Significance
Humans and our close evolutionary relatives respond differently to a large number of infections. Such differences are thought to result, at least in part, from interspecies differences in immune function. Here, we report on the whole-genome expression of blood leukocytes from four primate species responding to bacterial and viral stimulation. We show that apes mount a markedly stronger early transcriptional response to both viral and bacterial stimulation when compared to African and Asian monkeys. In addition, our findings suggest that apes activate a broader array of defense molecules that may be beneficial for early pathogen killing at the potential cost of increased energy expenditure and tissue damage. Our results provide insight into the evolution of immune responses in primates.
Keywords: pathogen-associated molecular patterns, primate evolution, early immune responses to infection, immunodeficiency viruses, gram-negative bacteria
Abstract
Despite their close genetic relatedness, apes and African and Asian monkeys (AAMs) differ in their susceptibility to severe bacterial and viral infections that are important causes of human disease. Such differences between humans and other primates are thought to be a result, at least in part, of interspecies differences in immune response to infection. However, because of the lack of comparative functional data across species, it remains unclear in what ways the immune systems of humans and other primates differ. Here, we report the whole-genome transcriptomic responses of ape species (human and chimpanzee) and AAMs (rhesus macaque and baboon) to bacterial and viral stimulation. We find stark differences in the responsiveness of these groups, with apes mounting a markedly stronger early transcriptional response to both viral and bacterial stimulation, altering the transcription of ∼40% more genes than AAMs. Additionally, we find that genes involved in the regulation of inflammatory and interferon responses show the most divergent early transcriptional responses across primates and that this divergence is attenuated over time. Finally, we find that relative to AAMs, apes engage a much less specific immune response to different classes of pathogens during the early hours of infection, up-regulating genes typical of anti-viral and anti-bacterial responses regardless of the nature of the stimulus. Overall, these findings suggest apes exhibit increased sensitivity to bacterial and viral immune stimulation, activating a broader array of defense molecules that may be beneficial for early pathogen killing at the potential cost of increased energy expenditure and tissue damage.
Despite being close evolutionary relatives, humans, chimpanzees, and African and Asian monkeys (AAMs) exhibit interspecies differences in sensitivity to and manifestation of certain bacterial and viral pathogens that are major causes of mortality in humans (e.g., HIV/AIDS, hepatitis C virus, and a broad range of commensal gram-negative bacteria commonly implicated in sepsis) (1 –5). Humans, for example, are highly sensitive to stimulation by the gram-negative bacterial-cell-wall component hexa-acylated lipopolysaccharide (LPS), miniscule amounts of which (2 to 4 μg/kg) can provoke inflammation, malaise, and fever, and a slightly higher dose, septic shock (15 μg/kg) (1, 6, 7). In contrast, baboons and macaques require doses nearly 10-fold higher in concentration to trigger similar symptoms (5, 8, 9). Pattern recognition receptors (PRRs) such as Toll-like receptors (TLRs) play a central role in the mediation of innate immune responses to pathogens (10). The limited number of studies comparing leukocyte function after stimulation with TLR-detected pathogen-associated molecules suggest that the differences in infectious disease susceptibility noted between apes and AAMs is, in part, the outcome of lineage-specific evolution of early innate immune-system regulation and signaling (11 –13). Indeed, innate immune components responsible for detecting pathogens, including TLRs that sense gram-negative bacteria and single-stranded RNA viruses, have been found to be under positive selection in primates (14, 15).
Despite stark differences in the manifestation of severe infections between apes and AAMs, there are few reports directly comparing the gene-expression response across species to bacterial and viral pathogens (11, 12). Furthermore, previous studies relied mainly on isolated cell types to characterize immune responses across primates (11, 16), which does not faithfully reflect the nature of the innate immune response that is a product of the interaction between several cell populations (17). To better understand the evolution of the primate immune system, this study compares the early responses of apes (humans and common chimpanzees) and AAMs (rhesus macaques and olive baboons) to bacterial and viral stimuli. Here, we report on the whole-genome expression of total blood leukocytes from these four primate species responding to bacterial and viral stimulation during the first 24 h of challenge. Our results show that apes and AAMs have diverged in sensitivity to specific microbial assaults, such that ape leukocyte responses favor robust antimicrobial power over pathogen specificity at the potential cost of increased energetic expenditure and bystander tissue damage.
Results
Evolutionary Relationships Explain Most of the Transcriptional Response Variation in Primates to Bacteria or Viral Stimulation.
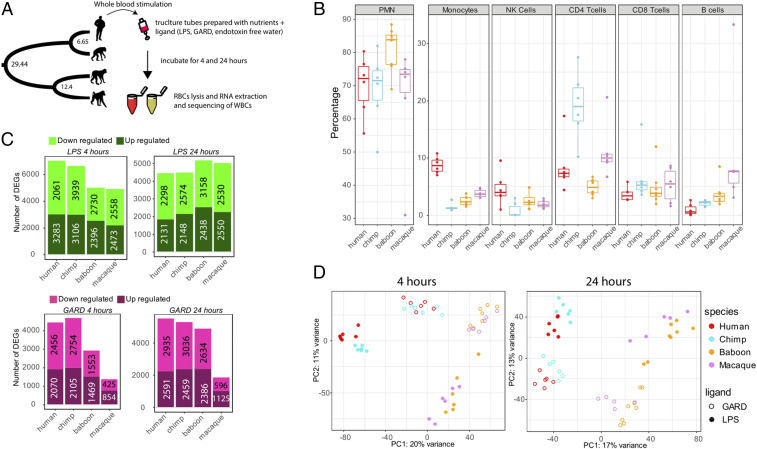
To assess differences in innate immune function between higher-order primates in as close an approximation to in vivo as possible, we challenged whole blood from humans (Homo sapiens; n = 6), common chimpanzees (Pan troglodytes; n = 6), rhesus macaques (Macaca mulattta; n = 6), and olive baboons (Papio anubis; n = 8) with bacterial or viral stimuli via venous draw directly into a media culture tube containing either LPS from Escherichia coli O111:B4, gardiquimod (GARD), a single-stranded RNA viral mimetic, or endotoxin-free water, as a negative control (Control). Both LPS and GARD were chosen as immune stimuli because they represent microbial components that are millions to billions of years old, shared across large groups of microorganisms including gram-negative bacterial and single-stranded RNA (ssRNA) viral pathogens, and are thought to be driving the evolutionary divergence of primate immune systems. Moreover, there are well-established differences between apes and AAMs in the manifestation of diseases mediated by such pathogens (e.g., immunodeficiency viruses, hepatitis C, and common commensal bacteria that cause gram-negative bacterial sepsis) (1, 3, 4). Blood was stimulated for 4 and 24 h before the total leukocytes were isolated and RNA extracted for RNA-sequencing (RNA-seq) (Fig. 1A ). Following quality control filtering, we analyzed 151 high-quality RNA-seq profiles across species and treatment combinations (see Materials and Methods and Dataset S1). We focus our comparative analyses on the expression levels of 14,140 one-to-one (1:1) orthologous genes, taking into account potential biases in expression estimates due to differences in mappability between species ( Materials and Methods ).
Fig. 1.
Characterizing innate immune response upon viral and bacterial stimulation of primate white blood cells. (A) Schematic representation of the study design. Whole-blood samples from humans, common chimpanzees, rhesus macaques, and olive baboons were stimulated with bacterial or viral stimuli via venous draw directly into a media culture tube containing either LPS, single-stranded RNA viral mimetic GARD, or endotoxin-free water, as a negative control (Control). At 4 and 24 h poststimulation, white blood cells were isolated, and RNA was extracted for RNA-seq. (B) Cell proportions of six populations of leukocytes for all species. Species are indicated on x-axis, and proportions of this population from total leukocytes are on y-axis. PMNs, polymorphonuclear cells; NK, Natural killer cells. (C) Number of differentially expressed genes (DEGs; FDR < 0.05) in response to LPS (Top) and GARD (Bottom) in each of the species at 4 and 24 h poststimulation. The exact number of up- and down-regulated genes in each condition in each species are indicated on the bar charts. (D) Principal component (PC) analysis performed on the log2 fold-change responses observed at 4 h post-LPS and GARD stimulation. PC1 primarily separates apes (human and chimpanzee) from AAM (macaque and baboon), and PC2 captures differences in immune response to bacterial or viral stimulation.
As whole blood contains a variety of leukocyte subtypes, we first characterized differences in total blood leukocyte composition between species using fluorescence-activated cell sorting (FACS). Leukocyte composition differs between species for all major subtypes measured, with the most notable differences an increase in the proportion of monocytes in humans (CD14+, P < 0.003) and helper T cells in chimpanzees (CD3+, CD4+; P = 0.0006 to 0.065), relative to other primates (Fig. 1B and Dataset S2). Using linear models that account for variation in cell composition, we next identified genes that respond to LPS and GARD in each of the species, at each of the time points (see Materials and Methods ). In all species, both treatments led to the up- or down-regulation of hundreds to thousands of genes (false discovery rate [FDR] < 0.05, Fig. 1C and Dataset S3). As expected, the transcriptional response to either stimulus was highly concordant across primates (Spearman’s r range 0.5 to 0.87 across all pairwise comparisons; SI Appendix, Fig. S1), with stronger correlations between closely related primates than between more distantly related pairs of species (e.g., at LPS 4 h Spearman’s r human versus chimpanzee = 0.84, human versus baboon = 0.50). Consistently, the first principal component (PC) of the log2 fold-change responses to both LPS and GARD accounted for ∼20% of the total variance in our dataset and separated apes (human and chimpanzee) from AAM (macaque and baboon) (t test; P < 1 × 10−10 for both 4 and 24 h, Fig. 1D ). The second PC captured differences in immune response to bacterial or viral stimulation (t test; P < 1 × 10−8 for 4 and 24 h; Fig. 1D ). We identified a set of 648 and 257 genes that early after stimulation (4 h) showed a consistently strong response across all species to LPS or GARD, respectively (defined as genes with |log2 FC| > 1 and FDR < 0.05 in all species, Dataset S4). These genes include most of the key transcription factors involved in the regulation of innate immune responses to bacterial (e.g., NFKB1/2) and viral pathogens (e.g., interferon regulatory factor [IRF]7/9), as well as several effector molecules involved in the regulation of inflammatory responses to infection (e.g., IL6, TNF-α, and IL1-β).
Stronger Early Innate Immune Response in Apes than in AAM.
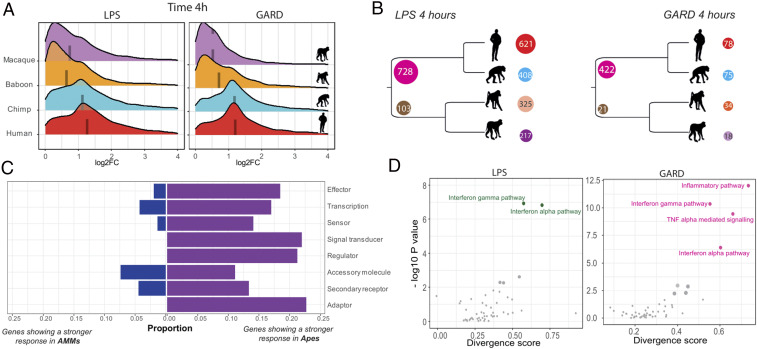
Next, we sought to characterize differences in immune responses across species. To do so, we first looked at overall differences in the magnitude of the transcriptional responses to LPS and GARD across species (see Materials and Methods ). We found that, at early time points, both ape species (human and chimpanzee) engage a much stronger transcriptional response to both stimuli as compared to rhesus and baboons (on average ∼twofold higher, Wilcoxon test P < 10−10, Fig. 2A ). Next, we identified genes for which the magnitude of the transcriptional response to LPS or GARD was significantly different between apes and AAM (FDR < 0.10 for all pairwise contrasts between an ape and an AAM species and an average |log2 FC| > 0.5). Hereafter, we refer to these genes as clade differentially responsive genes, or c-DRGs. We identified a total of 831 and 443 c-DRGs in the early response (4 h) to LPS and GARD, respectively (Fig. 2B and Dataset S5). Among c-DRGs, 83 to 92% showed a stronger response in apes as compared to AAM, consistent with the genome-wide pattern of an overall more robust transcriptional response to immune stimulation in apes. Importantly, the stronger response observed in apes is not explained by higher baseline expression levels of the receptors involved in the recognition of LPS (TLR4, CD14, LY96, and CASP4) and GARD (TLR8) ( SI Appendix, Fig. S2). Next, we focused our analyses on a manually curated list of 1,079 genes belonging to different modules of the innate immune system (18) and that were found to change gene expression in at least one of our experimental conditions, in at least one of the species from our dataset. These genes include sensors (n = 188), adaptors (n = 36), signal transducers (n = 209), transcription (factors) (n = 74), effector (molecules) (n = 115), accessory molecule (n = 54), and secondary receptors (n = 50). All modules show similar divergence between clades, with ∼15% of the genes within each module classified as c-DRGs with a stronger response in apes, as compared to less than 5% showing a significantly stronger response in AAM (Fig. 2C ).
Fig. 2.
Stronger early innate immune response in apes than monkeys. (A) For each combination of stimulus and time point, we show the distribution of the log2 fold changes (x-axis) among genes that respond to that treatment in at least one of the species. The median log2 fold-change responses in each species is represented by a dashed line. (B) Number of DRGs that are clade- or species-specific differently regulated genes at 4 h post-LPS (Left) and GARD (Right) stimulation. For c-DRG, we report the number of genes that show a stronger response in a specific clade at the beginning of the ancestral branch of the tree. For example, in response to LPS, we identified 831 c-DRGs from which 728 show a stronger response in apes and 103 in AAMs. For species-specific responsive genes, numbers are given in front of each species. The color codes for each species are red for human, cyan for chimp, orange for baboon, and violet for macaque. (C) Bar plots represent the proportions of different classes of innate immunity genes that are classified as c-DRGs with a higher response in apes (dark violet) or in AAMs (dark blue). (D) Scatter plot displaying total divergence scores of hallmark pathways for LPS (green) and GARD (pink) at 4 h stimulations. For a given pathway, the total divergence is given by divergence score (DS) on the x-axis and −log10 P values for each DS is on the y-axis. The pathways names, DS values, and corresponding P values are shown in Dataset S6. We highlight the pathways showing the most significant divergence scores for both the response to LPS and GARD.
To further characterize functional differences in immune regulation between apes and AAM, we devised a score of transcriptional divergence at the pathway level. We focused on the set of 50 “hallmark pathways,” which capture well-defined and curated biological states or processes (19). Briefly, for each gene in these pathways, a divergence score between apes and AAM was computed by calculating the average difference between the fold-change estimates between all pairs of species of the two clades, while taking into account variance in transcriptional response within each species. The pathway divergence score reflects the average divergence scores across all genes of a given pathway (see Materials and Methods for details). In the early response to LPS, the most divergent pathways between apes and AAM were “Interferon alpha response” and “Interferon gamma response” (P ≤ 0.01, Dataset S6), indicating that the regulation of interferon responses has significantly diverged since the separation between apes and AAM. In the early response to GARD, pathways directly related to the regulation of inflammatory responses, notably TNF-α signaling, were the most divergent (P ≤ 0.01) (Fig. 2D ). These results are consistent with recent finding showing that the transcriptional response of cytokines and chemokines to immune stimulation are among the most divergent across mammals (20). Of note, although many genes are differently expressed at baseline between apes and AAMs (at an FDR < 0.01, 871 and 890 at 4 and 24 h in culture, respectively; Dataset S7), pathways related to interferon responses and inflammation are among the least divergent in their baseline gene-expression levels between the two clades ( SI Appendix, Fig. S3). Moreover, gene ontology (GO) enrichment analysis did not reveal any immune-related term that is significantly enriched among genes differently expressed between apes and AAMs at baseline ( Dataset S8). Thus, we conclude that the differences in innate immune response identified between apes and AAMs are unlikely to be explained by baseline differences in the expression levels of inflammatory markers.
Species-Specific Immune Responses Reflect Unique Immune Regulation Mechanisms and Lineage-Specific Divergence.
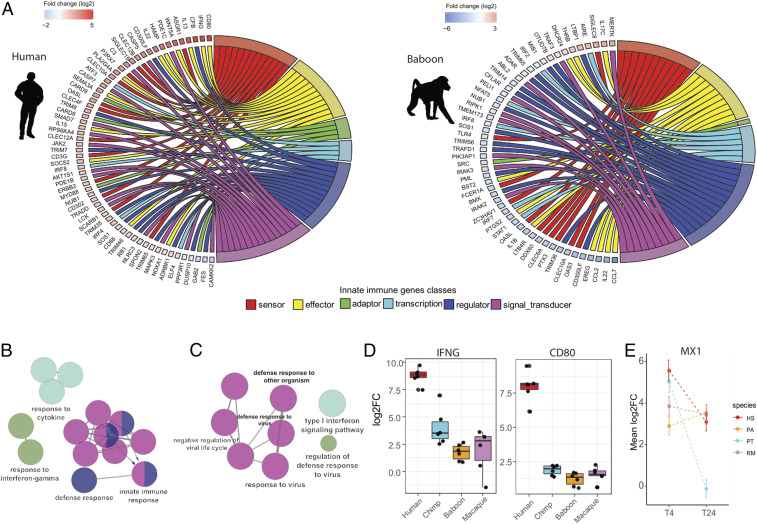
Next, we sought to identify genes that respond to immune stimulation in a species-specific fashion. These were characterized as genes for which the magnitude of the response to LPS or GARD in one species was significantly different to that observed in all other species (see Materials and Methods ). Across time points and immune stimulations, we identified a total of 980, 726, 425, and 655 species-specific responsive genes in human, chimpanzees, macaques, and baboons, respectively ( Dataset S9). Among baboon-specific responsive genes, the vast majority (69%) showed a weaker magnitude of the response to 4 h of LPS stimulation in baboons as compared to all other primates ( Dataset S9). GO enrichment analyses ( Dataset S10) revealed that these genes were enriched among defense response genes (FDR = 4.8 × 10−14) and a variety of other GO-associated immune terms (Fig. 3B ), including several key transcription factors (e.g., STAT1 and IRF7/9), major inflammatory cytokines (e.g., IL1B and CXCL8), and a number of genes directly involved in LPS sensing and recognition (adaptor molecules IRAK2, 3, and 4, and the primary LPS receptor, TLR4) (Fig. 3A and Dataset S9). The weaker response observed in baboons appears to be, at least in part, due to a higher baseline expression level of many of these innate immunity genes ( SI Appendix, Fig. S4). Baboons have been suggested to bear higher pathogen loads than apes because of their mating promiscuity, and so it is tempting to speculate that increased baseline might represent a mechanism of protection against frequent microbial infections (21, 22). In rhesus macaques, the other AAM species, genes showing a stronger response to LPS at both 4 and 24 h than that observed in all other species (n = 157, Dataset S9) were mostly enriched among genes involved in the regulation of inflammatory responses (FDR = 0.002, Dataset S10), including TREM2 a known suppressor of PI3K and NF-κ-B signaling in response to LPS.
Fig. 3.
Species-specific immune response reflects unique immune regulation mechanisms and lineage-specific divergence. (A) Circos plots showing different classes of innate immune genes (clustered using different color codes) classified as species-specific responsive at 4 h post-LPS stimulation in humans (Left) and baboons (Right). The log2FC key represent the average difference between species response versus all other species in which the positive (red color) and negative (blue) values indicate the magnitude of the stronger and weaker absolute response in this species versus all others, respectively. (B) GO enrichment analysis for genes showing a weaker response to LPS at 4 h in baboons as compared to all other species. (C) GO enrichment analysis for genes showing a weaker response to LPS at 24 h in chimpanzees as compared to all other species. For B and C only, top GO terms are presented. The full list of significant GO terms can be found in Dataset S10. (D) Boxplot represents the log2FC of IFN-γ and CD80 genes which, at 4 h post-LPS stimulation, were found to have a significantly stronger response in human than in other primates. (E) Estimates of the mean fold-changes response for MX1 (± SE) at the two time points across the four primate species studied.
Among chimpanzee-specific genes, the most notable GO enrichments were observed among genes showing a weaker response to LPS at 24 h relative to that observed in all other primates. These genes were significantly enriched for GO terms associated with viral defense mechanisms, including “response to virus,” or “type I interferon signaling pathway” (FDR < 1 × 10−9, Fig. 3C ). Further inspection of these genes revealed that the vast majority are strongly up-regulated at 4 h post-LPS stimulation—at similar levels to those observed in other species—but that chimpanzees have a unique ability to shutdown these genes at later time points. For example, the prototypic interferon-responsive gene MX1 is up-regulated by over fivefold in all primates at 4 h, but by 24 h, MX1 levels have revert to baseline uniquely in chimpanzees (Fig. 3E ), suggesting that chimpanzees are particularly divergent in the regulatory circuits associated with the control of viral responsive genes.
In contrast to the pattern observed for baboons, human-specific responses were associated with genes showing a stronger response to immune stimulation as compared to that observed in other primates. GO analyses revealed that these genes are overrepresented among terms related to the regulation of cytokine production involved in immune response (FDR = 0.045) and T-cell activation involved in immune response (FDR = 0.06) ( Dataset S10). Notable examples of human-specific responding genes include the canonical T-cell costimulatory molecule CD80 (average fivefold increase in response to both stimuli relative to other species) and IFN-γ, a cytokine central for protective immunity against a large number of infectious agents and the key determinant of the polarization of T cells toward a proinflammatory Th1 phenotype (23) (Fig. 3D ). The higher production of IFN-γ and CD80 in humans may mediate more effective killing of viral and bacterial pathogens. Furthermore, as these molecules are important regulators of cytokine production and T-cell activation, it also suggests significantly different regulation of T-cell responses (24, 25).
Regulatory Divergence Decreases as Infection Proceeds.
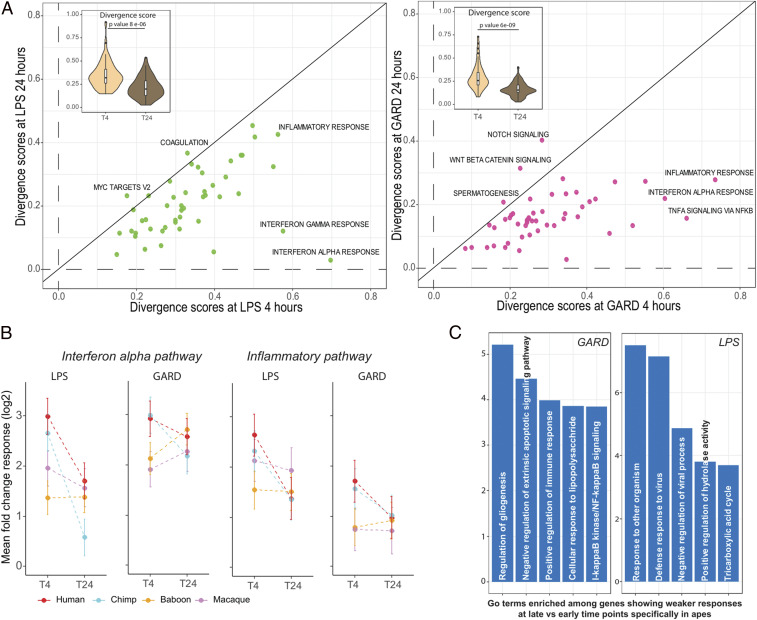
Next, we compared the transcriptional divergence between early (4 h) and late (24 h) immune responses. We observed a marked reduction in divergence scores at 24 h poststimulation of most hallmark pathways in the response to both LPS (P = 8 × 10−6) and GARD (P = 6 × 10−9) (Fig. 4A ). In LPS-stimulated cells, the most striking differences were observed for interferon-related pathways, which show a reduction in divergence score of ∼sixfold between the two time points. In GARD-stimulated cells, the largest reduction in divergence scores was observed among pathways related to the regulation of inflammatory responses (Fig. 4A ). These findings indicate that most transcriptional divergence in immune responses among primates occurs during the initial response to pathogens followed by an overall convergence to similar response levels at a later time point, specifically among genes involved in the regulation of inflammation and viral-associated interferon responses (Fig. 4B ). In apes (but not in AAMs), genes involved in the regulation of inflammation are strongly enriched among those for which the response to GARD significantly decreases at the later time point, whereas those decreasing in response to LPS are enriched for viral response genes (Fig. 4C and Dataset S11).
Fig. 4.
Divergence of immune response is reduced at later time point. (A) Scatter plots of divergence scores of hallmark pathways at early (x-axis) and later time points (y-axis) for LPS (green) and GARD (maroon) stimulations. The inset boxplots contrast the distribution of divergence scores among all pathways between the two time points. P values were obtained using Mann–Whitney U test. (B) Estimates of the mean response at the two time points for each species (± SE) across genes bellowing to the interferon alpha and inflammatory response hallmark pathways. (C) GO enrichment analysis for genes that showed significant decrease in response in apes only (FDR < 0.05 in apes and FDR > 0.05 in monkeys) for LPS and GARD. Top significant GO terms are given as indicated by −log10 P value on the x-axis.
Apes Engage a Less-Specific Innate Immune Response than AAM.
An aspect of innate immunity central to its success during microbial assault is its ability to recognize pathogens and initiate the most appropriate defense against them by type. The specificity of the innate immune response to infection is mediated by PRRs that detect the presence of danger signals via conserved molecular patterns associated with subtypes of pathogens and host damage (e.g., penta- and hexa-acylated LPS from gram-negative bacteria detected by TLR4-LY96 receptors) (26, 27). Signals of viral danger such as GARD are expected to activate a response mainly controlled by transcription factors prominent in antiviral defense such as Interferon Regulatory Factors (IRFs), which limit viral replication and dissemination through the up-regulation of interferons and interferon-regulated genes (28, 29). By contrast, recognition of gram-negative bacteria via cell-wall component LPS stimulates a broader array of cytokine responses that tends toward expression of proinflammatory cytokines regulated by transcription factors NF-κB and AP1 but can also include interferon expression regulated by transcription factors such as IRF3 and JAK-STAT (28, 30 –32)
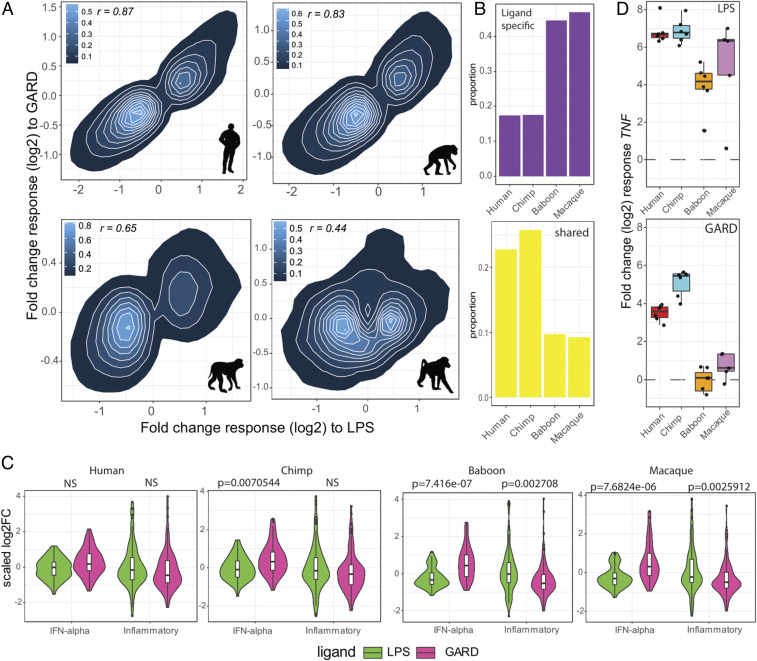
Two major lines of evidence indicate that early transcriptional immune responses are less specific in apes than in AAM. First, we found the transcriptional responses to LPS and GARD were more similar to each other in apes (humans r = 0.87, chimpanzees r = 0.83) than they were in baboons (r = 0.44) or macaques (r = 0.65) (Fig. 5A ). Accordingly, we found about three times more genes that respond uniquely to either LPS or GARD (i.e., “ligand specific” genes) in AAM as compared to apes (χ2 test; P = 2.2 × 10−16) (Fig. 5B , see Materials and Methods for details on the statistical model used to characterize ligand-specific and shared genes). The second piece of evidence comes from the nature of the genes that are differentially activated in response to LPS and GARD. The fact that apes show a higher correlation in GARD and LPS responses compared to AAMs predicts that they will tend to activate both antibacterial and antiviral defense mechanisms regardless of the nature of the stimuli. Supporting this notion, the genes that exhibited a stronger response in apes than in AAMs after stimulation with the viral mimetic GARD for 4 h were most significantly enriched genes involved in the regulation of inflammatory responses (P = 7.1 × 10−6; FDR = 0.008, Dataset S10), whereas genes involved in the response to viruses (GO term “response to virus”) were enriched upon bacterial LPS stimulation (P = 0.0023; FDR = 0.15, Dataset S10).
Fig. 5.
Apes engage a less-specific innate immune response than AAMs. (A) Correlation plots of the magnitude of the fold-change responses between LPS (x-axis) and GARD-stimulated cells (y-axis). For each of the species, we only include genes that were differentially expressed (FDR < 0.05) in response to at least one of the stimuli (N = 7,862, 7,874, 6,585, and 5,430 genes for human, chimp, baboon, and macaque, respectively). High correlation was found in apes (∼0.85), while modest correlation was found in baboon (0.44) and moderate in macaque (0.65). (B) Proportion of ligand-specific (i.e., genes that respond uniquely to either bacterial or viral stimuli) and shared genes (i.e., genes equally activated by both immune stimuli) across species. (C) Violin plots comparing (scaled) log2 fold-change responses to 4 h of LPS and GARD stimulation between genes belonging the hallmark pathways “Interferon (IFN) alpha” and “inflammatory response.” The P values shown have been Bonferroni corrected for the number of tests performed. NS, nonsignificant (i.e., P > 0.05). (D) Boxplots of the log2 fold-change response (y-axis) of TNF in response to LPS and GARD stimulations across primates.
To explore these differences in more detail, we focused on genes involved in the interferon alpha pathway (viral-associated response) or inflammatory response (bacterial-associated response). In AAMs, inflammatory response genes tended to be more strongly up-regulated in response to LPS compared to GARD (P ≤ 0.0027), suggesting that their transcription is particularly sensitive to receipt of a bacterial danger signal compared to a viral one. No significant differences in up-regulation of these same genes were noted between LPS and GARD cells in apes (Fig. 5C ). For example, the canonical proinflammatory cytokine TNF, which in macaques and baboons is strongly up-regulated only in response to LPS, is potently up-regulated in response to both stimuli in humans and chimpanzees (by over ∼fourfold, Fig. 5D ). Other examples of this pattern include the classical proinflammatory cytokines IL1A and IL1B ( SI Appendix, Fig. S5). Likewise, interferon-associated genes were more strongly up-regulated in response to GARD compared to LPS in AAM (P < = 7.7 × 10−6), while in apes these genes showed more concordant levels of up-regulation between stimuli (Fig. 5C ). Interferon-induced and potent antiviral genes, including MX1 and OAS1, were much more strongly up-regulated in response to GARD than to LPS in AAMs compared to apes ( SI Appendix, Fig. S5).
Discussion
Our study provides a genome-wide functional comparison of variation in innate immune responses between species belonging to two closely related clades of primates. Ape (human and chimpanzee) total blood leukocytes were significantly more responsive to bacterial and viral stimulation compared to total blood leukocytes obtained from AAM (rhesus macaques and baboons) during the early hours of challenge, mounting generally stronger and less-specific transcriptional responses. This increased response suggests apes maintain increased sensitivity to particular types of microbial assaults compared to AAM, a phenomenon likely to come with considerable energetic cost (1, 5). From an evolutionary standpoint, investment in increased sensitivity to pathogens can limit the negative effects of pathogen exposure on reproductive fitness. Humans and chimpanzees participate in a comparatively slower life history than rhesus macaque and olive baboon monkeys—they live decades longer, take longer to reach sexual maturity, nurture their young longer, and maintain a larger body size (33 –35). A long life at a large size increases risk of pathogen exposure, both in terms of number of exposures and absolute load, over the course of a life that will have long periods of time between the birth of offspring. A slow life history strategy can be concomitant with an increased risk in pathogen-mediated limitations to reproductive success, making a more substantial investment in robust early pathogen detection and elimination evolutionarily beneficial, compared to the ordinary metabolic costs of launching those responses (36, 37).
However, serious bystander tissue damage is a cost for immune protection during severe infections. Pathogen virulence may play a significant role in the evolution of high-energy low-specificity early immune responses. The primate genera in this study substantially differ in their evolutionary exposure to particular pathogens (e.g., dengue virus, immunodeficiency viruses, and Zika virus) (38 –41). Exposure to pathogens of high virulence may lead to a low cost–benefit ratio for primate hosts, since the reproductive and evolutionary benefit of a transiently demanding immune response outweighs its energetic and tissue costs (42, 43). Under this rubric, a robust but less-specific early response to pathogens is effective and beneficial most of the time. Any contribution that response might make to immunopathology in apes through potentially increased risk of sepsis or chronic inflammatory disease is evolutionarily negligible compared to the persistent risk of infection. Interestingly, among the most divergently responding pathways between apes and AAMs, several were associated with the regulation of interferon responses and responses to viruses. These findings are consistent with a growing body of literature that pathogens and, specifically, viruses have been important drivers of adaptive evolution in humans and other mammals (15, 44 –46).
Regardless of initial strength and divergence of transcriptional response to LPS and GARD, we show that the transcriptional activity of antiviral (interferon) and inflammatory pathways became attenuated over time and more similar between species. While acute-phase and early proinflammatory responses are typically countered by a later anti-inflammatory response to lessen host damage and maintain homeostasis, the dampening of this initial, powerful antimicrobial response here is profound over time (47). Remarkably, in apes the pathways that underwent the most pronounced attenuation after 24 h tended to be ones not expected to be strongly engaged in the response to the pathogen type in the experiment. For instance, the typically antiviral type-I IFN pathway response was found to be markedly reduced in apes after 24 h of bacterial but not viral stimulation. While the initial response of apes to immune stimulus is very strong, temporal regulation of responding pathways may reduce the energetic costs of such an immune strategy. What gene regulatory and immunological mechanisms are involved in such temporal regulation will require further investigation.
In conclusion, we show initial antibacterial and antiviral responses of apes to be highly correlated and strongly responsive when compared to close relatives AAMs. Apes appear to have adopted an immune strategy that emphasizes sterilization over specificity, strongly transcribing a greater number of genes in response to immune stimulation and releasing very similar immune transcriptomic “arsenals” regardless of pathogen type. This powerful response dramatically shifts during the opening hours of infection, to involve significantly fewer genes after 24 h, which may help limit bystander tissue damage. The energetically costly approach apes initiate in response to immune stimulation may be favored by this primate family’s adoption of slower life history with increased risk of pathogen exposure over reproductive life span or past pathogen exposure. The addition of more primate species, combined with the use of single-cell RNA sequencing methods, are important next steps to study the evolution of the immune system and more precisely map the immune cell types that contribute the most to divergence in immune response across primates.
Materials and Methods
Sample Collection and Blood Stimulation.
We measured innate immune responses on a panel of six humans, six chimpanzees, six rhesus macaques, and eight olive baboons (three females and three males for each species and four females and four males for baboon). Human samples were obtained via informed consent. This study was approved by the Research Ethics Board at the Centre Hospitalier Universitaire Sainte-Justine (Research Ethics Board approved protocol no. 3557). Nonhuman primate blood samples were humanely collected in accordance with the animal-subject regulatory standards of the Texas Biomedical Research Institute and Emory University Institutional Animal Care and Use Committees. These sample locations were chosen to help control for possible differences in genome expression stemming from care/management that was primate-center specific versus evolved differences in gene expression across the different primate lineages. Two more-distantly related monkey and ape species (rhesus macaque and chimpanzees) that were housed and under similar care at Yerkes National Regional Primate Center (Emory University) were sampled at that location, while their closest relatives (baboons for macaques and chimpanzees for humans) lived and were sampled at two different locations: the Texas Biomedical Research Institute and Montreal, Quebec. Chimpanzee samples were collected prior to the NIH ban on chimpanzee research.
All human and nonhuman primate subjects were broadly age matched and healthy. All individuals sampled were young adults to middle-aged adults, with all human subject between 22 to 55 y of age and all chimps 18 to 23 y of age. Baboons and macaques were between 7 to14 y of age, which corresponds to 28 to 55 human years, with the exception of two macaques of 3 and 20 y of age (∼12 and ∼80 human years) (48). The responses and differential cell counts of these individuals, however, were in keeping with other macaques. Subjects were screened to ensure they had no chronic illnesses/infections or history of cancer, free of anti-inflammatory medication, and reported or appeared [in the case of nonhuman primates (NHP)] healthy on the day of sampling. All rhesus macaques were specific-pathogen free (herpes B virus, simian immunodeficiency virus, simian betaretrovirus, and simian T-Cell lymphotrophic virus-1); chimpanzees were free of HIV and simian immunodeficiency virus, hepatitis B virus, and hepatitis C virus at the time of sampling. The individuals in this study had as similar a disease and health experience/status as is possible to know and control ( Dataset S1 provides information on the sex, age, and other features of our samples).
We drew 1 mL of whole blood from each animal directly into a TruCulture tube (Myriad RBM) that contained the following: 1) cell culture media only (“control”), 2) cell culture media plus 1 μg/mL ultra-pure LPS from the E. coli 0111:B4 strain (“LPS”), or 3) cell culture media plus 1 μg/mL GARD. Samples were incubated for 4 and 24 h at 37 °C. Following incubation, we separated the plasma and cellular fractions by centrifugation and lysed and discarded the red cells from the remaining cell pellet by applying red blood cell lysis buffer (RBC lysis solution, 5 Prime Inc.) for 10 min followed by centrifugation and washing with 1× PBS. The remaining white blood cells were lysed in Qiazol and frozen at −80 °C until library construction (Qiagen). To control for variation in cellular composition in downstream analyses, we used flow cytometry to quantify the proportions of leukocyte subtypes, accounting for polymorphonuclear (CD14dim/SSC-A > 100K/FSC-A > 100K/CD66+), classical monocytes (CD14+/CD16−), CD14+ intermediate monocytes (CD14+/CD16+), CD14-nonclassical monocytes (CD14−/CD16+), helper T cells (CD3+/CD4+), cytotoxic T cells (CD3+/CD8+), double positive T cells (CD3+/CD4+/CD8+), CD8− B cells (CD3−/CD20+/CD8−), CD8+ B cells (CD3−/CD20+/CD8+), natural killer T lymphocytes (CD3+/CD16+), and natural killer cells (for monkeys: CD3−/CD16+ in the lymphocyte scatter; for apes: CD3−/CD16+/CD56+ in the lymphocyte scatter). Samples for FACS were simultaneously cleared of red blood cells via lysis and fixed by application of BD FACS-lyse for 2 min, prior to washing with 1× PBS, and staining with fluorochrome conjugated monoclonal antibodies ( Dataset S12), before washing with 1× PBS and suspending in a 1× PBS and paraformadelhyde solution for analysis on the BD LSRFortessa platforms. Proportional analysis was completed in FlowJo X, using BD FACSBeads individually stained with the antibodies to calculate compensation.
RNA-Seq Data Generation.
Library construction.
Total RNA was isolated from cell lysate by phenol::chloroform extraction and spin column (miRNAeasy kit, Qiagen), quantified by spectrophotometry, and assessed for quality using the Agilent 2100 bioanalyzer (Agilent Technologies). Samples with no evidence of RNA degradation (Integrity number >8) were then used for RNA library development. Messenger RNA was isolated by magnetic bead and converted into RNA libraries using the Illumina TruSeq RNA Library preparation kit version 2 according to the manufacturer’s instructions (Illumina). Libraries were sequenced on a HiSEq. 2100, producing 151 transcriptomes, at 25 to 30 million reads per sample. The RNAseq data were deposited in the Gene Expression Omnibus database under accession no. GSE155918 (49).
Reads Mapping on 1:1 Orthologs.
Following sequencing, we trimmed Illumina adapter sequence from the ends of the reads and removed bases with quality scores <20 using Trim Galore (version 0.2.7). We used the software Spliced Transcripts Alignment to a Reference (STAR) to align the reads to an orthologous reference genome for all four species (50). We developed this genome using the XSAnno pipeline which combines whole-genome alignment, local alignment, and multiple filters to remove regions with difference in mappability between species (51). XSAnno pipeline identifies orthologous genes across two species using three major filters, namely, LiftOver to carry annotation of one species over the other, BLAT aligner to compare orthologous exons identity between the two species, and simNGS to identify exons that have different lengths between the species. We used the genome assemblies of hg19 for human, CHIMP2.1.4 for chimpanzee, MMUL 1.0 for macaque, and PapAnu2.0 for olive baboon species. We used human annotation as a reference. The pairwise alignment chains between human and each species were obtained from University of California, Santa Cruz genome browser (52). We used different thresholds to define orthologous regions between the two genomes to carry annotation from one species to another using AnnoConvert program that utilized LiftOver according to simulations using liftOverBlockSim PERL script from XSAnno pipeline (53). The values were 0.98, 0.92, and 0.91 for chimp, baboon, and macaque, respectively, that were used to assign -minMatch argument in AnnoConvert. The second step is using reciprocal whole-genome alignment using BLAT through BlatFilter software of the pipeline using annotations files generated previously (54). This step will filter exons that are highly divergent between the two species. The last filter is using simNGS to simulate reads for exons assuming they are not differentially expressed. Then, differential expression analysis is performed, and if exons are found to be differentially expressed, these will be filtered out as it reflects differential length of the exons between species.
Gene-expression estimates were obtained by summing the number of reads that mapped uniquely to each species-annotated genome using HTSeq-count (version 0.6.1) (55). After excluding samples that did not produce sequenceable libraries and postsequencing quality control, we analyzed read counts for 151 samples (Humans: 12 controls, 12 LPS, and 12 GARD; Chimpanzee: 12 controls, 12 LPS, and 12 GARD; Rhesus: 11 controls, 12 LPS, and 11 GARD; Baboons: 16 controls, 14 LPS, and 15 GARD; Dataset S1). We confirmed the identity of all samples based on genotype information derived from single nucleotide polymorphism calls made from the RNA-seq reads.
Read Normalization and Filtering Lowly Expressed Genes.
Prior to RNA-seq data analysis, we first filtered out genes that were very lowly or not detectably expressed in our samples. Specifically, we only kept genes whose expression was higher or equal to one count per million (CPM) in all the individuals from at least one species and one of the experimental conditions. This procedure yielded a total of 12,441 genes used for further analysis. Normalization for sequencing depth and library sizes was done using the trimmed mean of M-values method (56). We normalized the resulting read-count matrix using the function voom from the R-package limma to allow using linear models by limma package (57). The voom algorithm models mean-variance trend of logCPM for each gene and uses it to compute the variance as a weight of logCPM values. We then modeled the normalized expression values as a function of the different experimental factors in the study design such as species, ligand, and time points.
Statistical Analysis.
All statistical analysis was done on R version 3.6.2. Differential expression analysis was done using limma package version 3.34.9 (58). We employed linear regression to identify differentially expressed genes according to different questions asked by designing different models. We designated a model to test for differences of gene expression across species and treatments, ∼covariates + species + species:Time.point.stimulant. The arm Time.point.stimulant is the sample for each experimental condition, that is, LPS.4 h, LPS.24 h, GARD.4 h, and GARD.24 h. From this design, one can retrieve ligand responses in each species right away, while responses to ligands at 24 h are built from linear combinations, such as LPS.24 h to NC.24 h and GARD.24 h to NC.24 h. To take into account the paired structure of the data, with different samples coming from the same individuals, we used the duplicateCorrelation function. The used covariates are the different cell proportions collected by the FACS data. The cell proportions covariates are aimed to correct for the different proportion of white blood cells in different primate species since we conducted the transcriptomic characterization on all immune white blood cells. Genes with different magnitudes of response between clades, referred to as c-DRGs, were characterized. We established two filters to characterize significant c-DRGs in each treatment. First, we required the genes not to be differentially responsive to the treatment, even marginally, between within-clade species pairs (chimp versus human and baboon versus Macaque, showing FDR > 0.25). Second, we required that any pairwise comparison involving species from different clades to be significant at FDR < 0.1. Third, we also computed the average differences in responses between apes and AAMs. The absolute difference between the average response in apes versus AAMs is as follows:
And that contrast was required to be significant at FDR < 0.1, with genes featuring
being labeled ape specific and AAM specific for those for which
Species-specific DRGs were identified using pairwise comparisons at FDR < 0.01, consistent direction of expression in all contrasts, and systematic differences corresponding to stronger or weaker responses in the species of interest with respect to the any of the other three. Finally, we also required genes to show a logFC in response to the stimulus whose absolute differs in more than one log2FC with respect to the average of the other three animals. For humans, as an example, this means the following:
Ligand-specific genes in each species are genes that respond to one ligand (FDR < 0.05) but not to the other (FDR > 0.25), and whose responses to both ligands are in turn significantly different (FDR < 0.05). Shared genes are those whose responses to ligands are both significant (FDR < 0.05 in both) and, at the same time, not significantly different between them (FDR > 0.25).
Correction of multiple testing was done using FDR, as described by Benjamini-Hochberg (59).
Divergence Scores.
For each time point and stimulus, species were compared pairwise to retrieve the absolute differences between species’ responses to the stimulus under analysis. For the pair chimp versus human, for example, we can define the following:
Comparing these differences for pairs of animals within versus across clades, we obtained divergence scores as follows:
The analysis was conducted for all 50 hallmark pathways. We restrict the analysis in a given pathway to responsive genes (FDR < 0.05 in any species), whose average DS is reported. A P value for each DS of a given pathway was calculated by contrasting the DSs of genes of this specific pathway against the DSs of all responsive genes using Wilcoxon test.
We note that the fact that a given pathway is strongly responsive to immune activation does not necessarily translate into an elevated divergence score. The divergence score reflects how different (i.e., divergent) is the magnitude of the response between apes and AAMs, relative to differences in response within apes and AAMs. Thus, we could have a pathway that is strongly up- or down-regulated in response to immune stimulation and yet have a low divergence score. This could happen if 1) apes and AAMs respond to the stimulation with a similar magnitude, or 2) if the within-clade variance is larger than the between-clade variance in response.
Functional Characterization.
We conducted the functional characterization using GO enrichment implemented in the CluGO application (2.5.5) of Cytoscape (version 3.7.2) (60). The Benjamini-Hochberg method for multiple correction was used, and all orthologous genes, 12,441 genes, were used as a background. Default values were used for the rest of the parameters. FDR cutoff use was below 0.15.
Supplementary Material
Acknowledgments
We thank Steven Bosinger and Guido Silverstri of Yerkes Primate Center and Emory University for their assistance acquiring samples. We thank L.B.B.’s laboratory members for critical reading of the manuscript. We thank Calcul Québec and Compute Canada for providing access to the supercomputer Briaree from the University of Montreal. This work was supported by RGPIN/435917-2013 from the Natural Sciences and Engineering Research Council of Canada and R01-GM134376 from the National Institute of General Medical Sciences to L.B.B. J.F.B. is funded by NSF-BCS-1750675. The resources of the Southwest and Yerkes National Primate Research Centers are supported by NIH Grants P51-OD011133 and P51-OD011132, respectively, from the Office of Research Infrastructure Programs/Office of the Director.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. R.C.D. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2015855118/-/DCSupplemental.
Data Availability
The RNA-seq data generated in this study have been deposited in Gene Expression Omnibus (accession no. GSE155918) (49).
Change History
May 9, 2022: The Supporting Information section was updated.
References
- 1. Redl H., Bahrami S., Schlag G., Traber D. L., Clinical detection of LPS and animal models of endotoxemia. Immunobiology 187, 330–345 (1993). [DOI] [PubMed] [Google Scholar]
- 2. Munford R. S., Sensing gram-negative bacterial lipopolysaccharides: A human disease determinant? Infect. Immun. 76, 454–465 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Chahroudi A., Bosinger S. E., Vanderford T. H., Paiardini M., Silvestri G., Natural SIV hosts: Showing AIDS the door. Science 335, 1188–1193 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Sandmann L., Ploss A., Barriers of hepatitis C virus interspecies transmission. Virology 435, 70–80 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Vaure C., Liu Y., A comparative review of toll-like receptor 4 expression and functionality in different animal species. Front. Immunol. 5, 316 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Kumar A., et al., Experimental human endotoxemia is associated with depression of load-independent contractility indices: Prevention by the lipid a analogue E5531. Chest 126, 860–867 (2004). [DOI] [PubMed] [Google Scholar]
- 7. Taveira da Silva A. M., et al., Brief report: Shock and multiple-organ dysfunction after self-administration of Salmonella endotoxin. N. Engl. J. Med. 328, 1457–1460 (1993). [DOI] [PubMed] [Google Scholar]
- 8. Haudek S. B., et al., Lipopolysaccharide dose response in baboons. Shock 20, 431–436 (2003). [DOI] [PubMed] [Google Scholar]
- 9. Yin G. Q., et al., Endotoxic shock model with fluid resuscitation in Macaca mulatta. Lab. Anim. 39, 269–279 (2005). [DOI] [PubMed] [Google Scholar]
- 10. Akira S., Pathogen recognition by innate immunity and its signaling. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 85, 143–156 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Barreiro L. B., Marioni J. C., Blekhman R., Stephens M., Gilad Y., Functional comparison of innate immune signaling pathways in primates. PLoS Genet. 6, e1001249 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Brinkworth J. F., Pechenkina E. A., Silver J., Goyert S. M., Innate immune responses to TLR2 and TLR4 agonists differ between baboons, chimpanzees and humans. J. Med. Primatol. 41, 388–393 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Mandl J. N., et al., Divergent TLR7 and TLR9 signaling and type I interferon production distinguish pathogenic and nonpathogenic AIDS virus infections. Nat. Med. 14, 1077–1087 (2008). [DOI] [PubMed] [Google Scholar]
- 14. Wlasiuk G., Nachman M. W., Adaptation and constraint at Toll-like receptors in primates. Mol. Biol. Evol. 27, 2172–2186 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van der Lee R., Wiel L., van Dam T. J. P., Huynen M. A., Genome-scale detection of positive selection in nine primates predicts human-virus evolutionary conflicts. Nucleic Acids Res. 45, 10634–10648 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Danko C. G., et al., Dynamic evolution of regulatory element ensembles in primate CD4+ T cells. Nat. Ecol. Evol. 2, 537–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Rivera A., Siracusa M. C., Yap G. S., Gause W. C., Innate cell communication kick-starts pathogen-specific immunity. Nat. Immunol. 17, 356–363 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Deschamps M., et al., Genomic signatures of selective pressures and introgression from archaic hominins at human innate immunity genes. Am. J. Hum. Genet. 98, 5–21 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Liberzon A., et al., The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Hagai T., et al., Gene expression variability across cells and species shapes innate immunity. Nature 563, 197–202 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Nunn C. L., Gittleman J. L., Antonovics J., Promiscuity and the primate immune system. Science 290, 1168–1170 (2000). [DOI] [PubMed] [Google Scholar]
- 22. Nunn C. L., A comparative study of leukocyte counts and disease risk in primates. Evolution 56, 177–190 (2002). [DOI] [PubMed] [Google Scholar]
- 23. Bradley L. M., Dalton D. K., Croft M., A direct role for IFN-gamma in regulation of Th1 cell development. J. Immunol. 157, 1350–1358 (1996). [PubMed] [Google Scholar]
- 24. Kak G., Raza M., Tiwari B. K., Interferon-gamma (IFN-γ): Exploring its implications in infectious diseases. Biomol. Concepts 9, 64–79 (2018). [DOI] [PubMed] [Google Scholar]
- 25. Zha Z., et al., Interferon-γ is a master checkpoint regulator of cytokine-induced differentiation. Proc. Natl. Acad. Sci. U.S.A. 114, E6867–E6874 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Janeway C. A. Jr, Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb. Symp. Quant. Biol. 54, 1–13 (1989). [DOI] [PubMed] [Google Scholar]
- 27. Matzinger P., Tolerance, danger, and the extended family. Annu. Rev. Immunol. 12, 991–1045 (1994). [DOI] [PubMed] [Google Scholar]
- 28. Brubaker S. W., Bonham K. S., Zanoni I., Kagan J. C., Innate immune pattern recognition: A cell biological perspective. Annu. Rev. Immunol. 33, 257–290 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fitzgerald K. A., Kagan J. C., Toll-like receptors and the control of immunity. Cell 180, 1044–1066 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Kanevskiy L. M., Telford W. G., Sapozhnikov A. M., Kovalenko E. I., Lipopolysaccharide induces IFN-γ production in human NK cells. Front. Immunol. 4, 11 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Ghosh M., Subramani J., Rahman M. M., Shapiro L. H., CD13 restricts TLR4 endocytic signal transduction in inflammation. J. Immunol. 194, 4466–4476 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kagan J. C., et al., TRAM couples endocytosis of Toll-like receptor 4 to the induction of interferon-beta. Nat. Immunol. 9, 361–368 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Harvey P. H., Clutton-Brock T. H., Life history variation in primates. Evolution 39, 559–581 (1985). [DOI] [PubMed] [Google Scholar]
- 34. Ross C., Life history patterns and ecology and macaque species. Primates 33, 207–215 (1992). [Google Scholar]
- 35. Wich S. A., et al., Life history of wild Sumatran orangutans (Pongo abelii). J. Hum. Evol. 47, 385–398 (2004). [DOI] [PubMed] [Google Scholar]
- 36. Johnson P. T. J., et al., Living fast and dying of infection: Host life history drives interspecific variation in infection and disease risk. Ecol. Lett. 15, 235–242 (2012). [DOI] [PubMed] [Google Scholar]
- 37. Cronin J. P., Welsh M. E., Dekkers M. G., Abercrombie S. T., Mitchell C. E., Host physiological phenotype explains pathogen reservoir potential. Ecol. Lett. 13, 1221–1232 (2010). [DOI] [PubMed] [Google Scholar]
- 38. Buechler C. R., et al., Seroprevalence of Zika virus in wild African green monkeys and baboons. mSphere 2, e00392-16 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Gao F., et al., Origin of HIV-1 in the chimpanzee Pan troglodytes troglodytes. Nature 397, 436–441 (1999). [DOI] [PubMed] [Google Scholar]
- 40. Hirsch V. M., Olmsted R. A., Murphey-Corb M., Purcell R. H., Johnson P. R., An African primate lentivirus (SIVsm) closely related to HIV-2. Nature 339, 389–392 (1989). [DOI] [PubMed] [Google Scholar]
- 41. Vasilakis N., Cardosa J., Hanley K. A., Holmes E. C., Weaver S. C., Fever from the forest: Prospects for the continued emergence of sylvatic dengue virus and its impact on public health. Nat. Rev. Microbiol. 9, 532–541 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Okin D., Medzhitov R., Evolution of inflammatory diseases. Curr. Biol. 22, R733–R740 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Sorci G., Cornet S., Faivre B., Immune evasion, immunopathology and the regulation of the immune system. Pathogens 2, 71–91 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ito J., Gifford R. J., Sato K., Retroviruses drive the rapid evolution of mammalian APOBEC3 genes. Proc. Natl. Acad. Sci. U.S.A. 117, 610–618 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Enard D., Cai L., Gwennap C., Petrov D. A., Viruses are a dominant driver of protein adaptation in mammals. eLife 5, e12469 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Harrison G. F., et al., Natural selection contributed to immunological differences between hunter-gatherers and agriculturalists. Nat. Ecol. Evol. 3, 1253–1264 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Morris M. C., Gilliam E. A., Li L., Innate immune programing by endotoxin and its pathological consequences. Front. Immunol. 5, 680 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Mattison J. A., Vaughan K. L., An overview of nonhuman primates in aging research. Exp. Gerontol. 94, 41–45 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Fahmy Hawash M. B., et al., Primate innate immune responses to bacterial and viral pathogens reveals an evolutionary trade-off between strength and specificity. Gene Expression Omnibus. https://www-ncbi-nlm-nih-gov.ezproxy.u-pec.fr/geo/query/acc.cgi?acc=GSE155918. Deposited 8 August 2020. [DOI] [PMC free article] [PubMed]
- 50. Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Zhu Y., Li M., Sousa A. M., Sestan N., XSAnno: A framework for building ortholog models in cross-species transcriptome comparisons. BMC Genomics 15, 343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kent W. J., et al., The human genome browser at UCSC. Genome Res. 12, 996–1006 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Hinrichs A. S., et al., The UCSC genome browser database: Update 2006. Nucleic Acids Res. 34, D590–D598 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kent W. J., BLAT–the BLAST-like alignment tool. Genome Res. 12, 656–664 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Anders S., Pyl P. T., Huber W., HTSeq–A Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Robinson M. D., Oshlack A., A scaling normalization method for differential expression analysis of RNA-seq data. Genome Biol. 11, R25 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Law C. W., Chen Y., Shi W., Smyth G. K., voom: Precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol. 15, R29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Ritchie M. E., et al., Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 43, e47 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Benjamini Y., Hochberg Y., Controling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. Series B: Methodol. 57, 289–300 (1995). [Google Scholar]
- 60. Bindea G., et al., ClueGO: A Cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 25, 1091–1093 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The RNA-seq data generated in this study have been deposited in Gene Expression Omnibus (accession no. GSE155918) (49).