How autophagy in neurons is regulated by synaptic activity is largely unknown. Kulkarni et al. show that synaptic activity decreases the motility and increases the degradative function of autophagic vacuoles within dendrites but not axons.
Abstract
Macroautophagy (hereafter “autophagy”) is a lysosomal degradation pathway that is important for learning and memory, suggesting critical roles for autophagy at the neuronal synapse. Little is known, however, about the molecular details of how autophagy is regulated with synaptic activity. Here, we used live-cell confocal microscopy to define the autophagy pathway in primary hippocampal neurons under various paradigms of synaptic activity. We found that synaptic activity regulates the motility of autophagic vacuoles (AVs) in dendrites. Stimulation of synaptic activity dampens AV motility, whereas silencing synaptic activity induces AV motility. Activity-dependent effects on dendritic AV motility are local and reversible. Importantly, these effects are compartment specific, occurring in dendrites and not in axons. Most strikingly, synaptic activity increases the presence of degradative autolysosomes in dendrites and not in axons. On the basis of our findings, we propose a model whereby synaptic activity locally controls AV dynamics and function within dendrites that may regulate the synaptic proteome.
Introduction
Macroautophagy (hereafter “autophagy”) is a lysosomal degradation pathway that is important for neuronal development and survival (Kulkarni and Maday, 2018; Yamamoto and Yue, 2014). During autophagy, cellular components (e.g., proteins and organelles) are initially sequestered and packaged into autophagosome organelles. Autophagosomes then fuse with lysosomes to generate autolysosomes that degrade autophagic contents via lysosomal hydrolases. Breakdown products such as amino acids and lipids are exported from lysosomes to fuel new biosynthesis. In this way, autophagy can control the integrity and composition of the proteome and organelles.
Loss of autophagy can have a profound impact during neurodevelopment. Knockout of autophagy genes leads to defects in axon outgrowth and guidance and to the failure to form interhemispheric tracts in the brain (Dragich et al., 2016; Kannan et al., 2017; Stavoe et al., 2016; Yamaguchi et al., 2018). Autophagy deficits can impair synapse formation and development-based spine pruning (Shen and Ganetzky, 2009; Stavoe et al., 2016; Tang et al., 2014). Loss of autophagy can also elicit degeneration of established neuronal networks. Central nervous system– and Purkinje cell–specific deletion of key autophagy genes leads to a characteristic swelling of presynaptic terminals, followed by axon retraction and decay (Hara et al., 2006; Komatsu et al., 2006; Komatsu et al., 2007; Liang et al., 2010; Nishiyama et al., 2007; Yamaguchi et al., 2018; Zhao et al., 2015). Combined, autophagy serves essential roles in establishing and maintaining the proper connectivity of the nervous system.
Neuronal synapses are sites of high demand for protein and organelle quality control pathways. Neurons fire action potentials at rates that can reach up to ∼50–100 impulses per second for nearly a century of time. This activity renders the synaptic proteome vulnerable to stress and damage. Furthermore, the composition of the synaptic proteome is dynamic and remodeled to shape neuronal circuits, a foundational principle underlying learning and memory. In fact, loss of autophagy has been reported to cause deficits in learning and memory (Glatigny et al., 2019; Hylin et al., 2018; Pandey et al., 2020; Zhao et al., 2015). Moreover, stimulating autophagy can rescue age-related cognitive decline (Glatigny et al., 2019). These findings point toward critical roles for autophagy at synapses. Little is known, however, about the molecular underpinnings of autophagy in synaptic compartments.
We previously demonstrated that autophagosomes exhibit unique transport dynamics in axons as compared with dendrites (Maday and Holzbaur, 2014; Maday and Holzbaur, 2016; Maday et al., 2012), suggesting that there may be compartment-specific regulation of autophagy in neurons. We, along with other groups, found that in primary neurons, axonal autophagy follows a pathway of long-range unidirectional transport of autophagosomes that originate in the distal axon and travel to the soma for maturation into degradative compartments (Cheng et al., 2015; Hollenbeck, 1993; Lee et al., 2011; Maday and Holzbaur, 2014; Maday and Holzbaur, 2016; Maday et al., 2012; Wang et al., 2015; Yue, 2007). This pathway for axonal autophagy is conserved in intact nervous systems in vivo (Hill et al., 2019; Jin et al., 2018; Neisch et al., 2017; Soukup et al., 2016; Sung et al., 2016). By contrast, dendritic autophagosomes exhibit bidirectional and stationary movement (Maday and Holzbaur, 2014; Maday and Holzbaur, 2016). How the dynamics of autophagic vacuoles (AVs; i.e., autophagosomes and autolysosomes) are influenced by the activity state of the neuron and how alterations in these dynamics impact autophagic function are largely unknown.
Here, we used live-cell confocal microscopy to define how autophagy in dendritic and axonal compartments is regulated by synaptic activity in primary hippocampal neurons. We found that synaptic activity regulates the motility of AVs in dendrites. Stimulating synaptic activity dampens AV motility, whereas silencing synaptic activity induces AV motility. Activity-dependent effects on dendritic AV motility are local and reversible. Importantly, these effects are observed specifically within dendrites and not within axons, indicating compartment-specific regulation of AV dynamics with synaptic activity. Most strikingly, synaptic activity increases the presence of degradative autolysosomes in dendrites and not in axons. Our data suggest that synaptic activity may induce the maturation of dendritic AVs into proteolytically competent organelles. On the basis of our findings, we propose a model where synaptic activity locally regulates AV dynamics and degradative function within dendrites.
Results
Neuronal depolarization dampens AV motility specifically in dendrites and not in axons
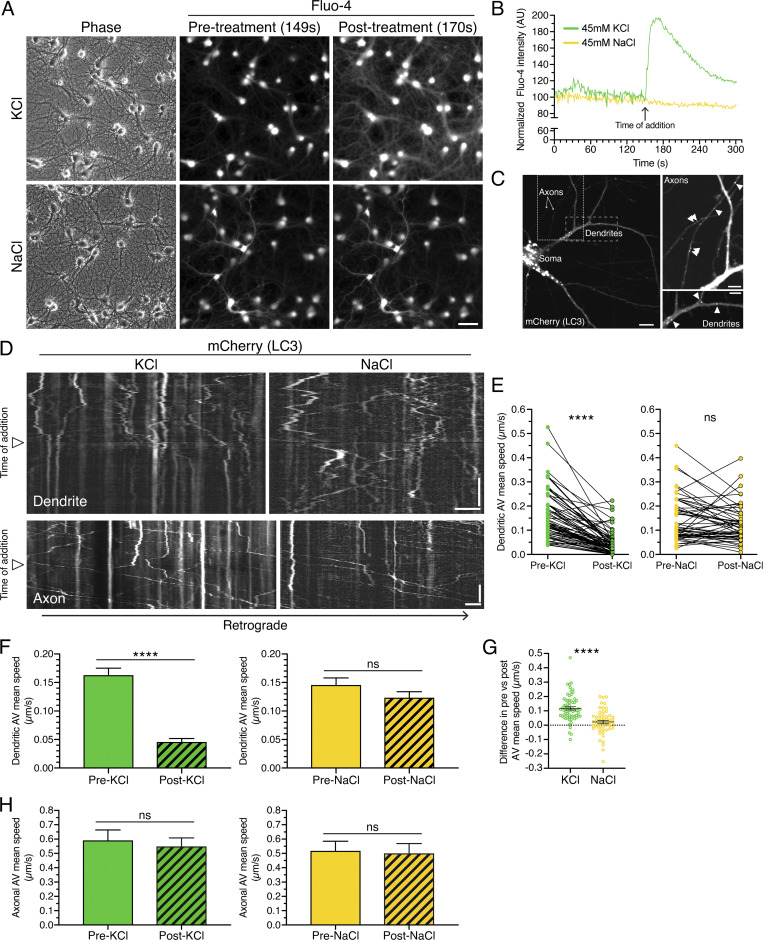
How autophagy is influenced by the activity state of the neuron is largely unknown. Here, we define how autophagy in dendritic and axonal compartments is regulated in response to synaptic activity in primary hippocampal neurons. To stimulate neuronal activity, we first treated rat hippocampal neurons with 45 mM KCl to induce depolarization (Wong et al., 2015). Neurons were loaded with 4 µM Fluo-4, a calcium-sensitive dye that fluoresces upon binding to calcium. We then performed live-cell imaging to measure intracellular calcium levels in real time. On-scope addition of KCl resulted in an immediate and dramatic increase in intracellular calcium, consistent with depolarization of the neurons (Fig. 1, A and B). As an osmotic control, we added 45 mM NaCl, which had no effect on calcium levels (Fig. 1, A and B). Thus, treatment with KCl, and not NaCl, results in depolarization of the neurons.
Figure 1.
Neuronal depolarization dampens AV motility specifically in dendrites and not in axons. (A) Rat hippocampal neurons (13 DIV) were loaded with the calcium indicator Fluo-4 and treated with KCl to depolarize the neurons or with NaCl as an osmotic control. Scale bar, 50 µm. (B) Quantitation of mean Fluo-4 fluorescence intensity before and after KCl or NaCl addition. (C) Distribution of mCherry-LC3–positive AVs in dendrites and axons of primary rat hippocampal neurons (13 DIV). Image with soma; scale bar, 10 µm. Magnified images of axons and dendrites with arrowheads denoting mCherry-LC3–positive puncta; scale bars, 5 µm. (D) Kymographs of LC3 motility along a dendrite or an axon of a rat hippocampal neuron. Retrograde direction is from left to right. Arrowheads denote time of KCl or NaCl addition. Horizontal bars, 5 µm. Vertical bars, 1 min. (E–G) Quantitation of AV mean speed in dendrites before and after KCl or NaCl addition (E, paired points represent a single AV before and after salt addition; F, mean ± SEM; G, difference in AV mean speed before and after salt addition; KCl, n = 68 AVs from 20 neurons from three independent experiments, 13–14 DIV; NaCl, n = 55 AVs from 20 neurons from three independent experiments, 13–14 DIV; E and F, paired t test; G, unpaired t test; ****, P ≤ 0.0001). (H) Quantitation of AV mean speed in axons before and after KCl or NaCl treatment (mean ± SEM; n = 56–125 AVs from 8–10 neurons from three independent experiments; 13–14 DIV; unpaired t test).
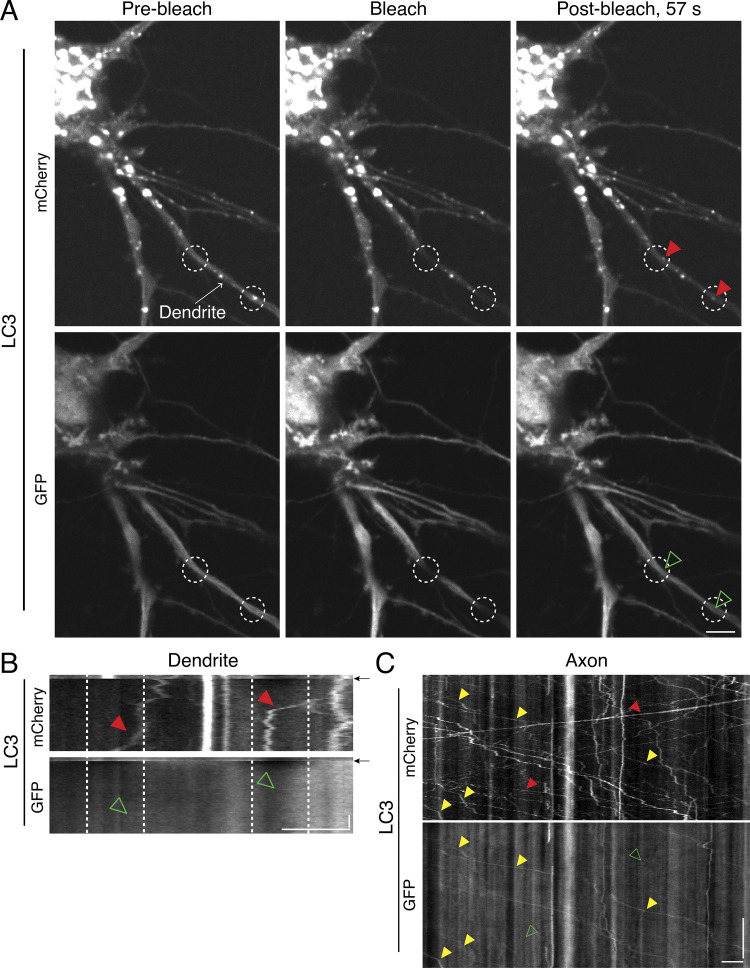
To examine the effects of stimulating neuronal activity on autophagy, we transfected rat hippocampal neurons with mCherry-GFP-LC3 (MAP1LC3/LC3; microtubule associated protein 1 light chain 3). LC3 is a well-characterized marker for autophagic organelles (Kabeya et al., 2000), and the dual-color LC3 reporter is a pH-sensitive marker that distinguishes the maturation state of organelles within the autophagy pathway (Kimura et al., 2007; Pankiv et al., 2007). Because mCherry fluorescence is more resistant than GFP to quenching in acidic environments, mCherry labels both immature autophagosomes and mature, fully acidified autolysosomes, while GFP labels only immature autophagosomes. However, we observed significant background in the GFP channel in dendrites that limited our ability to reliably quantify the GFP signal (Fig. S1). To reduce this background signal, we photobleached the GFP channel and determined the maturation state for those LC3 puncta that migrated into the bleached zone (Fig. S1, A and B). 15 of 15 LC3 puncta that moved into the bleached region were positive for mCherry only and lacked GFP fluorescence (Fig. S1, A and B). This result suggested a higher prevalence of autolysosomes in dendrites than in axons, where compartments copositive for mCherry and GFP are more readily detected (Fig. S1 C; Maday et al., 2012). Thus, the higher background with GFP in dendrites may be due to neurite-specific differences in microtubule density and/or microtubule binding (LC3 is a microtubule-binding protein; Mann and Hammarback, 1994). Furthermore, the longer lifetime and accumulation of mCherry in degradative compartments likely improves the signal-to-noise ratio for this fluorophore. Due to these limitations in GFP signal resolution in dendrites, we relied exclusively on the mCherry (LC3) signal to track autophagy. Because mCherry can label both autophagosomes and autolysosomes, we collectively refer to mCherry-LC3–positive organelles as AVs.
Figure S1.
Immature autophagic compartments are more readily detected in axons than in dendrites. (A and B) En face images (A) and associated kymographs (B) of mCherry-GFP-LC3 in dendrites of a hippocampal neuron. Dashed white regions denote location of photobleaching. Arrows in B indicate the time of photobleaching. Throughout the figure, AVs positive for mCherry and negative for GFP fluorescence are denoted with red arrowheads in the mCherry channel and open green arrowheads in the GFP channel. AVs copositive for both mCherry and GFP are denoted with yellow arrowheads in both channels. Horizontal bars, 5 µm. Vertical bar, 10 s. (C) Kymograph analysis of mCherry-GFP-LC3 motility in an axon of a hippocampal neuron. Horizontal bar, 5 µm. Vertical bar, 1 min.
To assess changes in autophagy in real time, we performed live-cell imaging to track the dynamics of mCherry (LC3)-positive AVs in each compartment of the neuron. For this experiment, we acquired time-lapse images for 5 min and performed on-scope addition of 45 mM KCl after 2.5 min of imaging. mCherry (LC3) marks AVs that are concentrated in the soma and also present within dendritic and axonal compartments (Fig. 1 C and insets). In dendrites, under basal conditions, mCherry (LC3)-positive AVs undergo bidirectional movement and oscillate within a confined region along the dendritic shaft (Fig. 1 D, before addition of treatment). Depolarization of neurons by addition of KCl resulted in a very striking and rapid decrease in AV motility within dendrites (Fig. 1 D). By contrast, AV motility in dendrites did not decrease with addition of NaCl, and AVs continued to undergo bidirectional movement after NaCl treatment (Fig. 1 D). To quantitate the effects of neuronal depolarization on dendritic AV motility, we tracked individual AVs before and after KCl addition and measured the mean speed (total distance traveled/total time traveled) for individual AVs. For this analysis, we focused on the population of AVs that exhibited motility before treatment addition by tracking only those AVs that traveled a total distance of ≥5 µm during the 2.5 min. We found that neuronal depolarization with KCl decreased the average AV mean speed in dendrites by ∼3.6-fold (Fig. 1, E and F). By contrast, we did not observe alterations in dendritic AV mean speed upon addition of NaCl (Fig. 1, E and F). To measure the change in AV speed upon addition of KCl or NaCl, for each individual AV, we subtracted the mean speed after salt addition from the paired mean speed before salt addition (Fig. 1 G). Treatment with KCl reduced the AV mean speed by an average of ∼0.12 µm/s, and this value was significantly larger than that measured for NaCl (Fig. 1 G). Indeed, the difference in mean speeds before and after NaCl treatment was near zero (∼0.02 µm/s; Fig. 1 G). Combined, these results indicate that neuronal activity dampens AV motility in dendrites.
Interestingly, we found that the effects of neuronal depolarization on AV motility are specific to dendrites. In axons, AV motility is biased toward long-distance unidirectional transport, in contrast to the short-range oscillatory movements exhibited by AVs in dendrites (Fig. 1 D). We previously demonstrated with GFP-LC3 tracking that autophagosomes exhibit a primarily retrograde motility (Maday and Holzbaur, 2014; Maday and Holzbaur, 2016; Maday et al., 2012). In addition to these retrograde tracks, mCherry (LC3) compartments also exhibit long-distance anterograde trajectories, emphasizing an additional population of mature AVs labeled with mCherry, consistent with our previous reports in dorsal root ganglion neurons (Maday et al., 2012). Addition of KCl (or NaCl), however, did not alter AV mean speeds in axons (Fig. 1 H). Thus, the activity-dependent decrease in AV mean speed is specific to dendrites and is not observed in axons, indicating compartment-specific regulation of AV dynamics.
Synaptic activity regulates AV motility specifically in dendrites and not in axons
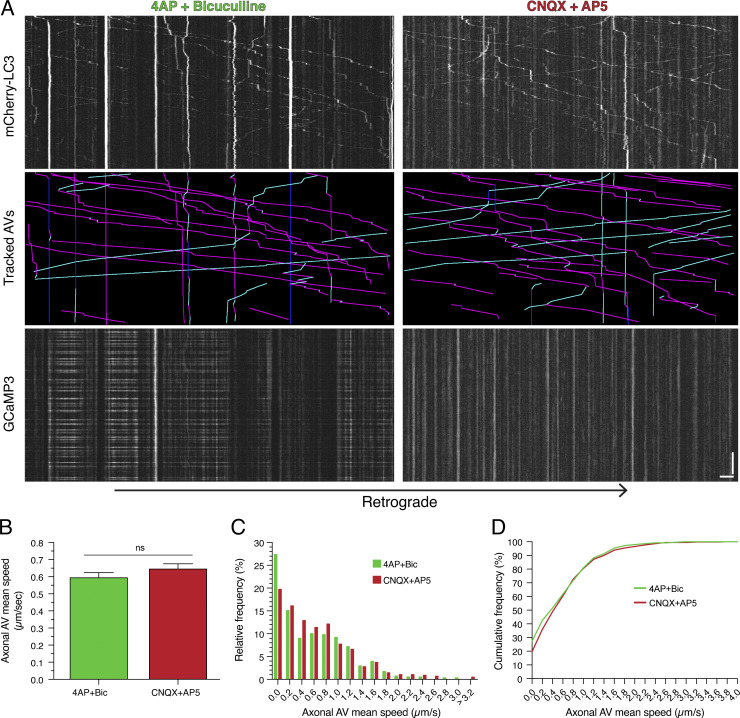
We next employed a pharmacological approach to modulate synaptic activity and assess effects on AV motility. To stimulate synaptic activity, we treated neurons with a cocktail of 50 µM 4-aminopyridine (4-AP) and 20 µM bicuculline (Bic), antagonists of voltage-gated potassium channels and inhibitory γ-aminobutyric acid A receptors, respectively (Hanus et al., 2014). To silence synaptic activity, we treated neurons with 50 µM 6-cyano-7-nitroquinoxaline-2,3-dione (CNQX) and 50 µM D-(−)-2-amino-5-phosphonopentanoic acid (AP5), antagonists of excitatory α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid (AMPA) and N-methyl-D-aspartate receptors, respectively (Hanus et al., 2014). Treatment with an equivalent volume of DMSO solvent served as a control to measure AV dynamics under the basal activity of the culture.
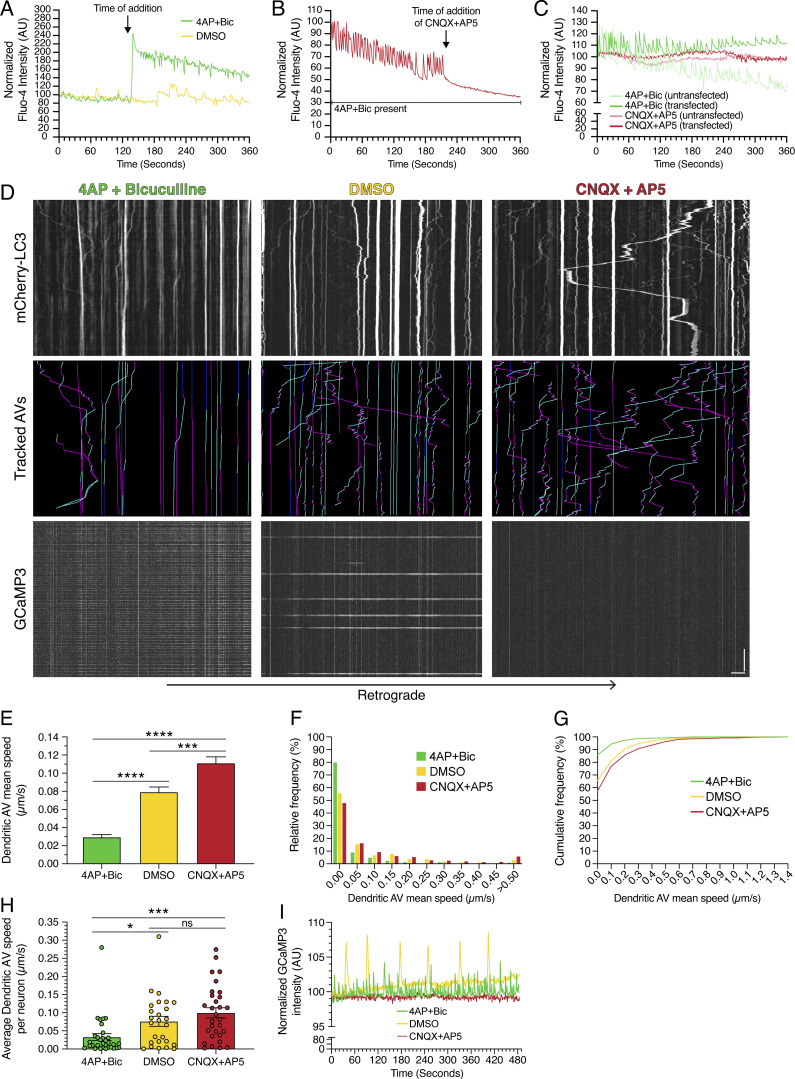
To validate the effectiveness of our drug treatments, we performed live-cell imaging to measure intracellular calcium dynamics using Fluo-4; oscillations in calcium levels serve as a proxy for the firing of action potentials. On-scope addition of 4-AP + Bic resulted in a dramatic increase in intracellular calcium, as evidenced by increased Fluo-4 signal relative to DMSO addition (Fig. 2 A). Furthermore, 4-AP + Bic treatment increased the frequency of calcium oscillations, consistent with increased firing of action potentials and stimulated synaptic activity (Fig. 2 A). Conversely, on-scope addition of CNQX + AP5 dampened calcium oscillations induced by 4-AP + Bic, consistent with silencing of synaptic activity (Fig. 2 B). Next, we transfected neurons with LC3 labeled with only mCherry (mCherry-LC3). We found that transfection of neurons to express mCherry-LC3 did not affect the response of neurons to the stimulation or silencing treatments (Fig. 2 C). Thus, our treatments establish paradigms of stimulated (4-AP + Bic) versus basal (DMSO) versus silenced (CNQX + AP5) synaptic activity, and mCherry-LC3 expression does not affect neuronal functionality.
Figure 2.
Synaptic activity regulates AV motility in dendrites. (A) Quantitation of mean Fluo-4 fluorescence intensity in rat hippocampal neurons (15 DIV) before and after on-scope addition of 4-AP + Bic or DMSO. (B) Quantitation of mean Fluo-4 fluorescence intensity in rat hippocampal neurons (25 DIV) treated with 4-AP + Bic before and after on-scope addition of CNQX + AP5. (C) Mean Fluo-4 intensity in rat hippocampal neurons (14 DIV) either transfected or untransfected with mCherry-LC3 and treated with 4-AP + Bic or CNQX + AP5 for 60 min. (D) Kymographs of mCherry-LC3, tracked AVs (magenta denotes retrograde, cyan denotes anterograde, and dark blue denotes stationary segments), and GCaMP3 from the same dendrite after 30 min of treatment in 4-AP + Bic, DMSO, or CNQX + AP5. Horizontal bar, 5 µm. Vertical bar, 1 min. (E–G) Quantitation of AV mean speed in dendrites of rat hippocampal neurons treated for 30 min in 4-AP + Bic, DMSO, or CNQX + AP5 (E, mean ± SEM; F, histogram of dendritic AV mean speed; G, cumulative frequency of dendritic AV mean speed; n = 629–731 AVs from 26–32 neurons from three to four independent experiments; 13–14 DIV; one-way ANOVA with Tukey’s post hoc test; ***, P ≤ 0.001; ****, P ≤ 0.0001). (H) Quantitation of average dendritic AV mean speed per neuron (mean ± SEM; n = 26–32 neurons from three to four independent experiments; 13–14 DIV; one-way ANOVA with Tukey’s post hoc test; *, P ≤ 0.05; ***, P ≤ 0.001). (I) Quantitation of GCaMP3 fluorescence intensity in the dendrite of a neuron treated with 4-AP + Bic, DMSO, or CNQX + AP5 for 30 min.
Next, we incubated neurons in each treatment condition (stimulated versus basal versus silenced synaptic activity) for 30 min and examined effects on AV dynamics. Consistent with the effects of KCl-induced neuronal depolarization, stimulating synaptic activity with 4-AP + Bic dampened AV motility in dendrites relative to the DMSO control (Fig. 2 D). With 4-AP + Bic treatment, we observed fewer motile AVs than with the DMSO control. Conversely, silencing synaptic activity with CNQX + AP5 induced AV motility in dendrites, as we observed a greater population of motile AVs than with the DMSO control (Fig. 2 D). These results suggest that synaptic activity and AV dynamics are inversely linked: Inducing synaptic activity dampens dendritic AV motility, whereas silencing synaptic activity increases dendritic AV motility.
To quantitate the effect of synaptic activity on AV motility, we measured the mean speed of individual mCherry-LC3–positive puncta under each paradigm of synaptic activity. The activity state of each neuron analyzed was confirmed by coexpression of the calcium sensor GCaMP3 to measure calcium oscillations. Treatment with 4-AP + Bic increased GCaMP3 oscillations by ∼10-fold relative to DMSO controls (mean rates of 0.28 versus 0.03 events/s), indicating induced synaptic activity (Fig. 2, D and I; and Fig. S2, C and D). Treatment with CNQX + AP5 eliminated GCaMP3 oscillations relative to DMSO controls (mean rate of 0.00 versus 0.03 events/s), indicating silenced synaptic activity (Fig. 2, D and I; and Fig. S2, C and D). Similar to KCl-mediated depolarization, stimulation of synaptic activity with 4-AP + Bic significantly reduced the average AV mean speed in dendrites by ∼2.7-fold relative to the DMSO control (Fig. 2 E). By contrast, silencing synaptic activity with CNQX + AP5 increased the mean speed of AVs in dendrites ∼1.4-fold relative to the DMSO control (Fig. 2 E). Thus, as synaptic activity decreased, the AV population shifted toward exhibiting higher mean speeds (Fig. 2, F and G). To evaluate the effects of activity on AV dynamics in individual neurons, we determined the average AV mean speed for each neuron analyzed (Fig. 2 H). Consistent with the effects observed by pooling individual AV mean speeds across neurons, 4-AP + Bic treatment decreased the average AV mean speed per neuron as compared with the DMSO control and neurons silenced with CNQX + AP5 (Fig. 2 H). Combined, our results indicate that the motility of AVs in dendritic compartments is regulated by the activity state of the neuron.
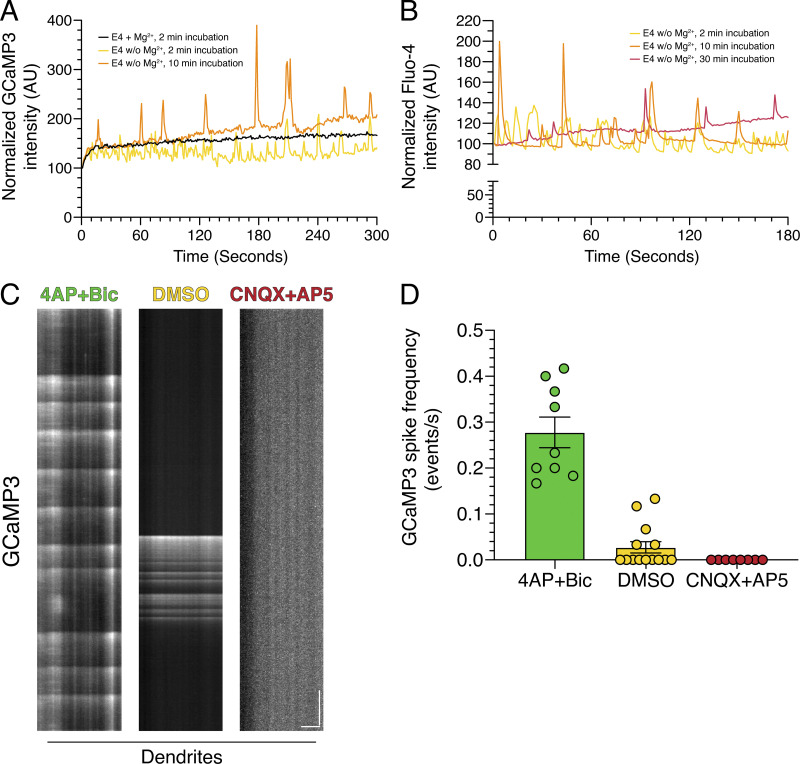
Figure S2.
Quantitation of calcium oscillations in rat hippocampal neurons. (A) Quantitation of mean GCaMP3 fluorescence in an individual neuron (14 DIV) as a function of time of incubation in E4 solution. (B) Quantitation of mean Fluo-4 fluorescence in a field of neurons (13 DIV) as a function of time of incubation in E4 solution. (C and D) High-speed imaging of GCaMP3 oscillations in dendrites from hippocampal neurons treated with 4-AP + Bic, DMSO, or CNQX + AP5 for 30 min. (C) Kymograph analysis of GCaMP3 in dendrites. Horizontal bar, 5 µm. Vertical bar, 5 s. (D) Corresponding quantitation of GCaMP3 spike frequency (mean ± SEM; n = 8–14 neurons from two independent experiments; 13–14 DIV).
To determine whether these effects are specific to dendrites or whether they also occur in axons, we measured AV mean speeds in axons. Similar to dendrites, calcium oscillations increased with 4-AP + Bic treatment and decreased with CNQX + AP5 treatment in axons (Fig. 3 A). However, unlike in dendrites, modulating synaptic activity did not alter the motility of AVs in axons (Fig. 3, A–D). Axonal AVs exhibited long-range movements with either 4-AP + Bic or CNQX + AP5 treatment, and no significant changes in AV mean speed were observed between stimulation or silencing conditions (Fig. 3, A–D). Thus, the dampening of AV motility with synaptic activity is compartment specific, occurring primarily in dendrites and not in axons.
Figure 3.
Synaptic activity does not alter AV motility in axons. (A) Kymographs of mCherry-LC3, tracked AVs (magenta denotes retrograde, cyan denotes anterograde, and dark blue denotes stationary segments), and GCaMP3 from the same axon after 60 min of treatment in 4-AP + Bic or CNQX + AP5. Horizontal bar, 5 µm. Vertical bar, 1 min. (B–D) Quantitation of AV mean speed in axons of rat hippocampal neurons treated with 4-AP + Bic or CNQX + AP5 for 60 min (B, mean ± SEM; C, histogram of axonal AV mean speed; D, cumulative frequency of axonal AV mean speed; n = 495–524 AVs from 21–23 neurons from three independent experiments, 13–14 DIV; unpaired t test).
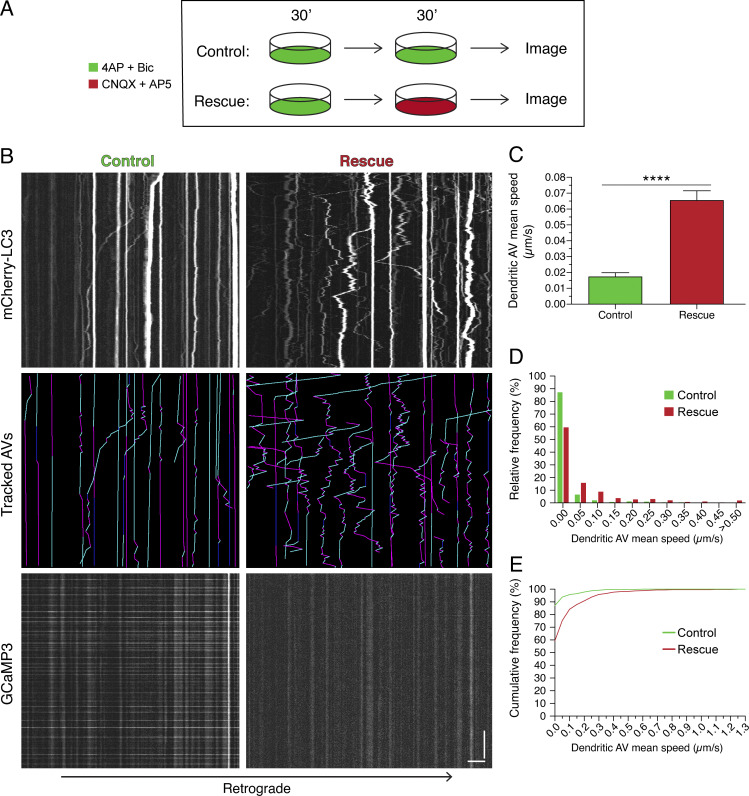
To determine whether the effects of synaptic stimulation on dendritic AV dynamics are reversible, we silenced neurons following a period of stimulation. For this experiment, neurons were first treated with 4-AP + Bic for 30 min to induce an activity-dependent decrease in dendritic AV motility (Fig. 4 A). Neurons were then treated for 30 min in either 4-AP + Bic (control sample) or CNQX + AP5 (rescue sample; Fig. 4 A). Neurons silenced with CNQX + AP5 exhibited increased AV motility as compared with neurons in 4-AP + Bic (Fig. 4 B). Silencing led to an increase in the average AV mean speed ∼3.7-fold relative to the stimulated control (Fig. 4, C–E) and ∼2.2-fold higher than the average AV mean speed observed after only 30 min of 4-AP + Bic treatment (Fig. 2 E). These results suggest that decreases in AV motility due to synaptic stimulation can be reversed by silencing neurons. Importantly, these findings also establish that activity-dependent dampening of AV dynamics is not due to cytotoxicity. Rather, we found that AV motility in dendrites is coordinated with the activity state of the neuron in a reversible manner.
Figure 4.
Activity-dependent dampening of AV motility in dendrites is reversed by silencing neurons. (A) Schematic of experimental paradigm. (B) Kymographs of mCherry-LC3, tracked AVs (magenta denotes retrograde, cyan denotes anterograde, and dark blue denotes stationary segments), and GCaMP3 from the same dendrite after the treatments described in A. Horizontal bar, 5 µm. Vertical bar, 1 min. (C–E) Quantitation of AV mean speed in rat hippocampal neurons treated with the experimental paradigm in A (C, mean ± SEM; D, histogram of dendritic AV mean speed; E, cumulative frequency of dendritic AV mean speed; n = 531–641 AVs from 25–26 neurons from three independent experiments; 13–14 DIV; unpaired t test; ****, P ≤ 0.0001).
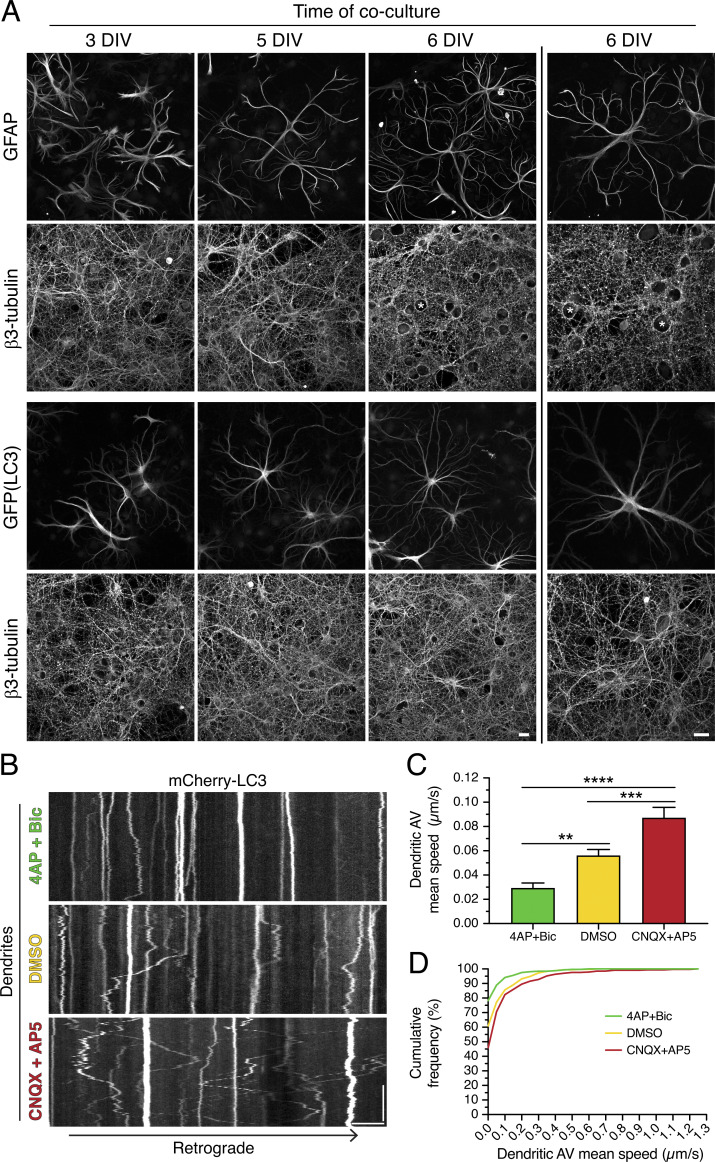
To determine whether synaptic activity regulates AV motility in neurons grown in a more complex environment, we cocultured primary neurons with astrocytes. For this experiment, we established a robust system for coculturing neurons and astrocytes to recapitulate intercellular connections present in vivo. Primary neurons (hippocampal or cortical) were grown for 7–8 d in vitro (DIV) to generate a monolayer of neurites onto which primary cortical astrocytes were plated. Neurons and astrocytes were then cocultured for up to 8 DIV. During this period of coculture, astrocytes transition from polygonal morphologies to elaborate starlike or “astral” morphologies reminiscent of structures observed in vivo (Fig. 5 A). To examine the effects of synaptic activity on dendritic AV motility, neurons were transfected with mCherry-LC3 on the day before astrocyte addition. After 7–8 DIV of coculture, samples were treated for 30 min in 4-AP + Bic, DMSO, or CNQX + AP5, and live-cell imaging was used to track AV motility in dendrites. Similar to monocultured neurons, cocultured neurons also exhibited an activity-dependent regulation of AV motility in dendrites (Fig. 5, B–D). Stimulation of synaptic activity dampened dendritic AV motility, and silencing synaptic activity increased dendritic AV motility (Fig. 5, B–D). Thus, this synaptic control of AV motility is a fundamental mechanism observed in neurons grown in monoculture and in coculture with astrocytes.
Figure 5.
Synaptic activity controls AV dynamics in dendrites of neurons cocultured with astrocytes. (A) Maximum projections of Z-stacks of rat hippocampal neurons (immunostained for neuron-specific β3-tubulin) cocultured with either wild-type mouse cortical astrocytes [immunostained for astrocyte-specific glial fibrillary acidic protein (GFAP)] or GFP-LC3–transgenic mouse cortical astrocytes (immunostained for GFP). Images to the left of the vertical line were taken with a 40× objective, and images to the right of the vertical line were taken with a 63× objective. Asterisks denote the location of neuronal cell bodies within the neurite meshwork. Scale bars, 20 µm. (B) Kymographs of mCherry-LC3 in dendrites of rat cortical neurons cocultured with mouse cortical astrocytes for 7–8 DIV (neurons are a total of 14–16 DIV) and treated for 30 min in 4-AP + Bic, DMSO, or CNQX + AP5. Retrograde direction is from left to right. Horizontal bar, 5 µm. Vertical bar, 1 min. (C and D) Quantitation of AV mean speed in dendrites of cortical neurons cocultured with cortical astrocytes for 7–8 DIV and treated for 30 min in 4-AP + Bic, DMSO, or CNQX + AP5 (C, mean ± SEM; D, cumulative frequency of dendritic AV mean speed; n = 323–368 AVs from 9–10 neurons from two independent experiments; neurons are 14–16 DIV; one-way ANOVA with Tukey’s post hoc test; **, P ≤ 0.01; ***, P ≤ 0.001; ****, P ≤ 0.0001).
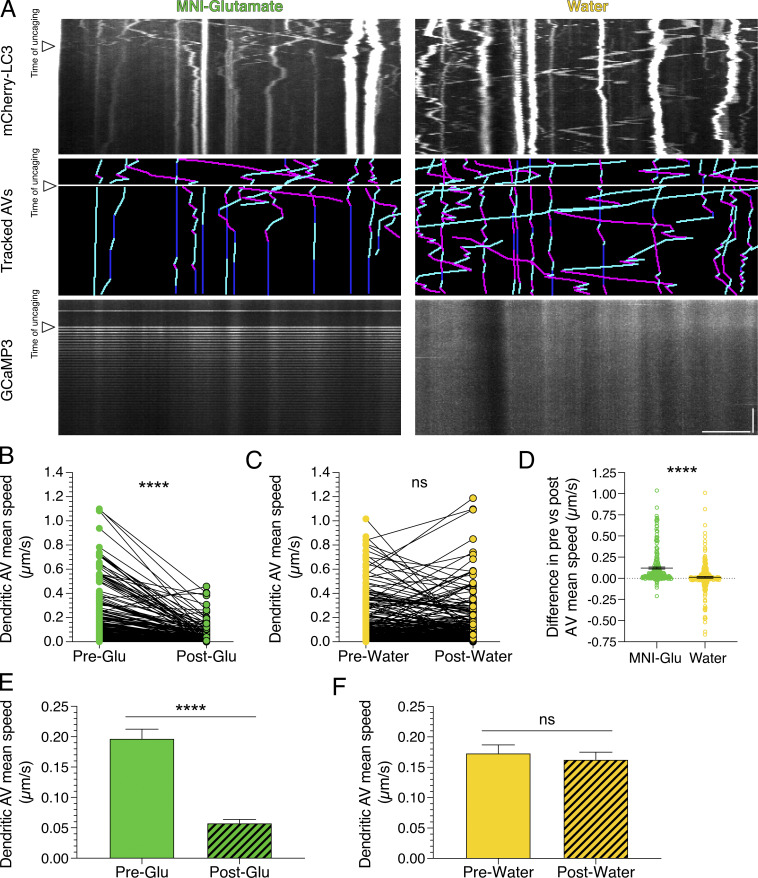
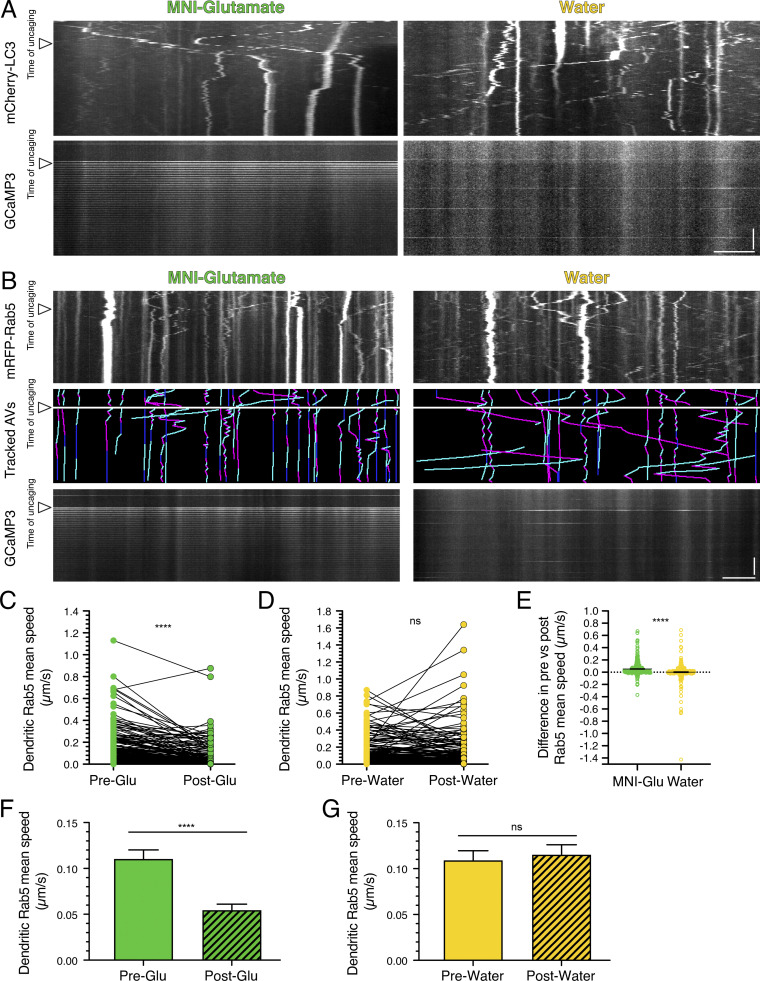
Next, we wanted to define whether the regulation of AV motility by synaptic activity is local. Thus, we locally stimulated synapses by photo-uncaging 4-methoxy-7-nitroindolinyl-caged-L-glutamate (MNI-glutamate). MNI-glutamate is a caged, or inactive, form of glutamate that, upon exposure to 405-nm light, releases glutamate to bind to its cognate receptors and stimulate glutamatergic signaling. When we photo-uncaged glutamate at 0.33 Hz for 40 pulses, GCaMP3 dendritic fluorescence spiked at every uncaging event, consistent with increased intracellular calcium in response to successful uncaging of the MNI-glutamate (Fig. 6 A). We did not observe rises in intracellular calcium levels during the uncaging pulses in the water controls that were exposed to the same laser conditions as samples with MNI-glutamate (Fig. 6 A). To measure the effects of glutamate release on AV motility, we tracked AVs before and during uncaging of MNI-glutamate. We observed a significant reduction in dendritic AV mean speed during glutamate uncaging (Fig. 6 B). We measured an ∼3.9-fold decrease in mean speed for AVs with paired tracks before and during treatment (Fig. 6 B) and an ∼3.4-fold decrease in mean speed for the total AVs that were tracked (Fig. 6 E). However, no change in dendritic AV mean speed was observed during the uncaging protocol in the water control (Fig. 6, C and F). Glutamate release reduced the AV mean speed by an average of ∼0.12 µm/s, and this value was significantly larger than that measured for the water control (Fig. 6 D); the difference in mean speeds before and during the water treatment was near zero (∼0.01 µm/s; Fig. 6 D). Importantly, these effects were not limited to hippocampal neurons. Glutamate uncaging also decreased AV motility in cortical neurons (Fig. S3 A). Combined, these results were strikingly similar to our previous findings of enhanced neuronal activity with KCl or 4-AP + Bic treatments on decreasing AV speeds (Figs. 1, 2, and 4). Our results with glutamate uncaging further indicate that the effects of synaptic activity on AV motility are rapid and can be controlled locally. Interestingly, we also found that the motility of early endosomes labeled with mRFP-Rab5 is also reduced in response to glutamate uncaging in dendrites (Fig. S3, B–G). Because synaptic activity regulates the motility of other dendritic organelles and degradative machinery (Bingol and Schuman, 2006; Hanus et al., 2014; MacAskill et al., 2009; Wang and Schwarz, 2009), these data suggest a core mechanism to regulate intracellular trafficking in a compartment-specific manner in neurons.
Figure 6.
Local stimulation of synaptic activity by uncaging MNI-glutamate (MNI-Glu) reduces AV motility in dendrites. (A) Kymographs of mCherry-LC3, tracked AVs (magenta denotes retrograde, cyan denotes anterograde, and dark blue denotes stationary segments), and GCaMP3 from the same dendrite before and after glutamate (Pre-Glu and Post-Glu, respectively) uncaging (arrowheads denote time of uncaging; water serves as a control). Horizontal bar, 5 µm. Vertical bar, 30 s. (B–F) Quantitation of AV mean speed in dendrites before and after uncaging. (B and C, paired points represent a single AV before and during uncaging; D, difference in AV mean speed before and after uncaging; MNI-glutamate, n = 226 AVs from 22 hippocampal neurons; water, n = 268 AVs from 25 hippocampal neurons; three independent experiments; 13–14 DIV; B and C, paired t test; D, unpaired t test; ****, P ≤ 0.0001). (E and F) Mean ± SEM of all AVs tracked, including those from paired analysis, before and after uncaging (E, n = 272 AVs in pre-uncaging and 277 AVs in post-uncaging from 22 neurons from three independent experiments; 13–14 DIV; F, n = 321 AVs in pretreatment and 346 AVs in post-treatment from 25 neurons from three independent experiments; 13–14 DIV; unpaired t test; ****, P ≤ 0.0001).
Figure S3.
Local activation of synapses by uncaging MNI-glutamate reduces dendritic AV motility in cortical neurons and early endosome motility in dendrites of hippocampal neurons. (A) Kymograph analysis of mCherry-LC3 and GCaMP3 in dendrites of cortical neurons before and after uncaging (arrowheads denote time of uncaging; water serves as a control). Horizontal bar, 5 µm. Vertical bar, 30 s. (B) Kymographs of mRFP-Rab5, tracked early endosomes (magenta denotes retrograde, cyan denotes anterograde, and dark blue denotes stationary segments), and GCaMP3 from the same dendrite before and after glutamate uncaging (arrowheads denote time of uncaging; water serves as a control). Horizontal bar, 5 µm. Vertical bar, 30 s. (C–G) Quantitation of Rab5 mean speed in dendrites from hippocampal neurons before and during uncaging (C and D, paired points represent a single early endosome before and after uncaging; E, difference in early endosome mean speed before and after uncaging; MNI-glutamate, n = 304 early endosomes from 21 hippocampal neurons; water, n = 313 early endosomes from 20 hippocampal neurons; three independent experiments; 13–14 DIV; C and D, paired t test; E, unpaired t test; ****, P ≤ 0.0001). (F and G) Mean ± SEM of all early endosomes tracked, including those from the paired analysis, before and after uncaging (F, n = 319 early endosomes in pre-uncaging and 340 early endosomes in post-uncaging from 21 neurons from three independent experiments; 13–14 DIV; G, n = 330 early endosomes in pretreatment and 363 early endosomes in post-treatment from 20 neurons from three independent experiments; 13–14 DIV; unpaired t test; ****, P ≤ 0.0001).
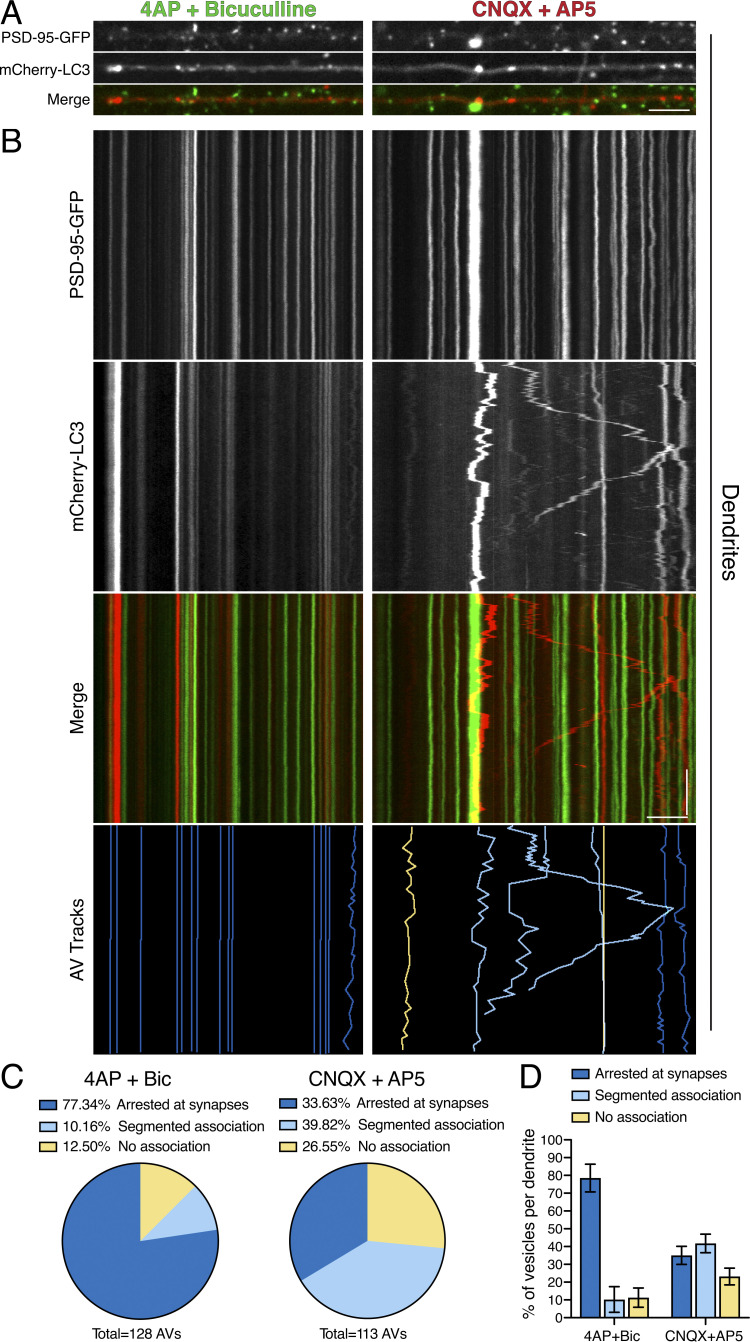
To determine whether AVs stop at synapses in response to activity, we performed dual-color imaging of mCherry-LC3 with the post-synaptic marker post-synaptic density protein 95 (PSD-95)–GFP under conditions of stimulated versus silenced synaptic activity (Fig. 7). We then tracked AVs and binned them into one of three categories based on their proximity and duration at PSD-95–positive puncta: (1) AVs that are “arrested” at synapses as defined by the localization of AVs within 0.75 µm on either side of a synapse for the entire duration of the AV track, (2) AVs with “segmented association” that localize within 0.75 µm on either side of a synapse for only a partial time of the AV track, and (3) AVs that exhibit “no association” with a post-synaptic compartment during its trajectory. We found that under conditions of stimulation of synaptic activity, ∼77% of AVs were arrested at or proximal to synapses (Fig. 7, A–D). This population, however, reduced to ∼34% with silenced synaptic activity, and we observed an increase in AVs that exhibit a segmented and transient association with synapses (Fig. 7, A–D). Furthermore, silencing synaptic activity doubled the percentage of AVs with no association with post-synaptic compartments (Fig. 7, A–D). Thus, we found that stimulating synaptic activity increases the persistent and sustained residence of AVs at or near synaptic compartments.
Figure 7.
Synaptic activity arrests AVs at or proximal to post-synaptic compartments. (A and B) En face images (A) and kymograph analysis (B) of PSD-95–GFP, mCherry-LC3, and tracked AVs (dark blue denotes AVs arrested at synapses, light blue denotes AVs with segmented association with synapses, and yellow denotes AVs with no association with synapses) in dendrites of hippocampal neurons. Horizontal bars in A and B, 5 µm. Vertical bar in B, 1 min. (C) Quantitation of individual AV association with synapses as a percentage of total AVs tracked (4-AP + Bic, n = 128 AVs from 10 neurons; CNQX + AP5, n = 113 AVs from 11 neurons; two independent experiments; 13–16 DIV). (D) Quantitation of percentage of AV association with synapses on a per dendrite basis (mean ± SEM; n = 10–11 neurons from two independent experiments; 13–16 DIV).
Synaptic activity increases degradative AVs in dendrites
Changes in organelle positioning and motility are often coupled with changes in their function (Korolchuk et al., 2011; Walton et al., 2018). Therefore, we proposed that because synaptic activity regulates AV motility, it may also regulate AV function. Because mCherry-LC3 labels immature, nondegradative autophagosomes as well as mature, degradative autolysosomes, we wanted to determine the maturation state of mCherry-LC3–positive AVs in dendrites and to define whether the nature of these compartments changes under paradigms of stimulated versus silenced synaptic activity.
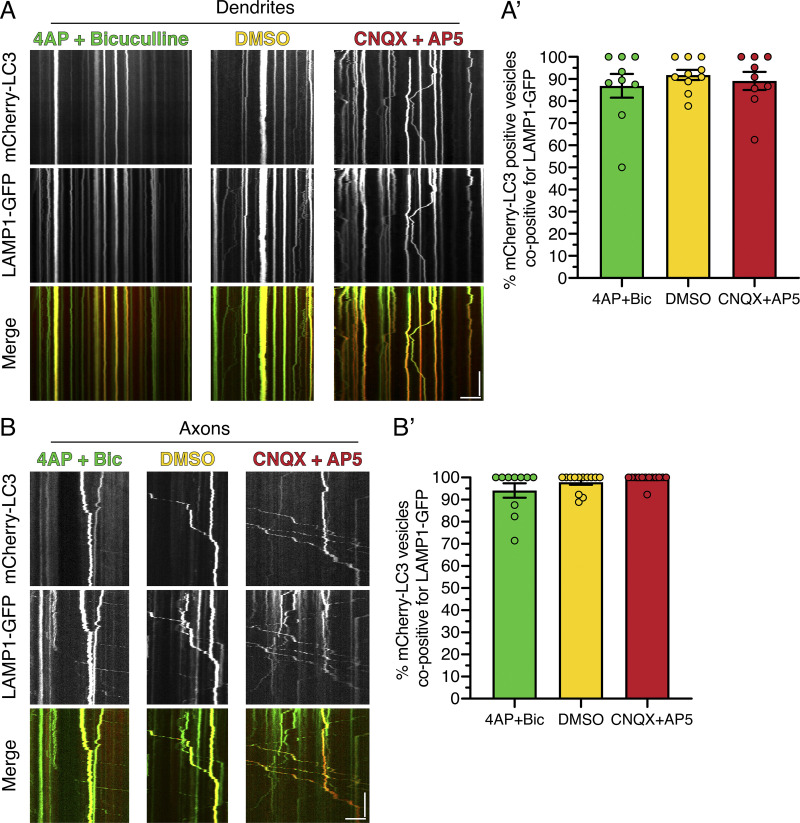
To begin to define the maturation state of AVs in dendrites under different paradigms of synaptic activity, we first measured colocalization of mCherry-LC3–positive AVs with lysosomal-associated membrane protein 1 (LAMP1)–GFP, a marker for late endosomes and lysosomes (Cheng et al., 2018; Yap et al., 2018). In the DMSO control, nearly all (∼90%) mCherry-LC3 puncta in dendrites were positive for LAMP1-GFP, and this value did not change with stimulating or silencing activity (Fig. S4, A and A′). Thus, we found that dendritic mCherry-LC3–positive compartments have largely acquired LAMP1, likely by fusion with late endosomes, and this step is not modulated with activity. Consistently, LAMP1-GFP dynamics in dendrites were regulated by activity similar to our observations for mCherry-LC3: dampened motility in 4-AP + Bic and increased motility in CNQX + AP5 (Fig. S4 A). Similar to dendrites, ∼95% of axonal mCherry-LC3–labeled compartments were copositive for LAMP1-GFP across activity paradigms (Fig. S4, B and B′), consistent with our previous reports under basal conditions (Maday and Holzbaur, 2016; Maday et al., 2012). These data suggest that mCherry-LC3–positive AVs in axons and dendrites have largely fused with late endosomes and or lysosomes and are predominantly amphisomes or autolysosomes. These results directed us to examine activity-dependent changes in downstream steps in AV maturation in dendrites.
Figure S4.
Nearly all AVs in dendrites and axons are positive for LAMP1. (A) Kymograph analysis of mCherry-LC3 and LAMP1-GFP in dendrites of hippocampal neurons treated with 4-AP + Bic, DMSO, or CNQX + AP5 for 30 min. Horizontal bar, 5 µm. Vertical bar, 1 min. (A’) Corresponding quantitation of the percentage colocalization between mCherry-LC3 and LAMP1-GFP in dendrites (mean ± SEM; n = 9–10 neurons from two independent experiments; 13–14 DIV). (B) Kymograph analysis of mCherry-LC3 and LAMP1-GFP in axons of hippocampal neurons treated with 4-AP + Bic, DMSO, or CNQX + AP5 for 30 min. Horizontal bar, 5 µm. Vertical bar, 1 min. (B’) Corresponding quantitation of the percentage colocalization between mCherry-LC3 and LAMP1-GFP in axons (mean ± SEM; n = 10–13 neurons from two independent experiments; 13–14 DIV).
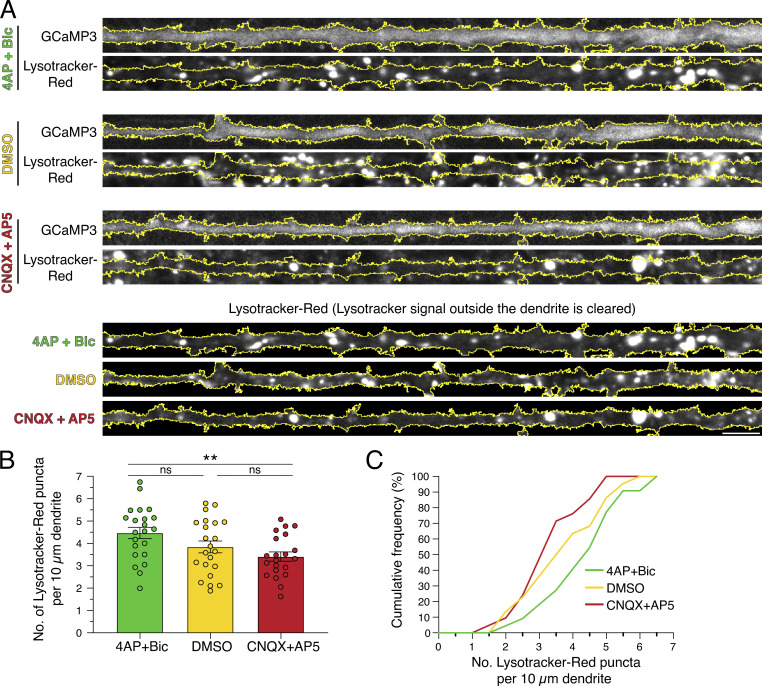
We next sought to define the effects of synaptic activity on the density of acidic organelles in dendrites. For this experiment, we treated neurons expressing GCaMP3 with 4-AP + Bic, DMSO, or CNQX + AP5 in the presence of Lysotracker-Red, a cell-permeant acidotropic dye (Fig. 8). Individual dendrites were outlined using GCaMP3 fluorescence, and the density of Lysotracker-Red–positive puncta was measured along the dendrite (Fig. 8, A–C). Interestingly, we observed a higher density of Lysotracker-Red puncta in dendrites treated with 4-AP + Bic than in those treated with CNQX + AP5 (Fig. 8, A–C). Thus, we found a greater number of acidic organelles in dendrites upon stimulating synaptic activity. Because lysosomal hydrolases are activated in acidic environments, our data suggest that synaptic activity may increase the presence of degradative organelles in dendrites.
Figure 8.
Synaptic activity increases the density of acidic organelles in dendrites. (A) Straightened images of GCaMP3 and Lysotracker-Red in dendrites from hippocampal neurons treated with 4-AP + Bic, DMSO, or CNQX + AP5 for 30 min. The soma is toward the right. Scale bar, 5 µm. (B and C) Corresponding quantitation of the number of Lysotracker-Red puncta per 10-µm dendrite (B, mean ± SEM; C, cumulative frequency; n = 21–22 neurons from three independent experiments; 13–14 DIV; one-way ANOVA with Tukey’s post hoc test; **, P ≤ 0.01).
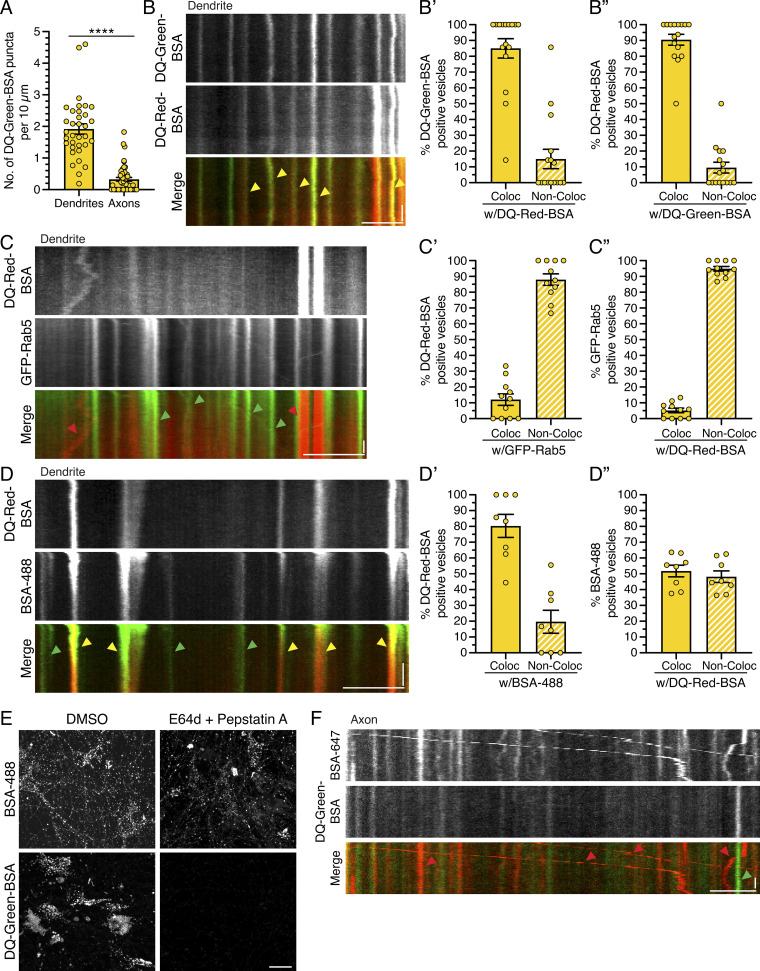
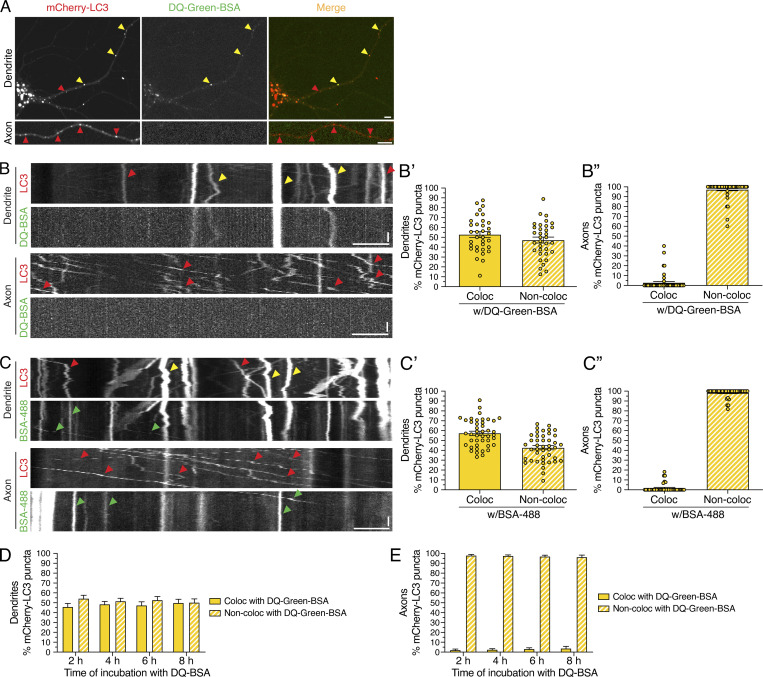
Next, we wanted to specifically label compartments that are proteolytically active. For this experiment, we incubated neurons with DQ-BSA, an endocytosed cargo that fluoresces only upon proteolytic cleavage. We used two different fluorescent conjugates, DQ-Green-BSA and DQ-Red-BSA, which exhibited ∼85–90% overlap and thus were used interchangeably (Fig. S5, B, B′, and B″). We found that DQ-Green-BSA–positive puncta were enriched in the soma, present within dendrites, and low in abundance in axons (Fig. 9, A and B; and Fig. S5 A), in accordance with results reported by Yap et al. (2018).
Figure S5.
DQ-BSA labels degradative compartments in the endolysosomal pathway in hippocampal neurons. (A) Quantitation of the density of DQ-Green-BSA–positive puncta in dendrites and axons (mean ± SEM; dendrites, n = 34 neurons; axons, n = 49 neurons from three independent experiments; 13–14 DIV; unpaired t test; ****, P ≤ 0.0001). (B) Kymograph analysis of DQ-Green-BSA and DQ-Red-BSA in dendrites. Yellow arrowheads denote puncta positive for both DQ-Green-BSA and DQ-Red-BSA. Horizontal bar, 5 µm. Vertical bar, 10 s. (B’ and B”) Corresponding quantitation of the percentage colocalization (Coloc) between DQ-Green-BSA and DQ-Red-BSA in dendrites (mean ± SEM; n = 16 neurons from three independent experiments; 13–15 DIV). (C) Kymograph analysis of DQ-Red-BSA and early endosomes labeled with GFP-Rab5 in dendrites. Red arrowheads denote puncta positive for only DQ-Red-BSA, and green arrowheads denote puncta positive for only GFP-Rab5. Horizontal bar, 5 µm. Vertical bar, 10 s. (C’ and C”) Corresponding quantitation of the percentage colocalization between DQ-Red-BSA and GFP-Rab5 in dendrites (mean ± SEM; n = 11 neurons from two independent experiments; 13–15 DIV). (D) Kymograph analysis of DQ-Red-BSA and endocytic cargo BSA-488 in dendrites. Yellow arrowheads denote puncta positive for both DQ-Red-BSA and BSA-488, and green arrowheads denote puncta positive for only BSA-488. Horizontal bar, 5 µm. Vertical bar, 10 s. (D’ and D”) Corresponding quantitation of the percentage colocalization between DQ-Red-BSA and BSA-488 in dendrites (mean ± SEM; n = 8 neurons from two independent experiments; 14 DIV). (E) Maximum projections of Z-stacks of rat hippocampal neurons treated with 100 µM E64d and 30 µM pepstatin A versus DMSO for 3 h. During the last hour of treatment, neurons were also incubated with BSA-488 or DQ-Green-BSA. Inhibition of protease activity reduces DQ-Green-BSA fluorescence but has minimal effect on BSA-488. Scale bar, 20 µm. (F) Kymograph analysis of BSA-647 and DQ-Green-BSA in the axon. Red arrowheads denote puncta positive for only BSA-647, and green arrowheads denote puncta positive for only DQ-Green-BSA. Horizontal bar, 5 µm. Vertical bar, 10 s.
Figure 9.
Degradative AVs labeled with DQ-BSA are detected more frequently in dendrites than in axons. (A) mCherry-LC3 and DQ-Green-BSA distribution in the soma, dendrite, and axon of a hippocampal neuron. Scale bars, 5 µm. In A–C, red arrowheads denote puncta positive for only mCherry-LC3, green arrowheads denote puncta positive for only BSA (either DQ-Green-BSA or BSA-488), and yellow arrowheads denote puncta positive for both LC3 and BSA. (B) Kymographs of mCherry-LC3 and DQ-Green-BSA motility in the dendrite and axon. Retrograde direction is from left to right. Horizontal bars, 5 µm. Vertical bars, 10 s. (B’) Quantitation of the percentage of mCherry-LC3–positive puncta that colocalize (Coloc) or do not colocalize (Non-Coloc) with DQ-Green-BSA in dendrites (mean ± SEM; n = 34 neurons from three independent experiments; 13–14 DIV). (B”) Quantitation of the percentage of mCherry-LC3–positive puncta that colocalize or do not colocalize with DQ-Green-BSA in axons (mean ± SEM; n = 49 neurons from three independent experiments; 13–14 DIV). (C) Kymographs of mCherry-LC3 and BSA-488 motility in the dendrite and axon. Horizontal bar, 5 µm. Vertical bar, 10 s. (C’) Quantitation of the percentage of mCherry-LC3 puncta positive for BSA-488 in dendrites (mean ± SEM; n = 43 hippocampal neurons from four independent experiments; 13 DIV). (C”) Quantitation of the percentage of mCherry-LC3 puncta positive for BSA-488 in axons (mean ± SEM; n = 40 hippocampal neurons from four independent experiments; 13 DIV). (D) Quantitation of the percentage of mCherry-LC3 AVs that are copositive for DQ-Green-BSA in dendrites as a function of time of incubation with DQ-BSA (mean ± SEM; n = 25–28 neurons from three independent experiments; 13 DIV). (E) Quantitation of the percentage of mCherry-LC3 AVs that are copositive for DQ-Green-BSA in axons as a function of time of incubation with DQ-BSA (mean ± SEM; n = 24–30 neurons from three independent experiments; 13 DIV).
We first validated the identity of the compartments labeled with DQ-BSA by performing dual-color imaging in dendrites with DQ-Red-BSA and various endocytic markers. To label early endosomes, we transfected neurons with GFP-Rab5 and measured overlap with DQ-Red-BSA. We observed that only ∼12% of DQ-Red-BSA–positive compartments in dendrites were copositive for GFP-Rab5, and only ∼5% of GFP-Rab5–positive compartments were copositive for DQ-Red-BSA (Fig. S5, C, C′, and C″). Thus, degradative compartments labeled with DQ-BSA are largely separate organelle populations from early endosomes. To label all compartments receiving endocytic input, we coincubated neurons with DQ-Red-BSA and the fluid-phase endocytic cargo BSA-488. We found that ∼80% of DQ-Red-BSA–positive compartments in dendrites were copositive for BSA-488 (Fig. S5, D and D′), indicating that most proteolytically active compartments receiving endocytic input during the incubation window colabel with both DQ-Red-BSA and BSA-488. With the converse analysis, we found that only ∼52% of BSA-488–positive compartments were copositive for DQ-Red-BSA (Fig. S5, D and D″). The remaining ∼48% of BSA-488–positive compartments likely represent early and late endosomes of low or no proteolytic activity. Last, we found that treatment with protease inhibitors E64d and pepstatin A eliminated fluorescence of DQ-Green-BSA but not of BSA-488 (Fig. S5 E), indicating that the loss in DQ-BSA fluorescence is due to a specific reduction in proteolytic processing rather than a block in endocytic flux. In total, we found that DQ-BSA labels degradative organelles within the endolysosomal pathway.
Do changes in synaptic activity affect the degradative nature of mCherry-LC3–positive AVs? To answer this question, we first needed to determine the percentage of mCherry-LC3–positive AVs that are degradative autolysosomes under basal conditions. For this experiment, we incubated neurons expressing mCherry-LC3 with DQ-Green-BSA and measured the percentage of AVs that were copositive for DQ-Green-BSA. We found that ∼51% of mCherry-LC3–positive AVs in dendrites were copositive for DQ-Green-BSA, suggesting that approximately half of dendritic AVs are degradative autolysosomes (Fig. 9, A, B, and B′). In contrast to dendrites, less than ∼3% of mCherry-LC3–positive AVs in axons were copositive for DQ-Green-BSA (Fig. 9 A, B, B″). Thus, we found a greater percentage of degradative autolysosomes, as labeled with DQ-BSA, in dendrites than in axons. Because DQ-BSA can label only those degradative compartments that are receiving endocytic input, we measured the percentage of mCherry-LC3–positive AVs that were copositive for the endocytic cargo BSA-488. We found that ∼57% of mCherry-LC3–positive AVs in dendrites were copositive for BSA-488 (Fig. 9, C and C′). By contrast, we find less than ∼2% of AVs in axons are positive for BSA-488 (Fig. 9, C and C″). The axon did indeed contain vesicles of endocytic origin, as labeled with BSA-488 (Fig. 9 C) or BSA-647 (Fig. S5 F). However, these endocytic compartments in axons were largely negative for mCherry-LC3 (Fig. 9 C) or DQ-BSA (Fig. S5 F), in contrast to the significant overlap observed in dendrites (Fig. 9, A–C; and Fig. S5 D, D′, and D″). Combined, these data highlight important differences in how the autophagic and endocytic pathways merge in dendritic versus axonal compartments. Specifically, autophagic organelles in dendritic compartments have fused with newly endocytosed cargo to a greater extent as compared to autophagic organelles in axons. Additionally, these compartment-specific differences in the overlap between mCherry-LC3 and endocytosed cargo were not due to insufficient labeling time. We performed a time-course analysis of 2, 4, 6, and 8 h of DQ-BSA uptake (Fig. 9, D and E) and found that longer incubation times with DQ-BSA did not alter the percentage of AVs copositive for DQ-BSA in either dendrites or axons (Fig. 9, D and E). Thus, the partial labeling of mCherry-LC3 compartments with DQ-BSA in dendrites and axons is not due to inadequate time for proteolytic processing of the DQ-BSA substrate.
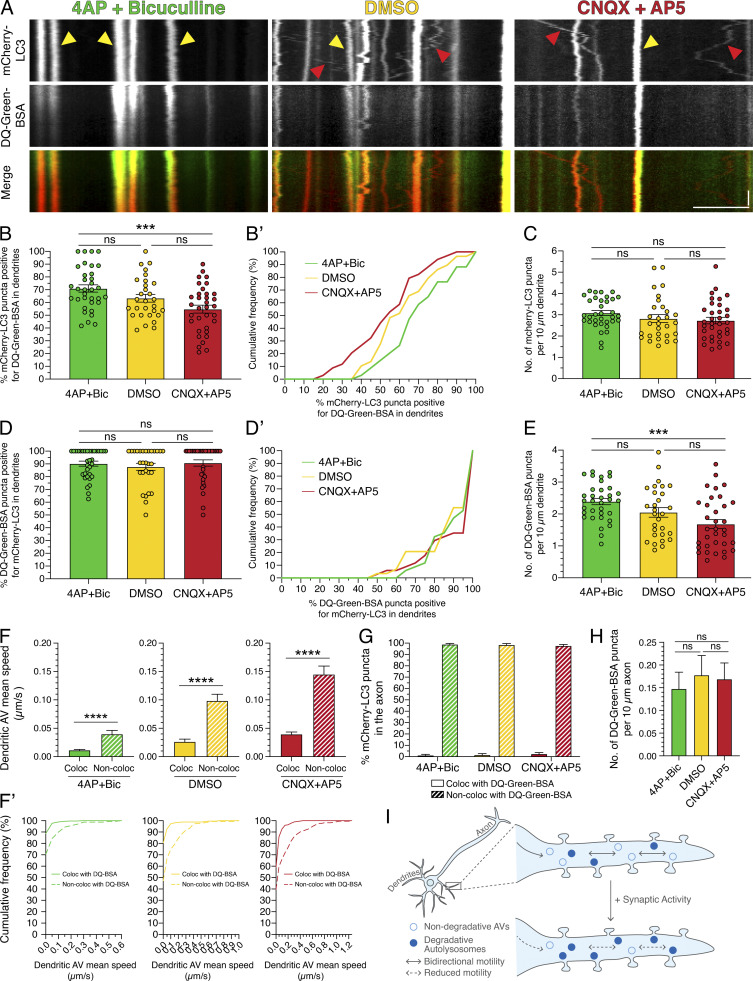
To determine whether the degradative state of dendritic AVs is regulated by synaptic activity, we measured the number of mCherry-LC3–positive AVs in dendrites that were copositive for DQ-Green-BSA under paradigms of stimulated versus silenced synaptic activity. In this experiment, neurons were preloaded for 2 h with DQ-BSA and then treated with modulators of synaptic activity. Consistent with our previous findings, mCherry-LC3 motility decreased with 4-AP + Bic treatment and increased with CNQX+AP5 treatment relative to the DMSO control (Fig. 10 A). Interestingly, we found that treatment with 4-AP + Bic increased the percentage of mCherry-LC3–positive AVs that were copositive for DQ-Green-BSA, while treatment with CNQX + AP5 reduced this degree of colocalization (Fig. 10, A, B, and B′). Thus, we found a higher percentage of degradative autolysosomes in dendrites with synaptic stimulation than with silencing. While the density of total mCherry-LC3 puncta in dendrites remained constant throughout activity paradigms (Fig. 10 C), the density of total DQ-Green-BSA puncta increased with synaptic activity (Fig. 10 E). Combined, these results indicate that synaptic stimulation increases the percentage of dendritic AVs that are degradative autolysosomes. These results are consistent with our previous observations that synaptic stimulation also increased the density of acidic organelles in dendrites (Fig. 8). Using the converse analysis, the percentage of DQ-Green-BSA–positive compartments that were copositive for mCherry-LC3 did not change across activity paradigms, suggesting that the percentage of degradative lysosomes that receive autophagic input remains constant (Fig. 10, D and D′).
Figure 10.
Synaptic activity increases degradative AVs in dendrites. (A) Kymographs of mCherry-LC3 and DQ-Green-BSA in dendrites of rat hippocampal neurons treated with 4-AP + Bic, DMSO, or CNQX + AP5 for 30 min. Yellow arrowheads denote puncta positive for both mCherry-LC3 and DQ-Green-BSA. Red arrowheads denote puncta positive for only mCherry-LC3. Retrograde direction is from left to right. Horizontal bar, 5 µm. Vertical bar, 10 s. (B and B’) Quantitation of the percentage of mCherry-LC3 puncta positive for DQ-Green-BSA in dendrites (B, mean ± SEM; B’, cumulative frequency; n = 29–34 neurons from four independent experiments; 15 DIV; one-way ANOVA with Tukey’s post hoc test; ***, P ≤ 0.001). (C) Number of mCherry-LC3 puncta per 10-µm dendrite (mean ± SEM; n = 29–34 neurons from four independent experiments; 15 DIV; one-way ANOVA with Tukey’s post hoc test). (D and D’) Quantitation of the percentage of DQ-Green-BSA puncta positive for mCherry-LC3 in dendrites (D, mean ± SEM; D’, cumulative frequency; n = 29–34 neurons from four independent experiments; 15 DIV; one-way ANOVA with Tukey’s post hoc test). (E) Number of DQ-Green-BSA puncta per 10-µm dendrite (mean ± SEM; n = 29–34 neurons from four independent experiments; 15 DIV; one-way ANOVA with Tukey’s post hoc test; ***, P ≤ 0.001). (F and F’) Mean speed of dendritic AVs that either colocalize or do not colocalize with DQ-Green-BSA (F, mean ± SEM; F’, cumulative frequency; n = 159–354 AVs from 29–34 neurons from four independent experiments; 15 DIV; unpaired t test; ****, P ≤ 0.0001). (G) Quantitation of the percentage of mCherry-LC3 puncta that colocalize or do not colocalize with DQ-Green-BSA puncta in axons treated with 4-AP + Bic, DMSO, or CNQX + AP5 for 30 min (mean ± SEM; n = 48–62 neurons from four independent experiments; 15 DIV). (H) Quantitation of the density of DQ-Green-BSA puncta in axons treated with 4-AP + Bic, DMSO, or CNQX + AP5 for 30 min (mean ± SEM; n = 46–62 neurons from four independent experiments; 15 DIV; one-way ANOVA with Tukey’s post hoc test). (I) Model for activity-dependent regulation of autophagy in dendrites. Under basal activity, AVs exhibit bidirectional movement in dendrites. Upon stimulation of synaptic activity, AV motility is dampened, and a population of degradative autolysosomes is increased.
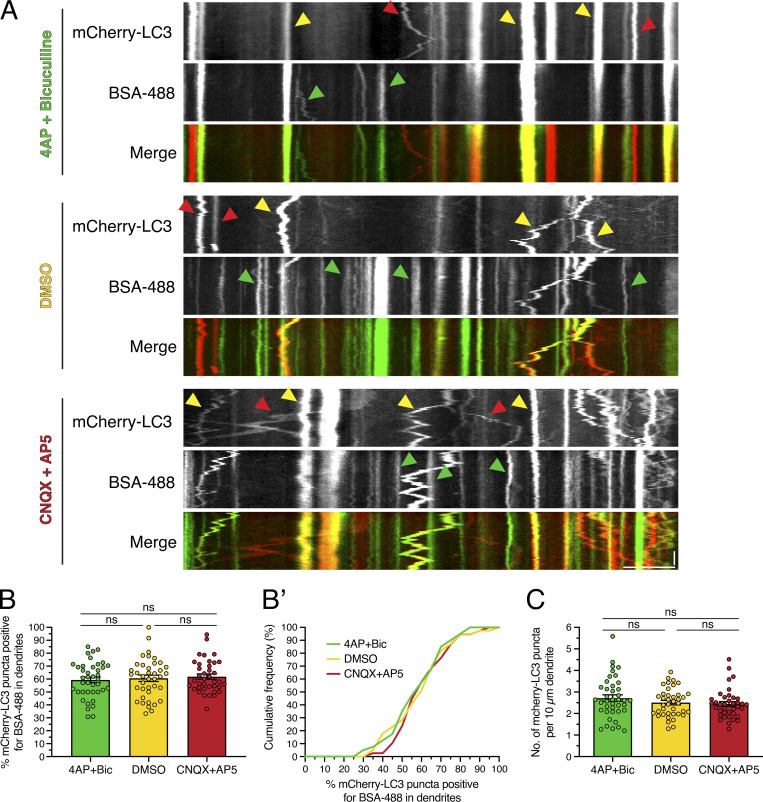
Our observations that synaptic activity increases the percentage of AVs marked with DQ-BSA could be attributed to a stimulation in either degradative activity or the delivery of endocytosed substrate (BSA) to autolysosomes that were already proteolytically competent. To distinguish between these possibilities, we measured the percentage of AVs receiving the endocytic cargo BSA-488 under the various paradigms of synaptic activity (Fig. S6). For this experiment, neurons were preloaded for 2 h with BSA-488 and then treated with modulators of synaptic activity. In contrast to our prior results with DQ-BSA, the percentage of mCherry-LC3–positive AVs that were copositive for BSA-488 did not change in response to modulating synaptic activity (Fig. S6, A, B, and B′). Consistent with our prior results, the density of mCherry-LC3 puncta in dendrites remained constant across the activity paradigms (Fig. S6 C). Thus, the activity-dependent increase in AVs labeled with DQ-BSA is likely not due to a stimulation in substrate delivery into autolysosomes; rather, these results suggest a stimulation in the presence of degradative compartments.
Figure S6.
Endocytic input to AVs does not change with synaptic activity. (A) Kymographs of mCherry-LC3 and BSA-488 in dendrites of rat hippocampal neurons treated with 4-AP + Bic, DMSO, or CNQX + AP5 for 30 min. Yellow arrowheads denote puncta positive for both mCherry-LC3 and BSA-488. Red arrowheads denote puncta positive for only mCherry-LC3. Green arrowheads denote puncta positive for only BSA-488. Retrograde direction is from left to right. Horizontal bar, 5 µm. Vertical bar, 10 s. (B and B’) Quantitation of the percentage of mCherry-LC3 puncta positive for BSA-488 in dendrites (B, mean ± SEM; B’, cumulative frequency; n = 38–40 neurons from four independent experiments; 13 DIV; one-way ANOVA with Tukey’s post hoc test). (C) Number of mCherry-LC3 puncta per 10-µm dendrite (mean ± SEM; n = 38–40 neurons from four independent experiments; 13 DIV; one-way ANOVA with Tukey’s post hoc test).
Strikingly, we observed these activity-dependent changes in AV maturation in only dendrites and not in axons. In axons, <3% of mCherry-LC3–positive AVs were copositive for DQ-Green-BSA in axons, and this value did not change in response to modulating synaptic activity (Fig. 10 G). Furthermore, the density of DQ-Green-BSA in axons also did not change across the activity paradigms (Fig. 10 H). Thus, the density of degradative organelles as labeled with DQ-BSA appears higher in dendrites than in axons and is specifically regulated with synaptic activity.
Last, we observed a pattern emerging between AV maturation state and dynamics. Across activity paradigms, mCherry-LC3–positive AVs that were copositive for DQ-Green-BSA exhibited lower mean speeds than mCherry-LC3 AVs that were negative for DQ-Green-BSA (Fig. 10, A, F, and F′). Thus, these data suggest that degradative AVs are less motile and that nondegradative AVs are more motile. Moreover, these observations suggest that AV transport and degradative function may be linked and that activity-dependent dampening of AV motility may be coupled with maturation into degradative organelles.
Discussion
How neuronal autophagy is regulated by synaptic activity is largely unexplored. Here, we defined the dynamics and degradative nature of autophagic organelles in compartments of the neuron (e.g., dendrites versus axons) in response to synaptic activity. We found that KCl-induced neuronal depolarization decreases AV motility in dendrites (Fig. 1). Furthermore, pharmacological manipulation of synaptic activity regulates AV motility in dendrites; stimulation of synaptic activity dampens AV motility, while silencing synaptic activity induces AV motility (Fig. 2). These effects occur in neurons grown in monoculture, as well as in neurons cocultured with astrocytes to recapitulate intercellular contacts observed in vivo (Fig. 5). Activity-dependent decreases in AV motility can be restored by silencing synaptic activity, suggesting that these effects are reversible and not due to cytotoxicity (Fig. 4). Most strikingly, synaptic activity does not dampen AV dynamics throughout the neuron. Rather, these effects are specific to dendrites and are not observed in axons (Figs. 1 and 3), indicating compartment-specific regulation of AV dynamics with synaptic activity. In fact, we also found that this process is regulated locally within dendrites because local stimulation of synaptic activity by uncaging MNI-glutamate is sufficient to reduce AV motility in dendrites (Fig. 6).
Because organelle motility and function can be linked (Korolchuk et al., 2011; Walton et al., 2018), we wanted to investigate the effects of synaptic activity on the degradative activity of autophagic organelles in dendrites. Thus, we needed to first characterize the maturation state of AVs under basal conditions. We found that ∼90% of mCherry-LC3–positive AVs in dendrites and axons are positive for the late endosomal/lysosomal marker LAMP1, suggesting that the vast majority of autophagic organelles in the neuron are either amphisomes or autolysosomes (Fig. S4). These results are consistent with prior work in axons showing that soon after autophagosomes form in the distal axon, they fuse with a LAMP1-positive compartment (Cheng et al., 2015; Lee et al., 2011; Maday and Holzbaur, 2016; Maday et al., 2012). To distinguish a population of AVs that are degradative autolysosomes, we incubated neurons with DQ-BSA, an endocytic substrate that fluoresces only upon proteolytic cleavage (Fig. S5). Under basal conditions, we found a higher proportion of degradative autolysosomes, as labeled with DQ-BSA, in dendrites than in axons (Fig. 9, A, B, B′, and B″). These compartment-specific differences are not due to the duration of incubation with DQ-BSA (Fig. 9, D and E; 2–8 h). Because DQ-BSA can label only those degradative compartments that are receiving endocytic input, we measured the percentage of mCherry-LC3–positive AVs that were copositive for the endocytic cargo BSA-488 and found a higher proportion of AVs positive for BSA-488 in dendrites than in axons (Fig. 9 C, C′, and C″). Thus, compared with axons, dendrites have a greater percentage of DQ-BSA labeled degradative autolysosomes due to compartment-specific differences in how autophagic organelles have fused with newly formed endosomes. Combined, these results reveal important differences in how the autophagy pathway intersects with the endolysosomal pathway in dendrites as compared with axons.
To determine the effects of synaptic activity on the degradative function of AVs, we pharmacologically stimulated or silenced synaptic activity and assessed changes in their proteolytic nature. Stimulating synaptic activity increased the percentage of dendritic AVs copositive for DQ-BSA (Fig. 10, A, B, and B′). This activity-induced increase in overlap between AVs and DQ-BSA is likely not due to an increase in endocytic delivery of the BSA substrate (Fig. S6). Interestingly, while the total number of AVs along the dendrite was unaltered upon changes in synaptic activity (Fig. 10 C), we did measure an increase in the presence of total DQ-BSA–labeled degradative compartments (Fig. 10 E). Consistent with this result, we observed that stimulating synaptic activity increased the density of acidic compartments relative to silencing within dendrites (Fig. 8). In sum, these observations suggest that dendritic AVs become more proteolytically active in response to stimulated synaptic activity. In contrast to dendrites, synaptic activity did not alter the percentage of AVs copositive for DQ-BSA or the density of total DQ-BSA–positive compartments, regardless of merging with autophagic compartments, in axons (Fig. 10, G and H). Thus, these data suggest that the activity-dependent increase in degradative autolysosomes is compartment specific, preferentially occurring in dendrites as compared with axons.
Combined, we propose a model whereby synaptic activity locally controls AV transport and degradative function in dendrites (Fig. 10 I). Our evidence suggests that synaptic activity may induce AV maturation into proteolytically active compartments in dendrites. Across all paradigms of synaptic activity, nondegradative AVs were more motile than degradative AVs (Fig. 10, F and F′), suggesting that AV motility and function may be correlated and that reduction in AV motility may facilitate maturation into proteolytically active compartments. We found that activity-dependent dampening of AV motility increases their residence time at or near post-synaptic compartments (Fig. 7). Thus, this process may facilitate local regulation of the synaptic proteome through degradation of post-synaptic components and/or through recycling of degradation products to fuel local protein synthesis. In total, our results suggest a more localized pathway for autophagy in dendrites, in contrast to the long-range pathway for autophagy that we previously defined in axons (Maday and Holzbaur, 2014; Maday and Holzbaur, 2016; Maday et al., 2012). While the mechanism underlying the increase in degradative activity of AVs in response to synaptic stimulation is yet to be unraveled, we note that increases in degradative capacity of lysosomal compartments have been observed in response to cellular signals in other settings (Trombetta et al., 2003). For example, during dendritic cell maturation, lysosomes become degradative through recruitment and activation of the vacuolar proton pump to promote processing of internalized antigens (Trombetta et al., 2003). Future studies will delineate the mechanism by which AVs become more degradative in dendrites in response to synaptic stimulation.
Autophagy may control the composition of the synaptic proteome through degradative, as well as biosynthetic, functions to impact the strength of a synapse. Autophagy has been shown to degrade post-synaptic components. In chemical models of long-term depression, autophagy facilitates the degradation of excitatory post-synaptic AMPA receptors (Shehata et al., 2012). Autophagy can also negatively regulate brain-derived neurotrophic factor–mediated long-term potentiation (LTP) by degrading post-synaptic scaffolding proteins (Nikoletopoulou et al., 2017). AVs may receive post-synaptic cargoes for degradation either by local fusion with endosomes or by inward budding of the autophagic membrane, similar to multivesicular body formation.
While most autophagy studies focus on cargo selection and elimination, another major function of autophagy is to liberate amino acids and lipids for new biosynthesis. Because we found that local formation of autophagosomes in dendrites is infrequent (Maday and Holzbaur, 2014), dendritic AVs may be sourced from the soma and preloaded with cargo. In this capacity, dendritic AVs may provide biosynthetic fuel, on demand in response to activity, for local translation at post-synaptic compartments, a way of creation by destruction. Consistent with this possibility, studies show that autophagy is required for LTP and LTP-induced dendritic spine growth (Glatigny et al., 2019; Zhao et al., 2015). In total, alterations in these pathways may provide a mechanistic basis for the learning and memory defects reported in autophagy-deficient models (Glatigny et al., 2019; Hylin et al., 2018; Nikoletopoulou et al., 2017; Pandey et al., 2020; Zhao et al., 2015).
Our observations regarding the impact of synaptic activity on autophagy are consistent with recent reports linking synaptic activity with lysosome function in post-synaptic compartments. Goo et al. reported that synaptic activity induces the localization of LAMP1-positive compartments to dendritic spines that may promote the degradation of AMPA receptors (Goo et al., 2017). Inhibition of lysosome function decreases dendritic spine density (Goo et al., 2017). Padamsey et al. demonstrated that synaptic activity induces exocytosis of lysosomes from dendrites, leading to remodeling of the extracellular matrix and enlargement of dendritic spines (Padamsey et al., 2017). We found significant overlap between LAMP1-labeled late endosomes/lysosomes and autophagic compartments in dendrites and axons (Fig. S4). We, along with other groups, previously reported that only newly formed autophagosomes in distal regions of the axon are negative for LAMP1 and mature into LAMP1-positive amphisomes during retrograde transport along the midaxon (Cheng et al., 2015; Lee et al., 2011; Maday and Holzbaur, 2016; Maday et al., 2012). In dendrites, we previously found very few immature autophagosomes as labeled with the acid-sensitive GFP-LC3 (Maday and Holzbaur, 2014), and our present study indicates a predominance of autolysosomes in dendrites, as evidenced with the mCherry-GFP-LC3 reporter (Fig. S1). Additionally, we found that ∼90% of degradative lysosomes labeled with DQ-BSA are copositive for LC3 (Fig. 10, D and D′), suggesting that nearly all DQ-BSA–labeled degradative lysosomes receive autophagic input. Consequently, it is challenging to untangle specific effects of synaptic activity on autophagic versus lysosomal maturation into degradative organelles. Future experiments will need to more specifically distinguish these possibilities.
Synaptic activity has been shown to regulate the distribution and function of other organelles and degradative machinery in dendrites. For example, neuronal activity recruits proteasomes to dendritic spines and stimulates their activity (Bingol and Schuman, 2006); activity-dependent proteasomal function results in new spine outgrowth (Hamilton et al., 2012). Synaptic activity also decreases the motility of post-ER secretory compartments (ER-Golgi intermediate compartments) in dendrites, resulting in increased appearance of receptors on the post-synaptic membrane (Hanus et al., 2014). In our study, we found that early endosomes labeled with Rab5 also arrest in response to synaptic activity (Fig. S3, B–G). Interestingly, mitochondrial movement is arrested in response to synaptic activity in both dendrites and axons via a mechanism that involves calcium-sensitive adaptor proteins that inactivate kinesin-based motility; synaptic mitochondria provide local ATP and calcium buffering (Chen and Sheng, 2013; MacAskill et al., 2009; Wang and Schwarz, 2009). Given the rapid effects of synaptic activity on AV dynamics, calcium may also regulate AV motility in dendrites. However, this mechanism of action is not applied in the axon, where AV motility is unaffected by increased calcium levels (Fig. 3). In total, activity-dependent effects on autophagy may be part of a broader mechanism to specifically localize biosynthetic and degradative organelles at post-synaptic compartments to impact synaptic function.
Most studies to date investigating synaptic autophagy have focused on autophagy in presynaptic compartments. Autophagy is important for the development as well as maintenance of presynaptic terminals (Komatsu et al., 2007; Nishiyama et al., 2007; Rudnick et al., 2017; Shen and Ganetzky, 2009; Stavoe et al., 2016; Yamaguchi et al., 2018). Studies have also shown that autophagy regulates presynaptic neurotransmission, potentially by controlling synaptic vesicle pool size (Hernandez et al., 2012; Rudnick et al., 2017). Axonal autophagosomes preferentially form in the distal axon and undergo retrograde movement toward the cell soma for degradation (Lee et al., 2011; Maday et al., 2012). Autophagosome biogenesis in the distal axon is regulated by several presynaptic proteins and can be induced with synaptic activity (Hill et al., 2019; Okerlund et al., 2017; Soukup et al., 2016). Wang et al. demonstrated that neuronal depolarization stimulates distal biogenesis and retrograde flux of autophagosomes from presynaptic terminals to the soma (Wang et al., 2015). Our study, however, provides new insights into the activity-dependent regulation of autophagic degradation in post-synaptic compartments, emphasizing the significance of degradative pathways on both sides of the synapse.
Here, we define a pathway for the activity-dependent regulation of autophagy in dendrites that is distinct from mechanisms previously described in the axon. This pathway may be particularly important in neurons with large receptive fields, as in the elaborate dendritic arbors of cerebellar Purkinje cells. Furthermore, alterations in this pathway may contribute to the preferential susceptibility of Purkinje cells to autophagy loss, a phenomenon observed across various mouse models deficient in key autophagy genes (Hara et al., 2006; Komatsu et al., 2006; Liang et al., 2010; Yamaguchi et al., 2018). Thus, compartment-specific roles for autophagy may be critical to enable the diverse demands and functions of different neuronal populations.
Materials and methods
Reagents
Constructs used included mCherry-EGFP-LC3B (gift from Terje Johansen, University of Tromsø, Tromsø, Norway), mCherry-LC3B (Maday and Holzbaur, 2016), GCaMP3 (Addgene; 22692), LAMP1-mGFP (Addgene; 34831), PSD-95–GFP (Addgene), GFP-Rab5 (gift from Marino Zerial, Max Planck Institute of Molecular Cell Biology and Genetics, Dresden, Germany), and mRFP-Rab5 (Addgene; 14437). BSA conjugates used included BSA-488 (Thermo Fisher Scientific/Molecular Probes; A13100), BSA-647 (Thermo Fisher Scientific/Molecular Probes; A34785), DQ-Green-BSA (Thermo Fisher Scientific/Molecular Probes; D12050), and DQ-Red-BSA (Thermo Fisher Scientific/Molecular Probes; D12051). Fluo-4 acetoxymethyl ester (Fluo-4 AM) was purchased from Thermo Fisher Scientific/Molecular Probes (F14201). Primary antibodies used for immunofluorescence included chicken anti-GFP (Aves Labs, Inc.; GFP1020), rabbit anti–glial fibrillary acidic protein (EMD Millipore; AB5804), and mouse anti-TUBB3/β3 tubulin (Novus Biologicals; MAB1195-SP). Secondary antibodies used for immunofluorescence included goat anti-chicken Alexa Fluor 488 (Jackson ImmunoResearch Laboratories; 103-545-155), goat anti-rabbit Alexa Fluor 488 (Thermo Fisher Scientific/Invitrogen; A11034), and goat anti-mouse Alexa Fluor 647 (Thermo Fisher Scientific/Invitrogen; A32728). Small molecules used included 4-AP (Thermo Fisher Scientific/Tocris Bioscience; 0940), Bic methiodide (Thermo Fisher Scientific/Tocris Bioscience; 2503), CNQX (Thermo Fisher Scientific/Tocris Bioscience; 0190), AP5 (Thermo Fisher Scientific/Tocris Bioscience; 0106), MNI-glutamate (Tocris Bioscience; 1490), E64d (Sigma-Aldrich; E8640), and pepstatin A (Sigma-Aldrich; P5318). Lipofectamine 2000 reagent (Thermo Fisher Scientific/Invitrogen; 11668-027) was used for transfecting neurons, following the manufacturer’s protocol.
Primary neuron culture
Rat hippocampal and rat cortical neurons were obtained from the Neurons R Us core facility at the University of Pennsylvania and as a gift from Dr. Kelly Jordan-Sciutto (University of Pennsylvania, Philadelphia, PA). Neurons were prepared from Sprague-Dawley rat embryos at gestational days 16–18. Neurons were plated at a density of 3,000,000 cells per 10-cm dish (∼4.7 × 104 cells/cm2) filled with eight 25-mm acid-washed glass coverslips coated with 1 mg/ml poly-L-lysine. Neurons were grown for 13–16 DIV in rat neuron maintenance medium (neurobasal medium supplemented with 2% B-27, 33 mM glucose, 100 U/ml penicillin, and 100 µg/ml streptomycin) at 37°C in a 5% CO2 incubator. After 3 DIV, 1.7 µM AraC (antimitotic drug, C6645; Sigma-Aldrich) was added, and every 7–8 DIV, ∼20% of medium volume was added.
Neuron–astrocyte coculture
Primary mouse cortical astrocytes (either GFP-LC3–transgenic or nontransgenic astrocytes) were isolated from GFP-LC3–transgenic mice [B6.Cg-Tg(CAG-EGFP/LC3)53Nmi/NmiRbrc; RIKEN BioResource Center] as we previously described (Dong et al., 2019; Kulkarni et al., 2020). Mouse cortical astrocytes were grown in glial media (DMEM supplemented with 10% FBS, 0.25% GlutaMAX, 100 U/ml penicillin, and 100 µg/ml streptomycin) on 10-cm tissue culture plates until ∼80–90% confluent. When the neuron culture was 7–8 DIV, mouse cortical astrocytes were dissociated from the 10-cm plate using 0.05% trypsin-EDTA. Upon dissociation from the 10-cm plate, trypsin was inactivated using glial media. Astrocytes were then resuspended in coculture media (neurobasal supplemented with 2% B-27, 1% G-5, 0.25% GlutaMAX, 100 U/ml penicillin, and 100 µg/ml streptomycin) and plated on the neuron culture and maintained at 37°C in a 5% CO2 incubator. For immunostain analysis, rat hippocampal neurons were plated at a density of 960,000 cells per 60-mm dish (∼4.7 × 104 cells/cm2) filled with 12 × 12–mm acid-washed glass coverslips coated with 1 mg/ml poly-L-lysine, onto which astrocytes were plated at a density of ∼6,600 cells/cm2. For live-cell imaging, rat cortical neurons were plated at a density of 3,000,000 cells per 10-cm dish (∼4.7 × 104 cells/cm2) filled with 8 × 25–mm acid-washed glass coverslips coated with 1 mg/ml poly-L-lysine, onto which astrocytes were plated at a density of ∼13,000–20,000 cells/cm2. After 2 d of coculture, 2 µM AraC was added to prevent cell division and promote maturation of the astrocytes. The coculture was maintained in the coculture medium for up to 8 d, with the day of astrocyte plating counted as day 0.
Live-cell imaging
Coverslips (25 mm) were sandwiched within a Chamlide CMB magnetic imaging chamber (BioVision Technologies). Live-cell imaging of organelle dynamics was performed on a BioVision spinning-disk confocal microscope system consisting of a Leica DMi8 inverted widefield microscope, a Yokagawa W1 spinning-disk confocal microscope, and a Photometrics Prime 95B scientific complementary metal–oxide–semiconductor camera. Images were acquired with VisiView software using a 63×/1.4 NA plan apochromat oil-immersion objective (with or without a 2× optical multiplier) or a 40×/1.3 NA plan apochromat oil-immersion objective and solid-state 405-, 488-, 561-, and 640-nm lasers for excitation. Fluo-4 imaging was performed on a Leica DMi8 inverted widefield microscope with a Hamamatsu Orca Flash 4.0 V2+ scientific complementary metal–oxide–semiconductor camera using a 20×/0.80 NA plan apochromat dry PH2 objective, FITC filter cube, and Leica software. Both microscopes are equipped with an environmental chamber at 37°C and adaptive focus control to maintain a constant focal plane during live-cell imaging.
Immunostaining of cocultured neurons and astrocytes
On days 3, 5, and 6 of the coculture, samples were fixed for 10 min in 4% PFA/4% sucrose in PBS (150 mM NaCl, 50 mM NaPO4, pH 7.4) previously warmed to 37°C. Cells were washed two times in PBS, permeabilized for 5 min in 0.1% Triton X-100 (Thermo Fisher Scientific; BP151-100) in PBS, washed two times in PBS, and then blocked for 1 h in PBS supplemented with 5% goat serum (Sigma-Aldrich; G9023) and 1% BSA (Thermo Fisher Scientific; BP1605-100). Cells were then incubated in primary antibody diluted in block solution for 1 h at room temperature, washed three times for 5 min each in PBS, and then incubated in secondary antibody diluted in block solution for 1 h at room temperature. Following incubation in secondary antibody, samples were washed three times for 5 min each in PBS. Samples were mounted in ProLong Gold and imaged with the BioVision spinning-disk confocal microscope system described above using the 40× or 63× objectives and using 488- and 640-nm lasers for excitation. Z-stacks spanning the entire depth of the field of neurons and astrocytes were obtained at 0.2-µm sections.
Photobleaching experiments
Primary rat hippocampal neurons were transfected with mCherry-GFP-LC3 at 7–8 DIV using Lipofectamine 2000. At 13–15 DIV, neurons were imaged at 1 frame per second for 1 min in E4 solution (150 mM NaCl, 3 mM KCl, 4 mM CaCl2, 15 mM glucose, 10 mM Hepes, pH 7.4) with the 63× objective and a 2× optical multiplier (total of 126× magnification). After a precount of 3 s, regions along the dendrite (as indicated in Fig. S1, A and B) were photobleached using the 488-nm laser. The dendrite was then imaged for an additional 57 s to observe the nature of vesicles moving into the photobleached region.
KCl and NaCl on-scope addition
Primary rat hippocampal neurons were transfected with mCherry-GFP-LC3 at 7–8 DIV using Lipofectamine 2000. At 13–14 DIV, neurons were imaged at 1 frame per second for 5 min in 500 µl of E4 solution (150 mM NaCl, 3 mM KCl, 4 mM CaCl2, 15 mM glucose, 10 mM Hepes, pH 7.4) with the 63× objective and a 2× optical multiplier (total of 126× magnification). Approximately halfway through the movie (∼149–150 s), 500 µl of 90 mM KCl or 90 mM NaCl diluted in E4 was added to the imaging chamber to reach a final concentration of 45 mM KCl or NaCl. Dendrites and axons were distinguished as we previously published (Maday and Holzbaur, 2014; Maday and Holzbaur, 2016), using morphological criteria established by Kaech and Banker (2006).
Pharmacological treatments to modulate synaptic activity
Primary rat hippocampal neurons were transfected with mCherry-LC3 and GCaMP3 (Figs. 2, 3, and 4), with mCherry-LC3 and PSD-95–GFP (Fig. 7), or with mCherry-LC3 and LAMP1-mGFP (Fig. S4) at 7–9 DIV using Lipofectamine 2000. At 13–15 DIV, neurons were treated with 50 µM 4-AP and 20 µM Bic (to stimulate synaptic activity), 50 µM CNQX and 50 µM AP5 (to silence synaptic activity), or an equal volume of DMSO solvent, in E4 solution for 30 min at 37°C in a 5% CO2 incubator. For the axon analysis in Fig. 3, samples were treated for 60 min at 37°C in a 5% CO2 incubator. Samples were imaged in the same treatment solution using the 63× objective and a 2× optical multiplier (total of 126× magnification) for a maximum of ∼30 min while on the microscope. For AV tracking in Figs. 2, 3, and 4, neurons were imaged at 1.4 frames per second for 7 min. For synapse association experiments in Fig. 7 and LAMP1 experiments in Fig. S4, neurons were imaged at 1 frame per second for 5 min. For the coculture experiments in Fig. 5, B–D, rat cortical neurons were transfected with mCherry-LC3 on 6–7 DIV 1 d before the addition of astrocytes. After 7–8 d of coculture (neurons are a total of 14–16 DIV), pharmacological treatments were performed as above, and neurons were imaged at 1 frame per second for 3 min.
For the rescue experiments in Fig. 4, mCherry-LC3 and GCaMP3 transfected neurons were first treated with 50 µM 4-AP and 20 µM Bic in E4 solution for 30 min at 37°C in a 5% CO2 incubator. One cohort (i.e., control samples) was treated with fresh 50 µM 4-AP and 20 µM Bic in E4 solution for an additional 30 min, whereas the other cohort (i.e., rescue samples) was treated with 50 µM CNQX and 50 µM AP5 in E4 solution for 30 min at 37°C in a 5% CO2 incubator. Neurons were imaged in the same treatment solution at 1.4 frames per second for 7 min with the 63× objective and a 2× optical multiplier (total of 126× magnification). Data were acquired for a maximum of ∼30 min while on the microscope.
Glutamate uncaging
Primary rat hippocampal neurons (Fig. 6 and Fig. S3, B–G) or primary rat cortical neurons (Fig. S3 A) were transfected with mCherry-LC3 and GCaMP3 (Figs. 6 and S3 A) or with mRFP-Rab5 and GCaMP3 (Fig. S3, B–G) at 6–8 DIV using Lipofectamine 2000. At 13–15 DIV, glutamate uncaging was performed. Neurons were first incubated for 10 min in E4 salt solution to equilibrate the neuronal field firing (described below). We then supplemented E4 with 0.5 mM MNI-glutamate or an equal volume of water as a solvent control. Neurons were imaged on the BioVision confocal spinning-disk system described above, with the 63× objective and a 2× optical multiplier (total of 126× magnification), and data from each coverslip were acquired for a maximum of ∼30 min while on the microscope. For each movie, neurons were imaged at 1 frame/s for 30 s before photo-uncaging to establish the baseline organelle motility. During the uncaging period, a region along the dendrite was exposed to the 405-nm laser at 0.33 Hz (one cycle every 3 s) for a total of 40 cycles over 2 min. Every pixel in the uncaged region received the 405-nm laser for 3 ms. Images were acquired after each cycle of photo-uncaging.
Fluo-4 AM and GCaMP3 imaging
In Fig. 1 B and Fig. 2, A–C, primary rat hippocampal neurons were incubated with 4 µM Fluo-4 AM for 20 min in rat neuron maintenance medium at 37°C in a 5% CO2 incubator. Neurons were then washed twice with rat neuron maintenance medium, transferred to E4 solution [or to Hibernate E (BrainBits) supplemented with 2% B-27 and 2 mM GlutaMAX for Fig. 2 A], and imaged at 1 frame per second for the indicated time. For imaging calcium dynamics in transfected versus untransfected neurons (Fig. 2 C), neurons were transferred to E4 solution containing 50 µM 4-AP and 20 µM Bic or 50 µM CNQX and 50 µM AP5 and incubated for 60 min at 37°C in a 5% CO2 incubator, followed by imaging in the same solution at 1 frame per second for 6 min.
To qualitatively assess changes in firing patterns in E4 solution (Fig. S2, A and B), we measured calcium oscillations by GCaMP3 and Fluo-4 fluorescence as a function of time of incubation in E4. In Fig. S2 A, primary rat hippocampal neurons were transfected with GCaMP3 at 8 DIV using Lipofectamine 2000. At 14 DIV, GCaMP3-transfected neurons were transferred to E4 solution and imaged at 2 and 10 min after E4 addition at 1 frame per second for 5 min. As a control, we confirmed the inhibitory effect of Mg2+ on neuronal firing in neurons incubated for 2 min in E4 with 2 mM Mg2+ (150 mM NaCl, 3 mM KCl, 2 mM CaCl2, 2 mM MgCl2, 15 mM glucose, 10 mM Hepes, pH 7.4). In Fig. S2 B, primary rat hippocampal neurons were preloaded with Fluo-4 as described above; transferred to E4 solution; and imaged at 2, 10, and 30 min after E4 addition at 1 frame/s for 3 min. The same coverslip was imaged for all three time points and returned to 37°C in a 5% CO2 incubator between incubation times. Combined, we found that calcium dynamics change with time of incubation in E4 solution. Initially, we observed sporadic and individual neuron events that transition to a synchronized and larger field firing of the entire culture with time in E4 (Fig. S2, A and B).
For high-speed imaging of calcium dynamics (Fig. S2, C and D), primary rat hippocampal neurons were transfected with GCaMP3 at 7–8 DIV using Lipofectamine 2000. At 13–14 DIV, neurons were treated with 50 µM 4-AP and 20 µM Bic, 50 µM CNQX and 50 µM AP5, or an equal volume of DMSO solvent, in E4 solution for 30 min at 37°C in a 5% CO2 incubator. Neurons were imaged in the same treatment solution at 10 frames per second for 1 min with the 63× objective.
BSA and DQ-BSA experiments
Primary rat hippocampal neurons were transfected with mCherry-LC3 at 7–8 DIV using Lipofectamine 2000. At 13–14 DIV, neurons were treated with 25 µg/ml BSA-488 or DQ-Green-BSA in rat neuron maintenance medium for 2 h (or 4, 6, and 8 h for the DQ-Green-BSA time-course experiments in Fig. 9, D and E) and rinsed twice with rat neuron maintenance medium. Neurons were either imaged in E4 solution immediately (as in Fig. 9) or treated with 50 µM 4-AP and 20 µM Bic, 50 µM CNQX and 50 µM AP5, or an equal volume of DMSO solvent, in E4 solution for 30 min at 37°C in a 5% CO2 incubator (as in Figs. 10 and S6) and imaged in the same treatment solution. Images were acquired at 1.4 frames per second for 42 s with the 63× objective and a 2× optical multiplier (126× total magnification); data were acquired for a maximum of 30 min on the microscope. For the dye validation experiments in Fig. S5, primary rat hippocampal neurons, untransfected or transfected with GFP-Rab5 at 7–8 DIV using Lipofectamine 2000, were treated at 13–15 DIV with 25 µg/ml DQ-Green/Red-BSA, BSA-488, or BSA-647 in rat neuron maintenance medium for 2 h and rinsed twice with rat neuron maintenance medium. Neurons were then imaged in E4 solution at 1–1.4 frames per second for 30–48 s with the 63× objective with a 2× multiplier (126× total magnification).
Treatment with protease inhibitors
Primary rat hippocampal neurons (23 DIV) were treated with 100 µM of E64d and 30 µM pepstatin A, or with an equivalent volume of DMSO as a solvent control, in rat neuron maintenance medium for 3 h. During the last hour of treatment, neurons were also treated with 25 µg/ml BSA-488 or DQ-Green-BSA in rat neuron maintenance medium. After treatment, neurons were washed twice in HibE imaging medium and imaged in HibE imaging medium. Z-stacks were obtained using the BioVision spinning-disk confocal microscope system described above by taking 0.2-µm sections using the 40× objective.
Lysotracker-Red experiments
Primary rat hippocampal neurons were transfected with GCaMP3 at 7–8 DIV using Lipofectamine 2000. At 13–14 DIV, neurons were treated with 100 nM Lysotracker-Red in rat neuron maintenance medium for 30 min. Following Lysotracker-Red treatment, neurons were treated with 50 µM 4-AP and 20 µM Bic, 50 µM CNQX and 50 µM AP5, or an equal volume of DMSO solvent, in E4 solution containing 100 nM Lysotracker-Red for 30 min at 37°C in a 5% CO2 incubator (Fig. 8). After treatment, neurons were washed twice with E4 solution containing 50 µM 4-AP and 20 µM Bic, 50 µM CNQX and 50 µM AP5, or an equal volume of DMSO solvent, and imaged in E4 solution containing activity modulators or DMSO control. Images were acquired at 1 frame per second for 3 min with the 63× objective and a 2× optical multiplier (126× total magnification); data were acquired for a maximum of ∼30 min on the microscope.
Image analysis
Quantitation of calcium oscillations using Fluo-4 AM and GCaMP3 imaging
For Fluo-4 imaging, the mean gray value of the entire field of view was determined for all images within a time series using Fiji software. Then, the mean gray value of an area within the field that did not contain any neurons or neurites, and represented background, was determined for all images within a time series. For each time point, the background value was subtracted from the corresponding field value. Each time point was then normalized to the first time point and expressed as a percentage (designated arbitrary units [AUs]). For GCaMP3 imaging, the mean gray value of an area within the somatodendritic region of a GCaMP3-expressing neuron was determined for all images within a time series. Each time point was then normalized to the first time point and expressed as a percentage (designated AUs). For Fig. S2, C and D, GCaMP3 spike frequency was calculated by generating kymographs in the somatodendritic region and counting the number of discrete firing events; data were plotted as the number of events/s.
Tracking AV dynamics with KCl and NaCl on-scope addition
Using the Kymoanalyzer plugin for Fiji (Neumann et al., 2017), kymographs were generated using line widths of 9 pixels for dendrites and 3 pixels for axons. In dendrites, kymographs were drawn >10 µm from the soma; kymographs ranged from ∼30 to ∼100 µm in length, measuring the late proximal to medial-distal region of the dendrite. For the dendrite analysis, only LC3-positive puncta that moved a total distance of 5 µm during the 2.5-min pretreatment window, and that could be tracked unambiguously for the entire duration of the movie, were manually tracked before and after treatment. Using the segmented line tool, individual tracks were manually traced by clicking on constant velocity segments. For each vesicle, the mean speed was measured by summing the individual segment lengths to get total distance traveled and dividing by the total time traveled. For the axon analysis, neighboring axons in the same field of view as analyzed dendrites were used for measurements. Because axonal AVs exhibit more processive movement than those in dendrites, individual AVs in the axon often do not remain within the field of view for the entire duration of the movie. Thus, we were unable to limit the tracking to only the same AVs before and after treatment as performed in the dendrite analysis. Rather, we tracked nearly all LC3 puncta visible in the axon throughout the movie.
Tracking AV and early endosome dynamics with modulators of synaptic activity
Using the KymoToolBox plugin for Fiji (Hangen et al., 2018), kymographs were generated using line widths of 10 pixels for dendrites and 3 pixels for axons. In dendrites, kymographs were drawn >10 µm from the soma; kymographs ranged from ∼30 to ∼100 µm in length, measuring the late proximal to medial-distal region of the dendrite. Only one dendrite was analyzed per neuron. In the axon, kymographs were drawn in the midaxon, at least 10 µm from the soma; directionality was confirmed by tracing the axon back to the soma. In both dendrite and axon kymograph analyses, nearly all AVs were tracked. To measure vesicle speeds, individual tracks were manually traced, using the segmented line tool, by clicking on constant velocity segments. For each vesicle tracked, the KymoToolBox plugin calculated a mean speed (total distance traveled divided by total time tracked). To generate an AV mean speed per neuron (Fig. 2 H), all the vesicles tracked from a single neuron were averaged to generate one value for AV mean speed. For glutamate uncaging experiments, movies were split into pre-uncaging (frames 1–29) and post-uncaging (frames 30–151), and kymographs were generated as described above. Nearly all AVs detected in the kymograph were manually tracked as above. Within the total population of AVs tracked, those AVs that could be traced in both the pre-uncaging and post-uncaging kymographs were identified, and their speeds were plotted separately and designated as “paired tracks.”
Colocalization measurements
Using the KymoToolBox plugin for Fiji (Hangen et al., 2018), kymographs were generated using line widths of 10 pixels for dendrites and 3 pixels for axons. In dendrites, kymographs were drawn >10 µm from the soma; kymographs ranged from ∼30 to ∼100 µm in length, measuring the late proximal to medial-distal region of the dendrite. Only one dendrite was analyzed per neuron. For the colocalization analysis in dendrites, only puncta present in the first frame of the movie and associated kymograph were included. Given the short-range movements exhibited by AVs in dendrites, puncta identified in the first frame account for >95% of dendritic AVs visible throughout the kymograph. Neighboring axons in the same field of view as analyzed dendrites were used for measurements in the axon analysis in Fig. 9; Fig. 10, G and H; and Fig. S4, B and B′. For the colocalization analysis in axons, because AVs in axons exhibit long-range movement and quickly move out of the field of view, nearly all vesicles detected throughout the movie and associated kymograph were included. For all colocalization measurements, both the movie and kymograph were consulted, and two markers were binned as colocalized if they overlapped and comigrated within the neurite. The number of AVs copositive for another marker (DQ-Green-BSA, BSA-488, or LAMP1-GFP) was expressed as a percentage of total AVs counted for each individual neurite. Similarly, the number of DQ-BSA puncta that were copositive for LC3 was expressed as a percentage of total DQ-BSA–positive puncta per neurite. The same calculations were performed for the markers in Fig. S5, B–D. The analysis in Fig. 10 for dendrites was performed blinded to the identity of the sample for three of four independent experiments. In Fig. 10, F and F′, the trajectories of the same AVs included in the colocalization and density analysis were manually traced by clicking on constant velocity segments. The KymoToolBox plugin calculated a mean speed (total distance traveled divided by total time tracked) for each vesicle tracked, and mean speeds were plotted separately on the basis of colocalization with DQ-BSA.
Quantitation of organelle density
For density measurements in either dendrites or axons, only puncta present in the first frame of the movie and associated kymograph were included in the analysis. The entire movie and associated kymograph were used, however, to resolve the number of individual puncta within clusters present in the first frame. AV and DQ-BSA density were measured as the number of discrete puncta, normalized for kymograph length (in µm) and plotted as the number of puncta per 10 µm of neurite. For quantitation of Lysotracker-Red puncta density (Fig. 8), the KymoToolBox plugin was used to generate straightened kymostacks with a width of 50 pixels using the criteria mentioned above for location along the dendrite. Because the GCaMP3 signal serves as a space filler for dendrites, we used the wand tool in Fiji to generate an outline of the dendrite based on the first frame of the GCaMP3 kymostack. This outline was overlaid onto the first frame of the respective kymostack for the Lysotracker-Red channel. Only discrete Lysotracker-Red puncta present inside the dendrite outline were counted; however, the entire movie was considered to determine whether a Lysotracker-Red punctum was contained within each dendrite of interest as opposed to a crossing axon or dendrite. The number of Lysotracker-Red puncta was normalized for kymostack total length (per µm) and plotted as the number of puncta per 10 µm of dendrite.
Quantitation of synapse association
Using the KymoToolBox plugin for Fiji (Hangen et al., 2018), kymographs were generated for the mCherry-LC3 and PSD-95–GFP channels using a line width of 10 pixels for dendrites, as described above. AV tracks were then manually traced by clicking on constant velocity segments in the mCherry-LC3 kymograph, and the AV tracks were then overlaid onto the PSD-95–GFP kymograph. Referencing both the movie and PSD-95 kymograph (with overlaid AV tracks), AVs were binned into one of three categories based on their proximity and duration at PSD-95–positive puncta: (1) “arrested” AVs, defined as AVs that localize within 0.75 µm on either side of a synapse for the entire duration of the AV track; (2) AVs with “segmented association,” where at least one segment of the AV track displays a sustained localization within 0.75 µm on either side of a synapse for ≥3 s; or (3) AVs that exhibit “no association” with a post-synaptic compartment during its trajectory, where the AV track did not have a sustained localization within 0.75 µm on either side of a synapse for ≥3 s. The number of AVs in each of these categories was plotted as a percentage of total AVs counted from either (1) a pool of all AVs combined across different neurons and experiments (Fig. 7 C) or (2) individual dendrites (Fig. 7 D).
Quantitation of dendritic AV maturation state by photobleaching analysis
The identity of the vesicles moving into the photobleached regions was assessed using kymographs generated in both red (mCherry) and green (GFP) channels using the KymoToolBox plugin for Fiji and a line width of 10 pixels for dendrites (Hangen et al., 2018). Vesicles were then binned either as copositive for GFP and mCherry or as positive for only mCherry.
Figure preparation and statistical analyses
All image measurements were obtained from the raw data. GraphPad Prism was used to plot graphs and perform statistical analyses; statistical tests are denoted within each figure legend. For data that do not appear normally distributed, the large n value (>20–25 data points) justifies a parametric test. For presentation of images, maximum and minimum gray values were adjusted linearly in Fiji, and images were assembled in Adobe Illustrator.
Online supplemental material
Fig. S1 shows that immature autophagic compartments are more readily detected in axons than in dendrites. Fig. S2 shows a quantitation of calcium oscillations in rat hippocampal neurons in response to various treatments. Fig. S3 A shows that local activation of synapses by uncaging MNI-glutamate also reduces dendritic AV motility in cortical neurons. Fig. S3, B–G, shows that local stimulation of synaptic activity by uncaging MNI-glutamate reduces early endosome motility in dendrites of hippocampal neurons. Fig. S4 shows that nearly all AVs in dendrites and axons are positive for LAMP1. Fig. S5 shows that DQ-BSA labels degradative compartments in the endolysosomal pathway in hippocampal neurons. Fig. S6 shows that endocytic input to AVs does not change with synaptic activity.
Acknowledgments
We thank Dr. Mary Putt in the Biostatistics and Bioinformatics Core at the Intellectual and Developmental Disabilities Research Center at the Children’s Hospital of Philadelphia for advice with statistical analyses; Dr. Kelly Jordan-Sciutto and her laboratory (University of Pennsylvania, Philadelphia, PA) for providing primary rat cortical and hippocampal neurons; and Dr. Kacy Cullen and Dr. John O’Donnell for advice with the neuron–glia coculture. We also thank members of the Maday laboratory for advice with experiments and preparation of the manuscript.
This work was supported by National Institutes of Health grants R01NS110716 and R00NS082619, the McCabe Fund Fellow Award, the University of Pennsylvania Alzheimer’s Disease Core Center, the Intellectual and Developmental Disabilities Research Center at the Children’s Hospital of Philadelphia and the University of Pennsylvania, and the Philadelphia Foundation (all to S. Maday).
The authors declare no competing financial interests.
Author contributions: Experiment conceptualization and design, V.V. Kulkarni and S. Maday; experiment execution, V.V. Kulkarni, J.B. Herr, and C. Miranda; data analysis and interpretation, V.V. Kulkarni, A. Anand, J.B. Herr, C. Miranda, M.C. Vogel, and S. Maday; writing – original draft of manuscript, V.V. Kulkarni and S. Maday; writing – review and editing of manuscript, V.V. Kulkarni, A. Anand, J.B. Herr, C. Miranda, M.C. Vogel, and S. Maday.
References
- Bingol, B., and Schuman E.M.. 2006. Activity-dependent dynamics and sequestration of proteasomes in dendritic spines. Nature. 441:1144–1148. 10.1038/nature04769 [DOI] [PubMed] [Google Scholar]
- Chen, Y., and Sheng Z.H.. 2013. Kinesin-1-syntaphilin coupling mediates activity-dependent regulation of axonal mitochondrial transport. J. Cell Biol. 202:351–364. 10.1083/jcb.201302040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X.T., Zhou B., Lin M.Y., Cai Q., and Sheng Z.H.. 2015. Axonal autophagosomes recruit dynein for retrograde transport through fusion with late endosomes. J. Cell Biol. 209:377–386. 10.1083/jcb.201412046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng, X.T., Xie Y.X., Zhou B., Huang N., Farfel-Becker T., and Sheng Z.H.. 2018. Characterization of LAMP1-labeled nondegradative lysosomal and endocytic compartments in neurons. J. Cell Biol. 217:3127–3139. 10.1083/jcb.201711083 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong, A., Kulkarni V.V., and Maday S.. 2019. Methods for imaging autophagosome dynamics in primary neurons. Methods Mol. Biol. 1880:243–256. 10.1007/978-1-4939-8873-0_16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dragich, J.M., Kuwajima T., Hirose-Ikeda M., Yoon M.S., Eenjes E., Bosco J.R., Fox L.M., Lystad A.H., Oo T.F., Yarygina O., et al. 2016. Autophagy linked FYVE (Alfy/WDFY3) is required for establishing neuronal connectivity in the mammalian brain. eLife. 5:e14810. 10.7554/eLife.14810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glatigny, M., Moriceau S., Rivagorda M., Ramos-Brossier M., Nascimbeni A.C., Lante F., Shanley M.R., Boudarene N., Rousseaud A., Friedman A.K., et al. 2019. Autophagy is required for memory formation and reverses age-related memory decline. Curr. Biol. 29:435–448.e8. 10.1016/j.cub.2018.12.021 [DOI] [PubMed] [Google Scholar]
- Goo, M.S., Sancho L., Slepak N., Boassa D., Deerinck T.J., Ellisman M.H., Bloodgood B.L., and Patrick G.N.. 2017. Activity-dependent trafficking of lysosomes in dendrites and dendritic spines. J. Cell Biol. 216:2499–2513. 10.1083/jcb.201704068 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hamilton, A.M., Oh W.C., Vega-Ramirez H., Stein I.S., Hell J.W., Patrick G.N., and Zito K.. 2012. Activity-dependent growth of new dendritic spines is regulated by the proteasome. Neuron. 74:1023–1030. 10.1016/j.neuron.2012.04.031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hangen, E., Cordelières F.P., Petersen J.D., Choquet D., and Coussen F.. 2018. Neuronal activity and intracellular calcium levels regulate intracellular transport of newly synthesized AMPAR. Cell Rep. 24:1001–1012.e3. 10.1016/j.celrep.2018.06.095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanus, C., Kochen L., Tom Dieck S., Racine V., Sibarita J.B., Schuman E.M., and Ehlers M.D.. 2014. Synaptic control of secretory trafficking in dendrites. Cell Rep. 7:1771–1778. 10.1016/j.celrep.2014.05.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hara, T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. 2006. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 441:885–889. 10.1038/nature04724 [DOI] [PubMed] [Google Scholar]
- Hernandez, D., Torres C.A., Setlik W., Cebrián C., Mosharov E.V., Tang G., Cheng H.C., Kholodilov N., Yarygina O., Burke R.E., et al. 2012. Regulation of presynaptic neurotransmission by macroautophagy. Neuron. 74:277–284. 10.1016/j.neuron.2012.02.020 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hill, S.E., Kauffman K.J., Krout M., Richmond J.E., Melia T.J., and Colón-Ramos D.A.. 2019. Maturation and clearance of autophagosomes in neurons depends on a specific cysteine protease isoform, ATG-4.2. Dev. Cell. 49:251–266.e8. 10.1016/j.devcel.2019.02.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hollenbeck, P.J. 1993. Products of endocytosis and autophagy are retrieved from axons by regulated retrograde organelle transport. J. Cell Biol. 121:305–315. 10.1083/jcb.121.2.305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hylin, M.J., Zhao J., Tangavelou K., Rozas N.S., Hood K.N., MacGowan J.S., Moore A.N., and Dash P.K.. 2018. A role for autophagy in long-term spatial memory formation in male rodents. J. Neurosci. Res. 96:416–426. 10.1002/jnr.24121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jin, E.J., Kiral F.R., Ozel M.N., Burchardt L.S., Osterland M., Epstein D., Wolfenberg H., Prohaska S., and Hiesinger P.R.. 2018. Live observation of two parallel membrane degradation pathways at axon terminals. Curr. Biol. 28:1027–1038.e4. 10.1016/j.cub.2018.02.032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kabeya, Y., Mizushima N., Ueno T., Yamamoto A., Kirisako T., Noda T., Kominami E., Ohsumi Y., and Yoshimori T.. 2000. LC3, a mammalian homologue of yeast Apg8p, is localized in autophagosome membranes after processing. EMBO J. 19:5720–5728. 10.1093/emboj/19.21.5720 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaech, S., and Banker G.. 2006. Culturing hippocampal neurons. Nat. Protoc. 1:2406–2415. 10.1038/nprot.2006.356 [DOI] [PubMed] [Google Scholar]
- Kannan, M., Bayam E., Wagner C., Rinaldi B., Kretz P.F., Tilly P., Roos M., McGillewie L., Bär S., Minocha S., et al. Sanger Mouse Genetics Project . 2017. WD40-repeat 47, a microtubule-associated protein, is essential for brain development and autophagy. Proc. Natl. Acad. Sci. USA. 114:E9308–E9317. 10.1073/pnas.1713625114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kimura, S., Noda T., and Yoshimori T.. 2007. Dissection of the autophagosome maturation process by a novel reporter protein, tandem fluorescent-tagged LC3. Autophagy. 3:452–460. 10.4161/auto.4451 [DOI] [PubMed] [Google Scholar]
- Komatsu, M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. 2006. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 441:880–884. 10.1038/nature04723 [DOI] [PubMed] [Google Scholar]
- Komatsu, M., Wang Q.J., Holstein G.R., Friedrich V.L. Jr., Iwata J., Kominami E., Chait B.T., Tanaka K., and Yue Z.. 2007. Essential role for autophagy protein Atg7 in the maintenance of axonal homeostasis and the prevention of axonal degeneration. Proc. Natl. Acad. Sci. USA. 104:14489–14494. 10.1073/pnas.0701311104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korolchuk, V.I., Saiki S., Lichtenberg M., Siddiqi F.H., Roberts E.A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F.M., et al. 2011. Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13:453–460. 10.1038/ncb2204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, V.V., and Maday S.. 2018. Compartment-specific dynamics and functions of autophagy in neurons. Dev. Neurobiol. 78:298–310. 10.1002/dneu.22562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kulkarni, A., Dong A., Kulkarni V.V., Chen J., Laxton O., Anand A., and Maday S.. 2020. Differential regulation of autophagy during metabolic stress in astrocytes and neurons. Autophagy. 16:1651–1667. 10.1080/15548627.2019.1703354 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee, S., Sato Y., and Nixon R.A.. 2011. Lysosomal proteolysis inhibition selectively disrupts axonal transport of degradative organelles and causes an Alzheimer’s-like axonal dystrophy. J. Neurosci. 31:7817–7830. 10.1523/JNEUROSCI.6412-10.2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang, C.C., Wang C., Peng X., Gan B., and Guan J.L.. 2010. Neural-specific deletion of FIP200 leads to cerebellar degeneration caused by increased neuronal death and axon degeneration. J. Biol. Chem. 285:3499–3509. 10.1074/jbc.M109.072389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- MacAskill, A.F., Rinholm J.E., Twelvetrees A.E., Arancibia-Carcamo I.L., Muir J., Fransson A., Aspenstrom P., Attwell D., and Kittler J.T.. 2009. Miro1 is a calcium sensor for glutamate receptor-dependent localization of mitochondria at synapses. Neuron. 61:541–555. 10.1016/j.neuron.2009.01.030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday, S., and Holzbaur E.L.. 2014. Autophagosome biogenesis in primary neurons follows an ordered and spatially regulated pathway. Dev. Cell. 30:71–85. 10.1016/j.devcel.2014.06.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday, S., and Holzbaur E.L.. 2016. Compartment-specific regulation of autophagy in primary neurons. J. Neurosci. 36:5933–5945. 10.1523/JNEUROSCI.4401-15.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maday, S., Wallace K.E., and Holzbaur E.L.. 2012. Autophagosomes initiate distally and mature during transport toward the cell soma in primary neurons. J. Cell Biol. 196:407–417. 10.1083/jcb.201106120 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann, S.S., and Hammarback J.A.. 1994. Molecular characterization of light chain 3. A microtubule binding subunit of MAP1A and MAP1B. J. Biol. Chem. 269:11492–11497. 10.1016/S0021-9258(19)78150-2 [DOI] [PubMed] [Google Scholar]
- Neisch, A.L., Neufeld T.P., and Hays T.S.. 2017. A STRIPAK complex mediates axonal transport of autophagosomes and dense core vesicles through PP2A regulation. J. Cell Biol. 216:441–461. 10.1083/jcb.201606082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann, S., Chassefeyre R., Campbell G.E., and Encalada S.E.. 2017. KymoAnalyzer: a software tool for the quantitative analysis of intracellular transport in neurons. Traffic. 18:71–88. 10.1111/tra.12456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikoletopoulou, V., Sidiropoulou K., Kallergi E., Dalezios Y., and Tavernarakis N.. 2017. Modulation of autophagy by BDNF underlies synaptic plasticity. Cell Metab. 26:230–242.e5. 10.1016/j.cmet.2017.06.005 [DOI] [PubMed] [Google Scholar]
- Nishiyama, J., Miura E., Mizushima N., Watanabe M., and Yuzaki M.. 2007. Aberrant membranes and double-membrane structures accumulate in the axons of Atg5-null Purkinje cells before neuronal death. Autophagy. 3:591–596. 10.4161/auto.4964 [DOI] [PubMed] [Google Scholar]
- Okerlund, N.D., Schneider K., Leal-Ortiz S., Montenegro-Venegas C., Kim S.A., Garner L.C., Waites C.L., Gundelfinger E.D., Reimer R.J., and Garner C.C.. 2017. Bassoon controls presynaptic autophagy through Atg5. Neuron. 93:897–913.e7. 10.1016/j.neuron.2017.01.026 [DOI] [PubMed] [Google Scholar]
- Padamsey, Z., McGuinness L., Bardo S.J., Reinhart M., Tong R., Hedegaard A., Hart M.L., and Emptage N.J.. 2017. Activity-dependent exocytosis of lysosomes regulates the structural plasticity of dendritic spines. Neuron. 93:132–146. 10.1016/j.neuron.2016.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pandey, K., Yu X.W., Steinmetz A., and Alberini C.M.. 2020. Autophagy coupled to translation is required for long-term memory. Autophagy. 10.1080/15548627.2020.1775393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pankiv, S., Clausen T.H., Lamark T., Brech A., Bruun J.A., Outzen H., Øvervatn A., Bjørkøy G., and Johansen T.. 2007. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 282:24131–24145. 10.1074/jbc.M702824200 [DOI] [PubMed] [Google Scholar]
- Rudnick, N.D., Griffey C.J., Guarnieri P., Gerbino V., Wang X., Piersaint J.A., Tapia J.C., Rich M.M., and Maniatis T.. 2017. Distinct roles for motor neuron autophagy early and late in the SOD1G93A mouse model of ALS. Proc. Natl. Acad. Sci. USA. 114:E8294–E8303. 10.1073/pnas.1704294114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shehata, M., Matsumura H., Okubo-Suzuki R., Ohkawa N., and Inokuchi K.. 2012. Neuronal stimulation induces autophagy in hippocampal neurons that is involved in AMPA receptor degradation after chemical long-term depression. J. Neurosci. 32:10413–10422. 10.1523/JNEUROSCI.4533-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shen, W., and Ganetzky B.. 2009. Autophagy promotes synapse development in Drosophila. J. Cell Biol. 187:71–79. 10.1083/jcb.200907109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soukup, S.F., Kuenen S., Vanhauwaert R., Manetsberger J., Hernández-Díaz S., Swerts J., Schoovaerts N., Vilain S., Gounko N.V., Vints K., et al. 2016. A LRRK2-dependent endophilinA phosphoswitch is critical for macroautophagy at presynaptic terminals. Neuron. 92:829–844. 10.1016/j.neuron.2016.09.037 [DOI] [PubMed] [Google Scholar]
- Stavoe, A.K., Hill S.E., Hall D.H., and Colón-Ramos D.A.. 2016. KIF1A/UNC-104 transports ATG-9 to regulate neurodevelopment and autophagy at synapses. Dev. Cell. 38:171–185. 10.1016/j.devcel.2016.06.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sung, H., Tandarich L.C., Nguyen K., and Hollenbeck P.J.. 2016. Compartmentalized regulation of parkin-mediated mitochondrial quality control in the Drosophila nervous system in vivo. J. Neurosci. 36:7375–7391. 10.1523/JNEUROSCI.0633-16.2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang, G., Gudsnuk K., Kuo S.H., Cotrina M.L., Rosoklija G., Sosunov A., Sonders M.S., Kanter E., Castagna C., Yamamoto A., et al. 2014. Loss of mTOR-dependent macroautophagy causes autistic-like synaptic pruning deficits. Neuron. 83:1131–1143. 10.1016/j.neuron.2014.07.040 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trombetta, E.S., Ebersold M., Garrett W., Pypaert M., and Mellman I.. 2003. Activation of lysosomal function during dendritic cell maturation. Science. 299:1400–1403. 10.1126/science.1080106 [DOI] [PubMed] [Google Scholar]
- Walton, Z.E., Patel C.H., Brooks R.C., Yu Y., Ibrahim-Hashim A., Riddle M., Porcu A., Jiang T., Ecker B.L., Tameire F., et al. 2018. Acid suspends the circadian clock in hypoxia through inhibition of mTOR. Cell. 174:72–87.e32. 10.1016/j.cell.2018.05.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, X., and Schwarz T.L.. 2009. The mechanism of Ca2+-dependent regulation of kinesin-mediated mitochondrial motility. Cell. 136:163–174. 10.1016/j.cell.2008.11.046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang, T., Martin S., Papadopulos A., Harper C.B., Mavlyutov T.A., Niranjan D., Glass N.R., Cooper-White J.J., Sibarita J.B., Choquet D., et al. 2015. Control of autophagosome axonal retrograde flux by presynaptic activity unveiled using botulinum neurotoxin type A. J. Neurosci. 35:6179–6194. 10.1523/JNEUROSCI.3757-14.2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong, Y.H., Lee C.M., Xie W., Cui B., and Poo M.M.. 2015. Activity-dependent BDNF release via endocytic pathways is regulated by synaptotagmin-6 and complexin. Proc. Natl. Acad. Sci. USA. 112:E4475–E4484. 10.1073/pnas.1511830112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, J., Suzuki C., Nanao T., Kakuta S., Ozawa K., Tanida I., Saitoh T., Sunabori T., Komatsu M., Tanaka K., et al. 2018. Atg9a deficiency causes axon-specific lesions including neuronal circuit dysgenesis. Autophagy. 14:764–777. 10.1080/15548627.2017.1314897 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto, A., and Yue Z.. 2014. Autophagy and its normal and pathogenic states in the brain. Annu. Rev. Neurosci. 37:55–78. 10.1146/annurev-neuro-071013-014149 [DOI] [PubMed] [Google Scholar]
- Yap, C.C., Digilio L., McMahon L.P., Garcia A.D.R., and Winckler B.. 2018. Degradation of dendritic cargos requires Rab7-dependent transport to somatic lysosomes. J. Cell Biol. 217:3141–3159. 10.1083/jcb.201711039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yue, Z. 2007. Regulation of neuronal autophagy in axon: implication of autophagy in axonal function and dysfunction/degeneration. Autophagy. 3:139–141. 10.4161/auto.3602 [DOI] [PubMed] [Google Scholar]
- Zhao, Y.G., Sun L., Miao G., Ji C., Zhao H., Sun H., Miao L., Yoshii S.R., Mizushima N., Wang X., et al. 2015. The autophagy gene Wdr45/Wipi4 regulates learning and memory function and axonal homeostasis. Autophagy. 11:881–890. 10.1080/15548627.2015.1047127 [DOI] [PMC free article] [PubMed] [Google Scholar]