Significance
Lipoylation is a posttranslational modification critical for the function of several metabolic enzymes. In the bacterial pathogen Staphylococcus aureus, lipoylation deficiency compromises growth and causes tissue-specific virulence defects. Perturbation of lipoylation causes attenuation in part due to disruption of the enzyme complex required for the synthesis of branched-chain fatty acids, an essential constituent of S. aureus membrane. S. aureus overcomes branched-chain fatty acid auxotrophy in the skin by acquiring host unsaturated fatty acids. This work underscores the adaptability of S. aureus when faced with nutrient scarcity and the relevance of lipoic acid sufficiency during infection. Our findings support the view that versatility in S. aureus membrane biogenesis must be considered when devising therapeutics.
Keywords: Staphylococcus aureus, branched-chain fatty acid, lipoic acid, membrane, virulence
Abstract
During infection, pathogenic microbes adapt to the nutritional milieu of the host through metabolic reprogramming and nutrient scavenging. For the bacterial pathogen Staphylococcus aureus, virulence in diverse infection sites is driven by the ability to scavenge myriad host nutrients, including lipoic acid, a cofactor required for the function of several critical metabolic enzyme complexes. S. aureus shuttles lipoic acid between these enzyme complexes via the amidotransferase, LipL. Here, we find that acquisition of lipoic acid, or its attachment via LipL to enzyme complexes required for the generation of acetyl-CoA and branched-chain fatty acids, is essential for bacteremia, yet dispensable for skin infection in mice. A lipL mutant is auxotrophic for carboxylic acid precursors required for synthesis of branched-chain fatty acids, an essential component of staphylococcal membrane lipids and the agent of membrane fluidity. However, the skin is devoid of branched-chain fatty acids. We showed that S. aureus instead scavenges host-derived unsaturated fatty acids from the skin using the secreted lipase, Geh, and the unsaturated fatty acid–binding protein, FakB2. Moreover, murine infections demonstrated the relevance of host lipid assimilation to staphylococcal survival. Altogether, these studies provide insight into an adaptive trait that bypasses de novo lipid synthesis to facilitate S. aureus persistence during superficial infection. The findings also reinforce the inherent challenges associated with targeting bacterial lipogenesis as an antibacterial strategy and support simultaneous inhibition of host fatty acid salvage during treatment.
The gram-positive bacterium Staphylococcus aureus is notorious for its capacity to cause widespread pathology in nearly every organ and tissue during infection (1). Much of this success can be attributed to the ability of S. aureus to extract essential nutrients from the host milieu (2). Hence, understanding the adaptive traits that allow S. aureus to exploit in situ resources for survival is critical to devising effectual treatments for staphylococcal diseases.
Lipoic acid is an organosulfur compound derived from an early-stage intermediate of fatty acid biosynthesis that is required for carbon shuttling in central metabolism via a redox-sensitive dithiolane ring at its distal end (3). In a previous study, we found that shuttling of lipoic acid to metabolic enzyme complexes by the amidotransferase, LipL, was paramount for S. aureus survival in a murine model of bacteremia but not skin infection (4). At minimum, LipL appears to be required for the transfer of lipoic acid to E2 subunits of the pyruvate dehydrogenase (PDH) and branched-chain α-ketoacid dehydrogenase (BCODH) complexes (4, 5). The absence of lipoic acid renders each complex nonfunctional (4, 5). The discrepancy in demand for lipoic acid attachment in these two infection sites suggests that the nutritional environment of the skin eases the requirement for lipoic acid–dependent metabolic processes, especially those that require PDH or BCODH complex activity. PDH is responsible for the generation of acetyl-CoA upon exit from glycolysis, while BCODH is essential for the synthesis of saturated branched-chain fatty acids (BCFAs), a major component of staphylococcal membrane phospholipids (6). Like unsaturated fatty acids (UFAs), BCFAs provide membrane fluidity to staphylococci, which neither synthesize UFAs nor encode a fatty acid (FA) desaturase that converts saturated fatty acids (SFAs) to unsaturated products (7). Since a lack of membrane fluidity triggers cell death as a result of lipid phase separation and protein segregation, bacterial survival hinges on the fine-tuning of membrane lipid composition to adapt to fluctuating environments (8).
Given the importance of de novo FA synthesis to ensure bacterial membrane integrity, enzymes that are involved in this metabolic process serve as attractive targets for antibacterial drug discovery. One such drug, triclosan, binds to the enoyl-acyl carrier protein reductase, FabI, a key enzyme in the type II FA synthesis (FASII) system found in archaea and bacteria (9). FASII restriction induces FA starvation and initiates the stringent response, where high concentrations of the alarmone (p)ppGpp inhibit synthesis of the FASII substrate malonyl-CoA (10). Nevertheless, mounting evidence suggests that S. aureus can resolve triclosan interference by incorporating host-derived UFAs into its membrane (11–13). During anti-FASII–adaptive outgrowth, dissipation of (p)ppGpp levels replenishes the malonyl-CoA pool and skews distribution of the FASII substrate to favor binding with FapR, a global repressor of phospholipid synthesis genes in S. aureus (10). Ultimately, this interaction sequesters FapR to allow expression of plsX and plsC, which encode enzymes that use host UFAs to synthesize phospholipids (10, 14). UFAs are commonly found esterified as triglycerides and cholesterol esters in vertebrates (15). In humans, both compounds form the lipid core of the five major groups of lipoproteins (chylomicron, VLDL, LDL, IDL, and HDL) that transport lipids throughout the body (15). Triglycerides are also the major constituents of adipocytes and exist to a lesser extent in nonadipose cells (16, 17). It has been suggested that S. aureus secretes the glycerol ester hydrolase Geh to liberate esterified FAs from triglycerides and LDL for use as FA substrates as a nutrient acquisition strategy, although this phenomenon has not yet been tested in vivo (18, 19).
Despite increasing understanding of FASII bypass by S. aureus in recent years, the overall mechanism of rescue continues to be investigated. Here, we draw upon insights derived from the study of S. aureus lipoic acid synthesis and acquisition to make connections between BCFA synthesis and bacterial survival during infection. We report that de novo BCFA synthesis by S. aureus is the primary enzymatic process that necessitates lipoic acid in vitro and during skin infection. However, the requirement for BCFA synthesis in the skin is bypassed through assimilation of UFAs from host tissue. We also provide genetic, in vivo, and chemical evidence, via treatment with the over-the-counter lipase inhibitor, orlistat, that suggests UFA acquisition depends on the secreted lipase, Geh, and the UFA binding protein, FakB2. Finally, we demonstrate that ablating both BCFA synthesis and exogenous FA uptake significantly improves staphylococcal infection outcome in mice.
Results
Tissue-Specific Requirement for Lipoic Acid during S. aureus Infection in Mice.
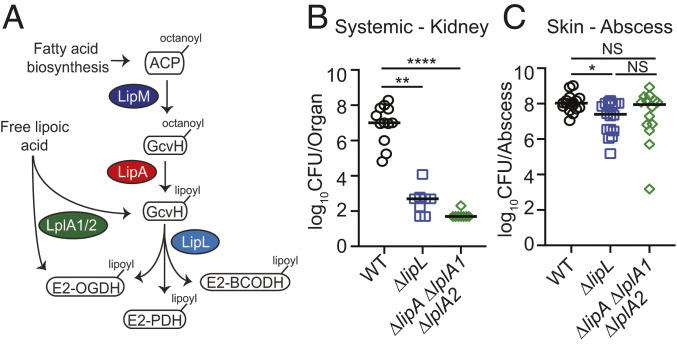
We previously characterized a putative S. aureus amidotransferase, LipL, and demonstrated that it is capable of shuttling lipoic acid between lipoyl-domain–containing proteins that require the cofactor for function (Fig. 1A) (4). Additionally, we showed that a ΔlipL mutant is highly attenuated for renal colonization in a murine sepsis model but exhibited a modest decrease in infectivity in a murine skin and soft tissue infection model (4). Therefore, we wondered if this difference in bacterial load reflects a distinct demand for lipoic acid between tissue sites. To this end, we infected mice intravenously or intradermally with either the wild-type (WT) strain, ΔlipL, or ΔlipA ΔlplA1 ΔlplA2 mutant strain that lacks the lipoyl synthase, LipA, and both lipoic acid salvage enzymes, LplA1 and LplA2 (5, 20). At 96 h postintravenous infection, kidneys of ΔlipL and ΔlipA ΔlplA1 ΔlplA2 mutant–infected animals had ∼10,000 to 100,000-fold fewer colony-forming units (CFU) than those infected with WT (Fig. 1B). At 120 h postintradermal infection, ∼fivefold fewer CFU were recovered from abscesses of ΔlipL, and no CFU difference was observed for ΔlipA ΔlplA1 ΔlplA2 mutant-infected animals compared to those infected with WT (Fig. 1C). Thus, a ΔlipA ΔlplA1 ΔlplA2 mutant is not attenuated, and a ΔlipL mutant is modestly attenuated in the skin, whereas both strains are markedly attenuated in the kidney, suggesting that S. aureus is largely refractory to lipoic acid deficiency in murine skin but not kidneys.
Fig. 1.
Differential requirement for lipoic acid by S. aureus during infection. (A) Lipoic acid synthesis and salvage pathways in S. aureus. De novo synthesis of lipoic acid by S. aureus requires the octanoyltransferase, LipM, and the lipoyl synthase, LipA. The amidotransferase, LipL, shuttles the lipoyl moiety to the E2 subunits of α-ketoacid dehydrogenase complexes that require the cofactor for function. S. aureus also encodes two salvage enzymes, LplA1 and LplA2, to scavenge free lipoic acid from the environment. ACP, acyl carrier protein; GcvH, H subunit glycine cleavage complex; E2-PDH, E2 subunit pyruvate dehydrogenase complex; E2-OGDH, E2 subunit 2-oxoglutarate dehydrogenase complex; E2-BCODH, branched-chain α-ketoacid dehydrogenase complex. (B) Bacterial burden (log10 CFU) in kidneys of mice at 96 h postinfection with 1 × 107 CFU WT (n = 12), ΔlipL (n = 8), and ΔlipA ΔlplA1 ΔlplA2 (n = 8) strains. (C) Bacterial burden (log10 CFU) in skin abscesses of mice at 120 h postinfection with 1 × 107 CFU WT (n = 16), ΔlipL (n = 16), and ΔlipA ΔlplA1 ΔlplA2 (n = 14) strains. P values were determined by a nonparametric one-way ANOVA (Kruskal–Wallis test) with Dunn’s posttest. *P < 0.05, **P < 0.01, ****P < 0.0001, and NS, not significant.
BCFA Auxotrophy Is Responsible for the Growth Defect of a Lipoic Acid–Deficient Strain.
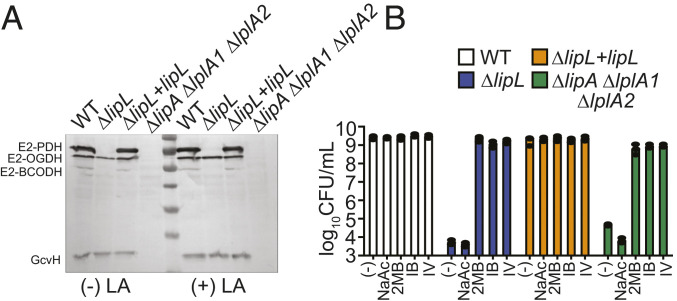
We surmised that lack of lipoylation on the E2 subunit of one or more metabolic enzyme complexes causes the reduced survival of the ΔlipL and ΔlipA ΔlplA1 ΔlplA2 mutant strains. To evaluate the lipoylation capabilities of these strains, we performed an anti-lipoic acid immunoblot on whole-cell lysates from each strain grown in lipoic acid bypass medium, which consists of Roswell Park Memorial Institute (RPMI) medium supplemented with sodium acetate (NaAc) and branched-chain carboxylic acids that are BCFA precursors (BCFAP). The supplemented precursors include 2-methylbutyric acid (2MB), isovaleric acid (IV), and isobutyric acid (IB) (21, 22). The ΔlipL mutant was unable to lipoylate E2-PDH and E2-BCODH, whereas the ΔlipA ΔlplA1 ΔlplA2 mutant could not lipoylate the E2 subunit of 2-oxoglutarate dehydrogenase and the glycine cleavage system H protein, GcvH, in addition to E2-PDH and E2-BCODH (Fig. 2A). Supplementation with exogenous lipoic acid or NaAc failed to rescue growth of ΔlipA ΔlplA1 ΔlplA2 and ΔlipL mutant strains in RPMI (Fig. 2B and 4, 5, 20). However, addition of any one BCFAP (2MB, IV, or IB) was sufficient to restore viability of both strains (Fig. 2B). Taken together, these observations suggest that the dearth of lipoyl-E2-BCODH, which is involved in BCFA synthesis, renders a lipoic acid–deficient strain nonviable in medium lacking this cofactor.
Fig. 2.
Lipoic acid deficiency causes BCFA auxotrophy in S. aureus. (A) In vivo lipoylation profiles of WT, ΔlipL, ΔlipL+lipL, and ΔlipA ΔlplA1 ΔlplA2 strains after 9 h subculture growth in RPMI + BCFAP [10 mM IB, 9 mM 2MB, 9 mM IV + 10 mM NaAc], with or without 5 μM lipoic acid (LA), were assessed by immunoblotting SDS-PAGE–resolved whole-cell lysates with rabbit α-lipoic acid antibody. Presented blot is representative of at least three independent experiments. (B) Growth (log10 CFU/mL) of WT, ΔlipL, ΔlipL+lipL, and ΔlipA ΔlplA1 ΔlplA2 strains after 24 h growth in RPMI only or RPMI + 10 mM NaAc or any one BCFAP (9 mM 2MB, 9 mM IV, or 10 mM IB).
Host-Derived UFAs Relieve Lipoic Acid Requirement of S. aureus.
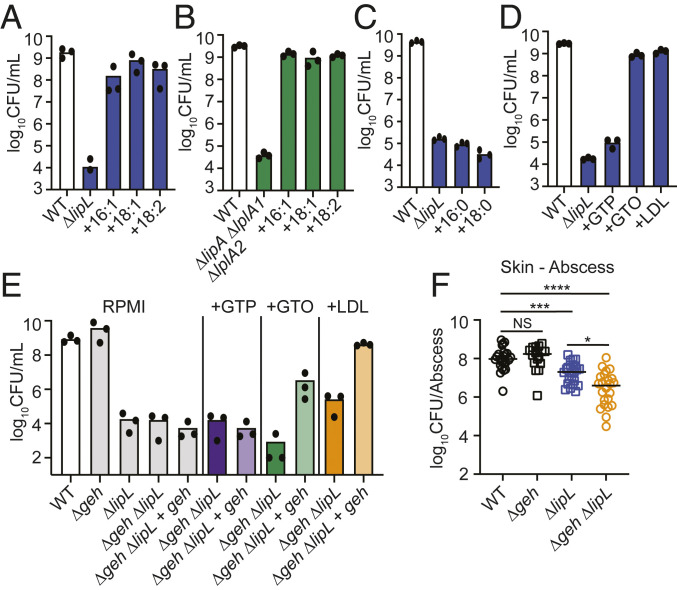
Our findings so far led us to hypothesize that the murine skin environment is enriched in certain nutrients that can restore viability to the ΔlipL and ΔlipA ΔlplA1 ΔlplA2 strains. Since these strains are auxotrophic for BCFA, which provides fluidity to staphylococcal membrane, we predict that the likely substrate is either BCFA or UFA. BCFAs are present in negligible quantities on murine skin, whereas UFAs, which contribute to membrane fluidity in many other bacteria including Escherichia coli, are of high abundance (23). Hence, to address whether UFAs can restore growth of a ΔlipL mutant in RPMI, we supplemented the medium with one of three UFAs [palmitoleic acid (16:1), oleic acid (18:1), or linoleic acid (18:2)] and evaluated growth of the mutant compared to the WT strain by quantifying CFUs. All three UFAs successfully restored growth of the ΔlipL mutant as well as the ΔlipA ΔlplA1 ΔlplA2 mutant to WT levels (Fig. 3 A and B). Conversely, supplementation with SFAs [palmitic acid (16:0) or stearic acid (18:0)] failed to reestablish bacterial growth (Fig. 3C). Considering most UFAs in the host originate from triglyceride stores and cholesterol esters (16, 17), we also assessed the ability of saturated and unsaturated triglycerides [glyceryl tripalmitate (GTP, 16:0) and glyceryl trioleate (GTO, 18:1)] or human low-density lipoprotein (LDL) to serve as FA substrates in RPMI. As expected, GTO and LDL but not the saturated triglyceride, GTP, successfully reestablished viability of these strains (Fig. 3D). The FA moieties from GTO and LDL are esterified to glycerol and glycerol/cholesterol, respectively, although some free FA may exist in purified human LDL. Therefore, we reasoned the acquisition of UFAs from GTO and LDL would require the major secreted lipase, Geh, to liberate free FAs before use. We generated a Δgeh ΔlipL double mutant and a Δgeh ΔlipL + geh complementation strain and monitored growth in RPMI or RPMI supplemented with GTP, GTO, and LDL. As predicted, the growth of the Δgeh ΔlipL double mutant was no longer rescued by supplementation with GTO or LDL, while complementation with geh partially restored growth in the presence of these substrates (Fig. 3E). To ascertain a role for Geh in liberating UFA to promote survival in vivo, we intradermally infected mice with WT, Δgeh, ΔlipL, and ΔlipL Δgeh strains and quantified CFU in abscesses after 120 h. A Δgeh mutant was not attenuated to any measurable degree in the skin after 120 h, whereas a ΔlipL mutant was modestly attenuated (Figs. 1C and 3F). Compared to the ΔlipL mutant, infection with ΔlipL Δgeh double mutant led to recovery of 10-fold fewer CFU. These results suggest that lipase activity is partially responsible for the bypass of lipoic acid requirement in the skin.
Fig. 3.
UFA substrates bypass requirement for lipoic acid by S. aureus. (A and B) 24 h subculture growth (log10 CFU/mL) of (A) ΔlipL and (B) ΔlipA ΔlplA1 ΔlplA2 strains in RPMI only or RPMI supplemented with 50 μM of each palmitoleic acid (16:1), oleic acid (18:1), or linoleic acid (18:2). (C and D) 24 h subculture growth (log10 CFU/mL) of ΔlipL strain in RPMI supplemented with (C) saturated FAs [50 μM palmitic acid (16:0) or 50 μM stearic acid (18:0)] or (D) covalently bound FA substrates (16.67 μM GTP, 16.67 μM GTO, or 0.34 mg/mL human LDL). (E) 24 h subculture growth (log10 CFU/mL) of Δgeh ΔlipL and Δgeh ΔlipL + lipL strains in RPMI supplemented with GTP, GTO, or LDL as indicated in (D). WT Δgeh, ΔlipL, Δgeh ΔlipL, and Δgeh ΔlipL + lipL strains were grown in RPMI without lipid supplementation as controls. (F) Bacterial burden (log10 CFU) in skin abscesses of mice at 120 h postinfection with 1 × 107 CFU WT (n = 24), Δgeh (n = 16), ΔlipL (n = 24), and Δgeh ΔlipL (n = 24) strains. P values were determined by a nonparametric one-way ANOVA (Kruskal–Wallis test) with Dunn’s posttest. *P < 0.05, ***P < 0.001, and ****P < 0.0001.
Liberation of UFAs Relieves BCFA Auxotrophy.
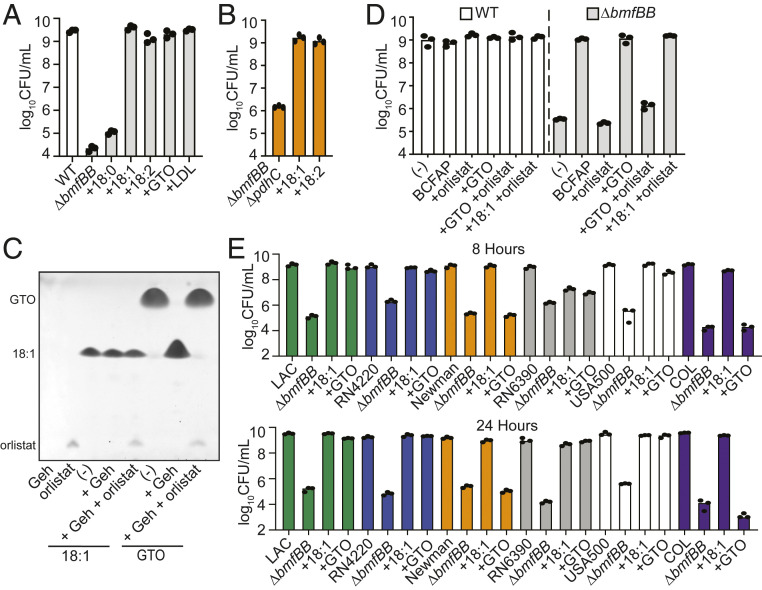
To confirm that uptake of UFAs by S. aureus sidesteps the requirement for BCFA synthesis, we generated a ΔbmfBB::kan mutant, which lacks the E2 subunit of BCODH, and tested the ability of the mutant strain to use free FAs (18:0, 18:1, and 18:2) as substrates for growth in RPMI. Similar to the ΔlipL mutant, the ΔbmfBB::kan mutant grew in RPMI supplemented with UFA but not SFA substrates (Fig. 4A). Deleting pdhC, which encodes E2-PDH, did not change the requirement of USFA for the growth of a ΔbmfBB::kan mutant (Fig. 4B). We also assessed the ability of the unsaturated triglyceride, GTO (18:1), or human LDL to serve as substrates for the ΔbmfBB::kan mutant in RPMI. Similar to the ΔlipL mutant, the ΔbmfBB::kan mutant grew in RPMI supplemented with GTO and LDL (Fig. 4A). To test the role of Geh in liberating 18:1 from GTO, we attempted to generate a ΔbmfBB Δgeh double mutant but were not successful after numerous attempts. Instead, we acquired an over-the-counter human pancreatic lipase inhibitor, orlistat, which was recently cocrystalized with S. aureus Geh (24) and tested its ability to inhibit Geh activity and prevent restoration of growth to the ΔbmfBB::kan mutant in the presence of GTO. When purified recombinant Geh was coincubated with orlistat and GTO, it was unable to hydrolyze the esterified 18:1 FA, whereas Geh alone was able to completely hydrolyze GTO (Fig. 4C). Consistent with its successful inhibition of Geh activity, orlistat prevented growth of a ΔbmfBB::kan mutant in the presence of GTO (Fig. 4D). To ascertain the relevance of strain-to-strain variability that has been proposed to affect UFA assimilation upon FASII inhibition (10–13, 25–28), we generated ΔbmfBB::kan mutations in the strains used in these studies as well as methicillin-resistant S. aureus strains that either harbor a WT geh allele or a naturally occurring phage disruption within the geh allele (SI Appendix, Table S1) and monitored growth in the presence of 18:1 and GTO. Our data indicate that ΔbmfBB::kan mutants of all strains were able to grow on 18:1 by 24 h, while ΔbmfBB::kan mutants in strains Newman and COL (both contain phage disruptions in geh) were unable to grow on GTO (Fig. 4D). RN6390 ΔbmfBB::kan was slower to adapt to 18:1 and GTO as evidenced by its limited growth at 8 h but reached a similar final CFU at 24 h, consistent with prior FASII inhibition studies (Fig. 4D). In sum, these observations demonstrate that host ester-linked UFAs are released by Geh-mediated ester hydrolysis and can functionally compensate for BCFA auxotrophy in strains with an intact geh allele.
Fig. 4.
UFA substrates rescue BCFA auxotrophy. (A) An 8 h subculture growth (log10 CFU/mL) of ΔbmfBB::kan strain in RPMI supplemented with different 18-carbon-long FA substrates [50 μM stearic acid (18:0), 50 μM oleic acid (18:1), 50 μM linoleic acid (18:2), or 16.67 μM GTO or 0.34 mg/mL human LDL]. (B) An 8 h subculture growth (log10 CFU/mL) of ΔbmfBB ΔpdhC::kan strain in RPMI only or RPMI supplemented with 50 μM 18:1 or 50 μM 18:2. (C) TLC of in vitro lipase activity assays containing 400 μM FA substrates [oleic acid (18:1) or GTO] in the presence or absence of 1 μM Geh ± 400 μM orlistat. (D) The effect of 50 μM orlistat on 8 h subculture growth (log10 CFU/mL) of WT and ΔbmfBB::kan strains in RPMI supplemented with 50 μM GTO or 50 μM 18:1. (E) An 8 and 24 h subculture growth (log10 CFU/mL) of the indicated ΔbmfBB::kan (ΔbmfBB) strains in RPMI supplemented with 50 μM oleic acid (18:1) or 16.67 μM GTO.
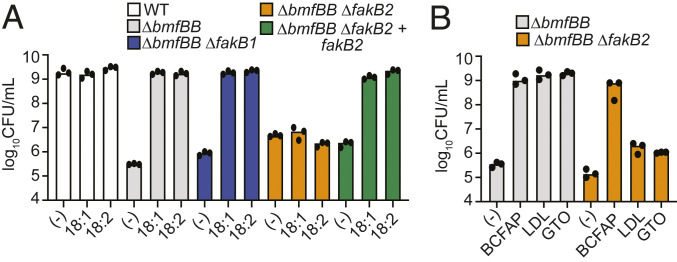
FakB2 Is Required for UFA-Dependent Growth of a BCFA Auxotroph.
S. aureus encodes two FA-binding proteins, FakB1 and FakB2 (29). FAs bound to either protein serve as substrates for the FA kinase FakA, which phosphorylates the FA acyl chains (29). These activated acyl-phosphates eventually get incorporated into membrane phospholipids (14). While both FA-binding proteins can bind SFAs, only FakB2 can bind UFAs (29, 30). Therefore, we predict that the ΔbmfBB::kan mutant requires FakB2 for UFA-dependent growth in RPMI. To determine the requirement of either FA-binding proteins for growth in RPMI supplemented with 18:1 or 18:2 UFA, we deleted fakB1 or fakB2 from the ΔbmfBB mutant strain. The ΔbmfBB::kan ΔfakB2 strain, but not the ΔbmfBB::kan ΔfakB1 strain, failed to replicate in RPMI supplemented with either of the aforementioned UFA substrates (Fig. 5A). This growth defect was reversed in the ΔbmfBB::kan ΔfakB2 + fakB2 strain (Fig. 5A). The ΔbmfBB::kan ΔfakB2 double mutant also failed to replicate in RPMI medium supplemented with GTO or LDL (Fig. 5B). Together, these findings underpin the role of FakB2 in scavenging exogenous UFAs to support growth of a BCFA auxotroph.
Fig. 5.
Use of UFA substrates requires FakB2. (A) An 8 h subculture growth (log10 CFU/mL) of WT, ΔbmfBB::kan, ΔbmfBB::kan ΔfakB1, ΔbmfBB::kan ΔfakB2, and ΔbmfBB::kan ΔfakB2 + fakB2 strains in RPMI only or RPMI supplemented with 50 μM oleic acid (18:1) or 50 μM linoleic acid (18:2). (B) An 8 h subculture growth (log10 CFU/mL) of ΔbmfBB::kan and ΔbmfBB::kan ΔfakB2 strains in RPMI supplemented with BCFAP (10 mM IB, 9 mM 2MB, 9 mM IV), 0.34 mg/mL human LDL, or 16.67 μM GTO.
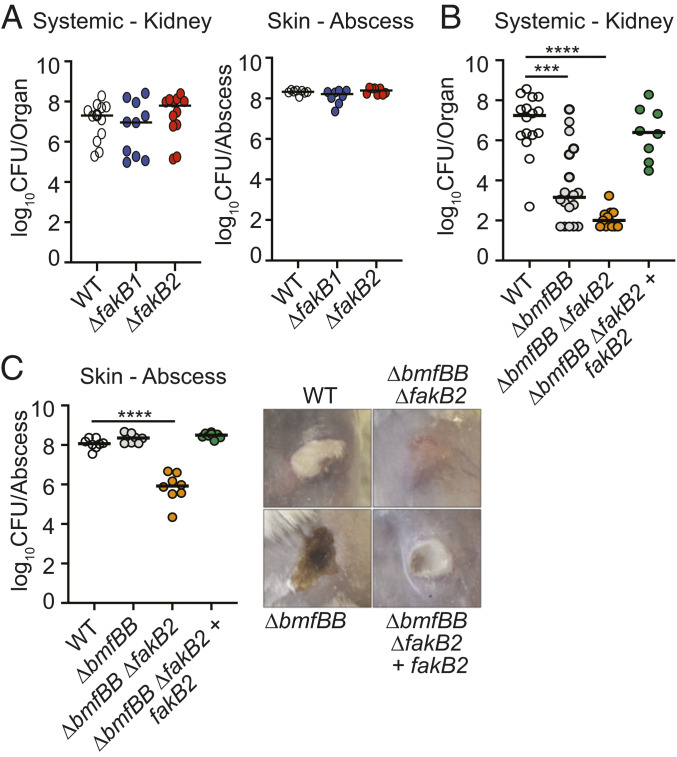
S. aureus Displays Tissue-Specific BCFA Dependency during Infection in Mice.
We showed that a ΔbmfBB::kan mutant is defective for growth in RPMI but that cell growth can be restored in a Geh- and FakB2-dependent manner by addition of UFAs and triglycerides containing UFAs. Given that a ΔbmfBB::kan mutant phenocopies a ΔlipL mutant for viability defects in broth culture, we surmised that a ΔbmfBB::kan mutant would also be compromised for systemic disease but not superficial skin infection in mice, as FakB2 would be critical for this survival advantage in the skin on account of UFA uptake. To this end, we initially infected mice intravenously or intradermally with WT, ΔfakB1, and ΔfakB2 mutant strains and monitored infection in the kidney (systemic) or superficial abscess (skin). No differences in recovered CFU were observed (Fig. 6A). We then infected mice intravenously or intradermally with WT, ΔbmfBB::kan, ΔbmfBB::kan ΔfakB2, and ΔbmfBB::kan ΔfakB2 + fakB2 strains. At 96 h postintravenous infection, kidneys of ΔbmfBB::kan mutant-infected animals had ∼10,000-fold fewer CFUs than those infected with WT, while renal CFUs of ΔbmfBB::kan ΔfakB2 mutant-infected animals were reduced an additional ∼10-fold (Fig. 6B). Complementation of the double mutant with fakB2 restored renal CFUs to nearly WT levels (Fig. 6B). At 120 h postintradermal infection, WT and ΔbmfBB::kan strains infected to similar levels (Fig. 6C). However, ∼100-fold fewer CFUs were recovered from the abscesses of ΔbmfBB::kan ΔfakB2 mutant-infected animals (Fig. 6C). Complementation of the double mutant with fakB2 restored CFUs to levels of WT and ΔbmfBB::kan strains (Fig. 6C). The differences in infection efficiency of these four strains were reflected in abscess severity at 96 h post infection (Fig. 6C). While we observed well-demarcated dermonecrotic abscesses in animals infected with WT, ΔbmfBB::kan, and ΔbmfBB::kan ΔfakB2 + fakB2 strains, infection with the ΔbmfBB::kan ΔfakB2 strain failed to induce abscess formation and only displayed minor redness at the site of injection (Fig. 6C). Thus, a ΔbmfBB::kan mutant mimics a ΔlipL mutant in terms of its differential infectivity in murine sepsis and skin disease models. These results also further cement the importance of FakB2 in supporting S. aureus infection when BCFA synthesis is hindered.
Fig. 6.
FakB2 is important for bypass of BCFA requirement during S. aureus infection. (A) Bacterial burden (log10 CFU) in kidneys of mice at 96 h postinfection with 1 × 107 CFU WT (n = 12), ΔfakB1 (n = 10), and ΔfakB2 (n = 12) strains; and bacterial burden (log10 CFU) in skin abscesses of mice at 120 h postinfection with 1 × 107 CFU WT (n = 8), ΔfakB1 (n = 8), and ΔfakB2 (n = 8) strains. (B) Bacterial burden (log10 CFU) in kidneys of mice at 96 h postinfection with 1 × 107 CFU WT (n = 16), ΔbmfBB::kan (n = 18), ΔbmfBB::kan ΔfakB2 (n = 12), and ΔbmfBB::kan ΔfakB2 + fakB2 (n = 8) strains. (C) Bacterial burden (log10 CFU) in skin abscesses of mice at 120 h postinfection with 1 × 107 CFU WT (n = 8), ΔbmfBB::kan (n = 8), ΔbmfBB::kan ΔfakB2 (n = 8), and ΔbmfBB::kan ΔfakB2 + fakB2 (n = 8) strains, and representative corresponding images of skin lesions. P values were determined by a nonparametric one-way ANOVA (Kruskal–Wallis test) with Dunn’s posttest. ***P < 0.001 and ****P < 0.0001.
Discussion
Evidence to support the link between lipoic acid metabolism and virulence of bacterial pathogens is growing (31). Indeed, we previously determined that lipoic acid synthesis and salvage supports S. aureus colonization during murine bloodstream infection (5) and restricts macrophage generation of reactive nitrogen and reactive oxygen species in a peritonitis infection model (32). We also found that autolysis-dependent release of S. aureus lipoyl E2-PDH suppresses macrophage activation by antagonizing Toll-like receptor (TLR) 1/2 signaling (33). Yet, the relative contributions of lipoic acid–dependent metabolic processes to S. aureus infection have not been explored. Here, we present evidence that BCODH, which converts transaminated branched-chain amino acids to precursors for BCFA synthesis, is arguably the most vital enzyme complex for optimal S. aureus colonization in mice during systemic disease but not skin and soft tissue infection. A similar tissue-specific requirement for LipL led us to determine that host-derived UFAs can rescue BCFA auxotrophy in the skin, and by extension, circumvent demand for lipoic acid for effective S. aureus infection.
We previously found that a S. aureus strain defective for lipoic acid salvage (ΔlplA1 ΔlplA2) is markedly attenuated in the kidneys of systemically infected mice (5). Therefore, lipoic acid salvage is instrumental for survival in this organ despite an intact de novo synthesis pathway. This is perhaps due to lower expression of lipoic acid synthesis enzymes or a higher demand for lipoyl proteins in the kidneys. Nevertheless, considering the findings in this study, we postulate that lipoic acid salvage is required in the kidney in order to augment BCODH activity since S. aureus appears unable to acquire sufficient UFAs from the renal milieu. However, our data do not rule out the alternative possibility that other lipoic acid–containing enzymes might also be required for survival in the kidney, namely E2-PDH. We are currently testing this hypothesis.
To make BCFA, BCODH-synthesized precursors in the form of short branched-chain acyl CoAs are elongated by FASII. Since this pathway is not found in mammals, which uses FASI (34), FASII enzymes were thought to be excellent targets for antibacterials. These include inhibitors of FabI (triclosan, AFN-1252, and MUT056399) (9, 35, 36), FabF (platensimycin and fasamycins A and B) (37, 38), and FabH (amycomicin) (39). While anti-FASII drugs remain potent antimycobacterial agents due to a dearth of exogenous very long chain FAs specific to members of this genus (40), Firmicutes, such as staphylococci and streptococci, were later found to be capable of exploiting host UFAs to evade FASII inhibition (11–13). Therefore, our findings further stress the sophisticated adaptive trait of acquiring a fail-safe system to maintain membrane homeostasis in the absence of BCFA.
In this study, we demonstrated that the free or triglyceride form of UFAs and human LDL were able to revitalize BCFA-deficient cells in broth medium. However, prior studies indicate that most UFAs exert potent antimicrobial activity on S. aureus by distorting membrane architecture (18, 41, 42). For example, while 18:1 can be directly incorporated into S. aureus phospholipids, allowing the bacterium to bypass FASII, 16:1 and 18:2 trigger membrane depolarization and release of low-molecular–weight proteins from cells (41). To counter these deleterious effects, S. aureus hydrates 16:1 and 18:2 to nontoxic hydroxy-16:0 and hydroxy-18:1, respectively, with the oleate hydratase, OhyA (43), or elongates 16:1 by two carbon units for assimilation into membrane lipids (41). Production of the surface protein IsdA by S. aureus also decreases its cellular hydrophobicity, in turn conferring resistance to the bactericidal actions of skin FAs (44). Moreover, S. aureus secretes FA-modifying enzymes that esterify free FAs with cholesterol or short-chain alcohols to inactivate these cytotoxic compounds (45). In addition, concomitant attachment of UFAs to lipoproteins renders S. aureus susceptible to the TLR2 immune response (46). Such mechanisms of detoxification are likely at play in our studies, allowing incorporation of less deleterious UFAs such as 18:1 and hydroxy-18:1 to promote survival in the skin.
Prior studies on adaptation to anti-FASII antibiotics highlighted strain-dependent variabilities in the assimilation of host FAs (10–13, 25–28). Using BCFA auxotroph mutants, we did not observe such variation after supplementation with free UFAs but did uncover growth defects upon GTO supplementation when using strains that harbor a phage disruption in the geh gene. Approximately 15% of sequenced isolates harbor this disruption, thus our current study implies the presence or absence of geh may have biologically meaningful consequences on nutritional adaptation in addition to previously described effects on immune function (47). RN6390, a derivative of strain 8325 (48, 49), was the only strain to exhibit a notable delay in UFA utilization upon deletion of bmfBB. This may be due to known genetic variations in this lineage that affect fakB1, among other regulatory and metabolic loci (10, 11, 49). Nevertheless, our findings argue that assimilation of free UFAs by S. aureus BCFA auxotrophs is likely conserved, whereas the ability to use triglycerides is predicted by the status of the geh gene.
Our combined approach of genetic and animal studies substantiates the model whereby FakB2 is responsible for binding exogenous UFAs, thus enabling S. aureus to tolerate anti-FASII treatment (11, 29, 30). We recapitulated the effects of this class of drugs by homing in on BCFA synthesis and genetically disrupting BCODH activity and determined that deletion of fakB2 abrogates the compensatory role of host UFAs in sustaining growth and infectivity of BCFA-deficient cells. It is notable that while chromosomal complementation of fakB2 in a ΔbmfBB::kan ΔfakB2 mutant restored colonization to WT levels during murine skin infection (Fig. 6C), the complemented mutant colonized the kidneys better than a ΔbmfBB mutant during systemic infection (Fig. 6B). We ascribe this outcome to the constitutive expression of complemented fakB2, which likely elevates incorporation of host UFAs and enhances renal infection. Since synthesis of 12-methyltetradecanoic acid (anteiso-15:0), the major BCFA species found in S. aureus membrane, requires BCODH, the ability of a ΔbmfBB mutant to grow in the presence of UFAs supports the view that the preferential occupancy of anteiso-15:0 at the sn-2 position of phosphatidylglycerol can be overcome to undermine anti-FASII efficacy (11–13, 27, 28, 50). In sum, the results of our studies emphasize the importance of the host environment in establishing an adaptive niche for survival and the synergistic potential of blocking host FA incorporation and de novo FA synthesis as a therapeutic approach for staphylococcal infections.
Materials and Methods
Bacterial Strains and Growth Conditions.
All bacterial strains used in this manuscript are listed in SI Appendix, Table S1. E. coli strains were routinely grown in Miller’s lysogeny broth (BD), while S. aureus strains were grown in either tryptic soy broth (BD) or in RPMI medium (Corning) supplemented with 1% casamino acids (VWR). When needed, cultures were supplemented with the following agents for selection: 100 μg ⋅ mL−1 ampicillin (GoldBio), 10 μg ⋅ mL−1 chloramphenicol (Amresco), 150 μg ⋅ mL−1 kanamycin (Amresco), and 0.1 mM cadmium chloride (Alfa Aesar). The following FA substrates were used for various assays throughout this study: 10 mM IB (Sigma-Aldrich), 9 mM 2-methylbutyric acid (Alfa Aesar), 9 mM IV (Sigma-Aldrich), 50 μM palmitic acid (MP Biomedicals), 50 μM palmitoleic acid (Sigma-Aldrich), 50 μM stearic acid (MP Biomedicals), 50 μM oleic acid (Sigma-Aldrich), 50 μM linoleic acid (VWR), 16.67 μM glyceryl tripalmitate (Sigma-Aldrich), 16.67 μM glyceryl trioleate (Sigma Aldrich), 0.34 μg/μL purified human LDL (Kalen Biomedical), and 50 μM orlistat (Alli, GlaxoSmithKline).
Murine Infection Models.
Murine systemic and skin and soft tissue infections were performed as previously described (5, 51) with WT, ΔlipL, Δgeh, ΔlipL Δgeh, ΔlipA ΔlplA1 ΔlplA2, ΔfakB1, ΔfakB2, ΔbmfBB::kan, ΔbmfBB::kan ΔfakB2, and ΔbmfBB::kan ΔfakB2 + fakB2 strains.
Determination of Protein Lipoylation.
Proteins from optical density (OD)-normalized whole-cell lysates were separated by sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) in 12% polyacrylamide gels at 120 V. Separated proteins were transferred from gels to 0.2 μm Immobilon polyvinylidene difluoride membranes (Millipore Sigma) at 100V for 30 min in a Criterion Blotter (Bio-Rad). After transfer, membranes were incubated for 1 h in Tris-buffered saline + 0.1% TWEEN 20 (Amresco) (TBST) supplemented with 5% bovine serum albumin (BSA) (GoldBio). Rabbit polyclonal α-lipoic acid antibody (Calbiochem) was added to the membranes at a 1:7,500 dilution followed by incubation for 1 h and three subsequent 15 min washes in ∼20 mL of TBST. Alkaline phosphatase (AP)-conjugated goat anti-rabbit IgG (H+L) (Invitrogen) was then added at a 1:5,000 dilution in 5% BSA in TBST for 45 min followed by three 15 min washes in ∼20 mL of TBST and another 5 min wash in AP Buffer (100 mM Tris pH 9.5, 100 mM NaCl, 5 mM MgCl2). Membranes were developed with 5-bromo-4-chloro-3-indoyl-phosphate/nitro blue tetrazolium color development substrate (GoldBio).
Generation of ΔbmfBB::kan and ΔbmfBB Strains.
The kanamycin resistance cassette, aphA3, was amplified from the plasmid pBTK (52) with primer pairs kanF-KasI and kanR-KasI (SI Appendix, Table S2), followed by subcloning into pIMAY harboring the ∼500-base-pair upstream and downstream regions of homology that flank the bmfBB gene using a KasI restriction site introduced between the two flanking regions (53). Mutagenesis was performed as previously described (5). An unmarked in-frame deletion of bmfBB was also generated with the same method but without using aphA3. Absence of any secondary mutations was verified by whole-genome sequencing.
Generation of ΔlipL Δgeh and ΔbmfBB ΔpdhC Strains.
Marked Δgeh::kan and ΔpdhC::kan mutations were transduced into ΔlipL and ΔbmfBB strains, respectively, as described previously (5), to generate ΔlipL Δgeh and ΔbmfBB ΔpdhC strains.
Generation of ΔlipL Δgeh + Geh Strains.
The pJC1112-geh construct (47) was transduced into the ΔlipL Δgeh strain as described previously (5).
Generation of ΔfakB1 and ΔfakB2 Strains.
A total of ∼500-base-pair regions of homology upstream and downstream of fakB1 and fakB2 genes were amplified from WT S. aureus genomic DNA (a list of primers is provided in SI Appendix, Table S2). The amplicons were used in a splicing by overlap extension (SOE) PCR to obtain the final amplicon, which was subcloned into the pIMAY plasmid (53). Mutagenesis was performed as previously described (5).
Generation of RN4220 ΔbmfBB::kan, RN6390 ΔbmfBB::kan, Newman ΔbmfBB::kan, COL ΔbmfBB::kan, BK2395 ΔbmfBB::kan, ΔbmfBB::kan ΔfakB1, and ΔbmfBB::kan ΔfakB2 Strains.
The ΔbmfBB::kan mutation was transduced into the indicated S. aureus strains, as described previously (5).
Generation of ΔbmfBB::kan ΔfakB2 + fakB2 Strain.
The ΔbmfBB::kan ΔfakB2 + fakB2 strain was generated using the pJC1111 plasmid (54). Using primers listed in SI Appendix, Table S2, the fakB2 gene was amplified from WT S. aureus genomic DNA, while the constitutive PHELP promoter was amplified from pIMAY plasmid (53). The resulting amplicons were subjected to SOE PCR to obtain PHELP-fakB2, which was cloned into pJC1111 at the PstI and SalI restriction sites. The recombinant construct was propagated in E. coli DH5-α and transformed into SaPI-1-integrase–expressing S. aureus (RN9011) to allow single-copy integration at the SaPI-1 site in the chromosome (54, 55). Bacteriophage Φ-11 was used to package the integrated complementation plasmid from RN9011 and was subsequently transduced into the ΔbmfBB::kan ΔfakB2 strain. ΔbmfBB::kan ΔfakB2 + fakB2 transductants were selected for based on cadmium chloride resistance and confirmed by PCR.
Bacterial Viability Assay.
S. aureus strains were grown in RPMI containing BCFAP and 10 mM NaAc (Sigma-Aldrich) overnight at 37 °C with shaking. Bacteria were washed three times and resuspended in the same volume of fresh RPMI before being subcultured 1:100 into 990 μL of the same medium supplemented with any one of aforementioned FA substrates in a 15 mL conical tube at 37 °C with shaking for 24 h. Bacterial viability was assessed by enumerating CFU on tryptic soy agar.
Purification of Geh-6xHis.
Geh-6xHis was purified as described previously (47).
Orlistat Activity Assay.
To assay the inhibitory activity of orlistat against Geh, 500 μL reactions were carried out at 37 °C in phosphate-buffered saline, with 400 μM FA substrate (oleic acid or glyceryl trioleate), 1 μM Geh-6xHis, and 400 μM orlistat for 6 h. A total of 5 μL of each reaction were applied onto a thin-layer chromatography (TLC) Silica gel 60 with concentrating zone 20 × 2.5 cm (Merck). The TLC plate was developed with hexane (Fisher):ethyl ether (VWR):acetic acid (Fisher) (80:20:2) as previously described (56).
Ethics Statement.
All experiments were performed following the ethical standards of the Institutional Biosafety Committee and the Institutional Animal Care and Use Committee (IACUC) at Loyola University Chicago Health Sciences Division. The institution is approved by Public Health Service (PHS; A3117-01 through 02/28/2022), is fully accredited by the Association for Assessment and Accreditation of Laboratory Animal Care International (000180, certification dated 11/17/2016), and is registered/licensed by the United States Department of Agriculture (USDA) (33-R-0024 through 08/24/2023). All animal experiments were performed in animal biosafety level 2 facilities with IACUC-approved protocols (IACUC #2017028) under the guidance of the Office of Laboratory Animal Welfare following USDA and PHS policy and guidelines on the humane care and use of laboratory animals.
Quantification and statistical analyses.
All experiments were repeated at least three independent times. For in vitro growth assays, data shown are representative experiments conducted in triplicate that were repeated on a minimum of three independent occasions. All statistical significance was analyzed using GraphPad Prism version 9.0 with statistical tests specified in the figure legends. All animal studies include combined data from at least two independent experiments.
Supplementary Material
Acknowledgments
We thank members of the laboratory for helpful discussions and for critically reviewing the manuscript. This study was supported by Grants NIH R01 AI120994 and a Burroughs Wellcome Fund Investigators in the Pathogenesis of Infectious Disease Award to F.A., AHA19POST34380259 to W.P.T., and AHA19PRE34400022 to X.C.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2022720118/-/DCSupplemental.
Data Availability
All study data are included in the article and/or SI Appendix.
References
- 1.Lu T., DeLeo F. R., “Pathogenesis of Staphylococcus aureus in humans” in Human Emerging and Re-Emerging Infections, Singh S. K., Ed. (John Wiley & Sons, Ltd, 2015), pp. 711–748. [Google Scholar]
- 2.Thomsen I. P., Liu G. Y., Targeting fundamental pathways to disrupt Staphylococcus aureus survival: Clinical implications of recent discoveries. JCI Insight 3, e98216 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cronan J. E., Assembly of lipoic acid on its cognate enzymes: An extraordinary and essential biosynthetic pathway. Microbiol. Mol. Biol. Rev. 80, 429–450 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Teoh W. P., Resko Z. J., Flury S., F. Alonzo, 3rd, Dynamic relay of protein-bound lipoic acid in Staphylococcus aureus. J. Bacteriol. 201, e00446-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zorzoli A., Grayczyk J. P., Alonzo F. 3rd, Staphylococcus aureus tissue infection during sepsis is supported by differential use of bacterial or host-derived lipoic acid. PLoS Pathog. 12, e1005933 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Singh V. K., et al., Roles of pyruvate dehydrogenase and branched-chain α-keto acid dehydrogenase in branched-chain membrane fatty acid levels and associated functions in Staphylococcus aureus. J. Med. Microbiol. 67, 570–578 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sen S., et al., Growth-environment dependent modulation of Staphylococcus aureus branched-chain to straight-chain fatty acid ratio and incorporation of unsaturated fatty acids. PLoS One 11, e0165300 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gohrbandt M., et al., Low membrane fluidity triggers lipid phase separation and protein segregation in vivo. bioRxiv [Preprint] (2019). https://www.biorxiv.org/content/10.1101/852160v1.full (Accessed 22 November 2019).
- 9.Heath R. J., et al., Mechanism of triclosan inhibition of bacterial fatty acid synthesis. J. Biol. Chem. 274, 11110–11114 (1999). [DOI] [PubMed] [Google Scholar]
- 10.Pathania A., et al., (p)ppGpp and malonyl-CoA set the pace for Staphylococcus aureus adaptation to FASII antibiotics and provide a basis for bi-therapy inhibition. bioRxiv [Preprint] (2020). https://www.biorxiv.org/content/10.1101/2020.03.26.007567v3 (Accessed 27 March 2020).
- 11.Kénanian G., et al., Permissive fatty acid incorporation promotes staphylococcal adaptation to FASII antibiotics in host environments. Cell Rep. 29, 3974–3982.e4 (2019). [DOI] [PubMed] [Google Scholar]
- 12.Brinster S., et al., Type II fatty acid synthesis is not a suitable antibiotic target for Gram-positive pathogens. Nature 458, 83–86 (2009). [DOI] [PubMed] [Google Scholar]
- 13.Morvan C., et al., Environmental fatty acids enable emergence of infectious Staphylococcus aureus resistant to FASII-targeted antimicrobials. Nat. Commun. 7, 12944 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons J. B., Frank M. W., Jackson P., Subramanian C., Rock C. O., Incorporation of extracellular fatty acids by a fatty acid kinase-dependent pathway in Staphylococcus aureus. Mol. Microbiol. 92, 234–245 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cox R. A., García-Palmieri M. R., “Cholesterol, triglycerides, and associated lipoproteins” in Clinical Methods: The History, Physical, and Laboratory Examinations, Walker H. K., Hall W. D., Hurst J. W., Eds. (Butterworths, ed. 3, 1990), pp. 153–160. [Google Scholar]
- 16.Frayn K. N., Tan G. D., Karpe F., Adipose tissue: A key target for diabetes pathophysiology and treatment? Horm. Metab. Res. 39, 739–742 (2007). [DOI] [PubMed] [Google Scholar]
- 17.Guilherme A., Virbasius J. V., Puri V., Czech M. P., Adipocyte dysfunctions linking obesity to insulin resistance and type 2 diabetes. Nat. Rev. Mol. Cell Biol. 9, 367–377 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cadieux B., Vijayakumaran V., Bernards M. A., McGavin M. J., Heinrichs D. E., Role of lipase from community-associated methicillin-resistant Staphylococcus aureus strain USA300 in hydrolyzing triglycerides into growth-inhibitory free fatty acids. J. Bacteriol. 196, 4044–4056 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Delekta P. C., Shook J. C., Lydic T. A., Mulks M. H., Hammer N. D., Staphylococcus aureus utilizes host-derived lipoprotein particles as sources of fatty acids. J. Bacteriol. 200, e00728-17 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laczkovich I., et al., Increased flexibility in the use of exogenous lipoic acid by Staphylococcus aureus. Mol. Microbiol. 109, 150–168 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Martin N., Lombardía E., Altabe S. G., de Mendoza D., Mansilla M. C., A lipA (yutB) mutant, encoding lipoic acid synthase, provides insight into the interplay between branched-chain and unsaturated fatty acid biosynthesis in Bacillus subtilis. J. Bacteriol. 191, 7447–7455 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin N., Christensen Q. H., Mansilla M. C., Cronan J. E., de Mendoza D., A novel two-gene requirement for the octanoyltransfer reaction of Bacillus subtilis lipoic acid biosynthesis. Mol. Microbiol. 80, 335–349 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wilkinson D. I., Karasek M. A., Skin lipids of a normal and mutant (asebic) mouse strain. J. Invest. Dermatol. 47, 449–455 (1966). [DOI] [PubMed] [Google Scholar]
- 24.Kitadokoro K., et al., Crystal structure of pathogenic Staphylococcus aureus lipase complex with the anti-obesity drug orlistat. Sci. Rep. 10, 5469 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Parsons J. B., Frank M. W., Rosch J. W., Rock C. O., Staphylococcus aureus fatty acid auxotrophs do not proliferate in mice. Antimicrob. Agents Chemother. 57, 5729–5732 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Balemans W., et al., Essentiality of FASII pathway for Staphylococcus aureus. Nature 463, E3–E3, discussion E4 (2010). [DOI] [PubMed] [Google Scholar]
- 27.Parsons J. B., Frank M. W., Subramanian C., Saenkham P., Rock C. O., Metabolic basis for the differential susceptibility of Gram-positive pathogens to fatty acid synthesis inhibitors. Proc. Natl. Acad. Sci. U.S.A. 108, 15378–15383 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Frank M. W., et al., Host fatty acid utilization by Staphylococcus aureus at the infection site. MBio 11, e00920-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Parsons J. B., et al., Identification of a two-component fatty acid kinase responsible for host fatty acid incorporation by Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 111, 10532–10537 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Cuypers M. G., et al., Acyl-chain selectivity and physiological roles of Staphylococcus aureus fatty acid-binding proteins. J. Biol. Chem. 294, 38–49 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Spalding M. D., Prigge S. T., Lipoic acid metabolism in microbial pathogens. Microbiol. Mol. Biol. Rev. 74, 200–228 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grayczyk J. P., F. Alonzo, 3rd, Staphylococcus aureus lipoic acid synthesis limits macrophage reactive oxygen and nitrogen species production to promote survival during infection. Infect. Immun. 87, e00344-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Grayczyk J. P., Harvey C. J., Laczkovich I., Alonzo F. 3rd, A Lipoylated metabolic protein released by Staphylococcus aureus suppresses macrophage activation. Cell Host Microbe 22, 678–687.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campbell J. W., Cronan J. E. Jr, Bacterial fatty acid biosynthesis: Targets for antibacterial drug discovery. Annu. Rev. Microbiol. 55, 305–332 (2001). [DOI] [PubMed] [Google Scholar]
- 35.Banevicius M. A., Kaplan N., Hafkin B., Nicolau D. P., Pharmacokinetics, pharmacodynamics and efficacy of novel FabI inhibitor AFN-1252 against MSSA and MRSA in the murine thigh infection model. J. Chemother. 25, 26–31 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Escaich S., et al., The MUT056399 inhibitor of FabI is a new antistaphylococcal compound. Antimicrob. Agents Chemother. 55, 4692–4697 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang J., et al., Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature 441, 358–361 (2006). [DOI] [PubMed] [Google Scholar]
- 38.Feng Z., Chakraborty D., Dewell S. B., Reddy B. V. B., Brady S. F., Environmental DNA-encoded antibiotics fasamycins A and B inhibit FabF in type II fatty acid biosynthesis. J. Am. Chem. Soc. 134, 2981–2987 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pishchany G., et al., Amycomicin is a potent and specific antibiotic discovered with a targeted interaction screen. Proc. Natl. Acad. Sci. U.S.A. 115, 10124–10129 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Takayama K., Wang C., Besra G. S., Pathway to synthesis and processing of mycolic acids in Mycobacterium tuberculosis. Clin. Microbiol. Rev. 18, 81–101 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Parsons J. B., Yao J., Frank M. W., Jackson P., Rock C. O., Membrane disruption by antimicrobial fatty acids releases low-molecular-weight proteins from Staphylococcus aureus. J. Bacteriol. 194, 5294–5304 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Butcher G. W., King G., Dyke K. G. H., Sensitivity of Staphylococcus aureus to unsaturated fatty acids. J. Gen. Microbiol. 94, 290–296 (1976). [DOI] [PubMed] [Google Scholar]
- 43.Subramanian C., Frank M. W., Batte J. L., Whaley S. G., Rock C. O., Oleate hydratase from Staphylococcus aureus protects against palmitoleic acid, the major antimicrobial fatty acid produced by mammalian skin. J. Biol. Chem. 294, 9285–9294 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Clarke S. R., et al., The Staphylococcus aureus surface protein IsdA mediates resistance to innate defenses of human skin. Cell Host Microbe 1, 199–212 (2007). [DOI] [PubMed] [Google Scholar]
- 45.Mortensen J. E., Shryock T. R., Kapral F. A., Modification of bactericidal fatty acids by an enzyme of Staphylococcus aureus. J. Med. Microbiol. 36, 293–298 (1992). [DOI] [PubMed] [Google Scholar]
- 46.Nguyen M. T., Hanzelmann D., Härtner T., Peschel A., Götz F., Skin-specific unsaturated fatty acids boost the Staphylococcus aureus innate immune response. Infect. Immun. 84, 205–215 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chen X., Alonzo F. 3rd, Bacterial lipolysis of immune-activating ligands promotes evasion of innate defenses. Proc. Natl. Acad. Sci. U.S.A. 116, 3764–3773 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Peng H. L., Novick R. P., Kreiswirth B., Kornblum J., Schlievert P., Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170, 4365–4372 (1988). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bæk K. T., et al., Genetic variation in the Staphylococcus aureus 8325 strain lineage revealed by whole-genome sequencing. PLoS One 8, e77122 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Hines Kelly M, et al., Lipidomic and Ultrastructural Characterization of the Cell Envelope of Staphylococcus aureus Grown in the Presence of Human Serum. mSphere 5 (3), 10.1128/mSphere.00339-20 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cosgriff C. J., White C. R., Teoh W. P., Grayczyk J. P., Alonzo F. 3rd, Control of Staphylococcus aureus quorum sensing by a membrane-embedded peptidase. Infect. Immun. 87, e00019-19 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fuller J. R., et al., Identification of a lactate-quinone oxidoreductase in Staphylococcus aureus that is essential for virulence. Front. Cell. Infect. Microbiol. 1, 19 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Monk I. R., Shah I. M., Xu M., Tan M.-W., Foster T. J., Transforming the untransformable: Application of direct transformation to manipulate genetically Staphylococcus aureus and Staphylococcus epidermidis. MBio 3, e00277-11 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Chen J., Yoong P., Ram G., Torres V. J., Novick R. P., Single-copy vectors for integration at the SaPI1 attachment site for Staphylococcus aureus. Plasmid 76, 1–7 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Green M. R., Sambrook J., Sambrook J., Molecular Cloning: A Laboratory Manual (Cold Spring Harbor Laboratory Press, ed. 4, 2012). [Google Scholar]
- 56.Khan H. A., Arif I. A., Williams J. B., Champagne A. M., Shobrak M., Skin lipids from Saudi Arabian birds. Saudi J. Biol. Sci. 21, 173–177 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All study data are included in the article and/or SI Appendix.