Graphical abstract
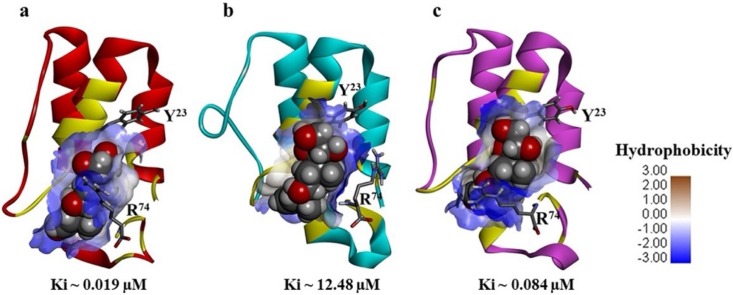
Illustrations of the C5a-prednisone complexes highlighting the range of estimated binding affinity.
Keywords: COVID19, Cytokine storm, C5a, Prednisone, Corticosteroids, Circular dichroism, Fluorescence, Molecular dynamics
Abstract
Complement system plays a dual role; physiological as well as pathophysiological. While physiological role protects the host, pathophysiological role can substantially harm the host, by triggering several hyper-inflammatory pathways, referred as “hypercytokinaemia”. Emerging clinical evidence suggests that exposure to severe acute respiratory syndrome coronavirus-2 (SARS-CoV2), tricks the complement to aberrantly activate the “hypercytokinaemia” loop, which significantly contributes to the severity of the COVID19. The pathophysiological response of the complement is usually amplified by the over production of potent chemoattractants and inflammatory modulators, like C3a and C5a. Therefore, it is logical that neutralizing the harmful effects of the inflammatory modulators of the complement system can be beneficial for the management of COVID19. While the hunt for safe and efficacious vaccines were underway, polypharmacology based combination therapies were fairly successful in reducing both the morbidity and mortality of COVID19 across the globe. Repurposing of small molecule drugs as “neutraligands” of C5a appears to be an alternative for modulating the hyper-inflammatory signals, triggered by the C5a-C5aR signaling axes. Thus, in the current study, few specific and non-specific immunomodulators (azithromycin, colchicine, famotidine, fluvoxamine, dexamethasone and prednisone) generally prescribed for prophylactic usage for management of COVID19 were subjected to computational and biophysical studies to probe whether any of the above drugs can act as “neutraligands”, by selectively binding to C5a over C3a. The data presented in this study indicates that corticosteroids, like prednisone can have potentially better selectively (Kd ∼ 0.38 μM) toward C5a than C3a, suggesting the positive modulatory role of C5a in the general success of the corticosteroid therapy in moderate to severe COVID19.
1. Introduction
COVID19 pandemic (Wiersinga et al., 2020) that has reshaped the world in many ways is caused by the orchestrated response of the immune system (Paces et al., 2020), triggered by the exposure and subsequent invasion of a virus, phylogenetically related to the broad class of coronavirus family (Hu et al., 2021), recently named as severe acute respiratory syndrome coronavirus-2 (SARS-CoV2). The function of the immune system is controlled by the active interplay between its two broad arms, (i) innate immunity (quick to respond) and (ii) adaptive immunity (slow to respond), which collectively provides lasting immunity to the hosts, against plethora of invading families of pathogens. The innate immunity arm of the immune system recruits the complement system (Freeley et al., 2016) -a cocktail of several proteins- as the first line of defence to protect the host from the initial assaults of the pathogen. However, increasing evidences strongly suggest that complement system also plays a pivotal role in controlling the adaptive immunity, by acting as a bridge between the two arms of the immune system (Lo and Woodruff, 2020). Thus, complement and immunity can be considered as both sides of the coin. Post detection of invading pathogens in the host body, controlled activation of the complement cascade is highly essential for restoring the normal physiology. Over stimulated complement cascade can significantly contribute toward several aggressive inflammation (Barrington et al., 2001) induced pathophysiological conditions, including multiple organ failure and death, through production of functionally diverse biological molecules (Schwartz and Suskind, 2020). Emerging clinical evidence suggests that in the process to fend off the SARS-CoV2 from the host body, the overwhelmed complement triggers the lethal “hypercytokinaemia” (Burgos-Blasco et al., 2020; Scala and Pacelli, 2020), which has been associated to contribute toward the complications, noted in patients with moderate to severe COVID19 (Badawi, 2020; Wang et al., 2020). In fact, it is observed that about 15 % of the patients severely infected with SARS-CoV2 have elevated levels of proinflammatory cytokines (Cugno et al., 2020; Guan et al., 2020) in the plasma and are highly susceptible to develop the severe acute respiratory distress syndrome (ARDS) (Holter et al., 2020). The hyper-inflammatory response of the overwhelmed complement is amplified by the over production of proinflammatory modulators, such as C3a (Coulthard and Woodruff, 2015) and C5a (Guo and Ward, 2005), among which the most potent one is the C5a, which has been strongly associated in aiding the “cytokine storm” (Chauhan et al., 2020; Mahmudpour et al., 2020; Mishra et al., 2020), mast cell degranulation (Ali, 2010) and enhancement of vascular permeability (Khan et al., 2013). In fact, several fold increase in the concentration of C5a (Carvelli et al., 2020; Prendecki et al., 2020), including the several cytokines have been observed in the plasma of moderate to severe COVID19 patients, compared to the healthy subjects. C5a post binding to C5aR, expressed in both myeloid and non-myeloid tissues promote the generation of reactive oxygen species (ROS) through oxidative burst, which contributes to the virus induced acute lung injury (ALI) and mortality (Wang et al., 2015). In addition, C5a-C5aR interaction promotes the adhesion of leukocytes, which subsequently transmigrate to the lung parenchyma. No wonder that respiratory distress is one of the prime contributors in the COVID19 related mortality (Carsana et al., 2020; Li and Ma, 2020). Further, overactivation of C5a-C5aR signaling axes can contribute toward endothelial dysfunction (Perico et al., 2021) and thus, even if lungs appear to be the first target, it can potentially affect the other organs (Wadman et al., 2020) due to the collision with other intertwined signaling axes (Chauhan et al., 2020). Therefore, it appears that neutralizing the harmful effects of the inflammatory modulators of the complement system (Risitano et al., 2020; Campbell and Kahwash, 2020) can actually be beneficial for the management of COVID19 (Huang et al., 2020). It is worth highlighting that clinical trials specifically targeting the complement proteins (Diurno et al., 2020; Giamarellos‐Bourboulis et al., 2020), such as C3 (trial id: NCT04395456), C5 (trial id: NCT04288713) and C5a (trial id: NCT04333420) with neutralizing antibodies have already been initiated with a hope to modulate the hyper-inflammatory role of complement in moderate to severe COVID19. Though, available data indicates the possible success of the approach (Mastellos et al., 2019), the comprehensive outcome of the above clinical studies are still awaited.
On the other hand, while the hunt for the safe and efficacious vaccines were underway, polypharmacology based combination therapies involving small molecule drugs were fairly successful in reducing both the morbidity and mortality of COVID19 across the globe. For example, synergistic combination of hydroxychloroquine / ivermectin (Patri and Fabbrocini, 2020), ruxolitinib / eculizumab (Giudice et al., 2020), nitazoxanide / azithromycin (Kelleni, 2020), and remdesivir / corticosteroids (Wang et al., 2021) have shown appreciable results in small to medium scale studies against COVID19. In addition, several small molecule drugs, such as azithromycin (Schwartz and Suskind, 2020; Pani et al., 2020), colchicine (Schlesinger et al., 2020), famotidine (Mather et al., 2020), fluvoxamine (Lenze et al., 2020), dexamethasone (Sterne et al., 2020; Ledford, 2020) and prednisone (Bartoletti et al., 2021) have also been subjected to small controlled studies as prophylactic to manage the COVID19 pandemic. Both azithromycin (macrolide) and colchicine have immunomodulatory effects and are known to downregulate proinflammatory cytokines, and chemokines respectively. Famotidine is a histamine receptor blocker used for treatment of peptic ulcer disease, whereas fluvoxamine is a selective serotonin reuptake inhibitor (SSRI), prescribed as an antidepressant for treatment of social anxiety disorder. Both dexamethasone and prednisone belongs to the corticosteroid class of drugs with potent anti-inflammatory activities (Rizk et al., 2020) and are generally prescribed for managing the “cytokine storm”, as they exerts antagonistic action on glucocorticoid receptors (Kadmiel and Cidlowski, 2013), a protein that shuttles between cytosol and nucleus, belonging to the broad class of nuclear receptor family. The publicly available results for the above drugs are generally encouraging in nature, though exact mechanism of action of these drugs in the context of COVID19 is not well understood so far. Interestingly, among all, corticosteroid class of drugs, such as dexamethasone and prednisone have demonstrated really appreciable results in large scale studies involving patients with moderate to severe COVID19 (Sterne et al., 2020; Bartoletti et al., 2021). It is worth mentioning that we have been pursuing the idea whether small molecule drugs can be repurposed to act as potential “neutraligands” of C5a (Mishra et al., 2020; Mishra and Rana, 2019; Mishra et al., 2021), as an alternative to the known antibodies, so that the amplification of the hyper-inflammatory response can be modulated in a dose dependent manner, under the disease settings. Our basic studies indicate that selective small molecule drugs can strongly bind to any one of three potential “hotspots” on C5a (Mishra et al., 2020; Mishra and Rana, 2019), triggering conformational alteration, which can significantly affect the interaction of C5a with C5aR and thus, can alleviate the strong inflammatory response. This approach has a strong potential similar to the small molecule protein degraders (Raina and Crews, 2017), which requires further exploration. Thus, in the current study, azithromycin, colchicine, famotidine, fluvoxamine, dexamethasone and prednisone (Fig. 1 ) were subjected to both computational and biophysical studies to probe whether the general success of any of the above drugs in managing COVID19 could be partially due to their selective neutralizing interactions with the complement proteins like C3a or C5a. The data presented in this study indicates that prednisone can have potentially better selectively toward C5a than C3a, which can act as a positive modulator in reducing the inflammatory response exerted by the immune system in COVID19. Prednisone’s dual action on C5a, including the glucocorticoid receptors (GRs) (Kadmiel and Cidlowski, 2013; Oakley and Cidlowski, 2013) could be the plausible reason behind the general success of the corticosteroid therapy in treatment of moderate to severe COVID19.
Fig. 1.
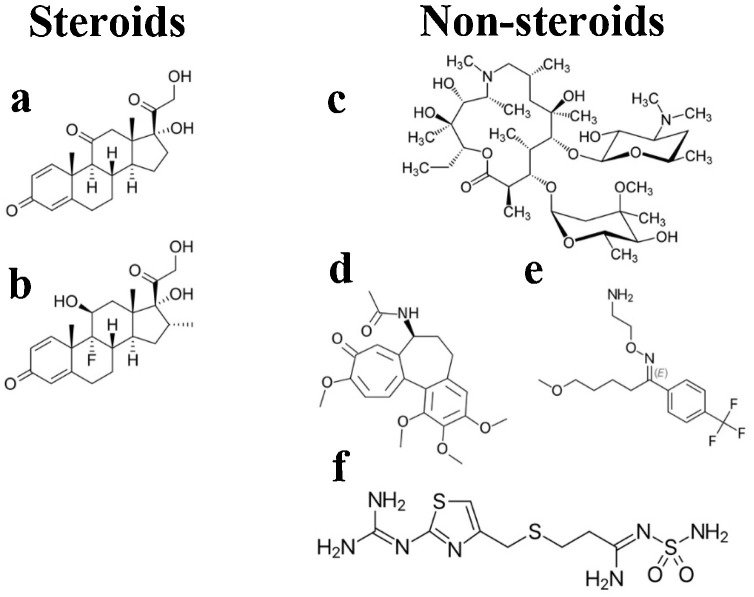
The steroid and non-steroid class of drugs screened against the various conformers of the complement proteins like C3a and C5a. (a) prednisone, (b) dexamethasone, (c) azithromycin, (d) colchicine, (e) fluvoxamine and (f) famotidine.
2. Material and methods
2.1. Computational methods and chemicals
PDB coordinates of the C3a (Bajic et al., 2013) (4HW5) and C5a (Zhang et al., 1997) (1KJS) were downloaded from www.rcsb.org. PyMOL (The PyMOL Molecular Graphics System, Version 1.1r1, Schrödinger, LLC) and Discovery studio (Accelrys) software were utilized for initial processing, visualization, analysis, and presentation of the protein structures. The initial coordinates of the drug molecules (a) prednisone (1H61) (Barna et al., 2001), (b) dexamethasone (3GN8) (Bridgham et al., 2009), (c) azithromycin (5UXD) (Pawlowski et al., 2018), (d) colchicine (5NKN) (Barkovskiy et al., 2019), (e) fluvoxamine (4MM9) (Wang et al., 2013) and (f) famotidine (6G3V) (Angeli et al., 2018) were respectively extracted from the www.rcsb.org. Subsequently, the topological parameter of all the drug molecules required for docking and molecular dynamics (MD) simulation were generated by using the PRODRG server (Schüttelkopf and Van Aalten, 2004). Prednisone’s parameters were appropriately edited to suit the gromos-96 43a1 force field built into GROMACS (Hess et al., 2008). Experimental and computational data were plotted in GraphPad Prism (version 6 for Windows, GraphPad Software, La Jolla California USA, www.graphpad.com). The R&D Systems provided the recombinant C5a protein. Sigma Aldrich supplied the prednisone molecule. Solvent accessible surface area (SASA) calculation for the fluorophores on C5a were performed by using the NACCESS program with a Ala-X-Ala template and solvent probe radius set to 1.4 Å.
2.2. Molecular dynamics (MD) studies
All the MD studies involving the proteins or protein complexed to drug molecule were carried out at 300 K in presence of explicit water. Prior to MD simulation, the system was subjected to energy minimization in a cubic box with appropriate periodic boundary, by recruiting the gromos-96 43a1 united atom force field built in to the GROMACS. Briefly, the side chains of Lys and Arg were positively charged, whereas the side chain of the Asp and Glu were negatively charged on both C3a and C5a proteins to roughly mimic the protonation state at the physiological pH. Both the systems were energy minimized to 100 kJ mol−1 nm− 1 tolerances with steepest descent, first in vacuum and then in presence of simple point charge (SPC) water molecules, as provided in GROMACS. The systems were solvated by using appropriate number of SPC water molecules covering the entire protein with solvent density set to the value corresponding to 1 atm at 300 K and further, the net charge of the systems were neutralized, by randomly placing requisite number of counter ions to the box. For example, the C3a system contained total 41,011 atoms with 13,390 solvent molecules and 7 chloride ions. Similarly, the C5a-prednisone system contained total 36,867 atoms with 12,031 solvent molecules and 7 chloride ions. Both the system were equilibrated through NVT (0.5 ns) and NPT (0.5 ns) conditions prior to MD studies. The C3a protein was subjected to 50 ns of production MD run, whereas the C5a-prednisone complex was subjected to 200 ns production MD run. Protein and solvents-ions were coupled independently to V-rescale bath at 300 K, to the coupling time constant 0.1 ps. Bonds were constrained with LINCS with order 4. Non-bonded pair list cut-off was 1.2 nm with a grid function. Numerical integrations were performed in step size of 2 fs and the coordinates were updated every 5 ps. Conformational clustering across the trajectory was performed every 50 ps with RMSD cut-off ≤ 1.5 Å, by recruiting the gromos fitting method, as defined in GROMACS. The trajectories were thoroughly analysed for both the systems, by recruiting the utility modules available within the GROMACS.
2.3. Automated docking studies
All the drug molecules were respectively subjected to automated docking, by recruiting the AutoDock (Morris et al., 2009) against various conformers of C3a and C5a. For example, all the drugs were screened against the entire surface area of energy minimized conformer of C3a, as well as against the most populated conformer of C3a evolved over 50 ns of MD. Similarly, the entire surface area of the most populated conformer of C5a, mono-glycosylated and tri-glycosylated C5a evolved over 50 ns of MD were subjected to scanning against all the drug molecules initially. Subsequently, the most populated conformers of C5a complexed to prednisone evolved over 200 ns of MD were further subjected to rigid docking to gauge the range for realistic binding affinities. Optimum grid dimension covering the entire protein surface along XYZ directions was achieved by using AutoGrid program. The Lamarckian genetic approach (LGA) was applied for a population size of 250 with the maximum number of generations set to 27,000. Structurally distinct conformational clusters of the drugs were ranked in terms of increasing energy. All the drug molecules were initially subjected to flexi dock, followed by rigid dock until the best bound conformer having the least binding energy with maximum intermolecular contacts was obtained.
2.4. Estimation of the binding free energies of the prednisone-C5a complex
The following equation: ΔGbinding = Gcomplex − (Gprotein − Gligand), implemented in the g_mmpbsa program was used for calculating the binding free energy of the prednisone complexed to C5a, by recruiting the Molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) method, as described elsewhere (Kumari et al., 2014). The free energy contribution of the prednisone was estimated by the following equation: G = EMM + Gsolv – TSsolute, where EMM (molecular mechanics energy) represents the summation of van der Waals and electrostatic, Gsolv represents the solvation energy contributed by both polar and non-polar solvation free energy, and TSsolute represents the temperature and entropy of the solute. Dielectric constant of solute and solvent were respectively fixed at 20 and 80 respectively for calculation of polar solvation energy. A value of 0.5 Å grid space was taken to calculate electrostatic energy. Probe radius was set to 1.4 Å to calculate the non-polar contribution to solvation free energy through SASA method. Finally, 200 conformers randomly selected from the first major cluster populated over the 200 ns of the MD trajectory were respectively used for calculating the average binding free energy for the prednisone.
2.5. Pilot biophysical studies
The fluorescence studies on C5a were performed in pure 1X PBS (pH ∼ 7.4) at 25 °C, by using the Cary Eclipse Fluorescence Spectrophotometer (Agilent Technologies) equipped with PCB 1500 Water Peltier System. The excitation and emission slit widths were set to 5 nm with excitation wavelength set at 280 nm and emission range set between 290 and 450 nm. The fluorescence spectra of 0.1 μM C5a were recorded both in presence and absence of 0–10 μM prednisone with an average of three scans and the background spectra of the buffer were appropriately subtracted. Prednisone with absorption maximum at 242 nm did not display significant fluorescence emission between 290−450 nm. The absorption spectra of prednisone was recorded by using nanodrop plates in a Spectramax ID3 plate reader. Binding affinity of prednisone toward C5a was estimated by fitting the normalized fluorescence of C5a against the prednisone doses, by recruiting the non-linear regression methods available in GraphPad Prism. The circular dichroism (CD) studies of C5a were carried on a Chirascan CD spectropolarimeter system in far-UV region at 25 °C. Each sample was subjected to minimum 3 scans with a time constant of 1 s and step size of 1 nm. The solvents and buffers used in the study were filtered and degassed by nitrogen bubbling method. The C5a was solubilized in 1X PBS (pH∼ 7.4) and the prednisone stock solutions were prepared in 1X PBS after solubilizing it in DMSO. The CD spectra of 0.1 μM C5a were recorded both in presence and absence of 1 μM prednisone. Similarly, CD spectra of 1 μM prednisone was also recorded and subsequently subtracted from the C5a + prednisone spectra. Samples were incubated for minimum 1 h at 4 °C prior to the CD studies. The molar ellipticity was converted to mean residue ellipticity [θMRE] after background subtraction, by using the methods described elsewhere (Greenfield, 2006).
3. Results
3.1. MD studies on C3a
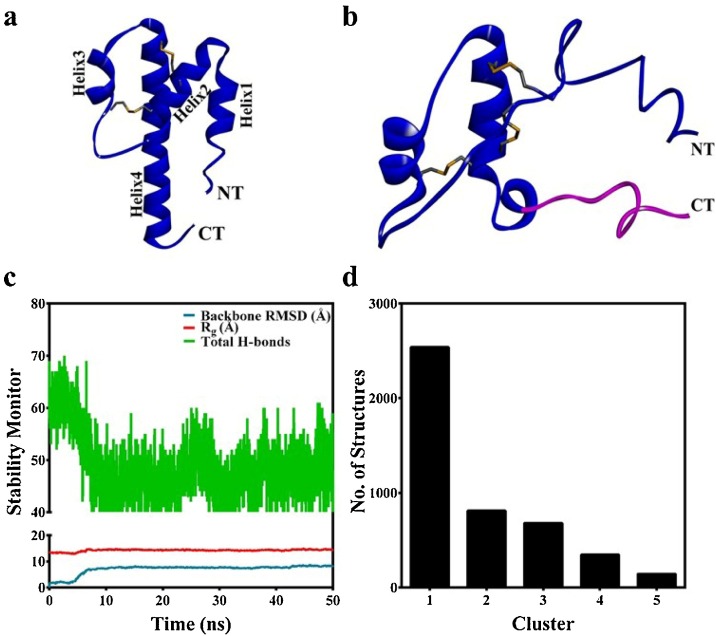
All the pathways of complement converge at C3, whose proteolytic cleavage produces the C3a anaphylatoxin (Fig. 2 a), a 77-aa polypeptide (Bajic et al., 2013) with close structural similarity with C5a. Interestingly, C3a does not have an unpaired cysteine as observed in case of C5a. Literature evidences that after reaching a critical concentration in the plasma, C3a can exert both pro-inflammatory and anti-inflammatory action on the surrounding cells (Coulthard and Woodruff, 2015). Though C3 concentration in plasma is usually high, anti-inflammatory action of C3a, in response to the proinflammatory role of C5a in COVID19 has not been clinically corelated to the elevated plasma concentration of C3a so far. Moreover, conformational integrity and dynamic behaviour of C3a are not postulated in the literature so far. Thus, the human C3a structure (4HW5) observed in solid was subjected to 50 ns MD simulation in presence of explicit water molecules to probe its conformational heterogeneity, prior to the screening of the drug molecules, illustrated in Fig. 1. Structural comparison indicates that the four helix bundle structure of C3a observed in solid (Fig. 2a) is not sustained in solution (Fig. 2b). The Helix2 on C3a becomes an extended loop in the major conformer of C3a evolved over 50 ns of MD. In addition, the length of the Helix4 is also shortened significantly, making the last 13 aa [Arg65-Arg77] of the C-terminus completely structurally unordered (Fig. 2b). This observation made in MD is in agreement with the conformational analysis data obtained from the synthetic peptides corresponding to Arg65-Arg77 region of C3a (Hugli and Erickson, 1977). Overall, the data presented in Fig. 2 indicates that C3a can be conformationally dynamic and perhaps prefers to remain as an intrinsically disordered protein (Oldfield and Dunker, 2014) in solution, after being detached from C3. Thus, it is important to consider this aspect of C3a, while performing both high throughput in silico or cell culture based drug screening studies (Rowley et al., 2020). More importantly, given the intrinsically disorder structure of C3a, the potent small molecule antagonist of C3aR receptors (Scully et al., 2010) should also be checked for their cross reactivity with C3a to further understand their biological implications.
Fig. 2.
(a) The energy minimized four helix bundle structure of C3a. (b) The central conformer of the first major cluster populated for C3a, over the 50 ns of MD at 300 K. The structurally unordered last 13 aa [Arg65-Arg77] of the C-terminus is highlighted in pink. (c) Monitoring the basic structural parameters of C3a over the duration of MD. The decrease in total number of hydrogen bonds indicates structurally unordered nature of C3a. (d) The number of major conformational clusters of C3a evolved over the duration of MD.
3.2. Screening of the drug molecules against the conformers of C3a
In the complement cascade, C3a is upstream of C5a and thus, the drug molecules presented in Fig. 1 were first subjected to screening against C3a, by recruiting automated docking studies. In this process, the entire surface area of the energy minimized structure (conformer 1) of C3a (Fig. 2a) and the major conformer of C3a (conformer 2) evolved during MD (Fig. 2b) were respectively screened against the drug molecules to find the potential binding sites with a cut-off for estimated Ki ≤ 10 μM. This was done to effectively probe the range of binding affinity, which can vary with conformational heterogeneity of C3a. The data summarized in Table 1 indicates that the steroid class of drugs, such as prednisone and dexamethasone demonstrated an estimated binding affinity ranging between 8−17 μM toward the conformers of C3a, whereas among the non-steroid drugs, only colchicine demonstrated an estimated binding affinity ranging between 6−23 μM toward the conformers of C3a. Thus, considering the general cut-off of 10 μM, beside colchicine, the steroid class of drugs, especially the prednisone appears to be the ideal candidate that can modulate the immune response, by acting on complement proteins like C3a.
Table 1.
Estimated binding affinity of the drugs screened against the major conformers of C3a. Drugs demonstrating ∼ Ki ≤ 10 μM are highlighted in bold.
| Drug | ∼ Ki (μM) / B. E. (kcal/mol) toward C3a |
|
|---|---|---|
| Conformer 1 | Conformer 2 | |
| Prednisone | 8.92 / -6.89 | 12.14 / -6.71 |
| Dexamethasone | 10.23 / -6.81 | 17.44 / -6.49 |
| Azithromycin | 4090 / -3.26 | 1020 / -4.08 |
| Colchicine | 23.32 / -6.32 | 6.17 / -7.11 |
| Fluvoxamine | 8040 / -2.83 | 815.95 / -4.21 |
| Famotidine | 352.95 / -4.71 | 508.21 / -4.49 |
3.3. Screening of the drug molecules against the conformers of C5a
C5a is the most potent proinflammatory polypeptide of the complement cascade, which has lasting effects on the surrounding tissues than any other complement protein. More importantly, C5a has been strongly associated with variety of inflammatory conditions, including the most recent COVID19 (Cugno et al., 2020). Thus, to find the most consistent drug molecule in the list, automated docking studies were also performed by recruiting three most populated conformers (Fig. S1), respectively evolved for unglycosylated (conformer 1), monoglycosylated (N-acetyl glucosamine attached to Asn64; conformer 2) and triglycosylated (N-acetyl glucosamine attached to Asn29, Asn30, and Asn64; conformer 3) C5a over 50 ns of MD (Mishra et al., 2021). The mono- and triglycosylated conformers of C5a were recruited to loosely mimic the heavily glycosylated natively activated state of C5a (Huber-Lang et al., 2002). The estimated binding affinity and energy observed for the drug molecules for the various conformers of C5a is presented in Table 2 . The data indicates that prednisone could bind to all the conformers of the C5a within a range of 0.8−6 μM, which is well within the hypothesized cut-off of estimated Ki ≤ 10 μM. Moreover, prednisone demonstrated binding at all the three hypothesized hotspots on C5a (Fig. S1). Similar observations has also been made in earlier in silico studies (Mishra et al., 2021) for trans-resveratrol, a phytoalexin known to demonstrate anti-inflammatory activities in both in vitro and in vivo studies (Issuree et al., 2009). It is interesting, as prednisone also demonstrated comparatively weaker binding affinity to C3a (Fig. S2). Apart from this, the next best molecule was dexamethasone, which demonstrated binding affinity in a range of 5−46 μM toward all the conformers of C5a. In comparison, colchicine demonstrated a binding affinity ranging between 3−89 μM toward all the conformers of C5a and thus, was not pursued further in the current study, including the azithromycin, famotidine and fluvoxamine (Fig. S3). Collectively, prednisone appears to be the most consistent drug molecule among the seven drug molecules that has been screened against C3a and C5a. Further, as prednisone demonstrated stronger binding affinity consistently toward C5a than C3a, it was subjected to further MD studies involving C5a.
Table 2.
Estimated binding affinity of the drugs screened against the major conformers of C5a. Drugs demonstrating ∼ Ki ≤ 10 μM are highlighted in bold.
| Drug | ∼ Ki (μM) / B. E. (kcal/mol) toward C5a |
||
|---|---|---|---|
| Conformer 1 | Conformer 2 | Conformer 3 | |
| Prednisone | 2.70 / -7.60 | 0.858 / -8.28 | 6.83 / -7.05 |
| Dexamethasone | 46.59 / -5.91 | 5.92 / -7.13 | 18.98 / -6.44 |
| Azithromycin | 380.53 / -4.67 | 1410 / -3.89 | 207.83 / -5.02 |
| Colchicine | 88.99 / -5.53 | 3.69 / -7.41 | 89.07 / -5.53 |
| Fluvoxamine | 558.98 / -4.44 | 735.51 / -4.27 | 117.01 / -5.36 |
| Famotidine | 26.16 / -6.25 | 146.26 / -5.23 | 118.11 / -5.36 |
3.4. Probing the structural stability of the C5a-prednisone complex
Among the three major conformers of C5a, prednisone demonstrated relatively strongest in silico affinity toward the monoglycosylated C5a and weakest affinity toward the triglycosylated C5a. Since, most of the biomolecular signaling studies reported in the field recruits the unglycosylated recombinant C5a, the prednisone complexed to unglycosylated C5a was subjected to MD studies in explicit water over 200 ns at 300 K. Further, this complex was preferred over others primarily for the ease of correlation with the subsequent pilot biophysical studies involving recombinant C5a.
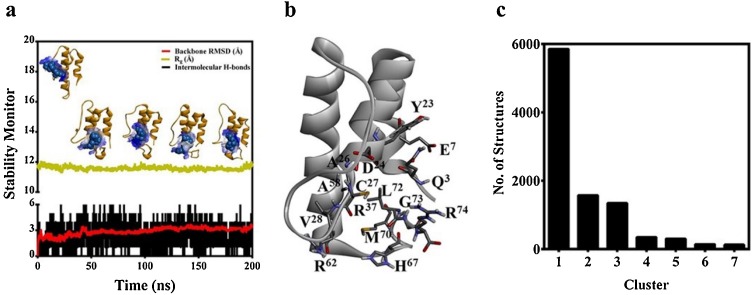
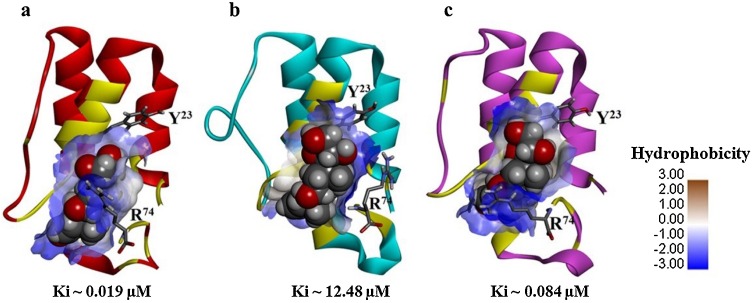
The data presented in Fig. 3 a suggests that prednisone could remain bound to the hotspot of C5a over the duration of the MD trajectory, indicating the physical viability of the strong intermolecular interactions existing between C5a and prednisone. The physical viability of the complex derives strength from average three number of intermolecular hydrogen bonds consistently sustained over the duration of MD (Fig. 3a). In addition to this, prednisone appears to be comfortably nestled inside the defined “hotspot” lined by several amino acids side chains that have both hydrophobic and hydrophilic properties (Fig. 3b). This is further supported by the central conformers (Fig. S4) of the C5a-prednisone complex, representing the first three major clusters (Fig. 3c), populated over the entire MD trajectory. The backbone RMSD of these central conformers vary between 0.98–2.35 Å from the unglycosylated structure of C5a (Fig S1a). In order to judge the more realistic range of binding affinity and to remove the conformational biasness from the docking results, prednisone was further subjected to guided docking involving the three central conformers (Fig. S4), which are illustrated as the representative C5a-prednisone complexes in Fig. 4 . The combined docking data indicates that prednisone may bind to recombinant human C5a with an estimated affinity ranging between 0.019−12 μM.
Fig. 3.
(a) Monitoring the various structural parameters for probing the physical viability of the C5a-prednisone complex at an interval of 50 ns over 200 ns of MD at 300 K. The prednisone molecule nestled inside the “hotspot” is presented in CPK model. (b) The representative amino acids lining the “hotspot” on C5a involved in binding the prednisone are highlighted in sticks. (c) The major conformational clusters of C5a evolved over the duration of MD, highlighting the number of structures in each cluster.
Fig. 4.
Illustrations of the C5a-prednisone complexes, highlighting the nanomolar to micromolar binding range of prednisone, nestled inside the defined “hotspot” of C5a. Prednisone highlighted in the CPK model is presented in complex with the respective central conformer of the (a) cluster 1, (b) cluster 2 and (c) cluster 3 of C5a evolved over the duration of MD. One of the fluorophore of C5a (Tyr 23) closest to the illustrated binding site is highlighted in stick. Prednisone binding interlocks the side chain of Arg74, presented in stick is known to be highly important for biomolecular signaling of C5a. The binding site of the C5a is rendered in typical “blue-white-brown” surface mode to indicate the contribution of both hydrophilic (blue) and hydrophobic (brown) side chains of different amino acids.
3.5. Estimating the binding free energy of the C5a-prednisone complex
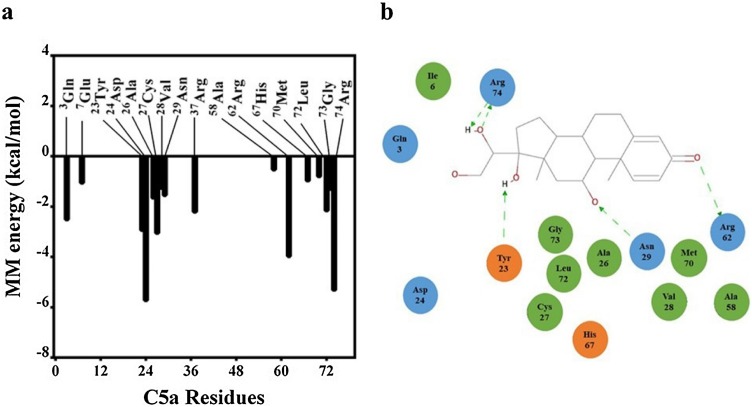
Molecular mechanics Poisson-Boltzmann surface area (MM-PBSA) calculation was performed to estimate the binding free energy of the C5a-prednisone complex. The calculation of the binding free energy of the complex involved 200 conformers selected randomly from the first major cluster populated over the entire 200 ns MD trajectory, by following the procedure detailed under the materials and methods. The binding free energy obtained for the complex (-60.89 ± 7.96 kcal/mol) suggests that prednisone strongly interacts with C5a, which can be positively correlated with the binding affinity estimated from the docking studies. The estimated free energy of binding is based on the van der Waals energy (-47.48 ± 5.37 kcal/mol), electrostatic energy (-23.79 ± 9.19 kcal/mol), polar solvation energy (14.14 ± 3.50 kcal/mol) and apolar energy (-3.76 ± 0.32 kcal/mol), as obtained for the C5a-prednisone complex. Further analysis (Fig. 5 a) suggests that several amino acids on C5a strongly contribute toward the overall free energy of binding of prednisone. Interestingly several C-terminal amino acids, responsible for downstream signaling of C5a, including the Arg74 strongly contributed toward the binding of prednisone.
Fig. 5.
(a) Decomposition of the MM energy plot highlighting the “hotspot” residues on C5a contributing toward the overall binding free energy of C5a-prednisone complex. (b) Schematic illustration of the intermolecular interaction observed in a representative C5a-prednisone complex. Amino acids involved in only hydrophobic interaction and the amino acids involved in both hydrophobic and electrostatic interactions are respectively coloured in green and blue. The amino acids coloured in orange are also involved in “alkyl-π” interactions. The green dotted arrows indicate the directionality of the hydrogen bonds.
3.6. Probing the interaction of prednisone with C5a by fluorescence and CD spectroscopy
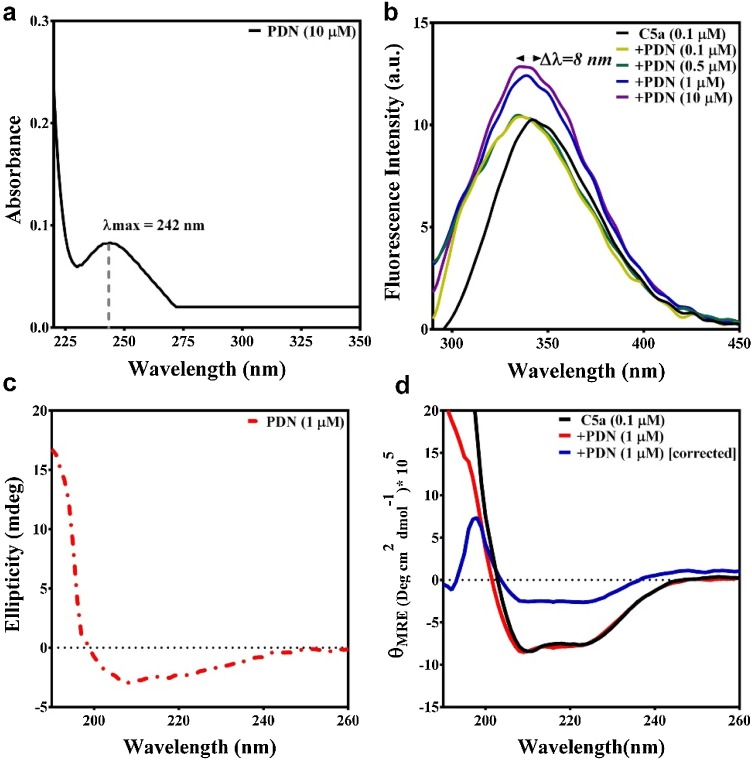
Pilot biophysical studies involving fluorescence and circular dichroism (CD) spectroscopy were performed further in PBS buffer to confirm the observations made in the computational studies. Prednisone with an absorption maximum (λmax) at 242 nm (Fig. 6 a) did not display any observable intrinsic fluorescence (Fig. 6b), while excited between 278−280 nm.
Fig. 6.
(a) Absorption spectra of prednisone (PDN), highlighting the absorption maximum in PBS buffer. (b) The effect of 0.1-10 μM prednisone (PDN) on the intrinsic fluorescence spectra of 0.1 μM C5a, highlighting the observed blueshift in the fluorescence emission maximum of C5a. (c) The signature CD pattern observed for 1 μM prednisone at 25 ℃ in PBS buffer. (d) Conformational perturbation observed for 0.1 μM C5a in response to 1 μM prednisone (PDN), suggesting the binding of prednisone to C5a.
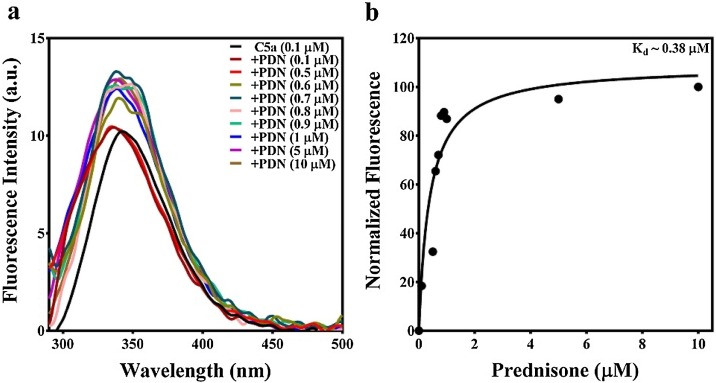
Since computational studies estimated that prednisone’s affinity (Fig. 4) toward C5a may be somewhere between 19 nM - 12 μM, the fluorescence of C5a was recorded both in presence and absence of 0−10 μM of prednisone. Interestingly, an appreciable blueshift (Δλ ∼ 8 nm) in the fluorescence emission maximum of C5a was noted in presence of 0−0.5 μM prednisone (Fig. 6b), though the intrinsic fluorescence intensity of C5a did not change significantly, suggesting changes in the microenvironment of the fluorophores of C5a, which is most likely due to the hydrophobic burial induced by the binding of prednisone. Further increase in concentration of prednisone (1−10 μM), significantly contributed toward the increase in the intrinsic fluorescence intensity of C5a (Fig. 6b), which almost saturated in presence of 1 μM prednisone, though minor changes in fluorescence intensity of C5a was also noted in presence of 5−10 μM prednisone. To probe this observation further, solvent accessible surface area (SASA) of the intrinsic fluorophores (Tyr 13 and Tyr 23) of C5a were calculated considering the energy minimized structure of free C5a as the reference. SASA calculation performed relative to Ala-X-Ala tripeptide indicates that the side chains of both Tyr 13 and Tyr 23 are highly buried and only 27 ± 2% of Tyr 13 and 30 ± 3% of Tyr 23 side chains are relatively exposed to the solvent. In comparison, the side chains of both Tyr 13 and Tyr 23, respectively demonstrated 18 ± 6% and 24 ± 4% relative solvent accessibility in C5a-prednisone complex, indicating further hydrophobic burial of the intrinsic fluorophores. Collectively, the data suggests that prednisone strongly binds to C5a, however further CD studies were performed to understand the overall effect of prednisone binding on the conformation of C5a. Since, 1 μM prednisone almost saturated the fluorescence signal of C5a, further CD studies were performed with 0.1 μM C5a both in presence and absence of 1 μM prednisone. C5a in the absence of prednisone produced the characteristic CD signature of helical bundle proteins. Similarly, prednisone being inherently chiral also demonstrated a characteristic CD signature in the far-UV region (Fig. 6c). As presented in Fig. 6d, the CD signature of the 0.1 μM C5a did not display any significant conformational changes in presence of 1 μM prednisone. Nevertheless, correcting the CD spectra for the contribution made by the 1 μM prednisone displayed the strong conformational changes in C5a, in agreement with the fluorescence studies. In addition, this observation is in sync with the earlier observations made for other neutraligands of C5a (Mishra and Rana, 2019; Mishra et al., 2021). Moreover, fluorescence titrations (Fig. 7 ) of 0−10 μM prednisone against 0.1 μM provided an estimated Kd ∼ 0.38 μM, confirming the high affinity binding interaction between prednisone and C5a.
Fig. 7.
(a) Fluorescence titration studies involving 0.1-10 μM prednisone (PDN) against 0.1 μM C5a. (b) Calculation of the binding affinity of prednisone from the normalized fluorescence data of C5a.
4. Discussion
Advanced clinical studies conducted so far to understand the pathophysiology of COVID19 suggest that it is a systemic syndrome, with elevated inflammatory markers in the serum, which gets exacerbated due to the strong hyperinflammatory response triggered by the host’s immune system, subsequently leading to the moderate to severe COVID19. The fine balance between the protective immunity and dysfunctional immune response decides recovery or death as one of the clinical outcome in case of COVID19. While both innate and adaptive arm of the immune system plays a cohesive role in providing lasting protective immunity leading to the recovery, the innate immunity is the fast acting arm of the first and second line of defence system built in to immune system, which provides non-specific protection against pathogens, by triggering both local and systemic inflammation. Complement system comes under the innate arm of the immunity, which acts as a bridge between innate and adaptive immunity. Thus, it is hypothesized that dysregulated complement can strongly contribute toward hyperinflammatory response, which can eventually modulate the immune system’s ability to clear pathogens from the host body. Indeed, the role of the complement in COVID19 is just beginning to unravel and no wonder that immunosuppression therapies (Rizk et al., 2020) targeting the excessive activation of the complement for managing severe COVID19 is being currently evaluated. Interestingly, several specific immunomodulators are also being tested for their anti-cytokine property for managing severe COVID19. In addition, corticosteroids like dexamethasone, methylprednisolone are also being tested as non-specific immunomodulators for managing COVID19. Beside, other drugs such as azithromycin, colchicine, hydroxychloroquine and several others are being used as prophylactic for their anti-inflammatory and immunomodulatory effects for managing the COVID19. It is noteworthy that C5a, is one of the most potent proinflammatory complement glycoprotein, whose overproduction in a dysregulated complement system (Java et al., 2020) has been associated with the lethal “hypercytokinaemia” and in fact C5a has been found to be elevated in patients with severe COVID19 (Carvelli et al., 2020). In this regard, our recent studies (Mishra et al., 2020) suggest that raloxifene, which is prescribed to alleviate the pain and inflammation in rheumatoid arthritis can be explored as a non-specific immunomodulator either as monotherapy or in combination with other synergistic drugs, as an alternative polytherapy for possible management of COVID19. In fact, raloxifene’s potential as an antiviral agent (Hong et al., 2021) has recently been explored and repurposing of raloxifene alone or in synergistic combination with nelfinavir (Schultz et al., 2021) for treatment of SARS-CoV2 infection is currently being explored. Among all non-specific immunomodulators being tested, corticosteroids have shown remarkable results in managing severe COVID19 (Sterne et al., 2020) so far across the globe. In fact, world health organization also endorses the benefit of corticosteroid use in patients with severe and critical COVID19. Corticosteroids (Liu et al., 2013) are in clinical practice for a very long time and are generally prescribed for effective treatment of various inflammatory and autoimmune diseases, such as chronic obstructive pulmonary disease (COPD) (de Jong et al., 2007), severe asthma, systemic lupus, rheumatoid arthritis etc. for their ability to act as both anti-inflammatory and immunosuppressive agents (Coutinho and Chapman, 2011). Both genomic and nongenomic mode of action contribute toward the overall effectiveness of the corticosteroid therapy (Stahn and Buttgereit, 2008). Under genomic mode of action, corticosteroids bind to cytoplasmic GRs, which further enters into nucleus and binds to glucocorticoid responsive element (GRE). This leads to the upregulation of anti-inflammatory genes and downregulation pro-inflammatory genes, which further exerts the anti-inflammatory and immunosuppressive effects. However, therapeutic benefit of non-genomic mode of action of corticosteroids is not completely clear and it appears from the current study that C5a could be a good target candidate for its non-genomic mode of action. Both dexamethasone and prednisone are corticosteroid class of drugs with identical backbone structure and the current study suggests that both of them can potentially interact with C5a, though prednisone appears to demonstrate higher affinity toward C5a.
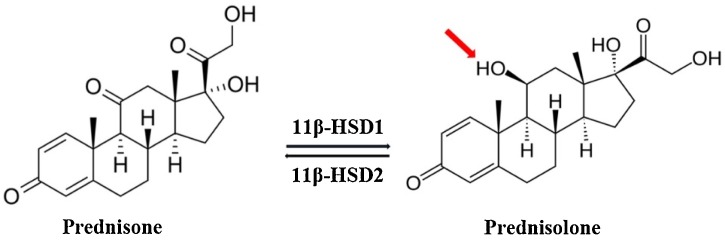
Prednisone is a prodrug, which gets rapidly metabolized to the bioactive prednisolone (Fig. 8 ) in presence of the 11 β-hydroxysteroid dehydrogenase enzymes, found to be expressed in both bone and synovial tissue, including macrophages (Raza et al., 2010). On the other hand, no such interconversion is known in case of dexamethasone, which has comparatively longer anti-inflammatory action than the prednisolone. However, given the minor structural differences between dexamethasone, prednisone and prednisolone, all are likely to bind C5a, which is upstream to GRs. Among all, dexamethasone is preferred, as it demonstrates lower mineral corticosteroid activity than the prednisolone derivatives and is less likely to contribute toward retention of sodium and fluids. On the other hand, low dose (35−70 mg / day/ 70 KG adult) of methylprednisolone is generally preferred for shorter duration to manage severe COVID19 with ARDS (Shang et al., 2020). In fact higher dose of the prednisolone (Edalatifard et al., 2020) has also been shown to reduce the “hypercytokinaemia”, observed in 15 % of patients with COVID19. In addition, prednisolone has also been shown to improve the condition of patients with severe COPD (de Jong et al., 2007). No wonder that collectively corticosteroid therapy (dexamethasone: 6 mg / day; or oral prednisone / prednisolone: 40 mg / day) (Farkas, 2020) have been shown to improve the survival rate in patients with severe COVID19. It is increasingly becoming clear that overactivated complement in the lungs, kidney, liver and heart has an undeniable role in the progression (Noris et al., 2020) of severe COVID19. It is also established that endothelial cells expresses the angiotensin converting enzyme 2 (ACE2) (Hamming et al., 2004), which acts as a gateway for the SARS-CoV2 infection of the cells (Hoffmann et al., 2020). Further, It is well known that C5a contributes significantly to the inflammation induced endothelial damage and virus induced acute lung injury (ALI) (Wang et al., 2015). In addition, it is believed that C5a could be an early biomarker for diagnosis of disease severity than the conventional biomarkers like C-reactive protein (Prendecki et al., 2020). Moreover, literature evidences that anti-complement protein products can be therapeutically valuable for management of COVID19 (Mastellos et al., 2019; Jodele and Köhl, 2020). Thus, there is a considerable evidence, which suggests that C5a may be a key protein contributing toward the lung pathology in patients with COVID19. The observation made in this study indicates that corticosteroid like prednisone can bind to C5a with Kd ∼ 0.38 μM, which is well within the therapeutic dose limit prescribed for managing the severe COVID19. It is noteworthy that synthetic corticosteroids have high oral bioavailability and are generally prescribed at a physiologically equivalent concentration. Interestingly, depending on the prescribed dose size, the total peak plasma concentration of corticosteroids can reach high micromolar range. However, most of the synthetic corticosteroids demonstrate ∼Kd 5−10 nM for the GRs (Andreae et al., 2001). The wide range of action demonstrated by the corticosteroids cannot be accounted by their sole interaction with the genomic targets. C5a is a potent glycoprotein of the complement system, known to contribute significantly toward the “cytokine storm” (Chauhan et al., 2020; Mahmudpour et al., 2020), as well as chronic inflammation (Guo and Ward, 2005). Further, the complex crosstalk between the complement, coagulation and cytokines is well known. C5a is upstream of glucocorticoid receptors and it is well known that clinicians/physicians usually prescribe the steroids at a concentration (high μM), which is way higher than what is required for completely blinding the GRs (low nM). Thus, it can be hypothesized that the excessive corticosteroids can contribute toward the specific non-genomic benefit, by binding to either plasma proteins or membrane receptors.
Fig. 8.
(a) Interconversion of prednisone to the bioactive prednisolone and vice-versa by the 11 β-hydroxysteroid dehydrogenase (11β-HSD) enzymes. The changed functional group is highlighted in prednisolone.
Under normal circumstances physiological concentration of C5a is tightly regulated, which synergistically function to saturate, desensitize and regenerate the C5aR in a defined signaling cycle to maintain the basal level activity. However, in response to stress, injury and infection, aberrant activation of the complement produces C5a at pathological concentration, which disrupts the synergistic signaling cycle of C5aR, and at a given point of time the number of C5a available can outnumber the number of C5aR available to saturate on the cell surface. Targeting the excessive C5a in a non-competitive manner under a disease setting like COVID19 can be really effective for controlling the hyperinflammatory response of C5aR. The bioavailability and half-life of the corticosteroids are generally very high and thus, the binding of corticosteroids to excessive C5a in the plasma at low μM concentration is technically feasible, which in principle can reduce the persistent saturation of C5aR, subsequently contributing toward the overall therapeutic benefit of the corticosteroids. It is noteworthy that currently the molecular complexes of C3a/C5a with their cognate receptors are not crystal clear. However, highly refined model structural complex of C5a bound to C5a receptor (C5aR) has already been described in the literature (Sahoo et al., 2018), which is most likely to be similar for C3a-C3aR system. It is evidenced that N-terminus of C5aR strongly interacts with bulk of C5a, which induces conformational changes in C5a, leading to the interaction of its C-terminus peptide with the extracellular surface of C5aR. This synergistic interaction is absolutely important for the formation of a biologically active structural complex between C5a and C5aR, which is required for initiation of the downstream signaling. A recent study suggests that blockage of C5a-C5aR can help COVID19 patients with ARDS (Carvelli et al., 2020), by reducing the excessive lung inflammation through controlled infiltration of the myeloid cells. The data presented in Figs. 4, 5 and 7 suggests that prednisone binding to C5a may be able to lock the C-terminus peptide of C5a, in addition to altering its biologically active conformation. The conformational altering induced by the binding of prednisone is likely to dampen the physiological affinity of C5a toward C5aR, in a non-competitive manner, which can eventually augment the disruption of a biologically active complex between C5a and C5aR (Mishra and Rana, 2019). A similar assumption can be made for C3a-C3aR system based on the data presented in Fig. S2. In addition, it can also be hypothesized that under immune inflammatory conditions, alteration in the biologically active conformer of C5a due to the binding of prednisone can reduce the excessive concentration of C5a available in the plasma post saturation of the C5aR, by triggering degradation similar to a targeted small molecule protein degrader, which can be helpful in controlling the undesired immune response in the body. As observed, the estimated affinity of the prednisone toward C5a is in sub-micromolar range compared to the native affinity of C5a toward C5aR, which is estimated to be in picomolar/sub-nanomolar range. The picomolar affinity of C5aR toward C5a is majorly contributed by the N-terminus peptide of C5aR binding to the bulk of C5a, as the signaling studies evidence that the peptide agonists based on the last ten amino acids of the C-terminus of C5a has a high micromolar affinity toward the extracellular surface (ECS) of C5aR. Small molecule based agonist/antagonist known for the C5aR basically target the ECS of C5aR, as technically such small molecules cannot compete off the N-terminus of C5aR binding to the bulk of C5a. Since, small molecule based ligands are known to block the ECS of C5aR to prevent the interaction of the C-terminus of C5a, then it can also be hypothesized that prednisone like small molecules can alternatively block the availability of the C-terminus of C5a (Figs. 3b and 5) for interaction with the ECS of C5aR. Considering the role of complement in the COVID19 and the data presented in this study, it appears that binding of corticosteroids can augment the neutralization of the excessive C5a under hyperinflammatory conditions, which may be contributing positively toward the general success of corticosteroid therapy in managing severe COVID19. However, further biomolecular signaling studies in appropriate system will be needed to rationalize this hypothesis.
5. Conclusion
The current study indicates that C5a could be one of the non-genomic target of corticosteroids and binding of dexamethasone / prednisone to C5a can potentially neutralize the hyperinflammatory signal triggered by the excessive C5a in the plasma, thereby amplifying the reported therapeutic benefits of the corticosteroid administrations, observed in patients with moderate to severe COVID19. It is understood that prolonged exposure to high dose corticosteroids can be generally immunosuppressive, as it can weaken the ability of immune system to fight pathogens, resulting unchecked pathogen replication and co-infections. Considering the immense importance of the immune system both in general physiology and severe pathology, further detailed studies are required not only to clearly understand the complement proteins role, but also to devise the timing for intervention in COVID19. Nevertheless, synergistic pairing of low dose corticosteroids with other drugs at an appropriately regulated regimen should further be explored vigorously in the clinical settings to harness the maximum therapeutic benefit, which may become essential in near future for effective management of severe COVID19. In addition, the current study also hints that design and synthesis of new chemical entities with exceptionally high binding affinity toward C5a should be further explored considering the established pathophysiological importance of the C5a under hyperinflammatory conditions.
Author contributions
A. D. conducted the biophysical experiments and prepared the figures. S. R. conducted the computational modelling and MD studies. A. D. and S. R. analysed the data. S. R. wrote the manuscript and conceived the idea for the project.
Declaration of Competing Interest
The authors report no declarations of interest.
Acknowledgements
This research is supported by the SERB (EMR/2016/000681). Use of supercomputing facility at IIT Delhi is highly appreciated. Usage of the analytical instrumentation facility of IIT Bhubaneswar is strongly acknowledged.
Footnotes
Supplementary material related to this article can be found, in the online version, at doi:https://doi.org/10.1016/j.compbiolchem.2021.107482.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- Ali H. Immunol. Lett. 2010;128:36–45. doi: 10.1016/j.imlet.2009.10.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andreae J., Tripmacher R., Weltrich R., Rohde W., Keitzer R., Wahn U., Paul K., Buttgereit F. Pediatr. Res. 2001;49:130–135. doi: 10.1203/00006450-200101000-00025. [DOI] [PubMed] [Google Scholar]
- Angeli A., Ferraroni M., Supuran C.T. ACS Med. Chem. Lett. 2018;9:1035–1038. doi: 10.1021/acsmedchemlett.8b00334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Badawi A. J. Inflamm. Res. 2020;13:255. doi: 10.2147/JIR.S259096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajic G., Yatime L., Klos A., Andersen G.R. Protein Sci. 2013;22:204–212. doi: 10.1002/pro.2200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkovskiy M., Ilyukhina E., Dauner M., Eichinger A., Skerra A. Biol. Chem. 2019;400:351–366. doi: 10.1515/hsz-2018-0342. [DOI] [PubMed] [Google Scholar]
- Barna T.M., Khan H., Bruce N.C., Barsukov I., Scrutton N.S., Moody P.C. J. Mol. Biol. 2001;310:433–447. doi: 10.1006/jmbi.2001.4779. [DOI] [PubMed] [Google Scholar]
- Barrington R., Zhang M., Fischer M., Carroll M.C. Immunol. Rev. 2001;180:5–15. doi: 10.1034/j.1600-065x.2001.1800101.x. [DOI] [PubMed] [Google Scholar]
- Bartoletti M., Marconi L., Scudeller L., Pancaldi L., Tedeschi S., Giannella M., Rinaldi M., Bussini L., Valentini I., Ferravante A.F., Potalivo A., Marchionni E., Fornaro G., Pascale R., Pasquini Z., Puoti M., Merli M., Barchiesi F., Volpato F., Rubin A., Saracino A., Tonetti T., Gaibani P., Ranieri V.M., Viale P., Cristini F., P.S. Group Clin. Microbiol. Infect. 2021;27:105–111. doi: 10.1016/j.cmi.2020.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridgham J.T., Ortlund E.A., Thornton J.W. Nature. 2009;461:515–519. doi: 10.1038/nature08249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burgos-Blasco B., Güemes-Villahoz N., Santiago J.L., Fernandez-Vigo J.I., Espino-Paisán L., Sarriá B., García-Feijoo J., Martinez-de-la-Casa J.M. Exp. Eye Res. 2020;200 doi: 10.1016/j.exer.2020.108253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell C.M., Kahwash R. Circulation. 2020;141:1739–1741. doi: 10.1161/CIRCULATIONAHA.120.047419. [DOI] [PubMed] [Google Scholar]
- Carsana L., Sonzogni A., Nasr A., Rossi R.S., Pellegrinelli A., Zerbi P., Rech R., Colombo R., Antinori S., Corbellino M., Galli M., Catena E., Tosoni A., Gianatti A., Nebuloni M. Lancet Infect. Dis. 2020;20:1135–1140. doi: 10.1016/S1473-3099(20)30434-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvelli J., Demaria O., Vély F., Batista L., Benmansour N.C., Fares J., Carpentier S., Thibult M.-L., Morel A., Remark R. Nature. 2020;588:146–150. [Google Scholar]
- Chauhan A.J., Wiffen L.J., Brown T.P. J. Thromb. Haemost. 2020;18:2110–2117. doi: 10.1111/jth.14981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coulthard L.G., Woodruff T.M. J. Immunol. 2015;194:3542–3548. doi: 10.4049/jimmunol.1403068. [DOI] [PubMed] [Google Scholar]
- Coutinho A.E., Chapman K.E. Mol. Cell. Endocrinol. 2011;335:2–13. doi: 10.1016/j.mce.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cugno M., Meroni P.L., Gualtierotti R., Griffini S., Grovetti E., Torri A., Panigada M., Aliberti S., Blasi F., Tedesco F. J. Allergy Clin. Immunol. 2020;146:215–217. doi: 10.1016/j.jaci.2020.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Jong Y.P., Uil S.M., Grotjohan H.P., Postma D.S., Kerstjens H.A., van den Berg J.W. Chest. 2007;132:1741–1747. doi: 10.1378/chest.07-0208. [DOI] [PubMed] [Google Scholar]
- Diurno F., Numis F.G., Porta G., Cirillo F., Maddaluno S., Ragozzino A., De Negri P., Di Gennaro C., Pagano A., Allegorico E., Bressy L., Bosso G., Ferrara A., Serra C., Montisci A., D’Amico M., Schiano Lo Morello S., Di Costanzo G., Tucci A.G., Marchetti P., Di Vincenzo U., Sorrentino I., Casciotta A., Fusco M., Buonerba C., Berretta M., Ceccarelli M., Nunnari G., Diessa Y., Cicala S., Facchini G. Eur. Rev. Med. Pharmacol. Sci. 2020;24:4040–4047. doi: 10.26355/eurrev_202004_20875. [DOI] [PubMed] [Google Scholar]
- Edalatifard M., Akhtari M., Salehi M., Naderi Z., Jamshidi A., Mostafaei S., Najafizadeh S.R., Farhadi E., Jalili N., Esfahani M. Eur. Respir. J. 2020;56 doi: 10.1183/13993003.02808-2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas J. 2020. The Internet Book of Critical Care. [Google Scholar]
- Freeley S., Kemper C., Le Friec G. Immunol. Rev. 2016;274:16–32. doi: 10.1111/imr.12472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giamarellos‐Bourboulis E., Argyropoulou M., Kanni T., Spyridopoulos T., Otto I., Zenker O., Guo R., Riedemann N. Br. J. Dermatol. 2020;183:176–178. doi: 10.1111/bjd.18877. [DOI] [PubMed] [Google Scholar]
- Giudice V., Pagliano P., Vatrella A., Masullo A., Poto S., Polverino B.M., Gammaldi R., Maglio A., Sellitto C., Vitale C., Serio B., Cuffa B., Borrelli A., Vecchione C., Filippelli A., Selleri C. Front. Pharmacol. 2020;11:857. doi: 10.3389/fphar.2020.00857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenfield N.J. Nat. Protoc. 2006;1:2876–2890. doi: 10.1038/nprot.2006.202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guan W.-j., Ni Z.-y., Hu Y., Liang W.-h., Ou C.-q., He J.-x., Liu L., Shan H., Lei C.-l., Hui D.S. N. Engl. J. Med. 2020;382:1708–1720. doi: 10.1056/NEJMoa2002032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo R.-F., Ward P.A. Annu. Rev. Immunol. 2005;23:821–852. doi: 10.1146/annurev.immunol.23.021704.115835. [DOI] [PubMed] [Google Scholar]
- Hamming I., Timens W., Bulthuis M., Lely A., Navis Gv., van Goor H. J. Pathol. 2004;203:631–637. doi: 10.1002/path.1570. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hess B., Kutzner C., van der Spoel D., Lindahl E. J. Chem. Theory Comput. 2008;4:435–447. doi: 10.1021/ct700301q. [DOI] [PubMed] [Google Scholar]
- Hoffmann M., Kleine-Weber H., Schroeder S., Krüger N., Herrler T., Erichsen S., Schiergens T.S., Herrler G., Wu N.-H., Nitsche A. Cell. 2020;181:271–280. doi: 10.1016/j.cell.2020.02.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holter J.C., Pischke S.E., de Boer E., Lind A., Jenum S., Holten A.R., Tonby K., Barratt-Due A., Sokolova M., Schjalm C. Proc. Natl. Acad. Sci. 2020;117:25018–25025. doi: 10.1073/pnas.2010540117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong S., Chang J., Jeong K., Lee W. J. Microbiol. 2021;59:124–131. doi: 10.1007/s12275-021-0617-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu B., Guo H., Zhou P., Shi Z.-L. Nat. Rev. Microbiol. 2021;19:141–154. doi: 10.1038/s41579-020-00459-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Q., Wu X., Zheng X., Luo S., Xu S., Weng J. Pharmacol. Res. 2020;159 doi: 10.1016/j.phrs.2020.105051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huber-Lang M., Younkin E.M., Sarma J.V., Riedemann N., McGuire S.R., Lu K.T., Kunkel R., Younger J.G., Zetoune F.S., Ward P.A. Am. J. Pathol. 2002;161:1849–1859. doi: 10.1016/S0002-9440(10)64461-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugli T.E., Erickson B.W. Proc. Natl. Acad. Sci. U. S. A. 1977;74:1826–1830. doi: 10.1073/pnas.74.5.1826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Issuree P.D., Pushparaj P.N., Pervaiz S., Melendez A.J. FASEB J. 2009;23:2412–2424. doi: 10.1096/fj.09-130542. [DOI] [PubMed] [Google Scholar]
- Java A., Apicelli A.J., Liszewski M.K., Coler-Reilly A., Atkinson J.P., Kim A.H., Kulkarni H.S. JCI Insight. 2020;5 doi: 10.1172/jci.insight.140711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jodele S., Köhl J. Br. J. Pharmacol. 2020:1–17. doi: 10.1111/bph.15187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kadmiel M., Cidlowski J.A. Trends Pharmacol. Sci. 2013;34:518–530. doi: 10.1016/j.tips.2013.07.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelleni M.T. Pharmacol. Res. 2020;157 doi: 10.1016/j.phrs.2020.104874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khan M.A., Maasch C., Vater A., Klussmann S., Morser J., Leung L.L., Atkinson C., Tomlinson S., Heeger P.S., Nicolls M.R. Proc. Natl. Acad. Sci. 2013;110:6061–6066. doi: 10.1073/pnas.1217991110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumari R., Kumar R., Consortium O.S.D.D., Lynn A. J. Chem. Inf. Model. 2014;54:1951–1962. doi: 10.1021/ci500020m. [DOI] [PubMed] [Google Scholar]
- Ledford H. Nature. 2020:469. doi: 10.1038/d41586-020-01824-5. [DOI] [PubMed] [Google Scholar]
- Lenze E.J., Mattar C., Zorumski C.F., Stevens A., Schweiger J., Nicol G.E., Miller J.P., Yang L., Yingling M., Avidan M.S., Reiersen A.M. JAMA. 2020;324:2292–2300. doi: 10.1001/jama.2020.22760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X., Ma X. Crit. Care. 2020;24:1–5. doi: 10.1186/s13054-020-02911-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu D., Ahmet A., Ward L., Krishnamoorthy P., Mandelcorn E.D., Leigh R., Brown J.P., Cohen A., Kim H. Allergy Asthma Clin. Immunol. 2013;9:30. doi: 10.1186/1710-1492-9-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lo M.W., Woodruff T.M. J. Leukoc. Biol. 2020;108:339–351. doi: 10.1002/JLB.3MIR0220-270R. [DOI] [PubMed] [Google Scholar]
- Mahmudpour M., Roozbeh J., Keshavarz M., Farrokhi S., Nabipour I. Cytokine. 2020 doi: 10.1016/j.cyto.2020.155151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mastellos D.C., Ricklin D., Lambris J.D. Nat. Rev. Drug Discov. 2019;18:707–729. doi: 10.1038/s41573-019-0031-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mather J.F., Seip R.L., McKay R.G. Am. J. Gastroenterol. 2020;115:1617–1623. doi: 10.14309/ajg.0000000000000832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R., Rana S. Bioorg. Med. Chem. 2019;27 doi: 10.1016/j.bmc.2019.115052. [DOI] [PubMed] [Google Scholar]
- Mishra R., Behera L.M., Rana S. J. Biomol. Struct. Dyn. 2020:1–13. doi: 10.1080/07391102.2020.1820381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mishra R., Das A., Rana S. J. Biomol. Struct. Dyn. 2021;39:1766–1780. doi: 10.1080/07391102.2020.1738958. [DOI] [PubMed] [Google Scholar]
- Morris G.M., Huey R., Lindstrom W., Sanner M.F., Belew R.K., Goodsell D.S., Olson A.J. J. Comput. Chem. 2009;30:2785–2791. doi: 10.1002/jcc.21256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noris M., Benigni A., Remuzzi G. Kidney Int. 2020;98:314–322. doi: 10.1016/j.kint.2020.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oakley R.H., Cidlowski J.A. J. Allergy Clin. Immunol. 2013;132:1033–1044. doi: 10.1016/j.jaci.2013.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oldfield C.J., Dunker A.K. Annu. Rev. Biochem. 2014;83:553–584. doi: 10.1146/annurev-biochem-072711-164947. [DOI] [PubMed] [Google Scholar]
- Paces J., Strizova Z., Smrz D., Cerny J. Physiol. Res. 2020;69:379–388. doi: 10.33549/physiolres.934492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pani A., Lauriola M., Romandini A., Scaglione F. Int. J. Antimicrob. Agents. 2020;56 doi: 10.1016/j.ijantimicag.2020.106053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patri A., Fabbrocini G. J. Am. Acad. Dermatol. 2020;82:e221. doi: 10.1016/j.jaad.2020.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pawlowski A.C., Stogios P.J., Koteva K., Skarina T., Evdokimova E., Savchenko A., Wright G.D. Nat. Commun. 2018;9:112. doi: 10.1038/s41467-017-02680-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perico L., Benigni A., Casiraghi F., Ng L.F.P., Renia L., Remuzzi G. Nat. Rev. Nephrol. 2021;17:46–64. doi: 10.1038/s41581-020-00357-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prendecki M., Clarke C., Medjeral-Thomas N., McAdoo S.P., Sandhu E., Peters J.E., Thomas D.C., Willicombe M., Botto M., Pickering M.C. Clin. Kidney J. 2020;13:889–896. doi: 10.1093/ckj/sfaa192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raina K., Crews C.M. Curr. Opin. Chem. Biol. 2017;39:46–53. doi: 10.1016/j.cbpa.2017.05.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Raza K., Hardy R., Cooper M.S. Rheumatology. 2010;49:2016–2023. doi: 10.1093/rheumatology/keq212. [DOI] [PubMed] [Google Scholar]
- Risitano A.M., Mastellos D.C., Huber-Lang M., Yancopoulou D., Garlanda C., Ciceri F., Lambris J.D. Nat. Rev. Immunol. 2020;20:343–344. doi: 10.1038/s41577-020-0320-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rizk J.G., Kalantar-Zadeh K., Mehra M.R., Lavie C.J., Rizk Y., Forthal D.N. Drugs. 2020;80:1267–1292. doi: 10.1007/s40265-020-01367-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rowley J.A., Reid R.C., Poon E.K.Y., Wu K.C., Lim J., Lohman R.J., Hamidon J.K., Yau M.K., Halili M.A., Durek T., Iyer A., Fairlie D.P. J. Med. Chem. 2020;63:529–541. doi: 10.1021/acs.jmedchem.9b00927. [DOI] [PubMed] [Google Scholar]
- Sahoo A.R., Mishra R., Rana S. Sci. Rep. 2018;8:2955. doi: 10.1038/s41598-018-21290-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scala S., Pacelli R. Front. Immunol. 2020;11:1201. doi: 10.3389/fimmu.2020.01201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlesinger N., Firestein B.L., Brunetti L. Curr. Pharmacol. Rep. 2020;6:137–145. doi: 10.1007/s40495-020-00225-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz B., Zaliani A., Ebeling C., Reinshagen J., Bojkova D., Lage-Rupprecht V., Karki R., Lukassen S., Gadiya Y., Ravindra N.G., Das S., Baksi S., Domingo-Fernández D., Lentzen M., Strivens M., Raschka T., Cinatl J., DeLong L.N., Gribbon P., Geisslinger G., Ciesek S., van Dijk D., Gardner S., Kodamullil A.T., Fröhlich H., Peitsch M., Jacobs M., Hoeng J., Eils R., Claussen C., Hofmann-Apitius M. bioRxiv. 2021 2020.2009.2023.308239. [Google Scholar]
- Schüttelkopf A.W., Van Aalten D.M. Acta Crystallogr. D Biol. Crystallogr. 2004;60:1355–1363. doi: 10.1107/S0907444904011679. [DOI] [PubMed] [Google Scholar]
- Schwartz R.A., Suskind R.M. Dermatol. Ther. 2020 doi: 10.1111/dth.13785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scully C.C., Blakeney J.S., Singh R., Hoang H.N., Abbenante G., Reid R.C., Fairlie D.P. J. Med. Chem. 2010;53:4938–4948. doi: 10.1021/jm1003705. [DOI] [PubMed] [Google Scholar]
- Shang L., Zhao J., Hu Y., Du R., Cao B. Lancet. 2020;395:683. doi: 10.1016/S0140-6736(20)30361-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stahn C., Buttgereit F. Nat. Clin. Pract. Rheumatol. 2008;4:525–533. doi: 10.1038/ncprheum0898. [DOI] [PubMed] [Google Scholar]
- Sterne J.A., Murthy S., Diaz J.V., Slutsky A.S., Villar J., Angus D.C., Annane D., Azevedo L.C.P., Berwanger O., Cavalcanti A.B. Jama. 2020;324:1330–1341. doi: 10.1001/jama.2020.17023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wadman M., Couzin-Frankel J., Kaiser J., Matacic C. Science. 2020;368:356–360. doi: 10.1126/science.368.6489.356. [DOI] [PubMed] [Google Scholar]
- Wang H., Goehring A., Wang K.H., Penmatsa A., Ressler R., Gouaux E. Nature. 2013;503:141–145. doi: 10.1038/nature12648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang R., Xiao H., Guo R., Li Y., Shen B. Emerg. Microbes Infect. 2015;4:e28. doi: 10.1038/emi.2015.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z., Yang X., Zhou Y., Sun J., Liu X., Zhang J., Mei X., Zhong J., Zhao J., Ran P. Am. J. Respir. Crit. Care Med. 2020;202:606–610. doi: 10.1164/rccm.202005-1701LE. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Y., Zhang D., Du G., Du R., Zhao J., Jin Y., Fu S., Gao L., Cheng Z., Lu Q., Hu Y., Luo G., Wang K., Lu Y., Li H., Wang S., Ruan S., Yang C., Mei C., Wang Y., Ding D., Wu F., Tang X., Ye X., Ye Y., Liu B., Yang J., Yin W., Wang A., Fan G., Zhou F., Liu Z., Gu X., Xu J., Shang L., Zhang Y., Cao L., Guo T., Wan Y., Qin H., Jiang Y., Jaki T., Hayden F.G., Horby P.W., Cao B., Wang C. Lancet. 2021;395:1569–1578. doi: 10.1016/S0140-6736(20)31022-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiersinga W.J., Rhodes A., Cheng A.C., Peacock S.J., Prescott H.C. JAMA. 2020;324:782–793. doi: 10.1001/jama.2020.12839. [DOI] [PubMed] [Google Scholar]
- Zhang X., Boyar W., Toth M.J., Wennogle L., Gonnella N.C. Proteins Struct. Funct. Bioinform. 1997;28:261–267. doi: 10.1002/(sici)1097-0134(199706)28:2<261::aid-prot13>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.