Significance
The trabecular meshwork is the major tissue regulating intraocular pressure (IOP). Malfunction of this tissue results in ocular hypertension, the major risk factor for glaucoma. How IOP is regulated is still unknown, but it is accepted that mechanosensation and mechanotransduction are critical regulators. In this study, we report that primary cilia (PC) is a mechanosensor for stretch-induced autophagy in TM cells and identified a reciprocal activation of AKT1 and SMAD2/3 as a signaling pathway regulating the activation of autophagy by PC. Most importantly, we showed that PC-mediated autophagy plays a critical role in IOP homeostasis. These findings tremendously contribute to our understanding of TM biology and identify new potential targets for therapeutic strategies.
Keywords: autophagy, primary cilia, glaucoma, trabecular meshwork, stretching
Abstract
Activation of autophagy is one of the responses elicited by high intraocular pressure (IOP) and mechanical stretch in trabecular meshwork (TM) cells. However, the mechanosensor and the molecular mechanisms by which autophagy is induced by mechanical stretch in these or other cell types is largely unknown. Here, we have investigated the mechanosensor and downstream signaling pathway that regulate cyclic mechanical stretch (CMS)-induced autophagy in TM cells. We report that primary cilia act as a mechanosensor for CMS-induced autophagy and identified a cross-regulatory talk between AKT1 and noncanonical SMAD2/3 signaling as critical components of primary cilia-mediated activation of autophagy by mechanical stretch. Furthermore, we demonstrated the physiological significance of our findings in ex vivo perfused eyes. Removal of primary cilia disrupted the homeostatic IOP compensatory response and prevented the increase in LC3-II protein levels in response to elevated pressure challenge, strongly supporting a role of primary cilia-mediated autophagy in regulating IOP homeostasis.
The trabecular meshwork (TM) is a pressure-sensitive tissue located in the anterior segment of the eye and key regulator of intraocular pressure (IOP). Malfunction of this tissue results in improper drainage of aqueous humor outflow, leading to ocular hypertension, the major risk factor for developing glaucoma (1–3). The TM consists of an irregular lattice of collagen beams lined by TM endothelial-like cells, followed by a zone of loose connective tissue containing TM cells, through which aqueous humor must pass before leaving the eye. Changes in pressure gradients and fluid flow associated with eye movement, circadian rhythm, or the ocular pulse cause small and high variations in IOP, which are translated in continuous cycles of tissue deformation and relaxation. Cells in the TM are known to be able to sense these deformations as mechanical forces and respond to them by eliciting a variety of responses, including reorganization of actin cytoskeleton, changes in gene expression, secretion of cytokines, modulation of matrix metalloproteinases, and extracellular-matrix remodeling (reviewed in ref. 4). It is believed that these mechanoresponses are critical regulators of IOP homeostasis; however, the mechanosensors and the downstream mechanotransduction signaling in TM have still not been characterized.
Our laboratory has identified the activation of macroautophagy (hereafter autophagy) as one of the responses elicited in TM cells following application of static or cyclic mechanical stretch (CMS) (5–7). Activation of autophagy was also observed quickly after pressure elevation in porcine perfused eyes (5), which prompted us to propose autophagy as a crucial physiological response to adapt to mechanical forces and maintain cellular homeostasis. The exact roles of autophagy in TM cells and tissue function are still under investigation. Most recently, we have shown the CMS-induced translocation of the autophagy marker LC3 (microtubule-associated protein 1 light chain 3) to the nuclear compartment, where it associates with the nucleolus and interacts with the ribophagy receptor NUFIP1 (nuclear FMR1 interacting protein 1), suggesting a potential role of CMS-induced autophagy in surveillance of the nucleolus activity (6). Furthermore, we have also recently provided direct evidence demonstrating autophagy as a regulator of TGF-β/SMAD-induced fibrogenesis in TM cells (8).
Autophagy is a fundamental process for degradation or recycling of intracellular components, which is essential to maintain cellular homeostasis. Autophagy occurs constitutively at basal levels, but it is quickly activated upon several types of stress, such as nutrient depletion, pathogen infections, drug treatment, accumulation of aggregated proteins and damaged organelles, and mechanical stress (7, 9–11). The molecular mechanisms by which cells recognize stress and regulate autophagic activity are very complex and differ based on the stimuli. A variety of components, for example, receptor tyrosine kinases, second messengers (Ca2+ or cAMP [cyclic adenosine monophosphate]), protein kinases, and downstream autophagy-related (ATG) complexes, participate in the regulation of autophagy (10). Among them, the best characterized is MTOR (mechanistic target of rapamycin kinase), a negative regulator of autophagy (10, 12, 13). Although MTOR acts as a core sensor in autophagic regulation, numerous studies have shown the MTOR-independent autophagy activation upon different stresses (10). Indeed, our own study and that of King et al. showed that the induction of autophagy in response to mechanical stress occurs in an MTOR-independent manner (5, 14). The mechanosensor and the downstream signaling pathway responsible for the activation of autophagy in response to stretching have still not been identified.
Primary cilium (PC) is a nonmotile cell-surface projection found almost ubiquitously in vertebrate cells, which acts as a cellular antenna that senses a wide variety of signals, including chemical and mechanical stimuli (15–19). PC plays a critical role in smell, sight, and mechanosensation. PC defects are associated with a number of human diseases termed ciliopathies. The most common feature in patients affected with ciliopathies include visual dysfunction (16). In particular, Lowe syndrome patients often develop ocular hypertension and glaucoma (20). Structurally, the PC consists of a microtubule-based core structure, called axoneme, and a basal body, which is a derivative of the centriole of centrosome from which axoneme is extended and surrounded by ciliary membrane (21). The ciliary membrane is a specialized domain extension of the plasma membrane enriched on signaling receptors and channels, including hedgehog (Hh) and Ca2+ pathways, which enables the PC to function as a signaling hub (16, 22, 23). Cargo trafficking into and out of the cilium is mediated by a specialized form of vesicle trafficking, named intraflagellar transport (IFT), that is composed of a multiprotein complex (16, 23).
Recent studies have demonstrated the reciprocal relationship between PC and autophagy. Autophagy has been shown to both positively and negatively regulate ciliogenesis. Under nutrient-rich conditions, basal autophagy inhibits cilia growth by limiting trafficking of PC components to the basal body through direct degradation of IFT20 (24). In contrast, nutrient starvation triggers the autophagic degradation of oral-facial-digital syndrome 1 and promotes cilia growth (24, 25). Conversely, functional PC are required for activation of autophagy in response to starvation and fluid flow. In both cases, autophagy was initiated by the recruitment of ATG16L to the basal body, suggesting that this event is a hallmark for PC-induced autophagy. Intriguingly, the signaling pathway mediating PC-induced autophagy activation differed based on the stimuli. While Hh/smoothened (SMO) was reported to mediate activation of autophagy in response to starvation (24), the LKB1–AMPK–MTOR signaling pathway was found to regulate PC-induced autophagy and cell volume in kidney epithelial cells under shear stress (26, 27). Whether PC are also involved in the regulation of autophagy triggered by mechanical stretching has not yet been explored.
The purpose of this study is to investigate a potential role of PC in stretch-induced autophagy in TM cells. Here, we report that PC acts as a mechanosensor for CMS-induced autophagy, and we identified AKT1 and SMAD2/3 as critical components of the signal mechanotransduction.
Results
The PC Is a Mechanosensor for CMS-Induced Autophagy in Human TM Cells.
Our laboratory previously showed the activation of autophagy with mechanical stress in cultured human TM cells (5–7). Here, we wanted to identify the mechanisms by which TM cells sense mechanical forces and activate autophagy. First, we compared the gene-expression profile of autophagy-deficient TM cells under nonstretched (NS) and stretched conditions (SI Appendix, Fig. S1A). For this, we knocked down the expression of the autophagy genes Atg5 and Atg7 via siRNA (small interfering RNA) (siAtg5/7), as reported in ref. 8. Scrambled siRNA was used as control (siCNT). At day two posttransfection, cells were subjected to CMS for 24 h. Changes in gene-expression profile were evaluated by gene-array analysis using Affymetrix Human Clariom D chips in conjunction with Partek Flow and Genomic Suite analysis software, as detailed in Materials and Methods. Complete array data sets have been deposited at the Gene Expression Omnibus (GSE122652 and GSE159793). Comparative analysis identified genes differentially expressed in autophagy-deficient cells under CMS compared to NS cultures (>1.5-fold, P < 0.05, SI Appendix, Fig. S1B). We used this data set of genes to further investigate which biological processes were most significantly affected by autophagy in the stretched cells, via gene ontology analysis. SI Appendix, Table S1 lists the top 10 biological process identified. Literature searches revealed that 47% of the up-regulated genes within this list participate in primary ciliogenesis (Table 1). This is particularly relevant for our studies given the reported role of PC as a mechanosensitive and pressure-sensitive organelle, which functions as a signaling hub. Based on this, we decided to focus on investigating the potential role of PC in stretch-induced autophagy.
Table 1.
Genes differentially expressed in autophagy-deficient TM cells subjected to CMS with a role in ciliogenesis
| Gene symbol | Fold change | P value | Ciliogenesis | Reference | |
| 1 | BIRC5 | 1.56899 | 0.0296575 | * | 28 |
| 2 | PLK4 | 1.57484 | 0.0051299 | * | 29 |
| 3 | SPAG5 | 1.60081 | 0.0303941 | ||
| 4 | CDKN3 | 1.64845 | 0.0167822 | ||
| 5 | UBE2C | 1.65023 | 0.0232858 | ||
| 6 | CENPE | 1.66315 | 0.0437133 | * | 28 |
| 7 | TYMS | 1.81371 | 0.0121627 | ||
| 8 | CDC20 | 1.92132 | 0.0077336 | * | 30 |
| 9 | CCNB1 | 1.94963 | 0.0184941 | * | 31 |
| 10 | NCAPH | 1.95889 | 0.0494857 | ||
| 11 | AURKA | 1.96758 | 0.0305636 | * | 32 |
| 12 | TPX2 | 2.05589 | 0.0366254 | ||
| 13 | ANLN | 2.05604 | 0.0339942 | ||
| 14 | PLK1 | 2.18769 | 0.0167943 | * | 32 |
| 15 | KIF23 | 2.198 | 0.0232678 | * | 28 |
| 16 | AURKB | 2.23499 | 0.0235399 | ||
| 17 | DLGAP5 | 2.38002 | 0.0202057 | ||
| 18 | NUSAP1 | 2.50417 | 0.0455912 | ||
| 19 | PRC1 | 2.64531 | 0.0273041 | * | 28 |
*Indicates a role in ciliogenesis.
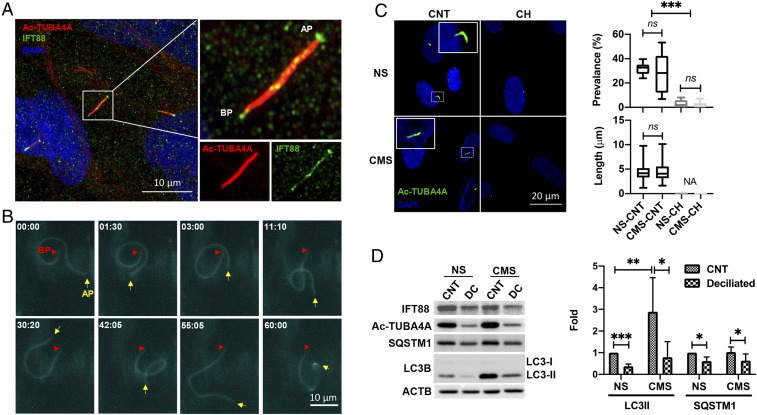
We first examined the presence of PC by immunocytochemistry and real-time, time-lapse live-cell imaging in TM cells. PC were identified as hair-like protrusions from the cell surface ∼4 to 6 µm in length, which coimmunostained by antibodies against acetylated-TUBA4A and the ciliary transport protein IFT88 (Fig. 1A and Movie S1). PC in live cells were observed by transfecting TM cells with p5HT6-mCherry, which expresses mCherry fluorescent protein at the ciliary membrane (33). Consistent with the results reported in another cell type (34), PC length in live cells is longer than that observed in fixed cells; cilia up to 10 to 20 µm in length were observed (Fig. 1B). Interestingly, time-lapse live-cell imaging showed that some portions of the PC move dynamically and bend even under NS normal cultured conditions (Fig. 1B and Movie S2), suggesting that PC are constantly sensing environmental changes to signal into the cell body.
Fig. 1.
PC is a mechanosensor for CMS-induced autophagy. (A) Representative immunocytochemical analysis of PC in TM cells. Acetylated TUBA4A (red fluorescence) and IFT88 (green fluorescence) antibodies were used to identify the PC. DAPI was used to stain nuclei. Images were acquired with super resolution confocal microscope and processed by using Fiji software. BP and AP represent basal and apical process, respectively. (B) Time-lapse live-cell imaging of PC in TM cells transfected with p5HT6-mCherry. Images were acquired with CELENA X High Content Imaging System equipped with the Ibidi stage-top incubation system and processed by using Fiji software. BP and AP are indicated by red arrowheads and yellow arrows, respectively. Captured time (minute : second) is represented in the upper left of each picture. (C) Prevalence and length of PC in NS and CMS cells disrupted by CH. Cells were treated in 4 mM CH for 3 d and subjected to CMS after CH removal. (Left) Images taken after 24 h in NS and CMS cells. Quantification analyses for the prevalence and length of PC were represented on the right. NS, nonstretch control; CMS, cyclic mechanical stretch; CH, chloral hydrate. Data are shown as the mean ± SD of three biological replicates (n = 10 for prevalence, n = 93 versus 55: NS versus CMS for length). NA, not applicable; ***P < 0.001; ns, not significant (ANOVA and Tukey’s post hoc test). (D) Protein levels of LC3-II and SQSTM1 in TM cells subjected to CMS (8% elongation, 24 h) with or without PC disruption, evaluated by WB. Band intensities were quantified by Image Lab touch software and normalized with β-actin (ACTB). Data are shown as the mean ± SD (n = 5 for LC3 II and 3 for SQSTM1). *P < 0.05; **P < 0.01; ***P < 0.001 (Student’s t test).
To investigate whether PC mediate CMS-induced autophagy in TM cells, we used two methods. We first attempted to block PC assembly by silencing IFT88 or IFT20 expression via siRNA. However, as shown in SI Appendix, Fig. S2, despite the reduced levels of the IFT88 and IFT20 proteins in the respective knockdown cultures, no significant decrease in acetylated tubulin was noted. Furthermore, immunofluorescence analysis revealed that PC were still present in these cells. Similar preservation of cilia structure with moderate knockdown expression of IFT20 and IFT88 has been previously reported in ref. 35. In view of this, we decided to chemically remove PC by treating the cells with the cilia disruptor chloral hydrate [CH, 4 mM (36)] for 3 d (37). Cells were then switched to fresh media to discard any potential effect of CH on autophagy or stretch response and subjected to CMS for 24 h. No detrimental effect on cell viability was observed (Movie S3). Efficient removal of PC by CH and the effect of CMS on PC prevalence and length was examined by immunocytochemistry. As seen in Fig. 1C, CH treatment significantly removed PC by over 90% (P < 0.001, n = 10, Fig. 1C). No significant changes were found in cilia prevalence (NS control [CNT] versus CMS CNT: 31.95 ± 5.17% versus 28.87 ± 16.72%, n = 10) or length (NS CNT versus CMS CNT: 4.17 ± 1.45 µm versus 4.40 ± 1.82 µm, n = 93 versus 55) at 24 h of CMS (Fig. 1C). Autophagy was evaluated by monitoring LC3-I to LC3-II lipidation in whole-cell lysates by Western blot (WB) analysis (5–7, 38). As shown in Fig. 1D, removal of PC significantly reduced CMS-induced LC3-II levels (0.23 ± 0.18 folds, P < 0.05, n = 5) compared to their nontreated control. A significant decrease in the protein levels of SQSTM1/p62 (sequestosome 1), another indicator of autophagy activity (38), was additionally found to be decreased (0.61 ± 0.19 and 0.63 ± 0.31 folds in NS and CMS, respectively, P < 0.05, n = 3) in deciliated cells.
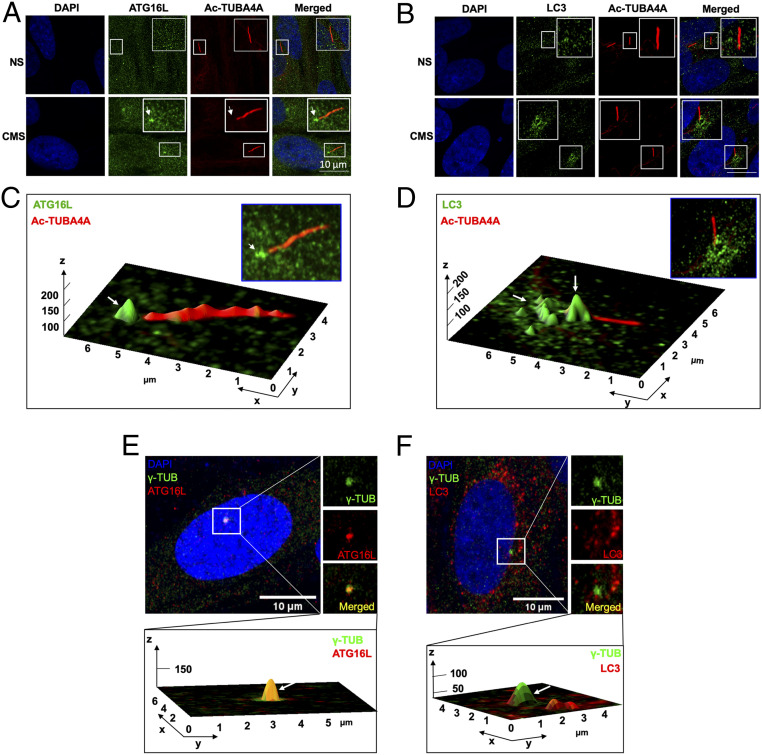
Recruitment of the autophagy protein ATG16L to the basal body of the PC has been reported to be required to initiate PC-dependent autophagy in response to starvation (24) and fluid flow (39), and it has been suggested to be one of the hallmarks for PC-induced autophagy. We examined by immunofluorescence in Airyscan confocal microscopy the recruitment of ATG16L and LC3 to the PC in CMS cells (Fig. 2). Both proteins, especially ATG16L, were found to be enriched at the basal body of cyclically stretched cells (Fig. 2 A–D). Furthermore, immunostaining with the basal cilia marker ɣ-tubulin showed 100% colocalization of Atg16L (Fig. 2E) and partial colocalization with LC3 (Fig. 2F). Altogether, these data very strongly indicate that PC mediate activation of autophagy in response to CMS in TM cells.
Fig. 2.
ATG16L and LC3 localize at the basal body of the PC upon CMS. (A and B) Representative immunostaining of ATG16L (A, green) and LC3 (B, green) in TM cells exposed for 24 h to CMS (8% elongation). PC were stained using acetylated TUBA4A (red); DAPI was used to stain nuclei. (C and D) Interactive three-dimensional (3D) surface plot analysis visualizing the accumulation of ATG16L (C, green) and LC3 (D, green) in the basal body of PC of the CMS cells (white arrows). (E and F) Representative immunostaining of ATG16L (A, red) and LC3 (B, red) with ɣ-tubulin (green), a maker for the basal body of PC, in TM cells exposed for 24 h to CMS (8% elongation). (Bottom) Interactive 3D surface plot analysis visualizing the colocalization of ATG16L or LC3 with ɣ-tubulin (white arrows).
Inhibition of Ca2+ Flux in PC Does Not Prevent CMS-Induced Autophagy in TM Cells.
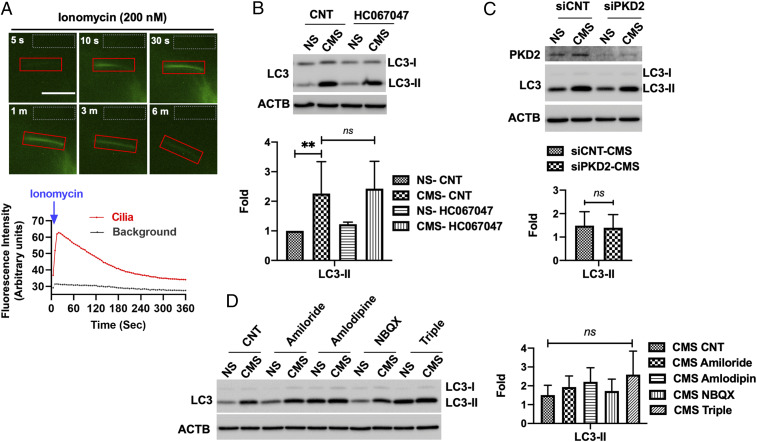
Next, we focused on the identification of the signaling pathway controlling CMS-induced autophagy via PC. A previous study reported that Ca2+ influx through the mechanosensitive channel TRPV4 regulates mechanosensation by PC in TM cells (40). Therefore, we investigated whether CMS-induced autophagy was mediated by a similar mechanism. First, we wanted to confirm that PC indeed mediate Ca2+ influx in TM cells, since this was not specifically tested in that report. For this, we transfected primary cultures of TM cells with p5HT6–G-GECO1.0, which expresses green fluorescent protein (GFP)-based Ca2+ indicator and target it to the cilia (34). Cells were then treated with 200 nM ionomycin, and Ca2+ influx in PC was monitored by live-cell time-lapse imaging. Faint GFP signals of 5HT6–G-GECO1.0 were observed in the majority of the PC at basal state. Upon stimulation with ionomcycin, GFP signals in the cilia started to increase reaching a maximal intensity at 30 s, after which it slowly decreased to basal levels at 6 min. (Fig. 3A and Movie S4). This data confirms that PC mediate Ca2+ influx in TM cells. We then evaluated the potential role of TRPV4-mediated Ca2+ influx in cyclic stretch-induced autophagy. TM cells were subjected to CMS in the presence or absence of HC067047 (10 µM), a TRPV4-specific inhibitor. As seen in Fig. 3B, inhibition of TRPV4-mediated Ca2+ influx did not prevent the increase in LC3-II by CMS.
Fig. 3.
Inhibition of Ca2+ influx in PC does not prevent CMS-induced autophagy in TM cells. (A) Time-lapse live-cell images of Ca2+ influx in PC of TM cells transfected with p5HT6–G-GECO1.0. Transfected cells were treated with ionomycin (200 nM), and the GFP fluorescence was monitored by the CELENA X live-cell imaging system for 10 min. The images were processed, and GFP intensity was quantified by Fiji software. PC-specific Ca2+ influx is shown in the red boxes. Area in gray boxes was used to measure background intensity of fluorescence. Quantitative analyses of alteration in GFP fluorescent intensities were represented in the lower panel. (B) Effect of HC067047 on CMS-induced autophagy. TM cells were subjected to CMS for 24 h in the absence or presence of HC067047 (10 µM). The expression levels of LC3 were measured in whole-cell lysates by WB. LC3-II band intensities were normalized by β-actin (ACTB). (C) Effect of PKD2 knockdown and (D) different types of calcium channel blockers on CMS-induced LC3-II levels, evaluated by WB. ACTB was used as a loading control. Graphs show fold changes in LC3-II levels calculated from the densitometric analysis of the bands. Data are shown as the mean ± SD (n = 3). **P < 0.01; ns, not significant (two-tailed unpaired Student’s t test or ANOVA and Tukey’s post hoc test).
We additionally explored the potential role of other functional transient receptor potential (TRP) Ca2+ channels and L-type Ca2+ channels known to be present at the PC (41) in PC-induced autophagy. The transient receptor cation channel polycystin 2 (PKD2) has been reported to regulate Ca2+ signaling and mediate PC-dependent autophagy in response to fluid flow in kidney epithelial cells (42). However, its knockdown via siRNA (siPKD2) did not block CMS-induced autophagy in TM cells (Fig. 3C) nor did any of the other Ca2+channel blockers tested [amiloride for T-type (43)], amlodipine for L-/N-type and NBQX for a specific Ca2+-permeable AMPA receptor antagonist (ref. 44, Fig. 3D). Together, these data suggest that CMS-induced autophagy is not primarily mediated by Ca2+ signaling in TM cells.
Noncanonical SMAD2/3 Signaling Pathway Regulates PC-Dependent CMS-Induced Autophagy.
Hh/SMO and LKB1–AMPK–MTOR signaling pathways have been identified to mediate starvation- and shear-stress–induced autophagy by PC in fibroblast (24, 45) and kidney epithelial cells (24, 26), respectively. We investigated the potential role of these pathways in regulating stretch-induced autophagy in TM cells (included as SI Appendix, Supplementary Materials and Methods). As shown in SI Appendix, Figs. S3 and S4, our data do not support Hh/SMO or LKB1 as mediators of PC-dependent activation of autophagy by CMS.
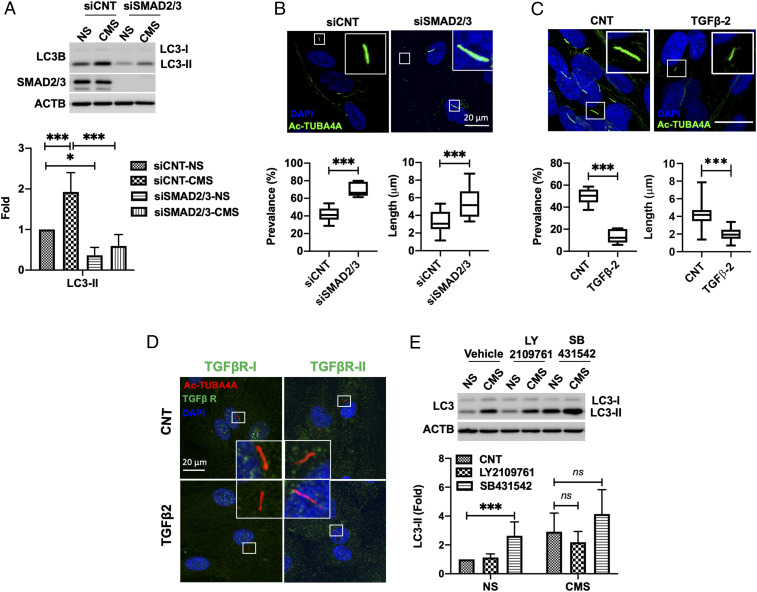
In a recent study, our laboratory has shown that down-regulation of SMAD2/3 decreases the constitutive and TGF-β–induced levels of LC3-II (8). We questioned whether SMAD2/3 could also regulate CMS-induced autophagy. For this, we transfected TM cells with siSMAD2/3 or siCNT and subjected them to CMS. LC3-II levels were quantified. As shown in Fig. 4A, silencing SMAD2/3 expression significantly abolished the increase in LC3-II caused by CMS (siCNT versus siSMAD2/3: 1.93 ± 0.47 versus 0.59 ± 0.28 folds, P < 0.01, n = 3). We then examined whether SMAD2/3 knockdown alters the prevalence and length of PC. Interestingly, both PC prevalence (siCNT versus siSMAD2/3: 41.64 ± 8.59% versus 70.03 ± 8.30%, P < 0.001, n = 6 versus 5) and length (siCNT versus siSMAD2/3: 3.21 ± 1.16 µm versus 5.37 ± 1.70 µm, P < 0.001, n = 22 versus 25) were significantly increased by SMAD2/3 knockdown (Fig. 4B). Moreover, TGF-β2 treatment (10 ng/mL) for 24 h significantly reduced the prevalence (CNT versus TGF-β2: 49.80 ± 7.59% versus 13.67 ± 6.37%, P < 0.001, n = 6 versus 5) and length (CNT versus TGF-β2: 4.18 ± 1.10 µm versus 2.02 ± 0.70 µm, P < 0.001, n = 140 versus 32) of PC in TM cells (Fig. 4C). Taken together, these data indicate that SMAD2/3 is a key component of the signaling pathway mediating PC-induced autophagy in response to CMS and suggest a role of TGF-β/SMAD in regulating ciliogenesis in TM cells.
Fig. 4.
Noncanonical SMAD2/3 signaling pathway regulates PC-dependent CMS-induced autophagy. (A) The effect of SMAD2/3 knockdown on CMS-induced autophagy. The expression levels of LC3 were measured in whole-cell lysates by WB. LC3-II band intensities were normalized by β-actin (ACTB) and fold changes calculated and graphed. Data are shown as the mean ± SD (n = 5). *P < 0.05; ***P < 0.001 (ANOVA). (B) Effect of SMAD2/3 knockdown on PC. PC were immunostained with anti-acetyl TUBA4A in siSMAD2/3-transfected TM cells. DAPI was used to stain nuclei. (Upper) Representative images of PC in each cell. The graphs in the lower panel show the quantification analysis for PC prevalence and length. Data are shown as the mean ± SD of three biological replicates (n = 6 versus 5 and 22 versus 25 in siCNT versus siSMAD2/3 for prevalence and length, respectively). ***P < 0.001 (two-tailed unpaired Student’s t test). (C) Effect of TGF-β-2 on PC. TM cells were treated with TGF-β-2 (10 ng/mL) for 24 h and PC analyzed with immunocytochemistry. Data are shown as the mean ± SD (n = 6 versus 5 and 140 versus 32 in CNT versus TGF-β-2 for prevalence and length, respectively). ***P < 0.001 (two-tailed unpaired Student’s t test). (D) TGF-βR-I and -II were not majorly localized to the PC in TM cells. Representative immunostaining of TGF-βRs (green) and PC (red) in CNT and TGF-β2 (10 ng/mL)–treated cells. DAPI was used to stain nuclei. (E) Effect of TGF-βRs inhibition on CMS-induced autophagy. TM cells were subjected to CMS (8% elongation, 24 h) in the presence or absence of 10 µM LY2109761 and SB431542. The expression levels of LC3 were quantified in whole-cell lysates by WB. LC3-II band intensities were normalized by ACTB and fold changes calculated. Data are shown as the mean ± SD (n = 3). *P < 0.05 (one-way ANOVA); ns, not significant.
SMAD2/3 is generally known to be phosphorylated and activated by TGF-β receptors (TGF-βR-I and -II) upon TGF-β stimulation (46). Some recent studies have suggested a PC-dependent TGF-β signaling regulated by the distinct spatial localization of TGF-βRs and SMAD components along the cilia structure (47, 48). Upon TGF-β stimulation, TGF-βRs located at the tip of the cilia travel to the ciliary base, where they are internalized within endosomes by clathrin-dependent endocytosis. TGF-βR–containing endosomes then activate SMAD2/3 at the ciliary base (41). We investigated whether the regulation of autophagy by SMAD2/3 is controlled by a PC-dependent TGF-β/SMAD pathway. We first examined by immunofluorescence whether TGF-βRs localize at the PC in TM cells. As shown in Fig. 4D, we did not find a clear localization pattern of the TGF-βRs at the PC in TM cells cultured in the absence or presence of TGF-β2 (10 ng/mL). We also tested the effect of inhibiting endocytosis on CMS-induced autophagy. For this, we treated TM cells with dynasore (100 and 200 µM), an inhibitor of clathrin-mediated endocytosis, and monitored endocytosis of DQ-BSA (bovine serum albumin) (SI Appendix, Fig. S5). While dynasore treatment inhibited the endocytosis of DQ-BSA (SI Appendix, Fig. S5A), it did not prevent the increase in LC3-II levels by CMS (SI Appendix, Fig. S5B).
We next investigated the contribution of the canonical TGF-β/SMAD pathway on PC-dependent CMS-induced autophagy. For this, we chemically inhibited TGF-βRs with LY2109761 (TGF-βR-I and -II dual inhibitor, 10 µM) or SB431542 (TGF-βR-I inhibitor, 10 µM). Both treatments effectively reduced the TGF-β–stimulated phosphorylation of SMAD2/3 (SI Appendix, Fig. S5C); however, chemical inhibition of TGF-βRs did not prevent the increase in LC3-II with CMS (Fig. 4E). Moreover, a further increase in LC3-II, rather than a decrease, was observed with SB431542 treatment in both CNT and CMS cells (Fig. 4E). We confirmed elevated autophagosome content with SB431542 in AdGFP-LC3–transduced TM cells. SB431542 treatment caused a dramatic increase in GFP-LC3 puncta size (SI Appendix, Fig. S5D, red arrows), as well as in the percentage of puncta-containing cells (CNT versus SB431542: 16.73 ± 5.12% versus 54.90 ± 6.57%, P < 0.001, n = 6, SI Appendix, Fig. S5D) compared to CNT. We additionally investigated whether CMS induces phosphorylation of SMAD2/3. Whereas TGF-β treatment (10 ng/mL) for 15 min clearly induces SMAD2/3 phosphorylation (pSMAD2/3), that was not observed with CMS application for 15 min or 1 h (SI Appendix, Fig. S5E). (Note, LC3-II levels are already increased after 1 h CMS.) Together, our results suggest the existence of an alternative signaling pathway to the canonical TGF-β/SMAD pathway that couples SMAD2/3 to PC to regulate CMS-induced autophagy in TM cells.
AKT1-SMAD2/3 Cross Talk Regulates CMS-Induced Autophagy via PC.
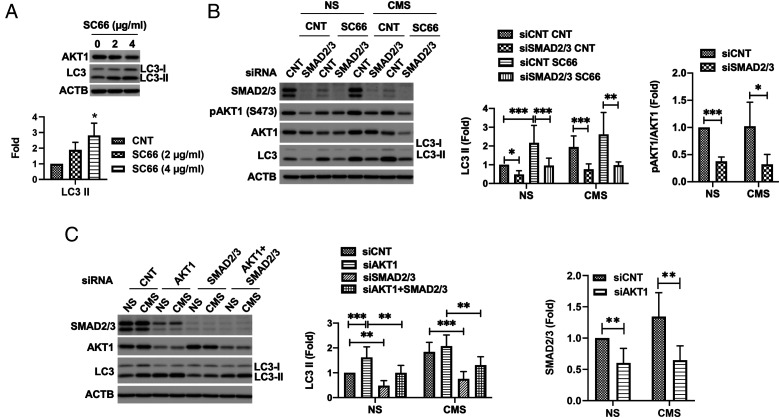
Our study and that from other research groups have reported mechanical stress-induced autophagy to occur in an MTOR-independent manner (5, 14). A recent work has implicated inhibition of AKT1 in the MTOR-independent activation of autophagy by trehalose (49). In view of this, we questioned whether AKT1 signaling could also regulate PC-dependent stretch-induced autophagy in TM cells. First, we tested whether AKT1 inhibition induces autophagy in TM cells. For this, we treated TM cells for 24 h with ∼2 to 4 µg/mL SC66 and evaluated LC3-II levels. SC66 is an allosteric inhibitor of AKT1 that facilitates its ubiquitination and deactivation by directly disrupting binding to phosphatidylinositol-3,4,5-triphosphate (PI-3,4,5-P3) thorough the pleckstrin homology (50). As seen in Fig. 5A, SC66 increased LC3-II levels in TM cells in a dose-dependent manner (P = 0.01, ANOVA, and n = 3). We then examined whether AKT1 inhibition-induced autophagy is regulated by SMAD2/3. To do this, we chemically (Fig. 5B) or genetically (Fig. 5C) inhibited AKT1 in SMAD2/3 knocked-down cells and evaluated LC3-II levels. As shown in Fig. 5B, the increase in LC3-II by SC66 treatment was significantly lowered by SMAD2/3 knock down in both NS and cyclically stretched cells (siCNT-SC66 versus siSMAD2/3-SC66: 2.17 ± 0.93 versus 0.96 ± 0.39 and 2.62 ± 1.17 versus 0.98 ± 0.17 folds in NS and CMS, respectively, P < 0.01, n = 3). Similarly, AKT1 knock down also resulted in increased LC3-II, which was lowered by SMAD2/3 knock down (Fig. 5C). Very intriguingly, knock down of SMAD2/3 or AKT1 reciprocally decreases AKT1 phosphorylation at S473 or SMAD2/3 protein level, respectively (Fig. 5 B and C), suggesting that AKT1 and SMAD2/3 reciprocally cross activate to regulate the induction of autophagy.
Fig. 5.
AKT1-SMAD2/3 cross talk regulates CMS-induced autophagy. (A) Autophagy induction by AKT inhibition in TM cells. TM cells were treated with ∼2 to 4 µg/mL SC66 for 24 h, and LC3 levels were evaluated by WB in the cells. LC3-II band intensities were normalized by β-actin (ACTB). Data are shown as the mean ± SD (n = 3). *P < 0.05 (one-way ANOVA). (B and C) Effect of SMAD2/3 knockdown on AKT inhibition-induced autophagy with SC66 (B) and siAKT1 (C). Data are shown as the mean ± SD (n = 3). *P < 0.05; **P < 0.01; ***P < 0.001 (ANOVA and Tukey’s post hoc test).
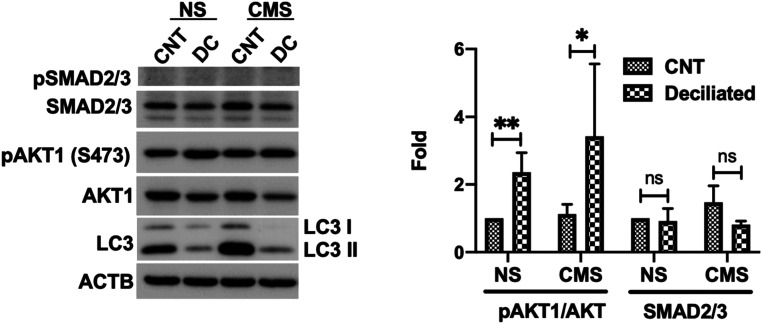
Finally, we examined whether PC directly control AKT1 or SMAD2/3 to regulate autophagy. For this, we treated TM cells with CH as detailed earlier; deciliated cells were then subjected to CMS, and the levels of pAKT and pSMAD2/3 were evaluated. No significant changes in SMAD2/3 protein levels were observed in deciliated cells either in NS or stretched conditions (Fig. 6). In contrast, deciliated cells showed higher levels of pAKT1/AKT in both NS (2.36 ± 0.57 folds, P < 0.01, and n = 6) and stretched (3.42 ± 2.14 folds, P < 0.05, and n = 6) conditions. These results suggest that AKT1 is directly regulated by PC to control induction of autophagy.
Fig. 6.
AKT phosphorylation is increased in deciliated cells. Cells were treated in 4 mM CH for 3 d and subjected to CMS after CH removal (8% elongation, 24 h). Protein levels of AKT1, pAKT1, SMAD2/3, and LC3 were evaluated by WB. Data are shown as the mean ± SD (n = 3). **P < 0.01; *P < 0.05 (two-tailed unpaired Student’s t test); ns, not significant.
PC-Mediated Autophagy Is Involved in IOP Homeostasis Regulation.
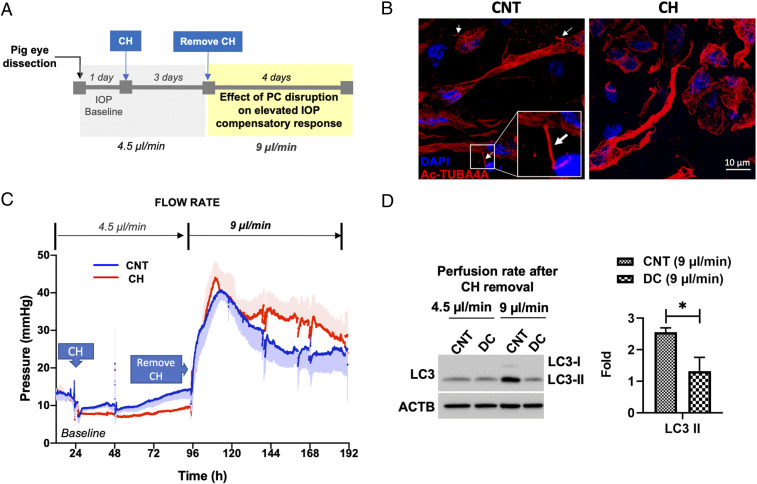
Next, we evaluated the physiological relevance of our findings. For this, we investigated the role of PC-mediated autophagy in IOP homeostasis in porcine eyes using the iPerfusion system. This system allows for continuous monitorization of outflow facility in enucleated eyes in response to treatment. A schematic diagram of the experimental outline is depicted in Fig. 7A. Anterior segments of enucleated eyes were perfused at a physiological flow rate of 4.5 µl/min. Once IOP baseline was established (∼24 h), eyes were perfused with either vehicle or CH (4 mM) for 3 d to remove PC (Fig. 7B). Perfusion media containing CH was replaced by fresh perfusion media, and flow rate was doubled to 9 µl/min and maintained as such for the length of the experiment. As expected, doubling flow rate led to an immediate increase in IOP, which gradually returned to baseline levels over the next 2 to 3 d, as part of the homeostatic IOP mechanism (51), by which outflow resistance is adjusted to restore normal pressure levels. Intriguingly, the IOP homeostatic response was retarded in the deciliated eyes compared to CNT eyes (Fig. 7C). We additionally evaluated the LC3-II protein levels in dissected perfused TM tissue by WB. Confirming our previous work (5), IOP elevation significantly increased LC3-II. Elevation in LC3-II in response to doubling flow rate was, however, not observed in the deciliated eyes (CNT versus DC: 2.55 ± 0.14 versus 1.31 ± 0.44 folds, P < 0.05, n = 3, Fig. 7D). Thick sections of perfused tissues showed preservation of outflow pathway tissue integrity in CH-treated eyes. (SI Appendix, Fig. S6). Together, these experiments very strongly support a role of PC-mediated autophagy in regulating IOP homeostasis.
Fig. 7.
PC-mediated autophagy is involved in IOP homeostasis regulation. (A) Schematic diagram of the experimental timeline. (B) Representative immunofluorescence analysis of PC in TM tissue perfused with CH. Acetylated TUBA4A (red fluorescence) was used to identify the PC. DAPI was used to stain nuclei. Images were acquired with confocal microscope and processed by using Fiji software. (C) Profile of IOP measurement in anterior segments of porcine eye perfused with CH and subjected to double flow rate. Data are presented as mean ± SD (solid lines ± shaded regions, n = 4). (D) LC3-II protein levels in whole-tissue lysates of dissected perfused TM tissues evaluated by WB. Data are shown as the mean ± SD (n = 3). *P < 0.05 (two-tailed unpaired Student’s t test). DC, deciliated.
Discussion
In this study, we report that PC is a mechanosensor for stretch-induced autophagy and that PC-mediated autophagy plays a critical role in IOP homeostasis. Moreover, we identified a reciprocal activation of AKT1 and SMAD2/3 as a signaling pathway regulating the activation of autophagy by PC. Furthermore, our data indicate TGF-β/SMAD2 as an important regulator of ciliogenesis in TM cells.
Despite its relevant role in mechanotransduction, the study of PC in TM cells has been very limited (35, 52, 53). Here, we show real-time live-cell imaging of TM cilia dynamics and Ca2+ flux in PC. Our gene-expression profile analysis highlighted genes involved in ciliogenesis in mechanically stretched TM cells deficient in autophagy. These exciting data were in line with an emerging body of evidence indicating a relationship between the PC and autophagy. PC have been shown to mediate autophagy induced by starvation (24) and flow-induced shear stress (26) to regulate energy status and cell volume, respectively. In our experiments, chemical removal of PC completely abolished CMS-induced autophagy, very strongly supporting a role of cilia as a mechanosensor for stretch-induced autophagy. As in the previous studies, we also observed the autophagy gene Atg16L and LC3 at the basal body of the PC in mechanically stretched cells, further confirming Atg16L recruitment to the basal body as a hallmark for PC-induced autophagy.
Interestingly, while Atg16L recruitment is shared in all cases for initiation of PC-mediated autophagy, the downstream signaling pathways seems to differ depending on the initial stimuli. The Hh/SMO signaling pathway was reported to regulate starvation-induced autophagy in fibroblasts (24, 26), and LKB1–AMPK–MTOR was linked to shear-stress–induced autophagy in kidney epithelial cells (24, 26). Our data shows that CMS does not induce autophagy through the Hh/SMO signaling pathway in PC. Similarly, LKB1 knockdown did not prevent the increase in LC3-II levels by CMS in TM cells. This was not unexpected given that, unlike starvation-induced autophagy, the induction of autophagy by mechanical stretch is independent of MTOR (5, 14). Thus, it is reasonable to speculate that the output results from the specific signaling components used upon different stimuli. With this in mind, we explored the role of Ca2+ signaling in stretch-induced autophagy, since Ca2+ influx through TRPV4 channels was reported to regulate mechanosensation by PC in TM cells (40). Although our data suggest a role of Ca2+ influx in regulating basal autophagy levels, it did not support a role of TRPV4 or other Ca2+ channels in the induction of autophagy by mechanical stretch.
Silencing SMAD2/3 via siRNA completely abolished the induction of autophagy by mechanical stress, suggesting that SMAD2/3 is a critical signaling component mediating CMS-induced autophagy. This was particularly interesting since some studies have suggested a PC-dependent TGF-β signaling, which is regulated by clathrin-dependent endocytosis of TGF-βRs and phosphorylation of SMAD at the ciliary base. Surprisingly, activation of autophagy by CMS was not prevented by treatment of cells with TGF-βR inhibitors (LY2109761 and SB431542). Furthermore, we did not detect phosphorylation of SMAD2/3 at the c terminal in stretched cells, even at time points in which LC3-II was already elevated. In addition, we did not observe clear localization of TGF-βRs at the PC, and inhibition of endocytosis with dynasore did not prevent the increase in LC3-II level by CMS. Together, these results indicated that canonical TGF-βR-I–mediated SMAD2/3 phosphorylation was not crucial to regulating CMS-induced autophagy in TM cells. In contrast, our data suggested a role of TGF-β/Smad in regulating ciliogenesis in TM cells. This is pathologically relevant, since TGF-β2 levels are elevated in the aqueous humor from patients with primary open-angle glaucoma (54–56).
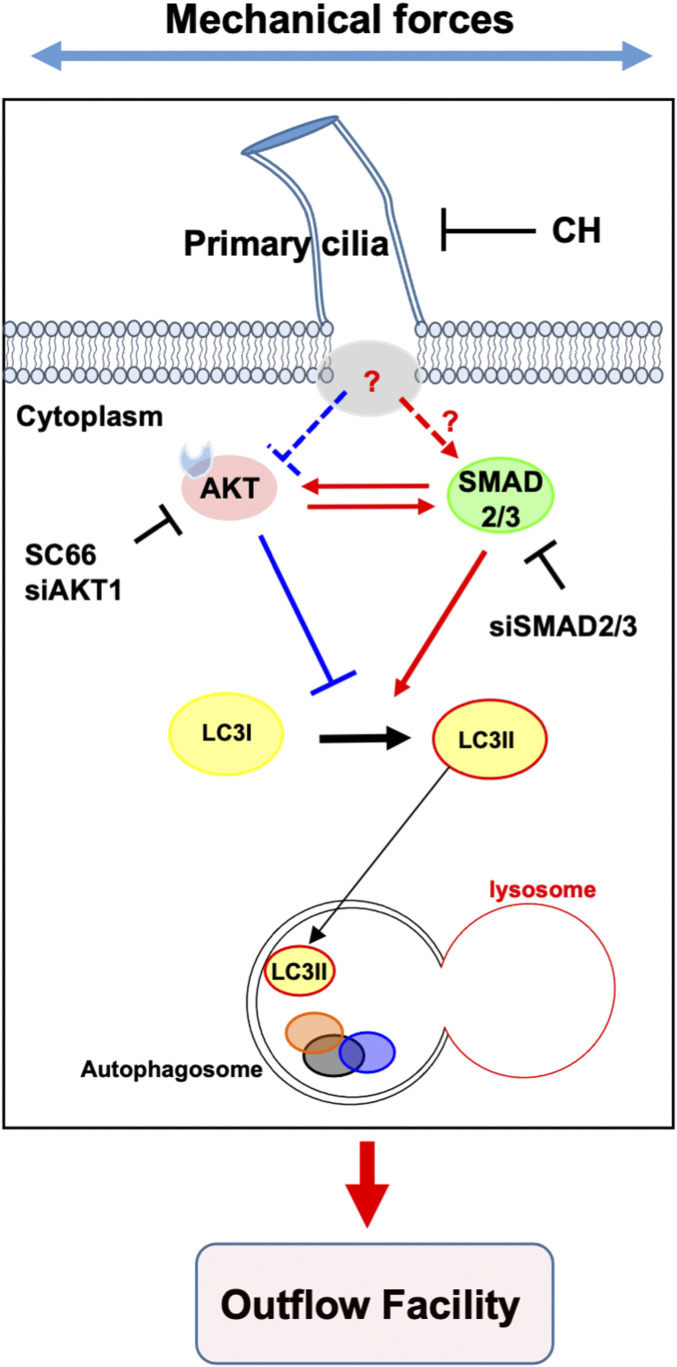
A very important finding in our work is the identification of a reciprocal activation between AKT1 and SMAD2/3 to regulate autophagy. Moreover, this reciprocal cross talk was additionally observed in primary-cultured human fibroblast cells (SI Appendix, Fig. S7). AKT1 negatively regulates autophagy in certain cancer cells (57, 58) and, most interestingly for our study, it has been recently shown to activate autophagy independently of MTOR (49). In line with these reports, our data showed that pharmacological inhibition or genetic knockdown of AKT1 induces autophagy in TM cells. Furthermore, the autophagy induction mechanism by SC66 or siAKT1 is SMAD2/3 dependent; silencing SMAD2/3 expression significantly lowered AKT1-increased LC3-II levels. AKT1 is a phosphatidylinositol-phosphate binding serine/threonine kinase, which is generally known as a key component of the phosphatidylinositol 3-kinase signaling pathway. AKT1 activity is regulated by phosphorylation (59). AKT1 phosphorylation by mechanical stretch has been reported in epidermal (60) and in porcine TM cells (61). In our experimental conditions, AKT1 and p-AKT1 protein levels varied among the different TM strains, and no statistical significance in pAKT1/AKT1 ratio with CMS was reached, although this was only evaluated at one time point. Interestingly, pAKT1/AKT1 ratio was found to be decreased in SMAD2/3 knocked-down cells. Moreover, knocking down AKT1 also reduced SMAD2/3 protein levels, indicating a cross talk between both signaling molecules. Furthermore, deciliated cells showed significant higher pAKT1/AKT1 levels, suggesting that AKT1-SMAD2/3 cross talk regulates PC-mediated CMS-induced autophagy. A model of our current hypothetical mechanism is represented in Fig. 8. Future studies will be directed at elucidating the molecular interaction between PC and AKT1-SMAD2/3 for the regulation of CMS-induced autophagy.
Fig. 8.
Hypothetical model of signaling pathway for PC-mediated CMS-induced autophagy. Under NS-CNT conditions, AKT1 and SMAD2/3 reciprocally activate each to maintain basal autophagy activity in TM cells. Upon mechanical stress, PC of TM cells sense the extracellular stress and transduce signal to regulate the cross talk between AKT1 and SMAD2/3, leading to autophagy induction to maintain IOP homeostasis. Solid lines represent pathway confirmed in this study, and dotted lines and question mark indicate a pathway that is not yet characterized.
Although the presence of PC in TM cells in situ has been documented in electron microscopy (EM) studies (62), its potential role as a sensory organ in outflow regulation, and its potential malfunction in glaucoma has hardly been explored, with just a couple of studies in the literature (40, 52), including the very recently published work by Prosseda et al., which reports a key role of phosphoinositides at the PC in IOP regulation in mice (52). Interestingly, patients affected with the ciliopathy Lowe syndrome are known to develop ocular hypertension and congenital glaucoma (20, 63). Similarly, defective aqueous humor outflow and elevated IOP is observed in a Lowe syndrome mouse model. Although clinical symptoms of Lowe syndrome typically manifest in the first year of life, whereas primary open-angle glaucoma (the most common form of the disease) develops in late adulthood, it provides a proof of concept of the relevance of PC in eye-pressure regulation.
Here, we demonstrated the physiological significance of our findings using the ex vivo perfused anterior segment system, which very accurately mimics outflow regulation in vivo. Very excitingly, our data clearly show that the compensatory pressure-lowering adaptive mechanism in response to elevated IOP is disrupted in eyes subjected to chemical removal of PC, compared to control eyes. Moreover, the CH-perfused TM failed to increase LC3-II protein levels in response to the elevated pressure challenge. These results very strongly indicate the role of PC-mediated autophagy in regulating IOP homeostasis. From a pathological point of view, previous work conducted in cultured TM cells isolated from glaucomatous eyes (64) and in glaucoma mouse models have revealed dysregulated autophagy in the TM of aging and hypertensive eyes (65–72). Future studies, specifically directed at evaluating PC in open-angle glaucoma models or human samples, will be conducted to establish a direct connection between dysregulated autophagy in glaucoma with defective ciliogenesis or cilia function and to investigate the AKT-SMAD2/3 cross talk.
Materials and Methods
Reagents.
Chemicals and other materials were obtained from the sources described in SI Appendix, Supplementary Materials and Methods.
Primary Human TM Cells Isolation, Maintenance, and Culture.
Primary human TM cells were isolated from dissected TM tissue from discarded corneal rims after surgical corneal transplantation at Duke University Eye Center and maintained as described previously (68). Each primary culture was obtained from one or two corneal rims (based on availability) from the same donor. Cells were passaged when confluent at a 1:2 ratio using growth media (10% fetal bovine serum [FBS], 1% nonessential amino acid, 1% penicillin/streptomycin [Pen/strep], and 0.1% gentamycin in Dulbecco's modified Eagle medium [DMEM]). Cells from passage four to eight were used in this study. Primary human TM cells were characterized by morphology and by the up-regulation of myocilin in response to dexamethasone treatment, in accordance to the consensus recommendations for TM cell isolation, characterization, and culture (69). The protocols involving the use of human tissue were approved under the Duke University Institutional Review Board (protocol no. 00050810) and were consistent with the tenets of the Declaration of Helsinki. Samples were deidentified prior to use in our study. Exogenous TGF-β2 (10 ng/mL) was added, when indicated, to the culture media.
CMS Application.
CMS was applied using the computer-controlled and vacuum-operated Flexcell FX-5000 Tension System (Flexcell International Corp.) as described previously (5, 6, 70). When indicated, an SMO agonist (SAG) (∼0.125 to 1 µM), cyclopamine (∼2.5 to 10 µM), HC067047 (10 µM), amiloride (100 µM), amlodipine (10 µM), NBQX (10 µM), LY2109761 (10 µM), SB431542 (10 µM), dynasore (100 or 200 µM), or SC66 (2 or 4 µg/mL) were added to the culture media 2 h prior to CMS.
Microarrays Analysis.
Microarray analysis was performed as described previously (8, 71). Gene ontology analysis of the differentially expressed genes was conducted with BiNGO version 3.04 (72), which is plugged in Cytoscape (73). The detailed procedure is in SI Appendix, Supplementary Materials and Methods. Array data were deposited at the Gene Expression Omnibus (National Center for Biotechnology; accession number: GSE122652 and GSE159793 for new deposit).
Transfection of Plasmids, siRNA, and Transduction of Adenovirus.
Plasmids for 5HT6-mCherry (33) (kindly provided by Dr. Takanari Inoue, Johns Hopkins University School of Medicine, Baltimore, MD) or 5HT6-G-GECO1.0 (34) (a gift from Dr. Takanari Inoue, Addgene plasmid no. 47499; www.addgene.org/47499/; RRID: Addgene_47499) were transfected using the Ca2+-phosphate method (74). siRNAs were transfected with Lipofectamine RNAiMAX Reagent as described previously (5). Adenovirus that express GFP–LC3 fusion protein (AdGFP-LC3; kindly provided by Dr. Wen-Xing Ding, University of Kansas Medical Center, Kansas City, KS) were transduced as described (6). More information is available in SI Appendix, Supplementary Materials and Methods.
Immunocyto and Histochemical Analyses.
Human TM cells and the anterior segments of the eye were fixed and immunostained using primary antibodies including anti-acetylated TUBA4A and anti-SMO (45). The complete information about antibodies used for immunostaining in this study and detailed methods are available in SI Appendix, Supplementary Materials and Methods. Images were acquired on a Nikon C2si confocal microscope (Nikon Instruments Inc.) or Zeiss 880 Airyscan Inverted Confocal microscope [Zeissand processed with Fiji (75)].
PC Prevalence and Length.
PC prevalence and length were measured as described (76, 77) with some modification. Briefly, for PC prevalence, multiple fields containing cells stained with Ac-TUBA4A and 4′,6-diamidino-2-phenylindole (DAPI) were randomly selected and imaged using a Nikon C2si confocal microscope with a ×40 1.3 numerical aperture (NA) or ×60 1.4 NA objective. Confocal z-stacks (0.2 µm step size) were generated containing the entire cell depth. The prevalence was determined as a percentage of total cells exhibiting PC (number of PC stained with Ac-TUBA4A)/ total cell (number of nucleus stained with DAPI) in a field of image. To evaluate cilia length, the same samples were imaged using a ×60 1.4 NA objective with the z-stacks images. Maximum intensity projection method was applied for the measuring of the prevalence and length as described (77) using Fiji.
Live-Cell Time-Lapse Imaging of PC Dynamics and PC-Ca2+ Influx.
Live-cell images of PC were captured using the CELENA X High Content Imaging System (Logos biosystems) equipped with high power light-emitting diode (LED) filters coupled with the Ibidi Perfusion System equipped with a CO2 and temperature-controlled stage-top incubation system (Ibidi). For live-cell time-lapse imaging of PC dynamics, human TM cells transfected with p5HT6-mCherry, as described earlier, were plated at 100% confluence onto µ-Slide VI 0.4 (Ibidi, 80606) in 30 µl volume and incubated for 72 h. PC dynamics was observed directly after the media change. For Ca2+ monitoring, cells grown on µ-Slide VI 0.4 were washed three times with Hanks balanced salt solution (HBSS), and media replaced with HBSS containing 200 nM ionomycin. Live-cell images were captured every 5 s. Fiji was used for image processing.
Whole-Cell Lysate Preparation and WB Analysis.
Whole-cell lysates preparation and WB were performed as described previously (6). Detailed information is available in SI Appendix, Supplementary Materials and Methods.
Outflow Facility Measurement in Porcine Anterior Segments.
Outflow facility in porcine eye was measured as described previously (78, 79) with some modification. Briefly, anterior segments were dissected from porcine eyes (four pairs) and perfused. Detailed procedure is available in SI Appendix, Supplementary Materials and Methods section. After initial perfusion to establish a stable outflow facility, CH (4 mM) was added to or removed from the anterior segment by exchanging the perfusion media (1% FBS, 0.05 mg/mL BSA, and 1% penicillin-streptomycin-glutamine) with or without the drug at a rate (1 mL/min) for ∼10 to 15 min, using the reservoir which is connected between the perfusion chamber and the pressure transducer, followed by a constant inflow rate (4.5 μL/min) for indicated time. After 3 d, CH was removed and double constant inflow rate (9 μL/min) was applied. The data were recorded and stored using the iPerfusion software system (80) based on Labview interface (National Instruments). The outflow facility from the acquired data were further calculated and processed with MATLAB software (Mathworks). After perfusion, TM tissue was either dissected and lysed in radioimmunoprecipitation assay (RIPA) buffer for WB analysis or fixed for histology.
Statistical Analyses.
All experimental procedures were repeated at least three times in independent experiments conducted with different TM primary cultures established from independent donors. Unless otherwise stated, n in figure legends represents biological replicates (corneal rims). Data were presented as the mean ± SD. Statistical analysis was done in GraphPad Prism software using two-tailed unpaired Student’s t test or one-way ANOVA with Tukey’s post hoc test. P < 0.05 was considered to be statistically significant.
Supplementary Material
Acknowledgments
We thank Dr. Takanari Inoue at Johns Hopkins University School of Medicine for kindly providing 5HT6-mCherry plasmid; Dr. Matthew P. Scott at Stanford University for generously providing anti-SMO antibody; Dr. Xiao-Ming Yin at Indiana University and Dr. Wen-Xing Ding at University of Kansas Medical Center for permission to use and providing AdGFP-LC3, respectively; Dr. W. Daniel Stamer and Dr. Fiona McDonnell at Duke University for kindly giving permission, training to use the iPerfusion system for measuring outflow facility in porcine eye, and providing primary human fibroblast cells; and Ying Hao at the Duke Eye Center EM unit for helping with histology samples preparation. Funding was provided by NIH, Eye Institute (EY026885, EY027733, and EY005722) and Unrestricted Research to Prevent Blindness Grant.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission. D.P. is a guest editor invited by the Editorial Board.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2021942118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or supporting information.
References
- 1.Weinreb R. N., Khaw P. T., Primary open-angle glaucoma. Lancet 363, 1711–1720 (2004). [DOI] [PubMed] [Google Scholar]
- 2.Zhang K., Zhang L., Weinreb R. N., Ophthalmic drug discovery: Novel targets and mechanisms for retinal diseases and glaucoma. Nat. Rev. Drug Discov. 11, 541–559 (2012). [DOI] [PubMed] [Google Scholar]
- 3.Weinreb R. N., et al., Primary open-angle glaucoma. Nat. Rev. Dis. Primers 2, 16067 (2016). [DOI] [PubMed] [Google Scholar]
- 4.WuDunn D., Mechanobiology of trabecular meshwork cells. Exp. Eye Res. 88, 718–723 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Porter K. M., Jeyabalan N., Liton P. B., MTOR-independent induction of autophagy in trabecular meshwork cells subjected to biaxial stretch. Biochim. Biophys. Acta 1843, 1054–1062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shim M. S., Nettesheim A., Hirt J., Liton P. B., The autophagic protein LC3 translocates to the nucleus and localizes in the nucleolus associated to NUFIP1 in response to cyclic mechanical stress. Autophagy 16, 1248–1261 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hirt J., Liton P. B., Autophagy and mechanotransduction in outflow pathway cells. Exp. Eye Res. 158, 146–153 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Nettesheim A., Shim M. S., Hirt J., Liton P. B., Transcriptome analysis reveals autophagy as regulator of TGFβ/Smad-induced fibrogenesis in trabecular meshwork cells. Sci. Rep. 9, 16092 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kroemer G., Mariño G., Levine B., Autophagy and the integrated stress response. Mol. Cell 40, 280–293 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sarkar S., Regulation of autophagy by mTOR-dependent and mTOR-independent pathways: Autophagy dysfunction in neurodegenerative diseases and therapeutic application of autophagy enhancers. Biochem. Soc. Trans. 41, 1103–1130 (2013). [DOI] [PubMed] [Google Scholar]
- 11.Mizushima N., Komatsu M., Autophagy: Renovation of cells and tissues. Cell 147, 728–741 (2011). [DOI] [PubMed] [Google Scholar]
- 12.Ganley I. G., et al., ULK1.ATG13.FIP200 complex mediates mTOR signaling and is essential for autophagy. J. Biol. Chem. 284, 12297–12305 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Liu G. Y., Sabatini D. M., mTOR at the nexus of nutrition, growth, ageing and disease. Nat. Rev. Mol. Cell Biol. 21, 183–203 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.King J. S., Veltman D. M., Insall R. H., The induction of autophagy by mechanical stress. Autophagy 7, 1490–1499 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Singla V., Reiter J. F., The primary cilium as the cell’s antenna: Signaling at a sensory organelle. Science 313, 629–633 (2006). [DOI] [PubMed] [Google Scholar]
- 16.May-Simera H., Nagel-Wolfrum K., Wolfrum U., Cilia–The sensory antennae in the eye. Prog. Retin. Eye Res. 60, 144–180 (2017). [DOI] [PubMed] [Google Scholar]
- 17.Satir P., Pedersen L. B., Christensen S. T., The primary cilium at a glance. J. Cell Sci. 123, 499–503 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orhon I., Dupont N., Pampliega O., Cuervo A. M., Codogno P., Autophagy and regulation of cilia function and assembly. Cell Death Differ. 22, 389–397 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gerdes J. M., Davis E. E., Katsanis N., The vertebrate primary cilium in development, homeostasis, and disease. Cell 137, 32–45 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Walton D. S., Katsavounidou G., Lowe C. U., Glaucoma with the oculocerebrorenal syndrome of Lowe. J. Glaucoma 14, 181–185 (2005). [DOI] [PubMed] [Google Scholar]
- 21.Ávalos Y., Peña-Oyarzun D., Budini M., Morselli E., Criollo A., New roles of the primary cilium in autophagy. BioMed Res. Int. 2017, 4367019 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Satir P., Satir B. H., The conserved ancestral signaling pathway from cilium to nucleus. J. Cell Sci. 132, jcs230441 (2019). [DOI] [PubMed] [Google Scholar]
- 23.Anvarian Z., Mykytyn K., Mukhopadhyay S., Pedersen L. B., Christensen S. T., Cellular signalling by primary cilia in development, organ function and disease. Nat. Rev. Nephrol. 15, 199–219 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pampliega O., et al., Functional interaction between autophagy and ciliogenesis. Nature 502, 194–200 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tang Z., et al., Autophagy promotes primary ciliogenesis by removing OFD1 from centriolar satellites. Nature 502, 254–257 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Orhon I., et al., Primary-cilium-dependent autophagy controls epithelial cell volume in response to fluid flow. Nat. Cell Biol. 18, 657–667 (2016). [DOI] [PubMed] [Google Scholar]
- 27.Zemirli N., et al., The primary cilium protein folliculin is part of the autophagy signaling pathway to regulate epithelial cell size in response to fluid flow. Cell Stress 3, 100–109 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith K. R., et al., A role for central spindle proteins in cilia structure and function. Cytoskeleton (Hoboken) 68, 112–124 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Coelho P. A., et al., Over-expression of Plk4 induces centrosome amplification, loss of primary cilia and associated tissue hyperplasia in the mouse. Open Biol. 5, 150209 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wang W., Wu T., Kirschner M. W., The master cell cycle regulator APC-Cdc20 regulates ciliary length and disassembly of the primary cilium. eLife 3, e03083 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spalluto C., Wilson D. I., Hearn T., Evidence for reciliation of RPE1 cells in late G1 phase, and ciliary localisation of cyclin B1. FEBS Open Bio 3, 334–340 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lee K. H., et al., Identification of a novel Wnt5a-CK1ε-Dvl2-Plk1-mediated primary cilia disassembly pathway. EMBO J. 31, 3104–3117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hong S. R., et al., Spatiotemporal manipulation of ciliary glutamylation reveals its roles in intraciliary trafficking and Hedgehog signaling. Nat. Commun. 9, 1732 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Su S., et al., Genetically encoded calcium indicator illuminates calcium dynamics in primary cilia. Nat. Methods 10, 1105–1107 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Follit J. A., Tuft R. A., Fogarty K. E., Pazour G. J., The intraflagellar transport protein IFT20 is associated with the Golgi complex and is required for cilia assembly. Mol. Biol. Cell 17, 3781–3792 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chakrabarti A., Schatten H., Mitchell K. D., Crosser M., Taylor M., Chloral hydrate alters the organization of the ciliary basal apparatus and cell organelles in sea urchin embryos. Cell Tissue Res. 293, 453–462 (1998). [DOI] [PubMed] [Google Scholar]
- 37.Kim C. H., Jeung E. B., Yoo Y. M., Combined fluid shear stress and melatonin enhances the ERK/Akt/mTOR signal in cilia-less MC3T3-E1 preosteoblast cells. Int. J. Mol. Sci. 19, 2929 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yoshii S. R., Mizushima N., Monitoring and measuring autophagy. Int. J. Mol. Sci. 18, 1865 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Boukhalfa A., et al., PI3KC2α-dependent and VPS34-independent generation of PI3P controls primary cilium-mediated autophagy in response to shear stress. Nat. Commun. 11, 294 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo N., et al., Primary cilia signaling mediates intraocular pressure sensation. Proc. Natl. Acad. Sci. U.S.A. 111, 12871–12876 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pala R., Alomari N., Nauli S. M., Primary cilium-dependent signaling mechanisms. Int. J. Mol. Sci. 18, 2272 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Nauli S. M., et al., Polycystins 1 and 2 mediate mechanosensation in the primary cilium of kidney cells. Nat. Genet. 33, 129–137 (2003). [DOI] [PubMed] [Google Scholar]
- 43.Tang C. M., Presser F., Morad M., Amiloride selectively blocks the low threshold (T) calcium channel. Science 240, 213–215 (1988). [DOI] [PubMed] [Google Scholar]
- 44.Hilton G. D., Bambrick L. L., Thompson S. M., McCarthy M. M., Estradiol modulation of kainic acid-induced calcium elevation in neonatal hippocampal neurons. Endocrinology 147, 1246–1255 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Rohatgi R., Milenkovic L., Scott M. P., Patched1 regulates hedgehog signaling at the primary cilium. Science 317, 372–376 (2007). [DOI] [PubMed] [Google Scholar]
- 46.Derynck R., Budi E. H., Specificity, versatility, and control of TGF-β family signaling. Sci. Signal. 12, eaav5183 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Clement C. A., et al., TGF-β signaling is associated with endocytosis at the pocket region of the primary cilium. Cell Rep. 3, 1806–1814 (2013). [DOI] [PubMed] [Google Scholar]
- 48.Pedersen L. B., Mogensen J. B., Christensen S. T., Endocytic control of cellular signaling at the primary cilium. Trends Biochem. Sci. 41, 784–797 (2016). [DOI] [PubMed] [Google Scholar]
- 49.Palmieri M., et al., mTORC1-independent TFEB activation via Akt inhibition promotes cellular clearance in neurodegenerative storage diseases. Nat. Commun. 8, 14338 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jo H., et al., Deactivation of Akt by a small molecule inhibitor targeting pleckstrin homology domain and facilitating Akt ubiquitination. Proc. Natl. Acad. Sci. U.S.A. 108, 6486–6491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Acott T. S., Vranka J. A., Keller K. E., Raghunathan V., Kelley M. J., Normal and glaucomatous outflow regulation. Prog. Retin. Eye Res., 100897 10.1016/j.preteyeres.2020.100897 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Prosseda P. P., et al., Optogenetic stimulation of phosphoinositides reveals a critical role of primary cilia in eye pressure regulation. Sci. Adv. 6, eaay8699 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Luo N., et al., Compensatory role of inositol 5-phosphatase INPP5B to OCRL in primary cilia formation in oculocerebrorenal syndrome of Lowe. PLoS One 8, e66727 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Inatani M., et al., Transforming growth factor-beta 2 levels in aqueous humor of glaucomatous eyes. Graefes Arch. Clin. Exp. Ophthalmol. 239, 109–113 (2001). [DOI] [PubMed] [Google Scholar]
- 55.Lütjen-Drecoll E., Morphological changes in glaucomatous eyes and the role of TGFbeta2 for the pathogenesis of the disease. Exp. Eye Res. 81, 1–4 (2005). [DOI] [PubMed] [Google Scholar]
- 56.Fuchshofer R., Tamm E. R., The role of TGF-β in the pathogenesis of primary open-angle glaucoma. Cell Tissue Res. 347, 279–290 (2012). [DOI] [PubMed] [Google Scholar]
- 57.Degtyarev M., et al., Akt inhibition promotes autophagy and sensitizes PTEN-null tumors to lysosomotropic agents. J. Cell Biol. 183, 101–116 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Wang R. C., et al., Akt-mediated regulation of autophagy and tumorigenesis through Beclin 1 phosphorylation. Science 338, 956–959 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Siess K. M., Leonard T. A., Lipid-dependent Akt-ivity: Where, when, and how. Biochem. Soc. Trans. 47, 897–908 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kippenberger S., et al., Mechanical stretch stimulates protein kinase B/Akt phosphorylation in epidermal cells via angiotensin II type 1 receptor and epidermal growth factor receptor. J. Biol. Chem. 280, 3060–3067 (2005). [DOI] [PubMed] [Google Scholar]
- 61.Bradley J. M., Kelley M. J., Rose A., Acott T. S., Signaling pathways used in trabecular matrix metalloproteinase response to mechanical stretch. Invest. Ophthalmol. Vis. Sci. 44, 5174–5181 (2003). [DOI] [PubMed] [Google Scholar]
- 62.Wickham M. G., Worthen D. M., Centrioles and cilia in the mesothelial cells of the pericanalicular region. Invest. Ophthalmol. 15, 499–502 (1976). [PubMed] [Google Scholar]
- 63.Attree O., et al., The Lowe’s oculocerebrorenal syndrome gene encodes a protein highly homologous to inositol polyphosphate-5-phosphatase. Nature 358, 239–242 (1992). [DOI] [PubMed] [Google Scholar]
- 64.Porter K., Hirt J., Stamer W. D., Liton P. B., Autophagic dysregulation in glaucomatous trabecular meshwork cells. Biochim. Biophys. Acta 1852, 379–385 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nettesheim A., et al., Autophagy in the aging and experimental ocular hypertensive mouse model. Invest. Ophthalmol. Vis. Sci. 61, 31 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Hirt J., Porter K., Dixon A., McKinnon S., Liton P. B., Contribution of autophagy to ocular hypertension and neurodegeneration in the DBA/2J spontaneous glaucoma mouse model. Cell Death Discov. 4, 14 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Zhu X., et al., Protective effects of rapamycin on Trabecular meshwork cells in glucocorticoid-induced glaucoma mice. Front. Pharmacol. 11, 1006 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Liton P. B., Li G., Luna C., Gonzalez P., Epstein D. L., Cross-talk between TGF-beta1 and IL-6 in human trabecular meshwork cells. Mol. Vis. 15, 326–334 (2009). [PMC free article] [PubMed] [Google Scholar]
- 69.Keller K. E., et al., Consensus recommendations for trabecular meshwork cell isolation, characterization and culture. Exp. Eye Res. 171, 164–173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Reina-Torres E., et al., VEGF as a paracrine regulator of conventional outflow facility. Invest. Ophthalmol. Vis. Sci. 58, 1899–1908 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Lee K. H., et al., Drosophila selenophosphate synthetase 1 regulates vitamin B6 metabolism: Prediction and confirmation. BMC Genomics 12, 426 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Maere S., Heymans K., Kuiper M., BiNGO: A Cytoscape plugin to assess overrepresentation of gene ontology categories in biological networks. Bioinformatics 21, 3448–3449 (2005). [DOI] [PubMed] [Google Scholar]
- 73.Shannon P., et al., Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 13, 2498–2504 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jiang M., Chen G., High Ca2+-phosphate transfection efficiency in low-density neuronal cultures. Nat. Protoc. 1, 695–700 (2006). [DOI] [PubMed] [Google Scholar]
- 75.Schindelin J., et al., Fiji: An open-source platform for biological-image analysis. Nat. Methods 9, 676–682 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Prodromou N. V., et al., Heat shock induces rapid resorption of primary cilia. J. Cell Sci. 125, 4297–4305 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Satir B. H., Pampliega O., Methods to study interactions between ciliogenesis and autophagy. Methods Mol. Biol. 1454, 53–67 (2016). [DOI] [PubMed] [Google Scholar]
- 78.McDonnell F., Dismuke W. M., Overby D. R., Stamer W. D., Pharmacological regulation of outflow resistance distal to Schlemm’s canal. Am. J. Physiol. Cell Physiol. 315, C44–C51 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Ramos R. F., Stamer W. D., Effects of cyclic intraocular pressure on conventional outflow facility. Invest. Ophthalmol. Vis. Sci. 49, 275–281 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sherwood J. M., Reina-Torres E., Bertrand J. A., Rowe B., Overby D. R., Measurement of outflow facility using iPerfusion. PLoS One 11, e0150694 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or supporting information.