Significance
Protein stability is central to the pathogenesis of several major human diseases and is the key to extensive research. Improved methods are needed for protein stability engineering. Here, we present a high-throughput screening strategy to stabilize proteins by linking their stabilities to the fluorescent readout of cells expressing an engineered bacterial enzyme. This strategy is generally applicable to proteins of different sources, sizes, and structural characteristics, including industrial-related enzymes and disease-related proteins. We also combined this strategy with deep mutational scanning to comprehensively understand how mutations shape the stability landscape of an important epigenetic enzyme. Our strategy expands the current toolbox of protein research and will also facilitate a variety of protein design.
Keywords: biosensor, protein stability, protein engineering, deep mutational scanning
Abstract
Protein stability affects the physiological functions of proteins and is also a desirable trait in many protein engineering tasks, yet improving protein stability is challenging because of limitations in methods for directly monitoring protein stability in cells. Here, we report an in vivo stability biosensor wherein a protein of interest (POI) is inserted into a microbial enzyme (CysGA) that catalyzes the formation of endogenous fluorescent compounds, thereby coupling POI stability to simple fluorescence readouts. We demonstrate the utility of the biosensor in directed evolution to obtain stabilized, less aggregation-prone variants of two POIs (including nonamyloidogenic variants of human islet amyloid polypeptide). Beyond engineering applications, we exploited our biosensor in deep mutational scanning for experimental delineation of the stability-related contributions of all residues throughout the catalytic domain of a histone H3K4 methyltransferase, thereby revealing its scientifically informative stability landscape. Thus, our highly accessible method for in vivo monitoring of the stability of diverse proteins will facilitate both basic research and applied protein engineering efforts.
Protein stability affects myriad aspects of biochemical and biological research and often appears as a challenge for the application of protein technologies. Most natural proteins are only marginally stable, having free energy values for unfolding as low as 5 to 10 kcal/mol, a level comparable to the energy needed to break only a few hydrogen bonds (1). Although these marginal stabilities enable proteins to be flexible and thereby support their diverse functions, there is a need for at least a minimal stability threshold to support adequately high enough protein folding efficiency for cell survival (2). Evolutionarily, this apparent tension has established a tight balance between increased functionality through accumulation of mutations and the ability to maintain an adequate level of stability (2). Because this balance is delicate, environmental and cellular disturbances—for example, elevated temperature or a limited pool of ligands—can often tip the balance and turn an active protein into a nonfunctional or misfolded, aggregated state (3).
It is increasingly appreciated that protein instability is often a major causative factor in human diseases (4). For example, destabilized mutations of the cellular tumor antigen p53 (5) or antioxidative superoxide dismutase 1 (SOD1) (6) are known to cause multiple human diseases. Misfolding or aggregation of specific proteins is also the hallmark of many neurodegenerative diseases, such as amyloid β peptide in Alzheimer's disease (7), α-synuclein in Parkinson’s disease (8), and polyglutamine in Huntington’s disease (9). Moreover, protein instability is very often a limiting factor in the development of protein technologies including biocatalysts, therapeutic proteins, and de novo protein design. Consider, for example, that natural enzymes cannot usually be directly deployed as biocatalysts; these tools must remain active under continuous stresses like high temperature, high ionic strength, and extreme pH during the industrial process (10) and must retain activity for days or even weeks in some cell-free applications (11). Similarly, therapeutic proteins often suffer from short half-lives in the human body and/or have a highly restricted shelf life (12). Finally, protein design based on the thermodynamic principles is often constrained by poor stability of the target: for example, 34% of the originally designed monomeric fluorescence-activating β-barrel structures were found to be insoluble, 37% were not expressed, and 7% were found to be toxic when expressed (13).
Regardless of widespread academic and industrial interest in stabilizing proteins, tools available for improving protein stability remain quite limited. Although computational stability design can be used to predict stabilizing mutations, its accuracy still needs to be substantially improved due to inadequacies in the quality of experimental results in public databases, in the accuracy of functional annotation information, and in the overall performance of the stability predicting algorithms themselves (14). Directed evolution represents another approach to obtain stabilized proteins, but a profound bottleneck for this approach is to establish high-throughput selection or screening strategies to rapidly monitor protein stability in vivo. The current widely used library-based display technologies rely on functional assays, which are by nature only indirect readouts of the stability of an analyte protein (15).
By contrast, protein stability biosensors offer a way to directly monitor protein stability in vivo (16–18). Although they have been successfully deployed in various protein stability evolution applications, the available technologies are not universal solutions (i.e., suitable for all proteins). As an example, our recent attempts to distinguish and evolve the in vivo stability of two members of the histone H3 lysine 4 (H3K4) methyltransferase family failed when using several well-established biosensors, including a green fluorescent protein (GFP)-based biosensor (16), an aminoglycoside 3″-adenylyltransferase–based biosensor (17), and a chloramphenicol acetyltransferase–based biosensor (18) (SI Appendix, Fig. S1). These failures in our own work highlight the need to expand the toolbox of high-throughput selection and screening strategies for the directed evolution of protein stability and served as the fundamental motivation for our work.
Here, we developed an enzyme-based fluorescent biosensor to monitor and evolve protein stability in vivo. Our strategy is based on insertion of a protein of interest (POI) between two halves of the Escherichia coli uroporphyrinogen-III methyltransferase CysGA protein (19), which catalyzes the formation of endogenous red fluorescent compounds. Linking protein folding to the activity of CysGA allows accurate and sensitive measurement of POI stability and solubility. Our biosensor does not require exogenous substrates or any prior structural knowledge or biophysical information about the POI, therefore engendering its use as a general screen for directed evolution of protein stability. We successfully applied our biosensor to identify stabilizing mutations of muscle acylphosphatase and nonamyloidogenic mutants of the human islet amyloid peptide. Combining this biosensor with deep mutational scanning, we systematically profiled the site-specific mutational tolerance and stability of MLL3SET, the catalytic domain of the H3K4 methyltransferase MLL3 (a member of the mixed lineage leukemia [MLL] family), therefore experimentally characterizing its stability landscape. At a fundamental level, the ability to dissect the molecular basis of protein stability allows the profile of residues that dictate stability to be generated and the stabilization hotspots in proteins to be mapped. Our study demonstrates the utility of our biosensor as a highly accessible, rapid, flexible, and robust tool for monitoring, evolving, and dissecting protein stability in vivo, allowing the improvement in the ability to engineer customized protein and a greater understanding of the relationship between sequence and stability.
Results
Conversion of CysGA into a Sandwiched Protein Stability Biosensor.
CysGA, the 256-residue C-terminal domain of siroheme synthase (CysG), is an S-adenosyl-l-methionine–dependent methyltransferase that converts uroporphyrinogen III into precorrin-2, an intermediate in tetrapyrrole biosynthesis in all organisms (20, 21). When CysGA is overexpressed in Escherichia coli, cells emit bright red fluorescence under ultraviolet (UV) light owing to the accumulation of both trimethylpyrrocorphin (a trimethylated product of precorrin-2) and sirohydrochlorin (the oxidation product of precorrin-2) (22) (SI Appendix, Fig. S2A). The fluorescence spectrum of cell extracts is characterized by an excitation maximum at 357 nm and an emission maximum at 620 nm (SI Appendix, Fig. S2B). This feature makes CysGA homologs useful as fluorescent reporters for gene expression, and such tools have been successfully implemented in bacteria, fission yeast, and cultured mammalian cells (23).
Since any fluorescence readout from CysGA depends on its enzymatic activity—which is closely related to the correct folding of CysGA—we envisioned that CysGA could be converted into a stability biosensor, specifically by coupling its capacity for proper folding to the stability of a given POI. To avoid defects with head-to-tail construction reported from previous biosensors (24), such as generating intact and active CysGA upon proteolytic cleavage of unstable POIs, we decided to create a tripartite, sandwich fusion biosensor comprising split CysGA on either side of the POI (Fig. 1A). The idea behind our design is that the POI can be inserted into a permissive site of CysGA so that the enzyme is physically separated into two segments. In theory, a stable, well-folded POI would bring the two CysGA segments into close proximity, potentially facilitating the reconstitution of CysGA’s enzymatic activity and thereby restoring the fluorescence readout function of the biosensor. In contrast, an unstable or misfolded POI would be vulnerable to aggregation and/or proteolysis, resulting in permanent separation of the two CysGA segments, thereby preventing the generation of any fluorescence readout.
Fig. 1.
Development of the CysGA tripartite system. (A) Schematic diagram of the CysGA tripartite system. The POI is inserted into the permissive site within the CysGA biosensor. If the POI is stable, then CysGA reassembles into its functional conformation and catalyzes formation of red fluorescent trimethylpyrrocorphin and sirohydrochlorin. In contrast, an unstable POI is prone to aggregation and/or proteolysis, resulting in elimination of functional CysGA and therefore weak or no fluorescence. (B) The Cpred score of each site was mapped onto the the crystal structure of CysGA from Salmonella typhimurium (PDB ID: 1PJS). In designing the CysGA biosensor, 10 candidate permissive sites for insertion were selected. (C) Schematic for fusion constructs generation. The POI is inserted into the permissive site via flexible, glycine–serine-rich linkers (sequences SSGSSG and GGGGSGGGGS). (D) Permissive site optimization based on relative fluorescence intensities for the POI Im7 (Escherichia coli) which was inserted at the 10 candidate permissive sites. POI insertion at the CysGA G364 site retained 80% of the fluorescence intensity of intact CysGA. P400 corresponds to Q400 in 1PJS. Data are the mean ± SD of three independent experiments.
A prerequisite for a tripartite CysGA biosensor is the identification of a suitable “permissive site” in CysGA that tolerates POI insertion while also successfully reconstituting CysGA’s enzymatic activity upon reassociation of its split segments. Pursuing this, we reasoned that sites which tolerate circular permutations may also tolerate insertions: both events disrupt the integrity of the structure of CysGA. In previously described circular permutations, the original N and C termini are fused, and new termini can be created at the designated site without substantially altering the original structure of the protein (25). We successfully employed a similar strategy in our previous development of a DsbA tripartite biosensor (26).
We first applied a web tool called Cpred (27) to predict sites permissive for circular permutation based on the crystal structure of CysGA from Salmonella typhimurium (20) (Protein Data Bank [PDB]: 1PJS), which shares 91% sequence identity with CysGA from E. coli. Among the CysGA sites with high probability scores, we selected 10 sites that are located at surface loops and identified their corresponding sites in E. coli CysGA (Fig. 1B and SI Appendix, Fig. S3 A and B). Next, to test their ability to tolerate insertions, we conducted in vivo assays wherein the immunity protein 7 (Im7)—an 86-residue α-helical model protein—was inserted into each of the 10 CysGA permissive candidates via two flexible glycine–serine linkers; we measured the fluorescence intensities of cells expressing each of these fusion constructs (Fig. 1 C and D). Although cells expressing most of the insertional constructs had very weak or no fluorescence, cells expressing the constructs with the Im7 POI inserted after residues V277 and G364 respectively maintained 60 and 80% of the fluorescence intensity of intact CysGA, indicating that these sites can tolerate the insertion of Im7. Given the known problems with solubility reported for circular permutation variants (28), it was encouraging that the G364 CysGA-Im7 fusions exhibited similar solubility as intact CysGA (SI Appendix, Fig. S3C). We selected G364 as the permissive site for further development of our CysGA biosensor.
Detection of Protein Stability In Vivo Using the CysGA Tripartite System.
To determine whether our engineered tripartite CysGA biosensor can be used to screen for stabilized protein variants, we first determined whether it has the ability to distinguish protein variants with a wide range of known thermodynamic stabilities. Our test proteins included seven variants of Im7, six variants of maltose binding protein (MBP), and six variants of human muscle acylphosphatase (AcP) as well as varying lengths of polyglutamine (polyQ) tracts (SI Appendix, Table S1). The first three proteins have been extensively used to study protein folding (29–31). PolyQ is a representative amyloidogenic protein, and abnormal polyQ expansions have been identified as a basis for cellular toxicity in at least eight neurodegenerative disorders, including Huntington’s disease (9).
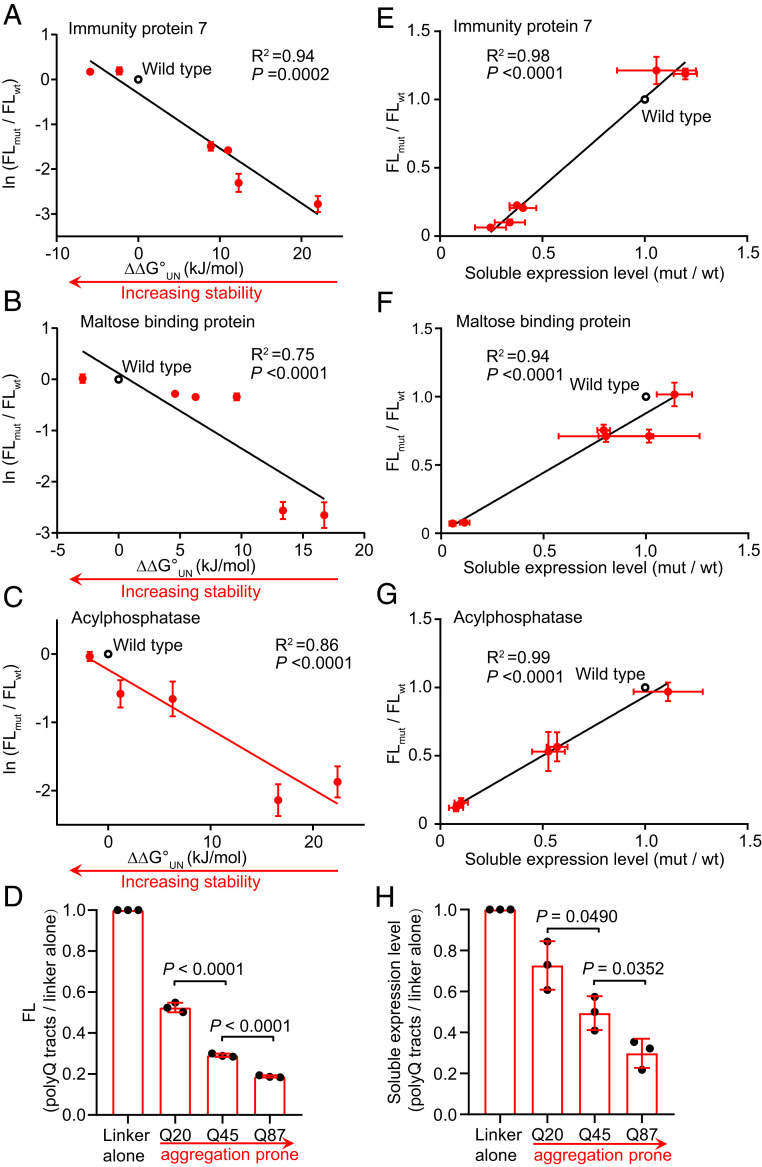
Upon insertion of these protein variants into CysGA after residue G364, we found a significant correlation between the fluorescence intensities of cells expressing different fusion constructs and the thermodynamic stabilities (ΔΔG°UN) for different variants of Im7, MBP, and AcP (Fig. 2 A–C). Numerous lines of evidence support that the soluble expression level of a protein is closely correlated with its thermodynamic stability (32, 33). To test whether the CysGA biosensor can also report protein solubility, we quantified the soluble amount of each tripartite fusion using Western blotting with an anti-CysGA antibody. We found a good correlation between the fluorescence intensities of strains expressing the tripartite fusions and the soluble amounts of each fusion protein for these nonamyloidogenic test proteins (Fig. 2 E–G).
Fig. 2.
Fluorescence intensities correlate with the stabilities and expression levels of different test proteins. (A–D) Protein stabilities correlate with fluorescence intensities (FL). CysGA-POI fusion variants containing insertion of POI sequences with known differential stabilities for Im7 (A, total of seven tested Im7 sequences), MBP (B, six sequences), AcP (C, six sequences), and polyQ (D, three sequences). The relative fluorescence intensities of cells expressing the various fusion proteins are plotted against ΔΔG°UN (or polyQ tract lengths) and normalized by the fluorescence intensities of cells expressing CysGA fused with wild-type POIs (or linker alone for polyQ tracts). A solubility-enhancing MBP tag was fused to the N terminus of CysGA-AcP fusions to facilitate their detection. (E–H) The soluble expression levels correlate well with fluorescence intensities for Im7 (E), MBP (F), AcP (G), and polyQ (H). Soluble expression levels, relative to wild-type POIs (or linker alone for polyQ tracts), are plotted against the relative fluorescence intensities. Data are the mean ± SD of three independent experiments. P values were determined by two-tailed unpaired Student’s t tests.
For polyQ, we observed a length-dependent decrease of the fluorescence intensities (Fig. 2D). It is widely accepted that the length of polyQ repeats is essential for disease’s onset (9). For Huntington’s disease, the threshold for protein aggregation and cellular toxicity has been speculated as around 30 to 40 glutamine residues. Consistent with previous observations (17), our CysGA-polyQ fusions displayed length-dependent aggregation behaviors in which polyQ45 and polyQ87 resulted in less soluble fusion proteins as compared to polyQ20 (Fig. 2H).
Collectively, these results indicate that our biosensor can display robust readouts of thermodynamic stabilities of proteins from diverse organisms with diverse sequences, various sizes, and distinct structural characteristics. Moreover, our biosensor has the ability to link the fluoresence readout to the aggregation propensity of test proteins and to distinguish amyloidogenicprotein variants.
Directed Evolution of the Stability of Human Muscle Acylphosphatase.
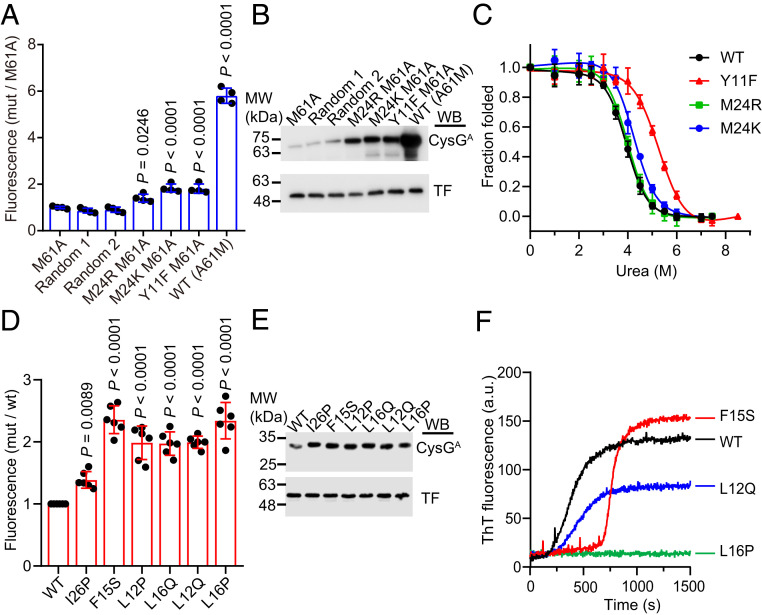
We next sought to extend our biosensor to the evolution of protein stability in vivo. Our procedure was quite straightforward: First, random mutations were introduced into the gene encoding a given POI for insertion between the two halves of CysGA. Then, the resulting E. coli transformants were induced for CysGA-POI expression on lysogeny broth (LB) agar plates, and colonies with enhanced fluorescence intensities detected under UV light were manually picked. As a proof of principle, we evolved the destabilized, M61A mutant of AcP, a slowly folding protein with a two-state behavior (31). AcPM61A resulted in nearly no fluorescence when inserted into the CysGA biosensor; this readily facilitated our screening because the background fluorescence was quite low. Then we used an error-prone PCR-based approach to create a plasmid library of ∼105 members with an average of 0.8 amino acid substitutions per AcP sequence. Following transformation of the library into the E. coli TransT1 strain, we screened 4% of the transformants (106) and picked 44 bright, red fluorescent colonies by naked eyes and then extracted and sequenced their plasmids. In each screen, there are about one to two bright colonies per 1,000 colonies. For the 44 colonies selected, three (6.8%) contain the A61M mutation that fully reversed to a wild-type sequence. Other abundant mutations include the known stabilizing mutation Y11F (31) identified in 29.5% of colonies and two previously uncharacterized mutations: M24K identified in 45% of colonies and M24R identified in 2.2% of colonies. We then introduced these four individual mutations into the acp M61A gene inserted in the tripartite biosensor system and retransformed the resulting plasmids into a fresh background strain. These transformants showed small but significant increases in the fluorescence compared to the control strain containing CysGA-AcPM61A (Fig. 3A). Western blot analysis also confirmed that these four mutations enhanced the soluble fraction of the CysGA-AcPM61A fusion protein (Fig. 3B).
Fig. 3.
Stabilization of AcP and human IAPP. (A) Relative fluorescence intensities of cells expressing CysGA fused with the indicated AcP mutants. A solubility-enhancing MBP tag was fused to the N terminus of CysGA-AcP fusions to facilitate their detection. Fluorescence intensities were normalized to that of cells expressing the CysGA fused with AcPM61A. Two randomly picked, unselected strains showed fluorescence similar to that of the unmutated control strain, indicating that the small but significant increase in fluorescence of our selected strains was not a stochastic event. (B) Soluble expression levels of CysGA fused with different AcP mutants. Soluble fractions of the cell lysates were analyzed by immunoblotting with antibodies against CysGA and a trigger factor (TF) protein that served as a loading control. (C) Equilibrium urea-induced unfolding of wild-type AcP and screened variants. The raw titration data were converted to the fraction of folded proteins. Equilibrium unfolding parameters of different AcP variants were determined via fitting of the denaturation curves to the equation as described in Materials and Methods. (D) Relative fluorescence intensities of cells expressing CysGA fused with different IAPP variants. All selected variants emitted stronger florescence intensities than the strain expressing CysGA fused with IAPPI26P, which was previously reported to display relatively weak aggregation. (E) Soluble expression levels of CysGA fused with different IAPP variants. (F) Thioflavin T assays for the previously uncharacterized IAPP mutants. Amyloid formation of IAPP was detected by measuring the fluorescence at an excitation wavelength of 440 nm and an emission wavelength of 485 nm. Data are the mean ± SD of four (A), three (C), or five (D) independent measurements. P values were determined by one-way ANOVA with a Dunnett’s multiple comparison test. Representative blots from three independent experiments are shown (B and E). One representative curve from three independent measurements is shown for each mutant (F). Uncropped Western blots are presented in SI Appendix, Fig. S12.
To test whether the two previously unreported mutations M24R and M24K exert any M61A-independent stabilizing effects, we purified the Y11F, M24R, and M24K mutants and determined their thermodynamic stabilities by equilibrium urea titration (Fig. 3C). Y11F stabilized AcP by 2.4 kJ/mol, which is comparable with the previously reported value of 1.8 kJ/mol (32). M24K stabilized AcP by 1.5 kJ/mol, but the M24R mutant showed similar stabilities as the wild-type protein, indicating that its stabilizing effect is specifically associated with the M61A mutation (Fig. 3C and Table 1). These data validate the ability of our CysGA tripartite system to be used as a screen for directed evolution experiments to select target proteins for increased stability.
Table 1.
Equilibrium unfolding parameters of AcP variants
| ΔG°UN (kJ ⋅ mol−1) | m (kJ ⋅ mol−1 ⋅ M−1) | Cm (M) | |
| Wild type | −22.5 ± 2.9 | 5.9 ± 0.7 | 3.94 ± 0.06 |
| Y11F | −24.9 ± 2.9 | 4.7 ± 0.5 | 5.25 ± 0.24 |
| M24K | −24.0 ± 3.1 | 5.5 ± 0.7 | 4.22 ± 0.08 |
| M24R | −22.7 ± 3.4 | 5.6 ± 0.9 | 3.98 ± 0.08 |
Values reported are the mean ± SEM. n = 3 independent samples.
Selection for Nonamyloidogenic Mutants of Islet Amyloid Polypeptide.
We then wondered whether amyloidogenic proteins could be similarly stabilized and solubilized using our biosensor. Human islet amyloid polypeptide (IAPP), a 37-amino-acid polypeptide, readily aggregates into oligomers or amyloid fibrils; such aggregation has been implicated as a hallmark of type 2 diabetes (34). Screening of a random mutagenesis library for IAPP inserted into CysGA identified five mutants with a fluorescence increase of at least twofold over wild-type IAPP (Fig. 3D). Indeed, the isolated mutants exhibited even higher fluorescence than the cells expressing CysGA fused with the nonamyloidogenic I26P mutant (35) in our test with the CysGA tripartite system. The soluble expression levels of these mutants and I26P in the tripartite fusion were also higher than that of wild-type IAPP (Fig. 3E). Two of these mutants (L12P and L16Q) have been reported to delay the aggregation of IAPP (36), supporting the validity of our screening. The effect of F15S is controversial: it was reported that small residue substitutions at F15, such as alanine or serine, may promote IAPP to associate into fibrils more rapidly (37). However, another study reported that F15A has higher solubility and reduced self-assembly potential compared to wild-type IAPP (38).
To confirm the ability of the newly isolated mutants (L12Q and L16P) and F15S to reduce aggregation, we synthesized these peptides and determined their aggregation profile by monitoring thioflavin T (ThT) binding, which emits strong fluorescence intensities upon association with amyloid fibrils. The L12Q mutation dramatically reduced the rate of fibril formation, and the L16P mutation almost completely inhibited fibril formation throughout the entire time course of measurement (Fig. 3F). For F15S, we observed a longer lag phase and higher final ThT fluorescence compared to wild-type IAPP. The initial lag phase is known to reflect the nucleation processes followed by a second, faster phase corresponding to fibril assembly (39). Therefore, the longer lag phase of F15S may reflect its ability to delay the nucleation process, whereas rapid fibril formation kinetics following the nucleation process probably allows more protomers to be incorporated into the final fibril. Alternatively, the fibers of F15S may have different morphology that allows them to bind more readily with ThT. These potentially distinct behaviors could help account for the controversial results for this mutant reported in the literature. Together, these results demonstrate the power of our CysGA tripartite system to evolve proteins with reduced aggregation propensity.
Deep Mutational Scanning Reveals Site-Specific Mutational Tolerance of the MLL3SET Protein.
Deep mutational scanning, which combines a genotype–phenotype platform with high-throughput DNA sequencing, has emerged as a powerful approach for dissecting protein properties in an unbiased manner (40). To demonstrate the applicability of our biosensor for analyzing the influence of each individual residue on the foldability/stability of an entire protein, we integrated our stability screening strategy with deep mutational scanning (SI Appendix, Fig. S4) and applied this hybrid approach to map the stability landscape of MLL3SET. In mammalian cells, MLL3 is responsible for monomethylation of histone H3 Lys4 (which regulates gene expression), and this function is mainly carried out through its catalytic SET domain (41). Although previous studies have suggested that the conformational stability of the MLL3 SET domain has a large impact on its catalytic activity (41), the lack of biochemical characterization for stabilized/destabilized MLL3SET mutants has precluded further mechanistic insights into this process. Thus, our delineation of the stability landscape of MLL3SET and obtaining its stable variants should facilitate deeper understanding of determinants of MLL3SET’s function and the development of chemical approaches to regulate MLL3SET’s function in vivo by adjusting its conformational stability.
We used error-prone PCR to construct a MLL3SET mutant library within the CysGA biosensor and obtained 106 members (the P0 library). Characterization of this library by deep sequencing revealed that it collectively harbors all substitutions that can be generated by single-nucleotide substitutions and that it ultimately samples 57.9% of the possible substitutions for each amino acid at every position (SI Appendix, Fig. S5). Additionally, the number of mutations in each MLL3SET sequence roughly conformed to the Poisson distribution with an average of five mutations, confirming as expected that these mutations were generated randomly in our error-prone PCR step (SI Appendix, Fig. S6A). We screened the P0 library based on fluorescence readouts of cells and initially isolated 1,374 clones (referred as the hit library, P1) exhibiting stronger fluorescence than the cells expressing the CysGA biosensor with wild-type MLL3SET (SI Appendix, Fig. S6B). Deep sequencing of the P1 library revealed that each sequence typically contains only one to two amino acid substitutions, and a large fraction of mutations decreased in frequency, suggesting that most mutations identified in the P0 library are not tolerant under our stability screening pressure (SI Appendix, Fig. S6 A and C).
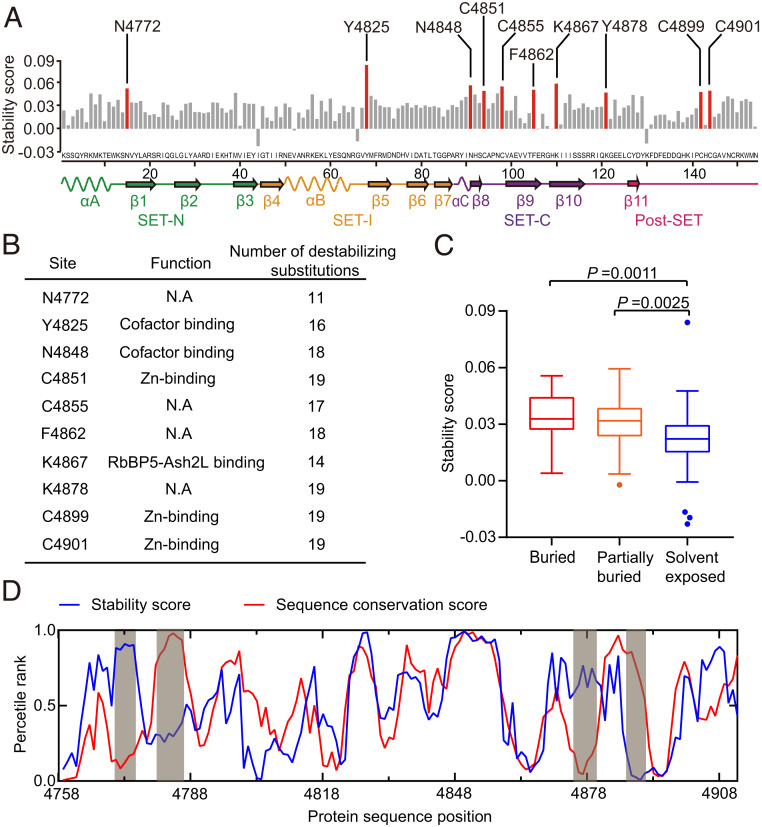
Frequency changes of wild-type amino acids in each site between the P0 and P1 libraries can reflect these residues’ differential contributions to the stability of MLL3SET. That is, residues which are intolerant of other amino acid substitutions can be assumed to somehow support protein stability, resulting in an increase of the frequency of the corresponding wild-type amino acids in the hit library. Seeking to characterize the contribution of each residue to stability, we defined a stability score (see Materials and Methods) based on our deep-sequencing data for the fold change in the frequency of the wild-type amino acid between the P0 and P1 libraries. We inferred that sites with a high stability score can likely only tolerate the wild-type amino acid at that particular position. In other word, in these sites, the wild-type amino acid is important for stability, and substitutions likely destabilized the protein. Then, we selected 10 top-ranking sites for further analyses (Fig. 4A). The wild-type residues at all of the 10 sites form extensive interaction networks with nearby residues and environmental water (SI Appendix, Fig. S7). Indeed, the FoldX (42) software predicted that, on average, 90% of the 19 possible substitutions at these sites are likely destabilizing mutations (Fig. 4B).
Fig. 4.
Site-specific mutational tolerance of the MLL3SET protein. (A) Stability score (defined as the frequency change of a wild-type amino acid before and after screening) for each site of MLL3SET. (B) Annotation of sites with high stability scores. The function of each site was experimentally confirmed in previous studies (41). The number of potential destabilizing mutations was predicted using the FoldX software. (C) Stability score of buried, partially buried, and solvent-exposed residues. Residues with relative solvent accessibility <0.05 were classified as buried, and residues with relative solvent accessibility >0.3 were classified as solvent exposed. Residues with relative solvent accessibility between 0.05 and 0.3 were classified as partially buried. P values were determined by two-tailed unpaired Student’s t tests. (D) Large-scale patterns of MLL3SET’s stability score and the TRX/MLL sequence conservation score. A moving average (five-site window) of the stability score and sequence conservation score is plotted over protein sequence positions. Percentile ranks are used to plot the two scores on the same axis. Data are the mean ± SD of four technical replicates.
It is generally believed that buried residues contribute more to protein stability than solvent-exposed residues because buried residues are often involved in protein core packing (43). Consistent with this idea, we found that buried residues are in general less tolerant to mutations than solvent-exposed residues (Fig. 4C). Of the 10 top-ranking sites, three (N4772, C4855, and C4901) are completely buried, and five (N4848, C4851, F4862, K4867, and C4899) are partially buried. Only two of the 10 top-ranking sites (Y4825 and K4878) are located on the surface of MLL3SET. Since half of these residues constitute the cofactor binding sites (Y4825 and N4848 for S-adenosyl-L-methionine [SAM] binding; C4851, C4899, and C4901 for Zn2+ binding), our findings suggest that cofactor binding in MLL3SET may largely contribute to its stability.
Residues important for stability are likely to be evolutionarily conserved (44). We defined a sequence conservation score (see Materials and Methods) based on the multiple sequence alignment of 744 SET domains of the TRX (trithorax)/MLL family. We found that the stability scores of each residue show a strong correlation with the calculated sequence conservation scores in most regions, despite four obvious local deviations (Fig. 4D). For the region of residues K4770-L4775 and R4874-G4879, we found that its conflicting low conservation score apparently results from sequence variation among different subcategories of the TRX/MLL family: members of the MLL3/MLL4 branch indeed contain similar sequences in this region (SI Appendix, Fig. S8). The other region comprising residues R4779-L4785 and D4885-F4890 have a low stability score and a high conservation score, which are known to contain residues important for SAM binding and the formation of the active center, respectively (41). Therefore, we suspected that selective pressures that made this region highly conserved may come from maintaining MLL’s physiological function rather than stability, so this region apparently illustrates an activity–stability tradeoff.
The Sequence-Stability Landscape of the MLL3SET Protein Facilitates Its Stability Evolution.
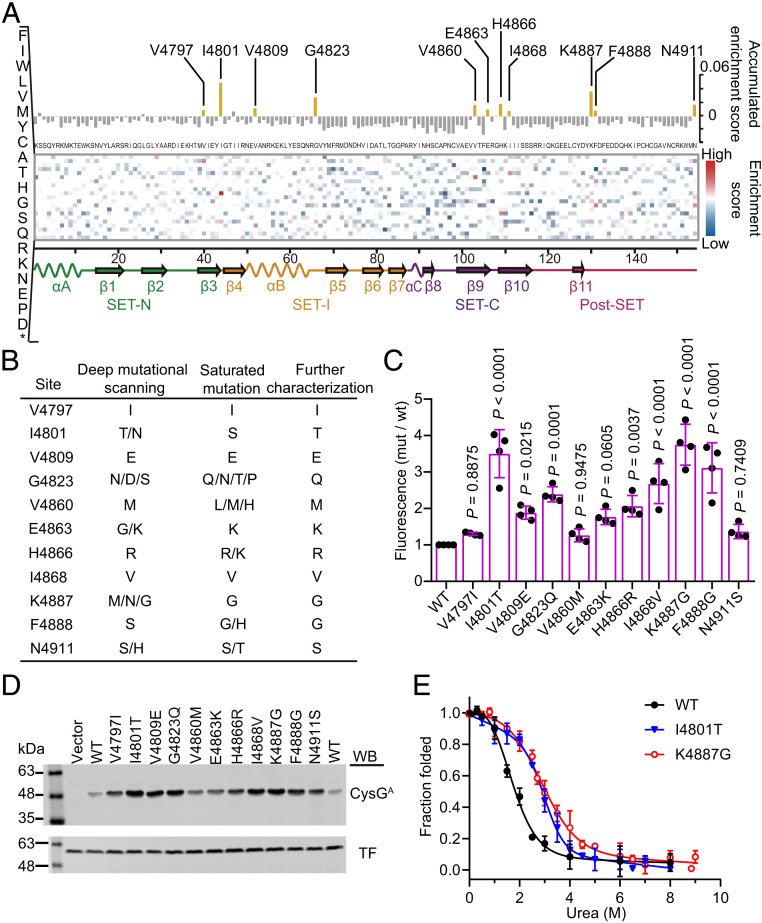
The large-scale mutational data generated by deep mutational scanning can also provide insights into another aspect of the sequence-stability landscape of MLL3SET: identification of stabilization hotspot residues [i.e., sites showing enrichment for mutations that are beneficial to MLL3SET stability; these stabilization hotspot residues can be inferred from frequency changes for each amino acid substitution before and after selection (45)]. We defined an enrichment score (see Materials and Methods) based on the frequencies of the corresponding substitution in the P0 and P1 libraries. In each site, substitutions with a high enrichment score are enriched after the stability screening. To analyze the effects of each amino acid substitution on the stability of MLL3SET, we organized our large-scale mutational dataset into a heat map that presents the enrichment score of every observed amino acid substitution at each site of the protein (Fig. 5A). In this map, most mutations showed a negative enrichment score, indicating that they are deleterious to MLL3SET stability. Only 9.1% of the mutations showed a positive enrichment score; these are mutations potentially beneficial for stability. Our results are consistent with observations from previous studies reporting that the large majority of mutations tend to reduce overall protein stability (45, 46).
Fig. 5.
The sequence-stability landscape of the MLL3SET protein. (A) Heat map of the enrichment scores for every observed amino acid substitution with the value for the accumulated enrichment score along the top, all possible mutations on the left axis, and the secondary structure elements displayed across the bottom. Unexamined amino acids and wild-type residues are colored in white. (B) Mutations enriched in the deep mutational scanning and identified from the saturation libraries. (C) Relative fluorescence intensities of cells expressing various fusion proteins inserted with the selected MLL3SET mutant variants. The fluorescence intensity of cells was normalized relative to cells expressing CysGA fused with wild-type MLL3SET. Data are the mean ± SD of four independent experiments. P values were determined by one-way ANOVA analysis with a Dunnett’s multiple comparison test. (D) Soluble expression level of the fusion protein containing the selected MLL3SET mutants. The soluble fraction of cell lysates was analyzed by immunoblotting with antibodies against CysGA and trigger factor (TF) protein that served as a loading control. The experiment was independently performed at least three times with similar results each time. Uncropped Western blots are presented in SI Appendix, Fig. S13. (E) Equilibrium urea-induced unfolding of wild-type MLL3SET and the selected variants. Data are the mean ± SD of three independent experiments.
Next, to identify stabilization hotspot residues and to compensate for the limitation of error-prone PCR to generate all possible single amino acid substitutions, we cataloged the enrichment score for every observed amino acid substitution at each site (Fig. 5A). Sites where some specific substitution can be enriched have high accumulated enrichment scores and are the potential hotspots for stabilization. On this basis, we selected the 11 top-ranking sites, generated complete saturation libraries for each, and screened those libraries using our CysGA tripartite system (SI Appendix, Fig. S9). This second round of screening uncovered 50% of the same potentially beneficial mutations as those from the deep mutational scanning; it also identified alternative substitutions in these sites (Fig. 5B). To rule out the effects of spontaneous genomic mutations that may also contribute to enhanced fluorescence in our screened cells, representative MLL3SET mutants were constructed, transformed into a fresh background strain, and verified for their ability to confer enhanced fluorescence (Fig. 5C). Western blotting of the fusion proteins also revealed that all of the mutants are more soluble than the wild-type fusion protein (Fig. 5D).
Seeking functional insights relating to the increased stabilization of the selected mutations, we used the FoldX force field and residue-interaction analysis based on the MLL3SET structure (PDB: 5F59) to assess the energy terms and possible interaction of each mutation (SI Appendix, Table S2 and Fig. S10). The V4797 and V4860 residues are buried in the structure, and the V4797I and V4860M mutations convert to larger hydrophobic side chains that apparently enhance hydrophobic interactions. The surface-exposed I4801T mutation may form extra hydrogen bonding with E4799 and the decrease in surface hydrophobicity, which is typically a benefit for protein stability (47). For V4809E and G4823Q, their contribution to stability may result from increased hydrogen bond formation. For H4866R, the arginine substitution apparently induces a relatively stronger electrostatic interaction with D4833 and D4832 to stabilize the protein. Finally, the N4911S, K4887G, and F4888G mutations may reduce entropy and/or Van der Waals’s torsional clashes. For the two mutations with the highest in vivo stability (I4801T and K4887G), we purified the recombinant MLL3SET variants and determined their thermodynamic stabilities using equilibrium denaturant titrations. Both mutants were more stable than wild-type MLL3SET, demonstrating 7.3 kJ/mol (I4801T) and 7.2 kJ/mol (K4887G) increases over ΔG°UN at 20 °C (Fig. 5E and Table 2). The fact that we obtained MLL3SET mutants with substantially improved stabilities through multiple library screens and site-specific mutational tolerance analysis clearly illustrates the utility of our platform combined with deep mutational scanning for selectively engineering highly stable proteins.
Table 2.
Equilibrium unfolding parameters of MLL3SET variants
| ΔG°UN (kJ ⋅ mol−1) | m (kJ ⋅ mol−1 ⋅ M−1) | Cm (M) | |
| Wild type | −6.7 ± 0.8 | 4.5 ± 0.9 | 1.39 ± 0.32 |
| I4801T | −14.0 ± 0.6 | 6.2 ± 1.2 | 3.03 ± 0.11 |
| K4887G | −13.9 ± 0.7 | 3.4 ± 0.6 | 2.81 ± 0.26 |
Values reported are the mean ± SEM. n = 3 independent samples.
Discussion
Protein stability often hinders the engineering of new or optimized protein functions. In this study, we developed a distinctive method for monitoring protein stability in vivo and applied this method for directed evolution of protein stability and for experimental delineation of the sequence-stability landscape. This method is based on the CysGA tripartite system consisting of a given POI and two segments of CysGA, which directly links the folding status of the POI to an easily detectable fluorescence phenotype. The method can be used with a wide range of POIs and is applicable to many proteins expressed in cytoplasm.
Many features of the CysGA protein render our biosensor to be a unique system. Given that almost all organisms can synthesis the tetrapyrrole intermediate and the CysGA substrate uroporphyrinogen III, our system should be applicable to a large diversity of experimental platforms and biological taxa without the extra introduction of substrate. Unlike methods based on mCherry or GFP that require oxygen to achieve maturation of their chromophores (48), our CysGA-based method does not rely on oxygen to produce of its fluorescent readout compound. Although preliminary, our initial testing of the tripartite CysGA system in anaerobic conditions does indicate that we can indeed monitor protein stability in this condition (SI Appendix, Fig. S11). Moreover, it was reported that a GFP-based reporter can suffer from a background autofluorescence of cells, commonly observed in the blue to green wavelength range (420 to 550 nm) overlapping with the emission spectra of GFP. This problem can be minimized with detection for red fluorescence using our CysGA biosensor.
Our design is conceptually similar to other tripartite fusion systems (49). Tripartite fusion design is advantageous compared to the “head-to-tail” design [e.g., the original GFP sensor (50)] in that it can avoid the false positive caused by proteolytic cleavage of a poorly folded POI and alternative translation because of frameshifting or the presence of internal cryptic ribosome-binding sites within the POI gene (24). Many of the available tripartite stability biosensors link cell viability to protein folding by engineering an antibiotic decomposing enzyme such as β-lactamase (51), aminoglycoside 3′-phosphotransferase (17), aminoglycoside 3″-adenylyltransferase (17), or nourseothricin acetyltransferase (17) or using an enzyme essential to cell growth under selective conditions such as orotate phosphoribosyl transferase (52) or DsbA (26). Linking protein folding to cell viability allows these biosensors to uncover rare stabilizing mutations, making them powerful selection techniques. For example, the β-lactamase tripartite reporter has been proved to be effective in identifying stabilized bovine pancreatic trypsin inhibitor variants (53), aggregation-resistant scFv sequences (54), inhibitors of IAPP aggregation (55), and in the custom tailoring of the cellular folding environment (26).
Unlike the β-lactamase sensor which functions in the periplasm of E. coli, our CysGA biosensor functions in the cytoplasm of E. coli, which is suitable for industrial-related and medically important proteins with multiple reduced cysteines (e.g., receptors with a cysteine-rich domain, thiol proteinases, and zinc finger proteins). In addition, while there may be many ways to gain intrinsic antibiotic resistance, the red fluorescent phenotype arising from CysGA activity cannot be complemented by any mutations from the E. coli chromosome to our best knowledge. Thus, screening based on cellular red fluorescence might be conceptually tighter than the selection based on antibiotic resistance.
As with any other folding reporters, our CysGA biosensor does have its certain limitations. First of all, once a certain stability threshold is reached, the majority of the POI is folded, and any additional increase in stability may not further enhance the foldability of the POI. In other words, the screening might be more beneficial when starting from a destabilizing protein than a highly stabilized one. Second, mutations other than those that affect the thermodynamic stability of the POI can also be enriched in the screening if they can affect CysGA activity, for example, those that impact the recognition by proteases or chaperones, or those changes the aggregation profile of CysGA when fused with it. Therefore, follow up inspection of the thermodynamic stability of the protein variants is always a necessary step.
We further envision that our current method can complement protein design efforts by offering a highly accessible and efficient means for evaluating the stability of designed proteins. Data for the stability landscape of a protein at single-residue resolution will enable systematic analyses of the relationships between sequence and stability, and large-scale studies can both help uncover general principles underlying protein stability and guide the rational design of protein activity while retaining adequate stability. We also inferred that residues with high stability scores apparently contribute to overall stability and may even serve as “global suppressors” which can buffer the effects of deleterious mutations (56). Accordingly, our data supports the recommendation that regions rich in such residues should almost certainly be retained during the construction of directed evolution libraries; we would anticipate that their retention should increase the robustness of the sequence against deleterious impacts from random mutations. Similar work using deep mutational scanning to assess protein stability will allow a deeper understanding of the fitness landscapes of proteins and generate a large number of training data sets that can be exploited by state-of-the-art technologies for protein design including machine learning (57).
In conclusion, we have developed a robust and reliable biosensor to monitor protein stability in vivo and demonstrated its application in directed evolution and deep mutational scanning. Again recalling the ubiquity of the CysGA substrate uroporphyrinogen III, we believe that our system can also be easily transferred to other advanced species and applied or engineered to detect more complicated proteins behaviors, such as screening for ligands that stabilize protein complexes. Thus, beyond illustrating successful engineering of stable proteins and revealing general insights about protein stability, our tripartite CysGA method will most likely facilitate protein stability–related research and engineering efforts in a wide diversity of living cells.
Materials and Methods
Detailed descriptions of all materials and methods are available in SI Appendix, Material and Methods. For the quantification of fluorescence, cells expressing the CysGA biosensor were resuspended in phosphate-buffered saline, and then the fluorescence of cells was measured using an automated plate reader (Synergy HTX Hybrid Reader, BioTek) by fixing the excitation filter at 380/20 nm and the emission filter at 600/40 nm. For the screening, the random libraries were transformed into E. coli and spread on LB plates containing 10 μM isopropyl β-D-1-thiogalactopyranoside and 200 μg/mL ampicillin. After incubation at 37 °C for 12 h, colonies exhibiting high fluorescence under the UV light were selected and sequenced.
Supplementary Material
Acknowledgments
We thank James C. A. Bardwell (University of Michigan) for helpful advice, Yong Chen (Chinese Academy of Sciences) for sharing plasmids and helpful suggestions on the study of MLL3SET, and Zixiao Xue and Yongxin Zheng (East China University of Science and Technology) for experimental support and helpful discussions. This work was supported by National Natural Science Foundation of China Grants 31870054, 31670802, and 31661143021 (to S.Q.), the Fundamental Research Funds for the Central Universities (Grant 22221818014 to S.Q.), and the Research Program of State Key Laboratory of Bioreactor Engineering (to S.Q.).
Footnotes
Competing interest statement: A patent application has been filed by East China University of Science and Technology for the technology disclosed in this publication.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2101618118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Goldenzweig A., Fleishman S. J., Principles of protein stability and their application in computational design. Annu. Rev. Biochem. 87, 105–129 (2018). [DOI] [PubMed] [Google Scholar]
- 2.Gershenson A., Gierasch L. M., Pastore A., Radford S. E., Energy landscapes of functional proteins are inherently risky. Nat. Chem. Biol. 10, 884–891 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stimple S. D., Smith M. D., Tessier P. M., Directed evolution methods for overcoming trade-offs between protein activity and stability. AIChE J. 66, e16814 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Clausen L., et al., Protein stability and degradation in health and disease. Adv. Protein Chem. Struct. Biol. 114, 61–83 (2019). [DOI] [PubMed] [Google Scholar]
- 5.Selivanova G., Wiman K. G., Reactivation of mutant p53: Molecular mechanisms and therapeutic potential. Oncogene 26, 2243–2254 (2007). [DOI] [PubMed] [Google Scholar]
- 6.Taylor J. P., Brown R. H. Jr., Cleveland D. W., Decoding ALS: From genes to mechanism. Nature 539, 197–206 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sun X., Chen W. D., Wang Y. D., β-Amyloid: The key peptide in the pathogenesis of Alzheimer’s disease. Front. Pharmacol. 6, 221 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stefanis L., α-Synuclein in Parkinson’s disease. Cold Spring Harb. Perspect. Med. 2, a009399 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gusella J. F., MacDonald M. E., Molecular genetics: Unmasking polyglutamine triggers in neurodegenerative disease. Nat. Rev. Neurosci. 1, 109–115 (2000). [DOI] [PubMed] [Google Scholar]
- 10.Silva C., Martins M., Jing S., Fu J., Cavaco-Paulo A., Practical insights on enzyme stabilization. Crit. Rev. Biotechnol. 38, 335–350 (2018). [DOI] [PubMed] [Google Scholar]
- 11.Moore S. J., MacDonald J. T., Freemont P. S., Cell-free synthetic biology for in vitro prototype engineering. Biochem. Soc. Trans. 45, 785–791 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kontermann R. E., Half-life extended biotherapeutics. Expert Opin. Biol. Ther. 16, 903–915 (2016). [DOI] [PubMed] [Google Scholar]
- 13.Dou J., et al., De novo design of a fluorescence-activating β-barrel. Nature 561, 485–491 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Magliery T. J., Protein stability: Computation, sequence statistics, and new experimental methods. Curr. Opin. Struct. Biol. 33, 161–168 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Galán A., et al., Library-based display technologies: Where do we stand? Mol. Biosyst. 12, 2342–2358 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Lindman S., Hernandez-Garcia A., Szczepankiewicz O., Frohm B., Linse S., In vivo protein stabilization based on fragment complementation and a split GFP system. Proc. Natl. Acad. Sci. U.S.A. 107, 19826–19831 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Malik A., Mueller-Schickert A., Bardwell J. C. A., Cytosolic selection systems to study protein stability. J. Bacteriol. 196, 4333–4343 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maxwell K. L., Mittermaier A. K., Forman-Kay J. D., Davidson A. R., A simple in vivo assay for increased protein solubility. Protein Sci. 8, 1908–1911 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Warren M. J., et al., Gene dissection demonstrates that the Escherichia coli cysG gene encodes a multifunctional protein. Biochem. J. 302, 837–844 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stroupe M. E., Leech H. K., Daniels D. S., Warren M. J., Getzoff E. D., CysG structure reveals tetrapyrrole-binding features and novel regulation of siroheme biosynthesis. Nat. Struct. Biol. 10, 1064–1073 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Bryant D. A., Hunter C. N., Warren M. J., Biosynthesis of the modified tetrapyrroles-the pigments of life. J. Biol. Chem. 295, 6888–6925 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Roessner C. A., Use of cobA and cysGA as red fluorescent indicators. Methods Mol. Biol. 183, 19–30 (2002). [DOI] [PubMed] [Google Scholar]
- 23.Stefan Wildt U., Deuschle, cobA, a red fluorescent transcriptional reporter for Escherichia coli, yeast, and mammalian cells. Nat. Biotechnol. 17, 1175–1178 (2000). [DOI] [PubMed] [Google Scholar]
- 24.Ren C., Wen X., Mencius J., Quan S., Selection and screening strategies in directed evolution to improve protein stability. Bioresour. Bioprocess. 6, 53 (2019). [Google Scholar]
- 25.Yu Y., Lutz S., Circular permutation: A different way to engineer enzyme structure and function. Trends Biotechnol. 29, 18–25 (2011). [DOI] [PubMed] [Google Scholar]
- 26.Quan S., et al., Genetic selection designed to stabilize proteins uncovers a chaperone called Spy. Nat. Struct. Mol. Biol. 18, 262–269 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lo W. C., Lee C. C., Lee C. Y., Lyu P. C., CPDB: A database of circular permutation in proteins. Nucleic Acids Res. 37, D328–D332 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ribeiro E. A. Jr., Ramos C. H., Circular permutation and deletion studies of myoglobin indicate that the correct position of its N-terminus is required for native stability and solubility but not for native-like heme binding and folding. Biochemistry 44, 4699–4709 (2005). [DOI] [PubMed] [Google Scholar]
- 29.Capaldi A. P., Kleanthous C., Radford S. E., Im7 folding mechanism: Misfolding on a path to the native state. Nat. Struct. Biol. 9, 209–216 (2002). [DOI] [PubMed] [Google Scholar]
- 30.Chun S. Y., Strobel S., Bassford P. Jr., Randall L. L., Folding of maltose-binding protein. Evidence for the identity of the rate-determining step in vivo and in vitro. J. Biol. Chem. 268, 20855–20862 (1993). [PubMed] [Google Scholar]
- 31.Chiti F., et al., Mutational analysis of acylphosphatase suggests the importance of topology and contact order in protein folding. Nat. Struct. Biol. 6, 1005–1009 (1999). [DOI] [PubMed] [Google Scholar]
- 32.Mayer S., Rüdiger S., Ang H. C., Joerger A. C., Fersht A. R., Correlation of levels of folded recombinant p53 in Escherichia coli with thermodynamic stability in vitro. J. Mol. Biol. 372, 268–276 (2007). [DOI] [PubMed] [Google Scholar]
- 33.Espargaró A., Sabaté R., Ventura S., Kinetic and thermodynamic stability of bacterial intracellular aggregates. FEBS Lett. 582, 3669–3673 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Akter R., et al., Islet amyloid polypeptide: Structure, function, and pathophysiology. J. Diabetes Res. 2016, 2798269 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Abedini A., Meng F., Raleigh D. P., A single-point mutation converts the highly amyloidogenic human islet amyloid polypeptide into a potent fibrillization inhibitor. J. Am. Chem. Soc. 129, 11300–11301 (2007). [DOI] [PubMed] [Google Scholar]
- 36.Fox A., et al., Selection for nonamyloidogenic mutants of islet amyloid polypeptide (IAPP) identifies an extended region for amyloidogenicity. Biochemistry 49, 7783–7789 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wiltzius J. J., Sievers S. A., Sawaya M. R., Eisenberg D., Atomic structures of IAPP (amylin) fusions suggest a mechanism for fibrillation and the role of insulin in the process. Protein Sci. 18, 1521–1530 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bakou M., et al., Key aromatic/hydrophobic amino acids controlling a cross-amyloid peptide interaction versus amyloid self-assembly. J. Biol. Chem. 292, 14587–14602 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Koo B. W., Miranker A. D., Contribution of the intrinsic disulfide to the assembly mechanism of islet amyloid. Protein Sci. 14, 231–239 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Fowler D. M., Fields S., Deep mutational scanning: A new style of protein science. Nat. Methods 11, 801–807 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Li Y., et al., Structural basis for activity regulation of MLL family methyltransferases. Nature 530, 447–452 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Delgado J., Radusky L. G., Cianferoni D., Serrano L., FoldX 5.0: Working with RNA, small molecules and a new graphical interface. Bioinformatics 35, 4168–4169 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou H., Zhou Y., Quantifying the effect of burial of amino acid residues on protein stability. Proteins 54, 315–322 (2004). [DOI] [PubMed] [Google Scholar]
- 44.Sternke M., Tripp K. W., Barrick D., Consensus sequence design as a general strategy to create hyperstable, biologically active proteins. Proc. Natl. Acad. Sci. U.S.A. 116, 11275–11284 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Romero P. A., Tran T. M., Abate A. R., Dissecting enzyme function with microfluidic-based deep mutational scanning. Proc. Natl. Acad. Sci. U.S.A. 112, 7159–7164 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Tokuriki N., Stricher F., Schymkowitz J., Serrano L., Tawfik D. S., The stability effects of protein mutations appear to be universally distributed. J. Mol. Biol. 369, 1318–1332 (2007). [DOI] [PubMed] [Google Scholar]
- 47.Pakula A. A., Sauer R. T., Reverse hydrophobic effects relieved by amino-acid substitutions at a protein surface. Nature 344, 363–364 (1990). [DOI] [PubMed] [Google Scholar]
- 48.Coralli C., Cemazar M., Kanthou C., Tozer G. M., Dachs G. U., Limitations of the reporter green fluorescent protein under simulated tumor conditions. Cancer Res. 61, 4784–4790 (2001). [PubMed] [Google Scholar]
- 49.Sachsenhauser V., Bardwell J. C., Directed evolution to improve protein folding in vivo. Curr. Opin. Struct. Biol. 48, 117–123 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Waldo G. S., Standish B. M., Berendzen J., Terwilliger T. C., Rapid protein-folding assay using green fluorescent protein. Nat. Biotechnol. 17, 691–695 (1999). [DOI] [PubMed] [Google Scholar]
- 51.Foit L., et al., Optimizing protein stability in vivo. Mol. Cell 36, 861–871 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Bjerre B., et al., Improving folding properties of computationally designed proteins. Protein Eng. Des. Sel. 32, 145–151 (2019). [DOI] [PubMed] [Google Scholar]
- 53.Foit L., et al., Genetic selection for enhanced folding in vivo targets the Cys14-Cys38 disulfide bond in bovine pancreatic trypsin inhibitor. Antioxid. Redox Signal. 14, 973–984 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ebo J. S., et al., An in vivo platform to select and evolve aggregation-resistant proteins. Nat. Commun. 11, 1816 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Saunders J. C., et al., An in vivo platform for identifying inhibitors of protein aggregation. Nat. Chem. Biol. 12, 94–101 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tokuriki N., Tawfik D. S., Stability effects of mutations and protein evolvability. Curr. Opin. Struct. Biol. 19, 596–604 (2009). [DOI] [PubMed] [Google Scholar]
- 57.Yang K. K., Wu Z., Arnold F. H., Machine-learning-guided directed evolution for protein engineering. Nat. Methods 16, 687–694 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.