Abstract
Rab32 coordinates a cell-intrinsic host defense mechanism that restricts the replication of intra-vacuolar pathogens such as Salmonella. Here we show that this mechanism requires aconitate decarboxylase 1 (IRG1), which synthesizes itaconate, a metabolite with antimicrobial activity. We find that Rab32 interacts with IRG1 upon Salmonella infection and facilitates the delivery of itaconate to the Salmonella-containing vacuole. Mice defective in IRG1 rescued the virulence defect of a S. Typhimurium mutant specifically defective in its ability to counter the Rab32 defense mechanism. These studies provide a link between a metabolite produced in the mitochondria after stimulation of innate immune receptors and a cell-autonomous defense mechanism that restricts the replication of an intracellular bacterial pathogen.
Keywords: innate immunity, bacterial pathogenesis, Salmonella pathogenesis, host defense, vesicle transport, Rab32, cell intrinsic immunity, Rab GTPases, mitochondria
Many cells are endowed with the capacity to control microbial invaders through cell-intrinsic defense mechanisms that synergize with the immune system to confer whole-body protection (1). Microbial pathogens respond to these host defense strategies by evolving virulence factors that prevent their detection or neutralize the effects of the antimicrobial mechanisms (2). Rab-family GTPases coordinate molecular transport across cellular compartments (3). A member of this family, Rab32, orchestrates a cell intrinsic host defense response that restricts the replication of intracellular bacterial pathogens including Salmonella Typhi (4–6). Salmonella Typhimurium, however, can neutralize this restriction mechanism with two effectors (SopD2 and GtgE) delivered by its type III protein secretion systems (7, 8). The mechanisms by which Rab32 controls bacterial replication are unknown. We hypothesized that Rab32 must control the delivery of an antimicrobial factor(s) to the Salmonella-containing vacuole (SCV). However, the nature of any potential factor(s) has remained elusive.
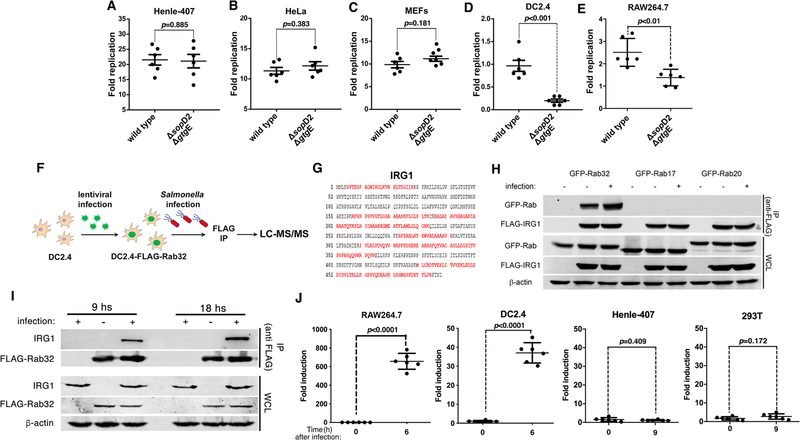
We searched for a cell line in which the Rab32-dependent restriction is robustly manifested by comparing the replication of wild-type S. Typhimurium with that of the ΔgtgE ΔsopD2 mutant, which is unable to neutralize it. We found no difference between the replication of the two strains in mouse embryo fibroblast, HeLa or Henle-407 cells (Fig. 1A–C), even after treatment with interferon (Fig. S1). By contrast, we observed significant differences in murine DC2.4 dendritic cells (Fig. 1D) and, to a lesser extent, in RAW264.7 macrophages (Fig. 1E). Thus, the Rab32 phenotype may be more robustly manifested in cells of hematopoietic origin.
Fig. 1. IRG1 interacts with Rab32 during Salmonella infection.
(A-E) The Rab32-associated pathogen restriction mechanism is manifested in myelocytic but not in epithelial cell lines. The ability of the S. Typhimurium ΔsopD2 ΔgtgE mutant strain to replicate within epithelial (Henle-407 and HeLa) or myelocytic (DC2.4 and RAW264.7) cell lines was evaluated by determining the CFU at different times after infection (MOI = 5). Fold replication represents the difference between the CFU at 1 and 9 hours post infection. Each circle represents the fold replication in each individual determination; the mean ± SEM of all the measurements and the p values of the indicated comparisons (two-sided Student’s t-test) are shown. (F-I) Rab32 interacts with IRG1 after Salmonella infection. DC2.4 cells expressing endogenous levels of FLAG-tagged Rab32 were infected with S. Typhimurium ΔsopD2 ΔgtgE (MOI = 30) and Rab32-interacting proteins were identified by affinity purification and LC—MS/MS analysis (F). The IRG1 peptides identified by the analyses are shown in red (G). (H and I) HEK293T cells transiently co-transfected with a plasmid expressing GFP-tagged Rab32, RAb17, or Rab20, along with a plasmid encoding FLAG-tagged IRG1 (H), or DC2.4 cells stably expressing FLAG-tagged Rab32 (I) were infected with S. Typhimurium ΔgtgE ΔsopD2 for 4 hours (MOI = 5). Cell lysates were then analyzed by immunoprecipitation with anti-FLAG and immunoblotting with anti-GFP, anti-FLAG, anti-IRG1, or anti-β-actin (as loading control) antibodies. IP: immunoprecipitates; WCL: whole-cell lysates. (J) Expression of IRG1 after Salmonella infection. The indicated cell lines were infected with S. Typhimurium ΔsopD2 ΔgtgE mutant strain (MOI = 5) and IRG1 mRNA levels were measured by qPCR 6 or 9 hours after infection. Each circle represents a single determination of the relative levels of IRG1 normalized to the levels of GAPDH; the mean ± SEM of all the measurements and p values of the indicated comparisons (two-sided Student’s t test) are shown.
We then searched for Rab32-interacting proteins in DC2.4 cells after infection with the S. Typhimurium ΔgtgE ΔsopD2 mutant strain at a time of infection that coincides with the recruitment of Rab32 to the SCV (4) (Fig. 1F). A prominent Rab32-interacting protein exclusively detected in infected cells was IRG1 (Fig. 1G, Table S1 and S2). This interaction was confirmed in cells expressing epitope-tagged versions of these proteins (Fig. 1H and Fig. S2) and in DC2.4 cells expressing endogenous IRG1 (Fig. 1I). The nucleotide state of Rab32 did not appear to affect its interaction with IRG1 (Fig. S3). However, the interaction was enhanced by the bacterial infection (Fig. 1H and 1I and Fig. S2). In addition, IRG1 expression was detected in Salmonella-infected or LPS-treated cells that showed the Rab32-restriction phenotype, but not in cells that did not (Fig. 1J and Fig. S4 and S5). Furthermore, when compared to the wild-type strain, the S. Typhimurium ΔgtgE ΔsopD2 mutant showed reduced intracellular replication in Henle-407 cells transiently expressing IRG1 (fig. S6). IRG1, which is highly expressed in mouse macrophages after stimulation of Toll-like receptors (9), converts cis-aconitate, a tricarboxylic acid cycle intermediate, to itaconic acid (10). By alkylating cysteine residues in the targeted molecules (11, 12), itaconic acid (or itaconate) inhibits methylmalonyl-CoA mutase (13), as well as isocitrate lyase (14) and succinate dehydrogenase (15), essential enzymes in the glyoxylate shunt pathway and the TCA cycle. These pathways are critical for the physiology and pathogenesis of Salmonella and Mycobacterium spp. (13, 16–20), which are susceptible to the Rab32-mediated defense mechanism (4, 7, 21). Furthermore, itaconic acid inhibits the growth of Mycobacterium spp., S. Typhimurium (10) and S. Typhi (Fig. S7).
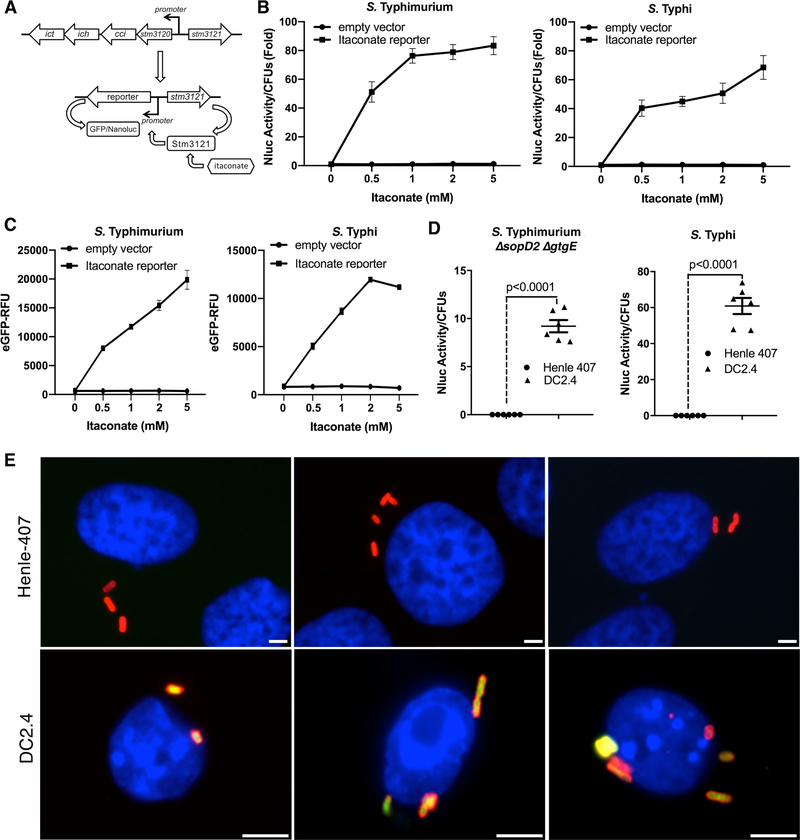
To investigate whether itaconic acid is delivered to the (SCV), we developed a biosensor to report the presence of itaconic acid in Salmonella. S. Typhimurium encodes a putative itaconate-degradation pathway (22), which is absent from S. Typhi (Fig. 2A). By analogy to similar systems in other bacteria (23), expression of this pathway is expected to be controlled by a transcriptional regulatory protein (STM3121), which directly senses itaconic acid (Fig. 2A). We constructed a transcriptional reporter in which the expression of the green fluorescent protein (GFP), or nanoluciferase was placed under the control of a promoter whose expression is directly controlled by STM3121 (Fig. 2A). The reporters responded to the presence of itaconic acid in a dose-dependent manner (Fig. 2B and 2C), maintaining a linear response up to itaconate concentrations of ~5–6 mM (Fig. S8). The reporter’s response was specific as addition of other metabolites or environmental stimuli did not result in a measurable transcriptional response (Fig. S9). We then tested whether itaconic acid was delivered to the SCV and whether the reporter strains could sense its presence within this environment. Infection of cells, which do not express IRG1 (Fig. 1), with Salmonella strains encoding the itaconate reporters, did not result in any measurable production of nanoluciferase (Fig. 2D and Fig. S10 and S11) or GFP (Fig. 2E and Fig. S12). By contrast, infection of cells that express IRG1 resulted in robust expression of the reporters (Fig. 2D, 2E, and Fig. S9, S10, and S11). Based on the dose response of the reporter (Fig. S8), the concentration of itaconate within the SCV is estimated to be ~6 mM, a concentration predicted to inhibit Salmonella growth (Fig. S7).
Fig. 2. Itaconate is delivered to the Salmonella-containing vacuole.
(A-C). Development of a biosensor to detect itaconate. (A) Chromosomal organization of the itaconate-degradation gene cluster in S. Typhimurium and diagram of the itaconate biosensor. (B and C) Effect of addition of itaconate on the biosensor transcriptional response. S. Typhimurium and S. Typhi strains carrying either the nanoluciferase or eGFP itaconate reporters were grown to an OD600 of 0.9 in the presence of different concentrations of itaconic acid (as indicated) and the levels of nanoluciferase or eGFP were determined. Values are the mean ± SD of three independent measurements. This experiment was repeated at least three times with equivalent results. (D and E) Detection of itaconate by intracellular Salmonella. DC2.4 or Henle-407 cells were infected with a S. Typhimurium ΔsopD2 ΔgtgE mutant (MOI=5) or S. Typhi (MOI=10) carrying a plasmid encoding a nanoluciferase-based itaconate biosensor. Eighteen hours after infection, the levels of nanoluciferase were measured in lysates of the infected cells (D). Each circle or triangle represents a single luciferase measurement; the mean ± SD and p values of the indicated comparisons (two-sided Student’s’s t test) are shown. This experiment was repeated at least three times with equivalent results. Alternatively, DC2.4 or Henle-407 cells were infected (MOI = 10) with S. Typhi carrying a plasmid encoding the eGFP-based itaconate biosensor (green). Eighteen hours after infection, cells were fixed, stained with DAPI (blue) to visualize nuclei, and stained with an anti-Salmonella LPS antibody along with Alexa 594-conjugated anti-rabbit antibody (red), and imaged under a fluorescence microscope (E). Scale bars: 5 μm.
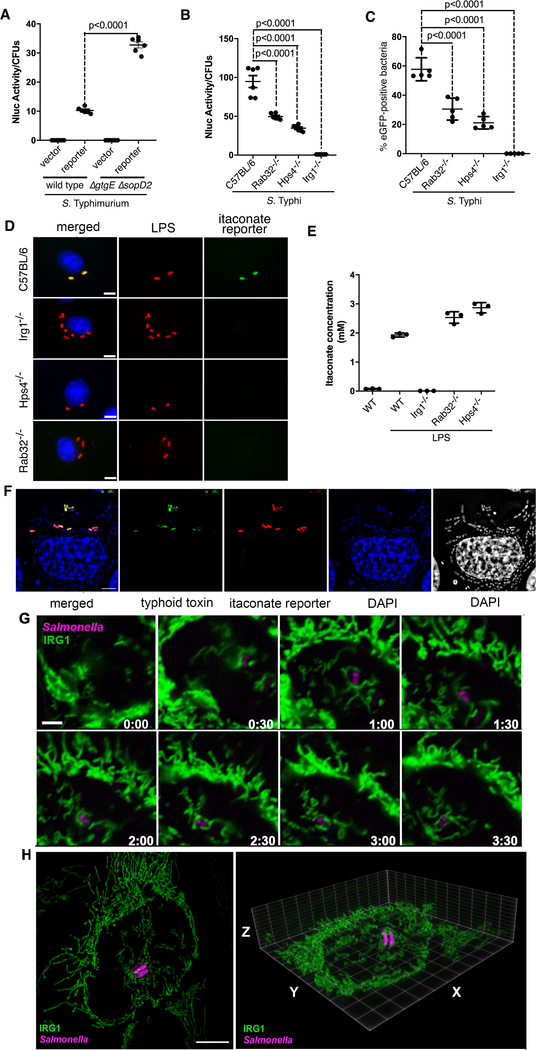
We then examined whether the Rab32 pathway influences the production or the delivery of itaconate to the SCV. We reasoned that if the Rab32 pathway influences the presence of itaconate in the SCV, wild-type S. Typhimurium should impair this process by the action of SopD2 and GtgE (4, 7). Consistent with this hypothesis, expression of the itaconate biosensor was detected at significantly reduced levels in DC2.4 or RAW264.7 cells infected with wild type bacteria in comparison to the ΔgtgE ΔsopD2 mutant strain (Fig. 3A and Fig. S13), despite equivalent levels of IRG1 expression in the infected cells (Fig. S14). We then compared the expression of the itaconate reporter in bone marrow derived macrophages (BMDMs) obtained from C57BL/6, IRG1−/−, BLOC3 deficient (Hsp4−/−) [the exchange factor for Rab32 (24)], or Rab32−/− mice, after infection with different Salmonella reporter strains. We found robust expression of the itaconate biosensor after infection of BMDMs obtained from C57BL/6 but not in BMDMs obtained from IRG1-deffective mice (Fig. 3B–D). Importantly, expression of the reporter was detected at significantly reduced levels in BMDMs obtained from Hsp4−/− or Rab32−/− mice. Expression of the reporter in Rab32−/− BMDMs was higher than in Hsp4−/− BMDMs, suggesting that in the absence of Rab32, the related Rab38 GTPase may partially compensate for its function (Fig. 3B–D). The levels of itaconate in BMDMs obtained from C57BL/6, IRG1−/−, Hsp4−/−, or Rab32−/− mice after stimulation with LPS were indistinguishable from one another (Fig. 3E and Table S3), indicating that the delivery of itaconate to the SCV, not its synthesis, is dependent on the Rab32—BLOC3 pathway.
Fig. 3. Rab32/BLOC3-dependent delivery of itaconate to the Salmonella-containing vacuole.
(A) Cultured DC2.4 cells were infected with wild-type or ΔgtgE ΔsopD2 S. Typhimurium strains (MOI = 5) encoding the luciferase-based itaconate biosensor and the levels of luciferase in cell lysates were measured 9 hours after infection. Each circle represents a single luciferase measurement; the mean ± SD and the p-values of the indicated comparisons (two-sided Student’s t-test) are shown. (B-D) Bone-marrow-derived macrophages (BMDMs) obtained from C57BL/6, Rab32−/−, Hsp4−/−, or IRG1−/− mice were infected with S. Typhi (MOI = 10) encoding the luciferase- or eGFP-based itaconate biosensors. Nine hours after infection, the levels of luciferase in cell lysates (B) or the number of cells expressing eGFP (C) were determined. Each circle in (B) represents a single luciferase measurement. Values in (C) represent the percentage of bacterial cells exhibiting fluorescence. A minimum of 200 cells in each condition was evaluated. The mean ± SD and p-values of the indicated comparisons (one-way Anova) are shown. Representative fields of BMDMs obtained from the indicated mouse lines infected with S. Typhi encoding the eGFP itaconate reporter (green) are shown. Cells were fixed, stained with DAPI (blue) to visualize nuclei and stained with an anti-Salmonella LPS antibody along with Alexa 594-conjugated anti-rabbit antibody (red) (D) (scale bar = 5 μm). (E) Itaconate levels in BMDMs obtained from the indicated mice before and after LPS treatment to induce the expression of IRG1. Values represent the mean ± SD of three independent measurements. (F) Expression of the itaconate reporter (red) by intravacuolar but not by cytosolic S. Typhi. HeLa cells transfected with a plasmid encoding FLAG-tagged IRG1 were infected by a S. Typhi strain encoding a mCherry itaconate reporter (red) and a pltB::GFP transcriptional reporter (green). PltB, a component of S. Typhi’s typhoid toxin, is exclusively produced by bacteria located within the SCV and therefore serves as a surrogate to report for intravacuolar (GFP positive) vs intra cytosolic (GFP negative) bacteria. Six hours after infection, cells were stained with DAPI (to visualize all bacteria) and examined under a fluorescence microscope (scale bars: 5 μm). (G) Live-cell fluorescence time-lapse microscopy of cultured HeLa cells stably expressing IRG1-GFP (green) infected with S. Typhimurium ΔgtgE ΔsopD2 mutant strain encoding an mCherry itaconate biosensor (magenta). Imaging was initiated 45 minutes after infection. The times (hours:min) after initiation of imaging are indicated in each frame (the entire sequence is shown in video S1; this experiment was conducted at least three independent times, imaging several independent positions in each experiment, with equivalent findings; see videos S2 and S3 for additional examples). (H) Snapshot of a 3-D rendering of 3D-SIM acquisitions of HeLa cells stably expressing IRG1-GFP (green) infected with S. Typhimurium ΔgtgE ΔsopD2 mutant strain encoding an mCherry itaconate biosensor (magenta) (videos of this and additional reconstructions can be found in videos S4–S7).
A proportion of the intracellular Salmonella breaks from the SCV to the cell cytosol where it can replicate at a faster rate (25). To investigate whether delivery of itaconate requires the integrity of the SCV, we examined the expression of the itaconate reporter in intravacuolar and cytoplasmic bacteria. Expression of S. Typhi’s typhoid toxin requires the environment of the SCV thus serving as a marker to distinguish intravacuolar vs intracytosolic bacteria (26). We found that all bacteria expressing the itaconate reporter were located within the SCV while no bacteria found within the cytosol showed expression of the reporter (Fig. 3F and Fig. S15).
The transport of mitochondria-originated products to other vesicular compartment is well documented (27, 28). As Rab32 is present in the mitochondria (29) and the SCV (4), this GTPase may aid the formation and/or delivery of itaconate and/or IRG1-transport carriers, or it may facilitate the tethering of the mitochondria with the SCV. We used live-cell time-lapse fluorescence microscopy to examine cells stably expressing GFP-tagged IRG1 that had been infected with Salmonella expressing an mCherry itaconate reporter. We found that in uninfected cells, IRG1 was uniformly distributed throughout the entire mitochondrial network (Fig. S16 and Table S4). After Salmonella infection, we observed many instances in which the IRG1-containing mitochondrial network repositioned to surround and make intimate contact with the SCV, a process that coincided with the activation of the itaconate biosensor in the intracellular bacteria (Fig. 3G, and videos S1–S3). Examination of the infected cells by two-color 3D super-resolution structured-illumination microscopy (3D-SIM) (30) revealed intimate contact between the IRG1-containing mitochondrial network and the SCV (Fig. 3H and videos S4–S7). These observations suggest a mechanism by which the close association between the IRG1-containing mitochondria and the SCV may facilitate their tethering and subsequent itaconate transport.
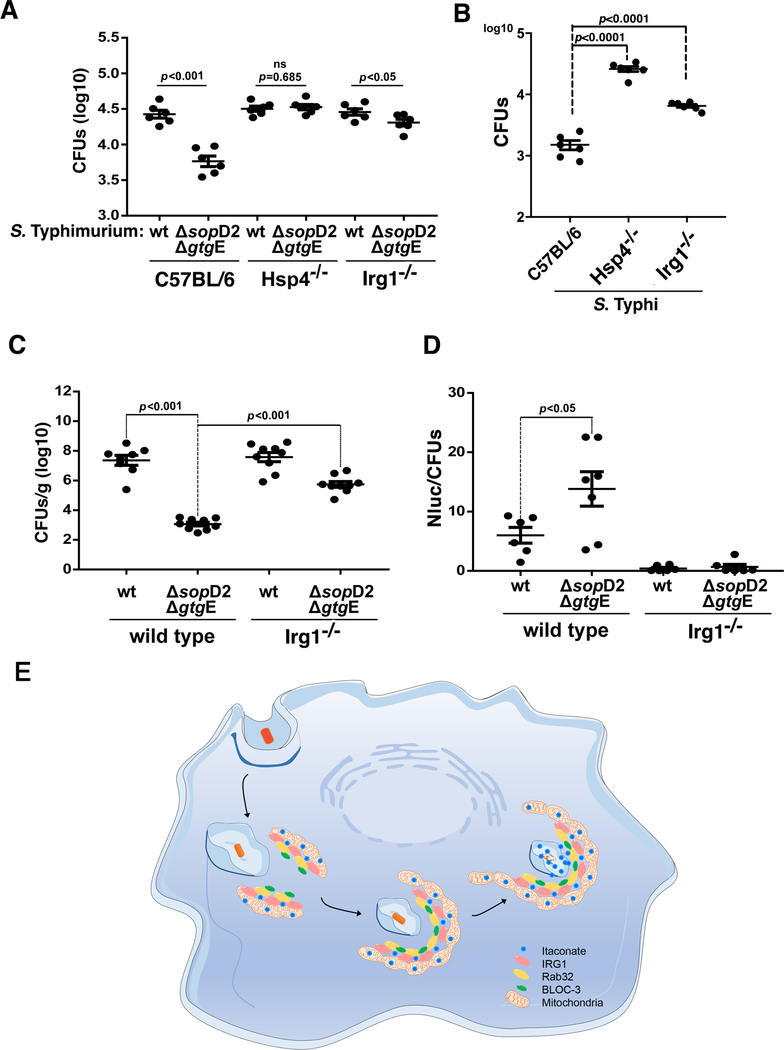
We compared the ability of wild-type S. Typhimurium or the ΔsopD2 ΔgtgE mutant to replicate within BMDMs obtained from C57BL/6, IRG1−/−, or Hsp4−/− mice. As previously shown (7), the S. Typhimurium ΔsopD2 ΔgtgE mutant exhibited reduced ability to replicate within C57BL/6 BMDMs, and this phenotype was rescued in BMDMs obtained from Hsp4−/− animals (Fig. 4A). Importantly, the replication-deficient phenotype was also rescued in BMDMs obtained from IRG1−/− mice, which allowed the replication of the S. Typhimurium ΔsopD2 ΔgtgE mutant to levels almost equivalent to those of wild type bacteria (Fig. 4A). The human-adapted pathogen S. Typhi is unable to replicate in mouse macrophages because the Rab32—BLOC3 pathway restricts its replication (4). As we have previously shown (4), S. Typhi was able to replicate in BMDMs from Hsp4−/− mice to levels almost equivalent to those of wild-type S. Typhimurium (Fig. 4B). Importantly, S. Typhi was able to replicate in BMDMs from IRG1 −/− mice although not to the same extent as to the levels observed in BMDMs from Hsp4−/− mice (Fig. 4B). This suggests that, in addition to itaconate, the Rab32—BLOC3 pathway may deliver additional antimicrobial factors. The S. Typhimurium ΔsopD2 ΔgtgE mutant exhibits significantly reduced mouse virulence when compared to the wild-type strain and the virulence defect can be reversed in BLOC3-defective mice (7). We therefore reasoned that if itaconate is an effector of this pathway, the virulence attenuation of the S. Typhimurium ΔsopD2 ΔgtgE mutant strain should be reversed in IRG1−/− mice. Consistent with this hypothesis, the virulence defect of the S. Typhimurium ΔsopD2 ΔgtgE mutant was significantly reversed in IRG1−/− mice (Fig. 4C). We also examined whether the deployment of the SopD2/GtgE effectors was able to blunt the delivery of itaconate to the SCV during infection. We found that twenty-four hours after infection, the itaconate reporter activity was almost undetectable in the spleens of animals infected with wild type S. Typhimurium. By contrast, significantly higher activity was detected in the spleens of animals infected with the ΔsopD2 ΔgtgE mutant strain (Fig. 4D).
Fig. 4. Susceptibility of IRG1-deficient mice to Salmonella infection.
(A and B) Bone-marrow-derived macrophages (BMDMs) obtained from C57BL/6 (WT), Hsp4−/−, or IRG1−/− mice were infected with wild-type S. Typhimurium (MOI=5), its ΔgtgE ΔsopD2 mutant derivative (MOI=5) (A), or wild-type S. Typhi (MOI=10) (B), and the number of CFU was determined 9 hours after infection. Each circle represents the CFU in independent measurements; the mean ± SEM of all the measurements and p-values of the indicated comparisons (two-sided Student’s t test) are shown. (C and D) C57BL/6 (wild-type) or IRG1−/− mice were intraperitoneally infected with wild-type or ΔgtgE ΔsopD2 S. Typhimurium (as indicated) (102 CFU). Five days after infection, bacterial loads in the spleen of the infected animals were determined (C). Alternatively, mice were intraperitoneally infected with the same strains (104 CFU) and the levels of luciferase activity in spleen lysates was quantified 24 hours after infection (D). Each circle in (C) represents the bacterial loads of the spleen of an individual animal, and in (D) represents the luciferase levels in the spleen of an individual animal normalized to the CFU. The mean ± SEM of all the determination and p-values of the indicated comparisons (two-sided Student’s t-test) are shown. (E) Model for the mechanism of Rab32—BLOC3-mediated itaconate delivery to the Salmonella-containing vacuole. Upon infection, the mitochondrial network repositions to surround the incoming bacteria, and the resulting close interaction between the mitochondria and the Salmonella-containing vacuole results in the Rab32—BLOC3 dependent delivery of itaconate, which is synthesized in the mitochondria by IRG1.
We have shown here that itaconate is an effector of the Rab32-dependent pathogen restriction pathway that limits the replication of Salmonella (Fig. 4E). In phagocytic cells, itaconate can also be delivered to vacuoles containing other bacteria (e.g. Escherichia coli) or avirulent S. Typhi lacking its two type III secretion systems (Fig. S17). We therefore hypothesized that this is a general mechanism of defense that may participate in the restriction of other vacuolar pathogenic bacteria. How itaconate inhibits bacterial growth is likely to be multi-factorial, exerting its function by altering bacterial metabolism through its ability to inhibit key metabolic enzymes. Although itaconate has also been reported to have modulatory activities over multicellular responses including inflammation (31), it is unlikely that those activities are central to the Rab32—BLOC3-mediated pathogen restriction mechanism, which involves the direct delivery of this metabolite to the bacterial-containing vacuole. These studies emphasize the critical role played by mitochondria in the control of microbial infections and the Rab32 pathway as a major link between this organelle and the compartments housing bacterial pathogens.
Supplementary Material
Acknowledgment
We thank Dr. M. Diamond and Dr. M. Artyomov for facilitating the Irg1−/− mouse. Funding: The Proteomics Resource of the WM Keck Foundation Biotechnology Resource Laboratory was partially supported by CTSA Grant Number UL1TR001863 from the National Center for Advancing Translational Sciences (NCATS) of the National Institutes of Health (NIH). This work was supported by NIH Grants R01AI114618 and R01AI055472 to J.G.
Footnotes
Competing interests: The authors declare no competing interests.
Data and materials availability:
all data are available in the main text, supplementary materials, and auxiliary files.
References and Notes
- 1.Randow F, MacMicking J, James L, Cellular self-defense: how cell-autonomous immunity protects against pathogens. Science 340, 701–706. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reddick L, Alto N, Bacteria fighting back: how pathogens target and subvert the host innate immune system. Mol Cell. 54, 321–328 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stenmark H, Rab GTPases as coordinators of vesicle traffic. Nat Rev Mol Cell Biol. 10, 513–525 (2009). [DOI] [PubMed] [Google Scholar]
- 4.Spanò S, Galán J, A Rab32-dependent pathway contributes to Salmonella typhi host restriction. Science 338, 960–963 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tang B, Rab32/38 and the xenophagic restriction of intracellular bacteria replication. Microbes Infect. 18, 595–603 (2016). [DOI] [PubMed] [Google Scholar]
- 6.Li Y et al. , Analysis of the Rab GTPase Interactome in Dendritic Cells Reveals Antimicrobial Functions of the Rab32 Complex in Bacterial Containment. Immunity 44, 422–437 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Spanò S, Gao X, Hannemann S, Lara-Tejero M, Galán J, A Bacterial Pathogen Targets a Host Rab-Family GTPase Defense Pathway with a GAP. Cell Host Microbe 19, 216–226 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Spano S, Liu X, Galan JE, Proteolytic targeting of Rab29 by an effector protein distinguishes the intracellular compartments of human-adapted and broad-host Salmonella. Proc Natl Acad Sci U S A 108, 18418–18423 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee C, Jenkins N, Gilbert D, Copeland N, O’Brien W, Cloning and analysis of gene regulation of a novel LPS-inducible cDNA. . Immunogenetics 41, 263–270 (1995). [DOI] [PubMed] [Google Scholar]
- 10.Michelucci A et al. , Immune-responsive gene 1 protein links metabolism to immunity by catalyzing itaconic acid production. Proc Natl Acad Sci U S A. 110, 7820–7825 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mills E et al. , Itaconate is an anti-inflammatory metabolite that activates Nrf2 via alkylation of KEAP1. Nature 556, 113–117 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Bambouskova M et al. , Electrophilic properties of itaconate and derivatives regulate the IκBζ-ATF3 inflammatory axis. Nature 556, 501–504 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ruetz M et al. , Itaconyl-CoA forms a stable biradical in methylmalonyl-CoA mutase and derails its activity and repair. Science 366, 589–593 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McFadden B, Purohit S, Itaconate, an isocitrate lyase-directed inhibitor in Pseudomonas indigofera. . J Bacteriol. 131, 136–144 (1977). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cordes T et al. , Immunoresponsive Gene 1 and Itaconate Inhibit Succinate Dehydrogenase to Modulate Intracellular Succinate Levels. J Biol Chem. 291, 14274–14284 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wilson R, Maloy S, Isolation and characterization of Salmonella typhimurium glyoxylate shunt mutants. J Bacteriol. 169, 3029–3034 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fang F, Libby S, Castor M, Fung A, Isocitrate lyase (AceA) is required for Salmonella persistence but not for acute lethal infection in mice. Infect Immun. 73, 2547–2549 (2005. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.McKinney J et al. , Persistence of Mycobacterium tuberculosis in macrophages and mice requires the glyoxylate shunt enzyme isocitrate lyase. Nature 406, 735–738 (2000). [DOI] [PubMed] [Google Scholar]
- 19.Mercado-Lubo R, Gauger E, Leatham M, Conway T, Cohen P, A Salmonella enterica serovar typhimurium succinate dehydrogenase/fumarate reductase double mutant is avirulent and immunogenic in BALB/c mice. Infect. Immun. 76, 1128–1134 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartman T et al. , Succinate dehydrogenase is the regulator of respiration in Mycobacterium tuberculosis. PLoS Pathog. 10, e1004510 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhang F et al. , Identification of two new loci at IL23R and RAB32 that influence susceptibility to leprosy. Nat Genet 43, 1247–1251 (2011). [DOI] [PubMed] [Google Scholar]
- 22.Sasikaran J, Ziemski M, Zadora P, Fleig A, Berg I, Bacterial itaconate degradation promotes pathogenicity. Nat. Chem. Biol. 10, 371–377 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Hanko E, Minton N, Malys N, A Transcription Factor-Based Biosensor for Detection of Itaconic Acid. ACS Synth Biol. 7, 1436–1446 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gerondopoulos A, Langemeyer L, Liang J, Linford A, Barr F, BLOC-3 mutated in Hermansky-Pudlak syndrome is a Rab32/38 guanine nucleotide exchange factor. Curr Biol. 22, 2135–2139 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Knodler L, Salmonella enterica: living a double life in epithelial cells. Curr Opin Microbiol. 23, 23–31 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Fowler C, Galán J, Decoding a Salmonella Typhi Regulatory Network that Controls Typhoid Toxin Expression within Human Cells. Cell Host Microbe 23, 65–76 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soto-Heredero G, Baixauli F, Mittelbrunn M, Interorganelle Communication between Mitochondria and the Endolysosomal System. Front. Cell Dev. Biol. 5, 95 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Abuaita B, Schultz T, O’Riordan M, Mitochondria-Derived Vesicles Deliver Antimicrobial Reactive Oxygen Species to Control Phagosome-Localized Staphylococcus aureus. Cell Host Microbe. 24, 625–636 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Alto N, Soderling J, Scott J, Rab32 is an A-kinase anchoring protein and participates in mitochondrial dynamics. J Cell Biol. 158, 659–668 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gustafsson M et al. , Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys J. 94, 4957–4970 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yu X, Zhang D, Zheng X, Tang C, Itaconate: an emerging determinant of inflammation in activated macrophages. Immunol Cell Biol. 97, 134–141 (2019). [DOI] [PubMed] [Google Scholar]
- 32.Gibson D et al. , Enzymatic assembly of DNA molecules up to several hundred kilobases. Nat Methods. 6, 343–345 (2009). [DOI] [PubMed] [Google Scholar]
- 33.Fowler C, Galán J, Decoding a Salmonella Typhi Regulatory Network that Controls Typhoid Toxin Expression within Human Cells. Cell Host Microbe 23, 65–76 (2018. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fitzsimmons L et al. , Zinc-dependent substrate-level phosphorylation powers Salmonella growth under nitrosative stress of the innate host response. PLoS Pathog. 14, e1007388 (2018. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fiolka R, Shao L, Rego E, Davidson M, Gustafsson M, Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. Proc Natl Acad Sci U S A. 109, 5311–5315 (2012. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schindelin J et al. , Fiji: an open-source platform for biological-image analysis. Nat Methods. 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lampropoulou V et al. , Itaconate Links Inhibition of Succinate Dehydrogenase with Macrophage Metabolic Remodeling and Regulation of Inflammation. Cell Metab. 24, 158–166 (2016. ). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
all data are available in the main text, supplementary materials, and auxiliary files.