Significance
The m7G cap is the canonical RNA cap in eukaryotes, but other noncanonical RNA caps exist, including the NAD+ cap. NAD captureSeq has been widely used to profile NAD+-capped RNAs (NAD-RNAs) in prokaryotes and eukaryotes. However, NAD captureSeq reacts at a low level with m7G-RNAs and introduces copper ions that cause RNA fragmentation, resulting in reduced sensitivity and loss of full-length sequence information. To address these issues, we developed the copper-free SPAAC-NAD-seq, which utilizes the strain-promoted azide–alkyne cycloaddition reaction to capture NAD-RNAs followed by high-throughput sequencing. Compared to NAD captureSeq, SPAAC-NAD-seq is more sensitive and retains full-length sequence information. This optimized technique provides a useful tool to profile NAD-RNAs in prokaryotes and, when combined with m7G-RNA depletion, in eukaryotes.
Keywords: NAD, NAD captureSeq, SPAAC-NAD-seq, NAD-RNA, m7G-RNA
Abstract
Nicotinamide adenine diphosphate (NAD+) is a novel messenger RNA 5′ cap in Escherichia coli, yeast, mammals, and Arabidopsis. Transcriptome-wide identification of NAD+-capped RNAs (NAD-RNAs) was accomplished through NAD captureSeq, which combines chemoenzymatic RNA enrichment with high-throughput sequencing. NAD-RNAs are enzymatically converted to alkyne-RNAs that are then biotinylated using a copper-catalyzed azide–alkyne cycloaddition (CuAAC) reaction. Originally applied to E. coli RNA, which lacks the m7G cap, NAD captureSeq was then applied to eukaryotes without extensive verification of its specificity for NAD-RNAs vs. m7G-capped RNAs (m7G-RNAs). In addition, the Cu2+ ion in the CuAAC reaction causes RNA fragmentation, leading to greatly reduced yield and loss of full-length sequence information. We developed an NAD-RNA capture scheme utilizing the copper-free, strain-promoted azide–alkyne cycloaddition reaction (SPAAC). We examined the specificity of CuAAC and SPAAC reactions toward NAD-RNAs and m7G-RNAs and found that both prefer the former, but also act on the latter. We demonstrated that SPAAC-NAD sequencing (SPAAC-NAD-seq), when combined with immunodepletion of m7G-RNAs, enables NAD-RNA identification with accuracy and sensitivity, leading to the discovery of new NAD-RNA profiles in Arabidopsis. Furthermore, SPAAC-NAD-seq retained full-length sequence information. Therefore, SPAAC-NAD-seq would enable specific and efficient discovery of NAD-RNAs in prokaryotes and, when combined with m7G-RNA depletion, in eukaryotes.
The modification of RNA species contributes to diverse biological processes by regulating gene expression (1). For messenger RNAs (mRNAs) in eukaryotes, one of the most predominant modifications is the 5′ m7G cap. Extensive studies over the past decades have revealed its wide-reaching roles in the life of an mRNA, such as transcript stability, polyadenylation, pre-mRNA splicing, mRNA export, and translation initiation (2, 3). In prokaryotes, it was long believed that RNAs contain a triphosphate at the 5′ end (4). Recently, this view was modified by the detection of nicotinamide adenine dinucleotide (NAD+), a redox cofactor, at the 5′ end of prokaryotic RNAs by high-performance liquid chromatography coupled to mass-spectrometry (HPLC-MS) (5). Being an ATP analog, NAD+ is used by RNA polymerases for incorporation into the RNA 5′ end (6, 7).
While affirming that bacterial RNAs possess an NAD+ cap, LC-MS does not supply sequence information on NAD+-capped RNAs (NAD-RNAs). In 2015, a robust new technique, known as NAD captureSeq, was developed for this purpose (8). Originally applied to Escherichia coli, NAD captureSeq combined chemoenzymatic reactions and next-generation sequencing to identify 44 annotated RNA species as NAD-RNAs, including both mRNAs and noncoding RNAs acting in cellular metabolism and stress responses (8). Following its demonstration in prokaryotes, NAD captureSeq was widely applied to eukaryotes, revealing thousands of NAD-RNAs in the transcriptomes of yeast, mammals, and plants (9–13). The widespread discovery of RNAs with this novel cap opened a new research field dedicated to deciphering the function of the NAD+ cap.
The methodology of NAD captureSeq includes an enzymatic reaction followed by click chemistry to specifically biotinylate NAD-RNAs for subsequent purification. First, adenosine diphosphate-ribosylcyclase (ADPRC) catalyzes a transglycosylation reaction of NAD+ to replace the nicotinamide moiety with an alkynyl alcohol. The alkyne product is further modified by click chemistry-mediated biotinylation through a copper-catalyzed azide–alkyne cycloaddition (CuAAC) reaction (14). For simplicity, we will refer to this original reaction as the CuAAC-NAD reaction. Finally, the biotinylated NAD-RNAs can be captured using streptavidin beads and subjected to high-throughput sequencing (8).
NAD captureSeq has been integral in expanding our knowledge of NAD-RNAs, but still suffers shortcomings. Most notably, the usage of copper ions during the biotinylation step induces RNA degradation that would prohibit the preservation of RNAs of low abundance, which may explain the enrichment of NAD-RNAs in noncoding transcripts (which may have secondary structures that promote stability) (8, 13, 15). Moreover, the fragmentation of RNA in the CuAAC-NAD reaction causes a loss of full-length sequence information, with reads being concentrated toward the 5′ ends of genes (10, 11). This is because the purification of 5′ biotinylated RNAs, which may have lost their 3′ portions due to copper ion-triggered cleavage, precedes library construction. Thus, the establishment of an optimized sequencing technology to avoid RNA fragmentation will pave the way for enhanced understanding of NAD-RNAs.
The strain-promoted azide–alkyne cycloaddition (SPAAC) reaction is an alternative approach for click chemistry that does not require a Cu2+ catalyst (16). This reaction is highly efficient and ultrafast because of the large amount of ring strain in the cyclooctyne (17). Due to the absence of exogenous metal catalysts, this reaction would reduce RNA degradation. ADPRC can catalyze the replacement of nicotinamide by various nucleophiles through transglycosylation (18), providing the possibility of formation of azide-functionalized NAD+. The SPAAC reaction would subsequently label this azide-functionalized NAD+ using cyclooctyne linked to a biotin group. For simplicity, we termed this approach the SPAAC-NAD reaction. The SPAAC-NAD reaction, in theory, may be able to enrich NAD-RNAs without RNA fragmentation to preserve the complete sequence and structural information of NAD-RNAs.
Other than the capability to enrich NAD-RNAs without fragmentation, the specificity of NAD-RNA capture is perhaps the most critical factor in accurate NAD-RNA identification. A plethora of chemical modifications to RNA exists, with ribosomal RNA (rRNA) and transfer RNA (tRNA) containing the largest number of diverse modifications (19). Therefore, nonspecific reactions involving some RNA modifications may occur during CuAAC- or SPAAC-NAD reactions, even without the addition of the ADPRC enzyme (ADPRC−) (11). This can be circumvented by defining significantly enriched transcripts between the ADPRC+ and ADPRC− NAD captureSeq samples as NAD-RNAs. Additionally, NAD+ is not the only substrate that can be catalyzed by ADPRC to produce a cyclic product in the absence of nucleophiles (20), which is a concern for specific NAD-RNA capture. Substrates such as bis-adenine dinucleotide (Ap2A) or bis-hypoxanthine dinucleotide (Hp2H) also show strong affinities for the ADPRC enzyme active site, implying that there may be other purine substrates of ADPRC. In prokaryotes like E. coli, where NAD captureSeq was originally applied, this imperfect specificity was less of a concern because RNAs are largely uncapped; however, in eukaryotes, the m7Gppp cap is the predominant cap structure and it may possess enzymatic and chemical reactivity (21, 22). In vitro assays showed that APDRC does have weak activity toward m7G-capped RNAs (m7G-RNAs) (9).
Here, we present an NAD-RNA profiling method, SPAAC-NAD-seq, which combines the ADPRC reaction with the copper-free SPAAC reaction for more efficient enrichment of intact NAD-RNAs followed by sequencing. We examined the specificities of the CuAAC- and SPAAC-NAD reactions and showed that they both act weakly on m7G-RNAs. Thus, we optimized NAD captureSeq and SPAAC-NAD-seq by incorporating an m7G-RNA depletion step with an m7G antibody. A large number of authentic NAD-RNA–producing genes from Arabidopsis was identified after eliminating competition from m7G-RNAs. In comparison to NAD captureSeq, SPAAC-NAD-seq exhibits high efficiency and no RNA degradation, providing a method to discover low-abundant NAD-RNAs as well as preserving full-length sequence information.
Results
Development of the SPAAC-NAD Reaction.
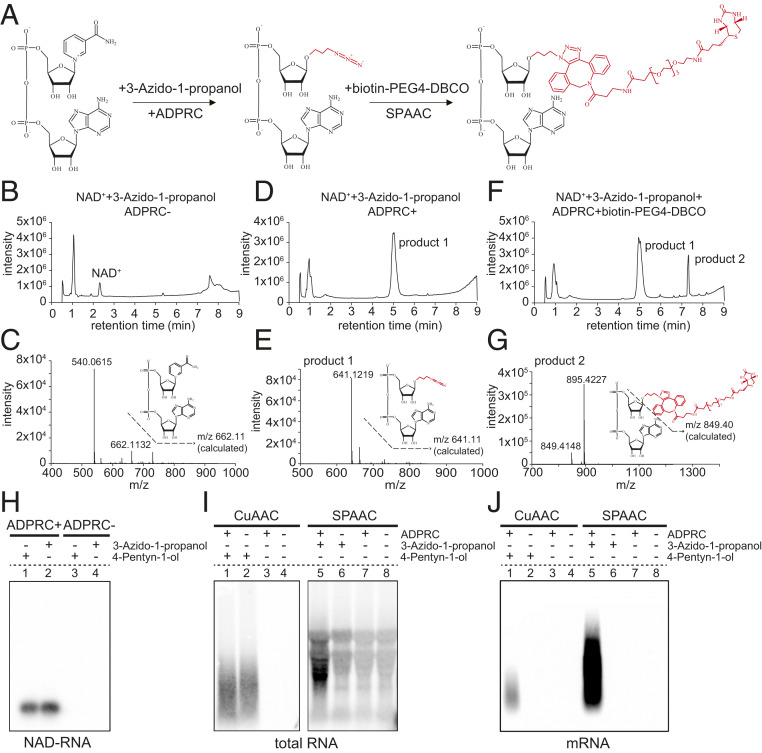
NAD captureSeq is the current method for global identification of NAD-RNAs in cells. Initially applied to E. coli (8), this method involves chemoenzymatic replacement of the nicotinamide moiety of NAD+ with 4-pentyn-1-ol by the ADPRC enzyme. Subsequently, the CuAAC reaction produces a biotinylated product through click chemistry (SI Appendix, Fig. S1 A–G). Unfortunately, the addition of copper ions in the reaction induces degradation of RNA. To circumvent the problem, we evaluated the potential use of the SPAAC reaction to biotinylate NAD-RNA (Fig. 1A). As in NAD captureSeq, the first step is the transglycosylation of NAD+ catalyzed by ADPRC. However, the alkyne is replaced by 3-azido-1-propanol, which is amenable to subsequent biotinylation using copper-free click chemistry. Untargeted HPLC showed that an NAD+ standard was completely converted to a new product in the presence of ADPRC, and MS confirmed that the nicotinamide moiety was replaced by 3-azido-1-propanol (Fig. 1 B–E). Subsequently, the azide moiety of this product reacted with the alkyne moiety of biotin-PEG4-DBCO through a SPAAC reaction, thus producing the final biotinylated NAD+-derived structure, which was confirmed by LC-MS (Fig. 1 F and G). In acknowledgment of the click chemistry reaction, we termed this method the SPAAC-NAD reaction.
Fig. 1.
The establishment and advantage of the SPAAC-NAD reaction. (A) Schematic diagram of the SPAAC-NAD reaction. ADPRC catalyzes the reaction replacing the nicotinamide moiety of NAD+ with 3-azido-1-propanol, followed by SPAAC-based biotinylation with biotin-PEG4-DBCO. (B) HPLC chromatogram of the control reaction in which NAD+ was incubated with 3-azido-1-propanol in the absence of ADPRC. The NAD+ peak is marked. (C) Mass spectrum of the NAD+ peak in B. A compound of mass 662.113 matches NAD+ in mass. (D) HPLC chromatogram of the ADPRC-catalyzed reaction of NAD+ with 3-azido-1-propanol. A product is found compared to B. (E) Mass spectrum of the “product 1” peak in D. A compound of mass 641.121 matches the expected product as diagramed. (F) HPLC chromatogram of the ADPRC-catalyzed reaction of NAD+ with 3-azido-1-propanol followed by SPAAC with biotin-PEG4-DBCO. Product 2 is formed as compared to D. (G) Mass spectrum of the “product 2” peak in F. The compound of mass 849.414 matches the moiety in red in the molecular diagram of product 2. (H–J) Gel blot assays showing NAD-RNA capture. CuAAC- or SPAAC-NAD reactions were performed with an in vitro-transcribed NAD-RNA (H), 100 μg of total Arabidopsis RNAs (I), or 1 μg of Arabidopsis mRNAs (J). RNAs were incubated with/without 3-azido-1-propanol or 4-pentyn-1-ol in the presence or absence of ADPRC. The RNAs were resolved in 2% agarose gel and transferred to a nylon N+ membrane. Biotin-labeled products were probed with streptavidin–horseradish peroxidase, and signals were detected with a chemiluminescent nucleic acid detection module kit.
To further test the feasibility of the SPAAC-NAD reaction to biotinylate NAD-RNAs, we subjected an in vitro-transcribed, 31-nucleotide (nt) NAD-RNA to the CuAAC- or SPAAC-NAD reaction. This was followed by incubation with streptavidin beads and the RNAs eluted from the beads were used to perform a gel blot assay, wherein biotin signal was detected. In the CuAAC- or SPAAC-NAD reaction, samples were processed in the presence or absence of ADPRC, where ADPRC− samples underwent all treatments except addition of the enzyme (Fig. 1H). The results suggested that the SPAAC-NAD reaction can successfully biotinylate NAD-RNA.
To compare the CuAAC- and SPAAC-NAD reactions on RNAs from cells, 100 μg of total RNA from Arabidopsis seedlings were subjected to the CuAAC- or SPAAC-NAD reaction. Consistent with our expectations, NAD-RNA captured by the SPAAC-NAD reaction showed intact RNA bands, while those captured by the CuAAC-NAD reaction were degraded to shorter fragments (Fig. 1I). In striking contrast to the weak smear of CuAAC-treated samples, SPAAC-treated samples showed much stronger biotin signals, implying that this method significantly increases the biotinylation efficiency of NAD-RNAs (Fig. 1I, compare lanes 1 and 5). However, in both methods, ADPRC− samples also produced signals (Fig. 1I, lanes 2 and 6) that probably represented RNAs containing unknown modifications that were able to react during the CuAAC- and SPAAC-NAD reactions, independently of the ADPRC enzyme. Nonspecific signals were also observed in SPAAC-treated samples that did not include 3-azido-1-propanol (Fig. 1I, lanes 7 and 8), suggesting that some unknown modifications in total RNAs can directly react with biotin-PEG4-DBCO used in the SPAAC-NAD reaction (Fig. 1I).
mRNAs usually contain low levels of RNA modifications; therefore, we isolated mRNAs from total RNAs and performed similar assays. The results showed that biotin signals were only detected in ADPRC+ samples (Fig. 1J, lanes 1 and 5), suggesting that the signals in ADPRC− samples from total RNA samples (Fig. 1I) were due to non-mRNA transcripts. Additionally, stronger signals with higher molecular weights were observed in SPAAC-treated samples compared to CuAAC-treated ones (Fig. 1J), suggesting higher biotinylation efficiency and less RNA degradation in the SPACC treatment.
Specificity of the SPAAC-NAD Reaction.
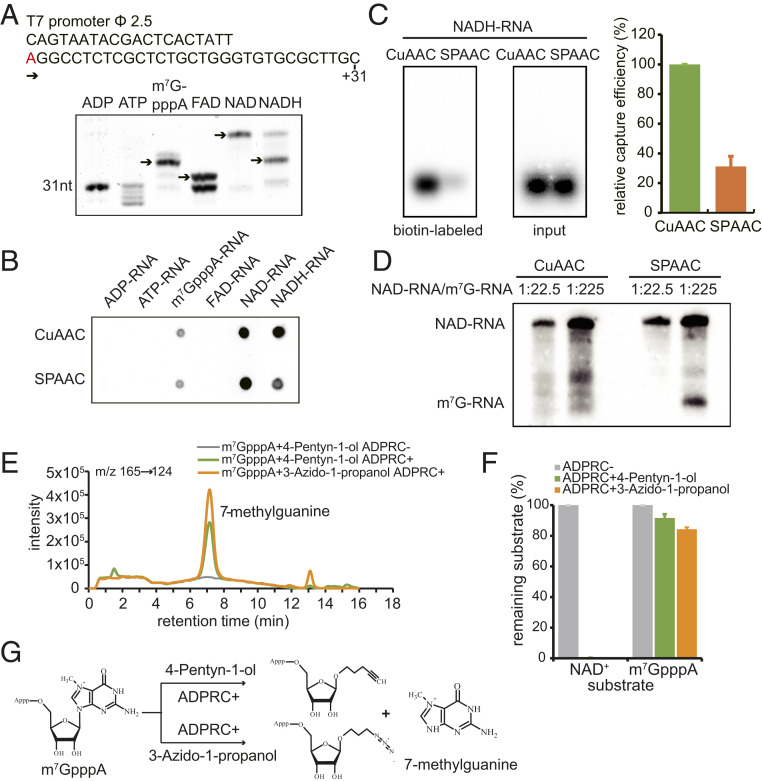
To examine the specificity of the CuAAC- and SPAAC-NAD reactions, RNA species capped with ATP, ADP, m7GpppA, FAD, NAD+, or NADH were tested. The same DNA sequence carrying a T7 promoter was used as a template for in vitro transcription to incorporate these 5′-end nucleotides (Fig. 2A). Boronate affinity electrophoresis was used to distinguish the RNAs with different 5′-end nucleotides (23). The boronic acid in the gel forms a covalent adduct with a vicinal diol on the ribose of each of the modified RNAs (NAD+, m7GpppA, FAD, NADH) causing lower mobility of these capped RNAs compared to the uncapped RNAs (ATP, ADP). Each cap also exhibits a different degree of migration retardation on boronate gels. Our results showed that RNAs with 5′-end caps all presented bands with relative electrophoretic mobilities consistent with those shown in previous studies (Fig. 2A) (24, 25). This suggested that various capped RNAs were successfully obtained for further use in CuAAC- and SPAAC-NAD reactions.
Fig. 2.
The specificity of CuAAC- and SPAAC-NAD reactions. (A) In vitro transcription with ADP, ATP, m7GpppA, FAD, NAD+, or NADH as the initiating nucleotide. (Top) Sequence of the DNA template with the T7 promoter. The “A” in red is the transcription start site. (Bottom) Boronate affinity electrophoresis of each in vitro-transcribed RNA with different 5′ end caps. The arrows indicate the properly capped species. (B) A dot blot assay to detect biotinylated RNAs after the in vitro-transcribed, 5′-end capped RNAs in A underwent CuAAC- or SPAAC-NAD reactions. The same amount of RNA was directly dotted onto a nylon N+ membrane and probed with streptavidin–horseradish peroxidase, and biotin signals were detected with the chemiluminescent nucleic acid detection module kit. (C) A gel blot assay of an in vitro-transcribed NADH-RNA after CuAAC- or SPAAC-NAD reactions. Biotin signals were detected as in B, and input RNA was detected with a radioactive probe. The intensities of bands in the gels were measured using ImageJ. The bar plot on the Right shows the relative capture efficiency of SPAAC- and CuAAC-NAD reactions as defined by the ratio between biotin signal intensity and input RNA level (with the CuAAC efficiency set as 1). Error bars represent SE from three independent replicates. (D) Comparison of reactivity between an NAD-RNA and an m7GpppA-RNA in CuAAC- or SPAAC-NAD reactions. The reactions were performed on mixtures of in vitro-transcribed NAD- and m7GpppA-RNAs at molar ratios 1:22.5 and 1:225. The RNAs were then resolved in a 12% PAGE gel and transferred to a nylon N+ membrane. Biotin signals were detected as in B. The lengths of the NAD-RNA and the m7GpppA-RNA were 70 and 31 nt, respectively. (E) HPLC ion chromatogram and MS/MS transitions (m/z 165→124) of the 7-methylguanine produced in the ADPRC-catalyzed reaction between m7GpppA and 3-azido-1-propanol or 4-pentyn-1-ol. Reaction of m7GpppA and 4-pentyn-1-ol without ADPRC was used as a control. (F) Substrates (m7GpppA or NAD+) remaining after the reaction with 3-azido-1-propanol or 4-pentyn-1-ol catalyzed by ADPRC. The percentage was calculated by the peak area of m7GpppA or NAD+ after the ADPRC reaction relative to the peak area in the ADPRC− control. The error bars represent SE calculated from three independent experiments. (G) Schematic diagram showing the proposed products of ADPRC-catalyzed transglycosylation with m7GpppA and 3-azido-1-propanol or 4-pentyn-1-ol.
According to the cap incorporation efficiency as determined by boronate affinity electrophoresis, the same amounts of each in vitro transcribed RNA were processed for CuAAC- or SPAAC-NAD reactions. A dot blot assay was used to detect biotinylated RNAs. Unfortunately, besides NAD-RNAs, both CuAAC- and SPAAC-NAD reactions also displayed the ability to react with m7GpppA- and NADH-RNAs (Fig. 2B), but the SPAAC-NAD reaction displayed only 30% of the NADH-RNA biotinylation efficiency when compared to the CuAAC-NAD reaction (Fig. 2C). Untargeted LC-MS analysis showed that NADH was catalyzed by ADPRC to produce the same transglycosylation product as NAD+ (SI Appendix, Fig. S2 A–G). Since NADH is the reduced form of, but otherwise shares a similar structure with, NAD+, it is not easy to differentiate NAD+- and NADH-capped RNAs. Thus, NAD-RNAs identified by CuAAC- or SPAAC-NAD reactions should actually be NAD(H)-RNAs.
Besides NADH-RNAs, m7G-RNAs also reacted in both methods (Fig. 2B). Although the signals were much weaker than for NAD-RNAs, the abundance of m7G-RNAs in vivo is much higher than that of NAD-RNAs. To better mimic the ratio of NAD-RNAs to m7G-RNAs in cells and compare the selectivity of each by both methods, an in vitro-transcribed, 31-nt m7G-RNA was mixed with a 70-nt NAD-RNA at molar ratios of 22.5:1 or 225:1. Subsequently, the CuAAC- or SPAAC-NAD reaction was performed. The gel blot revealed that the SPAAC-NAD reaction indeed exhibited activity on the m7G-RNA when it was at 225-fold the concentration of the NAD-RNA (Fig. 2D). The CuAAC reaction may have also acted on the m7G-RNA, but the RNA degradation activity precluded unequivocal identification of the m7G-RNA. We deduced that ADPRC may catalyze the same transglycosylation reaction with m7GpppA as with NAD+, allowing for nucleophilic attack of the same bond linking the base to the ribose in both m7GpppA and NAD+ (Fig. 2G). To test our hypothesis, the first step of the CuAAC- and SPAAC-NAD reactions was performed on m7GpppA and products were analyzed by targeted LC-MS. HPLC showed a new peak identified as 7-methylguanine in the ADPRC+ samples (Fig. 2E). This revealed that 7-methylguanine was replaced by 4-pentyn-1-ol or 3-azido-1-propanol in the presence of ADPRC (Fig. 2G). Only 8% and 16% of the m7GpppA substrate were consumed by CuAAC- and SPAAC-NAD reactions, respectively, while under the same conditions, NAD+ was completely consumed (Fig. 2F). Despite the lower m7GpppA reactivity, our findings suggest that m7G-RNAs likely produce false NAD-RNA signals in CuAAC and SPAAC-NAD reactions due to their high abundance in vivo.
Depletion of m7G-RNAs.
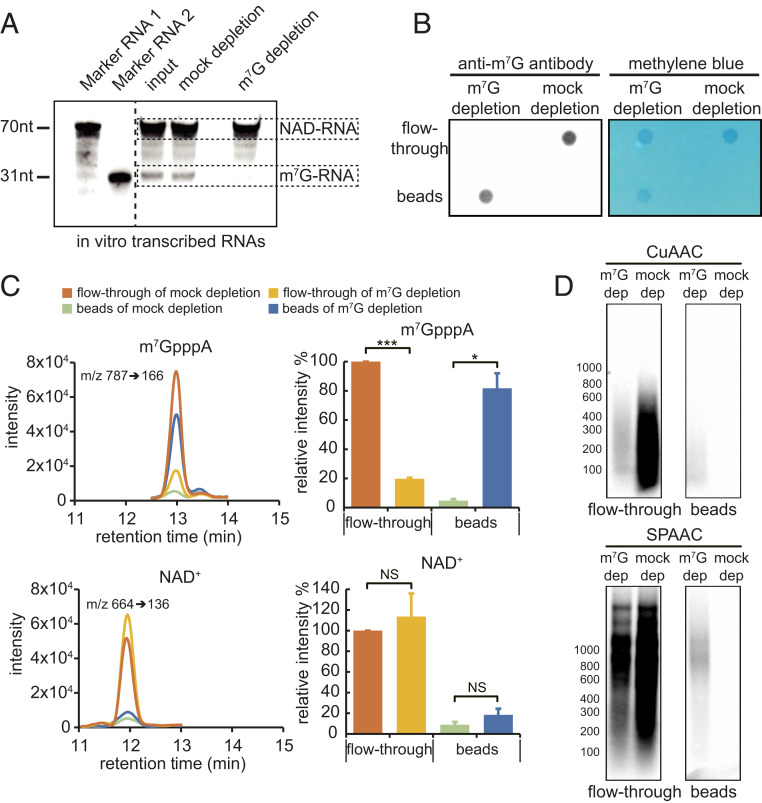
One strategy to remove m7G-RNAs is immunodepletion. We first examined the specificity of the anti-m7G antibody toward m7G- vs. NAD-RNAs. The antibody recognized an in vitro-transcribed m7G-RNA but not NAD-RNA (SI Appendix, Fig. S2H). Next, a mixture of in vitro-transcribed m7G- and NAD-RNAs at a molar ratio of 225:1 was treated without or with an anti-m7G antibody for immunodepletion, and the remaining RNA was subjected to the SPAAC reaction followed by gel blotting to detect biotinylated RNA. The results showed that biotin signals from the m7G-RNA were removed in the sample treated with the anti-m7G antibody, while NAD-RNA biotin signals were not affected (Fig. 3A). Next, we performed immunodepletion of m7G-RNAs using plant mRNAs. Poly(A) RNAs were either mock-treated or treated with the anti-m7G antibody, and RNAs in the flow-through or pulled down by the anti-m7G antibody on beads were isolated and then dot-blotted onto membrane. Methylene blue staining showed the presence of RNA in the flow-through in both treatments but only on beads in the anti-m7G treatment (Fig. 3B). Indeed, probing the blot with anti-m7G antibody showed that the RNAs on beads were m7G-RNAs (Fig. 3B). The lack of signal in the flow-through suggested that m7G-RNAs were almost completely removed by immunodepletion (Fig. 3B). To further confirm this, RNAs recovered from the flow-through and beads in both samples were subjected to nuclease P1 digestion followed by LC-MS detection. m7GpppA and NAD+ released from the RNAs by nuclease P1 were used as the indicator for m7G-RNAs and NAD-RNAs, respectively. Compared to the mock-depletion sample, m7GpppA levels decreased in the flow-through by more than 80% in the m7G-depletion sample (Fig. 3C). NAD+ levels in the flow-through were not significantly changed between mock-depletion and m7G-depletion samples (Fig. 3C). Therefore, after immunodepletion, the ratio of NAD-RNAs and m7G-RNAs was fivefold to sixfold that of the original ratio (Fig. 3C). Subsequently, RNAs from the flow-through were subjected to the CuAAC- or SPAAC-NAD reaction. A gel blot showed that signals from biotinylated products in the mock samples of both CuAAC- and SPAAC-NAD reactions were significantly stronger than those seen in the m7G-depletion samples, indicating that most of the signals in the mock samples were due to m7G-RNAs (Fig. 3D). RNAs from beads were also similarly examined. As expected, no biotinylated products were found in the mock samples, indicating that NAD-RNAs were not retained on beads (Fig. 3D). The weak signals found in the m7G-depletion samples could be due to reactivity of m7G-RNAs. Collectively, these data demonstrated that the abundance of NAD-RNAs in eukaryotic cells might be overestimated by the CuAAC- or SPAAC-NAD reaction.
Fig. 3.
CUAAC- and SPAAC-NAD reactions after m7G-RNA depletion. (A) A gel blot assay to evaluate the efficacy of m7G-RNA depletion. A mixture of in vitro-transcribed NAD-RNA (1 μg) and m7G-RNA (100 μg) (molar ratio, 1:225) was incubated with (m7G-depletion sample) or without the m7G antibody (mock-depletion sample). Protein G beads were then used to remove the m7G-RNA. Afterward, RNAs were subjected to the SPAAC-NAD reaction and biotin signals were detected with the chemiluminescent nucleic acid detection module kit after probing with streptavidin–horseradish peroxidase. The left two lanes (marker RNA 1 and marker RNA 2) are 70- and 31-nt NAD-RNAs after the SPAAC-NAD reaction; they are used as size markers. The input sample did not undergo mock or m7G-RNA depletion. (B) A dot blot assay demonstrating that m7G-RNAs in mRNAs were largely removed. Arabidopsis mRNA samples were treated with (m7G-depletion sample) or without the m7G antibody (mock-depletion sample). Protein G beads were then applied to specifically capture the m7G antibody. The mRNAs collected from the flow-through and beads were spotted onto a membrane and probed with the anti-m7G antibody or stained with methylene blue. (C) LC-MS analysis demonstrating that m7G-RNAs were largely removed from mRNAs but NAD-RNAs were not affected by immunodepletion. mRNA samples after m7G antibody treatment were subjected to nuclease P1 digestion and analyzed by HPLC-MS. Targeted detection of m7GpppA and NAD+ released by nuclease P1 indicates the amounts of m7G-RNAs and NAD-RNA, respectively. The bar plots show relative m7GpppA or NAD+ levels as calculated by the peak area among the samples. The error bars represent SE calculated from three independent experiments (two-tailed Student’s t test: *P < 0.05; ***P < 0.001; NS, not significant). (D) A gel blot assay of mRNAs that were pretreated with the anti-m7G antibody followed by CuAAC or SPAAC-NAD reactions. The anti-m7G antibody treatment and biotin signal detection were performed similarly to A. m7G dep, m7G depletion; mock dep, mock depletion.
Depletion of m7G-RNAs Promotes the Identification of Authentic, Low-Abundant NAD-RNAs.
Given that m7G-RNAs can be captured by both CuAAC- and SPAAC-NAD reactions, leading to false biotin signals during identification of NAD-RNAs, we exploited updated NAD-RNA sequencing approaches based on the original NAD captureSeq (8) and compared their applications in Arabidopsis. To avoid the nonspecific clickable reaction products in total RNAs (Fig. 1I), poly(A) RNAs, which displayed specific signals only in ADPRC+ samples (Fig. 1J), were used. To begin, anti-m7G antibody treatment was applied to poly(A) RNAs to remove m7G-RNAs. Samples treated with the anti-m7G antibody are referred to as the m7G-depletion samples. Subsequently, the CuAAC- or SPAAC-NAD reaction was performed. The biotinylated RNAs were captured by streptavidin beads, eluted, and subjected to RNA sequencing. Random primers were used for reverse transcription as copper-induced fragmentation of RNA in the CuAAC-NAD reaction would result in the loss of the poly(A) tail. We refer to the two approaches as CuAAC-NAD-seq, which is similar to NAD captureSeq, and SPAAC-NAD-seq hereafter.
In our previous study, ADPRC− samples, in which RNAs were not given the ADPRC enzyme, were deemed to be the background control for identifying NAD-RNAs (11). The transcripts in the ADPRC− samples represented nonspecific attachment to the beads. Due to their very low abundance and possible unequal representation of the transcriptome, transcripts in the ADPRC− samples are not ideal controls. Thus, we instead performed mRNA sequencing (mRNA-seq) on mock-depletion samples to reflect mRNA abundance, against which enriched NAD-RNAs were identified (Fig. 4A). Three biological replicates of m7G- and mock-depletion samples, both subjected to CuAAC- or SPAAC-NAD-seq, were performed (SI Appendix, Fig. S3A). The resulting libraries (mRNA-seq, CuAAC-NAD-seq, SPAAC-NAD-seq) each provided a minimum of ∼11 to 20 million uniquely mapped reads. Hierarchical clustering and principal component analysis revealed that the biological replicates of each library type clustered together, indicating the high quality and reproducibility of the sequencing data. The distance between the CuAAC- and SPAAC-NAD-seq of m7G-depletion samples implied that the identified NAD-RNAs differed by method (SI Appendix, Fig. S3 B and C).
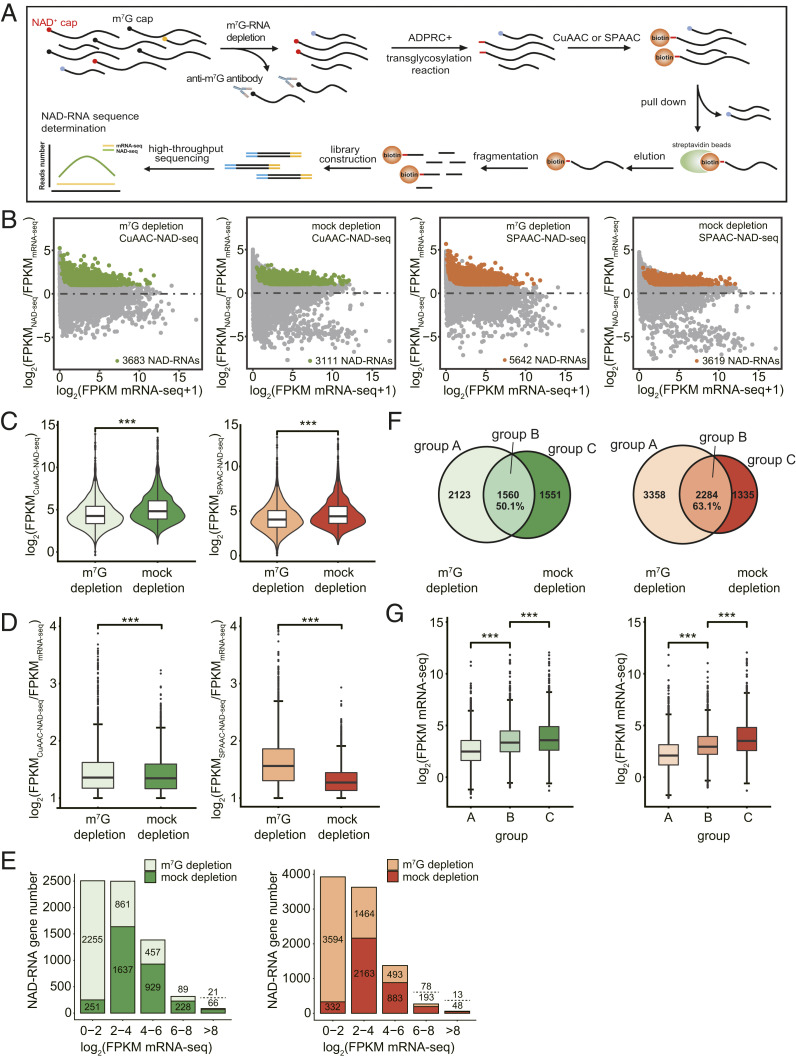
Fig. 4.
m7G-RNA depletion promotes the identification of low-abundant, authentic NAD-RNAs. (A) Schematic workflow of CuAAC- or SPAAC-NAD-seq. mRNAs were isolated and treated by the anti-m7G antibody for m7G-RNA depletion. CuAAC- or SPAAC-NAD reactions were subsequently performed to isolate NAD-RNAs. NAD-RNA libraries were constructed for high-throughput sequencing. NAD-RNAs were defined as ones with a twofold change in FPKM and adjusted P value < 0.05 in NAD-seq compared to mRNA-seq. (B) Scatter plots showing NAD-RNAs identified by CuAAC- and SPAAC-NAD-seq in m7G-depletion and mock-depletion samples. The log2 FPKM ratio between NAD-seq samples and mRNA-seq samples is plotted against the log2 FPKM of mRNA-seq. The green and orange dots represent the NAD-RNAs identified by CuAAC-NAD-seq or SPAAC-NAD-seq, respectively. (C) Violin plots showing the distribution of abundance (log2 FPKM) for all identified NAD-RNAs in m7G-depletion and mock-depletion samples by CuAAC-NAD-seq or SPAAC-NAD-seq. The center line in the “box” shows the median value. ***P < 0.001 by two-tailed Student’s t test. (D) The levels (log2[FPKMNAD-seq/FPKMmRNA-seq]) of all identified NAD-RNAs in m7G-depletion and mock-depletion samples by CuAAC-NAD-seq or SPAAC-NAD-seq. The center line in the “box” shows the median value. ***P < 0.001 by two-tailed Student’s t test. (E) The number of NAD-RNA genes identified in m7G-depletion and mock-depletion samples by CuAAC-NAD-seq (left plot) or SPAAC-NAD-seq (right plot). Genes were separated into five groups based on their mRNA-seq abundance (log2 FPKM). Numbers above and below the dotted line represent the numbers of NAD-RNA genes identified in the m7G-depletion and mock-depletion samples, respectively. (F) The overlap of NAD-RNAs in the m7G-depletion and mock-depletion samples as identified by CuAAC-NAD-seq (Venn diagram on the Left) or SPAAC-NAD-seq (Venn diagram on the Right). Numbers represent the number of NAD-RNAs in each group. The percentage is derived from the number of overlapped NAD-RNAs divided by the total number of NAD-RNAs in the mock-depletion sample. (G) The mRNA abundance (log2 FPKM mRNA-seq) for all identified NAD-RNA genes in each group (A, B, C) in F. The center line in the “box” shows the median value. ***P < 0.001 by two-tailed Student’s t test. The plots on the Left and Right represent results from CuAAC- and SPAAC-NAD-seq, respectively.
We then identified NAD-RNAs using fold change in NAD-seq vs. mRNA-seq ≥ 2 and adjusted P value < 0.05 as the criteria. In total, 3,683 and 3,111 NAD-RNAs were identified by CuAAC-NAD-seq in the m7G-depletion and mock-depletion samples, respectively. In total, 5,642 and 3,619 NAD-RNAs were identified by SPAAC-NAD-seq in the m7G-depletion and mock-depletion samples, respectively (Fig. 4B). Thus, after removing m7G-RNAs, more NAD-RNAs were discovered by both CuAAC- and SPAAC-NAD-seq. It is plausible that NAD-RNAs were captured more efficiently without competition from m7G-RNAs. The abundance (log2 fragments per kilobase of transcript per million mapped reads [FPKM]) of all identified NAD-RNAs in the mock-depletion sample was significantly higher than that in the m7G-depletion sample (Fig. 4C), which was consistent with our previous gel blot results (Fig. 3D) and was likely due to the extra biotinylated products stemming from m7G-RNAs in both the CuAAC- and SPAAC-NAD reactions. In striking contrast to the abundance levels, the FPKM fold change in NAD-seq vs. mRNA-seq, which represented the enrichment level of NAD-RNAs, was significantly lower in the mock-depletion samples than in the m7G-depletion samples (Fig. 4D). This suggested that not only were there more NAD-RNA species identified, but also that the levels of NAD-RNAs in the datasets were elevated after m7G-RNA depletion. Note that SPAAC-NAD-seq showed a much larger difference in the enrichment ratio between the m7G-depletion and mock-depletion samples than CuAAC-NAD-seq. This is consistent with our earlier assay that showed that in vitro-transcribed m7G-RNAs were more prone to biotinylation by the SPAAC-NAD reaction (Fig. 2D).
To understand whether there is a correlation between mRNA abundance and enrichment of NAD-RNAs, we grouped all NAD-RNAs based on total transcript abundance as determined by mRNA-seq. In m7G-depletion samples, the majority of NAD-RNAs belonged to the group of lowest abundance (log2 FPKM mRNA-seq < 2), whereas NAD-RNAs in mock-depletion samples tended to come from more abundant mRNAs (Fig. 4E). The substantial difference in this classification implicated that many of the low-abundant NAD-RNAs were overlooked in the mock-depletion samples. Therefore, when CuAAC- or SPAAC-NAD reactions were performed on a transcript pool that was free of m7G-RNAs, low-abundant NAD-RNAs may have acquired a higher chance to be captured and identified.
Specifically, in the m7G- and mock-depletion CuAAC-NAD-seq datasets, a total of 1,560 identified NAD-RNAs overlapped, which is represented by group B in Fig. 4F (Left). This suggests that, in the mock-depletion sample, nearly 50% of the identified NAD-RNAs were probably m7G-RNAs, as shown by group C. For NAD-RNAs identified in the m7G-depletion sample, 2,123 were novel (Fig. 4 F, Left). Consistent with our previous inference, NAD-RNAs that were only identified in the m7G-depletion samples, as seen in group A, exhibited significantly lower mRNA abundance, while m7G-RNAs in group C showed the highest mRNA levels (Fig. 4 G, Left). Similar results were observed in the SPAAC-NAD-seq data (Fig. 4 F and G, Right). These findings further underscore that m7G depletion promotes the identification of low-abundant and authentic NAD-RNAs.
SPAAC-NAD-Seq Preserves Full-Length NAD-RNAs.
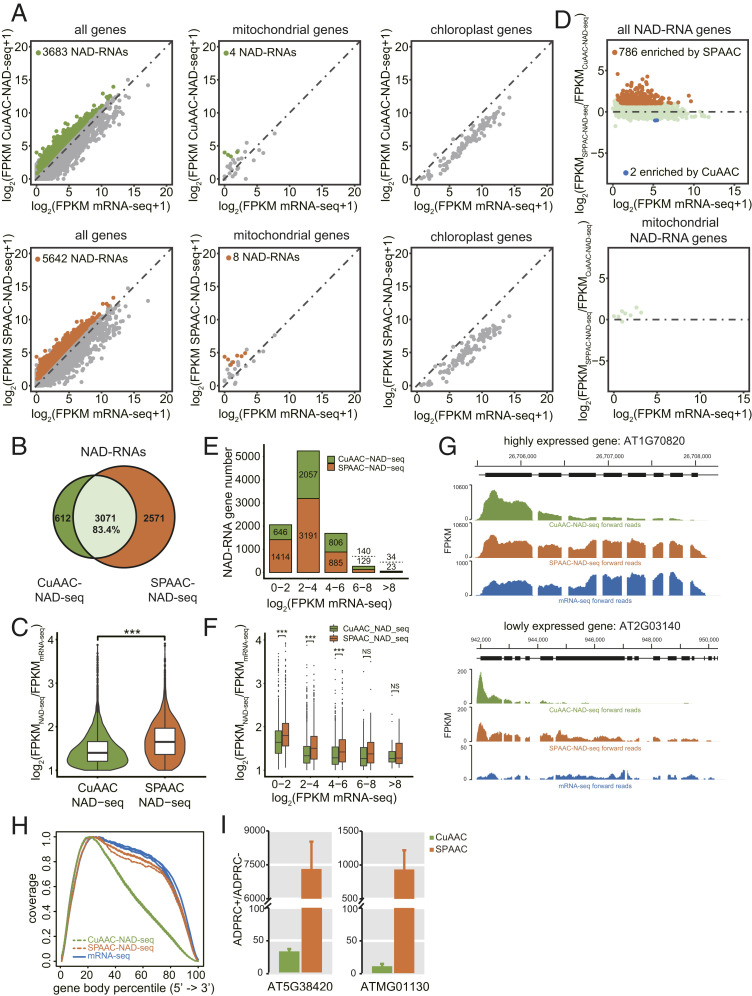
Gel blot assays with total RNA and mRNA both suggested that the SPAAC-NAD reaction enables the biotinylation of more NAD-RNAs than the CuAAC-NAD reaction (Fig. 1 I and J). Thus, to comprehensively compare both methods on a genomic scale, we analyzed the datasets from SPAAC- and CuAAC-NAD-seq of m7G-depletion samples. SPAAC-NAD-seq identified 5,642 NAD-RNAs, which was much more than CuAAC-NAD-seq, which only identified 3,683 NAD-RNAs (Fig. 5A). Additionally, SPAAC-NAD-seq identified 83.4% of the NAD-RNAs found by CuAAC-NAD-seq (Fig. 5B). In total, 2,571 additional NAD-RNAs were specifically discovered by SPAAC-NAD-seq (Fig. 5B). For the 3,071 NAD-RNAs identified by both methods, SPAAC-NAD-seq showed higher enrichment levels than CuAAC-NAD-seq (Fig. 5C). For reads that uniquely mapped to the mitochondrial genome (note that these reads did not map to the mitochondrial sequences that are present in the nuclear genome), eight NAD-RNAs were identified by SPAAC-NAD-seq, while four were identified by CuAAC-NAD-seq (Fig. 5A). Consistent with our previous findings (11), no chloroplast genes produced NAD-RNAs as determined by either CuAAC- or SPAAC-NAD-seq (Fig. 5A). Note that poly(A) RNAs were used as the starting material, which does not survey the entire mitochondrial or chloroplast transcriptomes as most transcripts in these organelles are expected to be nonpolyadenylated. When CuAAC- and SPAAC-NAD-seq were compared on all NAD-RNA genes, 786 NAD-RNAs showed significantly increased abundance in SPAAC-NAD-seq vs. CuAAC-NAD-seq (fold change ≥ 2 and adjusted P value < 0.05), while only 2 NAD-RNAs decreased in abundance significantly (Fig. 5D).
Fig. 5.
Profiling of NAD-RNAs by CuAAC and SPAAC-NAD-seq. (A) Scatter plots comparing log2 FPKM values between NAD-seq and mRNA-seq in m7G-depletion samples for all genes, mitochondrial genes, and chloroplast genes. The green and orange dots represent NAD-RNAs identified by CuAAC-NAD-seq and SPAAC-NAD-seq, respectively. (B) Scatter plots showing the relative levels of NAD-RNAs as determined by CuAAC-NAD-seq and SPAAC-NAD-seq. The log2 FPKM ratio between SPAAC-NAD-seq and CuAAC-NAD-seq for all identified NAD-RNAs was plotted against the log2 FPKM of mRNA-seq. The orange dots represent the NAD-RNAs enriched (fold change ≥ 2; adjusted P value < 0.05) in SPAAC-NAD-seq, and blue dots represent the NAD-RNAs enriched (fold change less than −2; adjusted P value < 0.05) in CuAAC-NAD-seq. (C) The overlap of NAD-RNAs identified by CuAAC-NAD-seq and SPAAC-NAD-seq in m7G-depletion samples. Numbers represent the numbers of NAD-RNAs. The percentage is derived from the number of overlapped NAD-RNAs divided by the total number of NAD-RNAs identified by CuAAC-NAD-seq. (D) Violin plots showing the levels (log2[FPKMNAD-seq/FPKMmRNA-seq]) of common NAD-RNAs identified by CuAAC-NAD-seq and SPAAC-NAD-seq in m7G-depletion samples. The center line in the “box” shows the median value. ***P value < 0.001 by two-tailed Student’s t test. (E) The numbers of NAD-RNA genes identified by CuAAC-NAD-seq or SPAAC-NAD-seq in m7G-depletion samples. Genes were separated into five groups based on their mRNA-seq abundance (log2 FPKM). Numbers above and below the dotted lines represent NAD-RNAs identified by CuAAC-NAD-seq and SPAAC-NAD-seq, respectively. (F) The levels (log2 [FPKMNAD-seq/FPKMmRNA-seq]) for all identified NAD-RNAs by CuAAC-NAD-seq or SPAAC-NAD-seq in m7G-depletion samples. Genes were separated into five groups based on their mRNA-seq abundance (log2 FPKM). The center line in the “box” shows the median value. NS denotes no significance; ***P < 0.001 by two-tailed Student’s t test. (G) Genome browser views of aligned reads from CuAAC-NAD-seq and SPAAC-NAD-seq in m7G-depletion samples and from mRNA-seq for two genes. Both genes were identified as NAD-RNAs by SPAAC-NAD-seq, but as non-NAD-RNAs by CuAAC-NAD-seq. The coding sequence is displayed as black bars. (H) Read distribution along gene bodies in CuAAC-NAD-seq, SPAAC-NAD-seq, and mRNA-seq in mock-depletion samples. Three lines of the same color represent biological replicates. (I) Quantification of transcript levels by qRT-PCR in ADPRC+ vs. ADPRC− samples after CuAAC or SPAAC-NAD reactions. The NAD-RNAs after both reactions were eluted from streptavidin beads, reverse transcribed by oligo(dT) primers, and then detected by PCR with gene-specific primers. Data are normalized to the ADPRC− control set as 1. Error bars represent SE. Three technical replicates from each of two biological replicates were performed.
To test whether both methods displayed different enrichment preferences for mRNAs with different abundances, we classified NAD-RNAs into five groups according to total mRNA levels and observed that, for low-abundant mRNAs, there was an increase in the NAD-RNAs identified by SPAAC-NAD-seq relative to CuAAC-NAD-seq (Fig. 5E). Indeed, for low-abundant mRNAs (log2 FPKM mRNA-seq < 6), the enrichment in SPAAC-NAD-seq was statistically significant (Fig. 5F). Collectively, these results demonstrate that SPAAC-NAD-seq possesses higher NAD-RNA enrichment efficiency. Also, it is capable of uncovering more NAD-RNAs that are otherwise overlooked.
To further elucidate the cause for the NAD-RNA enrichment difference between CuAAC- and SPAAC-NAD-seq, we analyzed the read distribution of two NAD-RNAs that were specifically identified by SPAAC-NAD-seq but deemed non-NAD-RNAs by CuAAC-NAD-seq (Fig. 5G). Relative to mRNA-seq, the reads were greatly enriched both in SPAAC- and CuAAC-NAD-seq, but were enriched in different regions. The former showed reads distributed along gene bodies, while the latter only showed read enrichment at the 5′ end. We then analyzed the global read distribution along gene bodies in each sample. It was apparent that a higher proportion of reads was enriched near the 5′ ends of genes in CuAAC-NAD-seq, while a near-uniform distribution along gene bodies was found in SPAAC-NAD-seq and mRNA-seq (Fig. 5H). This is likely due to RNA fragmentation caused by copper ions in the CuAAC reaction together with 5′-end capture. Consequently, the loss of read coverage may result in a low abundance value that precludes the identification of NAD-RNAs. To validate this hypothesis, qRT-PCR was performed for two NAD-RNA genes that were identified by both sequencing methods. The cDNA was reverse transcribed using an oligo(dT) primer. The SPAAC-NAD reaction showed a much higher enrichment ratio than the CuAAC-NAD reaction for the NAD-RNA genes (Fig. 5I). This is consistent with the notion that most of the NAD-RNAs were not polyadenylated after copper-induced cleavage. Therefore, to avoid underestimating the levels of NAD-RNAs, it is beneficial to use the copper-free SPAAC-NAD-seq to reveal full NAD-RNA profiles in cells.
Finally, to explore the biological processes that NAD-RNAs are involved in, we performed a Gene Ontology (GO) term analysis for 1,000 genes with FPKM mRNA-seq ≥ 5 and the highest FPKM fold change of NAD-seq vs. mRNA-seq. The highly enriched terms determined in the mock-depletion CuAAC-NAD-seq was consistent with our previous study (11, 12) and were mainly centered on the response to oxidative stress and cytokinin, or other processes such as photosynthesis (SI Appendix, Fig. S4). On the other hand, in the m7G-depletion CuAAC-NAD-seq, the terms with the highest enrichment differed from those in the mock-depletion samples. This suggests that some of the biological processes we previously detected may have been due to m7G-RNAs. After collecting more NAD-RNA genes through m7G-depletion SPAAC-NAD-seq, we found previously unidentified terms such as intracellular protein transport, starch biosynthetic processes, or responses to cadmium ion and bacterium defense (SI Appendix, Fig. S4). In conclusion, the SPAAC-NAD-seq on m7G-RNA-depleted mRNAs identified more NAD-RNA genes as compared to CuAAC-NAD-seq, supporting that this is a robust strategy for studies of NAD-RNAs.
Discussion
NAD+ is broadly present in diverse organisms and can serve as a noncanonical 5′-RNA cap (26). In Arabidopsis, thousands of NAD-RNAs were recently identified using NAD captureSeq that was originally developed for E. coli (11). This approach has also been used to identify NAD-RNAs in other eukaryotic species (9–11, 27). Based on the same enzymatic principle, NAD tagSeq appends an RNA tag to NAD-RNAs to enable single-molecule RNA sequencing (12). However, RNA degradation induced by copper ions is a serious issue for both NAD captureSeq and NAD tagSeq. CapZyme-seq, which combines enzymatic decapping of noncanonically capped 5′ ends with high-throughput sequencing, was developed to precisely determine the 5′ ends of RNAs with noncanonical caps (28). However, this method is unable to distinguish the NAD+ cap from other noncanonical caps. In this study, we developed SPAAC-NAD-seq, which in combination with m7G-RNA depletion, allows for sensitive and accurate identification of NAD-RNAs while retaining full-length sequence information.
The SPAAC-NAD reaction is based on the ADPRC-catalyzed transglycosylation in NAD captureSeq, but an azide replaces the alkyne in the ADPRC reaction to enable the subsequent SPAAC reaction in place of the CuAAC reaction (Fig. 1A). The dibenzocyclooctyne used in SPAAC can react with azide-functionalized compounds without the need for a copper catalyst, resulting in a stable triazole linkage. Therefore, the entire click chemistry reaction proceeds at a lower activation barrier and is not harmful to nucleic acids because of the absence of metal ions (14). In previous studies, most E. coli NAD-RNAs enriched by NAD captureSeq were mainly small RNAs (8). Also, in yeast, NAD captureSeq hardly displayed any full-length NAD-RNAs, and it was postulated that NAD-RNAs might be subjected to accelerated degradation at the 3′ end (13). However, the copper ions in the CuAAC-NAD reaction cause random RNA cleavage, leading to subsequent enrichment of 5′ RNA fragments after affinity purification, irrespective of the nature of the 3′ ends (29). Therefore, it was uncertain whether NAD-RNAs are actually 3′-truncated in vivo. Using SPAAC-NAD-seq, we observed that enriched reads were uniformly distributed along gene bodies (Fig. 5H). This is different from the CuAAC-NAD-seq pattern, where reads were preferentially mapped to the 5′ ends of genes (Fig. 5H), as reported previously (10, 11, 13). Thus, NAD-RNAs are full-length RNAs. The abundance of NAD-RNAs enriched in SPAAC-NAD-seq was significantly higher than in CuAAC-NAD-seq, probably for the following reasons. First, read abundance is higher because of uniform coverage along gene bodies. Furthermore, PEG4 in the SPAAC reaction has a large spacer that makes the compound more hydrophilic and allows for a greater distance between the reaction site with the NAD-azide and the biotin group (30). These features probably endow the SPAAC-NAD reaction a higher capture efficiency, leading to the identification of more NAD-RNAs that were overlooked previously. In the companion paper (31), we profiled NAD-RNAs from E. coli using NAD tagSeq II, a SPAAC-based, single-molecule NAD-RNA sequencing approach, and identified hundreds of NAD-RNAs including mRNAs with complete open reading frames.
The biological processes that the gene products of NAD-RNAs may be involved in differ between the mock-depletion and m7G-depletion samples generated through both CuAAC- and SPAAC-NAD-seq (SI Appendix, Fig. S4 and Datasets S6–S9). NAD captureSeq in Arabidopsis revealed GO terms such as photosynthesis and responses to oxidative stress or cytokinin (11, 12). This is highly consistent with our results in the mock-depletion samples by CuAAC-NAD-seq. In the m7G-depletion samples, these processes were not present or were present with less significance. It is probable that such GO terms were false due to contamination by m7G-RNAs. When performing SPAAC- or CuAAC-NAD-seq with m7G-depleted mRNAs, we found GO terms such as starch biosynthetic processes, intracellular protein transport, and responses to cadmium ions and bacteria. The information may guide future studies to understand the biological functions of NAD-RNAs. Notably, “response to water deprivation” and “response to ABA” are GO terms identified by SPAAC-NAD-seq (Datasets S7 and S9). This is consistent with a recent study showing that the hormone ABA remodels the NAD-RNA transcriptome (32).
SPAAC-NAD-seq will aid future studies on NAD-RNA metabolism and molecular functions. The RNA NAD+ cap can be removed by at least two classes of decapping enzymes, Nudix family proteins present in both prokaryotes and eukaryotes (8, 33–35) and DXO proteins found only in eukaryotes (9, 33, 36, 37). NAD-RNAs transfected into human cells and Arabidopsis protoplasts undergo DXO-dependent degradation (9, 37). In an Arabidopsis dxo mutant, NAD+-capped transcripts are channeled to RNA silencing for degradation (32, 36). NAD captureSeq was performed in yeast, human, and Arabidopsis dxo mutants to identify the in vivo targets of DXO (9, 13, 32). However, DXO and related decapping enzymes, such as yeast Rai1 or Dxo1, also show m7G cap-removing activity (38, 39). This, together with the reactivity of m7G-RNAs in the CuAAC reaction, may obscure the true set of NAD-RNA targets of DXO. Thus, removing m7G-RNAs is a critical step in profiling NAD-RNAs. Finally, in eukaryotes, translation initiation is primarily facilitated by the m7G cap. There are conflicting data on whether NAD-RNAs possess translatability (9, 11, 13). Our previous study using NAD captureSeq suggested that NAD-RNAs were enriched in the polysomal fractions undergoing active translation (11). Further NAD-RNA profiling with m7G-depleted polysomal RNAs should be performed to verify this conclusion. To date, research into NAD-RNAs in eukaryotes is still in its infancy. The establishment of SPAAC-NAD-seq and m7G-RNA depletion provides valuable tools for future NAD-RNA research.
Materials and Methods
Plant Materials.
The Columbia-0 (Col-0) ecotype of Arabidopsis thaliana was used in all experiments. Col-0 seeds were sown on solid 1/2 Murashige and Skoog medium with 1% sucrose and 0.7% agar. Plates were kept in the dark at 4 °C for 3 d for seed stratification and then were placed in a growth room with a 16/8-h light/dark photoperiod at 22 °C for 12 d. Seedlings were collected at 10 to 11 AM in this study.
CuAAC- and SPAAC-NAD Reactions.
CuAAC-NAD reactions were conducted following the two steps described by Cahová et al. (8). The procedure is described in detail in SI Appendix, Supplementary Methods. SPAAC-NAD reactions were conducted in two main steps. The first step was largely similar to the CuAAC-NAD reaction, with the only difference being that 3-azido-1-propanol (Sigma-Aldrich) replaced 4-pentyn-1-ol (Sigma-Aldrich). After the reaction, 2 μL of the mixture was diluted 200-fold in water for untargeted analysis by LC-MS. For the second step, 5 μL of the remaining mixture further underwent the SPAAC reaction on a 20-μL scale at 37 °C for 1 h with the addition of biotin-PEG4-DBCO (0.5 mM; Sigma-Aldrich) and 2 μL of 10× PBS buffer (pH 7.4). The reaction products were diluted 50-fold in water for untargeted analysis by LC-MS. The untargeted LC-MS conditions are described in detail in SI Appendix, Supplementary Methods.
Preparation of In Vitro Transcribed RNAs.
The template for in vitro transcription was generated by annealing complementary oligonucleotides (20 μM) such that the resulting template DNA carried the T7 class II promoter (ɸ2.5) and an adenosine at the transcription start site (primer sequences in SI Appendix, Table S1). In vitro transcription was performed in a 100-μL reaction using the T7 RNA polymerase under the conditions described in SI Appendix, Supplementary Methods. Quality of in vitro-transcribed RNAs was further analyzed by boronate affinity electrophoresis with a 7 M urea, 1× TBE, 12% polyacrylamide gel supplemented with 0.2% 3-acrylamidophenylboronic acid (Boron Molecular) (23).
CuAAC- and SPAAC-NAD Reactions with RNAs.
The amount of input RNAs varied between in vitro-transcribed RNAs (500 ng), total RNAs (100 μg), and mRNAs (1 μg) in CuAAC- and SPAAC-NAD reactions.
The CuAAC-NAD reaction was performed according to the original NAD captureSeq protocol (8) with minor modifications. The procedure is described in detail in SI Appendix, Supplementary Methods.
For the SPAAC-NAD reaction, RNAs were incubated with 10 μL of 3-azido-1-propanol (Sigma-Aldrich), ADPRC (0.85 μM, Sigma-Aldrich), Hepes (50 mM), MgCl2 (5 mM), and 100 units of RiboLock RNase Inhibitor (Thermo Fisher) on a 100-μL scale at 37 °C for 30 min. One hundred microliters of DEPC H2O was then added and acid phenol/chloroform extraction was performed to stop the reaction. RNAs were precipitated by ethanol, redissolved in 36 μL of 1× PBS buffer (pH 7.4), and then incubated with biotin-PEG4-DBCO (1 mM; Sigma-Aldrich) at 37 °C for 1 h. One hundred sixty microliters of DEPC H2O were then added, and acid phenol/chloroform extraction as well as ethanol precipitation were performed.
Dot Blot and Gel Blot Assays.
For dot blots, 1 μL of RNA after biotinylation by the CuAAC- or SPAAC-NAD reaction was denatured and spotted on a Hybond N+ membrane (GE healthcare). For gel blots, 10 μL of RNA after biotinylation by the CuAAC- or SPAAC-NAD reaction was denatured and resolved in a 2% denaturing agarose gel and transferred to a Hybond N+ membrane by the capillary method. The method to visualize biotinylated RNAs was described in detail in SI Appendix, Supplementary Methods.
Assessment of the Specificity of CuAAC and SPAAC Reactions toward NAD-RNA and m7G-RNA.
A 31-nt m7GpppA-RNA and a 70-nt NAD-RNA were produced by in vitro transcription and mixed together at molar ratios of 22.5:1 and 225:1 (10 μg/1 μg and 100 μg/1 μg). The mixture then underwent the CuAAC- or SPAAC-NAD reaction for biotinylation and was resolved in a 12% polyacrylamide urea gel. The RNAs were transferred to a Hybond N+ membrane by the semidry transfer method. The membrane was then soaked in a solution of 1.6 M 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide and 128 mM 1-methylinidazole (pH 8) at 65 °C for 1.5 h. Finally, the biotin-labeled nucleic acids on the membrane were probed by stabilized streptavidin-HRP and detected with the Chemiluminescent Nucleic Acid Detection Module Kit (Thermo Fisher) and imaged by ChemiDoc XRS+ camera (Bio-Rad).
Immunodepletion of m7G-RNAs.
m7G-RNAs were immunodepleted from mRNAs or a mixture of in vitro-transcribed NAD- and m7G-RNAs by using an anti-m7G antibody (RN017M; MBL). The procedure is described in detail in SI Appendix, Supplementary Methods. To examine the efficiency of m7G-RNA depletion, anti-m7G dot blot assays and nuclease P1 digestion followed by LC-MS detection were performed. Both assays are described in detail in SI Appendix, Supplementary Methods.
Sequencing Library Construction.
Total RNAs were extracted from three biological replicates of 12-d-old seedlings and subjected to mRNA isolation after DNase I treatment. Ten-microgram mRNAs were treated with an anti-m7G antibody to remove m7G-RNAs. Samples without the addition of the anti-m7G antibody were the mock-depletion samples. mRNAs in the supernatant were acid phenol/chloroform extracted, ethanol precipitated, and dissolved in DEPC H2O. Finally, the dissolved mRNAs were separated into three aliquots. One aliquot contained ∼150-ng mRNAs and was subjected to mRNA-seq library construction, while the other two aliquots each contained ∼3-μg mRNAs to be used for CuAAC- and SPAAC-NAD-seq. CuAAC- and SPAAC-NAD reactions were performed as described above, except that 100 μg of E. coli tRNA were added prior to the reactions to mitigate RNA loss during the NAD-RNA capture process. After CuAAC- or SPAAC-NAD reactions, RNAs were incubated at 25 °C for 1 h with 25 μL of magnetic MyOne streptavidin C1 beads, which were preblocked with 100 μg/mL acetylated bovine serum albumin in 100 μL of immobilization buffer containing NaCl (1 M), 10 mM Hepes (10 mM, pH 7), and ethylenediaminetetraacetic acid (5 mM). Beads were then washed five times with streptavidin wash buffer containing urea (8 mM), Tris⋅HCl (50 mM, pH 7.4), and Triton X-100 (0.25%), and three times with DEPC H2O. Biotinylated RNAs were finally eluted by incubating the beads with 20 μL of H2O at 80 °C for 5 min and subjected to library construction. NEBNext Ultra Directional RNA Library Prep Kit for Illumina (E7420; New England Biolabs) was used for subsequent mRNA-seq and NAD-seq library construction. First, the three batches of RNAs were incubated with the first Strand Synthesis Reaction Buffer and random primers at 94 °C for 8 min to fragment RNA. The fragmented and primed mRNAs were then subjected to first-strand cDNA synthesis. The remaining steps were performed according to the manufacturer’s instructions.
Sequencing and Data Analysis.
All prepared libraries were pooled and sequenced on the Illumina NovaSeq 6000 system (paired-end; 150 bp) at Novogene. RNA-seq data were analyzed using the pRNASeqTools pipeline (https://github.com/grubbybio/pRNASeqTools) (40). All reads were processed with cutadapt v1.14.0 with default parameters to remove the adapter sequences (AGATCGGAAGAGC), and reads longer than 20 bp were kept for downstream analysis. The remaining reads without de-duplication were mapped to the Arabidopsis genome (Araport 11) using STAR 2.7.3a (41) with the parameters “–alignIntronMax 5000–outSAMmultNmax 1–outFilterMultimapNmax 50–outFilterMismatchNoverLmax 0.1.” Mapped reads were counted by featureCounts v2.0.0 with the parameters “-p -B -C -O -s 0” (42). Transcript levels were quantified in fragments per kilobase per million total read counts manually. Sample hierarchical clustering was performed with the R function "hclust" using a matrix composed of all transcript levels of genes. NAD-RNAs were defined as fold change of transcript levels ≥ 2 and adjusted P value < 0.05 in NAD-seq samples compared to mRNA-seq samples using DESeq2 with default settings (43). The gene body coverage of NAD-seq reads was calculated using RSeQC (44) by counting the number of reads covering each nucleotide position; all transcripts were scaled to 100 bins. GO analysis was performed using the Database for Annotation, Visualization, and Integrated Discovery (45).
Supplementary Material
Acknowledgments
We thank Dr. Jay Kirkwood for training on LC-MS operations. This work was supported by grants from NIH (GM061146) to X.C., China Postdoctoral Science Foundation (2018M640720) and National Natural Science Foundation of China (31902051) to H.H., and the Research Grants Council of Hong Kong (C2009-19GF and AoE/M-403/16) to Y.X.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025595118/-/DCSupplemental.
Data Availability
The raw sequence data generated in this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (accession no. GSE162114). All other data are included in the article and/or supporting information.
References
- 1.Chen L. Q., Zhao W. S., Luo G. Z., Mapping and editing of nucleic acid modifications. Comput. Struct. Biotechnol. J. 18, 661–667 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ramanathan A., Robb G. B., Chan S. H., mRNA capping: Biological functions and applications. Nucleic Acids Res. 44, 7511–7526 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Galloway A., Cowling V. H., mRNA cap regulation in mammalian cell function and fate. Biochim. Biophys. Acta. Gene Regul. Mech. 1862, 270–279 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cannistraro V. J., Kennell D., The 5′ ends of RNA oligonucleotides in Escherichia coli and mRNA degradation. Eur. J. Biochem. 213, 285–293 (1993). [DOI] [PubMed] [Google Scholar]
- 5.Chen Y. G., Kowtoniuk W. E., Agarwal I., Shen Y., Liu D. R., LC/MS analysis of cellular RNA reveals NAD-linked RNA. Nat. Chem. Biol. 5, 879–881 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huang F., Efficient incorporation of CoA, NAD and FAD into RNA by in vitro transcription. Nucleic Acids Res. 31, e8 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bird J. G., et al., The mechanism of RNA 5′ capping with NAD+, NADH and desphospho-CoA. Nature 535, 444–447 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cahová H., Winz M. L., Höfer K., Nübel G., Jäschke A., NAD captureSeq indicates NAD as a bacterial cap for a subset of regulatory RNAs. Nature 519, 374–377 (2015). [DOI] [PubMed] [Google Scholar]
- 9.Jiao X., et al., 5′ end nicotinamide adenine dinucleotide cap in human cells promotes RNA decay through DXO-mediated deNADding. Cell 168, 1015–1027.e10 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Walters R. W., et al., Identification of NAD+ capped mRNAs in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. U.S.A. 114, 480–485 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang Y., et al., NAD+-capped RNAs are widespread in the Arabidopsis transcriptome and can probably be translated. Proc. Natl. Acad. Sci. U.S.A. 116, 12094–12102 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhang H., et al., NAD tagSeq reveals that NAD+-capped RNAs are mostly produced from a large number of protein-coding genes in Arabidopsis. Proc. Natl. Acad. Sci. U.S.A. 116, 12072–12077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang Y., et al., Extensive 5′-surveillance guards against non-canonical NAD-caps of nuclear mRNAs in yeast. Nat. Commun. 11, 5508 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rostovtsev V. V., Green L. G., Fokin V. V., Sharpless K. B., A stepwise huisgen cycloaddition process: Copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. Engl. 41, 2596–2599 (2002). [DOI] [PubMed] [Google Scholar]
- 15.Morales-Filloy H. G., et al., The 5′ NAD cap of RNAIII modulates toxin production in Staphylococcus aureus isolates. J. Bacteriol., 202 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jewett J. C., Sletten E. M., Bertozzi C. R., Rapid Cu-free click chemistry with readily synthesized biarylazacyclooctynones. J. Am. Chem. Soc. 132, 3688–3690 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Turner R. B., Jarrett A. D., Goebel P., Mallon B. J., Heats of hydrogenation. IX. Cyclic acetylenes and some miscellaneous olefins. J. Am. Chem. Soc. 95, 790–792 (1973). [Google Scholar]
- 18.Preugschat F., Tomberlin G. H., Porter D. J. T., The base exchange reaction of NAD+ glycohydrolase: Identification of novel heterocyclic alternative substrates. Arch. Biochem. Biophys. 479, 114–120 (2008). [DOI] [PubMed] [Google Scholar]
- 19.Roundtree I. A., Evans M. E., Pan T., He C., Dynamic RNA modifications in gene expression regulation. Cell 169, 1187–1200 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Migaud M. E., Pederick R. L., Bailey V. C., Potter B. V., Probing Aplysia californica adenosine 5′-diphosphate ribosyl cyclase for substrate binding requirements: Design of potent inhibitors. Biochemistry 38, 9105–9114 (1999). [DOI] [PubMed] [Google Scholar]
- 21.Holstein J. M., Stummer D., Rentmeister A., Enzymatic modification of 5′-capped RNA with a 4-vinylbenzyl group provides a platform for photoclick and inverse electron-demand Diels-Alder reaction. Chem. Sci. (Camb.) 6, 1362–1369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang L. S., et al., Transcriptome-wide mapping of internal N7-methylguanosine methylome in mammalian mRNA. Mol. Cell 74, 1304–1316.e8 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nübel G., Sorgenfrei F. A., Jäschke A., Boronate affinity electrophoresis for the purification and analysis of cofactor-modified RNAs. Methods 117, 14–20 (2017). [DOI] [PubMed] [Google Scholar]
- 24.Bird J. G., et al., Highly efficient 5′ capping of mitochondrial RNA with NAD+ and NADH by yeast and human mitochondrial RNA polymerase. eLife 7, e42179 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Luciano D. J., Belasco J. G., Analysis of RNA 5′ ends: Phosphate enumeration and cap characterization. Methods 155, 3–9 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kiledjian M., Eukaryotic RNA 5′-end NAD+ capping and deNADding. Trends Cell Biol. 28, 454–464 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Frindert J., et al., Identification, biosynthesis, and decapping of NAD-capped RNAs in B. subtilis. Cell Rep. 24, 1890–1901.e8 (2018). [DOI] [PubMed] [Google Scholar]
- 28.Vvedenskaya I. O., et al., CapZyme-seq comprehensively defines promoter-sequence determinants for RNA 5′ capping with NAD+. Mol. Cell 70, 553–564.e9 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu K., Lat P. K., Yu H. Z., Sen D., CLICK-17, a DNA enzyme that harnesses ultra-low concentrations of either Cu+ or Cu2+ to catalyze the azide-alkyne “click” reaction in water. Nucleic Acids Res. 48, 7356–7370 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Varasteh Z., et al., The effect of mini-PEG-based spacer length on binding and pharmacokinetic properties of a 68Ga-labeled NOTA-conjugated antagonistic analog of bombesin. Molecules 19, 10455–10472 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang H., et al., Use of NAD tagSeq II to identify growth-phase dependent alterations in E. coli RNA NAD+-capping. Proc. Natl. Acad. Sci. U.S.A. doi: 10.1073/pnas202026183118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu X., et al., Messenger RNA 5′ NAD+ capping is a dynamic regulatory epitranscriptome mark that is required for proper response to abscisic acid in Arabidopsis. Dev. Cell 56, 125–140.e6 (2021). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Mlynarska-Cieslak A., et al., Nicotinamide-containing Di- and trinucleotides as chemical tools for studies of NAD-capped RNAs. Org. Lett. 20, 7650–7655 (2018). [DOI] [PubMed] [Google Scholar]
- 34.Grudzien-Nogalska E., et al., Structural and mechanistic basis of mammalian Nudt12 RNA deNADding. Nat. Chem. Biol. 15, 575–582 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharma S., et al., Mammalian Nudix proteins cleave nucleotide metabolite caps on RNAs. Nucleic Acids Res. 48, 6788–6798 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kwasnik A., et al., Arabidopsis DXO1 links RNA turnover and chloroplast function independently of its enzymatic activity. Nucleic Acids Res. 47, 4751–4764 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pan S., et al., Arabidopsis DXO1 possesses deNADding and exonuclease activities and its mutation affects defense-related and photosynthetic gene expression. J. Integr. Plant Biol. 62, 967–983 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Jiao X., et al., Identification of a quality-control mechanism for mRNA 5′-end capping. Nature 467, 608–611 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chang J. H., et al., Dxo1 is a new type of eukaryotic enzyme with both decapping and 5′-3′ exoribonuclease activity. Nat. Struct. Mol. Biol. 19, 1011–1017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.You C., et al., FIERY1 promotes microRNA accumulation by suppressing rRNA-derived small interfering RNAs in Arabidopsis. Nat. Commun. 10, 4424 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liao Y., Smyth G. K., Shi W., featureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 30, 923–930 (2014). [DOI] [PubMed] [Google Scholar]
- 43.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wang L., Wang S., Li W., RSeQC: Quality control of RNA-seq experiments. Bioinformatics 28, 2184–2185 (2012). [DOI] [PubMed] [Google Scholar]
- 45.Dennis G. Jr, et al., DAVID: Database for Annotation, Visualization, and Integrated Discovery. Genome Biol. 4, 3 (2003). [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The raw sequence data generated in this study have been deposited in the National Center for Biotechnology Information Gene Expression Omnibus (GEO) database (accession no. GSE162114). All other data are included in the article and/or supporting information.