Significance
Harsh conditions in high elevations present strong stresses for organisms. Previous studies targeting phylogenetically distinct species revealed cases of diversified adaptations, but it remains largely unknown how common ancestry contributes to evolution of similar adaptations. Our study based on a species complex (snowfinches) living on Qinghai–Tibet Plateau shows that ancestral snowfinches had phenotypically evolved larger body size and genetically an accelerated selection on genes related to development and signaling. From this ancestral state of adaptation three descendants have undergone independent adaptive processes in response to the differences in selective pressures acting on them. A striking example is a DNA repair gene, DTL, in which nonsynonymous substitutions evolving in ancestor and descendants have led to different DNA damage repair kinetics.
Keywords: comparative genomics, high-elevation adaptations, common ancestry, snowfinches, DTL
Abstract
Species in a shared environment tend to evolve similar adaptations under the influence of their phylogenetic context. Using snowfinches, a monophyletic group of passerine birds (Passeridae), we study the relative roles of ancestral and species-specific adaptations to an extreme high-elevation environment, the Qinghai–Tibet Plateau. Our ancestral trait reconstruction shows that the ancestral snowfinch occupied high elevations and had a larger body mass than most nonsnowfinches in Passeridae. Subsequently, this phenotypic adaptation diversified in the descendant species. By comparing high-quality genomes from representatives of the three phylogenetic lineages, we find that about 95% of genes under positive selection in the descendant species are different from those in the ancestor. Consistently, the biological functions enriched for these species differ from those of their ancestor to various degrees (semantic similarity values ranging from 0.27 to 0.5), suggesting that the three descendant species have evolved divergently from the initial adaptation in their common ancestor. Using a functional assay to a highly selective gene, DTL, we demonstrate that the nonsynonymous substitutions in the ancestor and descendant species have improved the repair capacity of ultraviolet-induced DNA damage. The repair kinetics of the DTL gene shows a twofold to fourfold variation across the ancestor and the descendants. Collectively, this study reveals an exceptional case of adaptive evolution to high-elevation environments, an evolutionary process with an initial adaptation in the common ancestor followed by adaptive diversification of the descendant species.
Organisms living at high elevations are exposed to cold temperatures, low levels of oxygen, and strong ultraviolet (UV) radiation. These strong stresses drive drastic phenotypic adaptations, such as the increase of hemoglobin–oxygen affinity and metabolic rates, larger body size, and an enhanced UV tolerance (1–4). Despite the similarity in phenotypic adaptations, recent studies have demonstrated diversified and species-specific adaptations to high elevations at the genomic level (5–15). As the acquirement of similar genetic adaptations in a group of species is shaped by their phylogenetic relationship (16), it is an intriguing question whether a group of species sharing a common ancestry evolve different genetic adaptations to the high elevations.
The snowfinch species complex provides a unique opportunity to study this adaptive diversification within the context of common ancestry, i.e., how the adaptive processes of the descendant species differ from that of their common ancestor. Snowfinch is one of the few avian clades that have experienced an “in situ” radiation in extreme high-elevation environments, i.e., higher than 3,500 m above sea level (m a.s.l.) (17, 18). The Qinghai–Tibet Plateau (QTP), sometimes described as the “Third Pole” (19), houses six out of the total seven snowfinch species. Compared to most of their closest relatives among the Old World Sparrows (Passeridae), the snowfinches have a larger body size, darker plumage, and increased metabolic rate (20, 21). Previous studies suggested that snowfinches, consisting of the three genera of Montifringilla, Onychostruthus, and Pyrgilauda, form a monophyletic group (22–24). It is therefore possible that the observed similarities in phenotypic adaptations to high elevation are inherited from their common ancestor, assuming that the ancestral species had already adapted to this high-elevation environment. Furthermore, as the extant snowfinches differ somewhat in their morphology, niche utilization, and behavior (20, 21), they may also have evolved the differences in the adaptive characteristics after they split from their common ancestor. Under this adaptive scenario it is likely that the genetic adaptation of the snowfinches to high-elevation environments is an evolutionary process of an initial adaptation in the ancestor and a subsequent adaptive diversification in the descendant species. To our knowledge, this topic has not been explored previously in relation to high-elevation adaptation.
In this study, we generate three de novo genomes from representatives of the three lineages of snowfinches. We integrate phenotypic, genomic, and functional assay data to investigate the impact of the conditions of the ancestor (ancestral adaptation) on the adaptive evolution in the descendant species (species-specific adaptation). Our results show that the snowfinch ancestor has evolved relatively larger body size and an accelerated selection on genes related to development and cellular signaling. From these ancestral adaptive conditions, the three species of snowfinches have evolved species-specific adaptive strategies to high-elevation environments. Using a functional assay on a highly selected gene, denticleless E3 ubiquitin protein ligase homolog (DTL), we demonstrate that multiple nonsynonymous substitutions in the ancestor and the descendant species have increased the repair capacity of UV-induced DNA damage. Altogether, our results show an evolutionary process of an initial adaptation in the ancestor followed by adaptive diversification in the descendant species, which have concurrently generated similar but not identical evolutionary routes to reach the high-elevation adaptation.
Results
Generation of High-Quality Genomes of the Three Representative Lineages of Snowfinches.
To explore how snowfinches adapt to high-elevation environments, we sequenced the whole genomes of the representatives of three currently recognized major lineages within the group (Fig. 1A), the white-rumped snowfinch, Onychostruthus taczanowskii (hereafter referred to as taczan), the rufous-necked snowfinch, Pyrgilauda ruficollis (rufico), and the black-winged snowfinch, Montifringilla adamsi (adamsi). For each species, we generated more than 110× sequencing data with four to five different insert sizes (Table 1 and SI Appendix, SI Text 1 and Tables S1 and S2). We achieved high quality assemblies for all three species with scaffold N50 of ∼10MB and contig N50 of ∼0.5 MB (Table 1 and SI Appendix, SI Text 1 and Table S3). Combining methods of homology-based modeling, de novo gene prediction, and RNA-sequencing-based assemblies, we annotated around 15,000 protein-coding genes for each of the three species, 99.8% of which have homologs in the public protein databases (Table 1 and SI Appendix, SI Text 1 and Tables S4–S8). Using benchmarks against universal single-copy orthologs of birds (BUSCO, ref. 25), we estimated that the gene annotation is nearly complete (95%, SI Appendix, SI Text 1 and Tables S9–S11).
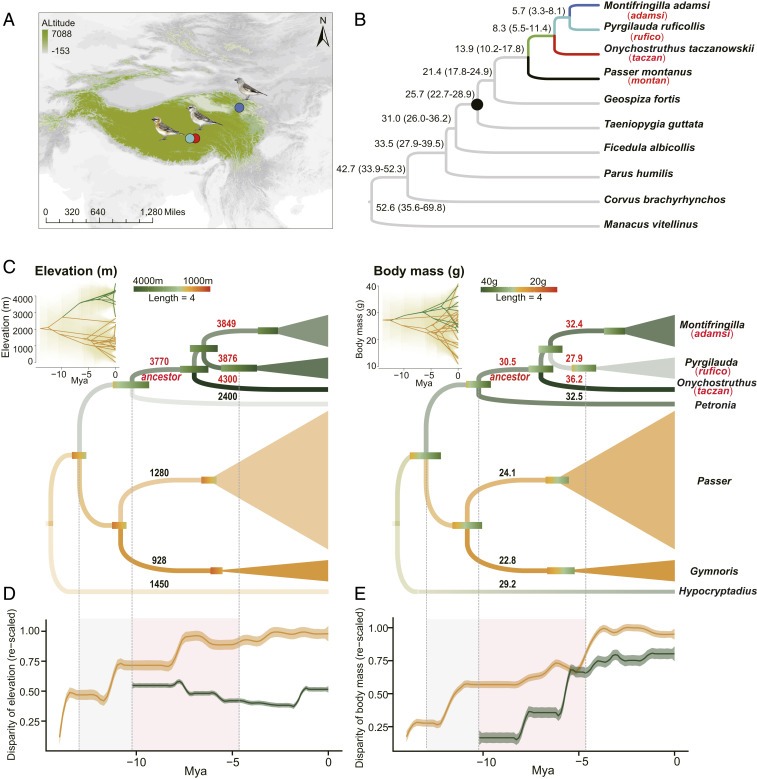
Fig. 1.
The phylogenomic tree and ancestral trait reconstruction of the snowfinches. (A) Sampling sites of three species of snowfinches used in this study. For (A) and (B), three snowfinch lineages are similarly color-coded. (B) The phylogenomic tree suggests that the snowfinches diverged from montan between 10 and 18 Ma. The black dot marks the calibration point (26.0 ± 3.0 Ma) in estimating divergence time. (C) Reconstruction of the ancestral traits for 27 species from Passeridae suggests that the ancestor of the snowfinches occupied a high-elevation environment (Left) and evolved a large body size (Right). The ancestral branch (ancestor), the representatives from three main lineages (adamsi, rufico, and taczan), and a lowland relative, the Eurasian tree sparrow (montan), are marked along the tree. Twenty-seven species in Passeridae are collapsed into seven genera. The point estimates and 95% CIs of traits reconstructed for common ancestors of given genera are shown in numerical values and bars. Note only the point estimates are shown for the genera that have a single species (i.e., Onychostruthus, Petronia, and Hypocryptadius). The size of the triangle is proportional to the number of species. Phenograms in the inset show the trait evolution of the 27 species (y axis) over time (x axis) based on ancestral trait reconstruction with 95% CIs shown as shades. (D and E) Accumulation of elevation disparity (D) and body mass disparity (E) through time for all species of Passeridae (yellow curve lines) and snowfinches (green curve lines). The gray and orange shaded areas indicate time ranges when snowfinches split from nonsnowfinches and snowfinches diversified, respectively. Note that the divergence time of snowfinches in Päckert et al. (24) (C, D, and E) slightly differs from our phylogenomic tree in B.
Table 1.
Basic statistics for genome sequencing, assembly, and annotation of snowfinch species
| Species | Common name | Sequence data (G) | Insert size | Scaffold N50, Mb | Contig N50, Mb | Gene no. |
| O. taczanowskii | White-rumped snowfinch | 152.6 | 170, 500, 800, 2,000, 5,000 | 9.07 | 0.67 | 15,585 |
| P. ruficollis | Rufous-necked snowfinch | 118.5 | 170, 500, 2,000, 5,000 | 9.60 | 0.51 | 15,206 |
| M. adamsi | Black-winged snowfinch | 139.7 | 170, 500, 2,000, 5,000 | 10.06 | 0.37 | 15,136 |
Snowfinches as a Monophyletic Group Emerged around 14 Ma.
Previous studies constructed the phylogeny of snowfinches based on a small number of genes (22–24). In this study, we corroborated the phylogenetic relationship of the three aforementioned phylogenetic lineages using whole genomic data. We selected seven phylogenetically representative oscine passerines as outgroups which have relatively well-annotated genomes available. We obtained a robust phylogenomic tree based on single-copy orthologous genes with 100% bootstrap values for all internal branches. This phylogenetic relationship is consistent with previously published results, supporting a monophyletic origin of three snowfinches (Fig. 1B). Given this robust phylogeny, we then estimated the time of divergence between the snowfinches and the other oscine passerines. Using the crown age of Passerida (26 ± 3 Ma, ref. 26) as a calibration point (Fig. 1B) we found that the ancestral snowfinch (ancestor) diverged from the most closely related lowland species, Passer montanus (Eurasian tree sparrow, montan) around 14 Ma (95% confidence interval 10.3 to 17.8 Ma), and the three major lineages diverged from one another between 5.7 and 8.3 Ma.
Ancestral Trait Reconstruction of Phenotypic Adaptation to a High-Elevation Environment.
We infer the ancestral elevation distribution of the snowfinches to determine whether the ancestral snowfinch had already inhabited high-elevation environments. To accurately infer the ancestral elevation, we collected midelevation records from the 27 extant species across seven genera of Passeridae and performed the ancestral trait reconstruction based on the species tree of Passeridae derived from Päckert et al. (24). Our results showed that the ancestor had likely already inhabited the QTP at an elevation of ∼3,770 m a.s.l, which is within the elevational range of many extant species of snowfinches (Fig. 1C). Most nonsnowfinch species of Passeridae, however, are distributed at elevation below 2,000 m a.s.l., with the exception of Petronia petronia (rock sparrow) at 2,400 m a.s.l. Consistently, we also found a remarkable division in elevational range at the time when the snowfinches split from other Passeridae species and a subsequent decreasing variation in elevation during the diversification of the snowfinches (Fig. 1D).
We then checked if ancestral snowfinch had evolved phenotypic characters specific to high-elevation animals. In general, birds at high elevation evolve a relatively larger body size than their low-elevation relatives (27). We thus examined the variation of body mass across the snowfinch phylogeny. The results of the ancestral trait reconstruction showed that the body mass of the ancestor had increased to ∼30 g, which is greater than that of most nonsnowfinch species of Passeridae but slightly smaller than that of the rock sparrow (Fig. 1C). During the diversification of snowfinches, both adamsi and taczan have increased their body mass whereas rufico has decreased its body mass compared to the ancestor (Fig. 1C). This result indicates an increased variation in body mass among snowfinches after their divergence from the common ancestor (Fig. 1E). Based on the time-scaled phylogeny, the results suggest that the ancestor began its adaptation to high-elevation environments around 14 Ma, and the descendant species have evolved divergent phenotypic adaptations during the speciation.
Positive Selection in the Common Ancestor and Descendant Species.
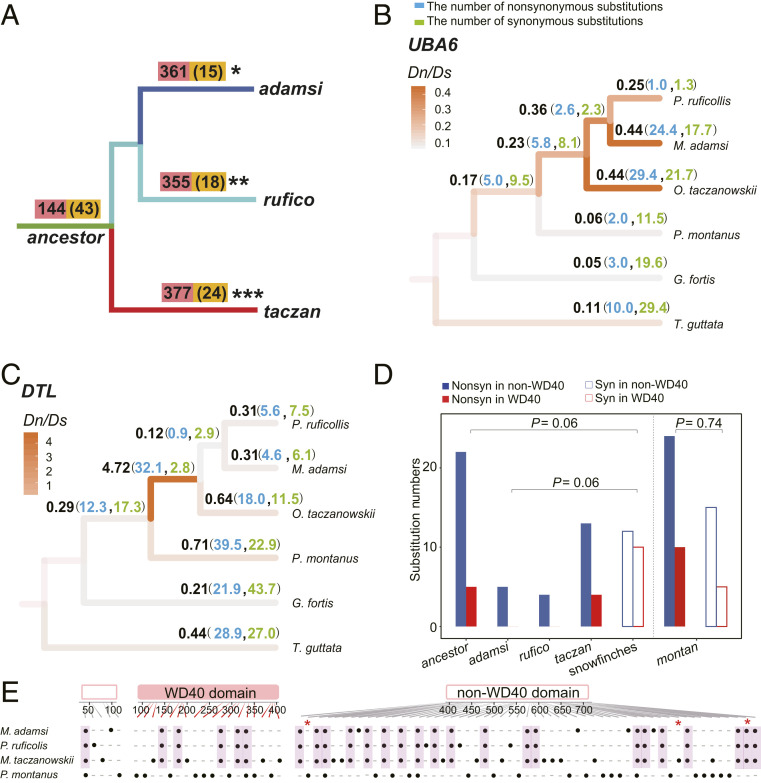
As the descendant snowfinches show diversification of phenotypic adaptations following an initial adaptation in the ancestor, we expect to find a similar trend at the genetic level. Using the branch-site model in PAML (28) we identified positively selected genes (PSGs) in the ancestor (144 PSGs) and the descendant species (355 to 377 PSGs; Fig. 2A and SI Appendix, Table S12). We found that ∼95% PSGs differ between the ancestor and the descendant species. However, we also found an excess of PSGs shared between the ancestor and the descendant snowfinches (one-sided binomial tests, false discovery rate [FDR]-adjusted P < 0.05; Fig. 2A). For example, the ubiquitin-like modifier activating enzyme 6 (UBA6), a gene activating ubiquitin activity (29), shows accelerated selection across snowfinch branches, with the ratio of nonsynonymous substitution rate and synonymous substitution rate (dN/dS) ranging from 0.23 to 0.44 as compared to ∼0.1 in other branches. It is worth noting that the dN/dS values are higher in adamsi and taczan (∼0.44) than in rufico (0.25; Fig. 2B), suggesting differences of selective strength among the descendant species. These results suggest that positive selection acting on the descendant species has driven divergent adaptations, demonstrated by the substantially different PSGs across species, although an appreciable number of genes (15 in adamsi to 24 in taczan; Fig. 2A) are under positive selection in both the ancestor and descendant species.
Fig. 2.
Positive selection analyses of snowfinches. (A) Numbers of PSGs in ancestor, taczan, rufico, and adamsi. Numbers of all PSGs are marked in red, and those shared between ancestor and all three descendant snowfinches (list on the ancestral branch) as well as between ancestor and each of the descendant snowfinches (list on the respective snowfinch branches) are marked in yellow and in brackets. We calculated the observed ratios of shared PSGs between ancestor and adamsi (0.04, 15/361), ancestor and rufico (0.05, 18/355), or ancestor and taczan (0.06, 24/377). We then tested these ratios against the expected ratio (0.02, 144/6,572 genes) using a binomial test. ***P < 0.001; **P < 0.01; *P < 0.05. (B and C) Evolutionary rates of two representative genes (B, UBA6 and C, DTL). dN/dS, dN and dS in each branch are shown by black, blue, and green, respectively. (D) Statistics of nonsynonymous and synonymous substitutions in the WD40 and non-WD40 regions of the ancestor and three descendant snowfinch DTLs. The reason that we merged all three descendant species is because each species consists of only a few synonymous substitutions. Nonsyn, nonsynonymous substitutions; syn, synonymous substitutions. (E) Distribution of the nonsynonymous substitutions of the snowfinch and montan DTLs in the WD40 and non-WD40 regions, which are marked by black dots. The nonsynonymous substitutions of ancestor DTL, i.e., those shared by three snowfinches, are highlighted by purple boxes. Asterisks show sites containing two nonsynonymous substitutions, which could not be drawn separately due to the space limit.
For the ancestor-only PSGs, which are genes showing an increase of dN/dS values only in the ancestral branch but not in the three descendant species, we hypothesize that the three descendant species may undergo a divergent adaptation by accumulating species-specific nonsynonymous substitutions. To test this hypothesis, we selected DTL, a gene related to DNA damage repair (30, 31), because DTL exhibits the strongest signal of positive selection in the ancestral branch (dN/dS, 4.72), higher than in any other branches (dN/dS, 0.12∼0.71; Fig. 2C). To examine which region is particularly subject to positive selection, we analyzed the spatial position of nonsynonymous substitutions across different functional domains in DTL using synonymous substitutions as neutral control. We found that the WD40 domain of DTL has been marginally depleted with nonsynonymous substitutions in the ancestor (5 substitutions in WD40 domain vs. 22 in non-WD40 domain region) compared to synonymous substitutions (10 vs. 12, Fisher’s exact test, FDR-adjusted P = 0.06; Fig. 2 D and E). This pattern differs from the positions of nonsynonymous and synonymous substitutions in WD40 and non-WD40 regions in montan (10 vs. 24 as compared to 5 vs. 15, FDR-adjusted P = 0.74; Fig. 2 D and E). As synonymous substitutions approximate the neutral mutation input, non-WD40 region of ancestor DTL seems to be the major target of positive selection. After the three descendant snowfinches split they evolved species-specific nonsynonymous substitutions in the non-WD40 region (4 vs. 22, FDR-adjusted P = 0.06; Fig. 2 D and E). DTL of taczan has a relatively large number (4 vs. 13), whereas adamsi and rufico DTLs have small numbers (rufico, 0 vs. 4; adamsi, 0 vs. 5). Thus, even for a gene that is highly selected in the ancestor, the descendant species have evolved varying numbers of nonsynonymous substitutions that probably lead to different functional dynamics.
Impact of DTL Nonsynonymous Substitutions in the Ancestral and Descendant Snowfinches on UV-Induced DNA Damage Repair.
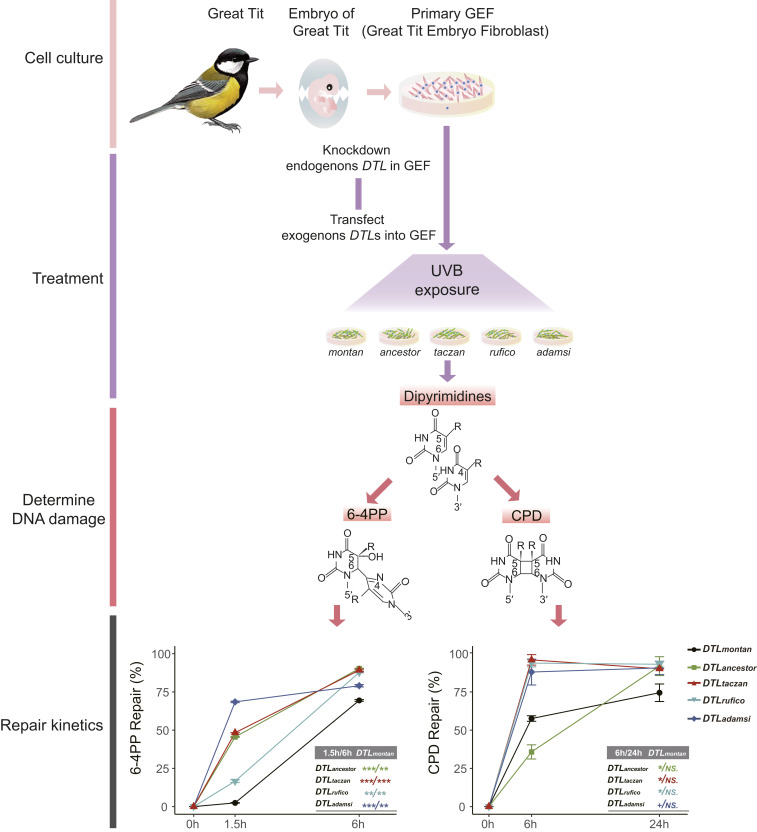
Because DTL is involved in DNA damage repair (30, 31), we examined whether the nonsynonymous substitutions in snowfinches affect repair capability of UV-induced DNA damage. We first predicted the functional effects of nonsynonymous substitutions using Provean (32) and Sift (33). Compared to those in montan, 6% (Provean) and 21% (Sift) substitutions potentially cause functional change and only one substitution (P506L) is predicted by both programs to be functionally relevant (SI Appendix, Table S13). Because the two programs give different results, we hypothesized that a cumulative effect of snowfinch-specific nonsynonymous substitutions could lead to more radical functional change. To examine this, we chemically synthesized the full-length protein-coding sequences of DTLs for the three descendant snowfinches (DTLtaczan, DTLrufico, and DTLadamsi) and their ancestor (DTLancestor). We also synthesized montan DTL (DTLmontan) as a control. We then cloned these DTL sequences into lentiviral vectors (Fig. 3 and SI Appendix, Text 2). We used our previously established embryo fibroblast cell line (GEF; Fig. 3) from a wild great tit (34) to test the repair function of DTL. The great tit is the closest relative to snowfinches with cell lines available. After knocking down the endogenous DTL expression in GEF (SI Appendix, Fig. S1), we overexpressed DTLtaczan, DTLrufico, DTLadamsi, DTLancestor, and DTLmontan via the lentiviral vectors. We exposed all five cell lines to UV irradiation and subsequently examined the repair kinetics of DTL (proportion of repair to damage) by measuring two representative UV-induced DNA lesions, pyrimidine (6-4) pyrimidone photoproduct (6-4PP) and cyclobutane pyrimidine dimers (CPD) (35).
Fig. 3.
A schematic representation of DTL’s functional experiment. The embryo fibroblast cells from the great tit (named GEF) were used. After knocking down endogenous DTL and transfecting exogenous DTLtaczan, DTLrufico, DTLadamsi, DTLancestor, and DTLmontan (SI Appendix, Fig. S1), we exposed GEFs to UVB irradiation and checked repair kinetics (percent repair) by measuring two UV-induced photoproducts, 6-4PP and CPD. For each curve, the bar of SE is shown based on three biological replicates. Only post hoc t tests between each of snowfinch DTLs and DTLmontan are shown. ***P < 0.001; **P < 0.01; *P ≤ 0.05; +P = 0.08; NS, not statistically significant. Statistical results of pairwise comparisons of snowfinch DTLs are shown in SI Appendix, Table S14.
In a two-way repeated measures ANOVA the two assays showed that snowfinch DTLs had a significant improvement of repair effects (6-4PP, P < 0.001; CPD, P < 0.001) and repair kinetics through time (6-4PP, P < 0.001; CPD, P < 0.001) as compared to montan DTL (Fig. 3). Specifically, at the time point of 1.5 h we found that 6-4PP repair capacity was significantly higher in the ancestral and descendant snowfinch DTLs (DTLtaczan, DTLrufico, DTLadamsi, and DTLancestor) than in DTLmontan (post hoc t test, FDR-adjusted P < 0.01; Fig. 3). The descendant and ancestral snowfinch DTLs differ in their repair kinetics (FDR-adjusted P < 0.05; Fig. 3 and SI Appendix, Table S14). DTLadamsi shows the greatest repair capability by repairing ∼65% DNA damage, whereas DTLrufico exhibits the smallest repair capability and only repairs ∼15% DNA damage. Both DTLancestor and DTLtaczan have an intermediate level of repair capability by repairing ∼45% DNA damage (FDR-adjusted P < 0.05). At the time point of 6 h all snowfinch DTLs had reached the repair levels of ∼80%, which are significantly higher than that of montan (∼60%, FDR-adjusted P < 0.05). DTLancestor and DTLtaczan show a similar repair kinetics (FDR-adjusted P value, nonsignificant), which significantly differs from that of DTLrufico and DTLadamsi (FDR-adjusted P < 0.05).
For the CPD assay, the three DTLs of the descendant snowfinches showed a similar repair kinetics and reached the peak of repair capability at the time point of 6 h (recovering ∼90% DNA damage, FDR-adjusted P values, nonsignificant). Their repair kinetics was significantly and marginally higher than that of DTLmontan (∼57%, FDR-adjusted P < 0.05 and P = 0.08). In contrast, DTLancestor only repairs about 40% damage, and its kinetics differs significantly from those of the descendant snowfinch DTLs (FDR-adjusted P < 0.05) and that of DTLmontan (FDR-adjusted P = 0.05). At the time point of 24 h, the repair kinetics of all snowfinch DTLs is higher than that of DTLmontan (∼95% vs. ∼74%), although the difference is not statistically significant after the multiple correction (Fig. 3 and SI Appendix, Table S14). Notably, DTLancestor shows lower efficiency of CPD repair at 6 h but higher efficiency at 24 h when compared to DTLmontan, and DTLancestor also has a constant higher efficiency of 6-4PP repair compared to DTLmontan. This pattern is likely a trade-off due to functional antagonism between 6-4PP and CPD repair or between early and late repair dynamics. Taken together, these results suggest that all snowfinch DTLs exhibit an improved repair capability of DNA damage, and the repair kinetics is different across the ancestor and the descendant species.
Comparison of Enriched Biological Processes between Descendant Species and Their Ancestor.
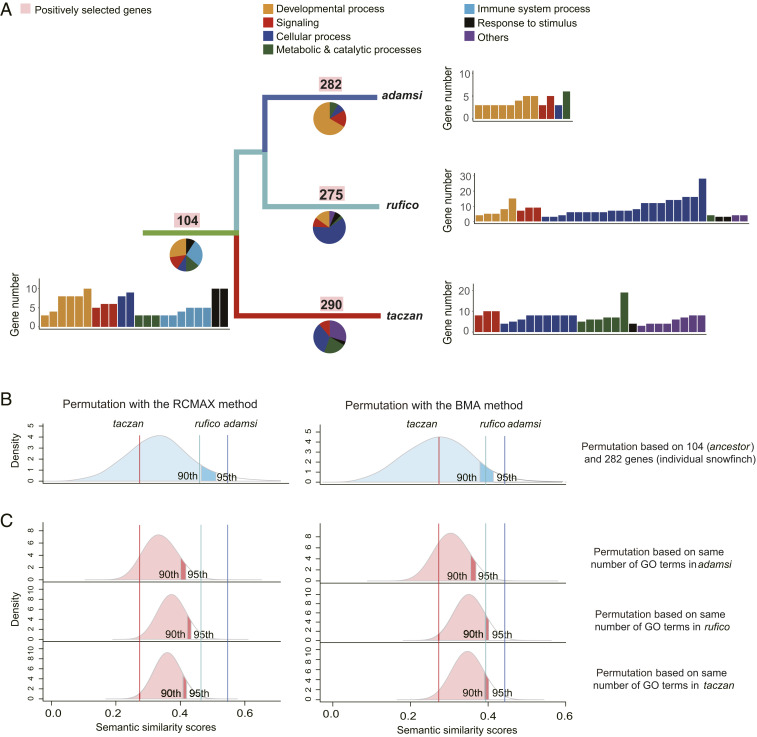
We tested whether the adaptive divergence observed in PSGs and DTL function in the descendant species also occurs in biological processes represented by Gene Ontology (GO) terms. We utilized GO annotations to analyze the functional enrichments of PSGs in the ancestor and the descendant species. We found that PSGs are generally related to development, signaling, and cellular processes (Fig. 4A). To quantify the differences between ancestor and descendant snowfinches in functional enrichments we used GOSemSim (36) to compute pairwise semantic similarity (SS) of GO annotations. We implemented two measurements (RCMAX and BMA) to examine the robustness of the results. A greater SS value indicates more similarity between the two sets of GO terms (37, 38). Our results show that the pairwise SS values are the highest between the ancestor and adamsi (0.547 and 0.446), intermediate between the ancestor and rufico (0.46 and 0.394), and lowest between the ancestor and taczan (0.274 and 0.273; Fig. 4B). This result indicates that the three descendant species exhibit different levels of similarity of GO terms to the ancestor.
Fig. 4.
Functional enrichment analyses of PSGs. (A) Distribution of overrepresented GO terms is shown as a pie chart below the branch. Numbers of PSGs applicable for enrichment analyses are shown above branches. Note that the number is smaller than the counterpart in Fig. 2A because some genes do not have a human ortholog and thus could not be included in the analyses. GO terms are grouped into seven major categories including developmental process (yellow), signaling (red), cellular process (dark blue), metabolic and catalytic processes (green), immune system process (light blue), response to stimulus (black), and others (purple). (B and C) The observed SS scores and permutated ones between ancestor and the descendant snowfinches based on genes (B) and GO terms (C). Two algorithms of summarization including RCMAX (Left) and BMA (Right) are used. Dark blue, light blue, and red vertical lines show the observed pairwise SS values between ancestor and each of the snowfinches (adamsi, rufico, and taczan, respectively). Blue (permutations based on same number of genes) and red (permutations based on same number of GO terms) shades show 10,000 permutated SS values with 95th and 90th percentiles labeled.
To test whether the observed SS values are statistically significant, we benchmarked the observed values against two permutations of the SS values calculated from 100 repeated samplings. We found that the observed SS values between adamsi and ancestor were higher than the 95th percentile value of the permutations (Fig. 4B), while the SS values between rufico and ancestor were higher than, or nearly equal to, the 90th percentile value. In contrast, the SS values between taczan and ancestor fall in the middle of the permutations, suggesting that they could be explained by random chance. We then generated a new permutation by sampling the same numbers of GO terms randomly. Using the new permutations, we obtained a similar trend (Fig. 4C). These results suggest that positive selection has driven different extents of adaptive divergence among the descendant species.
Discussion
High-elevation environments exert a strong selective pressure on organisms living there and thus provide an ideal natural setting for the study of adaptation. Using snowfinches, a species complex living at the QTP, we investigated the contribution of ancestral adaptation to the adaptive evolution in the descendant species. Our results show that some key features for the high-elevation adaptation, including the developmental process, cellular signaling, and DNA repair capacity, have already evolved in the common ancestor. After the initial adaptation in the ancestor, the descendant species have adapted divergently in response to local selective pressures and microhabitats unique to each species (20–22, 39), leading to a deviation of adaptations between the ancestor and each of its descendants. Altogether, this study demonstrates a case of high-elevation adaptation, in which the descendant species have evolved diversified and species-specific adaptations after a common adaptation established in the ancestor.
The snowfinches have evolved a large body size compared to their lowland relatives. This phenotypic change likely began in their common ancestor. In addition, we identify strong signals of positive selection on genes related to development, signaling and DNA repair in the ancestor. We further demonstrate the ancestral adaptation using a functional assay on a highly selected gene, DTL, where nonsynonymous substitutions of ancestral snowfinch have improved the repair capacity of UV-induced DNA damage. These adaptive characteristics are consistent with previous studies (5–14). Given the functional benefits of these characteristics, for example, a large body size can reduce heat loss with low surface-area-to-volume ratio (40), they are likely to be several common adaptive strategies for organisms to survive in a high-elevation environment with low oxygen levels, low temperatures, and strong UV radiation.
The common ancestry may partially constrain genetic adaptations in the descendant species, evidenced by the finding that a significant excess of PSGs (4 to 6%) shared between each of the descendant species and the common ancestor, and GO semantic similarity between ancestor and two descendants. This pattern could be explained by the evolution of a certain genetic makeup in the ancestor that had contributed to the acquirement of adaptive characteristics in the descendant species later on. This finding differs from what has been reported in previous genomic studies in the high-elevation animals, such as the Yak (Bos grunniens), Tibetan humans, ground tit (Parus humilis), and great tit (Parus major), as genes under positive selection have been rarely found to be shared across those species (5, 7, 41, 42). It is worthwhile to investigate how the common ancestry influences the adaptive evolution in more animal systems in the future.
Besides the inheritance of adaptive characteristics for high elevation from the common ancestor, the descendant species exhibit various extents of adaptive divergence compared to that of their common ancestor. These results suggest that the descendant species have evolved species-specific adaptive characteristics after they diversify. Since we find the GO terms enriched in adamsi and rufico show greater SS values than taczan, as compared to their common ancestor, this similarity seems evolved from their phylogenetic relatedness. However, we think this is less likely because of several lines of evidence. For example, the SS value between adamsi and rufico (0.28) is intermediate compared to the values between adamsi and taczan (0.20) and between taczan and rufico (0.34). Furthermore, adamsi and rufico do not exhibit more similarity when compared to taczan in terms of body mass (Fig. 1C) and DTL repair dynamics (Fig. 3). Moreover, our phylogenetic signal test does not find evidence for the influence of phylogenetic dependence on the evolution of the adaptive characteristics in the three descendant species (SI Appendix, Table S15). We therefore conclude that adaptive diversification of the three descendant species has evolved as a result of selection acting separately on each of them (20–22, 39).
Materials and Methods
Genome Sequencing, Assembly, and Annotation.
We collected samples of taczan and rufico from Naqu, Tibet (4,200 m a.s.l.) and of adamsi from Geermu, Qinghai (4,100 m a.s.l.; Fig. 1A). Our collection complies with the National Wildlife Conservation Law of China. Four to five paired-end short-read (2 × 150 bp) sequencing libraries with insert sizes of 170 bp, 500 bp, 800 bp, 2 kb, and 5 kb were constructed and sequenced on the Illumina HiSeq 2000 sequencing platform at BGI-Shenzhen. After the quality filtration, we assembled clean reads via SOAPdenovo (43) and SSPACE (44). We constructed gene sets by integrating de novo gene predictions, homology-based methods, and RNA-sequencing data-based methods. We annotated gene functions using Blastp based on their highest match to proteins in the UniProt database (release 2011-01), GO (45) and Kyoto Encyclopedia of Genes and Genomes database (release 58) (46). See SI Appendix, Text 1 for a detailed description of genome assembly and annotation.
Phylogenomic Tree Reconstruction.
We reconstructed a phylogenomic tree including three snowfinches and seven other oscine species. Both phylogenetic representativeness and genome assembly quality were considered to select the outgroups. We blasted all the protein-coding sequences from these species using TreeFam (47) with an E-value threshold of 10−7. We used Solar to concatenate high-scoring pair segments of each protein pair and used H-scores to evaluate the similarity among genes with a custom script. We identified gene families by clustering homologous gene sequences using Hcluster_sg (Version 0.5.0, https://github.com/douglasgscofield/hcluster). We identified the 784 best-to-best single-copy orthologous genes across all 10 species and used them for phylogenetic reconstruction. We calibrated the tree with the date of the clade Passerida (Fig. 1) (26), 26.0 ± 3.0 Ma. This clade includes three species of snowfinches, montan, the medium ground finch, Geospiza fortis, and the zebra finch, Taeniopygia guttata. We then estimated the divergence time by implementing a Bayesian relaxed clock model in Beast v.2 (48). We ran Markov chain Monte Carlo chains for 100 million generations (sampling once every 1,000 generations) with a relaxed lognormal distribution for the molecular clock model and assuming a birth–death speciation process for the tree prior. The gamma substitution model was applied. We checked for convergence and performance using Tracer v.1.5 (49) and accepted the results if the values of the estimated sample size were larger than 200, suggesting little autocorrelation between samples. We combined resulting trees in TreeAnnotator v.1.7.5 and visualized the consensus tree with the divergence dates in FigTree v.1.4.3 (50).
Ancestral Trait Reconstruction of Elevation and Body Mass.
We performed an ancestral trait reconstruction using a well-established phylogeny of Passeridae (24), which includes all 27 species from seven genera: Montifringilla, Onychostruthus, Pyrgilauda, Petronia, Gymnoris, Passer, and Hypocryptadius. We collected the elevation records from eBird (https://ebird.org/) and China Bird Reports (http://www.birdreport.cn). Body masses were curated from the National Zoological Museum, Institute of Zoology, Chinese Academy of Sciences, and from the Handbook of the Birds of the World (51). We inferred ancestral traits for midelevation and body mass using BayesTraits v.3.0.2 (52). We used a variable-rates model to account for variation in evolutionary rates across phylogenetic trees. We ran 110 million iterations to ensure parameter convergence by discarding the first 10 million iterations in the burn-in phase. We sampled parameters every 1,000 iterations and final parameters were estimated based on all samples. We summarized the results by calculating a mean rate-transformed tree based on all the trees scaled by the rate of change in the posterior samples. We estimated ancestral values for each trait using a maximum likelihood approach using the package Phytools (53). To account for unequal rates of trait evolution across the tree, we used the mean rate-transformed trees for each trait to estimate ancestral trait as described in Cooney et al. (54) and Venditti et al. (55). We used a 1-My time slice to estimate ancestral disparity through time using functional dispersion (FDisp) in package FD (56), which is a measurement of the average distance of all branches in the phylogeny to the centroid in a principal coordinates analysis (PCoA) of the given trait.
Positive Selection Analysis of Protein-Coding Genes.
Since the previous species tree including ten species only covers 784 orthologous genes, we performed positive selection analysis with six closely related species including taczan, rufico, adamsi, montan, medium ground finch, and zebra finch. Using Blast (57) and TreeFam (47), we obtained 6,572 single-copy best-to-best hits as orthologous genes, which were shared by all six species and used for positive selection analysis (SI Appendix, Fig. S2). We aligned the orthologous proteins using Mafft v.7.310 (58), which were reversely translated as codon-level alignment using Pal2nal v.14 (59). We used branch-site likelihood ratio tests in Codeml of the PAML package (v.4.9e) (28) to identify PSGs for ancestral branch and the branches leading to adamsi, rufico, and taczan, respectively. The branch-site model allows ω to vary both among sites in the protein and across branches on the tree and aims to detect positive selection affecting a few sites along particular lineages. Following http://abacus.gene.ucl.ac.uk/software/pamlDOC.pdf, we performed a branch-site model A test by comparing two models: the null model (using the settings model = 2, NSsites = 2, ω = 1) and the alternative model (model = 2, NSsites = 2). The likelihood ratio test has degrees of freedom =1. We used the F3 × 4 codon model of Goldman and Yang (60) to calculate the equilibrium codon frequencies from the average nucleotide frequencies at the three codon positions (CodonFreq = 2). We identified PSGs with multiple comparisons with Bonferroni–Holm correction at P < 0.05. We counted the shared PSGs between the ancestor and each of the descendant snowfinches and tested if the proportions of shared genes are significantly different from expected proportion (144/6,572) using a binomial test. FDR correction was used to control for multiple comparisons. For a few genes of interest, such as UBA6 and DTL, we used free-ratio model in Codeml to calculate dN/dS value on every branch. We tested whether the spatial position of nonsynonymous substitutions as compared to synonymous substitutions between WD40 and non-WD40 domain regions differs between snowfinch DTLs and montan DTL used Fisher’s exact test and FDR correction.
Functional Assays for Repair Capacity of DNA Damage to UV Irradiation.
The coding sequences were synthesized for ancestral snowfinch DTL (DTLancestor), descendant snowfinch DTLs (DTLtaczan, DTLrufico, and DTLadamsi), and montan DTL (DTLmontan). For each case, we cloned the DTL sequence into the pCDH-CMV-MCS-EF1-copGFP vector and packaged the vector into lentiviruses (Huaaobio). We then took the lentivirus to infect the great tit GEFs, whose endogenous DTL has been knocked down with short hairpin RNA targeting a conserved region (5′-GCACCAGCAAGCTCATCTTTA-3′; SI Appendix, Fig. S1). Applying a slot blot assay as described in Shah et al. (61), we treated GEF cells with UVB irradiation at 100 J/m2 for 2 min and subsequently harvested cells at 0, 1.5, and 6 h afterward for 6-4PP quantitation or at 0, 6, and 24 h post-UVB irradiation for CPD determination. To determine DNA repair kinetics, we calculated the percentage of repair as 1 minus the ratio between densities of the bands at specified times and that at the 0 time point (100% damage after 2 min of UVB exposure). We used two-way repeated measures ANOVA to investigate if time points of repair and different DTLs significantly influenced repair capacity of DNA damage. For each factor, we conducted a post hoc t test comparison with FDR correction.
We inferred ancestral DTL sequences using phylogenetic model of coding sequence in a maximum likelihood framework in PAML. The inferred ancestral sequences show alternative sequences during the inference process. For example, a topology with tree sparrow as outgroup was used to infer the ancestral sequence. The inferred sequence with the high likelihood (DTLancestor_1) differs from the sequence inferred from alternative topology using the three species of snowfinches (at two sites, L566P and H586N, DTLancestor). The ancestral sequence inference also identified two other sites with possible alternative variants (showing a probability of 0.3 to 0.4). Together the most likely (DTLancestor_1) and the least likely sequences (DTLancestor_2) differ at three sites (L566P, T472R, and Y605C; SI Appendix, Table S16).
We examined the functional difference among DTLancestor, DTLancestor_1, and DTLancestor_2 sequences using Provean (32) and PolyPhen-2 (62). We found that all these amino acid changes have negligible functional effects (SI Appendix, Table S16). We then carried out a functional experiment to test the repair capacities of the DNA damage of the three ancestral sequences using the 293T cell line. Both the CPD and 6-4PP functional assays show that the three ancestral sequences display similar repair capacities of DNA damage at all time points examined, except that DTLancestor_2 (the least likely sequence) shows a significantly lower repair capacity at the 24-h time point of CPD (SI Appendix, Fig. S3). Taken together, our computational and functional assay analyses show that the functionality of the three ancestral DTL sequences is roughly similar and stable to alternative amino acid state during the inference process.
Functional Enrichment Analyses and Semantic Similarity Permutation.
We performed functional enrichment analyses using Metascape (63), which has integrated the latest GO annotation. Specifically, we took advantage of the aforementioned 6,572 orthologs and compared snowfinch genes to zebra finch genes and then to their corresponding human orthologous based on Ensembl ortholog annotation (64). For 6,572 genes, 4,679 had orthologs in the human genome, which further reduced PSG numbers to 104 in ancestor and 276 to 290 in descendant species. We identified overrepresented biological process terms (P < 0.01 and enrichment fold > 2) in each group of these orthologs using 4,697 orthologs as a background gene set.
To evaluate the overall similarity of the functional enrichment patterns between the ancestor and each descendant species, we calculated pairwise SS values of the GO terms and examined SS values relative to random samples. We implemented the R package GoSemSim 3.10 (36) to calculate individual SS values. We applied two different algorithms, BMA and RCMAX, to check if results are robust when summarizing SS values of individual GO terms. To determine if the observed SS values significantly deviated from a random expectation, we performed two different permutations. First, we randomly repeated 100 times by sampling genes from the same gene pools (104 for ancestor and 282 for descendant species, median value of 276 to 290 PSGs of the three descendant species) from the pool of all 4,697 genes and performed analogous functional enrichment analysis for each random gene set followed by calculation of SS values. Second, we sampled 100 times of the same numbers of GO terms for ancestor and each species of snowfinches and calculate SS values. For both permutations, values greater than or equal to the 95th percentile values of the random samples were considered to be significant. Major conclusion is reproducible when more strict cutoffs (P < 0.005 and enrichment fold > 2) were used (SI Appendix, Fig. S4).
Phylogenetic Signal Test of the Adaptive Characteristics.
We tested whether there are significant phylogenetic signals on the adaptive characteristics of the three descendant species using Blomberg’s K (65) in Phytools (53), including the SS values, the numbers of the species-specific nonsynonymous substitutions of DTL, and the DNA damage repair kinetics.
Supplementary Material
Acknowledgments
We thank Tieshan Tang and Wei Li for discussions on the DTL functional experiment, Xiaohua Lei and Wei Wang for functional assay, Hejie Liu for illustrating the pictures of the three species of snowfinches, Xinhai Li for statistical analysis, and Shaoyuan Wu for valuable comments on the manuscript. This research was funded by the Strategic Priority Research Program of the Chinese Academy of Sciences of the Second Tibetan Plateau Scientific Expedition and Research (STEP) program (2019QZKK0304 to F.L.), the Strategic Priority Research Program of the Chinese Academy of Sciences (XDA19050202 to Y.Q.), Strategic Priority Research Program of the Chinese Academy of Sciences of the STEP program (2019QZKK0501 to Y.Q.), National Natural Science Foundation of China (NSFC32020103005 to Y.Q. 31771410 and 31970565 to Y.E.Z.), the Chinese Academy of Sciences (ZDBS-LY-SM005 to Y.E.Z.), and the Swedish Research Council (621-2017-3693 to P.G.P.E.). We acknowledge support from Science for Life Laboratory, the National Genomics Infrastructure, NGI, and Uppmax for providing assistance in massive parallel sequencing and computational infrastructure.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2012398118/-/DCSupplemental.
Data Availability
Sequencing data for the three species of the snowfinches have been deposited in the Sequence Read Archive under project number PRJNA417520.
References
- 1.Cheviron Z. A., et al., Integrating evolutionary and functional tests of adaptive hypotheses: A case study of altitudinal differentiation in hemoglobin function in an Andean sparrow, Zonotrichia capensis. Mol. Biol. Evol. 31, 2948–2962 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Storz J. F., Scott G. R., Life ascending: Mechanism and process in physiological adaptation to high-altitude hypoxia. Annu. Rev. Ecol. Evol. Syst. 50, 503–526 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.McGuire J. A., et al., Molecular phylogenetics and the diversification of hummingbirds. Curr. Biol. 24, 910–916 (2014). [DOI] [PubMed] [Google Scholar]
- 4.Lei F., et al., The feather microstructure of Passerine sparrows in China. J. Ornithol. 143, 205–213 (2002). [Google Scholar]
- 5.Qu Y., et al., Ground tit genome reveals avian adaptation to living at high altitudes in the Tibetan plateau. Nat. Commun. 4, 2071 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Li M., et al., Genomic analyses identify distinct patterns of selection in domesticated pigs and Tibetan wild boars. Nat. Genet. 45, 1431–1438 (2013). [DOI] [PubMed] [Google Scholar]
- 7.Qu Y., et al., Genetic responses to seasonal variation in altitudinal stress: Whole-genome resequencing of great tit in eastern Himalayas. Sci. Rep. 5, 14256 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li J. T., et al., Comparative genomic investigation of high-elevation adaptation in ectothermic snakes. Proc. Natl. Acad. Sci. U.S.A. 115, 8406–8411 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun Y. B., et al., Species groups distributed across elevational gradients reveal convergent and continuous genetic adaptation to high elevations. Proc. Natl. Acad. Sci. U.S.A. 115, E10634–E10641 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hao Y., et al., Comparative transcriptomics of 3 high-altitude passerine birds and their low-altitude relatives. Proc. Natl. Acad. Sci. U.S.A. 116, 11851–11856 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang M. S., et al., Genomic analyses reveal potential independent adaptation to high altitude in Tibetan Chicken. Mol. Biol. Evol. 32, 1880–1889 (2015). [DOI] [PubMed] [Google Scholar]
- 12.Yu L., et al., Genomic analysis of snub-nosed monkeys (Rhinopithecus) identifies genes and processes related to high-altitude adaptation. Nat. Genet. 48, 947–952 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Deng L., et al., Prioritizing natural selection signals from the deep-sequencing genomic data suggests multi-variant adaptation in Tibetan highlanders. Natl. Sci. Rev. 6, 1201–1222 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Qu Y., et al., Rapid phenotypic evolution with shallow genomic differentiation during early stages of high elevation adaptation in Eurasian Tree Sparrows. Natl. Sci. Rev. 7, 113–127 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barghi N., Hermisson J., Schlötterer C., Polygenic adaptation: A unifying framework to understand positive selection. Nat. Rev. Genet. 21, 769–781 (2020). [DOI] [PubMed] [Google Scholar]
- 16.Zhu X., et al., Divergent and parallel routes of biochemical adaptation in high-altitude passerine birds from the Qinghai-Tibet Plateau. Proc. Natl. Acad. Sci. U.S.A. 115, 1865–1870 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rosvold J., et al., Perennial ice and snow-covered land as important ecosystems for birds and mammals. J. Biogeogr. 43, 3–12 (2016). [Google Scholar]
- 18.Brambilla M., et al., Past and future impact of climate change on foraging habitat suitability in a high-alpine bird species: Management options to buffer against global warming effects. Biol. Conserv. 221, 209–218 (2018). [Google Scholar]
- 19.Wingfield J. C., et al., Organism-environment interactions in a changing world: A mechanistic approach. J. Ornithol. 152, S279–S288 (2011). [Google Scholar]
- 20.Deng H. L., Zhang X. A., Standard metabolic rate in several species of passerine birds in alpine meadow. Acta Zool. Sinica. 36, 377–384 (1990). [Google Scholar]
- 21.Gebauer A., Kaiser M., Biology and behavior of general Asiatic snow finches (Montifringilla) and mountain-steppe sparrows (Pyrgilauda). J. Ornithol. 135, 55–57 (1994). [Google Scholar]
- 22.Qu Y., et al., Molecular phylogenetic relationship of snow finch complex (genera Montifringilla, Pyrgilauda, and Onychostruthus) from the Tibetan plateau. Mol. Phylogenet. Evol. 40, 218–226 (2006). [DOI] [PubMed] [Google Scholar]
- 23.Burleigh J. G., Kimball R. T., Braun E. L., Building the avian tree of life using a large-scale, sparse supermatrix. Mol. Phylogenet. Evol. 84, 53–63 (2015). [DOI] [PubMed] [Google Scholar]
- 24.Päckert M., et al., “Into and Out of” the Qinghai-Tibet Plateau and the Himalayas: Centers of origin and diversification across five clades of Eurasian montane and alpine passerine birds. Ecol. Evol. 10, 9283–9300 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Simão F. A., Waterhouse R. M., Ioannidis P., Kriventseva E. V., Zdobnov E. M., BUSCO: Assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics 31, 3210–3212 (2015). [DOI] [PubMed] [Google Scholar]
- 26.Oliveros C. H., et al., Earth history and the passerine superradiation. Proc. Natl. Acad. Sci. U.S.A. 116, 7916–7925 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Schumm M., White A. E., Supriya K., Price T. D., Ecological limits as the drivers of bird species richness patterns along the East Himalayan elevational gradient. Am. Nat. 195, 802–817 (2020). [DOI] [PubMed] [Google Scholar]
- 28.Yang Z., PAML 4: Phylogenetic analysis by maximum likelihood. Mol. Biol. Evol. 24, 1586–1591 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Liu X., et al., Orthogonal ubiquitin transfer identifies ubiquitination substrates under differential control by the two ubiquitin activating enzymes. Nat. Commun. 8, 14286 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Abbas T., Dutta A., CRL4Cdt2: Master coordinator of cell cycle progression and genome stability. Cell Cycle 10, 241–249 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sansam C. L., et al., DTL/CDT2 is essential for both CDT1 regulation and the early G2/M checkpoint. Genes Dev. 20, 3117–3129 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi Y., Chan A. P., PROVEAN web server: A tool to predict the functional effect of amino acid substitutions and indels. Bioinformatics 31, 2745–2747 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ng P. C., Henikoff S., SIFT: Predicting amino acid changes that affect protein function. Nucleic Acids Res. 31, 3812–3814 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen X., et al., Mir-19b-3p regulates MAPK1 expression in embryonic fibroblasts from the Great tit (Parus major) under hypoxic conditions. Cell. Physiol. Biochem. 46, 546–560 (2018). [DOI] [PubMed] [Google Scholar]
- 35.Li J., et al., Dynamics and mechanism of repair of ultraviolet-induced (6-4) photoproduct by photolyase. Nature 466, 887–890 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yu G., et al., GOSemSim: An R package for measuring semantic similarity among GO terms and gene products. Bioinformatics 26, 976–978 (2010). [DOI] [PubMed] [Google Scholar]
- 37.Moon J. M., Capra J. A., Abbot P., Rokas A., Signatures of recent positive selection in enhancers across 41 human tissues. G3 (Bethesda) 9, 2761–2774 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hao Y., et al., Baby genomics: Tracing the evolutionary changes that gave rise to placentation. Genome Biol. Evol. 12, 35–47 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zeng X., Lu X., Interspecific dominance and asymmetric competition with respect to nesting habitats between two snowfinch species in a high-altitude extreme environment. Ecol. Res. 24, 607–616 (2009). [Google Scholar]
- 40.Meiri S., Dayan T., On the validity of Bergmann’s rule. J. Biogeogr. 30, 331–351 (2003). [Google Scholar]
- 41.Yi X., et al., Sequencing of 50 human exomes reveals adaptation to high altitude. Science 329, 75–78 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu Q., et al., The yak genome and adaptation to life at high altitude. Nat. Genet. 44, 946–949 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Li R., Li Y., Kristiansen K., Wang J., SOAP: Short oligonucleotide alignment program. Bioinformatics 24, 713–714 (2008). [DOI] [PubMed] [Google Scholar]
- 44.Boetzer M., Henkel C. V., Jansen H. J., Butler D., Pirovano W., Scaffolding pre-assembled contigs using SSPACE. Bioinformatics 27, 578–579 (2011). [DOI] [PubMed] [Google Scholar]
- 45.Ashburner M.et al.; The Gene Ontology Consortium , Gene Ontology: Tool for the unification of biology. Nat. Genet. 25, 25–29 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kanehisa M., Goto S., KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 28, 27–30 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H., et al., TreeFam: A curated database of phylogenetic trees of animal gene families. Nucleic Acids Res. 34, D572–D580 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bouckaert R., et al., BEAST 2: A software platform for Bayesian evolutionary analysis. PLOS Comput. Biol. 10, e1003537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rambaut A., Drummond A. J., Xie D., Baele G., Suchard M. A, Posterior Summarization in Bayesian Phylogenetics Using Tracer 1.7. Syst. Biol. 67, 901–904 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rambaut A., FigTree. tree.bio.ed.ac.uk/software/figtree/. Accessed 23 December 2014.
- 51.del Hoyo J., Elliott A., Sargatal J., Handbook of the Birds of the World (Lynx Edicions, 2008). [Google Scholar]
- 52.Mead A., Pagel M., BayesTraits. V3.0.2 manual. www.evolution.rdg.ac.uk/BayesTraits.html. Accessed 6 August 2020.
- 53.Revell L. J., Phytools: An R package for phylogenetic comparative biology (and other things). Methods Ecol. Evol. 3, 217–223 (2012). [Google Scholar]
- 54.Cooney C. R., et al., Mega-evolutionary dynamics of the adaptive radiation of birds. Nature 542, 344–347 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Venditti C., Meade A., Pagel M., Multiple routes to mammalian diversity. Nature 479, 393–396 (2011). [DOI] [PubMed] [Google Scholar]
- 56.Laliberté E., Legendre P., A distance-based framework for measuring functional diversity from multiple traits. Ecology 91, 299–305 (2010). [DOI] [PubMed] [Google Scholar]
- 57.Altschul S. F., Gish W., Miller W., Myers E. W., Lipman D. J., Basic local alignment search tool. J. Mol. Biol. 215, 403–410 (1990). [DOI] [PubMed] [Google Scholar]
- 58.Katoh K., Standley D. M., Kuma K., Miyata T., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Suyama M., Torrents D., P. Bork P. PAL2NAL: Robust conversion of protein sequence alignments into the corresponding codon alignments. Nucleic Acids Res. 34, W609–W612 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Goldman N., Yang Z., A codon-based model of nucleotide substitution for protein-coding DNA sequences. Mol. Biol. Evol. 11, 725–736 (1994). [DOI] [PubMed] [Google Scholar]
- 61.Shah P., Zhao B., Qiang L., He Y. Y., Phosphorylation of xeroderma pigmentosum group C regulates ultraviolet-induced DNA damage repair. Nucleic Acids Res. 46, 5050–5060 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Adzhubei I. A., et al., A method and server for predicting damaging missense mutations. Nat. Methods 7, 248–249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zhou Y., et al., Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat. Commun. 10, 1523 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Vilella A. J., et al., EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 19, 327–335 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Blomberg S. P., T. Garland, Jr, Ives A. R., Testing for phylogenetic signal in comparative data: Behavioral traits are more labile. Evolution 57, 717–745 (2003). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Sequencing data for the three species of the snowfinches have been deposited in the Sequence Read Archive under project number PRJNA417520.