Abstract
The inflammatory bowel diseases (IBD), comprising Crohn’s disease (CD) and ulcerative colitis (UC), are chronic, progressive, inflammatory conditions of the gastrointestinal tract. Imbalance in the gut microbial community, or dysbiosis, and the subsequent immune response, represent the critical relationship between genetic susceptibility, microbes, and environment factors, that result in IBD. Gastrointestinal pathogens – a common cause of dysbiosis – have been implicated as an environmental trigger in new onset IBD, as well as flare of existing IBD. In this article, we systematically review clinical data regarding the association between specific gastrointestinal pathogens and IBD. Numerous bacteria, viruses, fungi, and parasites have been implicated in the pathogenesis of IBD, and exacerbations of existing disease. In this article, we will also specifically discuss the less recognized microbes that have an inverse association with IBD, including certain bacterial pathogens, such as Helicobacter pylori, and parasites, such as Trichuris species. Future prospective and experimental studies are required to establish causality and clarify potential mechanisms of enteric pathogens in modifying the risk and course of IBD.
Keywords: Enteric infection, gastroenteritis, inflammatory bowel disease, pathogenesis, microbiome, flare, mucosal immunology
Background
The dynamic relationship between the gut microbiome on the host immune system is vital to maintaining physiologic homeostasis and human health. The host immune system establishes and modulates immunotolerance to the host microbial community, with the gut microbiome integrating environmental stimuli with genetic and immune inputs to influence metabolism, immunity, and response to pathogens.1,2 Several studies have identified and characterized pathways by which the gut microbiota impacts mucosal immunity. Indeed, dysregulation of this complex interaction is implicated across a wide spectrum of disease pathology, particularly immune-mediated diseases, and, not unexpectedly, particularly those involving the gastrointestinal tract.3,4
The inflammatory bowel diseases (IBD), comprising Crohn’s disease (CD) and ulcerative colitis (UC), are chronic, progressive, inflammatory conditions of the gastrointestinal tract affecting at least 0.4% of Europeans and North Americans, with a rising prevalence worldwide.5 IBD develops from a combination of genetic susceptibility and environmental factors that elicit a deleterious inflammatory response, with a disease course characterized by remitting and relapsing episodes of inflammation manifested as disease flares.5 Gastrointestinal microbial dysbiosis, defined as imbalance in the gut microbial community due to gain or loss of community members or changes in relative abundance of microbes, and the subsequent immune response represents the critical relationship between environment and genetics that results in the wide phenotypic range of IBD.1–4,6–9 Despite our increasing knowledge with respect to the relationship between the gut microbiome, dysbiosis, and the impact on mucosal immunity, much remains unknown regarding inciting environmental factors and underlying mechanisms that result in chronic, pathologic intestinal inflammation.
Enteric infections, leading culprits in gut microbial dysbiosis, are notoriously implicated as environmental triggers in new onset IBD or flare of existing IBD.8,10–14 However, enteric infections are not all the same. Indeed, some enteric infections are of no consequence, and perhaps even less recognized is the potential benefit of some gastrointestinal microbes, such as Helicobacter pylori and intestinal helminths, which have been associated inversely with risk of IBD. The ever-increasing health, societal, and economic burden of IBD worldwide highlights the urgent need to identify modifiable risk and protective factors implicated in IBD pathogenesis. We recently reviewed translational data implicating gastrointestinal pathogens in incident IBD.15 Thus, our primary objective was to conduct a comprehensive, clinical systematic review to define the association between specific gastrointestinal infections and (1) new onset IBD and (2) disease relapse, as well as provide an appraisal of specific studies to help with interpretation of current as well as future literature.
Methods
Search strategy
We searched electronic medical databases: PUBMED, Ovid, Scopus, ScienceDirect, CINAHL, LILACS, IMBIOMED, Scielo, IngentaConnect, Nature Publishing Group, and Cochrane database, from January 1990 to January 2020 for all English human and non-human studies assessing IBD and gastrointestinal infections. MeSH terms and/or text words included the broad terms inflammatory bowel disease, gastrointestinal infection, enteric infection. We also searched specific pathogens including bacteria (Campylobacter species, Helicobacter species, Mycobacterium avium paratuberculosis, Clostridioides difficile, Listeria monocytogenes, Yersinia enterocolitica, Salmonella species, Escherichia coli, Vibrio species, Plesiomonas shigelloides), viruses (Adenovirus, Astrovirus, Norovirus, Rotavirus, Sapovirus, cytomegalovirus, Epstein–Barr virus, human herpes virus 6), parasites (Trichuris species, Cryptosporidium species, Cyclospora cayetanensis, Entamoeba histolytica, Giardia lamblia), and fungi (Candida species, Aspergillus species, Cryptococcus neoformans), combined with “AND inflammatory bowel disease OR Crohn disease OR colitis, ulcerative OR indeterminate colitis.” We additionally reviewed the references of pertinent review articles and the studies meeting inclusion criteria for additional relevant articles.
Study selection
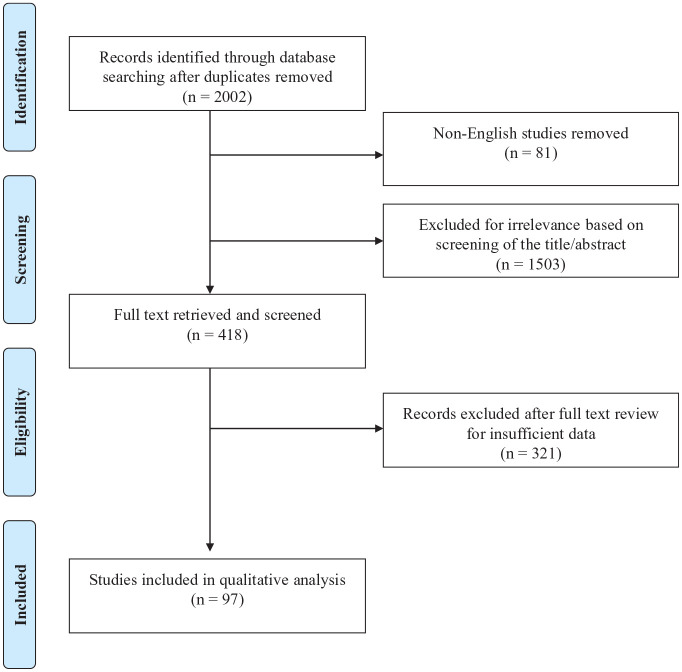
Experimental data, including specific gut microbiome and molecular changes associated with enteric infections in the absence of a diagnosis of new onset IBD or clinical disease flare is outside of the scope of this review. Clinical trials, cohort studies, and cross-sectional studies were eligible for inclusion. Case-series, review articles, and conference abstracts were excluded. Studies were included if they met the following inclusion criteria: (1) patients diagnosed with IBD [CD, UC, or IBD-undifferentiated (IBD-U)] according to standard diagnostic criteria; (2) diagnosis of gastrointestinal infection as defined by positive testing on any of the following modalities: histology, culture, serology, polymerase chain reaction (PCR) and/or other molecular technique as long as defined in the study; (3) documentation of criteria for disease flare qualification (e.g., clinical criteria, endoscopic criteria); (4) sufficient information provided to interpret or calculate comparative effect estimates; and (5) full-text available in English. We additionally documented the time course of gastrointestinal infection relative to incident IBD or disease flare, where available, such as in cohort studies. Of the 2002 unique studies identified, 418 studies were reviewed in full text, of which 97 met inclusion criteria for clinical studies of pathogens and IBD (Figure 1, Supplemental Table S1).
Figure 1.
Flowchart of study selection.
Gastrointestinal pathogens and increased risk of IBD
Bacteria
Many observational, cross-sectional or retrospective case-control and cohort studies link specific bacterial pathogens as potential triggers for new onset clinical IBD. Although no singular, causative microorganism or associated time course has been identified, several association studies implicate Campylobacter species, Salmonella species, enterohepatic Helicobacter species (EHS), Mycobacterium avium paratuberculosis (MAP), Clostridioides difficile, and Listeria monocytogenes (Table 1).
Table 1.
Specific pathogens associated incident IBD.
| Increased risk | Decreased risk | |
|---|---|---|
| Bacteria | Salmonella species | Helicobacter pylori |
| Escherichia coli | ||
| Yersinia enterocolitica | ||
| Campylobacter species | ||
| Enterohepatic Helicobacter species | ||
| Mycobacterium avium paratuberculosis | ||
| Clostridioides difficile | ||
| Listeria monocytogenes | ||
| Proteus mirabilis | ||
| Klebsiella pneumoniae | ||
| Citrobacter species | ||
| Viruses | Norovirus | |
| Cytomegalovirus | ||
| Epstein–Barr virus | ||
| Human herpes virus 3 | ||
| Human herpes virus 6 | ||
| Human herpes virus 8 | ||
| Measles virus | ||
| Mumps virus | ||
| Rubella virus | ||
| Rotavirus | ||
| Adenovirus | ||
| Fungi | Candida species | |
| Aspergillus species | ||
| Cryptococcus neoformans | ||
| Parasites | Amoeba/Entamoeba histolytica | Trichuris suis |
| Toxoplasma gondii | Hymenolepis diminuta | |
| Schistosoma species | ||
| Nector americanus |
IBD, inflammatory bowel disease.
Campylobacter species
In a nationwide cohort study from Denmark of 49,420 stool cultures positive for Campylobacter jejuni with 94,264,447 patient-years of follow up, both UC and CD were more common following an episode of gastroenteritis where the stool culture was positive for Campylobacter [UC IRR (incidence rate ratio) 2.6, 95% confidence interval (CI) 2.3–3.0; CD IRR 2.2, 95% CI 1.8–2.7] compared with patients without gastroenteritis. However, the incidence of UC and CD was significantly higher after a negative compared with a positive stool culture (UC IRR 8.2, 95% CI 8.0–8.5; CD IRR 6.4, 95% CI 6.1–6.7), suggesting possible detection bias.16 Despite the large sample size and duration of follow up, there may be significant selection or indication bias. Antimicrobial therapies administered contemporaneously were also not considered by the study investigators.16,17 Based on a more recent nationwide case-control study from Sweden of 480,721 patients, a diagnosis of Campylobacter species was associated with higher odds of UC [adjusted odds ratio (aOR), 1.86; 95% CI 1.32–2.61] and CD (aOR, 95% CI 1.87; 1.13–3.11) after adjusting for several factors including age, sex, birth year, place of residence, previous gastrointestinal surgery, autoimmune disease, and family history of IBD.18
Studies often analyze subset species as a composite genus group. This is particularly relevant for Campylobacter species, many of which are recognized as human pathogens and implicated in IBD pathogenesis, namely, Campylobacter jejuni, C. coli, C. ureolyticus, C. showae, and C. concisus. In particular, C. concisus, which has a virulence-associated restriction-modification system, has diverse pathogenic functions, including the ability to adhere to and invade host cells and secrete toxins.19 In a study of pediatric patients, C. concisus antibodies and DNA were detected with significantly higher frequency in patients with CD compared with patients without IBD.20,21 Among adults, one study demonstrated that C. concisus DNA was detected with significantly higher frequency in colonic biopsies of patients with CD (53%) and UC (77%) compared with adults without CD or UC (18% and 36%, respectively). These data have been confirmed in several other small studies using culture and PCR of intestinal biopsies from patients with CD and UC.22–25 Given the substantial genetic and functional diversity among C. concisus strains, further research is required.
There are some inconsistencies, as genus-specific PCR analyses have not demonstrated a significant difference between the prevalence of Campylobacter species in intestinal biopsies of patients with IBD versus those without IBD.26 In addition, in a broad meta-analysis comprising nine studies including 519 patients with IBD and 1133 non-IBD controls, C. concisus [pooled odds ratio (pOR) 3.76, 95% CI 1.46–9.70] and C. showae (pOR 2.39, 95% CI 1.11–5.18) were associated with increased odds of IBD.27 There were suggestive positive trends for C. hominis (pOR 1.58, 95% CI 0.91–2.75) and C. ureolyticus (pOR 2.34, 95% CI 0.77–7.16), while C. jejuni, C. rectus, and C. gracilis were not associated with odds of IBD, CD or UC.27
Salmonella species
In the same nationwide registry-based cohort study from Denmark with over 94 million patient-years of follow up, there were a total of 41,628 stool cultures that were positive for Salmonella species. An episode of gastroenteritis with stool culture positive for Salmonella species was significantly associated with increased risk of new-onset UC (IRR 3.0, 95% CI 2.6–3.4) and CD (IRR 2.2, 95% CI 1.7–2.7) compared with patients without gastroenteritis.16 The more recent nationwide case-control study from Sweden of 480,721 patients also cited above similarly demonstrated a positive association between a diagnosis of Salmonella and likelihood of IBD, albeit with effect estimates of slightly lower magnitude of effect (aOR for UC 1.49, 95% CI 1.15–1.94; aOR for CD 1.82, 95% CI 1.26–2.62).18
Enterohepatic Helicobacter species
Enterohepatic Helicobacter species (EHS) including Helicobacter fennelliae (H. fennelliae), H. cinaedi, and non-H. pylori-like (non-HPL) strains have been implicated in the development of clinical IBD. In the largest association study of Helicobacteraceae DNA and IBD, which included intestinal biopsies from 77 patients with CD and from 102 patients without IBD, non-HPL EHS were detected significantly more often in the intestinal biopsies of patients with CD compared with non-IBD controls (23% versus 12%, p = 0.04).28 A meta-analysis of nine case-control studies (545 patients with IBD and 524 non-IBD controls) confirmed these findings and demonstrated a pooled 2.6-fold higher odds (pOR 2.62, 95% CI 1.48–4.63) of IBD among individuals exposed versus non-exposed to non-HPL EPS.27 Notably, in this same study, a secondary meta-analysis of 15 studies totaling 906 patients with IBD and 758 patients without IBD demonstrated a statistically non-significant positive association between EHS and likelihood of IBD (pOR 1.51, 95% CI 0.95–2.41), as well as no difference in the odds of IBD among HPL-EHS exposed versus non-exposed (pOR 1.14, 95% CI 0.54–2.42).27 However, publication bias, delayed recognition of non-HPL-EHS, and heterogeneity may confound interpretation of these studies. The diagnostic tools used to detect non-HPL-EHS are likewise relevant, given that there is molecular overlap with HPL-EHS and Campylobacter species.
Mycobacterium avium paratuberculosis
There are over 65 studies analyzing the association between Mycobacterium avium paratuberculosis (MAP) and IBD, especially CD. Despite being one of the most frequently studied pathogens in IBD, the clinical relevance of MAP in IBD pathogenesis remains controversial.29 Part of the controversy derives from inconsistent methods of MAP detection, specifically culture techniques, which have many limitations including poor sensitivity, requirement for specialized media, and a long incubation time. By contrast, PCR-based testing allows for a more standardized method of detection and has now supplanted culture techniques. PCR-based testing of IS900 − a multicopy insertion element of MAP – is considered the most rigorous diagnostic method for MAP detection. Although a relatively new technique, the handful of studies that utilize these more refined PCR techniques support an association between MAP and CD.
In a cross-sectional study of 100 patients with CD, 100 with UC, and 100 controls without IBD, PCR testing was positive for the IS900 MAP insertion element in 52% of biopsies from patients with CD compared with 2% of UC and 5% of controls (p < 0.001).30 In a similarly designed study of 22 patients with CD, 20 with UC, 21 with aphthous ulcers of the terminal ileum, and 42 non-IBD controls, the prevalence of MAP based on IS900 assays was significantly higher in patients with CD compared with controls without IBD (29% versus 0%, p = 0.02).31 However, the prevalence of MAP was not significantly different between patients with UC (18%) or ileal aphthous ulcers (12%), although insufficient power due to small sample size is a consideration.31
Conversely, several studies utilizing culture and alternative PCR technologies (i.e., non IS900 PCR) have not demonstrated a significant association between MAP and CD.32–35 Collectively, these conflicting conclusions suggest that MAP may play a role in the pathogenesis or disease course of CD in a subset of individuals. However, optimal techniques for MAP identification and the causal role of MAP in IBD pathogenesis remain to be clarified.
Clostridioides difficile
There are numerous studies analyzing the association between C. difficile infection (CDI) and IBD. Very few studies, however, have analyzed CDI as a risk factor for new onset IBD, as it is challenging to discriminate reverse causality or the innocent bystander effect of CDI in IBD. In the aforementioned nationwide case-control study of 480,721 patients from Sweden, the largest study analyzing the association between CDI and IBD, a diagnosis of C. difficile was associated with higher odds of UC (aOR 4.02, 95% CI 2.94–5.49) and CD (aOR 4.25, 95% CI 2.79–6.47).18 One exploratory study of patients with CDI-associated diarrhea compared IgG-specific humoral immune response with C. difficile toxins A and B between 10 patients with concomitant IBD and 19 patients without IBD.36 While levels of circulating IgG antibodies to C. difficile toxins A and B were not significantly different between groups, IgG antibodies to C. difficile toxins A and B were detected on culture of large and small intestinal mucosal samples only in subjects with IBD.36 This finding is significant, as none of the patients with IBD had a history of clinical CDI, suggesting previous asymptomatic exposure to toxigenic C. difficile.36 High prevalence of toxigenic C. difficile carriage in patients with IBD is supported by several investigations,37–39 although the clinical impact is not clear.
While the reduced diversity of bacterial species in IBD may predispose to C. difficile colonization, emerging data also implicate genetic factors. In one study of 319 patients with UC, genetic risk alleles explained a larger portion of the variance in CDI risk than clinical factors such as antibiotic exposure, suggesting that host immunity may play an important role in susceptibility to CDI.40 These data provide some support that there may be a true contributory role of C. difficile in IBD pathogenesis, at least among genetically predisposed individuals.
Listeria monocytogenes
Few studies have examined the role of L. monocytogenes in IBD, either as a non-specific antigenic stimulus or as a specific pathogen trigger.41 In one non-comparative study of patients with CD, there was a positive antibody response to L. monocytogenes in 75% of intestinal and mesenteric lymph node specimens, with immunolabeled macrophages and giant cells distributed underneath ulcers, along fissures, around abscesses, within the lamina propria, in granulomas, and in the germinal centers of mesenteric lymph nodes.42 However, in a subsequent study of 274 colonoscopic biopsies, L. monocytogenes DNA was similarly detected in intestinal biopsies of both patients with IBD and in non-IBD controls. This underscores the widespread presence of this organism in the environment and brings into question the direct etiopathogenic role of L. monocytogenes in IBD pathogenesis.43
Yersinia species
In the aforementioned nationwide case-control study of 480,721 patients from Sweden, a diagnosis of Yersinia enterocolitica was associated with higher odds of CD (aOR 9.59, 95% CI 3.04–30.3), but not UC (aOR 2.61, 95% CI 0.87–7.83).18 In a case series of 54 intestinal resection specimens from 52 patients with CD, 40 normal bowel specimens, 30 cases of acute appendicitis, and 50 cases of other non-IBD active colitides, Yersinia DNA, including Y. pestis, Y. paratuberculosis, and Y. enterocolitica, was identified in 31% of CD specimens, but in 0% of the specimens obtained from the other groups.44 In a similarly designed study that tested colonic specimens from 77 children with Crohn’s colitis, 45 children with UC, and 10 children who underwent appendectomies, Yersinia species were detected significantly more often in specimens from children with CD compared with all other groups (9% versus 0%; p = 0.006).45 However, a recent cross-sectional study of 470 ileal samples from 262 CD patients and 76 non-IBD controls demonstrated discrepant findings, as Yersinia species were detected in 10% of CD patients and 12% of controls (p > 0.05).46 While none of these studies established causality, there is suggestion of shared genetic predisposition since loss-of-function mutations in Nucleotide Oligomerisation Domain 2 (NOD2) – a key gene of innate immunity – are associated both with CD as well as resistance to Yersinia. Some have hypothesized that some NOD2 mutations may have been selected for through evolution, specifically during past plague outbreaks.47
Viruses
Evidence for viruses as risk determinants of IBD among susceptible people is limited. The viruses most studied are those for which testing modalities are readily available. To this end, most studies to date have investigated species within the herpesvirus family, including cytomegalovirus (CMV), EBV, and human herpes virus 6 (HHV-6).
In one study of 79 IBD patients (47 UC and 32 CD) and 15 non-IBD controls, CMV and HHV-6 immunohistochemistry intensity and coexistence correlated with endoscopic disease severity, endoscopic activity, and number of immunosuppressive therapies.48 In another study of 84 patients with IBD and 115 non-IBD controls, EBV DNA load by immunohistochemistry in peripheral blood mononuclear cells was higher in patients with IBD, who consequently also showed high levels of lytic and latent EBV gene expression localized to proliferating B-lymphocytes; this was particularly pronounced among patients not responding to IBD therapy (p < 0.05).49 However, because there is nearly universal exposure to EBV by adulthood, strong conclusions from association studies of EBV and IBD, especially among adults, are limited at best. For example, one cross-sectional study of EBV seroprevalence in 263 IBD patients demonstrated that EBV seronegativity was similar to the general, with seropositivity approaching 100% after age 25 years, irrespective of IBD status.50 It is generally unclear why EBV-associated diseases such as mononucleosis and lymphoproliferative disorders occur in some individuals and not others. While not yet investigated, one possibility is that the timing of EBV exposure is relevant, such that EBV exposure in the early life years during the critical phases of immune development modifies IBD risk among genetically susceptible individuals.
With respect to HHVs more broadly, one study analyzed 41 patients with IBD, and reported that EBV, CMV, but not of herpes simplex virus (HSV)-1, HSV-2, Varicella zoster virus (VZV), HHV-6, HHV-7, or HHV-8, were more commonly identified in the colonic mucosa of individuals with versus without IBD (EBV: 54% versus 23%, p = 0.003; CMV: 24% versus 8%, p = 0.0001).51
Norovirus is one non-HHV that has been investigated as a risk determinant in IBD pathogenesis. However, data are limited and there is the consideration of selection bias as well given that norovirus is tested for in the setting of a clinical presentation of diarrhea. That being said, in the nationwide case-control study from Sweden already cited, a diagnosis of norovirus remained independently associated with higher odds of CD (aOR 3.19, 1.28–7.96), but not UC (aOR 1.74, 95% CI 0.85–3.57), after adjusting for several factors including sex, age, birth year, place of residence, previous gastrointestinal surgery, autoimmune disease, and family history of IBD.18
Based on these data, it remains unclear whether norovirus, CMV, EBV, and HHV-6 are involved in the pathogenesis of disease, associated with disease flares, complications, and response to therapy, or are simply innocent bystanders of active disease (or immunosuppression) and instead more reflect the baseline high prevalence in the general population.52 Well-designed prospective studies with adequate control populations are needed and will also help to define the impact of timing of exposure and other modifying factors.
Fungi
Technologic advancements have enabled deeper characterization of previously overlooked members of the gut microbiome that might also play a critical role in IBD pathogenesis and its disease course. Fungi fall into this category, but the same issues of causality and innocent bystander remain. Recent studies using advanced sequencing approaches demonstrate that fungal diversity is altered in ileal and colonic biopsies from patients with CD compared with healthy controls.53 Specifically, multiple studies have demonstrated varying proportions of gastrointestinal pathogenic Candida species, Malassezia species, Aspergillus species, and Cryptococcus neoformans.54–61 While studies on UC have been somewhat inconsistent, altered fungal diversity has still been reported.56,62,63 While the complex interplay and directionality between the gut mycobiome, fungal pathogens, and mucosal inflammation remain poorly understood, these findings do suggest specific fungi might be relevant in disease pathogenesis. Further studies are required, including studies of genetic and other susceptibility determinants.
Gastrointestinal pathogens and attenuated risk of IBD
Many population-based, cross-sectional, and retrospective studies have demonstrated a protective association between H. pylori or intestinal helminth colonization with IBD.
Bacteria
Helicobacter pylori
H. pylori is still the most common chronic bacterial infection worldwide, with over half the global population estimated to be colonized or infected64; however, the prevalence of H. pylori has been decreasing over the past several decades, due largely to industrialization, less crowding, and improved living conditions, as well as targeted eradication therapy. Decreases in H. pylori prevalence on a population level correspond to an increase in immune-mediated diseases, particularly CD, and is supported by many epidemiological studies. At least in endemic areas, H. pylori exposure most often occurs early in life, and, depending on host susceptibility factors and host-microbial interactions, might qualify as a “critical early exposure” given the downstream consequences of immunomodulatory effects and alterations of the gastric, oral, and colonic microbiome associated with H. pylori colonization/infection.65–67
To date, five meta-analyses of the association between H. pylori exposure and IBD have been published.27,68–71 The most recent and most comprehensive meta-analysis by Castano-Rodriguez and colleagues reported that, among 40 case-control studies with 6130 IBD cases and 74,659 non-IBD controls, H. pylori exposure was associated with a 57% lower odds of IBD (pOR 0.43, 95% CI 0.36–0.50), including CD (pOR 0.38, 95% CI 0.31–0.47), UC (pOR 0.53, 95% CI 0.44–0.65), and IBD-U (pOR 0.43, 95% CI 0.23–0.80). Regardless of age and geography, the inverse association was maintained, and was stronger for pediatric (pOR 0.24, 95% CI 0.14–0.43) versus adult (pOR 0.45, 95% CI 0.38–0.53) populations and Eastern (pOR 0.35, 95% CI 0.26–0.48) versus Western (pOR 0.46, 95% CI 0.38–0.55) populations, albeit not statistically significantly. The other meta-analyses, one of which included only Asian studies,68 reported consistent findings, with all meta-analyses demonstrating a significant inverse association between H. pylori and IBD that was generally more pronounced for CD versus UC. H. pylori strain-specific constituents might mediate, at least in part, this protective association, plausibly through immunomodulation. One recent meta-analysis of 1748 people (688 with CD, 272 with UC) found that, compared with CagA-negative H. pylori exposure or H. pylori non-exposure overall, exposure to CagA-positive H. pylori was associated with a significantly lower odds of IBD (pOR 0.31, 95% CI 0.21–0.44) and CD (pOR 0.23, 95% CI 0.15–0.35), with a suggestive trend for UC (pOR 0.66, 95% CI 0.34–1.27).71 Interestingly, there was no significant difference in the odds of IBD overall, CD, or UC between H. pylori exposed, CagA seronegative, and H. pylori non-exposed individuals.
It is important to emphasize that not all Helicobacter species are inversely associated with IBD. As detailed in the sections above, the meta-analysis by Castano-Rodriguez and colleagues also analyzed the association between EHS and IBD, stratified by non-HPL-EHS and HPL-EHS.
Parasites
Trichuris species
Helminths play an important immunoregulatory role in mucosal immunity and the gut microbiome. Similar to the epidemiology of H. pylori exposure, intestinal helminthic infections are more common in regions with higher population densities, less industrialization, and poorer water sanitation, which tend to also be regions of lower IBD prevalence. A handful of studies have suggested that the absence of intestinal helminths is associated with an increased likelihood of new onset IBD.72–74 Of the limited literature, the inverse association between Trichuris species and IBD is perhaps most described.75 One small case-control study of 151 South African patients with IBD and 219 controls without IBD reported that, after adjusting for age and sex, Trichuris helminth exposure versus non-exposure was associated with lower odds of IBD (aOR 0.2, 95% CI 0.1–0.4).76 These data, however, were based on self-reported infection early in life in a region where helminth infections are endemic. Indeed, this study also highlights the difficulty of conducting rigorous epidemiological and clinical studies in certain regions related to insufficient resources and infrastructure for exposure assessment and accurate case confirmation.
Few studies have sought to define mechanisms underlying these observations. Notably, based on the limited but biologically plausible evidence for a protective effect of intestinal helminths in IBD, several studies have evaluated the efficacy (and safety) of the pig whipworm Trichuris suis for the treatment of IBD with mixed results.77–82 Many confounders, measured and unmeasured, among other study limitations limit strong conclusions, however, and more data are needed.
Gastrointestinal pathogens in relapse of IBD
Gastrointestinal infections are commonly implicated in relapse (flares) of already established IBD (Table 2).83–88 There is considerable overlap between the clinical presentation of enteric infection and IBD flare, suggesting that these conditions may be mutually exclusive or co-exist, further complicating interpretation and causal determination. Current clinical guidelines recommend testing for C. difficile in all patients with IBD who have worsening or new onset diarrhea and testing for CMV in patients with severe active IBD, particularly if there is concomitant steroid use and disease refractory to medical therapy. Otherwise, there is no consensus for testing for other gastrointestinal pathogens in the setting of an IBD flare. The clinical significance of a positive result for non-C. difficile enteric infection in patients with active IBD is debated due to lack of robust outcomes-based evidence.
Table 2.
Specific pathogens associated flare of prevalent IBD.
| CD | UC | |
|---|---|---|
| Bacteria | + Campylobacter species | + Campylobacter species |
| + Clostridioides difficile | + Plesiomonas shigelloides | |
| + Enteroaggregative Escherichia coli | ||
| + Enteropathogenic Escherichia coli | ||
| + Clostridioides difficile | ||
| Viruses | + Norovirus | − Norovirus |
| + Cytomegalovirus | ||
| Parasites | − Giardia lambia | − Giardia lambia |
| − Cryptosporidium | − Cryptosporidium | |
| − Cyclospora cayetanensis | − Cyclospora cayetanensis | |
| − Entamoeba histolytica | − Entamoeba histolytica |
, Increased cross-sectional prevalence during flare compared to symptomatic patients without IBD.
, Decreased cross-sectional prevalence during flare compared to symptomatic patients without IBD.
CD, Crohn’s disease; IBD, inflammatory bowel disease; UC, ulcerative colitis.
Clostridioides difficile
Multiple studies have demonstrated consistently that, among patients with IBD, those who have confirmed CDI have more pronounced dysbiosis and significantly worse clinical outcomes, such as longer hospital stays, higher colectomy rates, higher recurrence rates, and increased mortality, compared with those without CDI.83,89–100 Moreover, established risk factors for CDI such as nosocomial acquisition, age, and recent antibiotic use may not be significant risk factors for CDI in patients with IBD.94,101–103
One study of 461 consecutive patients hospitalized for IBD flares, detected toxigenic C. difficile and non-toxigenic C. difficile in 35 (8%) and 10 (2%) hospitalized patients, respectively.104 In a similarly designed study that instead used multiplex PCR, of 214 patients with IBD who underwent 295 multiplex stool PCR tests for an IBD flare, toxigenic C. difficile was the most common pathogen identified (n = 38, 13%) among 79 (27%) tests where a pathogen was detected.83 Notwithstanding, there is controversy regarding the diagnosis and clinical relevance of C. difficile in IBD disease course. Based on the cross-sectional studies referenced above, patients with IBD have a high prevalence of asymptomatic C. difficile colonization, which similarly precludes major conclusions regarding the contribution of CDI to IBD flares.
Non-Clostridioides difficile enteric infections
The introduction of multiple PCR testing has allowed for more widespread detection of other enteric pathogens which may or may not be players in IBD flares. In the study cited above that included 214 patients with IBD who underwent 295 multiplex stool PCR tests during a flare, after C. difficile, the most commonly detected microbes were E. coli subtypes (8%) and viruses (5%).83 Patients who tested negative for an enteric infection were more likely to have IBD medications added or up-titrated (49% versus 29%, p = 0.027), suggesting that the results of enteric testing impacted IBD management.83 Focusing on culture data, other studies have demonstrated worse outcomes, including higher rates of colectomy and mortality, in patients with IBD flare complicated by Campylobacter and Salmonella.105,106
More recently, a cross-sectional study analyzed 577 patients with IBD (277 CD, 300 UC) flare and 8826 unmatched control subjects without IBD who had multiplex stool PCR tests ordered during a diarrheal illness. Compared with patients without IBD, patients with IBD were significantly less likely to test positive overall (CD 18.1%, UC 16.1%, no IBD 26.6%, p < 0.001), but it is important to note that not all microbes tested for with the stool PCR test are necessarily pathogenic (e.g., parasites).107 As such, there were some notable differences based on the specific pathogen. Compared with patients without IBD, patients with CD tested positive more often for norovirus (24.6% versus 17.4%, p = 0.05) and Campylobacter (13.1% versus 7.6%, p = 0.04), but less often for parasites, including Giardia lambia, Cryptosporidium, Cyclospora cayetanensis, and Entamoeba histolytica (0.8% versus 6.1%, p = 0.01).107 Compared with patients without IBD, patients with UC tested positive more often for Campylobacter (13.9% versus 7.6%, p = 0.01), Plesiomonas (2.6% versus 0.7%, p = 0.049), and E.coli subtypes (64.3% versus 47.6%, p < 0.001), including Enteroaggregative E.coli (EAEC; 20.9% versus 13.5%, p = 0.03), and Enteropathogenic E.coli (EPEC; 33.9% versus 22.1%, p = 0.004), but less often for parasites (0.9% versus 6.1%, p = 0.01) and norovirus (7.8% versus 17.4%, p = 0.02).107 When comparing IBD subtypes, patients with UC less often tested positive for viruses compared with patients with CD (16.5% versus 32.8%, p = 0.004), specifically norovirus (7.8% versus 24.6%, p < 0.001).107
There are some inconsistencies in the literature, though. In a study of enterovirus, norovirus G-I, norovirus G-II, rotavirus, astrovirus, and sapovirus RNA in stool samples of 33 children with IBD and 17 without IBD, viral RNA was detected only in children without IBD (3% versus 0%), although the frequency was overall low in both groups.108 In addition to limited power related to the very small sample size, this study was also limited in that both asymptomatic and symptomatic patients were included, possibly even further diluting the ability to detect a true difference. In a retrospective study of 1345 patients with IBD flare who underwent stool PCR or culture for non-C. difficile bacterial pathogens, only 25 tests (2%) were positive. Moreover, a higher proportion of patients with non-C. difficile bacterial infections were in remission of their IBD within 1 year compared with patients with C. difficile infection or non-infectious flare.109 This study, however, used mixed diagnostic methods and did not evaluate for enteric viruses.
Separately, CMV has been detected in approximately 30% cases of steroid-refractory IBD flares.110–112 Similar to many studies of other enteric infections, the variable study designs, study populations, diagnostic methods, and testing/indication biases, to name a few, compromise the ability to conduct rigorous analyses and discern true pathogenic role. CMV reactivation is common in patients with moderate to severe colonic disease especially with concomitant steroid use, with a reported prevalence of 5–17%, and as high as 25–30% among patients requiring colectomy for severe colitis.113 The clinical relevance of CMV infection as opposed to just a bystander detected in the setting of severe disease and immunosuppression is not clear, and most often depends on the clinical scenario. In one study of 69 patients with moderate-to-severe UC who were tested for CMV reactivation every 2 weeks for 8 weeks using the CMV antigenemia assay and serum quantitative real-time PCR assay, clinical outcomes, including rates of remission and colectomy, were not significantly different among the CMV reactivation-positive versus negative patients.114 Other serologic studies have demonstrated similar findings, arguing against CMV as a major pathogen in IBD relapse.115,116 However, latent or subclinical CMV does not correlate with CMV colitis which requires colonic tissue for diagnosis and these negative studies employed variable non-tissue diagnostics methods.
As with all gastrointestinal infections complicating flares of IBD, in the absence of serial, longitudinal, prospective testing and an appropriate reference group (e.g., patients without IBD with diarrhea or other GI symptoms), it is difficult to conclude that specific enteric pathogens are causative factors in disease flares or whether they represent a surrogate marker of disease severity or susceptibility to pathogen acquisition. In addition, very little is known regarding the influence of specific enteric pathogens on IBD outcomes and disease progression. Despite these and other limitations, further investigation would be valuable given the potential role of enteric pathogens in IBD incidence, disease flares, and complications.
Discussion
In summary, specific gastrointestinal infections including bacteria, viruses, parasites, and fungi, may modify the risk of developing IBD and trigger or complicate flares in patients with established IBD. Critical study design limitations and biases preclude the ability to definitively establish a direct causal relationship between gastrointestinal infection or colonization and new onset or flare of IBD. As such, the possibility of reverse causality, IBD increasing the risk of infection or colonization acquisition, and that these species are merely “innocent bystanders” remains unsettled and complicates our understanding of IBD. Specifically, C. difficile carriage occurs more often in the setting of reduced diversity of intestinal microbial species that occurs following antibiotic exposure, with reduced diversity predisposing to C. difficile colonization with or without clinical infection. This observation further complicates our understanding of its role in IBD pathogenesis and contributes to the reverse causality consideration, since reduced diversity might predispose to IBD, and reduced diversity also occurs as a result of IBD. Antibiotic exposures are likely also to modify or confound these associations. Nonetheless, clinical data described above have demonstrated a lower overall pathogen detection rate in patients with an exacerbation of existing IBD compared with non-IBD controls, the reasons for which are unclear. These data suggest that while patients with IBD have an impaired intestinal barrier with limited or dysfunctional antibacterial activity, this deficit does not directly translate into a broadly increased risk of gastrointestinal pathogen acquisition or infection. Thus, the above clinical data provide a wealth of insight and hypotheses for our understanding of factors driving the preclinical and clinical phases of IBD.
In addition to the above clinical associations, experimental and translational data offer a window into whether these clinical associations have mechanistic plausibility in IBD pathogenesis. Our group recently reviewed translational data implicating certain gastrointestinal pathogens in incident IBD15; herein, we highlight relevant mechanisms to provide context for the clinical evidence detailed above, but we encourage interested readers to refer to that more comprehensive review. In genetically susceptible individuals, specific pathogens and the associated downstream consequences might cause gut dysbiosis, exacerbate existing dysbiosis, induce immunological scar and deleteriously alter immune responses, or might activate immune responses via direct damage the intestinal mucosa; each of these mechanisms, alone and in concert, might trigger the clinical presentation of new IBD and flare of existing disease.15 Moreover, virulence mechanisms involved in microbial fitness can directly or indirectly damage the epithelial barrier, triggering the polarization of lymphocytes and myeloid cells towards an inflammatory state of activation in an attempt by the body to remove the offending pathogen.
A defective intestinal barrier is a major feature of IBD and infectious disease. A recent study demonstrated intestinal permeability precedes a diagnosis of CD, raising the possibility that a pathogen-induced disruption of the barrier is a contributing factor.117 Experimental systems, including those that seek to explain the link between certain genes and IBD risk, offer potential mechanisms. Abnormal microbial-sensing by pattern-recognition receptors, a compromised mucin barrier, impairment in autophagy, diminished functional antimicrobial activity, direct epithelial cell damage, and changes in tight junction permeability, all contribute to loss of intestinal barrier integrity.118 In the setting of reduced protective host-defense mechanisms and dysbiosis, there is greater commensal and pathogenic microbial contact with the intestinal epithelium, which is associated with aberrant mucosal immune responses that represent the hallmark of IBD. Conversely, pathogens such as Helicobacter and certain helminths, and their downstream products, might reduce dysbiosis and/or counteract inflammatory pathways, preventing IBD onset or flare of existing disease. The balance between pro- and anti-inflammatory microbes, and whether the individual is genetically predisposed to mounting a vigorous immune response, may determine the likelihood of developing IBD or triggering relapse of disease.
In terms of gastrointestinal pathogens and IBD onset, in addition to issues with establishing causality, many of the above studies analyzed subset species as a composite genus group. However, this may dilute the observed effect for species-specific pathogens and shroud clinically relevant differences. Other limiting factors include an inability to detect prior events, as PCR and culture techniques do not account for lifetime exposure to various gastrointestinal pathogens. The multitude of enteric infections, as well as the timing (e.g. early versus later in life), underlying genetic susceptibility, and change in the gut microbiome, rather than a singular pathogen, may be most relevant, particularly when considering the complexity of IBD pathogenesis. In fact, limited data has suggested gastroenteritis later in life is linked more strongly to an increased risk of IBD, or perhaps it is the lack of exposure earlier in life that may influence immune tolerance and increase the risk for subsequent IBD.18 Thus, timing of exposure to a gastrointestinal pathogen is important and more data are needed.
Overcoming selection bias in observational, associative studies investigating the role of gastrointestinal pathogens in IBD flares is also challenging, since testing for enteric infections is generally limited to those with an exacerbation in symptoms and is not routine for asymptomatic patients, thus limiting a true “control” population. Moreover, patients with IBD are more often tested for gastrointestinal pathogens compared with patients without IBD, since the identification of an enteric pathogen in a patient with IBD may directly impact clinical decision making to a greater degree.83,109 Although the lack of robust data, the heterogeneity of diagnostic methods, and inability to achieve complete confounder adjustment contribute to the inability to establish causality for most gastrointestinal pathogens in IBD flares, it is nevertheless conceivable that certain enteric pathogens may trigger relapse of clinical IBD or contribute to attenuated or failed responses to IBD therapies.
Experimental and translational studies provide complementary evidence for the role of enteric infections in IBD pathogenesis by delineating potential pathogenic and protective mechanisms. These mechanisms may be investigated for future preventative and therapeutic interventions in IBD. Further data incorporating deep sequencing technologies will continue to reveal the relationship between enteric pathogens and IBD. Well-designed prospective studies on carefully selected patients are required to evaluate the clinical implications of enteric pathogens on the risk and course of IBD, and how pathogens may contribute to the efficacy and safety of IBD therapies. A better understanding of these complex gene-microbe-environment interactions will advance our efforts toward IBD prevention and a more personalized approach to disease management.
Supplemental Material
Supplemental material, sj-docx-1-tag-10.1177_17562848211004493 for The role of gastrointestinal pathogens in inflammatory bowel disease: a systematic review by Jordan E. Axelrad, Ken H. Cadwell, Jean-Frederic Colombel and Shailja C. Shah in Therapeutic Advances in Gastroenterology
Footnotes
Authorship: Guarantor: Axelrad
Study concept and design: All co-authors.
Acquisition of data: JEA and SCS.
Writing first draft of the manuscript: JEA and SCS.
Critical revision of the manuscript for important intellectual content and approval of final version: All co-authors.
All authors approved the final version of the article, including the authorship list.
Conflict of interest: JEA reports receiving research grants from BioFire Diagnostics; consultancy fees or honorarium from BioFire Diagnostics and Janssen; and holds US patent 2012/0052124A1. KC reports receiving research funding from Pfizer, Takeda, and AbbVie; consultancy fees or honorarium from Puretech Health, Genentech, and AbbVie; and holds US patent 10,722,600 and provisional patent 62/935,035. JFC reports receiving research grants from AbbVie, Janssen Pharmaceuticals and Takeda; receiving payment for lectures from AbbVie, Amgen, Allergan, Inc. Ferring Pharmaceuticals, Shire, and Takeda; receiving consulting fees from AbbVie, Amgen, Arena Pharmaceuticals, Boehringer Ingelheim, Celgene Corporation, Celltrion, Eli Lilly, Enterome, Ferring Pharmaceuticals, Genentech, Janssen Pharmaceuticals, Landos, Ipsen, Medimmune, Merck, Novartis, Pfizer, Shire, Takeda, Tigenix, Viela bio; and holds stock options in Intestinal Biotech Development and Genfit.
Funding: The authors disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: JEA receives research support from the Crohn’s and Colitis Foundation, the Judith & Stewart Colton Center for Autoimmunity, Lyanne and Michael Saperstein, and the NIH NIDDK Diseases K23DK124570. KC has recently been supported by NIH grants DK093668, AI121244, HL123340, AI130945, AI140754, and DK124336; pilot awards from the NYU Cancer Center grant P30CA016087 and Judith & Stewart Colton Center of Autoimmunity; and Faculty Scholar grant from the Howard Hughes Medical Institute, Crohn’s & Colitis Foundation, Kenneth Rainin Foundation, and PATH award from the Burroughs Wellcome Fund. SCS supported by VA CDA: 5IK2CX002027.
ORCID iDs: Jordan E. Axelrad  https://orcid.org/0000-0003-1951-7790
https://orcid.org/0000-0003-1951-7790
Shailja C. Shah  https://orcid.org/0000-0002-2049-9959
https://orcid.org/0000-0002-2049-9959
Supplemental material: Supplemental material for this article is available online.
Contributor Information
Jordan E. Axelrad, Division of Gastroenterology, Department of Medicine, NYU Grossman School of Medicine, Inflammatory Bowel Disease Center, NYU Langone Health, 305 East 33rd Street, Lower Level, New York, NY 10016, USA.
Ken H. Cadwell, Division of Gastroenterology, Department of Medicine, NYU Grossman School of Medicine, New York, NY, USA Kimmel Center for Biology and Medicine at the Skirball Institute, NYU Grossman School of Medicine, New York, NY, USA; Department of Microbiology, NYU Grossman School of Medicine, New York, NY, USA.
Jean-Frederic Colombel, Division of Gastroenterology, Department of Medicine, Icahn School of Medicine at Mount Sinai, New York, NY, USA.
Shailja C. Shah, Section of Gastroenterology, Veterans Affairs Tennessee Valley Healthcare System, Nashville, TN San Diego Health System, La Jolla, CA, USA; Division of Gastroenterology, University of California, San Diego, La Jolla, CA, USA.
References
- 1. Thaiss CA, Zmora N, Levy M, et al. The microbiome and innate immunity. Nature 2016; 535: 65–74. [DOI] [PubMed] [Google Scholar]
- 2. Slack E, Hapfelmeier S, Stecher B, et al. Innate and adaptive immunity cooperate flexibly to maintain host-microbiota mutualism. Science 2009; 325: 617–620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Longman RS, Littman DR. The functional impact of the intestinal microbiome on mucosal immunity and systemic autoimmunity. Curr Opin Rheumatol 2015; 27: 381–387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Joeris T, Müller-Luda K, Agace WW, et al. Diversity and functions of intestinal mononuclear phagocytes. Mucosal Immunol 2017; 10: 845–864. [DOI] [PubMed] [Google Scholar]
- 5. Kaser A, Zeissig S, Blumberg RS. Inflammatory bowel disease. Annu Rev Immunol 2010; 28: 573–621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dubinsky M, Braun J. Diagnostic and prognostic microbial biomarkers in inflammatory bowel diseases. Gastroenterology 2015; 149: 1265–1274.e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Kostic AD, Xavier RJ, Gevers D. The microbiome in inflammatory bowel disease: current status and the future ahead. Gastroenterology 2014; 146: 1489–1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Yang WH, Heithoff DM, Aziz PV, et al. Recurrent infection progressively disables host protection against intestinal inflammation. Science 2017; 358: eaao5610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Khor B, Gardet A, Xavier RJ. Genetics and pathogenesis of inflammatory bowel disease. Nature 2011; 474: 307–317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Chow J, Tang H, Mazmanian SK. Pathobionts of the gastrointestinal microbiota and inflammatory disease. Curr Opin Immunol 2011; 23: 473–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Jostins L, Ripke S, Weersma RK, et al. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature 2012; 491: 119–124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Matsuzawa-Ishimoto Y, Shono Y, Gomez LE, et al. Autophagy protein ATG16L1 prevents necroptosis in the intestinal epithelium. J Exp Med 2017; 214: 3687–3705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Cadwell K, Patel KK, Maloney NS, et al. Virus-plus-susceptibility gene interaction determines Crohn’s disease gene Atg16L1 phenotypes in intestine. Cell 2010; 141: 1135–1145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Hand TW, Dos Santos LM, Bouladoux N, et al. Acute gastrointestinal infection induces long-lived microbiota-specific T cell responses. Science 2012; 337: 1553–1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Axelrad JE, Cadwell KH, Colombel J-F, et al. Systematic review: gastrointestinal infection and incident inflammatory bowel disease. Aliment Pharmacol Ther 2020; 51: 1222–1232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jess T, Simonsen J, Nielsen NM, et al. Enteric Salmonella or Campylobacter infections and the risk of inflammatory bowel disease. Gut 2011; 60: 318–324. [DOI] [PubMed] [Google Scholar]
- 17. Riddle MS, Porter CK. Detection bias and the association between inflammatory bowel disease and Salmonella and Campylobacter infection. Gut 2012; 61: 635. [DOI] [PubMed] [Google Scholar]
- 18. Axelrad JE, Olén O, Askling J, et al. Gastrointestinal infection increases odds of inflammatory bowel disease in a nationwide case-control study. Clin Gastroenterol Hepatol 2019; 17: 1311–1322.e7. [DOI] [PubMed] [Google Scholar]
- 19. Kaakoush NO, Mitchell HM, Man SM. Role of emerging Campylobacter species in inflammatory bowel diseases. Inflamm Bowel Dis 2014; 20: 2189–2197. [DOI] [PubMed] [Google Scholar]
- 20. Zhang L, Man SM, Day AS, et al. Detection and isolation of Campylobacter species other than C. Jejuni from children with Crohn’s disease. J Clin Microbiol 2009; 47: 453–455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Man SM, Zhang L, Day AS, et al. Campylobacter concisus and other Campylobacter species in children with newly diagnosed Crohn’s disease. Inflamm Bowel Dis 2010; 16: 1008–1016. [DOI] [PubMed] [Google Scholar]
- 22. Mukhopadhya I, Thomson JM, Hansen R, et al. Detection of Campylobacter concisus and other Campylobacter species in colonic biopsies from adults with ulcerative colitis. PLoS One 2011; 6: e21490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Kirk KF, Nielsen HL, Thorlacius-Ussing O, et al. Optimized cultivation of Campylobacter concisus from gut mucosal biopsies in inflammatory bowel disease. Gut Pathog 2016; 8: 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Deshpande NP, Kaakoush NO, Wilkins MR, et al. Comparative genomics of Campylobacter concisus isolates reveals genetic diversity and provides insights into disease association. BMC Genomics 2013; 14: 585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Kaakoush NO, Deshpande NP, Wilkins MR, et al. The pathogenic potential of Campylobacter concisus strains associated with chronic intestinal diseases. PLoS One 2011; 6: e29045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Hansen R, Berry SH, Mukhopadhya I, et al. The microaerophilic microbiota of de-novo paediatric inflammatory bowel disease: the BISCUIT study. PLoS One 2013; 8: e58825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Castaño-Rodríguez N, Kaakoush NO, Lee WS, et al. Dual role of Helicobacter and Campylobacter species in IBD: a systematic review and meta-analysis. Gut 2017; 66: 235–249. [DOI] [PubMed] [Google Scholar]
- 28. Kaakoush NO, Holmes J, Octavia S, et al. Detection of Helicobacteraceae in intestinal biopsies of children with Crohn’s disease. Helicobacter 2010; 15: 549–557. [DOI] [PubMed] [Google Scholar]
- 29. Naser SA, Sagramsingh SR, Naser AS, et al. Mycobacterium avium subspecies paratuberculosis causes Crohn’s disease in some inflammatory bowel disease patients. World J Gastroenterol 2014; 20: 7403–7415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Autschbach F, Eisold S, Hinz U, et al. High prevalence of Mycobacterium avium subspecies paratuberculosis IS900 DNA in gut tissues from individuals with Crohn’s disease. Gut 2005; 54: 944–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Timms VJ, Daskalopoulos G, Mitchell HM, et al. The association of Mycobacterium avium subsp. paratuberculosis with inflammatory bowel disease. PLoS One 2016; 11: e0148731. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Parrish NM, Radcliff RP, Brey BJ, et al. Absence of mycobacterium avium subsp. paratuberculosis in Crohn’s patients. Inflamm Bowel Dis 2009; 15: 558–565. [DOI] [PubMed] [Google Scholar]
- 33. Ricanek P, Lothe SM, Szpinda I, et al. Paucity of mycobacteria in mucosal bowel biopsies from adults and children with early inflammatory bowel disease. J Crohns Colitis 2010; 4: 561–566. [DOI] [PubMed] [Google Scholar]
- 34. Baksh FK, Finkelstein SD, Ariyanayagam-Baksh SM, et al. Absence of Mycobacterium avium subsp. paratuberculosis in the microdissected granulomas of Crohn’s disease. Mod Pathol 2004; 17: 1289–1294. [DOI] [PubMed] [Google Scholar]
- 35. Bernstein CN, Nayar G, Hamel A, et al. Study of animal-borne infections in the mucosas of patients with inflammatory bowel disease and population-based controls. J Clin Microbiol 2003; 41: 4986–4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Monaghan TM, Robins A, Knox A, et al. Circulating antibody and memory B-Cell responses to C. difficile toxins A B in patients with C. Difficile-associated diarrhoea, inflammatory bowel disease and cystic fibrosis. PLoS One 2013; 8: e74452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hourigan SK, Chirumamilla SR, Ross T, et al. Clostridium difficile carriage and serum antitoxin responses in children with inflammatory bowel disease. Inflamm Bowel Dis 2013; 19: 2744–2752. [DOI] [PubMed] [Google Scholar]
- 38. Clayton EM, Rea MC, Shanahan F, et al. The vexed relationship between Clostridium difficile and inflammatory bowel disease: an assessment of carriage in an outpatient setting among patients in remission. Am J Gastroenterol 2009; 104: 1162–1169. [DOI] [PubMed] [Google Scholar]
- 39. Monaghan TM, Cockayne A, Mahida YR. Pathogenesis of Clostridium difficile infection and its potential role in inflammatory bowel disease. Inflamm Bowel Dis 2015; 21: 1957–1966. [DOI] [PubMed] [Google Scholar]
- 40. Ananthakrishnan AN, Oxford EC, Nguyen DD, et al. Genetic risk factors for Clostridium difficile infection in ulcerative colitis. Aliment Pharmacol Ther 2013; 38: 522–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Miranda-Bautista J, Padilla-Suárez C, Bouza E, et al. Listeria monocytogenes infection in inflammatory bowel disease patients: case series and review of the literature. Eur J Gastroenterol Hepatol 2014; 26: 1247–1252. [DOI] [PubMed] [Google Scholar]
- 42. Liu Y, van Kruiningen HJ, West AB, et al. Immunocytochemical evidence of Listeria, Escherichia coli, and Streptococcus antigens in Crohn’s disease. Gastroenterology 1995; 108: 1396–1404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Chen W, Li D, Paulus B, et al. Detection of Listeria monocytogenes by polymerase chain reaction in intestinal mucosal biopsies from patients with inflammatory bowel disease and controls. J Gastroenterol Hepatol 2000; 15: 1145–1150. [DOI] [PubMed] [Google Scholar]
- 44. Lamps LW, Madhusudhan KT, Havens JM, et al. Pathogenic Yersinia DNA is detected in bowel and mesenteric lymph nodes from patients with Crohn’s disease. Am J Surg Pathol 2003; 27: 220–227. [DOI] [PubMed] [Google Scholar]
- 45. Leu SB, Shulman SC, Steelman CK, et al. Pathogenic Yersinia DNA in intestinal specimens of pediatric patients with Crohn’s disease. Fetal Pediatr Pathol 2013; 32: 367–370. [DOI] [PubMed] [Google Scholar]
- 46. Le Baut G, O’Brien C, Pavli P, et al. Prevalence of Yersinia species in the ileum of Crohn’s disease patients and controls. Front Cell Infect Microbiol 2018; 8: 336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dumay A, Gergaud O, Roy M, et al. Is Crohn disease the price to pay today for having survived to the Black Death? J Crohns Colitis 2019; 13: 1318–1322. [DOI] [PubMed] [Google Scholar]
- 48. Sipponen T, Turunen U, Lautenschlager I, et al. Human herpesvirus 6 and cytomegalovirus in ileocolonic mucosa in inflammatory bowel disease. Scand J Gastroenterol 2011; 46: 1324–1333. [DOI] [PubMed] [Google Scholar]
- 49. Sankaran-Walters S, Ransibrahmanakul K, Grishina I, et al. Epstein-Barr virus replication linked to B cell proliferation in inflamed areas of colonic mucosa of patients with inflammatory bowel disease. J Clin Virol 2011; 50: 31–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Linton MS, Kroeker K, Fedorak D, et al. Prevalence of Epstein-Barr Virus in a population of patients with inflammatory bowel disease: a prospective cohort study. Aliment Pharmacol Ther 2013; 38: 1248–1254. [DOI] [PubMed] [Google Scholar]
- 51. Shimada T, Nagata N, Okahara K, et al. PCR detection of human herpesviruses in colonic mucosa of individuals with inflammatory bowel disease: comparison with individuals with immunocompetency and HIV infection. PLoS One 2017; 12: e0184699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Norman JM, Handley SA, Baldridge MT, et al. Disease-specific alterations in the enteric virome in inflammatory bowel disease. Cell 2015; 160: 447–460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Miyoshi J, Sofia MA, Pierre JF. The evidence for fungus in Crohn’s disease pathogenesis. Clin J Gastroenterol 2018; 11: 449–456. [DOI] [PubMed] [Google Scholar]
- 54. Ott SJ, Kühbacher T, Musfeldt M, et al. Fungi and inflammatory bowel diseases: alterations of composition and diversity. Scand J Gastroenterol 2008; 43: 831–841. [DOI] [PubMed] [Google Scholar]
- 55. Li Q, Wang C, Tang C, et al. Dysbiosis of gut fungal microbiota is associated with mucosal inflammation in Crohn’s disease. J Clin Gastroenterol 2014; 48: 513–523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Sokol H, Leducq V, Aschard H, et al. Fungal microbiota dysbiosis in IBD. Gut 2017; 66: 1039–1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Liguori G, Lamas B, Richard ML, et al. Fungal dysbiosis in mucosa-associated microbiota of Crohn’s disease patients. J Crohns Colitis 2016; 10: 296–305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Richard ML, Lamas B, Liguori G, et al. Gut fungal microbiota: the Yin and Yang of inflammatory bowel disease. Inflamm Bowel Dis 2015; 21: 656–665. [DOI] [PubMed] [Google Scholar]
- 59. Lewis JD, Chen EZ, Baldassano RN, et al. Inflammation, antibiotics, and diet as environmental stressors of the gut microbiome in pediatric Crohn’s disease. Cell Host Microbe 2015; 18: 489–500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chehoud C, Albenberg LG, Judge C, et al. Fungal signature in the gut microbiota of pediatric patients with inflammatory bowel disease. Inflamm Bowel Dis 2015; 21: 1948–1956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Limon JJ, Tang J, Li D, et al. Malassezia is associated with Crohn’s disease and exacerbates colitis in mouse models. Cell Host Microbe 2019; 25: 377–388.e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Qiu X, Ma J, Jiao C, et al. Alterations in the mucosa-associated fungal microbiota in patients with ulcerative colitis. Oncotarget 2017; 8: 107577–107588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Mukherjee PK, Sendid B, Hoarau G, et al. Mycobiota in gastrointestinal diseases. Nat Rev Gastroenterol Hepatol 2015; 12: 77–87. [DOI] [PubMed] [Google Scholar]
- 64. Hooi JKY, Lai WY, Ng WK, et al. Global prevalence of Helicobacter pylori infection: systematic review and meta-analysis. Gastroenterology 2017; 153: 420–429. [DOI] [PubMed] [Google Scholar]
- 65. Benavides-Ward A, Vasquez-Achaya F, Silva-Caso W, et al. Helicobacter pylori and its relationship with variations of gut microbiota in asymptomatic children between 6 and 12 years. BMC Res Notes 2018; 11: 468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Bühling A, Radun D, Müller WA, et al. Influence of anti-Helicobacter triple-therapy with metronidazole, omeprazole and clarithromycin on intestinal microflora. Aliment Pharmacol Ther 2001; 15: 1445–1452. [DOI] [PubMed] [Google Scholar]
- 67. Shah SC. Friend or foe in IBD pathogenesis: not all infections are equal. Gastroenterology 2019; 157: 1441–1442. [DOI] [PubMed] [Google Scholar]
- 68. Wu X-W, Ji H-Z, Yang M-F, et al. Helicobacter pylori infection and inflammatory bowel disease in Asians: a meta-analysis. World J Gastroenterol 2015; 21: 4750–4756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Luther J, Dave M, Higgins PDR, et al. Association between Helicobacter pylori infection and inflammatory bowel disease: a meta-analysis and systematic review of the literature. Inflamm Bowel Dis 2010; 16: 1077–1084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Rokkas T, Gisbert JP, Niv Y, et al. The association between Helicobacter pylori infection and inflammatory bowel disease based on meta-analysis. United European Gastroenterol J 2015; 3: 539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Tepler A, Narula N, Peek RM, et al. Systematic review with meta-analysis: association between Helicobacter pylori CagA seropositivity and odds of inflammatory bowel disease. Aliment Pharmacol Ther 2019; 50: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Weinstock JV, Elliott DE. Helminths and the IBD hygiene hypothesis. Inflamm Bowel Dis 2009; 15: 128–133. [DOI] [PubMed] [Google Scholar]
- 73. Weinstock JV, Summers RW, Elliott DE, et al. The possible link between de-worming and the emergence of immunological disease. J Lab Clin Med 2002; 139: 334–338. [DOI] [PubMed] [Google Scholar]
- 74. Ramanan D, Bowcutt R, Lee SC, et al. Helminth infection promotes colonization resistance via type 2 immunity. Science 2016; 352: 608–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Wammes LJ, Mpairwe H, Elliott AM, et al. Helminth therapy or elimination: epidemiological, immunological, and clinical considerations. Lancet Infect Dis 2014; 14: 1150–1162. [DOI] [PubMed] [Google Scholar]
- 76. Chu KM, Watermeyer G, Shelly L, et al. Childhood helminth exposure is protective against inflammatory bowel disease: a case control study in South Africa. Inflamm Bowel Dis 2013; 19: 614–620. [DOI] [PubMed] [Google Scholar]
- 77. Sandborn WJ, Elliott DE, Weinstock J, et al. Randomised clinical trial: the safety and tolerability of Trichuris suis ova in patients with Crohn’s disease. Aliment Pharmacol Ther 2013; 38: 255–263. [DOI] [PubMed] [Google Scholar]
- 78. Summers RW, Elliott DE, Urban JF, et al. Trichuris suis therapy for active ulcerative colitis: a randomized controlled trial. Gastroenterology 2005; 128: 825–832. [DOI] [PubMed] [Google Scholar]
- 79. Schölmerich J, Fellermann K, Seibold FW, et al. A randomised, double-blind, placebo-controlled trial of Trichuris suis ova in active Crohn’s disease. J Crohns Colitis 2017; 11: 390–399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Garg SK, Croft AM, Bager P. Helminth therapy (worms) for induction of remission in inflammatory bowel disease. Cochrane Database Syst Rev 2014: CD009400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Huang X, Zeng L-R, Chen F-S, et al. Trichuris suis ova therapy in inflammatory bowel disease: a meta-analysis. Medicine 2018; 97: e12087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Summers RW, Elliott DE, Urban JF, et al. Trichuris suis therapy in Crohn’s disease. Gut 2005; 54: 87–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Axelrad JE, Joelson A, Nobel YR, et al. Enteric infection in relapse of inflammatory bowel disease: the utility of stool microbial PCR testing. Inflamm Bowel Dis 2017; 23: 1034–1039. [DOI] [PubMed] [Google Scholar]
- 84. Antonelli E, Baldoni M, Giovenali P, et al. Intestinal superinfections in patients with inflammatory bowel diseases. J Crohns Colitis 2012; 6: 154–159. [DOI] [PubMed] [Google Scholar]
- 85. Mylonaki M, Langmead L, Pantes A, et al. Enteric infection in relapse of inflammatory bowel disease: importance of microbiological examination of stool. Eur J Gastroenterol Hepatol 2004; 16: 775–778. [DOI] [PubMed] [Google Scholar]
- 86. Weber P, Koch M, Heizmann WR, et al. Microbic superinfection in relapse of inflammatory bowel disease. J Clin Gastroenterol 1992; 14: 302–308. [DOI] [PubMed] [Google Scholar]
- 87. Lobatón T, Domènech E. Bacterial intestinal superinfections in inflammatory bowel diseases beyond Clostridum difficile. Inflamm Bowel Dis 2016; 22: 1755–1762. [DOI] [PubMed] [Google Scholar]
- 88. Hong S, Zaki TA, Main M, et al. Comparative evaluation of conventional stool testing and multiplex molecular panel in outpatients with relapse of inflammatory bowel disease. Inflamm Bowel Dis. Epub ahead of print 2 January 2021. DOI: 10.1093/ibd/izaa336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Sokol H, Jegou S, McQuitty C, et al. Specificities of the intestinal microbiota in patients with inflammatory bowel disease and Clostridium difficile infection. Gut Microbes 2018; 9: 55–60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Axelrad JE, Shah BJ. Clostridium difficile Infection in inflammatory bowel disease: a nursing-based quality improvement strategy. J Healthc Qual 2016; 38: 283–289. [DOI] [PubMed] [Google Scholar]
- 91. Jodorkovsky D, Young Y, Abreu MT. Clinical outcomes of patients with ulcerative colitis and co-existing Clostridium difficile infection. Dig Dis Sci 2010; 55: 415–420. [DOI] [PubMed] [Google Scholar]
- 92. Issa M, Ananthakrishnan AN, Binion DG. Clostridium difficile and inflammatory bowel disease. Inflamm Bowel Dis 2008; 14: 1432–1442. [DOI] [PubMed] [Google Scholar]
- 93. Issa M, Vijayapal A, Graham MB, et al. Impact of Clostridium difficile on inflammatory bowel disease. Clin Gastroenterol Hepatol 2007; 5: 345–351. [DOI] [PubMed] [Google Scholar]
- 94. Ananthakrishnan AN, McGinley EL, Binion DG. Excess hospitalisation burden associated with Clostridium difficile in patients with inflammatory bowel disease. Gut 2008; 57: 205–210. [DOI] [PubMed] [Google Scholar]
- 95. Nguyen GC, Kaplan GG, Harris ML, et al. A national survey of the prevalence and impact of Clostridium difficile infection among hospitalized inflammatory bowel disease patients. Am J Gastroenterol 2008; 103: 1443–1450. [DOI] [PubMed] [Google Scholar]
- 96. Rodemann JF, Dubberke ER, Reske KA, et al. Incidence of Clostridium difficile infection in inflammatory bowel disease. Clin Gastroenterol Hepatol 2007; 5: 339–344. [DOI] [PubMed] [Google Scholar]
- 97. Kariv R, Navaneethan U, Venkatesh PGK, et al. Impact of Clostridium difficile infection in patients with ulcerative colitis. J Crohns Colitis 2011; 5: 34–40. [DOI] [PubMed] [Google Scholar]
- 98. Anderson A, Click B, Ramos-Rivers C, et al. Lasting impact of Clostridium difficile infection in inflammatory bowel disease: a propensity score matched analysis. Inflamm Bowel Dis 2017; 23: 2180–2188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Navaneethan U, Mukewar S, Venkatesh PGK, et al. Clostridium difficile infection is associated with worse long term outcome in patients with ulcerative colitis. J Crohns Colitis 2012; 6: 330–336. [DOI] [PubMed] [Google Scholar]
- 100. Murthy SK, Steinhart AH, Tinmouth J, et al. Impact of Clostridium difficile colitis on 5-year health outcomes in patients with ulcerative colitis. Aliment Pharmacol Ther 2012; 36: 1032–1039. [DOI] [PubMed] [Google Scholar]
- 101. Ananthakrishnan AN, McGinley EL, Saeian K, et al. Temporal trends in disease outcomes related to Clostridium difficile infection in patients with inflammatory bowel disease. Inflamm Bowel Dis 2011; 17: 976–983. [DOI] [PubMed] [Google Scholar]
- 102. Razik R, Rumman A, Bahreini Z, et al. Recurrence of Clostridium difficile infection in patients with inflammatory bowel disease: the RECIDIVISM study. Am J Gastroenterol 2016; 111: 1141–1146. [DOI] [PubMed] [Google Scholar]
- 103. Roy A, Lichtiger S. Clostridium difficile infection: a rarity in patients receiving chronic antibiotic treatment for Crohn’s disease. Inflamm Bowel Dis 2016; 22: 648–653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Sokol H, Lalande V, Landman C, et al. Clostridium difficile infection in acute flares of inflammatory bowel disease: a prospective study. Dig Liver Dis 2017; 49: 643–646. [DOI] [PubMed] [Google Scholar]
- 105. Alvarez-Lobos M, Pizarro DP, Palavecino CE, et al. Role of Salmonella enterica exposure in Chilean Crohn’s disease patients. World J Gastroenterol 2013; 19: 5855–5862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Arora Z, Mukewar S, Wu X, et al. Risk factors and clinical implication of superimposed Campylobacter jejuni infection in patients with underlying ulcerative colitis. Gastroenterol Rep (Oxf) 2016; 4: 287–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Axelrad JE, Joelson A, Green PHR, et al. Enteric infections are common in patients with flares of inflammatory bowel disease. Am J Gastroenterol 2018; 113: 1530–1539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Kolho K-L, Klemola P, Simonen-Tikka M-L, et al. Enteric viral pathogens in children with inflammatory bowel disease. J Med Virol 2012; 84: 345–347. [DOI] [PubMed] [Google Scholar]
- 109. Hanada Y, Khanna S, Loftus EV, et al. Non-Clostridium difficile bacterial infections are rare in patients with flares of inflammatory bowel disease. Clin Gastroenterol Hepatol 2018; 16: 528–533. [DOI] [PubMed] [Google Scholar]
- 110. Maher MM, Nassar MI. Acute cytomegalovirus infection is a risk factor in refractory and complicated inflammatory bowel disease. Dig Dis Sci 2009; 54: 2456–2462. [DOI] [PubMed] [Google Scholar]
- 111. Kandiel A, Lashner B. Cytomegalovirus colitis complicating inflammatory bowel disease. Am J Gastroenterol 2006; 101: 2857–2865. [DOI] [PubMed] [Google Scholar]
- 112. Cottone M, Pietrosi G, Martorana G, et al. Prevalence of cytomegalovirus infection in severe refractory ulcerative and Crohn’s colitis. Am J Gastroenterol 2001; 96: 773–775. [DOI] [PubMed] [Google Scholar]
- 113. Kojima T, Watanabe T, Hata K, et al. Cytomegalovirus infection in ulcerative colitis. Scand J Gastroenterol 2006; 41: 706–711. [DOI] [PubMed] [Google Scholar]
- 114. Matsuoka K, Iwao Y, Mori T, et al. Cytomegalovirus is frequently reactivated and disappears without antiviral agents in ulcerative colitis patients. Am J Gastroenterol 2007; 102: 331–337. [DOI] [PubMed] [Google Scholar]
- 115. Delvincourt M, Lopez A, Pillet S, et al. The impact of cytomegalovirus reactivation and its treatment on the course of inflammatory bowel disease. Aliment Pharmacol Ther 2014; 39: 712–720. [DOI] [PubMed] [Google Scholar]
- 116. Lévêque N, Brixi-Benmansour H, Reig T, et al. Low frequency of cytomegalovirus infection during exacerbations of inflammatory bowel diseases. J Med Virol 2010; 82: 1694–1700. [DOI] [PubMed] [Google Scholar]
- 117. Turpin W, Lee S-H, Raygoza Garay JA, et al. Increased intestinal permeability is associated with later development of Crohn’s disease. Gastroenterology 2020; 159: 2092–2100.e5. [DOI] [PubMed] [Google Scholar]
- 118. Nalle SC, Turner JR. Intestinal barrier loss as a critical pathogenic link between inflammatory bowel disease and graft-versus-host disease. Mucosal Immunol 2015; 8: 720–730. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental material, sj-docx-1-tag-10.1177_17562848211004493 for The role of gastrointestinal pathogens in inflammatory bowel disease: a systematic review by Jordan E. Axelrad, Ken H. Cadwell, Jean-Frederic Colombel and Shailja C. Shah in Therapeutic Advances in Gastroenterology