Significance
This study directly demonstrates virulence-associated features of Helicobacter suis infection using isolates obtained from human stomachs. H. suis is the second-most-prevalent Helicobacter species in the human stomach, but diagnostic tests for Helicobacter pylori often fail to diagnose H. suis. H. suis does not have orthologs of either cytotoxin-associated gene A (CagA) or vacuolating cytotoxin A (VacA), two major virulence factors in H. pylori, suggesting that the molecular pathogenicity of H. suis and clinical manifestations caused by H. suis infection are different from those caused by H. pylori. This study paves the way for epidemiological research aimed at identifying the causal relationships between H. suis and gastric diseases and developing diagnostic methods.
Keywords: Helicobacter suis, gastric diseases, zoonosis
Abstract
Helicobacter suis, a bacterial species naturally hosted by pigs, can colonize the human stomach in the context of gastric diseases such as gastric mucosa-associated lymphoid tissue (MALT) lymphoma. Because H. suis has been successfully isolated from pigs, but not from humans, evidence linking human H. suis infection to gastric diseases has remained incomplete. In this study, we successfully in vitro cultured H. suis directly from human stomachs. Unlike Helicobacter pylori, the viability of H. suis decreases significantly on neutral pH; therefore, we achieved this using a low-pH medium for transport of gastric biopsies. Ultimately, we isolated H. suis from three patients with gastric diseases, including gastric MALT lymphoma. Successful eradication of H. suis yielded significant improvements in endoscopic and histopathological findings. Oral infection of mice with H. suis clinical isolates elicited gastric and systemic inflammatory responses; in addition, progression of gastric mucosal metaplasia was observed 4 mo postinfection. Because H. suis could be isolated from the stomachs of infected mice, our findings satisfied Koch’s postulates. Although further prospective clinical studies are needed, H. suis, like H. pylori, is likely a gastric pathogen in humans. Furthermore, comparative genomic analysis of H. suis using complete genomes of clinical isolates revealed that the genome of each H. suis isolate contained highly plastic genomic regions encoding putative strain-specific virulence factors, including type IV secretion system–associated genes, and that H. suis isolates from humans and pigs were genetically very similar, suggesting possible pig-to-human transmission.
Non-Helicobacter pylori Helicobacters (NHPH), which have a corkscrew-like spiral morphology very different from that of H. pylori, have been known to be present in human stomachs since the 1980s (1). NHPH infection has been observed in patients with gastric diseases, including peptic ulcers, chronic gastritis, gastric cancers, and gastric mucosa-associated lymphoid tissue (MALT) lymphoma (2–4).
Because routine diagnostic methods based on the urease activity of H. pylori, such as the urea breath test and rapid urease test, often yield negative results for NHPH (5), and the bacteria is uncultivable, NHPH infection in individual patients could be missed. Thus, in the post-H. pylori era, NHPH could become an important gastric pathogen in people of all ages.
The primary human pathogenic NHPH species is Helicobacter suis, which is naturally hosted by pigs (6). H. suis infects 60 to 95% of livestock pigs, in which it causes gastritis, ulcers, and a reduced rate of weight gain (6, 7). Humans are frequently infected by H. pylori in childhood but rarely in adulthood (8); by contrast, the route and period of H. suis infection in humans is completely unknown. In some cases, H. suis was observed in patients with gastric diseases after H. pylori eradication therapy (9). It is not known whether only H. pylori was eradicated in individuals coinfected by H. pylori and H. suis or if instead H. suis arose after H. pylori eradication.
Successful isolation and in vitro culture of H. pylori has dramatically contributed to elucidating the causal relationships between bacterial infection and gastric diseases as well as the molecular and cellular mechanisms of gastric cancer pathogenesis (10, 11). Successful isolation and in vitro culture of H. suis from pig stomach were reported in 2008 (12), and animal infection experiments using H. suis isolates from pigs demonstrated its pathogenicity. In addition, previous reports strongly suggested that H. suis infection contributes to gastric diseases in humans (SI Appendix, Table S1). However, studies on H. suis have been hindered because H. suis has not been successfully isolated from humans. In other words, the first and third of Koch’s postulates (13) (“association of the microbe with symptoms and presence of the microbe at the site of infection” and “reproduction of the disease by inoculation of a susceptible host with the microbe”) have been partially achieved for H. suis, whereas the remaining two postulates (“isolation of the microbe from lesions” and “re-isolation of the microbe from experimentally infected individuals”) have not.
We developed a modified culture method for a human isolate, H. suis strain SNTW101c (14), which was isolated in 2008 from a patient with nodular gastritis and has been passaged in mouse stomach for 12 y by repeated inoculations of uninfected mice with gastric mucosal homogenates from infected mice (15). In this study, we succeeded in culturing H. suis directly from human gastric biopsies and demonstrated H. suis virulence in humans based on Koch’s postulates using an experimental animal model of H. suis infection. We also determined the complete genomes of H. suis isolates from humans and pigs and compared their genetic features.
Results
Isolation and Characterizations of H. suis from Humans and Pigs.
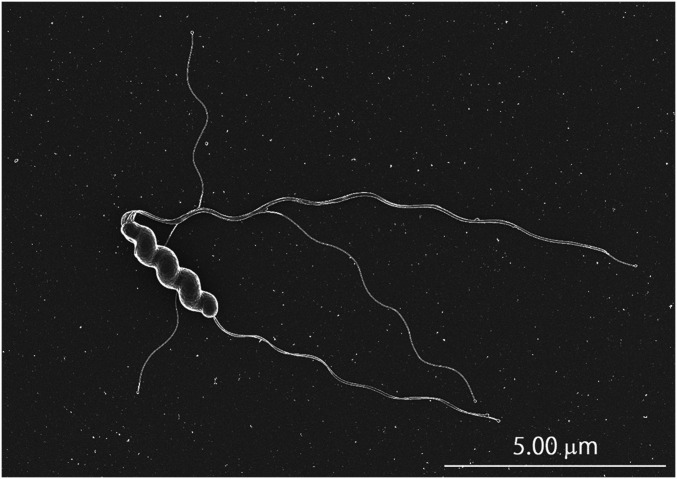
H. suis isolates were successfully cultured in vitro from patients A, B, and C, and named NHP19-4003, NHP19-4004, and NHP19-4022, respectively. H. suis isolates from slaughter pigs in Japan were named NHP19-0020 and NHP19-0033. In all cases, small clear colonies of H. suis on NHPH agar plates, smaller than those formed by H. pylori, appeared 8 to 10 d after inoculation, as shown in SI Appendix, Fig. S1. Scanning electron microscopy of H. suis isolates revealed a corkscrew-like spiral morphology with long bipolar flagella, which contribute to the motility of this bacterium (Fig. 1, https://www.youtube.com/watch?v=sHA4SSzu4eo&feature=youtu.be). Urease activities were detected in all isolates, although the rapid urease test and urea breath test were negative in all patients, in concordance with a previous report (SI Appendix, Table S1). All isolates were identified as H. suis by comparing average nucleotide identity (ANI) of whole-genome sequences of the isolates with H. suis strain HS5, the reference strain, which was originally isolated from a pig (accession no. NZ_ADHO01000000). ANI values of NHP19-4003, NHP4004, NHP19-4022, and NHP19-0020 were 99.64, 99.64, 99.74, and 99.64%, respectively.
Fig. 1.
Scanning electron micrograph of H. suis strain NHP19-4003 from a human patient.
In regard to antimicrobial susceptibility, none of the four H. suis isolates obtained in this study, as well as H. suis strain SNTW101c, were resistant to clarithromycin, levofloxacin, and oxacillin (Table 1). Minimum inhibitory concentrations (MICs) of amoxicillin in four of five isolates were 1.0 mg/L but were higher (16 mg/L) in one isolate, NHP19-4004. In that isolate, comparison of amino acid sequences of PBPA (PBP1), PBP2, and FtsI (PBP3), which are penicillin-binding proteins (PBPs) targeted by β-lactams, revealed the presence of one and three substitutions in the transpeptidase region (target region of β-lactams) in PBP2 and FtsI, respectively. The substitution at position 317 in PBP2 and the two substitutions at positions 532 and 547 were sterically close to the penicillin-binding motif in the predicted three-dimensional structures of PBPs (SI Appendix, Fig. S2). No specific substitutions were observed in the transpeptidase region of PBPA.
Table 1.
Antimicrobial susceptibility of the H. suis strain SNTW101c and H. suis strains obtained in this study
| Isolates | Origin | MIC (mg/L) of: | ||||||
| AMPC | OX | CAM | MINO | GM | LVFX | MTZ | ||
| SNTW101c | Human* | 1 | ≤0.25 | ≤0.25 | ≤2 | ≤4 | ≤0.5 | 16 |
| NHP19-4003 | Human | 1 | ≤0.25 | 0.5 | ≤2 | ≤4 | ≤0.5 | 8 |
| NHP19-4004 | Human | ≥16 | ≤0.25 | 0.5 | 8 | ≤4 | 1 | 16 |
| NHP19-4022 | Human | 1 | ≤0.25 | ≤0.25 | ≤2 | ≤4 | ≤0.5 | 16 |
| NHP19-0020 | Pig | 1 | ≤0.25 | ≤0.25 | 4 | ≤4 | ≤0.5 | 8 |
AMPC: Amoxicillin; OX: Oxacillin; CAM: Clarithromycin; MINO: Minocycline; GM: Gentamicin; LVFX: Levofloxacin; MTZ: Metronidazole.
Mouse-adapted.
Effects of H. suis Eradication on Gastric Diseases in Patients.
Endoscopic and histological data of patients pre- and posteradication were available for patient A and patient B.
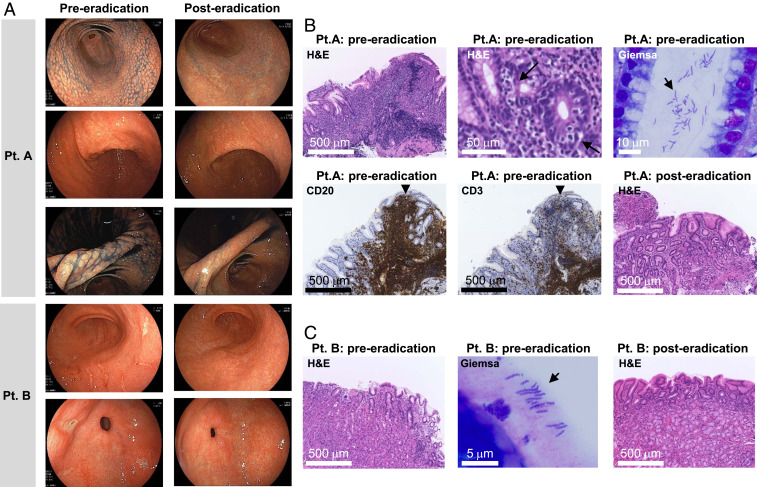
In patient A, thickened mucosae and nodular irregularities in the lesser curvature of the angle were observed endoscopically (Fig. 2A). Lymphocyte infiltration and formation of lymphoepithelial lesions were observed in biopsy tissues by hematoxylin–eosin (H&E) staining (Fig. 2B). Lymphocytes were CD20 positive but negative for CD3 (Fig. 2B), CD10, BCL6, and Cyclin D1 (SI Appendix, Fig. S3), as determined by immunohistochemistry. Patient A was diagnosed with H. suis–infected gastric MALT lymphoma with nodular gastritis and treated with the standard triple therapy for H. pylori: 750 mg amoxicillin, 400 mg clarithromycin, and 20 mg vonoprazan twice daily for 7 d. Successful eradication was confirmed by both PCR and culture 3 mo later. The gastric MALT lymphoma was confirmed to be in complete remission 9 mo later (Fig. 2 A and B).
Fig. 2.
Endoscopic and histological images from H. suis–infected patients. (A) Endoscopic images obtained from patient A and patient B pre- and posteradication. Patient A: Nodular gastritis from the antrum to the angle and the mucosal thickening in the angle of the stomach, endoscopically observed preeradication, improved 9 mo after eradication. Patient B: The open and linear multiple gastric ulcers observed before eradication disappeared. Histological examination of gastric biopsies obtained from patient A (B) and patient B (C) pre- and posteradication. Patient A: Preeradication, diffuse infiltration of lymphocytes and lymphoepithelial lesion (arrow) were observed by H&E staining. Immunostaining of infiltrating lymphocytes within the follicle was positive for CD20 and negative for CD3. Improvement of diffuse infiltration of lymphocytes and improvement of lymphoepithelial lesions were confirmed 3 mo after eradication. Patient B: Improvement of an infiltration of neutrophils observed by H&E staining pretreatment was confirmed posteradication. In both patients, bacteria with spiral morphology very different from that of H. pylori were observed by Giemsa staining.
Patient B was diagnosed with multiple H. suis–infected gastric ulcers by endoscopy (Fig. 2A) and histology (Fig. 2C) and was successfully treated with the same regimen as patient A (Fig. 2 A and C). Long and tightly coiled spiral microorganisms, distinct from H. pylori, were observed by Giemsa staining in patients A and B (Fig. 2 B and C).
Patient C was diagnosed with nodular gastritis and successfully treated with the same regimen as patient A.
Effects of H. suis Infections in Mice.
Clinical isolates of H. suis NHP19-4003 and NHP19-4004 and the mouse-adapted strain SNTW101c were used for experimental infection of mice. Because we knew that at least 3 mo were required to see the pathogenetic features in previous mouse infection experiments using the mouse-adapted strain TKY (16), we selected an infection duration of 4 mo for this study.
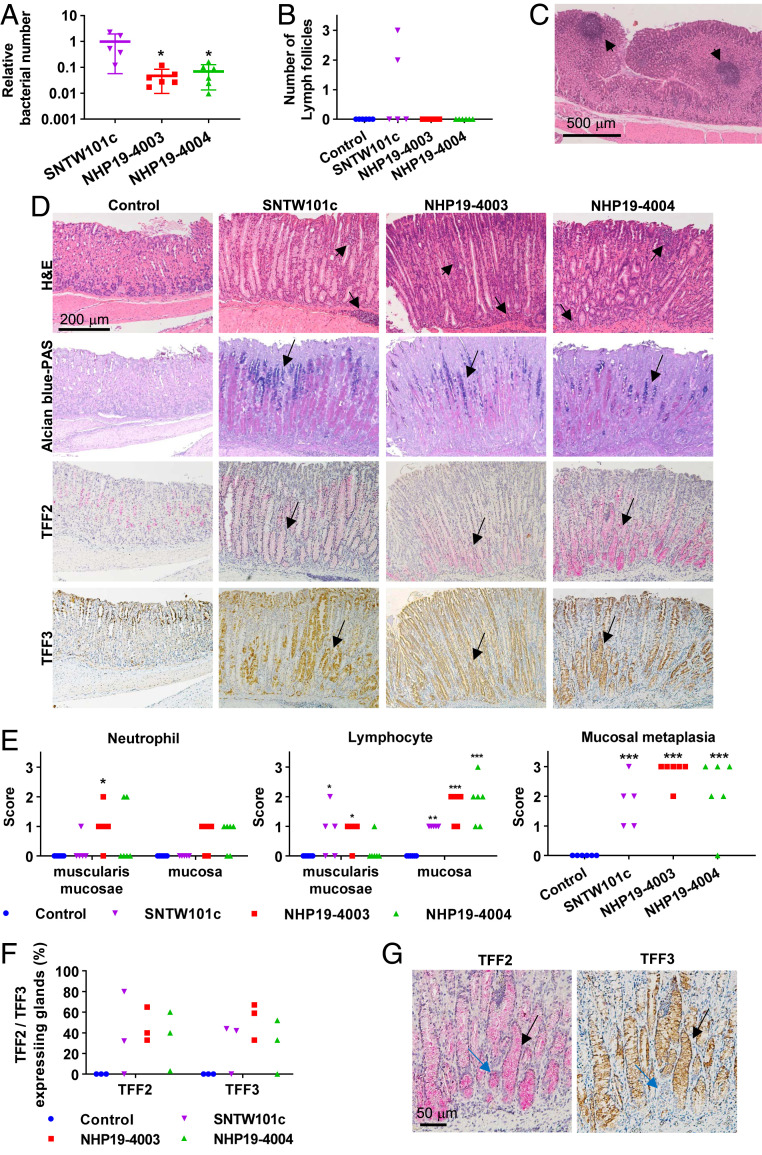
All isolates successfully infected the mice. The 4 mo after inoculation isolates were recovered from all mice. Comparison of bacterial numbers between isolates revealed that SNTW101c-infected mice contained 16 and 13 times as many bacteria as NHP19-4003– and NHP4004–infected mice, respectively (Fig. 3A). H. suis infection was further confirmed by serum H. suis–specific IgG measured by enzyme-linked immunosorbent assay (SI Appendix, Fig. S4).
Fig. 3.
Effects of H. suis infection in mice. (A) Relative bacterial number of H. suis in the mouse stomach. Bars indicate means ± SD. (B) Number of lymph follicles observed in stomach sections from H. suis–infected and control mice. (C) H&E staining of a lymph follicle (arrows) observed in mice infected with H. suis SNTW101c. (D) H&E, Alcian blue–PAS, TFF2, and TFF3 staining of stomach sections from H. suis–infected and control mice. Lymphocyte infiltration in mucosa and muscularis mucosae (arrows) was observed in H. suis–infected mice. Acidic mucus, stained blue with Alcian blue–PAS (arrows), was observed in H. suis–infected mice. Expression of TFF2 and TFF3 was observed at the bases of glands (arrows) in H. suis–infected mice. (E) Scoring of neutrophil and lymphocyte infiltration in muscularis mucosae and mucosa and mucosal metaplasia of stomach sections from H. suis–infected and control mice. Bars indicate means ± SD. (F) Percentage of glands expressing TFF2 or TFF3 in H. suis–infected and control mice. (G) TFF2- or TFF3-expressing glands of stomach from H. suis NHP19-4004–infected mice. Most glands expressed both TFF2 and TFF3 (black arrows), and rare glands (blue arrows) expressed only TFF2. In each of the H. suis–infected samples, the relative bacterial number of H. suis in the stomach of NHP19-4003– or NHP19-4004–infected mice was compared with that of SNTW101c–infected mice by one-way ANOVA with Dunnett’s test. Scores for cell infiltration observed in stomach sections in H. suis–infected mice were compared with those in control mice by two-way ANOVA with Sidak’s multiple comparisons test. Numbers of lymph follicles and scores for mucosal metaplasia in H. suis–infected mice were compared with those in controls by one-way ANOVA with Holm–Sidak’s multiple comparisons test. *P < 0.05, **P < 0.01, ***P < 0.001.
RANTES and TNF-α levels were both significantly higher in NHP19-4003–infected mice than in control mice, and RANTES level was significantly higher in NHP19-4004–infected mice than in control mice (SI Appendix, Fig. S5).
H&E staining of gastric mucosal tissue revealed lymphocyte infiltration in stomach mucosa of all H. suis–infected mice (Fig. 3 D and E). Significant neutrophil infiltration was observed in muscularis mucosae and/or mucosa of stomach in some NHP19-4003– and NHP19-4004–infected mice (Fig. 3 D and E). Lymphoid aggregates were observed in most H. suis–infected mice, and lymphoid follicles were especially prominent in SNTW101c-infected mice (Fig. 3 B and C). Alcian blue– periodic acid-Schiff (PAS) staining revealed mucosal metaplasia in almost all H. suis–infected mice, especially in NHP19-4003–infected mice (Fig. 3 D and E and SI Appendix, Fig. S6). Mucosal metaplasia was further evaluated by staining for the trefoil factor 2 (TFF2) and trefoil factor 3 (TFF3), representing spasmolytic polypeptide-expressing metaplasia (SPEM) and intestinal metaplasia (IM), respectively. Most H. suis–infected mice were positive for TFF2 and TFF3 and were confirmed to have late-stage SPEM or IM. TFF2 was coexpressed with TFF3 in most stomach glands of H. suis–infected mice (Fig. 3 D and G). The percentage of glands expressing TFF2 or TFF3 was calculated in three randomly selected mice in each group, revealing significant expression of TFF2 and TFF3 in all NHP19-4003–infected mice and two of three SNTW101c- and NHP19-4004–infected mice. By contrast, none of the control mice expressed TFF2 or TFF3 (Fig. 3F).
Genomic Comparisons of H. suis between Isolates from Humans and Pigs.
Genome sequences of H. suis isolates from humans and pigs (NHP19-4003, NHP19-4004, and NHP19-4022 from humans; and NHP19-0020 and NHP19-0033 from pigs) were determined by short- and long-read sequencing (SI Appendix, Table S2). The complete genomes of four isolates (NHP19-4003, NHP19-4004, NHP19-4022, and NHP19-0020) and the draft genome of NHPH190033 were determined. The complete genomes of the four isolates revealed that H. suis harbors plasmids carrying genes associated with DNA restriction–modification systems, as previously reported in SNTW101c (14). Importantly, H. suis was recovered from all stomachs of infected mice, and short-read sequencing revealed that their genomes were identical to those of the inoculated isolates (SI Appendix, Table S3), suggesting that H. suis infection was involved in the observed symptoms.
Based on multilocus sequence typing (MLST) analysis, NHP19-4003, NHP19-4004, and NHP19-4022 belonged to sequence types (STs), which were assigned as ST139, ST140, and ST141, respectively. Both NHP19-0020 and NHP19-0033 belonged to ST15, like the pig isolates B-V57 and 5Q from Belgium and Czech Republic, respectively. Phylogenetic trees constructed from a set of MLST data of 185 isolates, including five isolates in this study, revealed that all human isolates were scattered among the branches of pig isolates, suggesting that human and pig isolates are genetically very close (SI Appendix, Fig. S7).
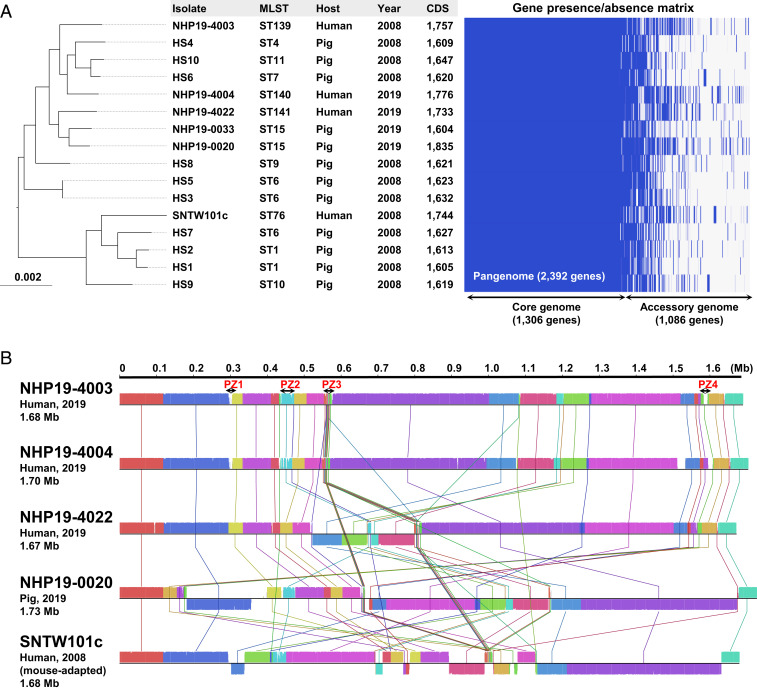
Genome-wide characterization and phylogenetic analysis of 16 genomes of H. suis, including 5 isolates from this study and 10 pig isolates from Belgium (17), further suggested a high genomic similarity between isolates from humans and pigs (Fig. 4A). Total numbers of coding sequences of H. suis were from 1,604 to 1,835 (the mean was 1,667). H. suis isolates shared a core genome of 1,306 genes, and therefore, each isolate possessed an accessory genome consisting of several hundred strain-specific genes, which resulted in a 2,392-gene pangenome (Fig. 4A). Comparative genomic analysis of five chromosomes of H. suis isolates (NHP19-4003, NHP19-4004, NHP19-4022, NHP19-0020, and SNTW101c) revealed that large recombination events have occurred in these genomes (Fig. 4B). Of note, each chromosome contained four nonconserved genomic regions, named plasticity zones (PZs) 1 to 4, which often contained genes associated with the type IV secretion system (T4SS), such as T4SS-specific ATPase genes virB4, virB11, and virD4, as in the cag pathogenicity island (cagPAI) in H. pylori (Fig. 4B and SI Appendix, Fig. S8).
Fig. 4.
Comparative analysis based on whole-genome data of H. suis isolates. (A) Pangenome analysis using genomes from the indicated 16 isolates. Phylogenetic trees generated using core genome alignments. Sequence type (ST), host, year of isolation, number of coding sequences, and gene presence/absence matrix are shown. Bar lengths represent the number of substitutions per site in the core genome. The pangenome of H. suis consisted of 2,392 genes, including 1,306 core genes (≥99.0% conserved in the isolates) and 1,086 accessory genes (<99.0% conserved in the isolates). (B) Linear sequence comparison of chromosomes of five isolates. Four genomic regions corresponding to PZs in NHP19-4003 are shown.
Discussion
NHPH in the human stomach was named “Helicobacter heilmanii” in acknowledgment of Konrad Heilmann’s work. In the 1990s, DNA sequencing of the 16S ribosomal RNA (rRNA) genes of NHPH from humans revealed that “H. heilmanii” included several species; however, the identification of individual species was difficult because they are genetically similar and unculturable. Later, H. suis was shown to be the most prevalent NHPH species in the human stomach (6). H. heilmannii is now identified as a species from cats (18). Culture of H. suis and H. heilmannii (from pig and cat, respectively) revealed that low pH is an important condition for culture in vitro (12, 18). Our preliminary results suggested that neutral pH has a significant effect on the viability of H. suis even after 1 h (https://www.youtube.com/watch?v=J68oZADtDj4&feature=youtu). Therefore, we used low-pH medium for the transport of gastric biopsies in this study, enabling us to isolate H. suis directly from human gastric biopsies.
Successful culture of H. suis from humans could help to answer many questions about the best therapy for the eradication of H. suis, the transmission route of H. suis, and the virulence factors contributing to H. suis pathogenicity in humans. H. suis can be effectively eradicated using a standard triple therapy, including amoxicillin and clarithromycin, that has also been used for the eradication of H. pylori infection (19, 20). Although clarithromycin resistance in H. pylori is the cause of most eradication failures in Japan (21), MICs of H. suis isolates obtained from humans demonstrated that three of the isolates were not resistant to clarithromycin. This is further supported by the fact that there are no mutations in the gene encoding 23S rRNA, the target protein of macrolides, corresponding to the position conferring clarithromycin resistance in H. pylori (22). Because low pH is necessary for H. suis growth, MICs of antimicrobial agents in H. suis tend to be higher than those in H. pylori, especially in the case of antimicrobial agents that degrade under acidic conditions. In this study, the MIC of amoxicillin in NHP19-4004 was high, possibly due to several mutations in PBP2 and FtsI. However, because the MIC of oxacillin in this strain was below 0.25 mg/L, these mutations are not critical for β-lactam resistance. Further studies are necessary to confirm a breakpoint for H. suis eradication therapy.
Comparative genomics of H. suis isolates from humans and pigs demonstrated that human isolates have a high genomic similarity to pig isolates (Fig. 4). H. suis has also been reported to infect monkeys (6, 23). A comparison of MLST data indicated that H. suis isolates from humans are less similar to isolates from monkeys than to isolates from pigs (SI Appendix, Fig. S7). Infection with Helicobacter spp. in the stomachs of slaughter pigs is highly prevalent (60 to 95%) in many countries (6, 7). In addition, H. suis infection has not been reported in animals other than pigs and monkeys (6). The H. suis–infected patients in this study were working in urban areas in Japan and had no direct contact with pigs or monkeys. Therefore, one possible route of infection is foodborne transmission, such as contaminated pig meat. Further analysis of H. suis from both pigs and humans will be necessary to identify the route of infection.
Infection of mice with H. suis clinical isolates NHP19-4003 and NHP19-4004, as well as the mouse-adapted strain SNYW101c, caused severe inflammatory responses in the gastric mucosa without exception. Mucosal metaplasia (SPEM and IM) characterized by the expression of TFF2 and TFF3, respectively, was observed in the stomachs of almost all H. suis–infected mice. Gastric IM is a preneoplastic gastric lesion that is also caused by the chronic infection of H. pylori in humans (11). Gastric SPEM is another type of TFF2-expressing metaplasia that is proposed to arise in chronic H. pylori infection in gerbils and has been detected in preneoplastic lesions in both humans and rodents (24, 25). SPEM has also been observed in H. pylori–infected mice, whereas IM has rarely been observed within 1 y of infection (25, 26). Observation of H. pylori infection in Mongolian gerbils suggests that SPEM likely represents a precursor of IM (24). In this study, we observed significant IM as well as SPEM in H. suis–infected mice, indicating that IM characterized by TFF3 expression in H. suis–infected mice may progress earlier than in H. pylori–infected mice. Although the effects of Helicobacter infection in rodents cannot be simply generalized to the effects in humans, as H. suis is the second-most prevalent Helicobacter species in human stomach, further investigations are needed to confirm the relationship between H. suis infection and gastric preneoplasia in human. In the case of H. pylori, two major virulence factors, cytotoxin-associated gene A (CagA) and vacuolating cytotoxin A (VacA), contribute significantly to pathogenesis. CagA, the only protein secreted via the T4SS encoded in cagPAI in H. pylori, elicits gastric epithelial proliferative and inflammatory responses (11). VacA is a toxin secreted via the autotransporter system in H. pylori and induces gastric epithelial cell death and T cell dysfunction (11). Although H. suis infection in mice was associated with gastric mucosal metaplasia even earlier than in H. pylori infection, H. suis does not possess orthologs of these major virulence factors of H. pylori, suggesting the contribution of other virulence factors common to H. suis and H. pylori. Common pathogenic factors, so-called pathogen-associated molecular patterns (PAMPs), include bacterial components such as lipopolysaccharide, peptidoglycan, and flagellins. H. pylori has three additional VacA paralogs, ImaA, VlpC, and FaaA, all of which are important for bacterial virulence; they are thought to have distinct functions (27). H. suis has a homolog of H. pylori VacA paralog-like autotransporter protein (gene product of NHP194003_11930 in NHP19-4003), which represents a candidate virulence factor. γ-glutamyl transpeptidase (GGT) is a membrane-associated enzyme that converts glutamine into glutamate and ammonia, as well as glutathione into glutamate and cysteinylglycin, and is involved in bacterial metabolism, gastric epithelial cell death, and T cell dysfunction during H. pylori infection (28). H. suis also has GGT, a potential virulence factor (29).
Nakamura et al. reported the formation of gastric MALT lymphoma in mice 6 mo after exposure to gastric mucus homogenate from mice infected with H. suis obtained from a cynomolgus monkey (23). In this study, infection of mice with a mouse-adapted strain of H. suis, SNTW101c, yielded a higher number of lymphoid follicles in the gastric mucosa than NHP19-4003 and NHP19-4004 infection (Fig. 3B). Bacterial load in the stomachs of SNTW101c-infected mice was more than 10-fold higher than those of NHP19-4003– and NHP19-4004–infected mice (Fig. 3A). Therefore, the differences in virulence-associated factors and PAMPs among H. suis strains could make a difference in the adaptive ability. We also observed differences in the progression of gastric mucosal metaplasia among mice infected with different H. suis strains: 4 mo after infection, gastric mucosal metaplasia was observed in all NHP19-4003–infected mice but not in all NHP19-4004–infected mice, even though bacterial loads did not differ (Fig. 3), suggesting that virulence differs among H. suis clinical isolates. Pangenomic comparison between H. suis isolates revealed that NHP19-4003 has 451 strain-specific genes in the accessory genome (Fig. 4A and https://dx.doi.org/10.17632/fvbvgz39f4.1). Although the function of most accessory genes is unknown, many T4SS-associated genes are included (SI Appendix, Fig. S8). Some types of H. pylori isolates have other T4SSs, termed tfs3 and tfs4, encoded in the PZs in their chromosomes independent of cagPAI (30, 31). The roles of tfs3 and tfs4 in H. pylori infection remain unknown, but some studies have suggested that the additional T4SSs are associated with bacterial virulence (30, 31). H. pylori cagPAI T4SS spontaneously secretes cytosolic metabolites, such as peptidoglycan and heptose 1,7-bisphosphate, that are recognized as PAMPs by host innate immune systems (32). Given that T4SS in the PZ is a strain-specific virulence factor for H. suis, and that activity is involved in the secretion of known and unknown PAMPs, leading to inflammatory responses, T4SS might affect the risks and outcomes of infection, such as gastric IM. Further analyses, including transcriptomics, proteomics, and metabolomics, will be required to identify the functions and impacts of accessory genes encoded in PZs during H. suis infection.
In conclusion, we successfully isolated H. suis from patients with gastric diseases, reconstituted bacterial pathogenicity using clinical isolates in laboratory mice, and recovered bacteria from the infected mice. Thus, we have certified the virulence of H. suis as a human gastric pathogen in accordance with Koch’s postulates. Further genomic epidemiological research on isolates from humans and animals will be useful for exploring the virulence of H. suis, its transmission to humans, and the diagnosis and treatment of H. suis–elicited gastric diseases. In any case, further research is needed to clarify how H. suis causes gastric disease in humans.
Materials and Methods
Subjects and Specimen Collection.
Gastric specimens were taken endoscopically from three H. pylori–negative patients: patient A (42-y-old female), patient B (52-y-old male), and patient C (45-y-old male). Ethical approvals were obtained from the ethics committee of Kyorin University School of Medicine and the medical research ethics committee of the National Institute of Infectious Diseases (NIID) for the use of human subjects. Written informed consent was obtained from all participants. H. pylori infection status in all patients was checked by urease breath test, rapid urease test, and pathological examination and confirmed to be negative by at least two tests. Gastric specimens from experimental pigs (11-wk-old), from a pig farm in Japan, were provided by the Nippon Institute for Biological Science in Tokyo, Japan in December of 2019. All gastric specimens from human subjects and pigs were transported to NIID using NHPH transport media for culture, as described below.
H. suis Isolation and MIC Measurement.
As described previously (12, 33), NHPH medium consisting of Brucella broth (Difco Becton Dickinson), 20% fetal bovine serum (Gibco), Skirrow supplement (Oxoid), Vitox supplement (Oxoid), and 0.05% HCl (FUJIFILM Wako; final pH is ∼5.0) was used for essentially all H. suis isolation procedures. NHPH transport media containing NHPH medium and 0.4% agar (Difco Becton Dickinson) was used for transportation of gastric biopsies. Transportation time from gastric biopsy collection to culture was about 6 h for patients A and B and 30 h for patient C. Homogenized gastric specimens were inoculated onto NHPH agar plates containing NHPH medium and 1.5% agar (Difco Becton Dickinson) and incubated under microaerobic conditions (5% O2, 12% CO2) in Forma Series II 3110 Water-Jacketed CO2 Incubators (Thermo Fisher Scientific). Single colonies on NHPH agar plates were subcultured on agar plates and further enriched by modified biphasic culture. For the enrichment, cells were cultured for 3 to 5 d in a biphasic NHPH medium consisting of slant NHPH medium agar and broth in a culture flask with gentle shaking (50 rpm). Bacterial number was observed microscopically, and the broth part of the biphasic culture was exchanged by centrifugation until the bacterial number reached ∼108 cells/mL All isolates were stocked in Brucella broth containing 30% glycerol at −80 °C for further experiments.
Susceptibility testing of H. suis isolates was performed by the broth microdilution method described in SI Appendix, Supplemental Materials and Methods. The MIC value was determined as the lowest concentration that inhibited bacterial growth. H. suis strain SNTW101c, recently isolated from mouse stomach, was also used in this study (14).
Genomic Sequencing and Comparative Genomic Analysis.
Genomic DNA from H. suis isolates was extracted using Genomic-tips 20/G and buffers (Qiagen) and sequenced on the MiniSeq platform (Illumina) using High Output Reagent Kit (300 cycles) and on the MinION platform (Oxford Nanopore Technologies [ONT]) using the R9.4.1 flow cell. The library for Illumina sequencing (paired-end, insert size of 500 to 900 bp) was prepared using Nextera XT DNA Library Prep Kit (Illumina), and the library for ONT sequencing was prepared using the Rapid Sequencing Kit (SQK-RAD004). ONT reads were base called with Guppy version 3.4.4 (https://github.com/joshwcomeau/guppy). For complete genome analysis, obtained Illumina and ONT reads were hybrid-assembled de novo using Unicycler version 0.4.8 (https://github.com/rrwick/unicycler). For draft genome analysis, Illumina reads were assembled de novo using Shovill version 1.0.4 (https://github.com/tseemann/shovill). The resultant genome sequences were annotated using the DNA Data Bank of Japan (DDBJ) Fast Annotation and Submission Tool (https://dfast.ddbj.nig.ac.jp/) (SI Appendix, Table S2). All genome sequences have been deposited at GenBank/European Molecular Biology Laboratory/DDBJ under BioProject number PRJDB8704.
Pangenome analysis of H. suis using the 5 genomes in this study and 11 publicly available genomes from the Pathosystems Resource Integration Center database (https://www.patricbrc.org/) was performed using Roary version 3.13.0 (https://sanger-pathogens.github.io/Roary/) with default settings (minimum identity for Protein Basic Local Alignment Search Tool: 95%) and visualized using Phandango version 1.3.0 (https://jameshadfield.github.io/phandango). Phylogenetic analysis based on the core genome alignments of H. suis from the Roary pipeline was performed using Randomized Axelerated Maximum Likelihood version 1.0.0 (https://github.com/stamatak/standard-RAxML) with 1,000 bootstraps and visualized using FigTree version 1.4.4 (http://tree.bio.ed.ac.uk/software/figtree/). Linear comparison of the chromosomal sequences of complete genomes of H. suis, including four genomes from this study, was performed using progressiveMauve version 2015_02_13.0 (http://darlinglab.org/mauve/mauve.html) with default settings. Genome sequencing and single-nucleotide variant/indel detection analysis of H. suis isolates recovered from infected mice are described in SI Appendix, Table S3).
Animal Experiments.
The 4-wk-old female C57BL/6NCrl mice (Charles River Japan) were infected intragastrically using a feeding needle with 1 × 108 colony-forming units of the indicated strain of H. suis. All mice were bred at the animal facility of the Kitasato Institute under an approved study protocol.
After 4 mo had passed following the infection, mice were killed, and the stomach and serum were collected from each mouse for evaluation. Mouse stomachs were cut along the greater curvature, rinsed with phosphate buffered saline (pH7.4) containing 0.01% (weight/volume) gelatin, and half of the stomach was subjected to histopathological examination. Longitudinal strips from the fundus, upper body, lower body, and antrum were fixed with 10% (wt/vol) neutral-buffered formalin (FUJIFILM Wako) and embedded in paraffin. Tissue sections ∼4 µm thick were prepared and mounted on glass slides. The sections were stained with H&E and Alcian blue–PAS. The images were captured with a Nano Zoomer XR (Hamamatsu Photonics). Histopathological scoring was performed as shown in SI Appendix, Table S4.
For analyzing metaplastic changes, we stained the sections with TFF2 for SPEM and TFF3 for IM. The sections were deparaffinized with HistoClear (National Diagnostics) and quenched with an alcohol series, and then endogenous peroxidase was blocked with 3% H2O2 in methanol for 3,3′-diaminobenzidine (DAB) staining. Antigen was retrieved by microwave heating for 20 min in an Immunosaver (Fujifilm Wako). For primary antibody conjugation, the section was blocked with serum and incubated overnight with anti-TFF2 antibody (1:100) [a kind gift from Sir Nicholas Wright (34)] or anti-TFF3 antibody (1:1,000) (UCLNT3, Thermo Fisher Scientific). The avidin-biotin complex (ABC) method (Vectastain ABC kit, Vector) was used to visualize staining. The Peroxidase–DAB system was used for TFF3 staining, and the Alkaline phosphatase–Vector Red system was used for TFF2 staining.
The other half of the stomach was homogenized in Brucella broth following treatment with 1% HCl for 45 min and then used for H. suis isolation (33) and DNA extraction. Relative H. suis count in mouse stomach was evaluated by the comparative ∆Ct method using probe-based qPCR targeting the gene encoding the H. pylori VacA-like autotransporter protein (NHP194003_11930 in NHP19-4003, SI Appendix, Supplemental Materials and Methods).
Serum cytokines, chemokines, and H. suis–specific IgG were measured by the methods shown in SI Appendix, Supplemental Materials and Methods.
All statistical analyses were performed using GraphPad Prism 8.4.1 (La Jolla) as shown in SI Appendix, Supplemental Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Akira Takeda (Takeda Hospital, Fukuoka, Japan) for providing gastric biopsies from a patient. We thank Dr. Shigetarou Mori (Department of Bacteriology II, NIID, Tokyo, Japan) for his advice on the experiments. We thank Hirotaka Kobayashi (Department of Pathology, NIID, Tokyo, Japan) for his support with scanning electron microscopic observation. We thank Midori Ozaki (Department of Pathology, NIID, Tokyo, Japan) and Miki Furuya (Department of Gastrointestinal Surgery, Graduate School of Medicine, The University of Tokyo, Tokyo, Japan) for their support with histological observations. We thank Sir Nicholas Wright for providing the anti-TFF2 antibody. We thank Dr. Tsutomu Tomida and Dr. Mitsuhiro Hashimoto (Timelapse Vision Inc., Saitama, Japan) for their support with microscopic observation of live bacteria. We thank Yuta Akaike (Nippon Institute for Biological Science, Tokyo, Japan) for his experimental support. We thank Dr. Masaru Usui (Rakuno Gakuen University, Hokkaido, Japan) for providing stool samples from domestic pigs. We thank Dr. Maya Watanabe (Inagi Municipal Hospital, Tokyo, Japan) for introducing a patient. We also thank Dr. Kiyotaka Nagahama (Department of Pathology, Kyorin University School of Medicine, Tokyo, Japan) for pathological consideration of patients’ gastric specimens. This work was supported by the Ministry of Education, Culture, Sports, Science and Technology/Japan Society for the Promotion of Science Grant Numbers JP19H03474 for (H. Matsui) and JP20K08365 for (K.T.), and Japan Agency for Medical Research and Development Grant Numbers JP20fk0108148 (E.R.) and JP20fk0108133 (M.S.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2026337118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Dent J. C., McNulty C. A., Uff J. C., Wilkinson S. P., Gear M. W., Spiral organisms in the gastric antrum. Lancet 2, 96 (1987). [DOI] [PubMed] [Google Scholar]
- 2.Debongnie J. C., et al., Gastric ulcers and Helicobacter heilmannii. Eur. J. Gastroenterol. Hepatol. 10, 251–254 (1998). [DOI] [PubMed] [Google Scholar]
- 3.Morgner A., Bayerdörffer E., Meining A., Stolte M., Kroher G., Helicobacter heilmannii and gastric cancer. Lancet 346, 511–512 (1995). [DOI] [PubMed] [Google Scholar]
- 4.Morgner A., et al., Helicobacter heilmannii-associated primary gastric low-grade MALT lymphoma: Complete remission after curing the infection. Gastroenterology 118, 821–828 (2000). [DOI] [PubMed] [Google Scholar]
- 5.Goji S., et al., Helicobacter suis-infected nodular gastritis and a review of diagnostic sensitivity for Helicobacter heilmannii-like organisms. Case Rep. Gastroenterol. 9, 179–187 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Haesebrouck F., et al., Gastric helicobacters in domestic animals and nonhuman primates and their significance for human health. Clin. Microbiol. Rev. 22, 202–223 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Foss D. L., et al., Identification of Helicobacter suis in pig-producing regions of the United States. J. Swine Health Prod. 21, 242–247 (2013). [Google Scholar]
- 8.Mendall M. A., et al., Childhood living conditions and Helicobacter pylori seropositivity in adult life. Lancet 339, 896–897 (1992). [DOI] [PubMed] [Google Scholar]
- 9.Blaecher C., et al., Significantly higher frequency of Helicobacter suis in patients with idiopathic parkinsonism than in control patients. Aliment. Pharmacol. Ther. 38, 1347–1353 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Warren J. R., Marshall B., Unidentified curved bacilli on gastric epithelium in active chronic gastritis. Lancet 1, 1273–1275 (1983). [PubMed] [Google Scholar]
- 11.Polk D. B., Peek R. M. Jr, Helicobacter pylori: Gastric cancer and beyond. Nat. Rev. Cancer 10, 403–414 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baele M., et al., Isolation and characterization of Helicobacter suis sp. nov. from pig stomachs. Int. J. Syst. Evol. Microbiol. 58, 1350–1358 (2008). [DOI] [PubMed] [Google Scholar]
- 13.Koch R., Ueber den augenblicklichen Stand der bakteriologischen Choleradiagnose. Z. Hyg. Infektionskr. 14, 319–338 (1893). [Google Scholar]
- 14.Rimbara E., et al., Complete genome sequence of Helicobacter suis strain SNTW101c, originally isolated from a patient with nodular gastritis. Microbiol. Resour. Announc. 9, e01340-19 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Matsui H., et al., Development of new PCR primers by comparative genomics for the detection of Helicobacter suis in gastric biopsy specimens. Helicobacter 19, 260–271 (2014). [DOI] [PubMed] [Google Scholar]
- 16.Matsui H., et al., Mouse models for assessing the protective efficacy of Lactobacillus gasseri SBT2055 against Helicobacter suis infection associated with the development of gastric mucosa-associated lymphoid tissue lymphoma. Helicobacter 20, 291–298 (2015). [DOI] [PubMed] [Google Scholar]
- 17.Vermoote M., et al., Genome sequence of Helicobacter suis supports its role in gastric pathology. Vet. Res. 42, 51 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Smet A., et al., Helicobacter heilmannii sp. nov., isolated from feline gastric mucosa. Int. J. Syst. Evol. Microbiol. 62, 299–306 (2012). Correction in: Int. J. Syst. Evol. Microbiol. 62 1016 (2012). [DOI] [PubMed] [Google Scholar]
- 19.Takigawa H., et al., Helicobacter suis infection is associated with nodular gastritis-like appearance of gastric mucosa-associated lymphoid tissue lymphoma. Cancer Med. 8, 4370–4379 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Goddard A. F., Logan R. P., Atherton J. C., Jenkins D., Spiller R. C., Healing of duodenal ulcer after eradication of Helicobacter heilmannii. Lancet 349, 1815–1816 (1997). [DOI] [PubMed] [Google Scholar]
- 21.Hoshiya S., et al., Relationship between eradication therapy and clarithromycin-resistant Helicobacter pylori in Japan. J. Gastroenterol. 35, 10–14 (2000). [PubMed] [Google Scholar]
- 22.Versalovic J., et al., Mutations in 23S rRNA are associated with clarithromycin resistance in Helicobacter pylori. Antimicrob. Agents Chemother. 40, 477–480 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nakamura M., et al., “Candidatus Helicobacter heilmannii” from a cynomolgus monkey induces gastric mucosa-associated lymphoid tissue lymphomas in C57BL/6 mice. Infect. Immun. 75, 1214–1222 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yoshizawa N., et al., Emergence of spasmolytic polypeptide-expressing metaplasia in Mongolian gerbils infected with Helicobacter pylori. Lab. Invest. 87, 1265–1276 (2007). [DOI] [PubMed] [Google Scholar]
- 25.Weis V. G., Goldenring J. R., Current understanding of SPEM and its standing in the preneoplastic process. Gastric Cancer 12, 189–197 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Weis V. G., et al., Heterogeneity in mouse spasmolytic polypeptide-expressing metaplasia lineages identifies markers of metaplastic progression. Gut 62, 1270–1279 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Foegeding N. J., Caston R. R., McClain M. S., Ohi M. D., Cover T. L., An overview of Helicobacter pylori VacA toxin biology. Toxins (Basel) 8, 173 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shibayama K., et al., Metabolism of glutamine and glutathione via gamma-glutamyltranspeptidase and glutamate transport in Helicobacter pylori: Possible significance in the pathophysiology of the organism. Mol. Microbiol. 64, 396–406 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Flahou B., et al., Gastric epithelial cell death caused by Helicobacter suis and Helicobacter pylori γ-glutamyl transpeptidase is mainly glutathione degradation-dependent. Cell. Microbiol. 13, 1933–1955 (2011). [DOI] [PubMed] [Google Scholar]
- 30.Kersulyte D., et al., Cluster of type IV secretion genes in Helicobacter pylori’s plasticity zone. J. Bacteriol. 185, 3764–3772 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Yuan X.-Y., Wang Y., Wang M.-Y., The type IV secretion system in Helicobacter pylori. Future Microbiol. 13, 1041–1054 (2018). [DOI] [PubMed] [Google Scholar]
- 32.Ying L., Ferrero R. L., Role of NOD1 and ALPK1/TIFA signalling in innate immunity against Helicobacter pylori infection. Curr. Top. Microbiol. Immunol. 421, 159–177 (2019). [DOI] [PubMed] [Google Scholar]
- 33.Blaecher C., et al., A novel isolation protocol and probe-based RT-PCR for diagnosis of gastric infections with the zoonotic pathogen Helicobacter suis. Helicobacter 22, 12369 (2017). [DOI] [PubMed] [Google Scholar]
- 34.Elia G., Chinery R., Hanby A. M., Poulsom R., Wright N. A., The production and characterization of a new monoclonal antibody to the trefoil peptide human spasmolytic polypeptide. Histochem. J. 26, 644–647 (1994). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.