Significance
Functionally relevant neuronal connections are often organized within discrete layers of neuropil to ensure proper connectivity and information processing. While layer-specific assembly of neuronal connectivity is a dynamic process involving stepwise interactions between different neuron types, the mechanisms underlying this critical developmental process are not well understood. Here, we investigate the role of the transcription factor dFezf in layer selection within the Drosophila visual system, which is important for synaptic specificity. Our findings show that dFezf functions as a transcriptional repressor governing the precise temporal expression pattern of downstream genes, including other transcription factors required for proper connectivity. Layer-specific assembly of neuronal connectivity in the fly visual system is thus orchestrated by precise, temporally controlled transcriptional cascades.
Keywords: growth cone, neural connectivity, dFezf, laminar organization, transcription
Abstract
The layered compartmentalization of synaptic connections, a common feature of nervous systems, underlies proper connectivity between neurons and enables parallel processing of neural information. However, the stepwise development of layered neuronal connections is not well understood. The medulla neuropil of the Drosophila visual system, which comprises 10 discrete layers (M1 to M10), where neural computations underlying distinct visual features are processed, serves as a model system for understanding layered synaptic connectivity. The first step in establishing layer-specific connectivity in the outer medulla (M1 to M6) is the innervation by lamina (L) neurons of one of two broad, primordial domains that will subsequently expand and transform into discrete layers. We previously found that the transcription factor dFezf cell-autonomously directs L3 lamina neurons to their proper primordial broad domain before they form synapses within the developing M3 layer. Here, we show that dFezf controls L3 broad domain selection through temporally precise transcriptional repression of the transcription factor slp1 (sloppy paired 1). In wild-type L3 neurons, slp1 is transiently expressed at a low level during broad domain selection. When dFezf is deleted, slp1 expression is up-regulated, and ablation of slp1 fully rescues the defect of broad domain selection in dFezf-null L3 neurons. Although the early, transient expression of slp1 is expendable for broad domain selection, it is surprisingly necessary for the subsequent L3 innervation of the M3 layer. DFezf thus functions as a transcriptional repressor to coordinate the temporal dynamics of a transcriptional cascade that orchestrates sequential steps of layer-specific synapse formation.
An organism’s ability to sense, adapt, and respond to its environment is dependent on precise neural connectivity. Compartmentalization of correct synaptic partners into discrete layers is thought to facilitate the establishment of proper connectivity during development. Layered organization of distinct neural connections may be advantageous for economizing wiring length (1), segregation of nonpartners (2), and enabling parallel processing of neural information (3–6). Despite the prominence of layers across brain regions, including the cerebral cortex (7) and spinal cord (8), as well as its conservation across species (9), the mechanisms underlying layer assembly of neural connections are not well understood.
The medulla neuropil of the Drosophila melanogaster optic lobe has a well-characterized layered structure rendering it an ideal system for defining molecular design principles underlying layer-specific synaptic connectivity (Fig. 1A). The medulla comprises neurites from over 100 morphologically distinct and genetically accessible neuron cell types, which form stereotyped connections within 1 or more of 10 discrete layers (M1 to M10).
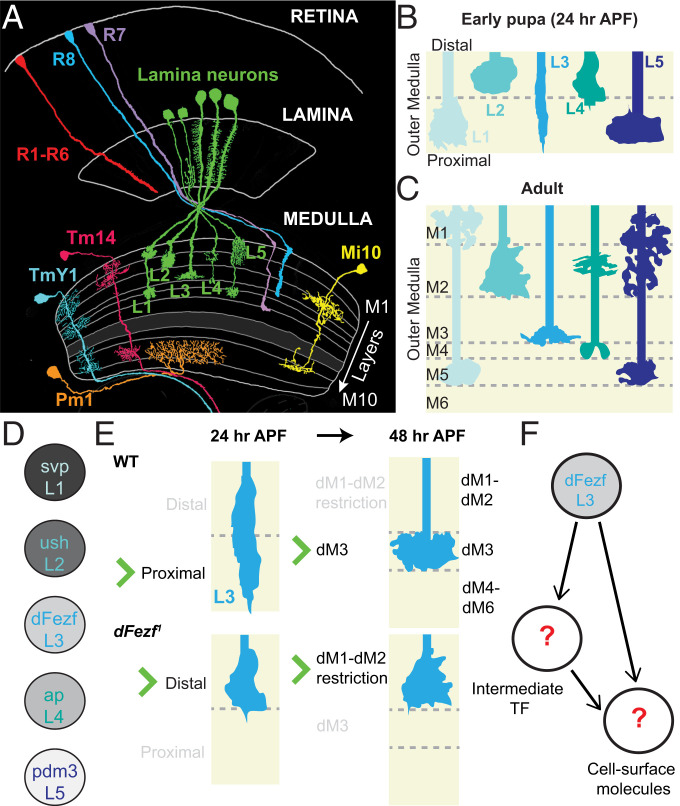
Fig. 1.
The D. melanogaster visual system and lamina neuron layer specificity. Schematics illustrating the various morphologies and layer-specific innervation patterns found throughout the fruit fly optic lobe, as well as a synopsis of the findings from Peng et al. (12). (A) Visual information is received by the retina and relayed to four downstream neuropil known as the lamina, medulla, lobula, and lobula plate (lobula and lobula plate are not shown in the diagram). The medulla neuropil consists of 10 layers (M1 to M10). The M1 to M6 layers constitute the outer medulla, while the M8 to M10 layers constitute the inner medulla, both of which are separated by the serpentine layer (M7). More than 100 morphologically distinct neurons innervate specific layers throughout the medulla, wherein they form stereotyped connections. Reprinted by permission from ref. 28, Springer Nature: Cell and Tissue Research, copyright (1989). (B and C) The L1 to L5 lamina neurons are major input neurons into the medulla that establish distinct innervation patterns throughout layers M1 to M5 of the outer medulla. (B) During early pupal development (24 h APF), L1 to L5 growth cones innervate the primordial outer medulla, which is partitioned into the distal and proximal broad domains at this stage. L1 to L5 establish primitive, overlapping targeting patterns before arborizing in unique patterns illustrated in C. (D) Each lamina neuron expresses a unique transcription factor, which we hypothesize confers the layer specificity for each cell type. (E) Summary of findings in Peng et al. (12). Growth cones of dFezf1 mutant L3 neurons are initially restricted to the distal domain and later terminate in superficial layers (dM1 to dM6 are the developing layers M1 to M6 observed in the adult [C]). We hypothesize dFezf regulates the expression of cell surface genes either directly or via intermediate regulators (F).
The medulla receives input directly from photoreceptors (R cells) R7 to R8 and lamina (L)1 to L5 monopolar lamina neurons. L1 to L5 neurons innervate specific layers throughout the outer medulla, which comprises the M1 to M6 layers (Fig. 1A). During early pupal development, the outer medulla is partitioned into two primordial, broad regions referred to as the distal and proximal domains (Fig. 1B). L1, L3, and L5 neuron growth cones become restricted to the proximal domain, while L2 and L4 neuron growth cones are restricted to the distal domain (10). As development proceeds, the outer medulla expands, and within this region, the M1 to M6 layers emerge along the distal-to-proximal axis. During this transition, lamina neurons undergo morphological maturation as they innervate their incipient layers, eventually adopting the unique shapes and synaptic contacts exhibited in the adult (Fig. 1C). The molecular mechanisms underlying growth cone selection of the distal or proximal domains (referred to as broad domain specificity), and the morphological transitions thereafter, are unclear.
Each lamina neuron type expresses one or more unique transcription factors, which may confer layer specificity of these neurons (11, 12) (Fig. 1D). Previously, we found that the transcription factor dFezf cell autonomously controls L3 neuron broad domain and layer specificity. L3 neurons are the only lamina neuron type that expresses dFezf. When dFezf is knocked out in L3 neurons, their growth cones are incorrectly restricted to the distal domain early in development, and later on their endings terminate in superficial layers M1 and M2 (Fig. 1E). We posited that dFezf directs L3 broad domain specificity by regulating expression of genes encoding cell surface molecules, either directly or through intermediate regulators such as other transcription factors (Fig. 1F). Consistent with this idea, we found that genes encoding cell surface proteins and transcription factors are two of the largest groups of differentially expressed genes in dFezf-null L3 neurons (12), setting the stage for identifying key molecular determinants of layer specificity in the medulla.
Here, we report that dFezf directs L3 broad domain specificity by functioning as a transcriptional repressor of the transcription factor slp1 (sloppy-paired 1). In wild-type (WT) L3 neurons, slp1 is transiently expressed at a low level during broad domain selection in early pupal development. In dFezf-null L3 neurons, slp1 expression is up-regulated, which causes L3 growth cones to improperly target the distal domain. Surprisingly, early expression of slp1 and another related transcription factor, slp2, although dispensable for broad domain selection, is required for subsequent innervation of the M3 layer by the L3 neuron. These findings thus indicate that dFezf functions as a transcriptional repressor to direct L3 broad domain specificity and ultimately layer specificity by orchestrating the precise temporal dynamics of an intricate transcriptional cascade.
Results
Identifying Candidate Molecular Determinants of L3 Broad Domain Specificity.
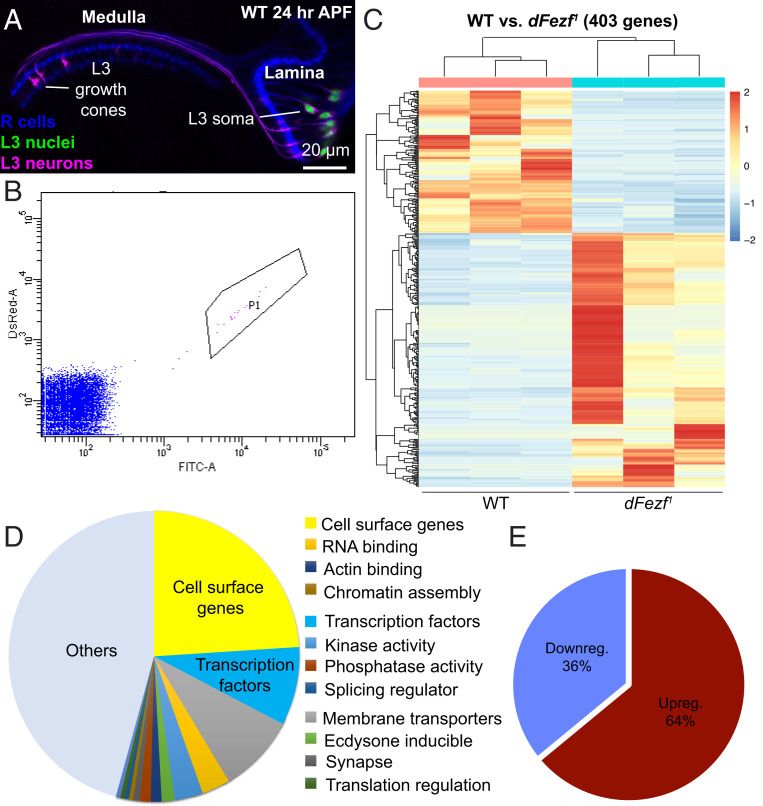
The transcription factor (TF) dFezf cell-autonomously directs L3 growth cones to the proximal domain of the outer medulla during early pupal development, a critical step in establishing L3 layer specificity (Fig. 1E). As a first step toward identifying relevant dFezf downstream genetic targets, we used RNA sequencing (RNA-seq) to generate transcriptomes of WT and dFezf1 L3 neurons at 24 h after puparium formation (APF), an early pupal stage of development near the initiation of lamina neuron broad domain selection (10) (Fig. 1B). Our previous RNA-seq dataset, generated at 40 h APF, and another dataset from a study examining WT lamina neuron transcriptomes at the same stage were used for comparison (11, 12). We used MARCM (Mosaic Analysis with a Repressible Cell Marker) (13) to generate single-cell clones of either WT or dFezf1 L3 neurons in an otherwise WT background. The L3 MARCM clones were fluorescently labeled with H2A-GFP (green) and myr-tdTOM (red) (Fig. 2A), which are nuclear and transmembrane reporters, respectively. Using this strategy, L3 MARCM clones were the only neurons labeled in the optic lobe. Cell-specific expression of fluorescent labels was verified by immunostaining and confocal microscopy (Fig. 2A). Fluorescently labeled WT and dFezf1 L3 MARCM clones were isolated by fluorescence-activated cell sorting (FACS) at 24 h APF (Fig. 2B), and complementary DNA (cDNA) libraries were then prepared from total messenger RNA (mRNA) and sequenced using an Illumina platform. Three biological replicates of each genotype were performed, and all the cDNA libraries were prepared and sequenced at the same time to minimize technical variability. Differential gene expression analysis identified 403 genes that were differentially expressed in WT and dFezf mutant clones (adjusted P < 0.05) (Fig. 2C and SI Appendix, Table S1). FlyMine enrichment analysis (14) showed that genes encoding cell surface proteins were significantly enriched in our dataset of differentially expressed (DE) genes (SI Appendix, Table S2) and represented the largest group of DE genes (99 out of 403 DE genes; Fig. 2D). Interestingly, TFs emerged as a second major DE class (36 out of 403; Fig. 2D), which is consistent with the possibility that dFezf regulates L3 broad domain specificity through the regulation of cell surface genes and/or intermediate TFs. Nearly two-thirds of the DE genes (258 out of 403 genes) were up-regulated in dFezf1 L3 neurons, while about one-third (145 out of 403 genes) were down-regulated (Fig. 2E), suggesting that dFezf may function primarily as a transcriptional repressor in directing L3 broad domain specificity.
Fig. 2.
RNA-seq identified 403 differentially expressed genes in dFezf1 mutant L3 neurons. We generated WT and dFezf1 transcriptomes of FACS-sorted L3 MARCM clones at 24 h APF. (A) Confocal image illustrating WT optic lobe at 24 h APF (at this stage, the lamina is located laterally to the medulla). L3 MARCM clones are pictured. L3 neuron somas and cell membranes are labeled with GFP (green) and tdTOM (magenta), respectively. R1 to R8 photoreceptors are also pictured in blue (mAB24B10). The sample was taken from the same crosses used to generate transcriptomes. (B) FACS plot illustrating selected cluster of GFP/tdTOM double-positive cells. These are L3 MARCM clones shown in A; 100,000 events are shown. (C) Heatmap illustrating up-regulated and down-regulated genes. Each column represents a different biological replicate of the genetic background indicated below. Each row represents the normalized expression of a differentially expressed gene. (D) Pie chart illustrating a representation of different gene types based on previously curated databases (11, 46). Cell surface genes and transcription factors are the two largest groups. (E) Pie chart illustrating the ratio of up- and down-regulated genes. The majority of differentially expressed genes are up-regulated.
DFezf Functions as a Transcriptional Repressor to Regulate L3 Broad Domain Specificity.
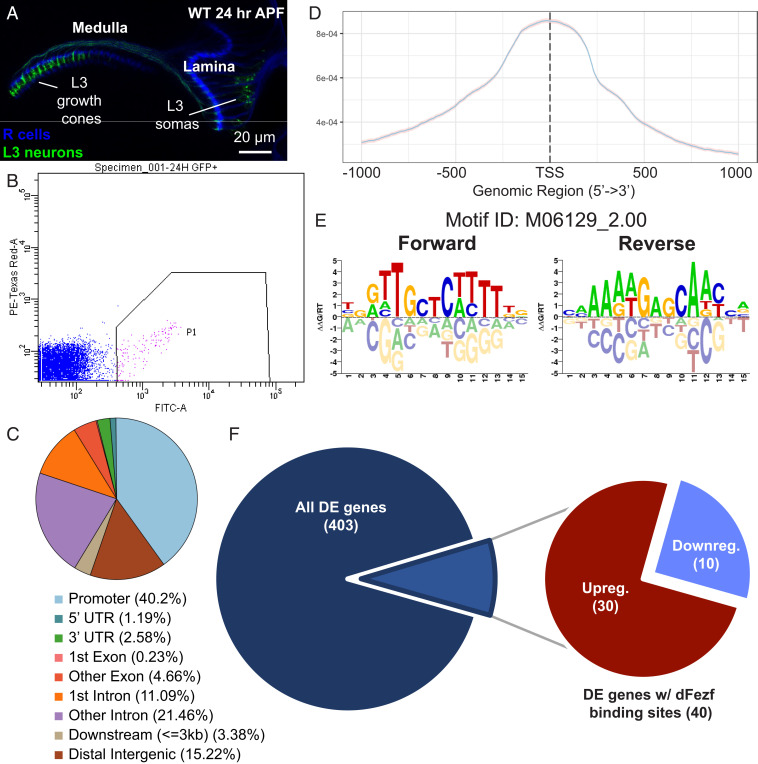
As a first step toward defining dFezf direct genetic targets, we performed ATAC-seq (assay for transposase-accessible chromatin with sequencing) (15) to identify the accessible genomic regions to which dFezf may directly bind in L3 neurons. We specifically labeled WT L3 neurons using GFP and isolated the fluorescently labeled cells by FACS at 24 h APF (Fig. 3 A and B), the same stage used for the RNA-seq experiments detailed above. DNA libraries were prepared from three biological replicates and sequenced at the same time using an Illumina platform. Overall, ∼23,000 peaks were called from the ATAC-seq data. About 40% of the peaks fall within gene promoter regions and are enriched in close proximity (i.e., within 2 kilobase pairs [kb]) of transcriptional start sites (TSSs) (Fig. 3 C and D) (see Materials and Methods for details regarding peak annotation). We then referred to our RNA-seq data at the same stage to identify the DE genes whose promoters are identified as accessible according to the ATAC-seq data. Next, matches to a dFezf binding motif (16) (Fig. 3E) were identified in the ATAC-seq peaks associated with DE genes. Genes identified with one or more putative dFezf binding sites in the accessible region were considered potential dFezf direct targets. Out of the 403 DE genes identified by RNA-seq, only 40 have putative dFezf binding sites in their open chromatin regions (Fig. 3F). We find that of these 40 genes, 13 are cell surface genes and 4 are TFs (SI Appendix, Table S3), further supporting a role for dFezf in directing L3 broad domain specificity through direct and/or indirect regulation of cell surface gene expression. Furthermore, 30 out of the 40 putative direct dFezf targets are up-regulated in dFezf1 mutant L3 neurons (Fig. 3F), again suggesting that dFezf may preferentially function as a transcriptional repressor in this context.
Fig. 3.
ATAC sequencing experiments suggest that dFezf may function as a transcriptional repressor. We FACS-sorted 24 h APF WT L3 neurons and sequenced the accessible genomic regions. We identified potential dFezf direct targets by scanning for matches to a dFezf-binding motif in the promoters of genes with dFezf-specific expression patterns using the RNA-seq data set from Fig. 2. (A) Confocal image illustrating WT optic lobe at 24 h APF (at this stage, the lamina is located laterally to the medulla). L3 neurons cell membranes are labeled in GFP (green). The youngest L3 neurons are unlabeled and therefore unseen, likely because these cells have yet to activate GAL4 expression. (B) FACS plot illustrating a cluster of GFP-positive L3 neurons; 100,000 events are shown. (C) Pie chart illustrating the representation of genomic region types that were found to be accessible. About 40% of the peaks fall within gene promoter regions. (D) The signal-density plot shows that there is a reasonable number of open regions identified within 2 kb of the TSS. (E) Sequence of the putative dFezf/erm DNA-binding motif. (F) Pie charts illustrating the portion of differentially expressed genes with putative dFezf-binding sites (Left) and ratio of up-regulated and down-regulated putative dFezf direct targets (Right).
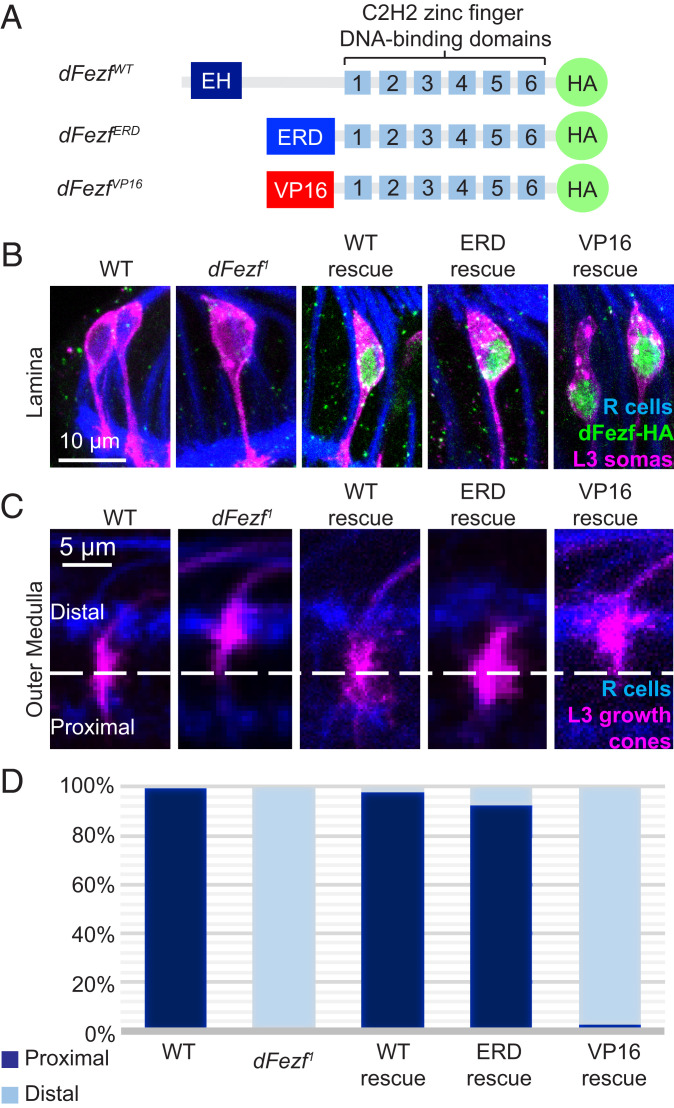
To test the idea that dFezf directs L3 growth cones to the proximal domain through transcriptional repression, we performed MARCM rescue experiments using 9-9-GAL4, an L3-specific GAL4 driver, and alternative UAS constructs encoding WT or modified forms of dFezf that either repress or activate transcription of target genes. WT dFezf (dFezfWT) contains an engrailed homology domain near its amino (N) terminus, which recruits the corepressor Groucho to repress gene transcription (17–19). The carboxyl (C) terminus of dFezfWT contains six C2H2 zinc finger DNA binding domains, which confer DNA sequence binding specificity of the TF (20). Previous studies generated UAS constructs encoding modified forms of dFezf in which the N terminus was truncated and replaced with either an engrailed transcriptional repressor domain (UAS-dFezfERD) or a VP16 transcriptional activation domain (UAS-dFezfVP16) (Fig. 4A); these dFezf chimeric variants thus function as either a repressor or activator, respectively (21). In each UAS construct, the encoded protein contains a hemagglutinin (HA) epitope tag at the C terminus (Fig. 4A), which allowed for immunohistochemical confirmation of protein expression (Fig. 4B).
Fig. 4.
DFezf functions as a transcriptional repressor in regulating L3 broad domain specificity. We performed MARCM rescue experiments using 9-9-GAL4, an L3-specific GAL4 driver, and UAS constructs encoding WT and chimeric forms of dFezf. (A) Schematic illustrating a simplified structure of UAS-dFezf constructs generated in the study by Janssens et al. (21). All three constructs contain six C2H2 zinc finger DNA-binding domains and an HA epitope tag at the C terminus. UAS-dFezfWT contains the WT dFezf structure with an engrailed homology domain (EH) at the N terminus. UAS-dFezfERD contains an ERD at the N terminus. UAS-dFezfVP16 contains a VP16 activation domain (VP16) at the N terminus. (B) Verification of UAS construct expression in MARCM rescue experiments in C. L3 somas are labeled in GFP (magenta). Anti-HA antibody was used to label dFezf-HA (green). R cells are labeled in blue (mAB24B10). Construct expression is confirmed for all three constructs used in this experiment. (C) Confocal images of L3 growth cone targeting at 24 h APF in MARCM rescue experiments using constructs illustrated in A. L3 growth cones are labeled with GFP (magenta). R cells are labeled in blue (mAB24B10). The UAS-dFezfWT and UAS-dFezfERD constructs rescue L3 broad domain specificity (i.e., targeting the proximal domain), while the UAS-dFezfVP16 construct does not. (D) Quantification of data from C.
For MARCM rescue experiments, we generated single-cell clones of WT and dFezf1 L3 neurons that express none or one of the UAS constructs (UAS construct activation is limited to MARCM clones in this experiment). The different genotypes are referred to as WT, dFezf1, WT rescue (dFezf1 + UAS-dFezfWT), engrailed repressor domain (ERD) rescue (dFezf1 + UAS-dFezfERD), and VP16 rescue (dFezf1 + UAS-dFezfVP16) (see Materials and Methods for complete genotypes). L3 broad domain specificity was assessed at 24 h APF (see Materials and Methods for details of proximal vs. distal designation). While WT L3 growth cones properly extended into the proximal domain (99.02%), dFezf1 L3 growth cones were restricted to the distal domain (99.08%; Fig. 4 C and D), consistent with our previous findings (12). The mistargeting phenotype was rescued by expressing the UAS-dFezfWT construct (98.8%) or the repressor-only UAS-dFezfERD construct (91.7%) but not the activator-only UAS-dFezfVP16 version (2.2%) (Fig. 4 C and D). These findings, which are consistent with the aforementioned ATAC-seq and RNA-seq findings, reveal that dFezf functions as a transcriptional repressor to control L3 broad domain specificity.
DFezf Cell-Autonomously Represses slp1 Expression to Direct L3 Broad Domain Specificity.
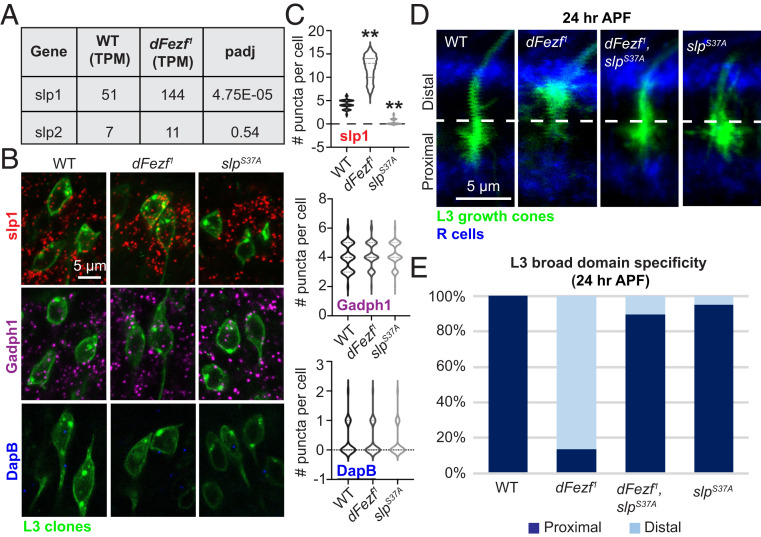
We next sought to identify downstream dFezf target(s) responsible for regulating L3 broad domain specificity. We focused on cell surface genes and TFs. Two complementary approaches interrogated genes up-regulated in dFezf1 mutant L3 neurons. In one approach, we misexpressed individual genes in WT L3 neurons using UAS constructs that were either newly generated or obtained from databases or other laboratories. In a second approach we used RNA interference (RNAi) or available null mutant alleles to ablate up-regulated genes in dFezf1 L3 neurons in an attempt to rescue L3 broad domain specificity. To examine down-regulated genes, we used RNAi and CRISPR/Cas9 (22) to ablate individual genes or combinations of genes that we suspected may function redundantly (e.g., if genes were members of the same gene family or were previously shown to interact with the same factors). We screened 42 out of 258 up-regulated DE genes and 38 out of 145 down-regulated DE genes (SI Appendix, Tables S4–S6). While this genetic screening strategy to identify downstream dFezf target genes did not reveal relevant genes encoding cell surface proteins, it did reveal that up-regulation of the TF sloppy-paired 1 (slp1) in dFezf1 mutants is responsible for the altered broad domain specificity of dFezf1 mutant L3 neurons. Slp1 is a forkhead family TF that, together with the related TF slp2, regulates a wide variety of developmental processes, including embryonic segmentation, heart formation, and visual system cell fate specification (23–25). Our RNA-seq data indicate that slp1/2 are both expressed at a low level in WT L3 neurons at 24 h APF (Fig. 5A), and our previous work showed that these TFs are not expressed at 40 h APF (12). When dFezf was knocked out in L3 neurons, expression of slp1 was significantly up-regulated, while slp2 expression was unchanged (Fig. 5A).
Fig. 5.
DFezf directs L3 broad domain specificity by repressing expression of slp1. We performed MARCM rescue experiments with null alleles for dFezf (dFezf1) and slp1/2 (slpS37A). (A) Chart indicating slp1 and slp2 expression levels in WT and dFezf1 mutant L3 neurons at 24 h APF from the dataset in Fig. 2. Both genes are lowly expressed in L3 neurons at this stage. Slp1 is significantly up-regulated when dFezf is knocked out. TPM, transcripts per million. (B) Confocal images illustrating RNA transcript expression in L3 MARCM clones. L3 cell bodies are labeled with GFP (green). Each row of images shows a different transcript: Slp1 (red), positive control Gapdh1 (magenta), and negative control DapB (blue). (All scale bars, 5 µm.) (C) Quantification of the corresponding data in B. Slp1 expression is significantly up-regulated in dFezf1 mutant L3 neurons and completely lost is slpS37A mutant L3 neurons. A two-tailed Student’s t test was performed. **P < 0.01. (D) Confocal images of L3 growth cone targeting at 24 h APF in MARCM experiments. L3 growth cones are labeled with GFP (green). R cells are labeled in blue (mAB24B10). DFezf1, slpS37A double-mutant L3 growth cones properly target the proximal domain. SlpS37A mutant L3 neurons do not exhibit growth cone-targeting defects. (E) Quantification of data in D.
To confirm a critical regulatory role of dFezf in controlling slp1 expression in L3 neurons, we next assessed slp1 mRNA levels by RNA in situ hybridization (RNAscope) in WT and dFezf1 L3 neurons. SlpS37A, a null allele in which both the slp1 and slp2 loci are deleted (26), was integrated into our dFezf1 MARCM strategy and used as a control to verify probe specificity (see below). To examine slp1 mRNA transcript levels, we first generated a GAL4 driver to label L3 neurons, as the previously used 9-9-GAL4 was not suitable for this experiment (see Materials and Methods and SI Appendix, Fig. S1). We modified the original dFezf BAC by replacing the majority of the dFezf coding region (all except the sequence encoding the first three amino acids) with that of GAL4 (SI Appendix, Fig. S1A), resulting in a GAL4 driver that is regulated by the endogenous regulatory sequence elements of dFezf (hereby named dFezf-GAL4). DFezf-GAL4 is only expressed in L3 neurons because dFezf is not expressed in any other lamina neuron type (SI Appendix, Fig. S1B).
We used the dFezf-GAL4 driver for RNAscope experiments (27) to assess slp1 mRNA levels in WT, dFezf1, and slpS37A L3 neurons. The findings confirmed that slp1 is indeed expressed in WT L3 neurons, with an average of 4.20 mRNA puncta detected per cell (n = 287; Fig. 5 B and C). In contrast, dFezf1 L3 neurons exhibited significantly up-regulated expression of slp1 (∼11.98 puncta per cell; n = 290), whereas slpS37A mutant L3 neurons, used as a control for probe specificity because slpS37A mutants lack both the slp1 and slp2 loci (26), showed only a background level of slp1 mRNA (∼0.39 puncta per cell; n = 308). Together with our RNA-seq data, these RNAscope findings indicate that dFezf functions cell-autonomously in L3 neurons to repress expression of slp1.
To determine if slp1 up-regulation is indeed responsible for the mistargeting phenotype observed in dFezf1 L3 neurons, we used MARCM to examine growth cone targeting of dFezf1, slpS37A double-mutant L3 neurons at 24 h APF. If aberrant slp1 expression in dFezf1 neurons is responsible for mistargeting of dFezf1 L3 neurons, then dFezf1, slpS37A double-mutant L3 growth cones should properly target the proximal domain. Indeed, the mistargeting phenotype of dFezf1 L3 growth cones was fully rescued by deletion of the slp1/2 genes (Fig. 5 D and E). Moreover, slpS37A single-mutant L3 growth cones also properly target the proximal domain (Fig. 5 D and E), indicating that the low expression level of slp1/2 observed in WT L3 neurons (Fig. 5A) is not required for L3 broad domain specificity. These findings support a model in which dFezf regulates L3 broad domain specificity through transcriptional repression of slp1.
Slp1/2 Are Required for L3 Growth Cone Distal Retraction.
After L3 growth cones reach the proximal domain, they segregate from other lamina neuron growth cones and innervate the developing M3 layer. This process involves distal retraction from the proximal edge of the outer medulla, and lateral extension within the developing M3 layer (Fig. 6A) (10). Since slp1/2 are expressed at low levels in WT L3 neurons at 24 h APF (Fig. 5A), we hypothesized that these TFs may play a role in M3 layer innervation by the L3 neuron. As slp1/2 are not necessary for L3 broad domain specificity (Fig. 5 D and E), we used the slpS37A allele to examine a putative role for early slp1/2 expression for subsequent steps of L3 growth cone targeting.
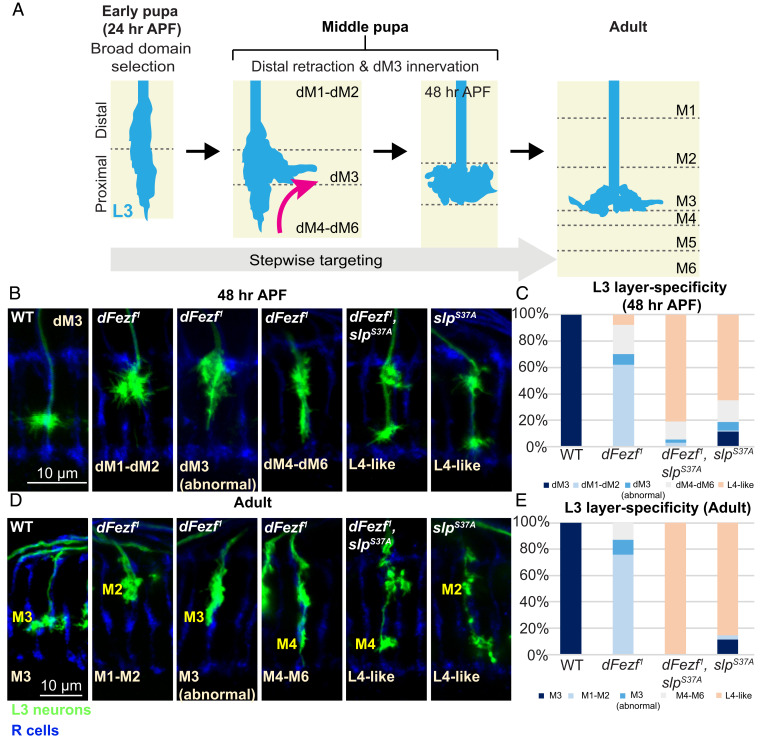
Fig. 6.
Slp1/2 are required for innervation of the M3 layer. In MARCM experiments, we examined L3 morphology and layer specificity during midpupal development (48 h APF) and adults. (A) Schematic illustrating stepwise innervation of the M3 layer by the L3 neuron. During early pupal development (24 h APF), dFezf directs L3 growth cones to the proximal domain (12). L3 neurons then undergo a distal retraction in which the growth cone retracts away from the proximal edge of the outer medulla and extends laterally (magenta arrow). This leads to confinement of the L3 growth cone within the developing M3 layer (dM3) and ultimately the M3-specific position of the L3 terminal in the adult. (B–E) Confocal images and quantification of L3 layer specificity in MARCM experiments at 48 h APF (B and C) and in the adult (D and E). L3 neurons are labeled with GFP (green). R cells are labeled in blue (mAB24B10). (B) At this stage, WT L3 growth cones are confined to the M3 layer. DFezf1 mutant L3 neurons predominantly innervate the distal neuropil (M1 and M2 developing layers). L3 neurons harboring the slpS37A mutation innervate more proximally than normal, suggesting a deficit in retraction. (C) Quantification of data from B. (D) L3 neuron-layer specificity observed in B is sustained in the adult stage. (E) Quantification of data from D.
We used MARCM to examine slpS37A mutant L3 growth cones at the time following broad domain selection. By midpupal development (48 h APF), the broad domains observed at 24 h APF have developed into the nascent layers of the outer medulla (Fig. 6A). By this stage, WT L3 terminals are confined to the M3 layer, while the majority of dFezf1 L3 growth cones terminate within the distal layers (M1 and M2), and a smaller population extend within the M4 to M6 layers (Fig. 6 B and C). These results are consistent with our previous findings (12). Surprisingly, the majority of slpS37A mutant L3 neurons exhibit an axon terminal extending and segregating into the M4 to M5 layers along with a second branch that invades the M1 and M2 layers (Fig. 6 B and C). The dFezf1, slpS37A double-mutant L3 neurons exhibited a similar targeting phenotype to the slpS37A mutant (Fig. 6 B and C). This targeting defect observed in slpS37A and dFezf1, slpS37A double mutants at 48 h APF persists into the adult stage (Fig. 6 D and E). Since the mutant allele used in this experiment is null for both slp1 and slp2, we cannot distinguish whether slp1, slp2, or both are involved in regulating L3 distal retraction during midpupal development. Nevertheless, these findings indicate that although slp1/2 are dispensable for the initial broad domain specificity of L3 neurons (Fig. 5 D and E), the early expression of at least one of these TFs is critical at a later stage for L3 growth cone innervation of the nascent M3 layer.
It is noteworthy that while the morphology of slpS37A and dFezf1, slpS37A mutant L3 neurons at 48 h APF are unique, the adult cells resemble the homologous L4 lamina neurons (28, 29). Therefore, to rule out the possibility that deleting slp1/2 unexpectedly causes activation of the dFezf-GAL4 driver in L4 neurons (and thus labeling those cells in the MARCM strategy), we used an antibody against dFezf to confirm the identity of the MARCM clones. Since the dFezf1 allele is a point mutation in the dFezf locus, the product of this mutant allele is a functionally defective protein that can be recognized by the dFezf antibody. The presence of dFezf antibody staining in cell bodies of the majority of the L3 MARCM clones confirmed that the cells labeled in this experiment are indeed L3 neurons (SI Appendix, Fig. S2). A small population of slpS37A and dFezf1, slpS37A double-mutant L3 neurons did not exhibit dFezf antibody staining (6.25 and 8.59%, respectively; SI Appendix, Fig. S2). Whether these few clones lacking dFezf antibody staining were L4 neurons is unclear. Nevertheless, these findings indicate that the majority of slpS37A single-mutant and dFezf1, slpS37A double-mutant neurons are indeed L3 neurons which have adopted an “L4-like” morphology (64.95 and 80.85%, respectively; Fig. 6 B and C). Therefore, dFezf functions in L3 neurons to control growth cone broad domain specificity through transcriptional repression of slp1 at an intermediate developmental stage, while earlier expression of slp1, slp2, or both, during a time prior to dFezf-mediated repression, mediates L3 distal growth cone retraction at a later, midpupal stage of development. Thus, precise temporal fidelity of slp gene transcription, coordinated by the transcriptional repressor dFezf, mediates distinct stages of L3 neuron targeting in the medulla neuropil.
Discussion
In this study, we show that transcription factor dFezf functions as a transcriptional repressor to coordinate a gene expression cascade that directs L3 broad domain specificity. DFezf cell-autonomously directs L3 growth cones to the proximal domain through transcriptional repression of slp1. Whether dFezf controls slp1 transcription directly or indirectly through intermediate TFs is presently unclear. Our ATAC-seq analysis did not reveal dFezf binding sites within 2 kb of the slp1 TSS, which may reflect an absence of dFezf binding to this region or a technical limitation of the assay. On the other hand, examination of the slp1 genomic region within 100 kb of the slp1 TSS revealed four putative dFezf binding sites, one of which resides within an ATAC-seq peak. Therefore, dFezf may bind to a slp1 distal enhancer to directly repress slp1 transcription. In vivo assays such as chromatin immunoprecipitation followed by sequencing (ChIP-seq) will be needed to determine whether dFezf directly binds to slp1 genomic enhancer regions in L3 neurons. It is also noteworthy that the vertebrate orthologs of dFezf (i.e., FEZF1/2) have been implicated in both activation and repression of gene expression (30–32). Interestingly, FEZF1 has recently been shown to direct layer-specific innervation in the mouse retina (33). Moving forward, it will be of interest to determine if Fezf family orthologs regulate layer assembly through conserved transcriptional cascades.
Which are the relevant genes downstream of dFezf and slp1 that determine L3 broad domain specificity? The most intuitive model is one in which dFezf-mediated repression of slp1 indirectly controls expression of cell surface molecules that mediate cell–cell interactions. As L3 neurons express over 300 genes encoding cell surface proteins (11), there may be functional redundancy among these genes in regulating broad domain specificity. This potential redundancy may account for our inability to identify relevant cell surface genes in our genetic screens. One caveat in our experimental approach is that without specific markers (e.g., specific antibodies), we were unable to verify altered expression levels of candidate genes following the various genetic perturbations. We attempted to circumvent this by testing multiple different RNAi constructs for the same gene in parallel, using multiple gRNA fragments to increase knockout efficiency of our CRISPR/Cas9 strategy and testing different UAS-cDNA constructs (SI Appendix, Tables S4–S6). However, none of these genetic manipulations of cell surface protein encoding genes recapitulated or rescued the dFezf1 mistargeting phenotype in L3 neurons. Our finding that slp1 is a downstream TF that governs L3 broad domain specificity suggests that identifying slp1 target genes will help guide efforts toward defining the relevant cell surface genes. In fact, putative slp1 binding sites are observed within 2 kb of the TSS of 42 of the genes screened in this study (SI Appendix, Table S7). Future work aimed at identifying relevant slp1 target genes will shed light on the molecular logic underlying layer-specific circuitry.
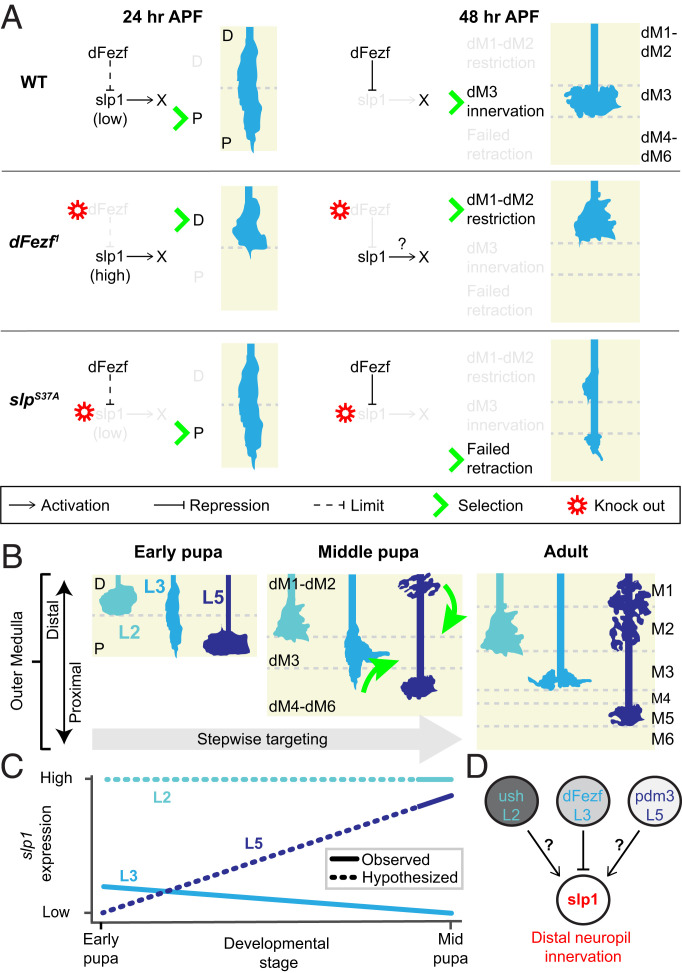
An unexpected finding from our study was that early expression of slp1 and/or slp2 plays a crucial role in L3 growth cone distal retraction, the step following broad domain selection (Fig. 7A). During this step, the L3 growth cone retracts distally and becomes confined to the nascent M3 layer. SlpS37A mutant L3 neurons, which lack both slp1/2 loci, exhibit a morphology that may reflect a retraction deficit (Fig. 6 B and D), suggesting that slp1/2 are necessary for distal retraction of the L3 neuron. Slp1/2 presumably accomplish this through the regulation of a downstream genetic program that is inconsequential during broad domain selection, but functionally relevant during M3 layer innervation after slp1/2 are repressed by dFezf.
Fig. 7.
Model for a transcriptional cascade regulating stepwise layer innervation. Schematics depicting hypothesized dFezf-slp1 cascade in L3 neurons and hypothesized function of slp1 function in other lamina neurons. (A) Observed L3-targeting defects in dFezf1 and slpS37A MARCM experiments. In WT L3 neurons (Top), dFezf limits slp1 expression, which directs L3 growth cones to the proximal domain (P) at 24 h APF. L3 growth cones are confined to the developing M3 layer (dM3) by 48 h APF. We hypothesize that a gene program downstream of slp1 (X) is necessary for L3 innervation of the M3 layer by 48 h APF (see below). When dFezf is knocked out (Middle), L3 growth cones are restricted to the distal domain (D) and terminate in distal layers (dM1 and dM2). When slp1 is knocked out (Bottom), L3 growth cones properly target the proximal domain, but fail to retract due to misregulation of gene program X. The possible role of slp2 in distal retraction is not depicted here (see Discussion). (B) Stepwise targeting of L2, L3, and L5 lamina neurons. During early pupal development (Left) lamina neurons target the distal (D) or proximal (P) domain. During midpupal development (Middle), lamina neurons either remain where initially positioned (e.g., L2) or undergo morphological changes (magenta arrows) as they innervate developing layers (dM1 to dM6). This process spans several hours, and the cartoons here are a general depiction of each transition. Ultimately, each lamina neuron establishes a unique arborization pattern throughout the outer medulla (Right). (C) Line graph illustrating observed and hypothesized changes in slp1 expression. We hypothesize that regulation of slp1 expression levels underlies broad domain specificity and morphology changes (depicted in B) of each lamina neuron. Solid lines represent observed slp1 expression pattern, while dashed lines represent hypothesized slp1 expression. Slp1 expression in lamina neurons is known from Tan et al. (11). Early pupal expression of slp1 expression in L3 neurons is presented here. Early pupal expression of slp1 in L2 was observed in M.Y.P.’s laboratory. (D) We speculate that slp1 may act downstream of each cell-specific TF as it does for dFezf.
A Temporally Precise Transcriptional Cascade and Lamina Neuron Target Selection.
Each lamina neuron arborizes in a unique manner within different neuropil regions along the distal-to-proximal axis. Lamina neurons first reach the primordial distal or proximal domains, thereafter arborizing within specific emerging layers during medulla expansion (Figs. 1 B and C and 7B). We envision that temporally regulated transcriptional cascades direct this modularized strategy of layer assembly. Although the processes of lamina neuron subtypes have distinct shapes, they exhibit partial overlap in their arborization patterns and laminar organization. For example, L2 and L5 neurons both innervate the nascent M2 layer (Fig. 7B). We hypothesize that lamina neurons achieve overlapping patterns of axon arborization via common or overlapping combinations of intermediate TFs that function downstream of cell type-specific TFs, such as dFezf (Fig. 7D) (11, 12). Our findings lead us to speculate that slp1 is one such intermediate TF that promotes innervation of the distal neuropil of multiple lamina neuron types. Consistent with this idea, during midpupal development, slp1 is normally highly expressed in L2 and L5 neurons, both of which arborize within the M2 layer (11, 12). Moreover, we previously found that dFezf misexpression in L5 neurons eliminates their M1 and M2 arbors (12), which we now speculate is due to dFezf-mediated repression of slp1. It will be interesting in future experiments to determine whether slp1 is necessary for L2 and L5 distal arborization patterns and whether changes in the levels of slp1 expression coincide with innervation of distal layers by these neurons (Fig. 7C).
In summary, our findings indicate that a unique and temporally precise transcriptional cascade orchestrates the stepwise nature of the L3 neuron’s laminar targeting and indeed the precision with which L3 synapse specificity within the medulla is achieved. We propose that each of the five lamina neuron subtypes utilizes a distinct transcriptional cascade, involving overlapping sets of TFs, to establish a unique molecular framework that coordinates their layer-specific patterns of connectivity within the medulla.
Materials and Methods
Fly Stocks and Husbandry.
All fly lines were raised at 25 °C and 60% relative humidity. The resources of fly stocks used in this study are listed in SI Appendix, Table S8. Genotypes of flies for each figure are listed in SI Appendix, Table S9. At least four brains for each genotype were examined for all the experiments.
Construction of 18B02 dFezf-GAL4.
The BAC with endogenous dFezf locus CH321-18B02 (BACPAC Resources) was modified using the recombineering protocol described previously (34). The entire coding region of dFezf (except the short sequence encoding the first three amino acids) was replaced by that of GAL4 (SI Appendix, Fig. S1A). The resulting DNA construct was inserted into the VK33 genomic site, resulting in the 18B02 dFezf-GAL4 strain.
Generation of CRISPR-Cas9 Knockout Flies.
gRNAs sequences are designed and evaluated using “DRSC Find CRISPRs” (https://www.flyrnai.org/crispr2/) and “flyCRISPR” (https://flycrispr.org/). For each gene, three gRNAs were selected to target genomic sequences that are near the translational start site and are conserved among fly species (https://evoprinter.ninds.nih.gov). Multiple gRNAs are assembled into pCFD6 plasmids (no. 73915; Addgene) using Gibson Assembly (E5510; New England BioLabs) to target single or a combination of genes. All gRNA constructs are inserted into attP40 site except for drp6 and dpr2 (VK14 site). Transgenic flies containing UAS-gRNAs allele were crossed with 9B08-Gal4/UAS-Cas9 flies to generate conditional knockout flies.
Immunohistochemistry and Microscopy.
Immunohistochemistry was performed as described previously (12). Fluorescence images were acquired using Zeiss LSM 800 Confocal Microscope and processed with Fiji ImageJ imaging software.
The primary antibodies used were as follows: chicken anti-GFP (1:1,000) (ab13970; Abcam), rabbit anti-HA (1:1,000) (3724S; Cell Signaling Technologies), rabbit anti-DsRed (1:200) (632496; Takara), and mouse anti-chaoptin (1:20) (24B10; Developmental Studies Hybridoma Bank). Rat anti-dFezf (1:50) was a gift from Dr. Claude Desplan (New York University, New York, NY).
The secondary antibodies used were as follows: Goat anti-chicken immune globulin (Ig) G (H + L) Alexa Fluor 488 (A-11039), goat anti-rabbit IgG (H + L) Alexa Fluor 568 (A-11011), goat anti-mouse IgG (H + L) Alexa Fluor 647 (A-21236), and goat anti-rat IgG (H + L) Alexa Fluor 568 (A-11077) were purchased from Thermo Fischer Scientific and used at a dilution of 1:500.
Assessment of Broad Domain Position.
Growth cone position within the outer medulla at 24 h APF was assessed based on previous characterization of the proximal and distal domains. As in previous studies, the lines drawn in this report to divide the outer medulla neuropil are based on the highly stereotyped positions of lamina neuron growth cones at this stage. L2 and L4 neurons are restricted to the distal half, while L1 and L5 are confined to the proximal half (10, 12). L3 broad domain specificity is a binary quality: Either the growth cone reaches the proximal domain or the bulk of the growth cone is restricted to the distal domain.
RNA-Seq Experiments.
The RNA-seq experiments were performed as described previously (12), except that the pupae were dissected at 24 h APF. Total RNA was purified with Qiagen RNeasy kit and used to make cDNA library by Smart-seq2 as previously described (35). The pooled libraries were sequenced in a single flow cell by NextSeq High Output.
ATAC-Seq.
The staging of pupae, dissection of fly brains, and cell dissociation were performed similarly as in the RNA-seq experiments. Upon dissociation, GFP-labeled L3 neurons were FACS sorted directly into 50 µL of lysis buffer (10 mM tris(hydroxymethyl)aminomethane hydrochloride (Tris⋅HCl) [pH 7.4], 10 mM NaCl, 3 mM MgCl2, and 0.1% octylphenoxy poly(ethyleneoxy)ethanol [IGEPAL CA-630]). Immediately after lysis, the nuclei were spun down at 800 × g for 10 min at 4 °C, and the supernatant was carefully removed. The pellet was resuspended in the transposase reaction mixture (25 µL of 2× Tagment DNA (TD) buffer [Illumina Nextera DNA Library Prep kit], 2.5 µL of Transposase [Illumina Nextera DNA Library Prep kit] and 22.5 µL of nuclease-free water) and incubated at 37 °C for 30 min. Following the transposition, the DNA sample was purified with a Qiagen MinElute kit. PCR amplification was performed in 50 µL of reaction (25 µL of 2× NEBnext PCR master mix; 5 µL each of Index 1 and Index 2 of choice [Illumina Nextera XT Index kit] and 15 µL of the purified DNA sample) using the following condition: 72 °C for 5 min, 98 °C for 30 s, followed by 12 cycles of 98 °C for 10 s, 63 °C for 30 s, and 72 °C for 1 min. The PCR product was purified with a Qiagen MinElute kit. A size-selection step was added by running the DNA samples in a 1% agarose gel together with a 100-base pair (bp) DNA ladder (New England BioLabs) and recovering the gel fragment between 100 bp and 1 kb. DNA was purified by gel extraction with Qiagen QG buffer and MinElute columns. DNA concentration was quantified by Qubit and sequenced by NextSeq High Output.
Sequencing Data Analysis.
RNA-seq.
All samples were processed using an RNA-seq pipeline implemented in the bcbio-nextgen project (https://bcbio-nextgen.readthedocs.org/en/latest/). Raw reads were examined for quality issues using FastQC (http://www.bioinformatics.babraham.ac.uk/projects/fastqc/) to ensure library generation and sequencing data were suitable for further analysis. Reads were aligned to the Ensembl build BDGP6 of the D. melanogaster genome using Spliced Transcripts Alignment to a Reference (STAR) (36). Quality of alignments was assessed by checking for evenness of coverage, ribosomal RNA content, genomic context of alignments, complexity, and other quality checks. Expression quantification was performed with Salmon (37) to identify transcript-level abundance estimates and then collapsed down to the gene level using the R Bioconductor package tximport (38). Differential expression was performed at the gene level using the R Bioconductor package DESeq2 (39). Default parameters were used with the exception of the Cook’s distance filter, which was turned off. Significant genes were identified using a false discovery rate (FDR) threshold of 0.05.
ATAC-seq.
Raw sequence reads were evaluated for quality using FASTQC. Reads were filtered and trimmed with Atropos (40). High quality reads were mapped to the Ensembl build BDGP6 of the D. melanogaster genome using Bowtie2 (41). To call the peaks we used MACS2 v2.1.1 (42) with parameters “-g dm–nomodel–nolambda–keep-dup all,” and peaks were filtered based on default settings and FDR < 0.05. Peaks were annotated to the nearest genes and to genomic elements such as TSSs, transcription termination sites, exons, and introns using the R Bioconductor package ChIPseeker (43). DNA sequences corresponding to ATAC-seq peaks located proximal to differentially expressed genes were scanned for matches to the dFezf/erm DNA-binding motif (CisBP ID M06129_2.00) (44) using the standard log likelihood scoring system (45) and a cutoff of 15.29 bits, which corresponds to 70% of the best possible score for that motif.
RNAscope.
Fly brains were dissected in cold Schneider’s medium and fixed in 4% paraformaldehyde in phosphate buffer saline (PBS) overnight at room temperature. After fixation, brains were washed for three times with phosphate-buffered saline (PBS) plus 0.1% Tween-20 (PBST) at room temperature for 10 min each. PBST was then replaced with PBS. Fixed brains were pretreated by incubation in Protease III (322340; ACDbio) for 5 min at room temperature, followed by rinsing for three times with PBS for 2 min each. RNAscope probes were preheated at 40 °C, and brains were incubated in probe solution overnight at 40 °C. All the subsequent washes were done with 1× Wash Buffer (diluted from 50× Wash Buffer stock; 310091; ACDbio) for three times and 5 min each. After probe incubation, brains were washed and subjected to amplification at 40 °C by consecutively incubating in the RNAscope Fluorescent Multiplex Detection Reagents (320851; ACDbio): Amp 1-FL for 30 min, Amp 2-FL for 15 min, Amp 3-FL for 30 min, and Amp 4-FL solution for 15 min, each followed by washing with 1× Wash Buffer. After amplification, brains were rinsed with PBS and mounted immediately in SlowFade mounting media. Confocal microscopy was used to acquire fluorescent images, which were then processed with ImageJ software; 287 to 308 cells were analyzed for each genotype in a blinded manner. The probes used in this study were either made to order or purchased as catalog probes from ACDbio, including RNAscope Probe-Dm-Slp1 (435501), RNAscope Probe-Dm-Gapdh1-C2 (made to order), and RNAscope 3-plex Negative Control Probe (320871).
Supplementary Material
Acknowledgments
This research was supported by NIH Grants R01 NS103905 (to M.Y.P.) and R01 NS099068 (to M.T.W.), the Lefler Center for the Study of Neurodegenerative Disorders (J.P. and D.D.G.), and HHMI (Gilliam Fellowship for Advanced Study [I.J.S.]). We thank M. Greenberg, A. Handler, C. Santiago, T. Schwarz, and T. Tan for valuable discussions and comments on the paper. D.D.G. is an investigator of HHMI. Finally, we acknowledge the late M.Y.P., who led us in this study. Matt’s passion for science was limitless and exhilarating. He gave us his best and inspired ours. Thank you, Matt.
Footnotes
The authors declare no competing interest.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025530118/-/DCSupplemental.
Data Availability
All raw data for RNA-seq and ATAC-seq have been deposited in the Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE163311) (47).
References
- 1.Wang I. E., Clandinin T. R., The influence of wiring economy on nervous system evolution. Curr. Biol. 26, R1101–R1108 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Baier H., Synaptic laminae in the visual system: Molecular mechanisms forming layers of perception. Annu. Rev. Cell Dev. Biol. 29, 385–416 (2013). [DOI] [PubMed] [Google Scholar]
- 3.Azeredo da Silveira R., Roska B., Cell types, circuits, computation. Curr. Opin. Neurobiol. 21, 664–671 (2011). [DOI] [PubMed] [Google Scholar]
- 4.Clark D. A., Demb J. B., Parallel computations in insect and mammalian visual motion processing. Curr. Biol. 26, R1062–R1072 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nassi J. J., Callaway E. M., Parallel processing strategies of the primate visual system. Nat. Rev. Neurosci. 10, 360–372 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Roska B., Werblin F., Vertical interactions across ten parallel, stacked representations in the mammalian retina. Nature 410, 583–587 (2001). [DOI] [PubMed] [Google Scholar]
- 7.Greig L. C., Woodworth M. B., Galazo M. J., Padmanabhan H., Macklis J. D., Molecular logic of neocortical projection neuron specification, development and diversity. Nat. Rev. Neurosci. 14, 755–769 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Todd A. J., Neuronal circuitry for pain processing in the dorsal horn. Nat. Rev. Neurosci. 11, 823–836 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sanes J. R., Zipursky S. L., Design principles of insect and vertebrate visual systems. Neuron 66, 15–36 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pecot M. Y., et al., Multiple interactions control synaptic layer specificity in the Drosophila visual system. Neuron 77, 299–310 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tan L., et al., Ig superfamily ligand and receptor pairs expressed in synaptic partners in Drosophila. Cell 163, 1756–1769 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peng J., et al., Drosophila Fezf coordinates laminar-specific connectivity through cell-intrinsic and cell-extrinsic mechanisms. eLife 7, e33962 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee T., Luo L., Mosaic analysis with a repressible cell marker for studies of gene function in neuronal morphogenesis. Neuron 22, 451–461 (1999). [DOI] [PubMed] [Google Scholar]
- 14.Lyne R., et al., FlyMine: An integrated database for Drosophila and Anopheles genomics. Genome Biol. 8, R129 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhu L. J., et al., FlyFactorSurvey: A database of Drosophila transcription factor binding specificities determined using the bacterial one-hybrid system. Nucleic Acids Res. 39, D111–D117 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tolkunova E. N., Fujioka M., Kobayashi M., Deka D., Jaynes J. B., Two distinct types of repression domain in engrailed: One interacts with the groucho corepressor and is preferentially active on integrated target genes. Mol. Cell. Biol. 18, 2804–2814 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Knust E., Tietze K., Campos-Ortega J. A., Molecular analysis of the neurogenic locus enhancer of split of Drosophila melanogaster. EMBO J. 6, 4113–4123 (1987). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goldstein R. E., et al., An eh1-like motif in odd-skipped mediates recruitment of Groucho and repression in vivo. Mol. Cell. Biol. 25, 10711–10720 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Razin S. V., Borunova V. V., Maksimenko O. G., Kantidze O. L., Cys2His2 zinc finger protein family: Classification, functions, and major members. Biochemistry (Mosc.) 77, 217–226 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Janssens D. H., et al., Earmuff restricts progenitor cell potential by attenuating the competence to respond to self-renewal factors. Development 141, 1036–1046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Port F., Bullock S. L., Augmenting CRISPR applications in Drosophila with tRNA-flanked sgRNAs. Nat. Methods 13, 852–854 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grossniklaus U., Pearson R. K., Gehring W. J., The Drosophila sloppy paired locus encodes two proteins involved in segmentation that show homology to mammalian transcription factors. Genes Dev. 6, 1030–1051 (1992). [DOI] [PubMed] [Google Scholar]
- 24.Park M., Wu X., Golden K., Axelrod J. D., Bodmer R., The wingless signaling pathway is directly involved in Drosophila heart development. Dev. Biol. 177, 104–116 (1996). [DOI] [PubMed] [Google Scholar]
- 25.Suzuki T., Kaido M., Takayama R., Sato M., A temporal mechanism that produces neuronal diversity in the Drosophila visual center. Dev. Biol. 380, 12–24 (2013). [DOI] [PubMed] [Google Scholar]
- 26.Sato A., Tomlinson A., Dorsal-ventral midline signaling in the developing Drosophila eye. Development 134, 659–667 (2007). [DOI] [PubMed] [Google Scholar]
- 27.Wang F., et al., RNAscope: A novel in situ RNA analysis platform for formalin-fixed, paraffin-embedded tissues. J. Mol. Diagn. 14, 22–29 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Fischbach K. F., Dittrich A. P. M., The optic lobe of Drosophila melanogaster. I. A Golgi analysis of wild-type structure. Cell Tissue Res. 258, 441–475 (1989). [Google Scholar]
- 29.Nern A., Zhu Y., Zipursky S. L., Local N-cadherin interactions mediate distinct steps in the targeting of lamina neurons. Neuron 58, 34–41 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hirata T., et al., Zinc-finger genes Fez and Fez-like function in the establishment of diencephalon subdivisions. Development 133, 3993–4004 (2006). [DOI] [PubMed] [Google Scholar]
- 31.Weng M., Golden K. L., Lee C. Y., dFezf/Earmuff maintains the restricted developmental potential of intermediate neural progenitors in Drosophila. Dev. Cell 18, 126–135 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang N., Dong Z., Guo S., Fezf2 regulates multilineage neuronal differentiation through activating basic helix-loop-helix and homeodomain genes in the zebrafish ventral forebrain. J. Neurosci. 32, 10940–10948 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Peng Y. R., et al., Binary fate choice between closely related interneuronal types is determined by a Fezf1-dependent postmitotic transcriptional switch. Neuron 105, 464–474.e6 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sharan S. K., Thomason L. C., Kuznetsov S. G., Court D. L., Recombineering: A homologous recombination-based method of genetic engineering. Nat. Protoc. 4, 206–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Picelli S., et al., Full-length RNA-seq from single cells using Smart-seq2. Nat. Protoc. 9, 171–181 (2014). [DOI] [PubMed] [Google Scholar]
- 36.Dobin A., et al., STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Patro R., Duggal G., Love M. I., Irizarry R. A., Kingsford C., Salmon provides fast and bias-aware quantification of transcript expression. Nat. Methods 14, 417–419 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Soneson C., Love M. I., Robinson M. D., Differential analyses for RNA-seq: Transcript-level estimates improve gene-level inferences. F1000 Res. 4, 1521 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Love M. I., Huber W., Anders S., Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Didion J. P., Martin M., Collins F. S., Atropos: Specific, sensitive, and speedy trimming of sequencing reads. PeerJ 5, e3720 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y., et al., Model-based analysis of ChIP-seq (MACS). Genome Biol. 9, R137 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Yu G., Wang L. G., He Q. Y., ChIPseeker: An R/bioconductor package for ChIP peak annotation, comparison and visualization. Bioinformatics 31, 2382–2383 (2015). [DOI] [PubMed] [Google Scholar]
- 44.Lambert S. A., et al., Similarity regression predicts evolution of transcription factor sequence specificity. Nat. Genet. 51, 981–989 (2019). [DOI] [PubMed] [Google Scholar]
- 45.Stormo G. D., DNA binding sites: Representation and discovery. Bioinformatics 16, 16–23 (2000). [DOI] [PubMed] [Google Scholar]
- 46.Kurusu M., et al., A screen of cell-surface molecules identifies leucine-rich repeat proteins as key mediators of synaptic target selection. Neuron 59, 972–985 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Peng J., Drosophila Fezf functions as a transcriptional repressor to direct layer specific synaptic connectivity in the fly visual system. Gene Expression Omnibus (GEO). https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE163311. Deposited 16 December 2020. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All raw data for RNA-seq and ATAC-seq have been deposited in the Gene Expression Omnibus, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE163311) (47).