Abstract
The NSP1– C terminal structure in complex with ribosome using cryo-EM is available now, and the N-terminal region structure in isolation is also deciphered in literature. However, as a reductionist approach, the conformation of NSP1– C terminal region (NSP1-CTR; amino acids 131–180) has not been studied in isolation. We found that NSP1-CTR conformation is disordered in an aqueous solution. Further, we examined the conformational propensity towards alpha-helical structure using trifluoroethanol, we observed induction of helical structure conformation using CD spectroscopy. Additionally, in SDS, NSP1-CTR shows a conformational change from disordered to ordered, possibly gaining alpha-helix in part. But in the presence of neutral lipid DOPC, a slight change in conformation is observed, which implies the possible role of hydrophobic interaction and electrostatic interaction on the conformational changes of NSP1. Fluorescence-based studies have shown a blue shift and fluorescence quenching in the presence of SDS, TFE, and lipid vesicles. In agreement with these results, fluorescence lifetime and fluorescence anisotropy decay suggest a change in conformational dynamics. The zeta potential studies further validated that the conformational dynamics are primarily because of hydrophobic interaction. These experimental studies were complemented through Molecular Dynamics (MD) simulations, which have shown a good correlation and testifies our experiments. We believe that the intrinsically disordered nature of the NSP1-CTR will have implications for enhanced molecular recognition feature properties of this IDR, which may add disorder to order transition and disorder-based binding promiscuity with its interacting proteins.
Keywords: SARS-CoV-2, NSP1, Hydrophobic interaction, Conformational dynamics
Graphical abstract
1. Introduction
Among four genera of coronaviruses (CoVs), alpha and beta can infect humans. CoVs have a single-stranded positive-sense RNA genome, of which the two-thirds part (~20 kb) is translated into two large polyproteins named pp1a and pp1ab. These two polyproteins encode sixteen non-structural proteins (NSP1–NSP16), which make up the viral replication-transcription complex (Romano et al., 2020; Giri et al., 2020). The exciting feature among the four CoVs genera is NSP1, present only in alpha- and beta-generates of CoVs (Narayanan et al., 2015). This feature may correlate with CoV’s pathogenicity as alpha and beta are highly pathogenic than the gamma and delta genera (Narayanan et al., 2015).
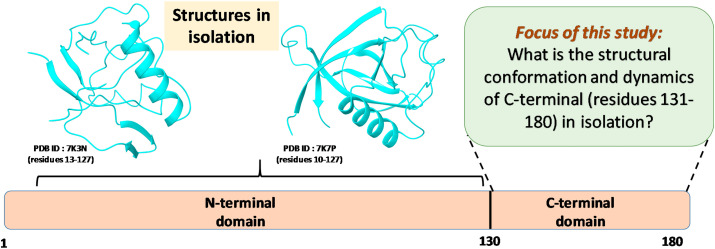
Moreover, the NSP1 size varies among different beta-CoVs lineages and shares different degrees of amino acid sequence similarity (Narayanan et al., 2015; Thoms et al., 2020). Despite the different amino acid compositions, the high structural similarity of NSP1 was found to regulate the same feature in all genera of alpha and beta-CoV (Shen et al., 2019). The NMR structure of SARS-CoV Nsp1 showed that N- and the C-terminal domains are disordered with a β-barrel fold in between (Almeida et al., 2007). There is no full-length NSP1 structure available to date. Recently two crystal structure of SARS-CoV-2 NSP1 corresponding to residues 10–127 (PDB ID: 7K7P) and 13–127 (PDB ID: 7K3N) have been reported, which suggest that residue 1–10 and 130–180 are highly flexible and cannot be crystallized (Fig. 1 ) (Semper et al., 2021; Clark et al., 2021). Also, the cryo-EM structure of SARS-CoV-2 NSP1-CTR has been reported in the presence of a binding partner (Thoms et al., 2020; Schubert et al., 2020). It is stated that the SARS-CoV NSP1 does not possess any enzymatic activity. The NSP1 C terminal region (NSP1-CTR) binds with the 40 S ribosomal subunit and blocks the host mRNA translation (Thoms et al., 2020; Schubert et al., 2020; Kamitani et al., 2009; Huang et al., 2011). Although the NSP1 N terminal domain has no role in the host translation shut down, it interacts with 5′ UTR of vRNA, lifts the NSP1-CTR-40 S inhibition, and allows the vRNA translation precisely (Banerjee et al., 2020; Shi et al., 2020). The NSP1 has been found to downregulate the host cell’s immune function through the RIG-I signaling pathway (Thoms et al., 2020; Tohya et al., 2009). SARS-CoV-2 NSP1, along with NSP3, NSP12, NSP13, NSP14, ORF3, ORF6, and M protein, is found to inhibit the Sendai virus-induced IFN-β promoter activation (Lei et al., 2020).
Fig. 1.
Literature-based schematic representation of domains of SARS-CoV-2 NSP1 depicting N- and C-terminal. The available 3D structures of the N-terminal domain using X-ray crystallography techniques are also shown.
Bioinformatics studies show intrinsically disordered proteins/regions (IDPs/IDRs) penetrance in viruses and all life domains from bacteria to eukaryotes (Ward et al., 2004; Xue et al., 2012; Gadhave et al., 2020; Kumar et al., 2020a; Giri et al., 2016). Recently, bioinformatics-based research by our group showed that NSP1 of SARS-CoV-2 regions (amino acids 99–179,137-145 and 172–179) is identified as a molecular recognition feature (MoRFs) regions (Giri et al., 2020). Generally, most IDPs are unfolded in solution and switch to a conformation when binding with particular ligands and undergo coupled folding and binding (Dunker et al., 1998; Wright and Dyson, 1999, 2009; Oldfield et al., 2005). The IDP’s capabilities to participate in multiple cellular signaling lies in their structural amenability, which can be related to the presence of MoRFs and their kinetics advantages (Kumar et al., 2020b, 2020c; Toto et al., 2014; Singh et al., 2018; Mishra et al., 2018). Therefore, IDPs’ molecular interactions are dynamic, which allows them to compete and exchange the binding partners in function regulating proteins.
The NSP1 majorly affects the host proteins, and the intracellular environment is highly crowded (Kumar et al., 2020b; Kuznetsova et al., 2014; Ellis, 2001). Therefore, the surrounding environment’s impact on the conformational behavior of NSP1-CTR needs to be addressed to assess its disorder-function paradigm, leading us to understand the interaction mechanistic with respective partners. Coronavirus NSP1 is also a membrane-associated protein that shuts off the host translation and immune evasion, thus facilitating viral replication (Narayanan et al., 2015). Therefore, it is essential to study the conformation of NSP-CTR in a membrane mimic environment or the presence of liposomes. Furthermore, given the earlier prediction that the C-terminal region of NSP1 has molecular recognition features, it is essential to investigate its folding propensity. Therefore, in the present study, structural conformations of SARS-CoV-2 NSP1 concerning C-terminal regions are explored in an organic solvent, membrane mimetic environment, and liposomes. The disorder to order transition of NSP1-CTR was found in these conditions. The current finding gives a more in-depth insight into disorder-order conformation in the respective environment, which will eventually help understand the broader aspect of NSP1 and its regions with binding partners still yet to be discovered.
2. Material and methods
2.1. Chemicals and reagents
The NSP1 peptide (residues 131–180), corresponding to the C-terminal regions with purity>82% was purchased from Gene script, USA. Organic solvents such as Trifluoroethanol (TFE) with ≥99% purity were purchased from Sigma-Aldrich. Lyophilized peptide was dissolved in ultra-pure water at a concentration of 1 mg/ml and calculated for its micromolar concentration, i.e., 184 μM, used as a stock for experimental working concentration.
2.2. Secondary structural analysis of NSP1 and insight into intrinsic disorder properties
The sequence of NSP1 C-terminal (residues 131–180) “NH2-AGGHSYGADLKSFDLGDELGTDPYEDFQENWNTKHSSGVTRELMRELNGG-COOH” was retrieved from the UniProt (ID: P0DTC1.1). The NSP1 (131–180 residues) were studied with the Clustal Omega tool to reveal their conserved sequences among coronaviruses and represented with ESPript 3.0. Further, as described earlier by our group, protein intrinsic disorder predictor PONDR® VSL2 was used to analyze the intrinsic disorder properties of NSP1-CTR (Giri et al., 2020; Kumar et al., 2020a; Giri et al., 2016; Kumar et al., 2020b; Bhardwaj et al. Giri; Obradovic et al., 2005). The secondary structure predisposition analysis for NSP1-CTR was performed with several different web-servers, namely pep2d, Jpred4, and PSIPRED (Buchan and Jones, 2019; Drozdetskiy et al., 2015; Singh et al., 2019).
2.3. Liposome preparation
The liposomes were prepared, as described earlier (Kumar et al., 2020b). Briefly, the neutral lipid DOPC (dioleoyl-phosphatidyl-ethanolamine) and negatively charged lipid DOPS (1,2-dioleoyl-sn-glycero-3-phospho-Lserine) were purchased from Avanti Polar Lipids (Alabaster, AL). The chloroform from the lipid solution was removed using a rotary evaporator at 40 °C, and the dry lipid films were hydrated in 50 mM phosphate buffer (pH 7.4). The final concentration of the DOPC and DOPS liposomes were 37.09 mM and 24.69 mM, respectively. The resulting suspension was freeze-thaw-vortex in liquid nitrogen and water at 60 °C, following which the lipids were extruded 25 times through the mini extruder (Avanti Polar Lipids, Inc. USA) through cut off filter of 100 nm polycarbonate membrane to prepare uniform Large unilamellar vesicles (LUV). The size of LUVs was determined by dynamic light scattering using Zetasizer Nano ZS (Malvern Instruments Ltd., UK). The size of DOPS and DOPC was 125 nm and 102 nm, respectively.
2.4. Circular dichroism spectroscopy
Circular dichroism spectrometer MOS500 (BioLogic) and JASCO machine (Jasco J-1500 CD spectrometer) were used for CD data recording. 20 μM peptide sample were prepared in 50 mM phosphate buffer, pH 7.0. The peptide was kept in organic solvents (TFE) with increasing concentration from 0 to 90%, and far-UV (190–240 nm) spectra were recorded in 1 mm quartz cuvette. Similarly, the peptide was assessed for structural changes in Sodium Dodecyl Sulfate (SDS) (below and above critical micellar concentration (CMC)) and in the presence of DOPS and DOPC LUVs (pH 7.4). All the spectra were recorded at a scan speed of 50 nm/min with a response time of 1s and 1 nm bandwidth and three technical repeats. The equivalent spectra of buffers were recorded and subtracted from the spectra of the test samples. The ellipticity with HT value of fewer than 600 V was considered to plot the spectra.
2.5. Fluorescence spectroscopy
We monitored the intrinsic Trp fluorescence intensity in NSP1-CTR. A 5 μM peptide in 50 mM sodium phosphate buffer (pH 7.0) was prepared with increasing TFE and SDS concentration and 20 μM peptide (pH 7.4) in DOPS and DOPC. Emission spectra were recorded from 300 to 500 nm at 295 nm excitation wavelength in a Horiba Fluorolog-3 spectrofluorometer. The individual negative blank was subtracted from each test sample (Kumar et al., 2020c).
2.6. Fluorescence lifetime measurement
Fluorescence lifetime measurements were carried out for the 5 μM peptide (pH 7.0) in increasing concentration of TFE and SDS, and 20 μM peptide (pH 7.4) in DOPC and DOPS by using TCSPC (Horiba Scientific Inc.) with pulsed LED sources. The excitation wavelength was set at 284 nm, and the emission wavelength was kept at 345 nm. A Ludox solution was used for the correct instrument response factor (IRF). Fluorescence decay data was analyzed through Data Analysis Software provided by Horiba Scientific. Decay data were fitted tri-exponentially decay function with a best fit chi-squared value approaching 1.0 to calculate the fluorescence lifetimes.
2.7. Time-Resolved Fluorescence Anisotropy Decay
Time-Resolved Fluorescence Anisotropy Decay was carried out for the 30 μM peptide (pH 7.4) in increasing SDS concentration by using TCSPC (Horiba Scientific Inc.) with pulsed LED sources. The measurement range of 200 ns, peak preset of 1000 counts was set up to monitor the decay with an emission wavelength of 345 nm. Ludox was used to monitor the prompt decay (at 284 nm wavelength) to correct the IRF. Fluorescence anisotropy measures the depolarization, which happens because it transfers to another molecule of a different orientation. Alternatively, molecular rotation is also caused by Brownian motion and the local environment, i.e., viscosity, size of the molecule, and molecular confinement affect the molecular motion (Lakowicz, 2006). Thus, a measurement of fluorescence anisotropy is useful in obtaining information concerning molecular size and mobility.
The time-resolved fluorescence anisotropy decay function, r(t) is given as:
| (1) |
where, and designate fluorescence decays obtained for parallel and perpendicular emission polarizations, respectively. G is the instrumental correction factor at the wavelength λ of emission. It is the ratio of efficiency of detection for vertically and horizontally polarized light.
The following equation can express the anisotropy decay curve
| (2) |
where, is the anisotropy at time , is the initial anisotropy and ranges from 0.4 (parallel transition dipoles) to - 0.2 (perpendicular dipoles), and are the amplitude associated with the rotational correlation time and , respectively.
2.8. Zeta potential measurement
NSP1 C-terminal interaction with DOPS and DOPC was studied for zeta potential measurements using Malvern Zetasizer Nano ZS device. Briefly, 200 μM of liposomes incubated with different concentrations of NSP1 peptide at 25 °C (no salt in any buffer) and measured for the charge in zeta potential. A total of 3 measurements with 50 runs each were obtained using disposable zeta cells with platinum gold-coated electrodes (Malvern). The electrophoretic mobility received was used for the zeta potential calculation with the help of the Smoluchowski equation.
2.9. Molecular dynamics (MD) simulation
The atomic-level dynamic and energetics could be understood through computer simulation for appropriate periods. Long simulations are helpful to explore structural properties and behavior with respect to time (Chong et al., 2017). We have utilized PepFold’s (Thévenet et al., 2012; Shen et al., 2014) web-server for constructing a 3D model for the C-terminal of NSP1. It applies a coarse-grained (CG) forcefield and performs up to 200 simulation runs to build an energy minimized structure. The resultant model was then prepared using Chimera by adding missing hydrogens and proper parameterization of asymmetrical residues (Pettersen et al., 2004).
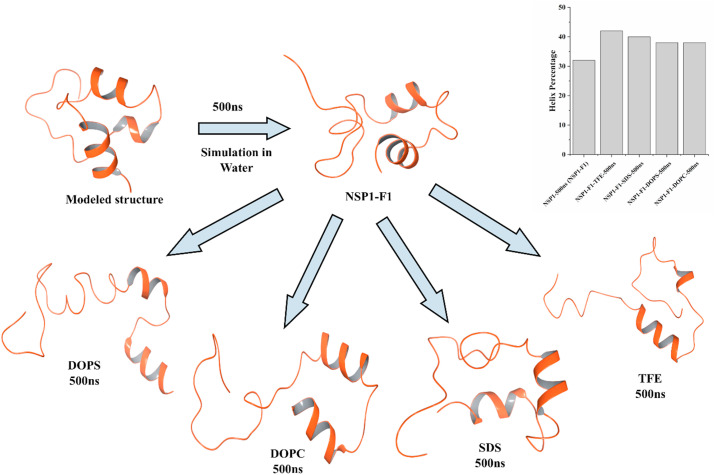
We used Gromacs v5, where simulation setup was built by placing the protein structure in a cubic box along with SPC water model, 0.15 M NaCl salt concentration. After solvation, the system was charge neutralized with counterions. The steepest descent method was used to attain an energy minimized simulation system until the system was converged within 1000 kJ/mol. Further, the equilibration of the system was done to optimize solvent in the environment. Using NVT and NPT ensembles within periodic boundary conditions for 100ps each, the system was equilibrated. The average temperature at 300 K and pressure at 1 bar were maintained using Nose-Hoover and Parrinello-Rahman coupling methods during the simulation. All bond-related constraints were solved using SHAKE algorithm. The final production run was performed for 500ns in our high-performing cluster at IIT Mandi. After analyzing the trajectory, the last frame (we named ‘NSP1–F1’ to avoid confusion) was chosen for further studies in different conditions.
Next, the structural conformations of the C-terminal of NSP1 (NSP1–F1) were analyzed in two different conditions of solvents: TFE (Trifluoroethanol; 8 M) and SDS (Sodium dodecyl sulfate) mixed with water in separate simulation runs. Using the above-described forcefield parameters and TFE and SDS molecules’ addition into the system, the simulation was performed for 500ns each. Further, to investigate the structural transitions upon lipid-mediated environments, we performed molecular dynamics simulation of frame NSP1–F1 in the presence of DOPS. Using previously described parameters for simulation in DOPS and DOPC (Saumya et al., 2020), we used CHARMM-GUI web interface (Jo et al., 2008) and inserted 300 molecules of DOPS and DOPC around protein structure in two separate systems. Following minimization and equilibration, the final production MD was performed for 500 ns? All trajectory analysis calculations were performed using Chimera, maestro, and Gromacs command for calculating the root mean square deviation (RMSD), root mean square fluctuation (RMSF), radius of gyration (Rg) for protein structure compactness, and solvent accessible surface area (SASA) for C-α atoms. The secondary structure component percentage was calculated using 2struc web server (Dp et al., 2010).
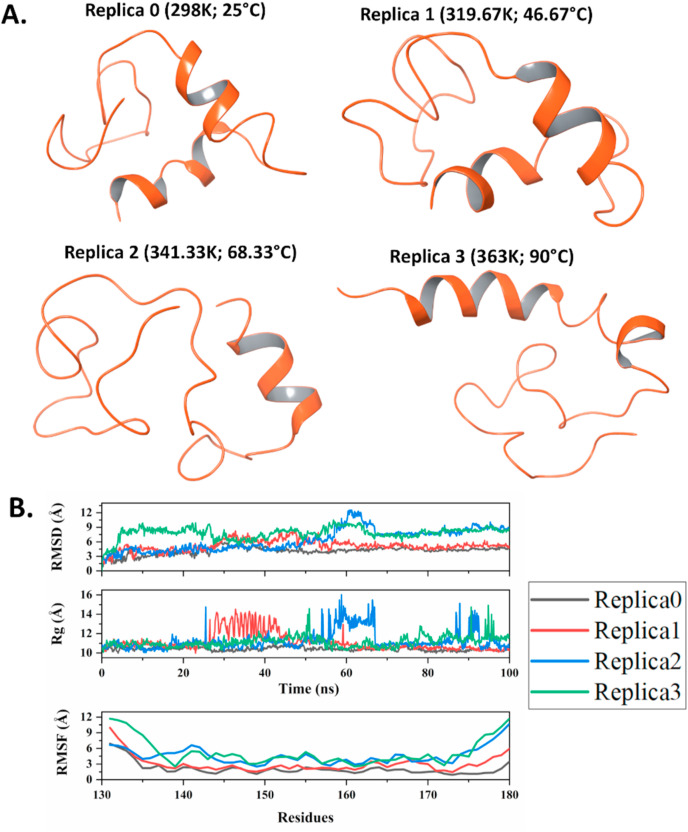
Additionally, to analyze the atomic level characteristics of NSP1–F1, we performed Replica Exchange Molecular Dynamics simulations on four different temperatures using our previously reported protocol (Kumar et al., 2020b). Using linear calculation mode of Desmond simulation package and experimental temperature limits, we selected four temperatures viz. 298 K (25 °C), 319.67 K (46.67 °C), 341.33 K (68.33 °C), and 363 K (90 °C). All these four replicas were allowed to exchange throughout the simulation period of 100 ns? The analyses of secondary structure content, RMSD, RMSF, and Rg were carried out using the Schrodinger Simulation Event Analysis module.
3. Results
3.1. Structural analysis of NSP1 C-terminal region and insight into intrinsic disorder properties
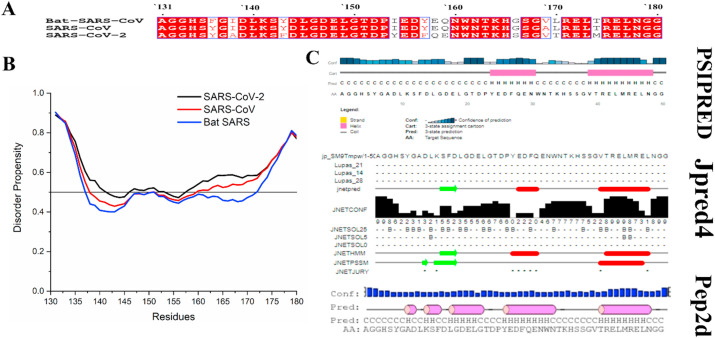
The SARS-CoV-2 C-terminal has few diverse amino acid residues, which indicated their evolutionary divergence from other coronaviruses (Fig. 2 B).
Fig. 2.
(A) The multiple sequence alignment of NSP1-CTR showing consensus sequences. The amino acids conserved in all the analyzed NSP1-CTR sequences are shown in a white with the red background and boxed in black. The residues shown in red with the white background are conserved in most analyzed sequences for their properties. (B) Disorder predisposition predictions of the NSP1-CTR (50 residues) using disorder predictor PONDR® VSL2. The interception at 0.5 scores on Y-axis shows the cut-off of disorderedness. Regions above 0.5 are considered as disordered. (C) Secondary structure predisposition predictions using servers PSIPRED, Jpred4, and Pep2D (C-Coil, E-Extended strand, B-Beta strand, and H-Helix).
The intrinsic disorder predisposition of NSP1 C-terminal was studied using commonly used VSL2 predictor of intrinsic disorder. It found that it is on the borderline between ordered and disordered nature, near the prediction cut-off of 0.5 for disorder prediction (Fig. 2C). Further, sequence-based secondary structure analysis was done by utilizing multiple web-servers, which clearly showed that the C-terminal part could acquire helical conformation. The PSIPRED has predicted potential for helical structure at the 154–160 and 169–178 region ( Fig. 2 D). Jpred4 has indicated the β-strand propensity at 141–144 region and helical propensity at155-158 and 169–177 regions. Pep2d predicted the very high α-helical tendency at multiple regions, which was 141–142, 145–149, 154–161, and 169–178 regions ( Fig. 2 D).
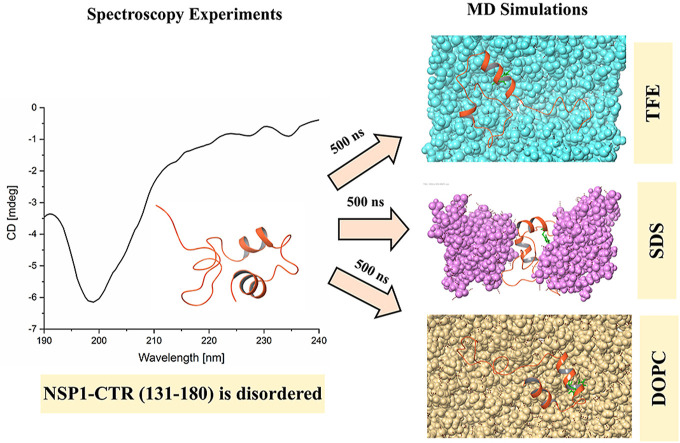
NSP1-CTR is disordered in isolation and has a folding propensity in the presence of organic solvents and lipids.
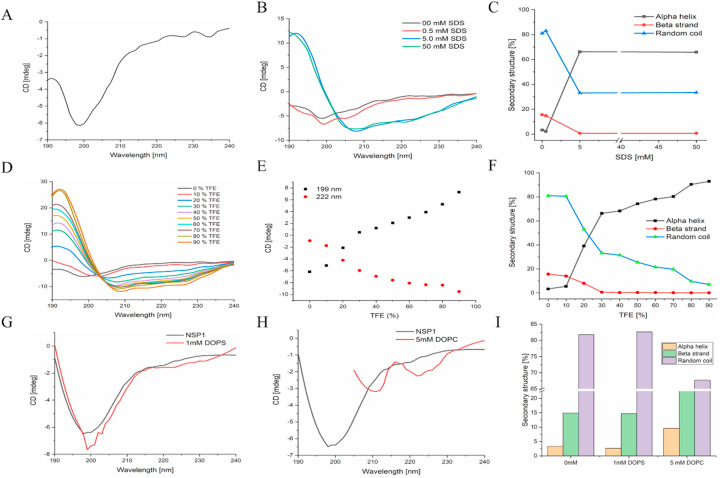
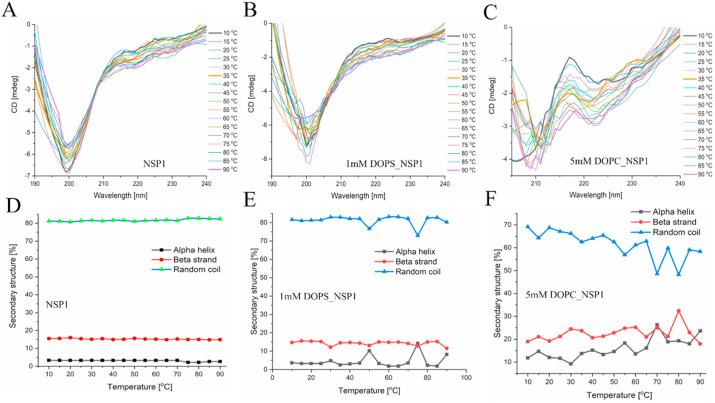
At a concentration of 20 μM in phosphate buffer (pH 7.0), the CD spectrum of NSP1-CTR was dominated by a single minimum (199 nm), which indicates that this region in isolation acquires random coil/disordered conformation ( Fig. 3 A). Therefore, in this study, we have used SDS, TFE, and liposomes for their hydrophobic/hydrophilic interface and charge mimic properties. CD Spectra showed the negative ellipticities at high SDS concentration at 208 and 222 nm, a typical characteristic of alpha-helical conformation. SDS’s effect on the conformation of NSP1-CTR was observed and found that SDS in its monomeric form (0.5 mM) doesn’t significantly impact the conformation of NSP1. However, SDS above critical micellar concentration (CMC; 5 mM and 50 mM) induce the helicity ( Fig. 3 B).
Fig. 3.
CD spectra showing effect of varying conditions on NSP1 C-terminal (A) 50 mM phosphate buffer (pH 7.0) (B) SDS micelle (D) TFE (G) DOPS and (H) DOPC. Panel (E) Ellipticity change at 198 nm and 222 nm in the presence of TFE. Panel (C, F, I) showing the secondary structure analysis of NSP1 in SDS, TFE, and liposomes, respectively.
Further, TFE was also found to induce helicity in the NSP1-CTR disorder system ( Fig. 3 D). The secondary structure analysis was performed using K2D3 indicates the gain in alpha-helical structure and loss in the random coil in the presence of SDS and TFE ( Fig. 3 C, E, F). In the presence of DOPS and DOPC there is no/partial change in the structural conformation that we can not say is either alpha-helical or disordered ( Fig. 3 G, H, I).
Further, the validation of experimental results was done through computational simulations. Firstly, the model built through the PEP-FOLD server was simulated in the presence of SPC water model using Gromos54A7 forcefield for 500 ns? Initially, the model constituted three helices (residues 145LGDEL149, 155EDFQE159, and 169VTRELMREL177) mainly at C-terminal. After the first simulation, the NSP1-CTR model started losing its structure and lost a significant amount of helix; designated as NSP1–F1 ( Fig. 4 ). The region with an unstable helix was found for residues 145–149, while the other two helices had insignificant fluctuations. As calculated using an online server 2struc, the simulated structure has lost its 10% helicity from 42% to 32%, as shown in Fig. 4. This indicates that the NSP1–C-terminal has less structure and more disorder character in isolation. The simulated model at 500 ns trajectory frame (NSP1–F1) was used for further simulation to study its structure ability. The first physiological condition was provided with 8 M trifluoroethanol in the simulation box, and the rest of the box was filled with water molecules. In the presence of an organic solvent, the NSP1–F1 structure showed increased helicity after 100ns up to 38% from 32% ( Fig. 4 ). Subsequently, after 300 ns, the trajectory frame shows 42% of helix present in the structure at residues 145–148, 155–163, and 171–177. However, the 500 ns frame has shown a little change at residues 171–177, while residues 145–148, 155–164 were intact. The MD simulation results are also purely correlated with the experimental observations. They suggest that in an organic solvent environment like TFE, the C-terminal of NSP1 has the propensity to gain structure.
Fig. 4.
MD Simulation of modeled structure in water and its last frame trajectory in SDS, TFE, DOPC, and DOPS. In the inset, bar graph representing helix percentage in modeled structure and frames of NSP1 obtained from MD simulation. Effect of temperature on the conformation of NSP1-CTR.
Further, MD simulation of NSP1–F1was performed in the presence of SDS and DOPC, and DOPS. In 500ns simulation time and the presence of nearly 60 molecules of SDS, the simulated frame or NSP1–F1 structure gained a slight increment in helical conformation at residues 155–161 and 170–176, with a total helicity of 40%. Similarly, DOPS has also shown a slight change in helical conformation at residues 159–161 and 170–178. In the DOPC lipid environment, the residues 155–163 and 170–178 have shown helical conformation. As calculated by 2struc server, the simulated structure in DOPS and DOPC has gained helicity from 32% to 38% and 38%, respectively, as shown in Fig. 4.
As depicted in Fig. 5 A, an increase in temperature leads to a partial change in the structural conformation of NSP1-CTR. Further, to know the impact of temperature on NSP1-CTR and lipid association, the effect of increasing temperature was studied ( Fig. 5 B, C). DOPC induced more contraction to this IDP system as compared to the contraction without the lipids. The secondary structure analysis using K2D3 also suggested a change in conformation in the presence of DOPC ( Fig. 5 D, E, F). The DOPS doesn’t enhance the contraction because both the peptide and lipid are negatively charged and may not be binding together. As observed in CD spectra without lipids, there was less contraction in NSP1 on increasing temperature, a similar trend was observed in Replica-exchange MD simulations. Based on Desmond simulation calculations, the helicity in NSP1–F1 at 298 K (replica 0) was 22.58%, while it got significantly decreased in subsequent replicas i.e., replica 1: 17.22; replica 2: 14.79; and replica 3: 14.05% (Fig. 6 A). The same can also be conferred from Fig. 6B, where the mean deviations and fluctuations of protein atoms are plotted in respect to the 100 ns simulation time. There were heavy fluctuations in high-temperature replicas 2 and 3 compared to low-temperature replicas (replica 0 and 1). The heavily fluctuating Rg values depict reduced compactness in NSP1 during simulations.
Fig. 5.
Effect of temperature on NSP1-CTR. Panel (A, D) showing the change in conformation and secondary structure in the presence of increasing temperature. Panel (B, E) and (C, F) showing the change in conformation and secondary structure in the presence of DOPS and DOPC, respectively.
Fig. 6.
Temperature-based Replica Exchange MD simulation: A. Snapshots of 100 ns frames from all replicas (replica 0 to 4) of four selected temperatures. B. RMSD, Rg, and RMSF analysis of all four replicas till 100 ns timescale. NSP1-CTR conformational change revealed by Trp fluorescence.
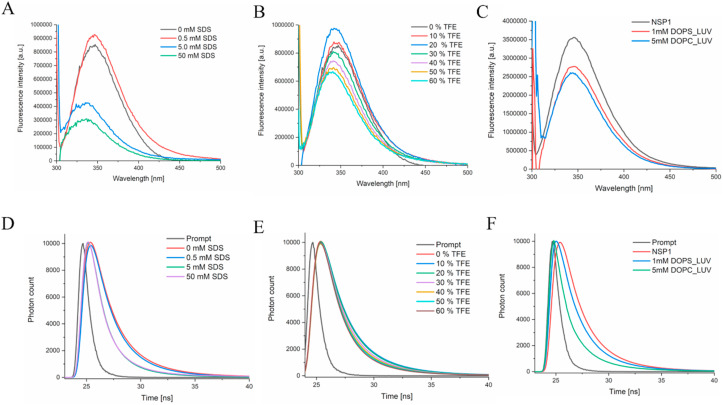
The NSP1-CTR has one Trp161, which showed emission maxima at 346 nm in phosphate buffer. With a rising concentration of SDS (above CMC), we observed decreased fluorescence intensity and a significant blue shift (emission maxima at 335 nm) ( Fig. 7 A). Similarly, the presence of TFE also reduces the fluorescence intensity with a blue shift (emission maxima at 339 nm) that may be due to increased non-polarity of the solvent ( Fig. 7 B). Further, NSP1-CTR interaction with the liposome (negatively charged liposome: DOPS; and neutral charged liposome: DOPC) reduces the fluorescence intensity with blue shift (emission maxima at 345 nm and 344 nm for DOPS and DOPC, respectively; see Fig. 7C). The average lifetime of fluorophore Trp in NSP1-CTR was observed to be 1.84 ns in phosphate buffer (pH 7.0), while 1.02 ns in the presence of 5 mM SDS, and 1.57 ns in 60% TFE (Table 1 and Fig. 7D and E). The average lifetime in the presence of DOPS and DOPC decreased to 0.634 ns and 0.0847 ns, respectively (Table 1 and Fig. 7F).
Fig. 7.
Tryptophan emission fluorescence analysis (A, B, C) and a comparison of fluorescence lifetime decay (D, E, F) curve of NSP1-CTR peptide in (A, D) SDS and (B, E) TFE and (C, F) Liposomes. In panel D the decay trace is of 0 mM and 0.5 mM of SDS is overlapping.
Table 1.
The average lifetime in different conditions after fitting data in three exponential terms. In the case of SDS and TFE the 5 μM peptide concentration is considered. For Liposomes, 20 μM peptide concentration is considered.
| Component | Concentration | Average lifetime (Nanosecond) | CHISEQ |
|---|---|---|---|
| NSP1-CTR only | 5 μM | 1.84 | 1.054 |
| SDS | 0.5 mM | 1.73 | 1.109 |
| 5 mM | 1.02 | 1.159 | |
| 50 mM | 1.02 | 1.345 | |
| TFE | 10% | 1.91 | 1.154 |
| 20% | 1.83 | 1.112 | |
| 30% | 1.72 | 1.097 | |
| 40% | 1.62 | 1.156 | |
| 50% | 1.61 | 1.152 | |
| 60% | 1.57 | 1.061 | |
| Liposome | 1 mM DOPS | 0.634 | 1.289 |
| 5 mM DOPC | 0.0847 | 1.088 |
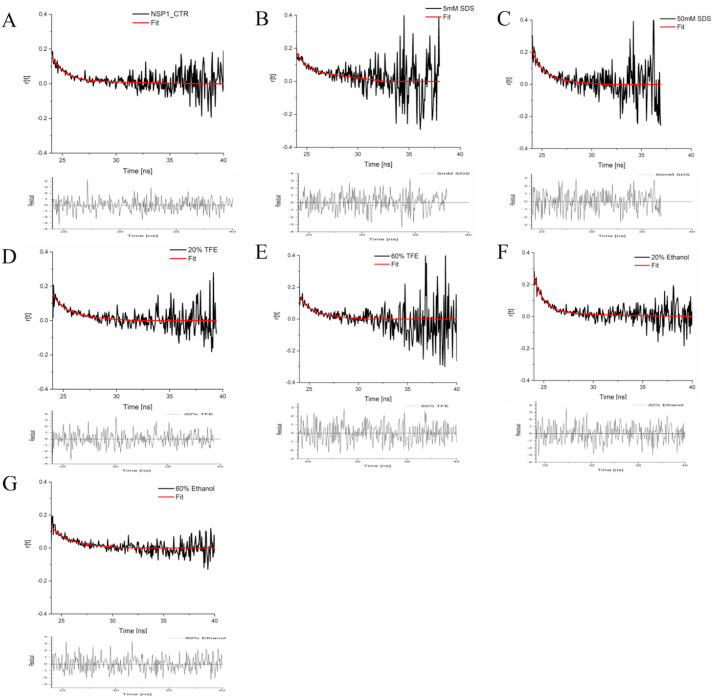
Fig. 8 shows the fluorescence anisotropy decay for the NSP1-CTR in the presence of SDS micelle, organic solvents like TFE, and ethanol. The decay curve was fitted with the bi-exponential decay function correlated with equation two described in the method section. The rotational correlation time θ1 (faster rotational correlation time) was observed around one ns in all conditions (Table 2 ). Here the θ1 represents the rotational correlation time of Trp161 residue motion. However, rotational correlation time θ2 (slower rotational correlation time) was observed beyond 200 ns at higher SDS concentration, TFE, and ethanol, representing the global conformational dynamics of NSP1-CTR. The slower rotational correlation times (θ2) for NSP1-CTR only is 16.94 ns, but it reduces to 4.09 ns, and 7.3 ns in 20% TFE and 20% ethanol, respectively. Further, in the presence of SDS (5 mM and 50 mM) and TFE (60%) where helical conformation was observed (monitored by CD; see Fig. 3), NSP1-CTR showed θ2 between 1300 and 1600 ns.
Fig. 8.
Decay of the fluorescence anisotropy of NSP1-CTR in the presence of (A) Buffer (B, C) SDS (D, E) TFE and (F, G) Ethanol. Surface charge distribution on LUVs in the presence of NSP1-CTR.
Table 2.
Anisotropy decay parameters for NSP1-CTR.
| Component | Concentration | θ1 (ns) | Relative amplitude (A1) | θ2 (ns) | Relative amplitude (A2) | Average rotational correlation time (ns) () |
|---|---|---|---|---|---|---|
| NSP1-CTR | 30 μM | 1.2 | 24.08 | 16.94 | 75.92 | 4.10 |
| SDS | 5 mM | 0.87 | 00 | 1610 | 100 | 1588 |
| 50 mM | 1.13 | 00 | 1384 | 100 | 1330 | |
| TFE | 20% | 0.90 | 20.36 | 4.09 | 79.64 | 2.38 |
| 60% | 1 | 00 | 1475 | 100 | 1452 | |
| Ethanol | 20% | 1.02 | 53.74 | 7.3 | 46.26 | 1.69 |
| 60% | 1.4 | 24.73 | 8.8 | 75.27 | 3.82 |
The zeta potential measurement was done to estimate the surface charge distribution of liposomes (DOPS LUV and DOPC LUV) upon the addition of the NSP1-CTR peptide. The results showed that there is a minor increase in the zeta potential towards the negative value for DOPS with NSP1 (−32.8 ± 3.15) than DOPS alone (−28.85 ± 0.21) while the minor change was found in the DOPC values (Table 3 ). The probable reason behind this might be the charge properties of the NSP1 CTR. The charge calculated for NSP1-CTR was −6, which thus cause electrostatic repulsion while interacting with DOPS.
Table 3.
Zeta potential measurement of liposome in the presence of NSP1 C-terminal region.
| Liposome | NSP1 concentration | Charge (mV) |
|---|---|---|
| DOPS 200 μM | 0 μM | −26.33 ± 2.35 |
| 5 μM | −28.85 ± 0.21 | |
| 20 μM | −32.8 ± 3.15 | |
| DOPC 200 μM | 0 μM | 1.86 ± 0.21 |
| 5 μM | 2.02 ± 0.15 | |
| 20 μM | 1.96 ± 0.38 |
3.2. Conformational dynamics of NSP1-CTR through MD simulation
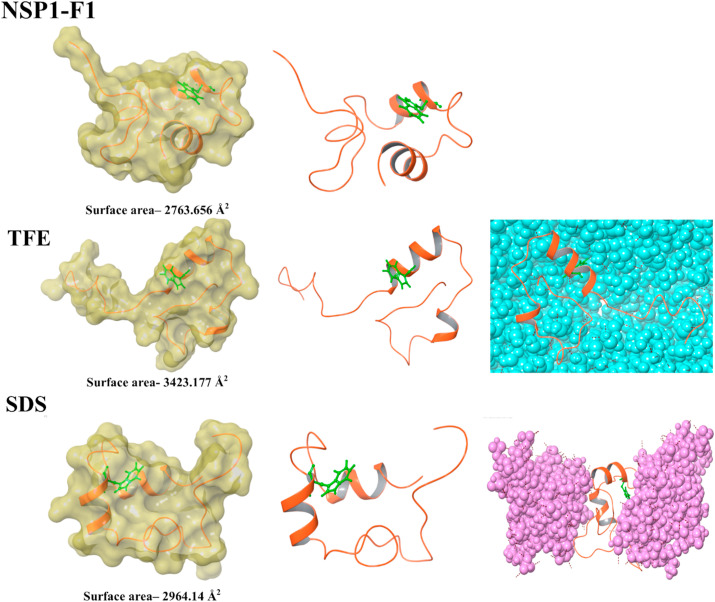
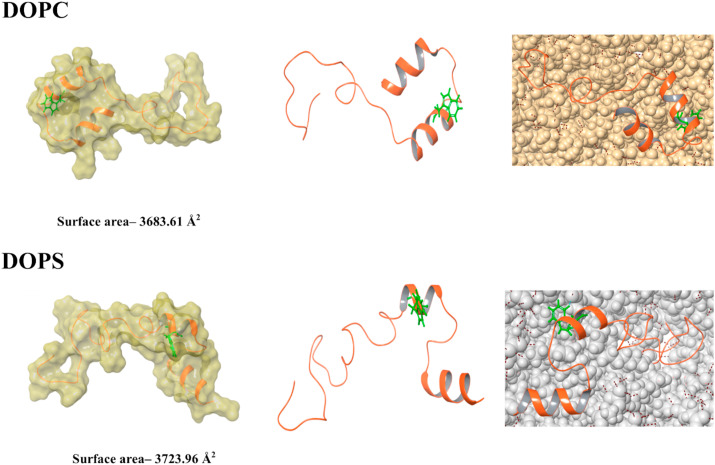
We have correlated the peptide’s conformational dynamics with its surrounding environment using CD spectroscopy and fluorescence spectroscopy. To complement our experiments, we further performed computational simulation. The NSP1–F1 simulation run has represented the change in conformation of NSP1-CTR in the presence of different environments i.e., TFE, SDS, and DOPS (Fig. 9A and B ). These changes may have occurred due to hydrophobic and electrostatic interactions that are likely to be formed during the simulation period. To analyze the change in conformation dynamics, we studied the surface properties and probed the Trp161 orientation in the respective environment with computational simulation (Fig. 9A and B ). The surface area on NSP1–F1 in water and biological mimic environment conditions showed changes related to its different conformations. The Trp161 has a different orientation in each situation, representing different conformation of NSP1-CTR (Fig. 9A and B). The NSP1–F1 structural dynamics corroborate with our experiments and strengthen the hypothesis of the interplay of multiple interactions for conformational dynamics of NSP1-CTR.
Fig. 9A.
Representation of structural conformations in MD simulation highlights the change in residues near to tryptophan and surface area (measured in Schrodinger’s maestro). Left panel: Surface representation of the final frame from the simulation. Middle panel: Final frames from the simulation. Right panel: TFE and SDS molecules are shown, which are surrounding the structure in MD.
Fig. 9B.
Representation of structural conformations in MD simulation highlights the change in residues near to tryptophan and surface area (measured in Schrodinger’s maestro). Left panel: Surface representation of the final frame from the simulation. Middle panel: Final frames from the simulation. Right panel: DOPC and DOPS molecules are shown, which are surrounding the structure in MD.
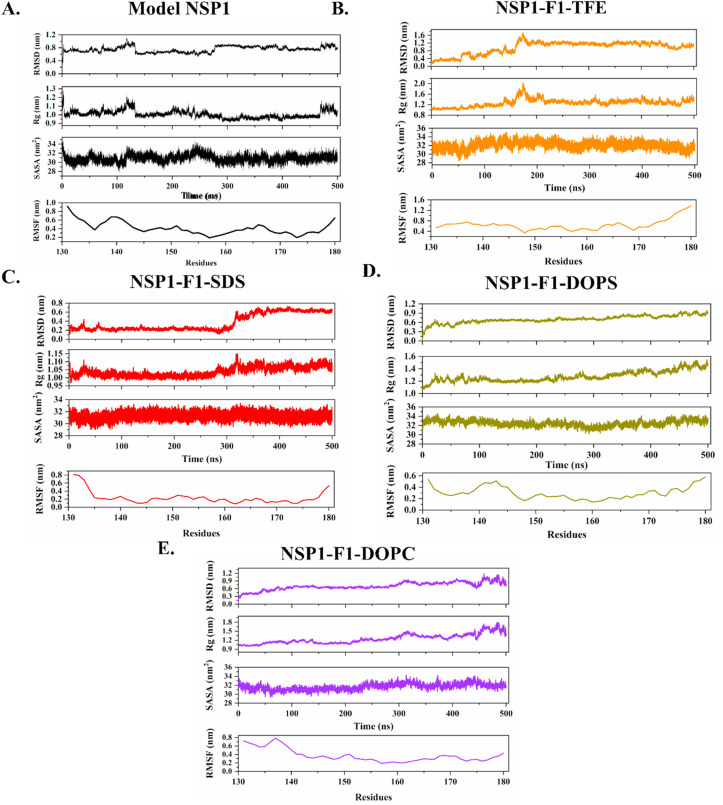
The time-dependent simulation frame analyses in Fig. 10 demonstrate these changes with RMSD, RMSF, Rg, and SASA values. First, the modeled structure of NSP1-CTR was simulated in water to check its conformation without any solvent condition, and its RMSD values were deviating initially (up to 0.8 nm till 125ns approx.) and then stabilized at 0.6 nm for the next 250 ns? The RMSD values remained same till 450 ns, but fluctuates in the last 50ns and get unstructured in some regions. These trends were also reflected in the other two time-dependent parameters (Rg and SASA). According to RMSF trend, the terminals’ residues fluctuate heavily, but nearby to C-terminal residues are found to be less fluctuating. In the presence of TFE, the radius of gyration and RMSD values show variations in the middle of 300ns simulation. During this period, the structural transitions occurred, and the region 145–148 got converted into helical conformation with one residue extra in the middle helix (Fig. 10B). After structure gain, the RMSF values decreased and illustrated that the structural transitions from unstructured to structured conformations are prominent. Further, in SDS’s presence, no such high fluctuations have been observed. However, secondary structure analysis predicted a gain of helicity (Fig. 4). Lastly, the CHARMM-GUI based lipid system also showed slight changes in structure as the RMSD and SASA values increased up to 100ns and then stabilized for the rest of the simulation period (Fig. 10).
Fig. 10.
Molecular Dynamics Simulation analysis of A. Model NSP1, B. NSP1–F1-TFE, C. NSP1–F1-SDS, D. NSP1–F1-DOPS, and E. NSP1–F1-DOPC. From up to down RMSD, Rg, SASA, and RMSF plots are shown.
4. Discussion
The SARS-CoV-2 NSP1-CTR among coronaviruses showed some significant substitution mutation, which may impact its folding propensity and binding affinity with respective partners. The NSP1-CTR was found to be disordered in an aqueous solution. The reason for peptide disorderedness in water has been described previously that the water makes hydrogen bonds with carbonyl group. Then, water oxygen starts to pair with N–H and disrupt the intramolecular hydrogen bonding (Rajan and Balaram, 1996). In comparison, TFE has a low dielectric constant and high dipole moment compared to water. The TFE directly interact with peptide and imparts hydrophobic effect. Irrespective of water, TFE makes stable hydrogen bonding with carbonyl groups of peptides and does not disrupt the intramolecular hydrogen bonding (Rajan and Balaram, 1996). Our finding revealed that the NSP1-CTR gain helical propensity in the presence of SDS and TFE.
The folding propensity of the disordered proteins was studied with organic solvents and lipids because their biological environment mimics properties (Uversky, 2009). In this context, we have used biological mimic environment conditions such as SDS micelles that provide a model for the hydrophobic and hydrophilic interface similar to lipid membranes (Popovic et al., 2007). According to the previously reported literature, the gain of the helical structure observed, where the interaction of disorder protein with SDS and TFE was used as a model system to mimic the biological environment such as hydrophobic nature (Kumar et al., 2020b, 2020c, 2020d; Saumya et al., 2020). Further, to elucidate the change in structure conformation, molecular dynamics studies have been performed. Previously, amyloid beta-peptide simulation studies showed that the peptide conformation changes at different concentrations of TFE (Jalili and Akhavan, 2009). Moreover, a model peptide and an antimicrobial peptide were also demonstrated in the co-presence of TFE with water, where the helical propensity of the structure was observed to be increased (Vymětal et al., 2016). Similar work by Jalili et al. has performed all atoms and coarse-grained simulation of Amyloid-beta (Aβ40) peptide in the presence of SDS. It has shown the formation of micelles around the peptide due to the self-assembly of molecules of SDS (S and A., 2010). The temperature has a significant impact on the protein’s backbone.
It is stated that the increase in the temperature leads to slight changes in the secondary structure of intrinsically disordered proteins, which is associated with hydrophobic interactions (Kumar et al., 2020b, 2020c). The contraction was observed in NSP1-CTR; it may be due to hydrophobic interaction collapse concerning secondary structure formation (Kumar et al., 2020b; Uversky and Fink, 2002). However, in the presence of DOPC, the changes in NSP1-CTR conformation were stable. This suggests that NSP1-CTR interacts with the neutrally charged liposome and acquires stable conformation, which in isolation was disordered. Previously, non-helical regions and hydrophilic chains in proteins were also responsible for the high-temperature mediated contraction in disordered proteins (Wuttke et al., 2014; Kjaergaard et al., 2010).
Further, we have measured the conformational dynamics and steric hindrance around the NSP1-CTR using Trp161 fluorophore in the SDS and organic solvent environment. The change in the fluorescence intensity of NSP1-CTR was due to conformational dynamics, which leads Trp residue to expose in a different environment. So, fluorescence studies suggest that the SDS, TFE, and liposome induce a conformational change in NSP1-CTR by imparting hydrophobic interaction and altering the hydrogen bonding. Tryptophan residue usually has a maximal fluorescence emission wavelength at 340–350 nm when exposed to water. In contrast, in our results, a blue shift was observed, which indicates buried Trp residue in the hydrophobic core of SDS micelle (Tulumello and Deber, 2009; Hou et al., 2020). This fluorescence-based study was further validated by the lifetime measurement of the Trp fluorescence. In our case, a fluorophore (Trp161), in response to excitation, spends time in the excited state, emits a photon, and returns to the ground state, which is denoted as its fluorescence lifetime. Any change in the physicochemical conditions leads to the change in fluorophore property and thus the fluorescence lifetime. Therefore, it can be seen that the lifetime decreases gradually in the presence of TFE. However, there is a sharp decrease in a lifetime in the presence of SDS and liposomes (both DOPC and DOPS).
We have also analyzed the NSP1-CTR conformational dynamics using Time-resolved fluorescence anisotropy decay measurements, which have been used previously for other IDPs (Karadi et al., 2020; Jain et al., 2016). The faster rotational correlation times (θ1) suggest a local motion of the fluorophore. In contrast, slower correlation times (θ2) are designated with the peptide’s segmental dynamics and the entire peptide’s global tumbling (Lakowicz, 2006; Jain et al., 2016). At a higher SDS concentration or TFE, a very long θ2 represents the prolonged global rotation of NSP1-CTR. At 20% TFE, the lower value of θ2 as compared to the higher concentration of TFE (60%) and in the absence of TFE may suggest an existence of a transition phase where partially folded NSP1-CTR dominated. The slow rotation in SDS case may be due to SDS’s self-aggregating nature, which forms micellar assemblies that increase the overall size of NSP1-CTR. SDS molecules’ amphiphilic nature allows them to self-aggregate and forms micellar assemblies (Panda et al., 2007; Maiti et al., 1997; Ulmer et al., 2005).
A similar type of approach has been employed previously to investigate the conformational dynamics of α-synuclein in the presence of SDS micelle using NMR that shows chemical entity of micelle doesn’t perturb the conformation change, but quantitative binding does (Ulmer et al., 2005). Overall, we may say that NSP1-CTR can acquire different structural conformation to perform functional roles in host cells, which need further experimental validation. Previously some evidence suggests that the two helical structures, alpha 1 and alpha 2 (Trp161, lie in between these two helical regions), acquired by NSP1-CTR when interacting with the 40 S ribosome subunit (Thoms et al., 2020). Thus, our study corroborates these results and found a change in structural conformation. Our group has recently demonstrated that p53 TAD2 was an IDR and evidenced a similar finding with an organic solvent, which relates its diverse conformations with different interacting partners (Kumar et al., 2020c). Further, to know the impact of hydrophobic interactions, we have studied the charge of liposomes and if any change in charge is observed in the presence of NSP1-CTR. The charge calculated for NSP1-CTR was −6, which thus cause electrostatic repulsion while interacting with DOPS. Previously it was shown that ionic interactions play an essential role in the interaction of the peptide (random coil to alpha-helical) with oppositely charged lipid vesicles (Saumya et al., 2020; Faustino et al., 2015). The CD data suggest that secondary structure in the presence of DOPS and DOPC remain partially affected. It has been reported earlier that alpha conformation solely is not the determining factor for the interaction between peptides and lipid vesicles, even though the headgroup of lipids may play an essential role in these interactions (Christiaens et al., 2002). The change in conformation of NSP1-CTR with neutrally charged liposome (no change in charge) further strengthens our hypothesis that hydrophobic interactions primarily play a role in the disorder-structure paradigm of this IDR system.
5. Conclusion
In summary, in this study, we have found that NSP1-CTR is an IDR in isolation, which is in line with previous discovery using Cryo-EM that the C-terminal is highly flexible in full length (NSP1) system. Further, we investigated the folding propensity in different environments and lipids and found conformational heterogeneity. The interplay of interaction forces is the driving force responsible for the protein-protein interactions and their associated functions. IDPs/IDRs binding promiscuity provides flexibility to the structural domain and functions independently with the number of partner ligands. The present study allowed us to understand the impact of hydrophobic interactions and electrostatic interaction, which agrees with the previous reports, where NSP1-CTR gains structural helicity after interaction with the 40 S and 80 S ribosomes (Thoms et al., 2020). The conformational dynamics of NSP1-CTR in solution could pave a path to understanding this disorder system’s structural ensemble. Further, some questions could be answered by the binding kinetics and folding studies of NSP1-CTR with ligand partner, which eventually helps us understand this peptide’s promiscuity and design a small molecular inhibitor.
Credit authorship contribution statement
RG and NG: study supervision. Amit Kumar and Ankur Kumar designed the experiment. Amit Kumar and Ankur Kumar conducted the experiment. Amit Kumar and PK acquisition and interpretation of computational data. Amit Kumar, Ankur Kumar, PK, and RG contributed to paper writing.
Declaration of competing interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.
Acknowledgments
All the authors would like to thank IIT Mandi for providing experimental and HPC facilities and Faculty Research Grant, SBS, IIT Mandi to RG. RG is thankful of DBT, Government of India (BT/11/IYBA/2018/06). AmK was supported by DBT, Government of India (BT/11/IYBA/2018/06). AnK was supported by fellowship from MHRD.
References
- Almeida M.S., Johnson M.A., Herrmann T., Geralt M., Wüthrich K. Novel β-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus. J. Virol. 2007;81:3151–3161. doi: 10.1128/JVI.01939-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A.K., Blanco M.R., Bruce E.A., Honson D.D., Chen L.M., Chow A., Bhat P., Ollikainen N., Quinodoz S.A., Loney C., Thai J., Miller Z.D., Lin A.E., Schmidt M.M., Stewart D.G., Goldfarb D., De Lorenzo G., Rihn S.J., Voorhees R.M., Botten J.W., Majumdar D., Guttman M. SARS-CoV-2 disrupts splicing, translation, and protein trafficking to suppress host defenses. Cell. 2020;183:1325–1339. doi: 10.1016/j.cell.2020.10.004. e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhardwaj, T., K.U. Saumya, P. Kumar, N. Sharma, K. Gadhave, V.N. Uversky, and R. Giri. Japanese encephalitis virus – exploring the dark proteome and disorder–function paradigm. FEBS J. n/a. [DOI] [PubMed]
- Buchan D.W.A., Jones D.T. The PSIPRED protein analysis workbench: 20 years on. Nucleic Acids Res. 2019;47:W402–W407. doi: 10.1093/nar/gkz297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chong S.-H., Chatterjee P., Ham S. Computer simulations of intrinsically disordered proteins. Annu. Rev. Phys. Chem. 2017;68:117–134. doi: 10.1146/annurev-physchem-052516-050843. [DOI] [PubMed] [Google Scholar]
- Christiaens B., Symoens S., Vanderheyden S., Engelborghs Y., Joliot A., Prochiantz A., Vandekerckhove J., Rosseneu M., Vanloo B. Tryptophan fluorescence study of the interaction of penetratin peptides with model membranes. Eur. J. Biochem. 2002;269:2918–2926. doi: 10.1046/j.1432-1033.2002.02963.x. [DOI] [PubMed] [Google Scholar]
- Clark L.K., Green T.J., Petit C.M. Structure of nonstructural protein 1 from SARS-CoV-2. J. Virol. 2021;95 doi: 10.1128/JVI.02019-20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dp K., Ba W., Rw J. 2Struc: the secondary structure server. Bioinforma. Oxf. Engl. 2010;26:2624–2625. doi: 10.1093/bioinformatics/btq480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drozdetskiy A., Cole C., Procter J., Barton G.J. JPred4: a protein secondary structure prediction server. Nucleic Acids Res. 2015;43:W389–W394. doi: 10.1093/nar/gkv332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A.K., Garner E., Guilliot S., Romero P., Albrecht K., Hart J., Obradovic Z., Kissinger C., Villafranca J.E. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac. Symp. Biocomput. Pac. Symp. Biocomput. 1998:473–484. [PubMed] [Google Scholar]
- Ellis R.J. Macromolecular crowding: obvious but underappreciated. Trends Biochem. Sci. 2001;26:597–604. doi: 10.1016/s0968-0004(01)01938-7. [DOI] [PubMed] [Google Scholar]
- Faustino A.F., Guerra G.M., Huber R.G., Hollmann A., Domingues M.M., Barbosa G.M., Enguita F.J., Bond P.J., Castanho M.A.R.B., Da Poian A.T., Almeida F.C.L., Santos N.C., Martins I.C. Understanding dengue virus capsid protein disordered N-terminus and pep14-23-based inhibition. ACS Chem. Biol. 2015;10:517–526. doi: 10.1021/cb500640t. [DOI] [PubMed] [Google Scholar]
- Gadhave K., Kumar P., Kapuganti S.K., Uversky V.N., Giri R. Unstructured biology of proteins from ubiquitin-proteasome system: roles in cancer and neurodegenerative diseases. Biomolecules. 2020;10:796. doi: 10.3390/biom10050796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri R., Kumar D., Sharma N., Uversky V.N. Intrinsically disordered side of the zika virus proteome. Front. Cell. Infect. Microbiol. 2016;6 doi: 10.3389/fcimb.2016.00144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giri R., Bhardwaj T., Shegane M., Gehi B.R., Kumar P., Gadhave K., Oldfield C.J., Uversky V.N. Understanding COVID-19 via comparative analysis of dark proteomes of SARS-CoV-2, human SARS and bat SARS-like coronaviruses. Cell. Mol. Life Sci. 2020 doi: 10.1007/s00018-020-03603-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hou H., He H., Wang Y. Effects of SDS on the activity and conformation of protein tyrosine phosphatase from thermus thermophilus HB27. Sci. Rep. 2020;10:3195. doi: 10.1038/s41598-020-60263-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang C., Lokugamage K.G., Rozovics J.M., Narayanan K., Semler B.L., Makino S. SARS coronavirus nsp1 protein induces template-dependent endonucleolytic cleavage of mRNAs: viral mRNAs are resistant to nsp1-induced RNA cleavage. PLoS Pathog. 2011;7 doi: 10.1371/journal.ppat.1002433. e1002433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N., Narang D., Bhasne K., Dalal V., Arya S., Bhattacharya M., Mukhopadhyay S. Direct observation of the intrinsic backbone torsional mobility of disordered proteins. Biophys. J. 2016;111:768–774. doi: 10.1016/j.bpj.2016.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jalili S., Akhavan M. A molecular dynamics simulation study OF conformational changes and solvation OF Aβ peptide IN trifluoroethanol and water. J. Theor. Comput. Chem. 2009:215–231. 08. [Google Scholar]
- Jo S., Kim T., Iyer V.G., Im W. CHARMM-GUI: a web-based graphical user interface for CHARMM. J. Comput. Chem. 2008;29:1859–1865. doi: 10.1002/jcc.20945. [DOI] [PubMed] [Google Scholar]
- Kamitani W., Huang C., Narayanan K., Lokugamage K.G., Makino S. A two-pronged strategy to suppress host protein synthesis by SARS coronavirus Nsp1 protein. Nat. Struct. Mol. Biol. 2009;16:1134–1140. doi: 10.1038/nsmb.1680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karadi K., Kapetanaki S.M., Raics K., Pecsi I., Kapronczai R., Fekete Z., Iuliano J.N., Collado J.T., Gil A.A., Orban J., Nyitrai M., Greetham G.M., Vos M.H., Tonge P.J., Meech S.R., Lukacs A. Functional dynamics of a single tryptophan residue in a BLUF protein revealed by fluorescence spectroscopy. Sci. Rep. 2020;10:2061. doi: 10.1038/s41598-020-59073-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kjaergaard M., Nørholm A.-B., Hendus‒Altenburger R., Pedersen S.F., Poulsen F.M., Kragelund B.B. Temperature-dependent structural changes in intrinsically disordered proteins: formation of α‒helices or loss of polyproline II? Protein Sci. 2010;19:1555–1564. doi: 10.1002/pro.435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar D., Singh A., Kumar P., Uversky V.N., Rao C.D., Giri R. Understanding the penetrance of intrinsic protein disorder in rotavirus proteome. Int. J. Biol. Macromol. 2020;144:892–908. doi: 10.1016/j.ijbiomac.2019.09.166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar A., Kumar P., Kumari S., Uversky V.N., Giri R. Folding and structural polymorphism of p53 C-terminal domain: one peptide with many conformations. Arch. Biochem. Biophys. 2020;684 doi: 10.1016/j.abb.2020.108342. 108342. [DOI] [PubMed] [Google Scholar]
- Kumar D., Mishra P.M., Gadhave K., Giri R. Conformational dynamics of p53 N-terminal TAD2 region under different solvent conditions. Arch. Biochem. Biophys. 2020;689 doi: 10.1016/j.abb.2020.108459. 108459. [DOI] [PubMed] [Google Scholar]
- Kumar A., Kumar P., Giri R. Zika virus NS4A cytosolic region (residues 1–48) is an intrinsically disordered domain and folds upon binding to lipids. Virology. 2020;550:27–36. doi: 10.1016/j.virol.2020.07.017. [DOI] [PubMed] [Google Scholar]
- Kuznetsova I.M., Turoverov K.K., Uversky V.N. What macromolecular crowding can do to a protein. Int. J. Mol. Sci. 2014;15:23090–23140. doi: 10.3390/ijms151223090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lakowicz J.R., editor. Time-Dependent Anisotropy Decays Principles of Fluorescence Spectroscopy. Springer US; Boston, MA: 2006. pp. 383–412. [Google Scholar]
- Lei X., Dong X., Ma R., Wang W., Xiao X., Tian Z., Wang C., Wang Y., Li L., Ren L., Guo F., Zhao Z., Zhou Z., Xiang Z., Wang J. Activation and evasion of type I interferon responses by SARS-CoV-2. Nat. Commun. 2020;11:3810. doi: 10.1038/s41467-020-17665-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maiti N.C., Krishna M.M.G., Britto P.J., Periasamy N. Fluorescence dynamics of dye probes in micelles. J. Phys. Chem. B. 1997;101:11051–11060. [Google Scholar]
- Mishra P.M., Uversky V.N., Giri R. Molecular recognition features in zika virus proteome. J. Mol. Biol. 2018;430:2372–2388. doi: 10.1016/j.jmb.2017.10.018. [DOI] [PubMed] [Google Scholar]
- Narayanan K., Ramirez S.I., Lokugamage K.G., Makino S. Coronavirus nonstructural protein 1: common and distinct functions in the regulation of host and viral gene expression. Virus Res. 2015;202:89–100. doi: 10.1016/j.virusres.2014.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obradovic Z., Peng K., Vucetic S., Radivojac P., Dunker A.K. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2005;61(Suppl. 7):176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- Oldfield C.J., Cheng Y., Cortese M.S., Romero P., Uversky V.N., Dunker A.K. Coupled folding and binding with α-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- Panda D., Khatua S., Datta A. Enhanced fluorescence of epicocconone in surfactant assemblies as a consequence of depth-dependent microviscosity. J. Phys. Chem. B. 2007;111:1648–1656. doi: 10.1021/jp065226o. [DOI] [PubMed] [Google Scholar]
- Pettersen E.F., Goddard T.D., Huang C.C., Couch G.S., Greenblatt D.M., Meng E.C., Ferrin T.E. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Popovic M., De Biasio A., Pintar A., Pongor S. The intracellular region of the Notch ligand Jagged-1 gains partial structure upon binding to synthetic membranes. FEBS J. 2007;274:5325–5336. doi: 10.1111/j.1742-4658.2007.06053.x. [DOI] [PubMed] [Google Scholar]
- Rajan R., Balaram P. A model for the interaction of trifluoroethanol with peptides and proteins. Int. J. Pept. Protein Res. 1996;48:328–336. doi: 10.1111/j.1399-3011.1996.tb00849.x. [DOI] [PubMed] [Google Scholar]
- Romano M., Ruggiero A., Squeglia F., Maga G., Berisio R. A structural view of SARS-CoV-2 RNA replication machinery: RNA synthesis, proofreading and final capping. Cells. 2020;9:1267. doi: 10.3390/cells9051267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- S J., A M. Study of the Alzheimer’s Aβ40 peptide in SDS micelles using molecular dynamics simulations. Biophys. Chem. 2010;153:179–186. doi: 10.1016/j.bpc.2010.11.007. [DOI] [PubMed] [Google Scholar]
- Saumya K.U., Kumar D., Kumar P., Giri R. Unlike dengue virus, the conserved 14–23 residues in N-terminal region of Zika virus capsid is not involved in lipid interactions. Biochim. Biophys. Acta BBA - Biomembr. 2020;1862 doi: 10.1016/j.bbamem.2020.183440. 183440. [DOI] [PubMed] [Google Scholar]
- Schubert K., Karousis E.D., Jomaa A., Scaiola A., Echeverria B., Gurzeler L.-A., Leibundgut M., Thiel V., Mühlemann O., Ban N. Author Correction: SARS-CoV-2 Nsp1 binds the ribosomal mRNA channel to inhibit translation. Nat. Struct. Mol. Biol. 2020;27:1094. doi: 10.1038/s41594-020-00533-x. 1094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Semper C., Watanabe N., Savchenko A. Structural characterization of nonstructural protein 1 from SARS-CoV-2. iScience. 2021;24 doi: 10.1016/j.isci.2020.101903. 101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen Y., Maupetit J., Derreumaux P., Tufféry P. Improved PEP-FOLD approach for peptide and miniprotein structure prediction. J. Chem. Theor. Comput. 2014;10:4745–4758. doi: 10.1021/ct500592m. [DOI] [PubMed] [Google Scholar]
- Shen Z., Wang G., Yang Y., Shi J., Fang L., Li F., Xiao S., Fu Z.F., Peng G. A conserved region of nonstructural protein 1 from alphacoronaviruses inhibits host gene expression and is critical for viral virulence. J. Biol. Chem. 2019;294:13606–13618. doi: 10.1074/jbc.RA119.009713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi M., Wang L., Fontana P., Vora S., Zhang Y., Fu T.-M., Lieberman J., Wu H. SARS-CoV-2 Nsp1 suppresses host but not viral translation through a bipartite mechanism. bioRxiv. 2020 2020.09.18.302901. [Google Scholar]
- Singh A., Kumar A., Uversky V.N., Giri R. Understanding the interactability of chikungunya virus proteins via molecular recognition feature analysis. RSC Adv. 2018;8:27293–27303. doi: 10.1039/c8ra04760j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh H., Singh S., Raghava G.P.S. 2019. Peptide Secondary Structure Prediction Using Evolutionary Information. bioRxiv. 558791. [Google Scholar]
- Thévenet P., Shen Y., Maupetit J., Guyon F., Derreumaux P., Tufféry P. PEP-FOLD: an updated de novo structure prediction server for both linear and disulfide bonded cyclic peptides. Nucleic Acids Res. 2012;40:W288–W293. doi: 10.1093/nar/gks419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thoms M., Buschauer R., Ameismeier M., Koepke L., Denk T., Hirschenberger M., Kratzat H., Hayn M., Mackens-Kiani T., Cheng J., Straub J.H., Stürzel C.M., Fröhlich T., Berninghausen O., Becker T., Kirchhoff F., Sparrer K.M.J., Beckmann R. Structural basis for translational shutdown and immune evasion by the Nsp1 protein of SARS-CoV-2. Science. 2020 doi: 10.1126/science.abc8665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tohya Y., Narayanan K., Kamitani W., Huang C., Lokugamage K., Makino S. Suppression of host gene expression by nsp1 proteins of group 2 bat coronaviruses. J. Virol. 2009;83:5282–5288. doi: 10.1128/JVI.02485-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Toto A., Giri R., Brunori M., Gianni S. The mechanism of binding of the KIX domain to the mixed lineage leukemia protein and its allosteric role in the recognition of c-Myb. Protein Sci. 2014;23:962–969. doi: 10.1002/pro.2480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tulumello D.V., Deber C.M. SDS micelles as a membrane-mimetic environment for transmembrane segments. Biochemistry. 2009;48:12096–12103. doi: 10.1021/bi9013819. [DOI] [PubMed] [Google Scholar]
- Ulmer T.S., Bax A., Cole N.B., Nussbaum R.L. Structure and dynamics of micelle-bound human alpha-synuclein. J. Biol. Chem. 2005;280:9595–9603. doi: 10.1074/jbc.M411805200. [DOI] [PubMed] [Google Scholar]
- Uversky V.N. Intrinsically disordered proteins and their environment: effects of strong denaturants, temperature, pH, counter ions, membranes, binding partners, osmolytes, and macromolecular crowding. Protein J. 2009;28:305–325. doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- Uversky V.N., Fink A.L. The chicken–egg scenario of protein folding revisited. FEBS Lett. 2002;515:79–83. doi: 10.1016/s0014-5793(02)02441-9. [DOI] [PubMed] [Google Scholar]
- Vymětal J., Bednárová L., Vondrášek J. Effect of TFE on the helical content of AK17 and HAL-1 peptides: theoretical insights into the mechanism of helix stabilization. J. Phys. Chem. B. 2016;120:1048–1059. doi: 10.1021/acs.jpcb.5b11228. [DOI] [PubMed] [Google Scholar]
- Ward J.J., Sodhi J.S., McGuffin L.J., Buxton B.F., Jones D.T. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J. Mol. Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J. Mol. Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- Wright P.E., Dyson H.J. Linking folding and binding. Curr. Opin. Struct. Biol. 2009;19:31–38. doi: 10.1016/j.sbi.2008.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wuttke R., Hofmann H., Nettels D., Borgia M.B., Mittal J., Best R.B., Schuler B. Temperature-dependent solvation modulates the dimensions of disordered proteins. Proc. Natl. Acad. Sci. Unit. States Am. 2014;111:5213–5218. doi: 10.1073/pnas.1313006111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue B., Dunker A.K., Uversky V.N. Orderly order in protein intrinsic disorder distribution: disorder in 3500 proteomes from viruses and the three domains of life. J. Biomol. Struct. Dyn. 2012;30:137–149. doi: 10.1080/07391102.2012.675145. [DOI] [PubMed] [Google Scholar]