Abstract
Biomarkers may be of value for the early detection of gastric cancer (GC) and the preoperative identification of tumor characteristics to guide treatment strategies. The present study analyzed the expression levels of phospholipids in plasma from patients with GC using liquid chromatography/electrospray ionization-mass spectrometry (LC/ESI-MS) to detect reliable biomarkers for GC. Furthermore, combining the results with a machine learning strategy, the present study attempted to establish a diagnostic system for GC. A total of 20 plasma samples from preoperative patients with GC and 16 plasma samples from tumor-free patients (controls) were selected from our biobank named ‘SHINGEN (Yamanashi Biobank of Gastroenterological Cancers)’, which includes a total of 1,592 plasma samples, and were analyzed by LC/ESI-MS. The obtained data were discriminated using a machine learning-based diagnostic algorithm, whose discriminant ability was confirmed through leave-one-out cross-validation. Using LC/ESI-MS, the levels of 236 lipid molecules were determined. Biomarker analysis revealed that a few lipids that were downregulated in the GC group could discriminate between the GC and control groups. Whole lipid composition analysis using partial least squares regression revealed good discrimination ability between the GC and control groups. Integrative analysis of all molecules using the aforementioned machine learning method exhibited a diagnostic accuracy of 94.4% (specificity, 93.8%; sensitivity, 95.0%). In conclusion, the outcomes of the present study suggested the potential future application of the aforementioned system in clinical settings. By accumulating more reliable data, the present system will be able to detect early-stage cancer and will be capable of predicting the efficacy of each therapeutic strategy.
Keywords: gastric cancer, lipidomics, mass spectrometry, machine learning, plasma
Introduction
Gastric cancer (GC) is the fifth most commonly diagnosed type of cancer worldwide, and the majority of patients with early GC are curable by appropriate treatments (1). Patients with advanced GC, however, have a poor prognosis, despite the progress achieved in various treatment strategies, including extended surgical resections and intensive chemotherapy, with or without the use of molecular targeted treatments. Preoperative chemotherapy has been demonstrated to be beneficial in certain subgroups of patients with GC (2,3). Therefore, early detection of this lethal disease is critical, and the preoperative identification of tumor characteristics can guide decision-making regarding the selection of the most appropriate treatment strategies. Toward these ends, various serum tumor markers have been established and utilized in clinical practice (4). Although they are widely used as supplementary information for diagnosis, their diagnostic accuracy, as well as their specificity and sensitivity for GC, have yet to be optimized (5).
Comprehensive molecular analyses have recently elucidated various genetic and epigenetic alterations in several types of cancer, and numerous studies in which circulating cell-free nucleic acids have been analyzed have reported the potential utility of blood molecular biomarkers (6). Blood biomarkers identified in the serum, plasma or other biological fluids derived from patients have several advantages, such as overcoming the undependability due to the tumor heterogeneities, and the feasibility of repeated sample collection. It has also been demonstrated that numerous metabolites are involved in carcinogenesis and cancer progression. In order to comprehensively identify and analyze the genetic and metabolic variations that are involved in carcinogenesis and cancer development, mass spectrometry (MS) techniques and devices are useful in the medical field (7). In addition, results of integrative metabolomic analyses using machine learning methods have been reported (8,9). It is expected that these technologies will become generally available, particularly for establishing a cancer diagnosis. Endoscopic examination, the gold standard for a definitive GC diagnosis, has revealed that a white opaque substance is a novel endoscopic finding in gastric neoplasms, indicating that there is intracellular accumulation of lipid droplets in GC (10,11). Therefore, a lipidomic approach to GC has attracted great research attention.
In recent clinical settings, not only surgical or endoscopic curative resection, but also perioperative combined treatments, such as neoadjuvant and/or adjuvant chemotherapies, are often practiced and have greatly affected survival outcomes (2,3). However, it is challenging to select the best treatment at the best time, as clinical situations vary constantly. Thus, in addition to achieving early cancer detection, it is necessary to identify and develop novel biomarkers for precision medicine to provide a highly effective and low-risk treatment to each individual patient. Various studies have been published on the expression of cancer-specific biomolecules in non-cancerous and cancerous tissues, including studies using MS (12–16). It is important to compare the differences between cancerous and non-cancerous tissues, or between patients with cancer and healthy volunteers (HVs) without any history of cancer, in order to identify cancer-specific molecules that can be useful in the early detection of cancer. In the future, personalized medicine will be more important in order to select the best treatment strategies for individual cases. Therefore, the present study conducted a comprehensive lipidomic approach using MS and a machine learning method to provide the basis of precision medicine for GC. Lipid metabolism is attracting increasing attention in tumor development and numerous other diseases. In particular, phospholipids have been reported to play important roles in various cancers (17–19). In addition, peripheral blood samples have a broad diagnostic utility due to their ease of use and accessibility, in contrast to tissue samples, which can be obtained only by surgical resection or biopsy with relatively highly invasiveness (20,21). Thus, methods such as liquid biopsy, which can be used to analyze primary tumors using body fluids, including blood, urine, digestive juice and cerebral spinal fluid, are an attractive research focus (22,23).
The present study analyzed the levels of phospholipids in plasma derived from patients with GC by using liquid chromatography/electrospray ionization-MS (LC/ESI-MS), a method which accurately identifies and quantifies lipid molecules. Furthermore, the present study aimed to establish a machine learning-based diagnostic algorithm for GC.
Materials and methods
Biobank establishment
A human biobank named ‘SHINGEN (Yamanashi Biobank of Gastroenterological Cancers)’, was established, which included frozen tissue, cell and fluid samples obtained from January 2018 until the present date. The tissue samples were derived from primary and metastatic malignant tumors, and from adjacent non-tumor tissues. Cellular and liquid components from the intraperitoneal lavage of patients collected during surgical procedures were obtained following centrifugation at 2,000 × g for 10 min at 4°C. Peripheral blood samples, including plasma and serum, were obtained from patients at various time points during clinical interventions or observations without treatment. For the preparation of the plasma samples, 5 ml peripheral whole blood was collected from each patient using vacuum blood sampling tubes containing the anticoagulant reagent EDTA (NP-EN0507; NIPRO Corp.). The tubes were immediately centrifuged at 2,000 × g for 10 min at 4°C. Serum samples were collected using vacuum blood sampling tubes containing separating agents and coagulation accelerators (VP-AS074K; Terumo Corp.). After incubation at 4°C for 30 min, the tubes were centrifuged following the aforementioned method. The plasma and serum samples obtained upon centrifugation were stored at −80°C until further analysis. Samples from patients with various digestive malignancies, such as GC, esophageal cancer, colorectal cancer, pancreatic cancer and hepatocellular carcinoma were included in the SHINGEN biobank. Furthermore, samples from patients who underwent subsequent surgical resection according to clinical guidelines after radical resection for their early cancer, were also included in this biobank. Of note, the majority of such patients had no residual malignancies at the time of surgery and/or blood sample collection.
Patient clinical information and ethical concerns
Patient clinical information, including pathological findings, was obtained from the electronic medical recording system at University of Yamanashi Hospital (Yamanashi, Japan). The pathological findings of GC were defined according to the Union for International Cancer Control classification of malignant tumors (8th edition) (24). This information was stored in the database of the biobank. The biological sample collection in the SHINGEN (#1665) biobank and the study design (#2192) were approved by the Ethics Committee of the University of Yamanashi, and the study was performed in accordance with the ethical standards of the Declaration of Helsinki and its later amendments (25). Written informed consent for the use of biological samples and clinical data was obtained from all the patients.
Inclusion and exclusion criteria
Between January 2018 and April 2020, a total of 1,592 blood plasma samples from patients who were treated at the First Department of Surgery at the University of Yamanashi Hospital, were collected prospectively in the SHINGEN biobank. A total of 910 plasma samples were obtained during the intra- or post-intervention period, and 682 were collected during the preoperative phase. Of these, 16 patient samples were set as the control group. These samples were obtained from patients who received additional resection after endoscopic resection for early tumors (12 patients with GC, 2 with esophageal cancer and 2 with colorectal cancer) and who were confirmed to have no residual malignancies based on their postoperative pathological findings. Of the remaining 666 plasma samples, 152 were from patients with GC and 514 from patients with other malignancies. After excluding patients with GC who had no lymph node metastasis based on their preoperative clinical findings and those who underwent non-radical resection with residual tumors, 20 patients with advanced GC were analyzed in the present study. The patient flow diagram is shown in Fig. S1.
LC/ESI-MS
A total of 10 µl human plasma was added into 990 µl 0.1% formic acid in methanol, and the sample solution was mixed using ThermoMixer C (Eppendorf) for 5 min at 4°C. After incubation on ice for 10 min, the sample solution was centrifuged at 15,000 × g for 10 min. The resultant supernatant was 2-fold diluted using 0.1% formic acid in methanol, and 300 µl diluted supernatant was applied into a LabTotal Vial (Shimadzu Corporation). The vial was inserted to the sample rack of the autosampler (Nexera X2 SIL-30AC; Shimadzu Corporation), and 3 µl sample was injected into the column for LC separation.
LC/ESI-MS was performed using the high-pressure LC installed LCMS-8060 (Shimadzu Corporation) system. To analyze the lipid components in human plasma, the LC/MS/MS Method Package for Phospholipid Profiling (Shimadzu Corporation) was used following the manufacturer's instructions. Kinetex C8 column (Kinetex C8, 150×2.1 mm i.d., 3.6-µm particle size; Phenomenex), mobile phase A (20 mM ammonium formate in water) and mobile phase B (acetonitrile: Isopropanol 1:1 v/v) were used for LC separation. The concentration of mobile phase B was programmed as 20% (0 min)-20% (1 min)-40% (2 min)-92.5% (25 min). The oven temperature was 45°C. Data processing and molecular identification/quantification were performed automatically by using the LabSolutions software (version 5.82 SP1; Shimadzu Corporation).
Statistical analysis
Age and tumor size were represented as the mean ± standard error of the mean. Statistical evaluation of the gender ratio between the two groups was performed using the χ2 test. The relative ion intensities of each molecule between the control and GC groups were calculated as the ratio of the mean values in each group. The variance of continuous values was confirmed by unpaired t-test. All statistical analyzes, receiver operating characteristic (ROC) graphs and box plots were performed using EZR (Saitama Medical Center, Jichi Medical University, Saitama, Japan), which is a graphical user interface for R (The R Foundation for Statistical Computing, Vienna, Austria) (26). Comparisons among groups were performed using partial least squares (PLS) regression according to a previously reported protocol (27). PLS regression is usually performed to compare the characteristics of the samples and to classify certain groups. In the present study, this method was used to determine the discriminability between the GC group and the control group before applying the machine learning approach.
Diagnostic algorithm
To construct the diagnostic algorithm of GC, logistic regression (LR), a type of machine learning method, was used for discriminant analysis. The expression levels (peak area in the chromatogram) of 536 lipid molecules obtained from each plasma sample were individually normalized by the median value. The normalized datasets of control and cancer were learned by LR, and blinded samples were classified as cancer or not. The cancer possibility was indicated as the probability value (0.0–1.0). The procedure and mathematical formulae used were those described in our previous study (27). The predictive accuracy of the LR classifier was evaluated by using a leave-one-out cross validation (LOOCV) procedure (28). These procedures are shown in Fig. S2.
Results
Patient characteristics
The clinicopathological characteristics of the patients are summarized in Table I. There were no differences in the age or sex of the patients between the two groups. The GC group included numerous patients with advanced GC who exhibited deep invasion, extensive lymph node metastasis and severe lymphovascular invasion. By contrast, tumor markers such as carcinoembryonic antigen (CEA) and carbohydrate antigen 19-9 (CA19-9) were not higher than the standard values in the majority of patients, despite being generally used as biomarkers of GC. Although three patients in the GC group had postoperative cancer recurrence during the follow-up period, none of the patients in the control group had residual malignancies or recurrences.
Table I.
Characteristics of controls and patients with GC.
| Variables | Control (n=16) | GC (n=20) | P-value |
|---|---|---|---|
| Comparison between control and GC | |||
| Age (mean ± SD) | 71.2±10.3 | 67.1±12.1 | 0.285 |
| Sex, n (male/female) | 9/7 | 11/9 | 0.999 |
| Cancer specific variables | |||
| Tumor size, mm (mean ± SD) | 66.4±49.9 | ||
| CEA, n (<5/≥5 ng/ml) | 17/3 | ||
| CA19-9, n (<37/≥37 U/ml) | 14/6 | ||
| T-factor, n (T1/2/3/4) | 2/3/8/7 | ||
| N-factor (N0/1/2/3) | 2/5/6/7 | ||
| M-factor, n (M0/1) | 19/1 | ||
| Stage, n (I/II/III/IV) | 0/10/9/1 | ||
| Lymphatic invasion, n (negative/positive) | 1/19 | ||
| Venous invasion, n (negative/positive) | 2/18 | ||
| Pathological subtypes, n (differentiated/undifferentiated) | 10/10 |
GC, gastric cancer; CEA, carcinoembryonic antigen; CA19-9, carbohydrate antigen 19-9.
Lipid biomarkers of GC
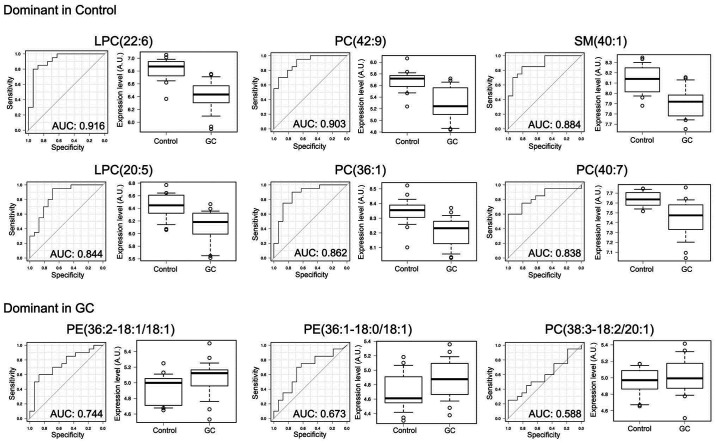
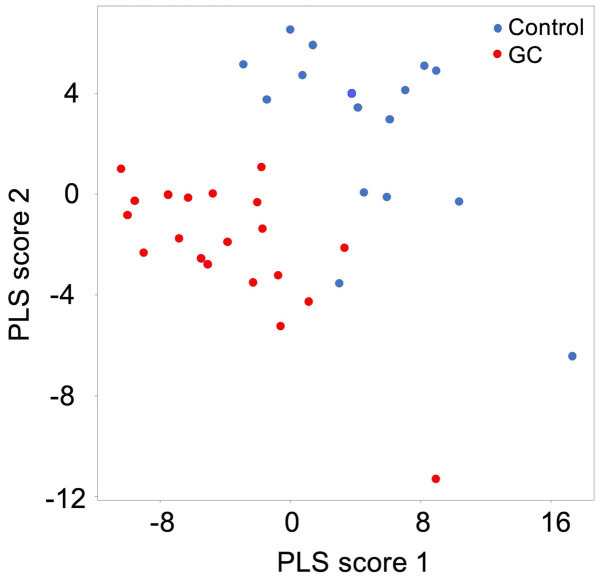
The simultaneous analysis of the plasma lipidome using LC/ESI-MS identified and quantified a total of 236 phospholipid molecules. The top ten molecules, which were up- or downregulated in the GC group, are listed in Table II. Of these, PE(36:2-18:1/18:1) showed the most remarkable upregulation in the GC group. By contrast, LPC(38:2) exhibited the most marked difference in relative expression in the control group, as a result of its suppression in the GC group. Overall, there was a downregulation trend for the ion intensities of the majority of molecules in GC plasma. The results of detailed analyses comparing the ion intensities between the GC and control groups, and the ROC curves with each area under the curve are shown in Fig. 1. These results suggested that certain lipid molecules, particularly the downregulated lipids, can be used as biomarkers for GC. In addition, the results of the PLS score plot indicated that the lipid composition of GC plasma is specifically changed, and can be used in discriminant analysis (Fig. 2).
Table II.
Candidate markers of phospholipids for GC.
| Candidate molecule | Relative expression levels (GC/control) | P-value (−log10) |
|---|---|---|
| Dominant in control | ||
| LPC(22:6) | 0.424 | 6.410 |
| PC(42:9) | 0.399 | 4.720 |
| SM(40:1) | 0.579 | 4.680 |
| LPC(20:5) | 0.502 | 3.970 |
| PC(36:1) | 0.732 | 3.850 |
| PC(40:7) | 0.680 | 3.600 |
| PC(40:1) | 0.329 | 3.590 |
| LPC(20:0) | 0.646 | 3.460 |
| LPC(18:2) | 0.644 | 3.290 |
| LPC(22:0) | 0.572 | 3.290 |
| Dominant in GC | ||
| PE(36:2-18:1/18:1) | 1.590 | 1.800 |
| PE(36:1-18:0/18:1) | 1.508 | 0.970 |
| PC(38:3-18:2/20:1) | 1.253 | 0.790 |
| PC(34:3-16:1/18:2) | 1.235 | 0.720 |
| PE(34:2-16:0/18:2) | 1.270 | 0.560 |
| PE(34:1) | 1.180 | 0.550 |
| PE(34:2) | 1.207 | 0.490 |
| PE(34:3) | 1.397 | 0.480 |
| PE(36:3) | 1.272 | 0.420 |
| PC(34:2) | 1.053 | 0.380 |
GC, gastric cancer; LPC, lysophosphatidylcholine; PC, phosphatidylcholine; SM, sphingomyelin; PE, phosphatidylethanolamine.
Figure 1.
Receiver operating characteristic curves and comparison of the expression levels for each lipid marker candidate for GC. The top six candidates dominant in control plasma and the top three in the GC group are shown. Each bracket shows the number of carbon and double bonds included. A.U., arbitrary unit; AUC, area under the curve; GC, gastric cancer; LPC, lysophosphatidylcholine; PC, phosphatidylcholine; SM, sphingomyelin; PE, phosphatidylethanolamine.
Figure 2.
Evaluation of distinguishability between the GC and control groups by PLS regression. Blue and red plots indicate the control and GC group, respectively. Scatter plots were depicted using PLS scores 1 and 2. PLS, partial least squares; GC, gastric cancer.
Discrimination of GC by machine learning
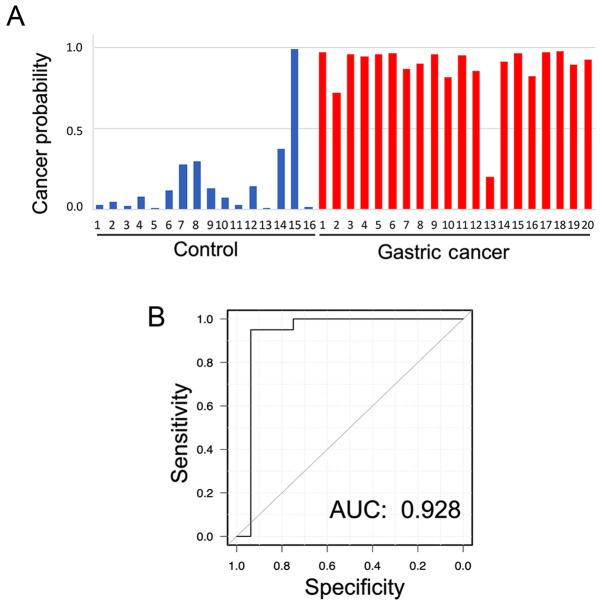
Fig. 3A and Table SI show the individual value of cancer probability in each plasma sample by using discriminant analysis with LOOCV. When a threshold of 0.5 was set for the probability of a sample being cancerous, each characteristic of each patient was distinguished. Although the correct characteristics of only one patient in each group could not be detected, a correct classification of subjects in the GC and control groups was achieved with an accuracy of 94.4% for almost all patients in both groups. The specificity and sensitivity were 93.8 and 95.0%, respectively. Furthermore, the area under the ROC curve was 0.928 (Fig. 3B).
Figure 3.
Results of discriminant analyses by machine learning for each patient and ROC curve. (A) Cancer probability in each patient. (B) ROC curve of probability by machine learning-based algorithm. The accuracy rate was 94.4%. The AUC was 0.928, and the specificity and sensitivity were 93.8 and 95.0%, respectively. AUC, area under the curve; ROC, receiver operating characteristic.
Discussion
The present study analyzed the expression levels of phospholipids, including phosphatidylcholine, sphingomyelins and phosphatidylethanolamine, in peripheral blood samples. Although these molecules are universally present in the whole body, certain plasma lipid molecules showed pronounced differences between the cancer-free controls and the patients with GC. However, the discrimination probabilities for each individual molecule were insufficient as an independent diagnostic tool. Therefore, an integrative analysis of all these molecules using a machine learning method was performed, aiming to establish more useful diagnostic systems with a higher accuracy. Compared with the results obtained using conventional GC markers, such as CEA and CA19-9, the results of integrative analysis showed a much higher sensitivity and specificity.
In discrimination analysis using LOOCV, correct results were not achieved for all the patients in each group. Regarding patient GC no. 13 (Fig. 3; Table SI), who was not determined as a correct cancerous characteristic, the patient had no specific characteristics observed besides pathological findings of T3N2M0 and non-elevation of both CEA and CA19-9. In the control group, patient control no. 15 (Fig. 3; Table SI), who did not display non-cancerous characteristics after discrimination, was after endoscopic resection for early surficial esophageal cancer. This could be one of the reasons for the inaccuracy of the results. Although further comparative analyses with strict setting of control specimens may be necessary to establish clinically useful diagnostic algorithms with a higher accuracy, our results suggest that, at least for advanced GC, using machine learning methods may be useful for the detection of GC.
To achieve significant accuracy in studies comparing patients with and without cancer, it is crucial to use the appropriate methodology for selecting the control specimens. Control specimens are frequently collected from ‘HVs’; however, there is often an age difference between patients with and without cancer, as the latter tend to be much younger than the former. This can significantly affect the results of the analysis, particularly in a lipidomic study, since the differences in the systemic metabolism associated with the patients' background and age are likely to be large (29). Previous studies have reported lipidomic approaches for cancer diagnosis using blood samples. For example, Guo et al (18) demonstrated that serum phospholipids were useful biomarkers for the different pathophysiological states of lung, gastric, intestinal and pancreatic cancer. Consistent with our results, the authors found that PC(34:2) was one of the six molecules that was increased in patients with GC. By contrast, Lee et al (19), reported that patients with GC had increased levels of LPC(18:2), and decreased levels of PC(34:2) and PE(36:3). One of the reasons for this discrepancy may be the control groups used in the different studies. In fact, in one report, the individuals in the control group were ~10 years younger than the patients in the GC group.
To prevent such a background bias, the present study used control plasma samples from patients who had underwent endoscopic resection for their early cancer months before the collection of biological samples for the SHINGEN biobank. For these patients, the clinical guidelines recommend additional surgical resection for their risk of lymph node metastases, and indeed this is commonly performed (30–32). However, the risk of metastases was generally low, and in the present study, there was no remaining cancer or metastasis in postoperative pathological findings. Therefore, in the present study, these cancer-free patients could be used as the control group in the different comparisons against the group of patients with advanced GC. By selecting this control group, the patients' background, including age and gender, were similar between the two groups. In addition, patients' status such as nutritional status, liver disorder or lipidemic disorder may affect the levels of plasma phospholipids. To investigate the effects of these factors, stratified analyses were performed for presence or absence of liver or lipidemic disorders. As a result, there were no obvious differences between patients with and without liver or lipidemic disorders in terms of specific phospholipid expression levels in the present study (data not shown). However, this should be further investigated in future studies using a large-scale cohort.
Concerning the control group settings, we previously performed the similar examinations to those described in the present study using plasma samples derived from HVs. Although each indicated molecule showed various expression levels in the HV group compared with those of the control and GC samples (Fig. S3), PLS analysis for the three groups (control, GC and HV) showed different characteristics for each group and good probability of discriminating each other (Fig. S4). This result means that amplification of the database to include HV samples should be considered for cancer screening during health checks, and that the integration analyses using machine learning methods presented in the current study contribute to discriminating each group. It appears to be reasonable that the cancer high-risk group was set as a control in the present study, and research including HV samples should be performed in future studies. To detect microresidual tumors or recurrent microtumors after surgery, comparison between pre- and postoperative conditions in the same patient is important, and this will be investigated in future studies.
To establish a novel diagnostic method for GC, previous studies have focused on the analysis of low molecular weight metabolites found in blood, serum or plasma, including lipids, primary metabolites and cell-free nucleic acids (33–36). During the preparation of serum, blood clots are formed by using serum separating agents and/or coagulation accelerators. In this process, numerous components in platelets, such as intracellular messengers/mediators, cell membrane and organelles, are released into the serum from disrupted platelets (37). The released components affect the MS results of the molecular composition of serum, particularly the phospholipid results. To avoid this problem, the plasma used in the present study was obtained by employing the anticoagulant agent EDTA and by centrifugation, without platelet disruption. This methodology may have contributed to the present results. Thus, the authors recommend the use of this methodology for similar studies in this field.
In summary, the novel cancer diagnosis approach employed in the present study may contribute to the development of relevant desirable biomarkers in the near future. The present study has certain limitations that need to be acknowledged. First, it is difficult to derive a definitive conclusion due to the small number of patients included in the study. Second, the patients in the control group, although they were cancer-free at the time of plasma collection and for months before sample collection, they had a previous cancer history. Despite certain limitations, the results of the present study strongly suggest that this new approach has a promising future as a diagnostic tool for GC. In conclusion, the present study suggests the diagnostic prospects of a plasma lipidomic and machine learning approach for GC. By accumulating more reliable data, this novel methodology may also be able to predict the efficacy of each therapy.
Supplementary Material
Acknowledgements
The authors would like to thank Ms. Arisa Ogihara (First Department of Surgery, Faculty of Medicine, University of Yamanashi, Yamanashi, Japan), Ms. Ayumi Manita (Department of Anatomy and Cell Biology, Faculty of Medicine, University of Yamanashi, Yamanashi, Japan) and Ms. Masumi Tanzawa (Department of Anatomy and Cell Biology, Faculty of Medicine, University of Yamanashi, Yamanashi, Japan) for their technical assistance. The authors would like to thank Shimadzu Corporation (Kyoto) (to which TM is affiliated) for lending the Mass Spectrometry instruments to ST.
Glossary
Abbreviations
- AUC
area under the curve
- CA19-9
carbohydrate antigen 19-9
- CEA
carcinoembryonic antigen
- EDTA
ethylenediamine tetraacetic acid
- GC
gastric cancer
- HV
healthy volunteer
- LC/ESI-MS
liquid chromatography/electrospray ionization-mass spectrometry
- LOOCV
leave-one-out cross validation
- LR
logistic regression
- MS
mass spectrometry
- PLS
partial least squares
- ROC
receiver operating characteristics
- SHINGEN
Yamanashi Biobank of Gastroenterological Cancers
- UICC
Union for International Cancer Control
Funding Statement
The present study was performed mainly with expenses grants of the University of Yamanashi, and partially supported by JSPS KAKENHI (grant nos. 17K10601 and 20K09031 to DI).
Funding
The present study was performed mainly with expenses grants of the University of Yamanashi, and partially supported by JSPS KAKENHI (grant nos. 17K10601 and 20K09031 to DI).
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
RS and KY performed the majority of the experiments and wrote the initial draft of the manuscript. RS, KY, KS, SF, HA, YK, TM, KO, TI, DI and ST designed the study and contributed to analysis and interpretation of data, and assisted in the preparation of the manuscript. RS, KY and TI were responsible for confirming the authenticity of all the raw data. All other authors have contributed to data collection and interpretation, and critically reviewed the manuscript. All authors approved the final version of the manuscript, and agree to be accountable for all aspects of the work in ensuring that questions related to the accuracy or integrity of any part of the work are appropriately investigated and resolved. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The biological sample collection in the SHINGEN (#1665) biobank and the study design (#2192) were approved by the Ethics Committee of the University of Yamanashi (Yamanashi, Japan). All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1964 and later versions. Written informed consent to be included in the study, or the equivalent, was obtained from all patients.
Patient consent for publication
Although detailed clinical data of individual patients were not included in the present study, consent for publication was obtained from all patients included in the present study.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Ferlay J, Colombet M, Soerjomataram I, Mathers C, Parkin DM, Piñeros M, Znaor A, Bray F. Estimating the global cancer incidence and mortality in 2018: GLOBOCAN sources and methods. Int J Cancer. 2019;144:1941–1953. doi: 10.1002/ijc.31937. [DOI] [PubMed] [Google Scholar]
- 2.Ito S, Sano T, Mizusawa J, Takahari D, Katayama H, Katai H, Kawashima Y, Kinoshita T, Terashima M, Nashimoto A, et al. A phase II study of preoperative chemotherapy with docetaxel, cisplatin, and S-1 followed by gastrectomy with D2 plus para-aortic lymph node dissection for gastric cancer with extensive lymph node metastasis: JCOG1002. Gastric Cancer. 2017;20:322–331. doi: 10.1007/s10120-016-0619-z. [DOI] [PubMed] [Google Scholar]
- 3.Sasako M, Sano T, Yamamoto S, Kurokawa Y, Nashimoto A, Kurita A, Hiratsuka M, Tsujinaka T, Kinoshita T, Arai K, et al. D2 lymphadenectomy alone or with para-aortic nodal dissection for gastric cancer. N Engl J Med. 2008;359:453–462. doi: 10.1056/NEJMoa0707035. [DOI] [PubMed] [Google Scholar]
- 4.Kim DH, Oh SJ, Oh CA, Choi MG, Noh JH, Sohn TS, Bae JM, Kim S. The relationships between perioperative CEA, CA 19-9, and CA 72-4 and recurrence in gastric cancer patients after curative radical gastrectomy. J Surg Oncol. 2011;104:585–591. doi: 10.1002/jso.21919. [DOI] [PubMed] [Google Scholar]
- 5.Shimada H, Noie T, Ohashi M, Oba K, Takahashi Y. Clinical significance of serum tumor markers for gastric cancer: A systematic review of literature by the Task Force of the Japanese Gastric Cancer Association. Gastric Cancer. 2014;17:26–33. doi: 10.1007/s10120-013-0259-5. [DOI] [PubMed] [Google Scholar]
- 6.Shoda K, Ichikawa D, Fujita Y, Masuda K, Hiramoto H, Hamada J, Arita T, Konishi H, Komatsu S, Shiozaki A, et al. Monitoring the HER2 copy number status in circulating tumor DNA by droplet digital PCR in patients with gastric cancer. Gastric Cancer. 2017;20:126–135. doi: 10.1007/s10120-016-0599-z. [DOI] [PubMed] [Google Scholar]
- 7.Vaysse PM, Heeren RMA, Porta T, Balluff B. Mass spectrometry imaging for clinical research-latest developments, applications, and current limitations. Analyst. 2017;142:2690–2712. doi: 10.1039/C7AN00565B. [DOI] [PubMed] [Google Scholar]
- 8.Zhou Z, Tu J, Zhu ZJ. Advancing the large-scale CCS database for metabolomics and lipidomics at the machine-learning era. Curr Opin Chem Biol. 2018;42:34–41. doi: 10.1016/j.cbpa.2017.10.033. [DOI] [PubMed] [Google Scholar]
- 9.Liebal UW, Phan ANT, Sudhakar M, Raman K, Blank LM. Machine learning applications for mass spectrometry-based metabolomics. Metabolites. 2020;10:243. doi: 10.3390/metabo10060243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Enjoji M, Kohjima M, Ohtsu K, Matsunaga K, Murata Y, Nakamuta M, Imamura K, Tanabe H, Iwashita A, Nagahama T, Yao K. Intracellular mechanisms underlying lipid accumulation (white opaque substance) in gastric epithelial neoplasms: A pilot study of expression profiles of lipid-metabolism-associated genes. J Gastroenterol Hepatol. 2016;31:776–781. doi: 10.1111/jgh.13216. [DOI] [PubMed] [Google Scholar]
- 11.Yao K, Iwashita A, Tanabe H, Nishimata N, Nagahama T, Maki S, Takaki Y, Hirai F, Hisabe T, Nishimura T, Matsui T. White opaque substance within superficial elevated gastric neoplasia as visualized by magnification endoscopy with narrow-band imaging: A new optical sign for differentiating between adenoma and carcinoma. Gastrointest Endosc. 2008;68:574–580. doi: 10.1016/j.gie.2008.04.011. [DOI] [PubMed] [Google Scholar]
- 12.de Carvalho AC, Kowalski LP, Campos AH, Soares FA, Carvalho AL, Vettore AL. Clinical significance of molecular alterations in histologically negative surgical margins of head and neck cancer patients. Oral Oncol. 2012;48:240–248. doi: 10.1016/j.oraloncology.2011.10.018. [DOI] [PubMed] [Google Scholar]
- 13.Xu J, Wang X, Xu P, Liu S, Teng F, Liu X, Zhu Q, Hua X, Gong Z, Jia X. Mass spectrometry-based peptidome profiling of human serous ovarian cancer tissues. Int J Biochem Cell Biol. 2019;107:53–61. doi: 10.1016/j.biocel.2018.12.004. [DOI] [PubMed] [Google Scholar]
- 14.Kerian KS, Jarmusch AK, Pirro V, Koch MO, Masterson TA, Cheng L, Cooks RG. Differentiation of prostate cancer from normal tissue in radical prostatectomy specimens by desorption electrospray ionization and touch spray ionization mass spectrometry. Analyst. 2015;140:1090–1098. doi: 10.1039/C4AN02039A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Iwano T, Yoshimura K, Inoue S, Odate T, Ogata K, Funatsu S, Tanihata H, Kondo T, Ichikawa D, Takeda S. Breast cancer diagnosis based on lipid profiling by probe electrospray ionization mass spectrometry. Br J Surg. 2020;107:632–635. doi: 10.1002/bjs.11613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ishii H, Saitoh M, Sakamoto K, Sakamoto K, Saigusa D, Kasai H, Ashizawa K, Miyazawa K, Takeda S, Masuyama K, Yoshimura K. Lipidome-based rapid diagnosis with machine learning for detection of TGF-β signalling activated area in head and neck cancer. Br J Cancer. 2020;122:995–1004. doi: 10.1038/s41416-020-0732-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Taguchi R, Nishijima M, Shimizu T. Basic analytical systems for lipidomics by mass spectrometry in Japan. Methods Enzymol. 2007;432:185–211. doi: 10.1016/S0076-6879(07)32008-9. [DOI] [PubMed] [Google Scholar]
- 18.Guo Y, Ren J, Li X, Liu X, Liu N, Wang Y, Li Z. Simultaneous quantification of serum multi-phospholipids as potential biomarkers for differentiating different pathophysiological states of lung, stomach, intestine, and pancreas. J Cancer. 2017;8:2191–2204. doi: 10.7150/jca.19128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee GB, Lee JC, Moon MH. Plasma lipid profile comparison of five different cancers by nanoflow ultrahigh performance liquid chromatography-tandem mass spectrometry. Anal Chim Acta. 2019;1063:117–126. doi: 10.1016/j.aca.2019.02.021. [DOI] [PubMed] [Google Scholar]
- 20.Yang M, Forbes ME, Bitting RL, O'Neill SS, Chou PC, Topaloglu U, Miller LD, Hawkins GA, Grant SC, DeYoung BR, et al. Incorporating blood-based liquid biopsy information into cancer staging: Time for a TNMB system? Ann Oncol. 2018;29:311–323. doi: 10.1093/annonc/mdx766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Vaidyanathan R, Soon RH, Zhang P, Jiang K, Lim CT. Cancer diagnosis: From tumor to liquid biopsy and beyond. Lab Chip. 2018;19:11–34. doi: 10.1039/c8lc00684a. [DOI] [PubMed] [Google Scholar]
- 22.Ponti G, Manfredini M, Tomasi A. Non-blood sources of cell-free DNA for cancer molecular profiling in clinical pathology and oncology. Crit Rev Oncol Hematol. 2019;141:36–42. doi: 10.1016/j.critrevonc.2019.06.005. [DOI] [PubMed] [Google Scholar]
- 23.Kaczor-Urbanowicz KE, Wei F, Rao SL, Kim J, Shin H, Cheng J, Tu M, Wong DTW, Kim Y. Clinical validity of saliva and novel technology for cancer detection. Biochim Biophys Acta Rev Cancer. 2019;1872:49–59. doi: 10.1016/j.bbcan.2019.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Union for International Cancer Control, corp-author. TNM classification of malignant tumours. In: Brierley JD, Gospodarowicz MK, Wittekind C, editors. 8th edition. John Wiley & Sons, Inc.; New York: 2017. [Google Scholar]
- 25.World Medical Association, corp-author. World medical association declaration of Helsinki: Ethical principles for medical research involving human subjects. JAMA. 2013;310:2191–2194. doi: 10.1001/jama.2013.281053. [DOI] [PubMed] [Google Scholar]
- 26.Kanda Y. Investigation of the freely available easy-to-use software ‘EZR’ for medical statistics. Bone Marrow Transplant. 2013;48:452–458. doi: 10.1038/bmt.2012.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Johno H, Yoshimura K, Mori Y, Kimura T, Niimi M, Yamada M, Tanigawa T, Fan J, Takeda S. Detection of potential new biomarkers of atherosclerosis by probe electrospray ionization mass spectrometry. Metabolomics. 2018;14:38. doi: 10.1007/s11306-018-1334-z. [DOI] [PubMed] [Google Scholar]
- 28.Bousquet O, Elisseeff A. Stability and generalization. J Mach Learn Res. 2002;2:499–526. [Google Scholar]
- 29.Singh R, Sharma S, Singh RK, Mahdi AA, Singh RK, Lee Gierke C, Cornelissen G. Effect of gender, age, diet and smoking status on chronomics of circulating plasma lipid components in healthy Indians. Clin Chim Acta. 2016;459:10–18. doi: 10.1016/j.cca.2016.05.010. [DOI] [PubMed] [Google Scholar]
- 30.Japanese Gastric Cancer Association, corp-author. Japanese gastric cancer treatment guidelines 2018 (5th edition) Gastric Cancer. 2021;24:1–21. doi: 10.1007/s10120-020-01042-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitagawa Y, Uno T, Oyama T, Kato K, Kato H, Kawakubo H, Kawamura O, Kusano M, Kuwano H, Takeuchi H, et al. Esophageal cancer practice guidelines 2017 edited by the Japan esophageal society: Part 2. Esophagus. 2019;16:25–43. doi: 10.1007/s10388-018-0641-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hashiguchi Y, Muro K, Saito Y, Ito Y, Ajioka Y, Hamaguchi T, Hasegawa K, Hotta K, Ishida H, Ishiguro M, et al. Japanese Society for Cancer of the Colon and Rectum (JSCCR) guidelines 2019 for the treatment of colorectal cancer. Int J Clin Oncol. 2020;25:1–42. doi: 10.1007/s10147-019-01485-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ros-Mazurczyk M, Jelonek K, Marczyk M, Binczyk F, Pietrowska M, Polanska J, Dziadziuszko R, Jassem J, Rzyman W, Widlak P. Serum lipid profile discriminates patients with early lung cancer from healthy controls. Lung Cancer. 2017;112:69–74. doi: 10.1016/j.lungcan.2017.07.036. [DOI] [PubMed] [Google Scholar]
- 34.Banales JM, Iñarrairaegui M, Arbelaiz A, Milkiewicz P, Muntané J, Muñoz-Bellvis L, La Casta A, Gonzalez LM, Arretxe E, Alonso C, et al. Serum metabolites as diagnostic biomarkers for cholangiocarcinoma, hepatocellular carcinoma, and primary sclerosing cholangitis. Hepatology. 2019;70:547–562. doi: 10.1002/hep.30319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Otandault A, Anker P, Al Amir Dache Z, Guillaumon V, Meddeb R, Pastor B, Pisareva E, Sanchez C, Tanos R, Tousch G, et al. Recent advances in circulating nucleic acids in oncology. Ann Oncol. 2019;30:374–384. doi: 10.1093/annonc/mdz031. [DOI] [PubMed] [Google Scholar]
- 36.Kahlert C. Liquid biopsy: Is there an advantage to analyzing circulating exosomal DNA compared to cfDNA or are they the same? Cancer Res. 2019;79:2462–2465. doi: 10.1158/0008-5472.CAN-19-0019. [DOI] [PubMed] [Google Scholar]
- 37.Provost P. The clinical significance of platelet microparticle-associated microRNAs. Clin Chem Lab Med. 2017;55:657–666. doi: 10.1515/cclm-2016-0895. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.