Significance
Amyotrophic lateral sclerosis (ALS) is a devastating neurodegenerative disease. Inhibition of RIPK1, a kinase which regulates cell death and neuroinflammation, has been efficacious in treating mouse models of ALS. RIPK1 inhibitors have reached phase 2 clinical trials in ALS patients. Here, we explore the impact of RIPK1 inhibition on microglial-mediated neuroinflammation in ALS mouse models. We find that there is a subclass of microglia which produces proinflammatory cytokines in a RIPK1-dependent manner, and that this population is reduced by RIPK1 inhibition. We believe that these inflammatory microglia are a hallmark of ALS which may be clinically relevant in advancing RIPK1 inhibitors as a potential ALS therapy.
Keywords: RIPK1, ALS, microglia, neuroinflammation, scRNAseq
Abstract
Microglial-derived inflammation has been linked to a broad range of neurodegenerative and neuropsychiatric conditions, including amyotrophic lateral sclerosis (ALS). Using single-cell RNA sequencing, a class of Disease-Associated Microglia (DAMs) have been characterized in neurodegeneration. However, the DAM phenotype alone is insufficient to explain the functional complexity of microglia, particularly with regard to regulating inflammation that is a hallmark of many neurodegenerative diseases. Here, we identify a subclass of microglia in mouse models of ALS which we term RIPK1-Regulated Inflammatory Microglia (RRIMs). RRIMs show significant up-regulation of classical proinflammatory pathways, including increased levels of Tnf and Il1b RNA and protein. We find that RRIMs are highly regulated by TNFα signaling and that the prevalence of these microglia can be suppressed by inhibiting receptor-interacting protein kinase 1 (RIPK1) activity downstream of the TNF receptor 1. These findings help to elucidate a mechanism by which RIPK1 kinase inhibition has been shown to provide therapeutic benefit in mouse models of ALS and may provide an additional biomarker for analysis in ongoing phase 2 clinical trials of RIPK1 inhibitors in ALS.
Neuroinflammation has been recognized as an important pathological mechanism underpinning many neurodegenerative and neuropsychiatric conditions, ranging from acute insults such as stroke, to neurodevelopmental alterations as in autism and schizophrenia, to chronic neurodegenerative disorders such as multiple sclerosis (MS), amyotrophic lateral sclerosis (ALS), Alzheimer’s Disease (AD), and Parkinson’s Disease (PD) (1). The presence of dysregulated microglia, a primary mediator of neuroinflammation, is a hallmark of neurodegenerative disorders (2). Microglia act as primary immune defenders of the central nervous system and play diverse roles, including engulfing and degrading extracellular aggregates and debris and promoting repair (3). When dysregulated, microglia can inappropriately prune neuronal circuits in development or generate a proinflammatory milieu to promote neurodegeneration (4).
Microglial “activation” has been broadly used to describe biochemical and morphological divergence from a homeostatic state. Emerging single-cell RNA sequencing (scRNAseq) and functional studies reveal that microglia assume many distinctive states that change over time. These states can be defined by unique RNA and protein markers, as well as by spatial localization within the brain and spinal cord (5, 6). For example, scRNAseq has identified Disease-Associated Microglia (DAMs) in mouse models and human samples of AD and ALS that do not reflect either an M1 or M2 state (7). DAMs have been shown to localize proximally to amyloid beta (Aβ) plaques in postmortem human AD brain samples, although their functional interactions with Aβ plaques and functional contributions to the pathology of AD remain unclear. Further studies have gone on to suggest that these DAMs may play a protective role in the context of disease, but also lack metabolic, engulfment, and digestive functions associated with microglia in healthy brains (8, 9). While the field has moved toward a better understanding of DAMs, the nature of the microglial activation states focused on the production of proinflammatory cytokines has yet to be deeply characterized using sophisticated sequencing approaches.
Neuroinflammation in PD, AD, and ALS has been shown to be strongly driven by TNFα signaling through TNF receptor 1 (TNFR1) and the NF-κB signaling pathway (10). However, clinical trials in MS using anti-TNFα antibodies or soluble TNF receptors to remove TNFα from the central nervous system (CNS) have exacerbated pathology as these approaches also block prosurvival signaling in neurons through TNFR2 (11). There remains significant interest in targeting neuroinflammation by selectively inhibiting inflammatory TNFR1 signaling while sparing TNFR2 (12). Receptor-interacting protein kinase 1 (RIPK1), a death-domain containing serine/threonine kinase, has an established role in selectively mediating deleterious mechanisms downstream of TNFR1 (13). RIPK1 kinase function is required for multiple forms of cell death, including necroptosis and RIPK1-dependent apoptosis, both of which have been linked to the death of CNS cells in response to inflammatory stimuli. RIPK1 kinase activity has also been shown to promote inflammatory responses under caspase-deficient conditions (14).
Mice carrying a RIPK1 D138N or K45A kinase dead knock-in mutation have been shown to develop normally but are resistant to TNFα-induced systemic inflammatory response (15, 16). We have developed a highly specific, blood brain barrier-permeable small molecule inhibitor of RIPK1, R-7-Cl-O-Necrostatin-1 (Nec-1s) (Kd = 3.1 nM) (17–19), which phenocopies the genetic inactivation seen with the D138N or K45A mutations in protecting against TNFα-mediated neuroinflammation (20–22). Further studies have gone on to assess the efficacy of Nec-1s and RIPK1 kinase inactivation in treating neuroinflammatory diseases and have demonstrated protective effects in the contexts of MS, ALS, and AD (20–22).
ALS is a devastating, terminal neurodegenerative disease affecting the motor neurons of the spinal cord and brainstem. In ALS pathogenesis, the myelinating oligodendrocytes surrounding the motor neurons die by necroptosis mediated by RIPK1 kinase activation (20). The loss of myelinating oligodendrocytes and subsequent death of motor neurons by Wallerian degeneration is an important driver of motor pathology. However, there is also clear evidence of microgliosis, microglial activation, and neuroinflammation in spinal cord tissue from postmortem ALS patients (23). In particular, both plasma and cerebrospinal fluid (CSF) from ALS patients show strong TNFα-mediated inflammation; however, it remains unclear which cells are responsible for producing and responding to TNFα and whether this is a therapeutically tractable approach to the treatment of ALS (24, 25).
There is substantial evidence supporting a proinflammatory role of RIPK1 kinase activity in the CNS of multiple animal models of ALS including SOD1G93A, Optn−/−, and TBK1+/− models (20, 26). In the CNS compartment, RIPK1 is highly expressed in microglia and astrocytes in healthy subjects, suggesting an important role for glial cells in perpetuating RIPK1-mediated disease pathology (27). Elevated production of proinflammatory cytokines in the spinal cords of Optn−/− mice can be inhibited upon blocking the kinase activity of RIPK1. Furthermore, RIPK1 drives the transcription of a large set of genes in Optn−/− microglia to promote inflammatory response. However, currently it is unclear if the activation of RIPK1 in microglia leads to a functional change in cellular state that can be defined at the molecular level.
Here, we investigate the impact of RIPK1 activation on the heterogeneity of microglia using multiple single-cell RNA sequencing approaches. We demonstrate the existence of a distinct inflammatory microglial subclass, named RIPK1-Regulated Inflammatory Microglia (RRIMs), which are enriched in and contribute to the inflammatory environment in the spinal cords of multiple mouse models of ALS. We demonstrate that these RRIMs show increased RIPK1 kinase-dependent production of proinflammatory cytokines, including Tnf and Il1b, at both the transcript and protein levels. Furthermore, we show that RRIMs are transcriptionally distinct from DAMs and may peak at different time points over the course of disease. Finally, we demonstrate that pharmacologic or genetic inhibition of RIPK1 kinase activity reduces levels of RRIMs in mouse models of ALS. Taken together, our data suggest that RRIMs may represent a microglial state that promotes RIPK1-mediated neuroinflammation in ALS.
Results
Elucidation of the Single-Cell Transcriptome in a Mouse Model of Familial ALS.
We performed scRNAseq using the Drop-Seq protocol on cells from the spinal cords of mice with mutations in SOD1, a commonly used model of familial ALS, compared with age- and sex-matched wild-type (WT) littermate controls (28, 29). Furthermore, within our group of SOD1G93A transgenic mice, we treated a subset of the mice beginning at 2.5 mo of age for 4 wk with necrostatin-1s (Nec-1s), a highly specific inhibitor of RIPK1 kinase activity, and the remaining SOD1G93A mice and WT controls with a vehicle control (Fig. 1A) (19, 20). We analyzed 4,329 single cells isolated from six adult male mice aged 3.5 mo including two WT vehicle-treated, two SOD1G93A vehicle-treated, and two SOD1G93A Nec-1s-treated using the Seurat package for unsupervised clustering (30, 31). This analysis identified clusters of 14 cell types, which were visualized by t-Distributed Stochastic Neighbor Embedding (tSNE) plotting (Fig. 1B). Cluster identities were determined using differential expression analysis of variable genes; canonical marker genes of each cluster were used to confirm cluster identity (SI Appendix, Fig. S1A) (27, 32, 33).
Fig. 1.
scRNAseq of spinal cords in SOD1G93A mouse model of ALS. (A) Schematic summarizing experimental design: whole spinal cords from 3.5-mo-old WT and SOD1G93A littermate male mice (n = 2 per group) treated with either a vehicle control or Nec-1s for 4 wk in drinking water were prepared using Drop-Seq and sequenced. (B) tSNE plot of cells isolated from all groups and identified based on differentially expressed genes generated using Seurat. n = 6 animals. (C) Proportion of each cell population across all samples. n = 6 animals. (D) tSNE plots of cells isolated plotted by group. n = 2 animals per group.
Consistent with findings across species that glia comprise the majority of cells in spinal cords, our data showed distinguishable populations of astrocytes (27%) and microglia (15%) as a proportion of total cells (Fig. 1C) (34). All cell populations were identified in all groups with the exception of skeletal muscle seen in the SOD1G93A vehicle-treated group, which may be the result of dissection due to the atrophied nature of the spinal cord in these animals (Fig. 1D and SI Appendix, Fig. S1B).
The degeneration of mature, myelinating oligodendrocytes in SOD1G93A mice and two models of MS (induced by experimental autoimmune encephalomyelitis and cuprizone demyelination) is driven in part by RIPK-dependent necroptosis (20, 22). To validate the integrity of our scRNAseq data, we examined whether we could observe these changes in captured cell populations. Consistent with published data, we found that SOD1G93A vehicle-treated mice had a significant reduction in myelinating oligodendrocytes as a proportion of total oligodendrocytes relative to WT littermates, which was protected by RIPK1 inhibition with Nec-1s (SI Appendix, Fig. S1C).
Differential expression analysis at the whole spinal cord level between groups yielded a surprisingly low number of differentially expressed genes relative to prior datasets comparing cell types with and without RIPK1 inhibition, which may be in part due to significant dropout effects seen with scRNAseq methodologies (SI Appendix, Fig. S2A) (20, 21). However, at both the gene and pathway level, as assessed during Gene Set Enrichment Analysis (GSEA) Gene Ontology (GO) pathways, there was evidence of differences in glial populations in SOD1G93A animals treated with Nec-1s (SI Appendix, Fig. S2B). Given prior work from our laboratory showing a role for RIPK1 kinase activity in mediating microglial inflammation in ALS, we focused subsequent analyses on understanding whether RIPK1 inhibition may impact microglial activation (20).
Identification of Altered Microglial States in SOD1G93A ALS.
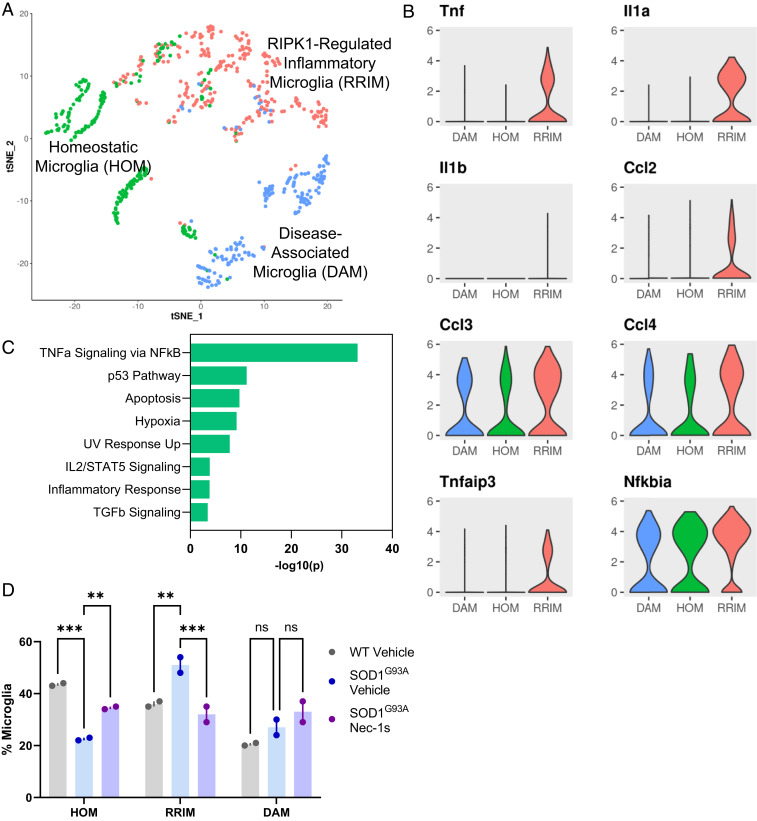
The cluster of microglia defined by high levels of Tmem119, Cx3cr1, Aif1, P2ry12, Csf1r, Trem2, and C1qa containing 654 cells (Fig. 1A) was extracted and reclustered in Seurat using semisupervised clustering; the resultant analysis showed three subclusters of microglia (Fig. 2A) (30, 31). One cluster was identified as DAMs based on significantly high expression of Apoe, Cd9, Trem2, Cd63, Cd52, and Ctsb as previously published (SI Appendix, Fig. S3A) (7). Another microglial cluster did not show marker genes associated with pathways beyond essential transcription, translational, and metabolic processes, which appeared to be Homeostatic Microglia (HOM).
Fig. 2.
RIPK1-regulated inflammatory microglia are up-regulated in SOD1G93A mice. (A) tSNE plot of microglia from all groups reclustered in Seurat using semisupervised clustering to identify three subpopulations, including previously identified DAM and HOM. n = 6 animals. (B) Violin plots of marker genes for the RRIM subcluster from all groups generated using Seurat. Values plotted are transcripts per million (TPM). n = 6 animals. (C) GO analysis of pathways enriched in marker genes (padj < 0.01, logFC >1.5) for RRIM subcluster analyzed using GSEA. (D) Proportion of RRIM cluster as a percentage of total microglia was calculated for each animal and quantified using two-way ANOVA with multiple comparison testing relative to SOD1G93A Vehicle group. n = 2 animals per group. **P < 0.01; ***P < 0.001; ns, not significant. Data are represented as mean ± SEM.
The third unidentified cluster of microglia showed differentially expressed marker genes associated with inflammatory and NF-κB-regulated pathways, including Tnf, Il1a, Il1b, Ccl2, Ccl3, Ccl4, Tnfaip3, and Nfkbia (Fig. 2B). Tnf, Il1a, and Il1b encode proinflammatory cytokines typically produced by immune cell populations during inflammatory responses in both autoimmune and neurodegenerative diseases. Increased levels of TNFα and IL1β have been reported in CSF and plasma from patients with SOD1 and C9ORF72 ALS mutations or sporadic ALS and may correlate negatively with patient survival (25). C-C motif chemokine ligand 2 (Ccl2, also known as monocyte chemoattractant protein 1 or Mcp2), C-C motif chemokine ligand 3 (Ccl3, also known as macrophage inflammatory protein 1-alpha or Mip1α), and C-C motif chemokine ligand 4 (Ccl4, also known as macrophage inflammatory protein 1-beta or Mip1β) are up-regulated by multiple types of antigen-presenting cells (APCs) in the context of inflammatory and autoimmune diseases to recruit and activate CD8+ T cells, and are also up-regulated in DAMs (5, 35). Tumor necrosis factor alpha-induced protein 3 (Tnfaip3, which encodes the protein A20) is induced by activation of the TNFα-induced NF-κB pathway and acts as a critical regulator of RIPK1 activation, cell death, and inflammation via its deubiquitinase activity (36, 37). Protein IκBα, encoded by Nfkbia, is transcribed and activated in response to multiple inflammatory stimuli and, depending on other cellular signals and posttranslational modifications, can either promote or inhibit NF-κB signaling (38, 39). Using GSEA to assess GO pathway enrichment of marker genes for this unidentified subcluster, TNFα signaling via the NF-κB pathway was highly enriched, along with canonical RIPK1-associated pathways such as cell death and inflammatory responses (Fig. 2C). Based on the expression of proinflammatory and NF-κB-regulated genes associated with RIPK1 kinase activity, we hypothesized that these cells may represent an inflammatory state and thus termed this subcluster RIPK1-Regulated Inflammatory Microglia (RRIMs) (Fig. 2A).
Assessing Proinflammatory Microglia in SOD1G93A ALS.
Given that RRIMs show the greatest up-regulation of TNFα signaling through the NF-κB pathway, we hypothesized that microglial polarization to the RRIM state would likely be impacted by RIPK1 inhibition, which has been shown to disrupt many signaling pathways through NF-κB (40). As hypothesized, relative to WT mice, SOD1G93A mice treated with vehicle had a significant increase in RRIMs, where this prevalence was reduced in SOD1G93A mice treated with Nec-1s (Fig. 2D and SI Appendix, Fig. S3B). Interestingly, while RIPK1 inhibition altered the levels of RRIMs in SOD1G93A mice, it did not seem to alter DAM prevalence, which was not significantly up-regulated in SOD1G93A mice relative to WT controls. We believe that this reflects the earlier time point of animals used in our experiments (3.5 mo old) relative to prior publications, which show a disease-progression-dependent increase in DAMs (7). While we have previously identified a role for RIPK1 regulating the expression of Cst7 in the endosomal/lysosomal degradation pathway within the DAM phenotype, we believe that RIPK1 is likely involved in perpetuating dysfunctional degradation within DAMs rather than inducing the initial formation of DAMs, which has increasingly been linked to the Apoe-Trem2 axis (8, 9, 21).
To validate the presence of RRIMs in an additional scRNAseq dataset with samples prepared using a different protocol, we analyzed the CD45High-sorted population from spinal cords of SODG93A mice at various ages published by Keren-Shaul et al. (7). Using both unsupervised and semisupervised clustering in Seurat, we were able to identify both RRIMs and DAMs from spinal cord microglia in the Keren-Shaul et al. (7) dataset, confirming that RRIMs are a distinct subcluster from DAMs (SI Appendix, Fig. S4A). The Keren-Shaul et al. dataset also allowed us to interrogate the dynamics of RRIMs and DAMs at two time points in disease progression: day 80 and day 135. The SODG93A mouse model of ALS starts to display motor dysfunction including hindlimb weakness beginning around day 90 and die or require euthanasia due to paralysis around day 130 (29, 41). Accordingly, the two time points represented in the dataset show a presymptomatic or early stage of the disease compared to a terminal stage. As was previously demonstrated, DAMs increase over the course of disease in the SODG93A model (as well as Alzheimer’s Disease) (7). We found that RRIMs were more up-regulated at the early stage of pathogenesis, representing 38% of total microglia in SOD1G93A animals at day 80, relative to terminal stage, where RRIMs represent only 18% of total microglia (SI Appendix, Fig. S4B). This led us to the hypothesis that perhaps DAMs could evolve from RRIMs as perhaps the published deficiencies in DAM metabolic fitness may mirror immune cell exhaustion as seen in the adaptive immune system (9, 21). Furthermore, the presence of plaques adjacent to these DAMs suggests that the induction of DAMs may be the result of a local need to remove misfolded proteins seen at later stages of disease (7). We used Monocle to computationally assess whether RRIMs may represent an intermediate “activated” state along the path to the “exhausted” DAM state (SI Appendix, Fig. S4C) (42–44). To our surprise, Monocle identified that both RRIMs and DAMs evolved from common homeostatic microglia. These results suggest that microglial polarization to DAMs or RRIMs originates from HOMs in response to varying stimuli at different stages of disease.
RRIMs Are Present in an Optn−/− Model of ALS.
To explore the significance of RRIMs in ALS more broadly, we used the optineurin knockout (Optn−/−) mouse model of ALS to explore microglial subclusters in disease using the inDrop scRNAseq protocol (20, 45, 46). Dysfunction of Optn, a ubiquitin-binding protein, has been shown in both familial and sporadic ALS, where it can mediate microglial- and RIPK1-mediated inflammation and pathology (20, 47). We enriched for CD45High cells from the spinal cords of Optn−/− mice treated with Vehicle or Nec-1s and OptnFl/Fl control animals treated with Vehicle for further analysis (Fig. 3A). We confirmed that this population was comprised predominantly of microglia, including 625 cells showing high levels of Tmem119 and Cx3cr1 (SI Appendix, Fig. S5A). Semisupervised clustering of microglia revealed three subclusters, including HOMs, DAMs, and RRIMs (Fig. 3B).
Fig. 3.
RRIMs are increased in the Optn−/− mouse model of ALS. (A) inDrop isolated spinal cords enriched for CD45+ cells of 3.5-mo-old OptnFl/Fl and Optn−/− male mice (n = 2 per group) treated with either a vehicle control or Nec-1s. tSNE plot of cells isolated from all groups and identified based on differentially expressed genes generated using Seurat. n = 6 animals. (B) tSNE plot of microglia from all Optn groups reclustered in Seurat using semisupervised clustering to identify four subpopulations, including HOM, DAM, and RRIM. n = 6 animals. (C) Violin plots of marker genes for RRIM subcluster from all Optn groups generated using Seurat. Values plotted are TPM. n = 6 animals. (D) GO analysis of pathways enriched in marker genes (padj < 0.01, logFC >1.5) for RRIM subcluster analyzed using GSEA. (E) Proportion of RRIM cluster as a percentage of total microglia were calculated for each animal and quantified using two-way ANOVA with multiple comparison testing relative to Optn−/− Vehicle group. n = 2 animals per group. *P < 0.05; ns, not significant. Data are represented as mean ± SEM. (F) SmartSeq2 prepared scRNAseq of FACS microglia from 3.5-mo-old Optn−/− untreated male mice (n = 2). tSNE plot of all cells generated using Seurat. (G) Volcano plot of transcriptional alterations between RRIM and HOM subclusters in SmartSeq2 Optn dataset. Significant genes indicated in red as logFC > 1.5 and −Log10P ≥ 10.
Consistent with our findings in SOD1G93A animals, RRIMs in the Optn dataset are defined by inflammatory cytokines and chemokines, markers of NF-κB pathway activation, as well as up-regulation of TNFα-driven inflammatory and cell-death-associated signaling pathways (Fig. 3 C and D). Similarly, Optn−/− mice also show a higher number of RRIMs compared to OptnFl/Fl controls, which is reduced by treatment with Nec-1s (Fig. 3E and SI Appendix, Fig. S5B). Optn−/− mice show spinal cord demyelination and axonal degeneration with weakness in the hind legs at 3.5 mo of age, but do not develop the severe phenotype including accumulation of misfolded SOD1 protein, loss of motor neurons, and paralysis seen in the SOD1G93A model (20). As we do not see evidence for increased levels of DAMs in Optn−/− mice irrespective of treatment, this may be due to a lack of protein aggregates in this model, which is different from what has been demonstrated in SOD1G93A mice and the APP/PS1 Alzheimer’s Disease mouse model where the induction of DAMs corresponds strongly to the presence and localization of misfolded proteins (7, 48).
While cytokines and chemokines, in particular Ib, show differential expression in ALS mouse models, these transcripts were particularly prone to dropout effects in our DropSeq and inDrop datasets. To more robustly capture transcriptional changes in cytokines and chemokines in RRIMs, we used a modified Smart-seq2 protocol to analyze microglia sorted from untreated Optn−/− mice using fluorescence-activated cell sorting (FACS) (SI Appendix, Fig. S6A) (49). Overall, there was much greater depth of gene expression in the Smart-seq2 dataset capturing on average 1,800 genes per cell, which corresponded well to expression patterns from the prior Optn−/− dataset (SI Appendix, Fig. S6B). Using semisupervised clustering, we found two subclusters corresponding to HOM and RRIMs, confirming the relative low levels of DAMs in Optn−/− relative to the SOD1G93A model (Fig. 3F). The Optn−/− Smart-seq2 dataset confirmed significant up-regulation of Tnf and Il1b in RRIMs relative to HOM and also captured significant increases in other inflammation-associated chemokines such as Ccl6 and Cxcl2 (Fig. 3G and SI Appendix, Fig. S6C).
In Situ Validation of RRIMs.
Several publications have identified microglial activation signatures induced by tissue digestion as is required for scRNAseq or FACS protocols (5, 50). Thus, it is critical to validate microglial immune activation transcriptional signatures seen in RRIMs using in situ methodologies that do not require tissue digestion. Using RNAscope in situ hybridization, we analyzed levels of individual Tnf and Apoe puncta in C1q+ microglia from the spinal cords of WT and SOD1G93A mice treated with Vehicle or Nec-1s as was previously described (Fig. 4A and SI Appendix, Fig. S7). Consistent with our sequencing data, C1q+ microglia in SOD1G93A mice treated with vehicle display higher levels of ApoeHigh TnfLow and ApoeLow TnfHigh cells relative to WT littermates, which may correspond to DAMs and RRIMs, respectively (Fig. 4C). Furthermore, SOD1G93A mice treated with Nec-1s showed decreased C1q+ ApoeLow TnfHigh RRIMs relative to those treated with vehicle, but no significant impact on C1q+ ApoeHigh TnfLow DAMs. To confirm the distinction between these two populations of cells, we cohybridized probes for Tnf and Ccl4, the former which should be restricted to RRIMs and the later which is increased in both RRIMs and DAMs. In SOD1G93A untreated mice, C1q+ Ccl4HighTnfHigh cells, representing 34% of analyzed cells, were distinct from C1q+ Ccl4HighTnfLow cells, which represented ∼14% of analyzed cells, but were not statistically different from WT littermates (SI Appendix, Fig. S8A). Conversely, cohybridization of probes for Tnf and Il1b, which should both be primarily in RRIMs and not DAMs, showed 39% C1q+ Il1bHigh TnfHigh cells and only 7% C1q+ cells that were either Il1bHigh TnfLow or Il1bLow TnfHigh in SOD1G93A mice (SI Appendix, Fig. S8B).
Fig. 4.
Validation of RRIMs in ALS mouse models using in situ hybridization. (A) Fresh-frozen spinal cords from 3.5-mo-old WT and SOD1G93A littermates treated with either vehicle or Nec-1s for 4 wk were sectioned and prepared using RNAscope with the indicated probes, followed by confocal imaging. Representative images from n = 3 mixed gender animals per group; n = 5 to 6 images from ventral horn (1 image per section). (Scale bar, 5 μm.) (B) Fresh-frozen spinal cords from 3.5-mo-old OptnFl/Fl, Optn−/−, and Optn−/−;Ripk1D138N animals were sectioned and prepared using RNAscope with the indicated probes, followed by confocal imaging. Representative images from n = 3 mixed gender animals per group; n = 5 to 6 images from ventral horn (1 image per section). (Scale bar, 5 μm.) (C) Quantification of (A) average of 14 microglia analyzed per image using CellProfiler. n = 3 mixed gender animals per group; n = 5 to 6 images from ventral horn (1 image per section). Two-way ANOVA with multiple comparison testing relative to SOD1G93A Vehicle group. *P < 0.05; **P < 0.01; ***P < 0.001; ns, not significant. Data are represented as mean ± SEM. (D) Quantification of (B) average of 17 microglia analyzed per image using CellProfiler. n = 3 mixed gender animals per group; n = 5 to 6 images from ventral horn (1 image per section). Two-way ANOVA with multiple comparison testing relative to Optn−/− Vehicle group. ****P < 0.0001; ns, not significant. Data are represented as mean ± SEM.
As genetic validation that induction of RRIMs relies specifically on RIPK1 kinase activation, we bred double-mutant mice harboring both Optn−/− and a RIPK1 kinase dead D138N mutation (15). Similar to the findings above with SOD1G93A mice, Optn−/− mice also showed increased C1q+ ApoeLow TnfHigh microglia corresponding to RRIMs, but minimal evidence of DAM-like cells, relative to OptnFl/Fl controls (Fig. 4 B and D). Similar to the sequencing data from Optn−/− mice treated with Nec-1s, double mutant Optn−/−;Ripk1D138N mice also showed a reduction in C1q+ ApoeLow TnfHigh microglia.
While changes in RIPK1 activation may impact cellular function at the transcriptional level, the gold standard biomarker for assessing RIPK1 activation is phosphorylation of S166 (51). To determine whether ALS mouse models showed RIPK1 activation in TNFα-producing inflammatory microglia, we coimmunostained pRIPK1 S166, TNFα, and IBA1, a microglial marker. Similar to the findings from in situ hybridization, immunofluorescent staining and quantification showed a significant increase in pRIPK1+TNFα+IBA1+ microglia in SOD1G93A mice treated with vehicle relative to WT littermates (Fig. 5A). We also wanted to examine whether at the protein level it was possible to distinguish RIPK1-activated microglia from DAMs and found that APOE+IBA1+ microglia were up-regulated in SOD1G93A mice irrespective of treatment, but that these microglia were distinct from pRIPK1+IBA1+ cells (SI Appendix, Fig. S9A). Consistent with initial findings from our SOD1G93A scRNAseq data and prior publications, pRIPK1 signal can be seen outside of IBA1+ microglia in SOD1G93A mice, in particular in Olig2+ myelinating oligodendrocytes (SI Appendix, Fig. S9B). In Optn−/− mice, we also found significant up-regulation of pRIPK1+TNFα+IBA1+ microglia corresponding to RRIMs relative to OptnFl/Fl controls, which were decreased in Optn−/−;Ripk1D138N double-mutant mice (Fig. 5B). Taken together, these experiments support the presence of RRIMs, a subclass of microglia with RIPK1-dependent activation of proinflammatory signatures in genetic models of ALS.
Fig. 5.
Validation of RRIMs in ALS mouse models using immunofluorescence. (A) Spinal cords from 4-mo-old WT and SOD1G93A littermates treated with either vehicle or Nec-1s for 4 wk were sectioned and immunostained for TNF, pS166 RIPK1, and IBA1. White arrow indicates representative TNF+pRIPK1+IBA1+ RRIM. n = 3 mixed gender animals per group; n = 5 to 6 images from ventral horn. One-way ANOVA with multiple comparison testing relative to SOD1G93A Vehicle group. **P < 0.01; ***P < 0.001. Data are represented as mean ± SEM. (Scale bar, 20 μm.) (B) Spinal cords from 3.5-mo-old OptnFl/Fl, Optn−/−, and Optn−/−;Ripk1D138N mice were sectioned and immunostained for TNF, pS166 RIPK1, and IBA1. White arrow indicates representative TNF+pRIPK1+IBA1+ RRIM. n = 3 mixed gender animals per group; n = 5 to 6 images from ventral horn. One-way ANOVA with multiple comparison testing relative to Optn−/− Vehicle group. ***P < 0.001. Data are represented as mean ± SEM. (Scale bar, 10 μm.)
Discussion
In this paper we use two mouse models of ALS to identify a subset of microglia, RRIMs. We demonstrate that these microglia express increased levels of proinflammatory cytokines, such as Tnf and Il1β, transcriptionally, in situ, and at the protein level. We confirm that RRIMs are up-regulated in the SOD1G93A and Optn−/− mouse models of ALS relative to WT and OptnFl/Fl controls, respectively. Of potential therapeutic relevance, we show that the prevalence of these microglia is governed by RIPK1 kinase activation, as inhibition of RIPK1 with Nec-1s or Ripk1D138N kinase dead mutation can restore RRIMs to wild-type levels. RRIMs are transcriptionally distinct from the previously identified DAMs and may occur at earlier time points in ALS pathogenesis relative to DAMs.
Collectively, based on our findings, we propose a model where early in disease pathogenesis TNFα signaling polarizes microglia to a RRIM state, increasing the proinflammatory milieu damaging the surrounding tissue. This increased production of inflammatory cytokines by RRIMs may contribute to the induction of “A1-like” reactive astrocytes observed in SOD1G93A mice, the death of myelinating oligodendrocytes, and subsequent demyelination and death of motor neurons in a RIPK1-dependent manner (20, 22, 52). While DAMs have been primarily associated with aggregates, RRIM levels are increased in the Optn−/− model, more so than in the SOD1G93A model. Optn−/− mice do not express a misfolded protein such as the misfolded SOD1 seen in SOD1G93A transgenic mice. Thus, induction of RRIMs may represent a distinct path in early stages of ALS which is independent of protein aggregation (7).
It remains unclear what the functional roles of RRIMs may be: Specifically, why do both transcriptomic and in situ analyses suggest that RRIM-like cells may exist in the homeostatic spinal cord at detectable levels? Microglia have been shown to assume M1-like, activated phenotypes in response to neurotrophic viruses, playing the same antiinfective, protective role of activated macrophages against pathogens in the periphery (53). However, it has been shown that activated microglia functionally remain poor antigen-presenting cells and are typically not the primary inducers of immune responses in conditions such as MS (54). As both RRIMs and "A1-like" reactive astrocytes appear to be regulated by TNFα-signaling pathways, we believe that RIPK1 inhibitors may offer a broad approach to reducing glial-mediated inflammation in chronic neurodegenerative diseases, such as ALS. Due to the significant reduction in inflammatory microglia seen downstream of Nec-1s treatment or Ripk1D138N kinase dead mutation in mouse models of ALS, we believe this helps to define RRIMs as a potential hallmark of RIPK1 activation for ongoing phase 2 clinical trials of RIPK1 inhibitors in ALS (51). While no direct markers of RIPK1 activation in the CNS, such as a PET tracer, are currently available, due to the inflammatory nature of RRIMs, it is possible that a pan-inflammatory biomarker like translocator protein (TSPO) PET imaging may capture changes in RRIM biology.
Materials and Methods
Animals.
OptnFl/Fl and Optn−/− mice were generated as described in ref. 20. Optn−/−;RIPK1D138N/D138N mice were generated by crossing Optn−/− mice with RIPK1D138N/D138N mice provided by Manolis Pasparakis of the University of Cologne, Germany, as described in ref. 15. SOD1G93A mice [B6SJL-Tg(SOD1G93A)1Gur/J; stock no: 002726] and C57BL/6 (B6) mice were obtained from Jackson Laboratory. All animals were maintained in a pathogen-free environment, and experiments on mice were conducted according to the protocols approved by the Harvard Medical School Animal Care Committee.
Nec-1s Administration.
The method of Nec-1s [R-5-((7-Chloro-1H-indol-3-yl)methyl)-3-methyl-2,4-imidazolidinedione] formulation and delivery was previously described in refs. 21 and 22. Custom synthesized Nec-1s was first dissolved in dimethylsulfoxide (50% wt/vol) and then transferred into 35% polyethylene glycol solution which was suspended in water containing 2% sucrose. Mice were provided with vehicle control or Nec-1s as drinking water ad libitum. Each mouse drank vehicle or Nec-1s containing water of about 5 to 10 mL/day (Nec-1s = 2.5 to 5 mg/day).
Tissue Preparation for Drop-Seq.
Mice were euthanized and perfused with phosphate-buffered saline (PBS). Spinal cords were removed and chopped with a razor blade in Accutase (Sigma). After 20 min of digestion, the spinal cords were homogenized with a 21-gauge needle and filtered through a 40-μM cell strainer. The pellet was resuspended in a gradient of OptiPrep and Hibernate A optimized for CNS isolation as described in ref. 55.
Tissue Preparation for inDrop.
Mice were euthanized and perfused with PBS. Spinal cords were removed and chopped with a razor blade followed by homogenization with a 19-gauge needle in Accutase (Sigma). After 10 min of digestion, the spinal cords were homogenized twice with a 21-gauge needle. The sample was centrifuged at room temperature for 5 min at 200 x g. The pellet was resuspended in PBS + 0.5% bovine serum albumin (BSA) (IgG and Protease Free from Jackson ImmunoResearch) and filtered through a 40-μM cell strainer. A gradient of 25% BSA was added on top, and the suspension was centrifuged for 10 min at 1,200 x g at room temperature. A top layer of myelin and supernatant was removed. The pellet was resuspended in an OptiPrep gradient for live/dead cell isolation as described in the OptiPrep Application Sheet C13. The sample was centrifuged for 20 min with the break off at 800 x g in room temperature. The top interface containing viable cells was transferred into a new tube, centrifuged for 5 min at 200 x g at room temperature. The pellet was resuspended in PBS + 0.5% BSA. Each sample was encapsulated and barcoded, and subsequently the complementary DNA (cDNA) library was prepared according to the protocols outlined in refs. 45 and 46.
Microglial Isolation for Smart-seq2.
Tissue was isolated and prepared for flow cytometry as described (21). The resulting pellet was blocked with Rat Anti-Mouse Fc (BD Biosciences) and then stained with CD11b, CD45, and CX3CR1 antibodies. Individual microglia (CD11bHigh CD45Intermediate CX3CR1High) were sorted into 96-well plates using a FACSAria III (BD Biosciences) and FACSDiva Software (BD Biosciences) and frozen at −80 °C. Smart-seq2 libraries were prepared with an optimized version of the protocol that was shared with us by Monika Kowalczyk and Aviv Regev of the Broad Institute (49).
RNA Sequencing.
All cDNA libraries were assessed for quality using a Bioanalyzer 2100 (Agilent). All samples were sequenced on a NextSEq. 550 (Illumina).
Computational Analysis.
Computational analyses were conducted using R (Version 3.6.1), Seurat (Version 2.3.4) (28, 30, 31), and Monocle 2 (Version 2.6.3) (42–44). GO analyses were conducted using the GO gene sets from the Broad Institute GSEA (56). Following publication of these findings, raw and processed datasets will be available on the National Center for Biotechnology Information Gene Expression Omnibus (GEO). The DropSeq raw sequencing dataset was processed using the data alignment tools described by the McCarroll Lab (28). The inDrop raw sequencing dataset was processed using the inDrop.py bioinformatics pipeline with support from the Harvard Chan Bioinformatics Core. The SmartSeq2 raw sequencing dataset was processed as previously described with support from Joshua Levin’s group at the Broad Institute (49). The DropSeq dataset was aligned to mm9 and inDrop, and SmartSeq2 datasets were aligned to mm10. Cells were filtered based on expressing between 200 and 2,000 genes. Single-cell data were normalized by the library size of each cell, and each gene was centered and normalized before undergoing principal component analysis and clustering. Unsupervised clustering was conducted including principal components with pval < 1e-03. Semisupervised clustering was conducted using known RIPK1 and DAM associated genes to define principal components followed by clustering. These genes were selected from published papers, specifically: Tnf, Apoe, Il1a, Il1b, Cst7, Spp1, Il10ra, Tnfaip3, Tnip1, Tnfaip2, Nfkbia, Ctsb, P2ry12, Aldoc, and Ccl2 (7, 57) Published data from Keren-Shaul et al. was obtained from GEO under accession no. GSE98969 (7). To conduct the Monocle Pseudotime analysis, the starting state was defined as the RRIM state based on Seurat-generated marker genes, and the end state was not specified.
Fluorescence In Situ Hybridization.
Animals were euthanized and perfused with PBS. Tissues were fresh frozen in Tissue-Tek OCT and rapidly cooled by submersion in 2-methylbutane (ThermoFisher). The lumbar regions of the spinal cord were dissected, and 16 μm spinal cord cross-sections were prepared on a Cryostat (Leica). Samples were prepared for RNAscope Fluorescent Multiplex Assay according to the manufacturer’s instructions with an overnight hybridization of catalog probes. Probes used were Mm-C1qa-C3 (ACD Bio, #441221-C3), Mm-Tnf-O1-C2 (ACD Bio, #844961-C2), Mm-Apoe (ACD Bio, #313271), Mm-Il1b (ACD Bio, #316891), and Mm-Ccl4 (ACD Bio, #421071). Five to six images per sample were randomly selected from the ventral horn of spinal cords. All images were collected with a Nikon Ti-E confocal microscope equipped with A1R scan head with spectral detector and resonant scanners; images were acquired with Nikon Elements Software (v4.11). For each image point, z-series optical sections were collected with a step size of 1 μm and then Z-stack projected with average intensity. Gamma, brightness, and contrast were adjusted on displayed images (identically for compared image sets) using FIJI software (ImageJ v1.53c). All image stacks except DAPI were converted into binary masks before analysis in CellProfiler. Quantification of fluorescence in situ hybridization data was conducted using a custom analysis pipeline built using the CellProfiler software developed by the Broad Institute as detailed in SI Appendix, Fig. S7 (58, 59).
Immunofluorescence.
Animals were euthanized and perfused with PBS followed by 100 mL 4% paraformaldehyde. Spinal cords (L2-L3) were dissected and immersed in 4% PFA for 30 min. Brain sections (30 μm) were prepared on a cryostat. For immunostaining, tissue sections were mounted and blocked with 10% normal goat serum and 1% BSA and then incubated with primary antibodies at 4 °C overnight. Four to six images per sample were randomly selected from ventral horn of spinal cords. All images were collected with a Nikon Ti-E confocal microscope equipped with A1R scan head with spectral detector and resonant scanners; images were acquired with Nikon Elements Software (v4.11). For each image point, z-series optical sections were collected with a step size of 1 μm and then Z-stack projected with maximum intensity. Gamma, brightness, and contrast were adjusted on displayed images (identically for compared image sets) using FIJI software.
Antibodies.
CD11b (BioLegend, #101235), CX3CR1 (BioLegend, #149007), CD45 (BioLegend, #103116 and #103125), CD16/CD32 Fc (BD Biosciences, #553142), pS166 RIPK1 (Biolynx, #BW60008), TNF (Thermo, #14–732-81), Apoe (Santa Cruz, #sc-390925), Olig2 (Santa Cruz, #sc-515947), and IBA1 (Abcam #ab48004). Alexa Fluor secondary antibodies against the various species were purchased from Life Technology and used according to the manufacturer’s instructions.
Supplementary Material
Acknowledgments
This work was supported in part by National Institute on Aging RF1-AG055521 (to J.Y.). We thank Dr. Manolis Pasparakis (University of Cologne) for the Ripk1D138N mice; the Wellcome Trust Sanger Institute for the Optntm1a(EUCOMM)Wtsi, which led to the generation of the Optn−/− mice; Dr. Jennifer Walters and staff at the Harvard Medical School Nikon microscope facility for assistance with fluorescence microscopy; Kris Holton (Harvard Medical School Research Computing group); the Harvard Chan Bioinformatics Core for assistance with computational efforts; the laboratory of Dr. Steve McCarroll for the development of, and assistance with, the Drop-Seq protocol; the Single Cell Core at Harvard Medical School and the laboratory of Dr. Allon Klein for the development of, and assistance with, the inDrop protocol; Dr. Aviv Regev and Dr. Monika Kowalczyk (Broad Institute) for sharing their modified Smart-seq2 protocol; and Dr. Samuel Marsh (Boston Children’s Hospital), Dr. Dimitry Ofengeim (Sanofi), and Dr. Thomas Sandmann (Denali Therapeutics) for helpful discussions. Graphics were created with BioRender.
Footnotes
Competing interest statement: J.Y. is a consultant for Denali Therapeutics and Sanofi. J.Y. and J.M.H. are co-authors on a 2018 Nomenclature review article.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.2025102118/-/DCSupplemental.
Data Availability
The scRNAseq data have been deposited in GEO under accession no. GSE167332.
References
- 1.Ransohoff R. M., How neuroinflammation contributes to neurodegeneration. Science 353, 777–783 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Salter M. W., Stevens B., Microglia emerge as central players in brain disease. Nat. Med. 23, 1018–1027 (2017). [DOI] [PubMed] [Google Scholar]
- 3.Tremblay M. E., et al., The role of microglia in the healthy brain. J. Neurosci. 31, 16064–16069 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wu Y., Dissing-Olesen L., MacVicar B. A., Stevens B., Microglia: Dynamic mediators of synapse development and plasticity. Trends Immunol. 36, 605–613 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hammond T. R., et al., Single-cell RNA sequencing of microglia throughout the mouse lifespan and in the injured brain reveals complex cell-state changes. Immunity 50, 253–271.e6 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li Q., et al., Developmental heterogeneity of microglia and brain myeloid cells revealed by deep single-cell RNA sequencing. Neuron 101, 207–223.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Keren-Shaul H., et al., A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169, 1276–1290.e17 (2017). [DOI] [PubMed] [Google Scholar]
- 8.Krasemann S., et al., The TREM2-APOE pathway drives the transcriptional phenotype of dysfunctional microglia in neurodegenerative diseases. Immunity 47, 566–581.e9 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ulland T. K., et al., TREM2 maintains microglial metabolic fitness in Alzheimer’s disease. Cell 170, 649–663.e13 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan J., Amin P., Ofengeim D., Necroptosis and RIPK1-mediated neuroinflammation in CNS diseases. Nat. Rev. Neurosci. 20, 19–33 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.The Lenercept Multiple Sclerosis Study Group and The University of British Columbia MS/MRI Analysis Group , TNF neutralization in MS: Results of a randomized, placebo-controlled multicenter study. Neurology 53, 457–465 (1999). [PubMed] [Google Scholar]
- 12.Tweedie D., Sambamurti K., Greig N. H., TNF-alpha inhibition as a treatment strategy for neurodegenerative disorders: New drug candidates and targets. Curr. Alzheimer Res. 4, 378–385 (2007). [DOI] [PubMed] [Google Scholar]
- 13.Ofengeim D., Yuan J., Regulation of RIP1 kinase signalling at the crossroads of inflammation and cell death. Nat. Rev. Mol. Cell Biol. 14, 727–736 (2013). [DOI] [PubMed] [Google Scholar]
- 14.Christofferson D. E., et al., A novel role for RIP1 kinase in mediating TNFα production. Cell Death Dis. 3, e320 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Polykratis A., et al., Cutting edge: RIPK1 kinase inactive mice are viable and protected from TNF-induced necroptosis in vivo. J. Immunol. 193, 1539–1543 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shutinoski B., et al., K45A mutation of RIPK1 results in poor necroptosis and cytokine signaling in macrophages, which impacts inflammatory responses in vivo. Cell Death Differ. 23, 1628–1637 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Degterev A., et al., Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat. Chem. Biol. 4, 313–321 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Degterev A., et al., Chemical inhibitor of nonapoptotic cell death with therapeutic potential for ischemic brain injury. Nat. Chem. Biol. 1, 112–119 (2005). [DOI] [PubMed] [Google Scholar]
- 19.Degterev A., Maki J. L., Yuan J., Activity and specificity of necrostatin-1, small-molecule inhibitor of RIP1 kinase. Cell Death Differ. 20, 366 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ito Y., et al., RIPK1 mediates axonal degeneration by promoting inflammation and necroptosis in ALS. Science 353, 603–608 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ofengeim D., et al., RIPK1 mediates a disease-associated microglial response in Alzheimer’s disease. Proc. Natl. Acad. Sci. U.S.A. 114, E8788–E8797 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ofengeim D., et al., Activation of necroptosis in multiple sclerosis. Cell Rep. 10, 1836–1849 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henkel J. S., et al., Presence of dendritic cells, MCP-1, and activated microglia/macrophages in amyotrophic lateral sclerosis spinal cord tissue. Ann. Neurol. 55, 221–235 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Cereda C., et al., TNF and sTNFR1/2 plasma levels in ALS patients. J. Neuroimmunol. 194, 123–131 (2008). [DOI] [PubMed] [Google Scholar]
- 25.Olesen M. N., et al., Inflammatory profiles relate to survival in subtypes of amyotrophic lateral sclerosis. Neurol. Neuroimmunol. Neuroinflamm. 7, e697 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu D., et al., TBK1 suppresses RIPK1-driven apoptosis and inflammation during development and in aging. Cell 174, 1477–1491.e19 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang Y., et al., An RNA-sequencing transcriptome and splicing database of glia, neurons, and vascular cells of the cerebral cortex. J. Neurosci. 34, 11929–11947 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Macosko E. Z., et al., Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gurney M. E., et al., Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–1775 (1994). [DOI] [PubMed] [Google Scholar]
- 30.Butler A., Hoffman P., Smibert P., Papalexi E., Satija R., Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat. Biotechnol. 36, 411–420 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Satija R., Farrell J. A., Gennert D., Schier A. F., Regev A., Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mancarci B. O., et al., Cross-laboratory analysis of brain cell type transcriptomes with applications to interpretation of bulk tissue data. eNeuro 4, ENEURO.0212-17.2017 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Heng T. S., Painter M. W.; Immunological Genome Project Consortium , The immunological genome project: Networks of gene expression in immune cells. Nat. Immunol. 9, 1091–1094 (2008). [DOI] [PubMed] [Google Scholar]
- 34.Bahney J., von Bartheld C. S., The cellular composition and glia-neuron ratio in the spinal cord of a human and a nonhuman primate: Comparison with other species and brain regions. Anat. Rec. (Hoboken) 301, 697–710 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Castellino F., et al., Chemokines enhance immunity by guiding naive CD8+ T cells to sites of CD4+ T cell-dendritic cell interaction. Nature 440, 890–895 (2006). [DOI] [PubMed] [Google Scholar]
- 36.Lee E. G., et al., Failure to regulate TNF-induced NF-kappaB and cell death responses in A20-deficient mice. Science 289, 2350–2354 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wertz I. E., et al., De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430, 694–699 (2004). [DOI] [PubMed] [Google Scholar]
- 38.Perkins N. D., Integrating cell-signalling pathways with NF-kappaB and IKK function. Nat. Rev. Mol. Cell Biol. 8, 49–62 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Hildebrand D. G., et al., IκBζ is a transcriptional key regulator of CCL2/MCP-1. J. Immunol. 190, 4812–4820 (2013). [DOI] [PubMed] [Google Scholar]
- 40.Zhu K., et al., Necroptosis promotes cell-autonomous activation of proinflammatory cytokine gene expression. Cell Death Dis. 9, 500 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chiu I. M., et al., A neurodegeneration-specific gene-expression signature of acutely isolated microglia from an amyotrophic lateral sclerosis mouse model. Cell Rep. 4, 385–401 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Qiu X., et al., Single-cell mRNA quantification and differential analysis with Census. Nat. Methods 14, 309–315 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Qiu X., et al., Reversed graph embedding resolves complex single-cell trajectories. Nat. Methods 14, 979–982 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trapnell C., et al., The dynamics and regulators of cell fate decisions are revealed by pseudotemporal ordering of single cells. Nat. Biotechnol. 32, 381–386 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zilionis R., et al., Single-cell barcoding and sequencing using droplet microfluidics. Nat. Protoc. 12, 44–73 (2017). [DOI] [PubMed] [Google Scholar]
- 46.Klein A. M., et al., Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Osawa T., et al., Optineurin in neurodegenerative diseases. Neuropathology 31, 569–574 (2011). [DOI] [PubMed] [Google Scholar]
- 48.Watanabe M., et al., Histological evidence of protein aggregation in mutant SOD1 transgenic mice and in amyotrophic lateral sclerosis neural tissues. Neurobiol. Dis. 8, 933–941 (2001). [DOI] [PubMed] [Google Scholar]
- 49.Ding J., et al., Systematic comparison of single-cell and single-nucleus RNA-sequencing methods. Nat. Biotechnol. 38, 737–746 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Marsh S. E., et al., Single cell sequencing reveals glial specific responses to tissue processing & enzymatic dissociation in mice and humans. bioRxiv [Preprint] (2020). 10.1101/2020.12.03.408542 (Accessed 3 December 2020). [DOI]
- 51.Mifflin L., Ofengeim D., Yuan J., Receptor-interacting protein kinase 1 (RIPK1) as a therapeutic target. Nat. Rev. Drug Discov. 19, 553–571 (2020). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Liddelow S. A., et al., Neurotoxic reactive astrocytes are induced by activated microglia. Nature 541, 481–487 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Klein R. S., et al., Neuroinflammation during RNA viral infections. Annu. Rev. Immunol. 37, 73–95 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wolf Y., et al., Microglial MHC class II is dispensable for experimental autoimmune encephalomyelitis and cuprizone-induced demyelination. Eur. J. Immunol. 48, 1308–1318 (2018). [DOI] [PubMed] [Google Scholar]
- 55.Brewer G. J., Torricelli J. R., Isolation and culture of adult neurons and neurospheres. Nat. Protoc. 2, 1490–1498 (2007). [DOI] [PubMed] [Google Scholar]
- 56.Subramanian A., et al., Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hitomi J., et al., Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135, 1311–1323 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Carpenter A. E., et al., CellProfiler: Image analysis software for identifying and quantifying cell phenotypes. Genome Biol. 7, R100 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Lamprecht M. R., Sabatini D. M., Carpenter A. E., CellProfiler: Free, versatile software for automated biological image analysis. Biotechniques 42, 71–75 (2007). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The scRNAseq data have been deposited in GEO under accession no. GSE167332.