Significance
Expression of keratin 17 (K17), an intermediate filament-forming cytoskeletal protein, is induced in a broad range of human carcinomas and has prognostic value for a growing number of tumor types. We report here that K17 is induced in response to DNA damage and that a nuclear pool of K17 regulates the immediate response to such damage, with an associated impact on cell survival. Impairing the nuclear pool of K17 markedly attenuates tumor initiation in mouse skin. Our finding adds a role for nuclear K17 in the DNA damage response (DDR) and further substantiates the significance of K17 expression and function during tumorigenesis.
Keywords: keratin, DNA damage, skin, tumor, intermediate filament
Abstract
High levels of the intermediate filament protein keratin 17 (K17) are associated with poor prognoses for several human carcinomas. Studies in mouse models have shown that K17 expression is positively associated with growth, survival, and inflammation in skin and that lack of K17 delays onset of tumorigenesis. K17 occurs in the nucleus of human and mouse tumor keratinocytes where it impacts chromatin architecture, gene expression, and cell proliferation. We report here that K17 is induced following DNA damage and promotes keratinocyte survival. The presence of nuclear K17 is required at an early stage of the double-stranded break (DSB) arm of the DNA damage and repair (DDR) cascade, consistent with its ability to associate with key DDR effectors, including γ-H2A.X, 53BP1, and DNA-PKcs. Mice lacking K17 or with attenuated K17 nuclear import showed curtailed initiation in a two-step skin carcinogenesis paradigm. The impact of nuclear-localized K17 on DDR and cell survival provides a basis for the link between K17 induction and poor clinical outcomes for several human carcinomas.
Keratin 17 (K17), a type I keratin intermediate filament protein, is robustly up-regulated in a broad array of epithelial tumors (1), and its expression at higher levels correlates with poor prognoses for breast (2), cervical (3), oral (4), ovarian (5), and pancreatic (6) cancers. In mouse models for tumorigenesis, genetic ablation of K17 delays the inception and/or attenuates tumor growth in several experimental paradigms, including Gli2tg- and HPV16tg-driven carcinomas in the skin (7, 8) and HPV16tg-driven cervical carcinoma (9). K17 occurs in the nucleus and interacts with autoimmune regulator (Aire) and hnRNPK, affecting tumor-associated inflammation (8, 10). Nuclear K17 regulates nuclear morphology and chromatin architecture and was identified to be associated with a large number of nuclear envelope and nucleoplasmic proteins involved in nuclear shape, chromatin organization, RNA processing, and gene expression (11). Nuclear K17 also facilitates degradation of cell cycle suppressor p27KIP1 during cervical cancer progression (12). Despite this significant progress, how K17 relates to tumorigenesis and whether it acts at the tumor initiation stage remain unclear.
Many classes of genetic alterations and associated molecular and functional changes have been recognized as hallmarks of the tumor state. Oncogenic driver mutations and genome instability each play essential roles in the inception and maintenance of the transformed state (13). Cells are normally equipped with a prominent array of mechanisms to sense and repair insults to their genome, and genomic instability is a direct consequence of faulty DNA damage response (DDR) (14). DNA repair processes protect the genome from endogenous or exogenous damage, ranging from single base alterations to broader lesions affecting multiple strands of DNA (15). Within the DDR cascade, single-stranded breaks (SSBs) or double-stranded breaks (DSBs) in DNA typically trigger distinct signaling cascades that allow cells to detect the type and extent of damage and then direct cell fate by either repairing the genetic insult or inducing apoptosis or senescence (16, 17). Given K17’s ability to directly impact keratinocyte survival and growth, the recent discovery of K17’s presence in the nucleus, and growing evidence that actin and other cytoskeletal proteins are involved in DNA damage-associated signaling (18, 19), we set out to investigate whether nuclear K17 regulates DDR as a component of early stage skin tumorigenesis.
Results
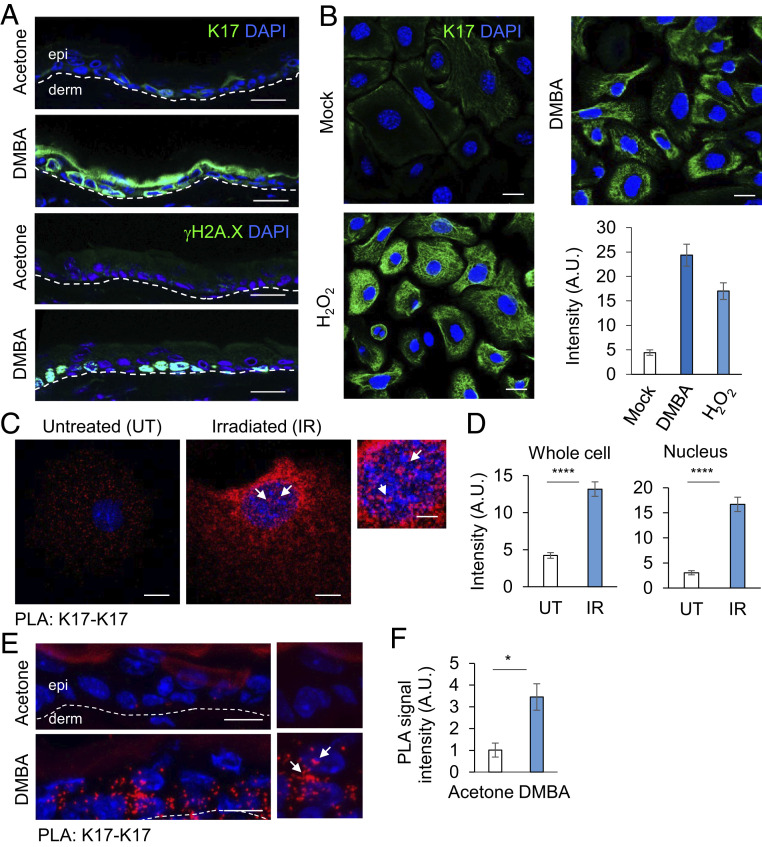
First, to evaluate whether K17 levels change in response to DNA damage, we treated mouse ear skin with 7,12-Dimethylbenz(a)anthracene (DMBA) (20 μg), a DNA damaging agent that forms DMBA-DNA adducts, or acetone (vehicle control). At 2 h after two topical treatments with DMBA, 24 h apart, K17 was robustly induced in a patchy fashion within the interfollicular epidermis (Fig. 1 A, Top), along with γH2A.X induction, a faithful readout for DNA damage (Fig. 1 A, Bottom). Skin keratinocytes from newborn mice in primary culture, treated with DMBA (1 μg, 12 h) or H2O2 (100 μM, 30 min) also showed increased K17 and γH2A.X levels by immunostaining (Fig. 1B and SI Appendix, Fig. S1 A and B), suggesting that K17 induction in response to DNA damage is keratinocyte-autonomous. Next, we used proximity ligation assays (PLAs) using two K17 antibodies raised in different species to evaluate whether the nuclear pool of K17 is sensitive to DNA damage. Fluorescence-based confocal imaging showed that a robust increase of the K17-K17 PLA signal occurs both in the cytoplasm and the nucleus of cultured keratinocytes from newborn mice after DNA damage using irradiation, 5 Gy X-ray (Fig. 1 C and D). This response to DDR may be K17-specific as there was no significant difference in the expression status of K14 (SI Appendix, Fig. S1 C and D), a highly related type 1 keratin (>83% sequence identical to K17). We performed subcellular fractionation of A431 human tumor keratinocytes in untreated and irradiated conditions and observed that there is a modest increase in nuclear K17 levels post DNA damage (SI Appendix, Fig. S1 E and F). We also treated 2-mo-old mice with DMBA or acetone on ear skin, as described above, and used PLA to evaluate nuclear and cytoplasmic K17 induction in vivo. K17 was induced as a result of DNA damage in the interfollicular epidermis, showing a presence in the nucleus along with cytoplasm (Fig. 1 E and F and SI Appendix, Fig. S1G).
Fig. 1.
K17 is induced during DNA damage. (A) K17 is induced in vivo upon DNA damage insult. 2-mo-old Krt17+/− mice were treated topically with 20 μg of DMBA dissolved in acetone on right ear and acetone only on left ear (two times, 24 h apart). Ear tissue was harvested 2 h after the second treatment, and immunostaining was performed for K17 or γH2A.X (green) expression status. (Scale bars: 20 μm.) Epidermis (epi) and dermis (derm) sides of the skin section are separated by a dashed line. n ≥ 3 mice per genotype. (B) K17 induction in mouse keratinocytes upon DNA damage. Mouse keratinocytes isolated from newborn pups in primary culture were treated with 1 μg of DMBA (12 h) or 100 μM H2O2 (30 min), fixed with 4% paraformaldehyde, permeabilized with 0.2% triton, and immunostained for K17 (green). (Scale bars: 20 μm.) Signal intensity per cell is quantified in the graph to the Right using ImageJ and Excel. Error bars are SEM. n = 3 replicates. (C) DNA damage induced K17 is both nuclear and cytoplasmic. Mouse keratinocytes isolated from newborn pups in primary culture were treated with 5 Gy X-ray irradiation and fixed after 3 h. PLA was performed using antibodies against K17 from two different species. The red dots indicate positive staining for K17. Krt17−/− cells were used as negative control. The representative data shown are single z-plane images in which DAPI (nuclei) of the cells are in focus to score for nuclear K17 punctae. (Scale bar: 10 μm.) The image on the Extreme Right is a zoomed section of the representative image for irradiated cells. (Scale bars: 5 μm.) (D) PLA: K17-K17 quantification. Single z-plane images were used to measure intensity through cell or only in DAPI stained nuclear region using ImageJ. Error bars are SEM. n = 3 replicates. (E) PLA for K17 induction post DMBA treatment in vivo. 2-mo-old mice were treated topically on the dorsal ear surface (20 μg of DMBA dissolved in acetone on right ear, acetone on left ear) two times, 24 h apart. Ear tissues were harvested 2 h after the second treatment and embedded in OCT. Tissue sections were processed for PLA using antibodies against K17 from two different species. The red dots indicate positive staining for K17. Krt17−/− mice sections were used as negative control. The representative images shown are single z-plane images in which most DAPI staining (nuclei) in the interfollicular epidermis is in focus to score for nuclear K17 punctae. (Scale bar: 20 μm.) n = 3 mice. (F) PLA: K17-K17 quantification in vivo. Maximum Intensity Projection images were generated using the Zen software, and, in every tissue section imaged, interfollicular epidermis was chosen as the ROI using ImageJ to measure fluorescence intensity. Error bars are SEM. White arrows in C and E denote areas of interest. A.U., arbitrary unit. *P < 0.05, ****P < 0.0001.
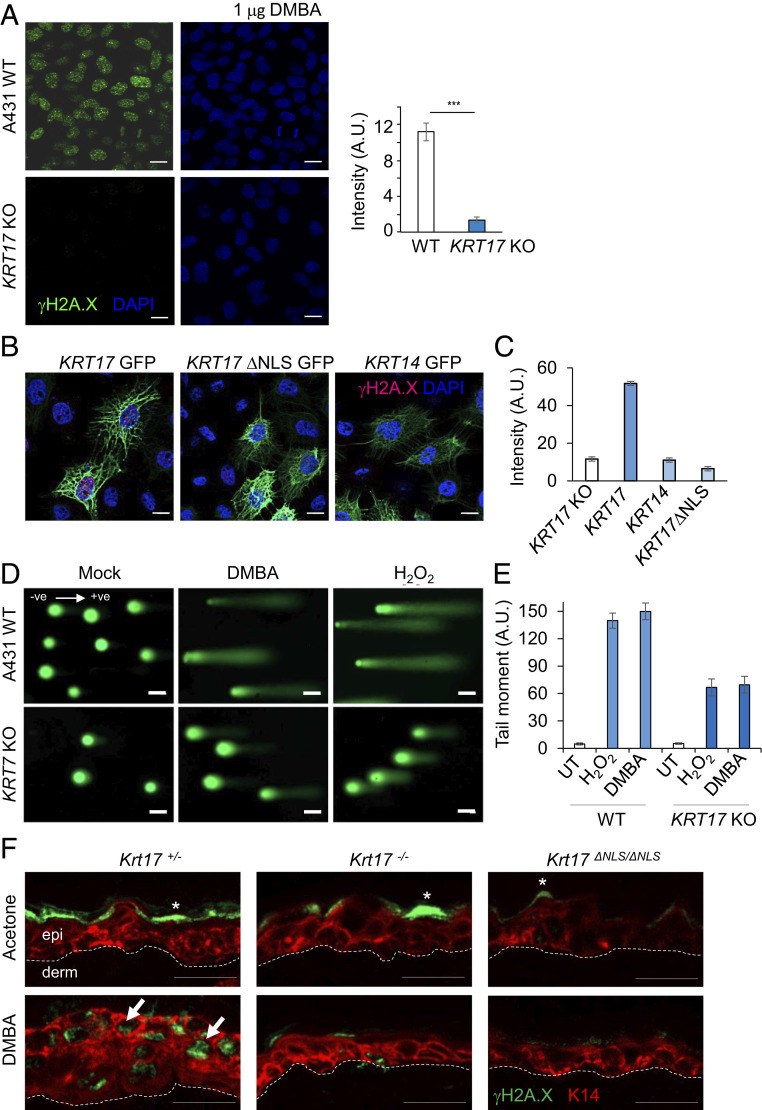
Complete loss of K17, or selective loss of its nuclear pool, markedly attenuated the extent of the DNA damage response. A431 human tumor keratinocytes, parental and KRT17 knockout (KO) (8), were treated with DMBA as described above. KRT17 KO A431 cells showed markedly reduced γH2A.X after treatment (Fig. 2A). This abnormal response to DNA damage was rescued, in a cell-autonomous fashion, by transfection-based reexpression of K17 in KRT17 KO A431 cells. By contrast, reexpression of a nuclear localization signal (NLS) mutant form of K17 (K399A, K400A) (8) or of K14 could not restore DMBA-induced γH2A.X levels in KRT17 KO A431 cells (Fig. 2 B and C). These findings suggest a direct correlation between nuclear K17 presence and DDR. We next performed alkaline comet assays, a gold standard approach to determine the extent of DNA fragmentation. Consistent with the observed K17-dependent reduction in γH2A.X levels in KRT17 KO A431 cells relative to wild-type (WT) parental cells following treatment with DMBA (12 h) or H2O2 (30 min), DNA damage-induced comet tails were shorter and less prominent in KRT17 KO A431 cells compared to WT parental cells (Fig. 2 D and E). The observed reductions in the DDR and in physical DNA fragmentation in KRT17 KO A431 cells in response to DMBA or H2O2 treatment suggest K17 involvement in the sensing of DNA damage, regulation of DDR kinetics, and/or chromatin organization to reduce accessibility to these damaging agents. Of note, nuclear K17 was recently reported to influence chromatin organization and associate with a number of chromatin binding proteins (11). To further evaluate the role of nuclear-localized K17 in skin tissue in situ, we exploited a mouse strain carrying K399A and K401A mutations at the Krt17 locus in its germline that renders the NLS sequence dysfunctional (Krt17ΔNLS/ΔNLS) (11). Krt17ΔNLS/ΔNLS mutant mice, along with mice null for Krt17 (Krt17−/−), failed to show γH2A.X induction in ear skin samples in response to DMBA treatment, compared to K17-expressing mice (Fig. 2F). These findings establish that the nuclear pool of K17 regulates DDR in both human and mouse keratinocytes.
Fig. 2.
Loss of K17 attenuates the DNA damage response. (A) γH2A.X induction is dependent on K17 after DNA damage. A431 skin tumor keratinocytes (both parental and KRT17 KO) were treated with 1 μg of DMBA (12 h) fixed and immunostained for γH2A.X. Signal intensity per cell is quantified in the graph at the Right using ImageJ and Excel. Error bars are SEM. n = 3 replicates. (Scale bars, 20 μm.) (B) Reexpression of K17, but not K17 lacking a nuclear localization sequence or of K14, restores the DDR in A431 tumor keratinocytes. KRT17 KO A431 skin tumor keratinocytes were transiently transfected with GFP-K17 or GFP-K17ΔNLS or GFP-K14, treated with 1 μg of DMBA (12 h), and fixed and immunostained for γH2A.X. Expression of GFP-K17, but not GFP-K17ΔNLS or GFP-K14, restored the ability of KRT17 KO cells to induce γH2A.X expression following DMBA treatment. (Scale bars: 20 μm.) (C) Signal intensity of γH2A.X per cell after reexpression of GFP-K17, GFP-K17ΔNLS, or GFP-K14 in KRT17 KO A431 skin tumor keratinocytes is quantified in the graph using ImageJ and Excel. Error bars are SEM. n = 3 replicates. (D) Comet Assay in response to 1 μg of DMBA (12 h) or 100 μM H2O2 (30 min) in A431 parental and KRT17 KO lines. Cells lacking K17 showed shorter comet tails. (Scale bars: 20 μm.) ve, direction of electric current ‘negative’ to ‘positive.’ (E) Tail moments from comet assay in Fig. 1D are quantified in the graph. Error bars are SEM. n = 3 replicates. UT, untreated. (F) K17 dependence on γH2A.X induction in vivo. The 2-mo-old Krt17+/−, Krt17−/−, and Krt17ΔNLS/ΔNLS mice were treated topically on the dorsal ear surface (20 μg of DMBA dissolved in acetone on right ear, acetone on left ear) two times, 24 h apart. Ear tissues were harvested 2 h after the second treatment and embedded in OCT. Tissue sections were immunostained for γH2A.X (green) and K14 (red). DAPI is shown in blue. Arrows indicate cells showing γH2A.X induction, and asterisks (*) indicate nonspecific staining. Induction of γH2A.X was attenuated in the absence of K17 as well as nuclear K17. (Scale bars: 20 μm.) n ≥ 3 mice per genotype.
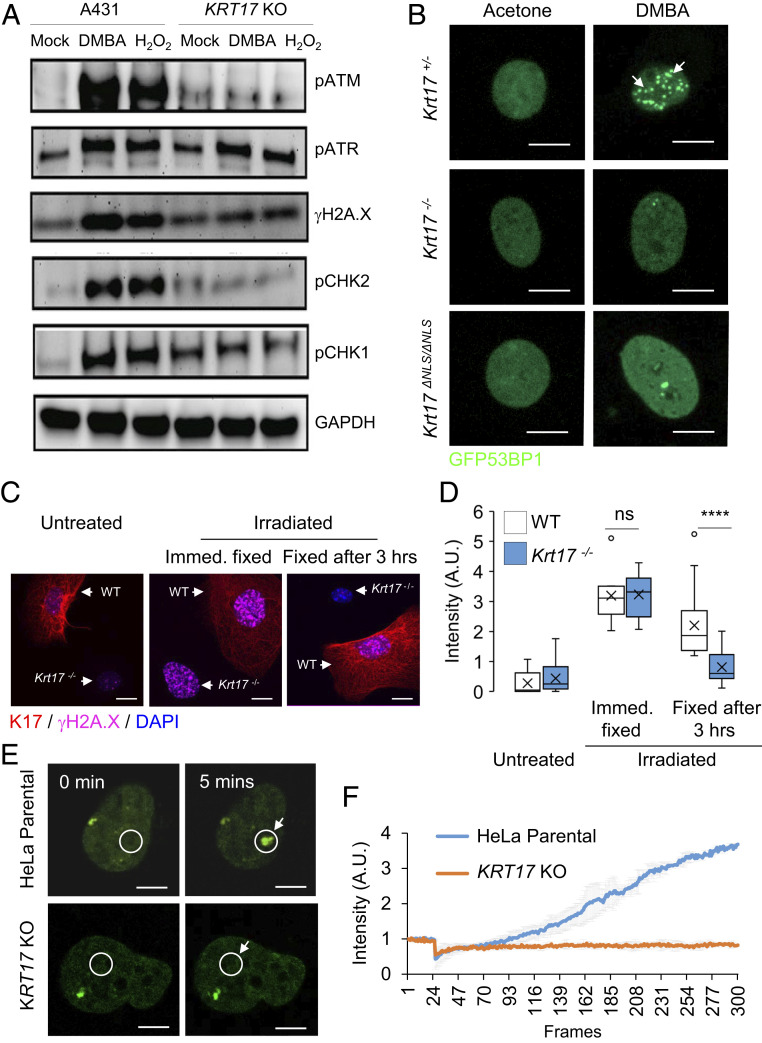
DDR consists of an elaborate homeostatic response that entails the engagement of several biochemical pathways and their molecular effectors (16). Next, we broadened the analysis of DDR effectors via Western blot analysis of total protein extracts from A431 human keratinocytes, parental and KRT17 KO, treated with DMBA or H2O2. We observed that, in addition to γH2A.X, the induction of key effectors of DSB DNA repair, phosphorylated ATM and CHK2 (pATM, pCHK2), were markedly diminished in KRT17 KO relative to parental cells (Fig. 3A and SI Appendix, Table S1). Key effectors of the repair of SSB, pATR, and pCHK1 were altered to a significantly lesser degree in the absence of K17 (Fig. 3A and SI Appendix, Table S1). Next, we focused on 53BP1, a faithful marker for DSBs involved in the nonhomologous end joining (NHEJ) pathway of DNA repair (20). A transfected GFP-53BP1 reporter in mouse keratinocytes in primary culture showed robust foci formation in response to DMBA treatment in Krt17+/− (control) keratinocytes, as expected (21), whereas this phenomenon was significantly reduced in Krt17−/− and Krt17ΔNLS/ΔNLS keratinocytes (Fig. 3B and SI Appendix, Fig. S2A). Similar findings were obtained when Krt17+/− and Krt17−/− keratinocytes were mixed and transfected with the reporter construct for the assay in which untreated cells were compared to damaged cells (SI Appendix, Fig. S2B). We next explored the dynamics of DSB recognition and repair utilizing irradiation as a DNA damage modality as it induces instant DSBs, allowing for kinetic analyses. Immediately after irradiation (X-ray; 5 Gy), there was γH2A.X staining in mouse keratinocytes in primary culture from Krt17+/− (control) and Krt17−/− mice. As soon as 3 h after irradiation, however, Krt17−/− keratinocytes showed significantly lesser γH2A.X staining whereas control Krt17+/− keratinocytes did not (Fig. 3 C and D). Moreover, the absence of K17 markedly reduced 53BP1 foci formation immediately after irradiation, suggesting that K17’s involvement occurs very early during DSBs (SI Appendix, Fig. S2C). Live cell imaging studies were next carried out to compare the fate of transfected GFP-53BP1 in KRT17-expressing and KRT17-null HeLa cells in real time after focal DNA damage induced by irradiation using a 405-nm laser under the microscope. In parental, KRT17-expressing HeLa cells, GFP-positive 53BP1 foci start to appear at the site of irradiation within seconds. Such accumulation of GFP-53BP1 did not occur within the same time scale in KRT17 KO HeLa cells (Fig. 3 E and F and Movies S1A and S1B). Virtually identical results were obtained when comparing Krt17+/− and Krt17−/− mouse skin keratinocytes in primary culture (SI Appendix, Fig. S2D). Krt17ΔNLS/ΔNLS keratinocytes also showed attenuated GFP-53BP1 foci formation (SI Appendix, Fig. S2D).
Fig. 3.
K17 impacts the regulation of key DNA damage response effectors. (A) K17 impacts the canonical DNA damage response pathway. Western blot analysis on total protein extracts from A431 parental and KRT17 KO cells for DDR-associated signaling proteins. (B) Mouse keratinocytes isolated from newborn pups, Krt17+/−, Krt17−/−, and Krt17ΔNLS/ΔNLS, were transfected with the GFP-53BP1 construct and treated with 1 μg of DMBA (12 h). Cells were fixed and imaged for 53BP1 foci formation. GFP-53BP1 is in green. Krt17+/− cells showed foci formation of 53BP1 post damage. (Scale bars: 10 μm.) n = 3 replicates. (C) Mouse keratinocytes isolated from WT and Krt17−/− newborn pups were mixed together and treated with 5 Gy X-ray irradiation and fixed either immediately or after 3 h to immunostain for γH2A.X. In the representative confocal images (maximum projection image of z stacks) K17 is red, γH2A.X is magenta, and DAPI is blue. Each representative image shows a single field containing both Krt17 expressing cell (red filament staining) and Krt17−/− cell (DAPI alone, no red filament staining). (Scale bars: 20 μm.) (D) Signal intensities of γH2A.X staining from Fig. 1C were quantified using ImageJ and Excel. Error bars are SEM. n = 3 replicates. (E) 53BP1 recruitment in response to laser-induced DSB visualized through live cell imaging requires K17. HeLa parental and KRT17 KO cells were transfected with the GFP-53BP1 construct and imaged under live conditions using a Zeiss LSM880 microscope. A 405-nm laser was used to create DSB at an ROI (white circle) and monitored for GFP-53BP1 foci formation during a course of 5 min. The Top shows the nucleus of a parental HeLa cell, and Bottom shows the nucleus of a HeLa KRT17 KO cell. (Scale bars: 5 μm.) (F) Signal intensities at the ROI during time lapse imaging represented in Fig. 1E were quantified using ImageJ and Excel for the graph. Error bars are SD for measurements across three single cell nuclei per genotype. White arrows in B, C, and E draw attention to area of interest. ns P > 0.05 and ****P < 0.0001.
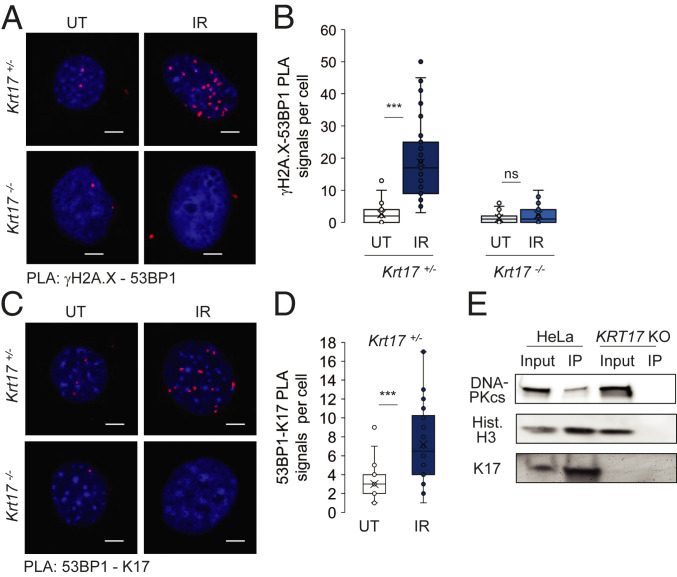
Further analysis on the dynamics of DDR foci using PLA showed that γH2A.X and 53BP1 were recruited to common intranuclear foci post DNA damage in Krt17+/− (control) keratinocytes, as expected, while this process was markedly lessened in Krt17−/− keratinocytes (Fig. 4 A and B). We also identified, using PLA, that 53BP1 and nuclear K17 were in proximity to a significantly higher degree following DNA damage, relative to baseline (Fig. 4 C and D). Additional findings further substantiate a link between K17 and the DSB arm of DDR. DNA-dependent protein kinase, catalytic subunit (DNA-PKcs), which is required for NHEJ (22), was identified as a nuclear K17 interacting protein (Fig. 4E). Of note, Aire, a transcriptional regulator previously identified to be a nuclear K17 interacting protein (8), is a DNA-PKcs interacting partner and is known to have a role during DNA repair and associated transcriptional regulations (23) (SI Appendix, Table S2). Other DDR proteins, like γH2A.X, were also identified to be coimmunoprecipitated with nuclear K17 (SI Appendix, Table S2). These findings establish a molecular paradigm where nuclear K17, either through direct or indirect interaction, is found within the same protein complex as several DDR effector proteins to facilitate an appropriate response. In the GFP-53BP1 reporter transfected cells, we were able to identify K17 as a protein associated with 53BP1 through immunoprecipitation (IP) using antibody against GFP, suggesting nuclear K17 is not only present in close proximity to 53BP1, as evident from PLA (Fig. 4C), but also is part of a complex involving 53BP1 at DDR foci (SI Appendix, Fig. S3A). Similar to previous outcomes on dysfunctional DDR in the absence of K17, the fate of phospho-KAP1, which is involved in chromatin decondensation during DSBs, was also abnormal (24). Cells lacking K17 showed pKAP1 staining prior to exposure to DNA damage, and the dynamics was also altered (SI Appendix, Fig. S3 B and C). These findings suggest that K17 plays a critical role, manifested very early on, in the DSB arm of the DDR program.
Fig. 4.
Keratin 17 regulates DDR foci formation. (A) PLA for γH2A.X and 53BP1 proximity status in mouse keratinocytes isolated from newborn pups in primary culture (Krt17+/− and Krt17−/−) before and after treatment with 5 Gy X-ray irradiation and fixed after 3 h. (Scale bars: 5 μm.) (B) Numbers of PLA signal dots per cell from PLA: γH2A.X-53BP1 represented in A were counted from maximum intensity projection images of z-stacked images. Error bars are SEM. n = 3 replicates. UT, untreated; IR, irradiated. (C) PLA for 53BP1 and K17 proximity status in mouse keratinocytes isolated from newborn pups in primary culture (Krt17+/−) before and after treatment with 5 Gy X-ray irradiation and fixed after 3 h. Krt17−/− cells were used as negative control. (Scale bars: 5 μm.) (D) Numbers of PLA signal dots per cell from PLA: 53BP1-K17 represented in C were counted from maximum intensity projection images of z-stacked images. Error bars are SEM. n = 3 replicates. UT, untreated; IR, irradiated. (E) Subcellular fractionation to collect nuclear lysate followed by K17 coimmunoprecipitation (co-IP) and Western blotting shows DNA-PKcs and nuclear K17 interaction. Hist-H3 serves as a positive control. Co-IP is not observed in KRT17 KO lysates. ***P < 0.001.
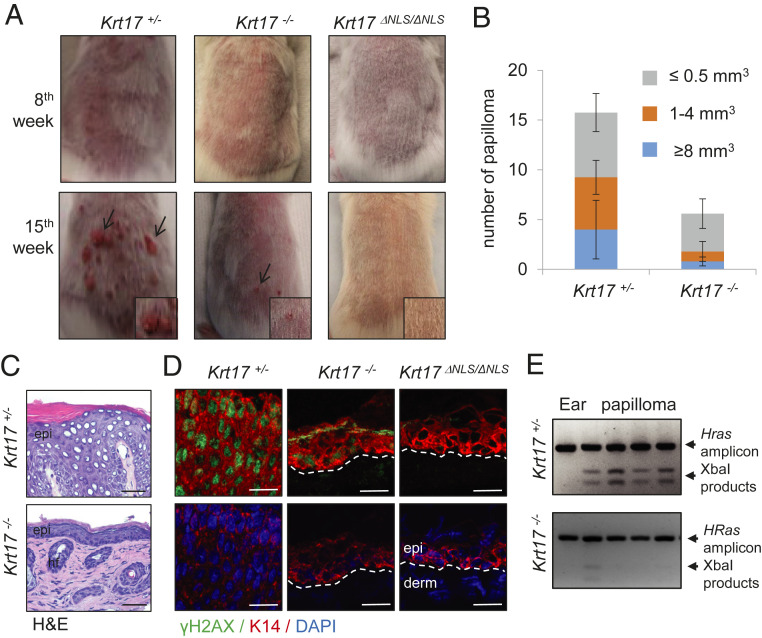
Given the observation of DDR dependence on K17 along with the known relation between K17 and tumor inception and growth in oncogene-driven mouse skin tumor models, we subjected Krt17+/− (control), Krt17−/− (null), and Krt17ΔNLS/ΔNLS mice to DMBA-TPA (7,12-dimethylbenz(a)anthracene and 12-O-tetradecanoylphorbol-13-acetate) two-step chemical skin carcinogenesis (25). Application of this protocol on the back skin of 8-wk-old mice induces papilloma formation, starting from ∼9 wk, in Krt17-expressing mice, whereas papilloma formation was reduced and delayed in Krt17−/− mice (Fig. 5 A and B). Remarkably, Krt17ΔNLS/ΔNLS mice failed to develop any papilloma during the 15-wk trial period (Fig. 5A). Follow-up analyses of papillomas at 15 wk showed the presence of significant epidermal thickening (Fig. 5C), along with robust γH2A.X expression in Krt17-expressing mice; again, Krt17−/− mice did not show these attributes (Fig. 5D). Analysis of genomic DNA (Methods) revealed a significantly higher frequency of Hras mutations in Krt17-expressing compared to Krt17−/− papilloma lesions (Fig. 5E), which was also confirmed by sequencing. These findings significantly extend previous studies involving Gli2tg (7) and HPV16tg mouse models (8) in establishing that absence of K17 during tumor initiation results in protection against skin tumorigenesis. Interestingly, similar tumor resistance to DMBA-TPA induced tumor development have been reported in other DDR-associated protein-deficient mice, like DNA-PK (26) and Rac1 (27). Possibly, the absence of K17 compromises DDR, resulting in the reduced survival of potential tumor-initiating cells and lesser papilloma formation, a notion that was addressed next.
Fig. 5.
Keratin 17 affects cancer initiation. (A) DMBA-TPA based two-step chemically induced skin carcinogenesis is dependent on the presence of K17. The 2-mo-old mice Krt17+/−, Krt17−/−, and Krt17ΔNLS/ΔNLS were subjected to DMBA-TPA two-step chemically-induced skin carcinogenesis and analyzed for papilloma formation on their back. n ≥ 3 mice per genotype. Arrows depict papilloma lesions and Inset shows zoomed image. (B) The presence of K17 affects number and size of the papilloma formed. The number of papillomas formed were counted for both Krt17+/− and Krt17−/−, and the size of the papilloma was measured and quantified. Error bars are SEM. (C) Hematoxylin/eosin (H&E) staining on the papillomas isolated from Krt17+/− and Krt17−/− mice after DMBA-TPA treatments. (Scale bars: 50 μm.) (D) Immunostaining of γH2A.X (green) and K14 (red) of papilloma/skin tissue after DMBA-TPA treatments from Krt17+/−, Krt17−/−, and Krt17ΔNLS/ΔNLS mice. (Scale bars: 20 μm.) (E) DMBA-induced HRas mutations showed higher frequency in Krt17-expressing mice. Genomic DNA was isolated from papillomas formed on Krt17+/− and Krt17−/− mice, and an HRas amplicon digestion assay was performed to identify the presence of mutations.
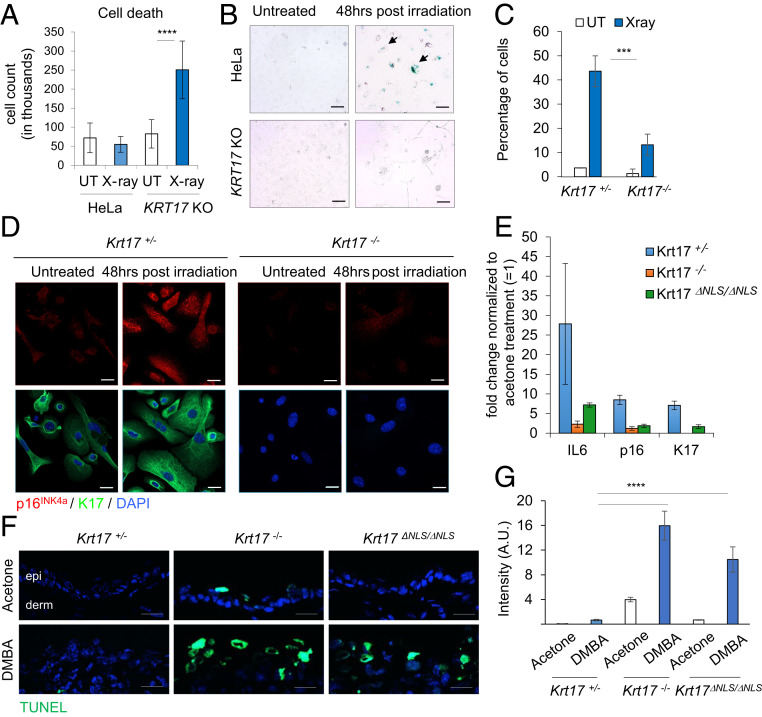
Analysis of the fate of keratinocytes following DNA damage confirmed that the presence of K17 is indeed critical for survival of damaged keratinocytes. A431 and HeLa cells, parental and KRT17 KO, were analyzed for cell viability using a trypan blue exclusion assay at 96 h after irradiation (X-ray; 5 Gy). Cells lacking K17 showed increased cell death relative to controls (Fig. 6A and SI Appendix, Fig. S4 A–C). We also performed terminal deoxynucleotidyltransferase-mediated dUTP nick end labeling (TUNEL) staining in primary cultures of skin keratinocytes from Krt17−/− and Krt17+/− controls, and Krt17ΔNLS/ΔNLS and WT controls, at 48 h after irradiation (X-ray; 5 Gy). Interestingly, significantly more TUNEL-positive keratinocytes were observed in Krt17−/− and Krt17ΔNLS/ΔNLS relative to their respective control (SI Appendix, Fig. S4 D–G), suggesting that a DDR response involving nuclear-localized K17 is critical for survival of cells with DNA damage. Next, we performed a senescence-associated beta galactosidase assay (28) on HeLa, parental, and KRT17 KO cells, as well as Krt17−/− and Krt17+/− mouse keratinocytes in primary culture, at 48 h after irradiation. This revealed that, while K17-expressing cells had the ability to enter senescence, cells lacking K17 showed impairment in this regard (Fig. 6 B and C). We also performed immunostaining for p16INK4a, a senescence marker (29), in Krt17−/− and Krt17+/− mouse keratinocytes subjected to the same protocol. K17-expressing keratinocytes showed robust p16INK4a expression compared to cells lacking K17 (Fig. 6D). Next, the role of K17 on cell fate determination in response to DNA damage was evaluated in vivo following DMBA treatment of 8-wk mouse ear skin (contralateral ear skin was treated with vehicle and used as internal reference). At 1 wk after two topical treatments, 24 h apart, Krt17+/− control skin showed a robust induction of senescence-associated markers p16INK4a and IL6 messenger RNAs (mRNAs), as determined via real time PCR analysis, whereas Krt17−/− and Krt17ΔNLS/ΔNLS mice did not (Fig. 6E). TUNEL staining on these ear samples showed that Krt17−/− and Krt17ΔNLS/ΔNLS skin showed increased cell death post-DNA damage, compared to Krt17+/− (control) mice (Fig. 6 F and G). These findings are consistent with the notion that the nuclear pool of K17 is required for keratinocyte survival after DNA damage in vitro and in vivo.
Fig. 6.
K17 affects cell survival through DDR. (A) K17 promotes survival post DNA damage. HeLa parental and KRT17 KO cells were treated with 5 Gy X-ray irradiation and harvested for trypan blue dye exclusion assay for cell viability after 96 h. The positively stained cells were a readout for dead cells quantified in the graph. Error bars are SEM. n = 3 replicates. ****P < 0.0001. (B) K17-expressing cells can proceed to senescence following persistent DDR. HeLa parental and KRT17 KO cells were treated with 5 Gy X-ray irradiation, and a senescence-associated beta galactosidase assay was performed after 48 h. Blue coloration within the cell shows positive staining. (Scale bars: 100 μm.) n = 3 replicates. Arrows draw attention to positively-stained cells. (C) Mouse keratinocytes isolated from Krt17+/− and Krt17−/− newborn pups were subjected to irradiation, and a senescence beta galactosidase assay was performed. Blue stained cells per field of image were calculated across experimental replicates (n = 3) and quantified. Error bars are SEM. ***P < 0.001. (D) Presence of K17 positively regulated p16INK4a induction. Mouse keratinocytes isolated from newborn pups in culture (Krt17+/− and Krt17−/−) were treated with 5 Gy X-ray irradiation and immunostained for p16INK4a after 48 h. (Scale bars: 20 μm.) n = 2 replicates. (E) Krt17-expressing mice showed induction of senescence markers in response to DNA damage. Two-mo-old mice (Krt17+/−, Krt17−/−, and Krt17ΔNLS/ΔNLS) were treated with 20 μg of DMBA dissolved in acetone on right ear and acetone on left ear two times, 24 h apart. Ear tissues were harvested 1 wk after first treatment, and RNA samples were isolated to perform real-time PCR for senescence-associated markers p16INK4a and IL6. Expression changes of IL6, p16INK4a, and K17 are normalized to βactin or GAPDH. Fold changes in expression of individual genes were quantified comparing acetone (= 1) to DMBA treatment for each genotype (n ≥ 2 mice per genotype). Error bars are SD. (F) TUNEL staining was performed on ear tissue sections 1 wk after acetone or DMBA treatment as described above on Krt17+/−, Krt17−/−, and Krt17ΔNLS/ΔNLS mice. (Scale bars: 20 μm.) n ≥ 2 mice per genotype. (G) TUNEL-positive cells (green) were quantified by measuring fluorescence intensity throughout interfollicular epidermis of each tissue section using ImageJ, and multiple regions per section were used. Error bars are SEM.
Discussion
We have here identified a role for the nuclear-localized pool of K17 in regulating DDR. Lack of K17, or of nuclear-localized K17, impairs keratinocyte survival post-DNA damage due to DDR dysregulation. Our findings clearly point to a preferential role for K17 in the repair of DSBs in DNA and are consistent with K17’s ability to interact or associate with a growing list of proteins involved in DDR, including Aire (8), hnRNPK (10), H2AX and γH2AX (11), DNA-PKcs, 53BP1 (this study), and chromatin architectural proteins like HMGN2 (11). A prosurvival role for K17 is consistent with, and extends, previous findings focused on hair follicles in mouse skin (30, 31). The findings reported here add to K17’s demonstrated ability to modulate inflammation-associated gene expression (7, 8, 10), cell proliferation (11, 12), and nuclear architecture (11) during stress and/or tumorigenesis and significantly extend the finding that K17 is subject to regulation by p53 after irradiation (32). Of note, we report here that Krt17ΔNLS/ΔNLS mice, which show impaired import of K17 to the nucleus but maintain a robust pool of K17-containing filaments in the cytoplasm of skin keratinocytes (11), are markedly less sensitive to DMBA-TPA–induced carcinogenesis, thus significantly extending studies involving Krt17-null mice (refs. 7–9 and this study).
At the cell and tissue levels, our findings are consistent with a model (SI Appendix, Fig. S5) that showcases the induction of K17 after DNA damage as an adaptation, akin to melanogenesis and pigmentation (33, 34), that promotes the survival of skin keratinocytes against additional exposures to such stresses. Speculating on the significance of our findings, we propose that the presence of nuclear-localized K17 skews the DDR response toward a quicker, and perhaps imperfect, resolution of double-stranded DNA breaks (e.g., via NHEJ-driven repair) (35), thereby promoting cell survival but also setting the stage for tumor initiation and subsequent growth if the tissue subsequently experiences tumor-promoting circumstances. While lacking in details (SI Appendix, Fig. S5), the model explains both the prooncogenic influence of K17 in a broad range of human carcinomas (Introduction) and the protection afforded by the complete loss of K17, or of its nuclear-localized form, in several experimental tumor paradigms in mice (refs. 7, 8 and this study). A previous study reported on the lesser sensitivity of Krt10-null mice to two-step chemical carcinogenesis in skin, correlating with a markedly enhanced turnover of keratinocytes (36). How the latter findings are related to the current study is unclear and warrants further examination.
At a molecular level, defining how the nuclear pool of K17 exerts a profound impact on DDR in keratinocytes of the epidermis awaits further investigation. In addition to further characterizing interactions between K17 and DDR effectors (see summary in SI Appendix, Table S2), improved biochemical and imaging-based tracking of nuclear-localized K17 represents a high priority toward defining the mechanisms underlying its role in DDR. K17 protein is dynamically modified, posttranslationally, via phosphorylation (37, 38), ubiquitylation (39), sumoylation (40), and likely other chemical groups, and this represents an ideal strategy to specify it for specific, context-dependent cellular roles (37, 38). Sumoylation is of particular interest as it occurs preferentially in the nucleus, is stimulated in the setting of DDR, and is known to play a role in nucleating the local formation of protein complexes (41, 42). Several DDR effector proteins associated with K17, including hnRNPK (43), H2AX (44), DNA-PKcs (45), and 53BP1 (42), undergo site-specific sumoylation after DNA damage as does K17 itself (40). K17 is differentially sumoylated at two sites, Lys15 and Lys399, after methyl methanesulfonate-induced DNA damage in HeLa cells (46), with the latter being part of its NLS (8, 12). Targeted follow-up studies on K17 sumoylation and other relevant posttranslational modifications may not only help elucidate the mechanisms that relate K17 to DDR, tumor initiation and, more broadly, the role(s) of K17 during tumorigenesis, but also understand how these elements play out in different populations of keratinocytes in skin and other tissues. Hair follicle keratinocytes, in particular, are of great interest since they are a frequent site of tumor initiation for nonmelanoma skin cancers (47, 48).
Finally, our findings expand the role of cytoskeletal proteins originally believed to be confined to the cytoplasm and recently shown to participate in DDR within the nucleus, as well as other intermediate filament proteins like synemin, which was recently reported to be involved in DNA damage response (49). Actin and actin-associated proteins, for instance, are required for several aspects of the response to DNA damage and, specifically, to DSB repair (20, 21, 50). Determining whether and how K17 and K17-dependent processes interface with the actin machinery in this setting represents yet another emerging issue of interest.
Methods
Cell Lines and Treatments.
HeLa and A431 cell lines were obtained from the American Type Culture Collection (ATCC). Cell lines were confirmed to be mycoplasma negative. The KRT17 knock out lines were generated in the P.A.C. laboratory using CRISPR/Cas9 technology and as previously described (8). Mouse epidermal keratinocytes were isolated from newborn mouse skin as described before (51). For DNA damaging purposes, cells in culture were treated with 1 μg of DMBA (D3254; Sigma-Aldrich) in acetone for 12 h before analysis or 100 μM H2O2 for 30 min. In addition to chemical agents, cells were irradiated to induce DNA damage. Irradiation was performed using an orthovoltage high energy X-ray machine (Irradiation Core Facility, University of Michigan) at a 5-Gy dose. X-ray exposure times were calculated based on dose rates provided by the Irradiation Core. Transfections were performed using FuGENE HD (E2311; Promega) at a 1:3 (DNA amount:FuGENE volume) ratio according to the manufacturer’s instructions or Nucleofector (Lonza) using transfection reagents and protocols provided by the manufacturer.
Mouse Models and Treatments.
Both Johns Hopkins University and University of Michigan Medical School’s Institutional Animal Use and Care Committee have approved all mouse experiments performed in this study. The Krt17+/− and Krt17−/− (30) and Krt17ΔNLS/ΔNLS (11) mouse lines were previously described. Acute DMBA treatments were performed on the ear skin of ∼8-wk-old mice; 20 μg of DMBA (dissolved in acetone) was used to treat one ear, and acetone was used as mock to treat the other ear of each mouse. Treatments were done twice, 24 h apart, and tissues were harvested 2 h after the second treatment. For the cell fate experiment, tissues were harvested 1 wk from the date of first treatment. For DMBA treatment on back skin of postnatal day 12 pups, back hair was shaved off, 40 μg of DMBA was used to do a one-time treatment, and tissues were harvested after 2 h. The two-step chemical carcinogenesis protocol, using DMBA and TPA (P1585; Sigma-Aldrich) treatments on the back skin of mice, was performed as described (25). Briefly, ∼8-wk-old Krt17+/−, Krt17−/−, and Krt17ΔNLS/ΔNLS mice (both male and female) were anesthetized using isoflurane, and their back hair was shaved using a hair clipper. Two days after shaving, mice were anesthetized and treated topically with single dose of 20 μg of DMBA in 200 μL of acetone on their back skin. At 7 d after the DMBA treatment, mice were treated topically with 2.0 μg per treatment of TPA in 200 μL of acetone on their back skin, twice weekly, for up to 15 wk. The papilloma burden was monitored during the entire 15-wk treatment duration. Skin tissue samples were harvest at 15 wk and embedded in optimal cutting temperature (OCT) compound. Tissue sections of 3 to 8 μm were obtained using a cryostat. For Hras Q61L mutation analysis, we isolated genomic DNA using TRIzol (Fisher Scientific) from each of the papillomas harvested from the mice skin, and XbaI digestion was performed on the HRas amplicon obtained by using HRas F 5′ AAGCCTGTTTTGCAGGA 3′ and Hras R 5′ GGTGGCTCACCTGTACTGATG 3′ primers. The samples post digestion were run on 3 to 4% agarose gel. Presence of the Q61L allele yields specific digestion products, as described previously (52). PCR products were subjected to DNA sequencing to confirm the presence of mutation.
Immunoblotting.
Protein lysates for immunoblotting were prepared in urea sample buffer (8 M deionized urea, 0.5% sodium dodecyl sulfate (SDS), 30 mM Tris, pH 6.8, 5% glycerol, and 5% β-mercaptoethanol) after collecting whole cell lysates from cultured cells. All samples were sheared using finger-gauged needles (22 1/2, 25 1/2, and 26 1∕2, in sequence) and subjected to a Bradford assay. All samples were loaded at 10 μg of protein per lane for immunoblotting. Proteins were resolved using either 10% or 12.5% SDS polyacrylamide gel electrophoresis and transferred to 0.45-μm nitrocellulose membranes (Bio-Rad, Hercules, CA) via a transblot turbo transfer or minitransblot cell system (Bio-Rad). Blots were blocked in 5% milk in tris-buffered saline, 0.1% Tween20 (TBST-T, wt/vol) for >1 h at room temperature (21 °C) and subsequently incubated overnight at 4 °C with primary antibodies. After rinses, secondary antibodies were applied for 1 h at room temperature (21 °C). Blots were developed using either ECL Select developing solution (GE Healthcare, Pittsburgh, PA) or SuperSignal West Pico chemiluminescent substrate (Thermo Scientific, Waltham, MA) and imaged using a FluorChem Q system (ProteinSimple, San Jose, CA).
Immunofluorescence.
For indirect immunofluorescence analysis, frozen sections of the aforementioned tissues were blocked for 1 h at room temperature in 5% normal goat serum and 1× phosphate-buffered saline (PBS). Primary antibodies were applied overnight at 4 °C. Cultured cell samples were fixed with 4% paraformaldehyde, permeabilized with 0.2% Triton X-100, and incubated with primary antibodies overnight at 4 °C and with secondary antibodies at room temperature with 1-h incubation time. Coverslips were mounted onto slides with FluoroSave reagent (345789-20ML; Calbiochem) and dried overnight. Both cultured cell samples and tissue sections were imaged using a Zeiss fluorescence microscope with Apotome attachment (Zeiss, Oberkochem, Germany) or a Zeiss LSM800 confocal microscope. Images for a given marker were acquired at the same exposure, pixel range, gamma values, and objective across all samples and genotype using the Zen software. All images were consistently adjusted for brightness and contrast and appropriately cropped for presentation purposes using Zen or ImageJ software.
Rescue Assay Using GFP-Tagged Keratin Constructs.
Previously reported constructs, GFP-K17, GFP-K14, and GFP-K17 ΔNLS (8, 11), were transiently transfected into KRT17 KO A431 cells for 48 to 72 h, followed by DMBA treatment as described before. Transfections were performed using FuGENE HD (E2311; Promega) at a 1:3 (DNA amount:FuGENE volume).
Proximity Ligation Assay.
Mouse keratinocytes were seeded on coverslips pretreated with collagen and cultured at 37 °C and 5% CO2 for 2 d before DNA damage treatment. Cells were washed with PBS and fixed using 4% paraformaldehyde solution (1× PBS) for 10 min at room temperature. After fixation, cells were washed with PBS and permeabilized with 0.2% Triton X-100 solution, diluted in PBS, for 5 min. Cells were rinsed with PBS and then blocked in 2.5% normal goat serum (in 1× PBS) for 45 min at 37 °C. Cells were then incubated with primary antibodies [1) rabbit polyclonal anti-K17 (53) and mouse monoclonal anti-K17 (sc-393002); 2) rabbit polyclonal anti-53BP1 (4937; Cell Signaling) and mouse monoclonal anti-K17 (sc-393002); and 3) rabbit polyclonal anti-53BP1 (4937; Cell Signaling) and mouse monoclonal anti-phospho-Histone H2A.X (Ser139) (05-636; Millipore Sigma)] diluted in the blocking buffer for 60 min at 37 °C. The manufacturer’s instructions from the Duolink PLA Fluorescence protocol (Millipore Sigma) were followed. PLA probes (DUO92002 and DUO92004; Millipore Sigma) and Duolink In Situ Detection Reagents Orange (DUO092007; Millipore Sigma) were used. Coverslips were mounted on microscope slides with mounting medium containing DAPI. Cells were imaged using a Zeiss LSM800 confocal microscope. For PLA on tissue sections, cryosectioned samples on glass slides were thawed, washed with PBS, and blocked using 5% NGS, followed by incubation with K17 antibodies and the same protocol as described above.
Live Imaging.
For live imaging analysis of 53BP1 foci formation, freshly isolated keratinocytes or cell lines were transfected with the GFP-tagged 53BP1 reporter construct (54) using the nucleofection method (Lonza) before plating in chamber slides. For keratinocytes, the chamber slides were coated with type I collagen (354236; Corning). HeLa and KRT17 KO HeLa cells were incubated in phenol red-free Dulbecco’s modified Eagle’s medium (DMEM) during imaging. Keratinocytes were incubated in phenol red-free epidermal culture media Cnt57 (Zenbio) during imaging. The cells were imaged on an LSM880 confocal microscope inside an imaging chamber equipped with 5% CO2 at 37 °C. Transfected cells with GFP-53BP1 expression throughout the nucleus were selected for imaging. Using Zen software, the time-lapse imaging was set up such that continuous capturing of the green fluorescent channel occurred at one frame per second rate, and, 10 s into the time lapse, a 405-nm laser was turned on at 100% laser power at a selected region of interest (ROI) within the nucleus of the cell chosen for imaging and turned off. This laser microirradiation induced DSBs as described previously (55). The imaging was continued for 300 s to capture the effect post microirradiation. The time-lapse videos were opened using ImageJ software, and the Time series Analyzer plugin was used to measure fluorescence intensity during the entire course of imaging. Microsoft excel was used to plot the graphs.
Alkaline Comet Assays.
Comet assays were conducted using the Comet Assay Kit (Trevigen) following the manufacturer’s instructions. Cells were collected and washed once with PBS, and 3 × 105 cells were combined with 1% molten LMAgarose at 37 °C at a ratio of 1:10 (vol/vol) and pipetted onto slides. Slides were then immersed in prechilled lysis buffer, followed by alkaline unwinding of chromatin. Alkaline electrophoresis of gelled slides was conducted at 30 V at 4 °C for 30 min. The DNA was visualized by SYBR gold staining, and images were taken with a Zeiss fluorescence microscope with Apotome attachment, and analyzed using the CometScore software (Tri-Tek, Sumerduck, VA).
K17 Coimmunoprecipitation.
HeLa cells (KRT17 WT and KRT17 KO) were lysed, fractionated, and immunoprecipitated (K17 IP, using rabbit polyclonal anti-K17 (11, 53). Prior to sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS/PAGE), inputs and IPs were denatured with 2× SDS sample buffer supplemented with 10% beta-mercaptoethanol and then heated at 95 °C for 10 min and run on a 4 to 15% SDS/PAGE gradient gel (456-1086; Bio-Rad). After gel electrophoresis, samples were transferred to a nitrocellulose membrane (162-0115; Bio-Rad). A 5% milk in Tris-buffered saline plus 0.1% Tween 20 (TBST) blocking solution was applied to the membrane for 60 min at room temperature. Primary antibodies used for immunoblotting, mouse monoclonal anti–DNA-PK (12311; Clone 3H6; Cell Signaling) and rabbit polyclonal anti-Histone H3 (9715; Cell Signaling), were diluted in 5% milk in TBST and incubated with the membranes overnight at 4 °C. Secondary antibodies used for immunoblotting, goat anti-mouse horseradish peroxidase (HRP) and goat anti-rabbit HRP, were diluted in 5% milk in TBST and incubated with the membranes for 45 min at room temperature prior to enhanced chemiluminescence (ECL) detection.
GFP-53BP1 and K17 Interaction Using IP.
A431 cells (KRT17 WT and KRT17 KO) were transfected with the GFP-tagged 53BP1 reporter construct as described before. After 48 h, untreated and cells cultured for 1 h after 10-Gy irradiation were lysed using NETN (150mM NaCl, 1mM EDTA – ethylenediaminetetraacetic acid, 20mM tris buffer pH 8.0, 0.5% v/v Nonidet P-40) lysis buffer. Dynabeads Protein G Immunoprecipitation kit (10007D; ThermoFisher Scientific) was used to pull down GFP-53BP1 and interacting partners using GFP antibody. For this, 10 μg of mouse monoclonal GFP antibody (9F9.F9; Novus Biologicals) and 500 μg of lysate were used. The lysate was allowed to interact with Dynabeads–GFP antibody complex in a cold room overnight. Prior to SDS/PAGE, inputs (30 μg per sample) and IPs (20 μL of eluted complex) were denatured with 2× SDS sample buffer supplemented with 10% beta-mercaptoethanol and then heated at 95 °C for 10 min and run on a 4 to 15% SDS/PAGE gradient gel (456-1086; Bio-Rad). Immunoblotting for K17 and endogenous 53BP1 (using rabbit polyclonal anti-53BP1) (A300-272A; Bethyl Laboratories) was performed as described before.
Antibodies and Reagents.
Primary antibodies used in this study were as follows: rabbit polyclonal antibodies against Krt17 (53), phospho-BRCA1 (S1524) (9009; Cell Signaling), Sam68 (SC-333; Santa Cruz), phospho-ATM (S1981) (5883; Cell Signaling), phospho-ATR (S428) (2853; Cell Signaling), γH2A.X (S139) (9718; Cell Signaling), phospho-CHK2 (T68) (2197; Cell Signaling), phospho-CHK1 (S345) (2341; Cell Signaling), DNA-PKcs (4602; Cell Signaling), Histone H3 (9715; Cell Signaling), phospho KAP-1 (S824) (A300-767A; Bethyl Laboratories Inc.), mouse monoclonal antibodies against Krt17 (E-4) (SC-393002; Santa Cruz), phospho-P53 (serine 15) (9286; Cell Signaling), p16INK4a (1E12E10; Thermo), mouse polyclonal antibody against glyceraldehyde-3-phosphate dehydrogenase (GAPDH) (SC-365062; Santa Cruz), and chicken polyclonal antibody against Krt14 (906001; Biolegend). All commercial antibodies were used according to the manufacturer’s recommendation. For TUNEL staining, Click-iT Plus TUNEL Assay for In Situ Apoptosis Detection, Alexa Fluor 488 dye (C10617; Invitrogen) was used. Manufacturer recommended protocol was followed. For senescence associated beta galactosidase staining, Senescence β Galactosidase staining kit (9860; Cell Signaling) was used, and the manufacturer’s instructions were followed for the assay.
Image Quantification and Statistical Analyses.
Signal intensity quantification for indirect immunofluorescence images was performed using ImageJ software. Most calculations were based on quantifying signal intensity of each individual cell within each field across biological replicates. Graphs represent the average, represented in arbitrary units comparing control to treated samples. Quantification for alkaline comet assays was performed using the CometScore software. The calculations were based on measuring tail lengths for each individual cell within the field of microscopy, at 10× magnification. Graphs accompanying alkaline comet assays represent the average tail lengths between each treatment condition and its respective genotype. For quantification of TUNEL-positive cells and SA-βgal–positive cells, number of cells stained positive, and total number of cells in multiple fields were counted. The percentage of positive cells was then calculated across biological replicates. Error bars on all histograms represent SE across biological replicates. The Student t test was done using Microsoft Excel to obtain P values. P values of <0.05 were considered statistically significant (*P < 0.05, **P < 0.01, ***P < 0.001, and ****P < 0.0001). For analysis of Western blot intensities shown in Fig. 3A and represented in SI Appendix, Table S1, statistical analysis was done using a web-based ANOVA calculator to compare WT versus K17 KO, DNA damage treated (DMBA and H2O2 together) groups for each DDR protein marker, and P values generated are mentioned.
Real-Time PCR Analysis for Gene Expression.
Tissue samples were homogenized, and RNA isolation was performed using TRIzol (Fisher Scientific) following the manufacturer’s protocol. Then, 2 μg of total RNA was used for complementary DNA (cDNA) conversion using iScript (Bio-Rad), which was directly used for quantitative Real-Time PCR analysis using the CFX 96 Real-time system (Bio-Rad). Primers used for amplification of specific genes are listed in SI Appendix, Table S3.
Supplementary Material
Acknowledgments
We thank P.A.C. laboratory members for advice and support; Beau Su and Caroline Shah for technical support; and Deepak Saini, Kenneth Kwan, Allan Balmain, Quan Tran, Fengyi Wan, Ritankar Majumdar, Song Chen, and Carole Parent for advice, protocols, and reagents. We thank the members of the Parent, Dlugosz, and Wong laboratories at the University of Michigan for comments and advice. This study was supported in part by NIH Grants AR044232 and CA160255 (to P.A.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2020150118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Karantza V., Keratins in health and cancer: More than mere epithelial cell markers. Oncogene 30, 127–138 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Merkin R. D., et al., Keratin 17 is overexpressed and predicts poor survival in estrogen receptor-negative/human epidermal growth factor receptor-2-negative breast cancer. Hum. Pathol. 62, 23–32 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Mockler D., et al., Keratin 17 is a prognostic biomarker in endocervical glandular neoplasia. Am. J. Clin. Pathol. 148, 264–273 (2017). [DOI] [PubMed] [Google Scholar]
- 4.Regenbogen E., et al., Elevated expression of keratin 17 in oropharyngeal squamous cell carcinoma is associated with decreased survival. Head Neck 40, 1788–1798 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Wang Y.-F., et al., Overexpression of keratin 17 is associated with poor prognosis in epithelial ovarian cancer. Tumour Biol. 34, 1685–1689 (2013). [DOI] [PubMed] [Google Scholar]
- 6.Roa-Peña L., et al., Keratin 17 identifies the most lethal molecular subtype of pancreatic cancer. Sci. Rep. 9, 11239 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Depianto D., Kerns M. L., Dlugosz A. A., Coulombe P. A., Keratin 17 promotes epithelial proliferation and tumor growth by polarizing the immune response in skin. Nat. Genet. 42, 910–914 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hobbs R. P., et al., Keratin-dependent regulation of Aire and gene expression in skin tumor keratinocytes. Nat. Genet. 47, 933–938 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hobbs R. P., Batazzi A. S., Han M. C., Coulombe P. A., Loss of keratin 17 induces tissue-specific cytokine polarization and cellular differentiation in HPV16-driven cervical tumorigenesis in vivo. Oncogene 35, 5653–5662 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chung B. M., et al., Regulation of C-X-C chemokine gene expression by keratin 17 and hnRNP K in skin tumor keratinocytes. J. Cell Biol. 208, 613–627 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Jacob J. T., et al., Keratin 17 regulates nuclear morphology and chromatin organization. J. Cell Sci. 133, jcs254094 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Escobar-Hoyos L. F., et al., Keratin-17 promotes p27KIP1nuclear export and degradation and offers potential prognostic utility. Cancer Res. 75, 3650–3662 (2015). [DOI] [PubMed] [Google Scholar]
- 13.Hanahan D., Weinberg R. A., Hallmarks of cancer: The next generation. Cell 144, 646–674 (2011). [DOI] [PubMed] [Google Scholar]
- 14.Giglia-Mari G., Zotter A., Vermeulen W., DNA damage response. Cold Spring Harb. Perspect. Biol. 3, a000745 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chatterjee N., Walker G. C., Mechanisms of DNA damage, repair, and mutagenesis. Environ. Mol. Mutagen. 58, 235–263 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.d’Adda di Fagagna F., Living on a break: Cellular senescence as a DNA-damage response. Nat. Rev. Cancer 8, 512–522 (2008). [DOI] [PubMed] [Google Scholar]
- 17.Nowsheen S., Yang E. S., The intersection between DNA damage response and cell death pathways. Exp. Oncol. 34, 243–254 (2012). [PMC free article] [PubMed] [Google Scholar]
- 18.Caridi C. P., Plessner M., Grosse R., Chiolo I., Nuclear actin filaments in DNA repair dynamics. Nat. Cell Biol. 21, 1068–1077 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hurst V., Shimada K., Gasser S. M., Nuclear actin and actin-binding proteins in DNA repair. Trends Cell Biol. 29, 462–476 (2019). [DOI] [PubMed] [Google Scholar]
- 20.Panier S., Boulton S. J., Double-strand break repair: 53BP1 comes into focus. Nat. Rev. Mol. Cell Biol. 15, 7–18 (2014). [DOI] [PubMed] [Google Scholar]
- 21.Zgheib O., Pataky K., Brugger J., Halazonetis T. D., An oligomerized 53BP1 tudor domain suffices for recognition of DNA double-strand breaks. Mol. Cell. Biol. 29, 1050–1058 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Davis A. J., Chen B. P. C., Chen D. J., DNA-PK: A dynamic enzyme in a versatile DSB repair pathway. DNA Repair (Amst.) 17, 21–29 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Abramson J., Goldfarb Y., AIRE: From promiscuous molecular partnerships to promiscuous gene expression. Eur. J. Immunol. 46, 22–33 (2016). [DOI] [PubMed] [Google Scholar]
- 24.Ziv Y., et al., Chromatin relaxation in response to DNA double-strand breaks is modulated by a novel ATM- and KAP-1 dependent pathway. Nat. Cell Biol. 8, 870–876 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Abel E. L., Angel J. M., Kiguchi K., DiGiovanni J., Multi-stage chemical carcinogenesis in mouse skin: Fundamentals and applications. Nat. Protoc. 4, 1350–1362 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kemp C. J., Vo K., Gurley K. E., Resistance to skin tumorigenesis in DNAPK-deficient SCID mice is not due to immunodeficiency but results from hypersensitivity to TPA-induced apoptosis. Carcinogenesis 20, 2051–2056 (1999). [DOI] [PubMed] [Google Scholar]
- 27.Deshmukh J., Pofahl R., Haase I., Epidermal Rac1 regulates the DNA damage response and protects from UV-light-induced keratinocyte apoptosis and skin carcinogenesis. Cell Death Dis. 8, e2664 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dimri G. P., et al., A biomarker that identifies senescent human cells in culture and in aging skin in vivo. Proc. Natl. Acad. Sci. U.S.A. 92, 9363–9367 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Itahana K., Campisi J., Dimri G. P., Methods to detect biomarkers of cellular senescence: The senescence-associated beta-galactosidase assay. Methods Mol. Biol. 371, 21–31 (2007). [DOI] [PubMed] [Google Scholar]
- 30.McGowan K. M., et al., Keratin 17 null mice exhibit age- and strain-dependent alopecia. Genes Dev. 16, 1412–1422 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tong X., Coulombe P. A., Keratin 17 modulates hair follicle cycling in a TNFalpha-dependent fashion. Genes Dev. 20, 1353–1364 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Liao C., et al., p53 is a direct transcriptional repressor of keratin 17: Lessons from a rat model of radiation dermatitis. J. Invest. Dermatol. 136, 680–689 (2016). [DOI] [PubMed] [Google Scholar]
- 33.Jablonski N. G., Chaplin G., Colloquium paper: Human skin pigmentation as an adaptation to UV radiation. Proc. Natl. Acad. Sci. U.S.A. 107 (suppl. 2), 8962–8968 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Agar N., Young A. R., Melanogenesis: A photoprotective response to DNA damage? Mutat. Res. 571, 121–132 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Chang H. H. Y., Pannunzio N. R., Adachi N., Lieber M. R., Non-homologous DNA end joining and alternative pathways to double-strand break repair. Nat. Rev. Mol. Cell Biol. 18, 495–506 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Reichelt J., Furstenberger G., Magin T. M., Loss of keratin 10 leads to mitogen-activated protein kinase (MAPK) activation, increased keratinocyte turnover, and decreased tumor formation in mice. J. Invest. Dermatol. 123, 973–981 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Kim S., Wong P., Coulombe P. A., A keratin cytoskeletal protein regulates protein synthesis and epithelial cell growth. Nature 441, 362–365 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Pan X., Kane L. A., Van Eyk J. E., Coulombe P. A., Type I keratin 17 protein is phosphorylated on serine 44 by p90 ribosomal protein S6 kinase 1 (RSK1) in a growth- and stress-dependent fashion. J. Biol. Chem. 286, 42403–42413 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yang L., et al., E3 ligase Trim21 ubiquitylates and stabilizes keratin 17 to induce STAT3 activation in psoriasis. J. Invest. Dermatol. 138, 2568–2577 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Hendriks I. A., Vertegaal A. C. O., A comprehensive compilation of SUMO proteomics. Nat. Rev. Mol. Cell Biol. 17, 581–595 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Müller S., Ledl A., Schmidt D., SUMO: A regulator of gene expression and genome integrity. Oncogene 23, 1998–2008 (2004). [DOI] [PubMed] [Google Scholar]
- 42.Galanty Y., et al., Mammalian SUMO E3-ligases PIAS1 and PIAS4 promote responses to DNA double-strand breaks. Nature 462, 935–939 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Lee S. W., et al., SUMOylation of hnRNP-K is required for p53-mediated cell-cycle arrest in response to DNA damage. EMBO J. 31, 4441–4452 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chen W.-T., et al., Systematic identification of functional residues in mammalian Histone H2AX. Mol. Cell. Biol. 33, 111–126 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Gao S.-S., et al., TIP60 K430 SUMOylation attenuates its interaction with DNA-PKcs in S-phase cells: Facilitating homologous recombination and emerging target for cancer therapy. Sci. Adv. 6, eaba7822 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hendriks I. A., Treffers L. W., Verlaan-de Vries M., Olsen J. V., Vertegaal A. C. O., SUMO-2 orchestrates chromatin modifiers in response to DNA damage. Cell Rep. 10, 1778–1791 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.E. H. Epstein, Jr, Mommy–Where do tumors come from? J. Clin. Invest. 121, 1681–1683 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Seykora J. T., Cotsarelis G., Keratin 15-positive stem cells give rise to basal cell carcinomas in irradiated Ptch1(+/-) mice. Cancer Cell 19, 5–6 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Deville S. S., et al., The intermediate filament synemin regulates non-homologous end joining in an ATM-dependent manner. Cancers (Basel) 12, 1717 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Serebryannyy L., de Lanerolle P., Nuclear actin: The new normal. Mutat. Res. 821, 111714 (2020). [DOI] [PubMed] [Google Scholar]
- 51.Wang F., Zieman A., Coulombe P. A., Skin keratins. Methods Enzymol. 568, 303–350 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nagase H., Mao J. H., Balmain A., Allele-specific Hras mutations and genetic alterations at tumor susceptibility loci in skin carcinomas from interspecific hybrid mice. Cancer Res. 63, 4849–4853 (2003). [PubMed] [Google Scholar]
- 53.McGowan K. M., Coulombe P. A., Onset of keratin 17 expression coincides with the definition of major epithelial lineages during skin development. J. Cell Biol. 143, 469–486 (1998). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Nair R. R., Bagheri M., Saini D. K., Temporally distinct roles of ATM and ROS in genotoxic-stress-dependent induction and maintenance of cellular senescence. J. Cell Sci. 128, 342–353 (2015). [DOI] [PubMed] [Google Scholar]
- 55.Holton N. W., Andrews J. F., Gassman N. R., Application of laser micro-irradiation for examination of single and double strand break repair in mammalian cells. J. Vis. Exp. 56265 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.