Significance
This work investigated the in vivo role of Paxbp1, a poorly studied nuclear protein, in regulating adult mouse muscle stem cells (MuSCs). By deleting Paxbp1 in adult mouse quiescent MuSCs, we found that Paxbp1-null MuSCs were unable to reenter the cell cycle to proliferate upon muscle injury and subsequently underwent apoptosis, resulting in a total failure in injury-induced muscle regeneration. Mechanistically, we found that Paxbp1 controls a late cell-growth checkpoint by targeting mTORC1. Loss of Paxbp1 in MuSCs led to increased levels of reactive oxygen species that in turn triggered p53 activation and induction of multiple p53 target genes, some of which contributed to cell-cycle arrest (e.g., Cdkn1a), apoptosis (e.g., Apaf1), and impaired mTORC1 signaling (e.g., Sesn2 and Ddit4).
Keywords: Paxbp1, muscle satellite cells, quiescence, ROS, cell growth
Abstract
Adult mouse muscle satellite cells (MuSCs) are quiescent in uninjured muscles. Upon muscle injury, MuSCs exit quiescence, reenter the cell cycle to proliferate and self-renew, and then differentiate and fuse to drive muscle regeneration. However, it remains poorly understood how MuSCs transition from quiescence to the cycling state. Here, we report that Pax3 and Pax7 binding protein 1 (Paxbp1) controls a key checkpoint during this critical transition. Deletion of Paxbp1 in adult MuSCs prevented them from reentering the cell cycle upon injury, resulting in a total regeneration failure. Mechanistically, we found an abnormal elevation of reactive oxygen species (ROS) in Paxbp1-null MuSCs, which induced p53 activation and impaired mTORC1 signaling, leading to defective cell growth, apoptosis, and failure in S-phase reentry. Deliberate ROS reduction partially rescued the cell-cycle reentry defect in mutant MuSCs. Our study reveals that Paxbp1 regulates a late cell-growth checkpoint essential for quiescent MuSCs to reenter the cell cycle upon activation.
In normal adult tissues of mammals, most cell types exist in one of the two postmitotic states: an irreversible terminally differentiated state as exemplified by mature neurons and myofibers of skeletal muscles, or a reversible quiescent state as exemplified by several somatic stem cells (e.g., hair follicle stem cells, muscle satellite cells, neural stem cells, and long-term hematopoietic stem cells) and naïve T cells (1–6). The quiescent cells (usually in G0 phase) can reenter the cell cycle upon injury or stimulation (6). As defects in quiescence regulation can lead to cell depletion, tissue degeneration, aging, or tumorigenesis (7), a better understanding of the underlying mechanisms is fully warranted.
In adult mammalian skeletal muscles, muscle satellite cells (MuSCs) uniquely express Pax7, a homeodomain and paired domain-containing transcription factor, and are sandwiched between the outer surface of the sarcolemma of individual myofibers and the surrounding basal lamina (5, 8–10). They are the predominant cell type responsible for injury-induced muscle regeneration. In noninjured homeostatic adult muscles, MuSCs are mostly quiescent with common characteristics of quiescent cells: small cell size with scarce cytoplasm, few mitochondria, and low metabolism (5, 6, 9–11). Upon muscle injury, quiescent MuSCs (QSCs) are rapidly activated to become activated satellite cells (ASCs) that are characterized by rapid induction of immediate-early genes (IEGs) including those encoding members of the AP1 family, expression of MyoD protein, an increase in cell size, and enhanced RNA and protein synthesis (5, 9, 10). Both in culture and in vivo, it takes QSCs an extended time (∼36 to 48 h) before they reenter S phase of the first cell cycle (12, 13), a period involving QSCs in a deep quiescent state transitioning to ASCs in varying degrees of shallow quiescent states (or more primed states). Once ASCs reenter the cell cycle, a transition commonly referred to as “quiescence exit,” they (now called myogenic precursor cells or myoblasts) can undergo both symmetric and asymmetric cell division to generate more proliferating myoblasts and to self-renew (14). Interestingly, it only takes ∼8 to 10 h for cycling myoblasts to complete subsequent cell cycles (12, 13). Myoblasts eventually differentiate and fuse with each other to complete muscle regeneration. Compared with QSCs, proliferating myoblasts and differentiating myocytes are known to undergo extensive metabolic reprogramming including reduced fatty acid oxidation, enhanced mitochondrial biogenesis, increased reactive oxygen species (ROS) production, and up-regulation of both glycolysis and oxidative phosphorylation (OXPHOS) (5, 13, 15–23). However, it remains unclear whether metabolic reprogramming is also a prerequisite for QSCs to reenter the cell cycle. Upon muscle injury, the initial transition of MuSCs from QSCs to ASCs, and then to S phase of the first cell cycle, is very critical, as any disruption during this period could potentially result in regeneration failure. Indeed, we and others recently found that the phosphatidylinositol 3-kinase (PI3K)-mediated signaling pathway involving both mTORC1 and FoxOs regulates an indispensable checkpoint for the early transition from QSCs to ASCs: MuSC-specific deletion of p110α (encoding the catalytic subunit of PI3K) prevented QSCs from transitioning to ASCs, while expression of a constitutively active p110α or deletion of Pten (encoding a well-established negative regulator of the PI3K pathway) drove spontaneous activation of QSCs in uninjured muscles, resulting in their gradual depletion (24, 25). In addition, in response to systemic signals from distant muscle injury, QSCs were found to transition to a reversible “primed” quiescent state (termed Galert) in an mTORC1-dependent manner with cells in Galert being metabolically more active and capable of reentering the first cell cycle faster than typical QSCs (13). These results indicate that the transition of adult MuSCs from quiescence to S-phase reentry is subject to active regulation. It remains unclear whether this early transition period is also regulated by other mechanisms.
Paxbp1 was previously identified by us as a nuclear Pax7-binding protein through yeast two-hybrid screening (26). It is conserved in Metazoa and is broadly expressed in multiple tissues and cell types including MuSCs. Mainly by cell culture-based assays, we previously found that Paxbp1 regulates the proliferation of Pax7-expressing myogenic precursor cells by bridging Pax7 with an H3K4 histone methyltransferase complex to promote the expression of Pax7 target genes including Cdc20 (26). However, it remained unclear whether Paxbp1 regulates adult MuSCs in vivo. Here, by genetically deleting Paxbp1 in adult mouse MuSCs in vivo, we found that Paxbp1-null MuSCs failed to reenter the cell cycle upon muscle injury and subsequently underwent apoptosis, resulting in a total failure of injury-induced muscle regeneration. Loss of Paxbp1 in MuSCs led to an abnormal elevation of ROS, which triggered p53 activation and impaired mTORC1 signaling. While the induction of many p53 target genes contributed to cell-cycle arrest and apoptosis, the impaired mTORC1 signaling compromised the late phase of cell growth before cell-cycle reentry. Collectively, these defects in Paxbp1-null MuSCs resulted in regeneration failure.
Results
Paxbp1 Was Indispensable for Injury-Induced Muscle Regeneration.
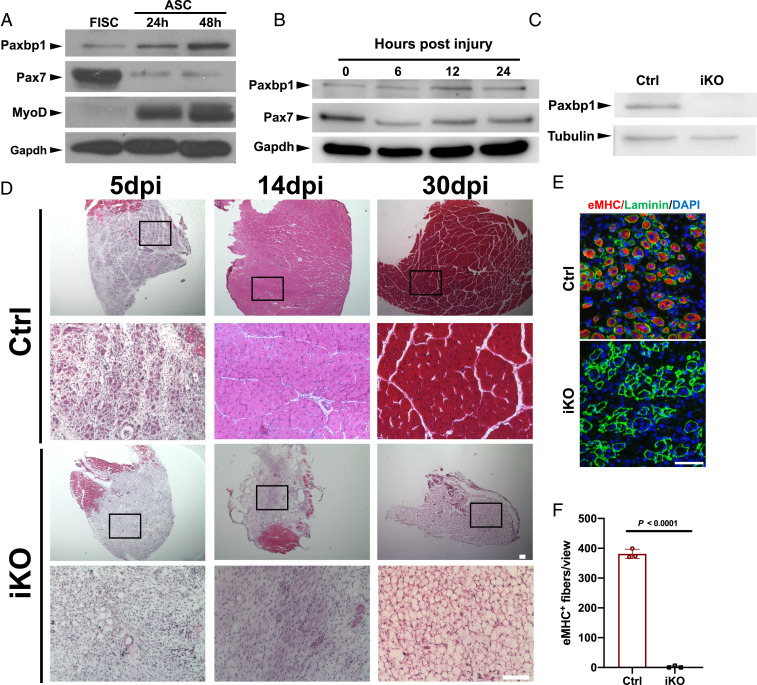
Previously, we showed that Paxbp1 could regulate myoblast proliferation mainly by cell culture-based studies (26). However, it was unclear whether Paxbp1 regulates adult MuSCs in vivo. To address this question, we first examined Paxbp1 messenger RNA (mRNA) and protein expression in adult MuSCs. Based on both public and our own RNA-sequencing (RNA-seq) data, Paxbp1 mRNA was expressed in both freshly isolated SCs (hereafter FISCs) from uninjured muscles and ASCs from injured and regenerating muscles (19, 24, 27, 28). Consistently, Paxbp1 protein was already expressed at low but detectable levels in FISCs (Fig. 1A). When FISCs were grown in culture, they could undergo spontaneous activation and reenter the cell cycle to become cycling myoblasts. In such settings, similar to MyoD, Paxbp1 protein levels further increased when MuSCs transitioned from a deeper quiescent state (FISCs) to a shallower one (after being grown in culture for 24 h), and then eventually to the cycling state (after being grown in culture for 48 h), a pattern opposite to that of Pax7 protein during this process. In addition, we also examined the protein expression patterns of both Paxbp1 and Pax7 in FISCs (without further culturing) from adult wild-type mice that were injured for 6, 12, and 24 h. Similar (but not identical) to the patterns seen in the cultured MuSCs, we found that Paxbp1 levels slightly increased, while Pax7 protein levels slightly decreased, in ASCs directly isolated from injured muscles (Fig. 1B). As Paxbp1 is broadly expressed in multiple tissues, to avoid potential embryonic lethality if Paxbp1 is to be deleted in the germline, we decided to conditionally delete Paxbp1 by generating a floxed Paxbp1 mouse strain (SI Appendix, Fig. S1A). The floxed allele enables deletion of exon 8 of Paxbp1 after Cre-mediated DNA recombination, which creates a premature stop codon (SI Appendix, Fig. S1A). We first generated a noninducible Pax7Cre; Paxbp1flox/flox mouse strain to conditionally delete Paxbp1 in Pax7-expressing cells during mouse development (29). No viable homozygous Paxbp1 knockout (KO) pups were found (SI Appendix, Fig. S1B), suggesting an indispensable role of Paxbp1 in mouse embryo development. To facilitate the study of Paxbp1 in adult MuSCs, we generated a tamoxifen (TMX)-inducible, MuSC-specific Paxbp1 KO strain (hereafter Paxbp1-iKO) by crossing the Paxbp1flox/flox mice with Pax7CreER mice (30). We also bred the R26RYFP/YFP allele into Paxbp1-iKO mice to facilitate MuSC tracking and sorting. Our TMX injection regimen achieved very efficient recombination in MuSCs: ∼90% of MuSCs (CD31−CD45−Sca1−Vcam1+) were YFP+ (SI Appendix, Fig. S1C), which indicates that yellow fluorescent protein (YFP) can be used as a surrogate marker for Pax7-expressing MuSCs. The efficient deletion of Paxbp1 at both mRNA and protein levels was further verified by RT-PCR and Western blot, respectively (Fig. 1C and SI Appendix, Fig. S1D). To assess the functions of MuSCs in vivo, we injected cardiotoxin (CTX) into the tibialis anterior (TA) muscles of Paxbp1-iKO mice and their heterozygous littermates (i.e., control with the following genotype: Pax7CreER/CreER; Paxbp1flox/+; R26RYFP/YFP) to induce muscle damage and then examined the extent of muscle regeneration. Five days postinjury (dpi), whereas numerous small regenerating myofibers (i.e., those with red color and round or oval shape with centrally localized dark nuclei) were observed on muscle sections from the control mice, such small regenerating myofibers were barely detectable on muscle sections from Paxbp1-iKO mice (Fig. 1 D, Left). Consistently, by staining for the embryonic myosin heavy chain (eMHC) that was only transiently expressed in regenerating myofibers between 4 and 6 dpi (31), we confirmed that eMHC was abundantly expressed in most of the regenerating myofibers from the control mice but not Paxbp1-iKO mice at 5 dpi (Fig. 1 E and F). By 14 dpi, whereas the damaged TA muscles were largely repaired in the control mice, no obvious regeneration was seen in damaged TA muscles from Paxbp1-iKO mice (Fig. 1 D, Middle). Moreover, this severe muscle regeneration defect was not resolved in Paxbp1-iKO mice even at 30 dpi (Fig. 1 D, Right), indicative of a total block in muscle regeneration in mutant mice. Consistently, the TA muscles that were dissected 30 dpi from Paxbp1-iKO mice were markedly paler and smaller (SI Appendix, Fig. S1E). By staining with oil red O or immunostaining for Perilipin 1 (a lipid droplet-associated protein enriched in adipocytes), we detected obvious adipocyte infiltration in regenerating muscles from Paxbp1-iKO mice at 30 dpi (SI Appendix, Fig. S1 F and G), which was commonly seen in injured muscles with defective muscle regeneration (32, 33). Collectively, our data demonstrated that Paxbp1 is indispensable for MuSCs to function during injury-induced muscle regeneration in vivo.
Fig. 1.
Paxbp1 was indispensable for muscle regeneration. (A–C) The expression of various proteins was examined by Western blot using equal amounts of soluble whole-cell lysates from (A) FISCs and ASCs cultured for 24 and 48 h; (B) MuSCs directly isolated from wild-type mice that were either noninjured or injured for various times as indicated; and (C) FISCs from control (Ctrl) and Paxbp1-iKO mice. (D) Representative images of regenerating TA muscles stained by hematoxylin & eosin at 5, 14, and 30 dpi in Ctrl and Paxbp1-iKO mice. The second and fourth rows are enlarged views from the corresponding images in the first and third rows, respectively. (E) Immunostaining for embryonic MHC (red) and laminin (green) on TA muscle cross-sections at 5 dpi from Ctrl and iKO mice. (F) Quantification of eMHC+ myofibers in E (n = 3 mice per group). In E, nuclei were counterstained with DAPI (blue). Data are presented as mean ± SD. (Scale bars, 100 µm [D] and 50 µm [E].)
Deletion of Paxbp1 Prevented MuSCs from Reentering the Cell Cycle upon Activation.
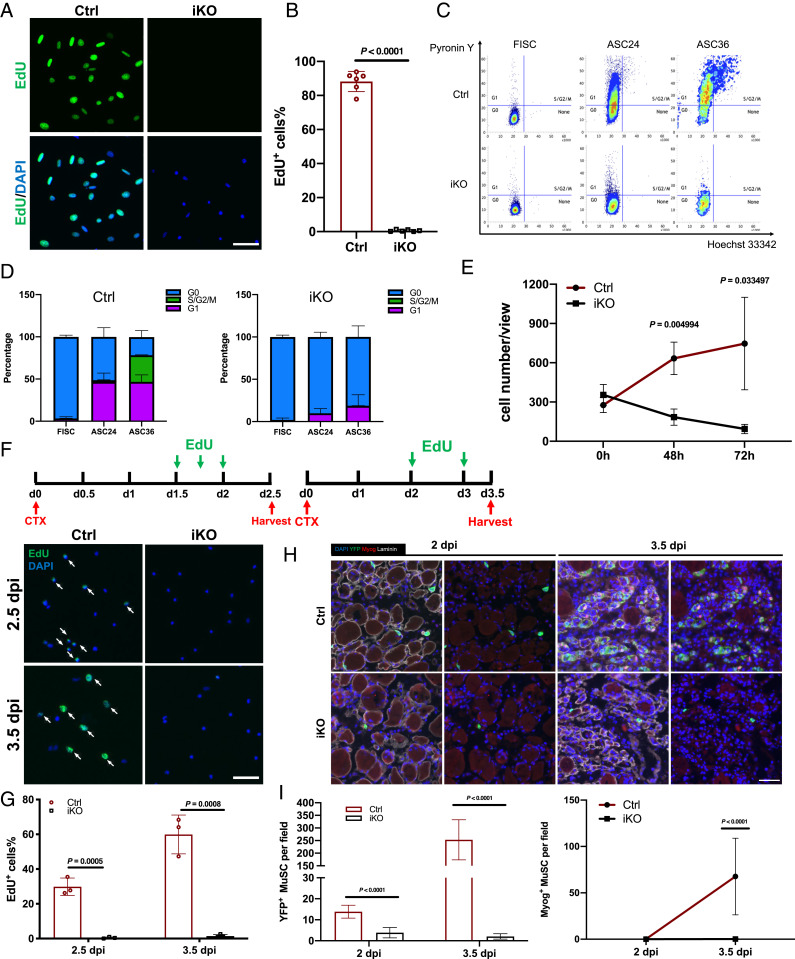
To further understand how Paxbp1 functions in MuSCs, we first investigated whether Paxbp1 regulates quiescence maintenance in adult MuSCs. After inducing Paxbp1 deletion in MuSCs from 2-mo-old adult mice by TMX injection, we monitored the number of YFP+ MuSCs by fluorescence-activated cell sorting (FACS) at different time points ranging from 1 to 5 mo after the last injection of TMX. We found that the number of MuSCs was comparable between control and Paxbp1-iKO mice (SI Appendix, Fig. S2A). Consistently, by immunostaining for either YFP or Pax7, we also detected similar numbers of MuSCs on uninjured TA muscle sections from both control and Paxbp1-iKO mice in which Paxbp1 was already deleted for more than 2 mo (SI Appendix, Fig. S2 B–E). Our data above indicated that Paxbp1 is not required for quiescence maintenance in adult MuSCs. Next, we examined whether Paxbp1 regulated the transition of MuSCs from QSCs to cell-cycle reentry upon activation. To do so, we first isolated MuSCs by FACS from control and Paxbp1-iKO mice, and then cultured them for 48 h in the presence of 5-ethynyl-2′-deoxyuridine (EdU) to identify cells that already reentered S phase of the cell cycle. While more than 80% of ASCs from control mice were EdU+ after 48 h in culture, few Paxbp1-null ASCs could incorporate EdU (Fig. 2 A and B). Next, we cultured freshly isolated individual myofibers in suspension in the presence of EdU for 48 h. In this setting, MuSCs still retained a part of their natural “niche” (i.e., SCs remained sandwiched between basal lamina and the surface of myofibers) where they could undergo activation, cell-cycle reentry and proliferation, and differentiation for a few days (34). Similarly, we found that most MuSCs attached to myofibers from control mice were already EdU+ by 48 h in culture, whereas few such EdU+ MuSCs were found on myofibers from Paxbp1-iKO mice (SI Appendix, Fig. S2 F and G). To further characterize MuSCs after activation, we also conducted flow cytometry-based cell-cycle analysis using FISCs grown in culture. For FISCs from control mice, nearly 50% of cells already entered G1 phase by 24 h in culture, and ∼80% were in G1 and S/G2/M by 36 h (Fig. 2 C and D). For FISCs from Paxbp1-iKO mice, less than 10% of the mutant cells entered G1 by 24 h and ∼20% by 36 h (Fig. 2 C and D). Importantly, none of the mutant cells entered S/G2/M by 36 h. To find out whether the cell-cycle reentry defect in Paxbp1-null MuSCs was merely a delay or a total block, we seeded similar numbers of FISCs from control and Paxbp1-iKO mice in culture for a longer time. By 72 h in culture, the control SCs already expanded more than twofold, whereas the number of Paxbp1-null SCs drastically decreased over the same period (Fig. 2E), indicative of cell death. By time-lapse video microscopy, we confirmed that Paxbp1-null MuSCs failed to divide and most were dead by ∼60 to 84 h in culture (Movies S1 and S2). To further investigate whether the loss of Paxbp1 prevented MuSCs from reentering the cell cycle in vivo, we first injured lower hindlimb muscles of control and Paxbp1-iKO mice with CTX to activate MuSCs followed by intraperitoneal EdU injection to label proliferating cells before killing the mice. At 2.5 and 3.5 dpi, YFP+ ASCs were isolated by FACS and stained for EdU. While ∼30% (at 2.5 dpi) and ∼60% (at 3.5 dpi) of control ASCs were EdU+, few EdU+ ASCs were detected from Paxbp1-iKO mice (Fig. 2 F and G). To exclude the possibility that defective expansion of the mutant MuSCs upon activation was caused by precocious differentiation, we first examined TA muscle sections from both control and Paxbp1-iKO mice that were injured for 2 and 3.5 d to look for YFP+ cells expressing myogenin (Myog), one of the earliest markers for myogenic differentiation. By 2 d after injury, some YFP+ cells were detected on TA muscle sections from the control mice, but no YFP+/Myog+ cells were found (Fig. 2 H and I). This was expected as most MuSCs had just reentered the first cell cycle by this time. By 3.5 d after injury, more YFP+ cells were detected due to SC activation and expansion, and many of them were YFP+/Myog+ (Fig. 2 H and I). By contrast, YFP+ cells were barely detected on muscle sections from Paxbp1-iKO mice that were injured for either 2 or 3.5 d (Fig. 2 H and I). Importantly, none of them were YFP+/Myog+. In addition, we also analyzed FISCs grown in culture. In cells from control mice, a few Myog+ cells started to appear after 48 h in culture and ∼60% Myog+ cells were detected after 72 h in culture (SI Appendix, Fig. S2 H and I). By contrast, no Myog+ cells were detected in SCs from Paxbp1-iKO mice after 24, 48, or 72 h in culture (SI Appendix, Fig. S2 H and I). Next, we assessed whether the cell-cycle reentry defect in Paxbp1-null MuSCs was cell-autonomous. By reexpressing Paxbp1 in Paxbp1-null FISCs using adenovirus, we found that more than 50% of the Paxbp1-null ASCs became EdU+ (SI Appendix, Fig. S2 J and K). Moreover, by time-lapse video microscopy, we showed that the cell-cycle defect in mutant ASCs was also obviously rescued with the reexpression of Paxbp1 (Movies S3 and S4), indicating that the cell-cycle reentry defect of Paxbp1-null MuSCs was indeed cell-autonomous. Taken together, our results demonstrated that Paxbp1 is indispensable for cell-cycle reentry upon MuSC activation.
Fig. 2.
Paxbp1 critically controlled cell-cycle reentry of quiescent MuSCs. (A) FISCs from Ctrl and Paxbp1-iKO mice were cultured for 48 h in the presence of EdU (10 µM) before fixation followed by staining for EdU. (B) Quantification of the percentage of EdU+ SCs over total sorted SCs (n = 6 mice per group). (C) FISCs from Ctrl and iKO mice with or without additional culturing were subjected to cell-cycle analysis using Pyronin Y and Hoechst 33342 double staining. Representative flow cytometry plots are shown. (D) Quantification of the data in C (n = 3 mice per group). (E) Similar numbers of FISCs from Ctrl and iKO mice were cultured for up to 3 d and growth curves were plotted (n = 3 mice per group). (F, Top) Experimental schematics. (F, Bottom) YFP+ SCs were sorted by FACS and immediately stained for EdU. White arrows indicate EdU+ MuSCs. (G) Quantification of the percentage of EdU+ SCs over total YFP+ SCs in F (n = 3 mice per group for each time point). (H) TA muscle sections from Ctrl and iKO mice that were injured for 2 and 3.5 d were subjected to immunostaining for YFP, Myog, and laminin. (I) Enumeration of YFP+ and Myog+ cells per field of view in H (cells in five different fields of view from each mouse were counted; n = 3 mice per group). Nuclei were counterstained with DAPI. Data are presented as mean ± SD. (Scale bars, 50 µm.)
Loss of Paxbp1 Induced Apoptosis during the Late Phase of Cell Growth without Affecting the Early Transition of QSCs to ASCs and the Early Phase of Cell Growth.
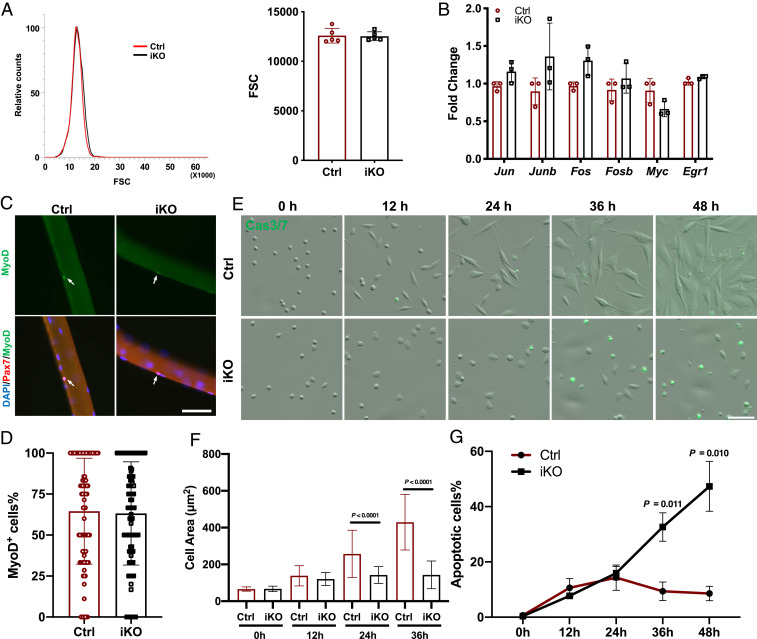
Given a severe blockage in cell-cycle reentry after Paxbp1 deletion in MuSCs, we next investigated when the defect occurred during the transition from QSCs to S phase of the first cell cycle. It became clear recently that FISCs are not bona fide “QSCs.” Instead, they are already partially activated, as evidenced by a rapid induction of multiple IEGs and slightly increased cell size even though such cells remain small and would take another 36 to 48 h before they enter S phase of the first cell cycle (24, 28, 35, 36). By flow cytometry analysis, we noticed that the average size of FISCs from Paxbp1-iKO mice was at least comparable to, if not slightly bigger than, that from the control mice (Fig. 3A), which was not the case for FISCs from p110α-iKO mice that failed to undergo early transition from QSCs to ASCs (24). By qRT-PCR, we also measured mRNA levels of multiple IEGs including those encoding different members of AP1 proteins, Myc, and Egr1 that are known to be rapidly induced in FISCs. Consistently, we found that these IEGs were all induced to similar levels in both control and Paxbp1-null FISCs (Fig. 3B). Next, we examined the protein expression of MyoD, a known early activation marker (10, 37), in SCs attached to intact myofibers that were freshly isolated and cultured for 8 h. By immunostaining, ∼60% of Pax7+ ASCs from both control and Paxbp1-iKO mice were MyoD+ (Fig. 3 C and D). Similarly, MyoD protein was also detected in ∼80% of FISCs from both control and Paxbp1-iKO mice after being grown in culture for 6 h (SI Appendix, Fig. S3A). We then monitored the gradual change in cell size during the course of early MuSC activation in culture by time-lapse video microscopy. As our previous results showed that Paxbp1-null MuSCs gradually underwent cell death in culture (Fig. 2E and Movies S1 and S2), here we added a fluorogenic substrate of caspase-3/7 to help determine the timing and nature of the cell death. As shown in Fig. 3 E–G and Movies S5 and S6, the size of the control MuSCs gradually increased in culture and peaked after 36 h. Interestingly, the size of the Paxbp1-null MuSCs also increased in culture for up to 24 h, but did not further increase beyond that point (Fig. 3 E and F). As for the timing and nature of the cell death, we found that the percentage of the fluorescent (i.e., apoptotic) cells was low and comparable between control and mutant ASCs that were cultured for no more than 24 h (Fig. 3 E and G). However, after 36 h in culture, multiple (∼30%) fluorescent cells started to appear in mutant cells only followed by obvious cell death, clearly revealing apoptosis as the nature of the cell death (Fig. 3G and Movies S5 and S6). In addition, after 24 h in culture, control ASCs became more spindle-shaped with obvious stress fibers and multiple filopodia, while Paxbp1-null ASCs appeared more rounded in shape with few stress fibers and filopodia (SI Appendix, Fig. S3B). Such abnormalities in the actin cytoskeleton may underlie the defective motility of Paxbp1-null ASCs in culture (Movie S2). Next, we examined the change in global RNA synthesis during early activation of MuSCs. Using 5-ethynyl-2′-uridine (EU) incorporation assays, we observed decreased EU incorporation (i.e., RNA synthesis) in mutant ASCs when compared with control ASCs after 24 h in culture (SI Appendix, Fig. S3 C and D). Collectively, our data above indicated that Paxbp1 is essential for the late phase of cell growth and cell survival after activation but not for the initial transition from QSCs to ASCs.
Fig. 3.
Loss of Paxbp1 impaired the cell growth of MuSCs without affecting the initial early activation. (A) Representative forward scatter (FSC) distribution of FISCs from a Ctrl and an iKO mouse (Left) and quantification of the mean FSC for two types of FISCs (Right) (n = 5 mice per group). (B) Measurement of relative mRNA expression of selected IEGs by qRT-PCR using FISCs from Ctrl and iKO mice (n = 3 mice per group). (C) Freshly isolated myofibers from Ctrl and iKO mice were cultured for 8 h before fixation followed by immunostaining for MyoD and Pax7. White arrows indicate ASCs on myofibers. (D) Quantification of the percentage of MyoD+ ASCs over Pax7+ ASCs in C; ∼90 fibers from n = 3 mice per group were used for quantification. (E) Live-cell imaging was conducted on FISCs from control and iKO mice grown in culture together with CellEvent Caspase-3/7 Green Detection Reagent for 72 h. Representative snapshots at the indicated time points are presented. (F) Quantification of the mean cell size in E by Fiji (ImageJ): At each time point, >100 cells from n = 3 mice per group were measured. (G) Quantification of the percentage of Caspase3/7-positive cells in E (n = 3 mice per group). Data are presented as mean ± SD. (Scale bars, 50 µm.)
Paxbp1 Regulated the Expression of Multiple Cell-Cycle and Anabolism-Related Genes in Activated SCs.
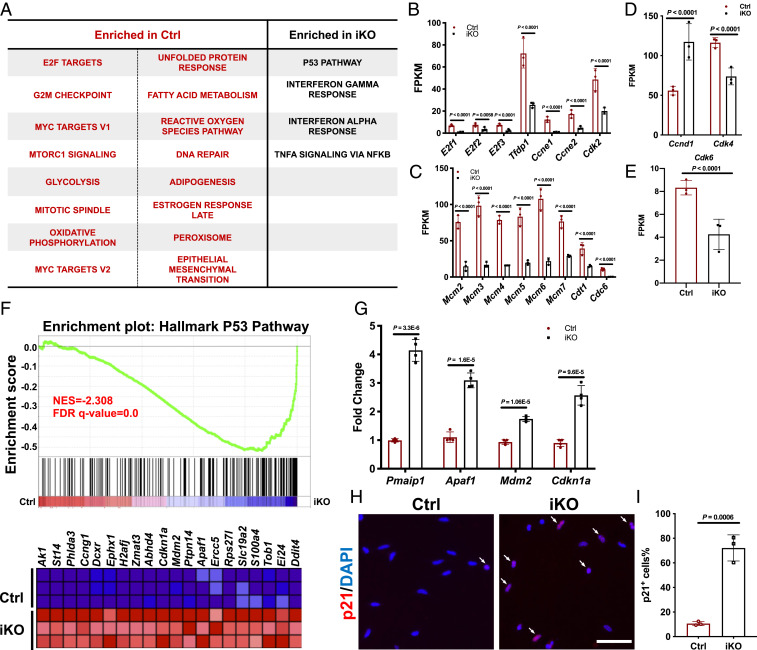
To better understand why the loss of Paxbp1 prevented MuSCs from reentering the cell cycle, we decided to examine the transcriptomes of control and Paxbp1-null MuSCs by RNA-seq. As the loss of Paxbp1 in QSCs did not affect either quiescence maintenance or the initial transition from QSCs to ASCs but compromised the late phase of cell growth after activation (Fig. 3 and SI Appendix, Fig. S2), we chose to examine ASCs 24 h after CTX-induced muscle injury, which we viewed as a suitable time point for comparison as both the mutant and control ASCs were activated but had not entered S phase yet (12, 13, 24). Moreover, based on our live-cell imaging study of FISCs in culture (Fig. 3G), we reasoned that extensive apoptosis would be less likely to occur in mutant ASCs at this time point in vivo. Selection of a later time point (e.g., 48 h after the injury) would have biased the differences toward either the cell cycle-related genes (as most control ASCs would have already entered the cell cycle to become cycling myoblasts) or apoptosis-related genes (as a large fraction of mutant ASCs would have already undergone apoptosis). To carry out RNA-seq, we freshly isolated YFP+ ASCs from control and Paxbp1-iKO mice by FACS without additional culturing, extracted the mRNA, and then subjected it to RNA-seq. Principal-component analysis revealed good reproducibility among biological triplicates and clear separation between control and Paxbp1-null ASCs (accession no. GSE141881) (SI Appendix, Fig. S4A). A large set of genes were differentially expressed between control and mutant ASCs (with ∼2,000 genes having fold change > 2 and Padj < 0.05) (SI Appendix, Fig. S4B). We then performed gene set enrichment analysis (GSEA) to identify dysregulated pathways or biological processes in mutant ASCs (38). Notably, many cell cycle-related terms, including “Myc targets,” “E2F targets,” “mitotic spindle,” as well as “G2M checkpoint,” were enriched in control ASCs (Fig. 4A), suggesting a requirement for control ASCs to induce the expression of multiple cell cycle-related genes long before they enter S phase and a failure of mutant ASCs in doing so. Consistently, Ki67, a prominent marker associated with proliferating but not quiescent cells (39), was undetectable in Paxbp1-null ASCs at both mRNA (mean FPKM [fragments per kilobase of transcript per million mapped reads] value 0.129) and protein levels (SI Appendix, Fig. S4 C and D). In particular, many genes involved in the G1/S transition including E2f1-3, Tfdp1 (encoding DP1), Ccne1/2 (encoding cyclin E1/2), and Cdk2 and those associated with DNA replication origin licensing (e.g., Cdc6, Cdt1, and Mcm2 to 7) were poorly expressed in mutant ASCs (Fig. 4 B–E). Among them, Cdc6 and Ccne1 were barely expressed in mutant ASCs (their mean FPKM values were 0.27 and 1.07, respectively), which could impede S-phase reentry from G0 as both are known to be indispensable for Mcm loading during replication origin licensing, a prerequisite for S-phase entry (40). Intriguingly, Ccnd1 (encoding cyclin D1) levels were around twofold higher in mutant ASCs than that of control (Fig. 4D). Moreover, both Cdk4 and Cdk6 were induced to similar extents in both control and mutant ASCs compared with their basal levels in FISCs, even though their mean FPKM values were slightly lower in mutant ASCs (Fig. 4 D and E). Collectively, our RNA-seq data supported our view that mutant ASCs were partially activated but likely arrested in late G0, or a “shallow” quiescent state (4, 5).
Fig. 4.
RNA-seq revealed dysregulated cell cycle-related genes and p53 target genes in Paxbp1-null ASCs. (A) Enriched terms (false discovery rate [FDR] < 0.05) in ASCs from Ctrl and iKO mice by GSEA using the Hallmark database are shown. (B–E) Based on RNA-seq data, the mean FPKM values of selected genes involved in G1/S transition (B, D, and E) and replication origin licensing (C) are shown (n = 3 mice per group). (F) GSEA plot showing many p53 target genes were up-regulated in Paxbp1-null ASCs. NES, normalized enrichment score. (G) The increased mRNA expression of selected p53 target genes in Paxbp1-null ASCs was validated by qRT-PCR using FACS-isolated ASCs 24 h postinjury (n = 4 mice per group). (H) p21 protein expression was examined in ASCs after 24 h in culture by immunostaining. White arrows indicate p21+ ASCs. (I) Quantification of the percentage of p21+ ASCs over total ASCs in H (n = 3 mice per group). Data are presented as mean ± SD. (Scale bar, 50 µm.)
Among the pathways enriched in Paxbp1-null ASCs, the term “p53 pathway” stood out (Fig. 4 A and F). The induction of many p53 target genes (e.g., Cdkn1a, Mdm2, Apaf1, and Pmaip1) in Paxbp1-null ASCs was further confirmed by qRT-PCR (Fig. 4G). Consistently, by immunostaining, we also detected abnormal induction of p21Cip1 protein (encoded by Cdkn1a) in mutant ASCs (Fig. 4 H and I). As the protein products of many p53 target genes are known to promote cell-cycle arrest (e.g., p21Cip1) and apoptosis (e.g., Apaf1 and Pmaip1), they certainly contributed to apoptosis in Paxbp1-null ASCs (Fig. 3 E and G).
A Further Increase in Paxbp1-Dependent mTORC1 Activity during the Late Phase of Cell Growth Promoted Mitochondrial Biogenesis and Anabolic Metabolism.
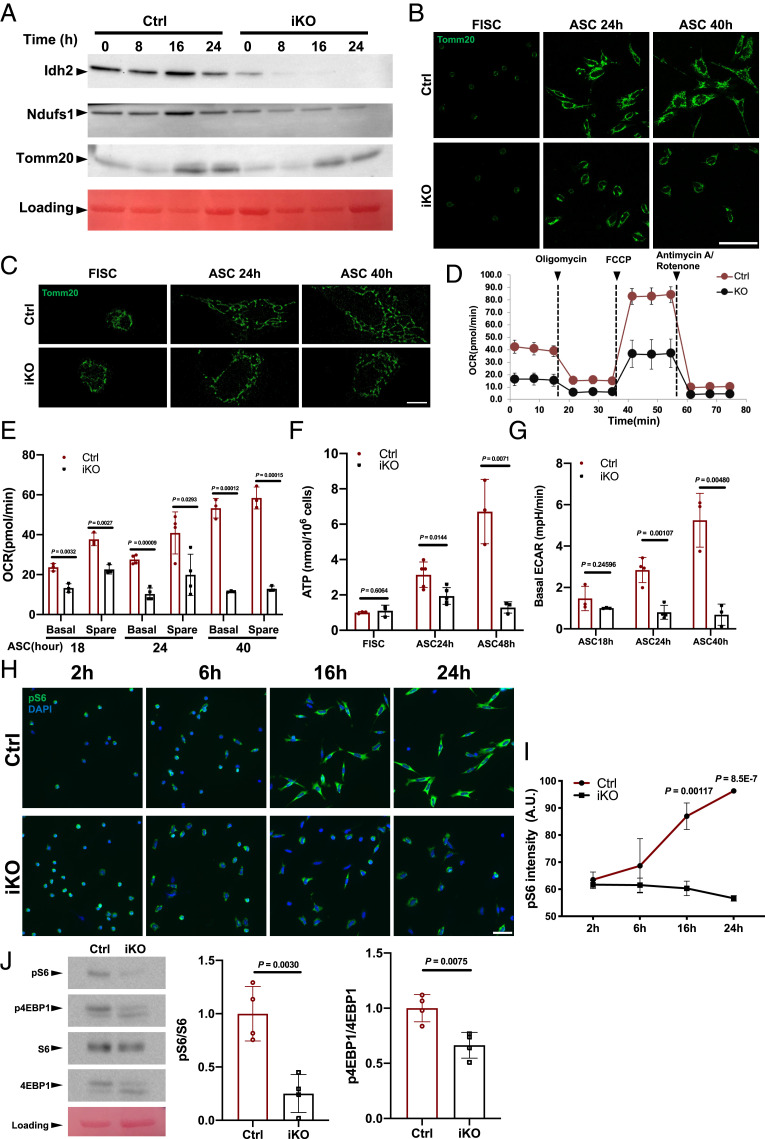
In addition to those cell cycle-related genes, the expression of many genes related to anabolic metabolism, including glycolysis, the tricarboxylic acid cycle, OXPHOS, fatty acid metabolism, and the pentose phosphate pathway (PPP), was also impaired in Paxbp1-null ASCs (Fig. 4A and SI Appendix, Fig. S4E). The reduced expression of selected metabolic genes (e.g., Pkm, Aco2, Cs, and Fh1) in mutant ASCs was further confirmed by qRT-PCR (SI Appendix, Fig. S4F). To functionally examine selected metabolic pathways in mutant ASCs, we first selected several mitochondria-associated genes (i.e., Idh2, Ndufs1, and Tomm20) whose mRNA levels were reduced in Paxbp1-null ASCs and then confirmed their reduced protein expression by Western blot (Fig. 5A). To assess mitochondrial biogenesis during MuSC activation, we used FISCs grown in culture and performed immunostaining for Tomm20, a protein specifically localized to the outer membrane of mitochondria. While few mitochondria could be detected in control FISCs, mitochondrial biogenesis became very obvious in control ASCs by 24 h in culture, with a further increase in control ASCs by 40 h in culture (Fig. 5B). Notably, mitochondrial biogenesis initially also occurred in Paxbp1-null ASCs by 24 h in culture, but at a reduced level compared with that in control ASCs (Fig. 5B). Importantly, there was no further increase in mitochondrial biogenesis in mutant ASCs by 40 h in culture, a pattern that mimicked the defective cell-size change in mutant cells during this period (Fig. 3 E and F). Moreover, by superresolution microscopy, we found that the mitochondria in control ASCs formed a more interconnected tubular network during activation, while they were largely fragmented and restricted to the perinuclear region in mutant ASCs (Fig. 5C). Next, we assessed the activity of the electron transport chain (ETC) in mitochondria and glycolysis. Using FISCs grown in culture, we measured the oxygen consumption rate (OCR), cellular adenosine triphosphate (ATP) levels, and basal glycolysis. We found that Paxbp1-null ASCs were already severely defective in ETC-dependent cellular respiration between 18 and 24 h in culture, a period before S-phase reentry (Fig. 5 D–F). Similar defects in glycolysis were also observed in mutant ASCs by 24 h in culture based on the measurement of the basal extracellular acidification rate (ECAR) (Fig. 5G). Although Paxbp1-null ASCs had dysfunctional OXPHOS and glycolysis, it was unclear whether such defects contributed to the failure of cell-cycle reentry in mutant ASCs. To address this issue, we treated FISCs from wild-type mice with various ETC inhibitors (i.e., oligomycin, FCCP, and antimycin A) or 2-deoxy-d-glucose (a glycolysis inhibitor), and then examined the EdU incorporation in culture. We found that inhibition of either ETC or glycolysis in FISCs potently blocked their cell-cycle reentry (SI Appendix, Fig. S5 A–D), suggesting that activation of both OXPHOS and glycolysis is a prerequisite for cell-cycle reentry in MuSCs.
Fig. 5.
Sharp increase in Paxbp1-dependent mTORC1 activity correlated with enhanced mitochondrial biogenesis and anabolic metabolism. (A) FISCs from Ctrl and iKO mice were cultured for various times as indicated. Cells were then collected and subjected to Western blot for the indicated mitochondrial proteins. Idh2, isocitrate dehydrogenase 2; Ndufs1, NADH-ubiquinone oxidoreductase Fe-S protein 1. (B and C) Representative confocal (B) and superresolution (C) microscopic images of Tomm20 immunostaining using FISCs and ASCs that were cultured for various times as indicated. (D) Representative OCR measured by Seahorse Mito Stress assays for ASCs after 24 h in culture. (E) Quantification of OCR for ASCs after 18, 24, and 40 h in culture (n = 3 mice per group for samples at 18 and 40 h; n = 4 mice per group for samples at 24 h). (F) Cellular ATP levels were measured in FISCs and ASCs that were cultured for 24 and 48 h (n = 3 mice per group for FISCs and samples at 48 h; n = 5 mice per group for samples at 24 h). (G) Quantification of ECAR for ASCs that were cultured for 18, 24, and 40 h (n = 3 mice per group for samples at 18 and 40 h; n = 4 mice per group for samples at 24 h). mpH/min, milli-pH/minute. (H) FISCs from Ctrl and iKO mice were cultured for the indicated times followed by immunostaining for pS6 (green). Nuclei were counterstained with DAPI (blue). (I) Quantification of the intensity of pS6 by Fiji (ImageJ) in H (n = 3 mice per group for each time point). A.U., arbitrary unit. (J) Representative Western blot (n = 4 mice per group) showing the levels of total protein and the phosphorylated forms for S6 and 4EBP1 in ASCs after 24 h in culture. Quantification data by densitometry are presented (Right). Data are presented as mean ± SD. (Scale bars, 50 µm [B and H] and 5 µm [C].)
To uncover the mechanisms responsible for defective anabolic pathways and mitochondrial biogenesis in Paxbp1-null ASCs, we turned to mTORC1, as it is a well-established master regulator capable of stimulating anabolic metabolism and mitochondrial biogenesis during cell growth (41, 42). Our GSEA indeed revealed that multiple genes implicated in “mTORC1 signaling” were down-regulated in mutant ASCs (SI Appendix, Fig. S5E). Consistently, when we treated FISCs from wild-type mice with Torin1 and Torin2, two potent mTORC1 inhibitors (43, 44), EdU incorporation was inhibited in both dose- and time-dependent manners: both drugs effectively inhibited EdU incorporation when added early (i.e., 2 h after culturing); they were still effective when added 18 h after culturing, but became less effective when added 24 h after culturing (SI Appendix, Fig. S5F). Next, we assessed the mTORC1 activity in cultured FISCs from both control and Paxbp1-iKO mice at various time points by immunostaining for the phosphorylated ribosomal protein S6 (pS6), a known substrate of mTORC1 and a sensitive marker for activated mTORC1 signaling (24, 42). As Paxbp1-null MuSCs could undergo early transition from QSCs to ASCs (Fig. 3), a known PI3K/mTORC1-dependent process (24), it was not surprising that most ASCs from both control and Paxbp1-iKO mice already became pS6+ by 2 h in culture (Fig. 5H). Unexpectedly, the intensity of the pS6 signal underwent a sharp increase in control ASCs by 16 to 24 h in culture, whereas such an increase was abolished in Paxbp1-null ASCs (Fig. 5 H and I). To further confirm this finding, we also assessed the activity of mTORC1 in ASCs by Western blot to monitor the levels of pS6 and phosphorylated 4EBP1 (p4EBP1), the latter being another well-characterized substrate of mTORC1 (42). Indeed, Paxbp1-null ASCs showed impaired mTORC1 activity as evidenced by reduced levels of pS6 and p4EBP1 (Fig. 5J). Collectively, our data indicated that a further increase in the Paxbp1-dependent mTORC1 activity promoted mitochondrial biogenesis and anabolic metabolism during the late phase of cell growth before QSCs reenter the cell cycle.
Paxbp1 Modulated mTORC1 Activity via Redox Regulation and p53.
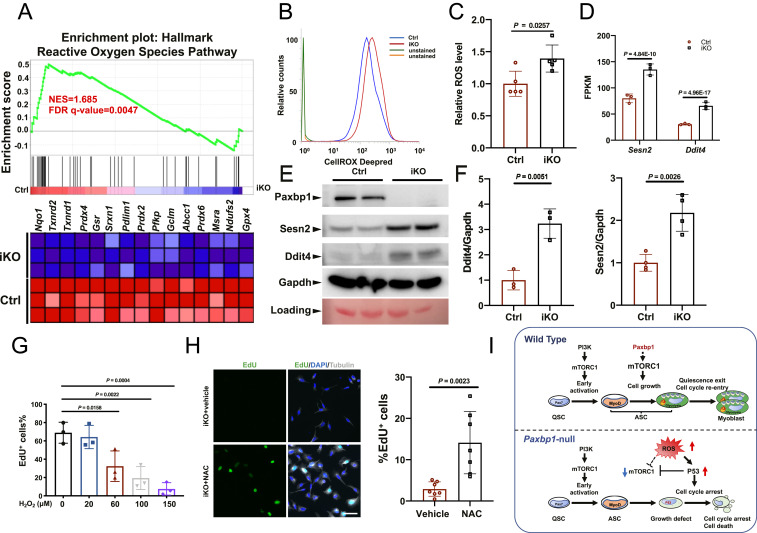
To further understand the mechanisms responsible for impaired mTORC1 activity in Paxbp1-null ASCs, we turned to our RNA-seq data and GSEA again. We noticed that many genes implicated in the “reactive oxygen species pathway” were down-regulated in Paxbp1-null ASCs (Figs. 4A and 6A). In particular, we noticed that many antioxidant genes including Prdx2, 3, 4, and 6, Txnrd1/2, Gsr, Srxn1, Gpx4, and Hmox1 were down-regulated in mutant ASCs (Fig. 6 A, Bottom). For selected antioxidant genes, we also further confirmed their down-regulation in freshly sorted ASCs from Paxbp1-iKO mice by qRT-PCR (SI Appendix, Fig. S6A). Such data suggested that the mutant ASCs could have elevated ROS levels. To test this hypothesis, we first cultured FISCs from both control and Paxbp1-iKO mice for 18 h and measured their ROS levels by flow cytometry. We found that ROS levels were indeed higher in mutant ASCs (SI Appendix, Fig. S6B). Importantly, we also measured ROS levels in freshly isolated ASCs (without additional culturing) from both control and Paxbp1-iKO mice that had been injured for 24 h and confirmed that ROS levels were indeed higher in Paxbp1-null ASCs (Fig. 6 B and C). As elevated ROS are known to activate p53 and to induce apoptosis (45), it is likely that induction of p53 target genes and apoptosis in Paxbp1-null ASCs was caused by elevated ROS. To test this hypothesis, we treated FISCs from wild-type mice with different doses of H2O2 to enhance intracellular ROS levels. We found that H2O2 treatment indeed led to induction of multiple p53 target genes and apoptosis (SI Appendix, Fig. S6 C and D). Among the p53-induced genes in mutant ASCs (Figs. 4F and 6D), Sesn2 and Ddit4 caught our attention as their protein products (Sestrin2 and Ddit4, respectively) were known to potently inhibit mTORC1 (46–48). Both Sesn2 and Ddit4 are also known to be induced by p53 in other cell types (49). By Western blot, we showed that both Sestrin2 and Ddit4 protein levels were indeed increased in mutant ASCs (Fig. 6 E and F). Consistently, we found that H2O2 treatment indeed impaired mTORC1 activities and inhibited EdU incorporation in ASCs from wild-type mice (Fig. 6G and SI Appendix, Fig. S6E). Thus, it appeared that elevated ROS were causally responsible for the cell-cycle reentry defect in Paxbp1-null ASCs. To test whether reduction of ROS levels could rescue the defect, we treated FISCs from Paxbp1-iKO mice with N-acetylcysteine (NAC), a widely used ROS scavenger (17, 50), and then measured EdU incorporation. We found that the NAC treatment partially rescued the cell-cycle reentry defect in mutant ASCs, with ∼15% of them becoming EdU+ after 60 h in culture (Fig. 6H). Taken together, our data supported the hypothesis that elevated ROS in Paxbp1-null ASCs increase p53 activity and impair mTORC1 activity, which promotes apoptosis and prevents ASCs from reentering the cell cycle (Fig. 6I).
Fig. 6.
Paxbp1-dependent redox regulation influenced the mTORC1 and p53 signaling and cell-cycle reentry of QSCs. (A) GSEA plot showing the defective ROS pathway in Paxbp1-null ASCs. (B) A representative flow cytometry plot showing increased ROS levels in Paxbp1-null ASCs directly isolated from Ctrl and iKO mice at 1 dpi. (C) Quantification of relative ROS levels in B (n = 5 mice per group). (D) Mean FPKM values for Sesn2 and Ddit4 based on our RNA-seq data. (E) A representative Western blot showing the protein levels of Sesn2 and Ddit4 in ASCs after 24 h in culture. (F) Quantification of data in E by densitometry (n ≥ 3). (G) FISCs from wild-type adult mice were cultured for 40 h with EdU in the absence or presence of different doses of H2O2 before fixation followed by staining for EdU. The percentage of EdU+ ASCs over total ASCs is shown (n = 3 independent experiments). (H) FISCs from iKO mice were cultured in the presence of EdU with or without NAC (5 mM) for 60 h before fixation followed by staining for EdU and tubulin. Quantification of the percentage of EdU+ ASCs over total ASCs (n = 7 independent experiments) is also shown. (I) Schematic diagrams summarizing key findings from our work. Data are presented as mean ± SD. (Scale bar, 50 µm.)
Discussion
Paxbp1 Controls a Late Cell-Growth Checkpoint Indispensable for Cell-Cycle Reentry during Early Activation of QSCs.
Quiescent cells generally take a longer time to reenter S phase of the first cell cycle compared with their cycling offspring after the latter complete the previous rounds of the cell cycle (40). The prolonged time required for quiescent cells to transition to the cycling state allows such cells to synthesize sufficient biomolecules for size increase and for subsequent cell division, and provides a regulatory window that could potentially involve multiple regulatory checkpoints. We and others recently identified an indispensable checkpoint for the early transition from QSCs to ASCs that is critically regulated by the PI3K-mTORC1/FoxO pathway: Loss of PI3K in QSCs prevents them from transitioning to ASCs upon injury-induced activation, whereas constitutive activation of PI3K in QSCs results in their spontaneous activation and quiescence exit in uninjured muscles and gradual depletion of QSCs (24, 25). Similar checkpoints for early activation and quiescence exit also operate in quiescent hematopoietic stem cells (HSCs) and naïve T cells (51, 52). Interestingly, unlike p110α (PI3K)-null QSCs, upon activation, Paxbp1-null QSCs could pass the initial PI3K-mediated checkpoint to transition to ASCs, express MyoD protein, and undergo partial mitochondrial biogenesis and size increase before displaying various cell growth-related defects between 16 and 24 h after the injury (Figs. 3, 4, and 5). This suggests the existence of another Paxbp1-dependent checkpoint for cell growth that functions beyond the initial checkpoint for the early transition from QSCs to ASCs (Fig. 6I). Failure in cell growth-associated anabolic pathways in other quiescent cell types also results in severe defects in their activation and cell-cycle reentry (11). For example, in hair follicle stem cells, inducible deletion of Ldha that encodes lactate dehydrogenase prevented their activation, while enhancing lactate production accelerated their activation (53). In naïve T cells, deletion of Cox10 that encodes a component of complex IV of the mitochondria ETC also impaired their cell-cycle reentry upon stimulation of T cell receptors (54). It remains to be tested whether the Paxbp1-dependent checkpoint for cell growth also operates during activation of other quiescent cells. Our work here also demonstrates that the severe muscle regeneration defect in Paxbp1-iKO mice is not caused by defective MuSC proliferation per se. Instead, it is caused by defective cell growth before QSCs reenter the cell cycle. mTOR is a well-established master regulator for cell growth in response to growth factors, nutrients, and cellular energy status by stimulating mitochondrial and ribosomal biogenesis and anabolic metabolism (42, 55, 56). Consistently, mTORC1 was shown to be indispensable for quiescent naïve T cells to increase in size and to reenter the cell cycle upon antigen stimulation (52). Intriguingly, we find that the PI3K-mTORC1 pathway is initially Paxbp1-independent in the early phase of cell growth in ASCs. By 16 to 24 h after injury, there is a further increase in the Paxbp1-dependent mTORC1 activity that drives the late phase of cell growth coupled with enhanced mitochondrial biogenesis and anabolic metabolism to promote cell-cycle reentry (Figs. 5 and 6I).
Cross-Talk among p53, mTORC1, and ROS.
Our RNA-seq data revealed that many known p53 target genes including p21Cip1, Mdm2, Apaf1, Pmaip1 (Noxa), Ercc5, Wig1, Sesn2, Ddit4, and Ei24 were induced in Paxbp1-null ASCs, which is indicative of enhanced p53 activity (Fig. 4). Induction of many p53 target genes is known to promote cell-cycle arrest (e.g., via p21Cip1) and apoptosis (e.g., via Apaf1 and Noxa), which match the phenotypes of the Paxbp1-null ASCs. Interestingly, mutant MuSCs lacking notchless homolog 1 (Nle1) were also found to have increased p53 activity and defective cell-cycle reentry upon activation (57). Unlike Paxbp1-null ASCs, Nle1-null ASCs did not undergo apoptosis in culture. As the mRNA levels of Nle1 did not change much in Paxbp1-null ASCs compared with that of control ASCs, it remains to be investigated whether Paxbp1 and Nle1 are mechanistically connected.
Increased p53 activity can inhibit mTORC1 via different mechanisms (49). In particular, Sestrin2 and Ddit4 (Redd1), the protein products of Sesn2 and Ddit4, respectively, that are induced by p53, are known to inhibit mTORC1 (46–48). Thus, the abnormal induction of Sesn2 and Ddit4 at both mRNA and protein levels in Paxbp1-null ASCs contributes to the reduced mTORC1 activity (Fig. 6). As a cytosolic leucine sensor, Sestrin2 represses mTORC1 by directly binding and repressing GATOR2 when cellular leucine levels are low (42). Consistently, based on our RNA-seq data, we found that Slc7A5 (encoding the leucine transporter), Slc7A1 (encoding the Arg transporter), Slc7A6 (encoding the cationic amino acid transporter), Slc1A4 (encoding the transporter for Ser, Thr, Cys, and Ala), and Slc38A1 (encoding the Gln transporter) were all down-regulated in Paxbp1-null ASCs, which supports our view that defective amino acid-sensing pathways also contribute to the reduced mTORC1 activity in mutant ASCs.
The increased p53 activity in Paxbp1-null ASCs is likely triggered by elevated ROS due to reduced expression of multiple antioxidant genes (e.g., Prdx 2, 3, 4, and 6, Gclm, Gsr, Hmox1, Srxn1, Sod1, Nqo1, Txn1, Txnrd1, and Txnrd2) in the mutant cells (Fig. 6) (45). Consistently, deliberate elevation of ROS levels in wild-type MuSCs by H2O2 treatment also resulted in induction of p53 target genes, increased apoptosis, impaired mTORC1 activity, and defective cell-cycle reentry (Fig. 6G and SI Appendix, Fig. S6 C–E). Moreover, the defective PPP in Paxbp1-null ASCs could also contribute to elevated ROS due to reduced NADPH production. NADPH is specifically utilized by glutathione reductase (encoded by Gsr) to convert the oxidized glutathione disulfide to the reduced glutathione, a major antioxidant in the cytosol. Although moderate levels of ROS promote cell signaling and quiescence exit in both MuSCs and HSCs (11, 17, 58, 59), excessive ROS can lead to cellular damage, p53 activation, cell-cycle arrest, and apoptosis (45, 60). In addition, ROS may also impair mTORC1 activity via p53-independent mechanisms (Fig. 6I) (61). A causal role for excessive ROS in Paxbp1-null MuSCs is further supported by our finding that treatment of mutant ASCs with a ROS scavenger (i.e., NAC) partially rescued their defects in S-phase reentry (Fig. 6H). The low rescue efficiency suggests that additional unknown factors also contribute to the cell-cycle reentry defects in mutant ASCs. Consistent with the mechanistic scheme we presented here (Fig. 6I), excessive ROS triggered by reduced glutathione levels in quiescent naïve T cells from Gclc (encoding the catalytic subunit of glutamate cysteine ligase)-null mice also led to reduced mTORC1 activity, defective metabolic reprogramming, and impaired cell proliferation upon stimulation (62). Moreover, abnormal accumulation of ROS was also detected in MuSCs with defective autophagy and mutant MuSCs lacking Pitx2/Pitx3 or Numb/Numbl, all of which promoted SC senescence and resulted in defective muscle regeneration (17, 50, 63). As the mRNA levels of Pitx2, Pitx3, Numb, and Numbl were not obviously affected in Paxbp1-null ASCs, it suggests that the defects of Paxbp1-null ASCs are not directly linked to these genes.
Connection with Pax7.
Paxbp1 was originally identified as a Pax7-binding protein in the mouse (26). Interestingly, two recent clinical studies reported homozygous mutations in human PAXBP1 and PAX7 that are linked to similar human genetic disorders with global developmental delay and myopathic hypotonia (64, 65). Are the defects of Paxbp1-null ASCs linked to Pax7? We found that both Pax7 mRNA and protein levels were not reduced in Paxbp1-null ASCs: The mRNA levels of Pax7 actually increased by approximately fourfold compared with that in control ASCs, while Pax7 protein levels were similar to (if not more than) that in control ASCs based on immunostaining. Functionally, loss of either Paxbp1 or Pax7 in adult MuSCs has distinct impacts on quiescence maintenance: While deletion of Paxbp1 did not affect the long-term maintenance of MuSCs in uninjured muscles (SI Appendix, Fig. S2), that of Pax7 led to gradual depletion of MuSCs (66). Nevertheless, our ongoing studies found that adult Pax7-null MuSCs were similarly defective in cell-cycle reentry upon activation, although such defects were less severe than that in Paxbp1-null MuSCs. Although both Paxbp1-null and Pax7-null MuSCs exhibit similar cell-cycle reentry defects upon activation, it remains to be rigorously tested whether the loss of Paxbp1 interferes with Pax7 functions in MuSCs. As Paxbp1 is conserved in Metazoa and expressed in many cell types and tissues that lack Pax7 expression, its cell growth-promoting role should not be solely dependent on Pax7 or limited to MuSCs. It is likely that Paxbp1 could also promote cell growth and cell-cycle reentry in other quiescent cells upon their activation.
Materials and Methods
Animals.
Paxbp1flox/flox mice were generated by the Model Animal Research Center at Nanjing University using the targeting vector of Paxbp1 (Paxbp1tm44128(L1L2_gt0)) purchased from the International Mouse Phenotyping Consortium with exon 8 of Paxbp1 flanked by loxP sites. Pax7CreERT2(Gaka) (stock 017763) and R26R-EYFP (stock 006148) mice were from The Jackson Laboratory. All the experiments were performed in accordance with protocols approved by the Animal Ethics Committee at the Hong Kong University of Science and Technology.
Isolation of Adult MuSCs by FACS.
This was performed essentially as described in Wang et al. (24).
RNA-Seq and Data Processing.
YFP+ ASCs (105) were isolated by FACS from control and Paxbp1-iKO mice 24 h after CTX-induced injury. Total RNA was extracted, and complementary DNA (cDNA) was synthesized and amplified by Smart-seq2 (67). Purified cDNA was fragmented followed by library construction. The paired-end library was sequenced by an Illumina NextSeq 500. Raw sequencing reads were mapped to the mouse reference genome (GRCm38/mm10) using STAR aligner (68). A raw count table was then generated with featureCounts (69), followed by analysis with the DESeq2 R package (70). Raw data were uploaded to the Gene Expression Omnibus (GEO) (accession no. GSE141881).
Time-Lapse Imaging and Apoptosis Detection.
FISCs were cultured in growth media (F10 with 10% horse serum) in the presence of 4 µM CellEvent Caspase-3/7 Green Detection Reagent for up to 72 h. Time-lapse imaging was then performed by a Zeiss Cell Discoverer 7 automated microscope for up to 72 h. Images were taken every 5 to 10 min and videos were generated using Zen blue software.
Supplementary Material
Acknowledgments
This work was supported by research grants from the Hong Kong Research Grant Council (GRF16101517, C6018-19G, AoE/M-09/12, AoE/M-604/16, and T13-605/18-W), the Center for Systems Biology & Human Health, the State Key Laboratory of Molecular Neuroscience at the Hong Kong University of Science and Technology, the Hong Kong Epigenome Project, and the Shenzhen Bay Laboratory (S201101002, Guangdong, China).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021093118/-/DCSupplemental.
Data Availability
The RNA-seq data reported in this article have been deposited in the GEO (accession no. GSE141881). Some study data are available in the supporting information.
References
- 1.Chapman N. M., Boothby M. R., Chi H., Metabolic coordination of T cell quiescence and activation. Nat. Rev. Immunol. 20, 55–70 (2020). [DOI] [PubMed] [Google Scholar]
- 2.Gonzales K. A. U., Fuchs E., Skin and its regenerative powers: An alliance between stem cells and their niche. Dev. Cell 43, 387–401 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Li L., Clevers H., Coexistence of quiescent and active adult stem cells in mammals. Science 327, 542–545 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Urbán N., Blomfield I. M., Guillemot F., Quiescence of adult mammalian neural stem cells: A highly regulated rest. Neuron 104, 834–848 (2019). [DOI] [PubMed] [Google Scholar]
- 5.van Velthoven C. T. J., Rando T. A., Stem cell quiescence: Dynamism, restraint, and cellular idling. Cell Stem Cell 24, 213–225 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marescal O., Cheeseman I. M., Cellular mechanisms and regulation of quiescence. Dev. Cell 55, 259–271 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ge Y., Fuchs E., Stretching the limits: From homeostasis to stem cell plasticity in wound healing and cancer. Nat. Rev. Genet. 19, 311–325 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Buckingham M., Relaix F., PAX3 and PAX7 as upstream regulators of myogenesis. Semin. Cell Dev. Biol. 44, 115–125 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Evano B., Tajbakhsh S., Skeletal muscle stem cells in comfort and stress. NPJ Regen. Med. 3, 24 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yin H., Price F., Rudnicki M. A., Satellite cells and the muscle stem cell niche. Physiol. Rev. 93, 23–67 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Coller H. A., The paradox of metabolism in quiescent stem cells. FEBS Lett. 593, 2817–2839 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rocheteau P., Gayraud-Morel B., Siegl-Cachedenier I., Blasco M. A., Tajbakhsh S., A subpopulation of adult skeletal muscle stem cells retains all template DNA strands after cell division. Cell 148, 112–125 (2012). [DOI] [PubMed] [Google Scholar]
- 13.Rodgers J. T., et al., mTORC1 controls the adaptive transition of quiescent stem cells from G0 to G(alert). Nature 510, 393–396 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Feige P., Brun C. E., Ritso M., Rudnicki M. A., Orienting muscle stem cells for regeneration in homeostasis, aging, and disease. Cell Stem Cell 23, 653–664 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen F., et al., YY1 regulates skeletal muscle regeneration through controlling metabolic reprogramming of satellite cells. EMBO J. 38, e99727 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dell’Orso S., et al., Single cell analysis of adult mouse skeletal muscle stem cells in homeostatic and regenerative conditions. Development 146, dev174177 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.L’honoré A., et al., The role of Pitx2 and Pitx3 in muscle stem cells gives new insights into P38α MAP kinase and redox regulation of muscle regeneration. eLife 7, e32991 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pala F., et al., Distinct metabolic states govern skeletal muscle stem cell fates during prenatal and postnatal myogenesis. J. Cell Sci. 131, jcs212977 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ryall J. G., et al., The NAD(+)-dependent SIRT1 deacetylase translates a metabolic switch into regulatory epigenetics in skeletal muscle stem cells. Cell Stem Cell 16, 171–183 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sala D., et al., The Stat3-Fam3a axis promotes muscle stem cell myogenic lineage progression by inducing mitochondrial respiration. Nat. Commun. 10, 1796 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tang A. H., Rando T. A., Induction of autophagy supports the bioenergetic demands of quiescent muscle stem cell activation. EMBO J. 33, 2782–2797 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Theret M., et al., AMPKα1-LDH pathway regulates muscle stem cell self-renewal by controlling metabolic homeostasis. EMBO J. 36, 1946–1962 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wüst S., et al., Metabolic maturation during muscle stem cell differentiation is achieved by miR-1/133a-mediated inhibition of the Dlk1-Dio3 mega gene cluster. Cell Metab. 27, 1026–1039.e6 (2018). [DOI] [PubMed] [Google Scholar]
- 24.Wang G., et al., p110α of PI3K is necessary and sufficient for quiescence exit in adult muscle satellite cells. EMBO J. 37, e98239 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yue F., et al., Pten is necessary for the quiescence and maintenance of adult muscle stem cells. Nat. Commun. 8, 14328 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Diao Y., et al., Pax3/7BP is a Pax7- and Pax3-binding protein that regulates the proliferation of muscle precursor cells by an epigenetic mechanism. Cell Stem Cell 11, 231–241 (2012). [DOI] [PubMed] [Google Scholar]
- 27.Liu L., et al., Chromatin modifications as determinants of muscle stem cell quiescence and chronological aging. Cell Rep. 4, 189–204 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Machado L., et al., In situ fixation redefines quiescence and early activation of skeletal muscle stem cells. Cell Rep. 21, 1982–1993 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Keller C., Hansen M. S., Coffin C. M., Capecchi M. R., Pax3:Fkhr interferes with embryonic Pax3 and Pax7 function: Implications for alveolar rhabdomyosarcoma cell of origin. Genes Dev. 18, 2608–2613 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Murphy M. M., Lawson J. A., Mathew S. J., Hutcheson D. A., Kardon G., Satellite cells, connective tissue fibroblasts and their interactions are crucial for muscle regeneration. Development 138, 3625–3637 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.d’Albis A., Couteaux R., Janmot C., Roulet A., Mira J. C., Regeneration after cardiotoxin injury of innervated and denervated slow and fast muscles of mammals. Myosin isoform analysis. Eur. J. Biochem. 174, 103–110 (1988). [DOI] [PubMed] [Google Scholar]
- 32.von Maltzahn J., Jones A. E., Parks R. J., Rudnicki M. A., Pax7 is critical for the normal function of satellite cells in adult skeletal muscle. Proc. Natl. Acad. Sci. U.S.A. 110, 16474–16479 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sambasivan R., et al., Pax7-expressing satellite cells are indispensable for adult skeletal muscle regeneration. Development 138, 3647–3656 (2011). [DOI] [PubMed] [Google Scholar]
- 34.Zammit P. S., et al., Muscle satellite cells adopt divergent fates: A mechanism for self-renewal? J. Cell Biol. 166, 347–357 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.van den Brink S. C., et al., Single-cell sequencing reveals dissociation-induced gene expression in tissue subpopulations. Nat. Methods 14, 935–936 (2017). [DOI] [PubMed] [Google Scholar]
- 36.van Velthoven C. T. J., de Morree A., Egner I. M., Brett J. O., Rando T. A., Transcriptional profiling of quiescent muscle stem cells in vivo. Cell Rep. 21, 1994–2004 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Cornelison D. D. W., Wold B. J., Single-cell analysis of regulatory gene expression in quiescent and activated mouse skeletal muscle satellite cells. Dev. Biol. 191, 270–283 (1997). [DOI] [PubMed] [Google Scholar]
- 38.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gerdes J., et al., Cell cycle analysis of a cell proliferation-associated human nuclear antigen defined by the monoclonal antibody Ki-67. J. Immunol. 133, 1710–1715 (1984). [PubMed] [Google Scholar]
- 40.Coller H. A., What’s taking so long? S-phase entry from quiescence versus proliferation. Nat. Rev. Mol. Cell Biol. 8, 667–670 (2007). [DOI] [PubMed] [Google Scholar]
- 41.González A., Hall M. N., Nutrient sensing and TOR signaling in yeast and mammals. EMBO J. 36, 397–408 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu G. Y., Sabatini D. M., mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liu Q., et al., Characterization of Torin2, an ATP-competitive inhibitor of mTOR, ATM, and ATR. Cancer Res. 73, 2574–2586 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Thoreen C. C., et al., An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Liu B., Chen Y., St Clair D. K., ROS and p53: A versatile partnership. Free Radic. Biol. Med. 44, 1529–1535 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Brugarolas J., et al., Regulation of mTOR function in response to hypoxia by REDD1 and the TSC1/TSC2 tumor suppressor complex. Genes Dev. 18, 2893–2904 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Budanov A. V., Karin M., p53 target genes sestrin1 and sestrin2 connect genotoxic stress and mTOR signaling. Cell 134, 451–460 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Segalés J., et al., Sestrin prevents atrophy of disused and aging muscles by integrating anabolic and catabolic signals. Nat. Commun. 11, 189 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Feng Z., Levine A. J., The regulation of energy metabolism and the IGF-1/mTOR pathways by the p53 protein. Trends Cell Biol. 20, 427–434 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Le Roux I., Konge J., Le Cam L., Flamant P., Tajbakhsh S., Numb is required to prevent p53-dependent senescence following skeletal muscle injury. Nat. Commun. 6, 8528 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang J., et al., PTEN maintains haematopoietic stem cells and acts in lineage choice and leukaemia prevention. Nature 441, 518–522 (2006). [DOI] [PubMed] [Google Scholar]
- 52.Chapman N. M., Chi H., Hallmarks of T-cell exit from quiescence. Cancer Immunol. Res. 6, 502–508 (2018). [DOI] [PubMed] [Google Scholar]
- 53.Flores A., et al., Lactate dehydrogenase activity drives hair follicle stem cell activation. Nat. Cell Biol. 19, 1017–1026 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Tan H., et al., Integrative proteomics and phosphoproteomics profiling reveals dynamic signaling networks and bioenergetics pathways underlying T cell activation. Immunity 46, 488–503 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hall M. N., TOR and paradigm change: Cell growth is controlled. Mol. Biol. Cell 27, 2804–2806 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Shimobayashi M., Hall M. N., Making new contacts: The mTOR network in metabolism and signalling crosstalk. Nat. Rev. Mol. Cell Biol. 15, 155–162 (2014). [DOI] [PubMed] [Google Scholar]
- 57.Gayraud-Morel B., Le Bouteiller M., Commere P.-H., Cohen-Tannoudji M., Tajbakhsh S., Notchless defines a stage-specific requirement for ribosome biogenesis during lineage progression in adult skeletal myogenesis. Development 145, dev162636 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Chen C., et al., TSC-mTOR maintains quiescence and function of hematopoietic stem cells by repressing mitochondrial biogenesis and reactive oxygen species. J. Exp. Med. 205, 2397–2408 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Reczek C. R., Chandel N. S., ROS-dependent signal transduction. Curr. Opin. Cell Biol. 33, 8–13 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Hamanaka R. B., Chandel N. S., Mitochondrial reactive oxygen species regulate cellular signaling and dictate biological outcomes. Trends Biochem. Sci. 35, 505–513 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Alexander A., et al., ATM signals to TSC2 in the cytoplasm to regulate mTORC1 in response to ROS. Proc. Natl. Acad. Sci. U.S.A. 107, 4153–4158 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Mak T. W., et al., Glutathione primes T cell metabolism for inflammation. Immunity 46, 675–689 (2017). [DOI] [PubMed] [Google Scholar]
- 63.García-Prat L., et al., Autophagy maintains stemness by preventing senescence. Nature 529, 37–42 (2016). [DOI] [PubMed] [Google Scholar]
- 64.Alharby E., et al., A homozygous potentially pathogenic variant in the PAXBP1 gene in a large family with global developmental delay and myopathic hypotonia. Clin. Genet. 92, 579–586 (2017). [DOI] [PubMed] [Google Scholar]
- 65.Proskorovski-Ohayon R., et al., PAX7 mutation in a syndrome of failure to thrive, hypotonia, and global neurodevelopmental delay. Hum. Mutat. 38, 1671–1683 (2017). [DOI] [PubMed] [Google Scholar]
- 66.Günther S., et al., Myf5-positive satellite cells contribute to Pax7-dependent long-term maintenance of adult muscle stem cells. Cell Stem Cell 13, 590–601 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Picelli S., et al., Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 68.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 70.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data reported in this article have been deposited in the GEO (accession no. GSE141881). Some study data are available in the supporting information.