Null mutations in secreted nuclease DNASE1L3 cause monogenic systemic lupus erythematosus (SLE). This study shows that many patients with sporadic SLE with nephritis manifest reduced DNASE1L3 activity, which is associated with blocking autoantibodies to the enzyme.
Abstract
Antibodies to double-stranded DNA (dsDNA) are prevalent in systemic lupus erythematosus (SLE), particularly in patients with lupus nephritis, yet the nature and regulation of antigenic cell-free DNA (cfDNA) are poorly understood. Null mutations in the secreted DNase DNASE1L3 cause human monogenic SLE with anti-dsDNA autoreactivity. We report that >50% of sporadic SLE patients with nephritis manifested reduced DNASE1L3 activity in circulation, which was associated with neutralizing autoantibodies to DNASE1L3. These patients had normal total plasma cfDNA levels but showed accumulation of cfDNA in circulating microparticles. Microparticle-associated cfDNA contained a higher fraction of longer polynucleosomal cfDNA fragments, which bound autoantibodies with higher affinity than mononucleosomal fragments. Autoantibodies to DNASE1L3-sensitive antigens on microparticles were prevalent in SLE nephritis patients and correlated with the accumulation of cfDNA in microparticles and with disease severity. DNASE1L3-sensitive antigens included DNA-associated proteins such as HMGB1. Our results reveal autoantibody-mediated impairment of DNASE1L3 activity as a common nongenetic mechanism facilitating anti-dsDNA autoreactivity in patients with severe sporadic SLE.
Introduction
The serological hallmark of systemic lupus erythematosus (SLE) is the production of antibodies (Abs) to nuclear antigens (Ags), with immune complex formation leading to systemic immune activation and tissue inflammation. Anti–double-stranded DNA (anti-dsDNA) IgG Abs can be found in up to 70–80% of SLE patients, of which ∼40% have high titers rarely seen in other autoimmune disorders (Rekvig, 2015; Tan et al., 1966). High titers of anti-dsDNA IgG correlate with disease severity (Banchereau et al., 2016) and often accompany disease flares, becoming especially evident when involving the kidney (Pisetsky, 2016; Yung and Chan, 2015). Despite the clinical relevance of anti-dsDNA Abs and numerous studies supporting the pathogenicity of this reactivity, the nature of antigenic DNA and the mechanisms that prevent its recognition by autoreactive B cells still remain largely elusive.
The development of anti-dsDNA Abs involves T cell help, making it unlikely that the “naked” protein-free dsDNA is the initiator of the response. In fact, Abs to chromatin often precede the onset of anti-dsDNA Abs, implicating protein-bound DNA in the initial breakdown of tolerance, followed by the spreading of autoreactivity toward naked dsDNA (Munroe et al., 2016). In turn, the antigenic chromatin may exist as small soluble fragments or comprise larger extracellular structures such as neutrophil extracellular traps (Garcia-Romo et al., 2011; Hakkim et al., 2010; Lood et al., 2016) and/or microparticles (MPs). Indeed, 1–2-µm-sized MPs that are derived from the cell membranes of apoptotic cells are found in the plasma of healthy subjects and SLE patients (Dieker et al., 2016; Nielsen et al., 2012). Moreover, MPs can carry apoptotic cell DNA and expose it on their surface (Casciola-Rosen et al., 1994; Radic et al., 2004; Ullal et al., 2011). This would effectively create an “ideal” source of antigenic DNA since it is accessible to autoreactive B cells, relatively resistant to degradation, and associated with proteins that can provide potential T cell epitopes (Beyer and Pisetsky, 2010). In addition to the physical form of DNA, the length of DNA is likely to influence its antigenicity. Anti-dsDNA Abs were proposed to bind longer fragments of naked dsDNA with higher avidity (Ali et al., 1985; Papalian et al., 1980), although this was shown to depend on assay conditions (Pisetsky and Reich, 1994). Overall, the precise physical form of cell-free DNA (cfDNA) that serves as an Ag in anti-dsDNA responses, and its relationship with DNA size, are not well understood.
The availability of self-DNA to autoreactive B cells can be limited by extracellular DNases that digest it (Elkon, 2018; Soni and Reizis, 2018). DNASE1L3 and DNASE1 are homologous extracellular DNases that are responsible for the majority of DNase activity in the circulation (Napirei et al., 2009). DNASE1L3 contains a unique positively charged C-terminal peptide that facilitates digestion of membrane- and/or protein-associated DNA, including intact chromatin (Napirei et al., 2005; Sisirak et al., 2016; Wilber et al., 2002). Homozygous null mutations in DNASE1L3 cause pediatric-onset familial SLE characterized by prominent anti-dsDNA Abs and renal involvement (Al-Mayouf et al., 2011; Batu et al., 2018; Belot, 2020; Carbonella et al., 2017; Ozçakar et al., 2013). In some of these cases, the disease initially manifested as hypocomplementemic urticarial vasculitis syndrome (HUVS; Jara et al., 2009) but almost invariably progressed to severe SLE with glomerulonephritis (Carbonella et al., 2017; Ozçakar et al., 2013). A coding polymorphism of DNASE1L3 that creates a hypofunctional protein DNASE1L3 R206C (Ueki et al., 2009) is associated with several seropositive autoimmune diseases, including SLE (Acosta-Herrera et al., 2019). Furthermore, DNASE1L3-deficient mice rapidly develop anti-chromatin and anti-dsDNA Abs followed by SLE-like disease with renal involvement (Sisirak et al., 2016; Soni et al., 2020; Weisenburger et al., 2018). Thus, DNASE1L3 represents a conserved extracellular DNase that maintains tolerance to self-DNA in a nonredundant manner. DNASE1L3 was shown to digest genomic self-DNA in apoptotic MPs (Sisirak et al., 2016), as well as reduce the level of polynucleosomal DNA fragments in the circulation (Chan et al., 2020; Serpas et al., 2019). However, it is unclear how these two activities are related to one another or relevant for the role of DNASE1L3 in preventing SLE. Moreover, the relevance of DNASE1L3 or its DNA substrates to sporadic SLE beyond the rare genetic DNASE1L3 deficiency has never been established.
This study addresses the relevance of DNASE1L3 and its DNA substrates to sporadic SLE, with an emphasis on lupus nephritis (LN). We show that patients with LN often manifest reduced activity of DNASE1L3 in the circulation, which is associated with auto-Abs to DNASE1L3 itself. Furthermore, reduced DNASE1L3 activity in LN patients is associated with cfDNA accumulation in MPs, as well as with autoreactivity toward DNA and DNA-associated proteins on the surface of MPs. In aggregate, these results establish DNASE1L3 as a novel self-Ag in patients with spontaneous SLE with renal involvement and the resulting deficiency of functional DNASE1L3 as a likely cause of cfDNA abnormalities that in turn facilitate anti-DNA responses.
Results
The activity of DNASE1L3 is reduced in SLE patients with renal involvement
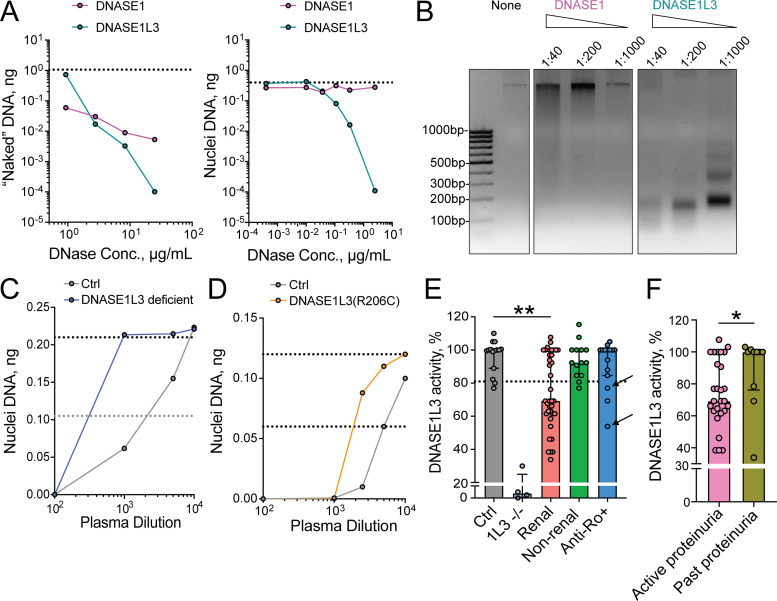
Initial experiments were done to establish a reliable assay for DNASE1L3 activity in the circulation. Unlike its homologue, DNASE1, DNASE1L3 efficiently digests native chromatin in intact nuclei (Napirei et al., 2005). Using purified recombinant DNASE1L3, we confirmed its superior ability, relative to DNASE1, to digest DNA in intact nuclei isolated from Jurkat T cells (Fig. 1 A). High concentrations of DNASE1 produced a smear pattern of nuclear DNA digest, consistent with inefficient random cleavage (Fig. 1 B). In contrast, even low concentrations of DNASE1L3 produced a characteristic nucleosomal ladder, suggesting efficient internucleosomal cleavage of chromatin (Fig. 1 B). The distinctive ability of DNASE1L3 to digest DNA in nuclei was used to measure its specific enzymatic activity in human plasma. Whereas control human plasma efficiently digested DNA in nuclei, plasma from DNASE1L3-deficient patients failed to do so in the same dilution range (Fig. 1 C). A lower dilution of plasma from DNASE1L3-deficient patients was able to digest nuclei in our assay, likely reflecting the activity of DNASE1 and/or nonspecific inhibition of detection (e.g., by plasma IgG; Sidstedt et al., 2018). Plasma from an SLE patient who was heterozygous for the hypomorphic R206C variant of DNASE1L3 also showed a noticeable reduction (Fig. 1 D), confirming the specificity and sensitivity of this assay. To measure the relative activity of DNASE1L3 in patient plasma, we set the 50% inhibitory concentration (IC50) value for pooled healthy control plasma (n = 15) at 100%, the value for DNASE1L3-deficient patient plasma at 0% and considered any activity below two standard deviations of the control (81%) as reduced.
Figure 1.
Reduced activity of DNASE1L3 in the plasma of patients with sporadic SLE. (A) Digestion of DNA substrates by human DNases. Recombinant human DNASE1 or recombinant DNASE1L3 was incubated with purified human genomic DNA (left) or intact nuclei from human Jurkat cells (right), and the remaining DNA was quantified by qPCR. Dashed lines indicate the amount of input DNA. Representative of three independent experiments. Conc., concentration. (B) Digestion of nuclear DNA by human DNases. Indicated dilutions of recombinant human DNASE1 or DNASE1L3 were incubated with intact nuclei from human Jurkat cells, and the resulting DNA was purified and analyzed by agarose gel electrophoresis along with the DNA size markers (bp). Representative of three independent experiments. (C and D) Validation of DNASE1L3 activity assay in human plasma. Serial dilutions of plasma were incubated with intact nuclei from human Jurkat cells, and the remaining DNA was quantified by qPCR. Shown are representative digestion curves for plasma from a DNASE1L3-deficient patient (C) or a patient with a hypomorphic DNASE1L3 (R206C) variant (D), along with plasma pooled from healthy controls (Ctrl). Dashed lines indicate 100% (top) and 50% (bottom) of input DNA. Representative of three independent experiments. (E) Plasma DNASE1L3 activity in SLE patients. DNASE1L3 activity was measured in the assay described in C and D and expressed as percentage of activity in pooled control plasma. Symbols represent individual subjects, and bars represent median ± interquartile range. Ctrl, healthy controls (n = 15); 1L3−/−, patients with genetic DNASE1L3 deficiency (n = 4); renal, “ever renal” SLE patients by ACR criteria (n = 37); nonrenal, “never renal” SLE patients by ACR criteria (n = 14); anti-Ro+, anti-Ro Ab–positive mothers of children with neonatal lupus (n = 15). Dashed line marks the threshold for reduced DNASE1L3 activity (81%); arrows mark the only two subjects in the anti-Ro+ cohort with renal SLE by ACR criteria. Statistical significance was determined by Kruskal–Wallis test followed by the Dunn’s multiple-comparison test; **, P < 0.01 (two tailed). (F) Plasma DNASE1L3 activity in SLE patients with active renal disease. DNASE1L3 activity was compared between patients with active proteinuria (UPCR > 0.5; n = 27) and those with resolved proteinuria (UCPR < 0.5; n = 10) from the “renal” SLE group in E. Symbols represent individual subjects, and bars represent median ± interquartile range. Statistical significance was determined by Mann–Whitney test; *, P < 0.05 (two tailed).
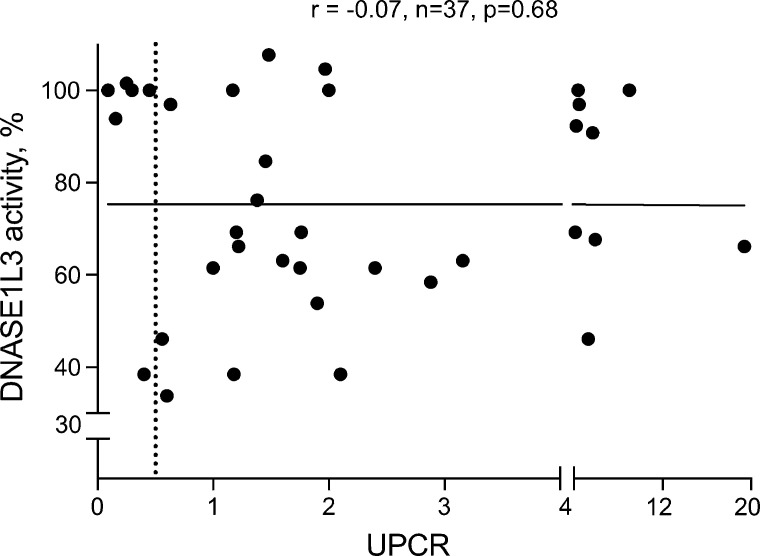
We then evaluated DNASE1L3 activity in several groups of SLE patients (patient demographics and average values for key readouts are provided in Table 1). Note that “renal SLE” indicates past or present renal disease and “LN” indicates renal SLE with persistent proteinuria (urine protein/creatinine ratio [UPCR] >0.5) accompanied by positive serologies (hypocomplementemia and/or elevated anti-dsDNA Abs) at the time of sampling. Average DNASE1L3 activity was not significantly decreased in 14 SLE patients without current or previous renal involvement compared with healthy controls (median activity 92%; Fig. 1 E). In contrast, 57% of patients with a history of renal disease (n = 37) showed a decrease in DNASE1L3 activity (median activity 69%) with a significant reduction compared with both healthy controls and SLE patients who never had renal disease (Fig. 1 E). Within the renal SLE group, DNASE1L3 activity was lower in those with active proteinuria than in those with resolved proteinuria (i.e., inactive renal disease) defined as UPCR <0.5 at the time of sampling (Fig. 1 F). Notably, plasma activity of DNASE1L3 did not correlate with the degree of proteinuria (Fig. S1), arguing against protein loss through the kidney as a cause of reduced DNASE1L3 activity. To further evaluate the clinical significance of reduced DNASE1L3 activity, we evaluated 15 mothers of children with heart block and high anti-Ro Abs, most of whom were clinically asymptomatic or had only minimal symptoms of SLE but did not meet formal classification criteria (Table 1). Only three showed a reduction in DNASE1L3 activity, two of whom were the only patients who progressed to SLE with renal disease (Fig. 1 E, arrows). Collectively, these data reveal a significant reduction of circulating DNASE1L3 activity in sporadic SLE patients with renal disease.
Table 1. Demographics and readouts in different study cohorts.
| Healthy control (n = 15) | DNASE1L3 deficient (n = 4) | SLE renal (n = 87) | SLE nonrenal (n = 33) | Anti-Ro+ (n = 45) | |
|---|---|---|---|---|---|
| Demographics | |||||
| Female, n (%) | 12 (80) | 2 (50) | 75 (87) | 31 (94) | 45 (100) |
| Age (yr), median (IQR) | 36 (23–59)a | 9 (7–14) | 3–7 (20-78) | 39 (27–80) | 35 (27–64) |
| Race/ethnicity, n (%) | |||||
| Hispanic/Latino | 0 | 0 | 25 (29) | 16 (48) | 0 |
| Non-Hispanic/Latino | 0 | 0 | 53 (61) | 17 (52) | 0 |
| White | 5 (33) | 4 (100) | 38 (44) | 23 (70) | 38 (84) |
| Black | 5 (33) | 0 | 27 (31) | 7 (21) | 1 (2) |
| Asian | 1 (7) | 0 | 21 (24) | 2 (6) | 3 (7) |
| Other | 1 (7) | 0 | 2 (2) | 1 (3) | 2 (4) |
| Not reported | 3 (20) | — | — | — | 1 (2) |
| Readoutsb | |||||
| DNASE1L3 activity, % | 100 (89–100.5, n = 15) | 0 (n = 4) | 68 (62–100, n = 37) | 92 (85–100, n = 14) | 100 (85–100, n = 15) |
| Anti-DNASE1L3 ELISA, OD | 0.15 (0.13–0.26, n = 15) | — | 0.40 (0.26–0.80, n = 40) | 0.30 (0.23–0.36, n = 18) | 0.19 (0.13–0.25, n = 18) |
| cfDNA total, ng/ml | 5.6 (3.2–8.4, n = 15) | 10.0 (9.0–17.6, n = 4) | 6.3 (3.4–11.8, n = 44) | 5.4 (2.6–10.2, n = 15) | 17.2 (6.5–26.5, n = 15) |
| MP cfDNA fraction, % | 23 (19–35, n = 15) | 70 (40–83, n = 4) | 62 (39–73, n = 44) | 33 (26–46, n = 15) | 49 (43–60, n = 15) |
| DNASE1L3-sensitive binding to MPsc | 0 (0%, n = 15) | 3 (75%, n = 4) | 50 (57%, n = 87) | 5 (15%, n = 33) | 3 (7%, n = 45) |
Continuous variables are given as median (interquartile range [IQR]), and categorical variables are given as number of cases (%).
The age of one control subject was unknown and was not included in the calculation.
Not all patients in every group were tested for all readouts. The n in each of the readouts refers to the number of patients from that group who were tested for this readout.
Figure S1.
DNASE1L3 activity does not correlate with renal function in patients with renal SLE. DNASE1L3 activity in plasma was measured as in Fig. 1, C–E, and is expressed as percentage of activity in healthy controls. Symbols represent DNASE1L3 activity and UPCR in individual patients with renal SLE. Dashed line indicates the threshold for proteinuria (UPCR >0.5). Results of Spearman correlation with a two-tailed P value are shown.
Reduced DNASE1L3 activity is associated with Abs to DNASE1L3
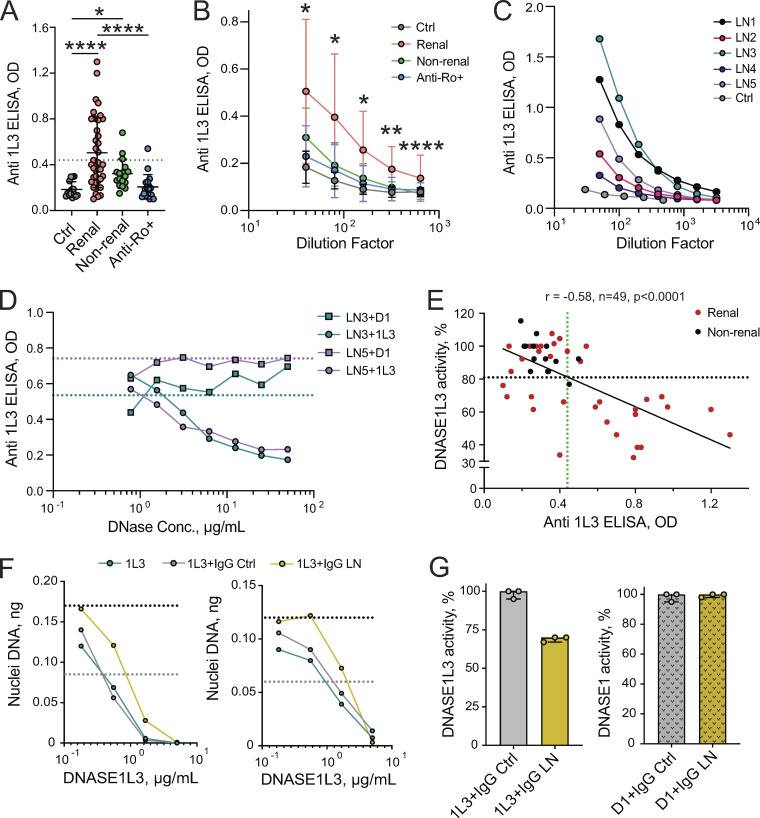
The known hypomorphic allelic variant of DNASE1L3, DNASE1L3 (R206C), is predominantly found in subjects of European ancestry, and indeed, only two patients in our SLE cohort carried this variant. However, reduced DNASE1L3 activity was observed in >50% of our renal SLE patients across all ethnic groups, prompting a search for a nongenetic cause. To test whether auto-Abs to DNASE1L3 might contribute to its decreased activity, we examined patient plasma by ELISA using purified DNASE1L3 as the Ag. No reactivity to DNASE1L3 was observed in plasma samples from healthy controls (Fig. 2, A–C) or DNASE1L3-deficient patients (data not shown). In contrast, 17 out of 40 (43%) evaluated patients with renal SLE showed a mean anti-DNASE1L3 Ab level greater than the threshold OD value of 0.44 (mean OD +2 SD of healthy controls; Fig. 2 A). Only 3 out of 17 (18%) plasma samples from nonrenal SLE patients and one anti-Ro+ mother with renal disease were positive in this assay (Fig. 2, A and B). The same plasma samples that showed strong reactivity to DNASE1L3 (Fig. 2 C) had minimal binding to the FLAG epitope present in this protein or immobilized DNASE1 (Fig. S2 A). To rule out any differences in immobilization between DNASE1 and DNASE1L3, we incubated plasma from SLE patients with high anti-DNASE1L3 titers with a concentration range of DNASE1 or DNASE1L3 and then tested the reactivity to plate-bound DNASE1L3 by ELISA. Representative examples in Fig. 2 D show that IgG binding was competed out by DNASE1L3, but not by DNASE1. Taken together, these data suggest a specific auto-Ab response predominantly directed toward DNASE1L3.
Figure 2.
Reduced DNASE1L3 activity is associated with neutralizing anti-DNASE1L3 Abs. (A–C) Anti-DNASE1L3 (1L3) Ab levels in the plasma of sporadic SLE patients as measured by ELISA. (A) Anti-DNASE1L3 (1L3) Ab OD values in individual patients at a single plasma dilution (1:40); bars represent mean ± SD. Patients included healthy controls (n = 15), renal (n = 40) and nonrenal SLE patients (n = 17) by ACR criteria, and anti-Ro Ab–positive mothers of children with neonatal lupus (Anti-Ro+, n = 18). Dashed line indicates the threshold for a positive anti-DNASE1L3 Ab (mean OD +2 SD of control). Statistical significance was determined by Kruskal–Wallis test followed by the Dunn’s multiple-comparison test; *, P < 0.05; ****, P <0.0001 (two tailed). (B) Average OD values ± SD for plasma dilutions (1:40–1:640) in the indicated groups of SLE patients and controls. Significance levels are given for the comparison between renal and nonrenal. Statistical significance was determined by Mann–Whitney test; *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (two tailed). (C) Dilution curves for healthy control plasma (Ctrl) and plasma from five SLE patients with LN (LN1–LN5). Representative of two independent experiments. (D) Cross-reactivity of anti-DNASE1L3 Abs to DNASE1 as measured in a competition assay. Recombinant DNASE1 (D1, squares) or DNASE1L3 (D1L3, circles) was incubated with plasma at a single dilution, and the binding of plasma IgG to DNASE1L3 was measured by ELISA. Shown are two representative patient samples (LN3 and LN5 from C). Dotted lines represent OD values in the absence of any competitor. Representative of two independent experiments. (E) Correlation between DNASE1L3 activity and anti-DNASE1L3 (1L3) Ab levels. Symbols represent individual patients with sporadic renal (red) or nonrenal (black) SLE. Dashed lines indicate thresholds for reduced DNASE1L3 activity (black) and positive anti-DNASE1L3 Ab (green). Results of Spearman correlation with a two-tailed P value are shown. (F and G) Inhibition of DNase activity by IgG from patients with anti-DNASE1L3 Abs. (F) Recombinant DNASE1L3 (1L3) was preincubated with purified IgG from plasma of a healthy control (Ctrl) or SLE patient with LN and used to digest DNA in native nuclei (left panel) or naked genomic DNA (right panel). Shown is the amount of remaining DNA after digestion with different concentrations of DNASE1L3. Dashed lines indicate 100% (black) and 50% (gray) of input DNA. Representative of two independent experiments. (G) Recombinant DNASE1L3 (1L3, left panel) or DNASE1 (right panel) was preincubated with purified IgG and used to digest naked DNA (as in Fig. 1 A). Shown is the relative enzymatic activity after preincubation with IgG from three controls or LN patients relative to the respective enzymes alone. Symbols represent IgG from individual patients; bars represent median ± interquartile range. Representative of two independent experiments.
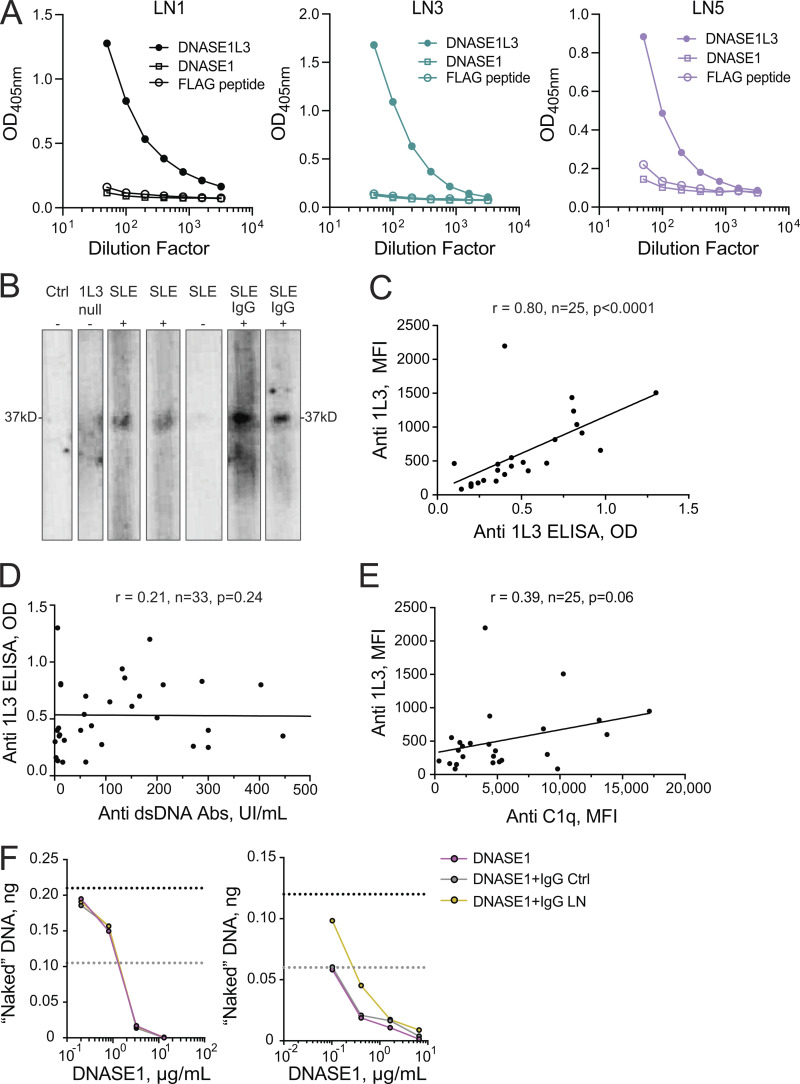
Figure S2.
Further characterization of auto-Abs to DNASE1L3 in SLE patients. (A) Abs to DNASE1, DNASE1L3, and FLAG peptide in the plasma of renal SLE patients (LN1, LN3, and LN5 from Fig. 2 C) as measured by ELISA. All Ags were plated at a concentration of 2 µg/ml. Representative of two independent experiments. (B) Ab binding to recombinant DNASE1L3 assessed by Western blot. Purified recombinant DNASE1L3 was analyzed by denaturing SDS-PAGE and probed by Western blot using plasma samples from a DNASE1L3-deficient patient (1L3 null), healthy control (Ctrl), or from SLE patients (SLE). The presence (+) or absence (−) of IgG binding to DNASE1L3 as assessed by ELISA is indicated. For sporadic SLE patients with a positive ELISA (SLE, +), both plasma and the purified IgG fraction (SLE IgG, +) were analyzed. Representative of three independent experiments. (C) Correlation between anti-DNASE1L3 (1L3) Ab levels assessed by plate-based ELISA (OD) and bead-based Ag array (mean fluorescence intensity [MFI]). Data points represent individual renal SLE patients. Results of Spearman correlation with a two-tailed P value are shown. (D) Correlation between anti-DNASE1L3 (1L3) Ab levels and anti-dsDNA Ab levels as assessed by ELISA. Symbols represent individual patients with SLE (combined renal and nonrenal). Results of Spearman correlation with a two-tailed P value are shown. (E) Correlation between anti-DNASE1L3 (1L3) Ab levels and anti-C1q Ab levels as assessed by respective bead array assays. Symbols represent individual patients with renal SLE. Results of Spearman correlation with a two-tailed P value are shown. (F) Inhibition of DNASE1 activity by IgG from patients with anti-DNASE1L3 Abs. IgG was purified from the plasma of a healthy control (Ctrl, gray) or LN patient with anti-DNASE1L3 Abs (LN, gold). Recombinant DNASE1 was preincubated with purified IgG at the same concentration as DNASE1L3 in Fig. 2 F (left panel) or at a 10-fold higher IgG concentration (right panel) and used to digest naked genomic Jurkat DNA. DNASE1 without preincubation with IgG (pink) was used as a control. Shown is the amount of remaining DNA after digestion with different concentrations of DNASE1. Dashed lines indicate 100% (black) and 50% (gray) of input DNA.
The binding of plasma IgG to recombinant DNASE1L3 was further confirmed by reducing SDS-PAGE followed by Western blot, in which the reactivity was observed only in plasma samples that showed anti-DNASE1L3 binding by ELISA (Fig. S2 B). In addition, DNASE1L3 was incorporated into a bead array that allows simultaneous analysis of reactivity to multiple Ags. Anti-DNASE1L3 Ab levels measured by bead array strongly correlated with those determined by ELISA (r = 0.80, P < 0.0001, n = 25; Fig. S2 C). C1q and dsDNA, Abs to which are closely linked to LN (Yung and Chan, 2015), were also included in the bead array to assess autoreactivity to these Ags. However, neither anti-C1q nor anti-dsDNA Ab levels showed a significant correlation with anti-DNASE1L3 reactivity (Fig. S2, D and E). Thus, Ab reactivity to DNASE1L3 in sporadic SLE could be demonstrated by three different methods and appears to be distinct from other autoreactivities.
Importantly, the levels of anti-DNASE1L3 Abs strongly correlated with reduced plasma activity of DNASE1L3 (Fig. 2 E). Moreover, positive anti-DNASE1L3 binding in ELISA reliably identified patients with DNASE1L3 activity below the threshold (Fig. 2 E, horizontal black dashed line; sensitivity, 71%; specificity, 89%; positive predictive value, 83%; negative predictive value, 81%). To verify that Abs to DNASE1L3 can inhibit its activity, we purified IgG from the plasma of three LN patients with positive anti-DNASE1L3 binding by ELISA (similar to LN1 and LN3 in Fig. 2 C). Preincubation of DNASE1L3 with IgG from these patients, but not with IgG from three healthy controls, decreased enzymatic activity by an average of 30% in our assay (Fig. 2 F and Fig. 2 G, left panel). Similar inhibitory effects were also observed when nuclear DNA and naked genomic DNA were used as DNA substrates (Fig. 2 F), suggesting that the Abs primarily target the core catalytic activity of DNASE1L3. In contrast, no inhibition of DNASE1 activity was observed with the same IgG concentration (Fig. S2 F and Fig. 2 G), and only minor inhibition was achieved at a 10-fold higher concentration (Fig. S2 F). Overall, these data suggest that the reduced DNASE1L3 activity in sporadic human SLE is caused primarily by specific Abs that develop against this protein.
DNASE1L3 controls the amount of DNA in MPs
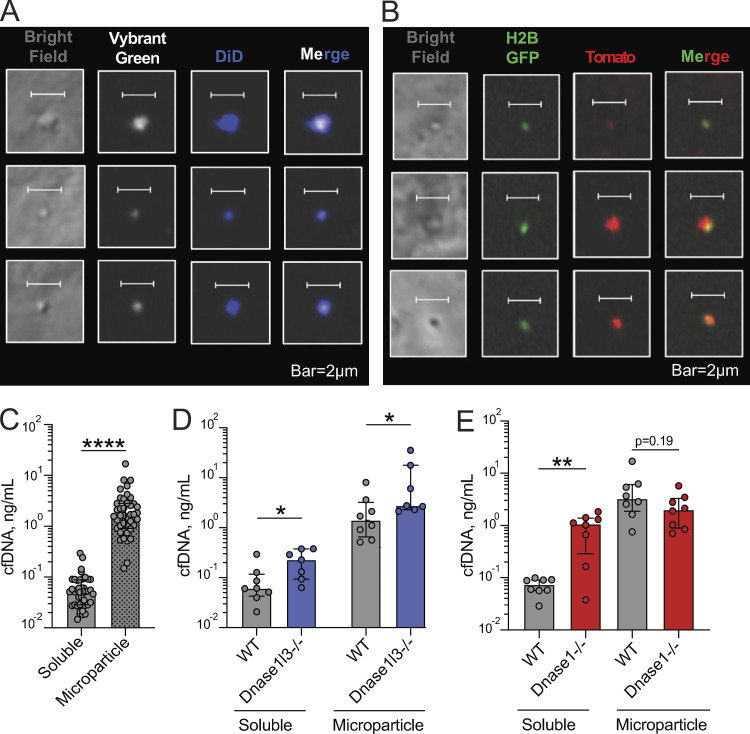
We next sought to determine the impact of reduced DNASE1L3 activity on cfDNA in sporadic SLE patients. Given that DNA in circulating MPs was proposed as a physiological substrate of DNASE1L3 (Sisirak et al., 2016), we sought to characterize MP-bound and soluble cfDNA in normal human plasma. Plasma from healthy donors was subjected to several centrifugation steps to isolate a MP fraction free of cell debris and platelets. Resulting MPs were then stained for plasma membrane and DNA, using a membrane-permeable dye, and examined by confocal microscopy. Fig. 3 A shows representative examples of 1–2-µm-sized particles composed of plasma membrane and containing dsDNA, suggesting that our centrifugation method indeed isolates human MPs with dsDNA. Similarly, a fraction of AnnexinV+ MP isolated from human plasma stained positive for DNA by flow cytometry (data not shown).
Figure 3.
DNASE1L3 controls the amount of cfDNA in circulating MPs. (A) The DNA content of MPs from the plasma of healthy human controls. MPs were stained with Vybrant DyeCycle Green for DNA (white) and Vybrant DiD for membrane (blue) and examined by confocal microscopy. Representative of three independent experiments. Scale bars, 2 µm. (B and C) cfDNA content of the MP fraction of human plasma. DNA was purified from the MP fraction, MP-depleted fraction (soluble), and total plasma and quantified by qPCR. Shown are concentrations of cfDNA adjusted to the original plasma volume purified from healthy controls (B; n = 15) or from DNASE1L3-deficient patients (C; n = 4). (D) The cfDNA content of the MP fraction, defined as a percentage of MP-associated cfDNA out of the sum of MP-associated and soluble cfDNA. Shown is the percentage of MP-associated cfDNA in healthy controls (Ctrl, n = 15); DNASE1L3-deficient patients (1L3−/−, n = 4); ever-renal SLE patients by ACR criteria (renal, n = 44); and never-renal SLE patients by ACR criteria (nonrenal, n = 15). Symbols represent individual patients, and bars represent median ± interquartile range. Statistical significance was determined by Kruskal–Wallis test followed by the Dunn’s multiple-comparison test; *, P < 0.05; ***, P < 0.001; ****, P < 0.0001 (two tailed). (E) Total concentration of cfDNA in the plasma of patient groups described in D. (F) Correlation of DNASE1L3 activity (Fig. 1 E) with the fraction of cfDNA in MP (panel D). Symbols represent individual patients with sporadic SLE. Renal patients are represented by red dots and nonrenal SLE patients by black dots, and the dashed line indicates the threshold for reduced DNASE1L3 activity. Results of Spearman correlation with a two-tailed P value are shown.
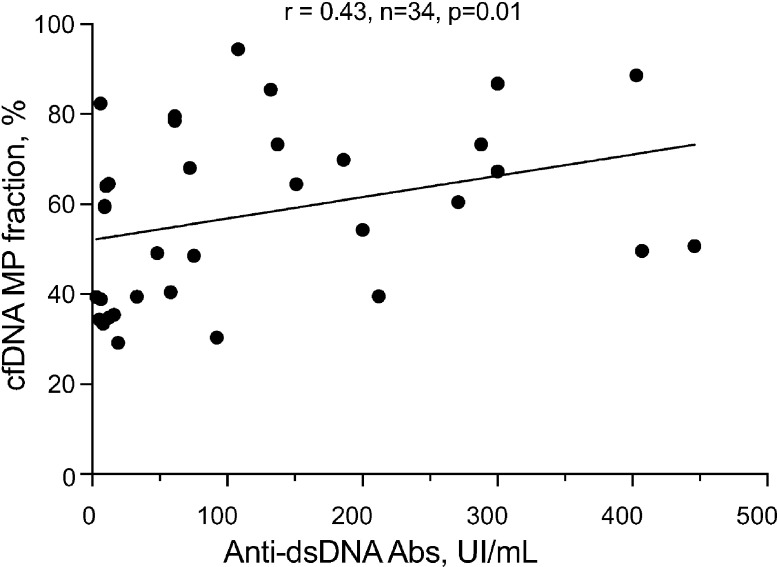
The fraction of MP-associated cfDNA was estimated by purifying DNA from the MP and MP-depleted fractions of 15 healthy controls and quantifying genomic DNA by quantitative PCR (qPCR) for Alu repeats in the genome. The concentration of MP-associated cfDNA ranged from 0.4–3.0 ng/ml plasma (Fig. 3 B), corresponding to 17–48% (median 23%) of total cfDNA in the circulation (Fig. 3 D). The three control samples with DNASE1L3 activity on or slightly below the threshold (Fig. 1 E) showed the fraction of MP-associated DNA (28–37%) that was slightly above the median. In contrast, the distribution of cfDNA was strongly skewed toward the MP fraction in three out of four DNASE1L3-deficient patients (Fig. 3 C), representing 31–86% (median 69%) of total cfDNA (Fig. 3 D). Of note, three of these four DNASE1L3-deficient patients were children diagnosed with HUVS before progression to SLE, suggesting that the observed cfDNA redistribution is not a consequence of severe SLE and/or anti-dsDNA reactivity. In our cohort of sporadic SLE patients, only those with renal SLE showed a significant increase in the MP cfDNA fraction (renal median, 62%; nonrenal median, 33%; Fig. 3 D), mirroring the changes observed with genetic DNASE1L3 deficiency. In contrast to the observed increase in MP-associated cfDNA, the total concentration of cfDNA was not significantly increased in any group, highlighting the specific effect of DNASE1L3 on cfDNA accumulation in MPs (Fig. 3 E). The concentration of soluble cfDNA was slightly decreased in renal SLE compared with controls (2.2 versus 5.0 ng/ml), but the difference was not significant (P = 0.08). Importantly, the fraction of MP-associated cfDNA showed a significant inverse correlation with DNASE1L3 activity (Fig. 3 F); thus, patients with reduced DNASE1L3 activity harbored on average 67% of cfDNA in the MP fraction. The fraction of MP cfDNA showed a significant correlation with anti-dsDNA Ab titers (Fig. S3); nevertheless, multiple patients with a high fraction of MP-associated cfDNA had low or absent anti-dsDNA Ab titers, ruling out the coincidental isolation of DNA-containing immune complexes with MPs. In summary, while a substantial fraction of total cfDNA in healthy humans is associated with MPs, the representation of this fraction is regulated by DNASE1L3 and is significantly increased in sporadic SLE patients with reduced plasma DNASE1L3 activity.
Figure S3.
Correlation between plasma cfDNA and anti-dsDNA Ab levels in sporadic SLE patients. Symbols represent individual patients with sporadic SLE; all patients with and without LN were included. Anti-dsDNA Ab levels were assessed by ELISA, and DNA concentrations were assessed by qPCR. Results of Spearman correlation with a two-tailed P value are shown.
The mouse model of DNASE1L3 deficiency was used to directly test the role of DNASE1L3 in the regulation of MP-associated DNA. The dual membrane/DNA staining protocol used for human MPs (Fig. 3 A) was used to confirm the presence of DNA-containing MPs in murine plasma (Fig. 4 A). DNA-carrying MPs in mice appeared slightly smaller than in humans, possibly reflecting differences between species and/or between isolation protocols. To visualize DNA-containing MPs in the absence of chemical dyes, we used dual reporter mice expressing a histone 2B (H2B) GFP fusion and membrane-targeted RFP tdTomato. All leukocytes in these mice were double positive for GFP and tdTomato by flow cytometry (data not shown), and their plasma MP fraction contained multiple tdTomato+ MPs carrying GFP (Fig. 4 B). Next, cfDNA was purified and measured in MPs and MP-depleted soluble fractions from the plasma of WT and Dnase1l3-deficient mice. To avoid potential secondary effects of anti-DNA Abs, only young male Dnase1l3−/− mice without anti-dsDNA reactivity were used. Unlike our findings in humans, the majority of total cfDNA in WT mice was contained within the MP fraction (Fig. 4 C). Importantly, Dnase1l3−/− mice showed a significantly increased DNA concentration in the MPs and soluble fractions compared with age- and sex-matched WT mice (Fig. 4 D). Conversely, Dnase1−/− mice harbored a normal amount of cfDNA in MPs but significantly more cfDNA in the soluble fraction compared with age- and sex-matched WT controls (Fig. 4 E). Thus, the loss of DNASE1L3, but not of its homologue, DNASE1, causes the accumulation of cfDNA in the MP fraction, implicating reduced DNASE1L3 activity in SLE patients as a likely cause of increased DNA load in MPs.
Figure 4.
DNASE1L3-mediated control of MP DNA is conserved in the mouse. (A) DNA-carrying MPs from the plasma of WT mice. MPs were stained with Vybrant DyeCycle Green for DNA (white) and Vybrant DiD for membrane (blue) and examined by confocal microscopy. Representative of three independent experiments. Scale bars, 2 µm. (B) Chromatin in plasma MPs visualized using genetically encoded fluorescent probes. MPs were isolated from the plasma of mice expressing a H2B-GFP fusion and membrane-targeted red fluorescent protein tdTomato and examined by confocal microscopy. Representative of two independent experiments. Scale bars, 2 µm. (C) Distribution of cfDNA in the plasma of WT mice. Shown are concentrations per original plasma volume of cfDNA in the MP fraction and in the MP-depleted (soluble) fraction of WT mice (n = 42). Symbols represent individual mice, and bars represent median ± interquartile range. Statistical significance was determined by Mann–Whitney test; ****, P < 0.0001 (two tailed). (D) Distribution of cfDNA in the plasma of age- and sex-matched WT and Dnase1l3-deficient mice. Shown are concentrations per original plasma volume of cfDNA in the MP fraction and in the MP-depleted (soluble) fraction of WT (n = 8) and Dnase1l3-deficient (n = 7) mice. Symbols represent individual mice, and bars represent median ± interquartile range. Statistical significance was determined by Mann–Whitney test; *, P < 0.05 (two tailed). (E) Distribution of cfDNA in the plasma of age- and sex-matched WT and Dnase1-deficient mice. Shown are concentrations per original plasma volume of cfDNA in the MP fraction and in the MP-depleted (soluble) fraction of WT (n = 8) and Dnase1-deficient (n = 8) mice. Symbols represent individual mice, and bars represent median ± interquartile range. Statistical significance was determined by Mann–Whitney test; **, P < 0.01 (two tailed).
MPs carry longer DNA fragments that are more immunogenic
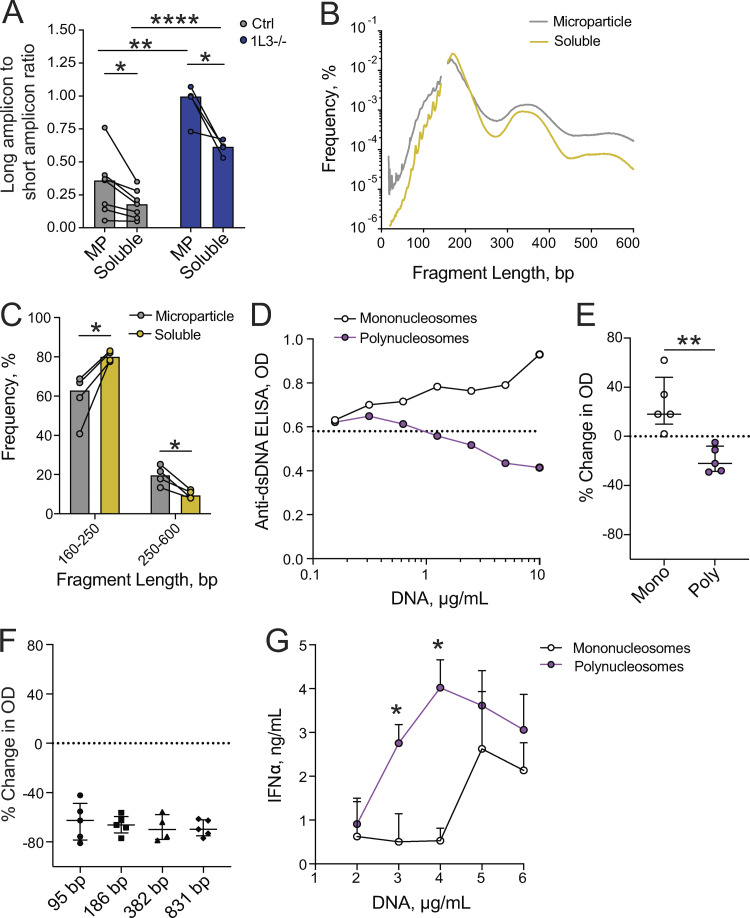
Given that a substantial fraction of human cfDNA is contained within the MP fraction, we tested whether this MP-associated DNA is qualitatively different from that in the soluble fraction. First, we estimated the length of cfDNA fragments in both fractions using the ratio of long to short amplicons recovered by qPCR (Jiang and Lo, 2016). This approach detected an increase in DNA length in the MP fraction compared with the soluble fraction of plasma from healthy controls (Fig. 5 A). Consistent with findings in the total cfDNA of DNASE1L3-deficient human patients (Chan et al., 2020), both MP-associated and soluble cfDNA from DNASE1L3-deficient patients was longer than in healthy controls (Fig. 5 A). To directly test the size distribution of MP-associated cfDNA in healthy controls, we sequenced it and computed the length of resulting fragments aligned to the genome (Jiang and Lo, 2016). No differences in genome coverage or in the end motif frequency were observed between MP-associated and soluble cfDNA (data not shown). However, the predominant DNA peak of mononucleosomal size (∼180 bp) was overrepresented in the soluble fraction, whereas larger DNA fragments in the di- to trinucleosomal range (250–600 bp) were significantly overrepresented in the MP fraction (Fig. 5, B and C). Thus, MP-associated cfDNA is enriched in longer polynucleosomal fragments compared with soluble cfDNA, and MPs from DNASE1L3-deficient patients show lengthening of their cfDNA cargo compared with healthy subjects.
Figure 5.
MPs carry longer polynucleosomal fragments with better antigenic properties. (A) Average length of cfDNA in the MP and MP-depleted (soluble) fractions of plasma from healthy controls (Ctrl; gray, n = 7) and DNASE1L3-deficient patients (1L3−/−; blue, n = 4) as determined by long to short amplicon ratio. Symbols represent individual subjects; bars represent mean. Unpaired samples (Ctrl versus 1L3−/−) were compared using unpaired Student's t test, and paired samples (MP versus soluble) were compared using paired Student's t test; *, P < 0.05; **, P < 0.01; ****, P < 0.0001 (two tailed). (B) Size distribution of cfDNA fragments in the MP (gray) and MP-depleted (soluble, gold) fractions of plasma from healthy controls as determined by sequencing. Lines represents averages of four healthy controls. (C) Fraction of cfDNA fragments of the indicated length ranges in the MP and MP-depleted (soluble) fractions of plasma from healthy controls as determined by sequencing. Symbols represent individual healthy subjects; bars represent mean. Samples were compared using paired Student's t test; *, P < 0.05 (two tailed). (D and E) The effect of chromatin size on binding to anti-dsDNA Abs in a competition assay using plasma of an SLE patient. D shows a representative titration of polynucleosomes (purple) and mononucleosomes (white); dashed line shows binding in the absence of competitor DNA. E shows the inhibition of binding by anti-dsDNA IgG from different SLE patients (n = 5) at the maximum inhibitor concentration (10 µg/ml). Symbols represent individual plasma samples, and bars represent median ± interquartile range. Statistical significance was determined by Mann–Whitney test; **, P < 0.01 (two tailed). (F) Inhibitory capacity of protein-free DNA fragments of the indicated lengths in bp on the binding by anti-dsDNA IgG at the maximum inhibitor concentration (10 µg/ml). Symbols represent individual SLE patients (n = 5), and bars represent median ± interquartile range. (G) The effect of chromatin size on interferon production by pDCs. Mononucleosomes (white) or polynucleosomes (purple) were incubated with antimicrobial peptide LL-37 and used to stimulate pDCs from healthy donors, and IFN-α was measured by ELISA after 24 h. Each point represents an average of two healthy donors, and bars represent SD of the mean. Data are representative of two independent experiments. Statistical significance was determined by Mann–Whitney test; *, P < 0.05 (two tailed).
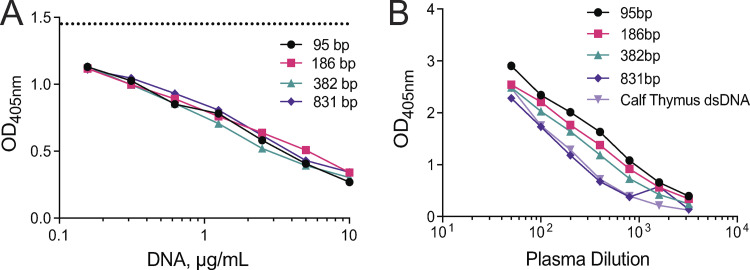
Given the observed enrichment of longer polynucleosomal fragments in MPs and the control of MP-associated DNA by DNASE1L3, we tested whether longer DNA fragments might represent more robust Ags. We incubated plasma from SLE patients with high anti-dsDNA titers with mononucleosomes or polynucleosomes and then tested the binding of plasma IgG to plate-bound DNA by ELISA. A representative example in Fig. 5 D shows that IgG binding to DNA was competed out by polynucleosomes, but not by mononucleosomes. This was observed using plasma samples from five patients (Fig. 5 E) and with purified IgG fractions from two of these patients (data not shown). In contrast, protein-free DNA fragments (95–831 bp) efficiently blocked IgG binding to DNA irrespective of their size (Fig. S4 A and Fig. 5 F). Moreover, IgG binding to the same fragments immobilized on a plate was slightly better for smaller fragments (Fig. S4 B) as observed previously (Pisetsky and Reich, 1994). The differences were not as prominent as observed by Pisetsky and Reich, likely because we used poly-lysine precoating to facilitate DNA binding to the plate. Thus, the impact of DNA length on auto-Ab binding is only apparent in solution and when the DNA is in its physiological chromatin state. In addition to serving as a self-Ag, cfDNA may induce innate immune responses such as the production of type I IFN (IFN-I) by plasmacytoid dendritic cells (pDCs), especially when in complexes with neutrophil-derived cationic peptides such as LL-37 (Garcia-Romo et al., 2011; Lande et al., 2011). We therefore incubated different concentrations of polynucleosomes or mononucleosomes with LL-37 and tested the ability of resulting complexes to activate pDCs from healthy donors. We found that polynucleosomes elicited IFN-I production at lower concentration than mononucleosomes (Fig. 5 G). Thus, longer chromatin fragments, such as those enriched in MPs, show better auto-Ab binding as well as better ability to elicit innate responses.
Figure S4.
The length of protein-free DNA does not impact binding of anti-dsDNA IgG from SLE patients. (A) The effect of DNA length on binding to anti-dsDNA Abs as measured in a competition assay. Plasma from SLE patients was incubated with increasing concentrations of PCR-amplified DNA products of different lengths (95 bp, 186 bp, 382 bp, and 831 bp), and the binding of IgG to calf thymus dsDNA was assessed by ELISA. Dashed line shows binding in the absence of competitor DNA. Shown is a representative example of the data quantified for five plasma samples (also used in Fig. 5 F). (B) The effect of DNA length on binding to anti-dsDNA Abs as measured by ELISA. Plate-bound PCR-amplified DNA products of different lengths (95 bp, 186 bp, 382 bp, and 831 bp) were used as Ag to assess anti-dsDNA Abs from plasma of SLE patients. Calf thymus dsDNA, used in standard anti-dsDNA measurements, was used as a control. Shown is a representative example of five patients tested. In both approaches, data are representative of two independent experiments.
DNASE1L3-sensitive MP Ags are targeted in SLE with renal involvement
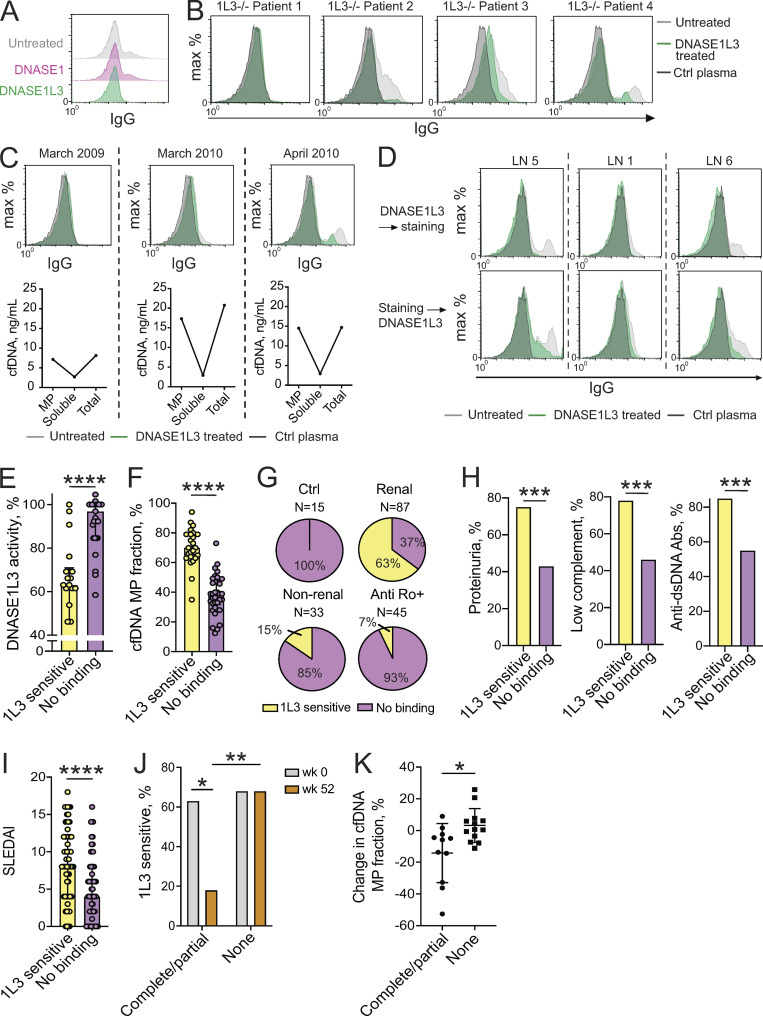
In view of reduced DNASE1L3 activity and the corresponding DNA accumulation in MPs of patients with renal SLE, we tested the functional consequences of these findings for autoreactivity in these patients. We have previously reported that DNASE1L3-deficient mice and ∼30% of human sporadic SLE patients harbored IgG binding to the surface of in vitro–generated MPs. The binding could be abolished by pretreatment of MPs with DNASE1L3, but not with DNASE1, suggesting Ab reactivity against DNASE1L3-sensitive Ags on the surface of MPs (Sisirak et al., 2016). We confirmed these observations (Fig. 6 A) and standardized the assay for an unbiased determination of DNASE1L3-sensitive binding of plasma IgG to MP Ags (referred to hereafter as “DNASE1L3-sensitive binding”). MPs are known to be heterogeneous (Dalli et al., 2013; Nielsen et al., 2011), thus the gating of MPs was deliberately broad to include all fractions below 1 µm in size; indeed, only a fraction of MP showed positive staining with IgG (Fig. 6, A–D). Among four patients with genetic DNASE1L3 deficiency, three manifested DNASE1L3-sensitive binding (Fig. 6 B). One DNASE1L3-deficient patient for whom longitudinal samples were available developed DNASE1L3-sensitive binding shortly before a severe flare of LN that required temporary dialysis. This was also accompanied by a marked increase of cfDNA in the MP fraction, but not in soluble cfDNA (Fig. 6 C).
Figure 6.
Reactivity to DNASE1L3-sensitive MP Ags is associated with reduced DNASE1L3 activity and renal SLE. (A–D) Auto-Ab binding to DNASE1L3-sensitive Ags on MPs. MPs from apoptotic Jurkat cells that were untreated or treated with human DNASE1 or DNASE1L3 were incubated with plasma from SLE patients or healthy control (Ctrl) plasma followed by anti-human IgG conjugates. (A) Histograms of IgG fluorescence using plasma from a SLE patient; representative of three independent experiments. (B) MP staining by plasma from patients with genetic DNASE1L3 deficiency (1L3−/−, n = 4) or from a healthy control (Ctrl). (C) MP binding by IgG from plasma of a DNASE1L3-deficient patient (patient 4 from B) who suffered a severe disease flare with renal failure in May 2010. Top row shows histograms of plasma IgG binding to MPs at the indicated dates. Bottom panel shows the concentration of cfDNA in MPs, the MP-depleted (soluble) plasma fraction, and total plasma at each date. (D) Auto-Ab binding to DNASE1L3-sensitive MP Ags in sporadic SLE patients with LN. Jurkat cell–derived MPs were treated with DNASE1L3 and stained with plasma (top row) or stained first and subsequently treated with DNASE1L3 (bottom row). Shown are representative histograms of IgG fluorescence in three LN patients with reduced DNASE1L3 activity and anti-DNASE1L3 Abs. (E) Relative DNASE1L3 activity in SLE patients with (n = 22) or without (n = 29) reactivity to DNASE1L3 (1L3)–sensitive Ags on MPs. Symbols represent individual patients, and bars represent median ± interquartile range. Statistical significance was determined by Mann–Whitney test; ****, P values < 0.0001 (two tailed). (F) The fraction of MP-associated cfDNA in sporadic SLE patients with (n = 25) or without (n = 34) reactivity to DNASE1L3-sensitive Ags on MPs. Symbols represent individual patients, and bars represent median ± interquartile range. Statistical significance was determined by Mann–Whitney test; ****, P < 0.0001 (two tailed). (G) Reactivity to DNASE1L3-sensitive Ags on MPs in the indicated patient groups. (H) Frequency of the indicated SLEDAI criteria in SLE patients (combined renal and nonrenal) with (n = 61) or without (n = 59) reactivity to DNASE1L3 (1L3)-sensitive Ags on MP. Fisher’s exact test was used to prepare contingency tables between the two groups; ***, P < 0.001 (two tailed). (I) SLEDAI index in renal and nonrenal SLE patients with (n = 61) or without (n = 59) reactivity to DNASE1L3 (1L3)-sensitive Ags on MPs. Symbols represent individual patients, and bars represent median ± interquartile range. Statistical significance was determined by Mann–Whitney test; ****, P < 0.0001 (two tailed). (J) Reactivity to DNASE1L3-sensitive Ags on MP in LN patients depending on treatment response. Shown is the percent of patients with DNASE1L3 (1L3)–sensitive IgG binding to MPs at baseline (week 0, gray) and after 52 wk of treatment (week 52, orange) for patients with complete (n = 8)/partial (n = 3) treatment response versus no response (n = 19). Fisher’s exact test was used to prepare contingency tables between the two groups; *, P < 0.05; **, P < 0.01 (two tailed). (K) Change in the fraction of MP-associated cfDNA in LN patients with a complete (n = 8)/partial (n = 3) or no treatment response (n = 13) at baseline and after 52 wk of treatment. Symbols represent individual subjects; bars represent mean ± SD. Statistical significance was determined by Mann–Whitney test; *, P < 0.05 (two tailed).
To test the prevalence of reactivity to DNASE1L3-sensitive Ags on MPs in sporadic SLE, we analyzed a well-characterized cohort of SLE patients (n = 120), many of whom were used in the initial description of Ab binding to DNASE1L3 described above. Of these patients, 87 (73%) had renal SLE and 62 (52%) had undergone longitudinal sampling. In total, 221 plasma samples were analyzed, revealing that 61 patients (51%) harbored DNASE1L3-sensitive binding during at least one sample collection. The clinical characteristics of sporadic SLE patients organized by the presence of DNASE1L3-sensitive binding are summarized in Table S1. As expected, no DNASE1L3-sensitive binding was observed in any of the healthy controls analyzed (n = 15). Binding of plasma IgG to MP Ags could be prevented or reversed by the treatment of MPs with DNASE1L3 before or after the incubation with patient plasma, respectively (Fig. 6 D). Importantly, patients with DNASE1L3-sensitive binding showed a significant decrease of DNASE1L3 activity in the plasma (Fig. 6 E) and a near-uniform increase in cfDNA in the MP fraction (Fig. 6 F). Furthermore, DNASE1L3-sensitive binding was strongly associated with renal involvement: 55 of 87 (63%) renal patients compared with 5 of 33 (15%) nonrenal SLE patients (P < 0.0001; Fig. 6 G). Moreover, patients with DNASE1L3-sensitive binding showed a significant enrichment in proteinuria, low complement (i.e., C3 and C4) levels, positive anti-dsDNA Abs (Fig. 6 H), and showed a higher median SLE disease activity index (SLEDAI) at the time of blood sampling (Fig. 6 I). In anti-Ro+ mothers, while 8 of 45 (18%) demonstrated IgG binding to MP Ags, this binding was sensitive to DNASE1L3 in only three patients (Fig. 6 G). Two of these were the only subjects with renal SLE, with one patient harboring Abs to DNASE1L3 (Fig. 2 A and Table 1).
To address whether DNASE1L3-sensitive binding tracks with clinical response in patients with active LN, samples were obtained from 30 patients at the time of renal biopsy and longitudinally over a 52-wk period. Clinical response was defined at 52 wk as complete (n = 8, UPCR <0.5), partial (n = 3, UPCR reduced by 50% from baseline but >0.5), or none (n = 19). Detailed information on patient characteristics organized by treatment response is given in Table S2 and characteristics for each individual patient included in this analysis are shown in Table S3. At baseline, there was no significant difference in the prevalence of DNASE1L3-sensitive binding between combined partial and complete responders (7/11, 63%) and nonresponders (13/19, 68%). However, at 52 wk, only 2/11 (18%) combined treatment responders still manifested DNASE1L3-sensitive binding, while this reactivity remained persistent in all nonresponders (13/19, 68%; Fig. 6 J). Additionally, at baseline, there was no difference in the fraction of cfDNA located in MPs, but combined treatment responders demonstrated a shift of cfDNA toward the MP-depleted plasma fraction, while the distribution of cfDNA remained unchanged in nonresponders (Fig. 6 K). Collectively, these data suggest that Abs to DNASE1L3-sensitive Ags on MPs inversely correlate with clinical response to treatment in patients with active LN.
Reactivity toward DNASE1L3-sensitive Ags includes DNA-associated proteins
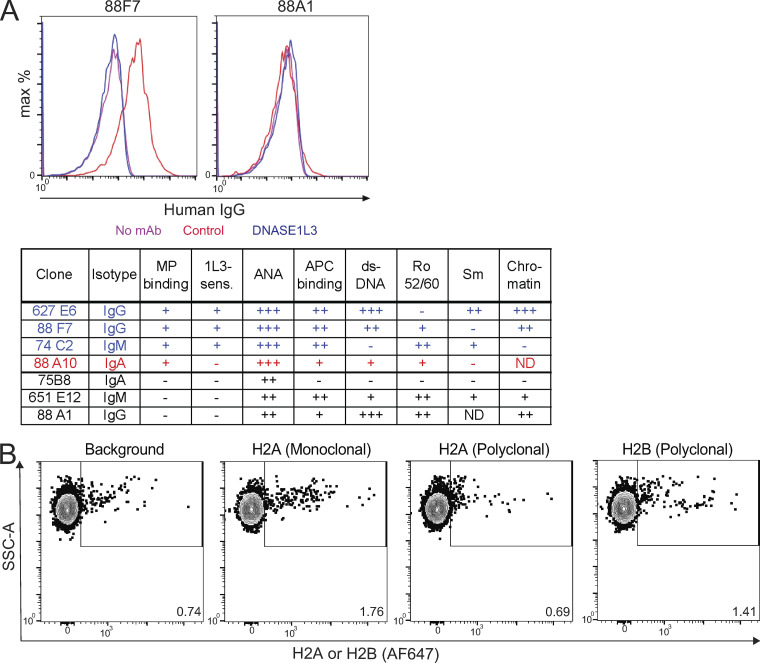
It was noted in evaluating the reactivity to DNASE1L3-sensitive MP Ags that this did not fully mirror anti-DNA Abs. Specifically, anti-dsDNA IgG was present in >80% of patients with this reactivity but also in >50% of patients without it (Fig. 6 H). Moreover, only one of the three DNASE1L3-deficient patients with DNASE1L3-sensitive binding (Fig. 6 B) had anti-dsDNA Abs as assessed by ELISA (data not shown). To further explore the relationship between Abs to dsDNA and MPs, we examined a panel of 9G4+ mAbs derived from SLE patients. Abs marked by the 9G4 idiotope are prominent among SLE-associated auto-Abs and bind multiple Ags, including DNA and apoptotic cell surface determinants (Pugh-Bernard et al., 2001; Richardson et al., 2013; Tipton et al., 2015). Out of 20 examined 9G4+ mAbs, three showed DNASE1L3-sensitive binding (Fig. S5 A). Two of those also bound dsDNA and chromatin as expected; however, one (74C2) did not. Conversely, other clones (e.g., 88A1) reacted with dsDNA and chromatin by ELISA but did not bind MPs (Fig. S5 A). Collectively, these data suggest that reactivity to DNASE1L3-sensitive MP Ags partially overlaps with, but is distinct from, anti-dsDNA Abs and likely includes additional antigenic targets associated with DNA.
Figure S5.
Further characterization of MP Ags and auto-Ab reactivity. (A) Binding of monoclonal Abs marked by the 9G4+ idiotope (9G4+ mAbs) to MP and nuclear Ags. Recombinant IgG mAb clones were generated based on auto-Ab sequences from SLE patients, and their reactivity to nuclear Ags, apoptotic cell surfaces, and DNASE1L3-sensitive Ags on MP was determined. Top panel shows the binding of representative clones to MP that were pretreated with control (red) or DNASE1L3-containing (blue) supernatants. Bottom panel lists the original isotype, binding to MP and whether it is DNASE1L3 (1L3) sensitive, and binding to nuclear/apoptotic cell Ags. (B) Exposure of H2A and H2B on the surface of Jurkat MPs assessed by flow cytometry. Untreated Jurkat MPs were stained in the absence (background) or presence of monoclonal or polyclonal Abs to H2A or a polyclonal Ab to H2B. Representative of three independent experiments. SSC-A, side scatter signal area.
To further characterize reactivity to MP Ags, we explored the recognition of MP proteins by IgG from SLE patients using lysates of MPs harvested from apoptotic Jurkat T cells. SLE plasma samples that had shown DNASE1L3-sensitive IgG binding to MPs by flow cytometry also demonstrated reactivity to multiple MP proteins by Western blot. Most prominently, reactivity to proteins of ∼10–15, 29, 35, 45, and 75 kD could be observed across multiple plasma samples (Fig. 7, A and B). We then took a candidate approach and explored the reactivity to histones and high-mobility group B1 (HMGB1) protein. High-mobility group proteins are structural transcription factors that can be incorporated into chromatin or released from activated or dying cells; HMGB1 in particular is an important mediator of inflammation and a common target of auto-Abs in SLE (Pisetsky, 2014a; Urbonaviciute et al., 2008; Wirestam et al., 2015). Both H2A/H2B and HMGB1 could be identified by Western blot in lysates of Jurkat cell MP, and bands of similar size (∼29 kD for HMGB1 and ∼13–15 kD for histones) were detected by SLE plasma samples with DNASE1L3-sensitive reactivity to MP Ags (Fig. 7 B). We next analyzed plasma IgG from Dnase1l3-deficient mice and observed IgG binding to proteins of similar size as those seen with human SLE plasma (Fig. 7 C). This reactivity was age dependent (Fig. 7 D), consistent with progressive autoreactivity and epitope spreading in older Dnase1l3-deficient mice (Sisirak et al., 2016).
Figure 7.
DNASE1L3-sensitive Ags on MP include DNA-associated proteins. (A) Human plasma IgG binding to proteins in MPs from Jurkat T cells. Plasma samples from healthy controls (Ctrl), DNASE1L3-deficient patients (1L3 null), sporadic SLE patients (SLE), or purified IgG from a healthy control (Ctrl IgG) and sporadic SLE patient (SLE IgG) were tested by Western blot. The presence (+) or absence (−) of IgG binding to DNASE1L3-sensitive MP Ags as assessed by flow cytometry is indicated. (B) Histones and HMGB1 as potential Ags on MPs as determined by Western blot. Plasma samples from patients with sporadic SLE were analyzed for IgG binding to MP proteins as in A. In parallel, MPs were probed by anti-H2A, anti-H2B, and anti-HMGB1 Abs. Representative of two independent experiments. (C and D) SLE-prone mouse plasma IgG binding to MP proteins as determined by Western blot. In C, plasma from a Dnase1l3-deficient mouse (1L3 KO mouse, dilutions 1:50, 1:150, and 1:450) and plasma from a patient with sporadic SLE (SLE human, dilutions 1:150, 1:500, and 1:1,500) were tested in parallel. In D, plasma from Dnase1l3-deficient (1L3 KO) mice before (3 mo) or after the onset of SLE-like disease (7–10 mo) was tested. Representative of two independent experiments. (E) DNASE1L3-sensitive exposure of HMGB1 and DNA on the surface of Jurkat MPs. MPs that were untreated or pretreated with recombinant human DNASE1 or DNASE1L3 were stained for HMGB1 (top) or DNA (Vybrant Green, bottom) and analyzed by flow cytometry. Representative of three independent experiments. SSC-A, side scatter signal area.
Although Western blot analysis confirmed the reactivity of patient auto-Abs with multiple protein Ags in MPs, it did not establish whether this reactivity is DNASE1L3 sensitive. To this end, we used flow cytometry to show that HMGB1 was exposed on the surface of Jurkat cell–derived MPs and that this exposure was sensitive to DNASE1L3, but not DNASE1 (Fig. 7 E). Staining for H2A and H2B did not yield a distinct positive MP population (Fig. S5 B), likely due to the blockade of histone epitopes by the associated nucleosomal DNA on the surface of MPs. Taken together, these data demonstrate that reactivity to DNASE1L3-sensitive Ags on MPs targets not only DNA but also multiple proteins, potentially including known SLE-associated self-Ags such as HMGB1 and histones.
Discussion
In this study, we explored the role of DNase activity in SLE, focusing specifically on DNASE1L3, the extracellular DNase implicated in monogenic SLE. It was reported that total DNase activity may be reduced in SLE patients (Martinez-Valle et al., 2009; Sallai et al., 2005), but no correlation with disease severity was observed, and the specific DNase was not identified. More recently, a decrease in total DNase activity was observed specifically in nephritis patients (Bruschi et al., 2020), although the protein levels of both DNASE1 and DNASE1L3 appeared normal. Using a novel functional assay that is specific for DNASE1L3, we found that more than half of SLE patients with renal involvement show reduced activity of DNASE1L3 in the circulation. Given that DNASE1L3 nonredundantly protects from SLE and that a partial reduction of its activity in DNASE1L3 (R206C) carriers is associated with autoimmunity, its specific reduction in severe sporadic SLE likely contributes to the pathogenesis.
Reduced DNASE1L3 activity strongly correlated with the appearance of activity-neutralizing Abs to DNASE1L3, suggesting that DNASE1L3 is blocked by this newly described auto-Ab. Indeed, the measurement of anti-DNASE1L3 Abs by ELISA provides a robust high-throughput assay to estimate DNASE1L3 activity in sporadic SLE patients in future studies. This mechanism does not rule out other possible scenarios such as reduced DNASE1L3 protein levels, a possibility that could not be explored here due to the lack of sufficiently specific protein detection reagents. Ab-mediated blockade is consistent with the results of Bruschi et al. (2020), who implicated inhibitory circulating proteins in the reduction of total DNase activity in renal SLE. Notably, auto-Abs preferentially bound DNASE1L3 compared with DNASE1 and thus appear distinct from previously described DNASE1 inhibitory substances (Hakkim et al., 2010) or polyreactive anti-DNASE1 Abs that also bind DNA (Puccetti et al., 1995; Yeh et al., 2003). The observed Ab-mediated inhibition of DNASE1L3 resembles a similar inhibition of the complement component C1Q. Like DNASE1L3, C1Q is required for self-tolerance, as null C1Q mutations in humans cause SLE with a near-complete penetrance (Leffler et al., 2014). In addition, C1Q can itself be targeted by a specific auto-Ab that reduces C1Q levels in HUVS (Jara et al., 2009) and is also found in sporadic SLE (Orbai et al., 2015). The inhibition of DNASE1L3 by specific auto-Abs appears to recapitulate its genetic deficiency by nongenetic means, emphasizing the critical role of DNASE1L3 in tolerance and its relevance for sporadic SLE.
Self-DNA that may activate innate or adaptive immunity in SLE may comprise inflammation-associated forms such as neutrophil extracellular traps and oxidized mitochondrial DNA (Caielli et al., 2016; Garcia-Romo et al., 2011; Lood et al., 2016) or cfDNA that exists in the steady state. Given the observed effects of DNASE1L3 on the latter (Serpas et al., 2019), we explored its physical nature in circulation, including the association with MPs. We confirmed the presence of DNA-carrying MPs in normal plasma and established that ∼25% and >90% of total cfDNA is MP associated in humans and mice, respectively. The reasons for this difference between the species are unknown but may involve the shorter time and route of blood circulation in mice, reducing the lifespan of soluble cfDNA. Importantly, MP-associated cfDNA was enriched in longer polynucleosomal DNA fragments, consistent with the detection of such fragments in MPs generated in vitro (Reich and Pisetsky, 2009). Fragments of naked DNA within the 100–4,000 bp size range were shown to bind anti-DNA Abs equally well in solution (Pisetsky and Reich, 1994). We confirmed these observations but showed that for nucleosome-bound DNA, longer polynucleosomal fragments bind auto-Abs with higher affinity. We also demonstrated the superior capacity of polynucleosomal fragments as inducers of IFN-I production. These data are consistent with the critical role of IFN-I in SLE (Elkon and Wiedeman, 2012) and with its contribution to autoreactivity and disease in DNASE1L3-deficient mice (Soni et al., 2020). Overall, our results highlight the abundance of MP-associated cfDNA and its potential as a self-Ag that requires tight control in vivo.
The MP-associated fraction of cfDNA was increased in DNASE1L3-deficient mice and humans and correlated with reduced DNASE1L3 activity in sporadic SLE patients with nephritis. This was in striking contrast to comparable levels of total plasma cfDNA, which did not show significant differences between patient groups in our study. Although increased total cfDNA has been reported in SLE patients compared with controls (Duvvuri and Lood, 2019), this difference showed no clear association with disease activity or with LN specifically (Atamaniuk et al., 2011; Xu et al., 2018). Sequencing of total cfDNA in SLE revealed certain abnormalities such as aberrant genome representation and shortening due to the presence of anti-dsDNA Abs, but no consistent features serving as biomarkers of the disease (Chan et al., 2014). Our results highlight the relevance of MP-associated cfDNA for SLE pathogenesis, given its striking increase in LN relative to healthy and non-LN patients. Moreover, they confirm DNASE1L3 as an essential and specific regulator of MP-associated cfDNA, which therefore represents the physiological substrate of this enzyme. They also help explain the lengthening of total cfDNA in the plasma of Dnase1l3-deficient mice (Serpas et al., 2019) and humans (Chan et al., 2020), which apparently reflects the accumulation of MP-associated polynucleosomal fragments. Thus, auto-Ab–mediated impairment of DNASE1L3 activity in renal SLE patients results in the accumulation of a self-Ag (i.e., MP-associated cfDNA and affiliated proteins).
Auto-Ab from SLE patients were shown to bind endogenous MPs (Nielsen et al., 2012) or in vitro–generated MPs (Ullal et al., 2011), yet the nature of bound Ag and clinical significance of this reactivity remained unclear. We report that Abs to MPs are prevalent in patients with SLE and target DNASE1L3-sensitive Ag on the MP surface. We also show that this Ab reactivity is associated with reduced DNASE1L3 activity and cfDNA accumulation into MPs, likely representing a consequence of these events. Unlike ELISA-based assays for Abs to DNA and other Ags, the binding to DNASE1L3-sensitive Ag on MPs is a physiological interaction that is unbiased with respect to a specific Ag. As such, it represents an integral readout that correlates but does not directly overlap with any single reactivity, including the canonical anti-dsDNA ELISA. This likely explains why reduced DNASE1L3 activity and/or DNASE1L3-targeting Abs did not correlate with anti-dsDNA (Fig. S2 D and data not shown) but were associated with reactivity to MPs (Fig. 6 E). Indeed, we demonstrate that the reactivity to MPs involves a set of protein Ag carried by MPs, with reactivity patterns shared among different patients and even with Dnase1l3-deficient mice. One established DNASE1L3-sensitive Ag is the nucleosome (Sisirak et al., 2016), consistent with its exposure on the MP surface (Ullal et al., 2011) and the presence of histones in MPs (this study); however, pure histones may not be targeted independently of the DNA. Another candidate DNASE1L3-sensitive Ag is HMGB1, a pleiotropic DNA-binding protein that is a common target of auto-Abs in SLE (Harris et al., 2012; Urbonaviciute et al., 2007). The exposure of HMGB1 on apoptotic MPs has been described (Pisetsky, 2014b), and here, we demonstrate that this phenomenon is controlled by DNASE1L3, presumably by digesting HMGB1–DNA complexes. Thus, by digesting DNA in MPs, DNASE1L3 controls the exposure of not only DNA but also multiple DNA-associated proteins, which are targeted by auto-Abs in SLE patients with reduced DNASE1L3 activity.
In conclusion, our data delineate the molecular basis of tolerogenic DNASE1L3 enzymatic activity and describe its impairment in some patients with SLE, most notably those with kidney involvement. Conceptually, our data reveal a novel mechanism of SLE pathogenesis based on auto-Abs targeting a critical “gatekeeper” of tolerance to self-DNA. The inhibition of DNASE1L3 activity by such auto-Abs recapitulates the genetic lesion of DNASE1L3-deficient patients by nongenetic means, emphasizing the key role of DNASE1L3 in sporadic SLE. Technically, our work introduces four novel readouts (DNASE1L3 activity, Abs to DNASE1L3, fraction of MP-associated cfDNA, and Abs to DNASE1L3-sensitive Ag) that correlate with each other and with active LN. Of these assays, IgG binding to DNASE1L3-sensitive Ag on MPs is a relatively simple, robust, high-throughput assay whose readout showed correlation with LN in both cross-sectional and longitudinal samples. Finally, the reduction of DNASE1L3 activity in SLE patients with nephritis warrants further exploration of DNASE1L3 as a potential therapeutic in this patient group. Our results provide an important caveat to such attempts by describing neutralizing Abs to DNASE1L3; nevertheless, the rationale for the therapeutic use of this secreted enzyme that is fundamentally linked to SLE pathogenesis appears compelling.
Materials and methods
Human subjects
The study population was composed of four subgroups: DNASE1L3-deficient patients (n = 4), the New York University (NYU) SLE cohort (n = 120), anti-Ro+ mothers of children with neonatal lupus (n = 45), and healthy controls (n = 15). The NYU SLE cohort was further stratified into SLE patients with (n = 87) and without (n = 33) renal involvement according to the Systemic Lupus Erythematosus International Cooperating Clinics criteria (Petri et al., 2012).
Sporadic SLE and healthy controls
Blood specimens were obtained from several sources. Patients with SLE and healthy controls older than 18 yr of age were enrolled in the NYU SAMPLE (Specimen and Matched Phenotype Linked Evaluation) Biorepository after signing informed consent approved by the NYU School of Medicine institutional review board. SLE was assigned using classification criteria as defined by one or more of the following: (1) 1997 Revised American College of Rheumatology (ACR; Hochberg, 1997; Tan et al., 1982), (2) Systemic Lupus Erythematosus International Cooperating Clinics (Petri et al., 2012), or (3) the most recent ACR/European League Against Rheumatism (Aringer et al., 2019). Plasma for the study of SLE patients was selected randomly to represent the full spectrum of clinical disease. All SLE patients were evaluated for the status of disease activity using a modification of the original Safety of Estrogens in Lupus National Assessment (SELENA)–SLEDAI (Petri et al., 2005) in which proteinuria was scored 4 points only when the UPCR increased by more than 0.5 from the previous visit. The modification applied in this study used the SLEDAI 2K definition of proteinuria in which a UPCR of >0.5 is always scored 4 points (Gladman et al., 2002). All other domains of the SELENA-SLEDAI were scored as originally defined. For the longitudinal follow up, a subset of SLE patients undergoing a kidney biopsy to evaluate a UPCR ratio of >0.5 (considered abnormal) was included in the study. A renal response at 52 wk was considered complete if the UPCR was <0.5 and a partial response if the baseline UPCR was reduced by 50% but did not resolve to <0.5.
Anti-Ro+ mothers of children with neonatal lupus
A second disease cohort evaluated were women enrolled in the Research Registry for Neonatal Lupus (RRNL; Buyon et al., 1998) after signing informed consent for participation approved by the NYU School of Medicine institutional review board. All participating mothers in the RRNL included in this study have a child with congenital heart block and/or characteristic rash and have Abs to at least one component of the Ro/La ribonucleoprotein complex, including 52-kD Ro, 60-kD Ro, or 48B/La, as confirmed by ELISA in the laboratory of R.M. Clancy and J.P. Buyon (Reed et al., 2012). Many of the women in the RRNL do not have any systemic disease and are only known to have anti-Ro Abs because of the workup for their child’s illness. Some mothers have an undifferentiated autoimmune disease, as they have insufficient criteria for SLE or Sjogren’s syndrome (Shiboski et al., 2017; Vitali et al., 2002) and less than half ever progress to SLE and/or Sjogren’s syndrome (Rivera et al., 2009). In this group of RRNL women, three had developed LN.
DNASE1L3-deficient patients
One patient with pediatric HUVS with renal involvement progressing to end-stage renal disease was diagnosed and treated at the Istituto Gaslini. Collection of patient samples was approved by the ethics committee of the Istituto Giannina Gaslini (approval BIOL 6/5/04). Molecular analysis by next-generation sequencing was performed in the Laboratory of Neurogenetics and Neuroinflammation at the Imagine Institute for Genetic Diseases, Paris. The patient was homozygous for the c.289-290delAC DNASE1L3 mutation, which was reported previously (Batu et al., 2018; Carbonella et al., 2017; Ozçakar et al., 2013) and is predicted to cause premature termination (p.Thr97Ilefs*2). Three patients with pediatric HUVS (age of onset, 9–14 yr) were diagnosed and treated at the Istituto di Ricovero e Cura a Carattere Scientifico Ospedale Pediatrico Bambino Gesù. Sample collection of patients from Ospedale Pediatrico Bambino Gesù was approved by the Institutional Ethical Committee number 1666_OPBG_2018. All patients had chronic urticarial vasculitis, hypocomplementemia, positive anti-C1Q Ab, negative anti-nuclear Abs, and negative anti-dsDNA by laboratory testing; one of the three showed positive anti-dsDNA by in-house ELISA. The patients were subjected to targeted resequencing on the Illumina NextSeq platform for genes associated with interferonopathies. All patients were homozygous for the c.290_291delCA DNASE1L3 mutation, which was reported recently (Belot, 2020) and is predicted to cause the same premature termination as the mutation above. All patients and/or their families signed informed consent for participation approved by the ethics committees at their respective institutions.
Animals
All animal studies were performed according to the investigator’s protocol approved by the Institutional Animal Care and Use Committee of NYU School of Medicine. Dnase1l3-deficient mice on C57BL/6 (B6) background (Dnase1l3LacZ/LacZ) were described previously (Sisirak et al., 2016). Mice carrying a null allele of Dnase1 (Dnase1tm1.1(KOMP)Vlcg) on B6 background were obtained from the Knockout Mouse Project, crossed with WT B6 mice and intercrossed to obtain Dnase1−/− mice. Mice with doxycycline-inducible ubiquitous expression of H2B-GFP fusion protein (B6;129S4-Gt(ROSA)26Sortm1(rtTA*M2)JaeCol1a1tm7(tetO-HIST1H2BJ/GFP)Jae/J; Foudi et al., 2009) and mice with constitutive ubiquitous expression of membrane-targeted tdTomato protein (B6.129(Cg)-Gt(ROSA)26Sortm4(ACTB-tdTomato,-EGFP)Luo/J; Muzumdar et al., 2007) were obtained from The Jackson Laboratory and intercrossed. The resulting mice heterozygous for both alleles were given oral doxycycline in water for at least 4–6 wk to induce the expression of H2B-GFP as described (Foudi et al., 2009). WT control mice of B6 background were bred in the same animal facility or purchased from Taconic and maintained in the same facility. All mice were age and sex matched when comparing groups. Male and female WT mice (57% female and 43% male) between 30 and 450 d of age (median 80 d) were analyzed (Fig. 4 C); no difference in the concentration or distribution of cfDNA was observed between mice of different age or sex. Dnase1l3-deficient males younger than 60 d were used in order to avoid any confounding effect of SLE-like disease that develops starting at 3 mo (Fig. 4 D). 1-yr-old Dnase1-deficient females were used in Fig. 4 E. These mice do not develop any autoreactivity at any age, and thus the age is not expected to impact our findings.
DNASE1L3 protein purification
Preliminary studies showed that the introduction of a 6-aa FLAG epitope into a predicted internal loop of the human full-length DNASE1L3 did not affect the activity of DNASE1L3, including its specific ability to digest chromatin. The resulting FLAG-DNASE1L3 open reading frame was cloned into a lentiviral vector, which was then used to transduce 293T cells followed by batch selection of stable transfectants. FLAG-DNASE1L3–expressing 293T cells were cultured in 150-mm dishes for 7 d, and the supernatants were incubated with magnetic anti-FLAG M2 beads (Sigma-Aldrich) overnight at 4°C. The beads were collected, washed, and incubated with 0.25 mg/ml FLAG peptide, and the eluted protein was quantified by SDS-PAGE with BSA as a standard.
Isolation of MPs
Human plasma
To obtain the MP fraction as well as the MP-depleted (soluble) fraction of human plasma, blood was collected in tubes containing EDTA (Becton Dickinson; 366643), and blood cells were removed by centrifugation at 1,400 g for 12 min at 4°C. A second centrifugation step (1,400 g for 12 min) was used to remove platelets. The resultant plasma (either fresh or stored at −80°C) was centrifuged at 22,000 g for 60 min to pellet the MP. The supernatant and MP pellet were used to measure cfDNA in the MP-depleted (soluble) and MP plasma fractions, respectively.
Murine plasma
Mice were anaesthetized by i.p. injection of 100 mg/kg ketamine/10 mg/kg xylazine in sterile PBS. Plasma was isolated by cardiac puncture using a 26G syringe and transferred into EDTA-coated tubes (Kent Scientific; 41.1395.105). Blood was centrifuged at 1,000 g for 10 min, and the supernatant was kept on ice. For DNA quantification, the plasma was spun a second time at 1,000 g for 10 min. For confocal imaging studies, the plasma was spun a second time at 10,000 g for 10 min to remove platelets. After the second spin, the supernatant was centrifuged at 21,130 g for 60 min at 4°C to pellet MPs. The supernatant and MP pellet were used to measure cfDNA in the MP-depleted (soluble) and MP plasma fractions, respectively.
Quantification of cfDNA
To extract cfDNA from the MP fraction and MP-depleted fraction, samples were centrifuged as described above. Circulating cfDNA was extracted from 50–400 µl human plasma or 150–200 µl murine plasma using the QIAamp DNA Blood Mini Kit (QIAGEN). The amount of DNA in the respective samples was measured by qPCR using intra-Alu human primers or inter-B1 mouse primers. All qPCRs were performed in 20 µl using FastStart Universal SYBR Green Master mix (Sigma-Aldrich 4913914001). Briefly, the method for intra-Alu human primers was as follows: initial denaturation of 12 min at 95°C followed by 45 amplification cycles of denaturation at 95°C for 20 s, annealing at 56°C for 1 min, and elongation at 72°C for 1 min (Walker et al., 2003). The mouse inter-B1 qPCR methods have been reported previously (Zhang et al., 2010). Briefly, each reaction was subjected to an initial denaturation of 8 min at 95°C followed by 40 amplification cycles of denaturation for 30s at 95°C, annealing for 40s at 55°C, and extension for 1 min at 72°C.
Estimation of cfDNA lengths by qPCR
Two sets of primers and the respective qPCR programs described by Lou et al. were used to amplify 76 bp (short) or 200 bp (long) of the human Alu repeat sequences (Lou et al., 2015). The concentration of cfDNA as determined by the 76-bp amplicons and 200-bp amplicons were calculated, and subsequently the ratio of long to short amplicon products was determined as a measure of cfDNA length in the MP fraction and MP-depleted (soluble) plasma fraction.
cfDNA sequencing in human patients
Plasma from four healthy controls (two males and two females) was isolated and centrifuged to separate MP and MP-depleted fractions as described above. DNA was purified from all samples using the QIAamp DSP DNA Blood Mini Kit (QIAGEN; 61104) and quantified by qPCR. DNA libraries were prepared using the KAPA HTP Library Preparation Kit (Roche) and purified using AMPure XP purification beads (Beckman Coulter) according to the manufacturer’s instructions. Adaptor-ligated libraries were analyzed on an Agilent 4200 TapeStation (Agilent Technologies) using the High Sensitivity D1000 ScreenTape System (Agilent Technologies) for quality control and quantified by Qubit (Invitrogen). The libraries were sequenced for 150 bp for each end in a paired-end format on a Hiseq 4000 or the equivalent Illumina sp300 flow cell.
DNA sequencing analysis
The adaptor sequences were trimmed using cutadapt (v1.18), and all of the reads from the sequencing experiment were mapped to the reference genome (hg19/GRCh37.75) using Bowtie2 (v2.2.4). Duplicate reads were removed using Picard tools (v.1.126), and the low-quality mapped reads (mapping quality <20) were removed from the analysis. The read per million normalized BigWig files were generated using BEDTools (v.2.17.0) and the bedGraphToBigWig tool (v.4). The insert sizes and their SDs were calculated using Picard tools and used to generate cfDNA length traces shown. Frequencies were calculated by dividing the number of reads from each insert size by the total number of reads acquired per sample. All downstream statistical analysis was performed in the R environment (v3.1.1). Sequence data have been deposited in the Genotypes and Phenotypes database hosted by the National Institutes of Health (BioProject accession no. PRJNA695527).
Assessment of DNASE1L3 activity
Digestion of DNA substrates with DNases
DNA substrates were incubated with different concentrations of human DNASE1 (Abcam; 73430) or human recombinant DNASE1L3 in the presence of 2 mM CaCl2/MnCl2 and a total volume of 10 µl for 15 min at 37°C. All dilutions of enzymes were prepared in HBSS (without magnesium and calcium) with 100 ng/ml molecular-grade BSA. Following digestion, the reactions were immediately kept on ice. The amount of remaining DNA was measured by qPCR using intra-Alu human primers as follows: initial denaturation of 10 min at 95°C followed by 45 amplification cycles of denaturation at 95°C for 15 s, annealing at 60°C for 15 s, and elongation at 72°C for 15 s. The following DNA substrates were used: 200 ng naked Jurkat DNA (Thermo Fisher) or nuclei purified from 15,000 Jurkat T cells as described previously (Nabbi and Riabowol, 2015). The digestion of nuclei was used to assess specific activity of plasma DNASE1L3, while the digestion of naked DNA was used to assess total plasma DNase activity. To assess whether plasma IgG could bind and inhibit enzymatic DNase activity, IgG was purified from healthy controls and three SLE patients with Abs to DNASE1L3 using Protein G beads. Different concentrations of DNases were preincubated with 0.1 mg/ml purified IgG at 37°C for 30 min. Preincubated DNase and untreated DNase were used to digest DNA substrates as described and the amount of remaining DNA was measured by qPCR. The DNase concentration at which 50% of the input DNA was digested (IC50) was established, and the percentage of DNase activity was compared with untreated DNase was calculated.
Quantitative DNASE1L3 activity assay
Several dilutions of human plasma were incubated with 15,000 Jurkat T cell–derived nuclei and 2 mM CaCl2/MnCl2 in a total volume of 10 µl for 10 min at 37°C. All plasma samples were diluted in HBSS (without magnesium and calcium) with 100 ng/ml molecular grade BSA. The remaining DNA was measured by qPCR using intra-Alu human primers as described above. The DNase concentration at which 50% of the input DNA was digested (IC50) was established, and the percentage of DNase activity was determined, considering the IC50 value for pooled healthy control plasma as 100% and the value for DNASE1L3-deficient patient plasma as 0%.
We have found that total reaction volume, digestion time, digestion solution, batch of nuclei, CaCl2/MnCl2 concentration, plasma diluent, and polymerase used for qPCR-based DNA quantification can affect the results of this assay. Detailed protocols and troubleshooting tips will be shared with all interested investigators upon request.
ELISA
Anti-DNase ELISA
All plate incubations were done in a hydration chamber unless otherwise specified. Nunc MaxiSorp plates (Invitrogen; 44–2404-21) were coated with 50 µl/well of 2 µg/ml recombinant human DNASE1 (Abcam; 73430), purified recombinant DNASE1L3, or FLAG peptide (Sigma-Aldrich) in PBS overnight at 4°C. Ag-coated plates were washed with PBS and blocked with 250 µl/well of PBS with 4% nonfat dry milk (NFDM) for 2–3 h at room temperature. Plates were then washed again three times with PBS and incubated with plasma diluted in PBS overnight at 4°C. Plates were washed three times with PBS containing 1% NFDM, and bound IgG was detected using goat anti-human IgG–alkaline phosphatase conjugate (Sigma-Aldrich; A1543-1ML) diluted 1:5,000 in PBS containing 1% NFDM. The plate was then washed three times with PBS containing 1% NFDM and developed using diethanolamine substrate buffer (Thermo Fisher) and para-nitrophenylphosphate phosphatase substrate tablets (Sigma-Aldrich).
Anti-dsDNA ELISA
Nunc MaxiSorp plates were coated with 0.01% poly-L lysine (Sigma-Aldrich) for 1 h at room temperature, washed with PBS, and coated with 50 µl/well of 10 µg/ml calf thymus DNA (Calbiochem) in PBS overnight at 4°C. Ag-coated plates were washed and then blocked with 200 µl/well of PBS containing 4% NFDM for 3 h at 4°C. Blocked plates were washed once with PBS + 1% NFDM, coated with diluted serum in PBS, and incubated overnight at 4°C. Unbound serum Abs were washed off with three washes of PBS + 1% NFDM. The bound IgG was detected using a goat anti-human IgG–alkaline phosphatase conjugate (Sigma-Aldrich; A1543) diluted 1:5,000 in PBS + 1% NFDM. Unbound secondary Ab was washed off with PBS+ 1% NFDM, and the plate was developed as described above.
Anti-dsDNA ELISA with DNA fragments of different length
Nunc MaxiSorp plates were coated with 0.01% poly-L lysine (Sigma-Aldrich) for 1 h at room temperature, washed with PBS, and coated with 50 µl/well of 10 µg/ml of DNA in PBS overnight at 4°C. DNA fragments of 95 bp, 186 bp, 382 bp, and 831 bp were prepared by PCR and purified using QIAquick Gel Extraction Kit (QIAGEN). Calf thymus DNA (Calbiochem) was used as a control. Ag-coated plates were washed and then blocked with 200 µl/well of PBS containing 4% NFDM for 3 h at 4°C. Blocked plates were washed once with PBS + 1% NFDM, coated with diluted serum in PBS, and incubated overnight at 4°C. Unbound serum Abs were washed off with three washes of PBS + 1% NFDM. The bound IgG was detected using a goat anti-human IgG–alkaline phosphatase conjugate (Sigma-Aldrich; A1543-1ML) diluted 1:5,000 in PBS + 1% NFDM. Unbound secondary Ab was washed off with PBS + 1% NFDM, and the plate was developed as described above.
Competition ELISA with nucleosomes
Anti-dsDNA plates were prepared as described above. During the plate blocking, plasma and inhibitor solutions were prepared separately in PBS. Each plasma sample was prepared at a dilution that yielded an OD405nm of 0.5–0.6 so that we could assess both increases and decreases in the OD. Different concentrations of purified HeLa mononucleosomes (EpiCypher; 16–0002) and purified HeLa polynucleosomes (EpiCypher; 16–0003) were prepared separately and mixed with an equal volume of diluted plasma. This solution was resuspended several times to mix, and 50 µl was immediately added to a freshly blocked dsDNA-coated plate. The plate was then processed as described above.
Competition ELISA with DNases
Anti-DNASE1L3 plates were prepared as described above. During the plate blocking, plasma and inhibitor solutions were prepared separately in PBS. Each plasma sample was prepared at a dilution that yielded a DNASE1L3 ELISA OD405nm of 0.5–0.6 so that we could assess both increases and decreases in the OD. Different concentrations of recombinant human DNASE1 (Abcam; 73430) or purified recombinant DNASE1L3 were prepared separately and mixed with an equal volume of diluted plasma. This solution was resuspended several times to mix, and 50 µl was immediately added to a freshly blocked plate coated with 2 µg/ml DNASE1L3. The plate was then processed as described above.
Competition ELISA with DNA fragments of different length
Anti-dsDNA plates were prepared as described above. During the plate blocking, plasma and inhibitor solutions were prepared separately in PBS. Each plasma sample was prepared at a dilution that yielded an OD405nm of 0.5–0.6 so that we could assess both increases and decreases in the OD. DNA fragments of 95 bp, 186 bp, 382 bp, and 831 bp were prepared by PCR and purified using QIAquick Gel Extraction Kit (QIAGEN). Different concentrations of PCR-amplified DNA fragments were prepared separately and mixed with an equal volume of diluted plasma. This solution was resuspended several times to mix, and 50 µl was immediately added to a freshly blocked dsDNA-coated plate. The plate was then processed as described above.
Western blot
IgG binding against Ags on MPs
MP proteins from 2–5 × 107 Jurkat-derived MPs were separated on a 4–12% Bis-tris Western blot gel (Invitrogen; NP0326BOX) in reducing conditions and transferred to a polyvinylidene fluoride microporous membrane (Sigma-Aldrich; PVH00005). Afterwards, the membrane was blocked in 5% BSA for 60 min at room temperature and incubated overnight with plasma diluted in blocking buffer. In some instances, incubation with plasma samples was performed using a multiplex blotting system (LI-COR Biosciences). After washing with TBS with 0.1% Tween-20, membranes were incubated with HRP-conjugated anti-human IgG Ab (Jackson Immunoresearch; 1:5,000) or anti-mouse IgG Ab (eBioscience; 1:1,000) diluted in blocking buffer at room temperature for 60 min. The immunoreactive bands were visualized using an Amersham Imager 680 blot and gel imager.
IgG binding to DNases
To assess IgG binding to DNases by Western blot, 2 µg recombinant DNASE1L3 (purified in our laboratory) or 2–40 µg recombinant DNASE1 (Abcam; 73430) was run on a 12.5% separating gel in reducing conditions, and Western blot was performed as above.
IgG binding to HMGB1 on MPs
MP proteins from Jurkat-derived MPs were separated on a 4–12% Bis-tris Western blot gel. A polyconal anti-HMGB1 Ab (Invitrogen; PA5-96160) was used as primary Ab (1:1,000 dilution) and detected with an HRP-conjugated anti-rabbit IgG Ab (Thermo Fisher; G-21234; 1:2,000 dilution).
IgG binding to histones on MPs
MP proteins from Jurkat-derived MPs were separated on a 4–12% Bis-tris Western blot gel. A polyconal anti-H2A Ab (Cell Signaling Technology; 2578S) or a polyconal anti-H2B Ab (Cell Signaling Technology; 8135S) was used as primary Ab (1:1,000 dilution) and detected with an HRP-conjugated anti-rabbit IgG Ab (Thermo Fisher; G-21234; 1:2,000 dilution).
Flow cytometry
Analysis of DNASE1L3-senstive IgG binding to MPs
MPs from Jurkat cells were generated as previously described (Sisirak et al., 2016; Ullal et al., 2011). Briefly, Jurkat cells were cultured in the presence of 1 µM staurosporine (Sigma-Aldrich; S440) overnight, harvested, and collected by centrifugation for 5 min at 400×g. The supernatants were collected and centrifuged at 22,000 g for 30 min at 4°C to pellet MPs. Surface staining of MPs was performed as follows: 105 MPs were mock-treated or treated with recombinant human DNASE1 (Abcam; 73430) or recombinant DNASE1L3. Where specified, MPs were treated with DNASE1 and DNASE1L3-containing supernatants, as described previously (Sisirak et al., 2016), rather than recombinant proteins. DNase-treated MPs were then stained with human plasma at 1:20 dilution in a total volume of 30 µl at 4°C for 90 min. Stained MPs were washed, pelleted by centrifugation at 22,000 g for 30 min at 4°C, and incubated with a secondary FITC-conjugated anti-human IgG Ab (Millipore; AP113F; 1:1,000). Samples were diluted to stop staining and acquired on an Attune NxT (Thermo Fisher) flow cytometer. Representative histograms were generated using FlowJo software (Tree Star).
To determine whether plasma IgG bound to the surface of native MPs, the respective histograms were subtracted from a control histogram. The control histogram was generated by merging histograms derived from staining native MPs with plasma from healthy controls (n = 10). To determine whether IgG binding to native MPs occurred in a DNASE1L3-sensitive manner, the histogram derived from staining DNASE1L3-treated MPs was subtracted from the histogram derived from staining native MPs for each plasma sample using the Overton subtraction method (Overton, 1988) as implemented in FSC Express (De Novo Software).
Analysis of MP Ags
3–4 × 105 MPs were mock-treated or treated with recombinant human DNASE1 (Abcam; 73430) or recombinant DNASE1L3. DNase-treated MPs were then diluted in HBSS without magnesium, calcium, or phenol red (Thermo Fisher; 14–175-095). They were centrifuged at 22,000 g for 30 min at 4°C to pellet MPs and resuspended in HBSS (for Vybrant Green staining) or PBS for other staining.
Vybrant Green
MPs were stained in 400 µl HBSS + Vybrant Green (Invitrogen; V35004, 1:10,000) for 15 min at 37°C. The sample was then kept on ice in the dark and run on an Attune NxT (Thermo Fisher) flow cytometer within 60 min.
HMGB1 or histones
MPs were stained in 50 µl PBS for 105 min at 4°C with HMGB1 (Invitrogen; PA5-96160, 1:50), H2A monoclonal (Cell Signaling Technology; 12349S, 1:50), H2A polyclonal (Cell Signaling Technology; 2578S, 1:50), or H2B (Cell Signaling Technology; 8135S, 1:50). After staining, samples were diluted with PBS and centrifuged at 22,000 g for 30 min at 4°C to pellet MPs. They were then stained with AF647-conjugated anti-rabbit IgG (Invitrogen; A21245, 1:500) for 45 min at 4°C. Samples were then diluted to 400 µl with PBS and run on an Attune NxT (Thermo Fisher) flow cytometer. MP gating was calibrated based on 1-µm sizing beads (Invitrogen).
Stimulation of human pDCs
Approximately 80 ml of blood was drawn from healthy controls into EDTA-coated tubes (BD Biosciences; 366643). The blood was then split equally between three 50-ml conical tubes and diluted to 40 ml with sterile PBS. 10 ml Ficoll-Paque (GE Healthcare; 17–1440-03) was added to the bottom of the tube using a glass Pasteur pipette. The cells were spun at 400 g for 22 min with low acceleration and no breaks at room temperature. Approximately 20 ml supernatant was aspirated, and the remaining top two layers were transferred into new conical tubes. The cells were spun at 200 g for 15 min at room temperature and washed three times with sterile wash buffer (PBS, 2% FCS, and 1 mM EDTA). The cells were then counted using an Accuri C6 flow cytometer (BD Biosciences) and diluted to a density of 50,000 cells/µl. pDCs were enriched from total peripheral blood mononuclear cells using EasySep Human Plasmacytoid DC Enrichment Kit (Stemcell; 19062A). After enrichment, the cells were immediately resuspended in full RPMI and transferred into a 96-well plate. 5,000 pDCs were added to each well in 180 µl of full RPMI, and Ag (e.g., mononucleosomes and polynucleosomes) was added in a volume of 20 µl for a total reaction volume of 200 µl. The cells were activated for 24 h at 37°C and 5% CO2. Following activation, the plate was spun at 1,300 rpm for 10 min. The supernatant was used immediately or stored at −80°C.
Ag preparation
Purified HeLa mononucleosomes (EpiCypher; 16–0002) and purified HeLa polynucleosomes (EpiCypher; 16–0003) were incubated with LL-37 (InvivoGen; tlrl-l37) for 30–40 min at room temperature in a 20-µl volume and then immediately added to purified pDCs. The pDC/Ag mixture was then resuspended several times without vortexing. LL-37 was incubated with DNA substrates at a concentration of 500 µg/ml, and this mixture was added to pDCs for a final concentration of 50 µg/ml. Mononucleosomes and polynucleosomes were incubated with LL-37 at concentrations between 60 and 20 µg/ml DNA, and the final concentrations after adding this mixture to pDCs were between 6 and 2 µg/ml. Full RPMI contained 10% FCS, 100 U/ml penicillin and 100 µg/ml streptomycin (Gibco; 15140122), 1× MEM nonessential amino acids solution (Gibco; 11140050), 2 mM L-glutamine (Gibco; 25030081), 1 mM sodium pyruvate (Gibco; 11360070).
Confocal microscopy of MPs
For human MPs, blood was drawn into EDTA-coated tubes (BD Biosciences; 366643) and centrifuged at 1,000 g for 10 min. The supernatant was then centrifuged at 10,000 g for 10 min to isolate platelet-free plasma. Platelet-free plasma was centrifuged at 21,130 g for 60 min at 4°C to pellet MPs. Murine MPs were isolated as described in the Isolation of MPs section of Materials and methods. MP pellets were resuspended in HBSS without magnesium, calcium, or phenol red (Thermo Fisher; 14–175-095), and 1–2 million MPs were used for staining. Samples were stained with Vybrant DiD Cell-labeling Solution (Invitrogen; V22887) and Vybrant Green (Invitrogen; V35004) in the dark at 37°C for 20 min. They were diluted to 1.5 ml with HBSS to stop the staining. The MPs were filtered through a 70-µm filter and spun at 21,130 g for 60 min at 4°C. The pellet was washed again with HBSS, filtered through a 70-µm filter, and spun again 21,130 g for 60 min at 4°C. The pellet was resuspended in 1 µl HBSS and transferred to a 1.5 coverslip (Corning; 2850–22). The sample was kept in a hydration chamber for 40 min and inverted onto a Poly-Prep slide (Sigma-Aldrich; P0425-72EA) containing one drop of Diamond Antifade Mountant (Thermo Fisher; P36961). Slides were dried for 60 min at room temperature and stored at 4°C until imaged. The imaging of H2B-GFP/tdTomato double reporter mice was done the same way but without any staining steps. Images were acquired using a Zeiss 880 confocal microscope with a 63× oil objective.
Primer sequences
The primer sequences were intra-alu human: (forward), 5′-TCACGCCTGTAATCCCAGCA-3′, and (reverse) 5′-AGCTGGGACTACAGGCGCCC-3′; for intra-Alu human 76 bp: (forward) 5′-AGACCATCCTGGCTAACACG-3′, and (reverse) 5′-GTTCACGCCATTCTCCTGC-3′; intra-Alu human 200 bp: (forward) 5′-AAAATTAGCCGGGCGTG-3′, and (reverse) 5′-AGACGGAGTCTCGCTCTGTC-3′; intra-Alu human 831 bp: (forward) 5′-AATGGCCAGTTTCCTGAGGATTGTC-3′, and (reverse) 5′-CCCTGGGCAGCAATAGTTAATG-3′; intra-Alu human 382 bp: (forward) 5′-AAGAGTGAAGACCCGTGTGC-3′, and (reverse) 5′-CCCCGTCTTCAAAGAACATT-3′; intra-alu human 186 bp: (forward) 5′-GAACCAGCTGATTACCCTGTTATCCCTAC-3′, and (reverse) 5′-CCCTGGGCAGCAATAGTTAATG-3′; intra-Alu human 95 bp: (forward) 5′-AAGAGTGAAGACCCGTGTGC-3′, and (reverse) 5′-CCAAGTTCACACTAGAGTCAAAGG-3′; and inter-B1 mouse: (forward) 5′-CCAGGACACCAGGGCTACAGAG-3′, and (reverse) 5′-CCCGAGTGCTGGGATTAAAG-3′.
Statistical analysis
Statistical analyses were performed using Prism software version 8 (GraphPad). Normal distribution of data were not assumed unless otherwise specified. Statistical significance between two unpaired experimental groups was determined by nonparametric Mann–Whitney test unless otherwise specified. For paired groups, normality was assessed using the Shapiro–Wilk test. If the data were normally distributed, a parametric paired t test was used. If the data were not normally distributed, a nonparametric Wilcoxon matched-pairs signed rank test was used. The nonparametric Kruskal–Wallis test followed by the Dunn’s multiple-comparison test was used for statistical analysis of more than two groups. Contingency tables were analyzed using Fisher’s exact test or χ2 test as appropriate. Correlation between two continuous variables was explored by Spearman correlation. All P values were two tailed, and differences were considered significant for P values < 0.05 (*), < 0.01 (**), < 0.001 (***), and < 0.0001 (****).
Online supplemental material
Fig. S1 shows the correlation of DNASE1L3 activity with renal function in SLE patients. Fig. S2 presents further characterization of auto-Abs to DNASE1L3 in SLE patients. Fig. S3 shows the correlation between the MP fraction of plasma cfDNA and anti-dsDNA Ab levels in SLE patients. Fig. S4 shows the impact of length of protein-free DNA on its binding to anti-dsDNA IgG. Fig. S5 further characterizes auto-Ab reactivity to MP Ags. Table S1 describes clinical characteristics of sporadic SLE patients with or without DNASE1L3-sensitive binding. Table S2 summarizes clinical characteristics of SLE patients organized by treatment response. Table S3 describes characteristics for each individual patient included in the analysis from Table S2.
Supplementary Material
shows a comparison of clinical characteristics between sporadic SLE patients with DNASE1L3-sensitive IgG binding to MP Ags versus patients with no binding.
summarizes clinical characteristics of sporadic SLE patients with LN organized by treatment response.
shows characteristics for each individual patient included in the analysis from Table S2.
Acknowledgments
This work was supported by National Institutes of Health grants AR071703 (B. Reizis and J.P. Buyon), AR070591 (B. Reizis, G.J. Silverman, J.P. Buyon, and R.M. Clancy), CA232666 (O.A. Perez), AI100853 (O.A. Perez, L. Serpas, and A. Rashidfarrokhi), AR069515 (L. Serpas), and GM136573 (L. Serpas). This work was also supported by the Lupus Research Alliance (B. Reizis), the Colton Center for Autoimmunity (B. Reizis and J.P. Buyon), and the German Research Foundation (J. Hartl).
Author contributions: J. Hartl, L. Serpas, A. Rashidfarrokhi, Y. Wang, O.A. Perez, B. Sally, V. Sisirak, C. Soni, and G.J. Silverman performed experiments and analyzed and interpreted results. A. Khodadadi-Jamayran, A. Tsirigos, and M.Y. Kim analyzed results. A.S. Chida and I. Sanz developed and provided reagents. I. Caiello, C. Bracaglia, S. Volpi, G.M. Ghiggeri, H.M. Belmont, R.M. Clancy, P.M. Izmirly, and J.P. Buyon obtained and provided patient samples. J.P. Buyon and B. Reizis conceived and supervised the project. J. Hartl, L. Serpas, and B. Reizis wrote the original manuscript. All authors reviewed and edited the manuscript.
References
- Acosta-Herrera, M., Kerick M., González-Serna D., Wijmenga C., Franke A., Gregersen P.K., Padyukov L., Worthington J., Vyse T.J., Alarcón-Riquelme M.E., et al. Scleroderma Genetics Consortium . 2019. Genome-wide meta-analysis reveals shared new loci in systemic seropositive rheumatic diseases. Ann. Rheum. Dis. 78:311–319. 10.1136/annrheumdis-2018-214127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Mayouf, S.M., Sunker A., Abdwani R., Abrawi S.A., Almurshedi F., Alhashmi N., Al Sonbul A., Sewairi W., Qari A., Abdallah E., et al. 2011. Loss-of-function variant in DNASE1L3 causes a familial form of systemic lupus erythematosus. Nat. Genet. 43:1186–1188. 10.1038/ng.975 [DOI] [PubMed] [Google Scholar]
- Ali, R., Dersimonian H., and Stollar B.D.. 1985. Binding of monoclonal anti-native DNA autoantibodies to DNA of varying size and conformation. Mol. Immunol. 22:1415–1422. 10.1016/0161-5890(85)90065-3 [DOI] [PubMed] [Google Scholar]
- Aringer, M., Costenbader K., Daikh D., Brinks R., Mosca M., Ramsey-Goldman R., Smolen J.S., Wofsy D., Boumpas D.T., Kamen D.L., et al. 2019. 2019 European League Against Rheumatism/American College of Rheumatology Classification Criteria for Systemic Lupus Erythematosus. Arthritis Rheumatol. 71:1400–1412. 10.1002/art.40930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Atamaniuk, J., Hsiao Y.Y., Mustak M., Bernhard D., Erlacher L., Fodinger M., Tiran B., and Stuhlmeier K.M.. 2011. Analysing cell-free plasma DNA and SLE disease activity. Eur. J. Clin. Invest. 41:579–583. 10.1111/j.1365-2362.2010.02435.x [DOI] [PubMed] [Google Scholar]
- Banchereau, R., Hong S., Cantarel B., Baldwin N., Baisch J., Edens M., Cepika A.M., Acs P., Turner J., Anguiano E., et al. 2016. Personalized Immunomonitoring Uncovers Molecular Networks that Stratify Lupus Patients. Cell. 165:1548–1550. 10.1016/j.cell.2016.05.057 [DOI] [PubMed] [Google Scholar]
- Batu, E.D., Koşukcu C., Taşkıran E., Sahin S., Akman S., Sözeri B., Ünsal E., Bilginer Y., Kasapcopur O., Alikaşifoğlu M., and Ozen S.. 2018. Whole Exome Sequencing in Early-onset Systemic Lupus Erythematosus. J. Rheumatol. 45:1671–1679. 10.3899/jrheum.171358 [DOI] [PubMed] [Google Scholar]
- Belot, A. 2020. Contribution of rare and predicted pathogenic gene variants to childhood-onset lupus: a large, genetic panel analysis of British and French cohorts. Lancet Rheumatol. 2:e99–e109. 10.1016/S2665-9913(19)30142-0 [DOI] [PubMed] [Google Scholar]
- Beyer, C., and Pisetsky D.S.. 2010. The role of microparticles in the pathogenesis of rheumatic diseases. Nat. Rev. Rheumatol. 6:21–29. 10.1038/nrrheum.2009.229 [DOI] [PubMed] [Google Scholar]
- Bruschi, M., Bonanni A., Petretto A., Vaglio A., Pratesi F., Santucci L., Migliorini P., Bertelli R., Galetti M., Belletti S., et al. 2020. Neutrophil Extracellular Traps Profiles in Patients with Incident Systemic Lupus Erythematosus and Lupus Nephritis. J. Rheumatol. 47:377–386. 10.3899/jrheum.181232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buyon, J.P., Hiebert R., Copel J., Craft J., Friedman D., Katholi M., Lee L.A., Provost T.T., Reichlin M., Rider L., et al. 1998. Autoimmune-associated congenital heart block: demographics, mortality, morbidity and recurrence rates obtained from a national neonatal lupus registry. J. Am. Coll. Cardiol. 31:1658–1666. 10.1016/S0735-1097(98)00161-2 [DOI] [PubMed] [Google Scholar]
- Caielli, S., Athale S., Domic B., Murat E., Chandra M., Banchereau R., Baisch J., Phelps K., Clayton S., Gong M., et al. 2016. Oxidized mitochondrial nucleoids released by neutrophils drive type I interferon production in human lupus. J. Exp. Med. 213:697–713. 10.1084/jem.20151876 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carbonella, A., Mancano G., Gremese E., Alkuraya F.S., Patel N., Gurrieri F., and Ferraccioli G.. 2017. An autosomal recessive DNASE1L3-related autoimmune disease with unusual clinical presentation mimicking systemic lupus erythematosus. Lupus. 26:768–772. 10.1177/0961203316676382 [DOI] [PubMed] [Google Scholar]
- Casciola-Rosen, L.A., Anhalt G., and Rosen A.. 1994. Autoantigens targeted in systemic lupus erythematosus are clustered in two populations of surface structures on apoptotic keratinocytes. J. Exp. Med. 179:1317–1330. 10.1084/jem.179.4.1317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, R.W., Jiang P., Peng X., Tam L.S., Liao G.J., Li E.K., Wong P.C., Sun H., Chan K.C., Chiu R.W., and Lo Y.M.. 2014. Plasma DNA aberrations in systemic lupus erythematosus revealed by genomic and methylomic sequencing. Proc. Natl. Acad. Sci. USA. 111:E5302–E5311. 10.1073/pnas.1421126111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan, R.W.Y., Serpas L., Ni M., Volpi S., Hiraki L.T., Tam L.S., Rashidfarrokhi A., Wong P.C.H., Tam L.H.P., Wang Y., et al. 2020. Plasma DNA Profile Associated with DNASE1L3 Gene Mutations: Clinical Observations, Relationships to Nuclease Substrate Preference, and In Vivo Correction. Am. J. Hum. Genet. 107:882–894. 10.1016/j.ajhg.2020.09.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalli, J., Montero-Melendez T., Norling L.V., Yin X., Hinds C., Haskard D., Mayr M., and Perretti M.. 2013. Heterogeneity in neutrophil microparticles reveals distinct proteome and functional properties. Mol. Cell. Proteomics. 12:2205–2219. 10.1074/mcp.M113.028589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dieker, J., Tel J., Pieterse E., Thielen A., Rother N., Bakker M., Fransen J., Dijkman H.B., Berden J.H., de Vries J.M., et al. 2016. Circulating Apoptotic Microparticles in Systemic Lupus Erythematosus Patients Drive the Activation of Dendritic Cell Subsets and Prime Neutrophils for NETosis. Arthritis Rheumatol. 68:462–472. 10.1002/art.39417 [DOI] [PubMed] [Google Scholar]
- Duvvuri, B., and Lood C.. 2019. Cell-Free DNA as a Biomarker in Autoimmune Rheumatic Diseases. Front. Immunol. 10:502. 10.3389/fimmu.2019.00502 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon, K.B. 2018. Review: Cell Death, Nucleic Acids, and Immunity: Inflammation Beyond the Grave. Arthritis Rheumatol. 70:805–816. 10.1002/art.40452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elkon, K.B., and Wiedeman A.. 2012. Type I IFN system in the development and manifestations of SLE. Curr. Opin. Rheumatol. 24:499–505. 10.1097/BOR.0b013e3283562c3e [DOI] [PubMed] [Google Scholar]
- Foudi, A., Hochedlinger K., Van Buren D., Schindler J.W., Jaenisch R., Carey V., and Hock H.. 2009. Analysis of histone 2B-GFP retention reveals slowly cycling hematopoietic stem cells. Nat. Biotechnol. 27:84–90. 10.1038/nbt.1517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Romo, G.S., Caielli S., Vega B., Connolly J., Allantaz F., Xu Z., Punaro M., Baisch J., Guiducci C., Coffman R.L., et al. 2011. Netting neutrophils are major inducers of type I IFN production in pediatric systemic lupus erythematosus. Sci. Transl. Med. 3:73ra20. 10.1126/scitranslmed.3001201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gladman, D.D., Ibañez D., and Urowitz M.B.. 2002. Systemic lupus erythematosus disease activity index 2000. J. Rheumatol. 29:288–291. [PubMed] [Google Scholar]
- Hakkim, A., Fürnrohr B.G., Amann K., Laube B., Abed U.A., Brinkmann V., Herrmann M., Voll R.E., and Zychlinsky A.. 2010. Impairment of neutrophil extracellular trap degradation is associated with lupus nephritis. Proc. Natl. Acad. Sci. USA. 107:9813–9818. 10.1073/pnas.0909927107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris, H.E., Andersson U., and Pisetsky D.S.. 2012. HMGB1: a multifunctional alarmin driving autoimmune and inflammatory disease. Nat. Rev. Rheumatol. 8:195–202. 10.1038/nrrheum.2011.222 [DOI] [PubMed] [Google Scholar]
- Hochberg, M.C. 1997. Updating the American College of Rheumatology revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 40:1725. 10.1002/art.1780400928 [DOI] [PubMed] [Google Scholar]
- Jara, L.J., Navarro C., Medina G., Vera-Lastra O., and Saavedra M.A.. 2009. Hypocomplementemic urticarial vasculitis syndrome. Curr. Rheumatol. Rep. 11:410–415. 10.1007/s11926-009-0060-y [DOI] [PubMed] [Google Scholar]
- Jiang, P., and Lo Y.M.D.. 2016. The Long and Short of Circulating Cell-Free DNA and the Ins and Outs of Molecular Diagnostics. Trends Genet. 32:360–371. 10.1016/j.tig.2016.03.009 [DOI] [PubMed] [Google Scholar]
- Lande, R., Ganguly D., Facchinetti V., Frasca L., Conrad C., Gregorio J., Meller S., Chamilos G., Sebasigari R., Riccieri V., et al. 2011. Neutrophils activate plasmacytoid dendritic cells by releasing self-DNA-peptide complexes in systemic lupus erythematosus. Sci. Transl. Med. 3:73ra19. 10.1126/scitranslmed.3001180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leffler, J., Bengtsson A.A., and Blom A.M.. 2014. The complement system in systemic lupus erythematosus: an update. Ann. Rheum. Dis. 73:1601–1606. 10.1136/annrheumdis-2014-205287 [DOI] [PubMed] [Google Scholar]
- Lood, C., Blanco L.P., Purmalek M.M., Carmona-Rivera C., De Ravin S.S., Smith C.K., Malech H.L., Ledbetter J.A., Elkon K.B., and Kaplan M.J.. 2016. Neutrophil extracellular traps enriched in oxidized mitochondrial DNA are interferogenic and contribute to lupus-like disease. Nat. Med. 22:146–153. 10.1038/nm.4027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lou, X., Hou Y., Liang D., Peng L., Chen H., Ma S., and Zhang L.. 2015. A novel Alu-based real-time PCR method for the quantitative detection of plasma circulating cell-free DNA: sensitivity and specificity for the diagnosis of myocardial infarction. Int. J. Mol. Med. 35:72–80. 10.3892/ijmm.2014.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez-Valle, F., Balada E., Ordi-Ros J., Bujan-Rivas S., Sellas-Fernandez A., and Vilardell-Tarres M.. 2009. DNase 1 activity in patients with systemic lupus erythematosus: relationship with epidemiological, clinical, immunological and therapeutical features. Lupus. 18:418–423. 10.1177/0961203308098189 [DOI] [PubMed] [Google Scholar]
- Munroe, M.E., Lu R., Zhao Y.D., Fife D.A., Robertson J.M., Guthridge J.M., Niewold T.B., Tsokos G.C., Keith M.P., Harley J.B., and James J.A.. 2016. Altered type II interferon precedes autoantibody accrual and elevated type I interferon activity prior to systemic lupus erythematosus classification. Ann. Rheum. Dis. 75:2014–2021. 10.1136/annrheumdis-2015-208140 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muzumdar, M.D., Tasic B., Miyamichi K., Li L., and Luo L.. 2007. A global double-fluorescent Cre reporter mouse. Genesis. 45:593–605. 10.1002/dvg.20335 [DOI] [PubMed] [Google Scholar]
- Nabbi, A., and Riabowol K.. 2015. Isolation of Nuclei. Cold Spring Harb. Protoc. 2015:731–734. 10.1101/pdb.top074583 [DOI] [PubMed] [Google Scholar]
- Napirei, M., Wulf S., Eulitz D., Mannherz H.G., and Kloeckl T.. 2005. Comparative characterization of rat deoxyribonuclease 1 (Dnase1) and murine deoxyribonuclease 1-like 3 (Dnase1l3). Biochem. J. 389:355–364. 10.1042/BJ20042124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Napirei, M., Ludwig S., Mezrhab J., Klöckl T., and Mannherz H.G.. 2009. Murine serum nucleases--contrasting effects of plasmin and heparin on the activities of DNase1 and DNase1-like 3 (DNase1l3). FEBS J. 276:1059–1073. 10.1111/j.1742-4658.2008.06849.x [DOI] [PubMed] [Google Scholar]
- Nielsen, C.T., Østergaard O., Johnsen C., Jacobsen S., and Heegaard N.H.. 2011. Distinct features of circulating microparticles and their relationship to clinical manifestations in systemic lupus erythematosus. Arthritis Rheum. 63:3067–3077. 10.1002/art.30499 [DOI] [PubMed] [Google Scholar]
- Nielsen, C.T., Østergaard O., Stener L., Iversen L.V., Truedsson L., Gullstrand B., Jacobsen S., and Heegaard N.H.. 2012. Increased IgG on cell-derived plasma microparticles in systemic lupus erythematosus is associated with autoantibodies and complement activation. Arthritis Rheum. 64:1227–1236. 10.1002/art.34381 [DOI] [PubMed] [Google Scholar]
- Orbai, A.M., Truedsson L., Sturfelt G., Nived O., Fang H., Alarcón G.S., Gordon C., Merrill J., Fortin P.R., Bruce I.N., et al. 2015. Anti-C1q antibodies in systemic lupus erythematosus. Lupus. 24:42–49. 10.1177/0961203314547791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Overton, W.R. 1988. Modified histogram subtraction technique for analysis of flow cytometry data. Cytometry. 9:619–626. 10.1002/cyto.990090617 [DOI] [PubMed] [Google Scholar]
- Ozçakar, Z.B., Foster J. II, Diaz-Horta O., Kasapcopur O., Fan Y.S., Yalçınkaya F., and Tekin M.. 2013. DNASE1L3 mutations in hypocomplementemic urticarial vasculitis syndrome. Arthritis Rheum. 65:2183–2189. 10.1002/art.38010 [DOI] [PubMed] [Google Scholar]
- Papalian, M., Lafer E., Wong R., and Stollar B.D.. 1980. Reaction of systemic lupus erythematosus antinative DNA antibodies with native DNA fragments from 20 to 1,200 base pairs. J. Clin. Invest. 65:469–477. 10.1172/JCI109690 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petri, M., Kim M.Y., Kalunian K.C., Grossman J., Hahn B.H., Sammaritano L.R., Lockshin M., Merrill J.T., Belmont H.M., Askanase A.D., et al. OC-SELENA Trial . 2005. Combined oral contraceptives in women with systemic lupus erythematosus. N. Engl. J. Med. 353:2550–2558. 10.1056/NEJMoa051135 [DOI] [PubMed] [Google Scholar]
- Petri, M., Orbai A.M., Alarcón G.S., Gordon C., Merrill J.T., Fortin P.R., Bruce I.N., Isenberg D., Wallace D.J., Nived O., et al. 2012. Derivation and validation of the Systemic Lupus International Collaborating Clinics classification criteria for systemic lupus erythematosus. Arthritis Rheum. 64:2677–2686. 10.1002/art.34473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisetsky, D.S. 2014a. The complex role of DNA, histones and HMGB1 in the pathogenesis of SLE. Autoimmunity. 47:487–493. 10.3109/08916934.2014.921811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisetsky, D.S. 2014b. The expression of HMGB1 on microparticles released during cell activation and cell death in vitro and in vivo. Mol. Med. 20:158–163. 10.2119/molmed.2014.00014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pisetsky, D.S. 2016. Anti-DNA antibodies--quintessential biomarkers of SLE. Nat. Rev. Rheumatol. 12:102–110. 10.1038/nrrheum.2015.151 [DOI] [PubMed] [Google Scholar]
- Pisetsky, D.S., and Reich C.F.. 1994. The influence of DNA size on the binding of anti-DNA antibodies in the solid and fluid phase. Clin. Immunol. Immunopathol. 72:350–356. 10.1006/clin.1994.1152 [DOI] [PubMed] [Google Scholar]
- Puccetti, A., Madaio M.P., Bellese G., and Migliorini P.. 1995. Anti-DNA antibodies bind to DNase I. J. Exp. Med. 181:1797–1804. 10.1084/jem.181.5.1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pugh-Bernard, A.E., Silverman G.J., Cappione A.J., Villano M.E., Ryan D.H., Insel R.A., and Sanz I.. 2001. Regulation of inherently autoreactive VH4-34 B cells in the maintenance of human B cell tolerance. J. Clin. Invest. 108:1061–1070. 10.1172/JCI200112462 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radic, M., Marion T., and Monestier M.. 2004. Nucleosomes are exposed at the cell surface in apoptosis. J. Immunol. 172:6692–6700. 10.4049/jimmunol.172.11.6692 [DOI] [PubMed] [Google Scholar]
- Reed, J.H., Clancy R.M., Lee K.H., Saxena A., Izmirly P.M., and Buyon J.P.. 2012. Umbilical cord blood levels of maternal antibodies reactive with p200 and full-length Ro 52 in the assessment of risk for cardiac manifestations of neonatal lupus. Arthritis Care Res. (Hoboken). 64:1373–1381. 10.1002/acr.21704 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich, C.F. III, and Pisetsky D.S.. 2009. The content of DNA and RNA in microparticles released by Jurkat and HL-60 cells undergoing in vitro apoptosis. Exp. Cell Res. 315:760–768. 10.1016/j.yexcr.2008.12.014 [DOI] [PubMed] [Google Scholar]
- Rekvig, O.P. 2015. The anti-DNA antibody: origin and impact, dogmas and controversies. Nat. Rev. Rheumatol. 11:530–540. 10.1038/nrrheum.2015.69 [DOI] [PubMed] [Google Scholar]
- Richardson, C., Chida A.S., Adlowitz D., Silver L., Fox E., Jenks S.A., Palmer E., Wang Y., Heimburg-Molinaro J., Li Q.Z., et al. 2013. Molecular basis of 9G4 B cell autoreactivity in human systemic lupus erythematosus. J. Immunol. 191:4926–4939. 10.4049/jimmunol.1202263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rivera, T.L., Izmirly P.M., Birnbaum B.K., Byrne P., Brauth J.B., Katholi M., Kim M.Y., Fischer J., Clancy R.M., and Buyon J.P.. 2009. Disease progression in mothers of children enrolled in the Research Registry for Neonatal Lupus. Ann. Rheum. Dis. 68:828–835. 10.1136/ard.2008.088054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallai, K., Nagy E., Derfalvy B., Müzes G., and Gergely P.. 2005. Antinucleosome antibodies and decreased deoxyribonuclease activity in sera of patients with systemic lupus erythematosus. Clin. Diagn. Lab. Immunol. 12:56–59. 10.1128/CDLI.12.1.56-59.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Serpas, L., Chan R.W.Y., Jiang P., Ni M., Sun K., Rashidfarrokhi A., Soni C., Sisirak V., Lee W.S., Cheng S.H., et al. 2019. Dnase1l3 deletion causes aberrations in length and end-motif frequencies in plasma DNA. Proc. Natl. Acad. Sci. USA. 116:641–649. 10.1073/pnas.1815031116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiboski, C.H., Shiboski S.C., Seror R., Criswell L.A., Labetoulle M., Lietman T.M., Rasmussen A., Scofield H., Vitali C., Bowman S.J., and Mariette X.. International Sjögren’s Syndrome Criteria Working Group . 2017. 2016 American College of Rheumatology/European League Against Rheumatism Classification Criteria for Primary Sjögren’s Syndrome: A Consensus and Data-Driven Methodology Involving Three International Patient Cohorts. Arthritis Rheumatol. 69:35–45. 10.1002/art.39859 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sidstedt, M., Hedman J., Romsos E.L., Waitara L., Wadsö L., Steffen C.R., Vallone P.M., and Rådström P.. 2018. Inhibition mechanisms of hemoglobin, immunoglobulin G, and whole blood in digital and real-time PCR. Anal. Bioanal. Chem. 410:2569–2583. 10.1007/s00216-018-0931-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sisirak, V., Sally B., D’Agati V., Martinez-Ortiz W., Özçakar Z.B., David J., Rashidfarrokhi A., Yeste A., Panea C., Chida A.S., et al. 2016. Digestion of Chromatin in Apoptotic Cell Microparticles Prevents Autoimmunity. Cell. 166:88–101. 10.1016/j.cell.2016.05.034 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni, C., and Reizis B.. 2018. DNA as a self-antigen: nature and regulation. Curr. Opin. Immunol. 55:31–37. 10.1016/j.coi.2018.09.009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soni, C., Perez O.A., Voss W.N., Pucella J.N., Serpas L., Mehl J., Ching K.L., Goike J., Georgiou G., Ippolito G.C., et al. 2020. Plasmacytoid Dendritic Cells and Type I Interferon Promote Extrafollicular B Cell Responses to Extracellular Self-DNA. Immunity. 52:1022–1038.e7. 10.1016/j.immuni.2020.04.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, E.M., Schur P.H., Carr R.I., and Kunkel H.G.. 1966. Deoxybonucleic acid (DNA) and antibodies to DNA in the serum of patients with systemic lupus erythematosus. J. Clin. Invest. 45:1732–1740. 10.1172/JCI105479 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan, E.M., Cohen A.S., Fries J.F., Masi A.T., McShane D.J., Rothfield N.F., Schaller J.G., Talal N., and Winchester R.J.. 1982. The 1982 revised criteria for the classification of systemic lupus erythematosus. Arthritis Rheum. 25:1271–1277. 10.1002/art.1780251101 [DOI] [PubMed] [Google Scholar]
- Tipton, C.M., Fucile C.F., Darce J., Chida A., Ichikawa T., Gregoretti I., Schieferl S., Hom J., Jenks S., Feldman R.J., et al. 2015. Diversity, cellular origin and autoreactivity of antibody-secreting cell population expansions in acute systemic lupus erythematosus. Nat. Immunol. 16:755–765. 10.1038/ni.3175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ueki, M., Takeshita H., Fujihara J., Iida R., Yuasa I., Kato H., Panduro A., Nakajima T., Kominato Y., and Yasuda T.. 2009. Caucasian-specific allele in non-synonymous single nucleotide polymorphisms of the gene encoding deoxyribonuclease I-like 3, potentially relevant to autoimmunity, produces an inactive enzyme. Clin. Chim. Acta. 407:20–24. 10.1016/j.cca.2009.06.022 [DOI] [PubMed] [Google Scholar]
- Ullal, A.J., Reich C.F. III, Clowse M., Criscione-Schreiber L.G., Tochacek M., Monestier M., and Pisetsky D.S.. 2011. Microparticles as antigenic targets of antibodies to DNA and nucleosomes in systemic lupus erythematosus. J. Autoimmun. 36:173–180. 10.1016/j.jaut.2011.02.001 [DOI] [PubMed] [Google Scholar]
- Urbonaviciute, V., Fürnrohr B.G., Weber C., Haslbeck M., Wilhelm S., Herrmann M., and Voll R.E.. 2007. Factors masking HMGB1 in human serum and plasma. J. Leukoc. Biol. 81:67–74. 10.1189/jlb.0306196 [DOI] [PubMed] [Google Scholar]
- Urbonaviciute, V., Fürnrohr B.G., Meister S., Munoz L., Heyder P., De Marchis F., Bianchi M.E., Kirschning C., Wagner H., Manfredi A.A., et al. 2008. Induction of inflammatory and immune responses by HMGB1-nucleosome complexes: implications for the pathogenesis of SLE. J. Exp. Med. 205:3007–3018. 10.1084/jem.20081165 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitali, C., Bombardieri S., Jonsson R., Moutsopoulos H.M., Alexander E.L., Carsons S.E., Daniels T.E., Fox P.C., Fox R.I., Kassan S.S., et al. European Study Group on Classification Criteria for Sjögren’s Syndrome . 2002. Classification criteria for Sjögren’s syndrome: a revised version of the European criteria proposed by the American-European Consensus Group. Ann. Rheum. Dis. 61:554–558. 10.1136/ard.61.6.554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker, J.A., Kilroy G.E., Xing J., Shewale J., Sinha S.K., and Batzer M.A.. 2003. Human DNA quantitation using Alu element-based polymerase chain reaction. Anal. Biochem. 315:122–128. 10.1016/S0003-2697(03)00081-2 [DOI] [PubMed] [Google Scholar]
- Weisenburger, T., von Neubeck B., Schneider A., Ebert N., Schreyer D., Acs A., and Winkler T.H.. 2018. Epistatic Interactions Between Mutations of Deoxyribonuclease 1-Like 3 and the Inhibitory Fc Gamma Receptor IIB Result in Very Early and Massive Autoantibodies Against Double-Stranded DNA. Front. Immunol. 9:1551. 10.3389/fimmu.2018.01551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilber, A., Lu M., and Schneider M.C.. 2002. Deoxyribonuclease I-like III is an inducible macrophage barrier to liposomal transfection. Mol. Ther. 6:35–42. 10.1006/mthe.2002.0625 [DOI] [PubMed] [Google Scholar]
- Wirestam, L., Schierbeck H., Skogh T., Gunnarsson I., Ottosson L., Erlandsson-Harris H., Wetterö J., and Sjöwall C.. 2015. Antibodies against High Mobility Group Box protein-1 (HMGB1) versus other anti-nuclear antibody fine-specificities and disease activity in systemic lupus erythematosus. Arthritis Res. Ther. 17:338. 10.1186/s13075-015-0856-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, Y., Song Y., Chang J., Zhou X., Qi Q., Tian X., Li M., Zeng X., Xu M., Zhang W., et al. 2018. High levels of circulating cell-free DNA are a biomarker of active SLE. Eur. J. Clin. Invest. 48:e13015. 10.1111/eci.13015 [DOI] [PubMed] [Google Scholar]
- Yeh, T.M., Chang H.C., Liang C.C., Wu J.J., and Liu M.F.. 2003. Deoxyribonuclease-inhibitory antibodies in systemic lupus erythematosus. J. Biomed. Sci. 10:544–551. 10.1007/BF02256116 [DOI] [PubMed] [Google Scholar]
- Yung, S., and Chan T.M.. 2015. Mechanisms of Kidney Injury in Lupus Nephritis - the Role of Anti-dsDNA Antibodies. Front. Immunol. 6:475. 10.3389/fimmu.2015.00475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, Z., Song L., Maurer K., Petri M.A., and Sullivan K.E.. 2010. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. 11:124–133. 10.1038/gene.2009.66 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
shows a comparison of clinical characteristics between sporadic SLE patients with DNASE1L3-sensitive IgG binding to MP Ags versus patients with no binding.
summarizes clinical characteristics of sporadic SLE patients with LN organized by treatment response.
shows characteristics for each individual patient included in the analysis from Table S2.