Abstract
Aims: TNF receptor-associated protein 1 (TRAP1), the mitochondrial paralog of the heat shock protein 90 (Hsp90) family of molecular chaperones, is required for neoplastic growth in several tumor cell models, where it inhibits succinate dehydrogenase (SDH) activity, thus favoring bioenergetic rewiring, maintenance of redox homeostasis, and orchestration of a hypoxia-inducible factor 1-alpha (HIF1α)-mediated pseudohypoxic program. Development of selective TRAP1 inhibitors is instrumental for targeted development of antineoplastic drugs, but it has been hampered up to now by the high degree of homology among catalytic pockets of Hsp90 family members. The vegetal derivative honokiol and its lipophilic bis-dichloroacetate ester, honokiol DCA (HDCA), are small-molecule compounds with antineoplastic activity. HDCA leads to oxidative stress and apoptosis in in vivo tumor models and displays an action that is functionally opposed to that of TRAP1, as it induces both SDH and the mitochondrial deacetylase sirtuin-3 (SIRT3), which further enhances SDH activity. We investigated whether HDCA could interact with TRAP1, inhibiting its chaperone function, and the effects of HDCA on tumor cells harboring TRAP1.
Results: An allosteric binding site in TRAP1 is able to host HDCA, which inhibits TRAP1 but not Hsp90 ATPase activity. In neoplastic cells, HDCA reverts TRAP1-dependent downregulation of SDH, decreases proliferation rate, increases mitochondrial superoxide levels, and abolishes tumorigenic growth.
Innovation: HDCA is a potential lead compound for the generation of antineoplastic approaches based on the allosteric inhibition of TRAP1 chaperone activity.
Conclusions: We have identified a selective TRAP1 inhibitor that can be used to better dissect TRAP1 biochemical functions and to tailor novel tumor-targeting strategies.
Keywords: TRAP1, honokiol, mitochondria, antineoplastic compound, molecular dynamics, neurofibromatosis type 1
Introduction
TNF receptor-associated protein 1 (TRAP1) is a molecular chaperone of the heat shock protein 90 (Hsp90) family that exerts important functions as a regulator of mitochondrial bioenergetics, contributing to the metabolic switch of tumor cells toward aerobic glycolysis (23), that is, enhanced glucose usage independently of oxygen availability paralleled by decreased oxidative phosphorylation (OXPHOS) (42). TRAP1 downregulates OXPHOS by inhibiting both cytochrome c oxidase, aka the respiratory complex IV, and succinate dehydrogenase (SDH), which is placed at the crossroad between OXPHOS and tricarboxylic acid cycle (34, 46). TRAP1 inhibition of SDH can establish a pseudohypoxic state by stabilizing hypoxia-inducible factor 1-alpha (HIF1α) in a succinate-dependent way (30, 34).
The transcriptional program coordinated by HIF1α increases invasiveness of tumor cells, elicits their proangiogenic capability, and further rewires metabolism (35), stepping up their malignancy. TRAP1 also shields neoplastic cells from oxidative insults and other death stimuli as it displays antioxidant activity, at least partially related to OXPHOS inhibition (14), and counteracts the mitochondrial permeability transition pore, a cell death-inducing mitochondrial channel (1).
In accord with these pro-neoplastic roles of TRAP1, its expression is upregulated in many cancer types (17, 30), where it correlates with progression, metastasis, and disease recurrence; whereas TRAP1 genetic ablation thwarts the growth of several tumor models (22, 23).
TRAP1 also acts as an energy rheostat and mitochondrial quality gatekeeper in neurons, astrocytes, and cardiomyocytes, and a protective function of the chaperone has been reported after ischemic damage (41, 44, 48); whereas functional impairment of TRAP1 occurs in models of Parkinsonism (3, 5, 12, 47). Therefore, compounds that selectively target TRAP1 would constitute useful tools in diverse clinical settings.
Innovation
The molecular chaperone TNF receptor-associated protein 1 (TRAP1) is the mitochondrial paralog of the heat shock protein 90 (Hsp90) family. TRAP1 plays a key role in the metabolic rewiring of tumor cells, and its genetic ablation hampers neoplastic growth. Selective TRAP1 inhibitors could allow the development of innovative antineoplastic strategies, but their identification is prevented by the similarity among ATP-binding pockets of Hsp90 family members. Here, we show that honokiol bis-dichloroacetate (HDCA) interacts with TRAP1 outside its ATP-binding pocket and inhibits it in a selective, allosteric way. HDCA rescues TRAP1-dependent inhibition of succinate dehydrogenase in tumor cells and abrogates their tumorigenicity, paving the way for a novel class of antitumor compounds.
TRAP1 functions as a dimer, and its chaperone activity requires sequential cycles of binding, hydrolysis, and release of ATP to provide the energy necessary for driving conformational changes in the two protomers and, in turn, to shape conformation and activity of substrates, termed clients (7, 18). Thus, targeting the ATP pocket of TRAP1 by small molecules constitutes an obvious strategy for inactivating its chaperone functions, and ATP-competitive inhibitors have demonstrated potent inhibitory activities. However, catalytic sites of the Hsp90 family proteins display a high degree of similarity, and current ATP-competitive compounds indiscriminately inhibit all these chaperones and their broad range of functions, encompassing regulation of client protein folding, trafficking and activity, assembly of multiprotein complexes, and tuning of signaling pathways (16, 33, 37).
This lack of selectivity elicits an overall cell toxicity, which has prevented further progress of these molecules into clinical use. Compounds targeting the paralogs of Hsp90 in a selective fashion are currently lacking and highly needed.
Honokiol (3′,5-di-2-propenyl-1,1′biphenol-2,4′-diol) is a small polyphenol compound extracted from the plant Magnolia grandiflora utilized in traditional Chinese medicine for its muscle relaxant, antioxidative, anti-inflammatory, antiallergic, and antibacterial activities, showing a long and safe record of usage (49), although the molecular mechanism(s) underlying its biological actions have never been elucidated. Recently, honokiol has emerged also as a promising anticancer agent in preclinical models, without any appreciable toxicity (36). However, solubility and bioavailability of honokiol are poor (43). To increase its lipophilicity and cellular uptake, honokiol was derivatized into the corresponding bis-dichloroacetate ester (HDCA), which upregulates the subunit B of SDH in melanoma cell models, where it exerts a remarkable pro-oxidant and antineoplastic function (2). Honokiol also activates the mitochondrial deacetylase sirtuin-3 (SIRT3) (28, 29), which increases SDH enzymatic activity (11).
Fine tuning of SDH activity is crucial in diverse models of tumor progression, as succinate can act as an oncometabolite when it accumulates downstream to SDH inhibition. Indeed, succinate inhibits a class of enzymes called dioxygenases, leading to the activation of several transcription and epigenetic pro-neoplastic programs, including those orchestrated by the transcription factor HIF1α (39). TRAP1 and HDCA exert opposite effects on SDH. We, therefore, examined the possibility that HDCA could act as a TRAP1 inhibitor, which could pave the way for the design of novel antineoplastic strategies.
Results
The structure of HDCA (Fig. 1A) is significantly different from that of all known ATP-competitive inhibitors of the Hsp90 chaperone family. Consequently, we evaluated the possibility that HDCA could act as an allosteric inhibitor of TRAP1, targeting binding sites that are distinct from its ATP-binding pocket. We previously identified a potential allosteric site in TRAP1 that is located in the straight protomer of the chaperone dimer and has the stereochemical features needed to host a small molecule (25), and we have recently found a set of compounds that inhibit TRAP1 by binding that region (32). The site was defined by characterizing the dynamic coordination of the ATP-binding site of each TRAP1 protomer with other residues of the protein by means of molecular dynamics (MD) simulations. We refer to the already published reference (25) for details.
FIG. 1.
Structure of HDCA and its complex with TRAP1 allosteric pocket. (A) Lewis structure of HDCA. (B) HDCA docked into the putative allosteric pockets. The protein residues making important contacts with the ligand are labeled. (C) The surface representation of the pocket, showing how HDCA fits into the allosteric site. Red, negatively charged areas; blue, positively charged areas. HDCA, honokiol bis-dichloroacetate; TRAP1, TNF receptor-associated protein 1.
The putative binding site of TRAP1 is characterized by a large positively charged area formed by the side chains of residue ARG341, LYS364, and ARG454 flanked by a negatively charged region due to residues GLU457 and ASP458 and, at the opposite side, by another negative region formed by residues GLU647 and GLU648. The two negatively charged regions are separated by a large hydrophobic one, formed by residues PRO361, MET363, PRO365, VAL370, LEU461, and PHE462 (Fig. 1B, C). In steric and sequence terms, this pocket is different from analogous allosteric pockets that we previously discovered (25).
To investigate the possibility for HDCA to bind this pocket, we carried out rigid and flexible computational docking experiments. Rigid docking on the representative structure of the most populated cluster obtained from the MD trajectory of TRAP1 could not find an optimal binding mode, as the target ligand does not fit into the receptor conformation, due to steric clashes.
In contrast, the use of an induced fit docking (IFD) protocol permitted structural reorganizations in the binding site that allowed HDCA to be optimally accommodated. The hydrophobic nature of the molecule fits well the large hydrophobic zone of the pocket where the carbonyl groups can also engage in additional hydrogen bonding with different residues of the protein and halogen bonding between the Cl atoms and the amino groups of the protein backbone. A pi-pi stacking interaction is frequently observed between the ligand and the TYR459 as well as a pi-cation interaction is seen with the residue forming the positive charged area of the binding pocket (Fig. 1B, C).
Overall, HDCA is able to exploit the potential interactions available in the putative binding pocket (Figs. 1B, C and 2). These models of HDCA/TRAP1 complexes may be used as a guide for a further derivatization of HDCA, to increase the selectivity for its putative binding pocket on TRAP1.
FIG. 2.
The three-dimensional structure of TRAP1. The elements of secondary structure are depicted as tubes (helices) or arrows (beta-sheets). The putative binding site is highlighted as a surface, colored according to the chemical properties of the constituting residues: blue, positively charged; red, negatively charged; white, hydrophobic.
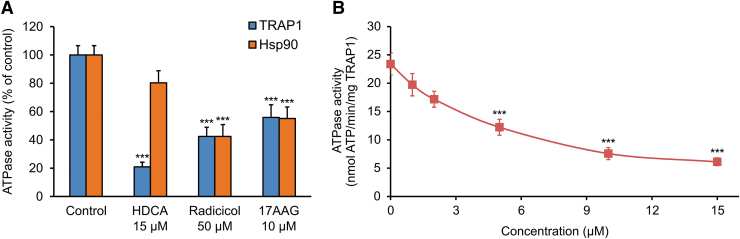
Given this possibility of interaction between HDCA and TRAP1, we measured whether HDCA could exert any effect on the chaperone function of TRAP1, assessing the ATPase activity of the recombinant human protein with an enzyme-coupled NADH absorbance assay (7, 19). In parallel, we carried out the same analysis on human recombinant Hsp90, to evaluate HDCA specificity. We found that HDCA causes a marked and concentration-dependent inhibition of TRAP1 ATPase activity (Fig. 3A, B), being even more effective than the wide-spectrum Hsp90 inhibitors 17-N-allylamino-17-demethoxygeldanamycin (17-AAG) and radicicol, which block ATP binding at the catalytic site of the Hsp90 family chaperones. Unlike radicicol and 17-AAG, the HDCA inhibitory effect was highly selective for TRAP1 over Hsp90, as HDCA could not inhibit Hsp90 activity (Fig. 3A).
FIG. 3.
HDCA inhibits TRAP1 but not Hsp90 ATPase activity. (A) Spectrophotometric quantification of the effect of HDCA on the ATPase activity of purified human TRAP1 or Hsp90α proteins (blue and orange bars, respectively). Wide-spectrum Hsp90 family inhibitors radicicol and 17-AAG were used as positive controls. (B) Dose-response analysis of the effect of HDCA on TRAP1 ATPase activity. Throughout the figure, data are presented as mean ± SEM (n = 4); ***p < 0.001 with an unpaired two-tailed Student's t test. 17-AAG, 17-N-allylamino-17-demethoxygeldanamycin; Hsp90, heat shock protein 90; SEM, standard error of the mean.
We have previously shown that TRAP1 down-modulates SDH enzymatic activity in several tumor cell types, and this blockade can be reversed either by silencing TRAP1 or by treatment with 17-AAG (34). TRAP1 chaperone activity is enhanced by extracellular signal–regulated kinases (ERK)-dependent phosphorylation in neoplastic cells endowed with hyperactivation of the Ras/ERK signaling pathway (22), as those related to the genetic syndrome neurofibromatosis type 1 (NF1) where expression of the Ras-GTPase-activating proteins (Ras-GAP) neurofibromin is ablated (31, 45). We, therefore, measured the succinate:coenzyme Q reductase (SQR) activity of SDH in NF1-related cells from either benign plexiform neurofibroma (PN) or malignant peripheral nerve sheath tumor (MPNST), as a readout to test the efficacy of HDCA inhibition of TRAP1 in tumor cells.
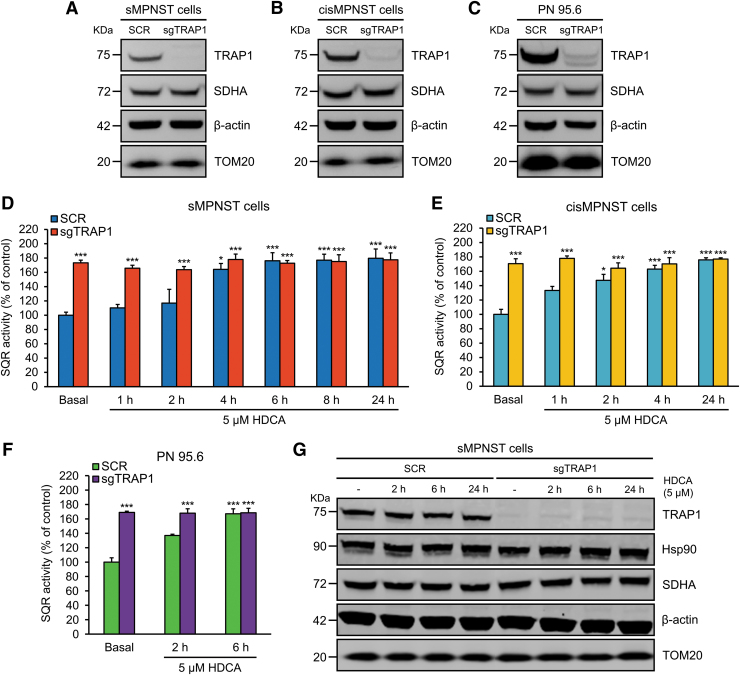
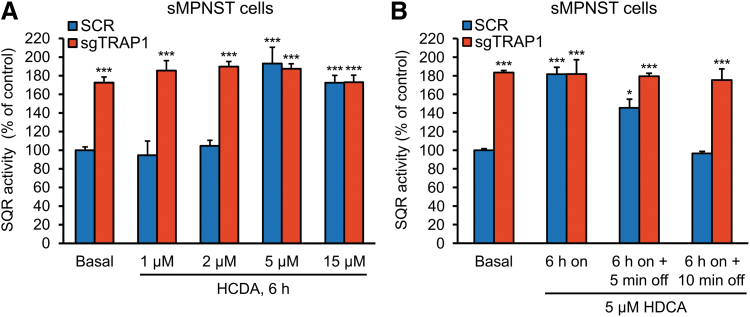
To address HDCA specificity, we used clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 technology to generate TRAP1 knock-out cells (Fig. 4A–C and Supplementary Fig. S1A, B). Genetic ablation of TRAP1 markedly increased the SQR activity of SDH in mouse sMPNST and cisMPNST cells, as well as in human plexiform neurofibroma PN95.6 cells (Fig. 4D–F). Treatment with HDCA upregulated SQR activity in all these cell models to the same levels achieved after knocking out TRAP1 expression (Fig. 4D–F) without changing either TRAP1 or SDH subunit A (SDHA) protein expression levels (Fig. 4G and Supplementary Fig. S2A, B). This effect of HDCA was fast and stable over time (Fig. 4D–F), occurred in a dose-dependent way (Fig. 5A), and was rapidly reversible after compound withdrawal (Fig. 5B). Importantly, TRAP1 knock-out cells were completely insensitive to HDCA treatment (Figs. 4D–F and 5A, B), indicating that HDCA enhances SDH activity as a consequence of TRAP1 inhibition.
FIG. 4.
HDCA reverts TRAP1-dependent inhibition of SDH activity. (A–C) Knocking out of TRAP1 expression by a single-guide RNA (sgTRAP1) by using a CRISPR/Cas9 approach (SCR: cells expressing a scrambled guide RNA) in sMPNST, cisMPNST, or PN 95.6 cells. TRAP1 ablation did not change expression levels of SDHA. (D–F) Spectrophotometric assessment of the SQR activity of SDH on whole cells treated with HDCA for the reported times. Data are shown as mean ± SEM (n > 3); ***p < 0.001; *p < 0.05 with one-way ANOVA with post hoc Bonferroni's test. (G) Western immunoblot analysis showing that TRAP1, Hsp90, and SDHA protein levels do not change after HDCA treatment. β-actin and TOM20 were used as cytosol and mitochondrial protein loading controls, respectively, in all Western immunoblots of the figure. ANOVA, analysis of variance; CRISPR, clustered regularly interspaced short palindromic repeat; MPNST, malignant peripheral nerve sheath tumor; PN, plexiform neurofibroma; SDH, succinate dehydrogenase; SDHA, SDH subunit A; SQR, succinate:coenzyme Q reductase.
FIG. 5.
HDCA reverts TRAP1-dependent inhibition of SDH in a concentration-dependent and reversible way. (A) Dose-response analysis of a 6-h cell exposure to the reported concentrations of HDCA. (B) Effect of a 6-h exposure of sMPNST cells to 5 μM HDCA (on) followed by compound withdrawal from medium (off) for the reported times. Data are shown as mean ± SEM (n > 3); ***p < 0.001; *p < 0.05 with one-way ANOVA with post hoc Bonferroni's test. Spectrophotometric assessments of SQR activity of SDH were carried out as in Figure 4. Cells are labeled as in Figure 4.
We then investigated whether HDCA treatment could display any TRAP1-related biological effect. HDCA did not increase mitochondrial depolarization (Fig. 6A), whereas it prompted a raise in mitochondrial superoxide levels, in agreement with previous studies (2) and independently of TRAP1 expression (Fig. 6B). No change in mitochondrial mass was caused either in wild-type or in TRAP1 knock-out cells by HDCA treatment (Fig. 6C). Genetic ablation of TRAP1 slightly slowed down the proliferation rate of sMPNST cells, and this effect was mimicked by HDCA treatment on TRAP1-expressing cells, whereas HDCA was completely ineffective on the proliferation of TRAP1 knock-out sMPNST cells (Fig. 6A).
FIG. 6.
Effect of HDCA on mitochondria, proliferation, and viability parameters. (A) Percentage of cells with polarized mitochondria measured by TMRM staining; (B) mitochondrial superoxide levels measured with MitoSOX Red; (C) mitochondrial mass assessed with the fluorescent dye NAO. In (A–C), analyses were carried out by cytofluorimetric inspections of scrambled (SCR) and TRAP1 knock-out (sgTRAP1) sMPNST cells treated with 5 μM HDCA for 24 h. Data are reported as mean ± SEM (n = 3); **p < 0.01; ***p < 0.001 against vehicle-treated SCR cells; §p < 0.05 against vehicle-treated sgTRAP1 cells with one-way ANOVA with post hoc Bonferroni's test. Cells are labeled as in Figure 4. (D) Effect of HDCA (5 μM, 96 h) on the proliferation rate of sMPNST cells. (E) Cell viability after treatment with 5 μM HDCA for 24 h under low serum conditions (1% FBS). On the left, representative traces of cytofluorimetric cell death analyses. Annexin-V and 7-AAD stainings are reported. Black: double negative, viable cells; green: Annexin V-FITC positive, early apoptotic cells; blue: Annexin V-FITC/7-AAD double positive, late apoptotic cells; red: 7-AAD positive, necrotic cells. The percentage of each cell subpopulation is indicated. On the right, bar graph quantification of cell viability is indicated. Data are reported as mean ± SEM (n = 3). Cells are labeled as in Figure 4. 7-AAD, 7-aminoactinomycin D; FBS, fetal bovine serum; NAO, 10-nonylacridine orange bromide; TMRM, tetramethylrhodamine methyl ester.
This lag in cell population growth was not caused by a higher propensity of cells to undergo apoptosis after genetic or pharmacological inhibition of TRAP1, as we could not detect any cell death either in TRAP1 knock-out or in HDCA-treated cells (Fig. 6B). Both genetic ablation and allosteric inhibition of TRAP1 could reduce intracellular succinate accumulation through reversion of TRAP1-dependent SDH inhibition. Therefore, it is possible to envision that the ensuing downregulation of succinate-driven epigenomic changes could decrease cell proliferation rate, as reported (6, 39).
We have previously observed that TRAP1-dependent downregulation of SDH activity is increased when tumor cells are exposed to stress conditions that resemble those found during neoplastic growth (34), and that TRAP1 silencing hampers tumorigenic growth of various cell models even when it does not affect in vitro proliferation (22, 34). Hence, we evaluated whether allosteric TRAP1 inhibition by HDCA might alter the tumorigenic capacity of neoplastic cells. As expected, sMPNST cells were tumorigenic, as they could overcome contact inhibition and form foci. TRAP1 genetic ablation nearly abolished foci formation, and this inhibition was recapitulated by treatment with HDCA (Fig. 7A, B). Notably, HDCA could block tumorigenic cell growth even when it was added to already formed foci (Fig. 7C, D).
FIG. 7.
Effect of TRAP1 inhibition on in vitro tumorigenesis. (A) Representative images of focus-forming assays performed on sMPNST cells grown for 9 days in the presence of the indicated concentrations of HDCA. (B) Bar graph quantification of the effect of TRAP1 inhibition, either by CRISPR/Cas9 knock-out or by administration of the reported concentrations of HDCA, on the growth of foci. Data are reported as mean ± SEM (n = 3); ***p < 0.001 with one-way ANOVA with post hoc Bonferroni's test. (C) Representative images of foci after 6 or 9 days of assay. Where indicated, 5 μM HDCA was added at the sixth experimental day and kept in the medium for the next 3 days. (D) Bar graph quantification of the effect of HDCA (5 μM, added as in C) on the growth of foci. Data are reported as mean ± SEM (n = 3); §§§p < 0.001 with an unpaired two-tailed Student's t test for the values obtained after 6 days of assay; ***p < 0.001 against basal values at the ninth experimental day with one-way ANOVA with post hoc Bonferroni's test. In (B, D), foci were quantified by using an integrated density parameter that englobes both their thickness and area. Cells are labeled as in Figure 4.
As honokiol is an activator of the mitochondrial deacetylase SIRT3 (28, 29), which enhances SDH activity (11), we investigated whether HDCA increases SQR activity of SDH via a SIRT3-dependent effect. We found that sMPNST cells harbor a very low level of SIRT3; as expected, its overexpression enhances SQR activity (Fig. 8A, B and Supplementary Fig. S3A, B). However, this effect was comparable to that of HDCA, which could further increase SQR activity in SIRT3-overexpressing cells (Fig. 8A, B). Taken together, these results suggest that HDCA has a double induction effect on the enzymatic activity of SDH: it inhibits the SDH inhibitor TRAP1 and augments the activity of the SDH enhancer SIRT3 (Fig. 8C).
FIG. 8.
Effect of HDCA on the SDH activity of SIRT3-overexpressing cells. (A) Western immunoblot showing SIRT3, TRAP1, and SDHA protein expression levels in sMPNST cells transfected either with a control pFUGW-GFP vector or with a pFUGW-SIRT3. β-actin was used as a protein loading control. (B) Spectrophotometric assessment of the SQR activity of SDH on cells treated with HDCA (5 μM, 6 h). Data are shown as mean ± SEM (n > 3); ***p < 0.001 against pFUGW-GFP-transfected cells; §§§p < 0.001 against HDCA-treated GFP cells with one-way ANOVA with post hoc Bonferroni's test. (C) Model of the HDCA effects on TRAP1 and SIRT3. SIRT3, sirtuin-3.
Discussion
In this work, we have reported the dynamic-based identification of an allosteric interaction between the vegetal derivative HDCA and the molecular chaperone TRAP1, and its effects on the biochemical activity of TRAP1 and on biological processes regulated by it. Our results demonstrate that unveiling specific allosteric domains of TRAP1, distal from its highly conserved ATP-binding site and characterized by structural and chemical features different from those of other Hsp90 family members (10, 25, 26), can disclose the possibility of detecting highly selective TRAP1 inhibitors [see also Sanchez-Martin et al. (32)]. Accordingly, HDCA displays a concentration-dependent inhibition of TRAP1 ATPase activity, while leaving cytosolic Hsp90 unperturbed.
By binding the allosteric site in the M-domain of TRAP1 (25), HDCA can potentially perturb the dynamic coupling mechanisms with the ATPase domain. Allostery is a key mechanism of fine conformational regulation of a protein: it can modulate its structural dynamic properties after binding of a ligand, altering reactivity toward a substrate at a distant catalytic site. This reverberates in a change of protein activity that affects the cell biochemical networks in which the protein is involved. The identification of specific allosteric ligands offers potent tools to investigate the complex biology of TRAP1, which probably interacts with a wide spectrum of clients in highly context-specific ways (23). This is particularly important in the neoplastic process, where TRAP1 contributes toward shaping the bioenergetic adaptations of mitochondria to the mutable environmental conditions encountered by the tumor (4, 17). The reported observation that in some specific neoplastic settings TRAP1 expression levels inversely correlate with tumor grade (30, 46) contrasts with its proposed pro-neoplastic role, and this further highlights the necessity of chemical entities that could selectively block the functions of TRAP1 for disentangling the complexity of its regulatory activities.
Importantly, TRAP1 activity can be subtly regulated by post-translational modifications, which influence downstream biological responses. Indeed, TRAP1 modulates cell tumorigenicity after interaction with the kinases Src and ERK1/2 (22, 46), or after its SIRT3-dependent deacetylation (27). In turn, TRAP1 stabilizes these kinases and deacetylases in mitochondria, even under stress conditions possibly related to specific tumor microenvironmental conditions. By selectively perturbing both TRAP1 and SIRT3 activity, HDCA could be used to study the biological meaning of this interaction in the native cellular milieu under specific conditions, such as metabolic stress or hypoxia, found in the growing tumor mass.
From the applicative point of view, blocking TRAP1 by HDCA without interfering with other Hsp90 paralogs can be a starting point for the development of further compounds with optimized pharmacological profiles. These molecules could be tested on pathological conditions that are strictly dependent on TRAP1, avoiding the problems connected with the Heat Shock Response observed on using large-spectrum Hsp90 inhibitors.
Honokiol and its derivative HDCA have shown remarkable biological effects, including their ability to tune cell redox homeostasis, OXPHOS function, and different signaling pathways, as well as the capacity to induce both apoptotic and necrotic cell death (15, 21, 38) and to act as antineoplastic compounds in in vitro and in vivo models (2, 8, 13, 36). Nonetheless, identification of honokiol intracellular interactors has remained elusive, hampering a thorough dissection of its mode(s) of action.
In this work, we demonstrate that the ATPase activity of the recombinant, purified human TRAP1 protein is inhibited by HDCA, indicating that TRAP1 is a molecular target of HDCA. This observation has implications for the choice of the pathophysiological models while assaying the biological effects of honokiol and derivatives. Moreover, it might be beneficial to conduct a further derivatization of HDCA, to optimize the distribution of the donor/acceptor features, with the aim of better adapting to the asymmetric nature of the TRAP1 target pocket that is schematically made of a positive charged area and a large hydrophobic zone separating two negatively charged regions. The ultimate goal could be the definition of molecular entities that are translatable to preclinical models of antineoplastic treatments.
Materials and Methods
Structural modeling of the complex between HDCA and TRAP1
Protein and ligand preparation
The coordinates for the protein were obtained from a cluster analysis of MD simulation trajectories from a previous work (25). The structure was prepared by using the Maestro software package. Hydrogen atoms were added, and a proper His tautomer was selected to maximize hydrogen bonding. All Asp, Glu, Arg, and Lys residues were left in their charged state. A brief minimization was performed by using the Protein Preparation module in Maestro with the “Refinement Only” option, where an all-atom constrained minimization allows the protein to relax. The minimization was terminated when the root mean square deviation reached a maximum value of 0.3 Å. Ligand was drawn in Maestro, and the appropriate bond orders and formal charges were manually adjusted. The ligand was subjected to a full minimization with the optimized potentials for liquid simulations (OPLS) force field, according to the Ligprep protocol. The minimized ligand was the starting point for the initial docking step.
Induced fit docking
All docking calculations were performed by using the Glide program of Schrodinger software release 2019-1. The first stage of the IFD protocol generates 20 initial poses of the complex using a softened-potential. The softened-potential consists of a potential energy function where the van der Waals radii are scaled by 0.5 (for receptor atoms with partial atomic charge q ≤ 0.25e and for ligand atoms with q ≤ 0.15e).
For each of the top 20 poses ranked according to GlideScore from the initial softened-potential docking, a protein refinement is performed with Prime. Prime uses the OPLS parameters and the Generalized Born implicit solvent model. A list of residues having at least one atom within 5 Å of an atom in any of the 20 ligand poses was generated, and all side chains of these residues underwent a conformational search and minimization. The low-energy solution identified with this step is further minimized to relax all residues in the list, both backbone and side chain, and the ligand. The complexes are ranked according to the Prime energy (molecular mechanics and solvation energy) and those within 30 kcal/mol in the lowest energy structure are passed through for a final round of Glide docking and scoring.
Finally, the minimized ligand used in the first docking run is re-docked by using Glide with default settings into the resulting 20 receptor structures from the protein refinement step. This iterative process produced a list of final poses, ordered by a score consisting of a linear combination of the total energy of the system (Prime energy, molecular mechanics force field in Variable-dielectric Surface Generalized Born solvent) and the energy between the ligand and the receptor, the Glide SP empirical scoring function, according to the following equation: GlideScore +0.05 × PrimeEnergy, which is used to rank the possible solutions.
Protein production and purification
Full-length recombinant human TRAP1 (without mitochondrial targeting sequence) was cloned into the pET151/D-TOPO bacterial expression plasmid (Cat. No. K15101; Thermo Fisher Scientific, Walthman, MA) as previously described (7). N-terminally His-tagged fusion TRAP1 was expressed in Escherichia coli BL21-AI cells (Cat. No. C607003; Thermo Fischer) grown in LB media (Cat. No. L3022; Sigma-Aldrich, St. Louis, MO) supplemented with 0.4% glucose (Cat. No. G8270; Sigma-Aldrich) by addition of 1 mM isopropyl β-D-1-thiogalactopyranoside (IPTG, Cat. No. V3955; Promega, Madison, WI) and 0.2% arabinose (Cat No. A3256; Sigma-Aldrich) at OD600 0.7–0.8 and then incubated under vigorous shaking for 20 h at 16°C.
After centrifugation, the pellet was resuspended in a binding buffer composed by 50 mM KH2PO4 (Cat. No. 5101; Merck, Darmstadt, Germany), pH 8.0, 500 mM NaCl (Cat No. S9888; Sigma-Aldrich), 20 mM imidazole (Cat. No. 56750; Sigma-Aldrich), and 3 mM β-mercaptoethanol (Cat No. M6250; Sigma-Aldrich). Samples were then sonicated in ice in the presence of 0.2 mM phenylmethyl sulfonyl fluoride (Cat. No. 78830; Sigma-Aldrich) and centrifuged at 20,000 g. The supernatant was incubated with Ni-NTA (Cat. No. 17-5318-01; GE Healthcare Biosciences, Pittsburgh, MA) under rotation for 1 h at 4°C and loaded onto a gravity column.
The His-tagged protein was eluted in a buffer containing 50 mM KH2PO4 pH 8.0, 300 mM NaCl, 500 mM imidazole, and 3 mM β-mercaptoethanol and dialyzed overnight in a buffer composed by 10 mM Tris-HCl (Cat. No. T3253; Sigma-Aldrich) pH 8.0, 200 mM NaCl, and 1 mM DTT (Cat. No. D0632; Sigma-Aldrich) supplemented with tobacco etch virus protease (Cat. No. T4455; Sigma-Aldrich) to cleave the N-terminal 6x-His-tag. The samples containing recombinant TRAP1 were then collected and stored at −80°C.
ATPase activity measurements
TRAP1 and Hsp90 ATPase assays were performed by using an ATP-regenerating system as described earlier (19). All experiments were carried out in a buffer composed by 50 mM Tris-HCl pH 7.4, 50 mM Sucrose (Cat. No. 84097; Sigma-Aldrich), 50 mM KCl (Cat. No. P3911; Sigma-Aldrich), 4 mM MgCl2 (Cat. No. 63068; Sigma-Aldrich), and 2 mM EGTA (Cat. No. E0396; Sigma-Aldrich) supplemented with 300 μM NADH (Cat. No. 10128023001; Sigma-Aldrich), 2 mM phosphoenolpyruvate (Cat. No. P7002; Sigma-Aldrich), 1.5 mU/mL l-lactate dehydrogenase (Cat. No. 427211; Sigma-Aldrich), 0.8 mU/mL pyruvate kinase (Cat. No. P1506; Sigma-Aldrich), and 450 ng human recombinant TRAP1 or Hsp90α (ADI-SPP-776-D; Enzo Life Sciences, Farmingdale, NY). Assays were started after addition of ATP (Cat. No. A26209; Sigma-Aldrich), 200 μM for TRAP1, and 1 mM for Hsp90α and the activity was recorded on a microplate spectrophotometer (Infinite® 200 PRO, Tecan, Switzerland) after NADH oxidation at 340 nm for 1 h at 37°C. Both Hsp90 family inhibitors, radicicol (Cat. No. sc-200620; Santa Cruz Biotechnology, Dallas, TX) and 17-AAG (Cat. No. A8476; Sigma-Aldrich), as well as HDCA were added 5 min before starting the recordings. HDCA was synthesized as previously described (2).
Cell culture and ablation of TRAP1 expression
All cells were cultured in Dulbecco's modified Eagle's medium (DMEM; Cat. No. 11965092; Thermo Fischer) supplemented with 10% fetal bovine serum (Cat. No. 26140079; Thermo Fischer), 2 mM glutamine (Cat. No. A2916801; Thermo Fischer), 1 mM sodium pyruvate (Cat. No. 11360070; Thermo Fischer), 100 μg/mL penicillin, and 100 μg/mL streptomycin (Cat. No. 15070063; Thermo Fischer) and kept at 37°C in a 5% CO2 humidified atmosphere. Human PN95.6 cells were immortalized from Nf1−/− primary Schwann cell cultures of NF1 patients (20). This cell line was provided by Dr. Margaret R. Wallace, University of Florida, College of Medicine, Gainesville, FL. Primary mouse MPNST cisMPNST cells were established from spontaneous MPNSTs arising in cis Nf1+/−;P53+/− mice (40), and sMPNST cells were derived from Nf1−/−;P53−/− skin precursors (24). Both MPNST cell types were provided by Dr. Lu Q. Le, University of Texas Southwestern Medical Center, Dallas, TX.
TRAP1 expression was stably knocked out with CRISPR/Cas9 technology by infecting cells with a lentivirus carrying single guides against either rodent or human TRAP1 genes (for mouse TRAP1: 5′-CACCGCGCCGAACTCCAGCCAGCGC-3′ and 5′-CACCGTTTGTGTGGGGCCCCTAAAC-3′; for human TRAP1: 5′-CACCGAAAGCTTCTTTGTCTCGGCC-3′ and 5′-CACCGCAGTTTTTCCAAGGCATCGC-3′). Scrambled single guides were always used as negative controls. Infected cells were selected in 1 μg/mL puromycin (Cat. No. P9620; Sigma-Aldrich). SIRT3 was overexpressed in TRAP1-expressing sMPNST cells by using a pFUGW vector. For these experiments, pFUGW-GFP was used as a negative control.
Western immunoblot assays
For Western immunoblots analyses, cells kept in standard culture conditions were lysed at 4°C in RIPA buffer composed by 150 mM NaCl, 50 mM Tris-HCl pH 8.0, 1% Nonidet P-40 (Cat. No. 74385; Sigma-Aldrich), 0.5% sodium deoxycholate (Cat. No. D5670; Sigma-Aldrich), and 0.1% sodium dodecyl sulfate (Cat. No. L3771; Sigma-Aldrich) in the presence of phosphatase (Cat. No. P5726; Sigma-Aldrich) and protease inhibitors (Cat. No. P8340; Sigma-Aldrich); proteins were then quantified by using a BCA Protein Assay Kit (Cat. No. 23227; Thermo Fisher).
Protein lysates were separated on 4–12% Bis-Tris NuPage gels (Life Technologies, Carlsbad, CA) and transferred onto nitrocellulose Hybond-C Extra membranes (Amersham, Uppsala, Sweden) following standard methods. Primary antibodies were incubated overnight at 4°C. Proteins were detected either with Immobilon® Western (Cat. No. WBKLS0500; Millipore, Darmstadt, Germany) after incubation with goat anti-mouse (Cat. No. 1706516; Bio-Rad laboratories, Hercules, CA) and anti-rabbit (Cat. No. 1706515; Bio-Rad Laboratories) horseradish peroxidase-conjugated secondary antibodies or with an Odyssey CLx Imaging System (LI-COR Biosciences, Lincoln, NE), after incubation with IRDye® 800CW goat (polyclonal) anti-rabbit (Cat. No. 926-68021; LI-COR Biosciences) or IRDye® 680LT goat (polyclonal) anti-mouse (Cat. No. 926-68020; LI-COR Biosciences) secondary antibodies. Anti-human TRAP1 (Cat. No. sc-73604), anti-SDHA (Cat. No. sc-166947), and anti β-actin (Cat. No. sc-47778) mouse monoclonal antibodies and anti-TOM20 (Cat. No. sc-11415) rabbit polyclonal antibody were all from Santa Cruz Biotechnologies; anti-rodent TRAP1 (Cat. No. 612344) and anti-Hsp90 (Cat. No. 610418) mouse monoclonal antibodies were from Becton Dickinson (Franklin Lakes, NJ).
Measurement of SQR activity of the SDH complex
To assay the SQR enzymatic activity of SDH, cells were collected and homogenized at 4°C in a buffer containing 25 mM KH2PO4 pH 7.2, 5 mM MgCl2, protease, and phosphatase inhibitors. Cell samples (40 μg protein per trace) were then preincubated for 10 min at 30°C in a buffer composed by 25 mM KH2PO4 pH 7.2, 5 mM MgCl2, 20 mM sodium succinate (Cat. No. 6624; Merck), and 10 μM alamethicin (Cat. No. A4665; Sigma-Aldrich). After the preincubation time, 5 mM sodium azide (Cat. No. S2002; Sigma-Aldrich), 5 μM antimycin A (Cat. No. 1782; Calbiochem, Darmstadt, Germany), 2 μM rotenone (Cat. No. R8875; Sigma-Aldrich), and 100 μM 2,6-dichlorophenolindophenol (DCPIP; Cat. No. D2908; Sigma-Aldrich) were added to the medium. The reaction started after the addition of 65 μM coenzyme Q1 (Cat. No. C7956; Sigma-Aldrich), and SQR activity was measured after the reduction of DCPIP at 600 nm (ɛ = 19.1 mM−1 cm−1) for 20 min at 30°C. Each measurement of SDH activity was normalized for protein amount.
Cell viability assays
Cell viability was quantified by flow cytometry analysis as previously described (9). MPNST cells were collected after treatment with vehicle or HDCA and incubated at 37°C in a buffer composed by 135 mM NaCl, 10 mM HEPES (Cat. No. H8651; Sigma-Aldrich), and 5 mM CaCl2 (Cat. No. C3306; Sigma-Aldrich). Cytofluorimetric recordings of phosphatidylserine exposure on the cell surface (increased FITC-conjugated Annexin-V staining) and loss of plasma membrane integrity (7-aminoactinomycin D [7-AAD] permeability and staining) were performed on an FACS Canto II flow cytometer (Becton Dickinson) and analyzed by using FACSDiva software.
Measurements of membrane potential, superoxide levels, and mass of mitochondria
Mitochondrial membrane potential was determined by using the fluorescent potentiometric dye tetramethylrhodamine methyl ester (TMRM; Cat. No. T668; Thermo Fisher). Cells were harvested in a phenol red-free DMEM containing 1 μM cyclosporine H (CsH; Cat No. AG-CN2-0447; Vinci-Biochem, Florence, Italy) to inhibit P-glycoproteins and were labeled with 20 nM TMRM at room temperature for 1 h. The protonophore carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone (FCCP, Cat No. C2920; Sigma-Aldrich) was used to dissipate the proton gradient as a positive control for depolarization of the mitochondrial membrane potential.
Mitochondrial superoxide levels were measured with the use of the fluorescence probe MitoSOX Red (2.5 μM, 2 h incubation; Cat No. M36008; Invitrogen, Carlsbad, CA) as previously reported (2, 13). The fluorescent dye 10-nonylacridine orange bromide (NAO, 200 nM, 30 min incubation; Cat. No. A7847; Sigma-Aldrich), which binds with high affinity to the negatively charged cardiolipin at the inner mitochondrial membrane, was used to quantify the mitochondrial mass. TMRM, MitoSOX Red, and NAO signals were recorded on an FACS Canto II flow cytometer and analyzed by using FACSDiva software.
Cell proliferation assays
To study the effect of HDCA on cell proliferation, sMPNST cells were seeded at a density of 1 × 104 cells per well in a 12-well plate and were allowed to attach overnight. Cells were then treated with either DMSO (Cat. No. 276855; Sigma-Aldrich) or HDCA at the indicated concentrations for 96 h. Counts were performed each day by using a Fuchs-Rosenthal chamber.
In vitro tumorigenesis assays
For focus-forming assays, cells were seeded in 12-well culture plates in DMEM medium with 10% fetal bovine serum. When cells reached confluence, serum concentration was reduced to 1%. HDCA was added at the indicated concentrations either immediately after changing serum concentration or 6 days after serum decrease. At the 10th day, foci appeared as thick masses; plates were then washed in PBS, fixed in methanol (Cat. No. 34860; Sigma-Aldrich) for 30 min, and foci stained with GIEMSA solution (Cat. No. 32884; Sigma-Aldrich) for 1 h. After washing in deionized water, surface area and thickness of foci were analyzed with ImageJ software and combined to obtain the integrated density parameter.
Statistics and reproducibility
A two-tailed, unpaired Student's t test was used to compare pairs of data groups. One-way analysis of variance followed by Bonferroni post hoc test was applied for multiple comparisons. In all figures, data were expressed as mean ± standard error of the mean. Statistical significance was calculated with Origin® 8 (OriginLab, Northampton, MA). Results with a p-value lower than 0.05 (***p < 0.001, **p < 0.01, *p < 0.05 compared with controls) were considered significant. Each experiment was repeated at least thrice.
Supplementary Material
Acknowledgments
The authors thank David Agard, University of California at San Francisco, CA, for providing the pET151/D-TOPO/TRAP1 plasmid; Danica Chen, University of California at Berkeley, CA, for providing the pFUGW-SIRT3 vector; and Elena Trevisan for invaluable technical support.
Abbreviations Used
- 17-AAG
17-N-allylamino-17-demethoxygeldanamycin
- 7-AAD
7-aminoactinomycin D
- ANOVA
analysis of variance
- CRISPR
clustered regularly interspaced short palindromic repeat
- DCPIP
2,6-dichlorophenolindophenol
- DMEM
Dulbecco's modified Eagle's medium
- ERK
extracellular signal–regulated kinases
- FBS
fetal bovine serum
- FCCP
carbonyl cyanide 4-(trifluoromethoxy)phenylhydrazone
- HDCA
honokiol bis-dichloroacetate
- HIF1α
hypoxia-inducible factor 1-alpha
- Hsp90
heat shock protein 90
- IFD
induced fit docking
- IPTG
isopropyl β-D-1-thiogalactopyranoside
- MD
molecular dynamics
- MPNST
malignant peripheral nerve sheath tumor
- NAO
10-nonylacridine orange bromide
- NF1
neurofibromatosis type 1
- Nf1
neurofibromin 1
- OPLS
optimized potentials for liquid simulations
- OXPHOS
oxidative phosphorylation
- PN
plexiform neurofibroma
- Ras-GAP
Ras-GTPase-activating proteins
- SDH
succinate dehydrogenase
- SDHA
SDH subunit A
- SEM
standard error of the mean
- SIRT3
sirtuin-3
- SQR
succinate:coenzyme Q reductase
- TMRM
tetramethylrhodamine methyl ester
- TRAP1
TNF receptor-associated protein 1
Author Disclosure Statement
No competing financial interests exist.
Funding Information
A.R. was supported by grants from the University of Padova, Neurofibromatosis Therapeutic Acceleration Program, Associazione Italiana Ricerca Cancro (AIRC grant IG 2017/20749), Children's Tumor Foundation Drug Discovery Initiative (Award Grant 2016A-05-009), Piano for Life Onlus and Linfa OdV. I.M. was recipient of a Young Investigator Award Grant from Children's Tumor Foundation. G.C. was supported by grants from Associazione Italiana Ricerca Cancro (AIRC grant IG 2017/20019) and from Neurofibromatosis Therapeutic Acceleration Program. J.L.A. was supported by grants from NIH (AR47901 grant) and the Reynolds Sarcoma foundation.
Supplementary Material
References
- 1. Bernardi P, Rasola A, Forte M, and Lippe G. The mitochondrial permeability transition pore: channel formation by F-ATP synthase, integration in signal transduction, and role in pathophysiology. Physiol Rev 95: 1111–1155, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Bonner MY, Karlsson I, Rodolfo M, Arnold RS, Vergani E, and Arbiser JL. Honokiol bis-dichloroacetate (Honokiol DCA) demonstrates activity in vemurafenib-resistant melanoma in vivo. Oncotarget 7: 12857–12868, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Butler EK, Voigt A, Lutz AK, Toegel JP, Gerhardt E, Karsten P, Falkenburger B, Reinartz A, Winklhofer KF, and Schulz JB. The mitochondrial chaperone protein TRAP1 mitigates alpha-Synuclein toxicity. PLoS Genet 8: e1002488, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Cannino G, Ciscato F, Masgras I, Sanchez-Martin C, and Rasola A. Metabolic plasticity of tumor cell mitochondria. Front Oncol 8: 333, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Costa AC, Loh SH, and Martins LM. Drosophila Trap1 protects against mitochondrial dysfunction in a PINK1/parkin model of Parkinson's disease. Cell Death Dis 4: e467, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dando I, Pozza ED, Ambrosini G, Torrens-Mas M, Butera G, Mullappilly N, Pacchiana R, Palmieri M, and Donadelli M. Oncometabolites in cancer aggressiveness and tumour repopulation. Biol Rev Camb Philos Soc 94: 1530–1546, 2019 [DOI] [PubMed] [Google Scholar]
- 7. Elnatan D, Betegon M, Liu Y, Ramelot T, Kennedy MA, and Agard DA. Symmetry broken and rebroken during the ATP hydrolysis cycle of the mitochondrial Hsp90 TRAP1. Elife 6: e25235, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fan Y, Xue W, Schachner M, and Zhao W. Honokiol eliminates glioma/glioblastoma stem cell-like cells via JAK-STAT3 signaling and inhibits tumor progression by targeting epidermal growth factor receptor. Cancers (Basel) 11: 22, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Fassetta M, D'Alessandro L, Coltella N, Di Renzo MF, and Rasola A. Hepatocyte growth factor installs a survival platform for colorectal cancer cell invasive growth and overcomes p38 MAPK-mediated apoptosis. Cell Signal 18: 1967–1976, 2006 [DOI] [PubMed] [Google Scholar]
- 10. Ferraro M, D'Annessa I, Moroni E, Morra G, Paladino A, Rinaldi S, Compostella F, and Colombo G. Allosteric modulators of HSP90 and HSP70: dynamics meets function through structure-based drug design. J Med Chem 62: 60–87, 2019 [DOI] [PubMed] [Google Scholar]
- 11. Finley LW, Haas W, Desquiret-Dumas V, Wallace DC, Procaccio V, Gygi SP, and Haigis MC. Succinate dehydrogenase is a direct target of sirtuin 3 deacetylase activity. PLoS One 6: e23295, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Fitzgerald JC, Zimprich A, Carvajal Berrio DA, Schindler KM, Maurer B, Schulte C, Bus C, Hauser AK, Kubler M, Lewin R, Bobbili DR, Schwarz LM, Vartholomaiou E, Brockmann K, Wust R, Madlung J, Nordheim A, Riess O, Martins LM, Glaab E, May P, Schenke-Layland K, Picard D, Sharma M, Gasser T, and Kruger R. Metformin reverses TRAP1 mutation-associated alterations in mitochondrial function in Parkinson's disease. Brain 140: 2444–2459, 2017 [DOI] [PubMed] [Google Scholar]
- 13. Fried LE and Arbiser JL. Honokiol, a multifunctional antiangiogenic and antitumor agent. Antioxid Redox Signal 11: 1139–1148, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Guzzo G, Sciacovelli M, Bernardi P, and Rasola A. Inhibition of succinate dehydrogenase by the mitochondrial chaperone TRAP1 has anti-oxidant and anti-apoptotic effects on tumor cells. Oncotarget 5: 11897–11908, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Huang K, Chen Y, Zhang R, Wu Y, Ma Y, Fang X, and Shen S. Honokiol induces apoptosis and autophagy via the ROS/ERK1/2 signaling pathway in human osteosarcoma cells in vitro and in vivo. Cell Death Dis 9: 157, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Karagoz GE and Rudiger SG. Hsp90 interaction with clients. Trends Biochem Sci 40: 117–125, 2015 [DOI] [PubMed] [Google Scholar]
- 17. Kowalik MA, Guzzo G, Morandi A, Perra A, Menegon S, Masgras I, Trevisan E, Angioni MM, Fornari F, Quagliata L, Ledda-Columbano GM, Gramantieri L, Terracciano L, Giordano S, Chiarugi P, Rasola A, and Columbano A. Metabolic reprogramming identifies the most aggressive lesions at early phases of hepatic carcinogenesis. Oncotarget 7: 32375–32393, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lavery LA, Partridge JR, Ramelot TA, Elnatan D, Kennedy MA, and Agard DA. Structural asymmetry in the closed state of mitochondrial Hsp90 (TRAP1) supports a two-step ATP hydrolysis mechanism. Mol Cell 53: 330–343, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Leskovar A, Wegele H, Werbeck ND, Buchner J, and Reinstein J. The ATPase cycle of the mitochondrial Hsp90 analog Trap1. J Biol Chem 283: 11677–11688, 2008 [DOI] [PubMed] [Google Scholar]
- 20. Li H, Chang LJ, Neubauer DR, Muir DF, and Wallace MR. Immortalization of human normal and NF1 neurofibroma Schwann cells. Lab Invest 96: 1105–1115, 2016 [DOI] [PubMed] [Google Scholar]
- 21. Li L, Han W, Gu Y, Qiu S, Lu Q, Jin J, Luo J, and Hu X. Honokiol induces a necrotic cell death through the mitochondrial permeability transition pore. Cancer Res 67: 4894–4903, 2007 [DOI] [PubMed] [Google Scholar]
- 22. Masgras I, Ciscato F, Brunati AM, Tibaldi E, Indraccolo S, Curtarello M, Chiara F, Cannino G, Papaleo E, Lambrughi M, Guzzo G, Gambalunga A, Pizzi M, Guzzardo V, Rugge M, Vuljan SE, Calabrese F, Bernardi P, and Rasola A. Absence of neurofibromin induces an oncogenic metabolic switch via mitochondrial ERK-mediated phosphorylation of the chaperone TRAP1. Cell Rep 18: 659–672, 2017 [DOI] [PubMed] [Google Scholar]
- 23. Masgras I, Sanchez-Martin C, Colombo G, and Rasola A. The chaperone TRAP1 as a modulator of the mitochondrial adaptations in cancer cells. Front Oncol 7: 58, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Mo W, Chen J, Patel A, Zhang L, Chau V, Li Y, Cho W, Lim K, Xu J, Lazar AJ, Creighton CJ, Bolshakov S, McKay RM, Lev D, Le LQ, and Parada LF. CXCR4/CXCL12 mediate autocrine cell- cycle progression in NF1-associated malignant peripheral nerve sheath tumors. Cell 152: 1077–1090, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Moroni E, Agard DA, and Colombo G. The structural asymmetry of mitochondrial Hsp90 (Trap1) determines fine tuning of functional dynamics. J Chem Theory Comput 14: 1033–1044, 2018 [DOI] [PubMed] [Google Scholar]
- 26. Morra G, Potestio R, Micheletti C, and Colombo G. Corresponding functional dynamics across the Hsp90 Chaperone family: insights from a multiscale analysis of MD simulations. PLoS Comput Biol 8: e1002433, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Park HK, Hong JH, Oh YT, Kim SS, Yin J, Lee AJ, Chae YC, Kim JH, Park SH, Park CK, Park MJ, Park JB, and Kang BH. Interplay between TRAP1 and Sirtuin-3 modulates mitochondrial respiration and oxidative stress to maintain stemness of glioma stem cells. Cancer Res 79: 1369–1382, 2019 [DOI] [PubMed] [Google Scholar]
- 28. Pillai VB, Kanwal A, Fang YH, Sharp WW, Samant S, Arbiser J, and Gupta MP. Honokiol, an activator of Sirtuin-3 (SIRT3) preserves mitochondria and protects the heart from doxorubicin-induced cardiomyopathy in mice. Oncotarget 8: 34082–34098, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Pillai VB, Samant S, Sundaresan NR, Raghuraman H, Kim G, Bonner MY, Arbiser JL, Walker DI, Jones DP, Gius D, and Gupta MP. Honokiol blocks and reverses cardiac hypertrophy in mice by activating mitochondrial Sirt3. Nat Commun 6: 6656, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rasola A, Neckers L, and Picard D. Mitochondrial oxidative phosphorylation TRAP(1)ped in tumor cells. Trends Cell Biol 24: 455–463, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ratner N and Miller SJ. A RASopathy gene commonly mutated in cancer: the neurofibromatosis type 1 tumour suppressor. Nat Rev Cancer 15: 290–301, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sanchez-Martin C, Moroni E, Ferraro M, Laquatra C, Cannino G, Masgras I, Negro A, Quadrelli P, Rasola A, and Colombo G. Rational design of allosteric and selective inhibitors of the molecular chaperone TRAP1. Cell Rep 31: 107531, 2020 [DOI] [PubMed] [Google Scholar]
- 33. Schopf FH, Biebl MM, and Buchner J. The HSP90 chaperone machinery. Nat Rev Mol Cell Biol 18: 345–360, 2017 [DOI] [PubMed] [Google Scholar]
- 34. Sciacovelli M, Guzzo G, Morello V, Frezza C, Zheng L, Nannini N, Calabrese F, Laudiero G, Esposito F, Landriscina M, Defilippi P, Bernardi P, and Rasola A. The mitochondrial chaperone TRAP1 promotes neoplastic growth by inhibiting succinate dehydrogenase. Cell Metab 17: 988–999, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Semenza GL. HIF-1 mediates metabolic responses to intratumoral hypoxia and oncogenic mutations. J Clin Invest 123: 3664–3671, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Song JM, Anandharaj A, Upadhyaya P, Kirtane AR, Kim JH, Hong KH, Panyam J, and Kassie F. Honokiol suppresses lung tumorigenesis by targeting EGFR and its downstream effectors. Oncotarget 7: 57752–57769, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Taipale M, Jarosz DF, and Lindquist S. HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat Rev Mol Cell Biol 11: 515–528, 2010 [DOI] [PubMed] [Google Scholar]
- 38. Tian W, Xu D, Han W, He H, Cai H, Chen H, Zhou M, Chen J, and Deng YC. Cyclophilin D modulates cell death transition from early apoptosis to programmed necrosis induced by honokiol. Int J Oncol 42: 1654–1663, 2013 [DOI] [PubMed] [Google Scholar]
- 39. van der Knaap JA and Verrijzer CP. Undercover: gene control by metabolites and metabolic enzymes. Genes Dev 30: 2345–2369, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Vogel KS, Klesse LJ, Velasco-Miguel S, Meyers K, Rushing EJ, and Parada LF. Mouse tumor model for neurofibromatosis type 1. Science 286: 2176–2179, 1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Voloboueva LA, Duan M, Ouyang Y, Emery JF, Stoy C, and Giffard RG. Overexpression of mitochondrial Hsp70/Hsp75 protects astrocytes against ischemic injury in vitro. J Cereb Blood Flow Metab 28: 1009–1016, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ward PS and Thompson CB. Metabolic reprogramming: a cancer hallmark even warburg did not anticipate. Cancer Cell 21: 297–308, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wu W, Wang L, Wang L, Zu Y, Wang S, Liu P, and Zhao X. Preparation of honokiol nanoparticles by liquid antisolvent precipitation technique, characterization, pharmacokinetics, and evaluation of inhibitory effect on HepG2 cells. Int J Nanomedicine 13: 5469–5483, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Xiang F, Huang YS, Shi XH, and Zhang Q. Mitochondrial chaperone tumour necrosis factor receptor-associated protein 1 protects cardiomyocytes from hypoxic injury by regulating mitochondrial permeability transition pore opening. FEBS J 277: 1929–1938, 2010 [DOI] [PubMed] [Google Scholar]
- 45. Yap YS, McPherson JR, Ong CK, Rozen SG, Teh BT, Lee AS, and Callen DF. The NF1 gene revisited—from bench to bedside. Oncotarget 5: 5873–5892, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Yoshida S, Tsutsumi S, Muhlebach G, Sourbier C, Lee MJ, Lee S, Vartholomaiou E, Tatokoro M, Beebe K, Miyajima N, Mohney RP, Chen Y, Hasumi H, Xu W, Fukushima H, Nakamura K, Koga F, Kihara K, Trepel J, Picard D, and Neckers L. Molecular chaperone TRAP1 regulates a metabolic switch between mitochondrial respiration and aerobic glycolysis. Proc Natl Acad Sci U S A 110: E1604–E1612, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhang L, Karsten P, Hamm S, Pogson JH, Muller-Rischart AK, Exner N, Haass C, Whitworth AJ, Winklhofer KF, Schulz JB, and Voigt A. TRAP1 rescues PINK1 loss-of-function phenotypes. Hum Mol Genet 22: 2829–2841, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang P, Lu Y, Yu D, Zhang D, and Hu W. TRAP1 provides protection against myocardial ischemia-reperfusion injury by ameliorating mitochondrial dysfunction. Cell Physiol Biochem 36: 2072–2082, 2015 [DOI] [PubMed] [Google Scholar]
- 49. Zhao X, Li F, Sun W, Gao L, Kim KS, Kim KT, Cai L, Zhang Z, and Zheng Y. Extracts of magnolia species-induced prevention of diabetic complications: a brief review. Int J Mol Sci 17: 1629, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.