Abstract
Cancer metastasis is the main cause of death in cancer patients, including patients with thyroid cancer (TC). TC is the most common malignant endocrine tumour. In the recent years, increasing evidence has demonstrated that circular RNAs (circRNAs) serve a significant role in the development of many types of human cancer. However, the function and underlying mechanism of circCCDC66 in TC remain unclear. The present study aimed to explore the role of circCCDC66 in TC. To do so, reverse transcription quantitative PCR was used to detect the expression level of circCCDC66. Cell viability, migratory and invasive abilities, and glucose consumption were evaluated by cell counting kit 8, Transwell and glucose consumption assays, respectively. The association between circCCDC66 or pyruvate dehydrogenase kinase 4 (PDK4) and miR-211-5p was verified by dual-luciferase reporter assay. The results demonstrated that circCCDC66 expression was significantly increased in TC tissues and cell lines. Furthermore, silencing circCCDC66 inhibited TC cell proliferation, migratory and invasive abilities and glycolysis in vitro. Further validation demonstrated that circCCDC66 directly interacted with the microRNA (miR) miR-211-5p. Subsequently, the activity of circCCDC66 was attenuated by miR-211-5p. In addition, the results demonstrated that circCCDC66 may promote papillary thyroid cancer progression by sponging miR-211-5p and increasing expression of PDK4. In conclusion, the present study demonstrated that circCCDC66 could promote TC cell proliferation, migratory and invasive abilities and invasion and glycolysis through the miR-211-5p/PDK4 axis. These findings suggested that targeting circCCDC66 may be considered as a promising therapeutic strategy for TC.
Keywords: circular RNA, circCCDC66, competitive endogenous RNA, miR-211-5p
Introduction
Circular RNAs (circRNAs) are non-coding RNAs with covalently closed loop structures formed by reverse splicing (1). Unlike linear RNAs, circRNAs have no exposed end and are not easily degraded by conventional intracellular degradation mechanisms. They are thus relatively stable in cells. Due to the limitations of technology and the low abundance of circRNAs in cells, circRNAs have not been well described (2). In the recent years, thanks to improvements in technology and algorithms, circRNAs have been implicated in numerous human diseases. For example, circSLC8A1 is downregulated in bladder cancer tissues and cell lines and its expression is associated with the pathological and histological stages of bladder cancer. In breast cancer, circTADA2As is a promising prognostic biomarker for patients with large triple-negative breast cancer (TNBC). Furthermore, the high expression of circRanGAP1 is closely related to the advanced TNM staging, lymph node metastasis and poor survival rate of patients with gastric cancer (3–5). Several studies have demonstrated that ncRNAs are differentially expressed in tumour and normal tissues and cells, including pediatric acute lymphoblastic leukemia and thyroid carcinoma (6,7). Some circRNAs have been reported to promote or inhibit cancer cell proliferation, migration and invasion. For example, Li et al (8) reported that circ-ITCH is expressed at lower levels in oesophageal carcinoma tissues compared with adjacent tissues, and that circ-ITCH can inhibit cell proliferation and tumorigenic ability by repressing the Wnt pathway. CircRNAs can bind to intracellular regulatory proteins, thereby affecting cellular metabolic activity. A study by Du et al (9) reported that circ-FOXO3 can interact with important regulators of cell cycle progression, including CDK6, p16, and p27, in order to regulate cell cycle progression of cancer cells.
Due to the expanded use of diagnostic imaging and surveillance, the prevalence of TC has risen worldwide (10). In 2013, 33,939 patients with TC (7,146 men and 26,793 women) were identified and the death toll was 4,974 (2,292 men and 2,682 women) in China (11). A steady increase has been observed in TC incidence worldwide, whereas the death rate has only slightly increased (12). According to previous studies, a number of genes and signaling pathways that play an important role in TC have been identified. Most patients with TC exhibit overactivated MAPK signalling, which plays an important role in the regulation of cell proliferation (13). The regulation of cellular metabolic pathways by circRNA is increasingly documented (14). For example, the circRNA circZFR was reported to promote the expression of C8orf4 in papillary TC (PTC) cells as a competitive endogenous RNA (ceRNA) of the microRNA (miR) miR-1261, thus regulating the carcinogenic effect of the miR-1261/C8orf4 axis (15).
A previous study by Hsiao et al (16) reported a novel carcinogenic function of circCCDC66 in colon cancer. The results demonstrated that circCCDC66 is highly upregulated in colorectal cancer and serves as a new cancer-related circRNA. Furthermore, the study showed that 88% of patients with colon cancer have high circCCDC66 expression, and that patients with higher circCCDC66 expression have a lower overall survival rate compared with patients with lower circCCDC66 expression. The dysregulation of circCCDC66 might therefore have a prognostic potential in human cancers. This study also reported that the suppression of circCCDC66 in cancer cells can inhibit tumour invasiveness in xenograft mouse models. In addition, circCCDC66 knockout results in decreased miRNA-targeted oncogene expression in circCCDC66-mediated sponge processes. Furthermore, the overexpression of circCCDC66 can promote cancer cell proliferation and colony formation, suggesting therefore that circCCDC66 might be considered as a critical driver of cancer aetiology. The overexpression of circCCDC66 can stimulate oncogenes by inhibiting miRNAs that are involved in the suppression of these oncogenes. circCCDC66 acts as a miRNA sponge, affecting the expression of miR-211, leading to the upregulation of SRCIN1 and promoting cell proliferation in non-small cell lung cancer (17). In osteosarcoma, circCCDC66 promotes the malignant phenotype of osteosarcoma by stimulating miR-338-3p to upregulate the expression of PTP1B (18). The present study aimed to investigate the effects of circCCDC66 on the proliferation, migratory and invasive abilities and glycolysis of TC cells.
Materials and methods
Human tissue samples
Samples of TC tissues and matched adjacent normal tissues were collected from nine patients with TC (age range, 32–69 years; mean age, 43.7±6.9 years; sex ratio, four men and 18 women) who underwent surgery at Qingdao Municipal Hospital. All subjects were recruited between April 2019 and December 2019. This study was approved by the Ethics Committee of Qingdao Municipal Hospital (approval no. 2019QMH126) and all patients provided informed consent.
Cell culture and transfection
The cell lines Nthy-ori 3-1, CAL62 and TPC1 were purchased from the American Type Culture Collection (ATCC). All cells were cultured in DMEM (Invitrogen; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS (Invitrogen; Thermo Fisher Scientific, Inc.) and were places at 37°C in a humidified incubator containing 5% CO2.
Cell transfection was carried out using Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to the manufacturers' instruction when cells in culture reached 60–80% confluence. Small interfering (si)RNAs targeting circCCDC66 (si-circCCDC66#1, 5′-CAAUUAGAGCAUCAGGAAA-3′; si-circCCDC66#2, 5′-GAGCAUCAGGAAACAGUAC-3′) and negative control (si-NC, 5′-UUCUCCGAACGUGUCACGUTT-3′) were purchased from Shanghai GenePharma Co., Ltd. miR-211-5p mimic and negative control were purchased from Guangzhou RiboBio Co., Ltd (cat. no. miR10000268-1-5). The transfection concentrations of oligonucleotides were as follows: si-NC, 40 nM; si-circCCDC66, 40 nM; miR-211-5p mimic, 50 nM; and miRNA control, 50 nM. Lipofectamine® 2000 Reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and si-RNAs or miR-mimic were diluted into serum-free DMEM medium, mixed together and incubated for 20 min at room temperature. This solution was subsequently was added to CAL62 and TPC1 cells and transfected for 4–6 h at 37°C in a humidified incubator containing 5% CO2.
Cell fractionation
The PARIS™ kit (cat. no. AM1921; Invitrogen; Thermo Fisher Scientific, Inc.) was used to extract cytoplasmic and nuclear RNA. Briefly, CAL62 and TPC1 cells were incubated with the lysis solution from the kit on ice for 10 min following two washes with PBS. Samples were collected, centrifuged at 500 × g for 30 min at 4°C, and cytoplasmic RNA was extracted from the supernatant, whereas nuclear RNA was extracted from the nuclear pellet. Samples were stored at −80°C until further use. GAPDH and U6 were used as the cytoplasmic RNA and nuclear RNA controls, respectively.
Online analysis of downstream target of circCCDC66
CircRNAs are known to exert their functions through sponging miRNAs and subsequently regulating downstream genes expression in cells (19,20). RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/index.html) was therefore used to predict the potential miRNA associated with circCCDC66. RegRNA2.0 is an integrated web server used to identify functional RNA motifs and sites. miRcode (http://www.mircode.org/mircode/) was subsequently used to predict the miRNA target. MiRcode provides the ‘whole transcriptome’ human miRNA target predictions based on the comprehensive GENCODE gene annotation.
Dual-luciferase reporter assay
To generate luciferase reporter expression constructs, full-length circCCDC66 or the 3′-UTR of PDK4 containing the expected binding sites was inserted into luciferase reporter DNA pGL3 vector (Promega Corporation). circCCDC66 mutant DNA or PDK4 3′-UTR mutant DNA was cloned into luciferase reporter DNA pGL3 vector. Plasmids were transfected with miRNAs in 293T cells (ATCC) using Lipofectamine® 2000 (Invitrogen; Thermo Fisher Scientific, Inc.) for 48 h. The measurement of luciferase activity was carried out with a Dual-Luciferase Reporter Assay System. The relative activity was normalized to Renilla luciferase activity.
Reverse transcription quantitative (RT-q) PCR
Total RNA was extracted from tissues or cells using TRIzol reagent (Invitrogen; Thermo Fisher Scientific, Inc.), which was then reverse transcribed into cDNA. The cDNA was generated using the Moloney Murine Leukemia Virus (M-MLV) first strand kit (Thermo Fisher Scientific, Inc.) or miRNA reverse transcription kit (Takara Biotechnology Co., Ltd.) according to the manufacturer's protocol. RT-qPCR reactions were conducted using a SYBR Green PCR Kit (Takara Biotechnology Co., Ltd). The thermocycling conditions are as follows: 95°C for 30 sec, followed by 40 cycles of 95°C for 5 sec and 60°C for 20 sec. All primers were purchased from Guangzhou RiboBio Co., Ltd. The relative expression levels were normalized to endogenous control and were expressed as 2−ΔΔCq (21). GAPDH and U6 were used as endogenous control. The sequences of the primers used were as follows: circCCDC66, forward 5′-TCTCTTGGACCCAGCTCAG-3′, reverse 5′-TGAATCAAAGTGCATTGCCC-3′; miR-211-5p, forward 5′-GATGCTGTAATGGATGATATGA-3′; reverse, 5′-ATTGGAACGATACAGAGAAGATT-3′; PDK4, forward 5′-GGAAGCATTGATCCTAACTGTGA-3′, reverse, 5′-GGTGAGAAGGAACATACACGATG-3′; GAPDH, forward 5′-GGAGCGAGATCCCTCCAAAAT-3′, reverse, 5′-GGCTGTTGTCATACTTCTCATGG-3′; and U6, forward 5′-GCTCGCTTCGGCAGCACA-3′ and reverse, 5′-AGGTATTCGCACCAGAGGA-3′.
Cell proliferation assay
CAL62 and TPC1 cells (2×103 cells/well) were seeded in a 96-well plate and five wells were used for technical replicates. Cell proliferation was assessed with a CCK-8 kit (Dojindo Molecular Technologies, Inc.) according to the manufacturers' protocol. The absorbance was measured at 450 nm on a microplate reader.
Cell migration and invasion assays
CAL62 and TPC1 cells (5×104 cells/well) were seeded in the upper chamber of a Transwell plate (Corning, Inc.) containing DMEM supplemented with 1% FBS, which was coated with (invasion assay) or without (migration assay) Matrigel mix (BD Biosciences). Medium containing 15% FBS was added to the lower chamber. After incubation for 48 h, methanol was used to fix the cells for 10 min at room temperature and cells were subsequently stained with DAPI for 10 min at room temperature. A fluorescent microscope (magnification, ×200) was used to count the number of cells in each treatment group (CAL62 cells transfected with si-circCCDC66, CAL62 cells transfected with si-NC, TPC1 cells transfected with si-circCCDC66 and TPC1 cells transfected with si-NC) and six areas were randomly selected to calculate the average value.
Detection of glucose consumption
The metabolism of malignant tumors is explained by the Warburg effect, which is the metabolic shift from oxidative phosphorylation (OXPHOS) to glycolysis in tumor cells (22). The effect of glycolysis on tumor growth is crucial. As previously described, glucose consumption was analysed. CAL62 and TPC1 cells (5×104 cells/well) were seeded into 24-well plates for 48 h. The cell supernatants were collected after 48 h of transfection. A glucose assay kit (cat. no. K606-100; BioVision, Inc.) was used to determine glucose levels in the supernatant according to the manufacturers' instructions.
Statistical analysis
Data from at least three independent experiments were analysed using SPSS 10.0 (SPSS, Inc.) and were presented as the means ± standard deviation. Normality of data was evaluated using Shapiro-Wilk normality test. Homoscedasticity of data was evaluated using F test. The differences between two groups were compared using Student's t-test. The differences among more than two groups were compared using one-way ANOVA followed by the Least Significance Difference post hoc test. P<0.05 was considered to indicate a statistically significant difference.
Results
circCCDC66 is upregulated in TC tissues
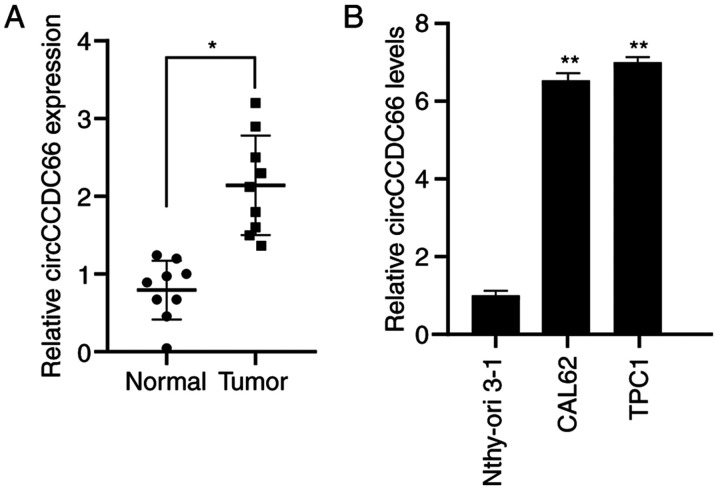
The results from RT-qPCR demonstrated that circCCDC66 was significantly overexpressed in tumour tissues compared with normal tissues (Fig. 1A). In addition, circCCDC66 expression levels were assessed in the TC cell lines TPC1 and CAL62 and the normal thyroid cell line Nthy-ori 3-1. The results showed that circCCDC66 was significantly overexpressed in TC cells compared with normal thyroid cells (Fig. 1B).
Figure 1.
circCCDC66 was upregulated in TC tissues and cell lines. (A) circCCDC66 expression was significantly increased in TC tissues compared with normal tissues. (B) circCCDC66 expression was significantly increased in TC cell lines CAL62 and TPC1 compared with normal cell line Nthy-ori 3-1. *P<0.05 and **P<0.01. TC, thyroid cancer.
circCCDC66 knockdown inhibits TC cell proliferation
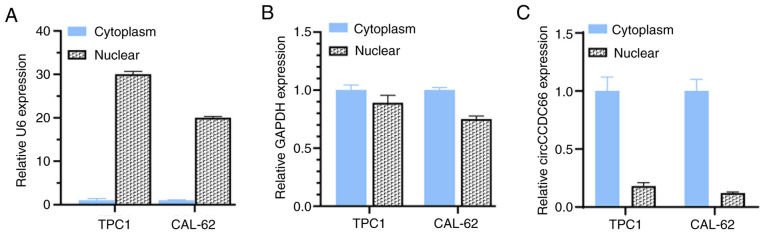
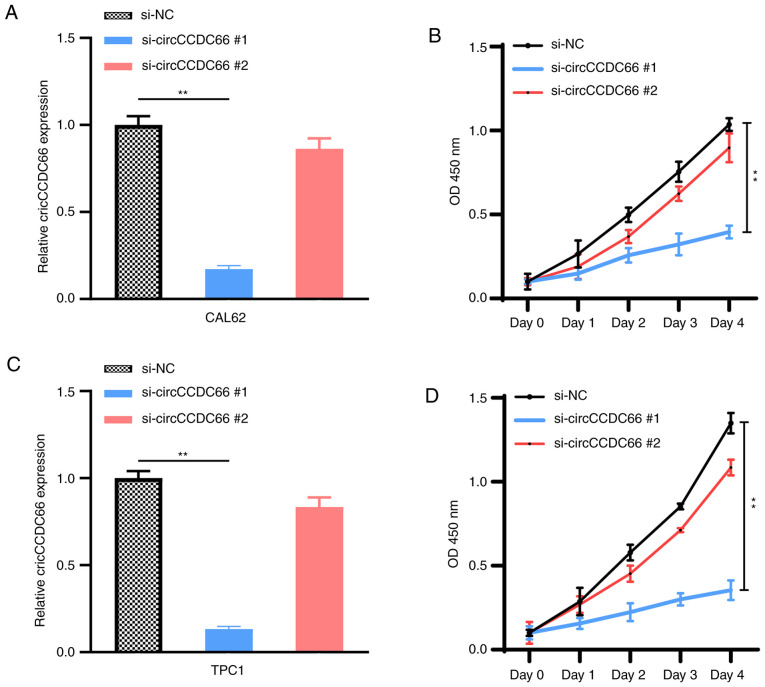
To investigate the biological function of circCCDC66 in TC cells, its subcellular localization was assessed in TPC1 and CAL62 cells. U6 in the nucleus and GAPDH in the cytoplasm were detected as nuclear and cytoplasmic markers (Fig. 2A and B). The results from RT-qPCR demonstrated that circCCDC66 was mainly located in the cytoplasm, similarly to GAPDH (Fig. 2C). TC cells (TPC1 and CAL62) were then transfected with si-circCCDC66#1 and si-circCCDC66#2 in order to knockdown circCCDC66 (Fig. 3A and C). The effect of circCCDC66 knockdown on TC cell proliferation was evaluated with a CCK-8 assay. Compared with control siRNA-transfected cells, the proliferation of TPC1 and CAL62 cells transfected with si-circCCDC66#1 was significantly decreased (Fig. 3B and D), suggesting that circCCDC66 knockdown could inhibit TC cell proliferation. Since the efficiency of knockdown and proliferation ability of si-circCCDC66#2 was not satisfactory, only si-circCCDC66#1 was used for subsequent experiments (represented as si-circCCDC66).
Figure 2.
circCCDC66 was mainly located in the cytoplasm. (A) In TPC1 and CAL62 cells, U6 was located in the nucleus according to RT-qPCR results. The expression level of U6 in cytoplasm was used as control. (B) GAPDH was mainly located in the cytoplasm in TPC1 and CAL62 cells according to RT-qPCR results. The expression level of GAPDH in cytoplasm was used as control. (C) circCCDC66 was mainly located in the cytoplasm according to RT-qPCR results. The expression level of circCCDC66 in cytoplasm was used as control. RT-qPCR, reverse transcription quantitative PCR.
Figure 3.
Knockdown of circCCDC66 inhibited thyroid cancer cell proliferation. (A,C) Following si-circCCDC66#1 was transfected into CAL62 and TPC1 cells, circCCDC66 expression was significantly decreased, which was not the case with si-circCCDC66#2. (B and D) Results from Cell Counting Kit-8 showed that the proliferation of CAL62 and TPC1 cells was significantly decreased following circCCDC66 silencing by si-circCCDC66#1, which was not the case with si-circCCDC66#2. **P<0.01. NC, negative control; OCD, optical density; si, silencing.
circCCDC66 silencing inhibits TC cell migratory and invasive abilities in vitro
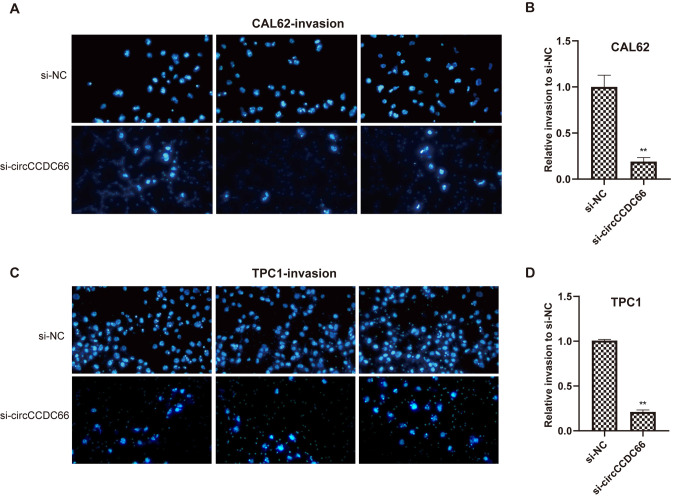
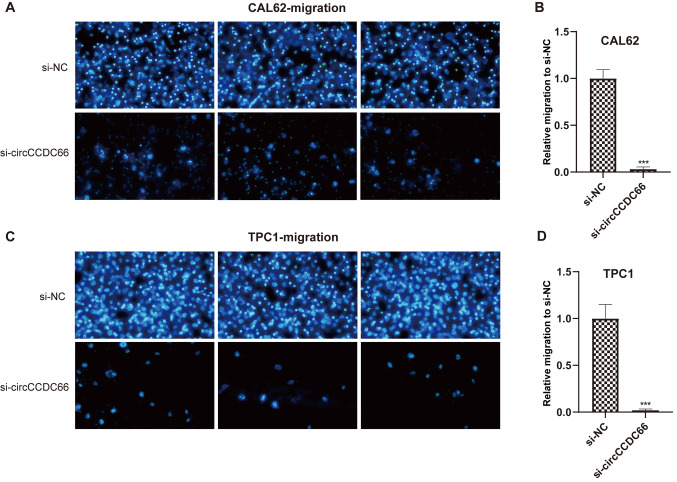
Whether circCCDC66 knockdown could affect the metastatic ability of the TC cells TPC1 and CAL62 was evaluated using Transwell assays. As presented in Figs. 4 and 5, circCCDC66 silencing resulted in a smaller number of migrating cells, suggesting that the knockdown of circCCDC66 inhibited TC cell migratory and invasive abilities.
Figure 4.
Knockdown of circCCDC66 inhibited TC cell invasive ability in vitro. (A and C) Number of nuclei decreased significantly after circCCDC66 silencing both in CAL62 and TPC1 cells. (B and D) Relative invasive ability of CAL62 and TPC1 cells following circCCDC66 knockdown was 3/4 lower than the control group. **P<0.01. NC, negative control; si, silencing.
Figure 5.
Knockdown of circCCDC66 inhibited TC cell migratory ability in vitro. (A and C) Number of nuclei decreased significantly after circCCDC66 silencing both in CAL62 and TPC1 cells. (B and D) Relative migratory ability of CAL62 and TPC1 cells following circCCDC66 knockdown was 19/20 lower than the control group. ***P<0.001. NC, negative control; si, silencing.
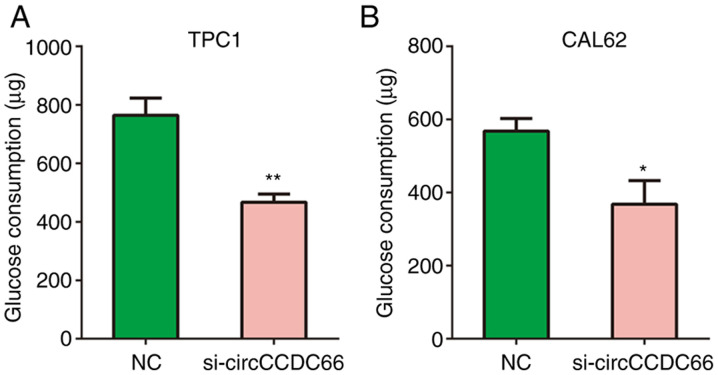
circCCDC66 knockdown inhibits TC glycolysis in vitro
Glycolysis serves a key role in cancer progression (23). Therefore, the effects of circCCDC66 knockdown on glycolytic metabolism in TPC1 and CAL62 cells were analysed in the present study. The results from Fig. 6A and B demonstrated that circCCDC66 knockdown significantly reduced the glucose consumption of TPC1 and CAL62 cells compared with control cells.
Figure 6.
Knockdown of circCCDC66 inhibited thyroid cancer cell glycolysis in vitro. (A and B) Compared with the negative control group, circCCDC66 knockdown significantly decreased the glucose consumption in (A) TCP1 and (B) CAL62 cells. *P<0.05 and **P<0.01. NC, negative control; si, silencing.
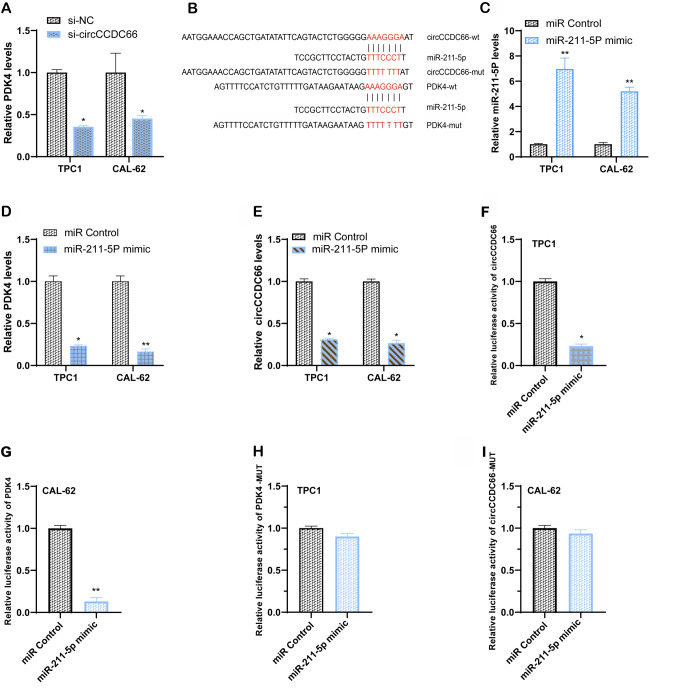
circCCDC66 serves as a sponge of miR-211-5p to promote PDK4 expression
To explore the mechanism by which circCCDC66 may serve important roles in TC cells, the mechanism by which circCCDC66 may regulate PDK4 expression was examined. The expression level of PDK4 was decreased following circCCDC66 silencing in TPC1 and CAL62 cells (Fig. 7A). circCCDC66 might act as a competing endogenous RNA and a sponge of miRNA and subsequently affect the expression of target genes. The results demonstrated that miR-211-5p and PDK4 were predicted as potential downstream targets associated with circCCDC66 function with RegRNA2.0 (http://regrna2.mbc.nctu.edu.tw/index.html) and miRcode (http://www.mircode.org/mircode/; Fig. 7B). Subsequently, the expression of PDK4 following miR-211-5p overexpression was determined. The results demonstrated that TC cells successfully transfected with miR-211-5p mimic (Fig. 7C) presented a lower PDK4 expression compared with control cells (Fig. 7D). Furthermore, the expression of circCCDC66 in both TC cell lines was inhibited following transfection with miR-211-5p-mimic (Fig. 7E).
Figure 7.
Expression of PDK4 was regulated by circCCDC66 via sponging miR-211-5p in thyroid cancer cells. (A) Compared with the control group, PDK4 expression level in circCCDC66 knockdown cells was decreased. (B) Schematic diagram of the wild-type or mutant-type binding sequence predicted on the online website. (C) Expression level of miR-211-5p was significantly upregulated in CAL62 and TPC1 cells following transfection with miR-211-5p-mimic. (D) Compared with the control group, PDK4 expression level was lower in cells overexpressing miR-211-5p. (E) Expression of circCCDC66 in TPC1 and CAL62 cells following transfection with miR-211-5p-mimic was decreased compared with control group. (F and G) miR-211-5p-mimic reduced the activity of circCCDC66 and PDK4 plasmid-transfected TPC1 and CAL62 cells according to results from luciferase reporter assay. (H and I) miR-211-5p-mimic had little effect on the activity of circCCDC66-MUT and PDK4-MUT plasmid-transfected TPC1 and CAL62 cells according to results from luciferase reporter assay. *P<0.05 and **P<0.01. MUT, mutant; miR, microRNA; PDK4, pyruvate dehydrogenase kinase 4.
In addition, to investigate the binding between miR-211-5p and circCCDC66 or the 3′-UTR of PDK4, a luciferase reporter assay was performed. The results demonstrated that a miR-211-5p-mimic could decrease the luciferase activity when wild-type circCCDC66 and wild-type 3′-UTR of PDK4 were expressed, which was not the case with mutant circCCDC66 and mutant 3′-UTR of PDK4 (Fig. 7F-I). These findings suggested that circCCDC66 may act as a sponge for miR-211-5p and promote PDK4 expression.
Discussion
High levels of metastasis and invasion are key characteristics of cancer cells (24). The present study revealed that circCCDC66 served a crucial stimulating role in TC cell proliferation and migratory and invasive abilities. The results demonstrated that circCCDC66 was expressed at higher levels in TC tissues compared with normal tissues and was also highly expressed in TC cells compared with normal cells. In addition, circCCDC66 knockdown inhibited the proliferation, glycolysis and migratory and invasive abilities of TC cells. Furthermore, circCCDC66 silencing decreased the expression of PDK4 by acting as a ceRNA and sponging miR-211-5p.
CircRNAs have been used as biomarkers for tumour detection due to their differential expression in tumour tissues and normal tissues (25). At the same time, the importance of numerous circRNAs in TC cells has also been reported. For example, Cui et al (26) performed a microarray analysis of circRNAs in TC and reported that 98 different circRNAs were abnormally regulated. The circRNA circZFR promotes the expression of C8orf4 by sponging miR-1261 and further promotes the proliferation and invasion of TC cells. The circRNA circ-ITCH inhibits cancer progression via the miR-22-3p/CBL/beta-catenin axis. However, the underlying mechanism of circCCDC66 in TC remains unclear.
The present study evaluated the role and underlying mechanism of circCCDC66 on the biological behaviour of TC cells. The results demonstrated that circCCDC66 was highly expressed in TC tissues and cells compared with normal thyroid tissues and cells, suggesting that circCCDC66 may promote the progression of TC. The results also demonstrated that the proliferation and migratory and invasive abilities of TC cells was significantly inhibited following circCCDC66 silencing. In addition, circCCDC66 was reported to bind to miR-211-5p and inhibit its expression. Subsequently, the overexpression of miR-211-5p in TC cells significant decreased the expression of PDK4 and circCCDC66. A luciferase assay demonstrated that miR-211-5p decreased the activity of pmiR-RB-reporter vectors containing PDK4 or circCCDC66 regions. These finding indicated that circCCDC66 may competitively inhibit the binding of miR-211-5p to PDK4, thereby affecting TC cell proliferation and invasiveness.
miR-211-5p has been found to be associated with metabolic activity in a variety of cancer cells. For example, the progression of TNBC is inhibited by miR-211-5p via targeting SET binding protein 1, suggesting that miR-211-5p might act as a potential prognostic biomarker and therapeutic target for TNBC (27). Furthermore, a previous study reported that the expression of snail family transcriptional repressor 1 (SNAI1) is decreased following miR-211-5p overexpression. Luciferase reporter assays showed that miR-211-5p can target the 3′-UTR of SNAI1. In addition, miR-211-5p reduces the weight of xenograft tumours and inhibits tumour metastasis in mice. These results indicate that the decrease in SNAIL1 expression mediated by miR-211-5p could help to inhibit the progression of renal cell carcinoma (28). Zinc finger E-box binding homeobox 2 (ZEB2) has also been reported to be a target of miR-211-5p and miR-211-5p serves an inhibitory role in hepatocellular carcinoma by inhibiting the expression of ZEB2 (29).
Pyruvate dehydrogenase kinase and phosphatase strictly regulate the pyruvate dehydrogenase complex through phosphorylation, thus regulating the irreversible reaction of pyruvate oxidative decarboxylation and affecting the glucose metabolism process and sugar homeostasis (30). Impaired glucose homeostasis is one of the risk factors for metabolic diseases, including obesity, type 2 diabetes and cancer (31). Previous studies reported that PDK4 is highly expressed in breast tumour tissues, and that higher PDK4 expression is associated with a worse prognosis in patients. A study reported that miR-211 can suppress PDK4 expression in breast cancer cells, while lower miR-211 expression levels are correlated with a worse overall survival, indicating that miR-211 and PDK4 expression levels are negatively correlated (32). In addition, the suppression of PDK4 by miR-211 can induce a dominant phenotype of oxidative phosphorylation, including decreased glucose and increased expression of key enzymes from pyruvate dehydrogenase and tricarboxylic acid cycle (33). These studies suggested that the effect of circCCDC66 on glycolysis might be mediated by PDK4 in TC cells.
The present study had several limitations. Although glucose consumption is a marker of glycolysis in cancer cells, it is not sufficient for evaluating glycolysis. Other experiment, including lactate production and oxygen consumption, should be performed. In future research, the signal pathways and metabolic pathways of circCCDC66 regulating glycolysis will be investigated. In addition, future investigation will further verify that PDK4 and miR-211-5p may affect the proliferation and migratory and invasive abilities of TC cells.
In conclusion, the present study demonstrated that circCCDC66 may promote TC cell proliferation, migratory and invasive abilities, and glycolysis in vitro by sponging miR-211-5p and increasing PDK4 expression. These findings suggested that targeting circCCDC66 may be considered as a promising therapeutic strategy for TC.
Acknowledgements
Not applicable.
Glossary
Abbreviations
- circRNA
circular RNA
- ceRNA
competitive endogenous RNA
- TC
thyroid cancer
- TNBC
triple-negative breast cancer
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
HR, ZS, CC and WM participated in the concept and design of the study. ZS and CC participated in data acquisition, analysis and interpretation. HR and WM participated in methodology design. ZS participated in sample collection. HR, ZS and WM participated in writing and revising the manuscript. All authors read and approved the final version.
Ethics approval and consent to participate
This study was approved by the Ethics Committee of the Qingdao Municipal Hospital (approval no. 2019QMH126) and all patients provided informed consent to participate to this study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Zhang JW, Long KR, Wang X, LI MZ, Ma JD. The research advance of circular RNA. Chin J Anim Veterinary Sci, 2016;11:2151–2158. (In Chinese) [Google Scholar]
- 2.Cai H, Li Y, Niringiyumukiza JD, Su P, Xiang W. Circular RNA involvement in aging: An emerging player with great potential. Mech Ageing Dev. 2019;178:16–24. doi: 10.1016/j.mad.2018.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Lu Q, Liu T, Feng H, Yang R, Zhao X, Chen W, Jiang B, Qin H, Guo X, Liu M, et al. Circular RNA circSLC8A1 acts as a sponge of miR-130b/miR-494 in suppressing bladder cancer progression via regulating PTEN. Mol Cancer. 2019;18:111. doi: 10.1186/s12943-019-1040-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Xu JZ, Shao CC, Wang XJ, Zhao X, Chen JQ, Ouyang YX, Feng J, Zhang F, Huang WH, Ying Q, et al. circTADA2As suppress breast cancer progression and metastasis via targeting miR-203a-3p/SOCS3 axis. Cell Death Dis. 2019;10:175. doi: 10.1038/s41419-019-1382-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lu J, Wang YH, Yoon C, Huang XY, Xu Y, Xie JW, Wang JB, Lin JX, Chen QY, Cao LL, et al. Circular RNA circ-RanGAP1 regulates VEGFA expression by targeting miR-877-3p to facilitate gastric cancer invasion and metastasis. Cancer Lett. 2020;471:38–48. doi: 10.1016/j.canlet.2019.11.038. [DOI] [PubMed] [Google Scholar]
- 6.Gaffo E, Boldrin E, Dal Molin A, Bresolin S, Bonizzato A, Trentin L, Frasson C, Debatin KM, Meyer LH, Te Kronnie G, Bortoluzzi S. Circular RNA differential expression in blood cell populations and exploration of circRNA deregulation in pediatric acute lymphoblastic leukemia. Sci Rep. 2019;9:14670. doi: 10.1038/s41598-019-50864-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Xiong X, Zhu H, Chen X. Low expression of long noncoding RNA CASC2 indicates a poor prognosis and promotes tumorigenesis in thyroid carcinoma. Biomed Pharmacother. 2017;93:391–397. doi: 10.1016/j.biopha.2017.06.063. [DOI] [PubMed] [Google Scholar]
- 8.Li S, Li Z, Guo F, Qin X, Liu B, Lei Z, Song Z, Sun L, Zhang HT, You J, Zhou Q. MiR-223 regulates migration and invasion by targeting Artemin in human esophageal carcinoma. J Biomed Sci. 2011;18:24. doi: 10.1186/1423-0127-18-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Du WW, Yang W, Liu E, Yang Z, Dhaliwal P, Yang BB. Foxo3 circular RNA retards cell cycle progression via forming ternary complexes with p21 and CDK2. Nucleic Acids Res. 2016;44:2846–2858. doi: 10.1093/nar/gkw027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Durante C, Grani G, Lamartina L, Filetti S, Mandel SJ, Cooper DS. The diagnosis and management of Thyroid Nodules: A review. JAMA. 2018;319:914–924. doi: 10.1001/jama.2018.0898. [DOI] [PubMed] [Google Scholar]
- 11.Cong S, Fang LW, Bao HL, Feng YJ, Wang N, Yin P, Li YC, Duan XN, Zhou MG. (Disease burden of thyroid cancer in the Chinese population, in 1990 and 2013) Zhonghua Liu Xing Bing Xue Za Zhi. 2016;37:773–777. doi: 10.3760/cma.j.issn.0254-6450.2016.06.006. [DOI] [PubMed] [Google Scholar]
- 12.Schmidt B, Davies L. Management of Differentiated Thyroid Cancer. Springer; Cham: 2017. The rising incidence of thyroid cancer: Contributions from healthcare practice and biologic risk factors; pp. 1–13. [DOI] [Google Scholar]
- 13.Murugan AK, Dong J, Xie J, Xing M. MEK1 mutations, but not ERK2 mutations, occur in melanomas and colon carcinomas, but none in thyroid carcinomas. Cell Cycle. 2009;8:2122–2124. doi: 10.4161/cc.8.13.8710. [DOI] [PubMed] [Google Scholar]
- 14.Gao Y, Wang J, Zheng Y, Zhang J, Chen S, Zhao F. Comprehensive identification of internal structure and alternative splicing events in circular RNAs. Nat Commun. 2016;7:12060. doi: 10.1038/ncomms12060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wei H, Pan L, Tao D, Li R. Circular RNA circZFR contributes to papillary thyroid cancer cell proliferation and invasion by sponging miR-1261 and facilitating C8orf4 expression. Biochem Biophys Res Commun. 2018;503:56–61. doi: 10.1016/j.bbrc.2018.05.174. [DOI] [PubMed] [Google Scholar]
- 16.Hsiao KY, Lin YC, Gupta SK, Chang N, Yen L, Sun HS, Tsai SJ. Noncoding effects of circular RNA CCDC66 promote colon cancer growth and metastasis. Cancer Res. 2017;77:2339–2350. doi: 10.1158/0008-5472.CAN-16-1883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hong W, Yu S, Zhuang Y, Zhang Q, Wang J, Gao X. SRCIN1 regulated by circCCDC66/miR-211 is upregulated and promotes cell proliferation in non-small-cell lung cancer. Biomed Res Int. 2020;2020:5307641. doi: 10.1155/2020/5307641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Xiang D, Li Y, Lin Y. Circular RNA circCCDC66 contributes to malignant phenotype of osteosarcoma by Sponging miR-338-3p to upregulate the expression of PTP1B. Biomed Res Int. 2020;2020:4637109. doi: 10.1155/2020/4637109. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 19.Wang M, Chen B, Ru Z, Cong L. CircRNA circ-ITCH suppresses papillary thyroid cancer progression through miR-22-3p/CBL/β-catenin pathway. Biochem Biophys Res Commun. 2018;504:283–288. doi: 10.1016/j.bbrc.2018.08.175. [DOI] [PubMed] [Google Scholar]
- 20.Xu H, Liu Y, Cheng P, Wang C, Liu Y, Zhou W, Xu Y, Ji G. CircRNA_0000392 promotes colorectal cancer progression through the miR-193a-5p/PIK3R3/AKT axis. J Exp Clin Cancer Res. 2020;39:283. doi: 10.1186/s13046-020-01799-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schmittgen TD, Livak KJ. Analyzing real-time PCR data by the comparative C(T) method. Nat Protoc. 2008;3:1101–1108. doi: 10.1038/nprot.2008.73. [DOI] [PubMed] [Google Scholar]
- 22.Nahm JH, Kim HM, Koo JS. Glycolysis-related protein expression in thyroid cancer. Tumour Biol. 2017;39:1010428317695922. doi: 10.1177/1010428317695922. [DOI] [PubMed] [Google Scholar]
- 23.Wang Y, Zhou X, Shan B, Han J, Wang F, Fan X, Lv Y, Chang L, Liu W. Downregulation of microRNA-33a promotes cyclin-dependent kinase 6, cyclin D1 and PIM1 expression and gastric cancer cell proliferation. Mol Med Rep. 2015;12:6491–6500. doi: 10.3892/mmr.2015.4296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim Y, Williams KC, Gavin CT, Jardine E, Chambers AF, Leong HS. Quantification of cancer cell extravasation in vivo. Nat Protoc. 2016;11:937–948. doi: 10.1038/nprot.2016.050. [DOI] [PubMed] [Google Scholar]
- 25.Wang KW, Dong M. Role of circular RNAs in gastric cancer: Recent advances and prospects. World J Gastrointest Oncol. 2019;11:459–469. doi: 10.4251/wjgo.v11.i6.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cui S, Qian Z, Chen Y, Li L, Li P, Ding H. Screening of up- and downregulation of circRNAs in HBV-related hepatocellular carcinoma by microarray. Oncol Lett. 2018;15:423–432. doi: 10.3892/ol.2017.7265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen LL, Zhang ZJ, Yi ZB, Li JJ. MicroRNA-211-5p suppresses tumour cell proliferation, invasion, migration and metastasis in triple-negative breast cancer by directly targeting SETBP1. Br J Cancer. 2017;117:78–88. doi: 10.1038/bjc.2017.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang K, Jin W, Jin P, Fei X, Wang X, Chen X. MiR-211-5p suppresses metastatic behavior by targeting SNAI1 in Renal cancer. Mol Cancer Res. 2017;15:448–456. doi: 10.1158/1541-7786.MCR-16-0288. [DOI] [PubMed] [Google Scholar]
- 29.Jiang G, Wen L, Deng W, Jian Z, Zheng H. Regulatory role of miR-211-5p in hepatocellular carcinoma metastasis by targeting ZEB2. Biomed Pharmacother. 2017;90:806–812. doi: 10.1016/j.biopha.2017.03.081. [DOI] [PubMed] [Google Scholar]
- 30.Shan C, Kang HB, Elf S, Xie J, Gu TL, Aguiar M, Lonning S, Hitosugi T, Chung TW, Arellano M, et al. Tyr-94 phosphorylation inhibits pyruvate dehydrogenase phosphatase 1 and promotes tumor growth. J Biol Chem. 2014;289:21413–21422. doi: 10.1074/jbc.M114.581124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sui W, Shi Z, Xue W, Ou M, Zhu Y, Chen J, Lin H, Liu F, Dai Y. Circular RNA and gene expression profiles in gastric cancer based on microarray chip technology. Oncol Rep. 2017;37:1804–1814. doi: 10.3892/or.2017.5415. [DOI] [PubMed] [Google Scholar]
- 32.Sun S, Zhao M, Han Y, Juanzi W, Peng W, Liu J. PDK4 mRNA Expression in Breast Cancer and Its Relationship with Prognosis. Cancer Res Prev Treat. 2018;45:73–76. [Google Scholar]
- 33.Mazar J, Richardson A, Qi F, Lee B, Duran A, Govindarajan S, Shelley J, Brill LM, Li JL, Han X, Moscat J, Perera RJ. MicroRNA-211 modulates energy metabolism in human melanoma cells by destabilizing HIF1-α and downregulating PDK4. AACR. 2014;74:S978. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Availability Statement
The datasets used and/or analysed during the current study are available from the corresponding author on reasonable request.