Significance
Mechanical force affects fundamental processes in biology like cell adhesion, tissue morphogenesis, and tumorigenesis. The molecular mechanisms by which cells utilize their protein machineries to sense and transduce mechanical forces remain to be elucidated. This study describes the application of neutron spin echo spectroscopy to reveal the activation of nanoscale motions of the actin-binding domain in the cadherin–catenin complex. The activated domain motion and an ensemble of domain configurations adopted by the mechanosensory actin-binding domain suggest the formation of an entropic trap in the cadherin–catenin complex. We hypothesize that mechanical tension facilitates the reduction in entropy of a mechanosensitive protein and thereby focuses the configuration space to specific conformations that are competent to bind the moving actin microfilament.
Keywords: cell adhesion, neutron spin echo spectroscopy, protein dynamics, mechanotransduction, catch bond
Abstract
As the core component of the adherens junction in cell–cell adhesion, the cadherin–catenin complex transduces mechanical tension between neighboring cells. Structural studies have shown that the cadherin–catenin complex exists as an ensemble of flexible conformations, with the actin-binding domain (ABD) of α-catenin adopting a variety of configurations. Here, we have determined the nanoscale protein domain dynamics of the cadherin–catenin complex using neutron spin echo spectroscopy (NSE), selective deuteration, and theoretical physics analyses. NSE reveals that, in the cadherin–catenin complex, the motion of the entire ABD becomes activated on nanosecond to submicrosecond timescales. By contrast, in the α-catenin homodimer, only the smaller disordered C-terminal tail of ABD is moving. Molecular dynamics (MD) simulations also show increased mobility of ABD in the cadherin–catenin complex, compared to the α-catenin homodimer. Biased MD simulations further reveal that the applied external forces promote the transition of ABD in the cadherin–catenin complex from an ensemble of diverse conformational states to specific states that resemble the actin-bound structure. The activated motion and an ensemble of flexible configurations of the mechanosensory ABD suggest the formation of an entropic trap in the cadherin–catenin complex, serving as negative allosteric regulation that impedes the complex from binding to actin under zero force. Mechanical tension facilitates the reduction in dynamics and narrows the conformational ensemble of ABD to specific configurations that are well suited to bind F-actin. Our results provide a protein dynamics and entropic explanation for the observed force-sensitive binding behavior of a mechanosensitive protein complex.
In multicellular organisms, the mechanical coupling of cell–cell adhesion or cell–matrix adhesion is crucial for embryonic morphogenesis and development (1, 2) and for tissue homeostasis, regeneration, and repair (3). The multiprotein complexes at the cell–cell or cell–matrix adhesion sites sense the fluctuations in tensile forces in the extracellular microenvironment and undergo structural and compositional changes to transform mechanical signals into biochemical signals (4). As a result, cells alter their intracellular signaling pathways, cytoskeletal organization, gene expression, and transcriptional programs to modulate cell migration, proliferation, and differentiation (5–7). Dysregulation of the cell–cell or cell–matrix mechanosensing or mechanotransduction complexes has been implicated in a number of pathological conditions such as cancer (8, 9) and in defective cardiovasculature (10) and neuronal development (11).
In cell–cell adhesion, the adherens junctions bridge the mechanical couplings between neighboring cells (12, 13). The core component of the adherens junction is composed of cadherin, the β-catenin and α-catenin complex, and the actomyosin complex. Cadherin is a transmembrane protein that employs its extracellular domain to form homophilic bonds with cadherin from a neighboring cell (14). The cytoplasmic domain of cadherin is attached to β-catenin, which in turn binds to α-catenin to form the cadherin–catenin complex. The cadherin–catenin complex interacts with the cytoskeletal actin microfilament in a mechanical force-dependent manner (15). The mechanical forces can be externally applied, such as during tissue movement or via the shearing force of blood flow in blood vessels, or forces can be generated inside the cell by the actomyosin assemblies. The actomyosin assemblies are composed of actin microfilaments and bundles and the attached myosin motors that can generate pulling forces on the actin microfilament, thereby causing tension within a filament as well as sliding movements between the filaments. In addition, the cadherin•β-catenin•α-catenin complex associates with p120-catenin, which stabilizes the cadherin–catenin complex at the cell membrane, and with other actin-binding proteins like vinculin, eplin, and zyxin (12, 16, 17). Because the cadherin•β-catenin•α-catenin complex physically couples the neighboring cells to the cytoskeleton, this complex can transduce mechanical forces between actomyosin assemblies and the cell surface cadherin (18). The mechanical coupling between the actomyosin and cadherin also acts in the reverse direction: Forces sensed and transmitted by cadherin are sent back to the catenin complex and to the actin cytoskeleton. The cadherin•β-catenin•α-catenin complex facilitates reciprocal and dynamic couplings of the actomyosin networks of neighboring cells and is crucial to maintain mechanical strength in quiescent tissues and also for collective cell movement during tissue morphogenesis and repair (19, 20).
The realization that the cadherin•β-catenin•α-catenin complex functions as a mechanical force sensor arises from a series of biochemical, structural, and single-molecule optical trap experiments (21–27). Besides participating in forming the cadherin•β-catenin•α-catenin complex, α-catenin also forms homodimers that are localized in the leading edge of migrating cells (21, 25, 28, 29). Biochemical studies find that the α-catenin homodimer binds to the actin filament robustly without the need of applied forces (21, 22, 30). However, when incorporated in the cadherin•β-catenin•α-catenin complex, α-catenin binds to the actin filament effectively only under mechanical tension, utilizing a mechanism called a catch bond (21, 24). Additionally, the recruitment of vinculin to the adherens junction by α-catenin is force dependent (31)—mechanical force is thought to be required to expose the cryptic vinculin-binding site buried in the M1 subdomain of α-catenin (32); see Fig. 1B for domain organization of α-catenin. α-Catenin is thus identified as a key mechanosensory component in the adherens junction complex. However, the mechanisms by which the cadherin–catenin complex enacts force-sensitive binding remain to be explained. Current structural models of the truncated actin-binding domain of α-catenin do not provide an explanation of why the α-catenin homodimer binds to actin microfilament under zero force, whereas the binding of actin filament by the cadherin–catenin complex is force sensitive. The development of physical models of motor-derived forces in cell–cell and cell–matrix interactions is an area of active research in mechanobiology (33–35).
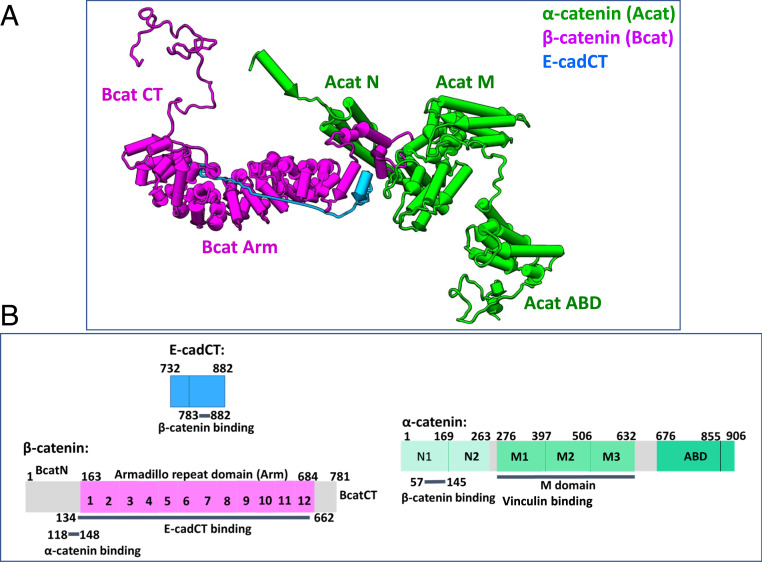
Fig. 1.
Structure and domain organizations of the ABE complex. (A) The model shown represents an ensemble-averaged structure obtained from small-angle X-ray and neutron scattering experiments (36). (B) Domain organizations and function of α-catenin, β-catenin, and E-cadherin cytoplasmic tail that form the ABE complex.
We have shown that the complex of E-cadherin cytoplasmic domain•β-catenin•α-catenin complex (hereafter called the ABE complex) displays an ensemble of flexible conformations, notably with the actin-binding domain of α-catenin adopting a variety of different configurations (36). The structural studies indicate that the cadherin–catenin complex populates thermodynamically reversible states and thus must possess a high degree of dynamics on a relatively flat free-energy landscape. To further understand the mechanism of mechanosensing by the ABE complex, we have determined the nanoscale protein domain motions in the ABE complex, using the relatively new technique of neutron spin echo spectroscopy (NSE). NSE belongs to a class of quasielastic neutron-scattering (QENS) techniques that determine the dynamics of matter (37–39). However, an NSE spectrometer measures dynamics at higher-energy resolution than the other QENS instruments such as the time-of-flight and backscattering spectrometers (40–42). An advanced NSE spectrometer is capable of determining the dynamics on nanometer length scales and on nanosecond to submicrosecond timescales in a protein (43–50). When combined with selective deuteration and theoretical physics analyses, NSE is uniquely suited to measure the dynamics of multidomain proteins and the multicomponent protein complex (44, 51).
Our NSE and selective deuteration experiments reveal that, in the ABE complex, the entire actin-binding domain (ABD) of α-catenin becomes more mobile than in the α-catenin homodimer, in which only the much smaller disordered C-terminal tail of the ABD is moving. Our molecular dynamics (MD) simulations also show increased mobility of the ABD as a part of the ABE complex, compared to that in the α-catenin homodimer. MD simulations further reveal that external forces can drive the conformational transition of the ABD in the ABE complex from an ensemble of diverse structures to specific states that resemble more the actin filament-bound structure of the ABD (52). Collectively, the study demonstrates that NSE can reveal which segment or domain in a protein or protein complex is likely to be a force sensor. The activated domain motion and an ensemble of flexible configurations adopted by the mechanosensory ABD suggest the formation of an entropic trap in the cadherin–catenin complex, which serves as the basis for negative allosteric regulation that impedes the ABE complex from binding to the actin microfilament. Mechanical tension facilitates the reduction in entropy of the cadherin–catenin complex and thereby focuses and narrows the configuration space to specific conformations that are competent to binding.
Results
NSE Spectra of Fully Hydrogenated and Selectively Deuterated ABE Complexes.
Fig. 1A is a representative structural model of the ABE complex obtained from small-angle X-ray and neutron scattering (36). Fig. 1B shows the domain organization and function of each domain in the ABE complex. The fully hydrogenated hAhBhE complex was reconstituted from hydrogenated α-catenin (hA) and hydrogenated β-catenin (hB) bound to hydrogenated E-cadherin cytoplasmic tail (hE), using the same method described in ref. 36. The selectively deuterated hAdBdE complex was reconstituted from the hydrogenated α-catenin, deuterated β-catenin (dB), and deuterated E-cadherin (dE). The selectively deuterated dAhBdE complex was formed from deuterated α-catenin (dA) bound to hydrogenated β-catenin and deuterated E-cadherin cytoplasmic tail (dE). The concentrations of the different protein complexes measured by NSE are shown in Materials and Methods. The dissociation constant Kd of β-catenin binding to E-cadherin cytoplasmic tail was determined to be 104 nM, and the Kd of α-catenin to the BE complex to be ∼200 nM (SI Appendix, Figs. S8 and S9A). Because the NSE experiments were performed at protein complex concentrations (of >50 μM) much higher than the dissociation constants of the ABE complex, the NSE data reported herein are from the discrete whole hydrogenated or selectively deuterated ABE complexes.
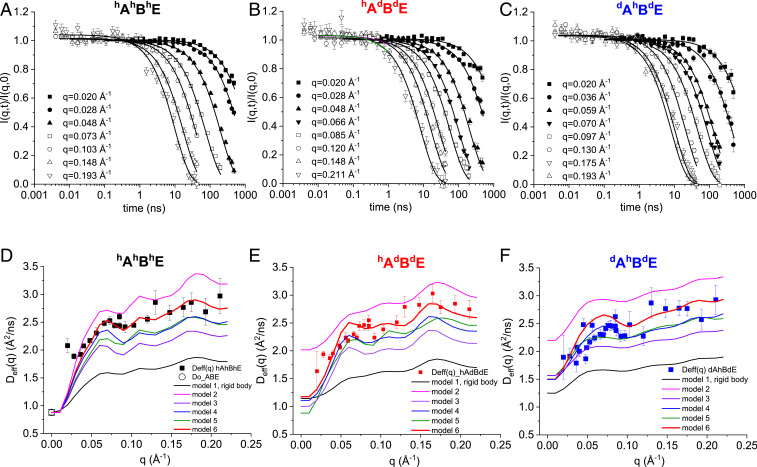
Fig. 2 A–C shows the representative intermediate scattering functions, I(q,t)/I(q,0) measured by NSE, of a fully hydrogenated hAhBhE, and the two selectively deuterated hAdBdE, and dAhBdE complexes. The complete NSE spectra of I(q,t)/I(q,0) at all q values for all three types of complexes are shown in SI Appendix, Figs. S1–S3. The initial slope fittings to I(q,t)/I(q,0) to obtain the decay rate Γ(q) and the effective diffusion constant Deff(q) are described in SI Appendix, Fig. S2 and Eq. S1. Fig. 2 D–F shows the Deff(q) of hAhBhE, hAdBdE, and dAhBdE, together with the center-of-mass diffusion constant Do of hAhBhE measured by dynamic light scattering (DLS); see SI Appendix, Figs. S3 and S4 for DLS experiments.
Fig. 2.
NSE data on hydrogenated and selectively deuterated ABE complex in D2O buffer. (A–C) Representative I(q,t)/I(q, 0) functions of the fully hydrogenated hAhBhE complex (A), of hydrogenated α-catenin in complex with deuterated β-catenin and deuterated E-cadherin cytoplasmic tail hAdBdE (B), and deuterated α-catenin in complex with hydrogenated β-catenin and deuterated E-cadherin cytoplasmic tail dAhBdE (C). (D–F) Experimental Deff(q) of hAhBhE (black squares) (D), hAdBdE (red squares) (E), and dAhBdE (blue squares) (F) from the initial slope fittings of I(q,t)/I(q,0) data. Black line is the calculated Deff(q) curve assuming the whole ABE moves as a single rigid body (model 1 in Fig. 3). Red line is model 6 that best fits the experimental data. Other colored lines are the different models depicted in Fig. 3. Note that the effective diffusion constant Deff(Q = 0) should be the same as the diffusion constant Do measured by DLS only for the fully hydrogenated complex, but this is not true for the selectively deuterated complexes (82).
The Entire ABD of α-Catenin Becomes Mobile within the ABE Complex, whereas Only the Disordered C-Terminal Tail of ABD Is Mobile in the α-Catenin Homodimer.
We developed a theoretical physics framework to interpret the experimentally measured Deff(q) and showed that Deff(q) allows us to test models of the mobility tensor (43–46, 53). The mobility tensor defines which domains of a protein complex move in concert with other domains. Our analysis rests upon an extension of the Akcasu–Gurol (AG) formula for Deff(q), generalized to include rigid body rotation (43) (SI Appendix, Fig. S2 and Eq. S2). The AG formula, in turn, is a sum rule for the initial slope in time of the density–density correlation function (54). This sum rule is a direct consequence of the fact that the associated Smoluchowski equation forms a Sturm–Liouville system (55).
The calculated Deff(q) will be different for each model of internal motion (as the mobility tensors are different), so by comparing the calculated Deff(q) for each model with the NSE data we can determine the nanoscale internal motion of the ABE complex. The strongest indication of internal motion comes from the magnitude of Deff(q). For example, in the simple case that a protein is composed of N identical domains that move independently, one has Deff(q→∞) = 2NDeff(0) (53). We have applied our framework to analyze the NSE data of the fully hydrogenated and selectively deuterated ABE complexes. We then determine which parts of the ABE complex are dynamically separated (i.e., which normal modes are active) on the nanosecond timescales accessible within the Fourier time of the NSE data; see SI Appendix, Fig. S2 for a more detailed description of the theoretical analyses.
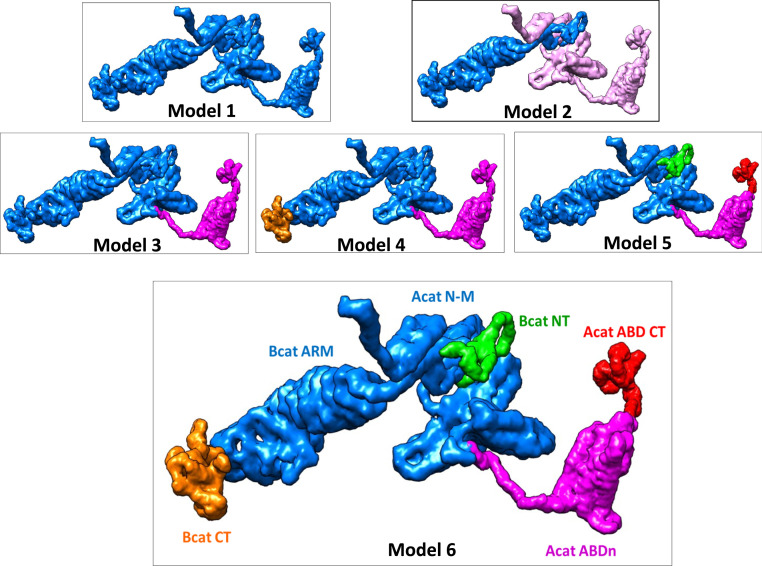
Fig. 3 shows six possible models of protein domain motions within the ABE complex, with respectively one, two, three, four, and five domains moving separately. The C-terminal tail of E-cadherin was omitted in the calculation because its scattering is only 8% of the total complex (Fig. 1B). The six models are as follows:
Model 1: Rigid body. The entire ABE complex is taken as a completely rigid body.
Model 2: Acat/Bcat, two moving segments. The ABE complex has α-catenin (light magenta) and β-catenin (blue) as two separately moving segments within the ABE complex.
Model 3: AcatABD/AcatN-M-Bcat, two moving segments (see Fig. 1B for domain organizations). The first moving segment is the entire ABD of α-catenin (magenta, amino acid residues 648 to 906), and the second segment is the combined α-catenin N and M domains (residues 19 to 647) plus the entire β-catenin protein (blue).
Model 4: AcatABD/AcatN-M-BcatNT-BcatARM/BcatCT, three moving segments. The ABE complex is parsed into three moving segments. The first segment is the entire ABD of α-catenin (amino acid residues 648 to 906, magenta). The second segment (blue), AcatN-M-BcatNT-BcatARM, is the central part of the ABE complex composed of the β-catenin N-terminal domain (BcatNT, residues 1 to 60), the α-catenin-binding domain, and the armadillo domain of β-catenin (BcatARM, residues 61 to 683), plus the α-catenin N and M domains (AcatN-M, residues 19 to 647). The third segment (orange) is the disordered C-terminal tail of β-catenin (BcatCT, residues 684 to 781).
Model 5: AcatABDct/AcatABDn/BcatNT/AcatN-M-BcatARM-BcatCT, four moving segments. The four segments are the ABD C-terminal tail of α-catenin (AcatABDct, residues 854 to 906, red), the N-terminal portion of ABD (AcatABDn, residues 648 to 853, magenta), BcatNT (green), and a fourth segment (blue) composed of the AcatN-M domains, BcatARM, and BcadCT.
Model 6: AcatABDct/AcatABDn/BcatNT/AcatN-M-BcatARM/BcatCT, five moving segments. The five segments are the α-catenin ABDct (red), AcatABDn (magenta), BcatNT (green), a fourth segment composed of AcatN-M domains and BcatARM (blue), and a fifth segment BcatCT (orange).
Fig. 3.
Models of moving segments within the ABE complex. Number of colors indicates the number of separately moving segments within the ABE complex. Model 6 shows the best agreement between theoretical calculations with NSE experimental data. See Fig. 1B for domain organization of α-catenin and β-catenin. Parsing α-catenin and β-catenin as separate moving segments results in Deff(q) that is significantly higher than the experimental data. SI Appendix, Fig. S6 shows that both the magnitude and q dependence of the calculated Deff(q) are sensitive to where the moving ABD domain is parsed in the linker region between M and ABD.
We first calculated Deff(q) for each of the above six models for the fully hydrogenated hAhBhE complex and compared the calculations with the NSE experimental data (Fig. 2D). The comparison obviously excludes the rigid body model (model 1) that has a significant lower Deff(q) than the experimental data. Model 2, parsing the domain in the middle of the complex, results in Deff(q) that is apparently higher than the experimental data, with distinction more pronounced in the selectively deuterated hAdBdE and dAhBdE complexes, Fig. 2 E and F. Further partitions of the domains of this two-segment model result in Deff(q) that is even higher than the experimental data (SI Appendix, Fig. S5). The comparisons suggest that there are no relative movements between β-catenin and the N-M domains of α-catenin on the timescale measured by NSE.
The motion of the entire ABD contributes most to the rise in Deff(q), Model 3. The calculations exhibit a better agreement with the NSE data when we include the motions of the disordered C-terminal tail of β-catenin (model 4) and the disordered N-terminal tail of β-catenin plus the disordered C-terminal tail of α-catenin ABD (models 5 and 6). The comparisons suggest that the five-segment mobility tensor model (model 6) exhibits the best concordance with the experimental Deff(q).
For a selectively deuterated protein complex, only the hydrogenated component contributes to the NSE measured I(q,t)/I(q,0) in D2O buffer. We previously showed that by combining NSE experiments with theoretical analyses on a selectively deuterated protein complex, one can highlight the moving hydrogenated subunit within a deuterated complex (44, 51). Here we calculated Deff(q) for the selectively deuterated hAdBdE complex, in which only the hydrogenated α-catenin hA is “visible” in D2O buffer (Fig. 2E), and for the dAhBdE complex in which only the hydrogenated hB is visible (Fig. 2F). Model 6 again shows better agreement with experiments than the other models. We also find that both the amplitude and the q dependence of the calculated Deff(q) are sensitive to the residue that defines the boundary of the moving ABD (SI Appendix, Fig. S6 A and B). This is particularly true for the selectively deuterated hAdBdE complex, because the moving ABD is in the hydrogenated subunit (SI Appendix, Fig. S6B). By contrast, Deff(q) is not sensitive to where a moving domain is parsed in the deuterated subunit (SI Appendix, Fig. S6 B and C). These analyses suggest that selective deuteration helps to refine the model of protein domain dynamics.
Note that Deff(q) at q = 0 for a partially deuterated sample need not be the same as the diffusion constant D0 measured by DLS. This is because the contribution to Deff(q = 0) of rotational diffusion is nonzero for a partially deuterated sample, since ΣI ri = 0 does not necessarily imply ΣI ri bi = 0, where r is the position of a scattering center and b is its scattering length (SI Appendix, Eq. S2) (51).
It is interesting to observe that the magnitudes of Deff(q) of the fully hydrogenated and selectively deuterated ABE complex are not significantly different. This is in contrast to the NSE data of another protein complex of NHERF1 bound to the ezrin FERM domain that we reported previously (44). The selectively deuterated hNHERF•dFERM complex has apparently higher Deff(q) than the fully hydrogenated NHERF1•FERM complex. The difference between these two complexes is due to the rather symmetric structural distribution of the deuterated and hydrogenated components in the ABE complex (Fig. 1A), whereas the deuterated component is located at the end of an elongated hNHERF1•dFERM. In both cases, the comparison with theoretical calculations serves to identify the active normal modes in the complex.
The above comparison between theoretical calculations and experiments proves that NSE is especially sensitive in pinpointing which segment is moving in a protein or protein complex. This is because Deff(q) depends strongly on the precise location of the domain boundaries and the number of dynamically active domains, as we show for the ABE complex (SI Appendix, Fig. S6), and demonstrated earlier for NHERF1 and for the α-catenin homodimer (53, 56).
To summarize the NSE results and analyses, the motions measured by NSE arise mainly from the movements of the entire ABD of α-catenin as a part of the ABE complex. The motions include the core of ABD, the disordered C-terminal tip of ABD, and the disordered N-terminal and C-terminal tails of β-catenin in the ABE complex. All these modes are active on the nanosecond timescales measured by NSE. By contrast our previous NSE study shows that in the α-catenin homodimer, only the C-terminal tip of ABD is active (56). Altogether, the analyses suggest that the entire ABD becomes activated in the ABD complex, while only the C-terminal disordered tip of the ABD is active in the α-catenin homodimer.
A Gain in Configurational Entropy upon Assembly of the ABE Complex.
The increased flexibility and domain motions of ABD in the ABE complex, compared to the α-catenin homodimer, imply a gain in entropy in the ABE complex. To estimate the entropy change upon forming the ABE complex, we measured the temperature dependence of Kd of α-catenin binding to the β-catenin•EcadCT (BE) complex using surface plasmon resonance (SI Appendix, Figs. S7–S9). Van ’t Hoff analysis indicates an increase in entropy (positive ΔS) and a decrease in enthalpy (negative ΔH) for the formation of ABE complex (SI Appendix, Fig. S9B and Table S2), which agree at least qualitatively with earlier isothermal titration calorimetry studies of the ABE complex (57, 58); see SI Appendix, Table S2 for comparison. The formation of the ABE complex is thus accompanied with a gain in entropy.
The above experimentally measured entropy change is the total change in entropy of forming the ABE complex, which includes the configuration entropy change of the binding partners, the entropy change of the solvent, and the entropy change associated with translational and rotational motion of binding partners and the complex (59). We used the change in Rg and phase volume to estimate the configurational entropy changes for the ABE complex (SI Appendix, Fig. S5 and Tables S1 and S2. The calculations show that both α-catenin and β-catenin have an increased configurational entropy as a part of the ABE complex.
Unbiased MD and Biased (Pulling) Simulations of the ABE Complex.
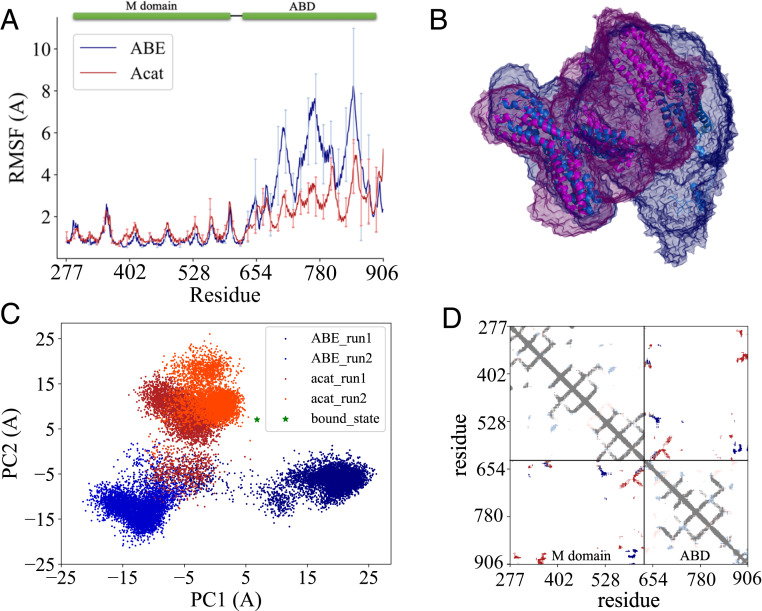
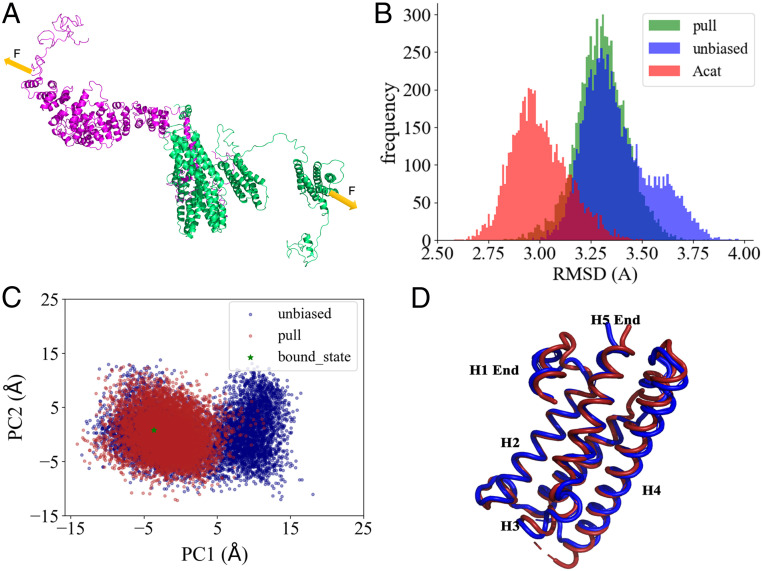
To provide insight into the conformational dynamics changes at residue levels, we performed MD simulations on both the ABE complex and the α-catenin homodimer; see SI Appendix, Fig. S6 for MD simulation methods. After equilibration, two production runs of 250 ns were collected for each system in an NPT ensemble at 300 K and 1 atm. The root-mean-square deviations (rmsds) of both systems rise quickly at the beginning of the simulations and reach a plateau of ∼25 and 20 Å for the ABE complex and the α-catenin dimer, respectively, after ∼100 ns simulation, suggesting that both systems undergo large-scale conformational transitions with the ABE complex being more dynamic than the α-catenin homodimer throughout the simulations (SI Appendix, Fig. S10). We further calculated the radius of gyration (Rg) of the M and ABD domains and the root-mean-square fluctuations (RMSFs) of ABD upon alignment of the M domain during the simulations; see Fig. 1 for α-catenin domain organization and function. Both the RMSF and Rg (SI Appendix, Figs. S10 and S11) profiles indicate that ABD in the ABE complex is more dynamic than in the α-catenin homodimer. As shown in Fig. 4B, the ABD explores greater space in the ABE complex than in the α-catenin dimer during the same simulation time (i.e., ∼250 ns).
Fig. 4.
MD simulation analyses of ABE and α-catenin dimer. (A) Comparison of RMSFs of M domain and ABD for ABE and α-catenin dimer during the simulations (from 150 to 250 ns). Error bars are SDs over different trajectories. (B) Space visited by M domain and ABD during the 250-ns simulations. ABE is in marine, and α-catenin dimer is in magenta. (C) Conformational landscape of ABD projected along the first two PC modes sampled in the simulations. Green star denotes the position of the actin filament-bound conformation. (D) Contact map of M domain and ABD. Dark blue indicates contacts shared by both ABE and α-catenin dimer. Red contacts exist only in ABE while light blue shows contacts only in α-catenin dimer.
Furthermore, we employed principal component analysis (PCA) (60) to reveal the most important motions in proteins during the simulations. Fig. 4C shows the projection of the MD trajectories onto the conformational landscape along the first two principal component (PC) modes of Cα atom fluctuations. It is evident that ABD explores two different regions in the ABE simulations but only one region in the α-catenin dimer simulations, supporting our result that ABD is more dynamic in the ABE complex than in the α-catenin dimer. Strikingly, in this principal component space, the ABD conformation in the α-catenin dimer is more similar to the actin filament-bound conformation of ABD than in the ABE complex (52), consistent with the experimental results that α-catenin alone can bind actin filament without the need of external forces. To understand the origin of the increased flexibility in the ABE complex, we inspected their structures during the simulations and found that the M and ABD domains form more contacts in the α-catenin dimer than in the ABE complex (Fig. 4D), likely due to the different orientation of the M domain in the two protein complexes. The more extensive contacts of ABD with the M domain seem to restrain the movement of ABD in the α-catenin dimer and make it less dynamic.
To shed light on how tensile forces promote the α-catenin to actin filament binding, we applied external forces to the ABE complex during the simulations to observe the potential structural and dynamics changes of ABD in response to the external perturbations. Ten-piconewton forces with the same magnitude and opposite directions were applied on two residues—one is Ala716 located in H2 of ABD and the other is Arg684 at the C terminus of the β-catenin Arm domain (Fig. 5A). We ran four independent 100-ns MD simulations with external forces. For comparison, we ran another four 100-ns unbiased simulations starting from the same structure as that in the biased simulations. To compare the ABD structures sampled in the simulations, we plotted the rmsd probability distributions of ABD in Fig. 5B. While two peaks appear in the unbiased simulations, the pulling simulations feature only one dominant peak and the peak is shifted slightly toward the actin filament-bound structure (52). We then carried out PCA on the simulation trajectories and projected them onto the conformational landscape along the first two PC modes. As shown in Fig. 5C, the ABD samples smaller (more restricted) space when external forces are applied, consistent with the rmsd profiles in Fig. 5B. In the direction of the first PC mode, two separate clusters are clearly shown in the unbiased simulations, but the right cluster disappears in the pulling simulations, indicating that the applied external forces can shift the system toward the left cluster or the binding favored conformations. To understand how external forces drive this conformation transition, we compared the ABD structures from both clusters (Fig. 5D). The major differences lie in the following regions: H1 end, loop between H3 and H4, loop between H4 and H5, and H5 end. It has been reported that H1 regulates the actin binding, and H4 and the C terminus form direct contact with the actin filament (52). Therefore, external forces can rearrange the binding interface, which will steer the ABD conformation shifting toward the strong bound states. Taken together, the simulation results confirm that ABD is more dynamic in the ABE complex and suggest that external forces can facilitate the transition of ABD from an ensemble of diverse structures to more specific structures that are primed for actin filament binding.
Fig. 5.
Biased MD simulation analyses of ABE under force. (A) Illustration of how external forces are applied in the simulations. (B) rmsd probability distributions of ABD during the pulling simulations of ABE and the unbiased simulations of ABE and α-catenin dimer. rmsd is calculated using the actin filament-bound structure as the reference. (C) Conformational landscape of ABD projected along the first two PC modes sampled in the pulling simulations. Green star denotes the position of the actin filament-bound conformation. (D) Two representative ABD structures from the left cluster (red) and the right cluster (blue), respectively, are shown.
Discussion
The NSE study reveals that, in the ABE complex, the entire ABD of α-catenin becomes mobile on nanosecond to submicrosecond timescales. This result stands in contrast to our earlier NSE experiments that showed that only the disordered C-terminal tip of the ABD is moving in the α-catenin homodimer (56). Together with our recent findings—that the ABE complex exists as an ensemble of domain configurations (36)—these studies demonstrate that the ABE complex is highly flexible, with activated ABD motions. It is counterintuitive and surprising that forming the ABE complex results in an increase in protein flexibility, activated ABD domain motion, and a gain in conformational entropy. This is because protein–protein or protein–ligand interactions are often accompanied by folding and reduced dynamics of the binding partners (61, 62). However, the increased domain motion and flexibility support a theoretical model that explains a negative allosteric regulation system (63, 64). This model postulates that, upon the binding of an effector, a negative allosteric regulation system experiences an increase in structural disorder and dynamic motion and therefore a gain in configurational entropy, which stabilizes the system and therefore impedes the system from binding to a target. The ABE complex is an exemplary paradigm of negative allosteric regulation: Upon binding to the effector β-catenin•Ecad, the affinity of α-catenin for the actin microfilament is reduced compared to α-catenin alone (22, 57). The enhanced ABD domain motion and increased configurational entropy in α-catenin as a part of the ABE complex are thus the source of negative allosteric regulation. The ABE complex is thus unusual, forming an entropic trap that serves as the basis for negative allosteric regulation under zero force conditions. We propose that the increased motion of the entire ABD domain is also the source of mechanosensitive binding.
Buckley et al. (24) demonstrated the force-sensitive binding of the ABE complex to the actin microfilament, and their results fitted a “two-state catch-bond” mechanism (24, 65). The catch-bond mechanism was proposed and shown experimentally to describe the unusual dissociation kinetics and bond lifetime of a ligand from an adhesion molecule whose bonds are already formed under force (66, 67). A signature of a catch bond is that the bond lifetime increases with force at low force, but decreases after a peak value is reached at intermediate force. The two-state catch-bond theory proposes the interconversion between a weak ligand-bound state and a strong ligand-bound state under force, which results in the nonmonotonic bond lifetime observed in experiments (65, 68, 69). The two-state catch-bond theories have been successful in explaining a number of mechanosensitive systems (65, 67, 70), besides the binding of the ABE complex to the actin filament (24).
We interpret our dynamics results in the context of association kinetics in the force-sensitive binding of ABE to the actin filament. Our results show enhanced ABD motion and an increase in configurational entropy of the ABE complex under zero force. The biased MD simulations show that the effect of force on the ABE complex is to focus ABD from an ensemble of conformations to a narrow conformational state that resembles the structure of ABD bound to the actin filament. The results suggest that the effect of force is to reduce the configurational entropy of ABD as a part of the ABE complex and thereby to increase the free energy of ABD as a reactant. Further, because when the ABE complex is under force, the conformation of ABD resembles that of the actin-bound state, force may also lower the free energy of the transition state of ABE to actin binding. Consequently, force favors the binding of ABD as a part of the ABE complex to the actin filament by raising free energy (reducing entropy) in the reactant and by lowering the energy in the transition state. Our results thus suggest an alternative (and possibly complementary) model to the “classical catch-bond” mechanisms: In our model, mechanical force facilitates binding by enhancing the association rate constant kon.
Another way to look at how protein domain motion and configuration entropy are employed by the ABE complex for mechanosensing is to consider the association kinetics of ABE binding to the actin filament in the framework of Kramers reaction kinetics (71), which is analogous to a hiker walking in a flat valley looking for a low pass through a tall mountain range. Once the hiker finds the pass, a thermal fluctuation will carry the hiker over the free-energy barrier of the pass to the next valley. The difficulty lies in locating the pass, which requires an extensive search through configuration space. An applied force serves to push the hiker toward the pass, guiding and focusing the hike, and thus speeding the search process. This simple analogy for the narrowing of configuration space by mechanical force underlies the use of an entropic trap by a mechanosensory protein for force-sensitive binding. We now elaborate on the basis for this hypothesis.
The differential activation of nanoscale domain motions of α-catenin in the homodimer and in the ABE complex, which are uniquely revealed by the NSE experiments, may serve to explain the different force-sensitive bindings to the actin filament by the α-catenin homodimer or by the truncated ABD domains and by α-catenin as a part of the ABE complex. Biochemical sedimentation experiments showed that the α-catenin homodimer or the ABD alone can readily bind to the actin filament, essentially without the need of an applied tensile force (21, 22, 29, 30, 56, 72). An optical tweezer experiment by Mei et al. (73) quantitatively showed that the intact C-terminal tip makes α-catenin ABD sensitive to 1-pN force (or 0.243 kBT/nm) for the binding of ABD to the actin filament, compared to a truncated ABD construct without this C-terminal tip. To put the NSE results in this context, the dynamic disordered C-terminal tip entropically obstructs the ABD core from binding to the actin filament and requires a small ∼1-pN force to narrow the configuration space of the C-terminal tip for the ABD binding to the actin filament. The binding of α-catenin homodimer to the actin filament likely enacts the same scenario, with only the C-terminal tip motion dynamically activated. The binding of α-catenin homodimer to the actin filament then requires only a slight or zero force in the in vitro sedimentation actin-binding assays. However, in the ABE complex, the entire ABD is dynamic and explores a larger configuration space. A relatively larger force must be applied to narrow the configurational space of the entire ABD to bind the actin filament than the C-terminal tip of ABD or the intact α-catenin homodimer. The optical tweezer experiment by Buckley et al. (24) shows that it takes a much stronger and physiologically relevant force of ∼10 pN (or 2.43 kBT/nm) to strengthen the binding of the ABE complex to the moving actin filaments that mimic the pulling force in the actomyosin bundles. NSE can reveal which segment or domain in a protein or protein complex is likely to be a force sensor.
The hypothesis that an entropic trap underlies mechanosensitive binding may also be found in other mechanosensitive systems. Indeed, the elasticity of the transition state (and thus the change of entropy under force) was identified as the source of mechanical stability (catch-bond behavior) in the unfolding of the titin immunoglobulin domains (74). In the case of the cadherin extracellular domain, force bends cadherin to a specific configuration and it then forms long-lived, force-induced hydrogen bonds that lock X dimers into tighter contact (75). In the inverse process, just as an applied force can reduce entropy, forces as high as 20 pN can be generated by entropic effects in chaperones (76). Thus, the differential activations of nanoscale domain motions revealed by NSE can serve to identify which segment in a protein or protein complex is likely to be a force sensor.
The purpose of this NSE study is to determine the domain-level dynamics of the intact ABE complex, which exhibits remarkable flexibility and domain motion on nanolength scales. The dynamic domain motions influence the association kinetics of the force-sensitive binding of ABE to actin filaments. High-resolution structural studies revealed that the isolated ABD of α-catenin is composed of a five-helix bundle H1–H5, with another helix H0 forming a cap at the N-terminal end of the five-helix bundle (26, 52, 73). Altogether, these studies find that H0 and H1 helices obstruct the H2–H5 bundle from strong binding to actin and that H0 and H1 helices are likely the source of a catch-bond behavior of ABD (26, 52, 73). However, the results on the truncated ABD do not explain why the α-catenin homodimer binds to the actin filament under zero force and why α-catenin in the ABE complex does not bind the actin filament without applied force. In the intact ABE complex, our simulations show that applied forces suppress fluctuations in the ABD, including the H0 and H1 regions and several loops connecting the helices in ABD (SI Appendix, Fig. S12). The likely scenario is that force also separates H0–H1 from the core H2–H5 helical bundle for strong actin binding. Thus, force promotes the association kinetics by overcoming the fluctuations of ABD and induces the catch-bond behavior of the dissociation kinetics by lowering the free energy of the strong bound state.
Conclusions
Using NSE, selective deuteration, theoretical physics analyses, and MD simulations, we revealed the activated motion of the α-catenin actin-binding domain within the cadherin–catenin complex. The primary effect of enhanced domain motion is an increase in entropy that prevents the cadherin–catenin complex from binding to the actin filament under zero force. This study thus illustrates an example of negative allosteric regulation that is accompanied by increased dynamics and structural disorder. Our NSE experiments and MD simulations suggest that the consequence of the mechanical force is to shift the ABE complex from a broad ensemble to a narrow population of conformations that is better suited to bind the actin microfilament. Thus, the activated nanoscale protein domain motions and the ensemble of flexible ABD conformations serve as an entropic trap, which underlies the force-sensitive binding behavior by the ABE complex. Nanoscale protein domain motion and entropic narrowing by mechanical force may be a common mechanism shared by other mechanosensitive proteins, as these proteins tend to be multidomain proteins, are very flexible, and show multiple domain configurations (14, 77–81). Currently, no experiment can simultaneously measure both force-sensitive binding and the molecular conformations and dynamics at sufficiently high temporal–spatial resolution. Future work should test this hypothesis.
Materials and Methods
Protein Purification and Protein Complex Reconstitution.
The bacterial expression and fast protein liquid chromatography (FPLC) purification of full-length human αE-catenin, human β-catenin, and the cytoplasmic domain of human E-cadherin (EcadCT) (residues 731 to 882) have been described previously (36). Deuterated αE-catenin, β-catenin, or EcadCT was grown in 85% D2O (vol/vol) M9 medium as described previously (51). Purification of deuterated proteins was the same as that of the hydrogenated proteins. Reconstitution of the hydrogenate hAhBhE and selectively deuterated dAhBdE and hAdBdE, complexes followed the same protocol described previously (36).
NSE Spectrometer and Experiments.
The NSE experiments were performed at the newly upgraded high-resolution IN15 NSE spectrometer at the Institut Laue-Langevin. We used three different wavelengths: 6, 10, and 13.5 Å covering 0.004- to 42-, 0.2- to 194-, and 0.55- to 477-ns time intervals. The 6-Å configuration used the extended time-range setup to capture eventual very fast motions at high Q. The covered Fourier time scales with the third power of the wavelength, but the incoming ux also drops roughly with the fourth power. The choice of the wavelength was made by optimizing the compromise between the resolution need and the incoming neutron flux. The beam monochromatization in each case was 15% full width at half-maximum as given by the neutron velocity selector. The samples were filled-in quartz cells with 4-mm sample thickness, and the temperature was controlled at 10.0 ± 0.1 °C or better. Instrumental resolution was measured from the standard Grafoil (GrafTech), which gives a strong, elastic, coherent small-angle scattering. The background was measured on the D2O buffer, and the sample spectra were corrected using the relative transmissions following the standard procedures. Details about protein sample preparation for NSE experiments and NSE data analyses are described in SI Appendix.
Supplementary Material
Acknowledgments
This research was funded by NSF Grant MCB-1817684 and by National Center for Research Resources Grant 2G12 RR003060 to City College of New York. We gratefully acknowledge the support of the Institut Laue-Langevin and the Science and Technology Facilities Council (United Kingdom) to carry out the NSE experiments.
ILL neutron beam time award: Neutron spin echo spectroscopy study of nanoscale dynamics of the α-catenin/β-catenin complex. (2019) ILL. DOI:10.5291/ILL-DATA.8-04-852
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025012118/-/DCSupplemental.
Data Availability
All study data are included in this article and/or SI Appendix.
References
- 1.Takeichi M., The cadherins: Cell-cell adhesion molecules controlling animal morphogenesis. Development 102, 639–655 (1988). [DOI] [PubMed] [Google Scholar]
- 2.Dewitz C., Duan X., Zampieri N., Organization of motor pools depends on the combined function of N-cadherin and type II cadherins. Development 146, dev180422 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pinheiro D., Bellaïche Y., Mechanical force-driven adherens junction remodeling and epithelial dynamics. Dev. Cell 47, 3–19 (2018). [DOI] [PubMed] [Google Scholar]
- 4.Iskratsch T., Wolfenson H., Sheetz M. P., Appreciating force and shape—the rise of mechanotransduction in cell biology. Nat. Rev. Mol. Cell Biol. 15, 825–833 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Benham-Pyle B. W., Pruitt B. L., Nelson W. J., Cell adhesion. Mechanical strain induces E-cadherin-dependent Yap1 and β-catenin activation to drive cell cycle entry. Science 348, 1024–1027 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Le H. Q., et al., Mechanical regulation of transcription controls Polycomb-mediated gene silencing during lineage commitment. Nat. Cell Biol. 18, 864–875 (2016). [DOI] [PubMed] [Google Scholar]
- 7.Rauskolb C., Sun S., Sun G., Pan Y., Irvine K. D., Cytoskeletal tension inhibits Hippo signaling through an Ajuba-Warts complex. Cell 158, 143–156 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mendonsa A. M., Na T. Y., Gumbiner B. M., E-cadherin in contact inhibition and cancer. Oncogene 37, 4769–4780 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cooper J., Giancotti F. G., Integrin signaling in cancer: Mechanotransduction, stemness, epithelial plasticity, and therapeutic resistance. Cancer Cell 35, 347–367 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Grimsley-Myers C. M., et al., VE-cadherin endocytosis controls vascular integrity and patterning during development. J. Cell Biol. 219, e201909081 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon K. J., et al., Zika-virus-encoded NS2A disrupts mammalian cortical neurogenesis by degrading adherens junction proteins. Cell Stem Cell 21, 349–358.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Takeichi M., Dynamic contacts: Rearranging adherens junctions to drive epithelial remodelling. Nat. Rev. Mol. Cell Biol. 15, 397–410 (2014). [DOI] [PubMed] [Google Scholar]
- 13.Mège R. M., Ishiyama N., Integration of cadherin adhesion and cytoskeleton at Adherens junctions. Cold Spring Harb. Perspect. Biol. 9, a028738 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brasch J., Harrison O. J., Honig B., Shapiro L., Thinking outside the cell: How cadherins drive adhesion. Trends Cell Biol. 22, 299–310 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yonemura S., Wada Y., Watanabe T., Nagafuchi A., Shibata M., Alpha-catenin as a tension transducer that induces adherens junction development. Nat. Cell Biol. 12, 533–542 (2010). [DOI] [PubMed] [Google Scholar]
- 16.Leckband D. E., de Rooij J., Cadherin adhesion and mechanotransduction. Annu. Rev. Cell Dev. Biol. 30, 291–315 (2014). [DOI] [PubMed] [Google Scholar]
- 17.Ishiyama N., et al., Dynamic and static interactions between p120 catenin and E-cadherin regulate the stability of cell-cell adhesion. Cell 141, 117–128 (2010). [DOI] [PubMed] [Google Scholar]
- 18.Huveneers S., de Rooij J., Mechanosensitive systems at the cadherin-F-actin interface. J. Cell Sci. 126, 403–413 (2013). [DOI] [PubMed] [Google Scholar]
- 19.Khalil A. A., de Rooij J., Cadherin mechanotransduction in leader-follower cell specification during collective migration. Exp. Cell Res. 376, 86–91 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Peyret G., et al., Sustained oscillations of epithelial cell sheets. Biophys. J. 117, 464–478 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Drees F., Pokutta S., Yamada S., Nelson W. J., Weis W. I., Alpha-catenin is a molecular switch that binds E-cadherin-beta-catenin and regulates actin-filament assembly. Cell 123, 903–915 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yamada S., Pokutta S., Drees F., Weis W. I., Nelson W. J., Deconstructing the cadherin-catenin-actin complex. Cell 123, 889–901 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Choi H. J., et al., αE-catenin is an autoinhibited molecule that coactivates vinculin. Proc. Natl. Acad. Sci. U.S.A. 109, 8576–8581 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Buckley C. D., et al., Cell adhesion. The minimal cadherin-catenin complex binds to actin filaments under force. Science 346, 1254211 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Rangarajan E. S., Izard T., Dimer asymmetry defines α-catenin interactions. Nat. Struct. Mol. Biol. 20, 188–193 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ishiyama N., et al., Force-dependent allostery of the α-catenin actin-binding domain controls adherens junction dynamics and functions. Nat. Commun. 9, 5121 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ishiyama N., et al., An autoinhibited structure of α-catenin and its implications for vinculin recruitment to adherens junctions. J. Biol. Chem. 288, 15913–15925 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pokutta S., Weis W. I., Structure of the dimerization and beta-catenin-binding region of alpha-catenin. Mol. Cell 5, 533–543 (2000). [DOI] [PubMed] [Google Scholar]
- 29.Wood M. N., et al., α-Catenin homodimers are recruited to phosphoinositide-activated membranes to promote adhesion. J. Cell Biol. 216, 3767–3783 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rimm D. L., Koslov E. R., Kebriaei P., Cianci C. D., Morrow J. S., Alpha 1(E)-catenin is an actin-binding and -bundling protein mediating the attachment of F-actin to the membrane adhesion complex. Proc. Natl. Acad. Sci. U.S.A. 92, 8813–8817 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huveneers S., et al., Vinculin associates with endothelial VE-cadherin junctions to control force-dependent remodeling. J. Cell Biol. 196, 641–652 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yao M., et al., Force-dependent conformational switch of α-catenin controls vinculin binding. Nat. Commun. 5, 4525 (2014). [DOI] [PubMed] [Google Scholar]
- 33.Chakrabarti S., Hinczewski M., Thirumalai D., Plasticity of hydrogen bond networks regulates mechanochemistry of cell adhesion complexes. Proc. Natl. Acad. Sci. U.S.A. 111, 9048–9053 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pegoraro A. F., Janmey P., Weitz D. A., Mechanical properties of the cytoskeleton and cells. Cold Spring Harb. Perspect. Biol. 9, a022038 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thomas W., Catch bonds in adhesion. Annu. Rev. Biomed. Eng. 10, 39–57 (2008). [DOI] [PubMed] [Google Scholar]
- 36.Bush M., et al., An ensemble of flexible conformations underlies mechanotransduction by the cadherin-catenin adhesion complex. Proc. Natl. Acad. Sci. U.S.A. 116, 21545–21555 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bee M., Quasielastic Neutron Scattering: Principles and Applications in Solid State Chemistry, Biology and Materials Science (Adam Hilger, Philadelphia, PA, 1988). [Google Scholar]
- 38.Smith J. C., Tan P., Petridis L., Hong L., Dynamic neutron scattering by biological systems. Annu. Rev. Biophys. 47, 335–354 (2018). [DOI] [PubMed] [Google Scholar]
- 39.Monkenbusch M., et al., Fast internal dynamics in alcohol dehydrogenase. J. Chem. Phys. 143, 075101 (2015). [DOI] [PubMed] [Google Scholar]
- 40.Ohl M., et al., The high-resolution neutron spin-echo spectrometer for the SNS with τ≥ 1μs. Physica B 350, 147–150 (2004). [Google Scholar]
- 41.Bu Z., Cook J., Callaway D. J., Dynamic regimes and correlated structural dynamics in native and denatured alpha-lactalbumin. J. Mol. Biol. 312, 865–873 (2001). [DOI] [PubMed] [Google Scholar]
- 42.Bu Z., et al., A view of dynamics changes in the molten globule-native folding step by quasielastic neutron scattering. J. Mol. Biol. 301, 525–536 (2000). [DOI] [PubMed] [Google Scholar]
- 43.Bu Z., Biehl R., Monkenbusch M., Richter D., Callaway D. J., Coupled protein domain motion in Taq polymerase revealed by neutron spin-echo spectroscopy. Proc. Natl. Acad. Sci. U.S.A. 102, 17646–17651 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Farago B., Li J., Cornilescu G., Callaway D. J., Bu Z., Activation of nanoscale allosteric protein domain motion revealed by neutron spin echo spectroscopy. Biophys. J. 99, 3473–3482 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Callaway D. J., Farago B., Bu Z., Nanoscale protein dynamics: A new frontier for neutron spin echo spectroscopy. Eur. Phys. J. E 36, 76 (2013). [DOI] [PubMed] [Google Scholar]
- 46.Callaway D. J. E., et al., Controllable activation of nanoscale dynamics in a disordered protein alters binding kinetics. J. Mol. Biol. 429, 987–998 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Balacescu L., et al., Transition between protein-like and polymer-like dynamic behavior: Internal friction in unfolded apomyoglobin depends on denaturing conditions. Sci. Rep. 10, 1570 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liu Y., Intermediate scattering function for macromolecules in solutions probed by neutron spin echo. Phys. Rev. E 95, 020501 (2017). [DOI] [PubMed] [Google Scholar]
- 49.Hong L., Smolin N., Smith J. C., de Gennes narrowing describes the relative motion of protein domains. Phys. Rev. Lett. 112, 158102 (2014). [DOI] [PubMed] [Google Scholar]
- 50.Gardner J. S., Ehlers G., Faraone A., García Sakai V., High-resolution neutron spectroscopy using backscattering and neutron spin-echo spectrometers in soft and hard condensed matter. Nat. Rev. Phys. 2, 103–116 (2020). [Google Scholar]
- 51.Callaway D. J., Bu Z., Essential strategies for revealing nanoscale protein dynamics by neutron spin echo spectroscopy. Methods Enzymol. 566, 253–270 (2016). [DOI] [PubMed] [Google Scholar]
- 52.Xu X. P., et al., Structural basis of αE-catenin-F-actin catch bond behavior. eLife 9, e60878 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Callaway D. J., Bu Z., Visualizing the nanoscale: Protein internal dynamics and neutron spin echo spectroscopy. Curr. Opin. Struct. Biol. 42, 1–5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Akcasu Z., Gurol H., Quasi-elastic scattering by dilute polymer-solutions. J. Polym. Sci. B Polym. Phys. 14, 1–10 (1976). [Google Scholar]
- 55.Doi M., Edwards S. F., The Theory of Polymer Dynamics (Oxford University Press, Oxford, UK, 1986). [Google Scholar]
- 56.Nicholl I. D., et al., α-Catenin structure and nanoscale dynamics in solution and in complex with F-actin. Biophys. J. 115, 642–654 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Pokutta S., Choi H. J., Ahlsen G., Hansen S. D., Weis W. I., Structural and thermodynamic characterization of cadherin·β-catenin·α-catenin complex formation. J. Biol. Chem. 289, 13589–13601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Terekhova K., et al., Binding partner- and force-promoted changes in αE-catenin conformation probed by native cysteine labeling. Sci. Rep. 9, 15375 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang Y., et al., Globally correlated conformational entropy underlies positive and negative cooperativity in a kinase’s enzymatic cycle. Nat. Commun. 10, 799 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.David C. C., Jacobs D. J., Principal component analysis: A method for determining the essential dynamics of proteins. Methods Mol. Biol. 1084, 193–226 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Wright P. E., Dyson H. J., Linking folding and binding. Curr. Opin. Struct. Biol. 19, 31–38 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rogers J. M., et al., Interplay between partner and ligand facilitates the folding and binding of an intrinsically disordered protein. Proc. Natl. Acad. Sci. U.S.A. 111, 15420–15425 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Williams D. H., Calderone C. T., O’Brien D. P., Zerella R., Changes in motion vs. bonding in positively vs. negatively cooperative interactions. Chem. Commun. (Camb.), 10.1039/B201428A (2002). [DOI] [PubMed] [Google Scholar]
- 64.Williams D. H., Stephens E., O’Brien D. P., Zhou M., Understanding noncovalent interactions: Ligand binding energy and catalytic efficiency from ligand-induced reductions in motion within receptors and enzymes. Angew. Chem. Int. Ed. Engl. 43, 6596–6616 (2004). [DOI] [PubMed] [Google Scholar]
- 65.Chakrabarti S., Hinczewski M., Thirumalai D., Phenomenological and microscopic theories for catch bonds. J. Struct. Biol. 197, 50–56 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Dembo M., Torney D. C., Saxman K., Hammer D., The reaction-limited kinetics of membrane-to-surface adhesion and detachment. Proc. R. Soc. Lond. B Biol. Sci. 234, 55–83 (1988). [DOI] [PubMed] [Google Scholar]
- 67.Marshall B. T., et al., Direct observation of catch bonds involving cell-adhesion molecules. Nature 423, 190–193 (2003). [DOI] [PubMed] [Google Scholar]
- 68.Barsegov V., Thirumalai D., Dynamics of unbinding of cell adhesion molecules: Transition from catch to slip bonds. Proc. Natl. Acad. Sci. U.S.A. 102, 1835–1839 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Thomas W. E., Vogel V., Sokurenko E., Biophysics of catch bonds. Annu. Rev. Biophys. 37, 399–416 (2008). [DOI] [PubMed] [Google Scholar]
- 70.Kong F., García A. J., Mould A. P., Humphries M. J., Zhu C., Demonstration of catch bonds between an integrin and its ligand. J. Cell Biol. 185, 1275–1284 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Howard J., Mechanics of Motor Proteins and the Cytoskeleton (Sinauer Associates, Sunderland, MA, 2001). [Google Scholar]
- 72.Koslov E. R., Maupin P., Pradhan D., Morrow J. S., Rimm D. L., Alpha-catenin can form asymmetric homodimeric complexes and/or heterodimeric complexes with beta-catenin. J. Biol. Chem. 272, 27301–27306 (1997). [DOI] [PubMed] [Google Scholar]
- 73.Mei L., et al., Molecular mechanism for direct actin force-sensing by α-catenin. eLife 9, e62514 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Yuan G., et al., Elasticity of the transition state leading to an unexpected mechanical stabilization of titin immunoglobulin domains. Angew. Chem. Int. Ed. Engl. 56, 5490–5493 (2017). [DOI] [PubMed] [Google Scholar]
- 75.Manibog K., Li H., Rakshit S., Sivasankar S., Resolving the molecular mechanism of cadherin catch bond formation. Nat. Commun. 5, 3941 (2014). [DOI] [PubMed] [Google Scholar]
- 76.De Los Rios P., Ben-Zvi A., Slutsky O., Azem A., Goloubinoff P., Hsp70 chaperones accelerate protein translocation and the unfolding of stable protein aggregates by entropic pulling. Proc. Natl. Acad. Sci. U.S.A. 103, 6166–6171 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Vogel V., Mechanotransduction involving multimodular proteins: Converting force into biochemical signals. Annu. Rev. Biophys. Biomol. Struct. 35, 459–488 (2006). [DOI] [PubMed] [Google Scholar]
- 78.Priest A. V., Shafraz O., Sivasankar S., Biophysical basis of cadherin mediated cell-cell adhesion. Exp. Cell Res. 358, 10–13 (2017). [DOI] [PubMed] [Google Scholar]
- 79.Springer T. A., Structural basis for selectin mechanochemistry. Proc. Natl. Acad. Sci. U.S.A. 106, 91–96 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Le Trong I., et al., Structural basis for mechanical force regulation of the adhesin FimH via finger trap-like beta sheet twisting. Cell 141, 645–655 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Su Y., et al., Relating conformation to function in integrin α5β1. Proc. Natl. Acad. Sci. U.S.A. 113, E3872–E3881 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Bu Z., Callaway D. J., Proteins move! Protein dynamics and long-range allostery in cell signaling. Adv. Protein Chem. Struct. Biol. 83, 163–221 (2011). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in this article and/or SI Appendix.