi: 10.1016/j.jgr.2015.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wilt T.J., Ishani A., Rutks I., MacDonald R. Phytotherapy for benign prostatic hyperplasia. Public Health Nutr. 2000;3:459–472. doi: 10.1017/s1368980000000549. [DOI] [PubMed] [Google Scholar]
- 20.Hirata H., Hinoda Y., Shahryari V., Deng G., Tanaka Y., Tabatabai Z.L. Genistein downregulates onco-miR-1260b and upregulates sFRP1 and Smad4 via demethylation and histone modification in prostate cancer cells. Br J Cancer. 2014;110:1645–1654. doi: 10.1038/bjc.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yoo J.H., Kwon H.C., Kim Y.J., Park J.H., Yang H.O. KG-135, enriched with selected ginsenosides, inhibits the proliferation of human prostate cancer cells in culture and inhibits xenograft growth in athymic mice. Cancer Lett. 2010;289:99–110. doi: 10.1016/j.canlet.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 22.Kim J.K., Kim J.Y., Kim H.J., Park K.G., Harris R.A., Cho W.J. Scoparone exerts anti-tumor activity against DU145 prostate cancer cells via inhibition of STAT3 activity. PLoS One. 2013;8 doi: 10.1371/journal.pone.0080391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pounis G., Tabolacci C., Costanzo S., Cordella M., Bonaccio M., Rago L. Reduction by coffee consumption of prostate cancer risk: evidence from the Moli-sani cohort and cellular models. Int J Cancer. 2017;141:72–82. doi: 10.1002/ijc.30720. [DOI] [PubMed] [Google Scholar]
- 24.Kim H.S., Lee H., Shin S.J., Beom S.H., Jung M., Bae S., Lee E.Y., Park K.H., Choi Y.Y., Son T. Complementary utility of targeted next-generation sequencing and immunohistochemistry panels as a screening platform to select targeted therapy for advanced gastric cancer. Oncotarget. 2017;8(24):38389–38398. doi: 10.18632/oncotarget.16409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Soares N.D., Machado C.L., Trindade B.B., Lima I.C., Gimba E.R., Teodoro A.J. Lycopene extracts from different tomato-based food products induce apoptosis in cultured human primary prostate cancer cells and regulate TP53, Bax and Bcl-2 transcript expression. Asian Pac J Cancer Prev. 2017;18:339–345. doi: 10.22034/APJCP.2017.18.2.339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhang H.Y., Cui J., Zhang Y., Wang Z.L., Chong T., Wang Z.M. Isoflavones and prostate cancer: a review of some critical issues. Chin Med J (Engl) 2016;129:341–347. doi: 10.4103/0366-6999.174488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y., Zhi F., Chen P., Zhao K., Xiang H., Mao Q. Green tea and the risk of prostate cancer: a systematic review and meta-analysis. Medicine (Baltimore) 2017;96 doi: 10.1097/MD.0000000000006426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lang A., Neuhaus J., Pfeiffenberger M., Schroder E., Ponomarev I., Weber Y. Optimization of a nonviral transfection system to evaluate Cox-2 controlled interleukin-4 expression for osteoarthritis gene therapy in vitro. J Gene Med. 2014;16:352–363. doi: 10.1002/jgm.2812. [DOI] [PubMed] [Google Scholar]
- 29.Li W., Yan M.H., Liu Y., Liu Z., Wang Z., Chen C. Ginsenoside Rg5 ameliorates cisplatin-induced nephrotoxicity in mice through inhibition of inflammation, oxidative stress, and apoptosis. Nutrients. 2016;8 doi: 10.3390/nu8090566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Han S.Y., Li H.X., Ma X., Zhang K., Ma Z.Z., Jiang Y. Evaluation of the anti-myocardial ischemia effect of individual and combined extracts of Panax notoginseng and Carthamus tinctorius in rats. J Ethnopharmacol. 2013;145:722–727. doi: 10.1016/j.jep.2012.11.036. [DOI] [PubMed] [Google Scholar]
- 31.Liang Q.L., Liang X.P., Wang Y.M., Xie Y.Y., Zhang R.L., Chen X. Effective components screening and anti-myocardial infarction mechanism study of the Chinese medicine NSLF6 based on “system to system” mode. J Transl Med. 2012;10:26. doi: 10.1186/1479-5876-10-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Attele Anoja S., Wu J.A., Yuan C.-S. Ginseng pharmacology Multiple constituents and multiple actions. Biochemical Pharmacology. 1999;58:1685–1693. doi: 10.1016/s0006-2952(99)00212-9. [DOI] [PubMed] [Google Scholar]
- 33.Schlag E.M., McIntosh M.S. Ginsenoside content and variation among and within American ginseng (Panax quinquefolius L.) populations. Phytochemistry. 2006;67:1510–1519. doi: 10.1016/j.phytochem.2006.05.028. [DOI] [PubMed] [Google Scholar]
- 34.Scholey A., Ossoukhova A., Owen L., Ibarra A., Pipingas A., He K. Effects of American ginseng (Panax quinquefolius) on neurocognitive function: an acute, randomised, double-blind, placebo-controlled, crossover study. Psychopharmacology (Berl) 2010;212:345–356. doi: 10.1007/s00213-010-1964-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chang Y.D., Smith J., Portman D., Kim R., Oberoi-Jassal R., Rajasekhara S. Single institute experience with methylphenidate and American Ginseng in cancer-related fatigue. Am J Hosp Palliat Care. 2018;35:144–150. doi: 10.1177/1049909117695733. [DOI] [PubMed] [Google Scholar]
- 36.Barton D.L., Liu H., Dakhil S.R., Linquist B., Sloan J.A., Nichols C.R. Wisconsin Ginseng (Panax quinquefolius) to improve cancer-related fatigue: a randomized, double-blind trial, N07C2. J Natl Cancer Inst. 2013;105:1230–1238. doi: 10.1093/jnci/djt181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Barton D.L., Soori G.S., Bauer B.A., Sloan J.A., Johnson P.A., Figueras C. Pilot study of Panax quinquefolius (American ginseng) to improve cancer-related fatigue: a randomized, double-blind, dose-finding evaluation: NCCTG trial N03CA. Support Care Cancer. 2010;18:179–187. doi: 10.1007/s00520-009-0642-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.de la Taille A., Hayek O.R., Burchardt M., Burchardt T., Katz A.E. Role of herbal compounds (PC-SPES) in hormone-refractory prostate cancer: two case reports. J Altern Complement Med. 2000;6:449–451. doi: 10.1089/acm.2000.6.449. [DOI] [PubMed] [Google Scholar]
- 39.Kim S.J., Kim A.K. Anti-breast cancer activity of fine black ginseng (Panax ginseng Meyer) and ginsenoside Rg5. J Ginseng Res. 2015;39:125–134. doi: 10.1016/j.jgr.2014.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Shan X., Fu Y.S., Aziz F., Wang X.Q., Yan Q., Liu J.W. Ginsenoside Rg3 inhibits melanoma cell proliferation through down-regulation of histone deacetylase 3 (HDAC3) and increase of p53 acetylation. PLoS One. 2014;9 doi: 10.1371/journal.pone.0115401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zhang Z., Du G.J., Wang C.Z., Wen X.D., Calway T., Li Z. Compound K, a ginsenoside metabolite, inhibits colon cancer growth via multiple pathways including p53-p21 interactions. Int J Mol Sci. 2013;14:2980–2995. doi: 10.3390/ijms14022980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen P., Luthria D., Harrington Pde B., Harnly J.M. Discrimination among Panax species using spectral fingerprinting. J AOAC Int. 2011;94:1411–1421. doi: 10.5740/jaoacint.10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Chen P., Harnly J.M., Harrington Pde B. Flow injection mass spectroscopic fingerprinting and multivariate analysis for differentiation of three Panax species. J AOAC Int. 2011;94:90–99. [PMC free article] [PubMed] [Google Scholar]
- 44.Jia L., Zhao Y. Current evaluation of the millennium phytomedicine--ginseng (I): etymology, pharmacognosy, phytochemistry, market and regulations. Curr Med Chem. 2009;16:2475–2484. doi: 10.2174/092986709788682146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Attele A.S., Wu J.A., Yuan C.S. Ginseng pharmacology: multiple constituents and multiple actions. Biochem Pharmacol. 1999;58:1685–1693. doi: 10.1016/s0006-2952(99)00212-9. [DOI] [PubMed] [Google Scholar]
- 46.Huang K.C. CRC Press LLC; 1999. The pharmacology of Chinese herbs. [Google Scholar]
- 47.Bostwick D.G., Burke H.B., Djakiew D., Euling S., Ho S.M., Landolph J. Human prostate cancer risk factors. Cancer. 2004;101:2371–2490. doi: 10.1002/cncr.20408. [DOI] [PubMed] [Google Scholar]
- 48.Zhou S.S., Xu J.D., Zhu H., Shen H., Xu J., Mao Q. Simultaneous determination of original, degraded ginsenosides and aglycones by ultra high performance liquid chromatography coupled with quadrupole time-of-flight mass spectrometry for quantitative evaluation of Du-Shen-Tang, the decoction of ginseng. Molecules. 2014;19:4083–4104. doi: 10.3390/molecules19044083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chen S W.Z., Huang Y., O'Barr S.A., Wong R.A., Yeung S., Chow M.S. Ginseng and anticancer drug combination to improve cancer. Evidence-Based Complementary and Alternative Medicine. 2014;2014 doi: 10.1155/2014/168940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Tang Y.C., Zhang Y., Zhou J., Zhi Q., Wu M.Y., Gong F.R. Ginsenoside Rg3 targets cancer stem cells and tumor angiogenesis to inhibit colorectal cancer progression in vivo. Int J Oncol. 2018;52:127–138. doi: 10.3892/ijo.2017.4183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Yang X., Zou J., Cai H., Huang X., Yang X., Guo D. Ginsenoside Rg3 inhibits colorectal tumor growth via down-regulation of C/EBPbeta/NF-kappaB signaling. Biomed Pharmacother. 2017;96:1240–1245. doi: 10.1016/j.biopha.2017.11.092. [DOI] [PubMed] [Google Scholar]
- 52.Jiang Z., Yang Y., Yang Y., Zhang Y., Yue Z., Pan Z. Ginsenoside Rg3 attenuates cisplatin resistance in lung cancer by downregulating PD-L1 and resuming immune. Biomed Pharmacother. 2017;96:378–383. doi: 10.1016/j.biopha.2017.09.129. [DOI] [PubMed] [Google Scholar]
- 53.Jiang J., Yuan Z., Sun Y., Bu Y., Li W., Fei Z. Ginsenoside Rg3 enhances the anti-proliferative activity of erlotinib in pancreatic cancer cell lines by downregulation of EGFR/PI3K/Akt signaling pathway. Biomed Pharmacother. 2017;96:619–625. doi: 10.1016/j.biopha.2017.10.043. [DOI] [PubMed] [Google Scholar]
- 54.Huang G., Khan I., Li X., Chen L., Leong W., Ho L.T. Ginsenosides Rb3 and Rd reduce polyps formation while reinstate the dysbiotic gut microbiota and the intestinal microenvironment in Apc(Min/+) mice. Sci Rep. 2017;7:12552. doi: 10.1038/s41598-017-12644-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu D., Liu T., Teng Y., Chen W., Zhao L., Li X. Ginsenoside Rb1 inhibits hypoxia-induced epithelial-mesenchymal transition in ovarian cancer cells by regulating microRNA-25. Exp Ther Med. 2017;14:2895–2902. doi: 10.3892/etm.2017.4889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Deng S., Wong C.K.C., Lai H.C., Wong A.S.T. Ginsenoside-Rb1 targets chemotherapy-resistant ovarian cancer stem cells via simultaneous inhibition of Wnt/beta-catenin signaling and epithelial-to-mesenchymal transition. Oncotarget. 2017;8:25897–25914. doi: 10.18632/oncotarget.13071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sivonova M.K., Vilckova M., Kliment J., Mahmood S., Jurečeková J., Dušenková S. Association of p53 and p21 polymorphisms with prostate cancer. Biomed Rep. 2015;3:707–714. doi: 10.3892/br.2015.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hemann M.T., Lowe S.W. The p53-Bcl-2 connection. Cell Death Differ. 2006;13:1256–1259. doi: 10.1038/sj.cdd.4401962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Paek A.L., Liu J.C., Loewer A., Forrester W.C., Lahav G. Cell-to-Cell variation in p53 dynamics leads to fractional killing. Cell. 2016;165:631–642. doi: 10.1016/j.cell.2016.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Catz S.D., Johnson J. BCL-2 in prostate cancer: a minireview. Apoptosis. 2003;8:29–37. doi: 10.1023/a:1021692801278. [DOI] [PubMed] [Google Scholar]
- 61.Quinn L.M., Richardson H. Bcl-2 in cell cycle regulation. Cell Cycle. 2014;3:6–8. [PubMed] [Google Scholar]
- 62.Levy D.E., Lee C.K. What does Stat3 do? J Clin Invest. 2002;109:1143–1148. doi: 10.1172/JCI15650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Pellecchia A., Pescucci C., De Lorenzo E., Luceri C., Passaro N., Sica M. Overexpression of ETV4 is oncogenic in prostate cells through promotion of both cell proliferation and epithelial to mesenchymal transition. Oncogenesis. 2012;1 doi: 10.1038/oncsis.2012.20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Kim Hogyoung, Datta A., Talwar Sudha, Saleem Sarmad N., Mondal Debasis, Abdel-Mageed Asim B. Estradiol-ERβ2 signaling axis confers growth and migration of CRPC cells through TMPRSS2-ETV5 gene fusion. Oncotarget. 2017;8 doi: 10.18632/oncotarget.11355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Leshem O., Madar S., Kogan-Sakin I., Kamer I., Goldstein I., Brosh R. TMPRSS2/ERG promotes epithelial to mesenchymal transition through the ZEB1/ZEB2 axis in a prostate cancer model. PLoS One. 2011;6 doi: 10.1371/journal.pone.0021650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Shen C., Oswald D., Phelps D., Cam H., Pelloski C.E., Pang Q. Regulation of FANCD2 by the mTOR pathway contributes to the resistance of cancer cells to DNA double-strand breaks. Cancer Res. 2013;73:3393–3401. doi: 10.1158/0008-5472.CAN-12-4282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bretz A.C., Gittler M.P., Charles J.P., Gremke N., Eckhardt I., Mernberger M. DeltaNp63 activates the Fanconi anemia DNA repair pathway and limits the efficacy of cisplatin treatment in squamous cell carcinoma. Nucleic Acids Res. 2016;44:3204–3218. doi: 10.1093/nar/gkw036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peverelli E., Giardino E., Treppiedi D., Locatelli M., Vaira V., Ferrero S. Dopamine receptor type 2 (DRD2) inhibits migration and invasion of human tumorous pituitary cells through ROCK-mediated cofilin inactivation. Cancer Lett. 2016;381:279–286. doi: 10.1016/j.canlet.2016.08.005. [DOI] [PubMed] [Google Scholar]
- 69.Jandaghi P., Najafabadi H.S., Bauer A.S., Papadakis A.I., Fassan M., Hall A. Expression of DRD2 is increased in human pancreatic ductal adenocarcinoma and inhibitors slow tumor growth in mice. Gastroenterology. 2016;151:1218–1231. doi: 10.1053/j.gastro.2016.08.040. [DOI] [PubMed] [Google Scholar]
- 70.O’Hurley G., Busch C., Fagerberg L., Hallstrom B.M., Stadler C., Tolf A. Analysis of the human prostate-specific proteome defined by transcriptomics and antibody-based profiling identifies TMEM79 and ACOXL as two putative, diagnostic markers in prostate cancer. PLoS One. 2015;10 doi: 10.1371/journal.pone.0133449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Nilsson S., Strang P., Aksnes A.K., Franzen L., Olivier P., Pecking A. A randomized, dose-response, multicenter phase II study of radium-223 chloride for the palliation of painful bone metastases in patients with castration-resistant prostate cancer. Eur J Cancer. 2012;48:678–686. doi: 10.1016/j.ejca.2011.12.023. [DOI] [PubMed] [Google Scholar]
- 72.Cai G., Laslett L.L., Aitken D., Halliday A., Pan F., Otahal P. Effect of zoledronic acid and denosumab in patients with low back pain and modic change: a proof-of-principle trial. J Bone Miner Res. 2018;33:773–782. doi: 10.1002/jbmr.3376. [DOI] [PubMed] [Google Scholar]
- 73.Lewiecki E.M. Safety and tolerability of denosumab for the treatment of postmenopausal osteoporosis. Drug Healthc Patient Saf. 2011;3:79–91. doi: 10.2147/DHPS.S7727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Beth-Tasdogan N.H., Mayer B., Hussein H., Zolk O. Interventions for managing medication-related osteonecrosis of the jaw. Cochrane Database Syst Rev. 2017;10 doi: 10.1002/14651858.CD012432.pub2. CD012432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kennel K.A., Drake M.T. Adverse effects of bisphosphonates: implications for osteoporosis management. Mayo Clin Proc. 2009;84:632–637. doi: 10.1016/S0025-6196(11)60752-0. quiz 638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Woo S.B., Hellstein J.W., Kalmar J.R. Narrative [corrected] review: bisphosphonates and osteonecrosis of the jaws. Ann Intern Med. 2006;144:753–761. doi: 10.7326/0003-4819-144-10-200605160-00009. [DOI] [PubMed] [Google Scholar]
- 77.Khosla S., Burr D., Cauley J., Dempster D.W., Ebeling P.R., Felsenberg D. Bisphosphonate-associated osteonecrosis of the jaw: report of a task force of the American Society for Bone and Mineral Research. J Bone Miner Res. 2007;22:1479–1491. doi: 10.1359/jbmr.0707onj. [DOI] [PubMed] [Google Scholar]
- 78.Gordaliza M. Natural products as leads to anticancer drugs. Clin Transl Oncol. 2007;9:767–776. doi: 10.1007/s12094-007-0138-9. [DOI] [PubMed] [Google Scholar]
- 79.Karikas G.A. Anticancer and chemopreventing natural products: some biochemical and therapeutic aspects. J BUON. 2010;15:627–638. [PubMed] [Google Scholar]
- 80.Kou N., Xue M., Yang L., Zang M.X., Qu H., Wang M.M. Panax quinquefolius saponins combined with dual antiplatelet drug therapy alleviate gastric mucosal injury and thrombogenesis through the COX/PG pathway in a rat model of acute myocardial infarction. PLoS One. 2018;13 doi: 10.1371/journal.pone.0194082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Shishtar E., Sievenpiper J.L., Djedovic V., Cozma A.I., Ha V., Jayalath V.H. The effect of ginseng (the genus panax) on glycemic control: a systematic review and meta-analysis of randomized controlled clinical trials. PLoS One. 2014;9 doi: 10.1371/journal.pone.0107391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim Y.S., Woo J.Y., Han C.K., Chang I.M. Safety analysis of panax ginseng in randomized clinical trials: a systematic review. Medicines (Basel) 2015;2:106–126. doi: 10.3390/medicines2020106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kim H.S., Lee E.H., Ko S.R., Choi K.J., Park J.H., Im D.S. Effects of ginsenosides Rg3 and Rh2 on the proliferation of prostate cancer cells. Arch Pharm Res. 2004;27:429–435. doi: 10.1007/BF02980085. [DOI] [PubMed] [Google Scholar]
- 84.Gioti K., Tenta R. Bioactive natural products against prostate cancer: mechanism of action and autophagic/apoptotic molecular pathways. Planta Med. 2015;81:543–562. doi: 10.1055/s-0035-1545845. [DOI] [PubMed] [Google Scholar]
- 85.Bae J.S., Park H.S., Park J.W., Li S.H., Chun Y.S. Red ginseng and 20(S)-Rg3 control testosterone-induced prostate hyperplasia by deregulating androgen receptor signaling. J Nat Med. 2012;66:476–485. doi: 10.1007/s11418-011-0609-8. [DOI] [PubMed] [Google Scholar]
- 86.Pan X.Y., Guo H., Han J., Hao F., An Y., Xu Y. Ginsenoside Rg3 attenuates cell migration via inhibition of aquaporin 1 expression in PC-3M prostate cancer cells. Eur J Pharmacol. 2012;683:27–34. doi: 10.1016/j.ejphar.2012.02.040. [DOI] [PubMed] [Google Scholar]
- 87.Liu J., Shimizu K., Yu H., Zhang C., Jin F., Kondo R. Stereospecificity of hydroxyl group at C-20 in antiproliferative action of ginsenoside Rh2 on prostate cancer cells. Fitoterapia. 2010;81:902–905. doi: 10.1016/j.fitote.2010.05.020. [DOI] [PubMed] [Google Scholar]
- 88.Fan T.P., Deal G., Koo H.L., Rees D., Sun H., Chen S. Future development of global regulations of Chinese herbal products. J Ethnopharmacol. 2012;140:568–586. doi: 10.1016/j.jep.2012.02.029. [DOI] [PubMed] [Google Scholar]
- 89.Xue M., Yang L., Shi D.Z., Radauer C., Breiteneder H., Ma Y. Qualitative analysis of xinyue capsules by high-performance liquid chromatography: preliminary evaluation of drug quality in a Sino-Austrian joint study. Chin J Integr Med. 2015;21:772–777. doi: 10.1007/s11655-015-2311-2. [DOI] [PubMed] [Google Scholar]
- 90.Justus C.R., Leffler N., Ruiz-Echevarria M., Yang L.V. In vitro cell migration and invasion assays. J Vis Exp. 2014 doi: 10.3791/51046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Piccolella M., Crippa V., Messi E., Tetel M.J., Poletti A. Modulators of estrogen receptor inhibit proliferation and migration of prostate cancer cells. Pharmacol Res. 2014;79:13–20. doi: 10.1016/j.phrs.2013.10.002. [DOI] [PubMed] [Google Scholar]
- 92.Gaudreau P.O., Stagg J., Soulieres D., Saad F. The present and future of biomarkers in prostate cancer: proteomics, genomics, and immunology advancements. Biomark Cancer. 2016;8:15–33. doi: 10.4137/BIC.S31802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Nguyen J.K., Magi-Galluzzi C. Unfavorable pathology, tissue biomarkers and genomic tests with clinical implications in prostate cancer management. Adv Anat Pathol. 2018;25:293–303. doi: 10.1097/PAP.0000000000000192. [DOI] [PubMed] [Google Scholar]
- 94.Bilani N., Bahmad H., Abou-Kheir W. Prostate cancer and aspirin use: synopsis of the proposed molecular mechanisms. Front Pharmacol. 2017;8:145. doi: 10.3389/fphar.2017.00145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Pietsch E.C., Humbey O., Murphy M.E. Polymorphisms in the p53 pathway. Oncogene. 2006;25:1602–1611. doi: 10.1038/sj.onc.1209367. [DOI] [PubMed] [Google Scholar]
- 96.Tyner A.L., Gartel A.L. The role of the cyclin-dependent kinase inhibitor p21. Molecular Cancer Therapeutics. 2002;1:639–649. [PubMed] [Google Scholar]
- 97.Willis S., Day C.L., Hinds M.G., Huang D.C. The Bcl-2-regulated apoptotic pathway. J Cell Sci. 2003;116:4053–4056. doi: 10.1242/jcs.00754. [DOI] [PubMed] [Google Scholar]
- 98.Kim S.K., Chung J.H., Lee B.C., Lee S.W., Lee K.H., Kim Y.O. Influence of panax ginseng on alpha-adrenergic receptor of benign prostatic hyperplasia. Int Neurourol J. 2014;18:179–186. doi: 10.5213/inj.2014.18.4.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Petros A.M., Gunasekera A., Xu N., Olejniczak E.T., Fesik S.W. Defining the p53 DNA-binding domain/Bcl-x(L)-binding interface using NMR. FEBS Lett. 2004;559:171–174. doi: 10.1016/S0014-5793(04)00059-6. [DOI] [PubMed] [Google Scholar]
- 100.Tomita Y., Marchenko N., Erster S., Nemajerova A., Dehner A., Klein C. WT p53, but not tumor-derived mutants, bind to Bcl2 via the DNA binding domain and induce mitochondrial permeabilization. J Biol Chem. 2006;281:8600–8606. doi: 10.1074/jbc.M507611200. [DOI] [PubMed] [Google Scholar]
- 101.Kang Jia, Chong S.J.F., Ooi Vignette Zi Qi, Vali Shireen, Kumar Ansu, Kapoor Shweta, Abbasi Taher, Hirpara Jayshree L., Loh Thomas, Goh Boon Cher. Overexpression of Bcl-2 induces STAT-3 activation via an increase in mitochondrial superoxide. Oncotarget. 2015;6(33) doi: 10.18632/oncotarget.5763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Niu G., Wright K.L., Ma Y., Wright G.M., Huang M., Irby R. Role of Stat3 in regulating p53 expression and function. Mol Cell Biol. 2005;25:7432–7440. doi: 10.1128/MCB.25.17.7432-7440.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Burdak-Rothkamm S., Rothkamm K., McClelland K., Al Rashid S.T., Prise K.M. BRCA1, FANCD2 and Chk1 are potential molecular targets for the modulation of a radiation-induced DNA damage response in bystander cells. Cancer Lett. 2015;356:454–461. doi: 10.1016/j.canlet.2014.09.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Fagerholm R., Sprott K., Heikkinen T., Bartkova J., Heikkilä P., Aittomäki K. Overabundant FANCD2, alone and combined with NQO1, is a sensitive marker of adverse prognosis in breast cancer. Ann Oncol. 2013;24:2780–2785. doi: 10.1093/annonc/mdt290. [DOI] [PubMed] [Google Scholar]
- 105.Taniguchi T., Garcia-Higuera I., Andreassen P.R., Gregory R.C., Grompe M., D’Andrea A.D. S-phase-specific interaction of the Fanconi anemia protein, FANCD2, with BRCA1 and RAD51. Blood. 2002;100:2414–2420. doi: 10.1182/blood-2002-01-0278. [DOI] [PubMed] [Google Scholar]
- 106.Wang H., He Z., Zhang C., Zhang L., Xu D. Transmembrane protein alignment and fold recognition based on predicted topology. PLoS One. 2013;8 doi: 10.1371/journal.pone.0069744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Klabunde T., Hessler G. Drug design strategies for targeting G-protein-coupled receptors. Chembiochem. 2002;3:928–944. doi: 10.1002/1439-7633(20021004)3:10<928::AID-CBIC928>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 108.Ng D.P., Poulsen B.E., Deber C.M. Membrane protein misassembly in disease. Biochim Biophys Acta. 2012;1818:1115–1122. doi: 10.1016/j.bbamem.2011.07.046. [DOI] [PubMed] [Google Scholar]
- 109.Hong M.G., Karlsson R., Magnusson P.K., Lewis M.R. A genome-wide assessment of variability in human serum metabolism. Hum Mutat. 2013;34:515–524. doi: 10.1002/humu.22267. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.