Significance
We show CD1d-restricted natural killer (NK)T cells have distinct metabolic profiles compared with CD4+ conventional T cells. Mature NKT cells have poor fatty acid oxidation and exhibit reduced mitochondrial respiratory reserve in the steady state. In addition, NKT cell development is more sensitive to alterations in mitochondrial electron transport chain function than conventional T cells. Using T cell-specific mitochondrial complex III ablation in mice, we further demonstrate that mitochondrial metabolism plays a crucial role in NKT cell development and function by modulating T cell receptor/interleukin-15 signaling and NFAT activity. Collectively, our data provide evidence for a critical role of mitochondrial metabolism in NKT cell development and activation, opening a new avenue for NKT cell-based immunotherapy by manipulating NKT cell metabolism.
Keywords: mitochondrial metabolism, NKT cells, T cell development, CD1, knockout mice
Abstract
Conventional T cell fate and function are determined by coordination between cellular signaling and mitochondrial metabolism. Invariant natural killer T (iNKT) cells are an important subset of “innate-like” T cells that exist in a preactivated effector state, and their dependence on mitochondrial metabolism has not been previously defined genetically or in vivo. Here, we show that mature iNKT cells have reduced mitochondrial respiratory reserve and iNKT cell development was highly sensitive to perturbation of mitochondrial function. Mice with T cell-specific ablation of Rieske iron-sulfur protein (RISP; T-Uqcrfs1−/−), an essential subunit of mitochondrial complex III, had a dramatic reduction of iNKT cells in the thymus and periphery, but no significant perturbation on the development of conventional T cells. The impaired development observed in T-Uqcrfs1−/− mice stems from a cell-autonomous defect in iNKT cells, resulting in a differentiation block at the early stages of iNKT cell development. Residual iNKT cells in T-Uqcrfs1−/− mice displayed increased apoptosis but retained the ability to proliferate in vivo, suggesting that their bioenergetic and biosynthetic demands were not compromised. However, they exhibited reduced expression of activation markers, decreased T cell receptor (TCR) signaling and impaired responses to TCR and interleukin-15 stimulation. Furthermore, knocking down RISP in mature iNKT cells diminished their cytokine production, correlating with reduced NFATc2 activity. Collectively, our data provide evidence for a critical role of mitochondrial metabolism in iNKT cell development and activation outside of its traditional role in supporting cellular bioenergetic demands.
Cellular metabolic pathways are interwoven with traditional signaling pathways to regulate the function and differentiation of T cells (1–3). Upon activation, effector T cells display a marked increase in glycolytic metabolism even in the presence of ample oxygen, termed aerobic glycolysis (4). We have previously shown that despite increased aerobic glycolysis, T cell activation depends on mitochondrial metabolism for generation of reactive oxygen species (ROS) for signaling (5). As activated T cells progress to a memory or regulatory phenotype, they preferentially oxidize fatty acids to support mitochondrial metabolism, and enhanced fatty acid oxidation (FAO) and spare respiratory capacity (SRC) are essential to maintenance of their phenotype (6, 7).
CD1d-restricted invariant natural killer T (iNKT) cells are a unique subset of lymphocytes that exhibit a preactivated phenotype with rapid effector responses (8, 9). iNKT cells are capable of producing large amount of proinflammatory and antiinflammatory cytokines thus have broad immunomodulatory roles (8–10). Given that these cells are poised for rapid proliferation and cytokine production, we hypothesized that coordination of cellular signaling with cellular metabolism will be especially critical for optimal iNKT function. In support of this hypothesis, several studies suggest that modulation of cellular metabolism affects iNKT cell development and function. iNKT cell development is diminished upon deletion of the miR-181 a1b1 cluster, which regulates phosphoinositide 3-kinase signaling and decreases aerobic glycolysis (11, 12). In addition, T cell-specific deletion of Raptor (a component of mTORC1), a metabolic regulator, leads to defects in iNKT cell development and function (13, 14). Loss of folliculin-interacting protein 1 (Fnip1), an adaptor protein that physically interacts with AMP-activated protein kinase, also results in defective NKT cell development, and interestingly conventional T cells develop normally (15). Furthermore, a number of studies targeting bioenergetics processes or related molecules, like alteration of glucose metabolism, mitochondrial-targeted antioxidant treatment, and receptor-interacting protein kinase 3-dependent activation of mitochondrial phosphatase, showed significant effects on iNKT cell ratio and function (16–19). A recent study showed that iNKT cells are less efficient in glucose uptake than CD4+ T cells. Furthermore, activated iNKT cells preferentially metabolize glucose by the pentose phosphate pathway and mitochondria, instead of converting into lactate, since high lactate environment is detrimental to their homeostasis and effector function (20).
In conventional lymphocytes, mitochondria clearly play a role in coordination of cell signaling and cell fate decisions outside of production of energy (5, 21–23). During T cell activation mitochondria localize at immune synapses that T cells form with antigen-presenting cells (22). T cell receptor (TCR) stimulation triggers mitochondrial ROS (mROS) production as well as mitochondrial ATP production that are released at the immune synapses and are critical for Ca2+ homeostasis and modulation of TCR-induced downstream signaling pathways (22). We previously showed that mice with T-cell–specific deletion of Rieske iron sulfur protein (RISP), a component of mitochondrial complex III of the mitochondrial electron transport chain (ETC), are defective in antigen-specific T cell activation due to deficiency of mROS required for cellular signaling (5). Several recent studies showed that ROS or factors that affect ROS production are also important in iNKT cell development and effector functions (24–27). In addition, inhibition of mitochondrial oxidative phosphorylation (OXPHOS) by oligomycin has been shown to decreased survival and cytokine production by splenic iNKT cells (20). However, the requirement of mitochondrial metabolism for iNKT cell development and function has not been previously defined genetically or in vivo.
Here we showed that iNKT cells have comparable basal mitochondrial oxygen consumption to conventional T cells but displayed lower SRC and FAO, which are thought to impart cells with mitochondrial reserve under stress. Using Uqcrfs1fl/fl;CD4-Cre+ (hereafter referred as T-Uqcrfs1−/−) mice, we showed that abrogation of mitochondrial metabolism resulted in a cell-autonomous defect in iNKT cell development in thymus and periphery. The iNKT cells were able to proliferate but exhibited impaired activation, suggesting that they were not lacking bioenergetically but rather had aberrant TCR signaling in vivo, leading to altered expression of downstream factors required for their terminal maturation. Accordingly, T-Uqcrfs1−/− iNKT cells displayed lower T-bet and CD122 levels and did not respond to interleukin (IL)-15 stimulation. Knockdown of RISP in mature iNKT cells also limited NFATc2 translocation to the nucleus. Collectively, our data highlighted an important role of mitochondrial metabolism in modulating TCR signaling in vivo and regulating iNKT cell development and function.
Results
iNKT Cells Are Metabolically Distinct from CD4+ Conventional T Cells.
While most conventional T cells circulate in the body in a naive resting state, NKT cells exhibit a preactivated effector phenotype (8). To determine whether this translates to distinct metabolic programs, we first compared mitochondrial oxygen consumption rate (OCR) between splenic iNKT cells and CD4+ conventional T cells. We found that splenic iNKT cells and CD4+ T cells had comparable rates of basal OCR; however, consistent with a previous study (28), splenic iNKT cells had markedly reduced mitochondrial SRC, as indicated by the lower increase of OCR following uncoupling with flurocarbonyl cyanide phenylhydrazone (FCCP) (Fig. 1 A and C). Given that iNKT cells are predominant in the liver, we further evaluated SRC in hepatic iNKT cells in comparison with hepatic CD4+ conventional T cells. Similar to splenic iNKT cells, hepatic iNKT cells had barely detectable SRC (Fig. 1 B and C). It is worth noting that hepatic CD4+ T cells exhibited lower SRC than splenic CD4+ T cells (Fig. 1C), indicating the unique tissue environment of the liver influenced metabolic profiles of resident T cells. However, the consistent patterns observed between splenic and hepatic iNKT cells suggest iNKT cells intrinsically possessed a lower SRC independent of organ distribution. SRC has previously been shown to be driven by FAO (4, 7). Therefore, we assessed ability to oxidize fatty acids by measuring OCR following treatment with the fatty acid palmitate-BSA. Both splenic and hepatic iNKT cells had significantly lower increase in OCR after treatment with palmitate-BSA compared to organ-matched CD4+ T cells and reduced sensitivity to etomoxir, an inhibitor of the rate-limiting step in FAO, suggesting reduced rates of FAO (Fig. 1 D and E). A reduction in SRC might be explained by a decrease in total mitochondrial mass. Consistent with previous reports (20, 28), we found iNKT cells had significantly lower mitochondrial mass compared to CD4+ T cells, as indicated by decreased MitoTracker Green staining and decreased relative mitochondrial DNA levels (Fig. 1 F, Upper, Fig. 1G, and SI Appendix, Fig. S1). iNKT cells also had lower mitochondrial membrane potential as shown by reduced tetramethylrhodamine, ethyl ester (TMRE) staining compared with CD4+ T cells (Fig. 1 F, Lower). Collectively, our data reveal that lower mitochondrial mass in iNKT cells is associated with reduced SRC and FAO, which might render them more susceptible or sensitive to mitochondrial injury or stress.
Fig. 1.
iNKT cells exhibit distinct metabolic profile. (A–C) OCR of purified iNKT cells and CD4+ T cells from the spleen and liver were measured under basal conditions and in response to oligomycin (oligo), FCCP, and rotenone plus antimycin A (Rot/AM). Real-time OCR levels (A and B) and SRC (percent maximum OCR after FCCP injection of baseline OCR) (C) of indicated cells are presented as mean + SEM (n = 3 to 6). (D) FAO of splenic iNKT and CD4+ T cells was detected by measuring the OCR level in response to substrate palmitate-BSA (Pal-BSA) and inhibitor etomoxir (Eto). (E) FAO of indicated cells was calculated as increase in OCR in response to palmitate-BSA and shown as mean + SEM (n = 6 for spleen and n = 3 for liver). (F) Cells from indicated organs of B6 mice were stained with CD1d/PBS57 tetramer and mAb against TCR-β, CD4, and CD8 followed by incubation with MitoTracker Green (MTG) or TMRE. Data are representative from seven experiments. (G) Relative mtDNA was quantifying the ratio of mitochondrial cytochrome C oxidase subunit 1 (MitoCO1) or NADH dehydrogenase (MitoND1) to nuclear DNA in iNKT cells and normalized to CD4+ T cells (n = 3). Data are shown as mean + SEM from three experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
iNKT Cell Development Is More Sensitive to Mitochondrial Perturbation than Conventional T Cell Development.
It has been previously reported that mitochondrial content and metabolic activity are dynamically regulated in iNKT cells (27, 29). Inhibition of mitochondrial OXPHOS block splenic iNKT cell proliferation and induces their apoptosis (20). To investigate the potential role of mitochondria in the development of NKT cells, we first set up thymic organ culture by culturing thymic lobes from 1-d-old B6 neonates in the presence of FCCP, a potent inhibitor of OXPHOS. In addition to inhibiting OXPHOS, FCCP impedes mitochondrial ROS diffusion into the cytosol by reducing mitochondrial membrane potential (5). In newborn thymic organ cultures, iNKT cells started appearing as early as day 2, reaching maximum frequency around day 8 (SI Appendix, Fig. S2A). Interestingly, FCCP treatment led to a significant reduction in iNKT cells in a dose-dependent manner but showed little effect on the development of CD4sp or CD8sp cells (Fig. 2 A–C). The impairment of iNKT cell development was not due to defects in survival or proliferation because low doses of FCCP (i.e., 2.5 μM and 5 μM) did not increase cell apoptosis or reduce cell proliferation (SI Appendix, Fig. S2 B and C). These results indicate iNKT cell development is more sensitive to alterations in mitochondrial ETC function than conventional T cells.
Fig. 2.
Disruption of mitochondrial metabolism affects iNKT cells development. Thymic lobes from 1-d-old B6 neonates were cultured in the presence of indicated doses of FCCP for 8 d. CD1d/PBS57 tetramer stained iNKT cells, CD4sp and CD8sp cells were detected by flow cytometry. (A) Representative dot plots from three independent experiments. (B) Bar graphs depict mean + SEM of percentages (Left) and total numbers (Right) of iNKT cells (n = 5). (C) Bar graphs depict mean + SEM of absolute numbers of CD4sp (Left) and CD8sp (Right) cells. *P < 0.05, **P < 0.01, and ***P < 0.001.
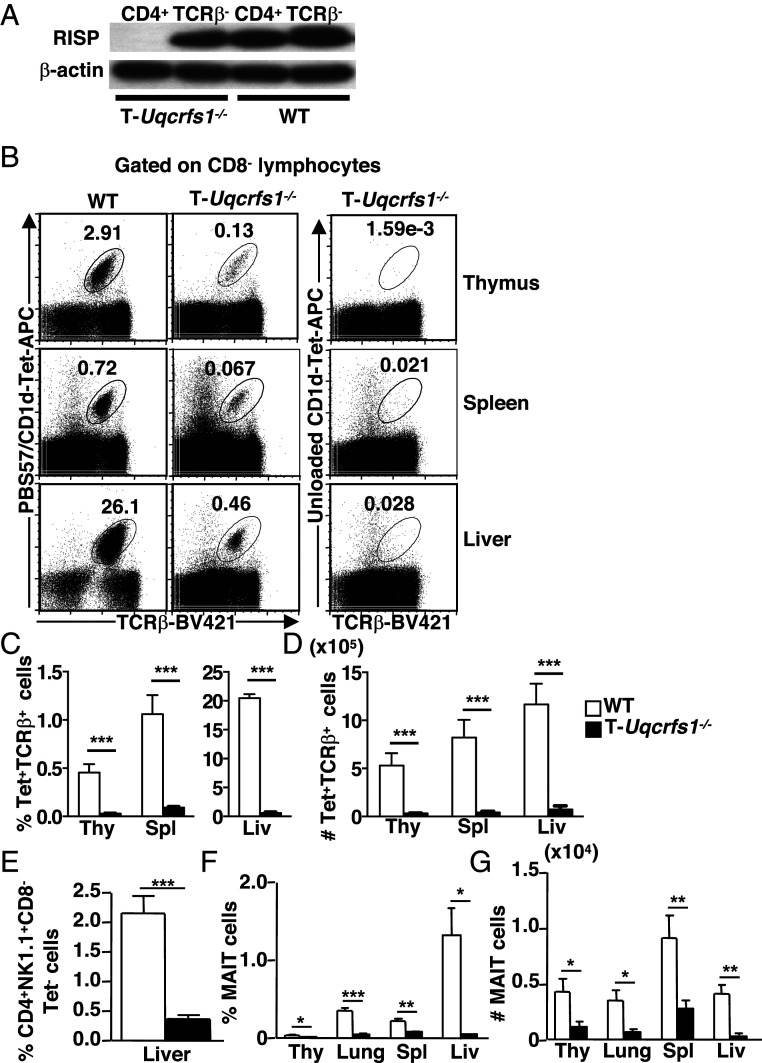
To further investigate the role of mitochondrial metabolism in iNKT cell development and function, we characterized iNKT cells in mice with a T cell-specific deletion of RISP (T-Uqcrfs1−/−) (5). RISP, a nuclear-encoded protein subunit of mitochondrial complex III, is required for transfer of electrons downstream of complex III as well as ROS production. Therefore, RISP-deficiency specifically affects mitochondrial production of ATP and complex III ROS (5). We backcrossed T-Uqcrfs1−/− mice into the B6 background and confirmed efficient ablation of RISP in T cells by immunoblotting (Fig. 3A). As reported previously, the total thymic cellularity and proportion of CD4sp, CD8sp, DN (CD4−CD8−), and DP (CD4+CD8+) was comparable between T-Uqcrfs1−/− mice and WT controls (5). However, both the frequency and absolute number of iNKT cells were drastically reduced in the thymus, spleen and liver of T-Uqcrfs1−/− mice (Fig. 3 B–D). In addition, we determined whether T-Uqcrfs1−/− mice had defects in other T cell subsets selected by DP thymocytes. Indeed, the percentage of CD1d-restricted type II NKT cells in the liver (defined as CD4+CD8−NK1.1+ CD1d/PBS57 tetramer− T cells) (30), was significantly reduced in T-Uqcrfs1−/− mice (Fig. 3E).
Fig. 3.
RISP-deficiency affects the development of iNKT cells, type II NKT cells, and MAIT cells. (A) Immunoblot of RISP protein in purified TCR-β+CD4+ and TCR-β− splenocytes from T-Uqcrfs1−/− and WT mice (n = 2). (B–D) Lymphocytes were stained with CD1d/PBS57 tetramer or unloaded CD1d tetramer and mAb against TCR-β, CD4, and CD8. CD8− cells were gated for flow cytometry analysis (B). Bar graphs depict mean + SEM of percentage (C) and number (D) of iNKT cells in indicated organs (n = 10). (E) Bar graph depicts mean + SEM of percentage of type II NKT cells in liver lymphocytes (n = 5). (F and G) Lymphocytes from indicated organs and mice were stained with anti–TCR-β and MR1/5OP-RU tetramer. Bar graph depicts mean + SEM of percentage (F) and number (G) of MAIT cells in indicated organs (n = 5). ***P < 0.001; **P < 0.01; *P < 0.05.
We also assessed the effect of RISP-deletion on the development of mucosal associated invariant T (MAIT) cells, another innate-like T cell population that exhibits an effector-memory phenotype similar to NKT cells (31, 32). We found that the frequency and total number of MAIT cells decreased significantly in the thymus and peripheral tissues of T-Uqcrfs1−/− mice (Fig. 3 F and G). In contrast, T-Uqcrfs1−/− mice have normal numbers of conventional CD4+ T cells in the spleen and liver. The total numbers of CD8+ T cells, in the spleen but not liver, was significantly reduced (SI Appendix, Fig. S3 A and B). In addition, T-Uqcrfs1−/− mice had normal number of Treg cells in the thymus while their numbers were modestly reduced in the spleen and liver (SI Appendix, Fig. S3 C–E). Thus, RISP ablation (and hence complex III dysfunction) has profound impacts on innate-like T cell development, but only minimally affects conventional T cell development.
Impaired iNKT Cell Development in T-Uqcrfs1−/− Mice Is Cell Intrinsic.
Having shown that iNKT cell numbers are decreased in T-Uqcfrs1−/− mice, we next questioned whether this defect was cell intrinsic or cell extrinsic. We found that the CD1d expression level on DP thymocytes, the cell type that mediates the positive selection of iNKT cells (9), is comparable between T-Uqcrfs1−/− and WT mice. Similar levels of CD1d expression were also observed in splenic B cells, T cells, and dendritic cells between these two groups (Fig. 4A). Furthermore, we found no difference in IL-2 production by iNKT cell hybridomas in responses to stimulation with either WT or T-Uqcrfs1−/− thymocytes in the presence or absence of α-GalCer (Fig. 4B), suggesting that RISP deletion did not affect the presentation of endogenous or exogenous lipid antigens.
Fig. 4.
Defective development of iNKT cells in T-Uqcrfs1−/− mice is cell intrinsic. (A) Cells were stained with anti-CD1d (filled histograms) or isotype control (open histograms). Representative histograms of CD1d expression on DP thymocytes and on splenic leukocyte subsets from three independent experiments. (B) IL-2 production by iNKT cell hybridomas cocultured with thymocytes in the absence or presence of α-GalCer, data shown as mean + SEM, figure generated from two experiments. (C and D) Lymphocytes from indicated organs of mixed BM chimeras were stained with CD1d/PBS57 tetramer and anti-CD45.1 for FACS analysis. (C) Numbers in each quadrant of representative dot plots show the percentage of tetramer+CD45.1+ iNKT cells in the indicated organs from two experiments. (D) Bar graph depicts the mean + SEM for the proportion of iNKT cells (n = 6). **P < 0.01; n.s., not significant.
To determine whether RISP-deficiency affects iNKT cell development via a cell-intrinsic mechanism, we generated mixed bone marrow (BM) chimeras by adoptive transfer of BM cells from CD45.1 congenic B6 (WT) and CD45.2 T-Uqcrfs1−/− mice at a 1:1 ratio into lethally irradiated Jα18−/− mice. We found that most of iNKT cells present in the Jα18−/− mice were derived from WT BM cells (Fig. 4 C and D). These data demonstrate that WT BM-derived cells were unable to rescue developmental defect of iNKT cells derived from the T-Uqcrfs1−/− BM. Collectively, these data show that impaired iNKT cell development observed in T-Uqcrfs1−/− mice stems from a cell-autonomous defect in iNKT cells.
RISP-Deficiency Affects iNKT Cell Maturation.
iNKT cells branch from the developmental pathway of conventional T cells at the DP thymocyte stage upon a productive Vα14Jα18 TCR-α gene rearrangement (9, 33). Transcriptome data revealed that T-Uqcrfs1−/− DP thymocytes had decreased expression of not only Uqcrfs1, but also several components of ETC, pointing to an overall decrease of the ETC functionality (SI Appendix, Fig. S4). To investigate how this defect affects the iNKT cell development, we first compared the Vα14Jα18 transcripts in total thymocytes and sorted DP thymocytes from WT and T-Uqcrfs1−/− mice, using qRT-PCR. The rearrangement of Vα14 to the distal Jα18 locus is known to require prolonged survival of DP thymocytes. Consistent with reduced iNKT cells in thymus, total thymocytes from T-Uqcrfs1−/− mice had lower Vα14Jα18 transcripts relative to WT counterparts. However, the levels of Vα14Jα18 transcripts in DP thymocytes were comparable between WT and T-Uqcrfs1−/− mice (Fig. 5A). Hence, RISP deficiency does not alter the number of thymic iNKT cell progenitors by reducing the lifespan of DP thymocytes.
Fig. 5.
Mitochondrial complex III deficiency impairs proper iNKT cell maturation. (A) Relative Vα14Jα18 expression in DP and total thymocytes from indicated mice. Data are representative of three experiments. (B and C) Thymocytes were stained with CD1d/PBS57 tetramer and mAb against TCR-β, CD24, CD44, and NK1.1. (B) Dot plots depict the gating strategy for iNKT cell developmental stages. (C) Bar graphs depict mean + SEM for the proportions and absolute numbers of iNKT stages (n = 6). (D–F) Thymocytes were stained with CD1d/PBS57 tetramer and mAb to TCR-β and CCR7 followed by intracellular staining with PLZF, T-bet, and RORγt (n = 6). (D) Dot plots depict the gating strategy for NKT1, NKT17, NKT2+preNKT (NKT2+Pre) subsets. (E) Bar graphs depict the for the percentages (Upper) and absolute numbers (Lower) of different NKT cell subsets. (F) The percentage of CCR7+ cells (mean + SEM) among PLZFhi iNKT cells. (G and H) Proportion of BrdU+ iNKT cells (mean + SEM) in indicated organs (G) and at different stages of iNKT maturation in thymus (n = 6). (I and J) Proportions of Annexin V+ iNKT cells (mean + SEM) in indicated organ (I) and at different stages of iNKT maturation in thymus (J) (n = 6). *P < 0.05, **P < 0.01, and ***P < 0.001.
Following positive selection, iNKT cells progress through conventionally characterized stages: stage 0 (CD24hiCD44−NK1.1−), stage 1 (CD24loCD44−NK1.1−), and stage 2 (CD24loCD44+NK1.1−) occur in the thymus, and terminal maturation to stage 3 (CD24loCD44+NK1.1+) occurs in the thymus or periphery (9, 10). In the thymus, we observed higher percentages of stages 0 to 2 iNKT cells in T-Uqcrfs1−/− mice compared to WT mice, while percentage of stage 3 iNKT cells were about fivefold lower (Fig. 5 B and C). Although ultimately T-Uqcrfs1−/− mice had significantly lower numbers of total thymic iNKT cells (Fig. 3D), they had comparable numbers of stage 0 iNKT cells. Differences were observed later, where T-Uqcrfs1−/− mice had significantly lower numbers of stages 1 to 3 iNKT cells compared to WT counterparts (Fig. 5C). These results indicate that RISP deletion-related mitochondrial dysfunction does not influence iNKT precursor selection (i.e., stage 0) but exerts its influence as early as the stage 0 to 1 transition, leading to significantly fewer mature iNKT cells.
When examining transcription factors, the expression of retinoic acid receptor-related orphan receptor γ (RORγt) and c-Myc, factors known to control iNKT cells development at a very early stage (34), was not significantly altered in T-Uqcrfs1−/− thymocytes (SI Appendix, Fig. S5 A and B). In addition, T-Uqcrfs1−/− thymic iNKT cells show no defect in the expression of promyelocytic leukemia zinc finger protein (PLZF) or early growth response 2 protein (Egr2) (SI Appendix, Fig. S5C), two transcription factors essential to induce a multipotent, unbiased effector program that precedes terminal differentiation of iNKT cells (35–37). Thus, the defect in iNKT cell maturation is PLZF and Egr2 independent. In the spleen and liver, total numbers of iNKT cells from all three stages also decreased dramatically in T-Uqcrfs1−/− mice (SI Appendix, Fig. S6 A and B). The increased ratio of stage 1 and stage 2 iNKT cells in T-Uqcrfs1−/− thymus and periphery (Fig. 5C and SI Appendix, Fig. S6 A and B) suggested that maturation of residual T-Uqcrfs1−/− iNKT cells was also impaired.
Based on transcription factor expression profiles and cytokine production, three different sublineages of iNKT cells, namely T-bet+ NKT1, GATA3+NKT2, and RORγt+ NKT17, have been described (10, 38). Due to significantly lower iNKT cell numbers in T-Uqcrfs1−/− mice, we found lower absolute numbers of all subsets detected by PLZF, RORγt, and T-bet expression (Fig. 5 D and E). A higher proportion of PLZFhi cells, enriched for NKT2 and NKT cell precursors (39), was found in T-Uqcrfs1−/− mice compared to WT littermates. Based on CCR7 expression, the NKT cell precursors (CCR7+) appear to be the major population of PLZFhi cells in T-Uqcrfs1−/− mice, while NKT2 cells (CCR7−) are the dominant PLZFhi cells in WT mice (Fig. 5F). Furthermore, the proportion of NKT1 (PLZFloT-bet+) was much lower in T-Uqcrfs1−/− mice compared to WT mice, suggesting complex III dysfunction led to a severe defect in interferon (IFN)-γ–producing NKT1 subsets (Fig. 5 D and E).
Residual iNKT Cells in T-Uqcrfs1−/− Mice Are Hyperproliferative but Undergo Enhanced Apoptosis.
Since iNKT cells have an intrathymic proliferation wave before acquiring maturity, we evaluated if defective maturation of T-Uqcrfs1−/− iNKT cells was due to impaired cell proliferation. Contrary to our expectation, we found higher bromodeoxyuridine (BrdU) incorporation in residual iNKT cells in the thymus, spleen, and liver of T-Uqcrfs1−/− mice (Fig. 5G), indicating T-Uqcrfs1−/− iNKT cells underwent hyperproliferation. In the thymus, T-Uqcrfs1−/− iNKT cells proliferated normally during stages 0 to 2 but exhibited increased proliferation during stage 3 compared with WT iNKT cells (Fig. 5H). We did not observe any difference in BrdU incorporation in conventional T cells (SI Appendix, Fig. S7A). The fact that RISP-deficiency does not reduce iNKT cell proliferation led us to examine whether it was affecting Uqcrfs1−/− iNKT cell survival. Indeed, we found iNKT cells in the thymus, spleen, and liver of T-Uqcrfs1−/− mice have higher percentage of Annexin V+ cells as compared with WT mice (Fig. 5I), indicating T-Uqcrfs1−/− iNKT cells are more prone to apoptosis. In thymic iNKT cells, the enhanced apoptosis in T-Uqcrfs1−/− iNKT cells was most profound at stage 2 (Fig. 5J). Consistent with previous reports, we did not detect significant differences in the survival of conventional T cells in T-Uqcrfs1−/− mice compared to WT counterparts (SI Appendix, Fig. S7B). Taken together, these results demonstrate that RISP deletion does not affect iNKT cell proliferation but leads to increased apoptosis of iNKT cells during their development and differentiation. However, impaired iNKT cell development in T-Uqcrfs1−/− mice might not be solely due to an intrinsic survival defect because transgenic overexpression of Bcl-xL in T-Uqcrfs1−/− mice does not restore iNKT cell population in either the thymus or the periphery (SI Appendix, Fig. S8).
RISP-Mediated Mitochondrial Metabolism Is Required for Optimal TCR Signaling.
Strong TCR signals are suggested to be crucial for iNKT cell positive selection and the early stages of iNKT development (40, 41). In T-Uqcrfs1−/− mice, we observed lower expression of Nur77, an orphan nuclear receptor used as a marker for TCR signaling strength (42), on CD4sp, CD8sp, and DP thymocytes as compared to WT counterparts (Fig. 6 A and B). This suggested that RISP-deficiency reduces the TCR signaling strength of developing thymocytes. T cell activation is known to elevate ROS and is associated with efflux of calcium ions from the endoplasmic reticulum (ER) into the cytosol (43). To further understand the effect of RISP-deficiency on TCR signaling strength, we examined the TCR-triggered calcium flux in DP thymocytes (containing iNKT cell precursors) from T-Uqcrfs1−/− and WT mice. DP thymocytes from T-Uqcrfs1−/− mice exhibited impaired TCR-driven ER calcium flux upon cross-linking of CD3. Moreover, after addition of Ca2+, store-operated Ca2+ influx in the T-Uqcrfs1−/− mice was also reduced (Fig. 6C). The magnitude of calcium flux in T-Uqcrfs1−/− mice was decreased by ∼30% both at TCR cross-linking and at Ca2+ addition stages (Fig. 6 D and E).
Fig. 6.
T-Uqcrfs1−/− iNKT cells have impaired TCR signaling and altered responsiveness to IL-15. (A) Expression of Nur77 in CD4sp, CD8sp, and DP thymocytes. (B) Bar graphs depict mean + SEM of Nur77 mean fluorescence intensity (MFI) (n = 6). (C) Representative recordings of Ca2+ flux in WT and T-Uqcrfs1−/− DP thymocytes measured before and after anti-CD3/anti-CD4 cross-linking in Ca2+ free DPBS, and after addition of 2 mM Ca2+ in DPBS. (D and E) Peak of calcium flux (ratios of Fluo 4/Flura red) after anti-CD3 cross-linking (D) and Ca2+ addition (E) (n = 5). (F) Expression of T-bet in thymic iNKT cells. (G) Bar graphs depict mean + SEM of T-bet MFI (n = 6). (H) Representative histograms show CD122 expression on thymic iNKT cells. (I) Bar graphs depict mean + SEM of CD122 MFI (n = 7). (J and K) Thymocytes were incubated in the presence or absence of IL-15. Surface staining with CD1d/PBS57 tetramer and anti–TCR-β were performed, followed by intracellular staining for pSTAT5. (J) Representative histograms show pSTAT5 level in iNKT cells. (K) Bar graphs depict mean + SEM of pSTAT5 MFI (n = 5). Data are representative of three experiments. *P < 0.05, **P < 0. 01, and ***P < 0.001.
It has been shown that TCR signaling strength distinctly regulates T-bet expression (44). T-bet up-regulates IL-15Rβ (CD122) and increases iNKT cell responsiveness to IL-15 signaling (45, 46), which are essential for iNKT cell terminal maturation and homeostasis (47, 48). Notably, expression of T-bet (Fig. 6 F and G) and CD122 (Fig. 6 H and I) were significantly reduced in T-Uqcrfs1−/− thymic iNKT cells. In addition, IL-15–induced phosphorylation of STAT5 was impaired in T-Uqcrfs1−/− iNKT cells (Fig. 6 J and K). Also, as a proof of principle, we found lower frequencies and total numbers of CD8αα intraepithelial lymphocytes (IELs) in T-Uqcrfs1−/− mice, the development of which is highly dependent on T-bet/IL-15 signaling (SI Appendix, Fig. S9). Considering the distinct roles of TCR and IL-15 signaling in different stages of iNKT cell development and maturation, we hypothesize that impaired TCR signaling may be responsible for the early block of iNKT cell development and T-bet/IL-15 signaling may be responsible for the defect in residual iNKT cell terminal maturation in T-Uqcrfs1−/− mice.
Residual iNKT Cells in T-Uqcrfs1−/− Mice Have Defective Responsiveness to TCR Stimulation.
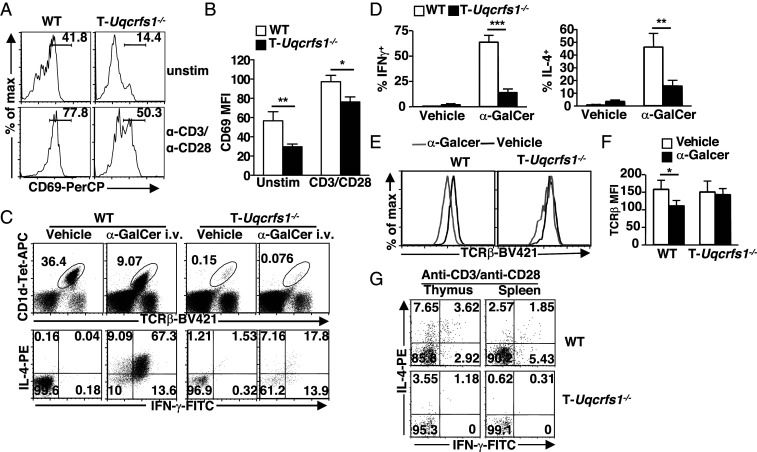
To assess whether RISP deletion affects iNKT cell activation and function, we examined CD69 expression and cytokine production in T-Uqcrfs1−/− iNKT cells. We found that residual T-Uqcrfs1−/− iNKT cells in the liver had lower constitutive and TCR-inducible CD69 expression (Fig. 7 A and B). Upon in vivo α-GalCer stimulation, T-Uqcrfs1−/− iNKT cells had a profound reduction in IFN-γ and IL-4 cytokine production compared with WT iNKT cells (Fig. 7 C and D). Similar to conventional T cells, iNKT cells down-regulate their TCR upon stimulation (49). We noticed that T-Uqcrfs1−/− iNKT cells had impaired TCR down-regulation while WT iNKT cells downmodulated their TCR normally upon α-GalCer stimulation (Fig. 7 E and F). This provided further evidence of impaired TCR signaling in T-Uqcrfs1−/− iNKT cells. Consistent with our in vivo observations, in vitro stimulation with anti-CD3/anti-CD28 resulted in significantly lower IFN-γ and IL-4 production by T-Uqcrfs1−/− iNKT cells compared to WT iNKT cells from the thymus and the spleen (Fig. 7G). As expected, T-Uqcrfs1−/− iNKT cells produced significantly reduced amounts of TCR-induced mROS as measured by MitoSox red (SI Appendix, Fig. S10 A and B). Consistent with our previous observation (5), splenic CD4+ T cells in T-Uqcrfs1−/− mice had lower TCR-induced mROS production (SI Appendix, Fig. S10 C and D) compared with WT controls. Thus, similar to conventional T cells, T-Uqcrfs1−/− iNKT cells were incapable of increasing mitochondrial complex III ROS upon activation, leading to impaired iNKT cell activation and function.
Fig. 7.
T-Uqcrfs1−/− iNKT cells have impaired activation and responsiveness to stimulation. (A) CD69 expression on iNKT cells ex vivo or upon anti-CD3/anti-CD28 stimulation. (B) Bar graphs depict mean + SEM of CD69 MFI (n = 5). (C–F) WT and T-Uqcrfs1−/− mice were injected intravenously with α-GalCer. Hepatic lymphocytes were harvested 1 h postinjection followed by flow cytometry analysis. (C) Represntative dot plots depict the percentages of iNKT cells (Upper) and IL-4– and IFN-γ–producing cells in iNKT cells. (D) Bar graphs show mean + SEM of IFN-γ+ and IL-4+ iNKT cells (n = 6). (E) Surface TCR-β expression on iNKT cells in α-GalCer (gray) and vehicle-injected mice (black). (F) Bar graphs depict mean + SEM of MFI of TCR-β (n = 6). (G) Dot plots show percentages of IL-4– and IFN-γ–producing cells in iNKT cells upon in vitro stimulation with anti-CD3/anti-CD28. Results shown are representative of three experiments. *P < 0.05, **P < 0.01, and ***P < 0.001.
Knockdown of Uqcrfs1 in Mature iNKT Cells Results in Impaired TCR-Induced Cytokine Production.
Since the periphery maturation of residual iNKT cells in T-Uqcrfs1−/− mice is impaired (SI Appendix, Fig. S6), we further investigated the role of RISP-dependent mitochondrial function in mature iNKT cells and CD4+ T cells that developed normally in a WT background. We performed nucleofection of splenocytes from Vα14Tg mice with small-interfering RNA (siRNA) against Uqcrfs1 or scrambled control (NC). We detected ∼50% reduction in Uqcrfs1 transcripts upon transfection with Uqcrfs1 siRNA compared to NC siRNA (Fig. 8A). The percentages and numbers of iNKT cells and CD4+ T cells (CD1d/PBS57 tetramer−) (Fig. 8 B and C) did not change significantly with ∼50% knockdown of Uqcrfs1 transcripts. However, anti-CD3 stimulated IFN-γ production was reduced by ∼40 to 50% in both iNKT cells and CD4+ T cells transfected with Uqcrfs1 siRNA compared with siNC-transfected counterparts (Fig. 8 D and E). Thus, RISP-mediated mitochondrial metabolism is essential for optimal TCR-induced cytokine production in mature iNKT and conventional CD4+ T cells.
Fig. 8.
Knockdown of Uqcrfs1 in mature iNKT cells results in impaired TCR-induced cytokine production. (A–E) Splenocytes from Vα14Tg mice were transfected with siNC and siUqcrfs1. After 3 d in culture, cells were stimulated with anti-CD3 for intracellular cytokine staining. (A) Knockdown efficiency of Uqcrfs1 was measured by qRT-PCR. (B and C) Percentage (B) and cell number (C) of iNKT cells and CD4+ T cells in siNC and siUqcrfs1-treated groups. (D and E) Bar graphs depict percentage of IFN-γ–producing iNKT cells and CD4+ T cells after anti-CD3 stimulation. (n = 3). (n.s., not significant. *P < 0.05 and **P < 0.01). (F–K) NKT cell hybridoma DN32.D3 cells were transduced with lentivirus encoding Uqcrfs1-targeting or NC shRNA. (F) Knockdown efficiency of Uqcrfs1 was confirmed by Western blot. (G) shUqcrfs1 and shNC-expressing DN32.D3 cells were stimulated with anti-CD3/anti-CD28 or α-GalCer for 48 h and IL-2 production was quantified by ELISA. (**P < 0.01). (H) Representative recordings of Ca2+ flux in shUqcrfs1 and shNC-expressing DN32.D3 cells. (I) Calcium flux of indicated cells after α-GalCer stimulation (n = 3). (J) Nuclear (N) and cytoplasmic (C) extracts were prepared from shNC and shUqcrfs1-expressing cells after stimulation with α-GalCer at the indicated time points and were immunoblotted for basal level and nuclear translocation of NFATc2. α-Tubulin and HDAC1 were served as loading control. (K) Bar graph of NFATc2 nuclear/cytoplasm ratio was normalized by the corresponding cells at 0 h. Data are representative of three experiments.
To further determine the mechanism by which Uqcrfs1 affects cytokine production by iNKT cells, we infected an iNKT cell hybridoma (DN32.D3) with lentiviruses encoding control or Uqcrfs1-targeting short-hairpin RNAs (shRNAs), and specific gene knockdown was verified by immunoblotting. Uqcrfs1 was successfully knockdown with shUqcrfs1-1 and shUqcrfs1-2 compared to shNC (Fig. 8F); further experiments were carried out with shUqcrfs1-2 (greater than 90% reduction of RISP expression). Consistent with Uqcrfs1 knockdown results in primary NKT cells, Uqcrfs1 knockdown in DN32.D3 cells significantly reduced anti-CD3 or α-GalCer–stimulated production of cytokine (IL-2) compared with control shNC-expressing DN32.D3 cells (Fig. 8G). In addition, calcium flux in shUqcrfs1-expressing DN32.D3 cells was significantly reduced compare to that of shNC-expressing cells upon α-GalCer stimulation (Fig. 8 H and I). NFATc2 (or NFAT1) is a key regulator for T cell function and development (50). Hypoxia, which paradoxically increases mROS production from complex III, induces NFATc2 nuclear translocation (51). Therefore, we examined nuclear translocation of NFATc2 to determine whether abrogating Uqcrfs1 activity and thus decreasing mROS production affected iNKT cells similarly to what is known in conventional CD4+ T cells (5). Nuclear translocation of NFATc2 was significantly increased in α-GalCer-treated control cells, but not detected in Uqcrfs1 knockdown cells. Interestingly, the abundance of NFATc2 in the cytoplasmic compartment was also decreased in Uqcrfs1 knockdown cells compared to control NKT cells (Fig. 8 J and K). These data suggest that mROS production from complex III may contribute to cytokine production in mature iNKT cells by regulating NFAT activity.
Discussions
T cells utilize glycolysis and OXHPOS in different ratios during activation, anergy, and development (52). The fine-tuning of metabolic programming throughout a T cell’s lifetime is crucial for proper effector function. iNKT cells are a unique subset of “innate-like” T cells, which constitutively express T cell activation markers and rapidly produce both Th1 and Th2 cytokines upon stimulation. Our study and others (20, 27, 28) suggest that irrespective of organs examined, iNKT cells have lower mitochondrial content and SRC that may be developmentally regulated during their thymic ontogeny (27). We provide further link between poor FAO and lower basal SRC in iNKT cells (Fig. 1). Importantly, through direct pharmacological (Fig. 2) and genetic (Fig. 3) ablation of mitochondrial function, we provide direct evidence that intact mitochondrial respiration is essential for iNKT cell development. Our data revealed the critical role of mitochondrial complex III-related mitochondrial metabolism in regulating iNKT cell development and function through modulation of TCR signaling strength, NFATc2 translocation and T-bet/IL-15 signaling (Figs. 6–8).
The reduced FAO in iNKT cells suggests that they lack the ability to utilize fat as energy source and rely on other carbon sources for their basal OXPHOS (Fig. 1 D and E). In CD8+ memory T cells, FAO that occurs in mitochondria has been proposed to play a critical role in supporting increased SRC, which is necessary for providing extra energy required for their long-term survival and rapid recall responses (7). Though iNKT cells have a “memory-like” phenotype, a single dose of α-GalCer induces a long-term anergic state of iNKT cells (53). In addition, iNKT cells have been shown to be anergic after certain microbial infections and antigen exposure (54, 55). It has been proposed that T cell anergy is partially due to an inability of the cell to broadly coordinate metabolic pathways for growth and function (52). Thus, we speculate that acquisition of an anergic phenotype in iNKT cells might be correlated with lack of SRC and utilization of FAO for energy. Recent studies showed that high concentration of etomoxir could impact mitochondrial respiration (56, 57) besides inhibiting CPT1, the rate-limiting enzyme FAO. Thus, further investigation of the effect of FAO on NKT cell survival, responsiveness or anergy would require the use of genetic tools.
Metabolic regulators have been shown to affect iNKT cell development by indirectly altering mitochondrial homeostasis (15, 26, 27, 29). For example, T cell-specific loss of Hippo/Mst signaling resulted in altered mitochondrial homeostasis, metabolism, and impaired iNKT cell development (27). Deletion of Fnip1 resulted in lower mitochondrial mass in iNKT cell precursors, but increased mROS production that was associated with their enhanced apoptosis (15). Far from simply being an agent of DNA damage, mROS generation is essential for cellular metabolism and survival, as well as intracellular signaling in T cells (58). Mitochondrial complex III is a crucial component for mROS generation (59). Studies in T-Uqcrfs−/− mice lacking mROS production from complex III in T cells demonstrated that conventional CD4+ T cells required mROS for NFAT nuclear translocation and subsequent production of IL-2 (5). Recent studies have also demonstrated that mice lacking RISP specifically in CD4+ Treg cells displayed fatal hyperinflammatory disease early in life due to their functional inability to limit inflammation (60). It is important to note that while RISP-deficiency impacts the activation and function of conventional CD4+, CD8+, and Treg cells, it does not affect the development of these T cell subsets. In contrast, our studies show a marked dependence of iNKT cells on intact mitochondrial function and mROS for proper thymic development and function. As such, iNKT cells from T-Uqcrfs1−/− mice are decreased in number, developmentally halted at the stage 0 to 1 transition, and functionally impaired. It is worth noting that the development of other innate-like T cells, such as type II NKT cells and MAIT cells, was also severely impeded in T-Uqcrfs1−/− mice. In T-Uqcrfs1−/− mice not only is mROS production reduced upon activation, but the activity of downstream ATP production (OXPHOS) is also inhibited. Thus, our data suggest a distinct requirement for mROS and OXPHOS in the development of innate-like T cells.
NKT cell development appears to be particularly sensitive to alterations in transcription factors, such as Ets-1 (61), PLZF (36, 62), Egr2 (37, 63), c-Myc (64, 65), RORγt (66), and T-bet (47, 67). This suggests that there are multiple checkpoints unique to iNKT cell differentiation. WT and RISP-deficient iNKT cells expressed comparable levels of PLZF, Egr2, c-Myc, and RORγt (SI Appendix, Fig. S5); however, the expression of T-bet was dramatically reduced in RISP-deficient iNKT cells (Fig. 6 F and G). It has been shown that high avidity TCR–MHC interactions induced low levels of T-bet expression in thymic IEL precursors that was bolstered by IL-15 signaling, which in turn regulated the differentiation and expansion of CD8αα IELs (46). iNKT cells share several developmental features with CD8αα IELs, such as agonist selection in the thymus and the requirement for T-bet– and IL-15–mediated differentiation and expansion after lineage commitment (42, 46). Thus, we hypothesized that differences in T-bet expression in iNKT cell precursors due to altered TCR signaling in T-Uqcrfs1−/− mice might be amplified over a period of time due to perturbations of the T-bet–driven, IL-15–dependent positive feed-back loop (45, 46). Consistent with this notion, we have shown that RISP-related mitochondrial metabolism may regulate multiple checkpoints in iNKT cell development by modulating TCR signaling strength and T-bet/IL-15 signaling.
First, defects in early iNKT cell development were correlated with decreased TCR signaling strength, as reflected by lower Nur77 expression and decreased calcium flux in DP thymocytes in T-Uqcrfs1−/− mice (Fig. 6). Second, Uqcrfs1−/− iNKT cells suffered high apoptosis during development and showed decreased IL-15 responsiveness (Figs. 5 I and J and 6 J and K). Third, functional iNKT cell development was disturbed in the T-Uqcrfs1−/− thymus, with T-bet+ NKT1 subset particularly affected (Fig. 5 D and E). Thus, our data suggest that RISP-dependent mitochondrial metabolism is essential for optimal TCR signaling strength and subsequent T-bet/IL-15 signaling to support iNKT cell survival, maturation, and function. Interestingly, a recent study showed that altered mitochondrial metabolism due to Mst1 deficiency could affect IL-15 signaling leading to enhanced apoptosis of stage 3 (T-bet–expressing) iNKT cells. However, this effect appears to be independent of aberration in TCR signaling strength (27).
A recent study found that iNKT cells have lower glucose uptake compared to CD4+ T cells, suggesting a quiescent phenotype in resting iNKT cells (20). However, activation and effector functions of iNKT cells were accompanied with enhanced OXPHOS (20) and iNKT cell proliferation, survival, and cytokine production were impaired upon OXPHOS inhibition. We had earlier reported that RISP-deficiency did not alter homeostatic proliferation of conventional T cell, but their antigen-induced expansion was affected (5). Our data and the report from Kumar et al. (20) imply that like conventional T cell OXPHOS is not required for homeostatic proliferation of iNKT cells but essential during TCR-induced proliferation and effector functions. However, intact mitochondrial metabolism is required for optimal TCR and IL-15 signaling, thus critical for integrating survival signals in iNKT cells during their development or effector functions. Furthermore, consistent with previous reports, we found that similar to conventional CD4+ T cells, mature iNKT cells require OXPHOS and complex III ROS for optimal TCR-induced cytokine production, supporting a signaling role of mitochondria in both T cell types. A previous report showed that ROS regulates the inflammatory function of iNKT cells through PLZF and high levels of ROS in the peripheral NKT cells was primarily produced from NADPH oxidase but not mitochondria (24). However, we found the residual iNKT cells in the spleen and liver of T-Uqcrfs1−/− mice produced reduced amounts of activation-induced mROS and had a profound reduction in cytokine production upon TCR stimulation (Fig. S10 and Fig. 7), supporting a role of mROS in the activation and function of iNKT cells. The decrease in cytokine production was likely due to the impairment of both basal expression and nuclear translocation of NFAT, as shown in the RISP-knockdown NKT cell hybridoma DN32.D3 cells (Fig. 8 J and K). More work is necessary to better understand the mechanism by which RISP regulates the expression of NFAT in NKT cells.
Immune dysfunctions and increased susceptibility to infections have been reported in patients with primary mitochondrial disorders due to ETC dysfunction (23). It will be interesting to study if mitochondrial diseases and associated immune deficiencies due to impaired ETC functions are related to defects in iNKT cell development and function. Furthermore, as discussed above, lower SRC in iNKT cells might be correlated with their enhanced propensity toward an anergic phenotype. It would be valuable to investigate if drugs can be developed to induce mitochondrial biogenesis and enhance SRC in iNKT cells and circumvent anergy, as this might have important implications for iNKT cell-based therapeutic research and development.
Materials and Methods
Mice.
B6, CD45.1 congenic B6 mice, Vα14-Jα18 transgenic (Vα14Tg) mice, TCR-α−/− mice, and Lckpr-Bcl-xL Tg mice were purchased from The Jackson Laboratory. Vα14Tg mice were crossed with TCR-α−/− to generate Vα14TgTCR-α−/− mice. Uqcrfs1fl/fl;CD4-Cre+ (T- Uqcrfs1−/−) and Uqcrfs1fl/fl on B6 background were generated as described previously (5). Jα18−/− mice on B6 background were provided by Luc Van Kaer, Vanderbilt University, Nashville, TN. All animal work was approved by the Northwestern University Institutional Animal Care and Use Committee.
Antibodies and Mitochondrial Probes.
The generation of CD1d-specific mAb 5C6 has been described previously (68). Allophycocyanin (APC) and Brilliant violet 421 (BV421) conjugated CD1d/PBS57-tetramers and APC conjugated MR1-5-OP-RU tetramers were obtained from the NIH tetramer core facility. Annexin V apoptosis detection kit and BrdU flow kit were purchased from BD Pharmingen. Fluorescently labeled antibodies against TCR-β, CD4, CD8α, CD8β, B220, CD24, CD44, NK1.1, CD69, CCR7, CD45.2, CD45.1, CD122, IL-4, IFN-γ, PLZF, T-bet, RORγt, Nur77, phosphor-STAT5, and biotinylated antibodies against CD3 and CD4 were purchased from BioLegend, BD, eBiosciences, or Cell Signaling technology. MitoTracker, TMRE for and MitoSox were purchased from ThermoFisher.
Surface Staining, Intracellular Staining, and Flow Cytometry Analysis.
Single-cell suspensions from various organs were prepared and surface stained as described previously (69, 70). For IL-15–induced STAT5 phosphorylation, thymocytes, were stained with CD1d/PBS57 tetramer and incubated with or without IL-15 (100 ng/mL) for 20 min. Cells were fixed with 4% paraformaldehyde (PFA) for 10 min at room temperature followed by methanol permeabilization on ice for 30 min. For staining of transcription factors, cells were surface stained first, fixed, and permeabilized with the Foxp3 staining buffer set (eBiosciences) and stained with relevant antibodies. For in vivo stimulation assay, mice were injected intravenously with 5 μg of α-GalCer. Liver leukocytes were isolated after 1 h and surface stained with CD1d/PBS57 tetramer and anti–TCR-β followed by fixation with 4% PFA. Cells were washed and permeabilized with 0.15% saponin, and then stained with anticytokine mAbs. For in vitro stimulation assays, lymphocytes were stimulated with plated bound anti-CD3 (5 μg/mL) and soluble anti-CD28 (5 μg/mL). Three hours later, monensin (0.01 mM) was added to each well and cells were incubated for another 3 h. Surface staining and intracellular cytokine staining were performed as above. For mitochondrial profiles detection, cells were incubated with 100 nM MitoTracker, 50 nM TMRE, or 5 μM MitoSox for 15 min in 37 °C incubator. Flow cytometry was performed with a FACS Canto II and analyzed using FlowJo software.
Metabolism Assays.
Splenic CD4+ T cells from B6 mice and iNKT cells from Vα14TgTCR-α−/− mice were stained with anti-CD4 and CD1d/PBS57-tetramer, respectively, and sorted using BD FACS Aria. Hepatic lymphocytes from Vα14TgTCR-α−/− mice and Jα18−/− mice were positively selected for CD4+ iNKT cells and CD4+ conventional T cells, respectively. Enriched cells were suspended in XF media (nonbuffered RPMI 1640 containing 20 mM glucose, 2 mM l-glutamine, and 1 mM sodium pyruvate) followed by measurement of OCR under basal conditions and in response to 200 μM etomoxir, 1 μM oligomycin, 1.5 μM FCCP, and 100 nM rotenone + 1 μM antimycin A (Sigma) using XF-96 Extracellular Flux Analyzer (Seahorse Biosciences).
Thymic Organ Cultures.
Thymic lobes were harvested from 1-d-old B6 neonates, placed on nitrocellulose filters (Millipore), and cultured in DMEM supplemented with 20% FCS, glutamine, sodium pyruvate, nonessential amino acids, 2-β mercaptoethanol, and antibiotics, in the presence or absence of indicated concentrations of FCCP at 37 °C for 8 d.
Immunoblotting.
Splenocytes from WT and T-Uqcrfs1−/− mice were stained with APC-anti mouse TCR-β, followed by anti-APC beads (Miltenyi). TCR-β+ and TCR-β− fractions were purified with a magnetic column. Immunoblotting was performed using antibody against RISP (MitoScience, Abcam) or α-tubulin (Santa Cruz) or β-tubulin (Calbiochem), as described previously (5).
Antigen Presentation Assay.
Thymocytes (5 × 105) isolated from WT and T-Uqcrfs1−/− mice were incubated with 5 × 104 iNKT cell hybridomas in the absence or presence of α-GalCer (100 ng/mL) for 24 h. Supernatants were collected and IL-2 production was quantified by ELISA.
Generation of BM Chimeras.
BM cells from WT (CD45.1) and T-Uqcrfs1−/− (CD45.2) mice were depleted of T cells with anti–Thy-1 and rabbit complement. Mixed BM cells (1 × 107 cells, 1:1 ratio) were injected intravenously into Jα18−/− recipient mice that were irradiated with 900 rad. Lymphocytes isolated from recipient mice were analyzed by flow cytometry 6 wk after transfer.
Real-Time PCR.
Splenic CD4+ T cells from B6 mice and iNKT cells from Vα14Tg mice were positively selected using MACS (Miltenyi). Genomic DNA and mitochondrial DNA were extracted with QIAamp DNA kit (Qiagen) for determining mt DNA/nuclear DNA ratio. Total RNA from thymocytes, sorted DP thymocytes or splenocytes transfected with siRNAs were extracted using RNeasy kit (Qiagen). Following reverse transcription, PCR was performed using SYBR Green PCR master mix with respective primers (SI Appendix, Table S1) and MyiQ real-time detection system (Bio-Rad). Each PCR was run in duplicate or triplicate and normalized to β-actin.
RNA-Sequencing Library Generation and Data Analysis.
RNA from FACS-sorted DP thymocytes was extracted with RNAeasy Mini Kit (Qiagen). Libraries were generated using the Illumina TruSeq preparation kit and sequenced on HiSeq 4000. Reads were analyzed through the Ceto pipeline (https://github.com/ebartom/NGSbartom) using STAR (71) for alignment on mm10 mouse genome, HTSEq. (72) for read counting, and edgeR (73) for differential expression analysis, and g:Profiler for gene enrichment analysis (74).
Proliferation and Apoptosis Assays.
Mice were injected intraperitonially with 100 μL of BrdU (10 mg/mL) and supplied with 0.8 mg/mL of BrdU in drinking water for 3 d. Thymocytes were enriched for iNKT cells by depleting CD8+ cells using anti-CD8α antibodies and rabbit complement, while splenocytes and liver leukocytes were enriched for iNKT cells using MACS negative selection (61). Enriched iNKT cells were surface stained with appropriate antibodies followed by staining with BrdU flow kit. To detect apoptotic cells, an annexin V-FITC labeling kit was used according to the manufacturer’s protocol.
Calcium Signaling.
Cells were incubated with Fluro-4 (4 μg/mL) and Fura Red (8 μg/mL; Molecular Probes) for 45 min in the dark. After washing with Hank’s buffer, cells were blocked with 2.4G2 and stained with anti-CD3–biotin, anti-CD4–biotin (RM4-4), anti-CD4–PE (GK1.5), and anti-CD8–BV510 (53-6.7) for 15 min. Cells were then washed and resuspended in Ca2+-free DPBS and placed in 37 °C incubator. Cross-linking with streptavidin and 2 mM Ca2+ solution was added at the indicated time points.
Transient Knockdown of Uqcrfs1 in iNKT Cells.
Splenocytes isolated from Vα14Tg mice were stimulated with plate bound anti-CD3 (5 μg/mL) and anti-CD28 (2 μg/mL) for 16 h. Nucleofection of stimulated splenocytes with siRNA targeting Uqcrfs1 or scrambled control (Integrated Technologies) was performed following the manufacturer’s instructions (Nucleofection Kit; Lonza). Transfected cells were cultured in complete RPMI supplemented with 200 U/mL IL-2 for additional 3 d followed by assessment of intracellular IFN-γ production.
Stable Knockdown of Uqcrfs1 in DN32.D3 NKT Cell Line.
DN32.D3 cells were transduced with lentivirus encoding Uqcrfs1-targeting or NC shRNA (Sigma). Stable knockdown cell line was generated through selection with 5 μg/mL puromycin-containing RPMI 1640 complete medium.
NFATc2 Nuclear Translocation.
DN32.D3 cell lines with stable knockdown of Uqcrfs1 were stimulated with α-GalCer (100 ng/mL) at 0, 1, 2, and 4 h. Isolation of nuclear and cytoplasmic fractions was performed according to the protocol of the Nuclear Extract Kit (Active Motif) followed by Western blotting. NFATc2 was detected by mAb 4G6-G5 (Santa Cruz). HDAC1 (Santa Cruz), and α-tubulin (Calbiochem) were used as a loading control for nuclear and cytoplasm fractions, respectively.
Statistical Analysis.
All statistical analyses were performed using Prism software. Comparisons for two groups were performed by Student’s t test. Comparisons for more than two groups were calculated by one-way or two-way ANOVA, followed by Fisher’s lease significant difference posttest. Results with a value of P < 0.05 were considered significant.
Supplementary Material
Acknowledgments
We thank Dr. Paul Schumacker for the Uqcrfs1fl/fl mice; Dr. Albert Bendelac for DN32.D3 NKT cell hybridoma; Dr. Sebastian Joyce for N38.2C12 natural killer T cell hybridoma; the NIH tetramer core facility for CD1d and MR1 tetramers; NUSeq Core for library construction and sequencing, and Northwestern University flow cytometry core facility for cell sorting services. This work was supported by NIH Grants R01 AI43407 and R01 AI057460 (to C.-R.W.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021385118/-/DCSupplemental.
Data Availability
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE157283). All other study data are included in the article and SI Appendix.
References
- 1.Geiger R., et al., L-arginine modulates T cell metabolism and enhances survival and anti-tumor activity. Cell 167, 829–842.e13 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Blagih J., et al., The energy sensor AMPK regulates T cell metabolic adaptation and effector responses in vivo. Immunity 42, 41–54 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Chang C. H., et al., Posttranscriptional control of T cell effector function by aerobic glycolysis. Cell 153, 1239–1251 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pearce E. L., Pearce E. J., Metabolic pathways in immune cell activation and quiescence. Immunity 38, 633–643 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sena L. A., et al., Mitochondria are required for antigen-specific T cell activation through reactive oxygen species signaling. Immunity 38, 225–236 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michalek R. D., et al., Cutting edge: Distinct glycolytic and lipid oxidative metabolic programs are essential for effector and regulatory CD4+ T cell subsets. J. Immunol. 186, 3299–3303 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.van der Windt G. J., et al., Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development. Immunity 36, 68–78 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brennan P. J., Brigl M., Brenner M. B., Invariant natural killer T cells: An innate activation scheme linked to diverse effector functions. Nat. Rev. Immunol. 13, 101–117 (2013). [DOI] [PubMed] [Google Scholar]
- 9.Godfrey D. I., Stankovic S., Baxter A. G., Raising the NKT cell family. Nat. Immunol. 11, 197–206 (2010). [DOI] [PubMed] [Google Scholar]
- 10.Constantinides M. G., Bendelac A., Transcriptional regulation of the NKT cell lineage. Curr. Opin. Immunol. 25, 161–167 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ziętara N., et al., Critical role for miR-181a/b-1 in agonist selection of invariant natural killer T cells. Proc. Natl. Acad. Sci. U.S.A. 110, 7407–7412 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Henao-Mejia J., et al., The microRNA miR-181 is a critical cellular metabolic rheostat essential for NKT cell ontogenesis and lymphocyte development and homeostasis. Immunity 38, 984–997 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Shin J., et al., Mechanistic target of rapamycin complex 1 is critical for invariant natural killer T-cell development and effector function. Proc. Natl. Acad. Sci. U.S.A. 111, E776–E783 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L., et al., Mammalian target of rapamycin complex 1 orchestrates invariant NKT cell differentiation and effector function. J. Immunol. 193, 1759–1765 (2014). [DOI] [PubMed] [Google Scholar]
- 15.Park H., Tsang M., Iritani B. M., Bevan M. J., Metabolic regulator Fnip1 is crucial for iNKT lymphocyte development. Proc. Natl. Acad. Sci. U.S.A. 111, 7066–7071 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Webb T. J., et al., Alterations in cellular metabolism modulate CD1d-mediated NKT-cell responses. Pathog. Dis. 74, ftw055 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bazhin A. V., et al., The novel mitochondria-targeted antioxidant SkQ1 modulates angiogenesis and inflammatory micromilieu in a murine orthotopic model of pancreatic cancer. Int. J. Cancer 139, 130–139 (2016). [DOI] [PubMed] [Google Scholar]
- 18.Kang Y. J., et al., Regulation of NKT cell-mediated immune responses to tumours and liver inflammation by mitochondrial PGAM5-Drp1 signalling. Nat. Commun. 6, 8371 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Finlay D. K., et al., Temporal differences in the dependency on phosphoinositide-dependent kinase 1 distinguish the development of invariant Valpha14 NKT cells and conventional T cells. J. Immunol. 185, 5973–5982 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar A., et al., Enhanced oxidative phosphorylation in NKT cells is essential for their survival and function. Proc. Natl. Acad. Sci. U.S.A. 116, 7439–7448 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Murphy M. P., Siegel R. M., Mitochondrial ROS fire up T cell activation. Immunity 38, 201–202 (2013). [DOI] [PubMed] [Google Scholar]
- 22.Ledderose C., et al., Mitochondria are gate-keepers of T cell function by producing the ATP that drives purinergic signaling. J. Biol. Chem. 289, 25936–25945 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kamiński M. M., Röth D., Krammer P. H., Gülow K., Mitochondria as oxidative signaling organelles in T-cell activation: Physiological role and pathological implications. Arch. Immunol. Ther. Exp. (Warsz.) 61, 367–384 (2013). [DOI] [PubMed] [Google Scholar]
- 24.Kim Y. H., Kumar A., Chang C. H., Pyaram K., Reactive oxygen species regulate the inflammatory function of NKT cells through promyelocytic leukemia zinc finger. J. Immunol. 199, 3478–3487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mak T. W., et al., Glutathione primes T cell metabolism for inflammation. Immunity 46, 1089–1090 (2017). [DOI] [PubMed] [Google Scholar]
- 26.Pyaram K., et al., Keap1-Nrf2 system plays an important role in invariant natural killer T cell development and homeostasis. Cell Rep. 27, 699–707.e4 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Raynor J. L., et al., Hippo/Mst signaling coordinates cellular quiescence with terminal maturation in iNKT cell development and fate decisions. J. Exp. Med. 217, e20191157 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fu S., et al., Immunometabolism regulates TCR recycling and iNKT cell functions. Sci. Signal. 12, eaau1788 (2019). [DOI] [PubMed] [Google Scholar]
- 29.Salio M., et al., Essential role for autophagy during invariant NKT cell development. Proc. Natl. Acad. Sci. U.S.A. 111, E5678–E5687 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhao J., Weng X., Bagchi S., Wang C. R., Polyclonal type II natural killer T cells require PLZF and SAP for their development and contribute to CpG-mediated antitumor response. Proc. Natl. Acad. Sci. U.S.A. 111, 2674–2679 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Garner L. C., Klenerman P., Provine N. M., Insights into mucosal-associated invariant T cell biology from studies of invariant natural killer T cells. Front. Immunol. 9, 1478 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Koay H. F., Godfrey D. I., Pellicci D. G., Development of mucosal-associated invariant T cells. Immunol. Cell Biol. 96, 598–606 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guo J., et al., Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat. Immunol. 3, 469–476 (2002). [DOI] [PubMed] [Google Scholar]
- 34.Egawa T., et al., Genetic evidence supporting selection of the Valpha14i NKT cell lineage from double-positive thymocyte precursors. Immunity 22, 705–716 (2005). [DOI] [PubMed] [Google Scholar]
- 35.McDonald B. D., Constantinides M. G., Bendelac A., Polarized effector programs for innate-like thymocytes. Nat. Immunol. 14, 1110–1111 (2013). [DOI] [PubMed] [Google Scholar]
- 36.Savage A. K., et al., The transcription factor PLZF directs the effector program of the NKT cell lineage. Immunity 29, 391–403 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Seiler M. P., et al., Elevated and sustained expression of the transcription factors Egr1 and Egr2 controls NKT lineage differentiation in response to TCR signaling. Nat. Immunol. 13, 264–271 (2012). Corrected in: Nat. Immunol.14, 413 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee Y. J., Holzapfel K. L., Zhu J., Jameson S. C., Hogquist K. A., Steady-state production of IL-4 modulates immunity in mouse strains and is determined by lineage diversity of iNKT cells. Nat. Immunol. 14, 1146–1154 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H., Hogquist K. A., CCR7 defines a precursor for murine iNKT cells in thymus and periphery. eLife 7, e34793 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Godfrey D. I., Pellicci D. G., Smyth M. J., Immunology. The elusive NKT cell antigen—Is the search over? Science 306, 1687–1689 (2004). [DOI] [PubMed] [Google Scholar]
- 41.Zhou D., et al., Lysosomal glycosphingolipid recognition by NKT cells. Science 306, 1786–1789 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Moran A. E., et al., T cell receptor signal strength in Treg and iNKT cell development demonstrated by a novel fluorescent reporter mouse. J. Exp. Med. 208, 1279–1289 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imboden J. B., Stobo J. D., Transmembrane signalling by the T cell antigen receptor. Perturbation of the T3-antigen receptor complex generates inositol phosphates and releases calcium ions from intracellular stores. J. Exp. Med. 161, 446–456 (1985). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Knudson K. M., Goplen N. P., Cunningham C. A., Daniels M. A., Teixeiro E., Low-affinity T cells are programmed to maintain normal primary responses but are impaired in their recall to low-affinity ligands. Cell Rep. 4, 554–565 (2013). [DOI] [PubMed] [Google Scholar]
- 45.Gordy L. E., et al., IL-15 regulates homeostasis and terminal maturation of NKT cells. J. Immunol. 187, 6335–6345 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Klose C. S., et al., The transcription factor T-bet is induced by IL-15 and thymic agonist selection and controls CD8αα(+) intraepithelial lymphocyte development. Immunity 41, 230–243 (2014). [DOI] [PubMed] [Google Scholar]
- 47.Townsend M. J., et al., T-bet regulates the terminal maturation and homeostasis of NK and Valpha14i NKT cells. Immunity 20, 477–494 (2004). [DOI] [PubMed] [Google Scholar]
- 48.Matsuda J. L., et al., Homeostasis of V alpha 14i NKT cells. Nat. Immunol. 3, 966–974 (2002). [DOI] [PubMed] [Google Scholar]
- 49.Wilson M. T., et al., The response of natural killer T cells to glycolipid antigens is characterized by surface receptor down-modulation and expansion. Proc. Natl. Acad. Sci. U.S.A. 100, 10913–10918 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Macian F., NFAT proteins: Key regulators of T-cell development and function. Nat. Rev. Immunol. 5, 472–484 (2005). [DOI] [PubMed] [Google Scholar]
- 51.Senavirathna L. K., et al., Hypoxia induces pulmonary fibroblast proliferation through NFAT signaling. Sci. Rep. 8, 2709 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacIver N. J., Michalek R. D., Rathmell J. C., Metabolic regulation of T lymphocytes. Annu. Rev. Immunol. 31, 259–283 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parekh V. V., et al., Glycolipid antigen induces long-term natural killer T cell anergy in mice. J. Clin. Invest. 115, 2572–2583 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kim S., et al., Impact of bacteria on the phenotype, functions, and therapeutic activities of invariant NKT cells in mice. J. Clin. Invest. 118, 2301–2315 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Braun N. A., et al., Development of spontaneous anergy in invariant natural killer T cells in a mouse model of dyslipidemia. Arterioscler. Thromb. Vasc. Biol. 30, 1758–1765 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Raud B., et al., Etomoxir actions on regulatory and memory T cells are independent of Cpt1a-mediated fatty acid oxidation. Cell Metab. 28, 504–515.e7 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Van den Bossche J., van der Windt G. J. W., Fatty acid oxidation in macrophages and T cells: Time for reassessment? Cell Metab. 28, 538–540 (2018). [DOI] [PubMed] [Google Scholar]
- 58.Weinberg S. E., Sena L. A., Chandel N. S., Mitochondria in the regulation of innate and adaptive immunity. Immunity 42, 406–417 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Turrens J. F., Mitochondrial formation of reactive oxygen species. J. Physiol. 552, 335–344 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Weinberg S. E., et al., Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 565, 495–499 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Choi H. J., et al., Differential requirements for the Ets transcription factor Elf-1 in the development of NKT cells and NK cells. Blood 117, 1880–1887 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kovalovsky D., et al., The BTB-zinc finger transcriptional regulator PLZF controls the development of invariant natural killer T cell effector functions. Nat. Immunol. 9, 1055–1064 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Lazarevic V., et al., The gene encoding early growth response 2, a target of the transcription factor NFAT, is required for the development and maturation of natural killer T cells. Nat. Immunol. 10, 306–313 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Dose M., et al., Intrathymic proliferation wave essential for Valpha14+ natural killer T cell development depends on c-Myc. Proc. Natl. Acad. Sci. U.S.A. 106, 8641–8646 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Mycko M. P., et al., Selective requirement for c-Myc at an early stage of V(alpha)14i NKT cell development. J. Immunol. 182, 4641–4648 (2009). [DOI] [PubMed] [Google Scholar]
- 66.Michel M. L., et al., Critical role of ROR-γt in a new thymic pathway leading to IL-17-producing invariant NKT cell differentiation. Proc. Natl. Acad. Sci. U.S.A. 105, 19845–19850 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Matsuda J. L., et al., T-bet concomitantly controls migration, survival, and effector functions during the development of Valpha14i NKT cells. Blood 107, 2797–2805 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Mandal M., et al., Tissue distribution, regulation and intracellular localization of murine CD1 molecules. Mol. Immunol. 35, 525–536 (1998). [DOI] [PubMed] [Google Scholar]
- 69.Goossens P. L., Jouin H., Marchal G., Milon G., Isolation and flow cytometric analysis of the free lymphomyeloid cells present in murine liver. J. Immunol. Methods 132, 137–144 (1990). [DOI] [PubMed] [Google Scholar]
- 70.Zimmer M. I., et al., A cell-type specific CD1d expression program modulates invariant NKT cell development and function. J. Immunol. 176, 1421–1430 (2006). [DOI] [PubMed] [Google Scholar]
- 71.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Anders S., Pyl P. T., Huber W., HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Robinson M. D., McCarthy D. J., Smyth G. K., edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26, 139–140 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Raudvere U., et al., g:Profiler: A web server for functional enrichment analysis and conversions of gene lists (2019 update). Nucleic Acids Res. 47, W191–W198 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo (accession no. GSE157283). All other study data are included in the article and SI Appendix.