Abstract
Seed germination and subsequent seedling establishment are important developmental processes that undergo extremely complex changes of physiological status and are precisely regulated at transcriptional and translational levels. Phytohormones including abscisic acid (ABA) and gibberellin (GA) are the critical signaling molecules that modulate the alteration from relative quiescent to a highly active state in seeds. Transcription factors such as ABA insensitive5 (ABI5) and DELLA domain-containing proteins play the central roles in response to ABA and GA, respectively, which antagonize each other during seed germination. Recent investigations have demonstrated that the regulations at translational and post-translational levels, especially post-translational modifications (PTMs), play a decisive role in seed germination. Specifically, phosphorylation and ubiquitination were shown to be involved in regulating the function of ABI5. In this review, we summarized the latest advancement on the function of PTMs involved in the regulation of seed germination, in which the PTMs for ABI5- and DELLA-containing proteins play the key roles. Meanwhile, the studies on PTM-based proteomics during seed germination and the crosstalk of different PTMs are also discussed. Hopefully, it will facilitate in obtaining a comprehensive understanding of the physiological functions of different PTMs in seed germination.
Keywords: post-translational modification, seed germination, phosphorylation, ubiquitylation, ABI5
Introduction
Seed germination is an indispensable event for initiating seedling establishment and plant growth for next generation, which presents as an intricate physiological process precisely regulated by endogenous and environmental cues (Mazer, 1999; Donohue et al., 2005; Finch-Savage and Leubner-Metzger, 2006; Holdsworth et al., 2008). Numerous genes affecting seed germination have been cloned, mainly including DOG1 (Nakabayashi et al., 2012; Graeber et al., 2014; Leubner-Metzger, 2014), CYP707A1/2 (Millar et al., 2006; Matakiadis et al., 2009; Shu et al., 2013), GA2oxs (Yamauchi et al., 2007), MYB96 (Lee H. G. et al., 2015; Lee K. et al., 2015), OsAP2-39 (Yaish et al., 2010), CHO1 (Yamagishi et al., 2009; Yano et al., 2009), WRKY41 (Ding et al., 2014), and ARF10/ARF16 (Liu et al., 2013). Almost all of these genes are involved in either abscisic acid (ABA) or gibberellic acid (GA) signaling, of which these two phytohormones play critical roles in seed germination through controlling the shift from seed dormancy to germination (Kucera et al., 2005; Finkelstein et al., 2008; Shu et al., 2016). The ABA molecule is recognized by its receptors PYRABACTIN RESISTANCE (PYR)/REGULATORY COMPONENT OF ABSCISIC ACID RECEPTOR (RCAR) (Ma et al., 2009; Miyazono et al., 2009; Nishimura et al., 2009; Santiago et al., 2009), and the ABA-bound receptors tightly combine with type 2C protein phosphatases (PP2Cs), resulting in the dissociation of SNF1-related kinases 2 (SnRK2s) from PP2C-SnRK2 complexes (Cutler et al., 2010). The released SnRK2s directly phosphorylate targeted transcription factors such as ABSCISIC ACID-INSENSITIVE5 (ABI5), ABI4, and ABI3 to mediate ABA responses (Kobayashi et al., 2005; Furihata et al., 2006; Fujii et al., 2007; Fujii and Zhu, 2009; Zhu et al., 2020). The mutant of ABA signaling related genes, such as biosynthetic genes aba deficient 1 (aba1) and nine-cis-epoxycarotenoid dioxygenase 6 (nced6) (Koornneef et al., 1982; Lefebvre et al., 2006), catabolic gene cyp707a2 (Matakiadis et al., 2009) and signal transduction genes abi3, abi4, and abi5 (Penfield et al., 2006; Park et al., 2011; Lim et al., 2013) in Arabidopsis had demonstrated that ABA could directly affect seed germination. GA, antagonizing with ABA, promotes seed germination (Holdsworth et al., 2008), which could be supported by the fact that GA-deficient mutants such as ga1 and ga2 fail to germinate (Lee et al., 2002; Shu et al., 2013). The balance between ABA and GA signal during seed germination is regulated by functional proteins such as GERMIN-LIKE PROTEIN 2-1 (OsGLP2-1), which binds to the promoters of ABI5 and GAMYB (Wang et al., 2020) and INDUCER OF CBF EXPRESSION1 (ICE1) to antagonize ABI5 and DELLA activity (Hu et al., 2019).
Besides ABA and GA, other phytohormones, such as jasmonate (JA) (Pan et al., 2020), ethylene (Jurdak et al., 2020), cytokinin (Wang et al., 2011), auxin (Liu et al., 2013), and brassinosteroids (BRs) (Hu and Yu, 2014), also regulate the process of seed germination in Arabidopsis. JA regulates seed germination through ABA signaling. JA ZIM-DOMAIN (JAZ) proteins could inhibit the expression of ABI3 and ABI5 (Pan et al., 2020), whereas, JAZ repressors could physically interact with ABI3 and activate ABA signaling. The effect of ethylene on seed germination depends on the reactive oxygen specifies (ROS) molecules produced by the mitochondrial electron transport chain through up-regulating AOX1a and ANAC013 in mitochondrial retrograde response complex (Jurdak et al., 2020). The mutant of cytokinin biosynthesis exhibited ABA insensitive phenotype during germination, and the cytokinin signal transducers and transcription repressors, type-A ARR4, ARR5, and ARR6, could physically interact with ABI5 to negatively regulate ABI5 expression (Wang et al., 2011). The mutation of auxin signaling or biosynthesis in Arabidopsis dramatically released seed dormancy, which recruits auxin response factors (ARF) 10 and 16 to control the expression of ABI3 during seed germination (Liu et al., 2013).
The synthesis, modification, localization, and degradation of proteins in the cells are critical for plants to survive from adverse environments, in which post-translational modifications (PTMs) of proteins increase the diversity of gene products and influence nearly every cellular process (Fulzele and Bennett, 2018). The prevalent PTMs mainly include phosphorylation (Mann et al., 2002; Ptacek et al., 2005; Thalassinos et al., 2008), ubiquitylation (Bennett et al., 2010; Xu et al., 2010; Kim et al., 2011), acetylation (Choudhary et al., 2009), glycosylation (Dell and Morris, 2001; Zhang et al., 2003), nitrosylation (Jaffrey et al., 2001), methylation (Clarke, 1993), and lipidation (Ichimura et al., 2000). With the development of analytical techniques, PTMs could be precisely detected at the global level or within a specific protein. For example, PHYTOCHROME INTERACTING FACTOR 3 (PIF3) is rapidly phosphorylated and degraded as a result of interaction between phytochrome B (phyB) and photo-activated PIF3, of which this process is needed for proper photomorphogenesis (Ni et al., 2013). Besides, the SUMOylated PIF3 in Lys13 residue also regulates the phyB abundant to affect plants’ photomorphogenesis (Bernula et al., 2020). PIF3 could be phosphorylated by multiple kinases such as GSK3-like kinase BRASSINOSTEROID-INSENSITIVE 2 (BIN2) and photo-regulatory protein kinases (PPKs), which is required for further ubiquitination of the proteins (Li et al., 2017; Xu et al., 2017).
Seed germination is an important physiological alteration from quiet dormant status to active seedling establishment, in which a large number of processes are reprogrammed. Han and Yang (2015) have summarized the progress in the molecular mechanisms of seed germination among different species, including morphological changes, cellular and its related structure recovery, metabolic variations, and transcription activation. However, more and more evidences have demonstrated that PTMs also play critical roles in regulating seed germination. In this paper, we reviewed the recent investigations on PTMs involved in seed germination (Figure 1), which will help to understand the molecular mechanism of seed germination.
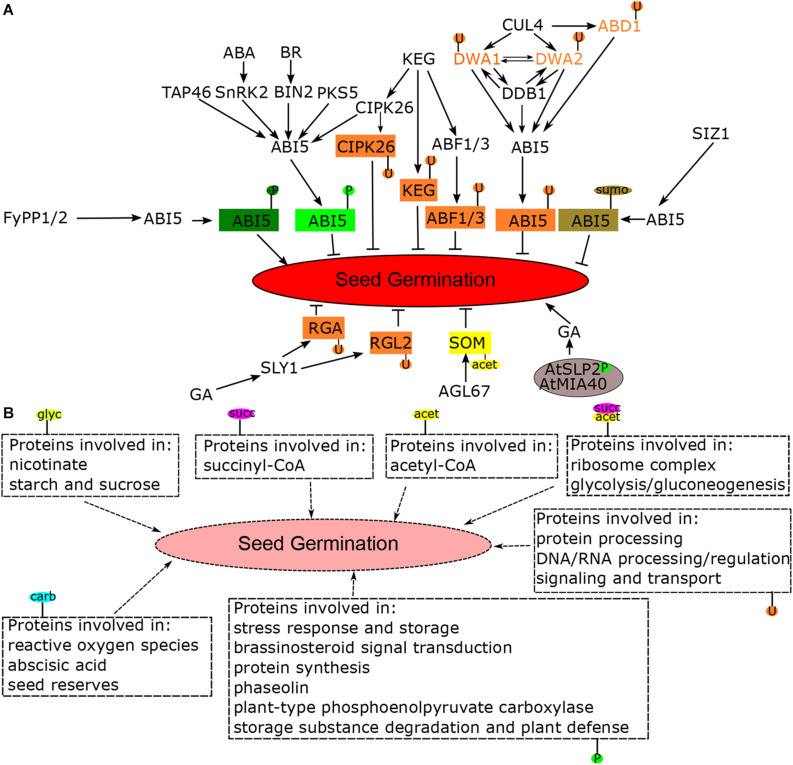
FIGURE 1.
The diverse post-translation modifications (PTMs) involved in seed germination. (A) The key genes regulated by PTMs involved in seed germination. (B) The PTMs identified by proteomics techniques involved in seed germination. The phytohormones GA and ABA function antagonistically with each other in regulating seed germination, of which the signal transduction is largely coordinated by PTMs. ABI5, a key transcription factor in response to ABA, suppresses the seed germination, and it is phosphorylated by different kinases such as SnRK2, BIN2, CIPK26, PKS5, and KEG. KEG also ubiquitinates ABI5, CIPK26, and ABF1/3 to directly inhibit seed germination. DWA1, DWA2, and DDB1 interact with each other forming a complex that elevates the level of ABI5, in which DWA1 and DWA2 are ubiquitinated by CUL4. CUL4 also ubiquitinated ABD1 targeted for ABI5 degradation through 26S proteasome. The phosphatase FyPP1/2 and SIZ1 dephosphorylated and sumoylated ABI5, respectively. GA facilitated the expression of SLY1, which ubiquitinated two DELLA containing proteins RGA and RGL2 to derepress for germination. MADS-box transcription factor AGL67 acetylated the promoter of zinc-finger protein SOM to promote its expression that inhibited seed germination, and AtMIA40 formed the complex with AtSLP2 to coordinate its phosphatase activity, which negatively regulated the GA-related process under seed germination. Furthermore, the PTMs including phosphorylation, ubiquitination, carbonylation, glycosylation, acetylation, and succinylation were also involved in multiple metabolism processes such as protein processing, ribosome complex, brassinosteroid signal transduction, and reactive oxygen species. GA, gibberellin; ABA, abscisic acid; ABI5, ABA insensitive 5; SnRK2s, SNF1-related kinases 2; BIN2, BRASSINOSTEROID-INSENSITIVE 2; CIPK26, Calcineurin B-like Interacting Protein Kinase 26; PKS5, SOS2-like protein kinase 5; KEG, KEEP ON GOING; ABF1/3: ABRE binding factor 1/3; DWA1/2, DWD hypersensitive to ABA1; CUL4, CULLIN4; DDB1, damaged DNA binding1; ABD1, ABA-hypersensitive DCAF1; SIZ1, SUMO E3 ligases; SLY1, F-box-containing proteins SLEEPY1; RGA, REPRESSOR OF ga1-3; RGL2, RGA-like 2; SOM, SOMNUS; AGL67, AGAMOUS-LIKE67; AtSLP2, Arabidopsis Shewanella-like protein phosphatase 2; AtMIA40, Arabidopsis mitochondrial oxidoreductase import and assembly protein 40; P, phosphorylation; U, ubiquitination; carb, carbonylation; glyc, glycosylation; acet, acetylation; succ, succinylation; sumo, sumoylation.
Phosphorylation Regulation of Seed Germination
Protein phosphorylation and dephosphorylation is an important switch for the activity of proteins, and more than 75% of eukaryotic proteins are potentially phosphorylated, which control almost all the biological processes (Sharma et al., 2014). ABI5 plays central roles in the ABA signaling pathway, and its phosphorylation regulation among ABA signaling transduction is well characterized, which directly influence the seed germination. In the presence of ABA, the protein kinase SnRK2 could phosphorylate ABI5 and promote its stability, through which seed germination was inhibited (Kobayashi et al., 2005; Fujii et al., 2007; Nakashima et al., 2009). The BR receptor BIN2 enhances the downstream signaling of ABA through phosphorylating ABI5 to further regulate the seed germination (Hu and Yu, 2014). SOS2-like protein kinase 5 (PKS5) also phosphorylates ABI5 at Ser-42 to regulate seed germination in Arabidopsis (Zhou et al., 2015). Calcineurin B-like Interacting Protein Kinase (CIPK) 26 not only interacts with ABI1, ABI2, and ABI5 in Arabidopsis, but also phosphorylates ABI5 in vitro, through which CIPK26 enhances the sensitivity of seed germination (Lyzenga et al., 2013). The Arabidopsis RING-ANK family protein, KEEP ON GOING (KEG), contains a kinase domain with phosphorylation activity, and negatively regulates ABA signaling through interacting with ABI5 (Stone et al., 2006). The mutation of two PROTEIN PHOSPHATASE6 (PP6) genes, FyPP1 and FyPP2, lead to the ABA hypersensitive phenotypes at seed germination and seedling growth in Arabidopsis, which directly interact with and dephosphorylate ABI5, acting antagonistically with SnRK2 kinases (Dai et al., 2013). Moreover, a protein phosphatase 2A-associated protein TAP46 could enhance the stability of ABI5 through binding to and phosphorylating ABI5 (Hu et al., 2014). These investigations collectively indicated that the phosphorylation and dephosphorylation of ABI5 is precisely regulated in the ABA-mediated signaling pathway, which further controls seed germination and seedling establishment. The recent investigation also showed that Arabidopsis Shewanella-like protein phosphatase 2 (AtSLP2) interacts with the mitochondrial oxidoreductase import and assembly protein 40 (AtMIA40) in the mitochondrial intermembrane space, which is required for the phosphatase activity of AtSLP2, and the complex of AtSLP2 and AtMIA40 negatively regulates the GA-related process during seed germination (Uhrig et al., 2017).
Besides the functional elucidation of phosphorylation in specific proteins, phosphoproteomics is a well way to globally identify the phosphorylated proteins in the tissues, through which numerous phosphorylation modified proteins involved in seed germination have been discovered. Systematical analysis of phosphorylated proteins in the embryo of germinating seeds at the early stage demonstrated that phosphorylation of proteins related to stress response and storage was gradually enhanced, and proteins involved in BR signal transduction were also phosphorylated while brassinolide treatment enhanced the ability of seed germination in rice, implying seed germination is possibly triggered by BR signal (Han et al.,2014a,b). A gel-free/label-free phosphoproteomics were conducted to detect phosphorylated nuclear proteins at the early stage of rice seed germination (Li M. et al., 2015). The results demonstrated that proteins related to protein synthesis were mainly phosphorylated with 29 proteins displaying significant changes in phosphorylation level over the period of imbibition. Phosphorylation analysis of phaseolin in the dormant and 4-day germinating bean seed of two different cultivars found that the phosphorylation levels of phaseolin were remarkably changed from the dormancy status to early germination stage (López-Pedrouso et al., 2014). The phosphorylation of multiple plant-type phosphoenolpyruvate carboxylase (PTPC) isoenzymes at their conserved N-terminal seryl site was also identified in sorghum during seed germination (Ruiz-Ballesta et al., 2016). Furthermore, the proteins involved in storage substance degradation and plant defense were also detected to be phosphorylated in wheat (Triticum aestivum L.) during seed germination (Dong et al., 2015), suggesting that protein phosphorylation participates in diverse metabolism processes in seed germination.
Ubiquitination Regulation of Seed Germination
Ubiquitination is another most prevalent PTMs in plants, which widely involves in various pivotal processes including protein turnover, genomic integrity, signaling processing among others (He et al., 2020a). GA is a key phytohormone that promotes seed germination with DELLA proteins being the most important repressors of GA signaling (Dill et al., 2001; Lee et al., 2002; Tyler et al., 2004; Davière and Achard, 2016). The GA receptor GID1 has a higher affinity for GA4, which also interacts with DELLA proteins (Nakajima et al., 2006). In the presence of GA, the F-box-containing protein SLEEPY1 (SLY1) directly interacts with the GA receptor and DELLA protein REPRESSOR OF ga1-3 (RGA) through their C-terminal GRAS domain, which mediates the ubiquitination and the subsequent degradation of RGA to promote the GA signaling pathway (Dill et al., 2001, 2004; Ueguchi-Tanaka et al., 2005). The RGA-like 2 (RGL2) is expressed during the imbibition period and plays a critical role in inhibiting seed germination, which is also degraded through F-box protein SLY1 mediated ubiquitination (Lee et al., 2002; Tyler et al., 2004; Piskurewicz et al., 2008). Other DELLA proteins, including GA insensitive (GAI), RGA and RGL1, enhance the function of RGL2, and the far-red light repressed the seed germination through stabilizing GAI, RGA, and RGL2 (Cao et al., 2005; Piskurewicz et al., 2009). These investigations illustrated that ubiquitination of DELLA proteins is the key step in derepressing the inhibition of GA-signaling involved in seed germination in Arabidopsis.
The level of ABI5 is also regulated by ubiquitination during seed germination. The RING-type E3 ligase KEG is needed to maintain the low level of ABI5, of which ABI5 is a substrate of KEG for ubiquitination (Liu and Stone, 2013). KEG is also self-ubiquitinated and degraded through the 26S proteasome system to increase the ABI5 level in response to ABA (Liu and Stone, 2010), and CIPK26 is also ubiquitinated by KEG and degraded through 26S proteasome (Lyzenga et al., 2013). Two ABI5-related transcription factor, ABRE binding factor 1 (ABF1) and ABF3, also regulate seed germination, and the detailed investigations demonstrated that KEG could directly interact with and ubiquitinate ABF1 and ABF3 to regulated their protein abundance (Finkelstein et al., 2005; Chen et al., 2013). The mutant of two Arabidopsis DWD proteins DWA1 (DWD hypersensitive to ABA1) and DWA2 that are substrate receptors of CULLIN4 (CUL4) E3 ubiquitin ligases displayed delayed germination, which were involved in ABA signal transduction (Lee et al., 2010). DWA1, DWA2 and Damaged DNA Binding1 (DDB1) can directly interact with each other to form CUL4-based complexes that target and mediate the ubiquitination of ABI5. The members of DDB1-CUL4–associated factors (DCAFs) family that bind to DDB1 are also the substrate receptors of CUL4. ABA-hypersensitive DCAF1 (ABD1) belongs to DCAF1 family that interacts with DDB1, and the loss of ABD1 could result in ABA-hypersensitivity phenotypes during germination and seedling growth (Seo et al., 2014). ABD1 directly interacts with ABI5 and the degradation of ABI5 by the 26S proteasome was also suppressed in the ABD1 mutant lines. Collectively, protein ubiquitination through E3 ubiquitin ligases is an important PTM regulating the ABI5 levels in ABA signaling during seed germination.
Furthermore, the high throughput ubiquitylome using K-ε-GG antibody enrichment integrated with mass spectrometry has been developed to identify a large amount of ubiquitinated proteins in tissues (Fulzele and Bennett, 2018). The PR-619-treated (deubiquitylase inhibitor) rice seed displayed a delayed germination, and analysis of ubiquitylated proteins at 0, 12, and 24 h after imbibition in rice embryos has demonstrated that 2,576 lysine sites in 1,171 proteins were ubiquitylated and the differentially ubiquitinated proteins were mainly involved in the categories of protein processing, DNA and RNA processing/regulation related, signaling and transport, indicating that ubiquitination is a regulator involved in the multifaceted process during seed germination (He et al., 2020b).
Other PTMS Regulation of Seed Germination
Phosphorylation and ubiquitination are the two prevalent PTMs that have been widely studied and shown to be involved in many physiological processes through regulating protein activity and levels. However, it has been shown that many PTMs other than these two, such as sumoylation, carbonylation, glycosylation, acetylation, and succinylation, are also involved in seed germination. Overexpression of small ubiquitin-related modifier 1/2 (AtSUMO1/2) exhibited increased levels of sumoylation and was less sensitive to ABA (Lois et al., 2003), while SUMO E3 ligases SIZ1 directly sumoylated ABI5 enhancing its stability in response to ABA in seed germination (Miura et al., 2009). MADS-box transcription factor AGAMOUS-LIKE67 (AGL67) recruits the histone mark reader EARLY BOLTING IN SHORT DAY (EBS) to form a complex that is necessary for histone H4K5 acetylation in the promoter of zinc-finger protein SOMNUS (SOM), which activates SOM expression, ultimately inhibiting seed germination under high-temperature stress (Li et al., 2020). The investigation of rice acetylated or succinylated embryonic proteins after 24 h imbibition using nano-LC-MS/MS has identified 699 acetylated sites from 389 proteins and 665 succinylated sites from 261 proteins, which covered nearly all aspects of cellular functions, with ribosome complex and glycolysis/gluconeogenesis related proteins being significantly enriched (He et al., 2016). The enzymes related to acetyl-CoA and succinyl-CoA metabolism were modified through acetylation or succinylation, respectively. The dynamic pattern of protein carbonylation in rice embryos during germination was also analyzed using sequential window acquisition of all theoretical fragment ion spectra (SWATH) method, and 288 carbonylated peptides corresponding to 144 proteins were identified, which mainly involved in maintaining the levels of reactive oxygen species, ABA and seed reserves (Zhang et al., 2016). The mutant of Arabidopsis UGT74F2, a glycosyltransferase, accumulates high levels of glycosylated nicotinate, and the mutant displayed decreased rates of seed germination, of which the germination drawback of the ugt74f2 mutant could be fully recovered by overexpressing UGT74F2 (Li W. et al., 2015). Moreover, N-glycosylation mapping of rice embryos during germination has discovered 242 glycosites from 191 unique proteins, and these N-glycosylated proteins were enriched in starch and sucrose metabolism pathway, which were predicted to interact with several BR signaling proteins, implying that N-glycosylation is involved in carbohydrate metabolism and BR signaling to regulate seed germination (Ying et al., 2017).
Crosstalk Among PTMS Involved in Seed Germination
The direct evidence of crosstalk regulation for PTMs involved in seed germination are from the modification of ABI5. As mentioned above, the protein levels of ABI5 could be precisely regulated through multiple PTMs. For example, protein kinase SnRK2, BIN2, PKS5 promote phosphorylation of ABI5 (Kobayashi et al., 2005; Fujii et al., 2007; Nakashima et al., 2009;, Hu and Yu 2014, Zhou et al., 2015), whereas protein phosphatase FyPP1 and FyPP2 lead to dephosphorylation of ABI5 (Dai et al., 2013). In addition, DWA1, DWA2, and ABD1 directly interact with and ubiquitinate ABI5 (Lee et al., 2010; Seo et al., 2014), and the SIZ1 sumoylates ABI5 during seed germination (Miura et al., 2009), KEG protein contains RING-HC and kinase domains, which function in ubiquitination and phosphorylation activity of ABI5 to control its protein level, respectively (Stone et al., 2006; Liu and Stone, 2010, 2013). KEG also ubiquitinated CIPK26 that phosphorylates ABI5. These results demonstrate the existence of crosstalk among phosphorylation, ubiquitination and sumoylation, with ABI5 being the node. Moreover, N-glycosylation and N-acetylation sites were predicted at the N terminus of ABI5 although experimental evidence is needed (Yu et al., 2015). Acetylation and succinylation analysis of germinated embryos in rice identified 133 common sites on 78 proteins modified by these two PTMs (He et al., 2016), implying the potential crosstalk between acetylation and succinylation during seed germination. Of the ubiquitylome in germinated rice embryo (He et al., 2020b), 88 proteins were also modified by phosphorylation (Han et al., 2014a) and 82 lysine residues in 49 proteins were also modified by acetylation (He et al., 2016), of which 12 proteins were modified by these three PTMs, indicating that co-modification occurred in these proteins. However, detailed investigations of specific proteins or sites are needed to clarify the molecular mechanism of how these PTMs co-regulated seed germination.
Concluding Remarks
A successful break of dormancy in seed to initiate germination is an irreplaceable process in the plant life cycle, and numerous efforts have been conducted to investigate the molecular mechanisms underlying the initiation of seed germination and seedling establishment at (post-)transcriptional and (post-)translational levels. This review presented here provides a relatively comprehensive summary of the PTMs involved in the regulation of seed germination, which mainly included phosphorylation, ubiquitination, sumoylation, carbonylation, glycosylation, acetylation, and succinylation. The PTMs on ABI5 provides a model that could be used to understand the stability and activity of specific proteins modified by different PTMs. However, numerous questions related to PTMs involved in seed germination still need to be investigated in the future. This is not only decided by the complexity of germinating processes but limited by the analytic techniques including the accuracy of identification and analysis for PTMs. Not only the protein functions are determined by the combination of multiple PTMs but also the crosstalk among PTM regulation of seed germination remains largely unknown. The summarization of PTMs presented here will promote the study of the molecular basis underlying seed germination especially for the processes regulated by PTMs. The future work will be mainly conducted to mine clues in regulation of seed germination as follows: (i) PTMs involved in the other hormones such as auxin and ethylene, except ABA and GA; (ii) whether phosphorylation or other PTMs are involved in GA signaling, because only ubiquitination was detected in DELLA proteins; (iii) the detailed mechanisms of PTMs such as sumoylation, carbonylation, glycosylation, acetylation, and succinylation identified through proteomics technique and how they regulate seed germination; and (iv) whether other PTMs are involved in seed germination since over 600 PTMs have been detected so far.
Author Contributions
FY and PY contributed to conceptualization and original draft preparation. PY, DH, and ML involved in review and editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors are grateful to Dr. Rebecca Njeri Damaris for her help in proofreading.
Footnotes
Funding. This work was supported by the National Natural Science Foundation of China (NSFC, No. 31271805) and the Young Science Foundation of Hubei University (Project 190804000005).
References
- Bennett E. J., Rush J., Gygi S. P., Harper J. W. (2010). Dynamics of cullin-RING ubiquitin ligase network revealed by systematic quantitative proteomics. Cell 143 951–965. 10.1016/j.cell.2010.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernula P., Pettkó-Szandtner A., Hajdu A., Kozma-Bognár L., Josse E. M., Ádám É, et al. (2020). SUMOylation of PHYTOCHROME INTERACTING FACTOR 3 promotes photomorphogenesis in Arabidopsis thaliana. New Phytol. 229 2050–2061. 10.1111/nph.17013 [DOI] [PubMed] [Google Scholar]
- Cao D., Hussain A., Cheng H., Peng J. (2005). Loss of function of four DELLA genes leads to light- and gibberellin-independent seed germination in Arabidopsis. Planta 223 105–113. 10.1007/s00425-005-0057-3 [DOI] [PubMed] [Google Scholar]
- Chen Y. T., Liu H., Stone S., Callis J. (2013). ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 75 965–976. 10.1111/tpj.12259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choudhary C., Kumar C., Gnad F., Nielsen M. L., Rehman M., Walther T. C., et al. (2009). Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325 834–840. 10.1126/science.1175371 [DOI] [PubMed] [Google Scholar]
- Clarke S. (1993). Protein methylation. Curr. Opin. Cell Biol. 5 977–983. [DOI] [PubMed] [Google Scholar]
- Cutler S. R., Rodriguez P. L., Finkelstein R. R., Abrams S. R. (2010). Abscisic acid: emergence of a core signaling network. Annu. Rev. Plant Biol. 61 651–679. 10.1146/annurev-arplant-042809-112122 [DOI] [PubMed] [Google Scholar]
- Dai M., Xue Q., Mccray T., Margavage K., Chen F., Lee J. H., et al. (2013). The PP6 phosphatase regulates ABI5 phosphorylation and abscisic acid signaling in Arabidopsis. Plant Cell 25 517–534. 10.1105/tpc.112.105767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davière J. M., Achard P. (2016). A pivotal role of DELLAs in regulating multiple hormone signals. Mol. Plant 9 10–20. 10.1016/j.molp.2015.09.011 [DOI] [PubMed] [Google Scholar]
- Dell A., Morris H. R. (2001). Glycoprotein structure determination by mass spectrometry. Science 291 2351–2356. 10.1126/science.1058890 [DOI] [PubMed] [Google Scholar]
- Dill A., Jung H. S., Sun T. P. (2001). The DELLA motif is essential for gibberellin-induced degradation of RGA. Proc. Natl. Acad. Sci. U.S.A. 98 14162–14167. 10.1073/pnas.251534098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dill A., Thomas S. G., Hu J., Steber C. M., Sun T. P. (2004). The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell 16 1392–1405. 10.1105/tpc.020958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ding Z. J., Yan J. Y., Li G. X., Wu Z. C., Zhang S. Q., Zheng S. J. (2014). WRKY41 controls Arabidopsis seed dormancy via direct regulation of ABI3 transcript levels not downstream of ABA. Plant J. 79 810–823. 10.1111/tpj.12597 [DOI] [PubMed] [Google Scholar]
- Dong K., Zhen S., Cheng Z., Cao H., Ge P., Yan Y. (2015). Proteomic analysis reveals key proteins and phosphoproteins upon seed germination of wheat (Triticum aestivum L.). Front. Plant Sci. 16:1017. 10.3389/fpls.2015.01017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donohue K., Dorn L., Griffith C., Kim E., Aguilera A., Polisetty C. R., et al. (2005). Environmental and genetic influences on the germination of Arabidopsis thaliana in the field. Evolution 59 740–757. 10.1554/04-419 [DOI] [PubMed] [Google Scholar]
- Finch-Savage W. E., Leubner-Metzger G. (2006). Seed dormancy and the control of germination. New Phytol. 171 501–523. [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Gampala S. S., Lynch T. J., Thomas T. L., Rock C. D. (2005). Redundant and distinct functions of the ABA response loci ABA-INSENSITIVE(ABI) 5 and ABRE-BINDING FACTOR (ABF) 3. Plant Mol. Biol. 59 253–267. 10.1007/s11103-005-8767-2 [DOI] [PubMed] [Google Scholar]
- Finkelstein R., Reeves W., Ariizumi T., Steber C. (2008). Molecular aspects of seed dormancy. Annu. Rev. Plant Biol. 59 387–415. [DOI] [PubMed] [Google Scholar]
- Fujii H., Zhu J. K. (2009). Arabidopsis mutant deficient in 3 abscisic acid-activated protein kinases reveals critical roles in growth, reproduction, and stress. Proc. Natl. Acad. Sci. U.S.A. 106 8380–8385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii H., Verslues P. E., Zhu J. K. (2007). Identification of two protein kinases required for abscisic acid regulation of seed germination, root growth, and gene expression in Arabidopsis. Plant Cell 19 485–494. 10.1105/tpc.106.048538 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulzele A., Bennett E. J. (2018). Ubiquitin diGLY proteomics as an approach to identify and quantify the ubiquitin-modified proteome. Methods Mol. Biol. 1844 363–384. 10.1007/978-1-4939-8706-1_23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Furihata T., Maruyama K., Fujita Y., Umezawa T., Yoshida R., Shinozaki K., et al. (2006). Abscisic acid-dependent multisite phosphorylation regulates the activity of a transcription activator AREB1. Proc. Natl. Acad. Sci. U.S.A. 103 1988–1993. 10.1073/pnas.0505667103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graeber K., Linkies A., Steinbrecher T., Mummenhoff K., Tarkowska D., Tureckova V., et al. (2014). DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. U.S.A. 111 E3571–E3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C., Yang P. (2015). Studies on the molecular mechanisms of seed germination. Proteomics 15 1671–1679. 10.1002/pmic.201400375 [DOI] [PubMed] [Google Scholar]
- Han C., Wang K., Yang P. (2014a). Gel-based comparative phosphoproteomic analysis on rice embryo during germination. Plant Cell Physiol. 55 1376–1394. 10.1093/pcp/pcu060 [DOI] [PubMed] [Google Scholar]
- Han C., Yang P., Sakata K., Komatsu S. (2014b). Quantitative proteomics reveals the role of protein phosphorylation in rice embryos during early stages of germination. J. Proteome Res. 13 1766–1782. 10.1021/pr401295c [DOI] [PubMed] [Google Scholar]
- He D., Damaris R. N., Li M., Khan I., Yang P. (2020a). Advances on plant ubiquitylome-from mechanism to application. Int. J. Mol. Sci. 21:7909. 10.3390/ijms21217909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He D., Li M., Damaris R. N., Bu C., Xue J., Yang P. (2020b). Quantitative ubiquitylomics approach for characterizing the dynamic change and extensive modulation of ubiquitylation in rice seed germination. Plant J. 101 1430–1447. 10.1111/tpj.14593 [DOI] [PubMed] [Google Scholar]
- He D., Wang Q., Li M., Damaris R. N., Yi X., Cheng Z., et al. (2016). Global proteome analyses of lysine Acetylation and Succinylation reveal the widespread involvement of both modification in metabolism in the embryo of germinating rice seed. J. Proteome Res. 15 879–890. 10.1021/acs.jproteome.5b00805 [DOI] [PubMed] [Google Scholar]
- Holdsworth M. J., Bentsink L., Soppe W. J. (2008). Molecular networks regulating Arabidopsis seed maturation, after-ripening, dormancy and germination. New Phytol. 179 33–54. 10.1111/j.1469-8137.2008.02437.x [DOI] [PubMed] [Google Scholar]
- Hu R., Zhu Y., Shen G., Zhang H. (2014). TAP46 plays a positive role in the ABSCISIC ACID INSENSITIVE5-regulated gene expression in Arabidopsis. Plant Physiol. 164 721–734. 10.1104/pp.113.233684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Yu D. (2014). BRASSINOSTEROID INSENSITIVE2 interacts with ABSCISIC ACID INSENSITIVE5 to mediate the antagonism of brassinosteroids to abscisic acid during seed germination in Arabidopsis. Plant Cell 26 4394–4408. 10.1105/tpc.114.130849 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu Y., Han X., Yang M., Zhang M., Pan J., Yu D. (2019). The transcription factor INDUCER OF CBF EXPRESSION1 interacts with ABSCISIC ACID INSENSITIVE5 and DELLA proteins to fine-tune Abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 31 1520–1538. 10.1105/tpc.18.00825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ichimura Y., Kirisako T., Takao T., Satomi Y. (2000). A ubiquitin-like system mediates protein lipidation. Nature 408:488. 10.1038/35044114 [DOI] [PubMed] [Google Scholar]
- Jaffrey S. R., Erdjument-Bromage H., Ferris C. D., Tempst P., Snyder S. H. (2001). Protein S-nitrosylation: a physiological signal for neuronal nitric oxide. Nat. Cell Biol. 3:193. 10.1038/35055104 [DOI] [PubMed] [Google Scholar]
- Jurdak R., Launay-Avon A., Paysant-Le Roux C., Bailly C. (2020). Retrograde signaling from the mitochondria to the nucleus translates the positive effect of ethylene on dormancy breaking of Arabidopsis thaliana seeds. New Phytol. 229 2192–2205. 10.1111/nph.16985 [DOI] [PubMed] [Google Scholar]
- Kim W., Bennett E. J., Huttlin E. L., Guo A., Li J., Possemato A., et al. (2011). Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol. Cell 44 325–340. 10.1016/j.molcel.2011.08.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kobayashi Y., Murata M., Minami H., Yamamoto S., Kagaya Y., Hobo T., et al. (2005). Abscisic acid activated SNRK2 protein kinases function in the gene-regulation pathway of ABA signal transduction by phosphorylating ABA response element-binding factors. Plant J. 44 939–949. 10.1111/j.1365-313x.2005.02583.x [DOI] [PubMed] [Google Scholar]
- Koornneef M., Jorna M. L., Brinkhorst-van der Swan D. L., Karssen C. M. (1982). The isolation of abscisic acid (ABA) deficient mutants by selection of induced revertants in non-germinating gibberellin sensitive lines of Arabidopsis thaliana (L.) Heynh. Theor. Appl. Genet. 61 385–393. 10.1007/bf00272861 [DOI] [PubMed] [Google Scholar]
- Kucera B., Cohn M. A., Leubner-Metzger G. (2005). Plant hormone interactions during seed dormancy release and germination. Seed Sci. Res. 15 281–307. 10.1079/ssr2005218 [DOI] [Google Scholar]
- Lee H. G., Lee K., Seo P. J. (2015). The Arabidopsis MYB96 transcription factor plays a role in seed dormancy. Plant Mol. Biol. 87 371–381. 10.1007/s11103-015-0283-4 [DOI] [PubMed] [Google Scholar]
- Lee J. H., Yoon H. J., Terzaghi W., Martinez C., Dai M., Li J., et al. (2010). DWA1 and DWA2, two Arabidopsis DWD protein components of CUL4-based E3 ligases, act together as negative regulators in ABA signal transduction. Plant Cell 22 1716–1732. 10.1105/tpc.109.073783 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee K., Lee H. G., Yoon S., Kim H. U., Seo P. J. (2015). The Arabidopsis MYB96 transcription factor is a positive regulator of ABI4 in the control of seed germination. Plant Physiol. 168 677–689. 10.1104/pp.15.00162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee S., Cheng H., King K. E., Wang W., He Y., Hussain A., et al. (2002). Gibberellin regulates Arabidopsis seed germination via RGL2, a GAI/RGA-like gene whose expression is up-regulated following imbibition. Genes Dev. 16 646–658. 10.1101/gad.969002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefebvre V., North H., Frey A., Sotta B., Seo M., Okamoto M., et al. (2006). Functional analysis of Arabidopsis NCED6 and NCED9 genes indicates that ABA synthesized in the endosperm is involved in the induction of seed dormancy. Plant J. 45 309–319. 10.1111/j.1365-313x.2005.02622.x [DOI] [PubMed] [Google Scholar]
- Leubner-Metzger G. (2014). DELAY OF GERMINATION 1 mediates a conserved coat-dormancy mechanism for the temperature- and gibberellin-dependent control of seed germination. Proc. Natl. Acad. Sci. U.S.A. 111 E3571–E3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li J., Zhu D., Deng X. W. (2017). Noncanonical role of Arabidopsis COP1/SPA complex in repressing BIN2-mediated PIF3 phosphorylation and degradation in darkness. Proc. Natl. Acad. Sci. U.S.A. 114 3539–3544. 10.1073/pnas.1700850114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li M., Yin X., Sakata K., Yang P., Komatsu S. (2015). Proteomic analysis of phosphoproteins in the rice nucleus during the early stage of seed germination. J. Proteome Res. 14 2884–2896. 10.1021/acs.jproteome.5b00215 [DOI] [PubMed] [Google Scholar]
- Li P., Zhang Q., He D., Zhou Y., Ni H., Tian D., et al. (2020). LIKE67 cooperates with the histone mark reader EBS to modulate seed germination under high temperature. Plant Physiol. 184 529–545. 10.1104/pp.20.00056 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li W., Zhang F., Chang Y., Zhao T., Schranz M. E., Wang G. (2015). Nicotinate O-glucosylation is an evolutionarily metabolic trait important for seed germination under stress conditions in Arabidopsis thaliana. Plant Cell 27 1907–1924. 10.1105/tpc.15.00223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S., Park J., Lee N., Jeong J., Toh S., Watanabe A., et al. (2013). ABA-insensitive3, ABA-insensitive5, and DELLAs Interact to activate the expression of SOMNUS and other high-temperature-inducible genes in imbibed seeds in Arabidopsis. Plant Cell 25 4863–4878. 10.1105/tpc.113.118604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S. L. (2010). Abscisic acid increases Arabidopsis ABI5 transcription factor levels by promoting KEG E3 ligase self-ubiquitination and proteasomal degradation. Plant Cell 22 2630–2641. 10.1105/tpc.110.076075 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu H., Stone S. L. (2013). Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 288 20267–20279. 10.1074/jbc.m113.465369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu X., Zhang H., Zhao Y., Feng Z., Li Q., Yang H. Q., et al. (2013). Auxin controls seed dormancy through stimulation of abscisic acid signaling by inducing ARF-mediated ABI3 activation in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 110 15485–15490. 10.1073/pnas.1304651110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lois L. M., Lima C. D., Chua N. H. (2003). Small ubiquitin-like modifier modulates abscisic acid signaling in Arabidopsis. Plant Cell 15 1347–1359. 10.1105/tpc.009902 [DOI] [PMC free article] [PubMed] [Google Scholar]
- López-Pedrouso M., Alonso J., Zapata C. (2014). Evidence for phosphorylation of the major seed storage protein of the common bean and its phosphorylation-dependent degradation during germination. Plant Mol. Biol. 84 415–428. 10.1007/s11103-013-0141-1 [DOI] [PubMed] [Google Scholar]
- Lyzenga W. J., Liu H., Schofield A., Muise-Hennessey A., Stone S. L. (2013). Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J. Exp. Bot. 64 2779–2791. 10.1093/jxb/ert123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Szostkiewicz I., Korte A., Moes D., Yang Y., Christmann A., et al. (2009). Regulators of PP2C phosphatase activity function as Abscisic acid sensors. Science 324 1064–1068. [DOI] [PubMed] [Google Scholar]
- Mann M., Ong S.-E., Grønborg M., Steen H., Jensen O. N., Pandey A. (2002). Analysis of protein phosphorylation using mass spectrometry: deciphering the phosphoproteome. Trends Biotechnol. 20 261–268. 10.1016/s0167-7799(02)01944-3 [DOI] [PubMed] [Google Scholar]
- Matakiadis T., Alboresi A., Jikumaru Y., Tatematsu K., Pichon O., Renou J. P., et al. (2009). The Arabidopsis abscisic acid catabolic gene CYP707A2 plays a key role in nitrate control of seed dormancy. Plant Physiol. 149 949–960. 10.1104/pp.108.126938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mazer S. J. (1999). Seeds-Ecology, biogeography, and evolution of dormancy and germination. Science 283:334. [Google Scholar]
- Millar A. A., Jacobsen J. V., Ross J. J., Helliwell C. A., Poole A. T., Scofield G., et al. (2006). Seed dormancy and ABA metabolism in Arabidopsis and barley: the role of ABA 8’-hydroxylase. Plant J. 45 942–954. 10.1111/j.1365-313x.2006.02659.x [DOI] [PubMed] [Google Scholar]
- Miura K., Lee J., Jin J. B., Yoo C. Y., Miura T., Hasegawa P. M. (2009). Sumoylation of ABI5 by the Arabidopsis SUMO E3 ligase SIZ1 negatively regulates abscisic acid signaling. Proc. Natl. Acad. Sci. U.S.A. 106 5418–5423. 10.1073/pnas.0811088106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyazono K., Miyakawa T., Sawano Y., Kubota K., Kang H. J., Asano A., et al. (2009). Structural basis of abscisic acid signalling. Nature 462 609–614. 10.1038/nature08583 [DOI] [PubMed] [Google Scholar]
- Nakabayashi K., Bartsch M., Xiang Y., Miatton E., Pellengahr S., Yano R., et al. (2012). The time required for dormancy release in Arabidopsis is determined by DELAY OF GERMINATION1 protein levels in freshly harvested seeds. Plant Cell 24 2826–2838. 10.1105/tpc.112.100214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakajima M., Shimada A., Takashi Y., Kim Y. C., Park S. H., Ueguchi-Tanaka M., et al. (2006). Identification and characterization of Arabidopsis gibberellin receptors. Plant J. 46 880–889. 10.1111/j.1365-313x.2006.02748.x [DOI] [PubMed] [Google Scholar]
- Nakashima K., Fujita Y., Kanamori N., Katagiri T., Umezawa T., Kidokoro S., et al. (2009). Three Arabidopsis SnRK2 protein kinases, SRK2D/SnRK2.2, SRK2E/SnRK2.6/OST1 and SRK2I/SnRK2.3, involved in ABA signaling are essential for the control of seed development and dormancy. Plant Cell Physiol. 50 1345–1363. 10.1093/pcp/pcp083 [DOI] [PubMed] [Google Scholar]
- Ni W., Xu S.-L., Chalkley R. J., Pham T. N. D., Guan S., Maltby D. A., et al. (2013). Multisite light-induced phosphorylation of the transcription factor PIF3 is necessary for both its rapid degradation and concomitant negative feedback modulation of photoreceptor phyB levels in Arabidopsis. Plant Cell 25 2679–2698. 10.1105/tpc.113.112342 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishimura N., Hitomi K., Arvai A. S., Rambo R. P., Hiotomi C., Cutler S. R., et al. (2009). Stuctural mechanism of absicsic acid binding and signaling by dimeric PYR1. Science 326 1373–1379. 10.1126/science.1181829 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan J., Hu Y., Wang H., Guo Q., Chen Y., Howe G. A., et al. (2020). Molecular mechanism underlying the synergetic effect of jasmonate on abscisic acid signaling during seed germination in Arabidopsis. Plant Cell 32 3846–3865. 10.1105/tpc.19.00838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Lee N., Kim W., Lim S., Choi G. (2011). ABI3 and PIL5 collaboratively activate the expression of SOMNUS by directly binding to its promoter in imbibed Arabidopsis seeds. Plant Cell 23 1404–1415. 10.1105/tpc.110.080721 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penfield S., Li Y., Gilday A. D., Graham S., Graham I. A. (2006). Arabidopsis ABA INSENSITIVE4 regulates lipid mobilization in the embryo and reveals repression of seed germination by the endosperm. Plant Cell 18 1887–1899. 10.1105/tpc.106.041277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Jikumaru Y., Kinoshita N., Nambara E., Kamiya Y., Lopez-Molina L. (2008). The gibberellic acid signaling repressor RGL2 inhibits Arabidopsis seed germination by stimulating abscisic acid synthesis and ABI5 activity. Plant Cell 20 2729–2745. 10.1105/tpc.108.061515 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piskurewicz U., Turecková V., Lacombe E., Lopez-Molina L. (2009). Far-red light inhibits germination through DELLA-dependent stimulation of ABA synthesis and ABI3 activity. EMBO J. 28 2259–2271. 10.1038/emboj.2009.170 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptacek J., Devgan G., Michaud G., Zhu H. (2005). Global analysis of protein phosphorylation in yeast. Nature 438:679. [DOI] [PubMed] [Google Scholar]
- Ruiz-Ballesta I., Baena G., Gandullo J., Wang L., She Y. M., Plaxton W. C., et al. (2016). New insights into the post-translational modification of multiple phosphoenolpyruvate carboxylase isoenzymes by phosphorylation and monoubiquitination during sorghum seed development and germination. J. Exp. Bot. 67 3523–3536. 10.1093/jxb/erw186 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santiago J., Dupeux F., Round A., Antoni R., Park S. Y., Jamin M., et al. (2009). The abscisic acid receptor PYR1 in complex with abscisic acid. Nature 462 665–668. 10.1038/nature08591 [DOI] [PubMed] [Google Scholar]
- Seo K. I., Lee J. H., Nezames C. D., Zhong S., Song E., Byun M. O., et al. (2014). ABD1 is an Arabidopsis DCAF substrate receptor for CUL4-DDB1-based E3 ligases that acts as a negative regulator of abscisic acid signaling. Plant Cell 26 695–711. 10.1105/tpc.113.119974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma K., D’Souza R. C., Tyanova S., Schaab C., Wi-Sniewski J. R., Cox J., et al. (2014). Ultradeep human phosphoproteome reveals a distinct regulatory nature of Tyr and Ser/Thr-based signaling. Cell Rep. 8 1583–1594. 10.1016/j.celrep.2014.07.036 [DOI] [PubMed] [Google Scholar]
- Shu K., Liu X. D., Xie Q., He Z. H. (2016). Two faces of one seed: hormonal regulation of dormancy and germination. Mol. Plant 9 34–45. 10.1016/j.molp.2015.08.010 [DOI] [PubMed] [Google Scholar]
- Shu K., Zhang H., Wang S., Chen M., Wu Y., Tang S., et al. (2013). ABI4 regulates primary seed dormancy by regulating the biogenesis of abscisic acid and gibberellins in Arabidopsis. PLoS Genet. 9:e1003577. 10.1371/journal.pgen.1003577 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stone S. L., Williams L. A., Farmer L. M., Vierstra R. D., Callis J. (2006). KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 18 3415–3428. 10.1105/tpc.106.046532 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thalassinos K., Grabenauer M., Slade S. E., Hilton G. R., Bowers M. T., Scrivens J. H. (2008). Characterization of phosphorylated peptides using traveling wave-based and drift cell ion mobility mass spectrometry. Anal. Chem. 81 248–254. 10.1021/ac801916h [DOI] [PubMed] [Google Scholar]
- Tyler L., Thomas S. G., Hu J., Dill A., Alonso J. M., Ecker J. R., et al. (2004). Della proteins and gibberellin-regulated seed germination and floral development in Arabidopsis. Plant Physiol. 135 1008–1019. 10.1104/pp.104.039578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueguchi-Tanaka M., Ashikari M., Nakajima M., Itoh H., Katoh E., Kobayashi M., et al. (2005). GIBBERELLIN INSENSITIVE DWARF1 encodes a soluble receptor for gibberellin. Nature 437 693–698. 10.1038/nature04028 [DOI] [PubMed] [Google Scholar]
- Uhrig R. G., Labandera A. M., Tang L. Y., Sieben N. A., Goudreault M., Yeung E., et al. (2017). Activation of mitochondrial protein phosphatase SLP2 by MIA40 regulates seed germination. Plant Physiol. 173 956–969. 10.1104/pp.16.01641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H., Zhang Y., Xiao N., Zhang G., Wang F., Chen X., et al. (2020). Rice GERMIN-LIKE PROTEIN 2-1 functions in seed dormancy under the control of abscisic acid and gibberellic acid signaling pathways. Plant Physiol. 183 1157–1170. 10.1104/pp.20.00253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Li L., Ye T., Zhao S., Liu Z., Feng Y. Q., et al. (2011). Cytokinin antagonizes ABA suppression to seed germination of Arabidopsis by down regulating ABI5 expression. Plant J. 68 249–261. 10.1111/j.1365-313x.2011.04683.x [DOI] [PubMed] [Google Scholar]
- Xu G., Paige J. S., Jaffrey S. R. (2010). Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat. Biotechnol. 28 868–873. 10.1038/nbt.1654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu S., Gonzalez-Grandio E., Chalkley R. J., Huhmer A. F. R., Burlingame A. L., Wang Z. Y., et al. (2017). PPKs mediate direct signal transfer from phytochrome photoreceptors to transcription factor PIF3. Nat. Commun. 8:15236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaish M. W., El-Kereamy A., Zhu T., Beatty P. H., Good A. G., Bi Y. M., et al. (2010). The APETALA-2-like transcription factor OsAP2-39 controls key interactions between abscisic acid and gibberellin in rice. PLoS Genet. 6:e1001098. 10.1371/journal.pgen.1001098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagishi K., Tatematsu K., Yano R., Preston J., Kitamura S., Takahashi H., et al. (2009). CHOTTO1, a double AP2 domain protein of Arabidopsis thaliana, regulates germination and seedling growth under excess supply of glucose and nitrate. Plant Cell Physiol. 50 330–340. 10.1093/pcp/pcn201 [DOI] [PubMed] [Google Scholar]
- Yamauchi Y., Takeda-Kamiya N., Hanada A., Ogawa M., Kuwahara A., Seo M., et al. (2007). Contribution of gibberellin deactivation by AtGA2ox2 to the suppression of germination of dark-imbibed Arabidopsis thaliana seeds. Plant Cell Physiol. 48 555–561. 10.1093/pcp/pcm023 [DOI] [PubMed] [Google Scholar]
- Yano R., Kanno Y., Jikumaru Y., Nakabayashi K., Kamiya Y., Nambara E. (2009). CHOTTO1, a putative double APETALA2 repeat transcription factor, is involved in abscisic acid-mediated repression of gibberellin biosynthesis during seed germination in Arabidopsis. Plant Physiol. 151 641–654. 10.1104/pp.109.142018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ying J., Zhao J., Hou Y., Wang Y., Qiu J., Li Z., et al. (2017). Mapping the N-linked glycosites of rice (Oryza sativa L.) germinating embryos. PLoS One 12:e0173853. 10.1371/journal.pone.0173853 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu F., Wu Y., Xie Q. (2015). Precise protein post-translational modifications modulate ABI5 activity. Trends Plant Sci. 20 569–575. 10.1016/j.tplants.2015.05.004 [DOI] [PubMed] [Google Scholar]
- Zhang H., He D., Yu J., Li M., Damaris R. N., Gupta R., et al. (2016). Analysis of dynamic protein carbonylation in rice embryo during germination through AP-SWATH. Proteomics 16 989–1000. 10.1002/pmic.201500248 [DOI] [PubMed] [Google Scholar]
- Zhang H., Xiao-jun L., Martin D. B., Aebersold R. (2003). Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 21:660. 10.1038/nbt827 [DOI] [PubMed] [Google Scholar]
- Zhou X., Hao H., Zhang Y., Bai Y., Zhu W., Qin Y., et al. (2015). SOS2-LIKE PROTEIN KINASE5, an SNF1-RELATED PROTEIN KINASE3-type protein kinase, is important for abscisic acid responses in Arabidopsis through phosphorylation of ABSCISIC ACID-INSENSITIVE5. Plant Physiol. 168 659–676. 10.1104/pp.114.255455 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhu Y., Hu X., Duan Y., Li S., Wang Y., Rehman A. U., et al. (2020). The Arabidopsis nodulin homeobox factor AtNDX Interacts with AtRING1A/B and negatively regulates abscisic acid signaling. Plant Cell 32 703–721. 10.1105/tpc.19.00604 [DOI] [PMC free article] [PubMed] [Google Scholar]