Highlights
-
•
ERK1/2 and ERK5 pathway inhibition induces mesenchymal to epithelial transition in breast cancer.
-
•
ERK5 is endogenously active and localized in the nucleus of TNBC cells.
-
•
Dual ERK5 and ERK1/2 or AKT inhibition is a relevant strategy to target breast cancer.
Keywords: ERK1/2, ERK5, Mesenchymal to epithelial transition (MET), Signaling, Breast cancer
Abbreviations: E-cadherin, epithelial cadherin; ERK, extracellular signal-regulated kinase; EMT, epithelial to mesenchymal transition; GFP, green fluorescent protein; MEK, mitogen-activated protein kinase kinase; MET, mesenchymal to epithelial transition; PDX, Patient-derived xenograft; RSK, ribosomal s6 kinase; TAMR, tamoxifen-resistant; TNBC, triple-negative breast cancer; ZEB, zinc-finger E-box binding homeobox
Abstract
The epithelial to mesenchymal transition (EMT) is characterized by a loss of cell polarity, a decrease in the epithelial cell marker E-cadherin, and an increase in mesenchymal markers including the zinc-finger E-box binding homeobox (ZEB1). The EMT is also associated with an increase in cell migration and anchorage-independent growth. Induction of a reversal of the EMT, a mesenchymal to epithelial transition (MET), is an emerging strategy being explored to attenuate the metastatic potential of aggressive cancer types, such as triple-negative breast cancers (TNBCs) and tamoxifen-resistant (TAMR) ER-positive breast cancers, which have a mesenchymal phenotype. Patients with these aggressive cancers have poor prognoses, quick relapse, and resistance to most chemotherapeutic drugs. Overexpression of extracellular signal-regulated kinase (ERK) 1/2 and ERK5 is associated with poor patient survival in breast cancer. Moreover, TNBC and tamoxifen resistant cancers are unresponsive to most targeted clinical therapies and there is a dire need for alternative therapies.
In the current study, we found that MAPK3, MAPK1, and MAPK7 gene expression correlated with EMT markers and poor overall survival in breast cancer patients using publicly available datasets. The effect of ERK1/2 and ERK5 pathway inhibition on MET was evaluated in MDA-MB-231, BT-549 TNBC cells, and tamoxifen-resistant MCF-7 breast cancer cells. Moreover, TU-BcX-4IC patient-derived primary TNBC cells were included to enhance the translational relevance of our study. We evaluated the effect of pharmacological inhibitors and lentivirus-induced activation or inhibition of the MEK1/2-ERK1/2 and MEK5-ERK5 pathways on cell morphology, E-cadherin, vimentin and ZEB1 expression. Additionally, the effects of pharmacological inhibition of trametinib and XMD8-92 on nuclear localization of ERK1/2 and ERK5, cell migration, proliferation, and spheroid formation were evaluated. Novel compounds that target the MEK1/2 and MEK5 pathways were used in combination with the AKT inhibitor ipatasertib to understand cell-specific responses to kinase inhibition. The results from this study will aid in the design of innovative therapeutic strategies that target cancer metastases.
1. Introduction
Metastases account for ~90% human deaths due to cancer [1]. The epithelial to mesenchymal transition (EMT), one of the first steps in metastases, leads to the loss of cell polarity, downregulation of E-cadherin, and upregulation of mesenchymal markers snail, zinc-finger E-box binding homeobox (ZEB1), and vimentin. EMT is also associated with drug resistance [2]. Additionally, cytoskeletal reorganization and loss of E-cadherin is an important step to initiate the epithelial to mesenchymal transition (EMT) and metastases [[3], [4], [5]]. Reversing the mesenchymal phenotype of cancer cells through activation of the mesenchymal to epithelial transition (MET) program is an emerging approach to attenuate the metastatic properties of cancer cells.
Most triple-negative breast cancer (TNBC) cells have a mesenchymal phenotype and show poor sensitivity to chemotherapy agents [6]. The loss of estrogen, progesterone hormone receptors, and human epidermal growth factor receptors (HER2) contributes to the aggressive state of TNBC and lack of targeted therapies [7]. Tamoxifen-resistance is associated with an induction of EMT in estrogen receptor (ER) positive MCF-7 cells [[8], [9]–10]. An increasing body of evidence suggests that activation of ERK1/2 and ERK5 signaling is a marker for node metastases and a predictor of poor responses to hormone therapy such as 4-OHT [[11], [12]–13]. Activation of intracellular signaling pathways, such as the ERK MAPK pathways, mediates tumorigenesis in TNBCs and tamoxifen resistant breast cancers [14,15]. ERK1/2 activation is known to mediate EMT in several cancer models [[16], [17], [18]–19]. Moreover, overexpression of the newest member of the MAPK family, ERK5, induces EMT and hormone-independent growth of breast cancer [13].
To identify the link between MAPK pathways and EMT, MAPK3 (ERK1), MAPK1(ERK2), and MAPK7 (ERK5) gene expression was correlated with EMT markers CDH1, ZEB1, or VIM in tumors derived from TNBC patients using publicly available datasets. Moreover, overall survival in patients with inflammatory breast cancer was plotted against ERK1, ERK2, or ERK5 gene expression using publicly available datasets. Although activation of the ERK1/2 and ERK5 pathways have been shown to mediate EMT, the effect of ERK1/2 and ERK5 inhibition on MET is poorly understood in cancer.
We hypothesize that inhibition of the ERK1/2 and ERK5 pathways is a relevant strategy to induce a MET in TNBCs. To test this hypothesis, we examined the effects the ERK1/2 and ERK5 pathways on the MET and nuclear localization of ERK5 using the pharmacological inhibitors trametinib (MEK1/2 inhibitor) and XMD8-92 (ERK5 inhibitor). Moreover, the effect of lentivirus-mediated activation or inhibition of ERK1/2 and ERK5 pathway components on the MET was examined. Cell morphology and protein expression of epithelial and mesenchymal markers, E-cadherin and ZEB1, respectively, were examined. Activation of ERK1/2, ERK5, and RSK, a downstream target of MAPK signaling, was evaluated. The effect of XMD8-92 and trametinib was evaluated on cell migration and cell proliferation in TNBC and TAMR breast cancer. Moreover, the crosstalk between MEK-ERK and PI3K-AKT pathway with respect to EMT was studied in a spheroid viability assay.
2. Material and methods
2.1. Cell culture
MDA-MB-231 and MDA-MB-231 VIM RFP cells (ATCC, Manassas, VA) were cultured in DMEM: F-12 and BT-549 and MCF-7 (ATCC) cells were cultured in RPMI media supplemented with 5% FBS (Gibco, Thermo Fisher Scientific, Waltham, MA), respectively. TUBcX-4IC patient-derived primary TNBC cells were generously provided by Burow lab. TU-BcX-4IC cells were cultured in DMEM:F-12 (1:1) media supplemented with 10% FBS and 1% Pen/Strep. TAMR-MCF-7 cells were cultured in phenol red free RPMI media supplemented with 5% charcoal-stripped FBS as previously described [20]. The cells were maintained at 37°C and 5% CO2 as per standard manufacturer's protocol.
2.2. Inhibitor treatment and EGF stimulation
Cells were cultured in a 6-well plate (250,000 cells/well) for 24 hrs. To examine kinase activity or inhibition, the cells were serum-starved for 18-24 h. The inhibitors XMD8-92 (Tocris, Minneapolis, MN) and trametinib (Selleckchem, Houston, TX) were added for 30 minutes prior to EGF (100ng/ml) stimulation for 15 minutes as previously described [21]. Cells were lysed and examined for kinase activation using standard western blot procedures.
2.3. Immunofluorescence assay
Cells were cultured in 96-well plates (5,000 cells/well). After 24 h of plating, treatments were added for 72 h. The media was removed, cells were washed with PBS and fixed with 4% paraformaldehyde for 10 minutes. The cells were washed and incubated with blocking buffer for 1 hour at room temperature. Primary antibodies (α-actinin, α-tubulin, ZEB1 and Ki67) were added at a dilution of 1:750 and the plate was incubated at 4°C overnight. The cells were washed with PBS three times at 5-minute intervals. Secondary antibodies (Goat anti-mouse Alexa Flour 488nm and goat anti-Rabbit Alexa Flour 555nm (1:1000, Invitrogen), counterstained with Hoechst (Fisher) were added for 1 hour at room temperature. Cells were washed with PBS three times at 5-minute intervals and the pictures were taken using the EVOS microscope (Thermo Fisher Scientific, Waltham, MA) at 10X magnification.
2.4. Migration assay
Cells were cultured in a 12-well plate (150,000 cells/ well). The compounds were added for 48 h and a scratch was made using 10 µl pipette tip. The underside of the plate was marked to denote the location of the initial wound. Cells were washed gently with 1x PBS to remove detached cells and debris. Treatments were added in fresh media, images were taken, and the plate was returned to the incubator for 24 h. The images were taken after 24 h from the time of scratch and the wound closure was calculated by the formula: (scratch at 24 h) – (scratch at 0 h) / (scratch at 0 h) X 100.
2.5. Lentivirus treatment
Lentivirus plasmids were a generous gift from Dr. Zhengui Xia (University of Seattle, Washington). Cells were cultured in a 12-well plate (150,000 cells/ well). The volume of required per well lentivirus (µL) was calculated as [(# of cells/well x desired multiplicity of infection (MOI)/viral titer (IU/µL)].. This volume of lentivirus was diluted in fresh media and 50% of media was replaced with the lentivirus-containing media. The infection efficiency was greater than 60% after 24 hours of treatment at MOI=1, as calculated by microscopic observation of the percentage of GFP-positive cells. The cells were infected with lentivirus at the MOI =1 for 96 h. Immunofluorescence staining and western blotting were performed to examine cell morphology, E-cadherin and ZEB1 protein expression or ERK1/2 and ERK5 activation.
2.6. Cell lysis and Western Blotting
Cells were cultured in a 6-well plate (250,000 cells/well) for 24 h. After 24 h, the inhibitors were added to the cells for 72 h to examine kinase activation and MET markers. The cells were lysed in ice-cold 1X cell lysis buffer (Cell Signaling Technology, Danvers, MA) buffer and 0.1 M PMSF. The lysates were centrifuged at 10,000 rpm for 10 minutes at 4°C. The supernatant was collected and stored at -80°C until further analyses. The lysates were denatured using β-mercaptoethanol. Bradford (Bio-Rad, Hercules, CA) protein assay was performed to determine the protein concentrations in the lysates. 30 µg of protein was loaded on 8% SDS-PAGE gels. The gels were transferred to nitrocellulose membranes. The membranes were incubated in casein blocking buffer at room temperature for 1 h. Primary antibodies ERK5, ERK1/2, ZEB1, pERK1/2 (phospho-p44/42), p-P90RSK (S380), RSK1/2/3, α-tubulin, and E-cadherin (Cell Signaling Technology) were added and the membranes were incubated at 4°C overnight. The membranes were washed in PBS-0.1% tween solution three times at 10-minute intervals. Secondary antibodies were added, and the membranes were incubated for 1 h and washed three times at 10-minute intervals at room temperature. The membranes were washed with PBS and scanned using an Odyssey (LI-CR, Lincoln, NE) imager at 700 and 800 nm wavelength. The blots were quantified using Image Studio Lite (LI-COR Biosciences).
2.7. Nuclear/Cytosolic fractionation
Cells were cultured in 6-well plates (500,000 cells/well) for 24 h. After 24 h, cells were treated with the kinase inhibitors for 72 h. The nuclear/ cytosolic fractionation was performed using standard manufacturer's instructions (Cell Biolabs, San Diego, CA). In brief, the medium was aspirated, and cells were washed with pre-chilled 1X PBS. DTT and Protease inhibitor cocktail was added to Cytosol Extraction Buffer (CEB). 100 µL CEB was added to cells for 10 minutes. Cells were scraped and collected in 1.5 mL pre-chilled microcentrifuge tubes. Cell lysis buffer was added for 5-15 minutes and the lysates were vortexed for 10 seconds. The lysates were centrifuged at 10,000 rpm for 10 minutes at 4°C. The supernatant (cytosolic fraction) was collected and stored at -80°C. The pellet was resuspended in CEB and lysis buffer was added for 10 minutes. The suspension was vortexed and centrifuged at 10,000 rpm for 10 minutes at 4°C. The supernatant was discarded, this step was performed to ensure clean separation. The pellet was resuspended in 40 µL nuclear extraction buffer (NEB) with DTT and protease inhibitors. The solution was incubated on ice for 30 minutes with occasional vortexing. The samples were centrifuged at 14,000 rpm for 30 minutes at 4°C. The supernatant (nuclear fraction) was stored at -80°C.
2.8. Spheroid assay
Cells were cultured in a 96-well plate (5000 cells/ well). After 24 h of plating, the spheroids were treated with different concentrations of the MAPK and/or AKT inhibitors and allowed to grow for 7 days. The pictures of spheroids were taken at the time of treatment and after 7 days of treatment. At the experimental endpoint, 10 µL Reliablue reagent (ATCC) was added to each well and the plate was returned to the incubator for 3 h. The fluorescence was measured at ex570/ em590 on a Synergy microplate reader (Biotek, Winooski, VT).
2.9. MTT cell viability assay
MTT (3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide) assay was performed to determine cell viability. Cells were seeded at a density of 5,000 per well in 96-well plates containing 90 μl of full media for 24 h and then treated with increasing concentrations of trametinib and/or XMD8-92 for 72 h. 10 μL of MTT (Acros, Cat. No. 298-93-1) solution (5 mg/ml in phosphate-buffered saline, PBS) was added to each well and the plate was incubated at 37°C for 3 h. After removal of the MTT solution from each well, 100 μl of DMSO was added to the wells for 10 min under agitation to dissolve the formazan crystals. Absorbance was measured at a wavelength of 570 nm.
2.10. Statistical analyses
Genomics data were analyzed using R2: Genomics analysis and visualization plaltform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). One-way or two-way analysis of variance (ANOVA) with Bonferroni post-hoc correction was applied to determine statistical significance across different concentrations of individual drugs compared to the control (DMSO or GFP) or to the individual drug where combination treatment was performed. GraphPad Prism version 7.03 for Windows (GraphPad Software La Jolla, California) was used for statistical analyses.
3. Results
3.1. ERK1, ERK2, and ERK5 expression correlates with EMT markers and is associated with poor patient survival in breast cancer
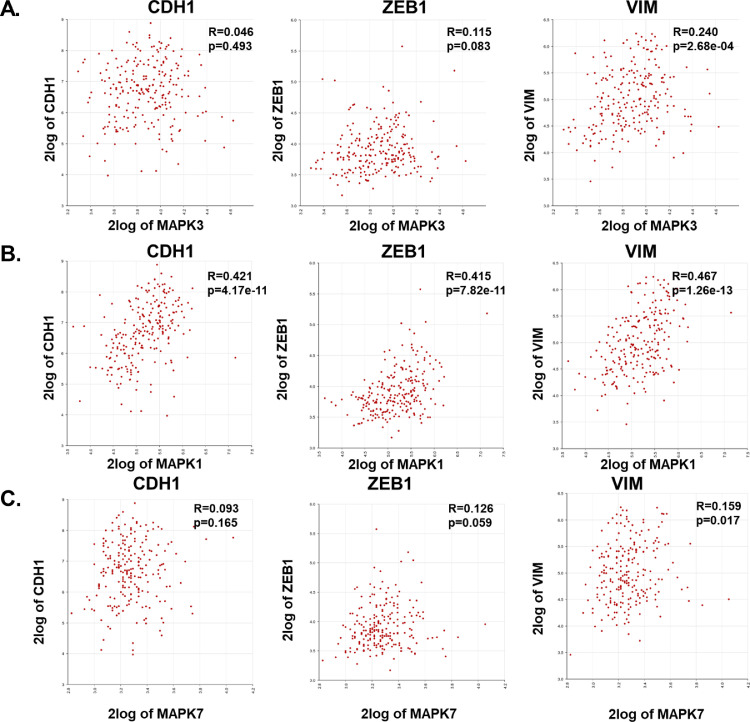
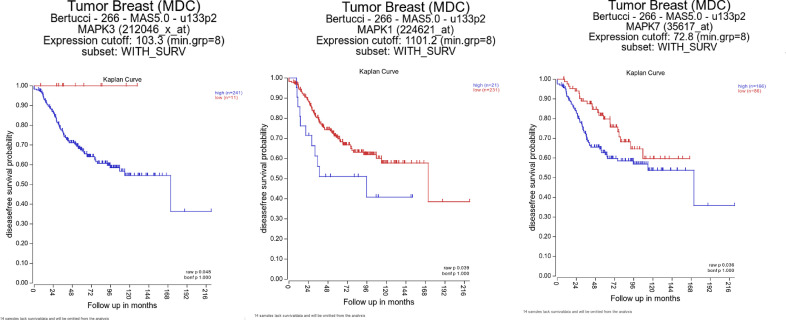
Since the effect of ERK1/2 and ERK5 pathways on EMT is less well-understood in TNBCs, we first used publicly available datasets from Purrington, K. S. and colleagues [22] to correlate MAPK3 (ERK1), MAPK1(ERK2), or MAPK7 (ERK5) gene expression with EMT markers CDH1 (E-cadherin), ZEB1, or vimentin in primary, invasive tumors derived from African-American TNBC patients (Fig. 1). There was a moderate to strong significant correlation between MAPK3, MAPK1, and MAPK7 with mesenchymal markers ZEB1 and vimentin. Moreover, we performed Kaplan-Meier survival analysis to examine relation between ERK1/2/5 expression and patient survival in inflammatory breast cancer using publicly available datasets from Bertucci and colleagues [23] MAPK3, MAPK1, or MAPK7 gene expression was found to be associated with poor patient survival in patients with inflammatory breast cancer (Fig. 2). Overall, these data suggest that ERK1, ERK2, and ERK5 are important therapeutic targets in breast cancer.
Fig. 1.
Correlation of ERK1, ERK2, or ERK5 with EMT markers in tumors derived from TNBC patients. Gene correlation between (A) MAPK3(ERK1), (B) MAPK1(ERK2), or (C) MAPK7 (ERK5) and EMT markers CDH1, ZEB1, or VIM was plotted using R2: Genomics analysis and visualization platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Datasets were exported from Tumor Breast (triple negative) - Purrington - 226 - rma_sketch - hugene21t.
Fig. 2.
MAPK3, MAPK1, and MAPK7 expression correlates with poor patient survival in breast cancer. Disease free survival was analyzed using R2: Genomics analysis and visualization platform (https://hgserver1.amc.nl/cgi-bin/r2/main.cgi). Datasets were exported from Tumor Breast (MDC) Bertucci - 266 - MAS5.0 - u133p2.
3.2. Pharmacological inhibition of the ERK1/2 and/or ERK5 pathways induces MET in TNBC and TAMR cells
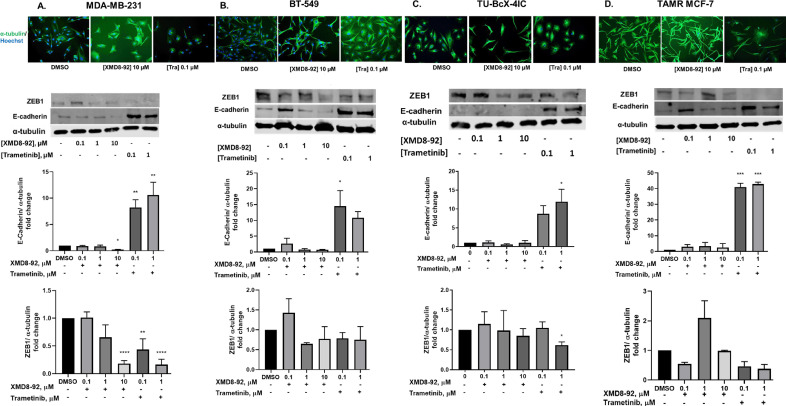
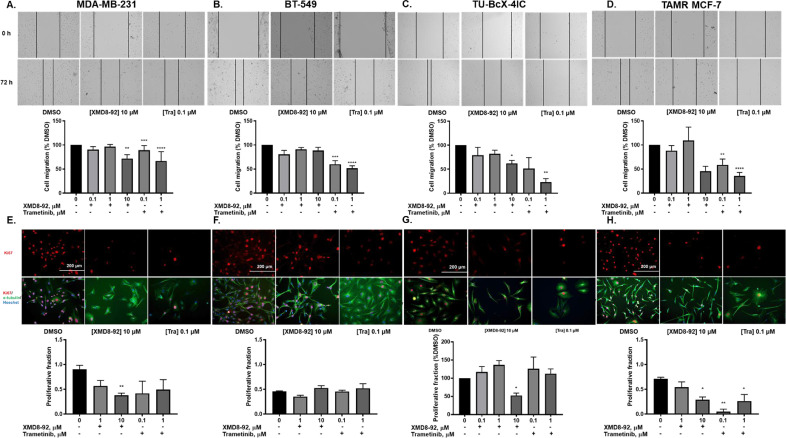
MDA-MB-231, BT-549, and TU-BcX-4IC TNBC cells were treated with increasing concentrations of XMD8-92 and trametinib for 72 hours. Moreover, the effects of XMD8-92 and trametinib were evaluated on MET and kinase signaling in tamoxifen-resistant (TAMR) MCF-7 ER-positive breast cancer cells, which have a mesenchymal phenotype. The generation of TAMR-MCF-7 cells in our lab and evidence of EMT have been previously reported [20]. Trametinib induced a morphological switch from mesenchymal to epithelial in all the cell lines, while XMD8-92 only induced this morphological change in MDA-MB-231 cells (Fig. 3).
Fig. 3.
ERK1/2 and ERK5 pathway inhibition induces MET in TNBC and TAMR MCF-7 cells. Cells were treated with XMD8-92 and trametinib at increasing concentrations for 72 hours. Cell morphology (20X magnification) and western blot analysis of EMT markers E-cadherin and ZEB1 in (A) MDA-MB-231 cells. (B) BT-549 cells (C) TU-BcX-4IC and (D) TAMR MCF-7 cells. Data represent the ± SEM of three different experiments for each inhibitor compared to DMSO control. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 vs DMSO control group determined by one-way ANOVA with the Bonferroni post hoc test.
In MDA-MB-231 cells, trametinib increased E-cadherin expression and decreased ZEB-1 expression, markers of epithelial and mesenchymal phenotypes, respectively. XMD8-92 decreased the expression of ZEB1 but had no effect on E-cadherin expression at low doses, and decreased E-cadherin expression at the highest dose in MDA-MB-231 cells (Fig. 3A). Treatment with trametinib significantly increased E-cadherin and led to a trending decrease in ZEB1 expression in BT-549 cells (Fig. 3B). Trametinib, but not XMD8-92 significantly decreased ZEB1 expression in TU-BcX-4IC cells. XMD8-92 did not alter cell morphology, E-cadherin, or ZEB1 expression in TAMR MCF-7 cells (Fig. 3C).
In order to examine the extent of MET induced by the inhibitors, we correlated the expression of E-cadherin to ZEB1. Treatment that induced E-cadherin expression by greater than 3-fold and decreased ZEB1 by greater than 0.3-fold was determined to induce a full MET switch whereas treatment that either induced 3-fold increase in E-cadherin expression or 0.3-fold was determined to induce a partial MET. Trametinib induced a full MET in MDA-MB-231 and TAMR MCF-7 cells at low and high doses whereas it induced a partial MET in BT-549 cells as noted by a significant increase in E-cadherin expression (Supplemental Figure 1). Therefore, we correlated WT-MCF-7 epithelial cells were included as a control to study EMT. We observed that treatment with XMD8-92 or trametinib did not alter cell morphology or E-cadherin expression in WT-MCF-7 cells (Supplemental figure 2A, B).
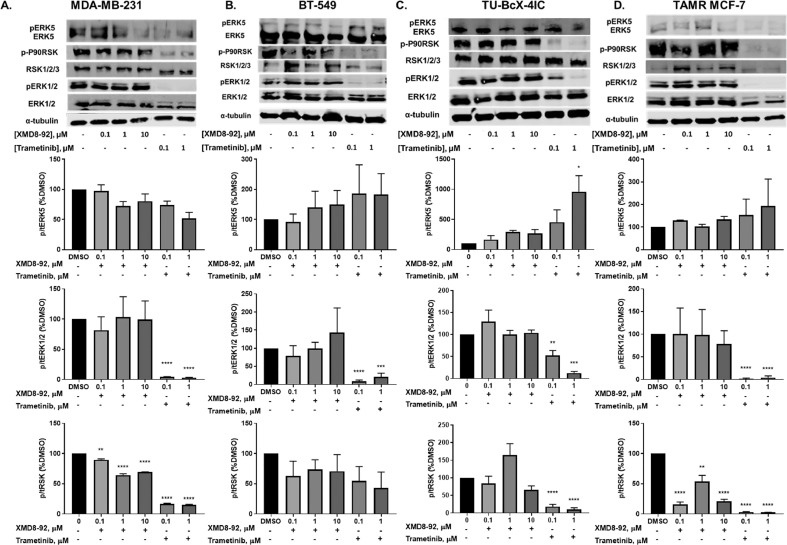
3.3. Trametinib and XMD8-92 differentially modulate ERK5 activation in breast cancer
The effects of XMD8-92 and trametinib were evaluated on ERK1/2, ERK5, and RSK activation in MDA-MB-231, BT-549, TU-BcX-4IC and TAMR MCF-7 cells at short time points (Supplemental figure 3) and after 72 hours of treatment (Fig. 4). At 72 hours, XMD8-92 decreased activation of RSK, a downstream target of ERK5 in MDA-MB-231 and TAMR MCF-7 cells but not in BT-549 and TU-BcX-4IC cells. As expected, trametinib significantly decreased ERK1/2 and/or RSK phosphorylation in MDA-MB-231, BT-549, TU-BcX-4IC and TAMR MCF-7 cells (Fig. 4). p-P90RSK protein expression was undetected in WT-MCF-7 cells (Supplemental figure 2C).
Fig. 4.
Western blot analysis of ERK5, ERK1/2, and RSK activation in TNBC cells. (A) MDA-MB-231, (B) BT-549, (C) TU-BcX-4IC, and (D) TAMR MCF-7 cells. Data represent the ± SEM of three different experiments for each inhibitor compared to DMSO control. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 vs DMSO control group determined by one-way ANOVA with the Bonferroni post hoc test.
Surprisingly, XMD8-92 did not decrease ERK5 activation at 72 hours in any model (Fig. 4). Therefore, ERK5 activation may be an early event that leads to alterations in cell signaling downstream at later time points. To examine this, cells were serum starved for 18-24 hours, treated with an inhibitor for 30 minutes, and then with epidermal growth factor (EGF) for 15 minutes. XMD8-92 decreased EGF-mediated ERK5 activation in MDA-MB-231 and TAMR-MCF-7 cells, but not in BT-549 or TU-BcX-4IC cells (Supplemental figure 3) which is consistent with the effects of XMD8-92 on RSK phosphorylation at 72 hours. Interestingly, XMD8-92 activated ERK1/2 in MDA-MB-231 cells compared to DMSO+EGF treatment control at short time points. This may be due a compensatory upregulation of ERK1/2 activity due to inhibition of ERK5 activation.
Trametinib significantly inhibited ERK1/2 activation at 72 hours in all cell types studied (Fig. 4). Interestingly, trametinib did not significantly decrease RSK phosphorylation in BT-549 cells (Fig. 4B). This may be because these cells are inherently dependent on alternative signaling pathways for RSK activation or EMT. Moreover, it is possible that AKT activation may mediate resistance to kinase inhibition in these cells, since the crosstalk between the MAPK and AKT pathways has been noted previously [24]. These data may explain why trametinib only partially induced MET in these cells. Trametinib also increased ERK5 phosphorylation in TU-BcX-4IC cells at 72 hours (Fig. 4C). Again, this may be a compensatory upregulation of ERK5 and/or indicate that ERK5 plays a lesser role in the EMT in these cells.
Trametinib significantly decreased ERK1/2, ERK5, and RSK phosphorylation in response to EGF stimulation in MDA-MB-231 cells in a dose-dependent manner at short time points (Supplemental figure 3A). However, trametinib significantly decreased ERK1/2 and RSK phosphorylation, but not ERK5 phosphorylation, in BT-549, TU-BcX-4IC, and TAMR MCF-7 cells (Supplemental figure 3B-D). These data indicate that trametinib may be a dual inhibitor of ERK1/2 and ERK5 pathways in MDA-MB-231 cells, but not in BT-549, TU-BcX-4IC, and TAMR MCF-7 cells.
3.4. Effects of XMD8-92 and trametinib on cell migration and proliferation in breast cancer
EMT is known to promote cell migration [[25], [26]]. XMD8-92 decreased cell migration in MDA-MB-231 and TAMR-MCF-7 cells at 10 μM concentration (Fig. 5A-D). Trametinib significantly decreased cell migration in TNBC and TAMR breast cancer cells (Fig. 5A-D). XMD8-92 produced a significant decrease in proliferative fraction of MDA-MB-231, TU-BcX-4IC, and TAMR-MCF-7 cells (Fig. 5E, G-H) but not in BT-549 cells (Fig. 5F). The immunostaining pictures for cell proliferation are similar to the morphology pictures shown in Fig. 3 with Ki67 added as the proliferation marker.
Fig. 5.
XMD8-92 and trametinib differentially decrease cell migration and proliferation in diverse breast cancer subtypes. (A) MDA-MB-231, (B) BT-549 cells, (C)TU-BcX-4IC, and (D) TAMR MCF-7 cells were treated with the kinase inhibitors and scratches were made after 48 hours of treatment. Cells were imaged at the time of scratch (0 h) and after 24 hours from the time of scratch (72 h) (20X magnification). Cell migration was measured as a percentage of DMSO control group. (E) MDA-MB-231, (F) BT-549, (G) TU-BcX-4IC and (H) TAMR MCF-7 cells were treated with XMD8-92 or trametinib for 72 hours (20X magnification). Proliferative fraction was evaluated as the number of Ki67 positive cells divided by the number of Hoechst positive cells. Data represent the ± SEM of three different experiments. *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 vs. DMSO control group determined by one-way ANOVA with the Bonferroni post hoc test.
3.5. Trametinib decreases nuclear ERK5 in MDA-MB-231, but not in BT-549 cells
ERK5 has a large C-terminal domain, which can facilitate its nuclear localization in response to growth factors or via autophosphorylation [27]. The location specific roles of ERK5 remain largely understudied. To understand differences in the signaling pathway across TNBC subtypes, we selected two models: MDA-MB-231 driven by mutations in RAF, leading to activation of the MEK-ERK pathway activation and BT-549 cells driven by loss of PTEN and subsequent increase in the PI3K-AKT pathway activation. MDA-MB-231 and BT-549 cells were treated with DMSO, 1 μM XMD, and 0.1 μM trametinib for 72 hours. ERK5 was found to be basally active and localized in the nucleus as well as the cytosol of MDA-MB-231 and BT-549 TNBC cells (Fig. 6). ERK1/2 was mainly localized in the cytosol of the TNBC cells and its activation or expression was not altered with XMD8-92. Trametinib did not decrease ERK5 activation but decreased the total expression in the nucleus as well as the cytosol of MDA-MB-231 cells, while it only decreased ERK5 activation in the cytosol of BT-549 cells (Fig. 6A, B). As expected, trametinib but not XMD8-92 significantly decreased ERK1/2 activation in the cytosol in MDA-MB-231 and BT-549 cells. Total ERK1/2 expression was not altered with either treatment in the cytosol of either cell line. We wanted to further evaluate the effects of dual ERK1/2 and ERK5 pathway activation or inhibition on EMT using lentivirus.
Fig. 6.
Effect of XMD8-92 and trametinib on ERK5 and ERK1/2 activation in the nucleus and cytoplasm. (A) MDA-MB-231 nuclear fraction (B) MDA-MB-231 cytosolic fraction (C) BT-549 nuclear fraction (D) BT-549 cytosolic fraction (72 h). *p<0.05; ***p<0.001 vs control group determined by one-way ANOVA with the Bonferroni post hoc test.
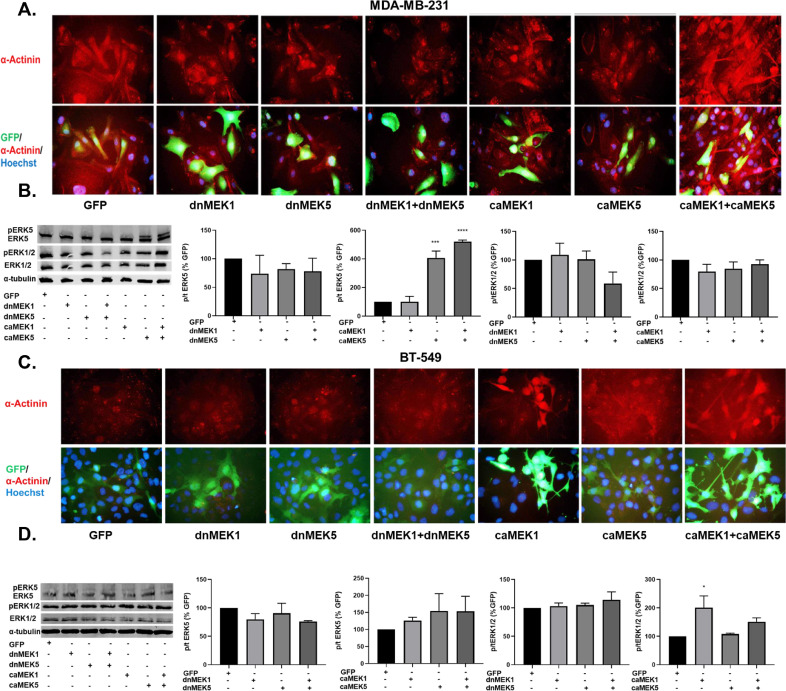
3.6. Diverse and converging roles of MEK1 and MEK5 on EMT and kinase activation in TNBC cells
To further examine the roles of the ERK1/2 and ERK5 pathways on MET and kinase activation, MDA-MB-231 and BT-549 cells were treated with dominant negative (dn) and constitutively active (ca) lentivirus vectors of MEK1 and MEK5 (generous gift from Dr. Zhengui Xia). The cells were transiently co-infected with GFP-tagged dnMEK1, dnMEK5, caMEK1, and/or caMEK5 lentivirus vectors as indicated for 96 hours. The morphology of infected cells was assessed via immunostaining for the cytoskeletal protein α-actinin (Fig. 7A). Cells that were infected with dnMEK1 and dnMEK5 alone or in combination displayed a phenotypic shift from a mesenchymal to epithelial. caMEK1, caMEK5, and caMEK5+caMEK1 treatments increased the mesenchymalization of MDA-MB-231 and BT-549 cells (Fig. 7A, C). caMEK5 and caMEK1+caMEK5 significantly increased ERK5 activation in MDA-MB-231 cells (Fig. 7B). There were no significant decreases or increases in ERK1/2 phosphorylation in MDA-MB-231 cells. dnMEK1 and dnMEK5 did not significantly decrease ERK5 or ERK1/2 in BT-549 cells; however, caMEK1 and caMEK1+caMEK5 groups significantly increased ERK1/2 activation in BT-549, but not MDA-MB-231 cells (Fig. 7D).
Fig. 7.
MEK1 and MEK5 activation mediates EMT in TNBC cells. (A, B) MDA-MB-231 and (C, D) BT-549 cells were treated with dnMEK5, dnMEK1, caMEK5, and caMEK1 alone and in combination as represented in the figure. The cells were incubated for 96 hours. Immunofluorescence staining for α-actinin was performed to assess the morphology (40X magnification). The effect on ERK1/2 and ERK5 activation was evaluated. *p<0.05; **p<0.01 vs GFP control group determined by one-way ANOVA with the Bonferroni post hoc test.
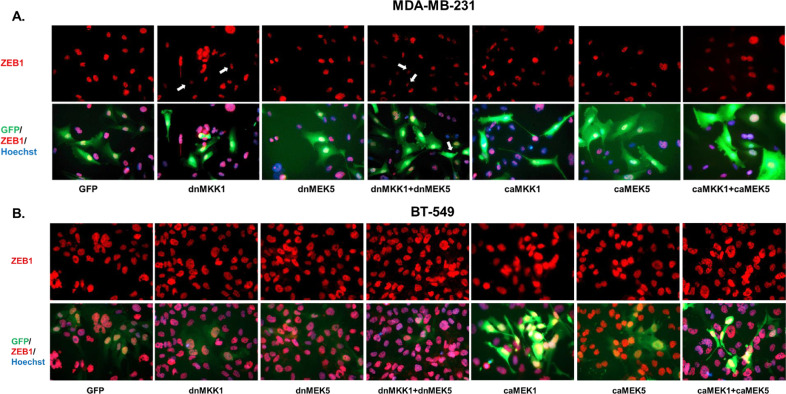
3.7. MEK1 and MEK5 pathways regulate ZEB1 expression in TNBC cells
Next, we wanted to examine cell-specific responses to MEK1/2 and MEK5 pathway inhibition and activation on EMT. Therefore, MDA-MB-231 and BT-549 cells infected with dnMEK1, dnMEK5, caMEK1, and caMEK5 lentivirus vectors alone and in combination were assessed for decreases and increases in ZEB1 expression by immunofluorescence. MDA-MB-231 cells that were infected with dnMEK1, dnMEK5, and dnMEK1+dnMEK5 vectors (GFP+ cells) had an attenuated ZEB1 expression (Fig. 8A). While cells infected with caMEK1, caMEK5, and caMEK1+caMEK5 groups had a more pronounced mesenchymal morphology compared to GFP control, there were no increases in ZEB1 expression (Fig. 8A). BT-549 cells that were infected with dnMEK1, dnMEK5, and dnMEK1+dnMEK5 groups appeared epithelial but had no reduction in ZEB1 expression (Fig. 8B). caMEK1, caMEK5, and caMEK1+caMEK5-infected cells had a more pronounced mesenchymal morphology and showed an increase in ZEB1 expression (Fig. 8B).
Fig. 8.
MEK1 and MEK5 activation mediates ZEB1 expression in TNBC cells. (A) MDA-MB-231 and (B) BT-549 cells were treated with dnMEK5, dnMEK1, caMEK5, and caMEK1 alone and in combination as represented in the figure. The cells were incubated for 96 hours. Immunofluorescence staining for ZEB1 was performed (40X magnification).
MDA-MB-231 and BT-549 cells were treated with 0.1 μM trametinib in the presence of GFP or caMEK5 lentivirus to examine the effects on cell morphology, E-cadherin, and ZEB1 expression. MDA-MB-231 cells treated with trametinib, which transitioned to an epithelial phenotype, had a reduction in ZEB1 expression. The reduction in ZEB1 was rescued in cells that were infected with caMEK5 as determined by immunofluorescence (Supplemental figure 4). While caMEK5 did not inhibit trametinib-mediated increases in E-cadherin expression (Supplemental Figure 5), it did reduce trametinib-mediated decrease in ZEB1 expression as determined by western blotting in MDA-MB-231 cells (Supplemental Figure 5A-B) but not in BT-549 cells (Supplemental Figure 5C-D). Putative signaling mechanisms driving EMT in TNBC are outlined (Supplemental Figure 5D, H).
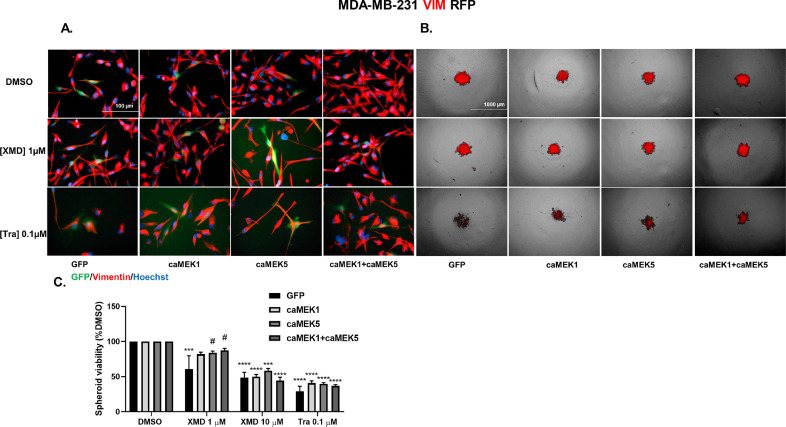
3.8. MEK1 and/or MEK5 activation reduces the ability of trametinib to decrease vimentin expression in MDA-MB-231 VIM RFP 2D and spheroid cultures
The effect of dual ERK1/2 and ERK5 pathway inhibition on MET was found to be most promising in MDA-MB-231 cells. Therefore, to strengthen the functional contribution of inhibiting the ERK1/2 and ERK5 pathways in MET, MDA-MB-231 VIM RFP cells, a new model for MET research, were infected with caMEK1 and/or caMEK5 in the presence of DMSO, trametinib, or XMD8-92. These cells have been transformed to constitutively express vimentin, a mesenchymal marker via CRISPR-knock-in system and serve as a good model to study MET. MET was examined via observing vimentin expression in 2D and spheroid cultures as well as spheroid viability after treatment with constitutively active MEK isoforms in the presence or absence of MAPK inhibitors. Treatment with caMEK1, caMEK5, and caMEK1+caMEK5 increased vimentin expression (Fig. 9A). While treatment with XMD8-92 alone did not reduce vimentin expression, treatment with trametinib moderately decreased vimentin expression, specifically in cells that underwent a MET as determined by examining morphology of GFP+ cells via microscopy (Fig. 9A). Treatment with constitutively active MEK1, MEK5, and MEK1+caMEK5 reduced the ability of trametinib to decrease vimentin expression.
Fig. 9.
MEK1 and/or MEK5 activation reduces the ability of XMD8-92 or trametinib to decrease spheroid viability or vimentin expression in MDA-MB-231 VIM RFP model. (A) MDA-MB-231 VIM RFP cells were treated with constitutively active MEK1, MEK5, and MEK1+MEK5 in the presence of DMSO, XMD8-92, or trametinib for 72 hours. The cells were fixed and stained with Hoechst. Images of Vimentin-, GFP-, and Hoechst-expressing cells were captured under 40X magnification using EVOS microscope (n=3, most representative image shown). (B) MDA-MB-231 VIM RFP cells were treated with constitutively active MEK1, MEK5, and MEK1+MEK5 in the presence of DMSO, XMD8-92, or trametinib for 96 hours. Images of spheroids under transmitted light and RFP channel were captured under 4X magnification using EVOS microscope (n=3, most representative image shown). (C) Spheroid viability was assessed after 7 days of treatment with the same groups. Data indicate ± SEM of experiments run in triplicate. ***p<0.001; ****p<0.0001 vs DMSO control group, #p<0.05; vs individual drug+GFP determined by two-way ANOVA with the Bonferroni post hoc test.
In spheroid culture, trametinib but not XMD8-92 reduced expression of vimentin at 96 hours, which was rescued in the presence of caMEK1, caMEK5, and caMEK1+caMEK5 groups (Fig. 9B). Spheroid viability was assessed after 7 days of treatment (Fig. 9C). There was no baseline difference in spheroid viability after treatment with caMEK1, caMEK5, or caMEK1+caMEK5 groups. This may indicate that the spheroid-forming ability of MDA-MB-231 VIM RFP cells is at its maximum and cannot be increased further. XMD8-92 significantly decreased spheroid viability at 1 and 10 µM concentrations. As expected, the reduction in spheroid viability at 1µM XMD8-92 concentration was rescued by co-treatment with caMEK5 or caMEK1+caMEK5 groups. The rescue effect by caMEK5 or caMEK1+caMEK5 groups was reversed in the presence of higher XMD8-92 concentration (10µM). While trametinib significantly decreased spheroid viability at 0.1µM concentration, these effects were not reversed in the presence of caMEK1, caMEK5, and caMEK1+caMEK5 groups (Fig. 9C). Pictures of vimentin-expressing spheroids at day 0, day 7, and evidence of lentivirus infection measured by examining GFP expression in spheroids are included in Supplemental Fig. 6.
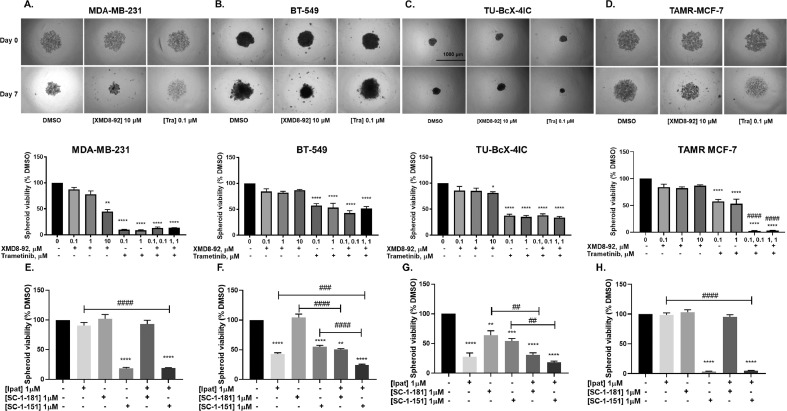
3.9. Effect of dual ERK1/2 and ERK5 pathway inhibition on spheroid formation and ipatasertib sensitivity in breast cancer
EMT is known to promote anchorage-independent growth [25,26]. We found that trametinib alone significantly decreased spheroid viability and/or cell viability in all breast cancer models and its effects were most pronounced in MDA-MB-231 cells (Fig. 10A-D). While XMD only decreased spheroid viability in MDA-MB-231 cells, Tra+XMD combination did not produce a greater effect on spheroid or cell viability compared to individual drugs in TNBC cells (Fig. 10 and Supplemental Fig. 7). However, in contrast to TNBC cells, Tra+XMD combination was effective in producing a synergistic inhibition of spheroid viability in TAMR MCF-7 cells (Fig. 10 and Supplemental Fig. 7). XMD8-92 did not significantly decrease spheroid viability in BT-549, TU-BcX-4IC, and TAMR-MCF-7 cells (Fig. 10B-D). The reduction in spheroid viability in response to trametinib was greater in MDA-MB-231 cells (~90%) compared to BT-549 (~40%), TU-BcX-4IC (~35%) or TAMR-MCF-7 cells (~40%). Moreover, the spheroids were treated with novel MAPK inhibitors SC-1-151 (dual MEK1/2 and MEK5 inhibitor) and SC-1-181 (MEK5 inhibitor) in combination with ipatasertib, an AKT inhibitor. Ipatasertib did not decrease spheroid viability but its effect was potentiated by MEK inhibitors in MDA-MB-231 and TAMR MCF-7 cells (Fig. 10A, C). Ipatasertib alone and in combination with MEK inhibitors synergistically decreased spheroid formation in BT-549 and TU-BcX-4IC cells (Fig. 10B, D).
Fig. 10.
Effect of ERK1/2 and ERK5 inhibition alone and together on spheroid viability and ipatasertib sensitivity in diverse breast cancer subtypes. The spheroids were treated with increasing concentrations of XMD8-92 and/or trametinib for 7 days. Pictures of spheroids were obtained before treatment and 7 days after treatment (4X magnification) (A) MDA-MB-231, (B) BT-549, and (C) TU-BcX-4IC, and (D) TAMR MCF-7 cells. Effect of SC-1-151 and SC-1-181 alone and in combination with ipatasertib on spheroid viability in (E) MDA-MB-231, (F) BT-549, (G) TU-BcX-4IC and (H) TAMR MCF-7 cells. Spheroid viability was assessed on day 7 after treatment. Data indicate ± SEM of experiments run in triplicate *p<0.05; **p<0.01; ***p<0.001; ****p<0.0001 vs DMSO control group, ##p<0.01; ###p<0.001; ####p<0.0001 vs individual drug determined by one-way ANOVA with the Bonferroni post hoc test.
4. Discussion
Mesenchymal cancer cells are migratory and invasive, leading to metastases. There are currently no effective treatments for metastases. Interestingly, activation of the ERK1/2 and ERK5 signaling pathways leads to an epithelial to mesenchymal transition (EMT) and poor patient survival, in several cancers, including TNBC and endocrine-resistant breast cancers [11–13, [28], [29], [30], [31], [32]–33]. Genomics data from our research indicates that MAPK3 and MAPK7 gene expression significantly correlated with mesenchymal marker VIM or ZEB1 but not with epithelial marker CDH1 (Fig. 1A and C). MAPK1 positively correlated with mesenchymal markers VIM and ZEB1 and epithelial marker CDH1 (E-cadherin), indicating that MAPK1 may mediate an intermediate epithelial/mesenchymal state where both epithelial and mesenchymal markers are co-expressed (Fig. 1B). These data suggest that ERK1/2 and ERK5 are relevant targets for treatment of TNBC. However, the relative roles of these pathways in inducing the MET in these cancers is unknown. Understanding of the tumor biology and response to therapy is further complicated due to crosstalk between the ERK1/2 and ERK5 pathways and different functions of ERK5 in the nucleus versus the cytosol. To our knowledge, this is the first study to examine the independent and overlapping roles of the ERK1/2 and ERK5 signaling cascades on MET, nuclear localization of ERK5, cell migration, proliferation, and spheroid formation in breast cancer.
In the current study, trametinib, a clinically relevant MEK1/2 inhibitor, and XMD8-92, an ERK5 inhibitor, induced a MET in MDA-MB-231 TNBC cells as shown by morphological characteristics, increased expression of E-cadherin, and/or decrease in ZEB1 (Fig. 3A). In BT-549, TU-BcX-4IC, and TAMR MCF-7 cells, treatment with trametinib, but not XMD8-92, resulted in an epithelial-like morphology (Fig. 3B-D). However, the morphological changes induced by trametinib in BT-549 cells were less pronounced than those in MDA-MB-231 cells, probably because trametinib inhibited ZEB1 in 231 cells, but not in BT-549 cells. The morphological changes induced by trametinib in the 231 cells were more pronounced than those induced by XMD8-92. This may be because trametinib increased E-cadherin expression and decreased ZEB1 expression, while XMD8-92 only reduced ZEB1 expression in 231 cells. Overall, these data suggest that inhibition of the ERK1/2 pathway alone is sufficient to induce a MET in BT-549, TU-BcX-4IC, and TAMR MCF-7 cells. Additionally, as trametinib consistently increased E-cadherin in TNBC and TAMR breast cancer models, E-cadherin may be used as a potential biomarker to predict MET induced by trametinib treatment in metastatic cancers. The inhibitors that induced a partial MET, as determined by ZEB1 and E-cadherin correlation, would have to be combined with additional EMT suppressors in future since a partial MET state is a predictor of metastases and poor patient survival [34,35].
Consistent with the effect on MET, trametinib decreased cell migration in TNBC and TAMR MCF-7 cells, suggesting that ERK1/2 inhibition is sufficient to decrease cell migration in these cells. At 10 μM concentration, XMD8-92 decreased cell migration only in MDA-MB-231 cells. This observation is consistent with reduction in ZEB1 and induction of MET following treatment with XMD8-92 in MDA-MB-231 cells. ERK1/2 and ERK5 activation are known to mediate cell proliferation by mediating G1-S transition during the cell cycle via distinct effects on cyclinD1 expression and activation [36]. Trametinib decreased cell proliferation by 80% in TAMR MCF-7 cells. However, in MDA-MB-231 cells, trametinib only decreased cell proliferation by ~50%. Although not statistically significant, this decrease in cell proliferation by trametinib may be biologically relevant. Moreover, MEK1/2 inhibition decreased colony formation by 20% and ERK5 inhibition did not decrease colony formation in MDA-MB-231 cells (data not shown). The effect of XMD8-92 on cell proliferation was evident only at the highest dose (10 μM) in MDA-MB-231 and TAMR MCF-7 cells. We have previously shown that effects of a high XMD8-92 dose could be recapitulated by the addition of AX15836 (ERK5 inhibitor) and CPI203 (BRD4 inhibitor) [24]. Therefore, at high doses, in addition to ERK5 inhibition, XMD8-92 may have off-target effects including inhibition of bromodomain (BRD)4 [37]. Trametinib or XMD8-92 did not decrease cell proliferation in BT-549 cells, which may indicate that alternative pathways mediate cell proliferation in these cells. As indicated above, BT-549 cells are PTEN mutant cells and may rely more on the AKT pathway for survival and proliferation.
XMD8-92 is a well-known ERK5 inhibitor [38]. We have seen previously that XMD8-92 inhibits ERK5 at as low as 3 or 5 μM concentration [24]. In this study, we have data that show that ERK5 activation by EGF is inhibited significantly in the presence of 1 μM XMD8-92 in MDA-MB-231 and TAMR-MCF-7 cells. Therefore, the lack of effect of XMD8-92 on ERK5 in BT-549 and TU-BcX-4IC cells may be due to compensatory increase in the AKT pathway or paradoxical activation and nuclear translocation of ERK5. Effect of trametinib was also less pronounced on RSK inhibition in these cells. It is also possible that inherently constitutively active AKT in BT-549 cells may be causing resistance to MAPK inhibitors. Above 10 μM concentration, XMD8-92 has toxic effects on cells as well as off-target effect on BRD4 and hence would confound data interpretation.
We observed that MDA-MB-231 cells were the most responsive to the effects of XMD8-92 and trametinib on MET, whereas BT-549 cells were the least responsive, which led us to characterize differences in cellular signaling between the two TNBC models. We wanted to characterize dual role of MEK1/2 and MEK5 pathways on EMT, nuclear localization of ERK5, and their relation to the PI3K-AKT pathway. We first examined the effects of trametinib and XMD on nuclear localization of ERK5. To further explore our hypothesis that inhibition of both the ERK1/2 and ERK5 pathways are necessary to induce the MET in TNBC, MDA-MB-231 and BT-549 cells were infected with dominant negative (dn) and constitutive active (ca) MEK1 and/or MEK5. We believe that this is the first study to examine nuclear localization of ERK5 in TNBC.
We found that the ERK5 inhibitor did not decrease nuclear ERK5 activation or total expression. This observation is consistent with a recent study, which has shown that ERK5 inhibitors that target the kinase domain were shown to activate the transcriptional activation domain (TAD) of ERK5, resulting in the nuclear localization and increased transcriptional activity of ERK5 [39]. This may explain why our data with respect to the effect of ERK5 inhibition on E-cadherin conflicts with studies that have shown that inhibition of ERK5 via knockdown or knockout enhances E-cadherin expression in several cancer models [40,41]. Since trametinib decreased nuclear ERK5, ERK1/2 activation may be a putative mechanism for the translocation of ERK5 into the nucleus in MDA-MB-231 cells. ERK1/2 has been shown previously to promote ERK5 translocation to the nucleus in response to growth factor stimulation [42]. However, constitutively active RAF may be responsible for constitutive ERK1/2 activation and subsequent translocation of ERK5 in the nucleus of MDA-MB-231 cells under unstimulated condition. We are currently investigating mechanisms for ERK5 nuclear translocation in BT-549 cells. These findings were further supported by Fig. 7B, where constitutively active MEK1 and MEK5 lentivirus vectors significantly increased ERK5 activation in MDA-MB-231 cells, but not in BT-549 cells.
Although phenotypic shifts were noted following infection of the ca or dn MEK, the morphological transitions were more pronounced when both pathways were activated or inhibited in MDA-MB-231 and BT-549 cells. This was supported by decrease in ZEB1 expression in MDA-MB-231 cells following dnMEK1 and/or MEK5 infection and increase in ZEB1 expression in BT-549 cells following caMEK1 and/or MEK5 infection (Fig. 8). It is possible that ZEB1 expression is maximum in MDA-MB-231 cells and could not be induced further. The more pronounced mesenchymal morphology following caMEK5 and caMEK1+caMEK5 infection may be a result of increase in vimentin expression (Fig. 7) or ERK5 activation (Fig. 7B) and its association with the actin cytoskeleton as previously described [18]. Moreover, ERK5 activation increased more significantly in caMEK1 + caMEK5 group vs GFP than caMEK5 group vs GFP (Fig. 7B), indicating that ERK5 may be activated by both MEK1 and MEK5 signaling. This observation supports the data that show that trametinib, a known MEK1/2 inhibitor, decreased ERK5 activation in MDA-MB-231 cells. We hypothesize that the effect of trametinib on ERK5 inhibition was mediated via MEK1/2 inhibition and not by direct binding of trametinib to MEK5 or ERK5. Since ERK1/2 and ERK5 share 50% sequence homology at the N-terminal domain, it is possible that MEK1/2 may phosphorylate ERK5 by direct binding. It is also possible that ERK1/2 may phosphorylate ERK5 at its C-terminal by direct interaction, as reported previously [42]. These data further support the conclusion that inhibition or activation of both pathways is necessary for the MET or EMT, respectively. These data, together with the effect of trametinib and XMD8-92 on morphology, suggest that inhibition of both the ERK1/2 and ERK5 pathways is necessary to induce a full MET in TNBC cells.
Reduction in trametinib-mediated decrease in ZEB1 expression in the presence of caMEK5 suggests that trametinib mediates its effect on MET via dual ERK1/2 and ERK5 inhibition. However, the trametinib-mediated increase in E-cadherin expression was not decreased by caMEK5, which may indicate that trametinib induces E-cadherin expression via ERK1/2 inhibition alone and not ERK5 inhibition (Supplemental Fig. 5). This observation further supports the data that suggest there is no change in E-cadherin expression following XMD8-92 treatment in MDA-MB-231 cells. While trametinib caused an overall decrease in ZEB1 expression in BT-549 cells with an epithelial morphology, caMEK5 did not decrease its ability to induce MET as determined by examining cell morphology, E-cadherin protein expression by western blotting, and ZEB1 expression via western blotting and immunofluorescence. This further supports that ERK1/2 inhibition but not ERK5 inhibition induces MET in BT-549 cells. The role of total ERK5 expression in modulating EMT in BT-549 cells needs to be further evaluated. We also observed that there was an overall reduction in ZEB1 expression in cells with an epithelial morphology following treatment with trametinib in both the TNBC cell lines and there was no cell population that had a complete loss of ZEB1. Such fine-tuning could be advantageous to avoid catastrophic side effects on healthy mesenchymal cells in the body, which depend on ZEB1 for their normal function.
The functional contribution of ERK1/2 and ERK5 pathways to vimentin expression in 2D cultures and spheroids was evaluated in MDA-MB-231 cells. Trametinib moderately decreased vimentin expression, in MDA-MB-231 VIM RFP cells in 2D culture (Fig. 9A), but completely inhibited vimentin expression in spheroid culture (Fig. 9B), indicating that treatment with trametinib may be specifically important in targeting mesenchymal and anoikis-resistant cells. Moreover, trametinib-mediated decrease in spheroid viability was not rescued by caMEK1, caMEK5, and caMEK1+caMEK5 groups at 7 day (Fig. 9C). This indicates that the reduction in vimentin expression in spheroids was not due to a decrease in spheroid viability. Since these effects of trametinib were partially reversed by co-treatment with constitutively active MEK1 and/or MEK5, ERK5, in addition to ERK1/2 may be co-regulated by the MEK1/2 pathway in MDA-MB-231 cells. XMD8-92 significantly decreased the spheroid viability at 1µM concentration, which was rescued by caMEK5 and caMEK1+caMEK5 groups indicating that ERK5 has a crucial role in regulating the survival of anoikis-resistant spheroids (Fig. 9C). While ERK1/2 activation has a greater role in regulating the EMT in spheroids, ERK5 activation regulates the survival of these anoikis-resistant spheroids.
As previously described, some single-agent inhibitors of MAPK pathway(s) led to an intermediate E/M state, which may be a predictor of metastases and poor prognosis. Therefore, combination strategy for MAPK inhibitors needs to be developed. We examined the effect of dual ERK1/2 and ERK5 pathway inhibition on spheroid viability, an assay representative of EMT. MAPK gene alterations, including ESR1 overamplification is common in endocrine-resistant breast cancer [43]. This explains why Tra and XMD combination produced greater inhibition of spheroid and cell viability compared to either drug alone in TAMR MCF-7 cells (Fig. 10 and Supplemental figure 7D).
As crosstalk between the ERK and AKT signaling pathways has been noted in TNBC,[24] the AKT pathway may be mediating the resistance to MAPK inhibitors in TNBC. Therefore, the effect of MAPK pathway inhibition in combination with AKT inhibition was evaluated on spheroid viability in TNBC and TAMR MCF-7 cells. We have previously shown that a novel, dual inhibitor (SC-1-151/compound 1) of MEK1/2 and MEK5, the upstream kinases of ERK1/2 and ERK5, respectively, was effective in inducing an mesenchymal to epithelial transition (MET) in triple negative and TAMR breast cancer [21,20]. SC-1-151-like effects on spheroid viability were recapitulated by Tra+XMD combination treatment in diverse breast cancer subtypes (Fig. 10). Consistent with the effects on MET, trametinib decreased spheroid formation in MDA-MB-231 cells to a greater extent compared to BT-549 or TAMR MCF-7 cells. This difference may be because trametinib, like SC-1-151, inhibits both ERK1/2 and ERK5 activation by EGF in MDA-MB-231 cells, but only inhibits the ERK1/2 activation in BT-549, TU-BcX-4IC, and TAMR MCF-7 cells. We found that dual ERK1/2 and ERK5 inhibition was sufficient to decrease spheroid formation in MDA-MB-231 and TAMR-MCF-7 cells whereas additional inhibition of AKT was necessary further inhibit spheroid formation in BT-549 and TU-BcX-4IC cells. The current study is the first to examine the distinct and overlapping roles of the ERK1/2 and ERK5 pathways on breast cancer MET and their relation to the AKT signaling with respect to EMT. Overall, the data from our research validates ERK1/2 and ERK5 as important therapeutic targets not only in triple-negative breast cancer but also in other aggressive forms of breast cancers such as inflammatory or tamoxifen-resistant breast cancer.
Declaration of Competing Interest
The authors have no conflicts of interests.
Acknowledgments
Acknowledgements
We would like to thank Dr. Zhengui Xia and Glen Abel (University of Washington, Seattle) for kindly sharing the dn and ca MEK1 and MEK5 plasmids.
Funding
This study was supported by the Department of Pharmacology at Duquesne University and NIH grant R15CA176496 awarded to Cavanaugh JE.
Footnotes
This article was transferred from the feeder journal Neoplasia before peer review, and the original manuscript No. is NEO-D-20-00490. Authors are told to contact OncologyOAtransfers@Elsevier.com for fully waiving their APC.
Supplementary material associated with this article can be found, in the online version, at doi:10.1016/j.tranon.2021.101021.
Appendix. Supplementary materials
References
- 1.Chaffer C.L., Weinberg R.A. A perspective on cancer cell metastasis. Science (New York, N.Y.) 2011;331(6024):1559–1564. doi: 10.1126/science.1203543. [DOI] [PubMed] [Google Scholar]
- 2.Thiery J.P., Sleeman J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006;7(2):131–142. doi: 10.1038/nrm1835. [DOI] [PubMed] [Google Scholar]
- 3.Kalluri R., Weinberg R.A. The basics of epithelial-mesenchymal transition. J. Clin. Invest. 2009;119(6):1420–1428. doi: 10.1172/JCI39104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oka H., Shiozaki H., Kobayashi K., Inoue M., Tahara H., Kobayashi T., Takatsuka Y., Matsuyoshi N., Hirano S., Takeichi M. Expression of E-cadherin cell adhesion molecules in human breast cancer tissues and its relationship to metastasis. Cancer Res. 1993;53(7):1696–1701. [PubMed] [Google Scholar]
- 5.Moll R., Mitze M., Frixen U.H., Birchmeier W. Differential loss of E-cadherin expression in infiltrating ductal and lobular breast carcinomas. Am. J. Pathol. 1993;143(6):1731–1742. [PMC free article] [PubMed] [Google Scholar]
- 6.Pal S.K., Childs B.H., Pegram M. Triple negative breast cancer: unmet medical needs. Breast Cancer Res. Treat. 2011;125(3):627–636. doi: 10.1007/s10549-010-1293-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.De Laurentiis M., Cianniello D., Caputo R., Stanzione B., Arpino G., Cinieri S., Lorusso V., De Placido S. Treatment of triple negative breast cancer (TNBC): current options and future perspectives. Cancer Treat. Rev. 2010;36(3):S80–S86. doi: 10.1016/S0305-7372(10)70025-6. Suppl. [DOI] [PubMed] [Google Scholar]
- 8.Chang M. Tamoxifen resistance in breast cancer. Biomol. Ther. (Seoul) 2012;20(3):256–267. doi: 10.4062/biomolther.2012.20.3.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hiscox S., Jiang W.G., Obermeier K., Taylor K., Morgan L., Burmi R., Barrow D., Nicholson R.I. Tamoxifen resistance in MCF7 cells promotes EMT-like behaviour and involves modulation of beta-catenin phosphorylation. Int. J. Cancer. 2006;118(2):290–301. doi: 10.1002/ijc.21355. [DOI] [PubMed] [Google Scholar]
- 10.Yuan J., Liu M., Yang L., Tu G., Zhu Q., Chen M., Cheng H., Luo H., Fu W., Li Z., Yang G. Acquisition of epithelial-mesenchymal transition phenotype in the tamoxifen-resistant breast cancer cell: a new role for G protein-coupled estrogen receptor in mediating tamoxifen resistance through cancer-associated fibroblast-derived fibronectin and β1-integrin signaling pathway in tumor cells. Breast Cancer Res. BCR. 2015;17(1):69. doi: 10.1186/s13058-015-0579-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gee J.M.W., Robertson J.F.R., Ellis I.O., Nicholson R.I. Phosphorylation of ERK1/2 mitogen-activated protein kinase is associated with poor response to anti-hormonal therapy and decreased patient survival in clinical breast cancer. Int. J. Cancer. 2001;95(4):247–254. doi: 10.1002/1097-0215(20010720)95:4<247::aid-ijc1042>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 12.Adeyinka A., Nui Y., Cherlet T., Snell L., Watson P.H., Murphy L.C. Activated mitogen-activated protein kinase expression during human breast tumorigenesis and breast cancer progression. Clin. Cancer Res. 2002;8(6):1747. [PubMed] [Google Scholar]
- 13.Antoon J.W., Martin E.C., Lai R., Salvo V.A., Tang Y., Nitzchke A.M., Elliott S., Nam S.Y., Xiong W., Rhodes L.V., Collins-Burow B., David O., Wang G., Shan B., Beckman B.S., Nephew K.P., Burow M.E. MEK5/ERK5 signaling suppresses estrogen receptor expression and promotes hormone-independent tumorigenesis. PloS one. 2013;8(8):e69291. doi: 10.1371/journal.pone.0069291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hoang V.T., Yan T.J., Cavanaugh J.E., Flaherty P.T., Beckman B.S., Burow M.E. Oncogenic signaling of MEK5-ERK5. Cancer Lett. 2017;392:51–59. doi: 10.1016/j.canlet.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ali S., Rasool M., Chaoudhry H.N., Pushparaj P., Jha P., Hafiz A., Mahfooz M., Abdus Sami G., Azhar Kamal M., Bashir S., Ali A., Sarwar Jamal M. Molecular mechanisms and mode of tamoxifen resistance in breast cancer. Bioinformation. 2016;12(3):135–139. doi: 10.6026/97320630012135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sinh N.D., Endo K., Miyazawa K., Saitoh M. Ets1 and ESE1 reciprocally regulate expression of ZEB1/ZEB2, dependent on ERK1/2 activity, in breast cancer cells. Cancer Sci. 2017;108(5):952–960. doi: 10.1111/cas.13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shin S., Dimitri C.A., Yoon S.-O., Dowdle W., Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Molecul. Cell. 2010;38(1):114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barros J.C., Marshall C.J. Activation of either ERK1/2 or ERK5 MAP kinase pathways can lead to disruption of the actin cytoskeleton. J. Cell Sci. 2005;118(8):1663. doi: 10.1242/jcs.02308. [DOI] [PubMed] [Google Scholar]
- 19.Buonato J.M., Lazzara M.J. ERK1/2 blockade prevents epithelial-mesenchymal transition in lung cancer cells and promotes their sensitivity to EGFR inhibition. Cancer Res. 2014;74(1):309–319. doi: 10.1158/0008-5472.CAN-12-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhatt A.B., Gupta M., Hoang V.T., Chakrabarty S., Wright T.D., Elliot S., Chopra I.K., Monlish D., Anna K., Burow M.E., Cavanaugh J.E., Flaherty P.T. Novel diphenylamine analogs induce mesenchymal to epithelial transition in triple negative breast cancer. Front. Oncol. 2019;9:672. doi: 10.3389/fonc.2019.00672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chakrabarty S., Monlish D.A., Gupta M., Wright T.D., Hoang V.T., Fedak M., Chopra I., Flaherty P.T., Madura J., Mannepelli S., Burow M.E., Cavanaugh J.E. Structure activity relationships of anthranilic acid-based compounds on cellular and in vivo mitogen activated protein kinase-5 signaling pathways. Bioorgan. Med. Chem. Lett. 2018;28(13):2294–2301. doi: 10.1016/j.bmcl.2018.05.029. [DOI] [PubMed] [Google Scholar]
- 22.Purrington K.S., Knight J., 3rd, Dyson G., Ali-Fehmi R., Schwartz A.G., Boerner J.L., Bandyopadhyay S. CLCA2 expression is associated with survival among African American women with triple negative breast cancer. PloS one. 2020;15(4) doi: 10.1371/journal.pone.0231712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bertucci F., Ueno N.T., Finetti P., Vermeulen P., Lucci A., Robertson F.M., Marsan M., Iwamoto T., Krishnamurthy S., Masuda H., Van Dam P., Woodward W.A., Cristofanilli M., Reuben J.M., Dirix L., Viens P., Symmans W.F., Birnbaum D., Van Laere S.J. Gene expression profiles of inflammatory breast cancer: correlation with response to neoadjuvant chemotherapy and metastasis-free survival. Ann. Oncol. 2014;25(2):358–365. doi: 10.1093/annonc/mdt496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wright T.D., Raybuck C., Bhatt A., Monlish D., Chakrabarty S., Wendekier K., Gartland N., Gupta M., Burow M.E., Flaherty P.T., Cavanaugh J.E. Pharmacological inhibition of the MEK5/ERK5 and PI3K/Akt signaling pathways synergistically reduces viability in triple-negative breast cancer. J. Cell. Biochem. 2019 doi: 10.1002/jcb.29350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cao Z., Livas T., Kyprianou N. Anoikis and EMT: Lethal "Liaisons" during cancer progression. Crit Rev Oncog. 2016;21(3-4):155–168. doi: 10.1615/CritRevOncog.2016016955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Mani S.A., Guo W., Liao M.J., Eaton E.N., Ayyanan A., Zhou A.Y., Brooks M., Reinhard F., Zhang C.C., Shipitsin M., Campbell L.L., Polyak K., Brisken C., Yang J., Weinberg R.A. The epithelial-mesenchymal transition generates cells with properties of stem cells. Cell. 2008;133(4):704–715. doi: 10.1016/j.cell.2008.03.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buschbeck M., Ullrich A. The unique C-terminal tail of the mitogen-activated protein kinase ERK5 regulates its activation and nuclear shuttling. J. Biol. Chem. 2005;280(4):2659–2667. doi: 10.1074/jbc.M412599200. [DOI] [PubMed] [Google Scholar]
- 28.Hoang V.T., Yan T.J., Cavanaugh J.E., Flaherty P.T., Beckman B.S., Burow M.E. Oncogenic signaling of MEK5-ERK5. Cancer Lett. 2017;392:51–59. doi: 10.1016/j.canlet.2017.01.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Olea-Flores M., Zuniga-Eulogio M.D., Mendoza-Catalan M.A., Rodriguez-Ruiz H.A., Castaneda-Saucedo E., Ortuno-Pineda C., Padilla-Benavides T., Navarro-Tito N. Extracellular-signal regulated kinase: a central molecule driving epithelial-mesenchymal transition in cancer. Int. J. Mol. Sci. 2019;(12):20. doi: 10.3390/ijms20122885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buonato J.M., Lazzara M.J. ERK1/2 blockade prevents epithelial–mesenchymal transition in lung cancer cells and promotes their sensitivity to EGFR inhibition. Cancer Res. 2014;74(1):309. doi: 10.1158/0008-5472.CAN-12-4721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith B.N., Burton L.J., Henderson V., Randle D.D., Morton D.J., Smith B.A., Taliaferro-Smith L., Nagappan P., Yates C., Zayzafoon M., Chung L.W., Odero-Marah V.A. Snail promotes epithelial mesenchymal transition in breast cancer cells in part via activation of nuclear ERK2. PloS one. 2014;9(8) doi: 10.1371/journal.pone.0104987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Miranda M., Rozali E., Khanna K.K., Al-Ejeh F. MEK5-ERK5 pathway associates with poor survival of breast cancer patients after systemic treatments. Oncoscience. 2015;2(2):99–101. doi: 10.18632/oncoscience.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pavan S., Meyer-Schaller N., Diepenbruck M., Kalathur R.K.R., Saxena M., Christofori G. A kinome-wide high-content siRNA screen identifies MEK5-ERK5 signaling as critical for breast cancer cell EMT and metastasis. Oncogene. 2018;37(31):4197–4213. doi: 10.1038/s41388-018-0270-8. [DOI] [PubMed] [Google Scholar]
- 34.Yang J., Antin P., Berx G., Blanpain C., Brabletz T., Bronner M., Campbell K., Cano A., Casanova J., Christofori G., Dedhar S., Derynck R., Ford H.L., Fuxe J., García de Herreros A., Goodall G.J., Hadjantonakis A.-K., Huang R.J.Y., Kalcheim C., Kalluri R., Kang Y., Khew-Goodall Y., Levine H., Liu J., Longmore G.D., Mani S.A., Massagué J., Mayor R., McClay D., Mostov K.E., Newgreen D.F., Nieto M.A., Puisieux A., Runyan R., Savagner P., Stanger B., Stemmler M.P., Takahashi Y., Takeichi M., Theveneau E., Thiery J.P., Thompson E.W., Weinberg R.A., Williams E.D., Xing J., Zhou B.P., Sheng G. On behalf of the, E. M. T. I. A., Guidelines and definitions for research on epithelial–mesenchymal transition. Nat. Rev. Mol. Cell Biol. 2020;21(6):341–352. doi: 10.1038/s41580-020-0237-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sinha D., Saha P., Samanta A., Bishayee A. Emerging concepts of hybrid epithelial-to-mesenchymal transition in cancer progression. Biomolecules. 2020;10(11):1561. doi: 10.3390/biom10111561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Nishimoto S., Nishida E. MAPK signalling: ERK5 versus ERK1/2. EMBO Rep. 2006;7(8):782–786. doi: 10.1038/sj.embor.7400755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin E.C.K., Amantea C.M., Nomanbhoy T.K., Weissig H., Ishiyama J., Hu Y., Sidique S., Li B., Kozarich J.W., Rosenblum J.S. ERK5 kinase activity is dispensable for cellular immune response and proliferation. Proc. Natl. Acad. Sci. 2016;113(42):11865. doi: 10.1073/pnas.1609019113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Deng X., Yang Q., Kwiatkowski N., Sim T., McDermott U., Settleman J.E., Lee J.-D., Gray N.S. Discovery of a benzo[e]pyrimido-[5,4-b][1,4]diazepin-6(11H)-one as a Potent and Selective Inhibitor of Big MAP Kinase 1. ACS Med. Chem. Lett. 2011;2(3):195–200. doi: 10.1021/ml100304b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lochhead P.A., Tucker J.A., Tatum N.J., Wang J., Oxley D., Kidger A.M., Johnson V.P., Cassidy M.A., Gray N.S., Noble M.E.M., Cook S.J. Paradoxical activation of the protein kinase-transcription factor ERK5 by ERK5 kinase inhibitors. Nat. Commun. 2020;11(1):1383. doi: 10.1038/s41467-020-15031-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Javaid S., Zhang J., Smolen G.A., Yu M., Wittner B.S., Singh A., Arora K.S., Madden M.W., Desai R., Zubrowski M.J., Schott B.J., Ting D.T., Stott S.L., Toner M., Maheswaran S., Shioda T., Ramaswamy S., Haber D.A. MAPK7 Regulates EMT Features and Modulates the Generation of CTCs. Mol. Cancer Res. MCR. 2015;13(5):934–943. doi: 10.1158/1541-7786.MCR-14-0604. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hoang V.T., Matossian M.D., Ucar D.A., Elliott S., La J., Wright M.K., Burks H.E., Perles A., Hossain F., King C.T., Browning V.E., Bursavich J., Fang F., Del Valle L., Bhatt A.B., Cavanaugh J.E., Flaherty P.T., Anbalagan M., Rowan B.G., Bratton M.R., Nephew K.P., Miele L., Collins-Burow B.M., Martin E.C., Burow M.E. ERK5 is required for tumor growth and maintenance through regulation of the extracellular matrix in triple negative breast cancer. Front. Oncol. 2020;10 doi: 10.3389/fonc.2020.01164. 1164-1164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Honda T., Obara Y., Yamauchi A., Couvillon A.D., Mason J.J., Ishii K., Nakahata N. Phosphorylation of ERK5 on Thr732 is associated with ERK5 nuclear localization and ERK5-dependent transcription. PloS one. 2015;10(2) doi: 10.1371/journal.pone.0117914. e0117914-e0117914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Razavi P., Chang M.T., Xu G., Bandlamudi C., Ross D.S., Vasan N., Cai Y., Bielski C.M., Donoghue M.T.A., Jonsson P., Penson A., Shen R., Pareja F., Kundra R., Middha S., Cheng M.L., Zehir A., Kandoth C., Patel R., Huberman K., Smyth L.M., Jhaveri K., Modi S., Traina T.A., Dang C., Zhang W., Weigelt B., Li B.T., Ladanyi M., Hyman D.M., Schultz N., Robson M.E., Hudis C., Brogi E., Viale A., Norton L., Dickler M.N., Berger M.F., Iacobuzio-Donahue C.A., Chandarlapaty S., Scaltriti M., Reis-Filho J.S., Solit D.B., Taylor B.S., Baselga J. The genomic landscape of endocrine-resistant advanced breast cancers. Cancer Cell. 2018;34(3):427–438. doi: 10.1016/j.ccell.2018.08.008. e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.