Abstract
BACKGROUND:
Carbapenems are the antibiotics of last-resort for the treatment of bacterial infections caused by multidrug-resistant organisms. The emergence of resistance is a critical and worrisome problem for clinicians and patients. Carbapenem-resistant Enterobacterales (CRE) are spreading globally, are associated with an increased frequency of reported outbreaks in many regions, and are becoming endemic in many others.
OBJECTIVES:
Determine the molecular epidemiology of CRE isolates from various regions of Saudi Arabia to identify the genes encoding resistance and their clones for a better understanding of the epidemio-logical origin and national spread.
DESIGN:
Multicenter, cross-sectional, laboratory-based study.
SETTING:
Samples were collected from 13 Ministry of Health tertiary-care hospitals from five different regions of Saudi Arabia.
METHODS:
Isolates were tested using the GeneXpert molecular platform to classify CRE.
MAIN OUTCOME MEASURES:
Prevalence of various types of CRE in Saudi Arabia.
SAMPLE SIZE:
519 carbapenem-resistant isolates.
RESULT:
Of 519 isolates, 440 (84.7%) were positive for CRE, with Klebsiella pneumoniae (410/456, 90%) being the most commonly isolated pathogen. The distribution of the CRE-positive K pneumoniae resistance genes was as follows: OXA-48 (n=292, 71.2%), NDM-1 (n=85, 20.7%), and NDM+OXA-48 (n=33, 8%). The highest percentage of a single blaOXA-48 gene was detected in the central and eastern regions (77%), while the blaNDM-gene was the predominant type in the northern region (27%). The southern regions showed the lowest percentages for harboring both blaOXA-48 and blaNDM genes (4%), while the western region isolates showed the highest percentage of harboring both genes (14%).
CONCLUSION:
The results illustrate the importance of molecular characterization of CRE isolates for patient care and infection prevention and control. Larger multicenter studies are needed to critically evaluate the risk factors and trends over time to understand the dynamics of spread and effective methods of control.
LIMITATIONS:
Lack of phenotypic susceptibility and clinical data.
CONFLICT OF INTEREST:
None.
INTRODUCTION
Antimicrobial resistance (AMR) is a major concern nationally and internationally. Major international efforts are ongoing by the World Health Organization to combat AMR through the initiation of a global action plan for antimicrobial resistance, the Global Antimicrobial Resistance Surveillance System (GLASS). The most recent reports of GLASS 2017 and 2018 showed that the prevalence rate of carbapenemase producers among Enterobacterales in Saudi Arabia range from 10–30%.1–3 Saudi Arabia became a leading member of these two programs in the Middle East. Continuous monitoring, molecular characterization, and identification of the source of these mechanisms of resistance are required to limit the spread of AMR.4,5 Globally, infections caused by gram-negative bacteria are associated with high rates of morbidity and mortality and cause many types of infections including bloodstream, urinary tract, respiratory tract and gastrointestinal tract infections.6 The emergence and dissemination of antibiotic resistance in gram-negative bacteria is complicating the treatment of serious nosocomial infections and is already creating bacterial species resistant to all currently available agents.7,8
Increasing degrees of resistance to major antimicrobials were first caused by extended-spectrum-β-lactamase (ESBL) producing gram-negative bacteria in the mid-2000's, and more recently by carbapenemase-producers, which are a huge clinical and public health concern worldwide; countries of the Gulf Cooperation Council (GCC) are no exception.9 Carbapenems are the last-resort antibiotics in the treatment of bacterial infections caused by multidrug-resistant organisms, and the emergence of resistance is an extremely critical and worrisome problem for clinicians and patients. The WHO considers carbapenem-resistant Enterobacterales to be a critical public health issue of global concern that requires immediate action.10 Resistance to carbapenems develops by several mechanisms; in Enterobacterales, this is mainly achieved by the acquisition of carbapenemases, resulting in carbapenem-resistant Enterobacterales (CRE). CRE are now spreading globally and are associated with an increased frequency of reported outbreaks in many regions; CRE are becoming endemic in some parts of the world.11–13 Other mechanisms of carbapenem resistance in Enterobacterales include permeability changes, AmpC- and ESBL acquisition/overexpression, and an efflux mechanism.14
There are five classes of carbapenemases found globally, which include three Ambler classes: KPC (Klebsiella pneumoniae carbapenemase) (class A); IMP (imipenem), NDM-1 (New Delhi metallo-β-lactamase), and VIM (Verona integron-encoded metallo-β-lactamase) (class B); and OXA-48-like (OXA: oxacillinase) carbapenemase (class D).12 Carbapenemase genes have very effective transmission abilities because they are encoded on mobile genetic elements, and their spread has been associated with both successful high-risk clones (e.g., KPC in Klebsiella pneumoniae clonal group ST258)15 and inter- and intra-species plasmid spread (e.g., blaKPC in pKpQIL and blaOXA-48-like in pOXA-48a).16 Additionally, these carbapenemase plasmids can harbor genes that encode resistance to other antibiotic classes like quinolones, polymyxin, and amino-glycosides that might be encoded on other plasmids or the chromosome, resulting in difficulty in finding other options to treat a patient infected with these organisms. These infections are associated with prolonged hospital stays and increased mortality.17–20 In this study, we aimed to determine the molecular characteristics of CREs isolated from various regions of Saudi Arabia to identify the genes and clones encoding resistance to better understand their epidemiological origin and spread.
METHODS
This multicenter, cross-sectional, laboratory-based study included Ministry of Health tertiary-care hospitals and laboratories selected from five different regions Saudi Arabia, which makes the data representative of the country. These hospital selections were based on special consideration, i.e., the number of beds, scope of services provided, and number of isolates obtained over one year starting from January to December, 2018. We included the first non-duplicate CRE isolated from each patient received from these hospitals, along with the demographic information on patients per the study protocol. All participating laboratories used the Clinical and Laboratory Standards Institute (CLSI) definition of CRE, screening by an automated system, and confirmation by E-test if the MIC was ≥1 for imipenem or meropenem. Molecular testing was performed using the Xpert Carba-R Assay (Cepheid, Sunnyvale, California, United States), a real-time polymerase chain reaction assay for the detection of blaKPC, blaNDM-1, blaVIM, blaIMP, and blaOXA-48 carbapenem resistance genes from bacterial isolates grown on blood agar or MacConkey agar.21 All data were analyzed using SPSS software (IBM Corp. released 2017, IBM SPSS Statistics for Windows, Version 25.0. Armonk, NY: IBM Corp.) to calculate percentages, chi-square values and P values. The respective institutional ethics and review boards of the included centers approved this study (Research Project No. E-20-4613).
RESULTS
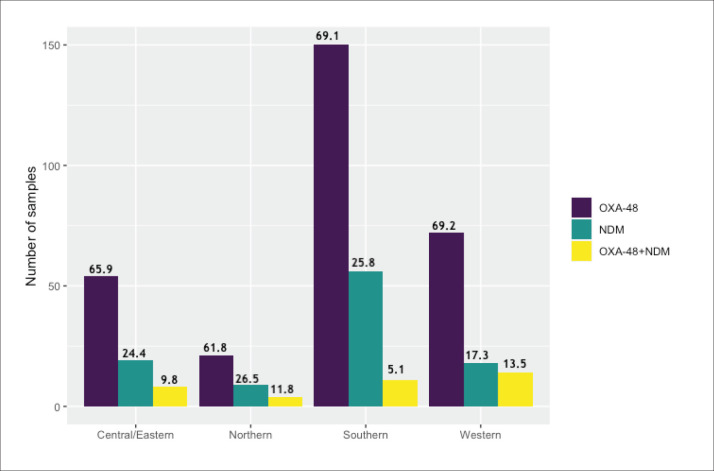
Of the 519 carbapenems-resistant isolates tested, 440 (84.7%) tested positive for CRE (Table 1). The rate of CRE was highest among K pneumoniae, with relatively few numbers of other gram-negative bacteria. All the three CRE-positive K oxytoca isolates were NDM-1 producers. Similarly, NDM-1 production was detected in 5/8 CRE-positive E cloacae, followed by 3 OXA-48 (n=3, 38%). IMP1, VIM, and KPC were not detected in any of these isolates. The anatomical source and classification of the 519 carbapenem-resistant Enterobacterales isolates are shown in Table 2. The distribution by region is shown in Figure 1.
Table 1.
Molecular characterization of the 519 carbapenem-resistant Enterobacterales isolates.
| Positive result | None Detected | Molecular classification | ||||
|---|---|---|---|---|---|---|
| NDM-1 | OXA-48 | NDM-1 +OXA-48 | VIM | |||
| Number of isolates (n=519) | 440/519 (85) | 79/519 (15) | 103/519 (20) | 298/519 (57) | 37/519 (7) | 2/519 (0.4) |
| K pneumoniae (n=456) | 410/456 (89.9) | 46/456 (10.1) | 85/410 (20.7) | 292/410 (71.2) | 33/410 (8) | – |
| K oxytoca (n=3) | 3/3 (100) | – | 3/3 (100) | – | – | – |
| E coli (n=21) | 13/21 (61.9) | 8/X (30) | 9/13 (69) | 1/13 (7.7) | 3/13 (23) | – |
| E cloacae (n=13) | 8/13 (62) | 5/13 (38) | 5/8 (63) | 3/8 (38) | – | – |
| Other gram-negative organismsa (n=26) | 26 (100) | 20/26 (76.9) | 1 (3.8) | 2 (7.7) | 1 (3.8) | 2 (7.7) |
Data are number (%).
Proteus mirabilis VIM and OXA-48 (n=2/8), Pseudomonas fluorescens VIM (n=1/1), Acinetobacter baumannii NDM (n=1/8), K ozaenae OXA-48 (n=1/1), Prevotella denticola NDM (n=1) and 1 OXA-48 (n=1), and Serratia marcescens (n=3) Morganella morganii (n=2) E aerogenes (n=1), Pseudomonas (n=1) negative for all genes.
Table 2.
The anatomical source of 519 carbapenem-resistant Enterobacterales isolates (n=519).
| Respiratory system | 136 (26.2) |
| Urine | 115 (22.2) |
| Tissue and wound | 103 (19.8) |
| Blood | 62 (11.9) |
| Swab | 32 (6.17) |
| Fluid | 17 (3.28) |
| Unknown | 54 (10.4) |
Data are number (%).
Figure 1. Participating regions and molecular classification of the 436 carbapenem-resistant Enterobacterales isolates by region (percentage for region shown above bars).
Tables 3 and 4 show the molecular classification of CRE-positive K pneumoniae across the different Saudi regions and hospitals participating in the study. The highest percentage of a single blaOXA-48 gene was detected in the central and eastern regions (77%), and the lowest percentage was detected in the northern region (61%). For the single blaNDM-1 gene the highest percentage was in the northern region (27%) and lowest in the central and eastern regions (15%). The southern region isolates showed the lowest percentage (4%) of both bla-OXA-48 and blaNDM-1 genes present together whereas the western region isolates showed the highest percentage harboring both genes (14%). This slight difference in the prevalence of carbapenemase types among regions was not statistically significant (chi-square=9.2854, df=6, P=.1582).
Table 3.
Molecular classification of 456 isolates of K pneumoniae submitted for molecular testing by region.
| Regions | K pneumoniae received (n=456) | K pneumoniae positive (n=410) | Single NDM-1 (n=85) | Single OXA-48 (n=292) | NDM-1+OXA-48 (n=33) |
|---|---|---|---|---|---|
| Northern | 35 | 33 | 9 (27) | 20 (61) | 4 (12) |
| Central and Eastern | 90 | 78 | 12 (15) | 60 (77) | 6 (7) |
| Western | 111 | 94 | 15 (16) | 65 (69) | 14 (14) |
| Southern | 220 | 205 | 49 (24) | 147 (72) | 9 (4) |
| Mean number (%) for all regions | 114 | 102.5 | 21.1 (20.5) | 73 (71.2) | 8.25 (8.1) |
Data are number (%). Chi-square=9.2854, df=6, P=.1582 for comparison of genetic types (shaded light blue).
Table 4.
Participating hospitals and molecular classification of 456 isolates of K pneumoniae.
| Region | Participating hospitals | K pneumoniae received (n=456) | K pneumoniae positive (n=410) | NDM-1 detected (n=85) | OXA48 detected (n=292) | NDM-1 +OXA48 detected (n=33) |
|---|---|---|---|---|---|---|
| Central | King Salman Bin Abdulaziz Hospital, Riyadh (n=54) | 47 | 45 (96) | 5 (11) | 40 (89) | 0 (0) |
| Central | King Khalid Hospital and Prince Sultan Center for Health Service, Al Kharj (n=24) | 17 | 9 (53) | 3 (33) | 6 (67) | 0 (0) |
| Central | King Fahd Specialist Hospital, Buraydah (n=22) | 20 | 18 (90) | 4 (22) | 8 (44) | 6 (33) |
| East | Regional Laboratory and Blood Bank, Microbiology Department, Dammam (n=7) | 6 | 6 (100) | 0 (0) | 6 (100) | 0 (0) |
| North | Gurayat General Hospital (n=23) | 23 | 22 (96) | 7 (32) | 11 (50) | 4 (18) |
| North | Hael General Hospital (n=20) | 6 | 5 (83) | 0 (0) | 5 (100) | 0 (0) |
| North | Arar Central Hospital (n=6) | 6 | 6 (100) | 2 (33) | 4 (67) | 0 (0) |
| South | Asir Hospital (n=222) | 208 | 193 (93) | 49 (25) | 137 (71) | 7 (4) |
| South | King Fahd Hospital, Al Baha, (n=12) | 12 | 12 (100) | 0 (0) | 10 (83) | 2 (17) |
| West | King Faisal Medical Complex, Taif (n=15) | 15 | 12 (80) | 2 (17) | 8 (67) | 2 (17) |
| West | King Abdulaziz Specialist Hospital, Taif (n=8) | 8 | 8 (100) | 1 (13) | 7 (88) | 0 (0) |
| West | King Fahad Hospital, Madina Munawara (n=95) | 83 | 70 (84) | 11 (16) | 49 (70) | 10 (!4) |
| West | Hera General Hospital (n=11) | 5 | 4 (80) | 1 (25) | 1 (25) | 2 (50) |
Data are number (%).
DISCUSSION
Over the past ten years, carbapenemase-producing Enterobacterales (CRE) have been increasingly reported worldwide,22–24 and their dissemination has been highlighted as a significant global public health threat. Enterobacterales that produce carbapenemases commonly have so few treatment options that their spread threatens our system of modern healthcare. Recent GLASS data have shown an increase in the rate of carbapenem-resistant K pneumoniae isolated in 2017 and 2018 from different infection sites including urine isolates (increased from 15% to 22%) and blood isolates (increased from 30% to 35%).25
Since the discovery and early isolation of the first metallo-β-lactamase from a urinary tract infection in a Swedish patient who had traveled to New Delhi in 2008,26 there has been a dramatic spread and global dissemination of this carbapenemase-producing gene (named the New Delhi metallo-β-lactamase) worldwide, with several reports showing variability in the prevalence of NDM-1 and other types of carbapenemases in Enterobacterales across different countries. Before long, CRE became endemic to Asia, northern European and Pacific regions, the UK, and the Middle East.27–30 The other types of carbapenemases in Enterobacterales common to the Middle East and Arabian countries are OXA-48 and OXA-48-like (i.e., OXA-163 and OXA-181) carbapenemases with a prevalence range of 2–10%.30–33
In the current study, the mean percentages of genes encoding for carbapenemase-producing K pneumoniae were consistent with previous studies: the single OXA-48 type was the most common (71.2%) followed by NDM-1 (20.5%), and the least common isolates were those harboring both OXA-48 and NDM-1, across all study centers. Two earlier studies from central Saudi Arabia reported the following: the first one performed on 71 CRE isolates showed that OXA-48 was the most common type (67.6%), followed by NDM-1 (12.7%) and both (8.5%),34 The second study performed on 60 CRE isolates which showed that OXA-48 was detected in 78.3% and NDM-1 in 20%.35 Another more recent study from the same region performed on 31 CREs isolated from three hospitals showed that OXA-48 and NDM-1 accounted for 58% and 42%, respectively.36 In 2015, another study on a smaller number of isolates from the two largest hospitals in the southern region detected 81.5% OXA-48 and 7.4% NDM-1, respectively.37 Recently several studies reported the appearance of the KPC type in the Central and Western regions of Saudi Arabia, the GCC and other Middle Eastern countries.30,38–41
In this study, some of the isolates tested negative for carbapenemases (n=79 [15.4%]) and these isolates might harbor other carbapenemases enzymes not included in Xpert Carba-R Assay like OXA-48-like carbapenemase or they might contain other mechanisms for carbapenems resistance including the diminishment of outer membrane permeability due to decreased expression or loss of OprD porin expression, efflux pumps, or AmpC ?-lactamases.42 There was a slight variability in the percentage of CRE encoding genes between regions, which could be explained by the circulating strain causing outbreaks in that specific region during the course of the study. The variability among studies can also be explained by the inclusion criteria, types and number of patients, and the geographic location of the study.
In the GCC, a molecular classification of 45 CRE isolates found that the blaOXA-48 gene was detected in 56.4%, blaNDM-1 gene in 25.8%, and both blaOXA-48 and blaNDM-1 genes were detected in 9.7% of the isolates.30 In the Gulf region, the most prevalent carbapenemase classes are NDM-1 and OXA-48, and these genes are carried on plasmid replicon type Inc L/M, Tn1999, IS1999 and rarely IncA/C in OXA-48-like. A mix of IncR, IncFII, were responsible for a spread of NDM-1.43 These plasmids are mobile elements and facilitate the horizontal transmission of these resistance mechanisms among patients in hospitals, especially those with a risk factor for colonization and infection with CRE.44
In the current study, nearly 10% of carbapenem-resistant K pneumoniae isolates were negative for all genes encoding the CRE type on the Xpert Carba-R Assay, which is most likely indicative of other mechanisms of resistance like AmpC hyper-production with efflux, loss of porins, or outer membrane protein mutations.45
Recently, the rate of colonization in hospitalized patients with CRE has been confirmed to be in the range of 2–13.5%, especially among high-risk groups such as patients in ICUs and patients on multiple antibiotics, which can lead to the spread of this organism in health-care facilities, longer hospital stays, and a higher rate of morbidity and mortality.46–49 Asymptomatic carriage of CREs has been documented in patients admitted to the hospital; in a survey of 1806 hospitals, 299 (16.5%) patients showed colonization with CRE, which progressed to clinical infection.50 This might be due to widespread CRE contamination of the environment, especially drinking and sewage water, which was shown earlier in a study in India.51
Various laboratory methods including both phenotypic and genotypic methods, have been used to screen patients for the carriage of CRE. The implementation varies from one clinical microbiologist to another and depends on the available resources; however, the molecular methods are rapid, sensitive, and specific, and have been made more affordable in recent years.52 To prove transmission and document an outbreak of CRE, molecular typing using whole-genome sequencing can be performed to determine if the strains of the outbreak are related to the same clone or sequence type or not. The majority of OXA-48 are related to ST147, ST11, ST101, ST405, and ST395 while NDM-1 are related to ST101, ST11.53
This study is an initial surveillance study to determine the most common CRE types nationwide. Our future plan is to perform molecular sequencing to identify the ST and plasmids associated with the spread and identify whether differences exist between different geographic regions. We have not included the phenotypic susceptibility testing and minimal inhibitory concentration of these isolates to carbapenems because of inconsistency in the methods of testing and reporting in these hospitals. Despite that, our study provides valuable epidemiological data and likely, good guidance for future national antimicrobial guidelines and use of new ?-lactamase inhibitor combinations, which have variable antibacterial activity depending on the molecular type of CRE; for example ceftazidime/avibactam has higher activity against OXA-48-like CRE type and the addition of aztreonam is required to treat NDM-1 CRE types.54–55
Funding Statement
None.
REFERENCES
- 1.World Health Organization (WHO). Global action plan for antimicrobial resistance. 2015. [cited 2020 Nov 23]. Available from: http://apps.who.int/iris/bitstream/10665/193736/1/9789241509763_eng.pdf?ua=1. [DOI] [PubMed]
- 2.World Health Organization (WHO). Global Antimicrobial Resistance Surveil-lance System Manual for Early Implementation. 2015. [cited 2020 Nov 23]. Available from: http://apps.who.int/iris/bitstream/10665/188783/1/9789241549400_eng.pdf
- 3.World Health Organization (WHO). Global Antimicrobial Resistance Surveillance System (GLASS) Report Early implementation 2017-2018. Pages 182-185. [Cited 2020 Dec 25]. Available from: https://www.who.int/glass/resources/publications/early-implementation-report-2017-2018/en/
- 4.Liu YY, Wang Y, Walsh TR, Yi LX, Zhang R, Spencer J, et al. Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect Dis. 2016;16(2):161-8. [DOI] [PubMed] [Google Scholar]
- 5.Clinical and Laboratory Standards Institute (CLSI). Performance Standards for Antimicrobial Susceptibility Testing. 28th ed. CLSI supplement M100. Wayne, PA: Clinical and Laboratory Standards Institute; 2018. [Cited 2020 Dec 26]. Available from: http://file.qums.ac.ir/repository/mmrc/CLSI-2018-M100-S28.pdf [Google Scholar]
- 6.Morris S, Cerceo E. Trends, Epidemiology, and Management of Multi-Drug Resistant Gram-Negative Bacterial Infections in the Hospitalized Setting. Antibiotics (Basel). 2020;9(4):196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Karakonstantis S, Kritsotakis EI, Gikas A. Treatment options for K. pneumoniae, P. aeruginosa and A. baumannii co-resistant to carbapenems, aminoglycosides, polymyxins and tigecycline: an approach based on the mechanisms of resistance to carbapenems. Infection. 2020:1–17. [DOI] [PMC free article] [PubMed]
- 8.Potter RF, D’Souza AW, Dantas G. The rapid spread of carbapenem-resistant Enterobacteriacea. Drug Res Updates. 2016;29:30-46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Moghnieh RA, Kanafani ZA, Tabaja HZ, Sharara SL, Awad LS, Kanj SS. Epidemiology of common resistant bacterial pathogens in the countries of the Arab League. The Lancet Infect Dis. 2018;18(12):E379-E394. [DOI] [PubMed] [Google Scholar]
- 10.World Health Organization (WHO). Antimicrobial Resistance: Global Report on Surveillance. 2014. [cited 2020 Nov 23]. Available from: http://apps.who.int/iris/bitstream/10665/112642/1/9789241564748_eng.pdf.
- 11.Nordmann P. Carbapenemase-producing Enterobacteriacea: overview of a major public health challenge. Med Mal Infect. 2014;44:51-6. [DOI] [PubMed] [Google Scholar]
- 12.Cui X, Zhang H, Du H. Carbapenemases in Enterobacteriaceae: Detection and Antimicrobial Therapy. Front Microbiol. 2019;10:1823. doi: 10.3389/fmicb.2019.01823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cantón R, Akóva M, Carmeli Y, Giske CG, Glupczynski Y, Gniadkowski M, et al. Rapid evolution and spread of carbapenemases among Enterobacteriacea in Europe. Clin Microbiol Infect. 2012;18(5):413-31. [DOI] [PubMed] [Google Scholar]
- 14.Codjoe FS, Donkor ES. Carbapenem resistance: A review. Med Sci (Basel). 2017;6(1):1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chen L, Mathema B, Chavda KD, DeLeo FR, Bonomo RA, Kreiswirth BN. Carbapenemase-producing Klebsiella pneumoniae: molecular and genetic decoding. Trends Microbiol. 2014;22(12):686-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Poirel L, Bonnin RA, Nordmann P. Genetic features of the widespread plasmid coding for the carbapenemase OXA-48. Antimicrob Agents Chemother. 2012;56:559-62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Villa L, Poirel L, Nordmann P, Carta C, Carattoli A. Complete sequencing of an IncH plasmid carrying the blaNDM-1, blaCTXM-15 and qnrB1 genes. J Antimicrob Che-mother. 2012;67(7):1645-50. [DOI] [PubMed] [Google Scholar]
- 18.Tzouvelekis LS, Markogiannakis A, Psichogiou M, Tassios PT, Daikos GL. Carbapenemases in Klebsiella pneumoniae and other Enterobacteriacea: an evolving crisis of global dimensions. Clin Microbiol Rev. 2012;25(4):682-707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3:15-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zowawi H, Forde B, Alfaresi M, Alzarouni A, Farahat Y, Chong TM, et al. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep 2015;5:15082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Traczewski MM, Carretto E, Canton R, Moore NM. Multicenter Evaluation of the Xpert Carba-R assay for detection of carbapenemase genes in gram-negative isolates. J Clin Microbiol. 2018;56(8):e00272-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nordmann P, Naas T, Poirel L. Global spread of Carbapenemase-producing Enterobacteriacea. Emerg Infect Dis. 2011;17(10):1791-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Queenan AM, Bush K. Carbapenemases: the versatile beta-lactamases. Clin Microbiol Rev. 2007;20(3):440-58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Nordmann P, Poirel L. The difficult-to-control spread of carbapenemase producers among Enterobacteriacea worldwide. Clin Microbiol Infect. 2014;20(9):821-30. [DOI] [PubMed] [Google Scholar]
- 25.World Health Organization (WHO). Global Antimicrobial Resistance Surveil-lance System (GLASS) Report Early implementation 2017-2018. Pages 182-185. 2018. [cited 2020 Nov 23]. Available from: https://apps.who.int/iris/bitstream/handle/10665/279656/9789241515061-eng.pdf?ua=1
- 26.Yong D, Toleman MA, Giske CG, Cho HS, Sundman K, Lee K, et al. Characterization of a new metallo-beta-lactamase gene, bla(NDM-1), and a novel erythromycin esterase gene carried on a unique genetic structure in Klebsiella pneumoniae sequence type 14 from India. Antimicrob Agents Che-mother. 2009; 53:5046-54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.National Institute of Allergy and Infectious Diseases (NIAID). NIAID's Antibacterial Resistance Program: current status and future directions. 2014. [cited 2020 Nov 23]. Available from: https://www.niaid.nih.gov/sites/default/files/arstrategicplan2014 [[AUTHOR: link 404]]
- 28.Thaden JT, Lewis SS, Hazen KC, Huslage K, Fowler VG Jr, Moehring RW, et al. Rising rates of carbapenem-resistant Enterobacteriacea in community hospitals: a mixed-methods review of epidemiology and microbiology practices in a network of community hospitals in the southeastern United States. Infect Control Hosp Epidemiol. 2014;35:978-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bush K. Proliferation and significance of clinically relevant β-lactamases. Ann NY Acad Sci. 2013;1277:84-90. [DOI] [PubMed] [Google Scholar]
- 30.Zowawi HM, Sartor AL, Balkhy HH, Walsh TR, AlJohani SM, AlJindan RY, et al. Molecular characterization of carbapenemase-producing Escherichia coli and Klebsiella pneumoniae in the countries of the Gulf Cooperation Council: dominance of OXA-48 and NDM producers. Antimicrob Agents Chemother. 2014;58:3085-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wartiti MA, Bahmani FZ, Elouennass M, Benouda A. Prevalence of carbapenemase-producing Enterobacteriacea in a University Hospital in Rabat, Morocco: a 19-months prospective study. Int Arab J Antimicrob Agents. 2012;2(3:4):1-6. [Google Scholar]
- 32.Wadi J, Haloub N, AlAhmad MA, Samara A, Romman A. Prevalence of meropenem susceptibility among Gram-negative pathogens isolated from intensive care units in Jordan. Int Arab J Antimicrob Agents. 2011;1(1:3):1- 8. [Google Scholar]
- 33.Kattan R, Liddawi R, Ghneim R, Siryani I, Al-Dawodi R, Abu-Diab A, et al. Emergence of Klebsiella pneumoniae carbapenemase (blaKPC-2) in members of the Enterobacteriacea family in Palestine. Int Arab J Antimicrob Agents. 2012;2(2):4. [Google Scholar]
- 34.Zaman T, Alrodayyan M, Albladi M, Aldrees M, Siddique MI, Aljohani S, et al. Clonal diversity and genetic profiling of antibiotic resistance among multidrug carbapenem-resistant Klebsiella pneumoniae isolates from a tertiary care hospital in Saudi Arabia. BMC Infect Dis. 2018;18(1):205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shibl A, Al-Agamy M, Memish Z, Senok A, Khader SA, Assiri A. The emergence of OXA-48- and NDM-1-positive Klebsiella pneumoniae in Riyadh, Saudi Arabia. Int J Infect Dis. 2013;17:e1130–33. [DOI] [PubMed] [Google Scholar]
- 36.Al-Agamy MH, Aljallala A, Radwana HH, Shibl AM. Characterization of carbapenemases, ESBLs, and plasmid-mediated quino-lone determinants in carbapenem-insensitive Escherichia coli and Klebsiella pneumoniae in Riyadh hospitals. J Infect Public Health. 2018;11:64-8. [DOI] [PubMed] [Google Scholar]
- 37.Al-Zahrani IA, Alasiri BA. The emergence of carbapenem-resistant Klebsiella pneumoniae isolates producing OXA-48 and NDM in the Southern (Asir) province, Saudi Arabia. Saudi Med J. 2018;39(1):23–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Khan MA, Mohamed AM, Faiz A, Ahmad J. Enterobacterial infection in Saudi Arabia: First record of Klebsiella pneumoniae with triple carbapenemase genes resistance. J Infect Dev Ctries. 2019;13(4):334-341. [DOI] [PubMed] [Google Scholar]
- 39.Hala S, Antony CP, Alshehri M, Althaqafi AO, Alsaedi A, Mufti A, et al. First report of Klebsiella quasipneumoniae harboring blaKPC-2 in Saudi Arabia. Antimicrob Res Infect Control. 2019;8(203). 10.1186/s13756-019-0653-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Alghoribi MF, Binkhamis K, Alswaji AA, Alhijji A, Alsharidi A, Balkhy HH, et al. Genomic analysis of the first KPC-producing Klebsiella pneumoniae isolated from a patient in Riyadh: A new public health concern in Saudi Arabia. J Infect Public Health. 2020. Apr;13(4):647-650. [DOI] [PubMed] [Google Scholar]
- 41.Al Salman J, Al Dabal L, Bassetti M, Alfouzan WA, Al Maslamani M, Alraddadi B, et al. Management of infections caused by WHO critical priority Gram-negative pathogens in Arab countries of the Middle East: a consensus paper. Int J Antimicrob Agents. 2020;56(4):106104. doi: 10.1016/j.ijantimicag.2020.106104. [DOI] [PubMed] [Google Scholar]
- 42.Meletis G. Carbapenem resistance: overview of the problem and future perspectives. Ther Adv Infect Dis. 2016;3(1):15-21. doi: 10.1177/2049936115621709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Baraniak A, Izdebski R, Fiett J, Gawryszewska I, Bojarska K, Herda M, et al. NDM-producing Enterobacteriacea in Poland, 2012–14: interregional outbreak of Klebsiella pneumoniae ST11 and sporadic cases. J Antimicrob Chemother. 2016;71(1):85-91. [DOI] [PubMed] [Google Scholar]
- 44.Tischendorf J, de Avilab RA, Safdar N. Risk of infection following colonization with carbapenem resistant Enterobactericeae: A systematic review. Am J Infect Control. 2016; 44(5):539-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Papp-Wallace KM, Endimiani A, Tara-cila MA, Bonomo RA. Carbapenems: Past, Present, and Future. Antimicrob Agents Chemother. 2011;55(11):4943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kumarasamy KK, Toleman MA, Walsh TR, Bagaria J, Butt F, Balakrishnan R, et al. Emergence of a new antibiotic resistance mechanism in India, Pakistan, and the UK: a molecular, biological, and epidemiological study. Lancet Infect Dis. 2010;10:597-602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Bharadwaj R, Joshi S, Dohe V, Gaikwad V, Kulkarni G, Shouche Y. Prevalence of New Delhi metallo-beta-lactamase (NDM-1)-positive bacteria in a tertiary care centre in Pune, India. Int J Antimicrob Agents. 2012;39:265-6. [DOI] [PubMed] [Google Scholar]
- 48.Soman R, Rodrigues C. New Delhi metallo 1: have carbapenems met their doom? Clin Infect Dis. 2010; 51:1222. 53. [DOI] [PubMed] [Google Scholar]
- 49.Perry JD, Naqvi SH, Mirza IA, Alizai SA, Hussain A, Ghirardi S, et al. Prevalence of faecal carriage of Enterobacteriacea with NDM-1 carbapenemase at military hospitals in Pakistan, and evaluation of two chromogenic media. J Antimicrob Chemother. 2011;66:2288-94. [DOI] [PubMed] [Google Scholar]
- 50.Tischendorf J, de Avila RA, Safdar N. Risk of infection following colonization with carbapenem resistant Enterobacteriacea: a systematic review. Am J Infect Control. 2016;44:539-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Walsh TR, Weeks J, Livermore DM, Toleman MA. Dissemination of NDM-1 positive bacteria in the New Delhi environment and its implications for human health: an environmental point prevalence study. Lancet Infect Dis. 2011; 11:355-62. [DOI] [PubMed] [Google Scholar]
- 52.Viau R, Frank KM, Jacobs MR, Wilson B, Kaye K, Donskey CJ, et al. Intestinal Carriage of Carbapenemase-Producing Organisms: Current Status of Surveillance Methods. Clin Microbiol Rev. 2016;29(1):1-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Logan LK, Weinstein RA. The Epidemiology of Carbapenem-Resistant Enterobacteriacea: The Impact and Evolution of a Global Menace. J Infect Dis. 2017;215(1):S28-S36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Aktaş Z, Kayacan C, Oncul O. In vitro activity of avibactam (NXL104) in combination with β-lactams against gram-negative bacteria, including OXA48 β-lactamase-producing Klebsiella pneumoniae. Int J Antimicrob Agents. 2012; 39:86-9. [DOI] [PubMed] [Google Scholar]
- 55.Shaw E, Rombauts A, Tubau F, Padullés A, Càmara J, Lozano T, et al. Clinical outcomes after combination treatment with ceftazidime/avibactam and aztreonam for NDM-1/OXA-48/CTX-M-15-producing Klebsiella pneumoniae infection. J Antimicrob Chemother. 2018;73(4):1104-1106. [DOI] [PubMed] [Google Scholar]