Significance
Hepadnaviruses, including hepatitis B virus (HBV) as a major human pathogen, are small, enveloped DNA viruses, which replicate by reverse transcription of an RNA intermediate. Unlike retroviruses, which use tRNA as primer for reverse transcription, hepadnaviruses employ a unique protein-priming mechanism involving de novo synthesis of a DNA primer covalently attached to the viral polymerase. Here, we show that this mechanism is highly conserved on the molecular level in distantly related non-enveloped fish viruses which diverged from ancestral hepadnaviruses more than 400 Mya. The exceptional level of conservation and the absence of known homologous cellular mechanisms renders HBV protein priming a promising, yet unexplored, antiviral target for the development of novel therapeutics against this highly relevant pathogen.
Keywords: protein priming, initiation of reverse transcription, HBV replication mechanism, HBV long-term evolution, paleovirology
Abstract
Hepadnaviruses, with the human hepatitis B virus as prototype, are small, enveloped hepatotropic DNA viruses which replicate by reverse transcription of an RNA intermediate. Replication is initiated by a unique protein-priming mechanism whereby a hydroxy amino acid side chain of the terminal protein (TP) domain of the viral polymerase (P) is extended into a short DNA oligonucleotide, which subsequently serves as primer for first-strand synthesis. A key component in the priming of reverse transcription is the viral RNA element epsilon, which contains the replication origin and serves as a template for DNA primer synthesis. Here, we show that recently discovered non-enveloped fish viruses, termed nackednaviruses [C. Lauber et al., Cell Host Microbe 22, 387–399 (2017)], employ a fundamentally similar replication mechanism despite their huge phylogenetic distance and major differences in genome organization and viral lifestyle. In vitro cross-priming studies revealed that few strategic nucleotide substitutions in epsilon enable site-specific protein priming by heterologous P proteins, demonstrating that epsilon is functionally conserved since the two virus families diverged more than 400 Mya. In addition, other cis elements crucial for the hepadnavirus-typical replication of pregenomic RNA into relaxed circular double-stranded DNA were identified at conserved positions in the nackednavirus genomes. Hence, the replication mode of both hepadnaviruses and nackednaviruses was already established in their Paleozoic common ancestor, making it a truly ancient and evolutionary robust principle of genome replication that is more widespread than previously thought.
Hepatitis B virus (HBV) is a major human pathogen. Globally, ∼250 million chronically HBV infected people are at an increased risk of developing liver cirrhosis and hepatocellular carcinoma, accounting for close to 900,000 deaths annually (1). HBV is the prototypic member of the Hepadnaviridae, a family of small, enveloped DNA viruses from mammals (genus Orthohepadnavirus), birds (Avihepadnavirus), reptiles and amphibians (Herpetohepadnavirus) (2), and fishes (Para- and Metahepadnavirus) (2, 3). About 3 kb in size, hepadnavirus genomes are among the smallest of all animal viruses, containing just four extensively overlapping genes. Genome replication occurs inside viral capsids by reverse transcription of a terminally redundant pregenomic RNA (pgRNA) into a partially double-stranded (ds), noncovalently closed, relaxed circular DNA (rcDNA) (4, 5). Upon infection, the rcDNA is converted into covalently closed circular DNA (cccDNA) that, as an episomal minichromosome, serves as a template for viral transcripts and establishes viral persistence (6).
The viral polymerase P comprises reverse transcriptase (RT) and RNase H (RH) domains with weak homology to retroviral replicases (7) plus a unique terminal protein (TP) domain essential for the unusual mode of reverse transcription initiation. The replication origin for first-strand DNA synthesis is contained within the structured 5′-proximal pgRNA element epsilon (ε) (8-10). ε serves as a specific entry site for P and directs copackaging of pgRNA and P into capsids (11, 12). Unlike the host transfer RNA priming employed by retroviruses, hepadnaviral reverse transcription is initiated by de novo synthesis of a short DNA primer that is copied from ε and, upon transfer to a complementary 3′-proximal site, is elongated into full-length minus-strand DNA [(−)DNA] (9, 10, 13, 14) (SI Appendix, Fig. S1A). Peculiar to this initiation process is the covalent attachment of the first nucleotide (nt) to P itself, mediated by an auto-nucleotidylation mechanism that, beyond the cognate εRNA, requires the DNA polymerase activity of RT and a specific tyrosine residue of TP as an acceptor (15–17). This protein-priming reaction links P to the nascent (−)DNA via a tyrosyl-DNA phophodiester bond, which is preserved through viral DNA maturation. As (−)DNA synthesis proceeds, the RH activity degrades the pgRNA template, leaving a short 5′ RNA fragment comprising direct repeat 1 (DR1), which, upon transfer to DR2, serves as primer for (+)DNA synthesis (18). A further template switch of the nascent (+)DNA between the terminally redundant ends of the (−)DNA template accomplishes circularization, and (+)DNA elongation establishes the mature rcDNA genome (4).
For decades, the hepadnaviral replication mechanism and the distinctive TP domain were considered unique for hepadnaviruses, and consequently, their origin and evolution remained enigmatic. Recently, we discovered a family of non-enveloped fish viruses, termed nackednaviruses (2); they contain small circular dsDNA genomes of HBV genome-like size inside icosahedral capsids that are structurally related to those of hepadnaviruses (2). Importantly, although nackednaviruses lack envelope protein genes, they encode P-like reverse transcriptases with sequence homology to the conserved regions of the hepadnaviral P protein domains TP, RT, and RH (2). Moreover, these nackednaviral P proteins possess TP-, RT- and RNA-dependent auto-guanylylation activity (2) (SI Appendix, Fig. S2), reminescent of hepadnaviral protein priming, although further mechanistic details of nackednaviral reverse transcription including the location and nature of the replication origin are unknown. In fact, phylogenetic reconstructions date the common evolutionary origin 430 Mya before the rise of tetrapods (2). While nackednaviruses retained their envelope-less lifestyle, hepadnaviruses evolved an envelope protein gene de novo by overprinting of the P open reading frame and coevolved with their hosts over geological eras during tetrapod evolution (2). This unique phylogenetic relation between the two distantly related virus families provides an unprecedented opportunity to study the origin and long-term evolution of the peculiar hepadnaviral reverse transcription mechanism.
In this work, we examined the nackednaviral replication mechanism using biochemical and bioinformatic methods. We show that nackednaviruses encode ε-like RNA elements, which serve as origin for TP-primed reverse transcription. Nackedna- and hepadnaviral ε elements and P proteins are functionally highly conserved, and two nucleotide substitutions in ε were sufficient to enable heterologous P protein-priming activity. Moreover, identification in nackednaviral genomes of further, positionally conserved cis elements crucial for hepadnaviral replication indicates that the replication mechanism as a whole is shared by both virus families. In conclusion, our data show that hepadnaviral reverse transcription is a primordial, evolutionary robust principle of nucleic acid primer independent genome replication whose origin is likely much deeper than the estimated phylogenetic divergence of nackedna- and hepadnaviruses >400 Mya.
Results
Nackednaviral P Protein Initiates Reverse Transcription by Protein Priming at an ε-like RNA Element.
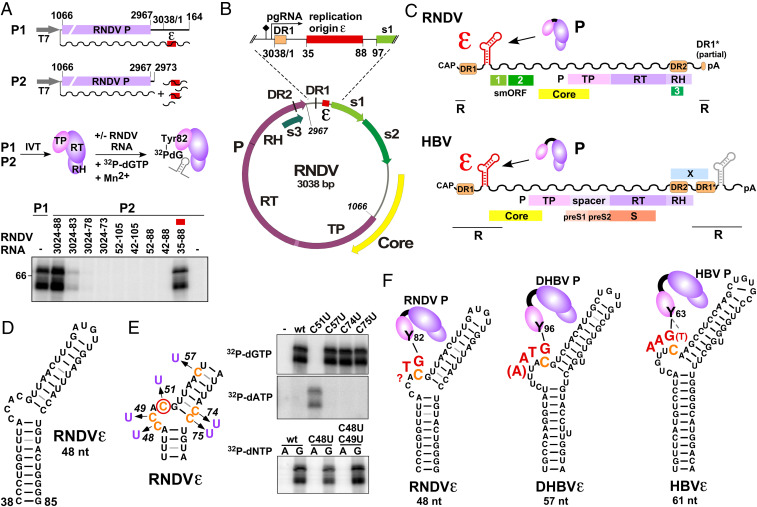
To gain insight into the replication mechanism of nackednaviruses, we mapped RNA elements promoting auto-guanylylation of in vitro translated P protein of the prototypic rockfish nackednavirus (RNDV). Initially, we expressed P from the circularly permuted construct P1 in which P represents the most upstream ORF, followed by 235 nt (positions 2,967 to 164) spanning the 3′ and 5′ untranslated regions (UTR) of the predicted RNDV pgRNA (Fig. 1A). Using a Mn2+-supplemented assay buffer previously shown to enhance duck HBV (DHBV) P in vitro priming efficiency (19), we observed robust RNDV P auto-guanylylation activity upon provision of [α-32P]-dGTP. In contrast, deletion of the UTR sequences (construct P2) completely abolished guanylylation. However, guanylylation was rescued by adding specific UTR RNA fragments in trans, indicating the presence of an essential RNA element between genome positions 2,973 and 164. Using an iterative approach, we identified a sequence of about 50 nt between genome positions 35 to 88 as necessary and sufficient for guanylylation activity, which we termed RNDVε. Guanylylation was RNDVε dose dependent and reached half-maximal activity at about 20 nM (SI Appendix, Fig. S3), in line with the reported dissociation constant for the DHBV ε-P interaction (12). On the predicted pgRNA, RNDVε is located in the 5′ UTR about 40 nt downstream of the 5′ end (Fig. 1B) and is therefore similarly positioned as hepadnaviral ε elements (Fig. 1C). Despite its substantially different sequence, RNDVε can adopt a hepadnaviral ε-like stem-bulge–stem-loop structure (Fig. 1D), although the upper stem comprises mostly weak base pairs. RNDV P auto-nucleotidylation was highly selective for guanosine, arguing for a templated reaction (SI Appendix, Fig. S4). To analyze whether that template is located within RNDVε, individual C residues were mutated to U, and guanylylation versus adenylylation activities were investigated (Fig. 1E). All mutants except C51U showed wild-type (wt)-like guanylylation without detectable adenylylation, whereas mutant C51U completely abolished guanylylation but supported adenylylation. Hence, RNDV P guanylylation is specifically templated by RNDVε C51, strongly suggesting this residue as the origin of replication. C51 maps to the 3′ position of the ε bulge and therefore is in the same structural context as the initiation sites of HBV and DHBV (Fig. 1F). Furthermore, guanosine initiation appears to be a common principle in both virus families (10, 20, 21). Hence, our data reveal intriguing similarities in the initiation of reverse transcription by nackedna- and hepadnaviruses despite their huge phylogenetic distance.
Fig. 1.
Identification and characterization of the RNDV replication origin RNDVε. (A) Mapping of RNDVε (red box). RNDV P was expressed in a coupled in vitro transcription/translation (IVT) system from construct P1 or from P2 complemented with the indicated RNDV UTR RNA fragments (wavy lines, RNA transcripts; T7, T7 promoter). Priming assays were performed, and guanylylation of P was assessed by SDS polyacrylamide gel electrophoresis and phosphoimaging. The top band represents the 72 kDa full-length RNDV P, the band below a truncated yet functional RNDV P translation product. Numbers refer to genomic nt positions as previously defined (2). (B) Position of RNDVε in the RNDV genome. The predicted pgRNA start site and the polyA signal are indicated by arrow and diamond, respectively; s1, s2, and s3 denote nackednavirus specific small ORFs (smORFs) of unknown function. (C) The 5′-proximal position of ε on the pgRNA is conserved between RNDV and HBV. pgRNAs are depicted as wavy lines; terminal redundancy (R), polyA tail (pA). HBV contains a second copy of ε in the 3′ redundancy (gray) dispensible for replication (4). (D) Secondary structure of RNDVε predicted by MC-Fold. (E) The RNDV reverse transcription initiation site maps to RNDVε nt C51. RNDV P was expressed from P2, and nucleotidylation of P was assessed using the indicated C to U point mutants of RNDVε plus [α-32P]dGTP or [α-32P]dATP as substrate. (F) (−)DNA primer (red) and structural context of the primer template of RNDV, DHBV, and HBV. The Tyr residue (Y) of TP, covalently attached to the initiating G residue, is indicated, and the templating C is highlighted in orange. The length of the RNDV primer has not been determined, but the limited complementarity to the putative primer transfer site suggests it does not exceed two nt. HBV possesses a potential alternative initiation site (dashed line to bracketed T).
Conserved Features of Hepadna- and Nackednaviral ε Elements.
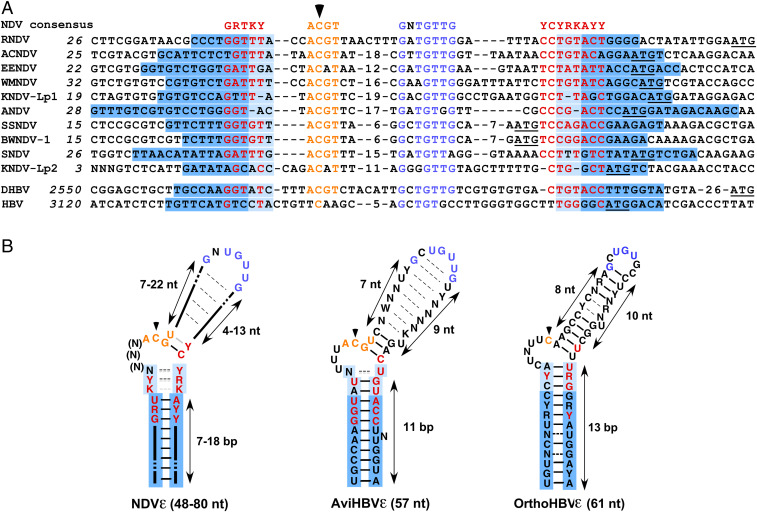
By in silico analyses, we next could identify ε elements in all known nackednaviruses at homologous genomic positions (Fig. 2A). Structurally, a lower stem of variable length (7 to 18 base pair [bp]) is common to all nackednaviral ε elements (Fig. 2B; for individual structures see SI Appendix, Fig. S5). In contrast, the apical regions vary in length from 24 to 43 nt and are structurally not well defined, except for a conserved potential G-C bulge closing bp. This finding resembles the structural variability in the upper stem of avian hepadnavirus ε elements, which is functionally explained by the common priming-active structure the different RNAs adopt upon productive binding to P (22). The low overall sequence similarity between individual nackednavirus ε elements reflects their high degree of diversification, but two regions are highly conserved: 1) a 5′-ACGU motif encompassing the C51 initiation site, suggesting that (–)DNA initiation by 5′-GT (as depicted for RNDV in Fig. 1F) is absolutely conserved, and 2) the motif GNUGUUG in the apical region crucial for priming (SI Appendix, Fig. S6). Strikingly, in avihepadnaviruses, the same two motifs are strictly conserved (Fig. 2B) and functionally essential (10, 12, 23), revealing an unexpectedly high similarity between the two divergent virus families. The still higher sequence similarity between avian HBVε and NDVε than between avian and mammalian HBVε (Fig. 2B) supports the hypothesis that nackedna- and avihepadnaviral ε elements are plesiomorph, whereas mammalian ε elements are apomorph, in line with avihepadnaviruses being more ancestral than orthohepadnaviruses. This conclusion is consistent with P sequence homology and the pattern of indels in P (2) and with the higher complexity of mammalian HBV genomes carrying an additional X gene.
Fig. 2.
Comparison of nackednaviral and hepadnaviral ε elements. (A) Alignment of nackednaviral and hepadnaviral ε sequences. Colored letters indicate conserved sequence elements in lower stem (red), initiation region (orange), and apical region (blue). Base-paired lower stem regions are shaded in blue, and the variably base-paired tip of the lower stem is in light blue. A black arrowhead denotes the intiation site. The smORF1 start codons of nackednaviruses and the core start codons of HBV and DHBV are underlined. Internal numbers denote nt not depicted in the figure. Italic numbers refer to genomic nt positions (2). African cichlid nackednavirus (ACNDV), European eel nackednavirus (EENDV), Western mosquitofish nackednavirus (WMNDV), Lucania parva killifish nackednavirus-1 and -2 (KNDV-Lp1, KNDV-Lp2), Astatotilapia nackednavirus (ANDV), sockeye salmon nackednavirus (SSNDV), baby whale nackednavirus-1 (BWNDV-1), and stickleback nackednavirus (SNDV). (B) Consensus ε structures of nackednaviruses (Left), avihepadnaviruses (Middle), and orthohepadnaviruses (Right). Dashed lines between nt indicate nonconserved, facultative bp, and gray lines indicate potential noncanonical bp.
Cross-Priming Experiments Reveal Functional Conservation of Hepadna- and Nackednaviral ε Elements.
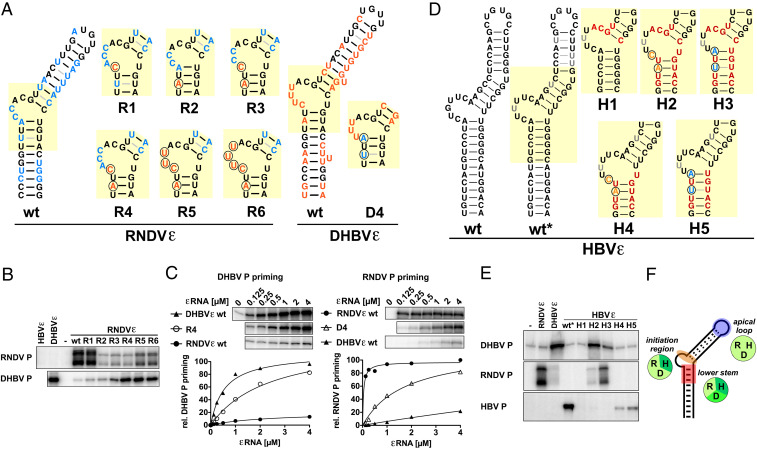
The presence of conserved subelements in nackedna- and hepadnaviral ε elements, despite their divergent sequence context, prompted us to investigate the functional distance between RNDVε, DHBVε, and HBVε by cross-priming assays. To identify P-ε specificity determinants, we examined priming activities of chimeric ε variants with different P proteins. For both HBV and DHBV, such determinants for P binding and priming have been described in the apical loop and in the region surrounding the central bulge (12, 24, 25), although HBVε is not a productive substrate for DHBV P and vice versa (20, 26). As the apical loop motif is nearly identical in nackedna- and avihepadnaviral ε elements, we first focused on the more variable central region to compare RNDVε with DHBVε. In variants R1 to R6, RNDVε-specific nt were gradually replaced by up to five of the corresponding DHBVε nt (Fig. 3A), and P guanylylation activity was tested in standard priming buffer containing Mn2+ (Fig. 3B) and, to control for potential Mn2+-mediated impacts, also under Mg2+-only conditions (SI Appendix, Fig. S7). As expected, wt RNDVε did not support substantial guanylylation of DHBV P. However, two nt mutations in the tip of the lower stem (variant R4) were sufficient to gain strong, Mn2+-independent DHBV P guanylylation accompanied by a substantial drop in cognate RNDV P activity. In the sole presence of Mg2+ (SI Appendix, Fig. S7), virtually the same patterns were seen, except that the weak RNDV P signals with variant RNAs R2 to R6 remained undetectable. Reciprocal mutations in DHBVε (variant D4) enabled RNDV P guanylylation in a similar manner, corroborating these findings (Fig. 3C). Hence, the barrier for virus-specific priming activity between DHBV and RNDV P is defined by only two key nt in ε, revealing a surprisingly high level of conservation between both virus families.
Fig. 3.
Functional adaptation of nackedna- and hepadnaviral ε elements to heterologous P proteins. Few strategic nt exchanges in ε enable cross-species P protein priming. (A) RNDV/DHBV chimeric ε variants. Substituted nt are encircled. RNDV-specific nt are in blue, and DHBV-specific nt are in orange. Note that the RNDVε variant R4 and the DHBVε variant D4 contain reciprocal substitutions at homologous positions. (B) RNDV and DHBV P guanylylation activities of RNDVε variants R1 to R6. P proteins were expressed from ε-deficient templates, and priming assays were performed in the presence of the indicated ε variants (1 µM) as described in Fig. 1A. (C) Guanylylation activities of DHBV P adapted RNDVε variant R4 (Left) and RNDV P adapted DHBVε variant D4 (Right) at different ε concentrations. Maximal priming activity of the cognate ε was set to 100%. Only the upper/full-length band of RNDV P is shown. (D) Chimeric ε variants for adaptation of HBVε to DHBV and RNDV P. DHBV-specific nt are in orange, RNDV-specific nt are in blue, and nt identical between DHBV and RNDV are in red. Note that HBVε wt* contains structure destabilizing mutations (gray) essential for HBV P in vitro priming activity which do not affect P specificity. (E) DHBV, RNDV, and HBV P guanylylation activities of HBVε variants H1 to H5 (performed as described in B). (F) Compatibility of ε determinants for priming of reverse transcription between nackedna- and hepadnaviruses. Cross-functionality of heterologous pairs of P and ε is indicated by identical background color of circular icons. RNDV (R), DHBV (D), HBV (H).
Inclusion of HBVε in this functional comparison was initially hampered by a lack of priming activity of in vitro translated HBV P (27). This block could be overcome by using a truncated HBV P (amino acid sequence in SI Appendix) in combination with a novel HBVε variant wt* (Fig. 3D and SI Appendix, Fig. S8). Wt* contains a few structure-destabilizing mutations in the upper stem, which facilitate essential structural rearrangements in functional ε-P priming complexes (22, 28). As intended, HBVε wt* supported guanylylation of HBV P but not DHBV P or RNDV P (Fig. 3E). Replacing just the tip of the lower stem of HBVε wt* by DHBV or RNDV sequences strongly reduced HBV P guanylylation (variants H4 and H5) but did not elicit substantial DHBV or RNDV P activity. However, additional sequence adjustments in the central initiation region enabled robust guanylylation of either DHBV P (H2) or RNDV P (H3), depending on the cognate P specificity determinant in the lower stem, and, as in variant H1, they completely ablated detectable HBV P priming. Notably, fully congruent results were obtained using FLAG-tagged full-length HBV P protein from transfected mammalian cells with authentic wt HBVε and the fully wt HBVε-based variants H1′ to H5′ in Mn2+-free buffer (SI Appendix, Fig. S9). Hence, the initiation region as well as the tip of the lower stem contribute to P discrimination, and the priming relevant features of the apical loop are largely conserved among nackedna- and hepadnaviruses (Fig. 3F). In sum, our data demonstrate that key features of the εRNA elements and their replication initiation-relevant interactions with P proteins are structurally and mechanistically highly similar in nackedna- and hepadnaviruses.
Bioinformatic Reconstruction of Postpriming Events in Nackednavirus Genome Replication.
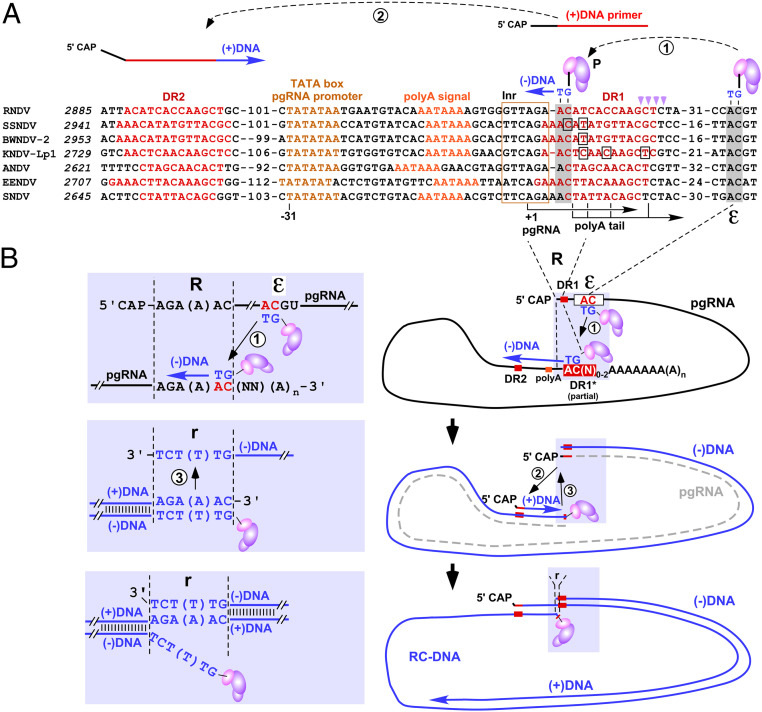
To derive a model for the entire nackednavirus replication mechanism, we screened NDV genomes in silico for further cis elements, which are known to be critical in HBV replication (Fig. 4A). Nackednaviral pgRNA promoters contain TATA boxes and initiator elements, allowing prediction of pgRNA initiation sites (29, 30). Together with the position of the pgRNA polyA signals, this implies that transcription from a circular genome produces pgRNAs with short terminal redundancies of about 10 nt (Fig. 4B). These predictions were directly confirmed by the mapping of primary transcriptome shotgun sequencing reads representing the pgRNA termini (SI Appendix, Fig. S10). In hepadnaviruses, the polyA signal shifted to a position downstream of ε, resulting in a larger terminal redundancy that includes a second copy of ε (Fig. 1C and SI Appendix, Fig. S1). Notably, artificial back shifting of the HBV pA signal to a nackednavirus-similar position did not impair HBV replication (SI Appendix, Fig. S11). To copy the entire genome without loss of genetic information, the synthesized primer needs to be transferred to a complementary site in the 3′ redundancy. Conservation of both the 5′-AC initiation motif in ε and an AC motif in the pgRNA’s 3′ redundancy suggests that nackednaviral reverse transcription is initiated by transfer of the 5′ε templated 5′-GT primer to this complementary 3′-proximal site (Fig. 4 A and B). Its genomic position, a few nt downstream of the pgRNA start and at the 5′ border of DR1, is identical to the position of primer transfer sites in hepadnaviruses, further supporting our model (9, 10). Accordingly, nackedna- and hepadnaviral pgRNAs are both reverse transcribed into (−)DNAs containing similarly sized (<10 nt) terminal redundancies, essential for subsequent circularization and rcDNA synthesis. Complementarity between primer and transfer site is limited to two nt in 11 out of 12 nackednavirus genomes, arguing for a primer size of only two nt versus 3 to 4 nt in hepadnaviruses (Fig. 1F). The specificity of the transfer of such extremely short primers to the polyA proximal transfer site may be facilitated by the polyA tail and/or polyA binding proteins as described for picornaviruses (31), which initiate replication of their RNA genome by a two nt RNA primer copied from an internal stem-loop upon transfer to the polyA tail (32). (+)DNA synthesis and genome circularization of hepadnaviruses require two direct repeat elements (DR1 and DR2). Nackednaviruses contain DRs of 10 to 15 nt at homologous positions (Figs. 1C and 4A). Hepadnaviral (+)DNA is primed by a pgRNA-derived, DR1-containing oligoribonucleotide processed by the RH activity of P at 15 to 18 nt downstream of the 5′ end of pgRNA (18). The DR1 part renders the oligoribonucleotide competent for (+)DNA priming at DR2. The 3′ borders of nackednaviral DR1 are analogously located 15 to 17 nt downstream of the predicted pgRNA start sites (Fig. 4A). Nackednaviral DR2 elements are positioned 131 to 141 nt from the 5′ end of (−)DNA, similar to hepadnaviruses (HBV: 265 nt, DHBV: 49 nt). In both virus families, DR2 overlaps with the C-terminal region of the P ORF. Finally, the terminal redundancy of nackednaviral (−)DNA enables hepadnavirus-like genome circularization by terminal template switching during (+)DNA synthesis and eventually the formation of hepadnavirus-like rcDNA (Fig. 4B). The high conservation of all these features strongly supports a common functionality.
Fig. 4.
Model of nackednaviral replication. (A) Alignment of replication relevant cis elements of nackednaviruses. Italics indicate genomic positions. Internal numbers denote nt not depicted in the figure. The conserved consensus initiator element (30) (Inr, boxed in gold) harbors the predicted pgRNA initiation site (+1) located at a canonical distance of 31 nt to the conserved TATA box of the pgRNA promoter. Boxed nt indicate mapped polyA attachment sites. Template switches are indicated by encircled numbers. The GT primer template in ε and the primer transfer site are shaded in gray. Magenta arrowheads mark RNase H cleavage sites for (+) strand primer generation. (B) Model for hepadnavirus-like reverse transcription of nackednaviral pgRNA into rcDNA. A TP-linked GT primer is copied from ε (white box) and transferred to a conserved AC 3′ element (red) in the terminal redundancy (R). The entire pgRNA is copied into (−)DNA by primer elongation, and the pgRNA is degraded (dashed gray line) by P’s RH activity, leaving a short RNA oligo (red line), which serves as (+)DNA primer upon transfer to complementary DR2. A third template switch from the 5′ to the 3′ end of (−)DNA fostered by the short terminal redundancy (r) of five to six nt (detailed in shaded inset, hepadnaviruses eight to nine nt) accomplishes circularization, and (+)DNA elongation generates HBV-like rcDNA. Note that the RT domain of P linked to the (−)DNA is engaged in DNA synthesis and associated with the 3′ end of nascent DNA, which is not depicted in the figure.
Discussion
Our study suggests a surprisingly high concordance in the replication mechanisms of hepadna- and nackednaviruses and therefore firmly corroborates the proposed phylogenetic relation between both virus families (2), yet the functions of the putative replication elements in nackednaviruses, other than ε, remain to be established experimentally. Furthermore, our data imply that the HBV-like reverse transcription mechanism was already well established in the last common ancestor (LCA) of hepadna- and nackednaviruses and therefore has been preserved for more than 400 My. Such an LCA would have contained core and P genes whose products, in hepadnaviruses, are the only viral proteins required for intracellular genome amplification (4, 33), plus the cis elements ε, DR1, and DR2, which, as shown here, are common to both hepadna- and nackednaviruses. Further unraveling the origin of the LCA is hampered by the limitation of known gene homologs to the RT and RH domains of P protein. Phylogenetic comparisons of these two domains suggest that hepadnaviruses constitute a very basal lineage within the diverse group of long terminal repeat retroelements (LTR-RE), which also include Retroviridae (7, 34, 35). However, because all other proteins plus the entire replication mechanism of LTR-REs are fundamentally different from those of hepadnaviruses (36), we consider it very unlikely that the hepadna-/nackednaviral lineage derived from LTR-REs by secondary loss of LTR-RE–specific genes. As an alternative parsimonious hypothesis, we propose that the LCA evolved from a primordial ε-P(TP-RT-RH)-DR1/2-containing retroplasmid capable of self-sustained replication similar to, but mechanistically distinct from, known retroplasmids (37, 38). Such retroplasmids could have developed into viruses by de novo evolution of the core or by acquisition of a preexisting proto-core gene through horizontal gene transfer (39, 40). In this scenario, the evolution of HBV ancestors would have been independent from retrovirus-like obligate host genome integration. Indeed, there is no evidence for genomically integrated primordial TP-containing P sequences that predate the split between the nackedna- and hepadnaviral lineages.
However, further insights into the evolutionary origin of protein-primed reverse transcription are inevitably linked to the unraveling the enigmatic origin of TP and ε. Although several other viruses and plasmids employ protein priming (41), the respective genome-linked proteins share no sequence homology with TP proteins, suggesting independent origins. Likewise, to date, no significant homology of TP to any extant cellular protein has been reported. Hence, it appears most likely that the hepadnaviral reverse transcription mechanism has evolved de novo in a primordial replicon as a unique strategy for replication of small circular dsDNAs independent of nucleic acid primers and host genome integration.
While such early evolution considerations leave room for speculation, the observed long-term, nucleotide-level conservation of functionally crucial signature sequences within the ε elements of hepadna- and nackednaviruses indicates a low viral escape potential toward interference with a productive P-ε interaction. This scenario renders the P-ε complex a promising target for the development of novel anti-HBV compounds aimed to inhibit protein priming and packaging of pgRNA into nucleocapsids. Moreover, because hepadnaviral protein priming appears to be mechanistically distinct from all known enzymatic activities of the host, such drugs can be expected to pose a very low risk for unwanted off-target effects and finally may contribute to combat currently uncurable chronic hepatitis B.
Materials and Methods
Virus Sequences.
Accession numbers of all virus sequences used in this study are given in SI Appendix.
Priming/Guanylylation Assay.
Sequences and technical information for all P protein expression plasmids are provided in SI Appendix. RNDV and DHBV P proteins were expressed by in vitro translation in rabbit reticulocyte lysate using the TNT T7 Quick Coupled Transcription/Translation System (Promega) according to manufacturer’s instructions. After incubation for 1 h at 30 °C, the samples were split into aliquots of 9 µl, and 1 µl of an appropriately concentrated stock solution of εRNA was added. Unless stated otherwise, the final concentration of εRNA was 1 µM. Samples were incubated for 30 min at 30 °C plus 30 min at 23 °C, mixed with 5 µl of priming buffer (50 mM Tris HCl [pH 8.0], 50 mM NaCl, 6 mM MnCl2, and 2 µCi [α-32P]dGTP [3,000 Ci/mmol]), and serially incubated at 23 °C and 37 °C for 30 min each. Reactions were stopped by adding three volumes of sodium dodecyl sulfate (SDS) sample buffer, heat denatured at 100 °C for 5 min, and analyzed by SDS polyacrylamide gel electrophoresis. Gels were vacuum dried and primed P protein was detected by phosphorimaging (Typhoon FLA 7000; GE Healthcare) and quantified using ImageQuant TL software (GE Healthcare). The priming of DHBV P by RNDV/DHBVε chimeras (Fig. 3B) was examined using DHBV miniPTEV expressed in Escherichia coli and refolded from purified and solubilized inclusion bodies essentially as previously described (19). εRNAs were added to freshly refolded miniPTEV, and the samples were incubated for 1 h at 23 °C followed by priming assay using [α-32P]dGTP as described above. Homemade wheat germ extract (kind gift of A. Böckmann, University of Lyon, France) for HBV P expression was prepared from wheat seeds (42, 43). In vitro translation using a bilayer method was performed essentially as previously described (43, 44). εRNA was added cotranslationally to the feeding buffer, and samples were incubated for 16 h at 22 °C, and aliquots of 10 µl were subjected to priming assay using [α-32P]dGTP as described above.
εRNA Synthesis.
εRNAs were generated by T3 or T7 promoter driven in vitro transcription from linearized plasmids or PCR products using T3 RNA polymerase (New England Biolabs) or Ampliscribe T7 High Yield Transcription Kit (Lucigen) according to manufacturer’s recommendations. Details on template DNA generation and sequences of εRNA transcripts are listed in SI Appendix, Tables S1 and S2. RNA concentrations were determined photometrically. Transcript integrity was verified by gel electrophoresis and ethidium bromide staining.
RNDVε Secondary Structure.
The secondary structure model of RNDVε was obtained by the calculation of the minimum free energy state of RNDV nt 38 to 85 by MC-Fold (45), forcing the guanylylation template C51 to be unpaired. The model ranks within the top five predicted structures of unconstraint calculations (difference in free energy <5%), which is typical for biologically active and experimentally confirmed structures (45).
Identification of Nackednaviral ε Elements.
Because of low sequence conservation and frequent indels, linear computational alignment with the sequence of the experimentally confirmed RNDVε failed to identify ε elements in other nackednaviruses. Instead, MC-Fold (45) was used for scanning the 5′-proximal region of pgRNA for potential stem structures with a variable window size ranging from 50 to 80 nt. Stem regions plus the intervening sequences were aligned using the multiple sequence alignment tool multiple sequence comparison by log-expectation (MUSCLE; https://www.ebi.ac.uk/Tools/msa/muscle/), revealing the highly conserved motifs in the initiation region and apical loop region. The primary computed alignment was further edited by visual inspection to yield the alignment given in Fig. 3A.
ε Element Consensus Sequences.
Avi- and orthohepadnaviral ε consensus sequences were derived from alignments of 18 and 61, respectively, sequences representing the full spectrum of sequence variability (for accession numbers see SI Appendix). Alignment data were used to generate consensus structures shown in Fig. 2B.
Identification of Transcription- and Replication-Relevant Cis Elements.
For SSNDV and KNDV-Lp-1, the pgRNA transcription start and polyA addition sites could be directly identified in high coverage transcriptome sequencing experiments of sockeye salmon (SRX265390: SRR827572, SRR827573; SRX265393: SRR827512, SRR827513) and Lucania parva killifish (SRX340836: SRR958778) tissues, respectively (2). For visualization, all primary sequence reads covering the 5′ copy of DR1 and adjacent regions were aligned to the derived viral cccDNA sequence using MUSCLE (SI Appendix, Fig. S6). These data were used to identify for each analyzed nackednavirus the pgRNA’s promoter (TATA box) and polyA signal by alignment of genomic sequences proximal to the pgRNA’s termini using MUSCLE and further manual refinement (Fig. 4A). Direct repeats were identified by a homology search within each genome using the pgRNA UTRs as seed sequences.
Supplementary Material
Acknowledgments
We are grateful to Anja Böckmann (University of Lyon, Lyon, France) for providing wheat germ extracts, in vitro translation reagents, and plasmid pEU-E01-MCS and to Rainer Rothe (University of Lyon, Lyon, France) for technical advice on wheat germ expression. We thank Peter Zimmermann for providing data on HBV deletion variants and Ida Wingert for excellent technical assistance. Part of this work was funded by Infect-ERA project hepBccc (#031A508) to M.N.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022373118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Revill P. A.et al.; Members of the ICE-HBV Working Groups; ICE-HBV Stakeholders Group Chairs; ICE-HBV Senior Advisors , A global scientific strategy to cure hepatitis B. Lancet Gastroenterol. Hepatol. 4, 545–558 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lauber C., et al., Deciphering the origin and evolution of hepatitis B viruses by means of a family of non-enveloped fish viruses. Cell Host Microbe 22, 387–399.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Magnius L.et al.; Ictv Report Consortium , ICTV virus taxonomy profile: Hepadnaviridae. J. Gen. Virol. 101, 571–572 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beck J., Nassal M., Hepatitis B virus replication. World J. Gastroenterol. 13, 48–64 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nassal M., Hepatitis B viruses: Reverse transcription a different way. Virus Res. 134, 235–249 (2008). [DOI] [PubMed] [Google Scholar]
- 6.Nassal M., HBV cccDNA: Viral persistence reservoir and key obstacle for a cure of chronic hepatitis B. Gut 64, 1972–1984 (2015). [DOI] [PubMed] [Google Scholar]
- 7.Xiong Y., Eickbush T. H., Origin and evolution of retroelements based upon their reverse transcriptase sequences. EMBO J. 9, 3353–3362 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Knaus T., Nassal M., The encapsidation signal on the hepatitis B virus RNA pregenome forms a stem-loop structure that is critical for its function. Nucleic Acids Res. 21, 3967–3975 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nassal M., Rieger A., A bulged region of the hepatitis B virus RNA encapsidation signal contains the replication origin for discontinuous first-strand DNA synthesis. J. Virol. 70, 2764–2773 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang G. H., Seeger C., Novel mechanism for reverse transcription in hepatitis B viruses. J. Virol. 67, 6507–6512 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bartenschlager R., Schaller H., Hepadnaviral assembly is initiated by polymerase binding to the encapsidation signal in the viral RNA genome. EMBO J. 11, 3413–3420 (1992). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pollack J. R., Ganem D., Site-specific RNA binding by a hepatitis B virus reverse transcriptase initiates two distinct reactions: RNA packaging and DNA synthesis. J. Virol. 68, 5579–5587 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rieger A., Nassal M., Specific hepatitis B virus minus-strand DNA synthesis requires only the 5′ encapsidation signal and the 3′-proximal direct repeat DR1. J. Virol. 70, 585–589 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tavis J. E., Perri S., Ganem D., Hepadnavirus reverse transcription initiates within the stem-loop of the RNA packaging signal and employs a novel strand transfer. J. Virol. 68, 3536–3543 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lanford R. E., Notvall L., Lee H., Beames B., Transcomplementation of nucleotide priming and reverse transcription between independently expressed TP and RT domains of the hepatitis B virus reverse transcriptase. J. Virol. 71, 2996–3004 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weber M., et al., Hepadnavirus P protein utilizes a tyrosine residue in the TP domain to prime reverse transcription. J. Virol. 68, 2994–2999 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zoulim F., Seeger C., Reverse transcription in hepatitis B viruses is primed by a tyrosine residue of the polymerase. J. Virol. 68, 6–13 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Loeb D. D., Hirsch R. C., Ganem D., Sequence-independent RNA cleavages generate the primers for plus strand DNA synthesis in hepatitis B viruses: Implications for other reverse transcribing elements. EMBO J. 10, 3533–3540 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Beck J., Nassal M., A Tyr residue in the reverse transcriptase domain can mimic the protein-priming Tyr residue in the terminal protein domain of a hepadnavirus P protein. J. Virol. 85, 7742–7753 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jones S. A., Boregowda R., Spratt T. E., Hu J., In vitro epsilon RNA-dependent protein priming activity of human hepatitis B virus polymerase. J. Virol. 86, 5134–5150 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang G. H., Seeger C., The reverse transcriptase of hepatitis B virus acts as a protein primer for viral DNA synthesis. Cell 71, 663–670 (1992). [DOI] [PubMed] [Google Scholar]
- 22.Beck J., Nassal M., Formation of a functional hepatitis B virus replication initiation complex involves a major structural alteration in the RNA template. Mol. Cell. Biol. 18, 6265–6272 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gajer M., et al., Few basepairing-independent motifs in the apical half of the avian HBV ε RNA stem-loop determine site-specific initiation of protein-priming. Sci. Rep. 7, 7120 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Beck J., Nassal M., Sequence- and structure-specific determinants in the interaction between the RNA encapsidation signal and reverse transcriptase of avian hepatitis B viruses. J. Virol. 71, 4971–4980 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schaaf S. G., Beck J., Nassal M., A small 2′-OH- and base-dependent recognition element downstream of the initiation site in the RNA encapsidation signal is essential for hepatitis B virus replication initiation. J. Biol. Chem. 274, 37787–37794 (1999). [DOI] [PubMed] [Google Scholar]
- 26.Wang G. H., Zoulim F., Leber E. H., Kitson J., Seeger C., Role of RNA in enzymatic activity of the reverse transcriptase of hepatitis B viruses. J. Virol. 68, 8437–8442 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hu J., Boyer M., Hepatitis B virus reverse transcriptase and epsilon RNA sequences required for specific interaction in vitro. J. Virol. 80, 2141–2150 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dörnbrack K., “In vitro Rekonstitution der Replikationsinitiation des humanen Hepatitis B Virus,” PhD thesis, University of Freiburg, Germany (2014).
- 29.Juven-Gershon T., Hsu J. Y., Theisen J. W., Kadonaga J. T., The RNA polymerase II core promoter - the gateway to transcription. Curr. Opin. Cell Biol. 20, 253–259 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vo Ngoc L., Cassidy C. J., Huang C. Y., Duttke S. H., Kadonaga J. T., The human initiator is a distinct and abundant element that is precisely positioned in focused core promoters. Genes Dev. 31, 6–11 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ogram S. A., Flanegan J. B., Non-template functions of viral RNA in picornavirus replication. Curr. Opin. Virol. 1, 339–346 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Paul A. V., Wimmer E., Initiation of protein-primed picornavirus RNA synthesis. Virus Res. 206, 12–26 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Summers J., Smith P. M., Horwich A. L., Hepadnavirus envelope proteins regulate covalently closed circular DNA amplification. J. Virol. 64, 2819–2824 (1990). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Krupovic M., Koonin E. V., Homologous capsid proteins testify to the common ancestry of retroviruses, caulimoviruses, pseudoviruses, and metaviruses. J. Virol. 91, e00210–e00217 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Miller R. H., Robinson W. S., Common evolutionary origin of hepatitis B virus and retroviruses. Proc. Natl. Acad. Sci. U.S.A. 83, 2531–2535 (1986). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Krupovic M., et al., Ortervirales: New virus order unifying five families of reverse-transcribing viruses. J. Virol. 92, e00515–e00518 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Galligan J. T., Marchetti S. E., Kennell J. C., Reverse transcription of the pFOXC mitochondrial retroplasmids of Fusarium oxysporum is protein primed. Mob. DNA 2, 1 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang H., Lambowitz A. M., The Mauriceville plasmid reverse transcriptase can initiate cDNA synthesis de novo and may be related to reverse transcriptase and DNA polymerase progenitor. Cell 75, 1071–1081 (1993). [DOI] [PubMed] [Google Scholar]
- 39.Krupovic M., Dolja V. V., Koonin E. V., Origin of viruses: Primordial replicators recruiting capsids from hosts. Nat. Rev. Microbiol. 17, 449–458 (2019). [DOI] [PubMed] [Google Scholar]
- 40.Koonin E. V., Dolja V. V., Virus world as an evolutionary network of viruses and capsidless selfish elements. Microbiol. Mol. Biol. Rev. 78, 278–303 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Salas M., Protein-priming of DNA replication. Annu. Rev. Biochem. 60, 39–71 (1991). [DOI] [PubMed] [Google Scholar]
- 42.Fogeron M. L., Badillo A., Penin F., Böckmann A., Wheat germ cell-free overexpression for the production of membrane proteins. Methods Mol. Biol. 1635, 91–108 (2017). [DOI] [PubMed] [Google Scholar]
- 43.Takai K., Sawasaki T., Endo Y., Practical cell-free protein synthesis system using purified wheat embryos. Nat. Protoc. 5, 227–238 (2010). [DOI] [PubMed] [Google Scholar]
- 44.David G., et al., Phosphorylation and alternative translation on wheat germ cell-free protein synthesis of the DHBV large envelope protein. Front. Mol. Biosci. 6, 138 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Parisien M., Major F., The MC-Fold and MC-Sym pipeline infers RNA structure from sequence data. Nature 452, 51–55 (2008). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.